Abstract
A current focus in psychiatric genetics is detection of multiple common risk alleles through very large GWAS analyses. Yet families do exist, albeit rare, that have multiple affected members who are presumed to have a similar inherited cause to their illnesses. We hypothesized that within some of these families there may be rare highly penetrant mutations that segregate with illness. In this exploratory study, the genomes of ninety individuals across nine families were sequenced. Each family included a minimum of three available relatives affected with a psychotic illness and three available unaffected relatives. Twenty-six variants were identified that are private to a family, alter protein sequence, and are transmitted to all affected individuals within the family. In one family, seven siblings with schizophrenia spectrum disorders each carry a novel private missense variant within the SHANK2 gene. This variant lies within the consensus SH3 protein-binding motif by which SHANK2 may interact with post-synaptic glutamate receptors. In another family, four affected siblings and their unaffected mother each carry a novel private missense variant in the SMARCA1 gene on the X chromosome. Both variants represent candidates that may be causal for psychotic disorders when considered in the context of their transmission pattern and known gene and disease biology.
Introduction
Recently, considerable advances have been made in the understanding of the genetics of psychiatric disorders, particularly schizophrenia. A Genome-Wide Association Study (GWAS) meta-analysis of 39,989 individuals with schizophrenia identified 108 loci with risk alleles for schizophrenia1. Notably, the risk alleles were all of modest effect, supporting a model of a highly polygenic disease characterized by small cumulative effects of a large number of common risk alleles. This effort was complemented by two recent exome sequencing studies: one searching for rare variants among 2,536 individuals with schizophrenia2, and the other searching for de novo causative mutations among 623 trios3. The rare variant and de novo studies did not implicate any specific gene, but supported the role of synaptic pathways and the immune system in the etiology of the disease.
Collectively, these studies suggest that a large number of genes can contribute to disease risk and that no particular gene is specifically required or strongly over-represented among the risk loci. While many instances of schizophrenia may be polygenic (i.e., resulting from cumulative effect of modest-effect alleles), a subset may arise from single rare high-penetrance variants, possibly drawn from the large pool of genes influencing neurodevelopmental pathways. For example, several rare Copy Number Variants (CNVs) with a strong contribution to disease risk have been identified4, 5, a risk possibly amplified by somatic copy number variation in the brain6. The high heritability ascertained from twin studies, however, suggests a degree of schizophrenia heritability that is not yet accounted for7. Here, we extend the search for rare highly-penetrant causative variants by focusing on families with a high prevalence of schizophrenia, as they may be more likely to harbor such variants. This approach has limitations, but can serve to suggest candidate variants that may be of particular value in the development of experimental models for schizophrenia and drug target discovery. The results reported here were obtained from whole genome sequencing of 83 individuals in 9 families. Variants identified include family private candidate variants in the SHANK2 post-synaptic density scaffolding protein and the SMARCA1 transcriptional regulator that might reasonably be expected to disrupt neuronal development or signaling.
Methods
Sample acquisition
Families with at least three members affected with schizophrenia were identified through advertisements placed primarily in local support group newsletters throughout the USA. The National Alliance for Mental Illness Chapters, initially in the Boston area and then throughout the USA, were approached for referrals and to advertise this study in their local newsletters. Several years ago, LE DeLisi also identified and evaluated families in a similar manner, and cell lines from these families were stored at the Coriell Institute in Camden, New Jersey. For the current analyses, five families’ cell lines (pedigrees p1250, p1271, p1274, p1333, and pSB285) were obtained from the Coriell Institute collection. Whole blood was obtained from an additional four families (pedigrees pVA02, pVA03, pVA04, and pVA07) from the later collection initiated in 2013. IRB approval was obtained at each institution where the data were gathered, and the overall study is currently approved by the VA Boston Healthcare System local IRB. All individuals signed written informed consent for their blood sample to be used for finding genes for risk for schizophrenia. All samples and corresponding clinical information were coded using both family and unique individual codes to mask identities. All individuals were interviewed using the Diagnostic Interview for Genetic Studies (http://www.nimhgenetics.org/interviews/digs_2.0/digs2.0.pdf), and diagnoses were made based on DSM-IV criteria by two independent investigators and consensus with a third if necessary.
