Abstract
The expression of biliary/progenitor markers by hepatocellular carcinoma (HCC) is often associated with poor prognosis and stem cell-like behaviors of tumor cells. Hepatocellular adenomas (HCA) also often express biliary/progenitor markers and frequently act as precursor lesions for HCC. However, the cell of origin of HCA and HCC that expresses these markers still remains unclear. Therefore, to evaluate if mature hepatocytes give rise to HCA and HCC tumors, and to understand the molecular pathways involved in tumorigenesis, we lineage-labeled hepatocytes by injecting adeno-associated virus (AAV) containing thyroxine-binding globulin (TBG) promoter driven-Cre into RosaYFP mice. Yellow fluorescent protein (YFP) was present in more than 96% of hepatocytes before exposure to carcinogens. We treated AAV-TBG-Cre;RosaYFP mice with diethylnitrosamine (DEN) followed by multiple injections of carbon tetrachloride (CCl4) to induce carcinogenesis and fibrosis, and found that HCA and HCC nodules were YFP+ lineage-labeled and also positive for osteopontin (Opn), SRY (sex determining region Y)-box 9 (Sox9), and epithelial cell adhesion molecule (EpCAM), and enriched for transcripts of biliary/progenitor markers such as Prom1, Cd44, and Dlk1. Next, we performed the converse experiment and lineage-labeled Foxl1-positive hepatic progenitor cells simultaneously with exposure to carcinogens. None of the tumor nodules expressed YFP, indicating that Foxl1-expressing cells are not the cell of origin for hepatotoxin-induced liver tumors. We confirmed that HCA and HCC cells are derived from mature hepatocytes and not from Foxl1-Cre-marked cells in a second model of toxin-induced hepatic neoplasia, using DEN and 3,3’,5,5’-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP).
Conclusion
Our results indicate that hepatocytes are the cell of origin of HCA and HCC in DEN/CCl4 and DEN/TCPOBOP-induced liver tumors.
Keywords: Cancer-initiating cells, hepatocytes, adult hepatic progenitor cells, liver cancer, hepatocellular carcinoma, hepatocellular adenoma, lineage labeling, Foxl1
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer. Risk factors associated with liver cancer include viral hepatitis type B and C, alcohol, non-alcoholic fatty liver disease (NAFLD), diabetes, environmental toxin exposure, and obesity (1, 2). It has been estimated that more than 33,000 new cases of liver cancer are diagnosed in the United States yearly, and that 80% of these are HCC (3). Therefore, it is critical to understand the cellular mechanisms of HCC pathogenesis in order to develop effective anti-cancer therapy.
Human HCC cells frequently express markers of biliary/progenitor cells that are absent from normal hepatocytes, including osteopontin (OPN), CD133 (encoded by PROM1), CD44, delta-like 1 homolog (DLK1), SRY (sex determining region Y)-box 9 (SOX9), and EpCAM (epithelial cell adhesion molecule) (4-6). Expression of progenitor markers by HCC has been linked to aggressive behavior of tumors and poor prognosis (4, 7), highlighting the importance of understanding the cell of origin of these cancers. It also has been reported that biliary/progenitor markers are expressed in hepatocellular adenomas (HCA), benign tumors with the risk for malignant transformation (8-10). Hepatic progenitor cells (HPCs), the biliary epithelial component of ductular reactions, have been proposed to be the cell of origin of HCC based on correlative data (11-15). Surprisingly, the cellular origin of HCA and HCC nodules (referred as to “tumor nodules” in this paper) that are positive for biliary/progenitor markers has not been fully addressed experimentally.
Mature hepatocytes acquire markers of biliary/progenitor cells in response to injury (16), which led us to hypothesize that mature hepatocytes give rise to tumor nodules positive for progenitor markers. Therefore, to test our hypothesis, we lineage-traced mature hepatocytes using adeno-associated virus (AAV)-based genetic labeling (16, 17). We included HCA nodules as well as HCC nodules in our analyses to gain insight into cellular mechanisms of hepatocyte de-differentiation at the early stages of tumor progression. It has been well documented that HCA is a neoplastic lesion that progresses to HCC in mouse models (18-20), and that there is an increased risk of malignant transformation in a subset of HCA in human (8, 9).
We employed two different combinations of hepatotoxins that induce progenitor marker-expressing HCA and HCC nodules accompanied by ductular reactions and fibrosis. In addition, we also lineage-traced cells that express the winged helix transcription factor forkhead box protein (Foxl1) using Foxl1-Cre;RosaYFP mice, because our previous studies indicated that Foxl1 is a marker for HPCs (21-23). We also investigated whether tumor nodules express c-myc and components of the Wnt, Notch, and Hippo signaling pathways, key regulators of hepatic cell specification and tumorigenesis (24-29). Our study clarifies the long-debated cellular origin of tumor cells that express progenitor markers by tracing hepatocytes to tumor nodules in two mouse models of toxin-induced HCA and HCC.
