Abstract
Botulinum neurotoxin serotype A1 (BoNT/A1) is one of the most dangerous potential bioterrorism agents, and exerts its action by invading motoneurons. It is also a licensed drug widely used for medical and cosmetic applications. Here we report a 2.0 Å resolution crystal structure of BoNT/A1 receptor-binding domain in complex with its neuronal receptor, the glycosylated human SV2C. We find that the neuronal tropism of BoNT/A1 requires recognition of both the peptide moiety and an N-linked glycan on SV2. This N-glycan—conserved in all SV2 isoforms across vertebrates—is essential for BoNT/A1 binding to neurons and its potent neurotoxicity. The glycan-binding interface on SV2 is targeted by a human BoNT/A1-neutralizing antibody currently licensed as an anti-botulism drug. Our studies reveal a new paradigm of host-pathogen interactions, in which pathogens exploit conserved host post-translational modifications to achieve highly specific receptor binding while also tolerating genetic changes across multiple isoforms of receptors.
BoNT/A1 is one of the seven major serotypes of BoNT (termed BoNT/A–G). According to a well-accepted dual-receptor model, the extreme potency of BoNT/A1 targeting motoneurons is mediated by its receptor-binding domain (HCA), which synergistically binds to host protein receptors and gangliosides on the neuronal surface at neuromuscular junctions 1–3. The synaptic vesicle glycoprotein 2 (SV2), a family of 12-transmembrane domain proteins that have three isoforms (SV2A, 2B, and 2C) in mammals, are protein receptors for BoNT/A1 4,5, as well as for BoNT/E 6, BoNT/D 7, and potentially BoNT/F 8,9.
We have previously mapped the BoNT/A1-binding site to the fourth luminal domain of SV2 (SV2-L4) 4,5. A crystal structure of HCA in complex with the recombinant human SV2C-L4 expressed in E. coli (referred to as bSV2C with b indicating bacterial expression) has been reported recently 10. It shows that HCA–bSV2C recognition relies mostly on backbone-to-backbone interactions within a small interface (~596 Å2), mediated by two β-strands in HCA and one open edge of the quadrilateral β-helices of bSV2C 10. This binding mode is in sharp contrast to BoNT/B, which recognizes its receptors synaptotagmin-I/II (Syt-I/II) through an extensive side-chain mediated protein–protein interaction network that ensures high binding affinity and specificity towards Syt-I/II 11,12. So, how could BoNT/A1 possibly achieve extreme efficacy of targeting neurons using mostly backbone-mediated interactions for receptor recognition?
To better understand the molecular mechanism underlying BoNT/A1’s extraordinary neuronal tropism, we determined the crystal structures of HCA in complex with rat bSV2C-L4 and the physiologically more relevant glycosylated human SV2C-L4. We found that BoNT/A1 recognizes two distinct structural elements on SV2C: the protein moiety and an N-linked glycan that is conserved in all known SV2 homologs across vertebrates. Further biophysical, cellular and functional studies demonstrated that SV2 glycans are essential for BoNT/A1 binding to neuron and its extreme toxicity at its physiological site of action, the motor nerve terminals. Moreover, we found that the glycan-binding site of BoNT/A1 is also the target of a potent human neutralizing antibody, suggesting the potential for SV2 glycan as a novel target for developing BoNT inhibitors.
RESULTS
The crystal structure of HCA in complex with rat bSV2C
Amino acid sequence analyses showed that even the few residues that mediate side-chain interactions in the HCA and human bSV2C complex are not strictly conserved in SV2A and SV2B, or even SV2C from other species (e.g. rodents) (Supplementary Note 1). To gain a better insight into how BoNT/A1 can recognize SV2C from different species, we determined the crystal structure of HCA in complex with rat SV2C-L4 expressed in E. coli (Table 1). The structure of the rat bSV2C–HCA complex is virtually identical to that of the human bSV2C complex [root mean square deviation (RMSD) ~0.70 Å over 496 aligned Cα pairs]. Two major differences are observed. First, HCA-R1294 forms hydrogen bonds with S519, C520, T521, and D539 of rat bSV2C (Supplementary Fig. 1), which are not observed in the structure of the human bSV2C–HCA complex probably due to the different crystal packing modes 10. Interestingly, R1294 only exists in two of the eight BoNT/A subtypes currently known (BoNT/A1 and A4). Second, a cation-π stacking interaction between BoNT/A1-R1156—a residue exclusively existing in subtype BoNT/A1—and human SV2C-F563, previously thought to be critical for BoNT/A1–SV2C recognition 10, does not exist in the rat bSV2C–HCA complex because rat SV2C has a leucine (L563) in the place of human SV2C-F563. Leucine is also the homologous residue on SV2A and SV2B in both humans and rodents (Supplementary Note 1). These findings suggest that the side-chain mediated interactions may vary substantially among different BoNT/A subtypes and SV2 isoforms, thus unlikely provide sufficient binding specificity and affinity between them. Therefore, some crucial BoNT/A–SV2 interactions are missing in the crystal structures of HCA–bSV2C complexes described here and previously 10.
Table 1.
Data collection and refinement statistics
| HCA-bSV2C (PDB code: 5JMC) | HCA-gSV2C (PDB code: 5JLV) | |
|---|---|---|
| Data collection | ||
| Space group | P 1 21 1 | C 1 2 1 |
| Cell dimensions | ||
| a, b, c (Å) | 88.66, 143.99, 110.92 | 109.00, 111.85, 126.25 |
| α, β, γ (°) | 90, 93.6, 90 | 90, 101.3, 90 |
| Resolution (Å) | 87.76–2.64 (2.73–2.64)a | 123.81–2.00 (2.03–2.00) |
| Rmeas | 0.172 (1.454) | 0.159 (0.889) |
| I/σ(I) | 9.09 (1.43) | 6.7 (2.1) |
| CC1/2 | 0.991 (0.599) | 0.984 (0.722) |
| Completeness (%) | 99.4 (99.2) | 99.6 (99.9) |
| Redundancy | 3.4 (3.1) | 2.9 (2.9) |
| Refinement | ||
| Resolution (Å) | 87.76–2.64 | 123.81–2.00 |
| No. reflections | 80,958 | 99,659 |
| Rwork / Rfree | 0.239 / 0.275 | 0.175 / 0.216 |
| No. atoms | ||
| Protein | 16,825 | 8,641 |
| Ligand | – | 128 |
| Water | 162 | 659 |
| B factors | ||
| Protein | 58.50 | 44.50 |
| Ligand | – | 58.70 |
| Water | 55.40 | 51.50 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.012 | 0.009 |
| Bond angles (°) | 1.22 | 1.01 |
One crystal was used for each structure.
Values in parentheses are for highest-resolution shell.