Sample ancestry
Ancestry of individuals was assessed using iAdmix software (v0.2)8 with the bundled HapMap 39 population allele frequencies. All individuals most closely resembled the CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) and TSI (Toscani in Italia) populations, with the sum of the remaining population frequencies never exceeding 6% in any individual. Individuals in the pVA07 pedigree were most similar to TSI (68-78%), while the other individuals were most similar to CEU, with the exception of pVA04 individual 4, who was calculated to be 46% CEU, 51% TSI, 2% JPT (Japanese in Tokyo, Japan). When interviewed, these families all described their ancestry as solely European.
Sample preparation and sequencing
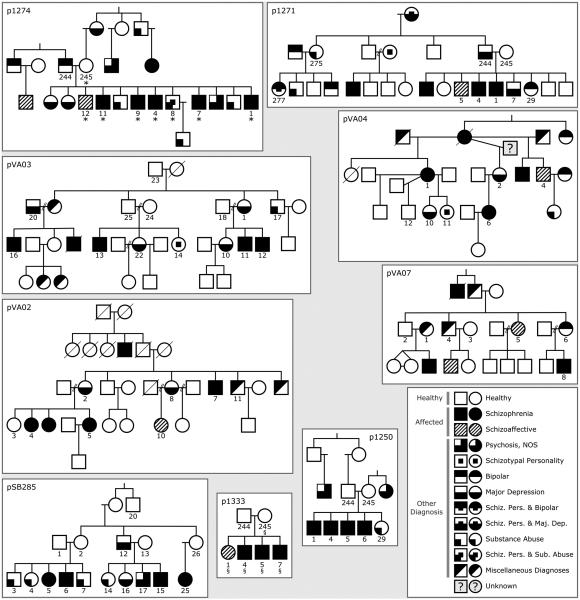
Cryopreserved B-lymphoblastoid cell lines were obtained from the Coriell Institute for Medical Research (Camden, NJ). Cells were thawed and grown in RPMI 1640 base medium supplemented with 15% fetal bovine serum in a T-75 flask, and placed in a humidified growth chamber at 37°C and 5% CO2. Cells were harvested at a density of 5×106 to 1×107 cells. Genomic DNA was isolated from cells using the Gentra Puregene cell kit (Qiagen, Inc.). Cell pellets were sent to Expression Analysis (Durham, NC) for genomic DNA isolation. Genomic DNA libraries were prepared and sequenced by deCODE Genetics in Iceland (Illumina 2×150 bp; 340-450 million reads/sample; 20-45x median CDS coverage). A total of nine families and 90 DNA samples were sequenced. After sequencing, QC analyses were performed and 83 samples remained (Figure 1; Supplementary Methods). For the purposes of reproducing our results, raw sequencing data are available on request.
Figure 1. Pedigrees selected for whole genome sequencing.
Numbered individuals were sequenced. The numbers are arbitrary identifiers and are unique within each family. *Individuals with SHANK2 Variant. §Individuals with SMARCA1 variant. Within the diagnostic key, "Miscellaneous Diagnoses" represents diagnoses not otherwise mentioned, such as "anxiety disorder", anorexia, and non-schizophrenia-related personality disorders. However, no family members were diagnosed with autism or mental retardation. The diagnostic key also delineates the grouping of diagnoses into classes designated “healthy”, “affected”, and “other diagnosis”.
Sequence analysis
Detailed documentation of sequence analysis and quality control measures are provided in the Supplementary Methods. In brief, SNP and INDEL discovery was performed in adherence to the May 2015 version of the Genome Analysis Toolkit (GATK)10 Best Practices recommendations11, 12. Reads were aligned to the hg19 reference genome using BWA13, and then GATK was used to perform base quality score recalibration, indel realignment, duplicate removal, variant discovery across the full sample set, and filtering using variant quality score recalibration. Copy number variants were discovered using Genome STRiP (v2.00.1533)14, and other structural variants were discovered using Lumpy (v0.2.11)15. Variants were annotated using the Ensembl Variant Effect Predictor16, and SNPs and INDELs were then categorized according to rarity using variant calls and frequency annotations obtained from dbSNP (build v142)17, 1000 genomes (release v5a)18, and the Exome Aggregation Consortium (ExAC; Cambridge, MA; release r0.3; http://exac.broadinstitute.org). Annotation and manipulation of the VCF format variant calls utilized custom Java code and the Picard-tools Java library (v1.128; http://broadinstitute.github.io/picard/). We have focused this study on private and very rare variants within individual families that statistically represent very small numbers, and thus were not able to describe an odds ratio or a statistical probability of the null hypothesis. Seven sequenced samples were omitted from the analysis due to QC issues (see Supplementary Methods). Variant calls for both SHANK2 and SMARCA1 were verified with Sanger sequencing for each member of pedigrees p1274 and p1333. The two families served as negative controls for one another, as the SHANK2 variant was specific to pedigree p1274 and the SMARCA1 variant was specific to pedigree p1333.