Materials and Methods
Mice
For lineage-tracing of hepatocytes, 6-day-old RosaYFP (Rosa26loxP-stop-loxP-YFP) reporter mice were injected with serotype 8 AAV-thyroxine-binding globulin (TBG)-Cre (4×1010 gene copies per mouse, intraperitoneally) (University of Pennsylvania Vector Core) (16, 17). For lineage tracing of Foxl1-expressing cells, Foxl1-Cre mice (30) were crossed to RosaYFP reporter mice (31). Two different strategies were used to induce HCC as described previously (31, 32). First, 15-day-old mice were injected with DEN (25 mg/kg body weight, intraperitoneally, Sigma-Aldrich, St. Louis, MO). Beginning at 29 days, mice were injected with CCl4 (0.5 mg/kg body weight, intraperitoneally, Sigma-Aldrich, St. Louis, MO) weekly for 14 to 21 weeks (32). Tissues were harvested one to eight weeks after the last injection. Second, 15-day-old mice were injected with DEN (20 mg/kg body weight, intraperitoneally). Beginning at 29 days, mice were injected with TCPOBOP (3 mg/kg body weight, intraperitoneally, Sigma-Aldrich, St. Louis, MO) biweekly for 16-26 weeks (33). Tissues were harvested two to eight weeks after the last injection. All protocols were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Histology and Cell Counting
HCA and HCC nodules were identified by a board-certified veterinary anatomic pathologist based on histomorphology of H&E-stained sections according to published guidelines (19). Co-localization analysis for hepatocyte, biliary, and/or progenitor cell markers, as well as yellow fluorescent protein (YFP) on stained sections was performed as described (34). Briefly, liver lobes were fixed in 4% paraformaldehyde for 24 hours at 4°C and embedded in paraffin. Slides (5 μm sections) were subjected to antigen retrieval using a 2100 Retriever (Electron Microscopy Sciences, Hatfield, PA). Slides were incubated with primary antibodies diluted in CAS-Block (Life Technologies, Grand Island, NY) overnight at 4°C and then incubated with appropriate secondary antibodies diluted in CAS-Block for 2 hours at room temperature. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei. For analysis of the ductular response, ten random pictures centered on the portal triad were taken for each section. For measurement of lineage labeling efficiency, approximately 1,300 cells were counted per mouse. For analysis and quantification of tumor nodules, serial sections were stained by immunofluorescence or immunohistochemisty as described (21). High-resolution slide scan images were obtained using a light microscopy (Keyence BZ-X700, Japan). Image J Software was used for analyses (35). The following antibodies were used: HNF4α (PP-H1415-00, R&D Systems, Minneapolis, MN); YFP (ab6673, Abcam, Cambridge, MA and GFP-1210, Aves Labs, Tigard, OR); Opn (AF808, R&DSystems, Minneapolis, MN); EpCAM (ab71916, Abcam, Cambridge, MA), Sox9 (AB5535, Millipore, Norwood, OH), vimentin (5741, Cell Signaling Technologies, Danvers, MA), Yap1 (4912, Cell Signaling Technologies, Danvers, MA), AFP (sc8108, Santa Cruz Biotech, CA). The CK19 antibody was a kind gift from Dr. Joshua R Friedman (University of Pennsylvania).
RNA Extraction and Quantitative Reverse Transcription Polymerase chain reaction (qRT-PCR)
Total RNA was extracted from liver samples using the PerfectPure RNA Tissue Kit (5 PRIME, Gaithersburg, MD) based on the manufacturer's protocol. Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) was used for generating cDNA. PCR reactions were performed using SYBRGreen QPCR Master Mix (Agilent Technologies, Santa Clara, CA) on Mx3000 PCR cycler (Agilent Technologies, Santa Clara, CA). Reactions were performed in triplicate and normalized relative to the ROX reference dye, and median cycle threshold values were used for subsequent analyses. TATA-box binding protein was used to normalize mRNA expression. The following primers were used: Col1a1(F): GTGGACGGCTGCACGAGTCA; Col1a1(R): GGCTGGGTGGGAGGGAACCA; Mmp2(F): GGACCTGCAGGGTGGTGGTCAT; Mmp2(R): TAGGGCCCGTGGGAACAGGG; Timp1(F): ACCAGAGCAGATACCATGATGGCCC; Timp1(R): TGGGGTGGGGCACAGCTACA; Prom1(F): GAAAAGTTGCTCTGCGAACC; Prom1(R): TCTCAAGCTGAAAAGCAGCA; Cd44(F): TGACCCGTTGTGCTGTGATCCT; Cd44(R): GCACAAAAGGGACTGAAGCTTGCC; Dlk1(F): GTGGCCATCGTCTTTCTCAAC; Dlk1(R): GATGATATTGACCGCCAGCTC; Sox9(F): GAGGTTTCAGATGCAGTGAGGA; Sox9(R): TGTCACAACACACGCACACA; Myc (F):CTCCACTCACCAGCACAACT; Myc(R): CTGTCCAACTTGGCCCTCTT; Ctnnb1(F):TGACACCTCCCAAGTCCTTT; Ctnnb1(R): CATGCCCTCATCTAGCGTCT; Notch1(F): CCTCATGATTGCCTCCTGCAGTGG ; Notch1(R): CAGGATCAGTGGAGTTGTGCCATCATGCAT; Notch2(F):CAGCAGACTGGATGAACCGT; Notch2(R): GAAAGTCACGATGGGAGGCA; Hes1(F): AGAGGCTGCCAAGGTTTTTG; Hes1(R): TCCCACTGTTGCTGGTGTAGA. Tbp primers were as described (21).
Statistical Analysis
Student's t-test was used to determine the significance of differences between two groups. A p-value < 0.05 was considered to be statistically significant.
Results
Hepatocytes Give Rise to Tumor Cells in the DEN/CCl4-Induced Tumor Model
We asked whether established mature hepatocytes are the major source of hepatotoxin-induced tumor nodules. In RosaYFP mice, expression of yellow fluorescent protein (YFP) is blocked by a transcriptional stop sequences flanked by loxP sites (31). Expression of Cre-recombinase allows for permanent labeling of the Cre-expressing cells and all its descendants with YFP. Injection of an AAV serotype with a high tropism for hepatocytes that expresses Cre recombinase under the control of the hepatocyte-specific thyroxine-binding globulin promoter (AAV-TBG-Cre) (16, 17) into RosaYFP mice resulted in efficient labeling of nearly all hepatocytes (96.9 ± 0.7%, n = 3) with YFP (Fig. 1A-C). Other hepatic cells, including CK19-expressing cholangiocytes and vimentin-expressing mesenchymal cells remained YFP-negative, confirming the specificity of this labeling strategy (Fig. 1B,C).