SV2 glycosylation is crucial for BoNT/A1 binding to neurons
Native SV2s are glycosylated in neurons 13, and one of the three N-linked glycosylation motifs in L4—conserved in all SV2 isoforms across vertebrates—is located at the center of the BoNT/A1-binding interface of SV2 (e.g. N573 in SV2A, N516 in SV2B, and N559 in SV2C for human) (Supplementary Note 1). To explore the functional role of this N-linked glycan, we utilized a molecule replacement approach to express either wild type (WT) SV2A–C, or the corresponding deglycosylation mutants (SV2A-N573A, SV2B-N516A, and SV2C-N559A) in hippocampal/cortical neurons cultured from SV2A/SV2B double knockout (KO) mice. Most hippocampal/cortical neurons do not express SV2C 13, and thus these neurons cultured from SV2A/SV2B double KO mice serve as an SV2-null neuron model. These deglycosylation mutations of SV2 do not affect protein–protein interactions with BoNT/A1, as e.g. bSV2C-N559A maintained a WT-like binding to HCA 10.
The deglycosylation mutants of SV2A, 2B, and 2C all showed a lower molecular weight compared to WT SV2s, confirming that this asparagine residue is indeed glycosylated in neurons (Fig. 1a,b and Supplementary Fig. 2). The expression level of SV2A-N573A was comparable to the WT SV2A, but there was a drastic reduction of SV2B-N516A expression and a mild reduction of SV2C-N559A expression (Supplementary Fig. 2). It suggests that glycosylation at this site is crucial for folding and/or stability of SV2B and to a lesser degree for SV2C—a known function of N-linked glycans 14.
Figure 1.
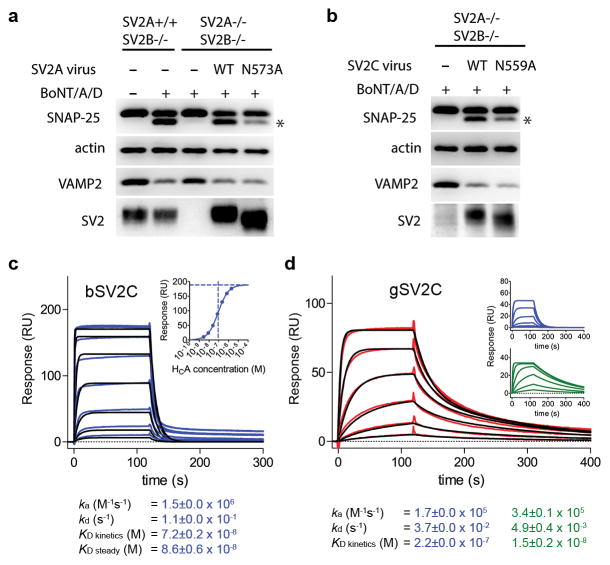
SV2 glycosylation is critical for BoNT/A1 binding and entry into neurons. (a,b) Neurons that express either the WT or the deglycosylation mutants of SV2A (a) or SV2C (b) were simultaneously exposed to BoNT/A1 (1 nM) and BoNT/D (0.1 nM). Cell lysates were harvested and subjected to immunoblot analysis using mouse monoclonal antibodies against SNAP-25 (Cl 71.2), VAMP2 (Cl 69.1), and SV2 (pan-SV2). BoNT/D served as an internal control to confirm that SV2 mutants still sorted and localized correctly. Cleavage of SNAP-25 by BoNT/A1 generates a smaller fragment that is marked by an asterisk. Actin served as a loading control. The uncropped images of the blots are shown in Supplementary Data Set 1. (c) Surface plasmon resonance sensorgrams of HCA binding to immobilized human bSV2C (blue) overlaid with a fit of 1:1 binding model (black). Since equilibrium binding was reached for all conditions tested, a steady state affinity was also determined (insert). (d) HCA binding to immobilized human gSV2C (red) was best fit to a heterogeneous binding model (black), whereas a 1:1 binding model was inapplicable. Insert: individual contributions of the two binding events observed in the heterogeneous interaction (blue and green). Shown values represent the mean ± S.D. (2 and 3 independent experiments for bSV2C and gSV2C, respectively).
We thus focused on SV2A-N573A and SV2C-N559A, and examined SV2-mediated toxin entry for BoNT/A1 and BoNT/D by analyzing the cleavage of synaptosomal-associated protein of 25 kDa (SNAP-25, the substrate of BoNT/A1) and synaptobrevin/vesicle-associated membrane protein 2 (VAMP2, the substrate of BoNT/D) after toxin exposure. BoNT/D serves here as an internal control for regular trafficking and sorting of SV2 mutants, as BoNT/D uses SV2s as receptors independent to N-glycosylation 7. Furthermore, we have previously shown that BoNT/B that does not use SV2 as its receptor binds and enters the WT and SV2 KO neurons at similar levels 4,6. We found that the SV2 KO neurons did not show BoNT/A1 entry at the conditions tested, which is likely due to BoNT/A1’s low affinity for the ganglioside receptor that is unable to allow cell entry in the absence of SV2 15,16. Expression of SV2A-N573A and SV2C-N559A mediated substantially less entry of BoNT/A1 compared to the WT SV2s, whereas BoNT/D entry was not affected (Fig. 1a,b). These results suggest that SV2 glycosylation at this strictly conserved site clearly contributes to BoNT/A1 binding and entry into neurons.
The SV2C glycans greatly enhance HCA binding
Glycosylation is a common and highly diverse post-translational protein modification that profoundly alters the protein behavior 14. Do the N-glycans contribute directly to BoNT/A1 binding? In this regard, we first carried out surface plasmon resonance (SPR) to examine how HCA binds to the human bSV2C. We found that the binding displays a fast association rate (ka: 1.5 × 106 M−1s−1) and a fast dissociation rate (kd: 0.11 s−1) (Fig. 1c), and the overall dissociation constant (KD) of ~86 nM was comparable to a previously reported KD of 260 nM determined by fluorescence anisotropy experiment 10. The nature of this transient interaction is consistent with the pattern of backbone-to-backbone interactions revealed in the crystal structure, but difficult to reconcile with the extreme specificity of BoNT/A1 toward neurons.
We then expressed the human SV2C-L4 (residues V473–T567) as a secreted protein using human embryonic kidney 293 cells (HEK293), to mimic the physiologically relevant glycosylated receptor. HEK293 cells have been widely used to produce glycoproteins with human glycosylation patterns 17. The resulting protein (referred to as gSV2C, g stands for glycosylation) is glycosylated as evidenced by the appearance of multiple bands on SDS-PAGE that are bigger than its peptide mass and represent heterogeneous glycoforms. We characterized binding of HCA to gSV2C by SPR, which revealed two binding components (Fig. 1d). A transient low affinity binding (KD ~220 nM) closely resembles the binding of HCA to bSV2C, which is likely due to heterogeneous glycosylation of SV2C under over-expression conditions 18. Notably, a high affinity binding (KD ~15 nM) of gSV2C displays a ~22-fold slower dissociation rate and a ~4-fold slower association rate compared to bSV2C. The slightly decreased association rate of gSV2C likely stems from restricted carbohydrate flexibility upon toxin binding, a physiological event that BoNT/A1 will encounter in vivo because the neuronal SV2s are glycosylated. Therefore, the relatively fast association rate displayed by bSV2C artificially increases the binding affinity of bSV2C. Taken together, these data demonstrate that the glycans of gSV2C stabilize the HCA–gSV2C complex by markedly decreasing the dissociation rate.