Code Availability
The code and precise parameters used to run the DNA-Seq alignment and variant calling pipeline are described in detail in the Supplementary Methods. Custom code – used primarily for binning variants by rarity and quality control – is not provided, but is described in sufficient detail in the Supplementary Methods to reproduce the analysis.
Results
The total number of sequenced individuals remaining after application of quality control measures is shown for each pedigree in Table 1. Individuals diagnosed as schizophrenia or schizoaffective were classified as “affected” and served as the basis for candidate variant identification. Although we recognize that some mutations may cross diagnostic boundaries and also contribute to affective disorders, autism, and mental retardation, we chose not to include any affective disorders as "affected" in this initial exploratory search. No individuals in our families had diagnoses of autism or mental retardation. The analysis focused on variants that were (1) predicted to alter protein sequence, (2) present in all sequenced affected individuals, and (3) unique to that pedigree. These variants were classified as family private, and all 26 such variants are listed in Table 2. None of these variants are homozygous in the sequenced individuals, and none overlap with the 108 association loci identified by the recent large meta-GWAS study1 or the set of de novo mutations in the recent trio exome-seq study3. To address the possibility that the uncommonly high disease incidence in our ascertained families simply resulted from unusual concentrations of common risk alleles, all sequenced individuals were analyzed for presence of common protective or risk alleles at index SNPs identified as markers of schizophrenia-associated loci defined with GWAS (see Supplementary Methods). Every individual carried a close balance of “risk” and “protective” common variants, and within each family the proportion of risk and common variants was the same in diseased and healthy individuals, making common variation an unlikely source of disease. For extended analysis, we also considered more relaxed selection criteria, such as including variants with low population-level allele frequencies and instances in which a variant is found in all but one affected individual (see Supplementary Table for extended list of variants and annotations). It is entirely possible that variants that segregate still less strongly with phenotype could contribute or predispose to disease. The filtering we describe below is of necessity somewhat arbitrary, and a full catalog of rare coding variants is provided in the Supplementary Table.
Table 1.
Counts of sequenced individuals and variants by family, disease status, and variant classification.
| # Sequenced | # Variants in all Affected |
||||
|---|---|---|---|---|---|
|
|
|||||
| Pedigree | Affected† | Other Diagnosis‡ |
Healthy | Family Private | Rare |
| p1250 | 4 | 1 | 2 | 2 | 12 |
|
| |||||
| p1271 | 3 | 5 | 1 | 9 | 29 |
|
| |||||
| p1274 | 6 | 2 | 1 | 4 | 9 |
|
| |||||
| p1333 | 4 | 0 | 2 | 4 | 16 |
|
| |||||
| pSB285 | 4 | 7 | 5 | 0 | 2 |
|
| |||||
| pVA02 | 4 | 3 | 1 | 1 | 1 |
|
| |||||
| pVA03 | 4 | 6 | 4 | 0 | 0 |
|
| |||||
| pVA04 | 3 | 3 | 1 | 3 | 6 |
|
| |||||
| pVA07 | 2 | 3 | 2 | 3 | 61 |
Diagnosis of ‘schizophrenia’ or ‘schizoaffective’
Diagnosis other than ‘healthy’, ‘schizophrenia’, or ‘schizoaffective’
Table 2.