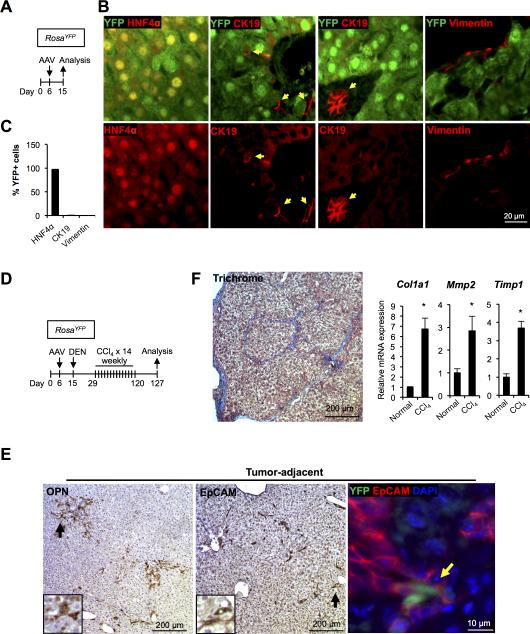
Figure 1. DEN and CCl4 treatment leads to ductular reactions and fibrosis.
(A) Schema of the treatment paradigm to test the AAV-TBG-Cre (AAV)-lineage labeling strategy. RosaYFP mice were injected with AAV-TBG-Cre 6 days after birth. (B) AAV-TBG-Cre efficiently labels hepatocytes in RosaYFP mice. Almost all hepatocytes (> 96%) were selectively marked by YFP at day 15, while Ck19+ cholangiocytes and vimentin+ mesenchymal cells are unlabeled. Red arrows: CK19+YFP− cells. (C) Quantification of YFP+ cells in the three cellular compartments analyzed. Error bars represent the standard error of the mean (n = 3-5 per group). (D) Schema of the treatment paradigm to induce liver tumors. RosaYFP mice were injected with AAV-TBG-Cre at day 6 and with DEN at day 15, and received fourteen weekly injection of CCl4. (E) Ductular reactions were detected in tumor-adjacent areas at the end of the treatment period by staining for Opn and Epcam. Inset: higher magnification of the area marked by the black arrow. A small fraction (6.1%) of Epcam+ cells were YFP+ (Yellow arrow). (F) Bridging fibrosis was detected at the end of treatment period by trichrome staining (left panel). Quantitative RT-PCR analyses revealed higher mRNA levels of fibrotic markers in the liver of mice treated with DEN/CCl4 as compared to normal chow-fed mice (n = 3-4 per each group). *P < 0.05 (right panel).
For the induction of HCC, we employed the recently developed paradigm in which a one-time administration of the mutagen diethylnitrosamine (DEN) is combined with multiple injections of the hepatotoxin carbon tetrachloride (CCl4) (Fig. 1D) (32). This model of chemical carcinogenesis is thought to approximate human pathophysiology because it induces chronic inflammation, elevation of endotoxin levels, and fibrosis in the liver, creating an environment permissive for the development of cancer (32, 36). As expected, treatment of mice led to the activation of ductular reactions positive for biliary markers Opn and EpCAM (Fig. 1E), and development of bridging fibrosis as well as up-regulation of markers of fibrosis (Fig. 1F). A small number of EpCAM-positive cells within ductular reactions were labeled with YFP (8.7 ± 6.1%, n = 4) (Fig. 1E, right panel), indicating a hepatocyte origin. Our finding is in line with two recent studies reporting that ductular reactions mostly but not exclusively originate from biliary cells in mouse models of liver cancer (37, 38).
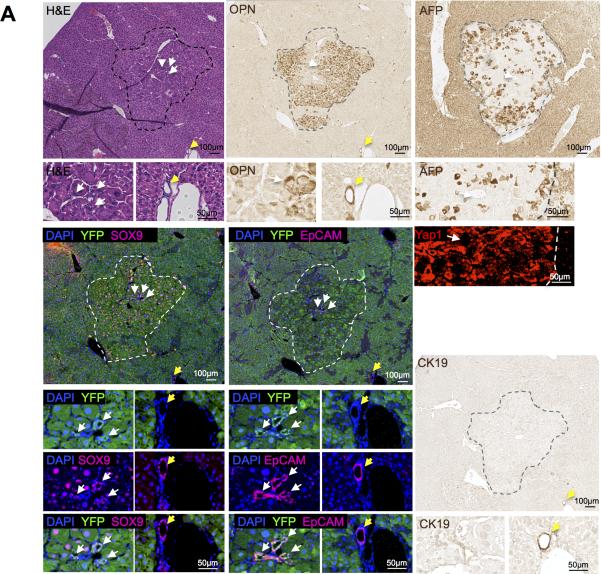
At the end of the treatment with DEN/CCl4, we detected multiple nodules with the morphology of HCA and HCC, all of which contained AFP and Opn-positive cells (Table 1; Fig. 2, S1). OPN is a frequently used marker of human HCC (39-41), although it is also found in cholangiocytes and HPCs (42, 43). All of the HCC tumor nodules were YFP-positive (Table 1; Fig. 2), and 26 of 27 HCA nodules were YFP-positive (96.2%) (Table 1; Fig. S1), indicating that tumor cell positivity for YFP was in line with the observed efficiency of labeling of hepatocytes with AAV. In addition, we confirmed that most of the HCA and HCC nodules contained cells marked with Sox9 in tumor parenchymal cells or in tumor-associated duct structures, or in both, and that cells expressing Sox9 in the tumors were also YFP-positive (Figure 2). SOX9 is a progenitor cell marker and is associated with tumor progression of human HCC (4). Similarly, EpCAM, a marker of an HCC subtype with progenitor features (6), was present in tumor-associated ductular structures that were YFP-positive in HCCs and in HCAs (Fig. 2, S1). Peri-tumoral ductular structures were positive for Opn, Sox9, EpCAM, and CK19, but were negative for YFP (Figure 2). We also investigated whether tumor nodules express Yap1, a component of the Hippo signaling pathway that is involved in re-programming of hepatocytes into cells with progenitor characteristics (28, 29). Yap1 was present in nuclei in both tumor and non-tumor tissues in HCAs and HCCs, with marked Yap1 cytoplasmic expression in the HCCs (Fig. 2, S1). AFP, a tumor marker and marker of hepatoblasts, was expressed in tumor and non-tumor cells, which is likely a reflection of the injury induced by CCl4 (44), though many of the tumors had darker staining than the surrounding tissues (Fig. 2, S1). CK19, a marker of cholangiocytes and of hepatoblasts (15), was expressed in peri-tumoral bile ducts, but was absent from the tumors in the DEN/CCl4 model (Fig. 2, S1).