The crystal structure of HCA in complex with human gSV2C
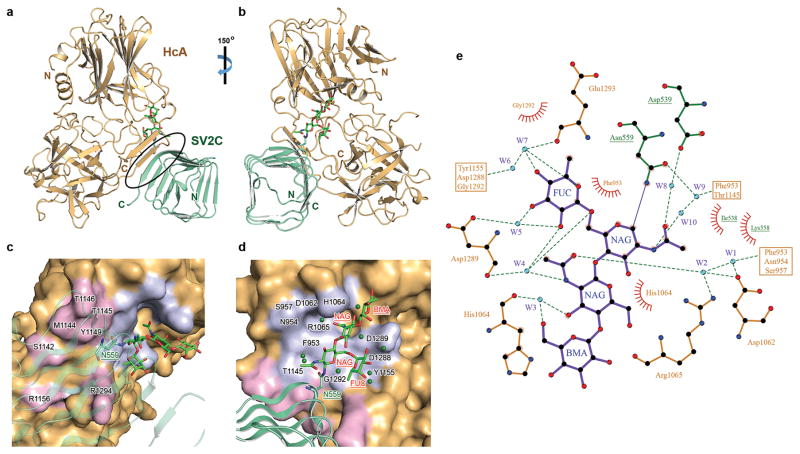
We next determined the crystal structure of HCA in complex with human gSV2C at 2.0 Å resolution (Table 1). The overall architecture of the HCA–gSV2C complex is similar to that of the bSV2C complex (RMSD ~0.88 Å over 489 aligned Cα pairs). A complex-type N-linked glycan attached to gSV2C-N559 was observed with clear electron densities for the quadruple-saccharide core made up of two N-acetylglucosamine (NAG), a mannose (BMA), and a fucose (FUC) (Fig. 2 and Supplementary Fig. 3). There are two other putative N-linked glycosylation sites (N484 and N534) that are localized on SV2C-L4 and also conserved on SV2A and SV2B (Supplementary Note 1). Only one NAG could be resolved in the electron density for the N534 glycan likely due to its high mobility, whereas no sugar could be identified at N484. SV2C N484 and N534 are located far away from BoNT/A1-binding interface, and thus unlikely to directly participate in toxin binding.
Figure 2.
Structure of HCA in complex with human gSV2C. (a) Cartoon representation of the complex whereas HCA is gold and gSV2C is green. The black oval highlights the interacting β-strands between them. gSV2C-N559 and the attached N-linked glycan are shown in stick models. (b) A ~150° rotation of the complex about a vertical axis. (c,d) Close-up views of the protein–protein and protein–glycan association interfaces between HCA and gSV2C. HCA is in surface representation (gold), whereas HCA residues that directly interact with the peptide moiety of gSV2C or the N559 glycan are colored purple and light blue, respectively. Well-defined water molecules that mediate HCA–glycan binding are shown as green spheres. (e) Extensive interactions between gSV2C-N559 glycan and HCA. The plots were generated using LIGPLOT 36. HCA and gSV2C residues are labeled brown and green, respectively. Hydrogen bonds are indicated by dashed green lines. Key hydrogen bonding distances are listed in Supplementary Table 1. Residues involved in hydrophobic interactions are represented by an arc with spokes radiating towards the binding partners they contact.
Remarkably the N559-glycan directly interacts with HCA through a network of hydrogen bonds and van der Waals contacts. The most prominent are stacking interactions between residues F953 and H1064 of HCA and the hydrophobic faces of the two NAG. In addition, ten well-defined water molecules act as molecular “glue” in the glycan–HCA interface to further strengthen the interactions (Fig. 2d,e and Supplementary Table 1). These interactions almost double the contact area between HCA and SV2C from 557 to 925 Å2. There are also weak electron densities beyond the mannose, suggesting that there might be more extensive HCA–glycan interactions that were not resolved, possibly due to the inherent flexibility of glycans and heterogeneous glycosylation. This structure unambiguously reveals that the N559-glycan of SV2C is recognized directly by BoNT/A1 as an integral part of the toxin-binding site.
SV2 glycan is critical for BoNT/A1 binding to neurons
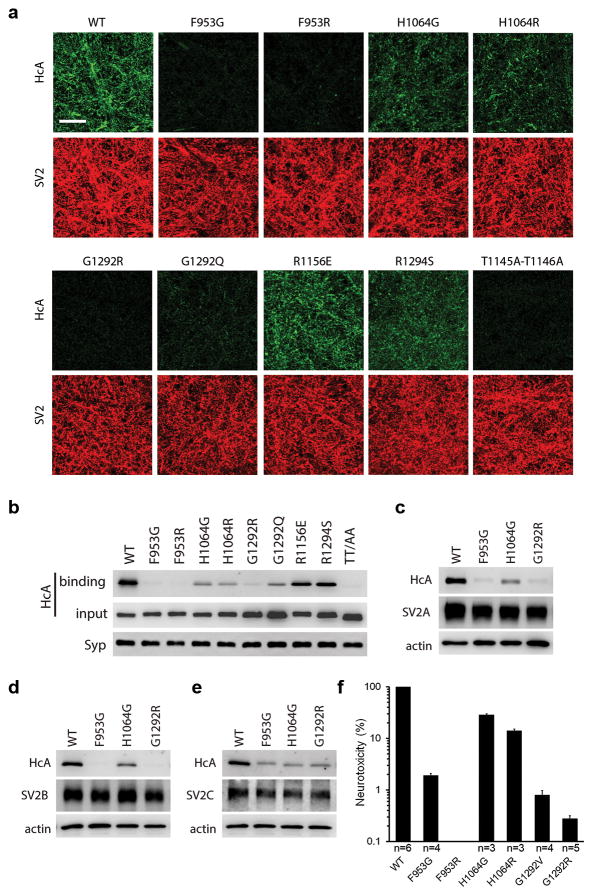
We then sought to understand the functional role of protein–glycan interactions in BoNT/A1–SV2 recognition. Based on the crystal structure, we designed a set of single-site mutations on HCA that selectively disrupt glycan binding: F953G and H1064G abolish the critical stacking of aromatic side chains against the sugar rings, and F953R, H1064R, G1292Q, and G1292R cause clashes between their bulky side chains and the N559-glycan. None of these mutations affected HCA folding and stability as verified by thermal denaturation experiments, nor did they affect HCA binding to bSV2C based on pull-down and SPR studies. As expected, these mutants markedly decreased HCA binding to gSV2C, strongly supporting the direct involvement of N559-glycan of SV2C in BoNT/A1 binding (Supplementary Figs. 4 and 5a,b,c). Interestingly, SPR studies showed that the glycan-binding deficient HCA-F953G binds to gSV2C much weaker than bSV2C (KD: ~760 nM vs. ~160 nM) (Supplementary Fig. 5d,e). Therefore, the native neuronal SV2s that are always glycosylated initially impose steric hindrances for BoNT/A1 recognition. But BoNT/A1 manages to overcome this obstacle by directly employing SV2 glycans to strengthen binding.