“Family private” variants present in all affected individuals.
| Pedigree | Gene | Affected † |
Healthy | Other Diagnosis ‡ |
Genomic Position | Variant | Impact§ |
|---|---|---|---|---|---|---|---|
| p1250 | KIT | 4 | 1 of 2 | 1 of 1 | 4q12:55,603,415 T > C | I924T | Neutral (0.86) |
|
| |||||||
| p1250 | HEXA | 4 | 1 of 2 | 1 of 1 | 15q23:72,638,995 C > T | M401I | Neutral (0.76) |
|
| |||||||
| p1271 | ZMYM6 | 3 | 1 of 1 | 2 of 5 | 1p34.3:35,453,774 A > C | L970R | Deleterious (1) |
|
| |||||||
| p1271 | SPAG17 | 3 | 1 of 1 | 1 of 5 | 1p12:118,509,369 G > C | A2132G | Deleterious (1) |
|
| |||||||
| p1271 | TCP1 | 3 | 0 of 1 | 2 of 5 | 6q25.3:160,201,497 C > G | D359H | Deleterious (0.99) |
|
| |||||||
| p1271 | STAG3 | 3 | 1 of 1 | 1 of 5 | 7q22.1:99,794,880 T > G | I348S | Deleterious (1) |
|
| |||||||
| p1271 | STK33 | 3 | 0 of 1 | 3 of 5 | 11p15.4:8,588,781 TCTGGTTCAGAGCTCA > T |
<splice> | n/a |
|
| |||||||
| p1271 | DDX55 | 3 | 1 of 1 | 1 of 5 | 12q24.31:124,086,789 A > G | M32V | Neutral (0.93) |
|
| |||||||
| p1271 | NTRK3 | 3 | 0 of 1 | 2 of 5 | 15q25.3:88,472,466 C > G | D697H | Deleterious (1) |
|
| |||||||
| p1271 | GREB1L | 3 | 1 of 1 | 1 of 5 | 18q11.2:19,093,839 A > G | N1598S | Deleterious (1) |
|
| |||||||
| p1271 | CEACAM16 | 3 | 0 of 1 | 3 of 5 | 19q13.32:45,211,144 G > A | A318T | Neutral (0.79) |
|
| |||||||
| p1274 | STIP1 | 6 | 1 of 1 | 1 of 2 | 11q13.1:63,963,130 C > G | P173A | Neutral (0.97) |
|
| |||||||
| p1274 | SHANK2 | 6 | 1 of 1 | 1 of 2 | 11q13.4:70,644,595 G > A | A578V | Deleterious (1) |
|
| |||||||
| p1274 |
RP11-
796G6.2 |
6 | 0 of 1 | 2 of 2 | 14q32.31:102,198,574 G > A | W112* | n/a |
|
| |||||||
| p1274 | DEF8 | 6 | 0 of 1 | 1 of 2 | 16q24.3:90,030,588 G > A | R399Q | n/a Deleterious (1) |
|
| |||||||
| p1333 | RNASEL | 4 | 1 of 2 | 0 of 0 | 1q25.3:182,545,525 C > G | <splice> | n/a |
|
| |||||||
| p1333 | KIAA1524 | 4 | 1 of 2 | 0 of 0 | 3q13.13:108,301,916 T > C | T89A | Neutral (0.85) |
|
| |||||||
| p1333 | AVEN | 4 | 1 of 2 | 0 of 0 | 15q14:34,331,202 C > T | G16S | Deleterious (1) |
|
| |||||||
| p1333 | SMARCA1 | 4 | 1 of 2 | 0 of 0 | Xq25:128,638,728 C > T | V384M | Deleterious (1) |
|
| |||||||
| pVA02 | TGM6 | 4 | 0 of 1 | 2 of 3 | 20p13:2,384,113 G > A | V354I | Deleterious (0.98) |
|
| |||||||
| pVA04 | HSD17B4 | 4 | 0 of 1 | 2 of 3 | 5q23.1:118,835,147 G > C | D395H | Neutral (0.86) |
|
| |||||||
| pVA04 | MAN1A1 | 4 | 1 of 1 | 3 of 3 | 6q22.31:119,670,217 C > T | G5D | Neutral (0.87) |
|
| |||||||
| pVA04 | ALPK3 | 4 | 0 of 1 | 2 of 3 | 15q25.3:85,400,551 G > | S1063N | Deleterious (0.99) |
|
| |||||||
| pVA07 | BEND7 | 2 | 1 of 2 | 2 of 3 | 10p13:13,534,853 G > T | P147T | Neutral (0.92) |
|
| |||||||
| pVA07 | WDFY4 | 2 | 1 of 2 | 2 of 3 | 10q11.23:49,982,576 T > A | V876D | Deleterious (1) |
|
| |||||||
| pVA07 | WDR25 | 2 | 1 of 2 | 2 of 3 | 14q32.2:100,992,319 G > A | R405H | Deleterious (1) |
In order to find rare variants that may be causal for schizophrenia, we further evaluated the 26 “family private” SNPs and INDELs in the context of supporting data, including predicted deleterious impact, transmission genetics, presence in individuals with other diagnoses (i.e. non-healthy and non-affected), and functional characterization. First, we considered the three potential disruptive variants. Family p1271 harbors the only non-SNP variant in our candidate variant set, a 15bp deletion that disrupts the splice donor site of the serine/threonine kinase STK33. However, the impacted splice site joins two untranslated 5’ exons in a non-canonical splice variant of dubious validity. Family p1333 harbors a splice acceptor mutation in RNASEL (ribonuclease L), present in all four affected children and transmitted through the healthy father. RNASEL is involved in interferon antiviral response19, but no obvious connection to schizophrenia or neuronal biology has been reported. The final disruptive variant is a stop gain variant in the RP11-796G6.2 gene of Family p1274, but this gene appears to have been reclassified as a lincRNA in Ensembl GRCh38 (release 80)20.