| Treatment | DEN/CCl4 | DEN/TCPOBOP | ||
|---|---|---|---|---|
| Cre Driver | AAV-TBG-Cre | Foxl1-Cre | AAV-TBG-Cre | Foxl1-Cre |
| Labeled cells | Hepatocytes | HPCs | Hepatocytes | HPCs |
| Number of mice | 9 | 9 | 11 | 9 |
| Number of HCAs analyzed | 27 | 6 | 7 | 17 |
| Number of YFP+ HCAs | 26 | 0 | 7 | 0 |
| Number of HCCs analyzed | 3 | 0 | 1 | 8 |
| Number of YFP+ HCCs | 3 | 0 | 1 | 0 |
| Number of HCCs with Sox9+ tumor-associated duct structures | 3 | 0 | 0 | 8 |
| Number of HCCs with Sox9+ parenchymal cells | 3 | 0 | 1 | 6 |
| Number of OPN+ HCCs | 3 | 0 | 1 | 8 |
| Number of Yap1+ HCCs | 3 | 0 | 1 | 8 |
| Number of HCCs with EpCAM+ tumor-associated duct structures | 3 | 0 | 0 | 7 |
| Number of HCCs with EpCAM+ parenchymal cells | 0 | 0 | 1 | 4 |
| Number of CK19+ HCC nodules | 0 | 0 | 0 | 7 |
| Number of HCCs with nuclear β-catenin-positive parenchymal cells | 0 | 0 | 1 | 7 |
| Number of Afp+ HCC nodules | 3 | 0 | 1 | 8 |
Figure 2. Lineage tracing demonstrates that HCC originates from hepatocytes when carcinogenesis is induced by DEN and CCl4 treatment.
Serial sections of a representative HCC stained for H&E, Opn, AFP, YFP/Sox9, YFP/Epcam, Yap1, and CK19. Dotted lines indicate tumor margins. White arrows point to tumor-associated ductular structures. Yellow arrows mark extra-tumoral bile ducts.
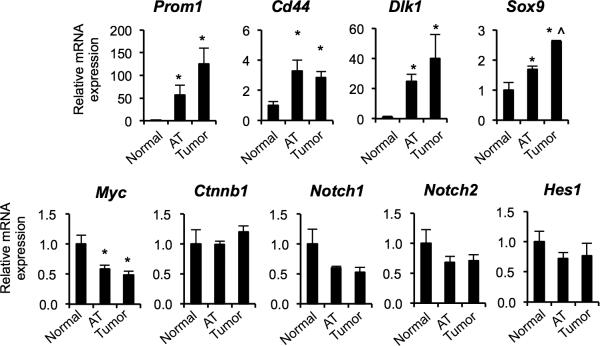
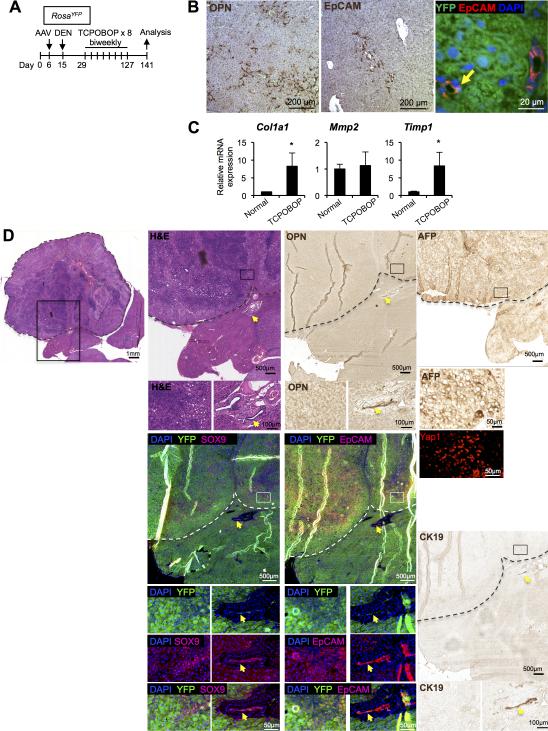
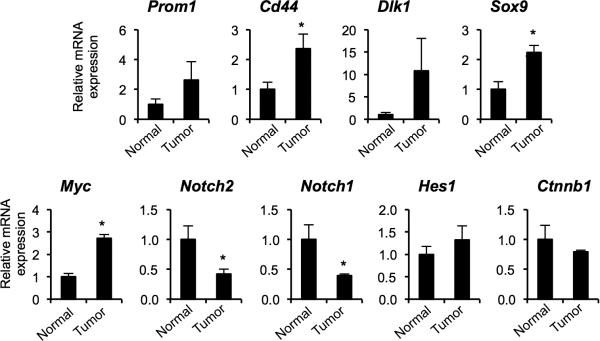
Quantitative PCR analysis indicated that tumor nodules express significantly higher mRNA levels of the markers of biliary/progenitor cells including Prom1, Cd44, Dlk1, and Sox9 than the normal liver (Fig. 3). While Sox9 mRNA levels were significantly higher in tumor nodules compared to tumor-adjacent areas, other markers were expressed in tumor-adjacent areas at levels slightly lower or comparable to tumor nodules. The similar levels of expression in tumor and tumor-surrounding areas is consistent with hepatotoxin-induced ductular reactions in tumor surrounding areas, and also with de-differentiation of mature hepatocytes. Both ductular reactions and tumors came to express progenitor cell and ductal cell markers. Our results demonstrate that mature hepatocytes give rise to tumor nodules positive for progenitor markers in the DEN/CCl4 model.