We examined how these HCA mutants bind to native SV2s in neurons. First, we analyzed binding of HCA to endogenous SV2A and SV2B using cultured rat hippocampal/cortical neuron 13 (Fig. 3a,b). Mutations F953G, F953R, G1292Q, and G1292R largely abolished binding of HCA to neurons; H1064G and H1064R also drastically reduced binding, suggesting that protein–glycan interactions are essential for BoNT/A1 binding to native SV2s on neurons. We further examined neurons that exclusively expressed SV2A, 2B, or 2C: hippocampal/cortical neurons cultured from SV2A(+/+)SV2B(−/−) mice served as neurons that only express SV2A, and neurons that only express SV2B or SV2C were created by infecting neurons cultured from SV2A(−/−)SV2B(−/−) mice with lentiviruses that express SV2B or SV2C, respectively. Mutating glycan-binding residues on HCA (e.g. F953G, G1292R, and H1064G) reduced HCA binding in all cases tested, demonstrating that BoNT/A1–glycan interactions are conserved and essential for all three SV2 isoforms (Fig. 3c,d,e and Supplementary Fig. 6).
Figure 3.
Site-directed mutagenesis analysis of the SV2-binding site on HCA. (a,b) Binding of HCA variants (100 nM) to rat hippocampal/cortical neurons was analyzed by immunostaining (a) or immunoblot (b). Scale bar, 20 μm. Synaptophysin (Syp) served as a loading control. (c,d,e) Binding of HCA variants (100 nM) to neurons that express individual SV2A, 2B, or 2C was analyzed by immunoblot. The results of immunostaining analysis are shown in Supplementary Fig. 8. The uncropped images of the blots are shown in Supplementary Data Set 1. (f) Glycan binding-deficient BoNT/A1 displays drastic decreased neurotoxicity as examined by the mouse phrenic nerve (MPN) assay. BoNT/A1 F953R mutant showed no detectable toxicity even at the maximal concentration tested (200 nM), indicating a larger than 106-fold reduction on toxicity. Shown are mean ± S.D. of 3–6 technical replicates. The source data are shown in Source Data Figure 3f.
SV2 glycan allows to tolerate genetic changes on BoNT/A
Complementing our studies on protein–glycan interactions, we also examine the contribution of side-chain-mediated protein–protein interactions to SV2 recognition, which could vary substantially among the eight different BoNT/A subtypes (Supplementary Note 2). In this regard, residues R1156 and R1294 of BoNT/A1 were mutated to their counterparts in other BoNT/A subtypes (e.g. R1156E and R1294S), which disrupt their interactions with the peptide moiety of SV2C but without affecting glycan binding. We used the double mutant HCA-T1145A-T1146A as a control 10. These two threonine residues, located at the core of protein–protein interface, are conserved in all BoNT/A subtypes (Supplementary Note 2). We found that binding of HCA-T1145A-T1146A to cultured rat hippocampal/cortical neurons was abolished (Fig. 3a,b). This is consistent with an earlier study showing that HCA-T1145A-T1146A can no longer bind to bSV2C 10. Interestingly, we found that HCA-R1156E and R1294S still bound substantially to neurons (Fig. 3a,b), even though they showed decreased binding to both human bSV2C and gSV2C in vitro (Supplementary Fig. 5a,b,c). These data suggest that loss of side-chain-mediated interactions at R1156 and R1294 of BoNT/A1 is tolerated on neuronal surfaces likely due to the presence of SV2 glycan and co-receptor gangliosides.
Glycan-binding is essential for BoNT/A1’s extreme potency
To further establish the physiological relevance of protein–glycan interactions, we produced full-length BoNT/A1 containing single-site glycan-binding deficient mutations (F953G, F953R, H1064G, H1064R, G1292V, or G1292R) and examined their neurotoxicity at motor nerve terminals using an ex vivo mouse phrenic nerve hemi-diaphragm (MPN) assay (Fig. 3f) 19. Remarkably, all these mutations drastically reduced the potency of BoNT/A1. BoNT/A1-F953R had no detectable toxicity even at the maximal concentration tested (200 nM), which reflects a larger than 106-fold reduction of neurotoxicity even though it displayed fully functional Zn2+-endoprotease activity in vitro. Mutation G1292R also severely reduced the toxicity by 350-fold 20. Mutations H1064G and H1064R displayed 3- and 7-fold reduction, respectively. These data demonstrate that glycan-binding is essential for the extreme potency of BoNT/A1 at the motor nerve terminals, its physiological site of action.
The SV2 glycan-binding mode is conserved in BoNT/HA
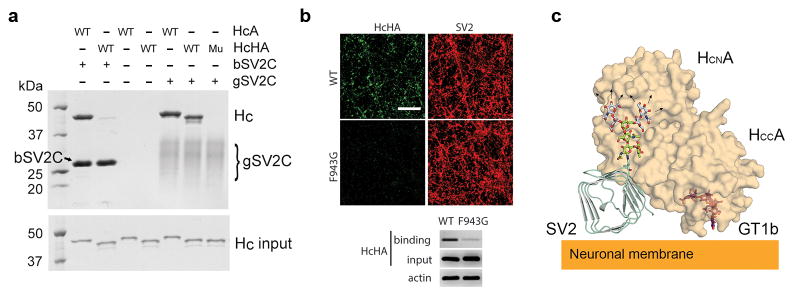
We next examined whether glycan binding is conserved in natural variants of BoNT/A1, as BoNT genes are actively evolved and at least 40 different subtypes of BoNT have been reported 21. In this regard, we focused on a newly reported mosaic toxin type HA (BoNT/HA, also known as BoNT/FA), which has a hybrid-like structure including a BoNT/A1-like HC 22–26. HCHA is highly similar to HCA (~83% identical) (Supplementary Note 2). Sequence alignment revealed that most of the glycan-binding residues, such as F953 and H1064, are conserved in HCA and HCHA, but residues R1156 and R1294 of HCA are changed to M1148 and S1286 in HCHA. HCHA showed much weaker binding to human bSV2C as compared to HCA, whereas its binding to human gSV2C was comparable to that of HCA (Fig. 4a). It suggests that the loss of side-chain interactions due to genetic changes of M1148 and S1286 in HCHA is compensated by SV2C glycan. In contrast, HCHA-F943G (equivalent to HCA-F953G) failed to bind to gSV2C based on a pull-down assay, nor did it bind to cortical neurons, likely due to the disruption of glycan binding (Fig. 4a,b). These data confirm that a similar glycan-binding mode is conserved in HCHA and is critical for its binding to SV2s on neurons.
Figure 4.
The SV2 glycan binding mode is conserved in BoNT/HA. (a) The SV2C glycan plays an indispensable role for HCHA binding as demonstrated by a pull-down assay using human bSV2C (with a SUMO tag) or gSV2C as baits. “Mu” is HCHA-F943G mutant. (b) The F943G mutation dramatically decreased HCHA binding to rat cortical neurons as analyzed by immunostaining and immunoblot. The uncropped images of the gels and blots are shown in Supplementary Data Set 1. (c) Proposed model for simultaneous binding of BoNT/A1 to two neuronal surface receptors: glycosylated SV2 and ganglioside. GT1b is modeled based on the structure of a GT1b-bound HCA (PDB code 2VU9) 37. A representative complex type N-linked glycan is modeled based on the structure of a glycan of human IgG1 (PDB code 3AVE) 38. The glycan core that is observed in the HCA–gSV2C complex is colored green, whereas the remaining carbohydrates (mannose and NAG colored gray) could potentially extend to the N-terminal sub-domain of HCA (HCNA).