The missense SNPs predicted to have deleterious impact on protein function (Table 2; column labeled ‘Impact’) were also examined. Family p1271 harbors six such variants. Four were dismissed (ZMYM6, SPAG17, STAG3, and GREB1L) as inconsistent with our transmission hypothesis, as they were all transmitted from the married-in mother (individual 245; also the case for the neutral variant in DDX55). The remaining two variants impact the genes TCP1 and NTRK3. Connections between TCP1 and schizophrenia are tenuous. TCP1 is a molecular chaperone thought to be associated with schizophrenia in a Han Chinese population21, but a follow-up attempt to replicate the result failed22. TCP1 was also reported to be significantly downregulated in a small comparison of schizophrenic and healthy postmortem brain samples23. NTRK3 is a neurotrophic tyrosine kinase receptor with reported associations with bipolar disorder24, 25 and schizophrenia26.
Family p1274 harbors two missense SNPs that are predicted to be deleterious. The first, in the gene DEF8, was transmitted through the bipolar father, and the second, in SHANK2, was transmitted through the healthy mother. DEF8 is very poorly characterized, but does exhibit elevated expression in the brain relative to most other normal tissues (GTEx consortium RNA-Seq dataset27). SHANK2 is a post-synaptic scaffolding protein at glutamatergic synapses with several connections to schizophrenia, and the A578V variant localizes to the SH3 consensus protein-binding domain28-31.
Family p1333 also harbors two potentially deleterious missense SNPs, located in the genes AVEN and SMARCA1. AVEN is a caspase activation inhibitor 32 with a possible schizophrenia association reported in a small fine-mapping study of chromosomal region 15q13-q1433. SMARCA1 resides on the X chromosome, and thus exists in a hemizygous state in the three affected males and a heterozygous state subject to X inactivation in the unaffected mother and schizoaffective daughter. SMARCA1 is a member of the SWI/SNF family of chromatin remodeling genes and has multiple connections to neurogenesis and schizophrenia (see Discussion).
The two candidate genes in family pVA07, WDFY4 and WDR25, are both poorly characterized. Analysis of this family was compromised by QC issues that reduced the number of sequenced affected individuals to two. In family pVA04, a single family private potentially deleterious variant in the gene ALPK3 was identified. This kinase has no known connection to neurobiology. Finally, in family pVA02, the gene TGM6 was identified as a potential candidate. TGM6, a member of the transglutaminase family, may play a role in neurogenesis34 and has been implicated in dominant spinocerebellar ataxias35.
Existing algorithms for discovery and analysis of structural variants in whole genome sequence are far less robust than those for SNPs and small INDELs. Preliminary attempts to identify structural variants yielded numerous probable false positives (see Supplementary Methods), and were likely further compromised by false negatives and inexact breakpoint localization. However, one interesting family private chr6:chr11 translocation with strong support from reads spanning the predicted breakpoint was observed. This structural variant was transmitted from the healthy mother to all affected children in family p1333 and disrupts the TEAD1 gene, which encodes a transcriptional activator.