Figure 3. Tumors induced by DEN and CCl4 treatment express progenitor markers.
Quantitative RT-PCR analyses for mRNA levels of progenitor markers in the tumor nodules as compared to tumor-adjacent areas (AT) and the liver of normal chowfed mice (n = 4 mice per group). Note that we pooled HCA and HCC tumor nodules isolated from the same mouse, 6 nodules per mouse. *P < 0.05, AT versus normal and tumors versus normal; ^P < 0.05, tumors vs. AT. Error bars represent the standard error of the mean.
To further understand which signaling molecules are associated with de-differentiation of hepatocytes and tumor formation, we investigated expression of Myc, Ctnnb1, Notch1, Notch2, and Hes1 on the transcript level (Fig. 3). It has been demonstrated that activation of c-myc is required to reprogram mature hepatocytes into cancer stem cells (45). Activation of Wnt signaling has been suggested as a marker of malignant transformation of HCA into HCC (9). Activation of Notch signaling has been associated with HCC formation in mice (46). Furthermore, the Wnt and Notch signaling pathways are key regulators of hepatic cell specification (24, 26). Surprisingly, the levels of Myc mRNA were down-regulated in DEN/CCl4-induced tumors compared to normal liver, and the transcript levels of Ctnnb1, Notch1, Notch2, and Hes1 remained unchanged.
Foxl1-Cre-Marked Cells Do Not Contribute to Tumors in the DEN/CCl4 Model
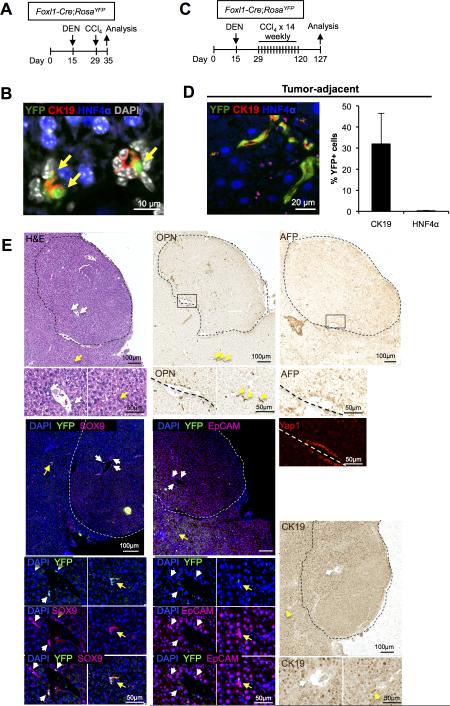
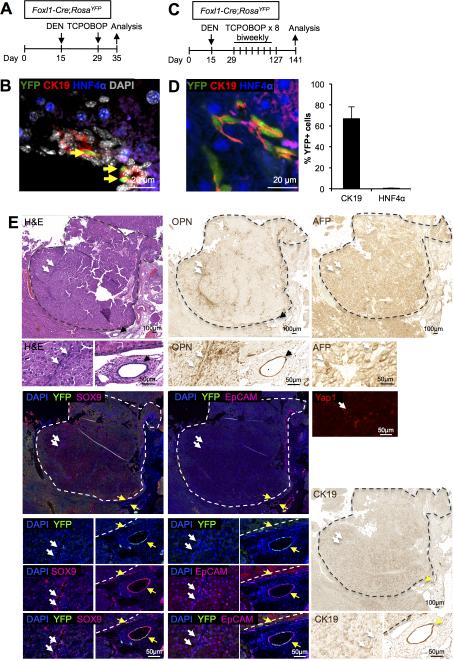
We have previously demonstrated that Foxl1+ cells appear in the portal triad following certain types of liver injury, including bile duct ligation and with the choline-deficient, ethionine-supplemented (CDE) diet (21, 34). Foxl1-Cre labeled cells express markers of biliary/progenitor cells and contribute to parenchymal regeneration (23, 34). Therefore, we asked whether Foxl1+ HPCs contribute to DEN/CCl4-induced tumor nodules. To follow Foxl1+ HPCs, we employed Foxl1-Cre;RosaYFP mice in which Foxl1-expressing cells as well as their descendants are marked by YFP (23, 30, 31). To test whether the administration of DEN and CCl4 activates Foxl1-Cre expression in the liver prior to tumorigenesis, we treated Foxl1-Cre;RosaYFP mice with DEN on postnatal day 15, and once with CCl4 on postnatal day 29, and harvested the liver one week later (Fig.4A). We detected Ck19+YFP+ cells in the portal triad (Fig.4B), indicating that these hepatotoxins activate the Foxl1+ progenitor cell compartment prior to the induction of liver tumors. Next, to induce tumors in the mouse liver, Foxl1-Cre;RosaYFP mice received DEN followed by up to 21 weekly injections of CCl4 (Fig.4C). At the end of the treatment with DEN/CCl4, Foxl1-Cre-labeled cells were confined to non-tumor areas, and YFP expression was detected in 32% of all Ck19+ cells (which include both cholangiocytes and HPCs), but only in 0.2% of HNF4α+ hepatocytes (Fig. 4D). We detected six HCA nodules that contained cells expressing Opn, AFP, Sox9, and EpCAM, similar to what was seen in the hepatocyte lineage-tracing model, except that in this case the tumor cells were all YFP-negative (Fig. 4E). None of the tumors in this experimental group were classified as HCC by histopathological criteria, but the staining pattern of tumor markers was similar between HCAs and HCCs (Fig. 4E). These results indicate that Foxl1+ cells are not tumor-initiating cells in this model of hepatic tumorigenesis.
Figure 4. Foxl1-Cre-marked HPCs are not tumor-initiating cells in the DEN/CCl4 hepatotoxin–induced tumor model.