Furthermore, we found that the SV2 glycan-binding residues are largely conserved in seven out of the eight BoNT/A subtypes (BoNT/A1–A3 and A5–A8) that have been identified up to date (Supplementary Note 2) 27. Notably, BoNT/A4 is the only one that has an arginine residue (R1292) at the equivalent position of BoNT/A1-G1292. Since BoNT/A1-G1292R mutant drastically decreases BoNT/A1 toxicity by blocking its binding to SV2 glycan (Fig. 3 and Supplementary Figs. 5,6) 20, we suggest that this is a major reason that causes the ~1,000-fold reduced biological activity of BoNT/A4 compared to BoNT/A1 28. Taken together, our findings suggest that BoNT/A1 and variants utilizes the genetically invariable carbohydrates as surrogate amino acids to engage the host receptors. Synergistic binding to two distinct cell surface receptors, SV2 (including its peptide and glycan moieties) and ganglioside, provides a plausible explanation for the extreme potency of BoNT/A and its remarkable specificity for nerve terminals (Fig. 4c).
A new strategy for developing antibodies against BoNT/A
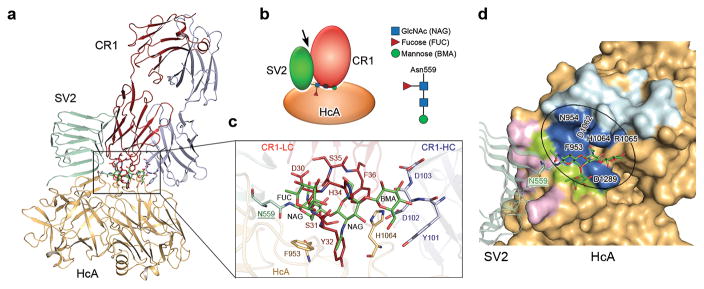
The novel glycan receptor for BoNT/A presents a promising target for developing therapeutic toxin inhibitors. Remarkably, we found that a BoNT/A1-neutralizing human monoclonal antibody family CR2/CR1, which is currently in clinical trial 29,30, directly targets the glycan-binding site on HCA with its first antigen-binding loop of the light chain variable region occupying the glycan-binding site (Fig. 5a). The key epitopes for CR2/CR1 include precisely residues F953 and H1064 29. Interestingly, residue F36 of CR2/CR1 uses a π-stacking interaction to bind BoNT/A1-H1064, mimicking its interactions with SV2C N559-glycan. CR2/CR1 also blocks binding of BoNT/A1 to bSV2C 20, but this is due to the large size of CR2/CR1 causing side-to-side clash with bSV2C, as CR2/CR1 and bSV2C have non-overlapping binding sites on HCA (Fig. 5b,c,d). Together our data reveal that the strong neutralization potency of CR2/CR1 is empowered by simultaneously blocking BoNT/A1 binding to the glycan and peptide moieties of SV2.
Figure 5.
The SV2 glycan-binding site on BoNT/A1 is the target for the neutralizing antibody CR1. (a,b) BoNT/A1-neutralizing therapeutic antibody CR1 (PDB code: 2NYY) 29 occupies the SV2C glycan-binding site on HCA, but does not affect HCA’s SV2C peptide-binding site. An arrow in the schematic diagram indicates the side-to-side clash between SV2C and CR1. (c) A close-up view of the interface. gSV2C-N559 glycan (green), HCA-F953 and -H1064 (gold), residues 30–36 of CR1 ligand chain (LC, red), and residues 101–103 of CR1 heavy chain (HC, gray) are shown in stick models. (d) Residues of HCA that exclusively bind SV2C peptide, N559 glycan or CR1 are colored in purple, green or cyan, respectively, while the HCA residues that are contacted by both N559 glycan and CR1 are in blue.
DISCUSSION
Host cell-surface glycans are crucial for pathogen recognition. For example, influenza hemagglutinins use carbohydrates in determining the host range (e.g. swine, avian or human) 31. Our results reveal a novel host-recognition strategy, by which pathogens simultaneously recognize a protein segment and the neighboring glycans as a composite binding site. This unique strategy uses a conserved post-translational modification as an evolutionarily static recognition site, in addition to protein–protein interactions that encode the location and specificity information. Together, it provides a powerful solution to address the competing needs of achieving highly specific binding while also tolerating residue changes in receptors across multiple isoforms and species variants.
Intriguingly, a similar strategy may be utilized by some important broad-neutralizing human antibodies, which are capable of neutralizing multiple serotypes of targeted viruses, such as dengue viruses and immunodeficiency virus-1 (HIV-1), by simultaneously recognizing the protein components and the highly conserved glycans on virus proteins 32–34. A similar model was also recently demonstrated for Notch1 receptor binding with its ligand Delta like 4 (DLL4) 35, where DLL4 binds to the fucose and glucose at the base of an O-glycosylation site located within the protein–protein interface. Therefore, our findings offer an important strategy for engineering ligand–receptor interactions and broadly neutralizing antibodies for therapeutic applications.
METHODS
No statistical method was used to predetermine sample size. The experiments were not randomized and were not performed with blinding to the conditions of the experiments.
Construct design and cloning
The gene of HCA (residues N872–L1296) was cloned into expression vector pQE30 with an N-terminal 6xHis-tag and a PreScission protease cleavage site. HCHA (residues E860–L1286) was cloned into pGEX-4T-2 vector that has a thrombin cleavage site following GST. The core region of human SV2C-L4 (residues V473–T567) was cloned into two different vectors: pET28a vector for E. coli expression and a pcDNA vector for mammalian cell expression. For E. coli expression, a 6xHis/SUMO (Saccharomyces cerevisiae Smt3p) tag was introduced to the N-terminus of SV2C-L4 to facilitate protein expression and purification (SUMO-bSV2C). For mammalian cell expression, a human IL2 signal sequence (MYRMQLLSCIALSLALVTNS), a 9xHis-tag, and a Factor Xa cleavage site were added to the N-terminus of SV2C-L4 (gSV2C). A second SV2C mammalian expression construct was made based on gSV2C by inserting SUMO between the Factor Xa site and SV2C (SUMO-gSV2C). Rat SV2C-L4 (residues P455–Y577) was covalently linked to the C-terminus of HCA through a peptide linker composed of a thrombin cleavage site (LVPRGS) and a PreScission protease cleavage site (LEVLFQGP), and the covalently linked HCA and rat bSV2C (HCA–bSV2C) was cloned into pET28a vector with an N-terminal 6xHis-tag and a thrombin cleavage site. All HCA and HCHA mutations were generated using QuikChange site-directed mutagenesis (Agilent). All pH6tBoNTA mutants were prepared using the GeneTailor method (Invitrogen GmbH, Karlsruhe, Germany) employing suitable primers and pH6tBoNTA as template DNA 20.