Discussion
Through whole genome sequencing of multiplex families, 25 SNPs and one small deletion were identified that potentially alter or disrupt protein sequence, are private to a single family, and are observed in all sequenced affected individuals. This study was not intended as an exhaustive attempt to assign a genetic basis for disease to each pedigree, but rather as an exploratory search designed to identify rare instances in which the schizophrenia spectrum phenotype might result from a single high penetrance variant. This approach has yielded several variants with strong contextual support in the form of pre-existing mechanistic and genetic data, among which the SHANK2 variant in family p1274 is perhaps the most compelling. Rare predicted-damaging variants segregating with disease but without such contextual support were also found (Table 2), and from this study alone we do not exclude these variants as playing a role in disease. If relevant to disease, such variants may play an undiscovered role in neuronal development or signaling.
Along with gene family members SHANK1 and SHANK3, SHANK2 is an integral part of the post-synaptic architecture of glutamatergic synapses. Recently, Peykov and colleagues36 reported a significant enrichment of rare and common SHANK2 variants in schizophrenic individuals and demonstrated that some variants impaired synaptic clustering in hippocampal neurons. Unlike the variants in the Peykov study, the A578V SHANK2 variant identified in this study lies within the SH3 domain and thus might result in an even more severe phenotype. The Peykov study variants do share one feature with the A578V variant: the observed transmission was from heterozygous unaffected mother to heterozygous affected son (for all five variants that the study was able to trace). While this trend might arise by chance, it is also possible that the transmission from healthy mother to seven sons observed in family p1274 reflects an as yet uncharacterized sex-dependent influence on developmental pathways.
SHANK2 has also previously been reported as being associated with autism, intellectual disability, and other neuropsychiatric disorders37, 38, as has SHANK139. Interestingly, one familial autism-associated variant included deletion of the SH3 domain, and a population study found overrepresentation of missense variant S557N within SH3 in autism probands40. It is unknown how different variants in SHANK2, even within the SH3 domain, might predispose differently to autism, intellectual disability, or schizophrenia - clinically distinct syndromes that in some instances are enriched for the same or similar genetic associations. However, these variants offer tools for further study. Within the SHANK family, SHANK3 also has been implicated in schizophrenia via rare missense and stop-gain variants41, 42, and a noncoding variant near SHANK1 has been linked to mild cognitive deficits in schizophrenia43.
While SHANK2 represents a compelling candidate for a high penetrance variant, it alone cannot fully explain disease transmission within the p1274 family. The bipolar father (individual 244; Figure 1) does not harbor the SHANK2 variant, yet he also has a bipolar brother and schizoaffective nephew. It is possible that the family private DEF8 variant, which was transmitted through the father, contributes to the phenotype. However this gene is very poorly characterized and lacks the preponderance of connections to synaptic function and neurodevelopmental disorders of the SHANK2 variant. More broadly, this ambiguity reflects the limitations of the multiplex variant analysis, which involves too few samples to power statistical genetic approaches. In many instances the families might have been enriched for high background levels of variants of low effect size, and in others high-penetrance variants might have escaped notice because they resided in regulatory regions, involved structural variants that evaded detection, or were obscured by our stringent selection criteria.
Several of the other families also harbored variants that represent compelling candidates, though perhaps none as compelling as SHANK2. Multiple variants occur in genes that have previously been reported as having some connection to neurodevelopment or schizophrenia, including NTRK3, SMARCA1, TCP1, and TGM6. However, while interesting, these associations should be viewed with caution, as the number of genes that have some such link within the vast body of literature is no doubt quite large. SMARCA1 is worthy of special note, as similar to the SHANK family, the SMARCA protein family has multiple plausible connections to schizophrenia and brain function. SMARCA1 has been implicated in neurogenesis in several mouse studies44-46, and in humans a loss of function mutation was recently identified in an individual suffering from microcephaly and intellectual disability47. Furthermore, gene family member SMARCA2 has previously been implicated in schizophrenia48, 49. The SMARCA1 variant in family p1333 is X-linked, and was the only gene in our list of top candidates (Table 2) that had no wild-type allele expressed in the affected individuals (with the possible exception of the affected daughter, subject to X-inactivation state).