(A) Schema of the treatment paradigm to label HPCs prior to the induction of liver tumors. (B) Ck19+HNF4α−YFP+ HPCs (yellow arrows) derived from Foxl1-Cre-expressing cells are clearly visible in the portal tract. (C) Schema of the treatment paradigm to induce liver tumors. (D) Ck19+HNF4α−YFP+ cells were detected in non-tumor areas at the end of treatment period (left panel). Quantification of the percentage of YFP+Ck19+ or YFP+HNF4α+ cells in DEN/CCl4-treated mice (n = 4 per each group) (right panel). Error bars represent the standard error of the mean. (E) YFP-expressing cells are absent from HCA tumor nodules. Serial sections of a representative HCA stained for H&E, Opn, AFP, YFP/Sox9, YFP/Epcam, Yap1, and CK19 are shown. Dotted lines indicate the margins of the tumor. White arrows point to tumor-associated ductular structures. Yellow arrows mark extra-tumoral bile ducts.
Hepatocytes but Not Foxl1-Cre-Marked HPCs Give Rise to Tumors in the DEN/TCPOBOP Model of Liver Tumorigenesis
Having established that hepatocytes are the cells of origin of tumor cells in the DEN/CCl4 model, we tested another tumorigenesis paradigm, the combination of DEN with 3,3’,5,5’-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP) (33). TCPOBOP is an agonist for the constitutive androstane receptor, and promotes hepatocarcinogenesis through multiple mechanisms including activation of the c-Myc-FoxM1 pathway and upregulation of anti-apoptotic proteins (47-49). To lineage-trace hepatocytes, we injected AAV-TBG-Cre into RosaYFP mice before hepatotoxin treatment (Fig. 5A). Treatment of mice led to activation of the ductular reaction as indicated by Opn and EpCAM expression (Fig. 5B), as well as up-regulation of markers of fibrosis (Fig. 5C), although the fibrosis was not extensive enough to cause bridging of fibrous septa (data not shown). Consistent with our results in the DEN/CCl4 model, only a small fraction (fewer than 2%) of EpCAM-positive cells within ductular reactions were labeled by YFP (1.6 ± 1.0%, n = 4) (Fig. 5B, right panel).
Figure 5. HCC induced by DEN and TCPOBOP treatment originates from hepatocytes.
(A) Schema of the treatment paradigm to induce liver tumors. (B) Ductular reactions were detected in tumor-adjacent areas at the end of treatment period by staining for Opn and Epcam. Yellow arrow: Epcam+YFP+ cells. (C) Quantitative RTPCR analyses revealed higher mRNA levels of fibrotic markers in the liver of mice treated with DEN/TCPOBOP as compared to normal chow-fed mice (n = 3-4 per each group). *P < 0.05. Error bars represent the standard error of the mean. (D) Serial sections of an HCC stained for H&E, Opn, AFP, YFP/Sox9, YFP/Epcam, Yap1, and CK19 are shown. Dotted lines indicate the tumor margin. Yellow arrows mark extra-tumoral bile ducts.
Next, to induce tumor formation, we treated AAV-TBG-Cre-injected RosaYFP mice with DEN followed by biweekly injections of TCPOBOP for up to 26 weeks. Toxin treatment led to development of HCA and HCC nodules in both AAV-TBG-Cre-treated and Foxl1-Cre-marked groups (Table 1). In AAV-TBG-Cre-treated mice, HCAs and an HCC induced by DEN/TCPOBOP treatment were positive for AFP, Opn, EpCAM, Yap1 (Fig. 5D, S2). As shown in Figures 5D, S2, and Table 1, the HCAs and HCC were YFP-positive, demonstrating that the tumors had originated from mature hepatocytes in the DEN/TCPOBOP model. Our quantitative RT-PCR analyses indicated that tumor nodules were enriched for biliary progenitor markers as well as for c-myc transcripts (Fig. 6). The levels of Ctnnb1 and Hes1 remained unchanged, while expression of Notch1 and Notch2 was down-regulated in tumor nodules.
Figure 6. Tumors induced by DEN and TCPOBOP treatment express progenitor markers.
Quantitative RT-PCR analyses revealed higher mRNA levels of progenitor markers in the tumor nodules as compared to the liver of normal chow-fed mice (n = 3-4 mice per group). Note that we pooled HCA and HCC tumor nodules isolated from the same mouse, 6 nodules per mouse. *P < 0.05, tumors versus normal. Error bars represent the standard error of the mean.
We then asked whether Foxl1+ HPCs contribute to DEN/TCPOBOP-induced tumor formation. First, to test whether the administration of DEN and TCPOBOP activates Foxl1 expression in the liver prior to tumor formation, we treated Foxl1-Cre;RosaYFP mice with DEN 15 days after birth, and once with TCPOBOP on postnatal day 29, and harvested the liver 1 week later (Fig.7A). The presence of Ck19+YFP+ cells in the portal triad at this early time point (Fig. 7B) indicated that the DEN/TCPOBOP protocol activates the Foxl1+ progenitor cell compartment prior to the induction of HCA/HCC. Next, to induce tumor formation, we treated Foxl1-Cre;RosaYFP mice with DEN followed by biweekly injections of TCPOBOP for up to 26 weeks (Fig. 7C). YFP expression was detected in Ck19+ cells in non-tumor areas (Fig. 7D). HCA and HCC nodules expressed AFP, Opn, Sox9, EpCAM, Yap1 (Table 1, Fig. 7E, S3). In contrast to the DEN/CCl4 model, the DEN/TCPOBOP condition led to CK19-positive tumor and non-tumor parenchymal cells, albeit at lower levels than at the bile ducts (Fig. 7E, Table 1). Despite the presence of multiple YFP+ cells in tumor-adjacent areas, all tumor nodules remained YFP-negative, indicating that Foxl1+ cells are not a source of tumor nodules in this model (Table 1, Fig. 7E,F).
Figure 7. Foxl1-Cre-marked HPCs are not tumor-initiating cells in the DEN/TCPOBOP hepatotoxin–induced HCC model.