Protein expression and purification
HCA, HCHA, SUMO-bSV2C, and HCA–bSV2C were expressed in E. coli strain BL21-Star (DE3) (Invitrogen). Bacteria were cultured at 37°C in LB medium containing appropriate selecting antibiotics. The temperature was reduced to 18°C when OD600 reached 0.4. Expression was induced with 0.2 mM IPTG (isopropyl-b-D-thiogalactopyranoside) when OD600 reached 0.7, and continued at 18°C for ~16 hours. The cells were harvested by centrifugation and stored at −80°C until use.
Wild-type and mutated recombinant full-length neurotoxin H6tBoNTA were produced under biosafety level 2 containment (project number GAA A/Z 40654/3/123) utilizing the E. coli strain M15pREP4 (Qiagen, Hilden, Germany) during 16 h of induction at 22°C in the presence of 0.2 mM IPTG, and were purified on Co2+-Talon matrix (Takara Bio Europe S.A.S., France). Full-length neurotoxins were eluted using 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 250 mM imidazole, subjected to size-exclusion chromatography (SEC; Superdex-200 16/60 column, GE Healthcare, Germany) in 100 mM Tris-HCl, pH 8.0, and 150 mM NaCl, frozen in liquid nitrogen and kept at −70°C.
The His-tagged proteins (HCA, SUMO-bSV2C and HCA–bSV2C) were purified using Ni2+-NTA (nitrilotriacetic acid, Qiagen, Hilden, Germany) affinity resins in a buffer containing 50 mM Tris, pH 8.0, 400 mM NaCl, and 40 mM imidazole. The proteins were eluted with a high-imidazole buffer (50 mM Tris, pH 8.0, 400 mM NaCl, and 300 mM imidazole) and then dialyzed at 4°C against a buffer containing 20 mM HEPES, pH 7.5, and 150 mM NaCl. The His-tag of HCA and the His/SUMO-tag of SUMO-bSV2C were cleaved by PreScission and SUMO proteases, respectively. For the covalently linked HCA–bSV2C, both thrombin and PreScission protease were used to cut the linker between the two proteins in order to avoid potential conformational constraint. GST-HCHA fusion protein was purified using glutathione Sepharose 4B affinity resin (GE Healthcare) in a buffer containing 20 mM HEPES, pH 7.5, and 150 mM NaCl. HCHA was then released from the resins by on-column cleavage using thrombin.
Tag-cleaved HCA and HCHA were further purified by MonoS ion-exchange chromatography (GE Healthcare) in a buffer containing 50 mM MES, pH 6.0, and eluted with a NaCl gradient. The peak fractions were then subjected to Superdex-200 SEC (GE Healthcare) in a buffer containing 20 mM sodium phosphate, pH 6.0, and 50 mM NaCl. SUMO-bSV2C (His/SUMO-tag cleaved or un-cleaved) and the cleaved HCA–bSV2C were further purified by Superdex-200 SEC in a buffer containing 20 mM HEPES, pH 7.5, and 150 mM NaCl. The HCA–bSV2C complex was concentrated to ~2 mg/ml for crystallization.
SUMO-gSV2C and gSV2C were expressed and secreted from HEK 293 cells (BioLegend) and purified directly from cell culture media using Ni2+-NTA. The proteins were eluted from the resins with high concentration of imidazole and dialyzed against a buffer containing 50 mM Tris, pH 8.0, and 400 mM NaCl. gSV2C was then mixed with the purified HCA at a molar ratio of ~1:2, and the HCA–gSV2C complex was isolated by Ni2+-NTA resins. After dialysis against a buffer containing 20 mM HEPES, pH 7.5, and 150 mM NaCl, the complex was further purified by Superdex-200 SEC using the same buffer. The complex was concentrated to ~10 mg/ml for crystallization.
Crystallization
Initial crystallization screens were carried out using a Gryphon crystallization robot (Art Robbins Instruments) with high-throughput crystallization screening kits (Hampton Research and Qiagen). Extensive manual optimization was then performed at 20°C using the hanging-drop vapor-diffusion method when proteins were mixed with reservoir solutions in 1:1 ratio. The HCA–bSV2C complex was initially crystallized in a condition containing 100 mM sodium cacodylate, pH 6.5, 13% polyethylene glycol (PEG) 3,350, and 200 mM NaCl. The best crystals were obtained in the presence of 0.7% (v/v) 1-butanol, which was identified using an additive screen kit (Hampton Research). The crystals were cryo-protected in the original mother liquor supplemented with 20% (v/v) glycerol and flash-frozen in liquid nitrogen. The HCA–gSV2C complex was originally crystallized as thin plates in a condition composed of 100 mM sodium acetate, pH 4.6, 20% PEG 3,350, and 200 mM ammonium phosphate monobasic. These crystals diffracted poorly. After extensive additive screening and optimization, the best crystals were obtained in the presence of 4% (w/v) polypropylene glycol P 400. The crystals were cryo-protected in the reservoir solution supplemented with 20% (v/v) ethylene glycol and flash-frozen in liquid nitrogen.
Data collection and structure determination
The X-ray diffraction data were collected at 100 K at the NE-CAT beamline 24-ID, Advanced Photon Source (APS). The data were processed with XDS 39. The structure of HCA–bSV2C was determined by molecular replacement software Phaser 40 using the structure of HCA (PDB code 3FUO) 9 as the search model. The structural model of bSV2C was manually built, and the structural modeling and refinement were carried out iteratively using COOT 41 and Refmac from the CCP4 suite 42. This structure was later used as the search model to determine the structure of the HCA–gSV2C complex. All the refinement progress was monitored with the free R value using a 5% randomly selected test set 43. The structures were validated through the MolProbity web server 44 and showed excellent stereochemistry. Data collection and structural refinement statistics are listed in Table 1. All structure figures were prepared with PyMol (http://www.pymol.org).
Pull-down assay
The pull-down assay was performed using Ni2+-NTA resins in 1 ml buffer containing 50 mM Tris, pH 8.0, 400 mM NaCl, 10 mM imidazole, and 0.1% Tween-20. SUMO-bSV2C or gSV2C served as the bait while HCA or HCHA variants were the preys. SV2C was pre-incubated with Ni2+-NTA resins at 4°C for 1 hour, and the unbound protein was washed away. The resins were then divided into small aliquots (~5 μg of bait) and mixed with the preys (~30 μg). The pull-down assay was carried out at 4°C for ~1.5 hours. The resins were washed twice, and the bound proteins were released from resins using 300 mM imidazole.
Protein melting assay
The thermal stability of HCA or HCHA variants was measured using a fluorescence-based thermal shift assay on a StepOne real-time PCR machine (Life Technologies). The protein (~5 μM) was mixed with the fluorescent dye SYPRO Orange (Sigma-Aldrich) immediately before the experiment. The samples were heated from 25°C to 90°C in ~45 min. The midpoint of the protein-melting curve (Tm) was determined using the analysis software provided by the instrument manufacturer. The data obtained from three independent experiments were averaged to generate the bar graph.
Surface plasmon resonance (SPR)
Binding kinetics and affinity were determined on a Biacore X100 unit (GE Healthcare) at 25°C using HBS-EP+ (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20) as running buffer at a flow rate of 30 μL/min. Using standard EDC/NHS amine coupling chemistry, SUMO-gSV2C or SUMO-bSV2C were coupled on flow cell (Fc) 2 of a CM5 sensor chip (GE Healthcare) to a surface density of ~83 resonance units (RUs) or ~105 RUs, respectively. Control Fc1 was blank immobilized by EDC/NHS activation before blocking with 1 M ethanolamine (GE Healthcare).