While functional effects of these variants are untested, and this study was not designed to conclusively identify high penetrance disease causing variants, we have identified several plausible candidates, including two particularly compelling candidates in SHANK2 and SMARCA1. Thus, the multiplex pedigree sequencing strategy can be considered as a useful complement to the larger sample size GWAS and rare variant studies that don't examine individuals from the same family. The observations from the pedigrees may make valuable contributions towards understanding the biology of specific genes and biological pathways implicated in schizophrenia.
The identification of multiplex families for genetic studies, however, is difficult, not because they are rare, but rather because the affected individuals are frequently unavailable for study due to suicide, family disruption, and the nature of the illness itself. While it is known that family studies as a whole have calculated morbid risks for schizophrenia as only 8-10 percent to specific first degree relatives50, other studies surveying family history in general in people with schizophrenia find a much higher percentage of positive history51. Multiplex families are potentially enriched for extreme and high penetrance phenotypes, yielding variants that are experimentally useful for future studies. For example, mice with disrupted SHANK2 are viable and show behavioral abnormalities possibly correctable pharmacologically52-54, which is encouraging for potential use of the SHANK2 pathway to generate tools that reflect molecular and cellular mechanisms of schizophrenia.
Supplementary Material
Acknowledgements
The pedigree collections were partially supported by NIMH from 1992-1999 (MHR01 44245) and more recently by Amgen 2013-present.
Footnotes
Conflicts of Interest
Dr. DeLisi has received funds from Amgen for clinical evaluations of multiplex families. All other co-authors are employees of Amgen. The company, however, did not influence the study design, analyses, or interpretation of the results presented in this manuscript.
Supplementary information is available at Molecular Psychiatry’s website.
References
- 1.Schizophrenia Working Group of the Psychiatric Genomics C Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pocklington AJ, Rees E, Walters JT, Han J, Kavanagh DH, Chambert KD, et al. Novel Findings from CNVs Implicate Inhibitory and Excitatory Signaling Complexes in Schizophrenia. Neuron. 2015;86(5):1203–1214. doi: 10.1016/j.neuron.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL, et al. Analysis of copy number variations at 15 schizophrenia-associated loci. The British journal of psychiatry : the journal of mental science. 2014;204(2):108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, et al. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of general psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 8.Bansal V, Libiger O. Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC bioinformatics. 2015;16:4. doi: 10.1186/s12859-014-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International HapMap C The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al] 2013;11(1110):11 10 11–11 10 33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, et al. Large multiallelic copy number variations in humans. Nature genetics. 2015;47(3):296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome biology. 2014;15(6):R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics (Oxford, England) 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. The EMBO journal. 1993;12(8):3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2014. Nucleic acids research. 2014;42:D749–755. doi: 10.1093/nar/gkt1196. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang MS, Yu L, Guo TW, Zhu SM, Liu HJ, Shi YY, et al. Evidence for association between single nucleotide polymorphisms in T complex protein 1 gene and schizophrenia in the Chinese Han population. Journal of medical genetics. 2004;41(5):e63. doi: 10.1136/jmg.2003.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang W, Shi Y, Feng G, Yan L, Xing Y, Zhu S, et al. Family-based association studies of the TCP1 gene and schizophrenia in the Chinese Han population. Journal of neural transmission. 2006;113(10):1537–1543. doi: 10.1007/s00702-005-0419-9. [DOI] [PubMed] [Google Scholar]
- 23.Chu TT, Liu Y. An integrated genomic analysis of gene-function correlation on schizophrenia susceptibility genes. Journal of human genetics. 2010;55(5):285–292. doi: 10.1038/jhg.2010.24. [DOI] [PubMed] [Google Scholar]