(A) Schema of the treatment paradigm to label HPCs prior to the induction of HCC. (B) Ck19+HNF4α−YFP+ HPCs (yellow arrows) in the portal tract are clearly visible, indicating activation of the Foxl1-Cre promoter. (C) Schema of the treatment paradigm to induce liver tumors. (D) Ck19+HNF4α−YFP+ cells were detected in non-tumor areas at the end of the treatment period (left panel). Quantification of the percentage of YFP+Ck19+ or YFP+HNF4α+ cells in DEN/TCPOBOP-treated mice (n = 6 per each group) (right panel). Error bars represent the standard error of the mean. (E) YFP-expressing cells are absent from HCA tumor nodules. Serial sections of a representative tumor nodule stained for H&E, Opn, and YFP/Sox9 are shown. (F) YFP-expressing cells are absent from HCC tumor nodules. Serial sections of a representative HCC stained for H&E, Opn, AFP, YFP/Sox9, YFP/Epcam, Yap1, and CK19 are shown. Dotted lines indicate tumor margins. White arrows point to tumor-associated ductular structures. Yellow arrows mark extra-tumoral bile ducts.
Discussion
Identifying the cell-of-origin of HCC is crucial to understanding tumorigenesis in the liver. However, which types of liver cells give rise to HCC is still a subject of debate. Hepatocellular carcinomas frequently express biliary/progenitor markers (7, 50, 51), which are associated with poor prognosis, and stem cell-like features such as ability to self-renew and differentiate (4, 7). These findings have led to the notion that the cell-of-origin of HCC might be a progenitor or biliary epithelial cell. Indeed, Holczbauer and colleagues demonstrated that oncogenic H-Ras and SV40LT transform HPCs, hepatoblasts, and mature hepatocytes into cells that express biliary/progenitor markers and give rise to tumors upon transplantation, indicating that a strong oncogenic stimulus can transform normal cells regardless of their lineage (45). However, two independent studies demonstrated that endogenous biliary lineages do not contribute to HCC in mouse models of liver cancer using lineage tracing (37, 38), and that wild-type HPCs do not give rise to HCC upon transplantation (52), putting the progenitor hypothesis of HCC origin into question.
On the other hand, increasing evidence suggests a hepatocyte origin of HCC. Multiple studies demonstrated the remarkable plasticity of mature hepatocytes (16, 53, 54). Furthermore, He and colleagues identified HCC progenitor cells (HcPCs) that express biliary/progenitor markers such as Sox9 and Cd44 in foci of altered hepatocytes of DEN-treated mouse liver (5). Although the authors did not perform lineage-tracing experiments to identify the cell-of-origin of HcPCs, correlative evidence strongly suggested that mature hepatocytes are the cells of origin of DEN-induced HcPCs. Based on these reports, we hypothesized that mature hepatocytes give rise to HCC, at least in models that include exposure to hepatotoxins, even if they come to express biliary/progenitor markers. Our results clearly demonstrate that in two different mouse models of hepatotoxin-induced liver tumorigenesis, hepatocytes are the cellular source for HCA or HCC that are positive for biliary/progenitor markers.
Two recent studies identified novel cell populations that contribute to parenchymal regeneration – hybrid periportal hepatocytes and Axin2+ pericentral cells (52, 55). While hybrid periportal hepatocytes did not give rise to HCC in three different mouse models of liver cancer, the tumorigenic potential of Axin2+ cells has not yet been evaluated. In the current study, AAV labeled both periportal and pericentral hepatocytes. Further studies using alternative labeling strategies will be required to identify the subpopulation of hepatocytes that contributes to tumorigenesis.
Using a complementary approach, we investigated whether Foxl1+ hepatic progenitor cells can give rise to tumor cells, because a subset of biliary cells within ductular reactions marked by Foxl1-Cre become HPCs that possess the ability to self-renew and differentiate towards both cholangiocyte and hepatocyte lineages (21, 23, 34). Foxl1+ marked HPCs express the progenitor/biliary markers EpCAM, Sox9, Prom1, and Cd44 (23). Our Foxl1-Cre lineage labeling is constitutive, marking cells with YFP whenever Foxl1 is expressed during development or injury. Thus, if we had found that liver tumors in Foxl1-Cre;RosaYFP mice express YFP, we would not have been able to state conclusively that these tumors arose from HPCs, as the Foxl1 promoter might have been activated in a hepatocytes, mesenchymal cells, or other potential tumor precursor cells at any time during toxin exposure and tumorigenesis. However, the fact that all HCA and HCC nodules remained YFP-negative in Foxl1-Cre;RosaYFP mice indicates that Foxl1-expressing cells do not contribute to HCA/HCC nodules. We conclude that Foxl1-Cre-marked HPCs are not tumor-initiating cells in two models of hepatotoxin-induced tumorigenesis.
Our findings do not rule out the possible contribution of different stem/progenitor populations within the liver to tumor cells, as the Foxl1-Cre transgene does not mark all Ck19-positive cells. However, the fact that AAV-TBG-Cre labels more than 96% of HCAs and all HCC tumor nodules suggests that the contribution of non-hepatocyte cells to DEN/CCl4 or DEN/TCPOBOP-induced tumors is minimal. Our results are consistent with a recent report demonstrating that cells expressing Hnf1β, a transcription factor specific to biliary cells including ductular reactions, do not contribute to HCC (37). Furthermore, biliary cells marked with Opn-iCreERT2 also do not contribute to HCC, further supporting our results (38). Thus, the biological significance of the expansion of ductular reactions during tumor development in models of chemically-induced liver cancer appears limited.
Finally, we note that our study does not rule out the potential contribution of Foxl1+ HPCs to tumorigenesis in other models of HCC. The hepatotoxin DEN is metabolically activated in hepatocytes (5), implying that Foxl1+HPCs may not have been exposed to genotoxic stress in our mouse models. Nevertheless, our results indicate that mature hepatocytes can give rise to tumor cells positive for biliary/progenitor markers despite the presence of ductular reactions in tumor-adjacent areas, thus indicating that expression of biliary/progenitor markers by HCC on its own does not support the progenitor hypothesis of HCC origin. In conclusion, our study demonstrates that mature hepatocytes are the cell of origin in two mouse models of liver tumors induced by hepatotoxin treatment. An important question for the future is to determine why HCCs that express progenitor cell markers have a worse prognosis than those that do not.