For kinetic measurements, HCA was injected in 1:3 dilution series ranging from 1,200 nM to 4.94 nM for gSV2C or 2,000 nM to 2.74 nM for bSV2C. Each measurement was started and ended with injection of the highest analyte concentration to ensure retained binding capacity. Association was monitored for 120 seconds by analyte injection followed by 300-second injections of running buffer to monitor binding dissociation. Between measurements, the surface was regenerated by 60 second injections of 10 mM glycine-HCl (pH 1.7) at 10 μL/min. Binding kinetics were determined by fitting the double referenced 45 binding curves using the heterogeneous (gSV2C) or the 1:1 Langmuir binding models (bSV2C) with global Rmax and RI set to zero (Biacore Evaluation Software 2.01). Due to the highly transient interaction of HCA with bSV2C, equilibrium binding was reached for all HCA concentrations tested. Therefore, steady state affinity was determined for bSV2C by fitting a four-parametric Hill equation to the binding responses 65 seconds after analyte injection over log-transformed HCA-concentrations using Prism 5.04 (GraphPad) with bottom constraint set to zero. The good agreement between the binding affinity determined by kinetic analysis and the steady state affinity proved that kinetic binding rates for bSV2C were reliable despite being close to the measurement limits of the instrument. Kinetic binding rates were determined by n = 2 (bSV2C) or n = 3 (gSV2C) independent experiments. Shown values represent the mean ± standard deviation.
To compare the interaction of HCA WT and its mutants with bSV2C (511 RUs) and separately with gSV2C (479 RUs), the reactions were performed in the “interactive manual run mode”. The HCA variants were injected at concentrations of 0.01, 1.0, 10, 25, 50, 75, 100 and 200 nM, each in triplicate, and at a flow-rate of 10 μL/min. The chip surface was regenerated by two injections of glycine-HCl (10 mM, pH 2.0, 30 sec contact time) allowing the removal of the analyte without changing the activity of the immobilized ligand. This was confirmed by the equal responses obtained from the binding assays before and after regeneration. Results were plotted as response (RU) versus concentration of HCA using Prism 6 (GraphPad Software Inc., La Jolla, CA).
Cell biology materials and constructs
SV2A and SV2B knockout mice were obtained from the Jackson Laboratory. Rat cDNAs encoding SV2A, SV2B, and SV2C were generously provided by R. Janz (Houston, TX). They were cloned into a lentiviral vector (Lox-Syn-Syn) as we previously described 7. This vector contains two separate neuronal-specific synapsin promoters. One promoter drives expression of SV2 and the other one drives expression of GFP as a marker. Human cDNA encoding SV2C was obtained from PlasmID repository of Harvard Medical School. Human monoclonal antibody against HCA (RAZ-1) was generously provided by J. Marks (San Francisco, CA). Mouse monoclonal antibodies against VAMP2 (Cl 69.1), SNAP-25 (Cl 71.2), Syp (Cl 7.2) were generously provided by E. Chapman (Madison, WI) and are available from Synaptic Systems (Göttingen, Germany). SV2 (pan-SV2) was also provided by E. Chapman (Madison, WI) and it is available from The Developmental Studies Hybridoma Bank (AB2315387, Iowa City, IA). The following antibodies were purchased from indicated vendors: mouse monoclonal antibody against actin (Sigma, A2228); rabbit polyclonal antibody against synapsin (Millipore, Ab1543p); chicken polyclonal antibody against GFP (Aves lab, GFP-1020). Purified BoNT/A1 of Hall-A strain and BoNT/D of D1873 strain were generously provided by E. Johnson (Madison, WI).
Neuron culture and lentivirus transduction
Rat hippocampal/cortical neurons were prepared from E18–19 embryos. Mouse SV2A/B double knockout neurons were prepared from postnatal day 1 pups as previously described 7. Dissected hippocampi and cortex were dissociated with papain following manufacture instructions (Worthington Biochemical, NJ). Cells were plated on poly-D-lysine coated coverslips. Experiments were carried out generally using DIV (days in vitro) 13–15 neurons. Lentiviruses were prepared as described previously using HEK293FT cells 7. Viruses were added to neurons at DIV5.
HCA and HCHA binding to neurons
Neurons were exposed to 100 nM HCA or HCHA in high K+ buffer, which contains (mM): NaCl 140, KCl 3, KH2PO4 1.5, Na2HPO4 8, MgCl2 0.5, and CaCl2 1, for 5 minutes at 37°C. Cells were then washed three times with phosphate-buffered saline (PBS). Binding of HCA or HCHA was examined using two complementary approaches. (1) Immunostaining: neurons were fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100 in PBS solution. Images were collected using a Leica TCS SP8 confocal microscope with a 40X oil objective. At least three representative images were collected per condition. (2) Immunoblot analysis: neurons were harvested in a lysis buffer (PBS with 1% Triton X-100, 0.05% SDS and protease inhibitor cocktail (Roche, CA), 100 μL per one well of 24-well plates). Lysates were centrifuged for 10 minutes at 4°C and the supernatants were subjected to SDS-PAGE and western blot analysis. Binding of HCA or HCHA was detected using a monoclonal human anti-HCA antibody (RAZ-1), which recognizes both HCA and HCHA. It also recognizes mutants of HCA and HCHA examined in this study with similar sensitivity (“input” lanes in Figs. 3b and 4b). All experiments were repeated three times independently.
Entry of BoNTs into neurons
Neurons were exposed to 1 nM BoNT/A1 and 0.1 nM BoNT/D in high K+ buffer for 5 minutes at 37°C. Cells were washed three times with PBS and further incubated in toxin-free media for 8 hrs. Neuron lysates were then harvested and subjected to immunoblot analysis, detecting cleavage of toxin substrate SNAP-25 (for BoNT/A1) and VAMP2 (for BoNT/D). Cleavage of SNAP-25 by BoNT/A1 generates a smaller fragment that can be detected on immunoblot. Cleavage of VAMP2 by BoNT/D resulted in a loss of immunoblot signal of VAMP2. All experiments were repeated three times independently.
Mouse phrenic nerve hemidiaphragm (MPN) assay
The MPN assay was performed as described previously employing 20–30 g NMRI mice (Janvier SA, France) 19,20. According to §4 Abs. 3 (killing of animals for scientific purposes, German animal protection law (TSchG)) animals were sacrificed by trained personnel before dissection of organs and its number reported yearly to the animal welfare officer of the Central Animal Laboratory and to the local authority, Veterinäramt Hannover. First, mice were euthanized by CO2 anaesthesia and subsequent exsanguination via an incision of the ventral aspect of the throat. The chest of the cadaver was opened, the phrenic nerve hemidiaphragm tissue was explanted and placed into an organ bath. The phrenic nerve was continuously stimulated at 5–25 mA with a frequency of 1 Hz and with a 0.1 ms pulse duration. Isometric contractions were transformed using a force transducer and recorded with VitroDat Online software (FMI GmbH, Germany). The time required to decrease the amplitude to 50% of the starting value (paralytic half-time) was determined. To allow comparison of the altered neurotoxicity of mutants with H6tBoNTA wild-type, a power function (y(H6tBoNTA; 10, 30, 80 pM) = 139.6x−0.1957, R2 = 0.9991) was fitted to a concentration-response-curve consisting of three concentrations determined in 3–6 technical replicates. Resulting paralytic half-times of the H6tBoNTA mutants were converted to concentrations of the wild-type employing the above power functions and finally expressed as relative neurotoxicity.