- 24.Athanasiu L, Mattingsdal M, Melle I, Inderhaug E, Lien T, Agartz I, et al. Intron 12 in NTRK3 is associated with bipolar disorder. Psychiatry research. 2011;185(3):358–362. doi: 10.1016/j.psychres.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Nurnberger JI, Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA psychiatry. 2014;71(6):657–664. doi: 10.1001/jamapsychiatry.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otnaess MK, Djurovic S, Rimol LM, Kulle B, Kahler AK, Jonsson EG, et al. Evidence for a possible association of neurotrophin receptor (NTRK-3) gene polymorphisms with hippocampal function and schizophrenia. Neurobiology of disease. 2009;34(3):518–524. doi: 10.1016/j.nbd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 27.The Genotype-Tissue Expression (GTEx) project Nature genetics. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. The Journal of biological chemistry. 1999;274(41):29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- 29.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23(3):569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 30.Pawson T, Schlessingert J. SH2 and SH3 domains. Current biology : CB. 1993;3(7):434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 31.Sheng M, Kim E. The Shank family of scaffold proteins. Journal of cell science. 2000;113:1851–1856. doi: 10.1242/jcs.113.11.1851. Pt 11. [DOI] [PubMed] [Google Scholar]
- 32.Chau BN, Cheng EH, Kerr DA, Hardwick JM. Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Molecular cell. 2000;6(1):31–40. [PubMed] [Google Scholar]
- 33.Stephens SH, Franks A, Berger R, Palionyte M, Fingerlin TE, Wagner B, et al. Multiple genes in the 15q13-q14 chromosomal region are associated with schizophrenia. Psychiatric genetics. 2012;22(1):1–14. doi: 10.1097/YPG.0b013e32834c0c33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas H, Beck K, Adamczyk M, Aeschlimann P, Langley M, Oita RC, et al. Transglutaminase 6: a protein associated with central nervous system development and motor function. Amino acids. 2013;44(1):161–177. doi: 10.1007/s00726-011-1091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain : a journal of neurology. 2010;133(Pt 12):3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 36.Peykov S, Berkel S, Schoen M, Weiss K, Degenhardt F, Strohmaier J, et al. Identification and functional characterization of rare SHANK2 variants in schizophrenia. Molecular psychiatry. 2015 doi: 10.1038/mp.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nature genetics. 2010;42(6):489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 38.Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P, et al. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Human molecular genetics. 2012;21(2):344–357. doi: 10.1093/hmg/ddr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S, et al. SHANK1 Deletions in Males with Autism Spectrum Disorder. American journal of human genetics. 2012;90(5):879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS genetics. 2012;8(2):e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature genetics. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(17):7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lennertz L, Wagner M, Wolwer W, Schuhmacher A, Frommann I, Berning J, et al. A promoter variant of SHANK1 affects auditory working memory in schizophrenia patients and in subjects clinically at risk for psychosis. European archives of psychiatry and clinical neuroscience. 2012;262(2):117–124. doi: 10.1007/s00406-011-0233-3. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez-Saavedra M, De Repentigny Y, Lagali PS, Raghu Ram EV, Yan K, Hashem E, et al. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nature communications. 2014;5:4181. doi: 10.1038/ncomms5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metzakopian E, Bouhali K, Alvarez-Saavedra M, Whitsett JA, Picketts DJ, Ang SL. Genome-wide characterisation of Foxa1 binding sites reveals several mechanisms for regulating neuronal differentiation in midbrain dopamine cells. Development. 2015;142(7):1315–1324. doi: 10.1242/dev.115808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yip DJ, Corcoran CP, Alvarez-Saavedra M, DeMaria A, Rennick S, Mears AJ, et al. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Developmental cell. 2012;22(4):871–878. doi: 10.1016/j.devcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, et al. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88(3):499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Human molecular genetics. 2009;18(13):2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- 49.Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H, et al. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Human molecular genetics. 2010;19(14):2841–2857. doi: 10.1093/hmg/ddq184. [DOI] [PubMed] [Google Scholar]
- 50.Gershon ES, DeLisi LE, Hamovit J, Nurnberger JI, Jr., Maxwell ME, Schreiber J, et al. A controlled family study of chronic psychoses. Schizophrenia and schizoaffective disorder. Archives of general psychiatry. 1988;45(4):328–336. doi: 10.1001/archpsyc.1988.01800280038006. [DOI] [PubMed] [Google Scholar]
- 51.Proal AC, Fleming J, Galvez-Buccollini JA, Delisi LE. A controlled family study of cannabis users with and without psychosis. Schizophrenia research. 2014;152(1):283–288. doi: 10.1016/j.schres.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78(1):8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486(7402):256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 54.Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486(7402):261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- 55.Lopes MC, Joyce C, Ritchie GR, John SL, Cunningham F, Asimit J, et al. A combined functional annotation score for non-synonymous variants. Human heredity. 2012;73(1):47–51. doi: 10.1159/000334984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 57.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.