Supplementary Material
Acknowledgments
This work was partly supported by National Institute of Allergy and Infectious Diseases (NIAID) grants R01AI091823 and R21AI123920 to R.J. and R01AI096169 to M.K; by National Institute of Neurological Disorders and Stroke (NINDS) Grant R01NS080833 to M.D.; and by the Bundesministerium für Bildung und Forschung grants FK031A212A to A.R. and FK031A212B to Brigitte G. Dorner (RKI). NE-CAT at the Advanced Photon Source (APS) is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
Accession codes. Atomic coordinates and structure factors for the HCA–bSV2C and HCA–gSV2C complexes have been deposited in the Protein Data Bank under accession codes 5JMC and 5JLV, respectively.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
G.Y. and S.M. performed the cloning and mutagenesis. G.Y., K.L. and R.J. carried out the protein expression, purification, characterization and crystallographic studies. K.P. collected the X-ray diffraction data. S.Z. and M.D. performed all experiments on cultured neurons. A.R. and S.M. generated the full-length BoNT/A1 mutants and performed the MPN assay with support of Nadja Krez. Dr. Jasmin Weisemann cloned HCHA. D.S., K.B. and M.K. performed the SPR studies. R.J., M.D. and A.R. wrote the manuscript with input from other authors.
References
- 1.Montecucco C. How do tetanus and botulinum neurotoxins bind to neuronal membranes? Trends Biochem Sci. 1986;11:314–17. [Google Scholar]
- 2.Montecucco C, Rossetto O, Schiavo G. Presynaptic receptor arrays for clostridial neurotoxins. Trends Microbiol. 2004;12:442–6. doi: 10.1016/j.tim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Rummel A. Double Receptor Anchorage of Botulinum Neurotoxins Accounts for their Exquisite Neurospecificity. Curr Top Microbiol Immunol. 2013;364:61–90. doi: 10.1007/978-3-642-33570-9_4. [DOI] [PubMed] [Google Scholar]
- 4.Dong M, et al. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–6. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 5.Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 2006;580:2011–4. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 6.Dong M, et al. Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol Biol Cell. 2008;19:5226–37. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng L, Tepp WH, Johnson EA, Dong M. Botulinum neurotoxin D uses synaptic vesicle protein SV2 and gangliosides as receptors. PLoS Pathog. 2011;7:e1002008. doi: 10.1371/journal.ppat.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rummel A, et al. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–54. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- 9.Fu Z, Chen C, Barbieri JT, Kim JJ, Baldwin MR. Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F. Biochemistry. 2009;48:5631–41. doi: 10.1021/bi9002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benoit RM, et al. Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A. Nature. 2014;505:108–11. doi: 10.1038/nature12732. [DOI] [PubMed] [Google Scholar]
- 11.Chai Q, et al. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 12.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–5. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 13.Janz R, Sudhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience. 1999;94:1279–90. doi: 10.1016/s0306-4522(99)00370-x. [DOI] [PubMed] [Google Scholar]
- 14.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–9. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 15.Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol Microbiol. 2004;51:631–43. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- 16.Pirazzini M, Rossetto O, Bolognese P, Shone CC, Montecucco C. Double anchorage to the membrane and intact inter-chain disulfide bond are required for the low pH induced entry of tetanus and botulinum neurotoxins into neurons. Cell Microbiol. 2011;13:1731–43. doi: 10.1111/j.1462-5822.2011.01654.x. [DOI] [PubMed] [Google Scholar]
- 17.Swiech K, Picanco-Castro V, Covas DT. Human cells: new platform for recombinant therapeutic protein production. Protein Expr Purif. 2012;84:147–53. doi: 10.1016/j.pep.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 18.Croset A, et al. Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. J Biotechnol. 2012;161:336–48. doi: 10.1016/j.jbiotec.2012.06.038. [DOI] [PubMed] [Google Scholar]
- 19.Bigalke H, Rummel A. Botulinum Neurotoxins: Qualitative and Quantitative Analysis Using the Mouse Phrenic Nerve Hemidiaphragm Assay (MPN) Toxins (Basel) 2015;7:4895–905. doi: 10.3390/toxins7124855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strotmeier J, et al. Identification of the synaptic vesicle glycoprotein 2 receptor binding site in botulinum neurotoxin A. FEBS Lett. 2014;588:1087–93. doi: 10.1016/j.febslet.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossetto O, Pirazzini M, Montecucco C. Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat Rev Microbiol. 2014;12:535–49. doi: 10.1038/nrmicro3295. [DOI] [PubMed] [Google Scholar]
- 22.Barash JR, Arnon SS. A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J Infect Dis. 2014;209:183–91. doi: 10.1093/infdis/jit449. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Escalona N, et al. Draft Genome Sequence of Bivalent Clostridium botulinum Strain IBCA10-7060, Encoding Botulinum Neurotoxin B and a New FA Mosaic Type. Genome Announc. 2014;2 doi: 10.1128/genomeA.01275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalb SR, et al. Functional characterization of botulinum neurotoxin serotype H as a hybrid of known serotypes F and A (BoNT F/A) Anal Chem. 2015;87:3911–7. doi: 10.1021/ac504716v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maslanka SE, et al. A Novel Botulinum Toxin, Previously Reported as Serotype H, has a Hybrid Structure of Known Serotypes A and F that is Neutralized with Serotype A Antitoxin. J Infect Dis. 2015 doi: 10.1093/infdis/jiv327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Y, et al. Immunological Characterization and Neutralizing Ability of Monoclonal Antibodies Directed Against Botulinum Neurotoxin Type H. J Infect Dis. 2016 doi: 10.1093/infdis/jiv770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kull S, et al. Isolation and functional characterization of the novel Clostridium botulinum neurotoxin A8 subtype. PLoS One. 2015;10:e0116381. doi: 10.1371/journal.pone.0116381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitemarsh RC, et al. Characterization of botulinum neurotoxin A subtypes 1 through 5 by investigation of activities in mice, in neuronal cell cultures, and in vitro. Infect Immun. 2013;81:3894–902. doi: 10.1128/IAI.00536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Rodriguez C, et al. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–16. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 30.Nayak SU, et al. Safety and pharmacokinetics of XOMA 3AB, a novel mixture of three monoclonal antibodies against botulinum toxin A. Antimicrob Agents Chemother. 2014;58:5047–53. doi: 10.1128/AAC.02830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 32.Rouvinski A, et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015 doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 33.Garces F, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong L, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luca VC, et al. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–53. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–86. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 37.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumiya S, et al. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol. 2007;368:767–79. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–5. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–84. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.