Supplementary Material
Acknowledgements
We thank Tia Bernard-Banks (University of Pennsylvania) for managing the mouse colony, Dr. Joshua R Friedman (University of Pennsylvania) for providing the anti-Ck19 antibody.
Financial Support
This work was supported by the American Association for the Study of Liver Diseases/American Liver Foundation Liver Scholar Award (S.S.) and the National Institutes of Health through T32DK007066 (K.J.W.) and R01-DK102667 (K.H.K.). This study was facilitated by the Molecular Pathology and Imaging Core of the Penn Center for Molecular Studies in Digestive and Liver Disease (P30-DK50306).
List of Abbreviations
- AFP
alpha fetoprotein
- AAV
adeno-associated virus
- CCl4
carbon tetrachloride
- CK19
cytokeratin 19
- Ctnnb1
beta-catenin
- DEN
diethylnitrosamine
- Dlk1
delta-like 1 homolog
- EpCAM
epithelial cell adhesion molecule
- Foxl1
forkhead box protein L1
- HCA
hepatocellular adenoma
- HCC
hepatocellular carcinoma
- HNF4α
hepatocyte nuclear factor 4 alpha
- HPCs
hepatic progenitor cells
- Opn
osteopontin
- Sox9
SRY (sex determining region Y)-box 9
- TBG
thyroxine-binding globulin
- TCPOBOP
tetrachloro-1,4-bis(pyridyloxy)benzene
- Yap1
yes-associated protein 1
- YFP
yellow fluorescent protein
References
- 1.Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109:542–553. doi: 10.1038/ajg.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: Where are we? Where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Cancer Society . Cancer Facts & Figures 2014. American Cancer Society; Atlanta, Georgia, USA: 2014. [Google Scholar]
- 4.Guo X, Xiong L, Sun T, Peng R, Zou L, Zhu H, Zhang J, et al. Expression features of SOX9 associate with tumor progression and poor prognosis of hepatocellular carcinoma. Diagn Pathol. 2012;7:44. doi: 10.1186/1746-1596-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–396. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 7.Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, Llovet JM. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farges O, Ferreira N, Dokmak S, Belghiti J, Bedossa P, Paradis V. Changing trends in malignant transformation of hepatocellular adenoma. Gut. 2011;60:85–89. doi: 10.1136/gut.2010.222109. [DOI] [PubMed] [Google Scholar]
- 9.Liau SS, Qureshi MS, Praseedom R, Huguet E. Molecular pathogenesis of hepatic adenomas and its implications for surgical management. J Gastrointest Surg. 2013;17:1869–1882. doi: 10.1007/s11605-013-2274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Libbrecht L, De Vos R, Cassiman D, Desmet V, Aerts R, Roskams T. Hepatic progenitor cells in hepatocellular adenomas. Am J Surg Pathol. 2001;25:1388–1396. doi: 10.1097/00000478-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Kung JW, Currie IS, Forbes SJ, Ross JA. Liver development, regeneration, and carcinogenesis. J Biomed Biotechnol. 2010;2010:984248. doi: 10.1155/2010/984248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee TK, Castilho A, Ma S, Ng IO. Liver cancer stem cells: implications for a new therapeutic target. Liver Int. 2009;29:955–965. doi: 10.1111/j.1478-3231.2009.02040.x. [DOI] [PubMed] [Google Scholar]
- 13.Sell S. Cellular origin of hepatocellular carcinomas. Semin Cell Dev Biol. 2002;13:419–424. doi: 10.1016/s1084952102001295. [DOI] [PubMed] [Google Scholar]
- 14.Song K, Wu J, Jiang C. Dysregulation of signaling pathways and putative biomarkers in liver cancer stem cells (Review). Oncol Rep. 2013;29:3–12. doi: 10.3892/or.2012.2082. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911–1918. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Wang H, Bell P, McMenamin D, Wilson JM. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum Gene Ther. 2012;23:533–539. doi: 10.1089/hum.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang JJ, Weghorst CM, Henneman JR, Devor DE, Ward JM. Progressive atypia in spontaneous and N-nitrosodiethylamine-induced hepatocellular adenomas of C3H/HeNCr mice. Carcinogenesis. 1992;13:1541–1547. doi: 10.1093/carcin/13.9.1541. [DOI] [PubMed] [Google Scholar]
- 19.Thoolen B, Maronpot RR, Harada T, Nyska A, Rousseaux C, Nolte T, Malarkey DE, et al. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol. 2010;38:5S–81S. doi: 10.1177/0192623310386499. [DOI] [PubMed] [Google Scholar]
- 20.Uehara T, Ainslie GR, Kutanzi K, Pogribny IP, Muskhelishvili L, Izawa T, Yamate J, et al. Molecular mechanisms of fibrosis-associated promotion of liver carcinogenesis. Toxicol Sci. 2013;132:53–63. doi: 10.1093/toxsci/kfs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sackett SD, Li Z, Hurtt R, Gao Y, Wells RG, Brondell K, Kaestner KH, et al. Foxl1 is a marker of bipotential hepatic progenitor cells in mice. Hepatology. 2009;49:920–929. doi: 10.1002/hep.22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin S, Kaestner KH. The origin, biology, and therapeutic potential of facultative adult hepatic progenitor cells. Curr Top Dev Biol. 2014;107:269–292. doi: 10.1016/B978-0-12-416022-4.00010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S, Walton G, Aoki R, Brondell K, Schug J, Fox A, Smirnova O, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15:653–667. doi: 10.1038/nrc4017. [DOI] [PubMed] [Google Scholar]
- 26.Spee B, Carpino G, Schotanus BA, Katoonizadeh A, Vander Borght S, Gaudio E, Roskams T. Characterisation of the liver progenitor cell niche in liver diseases: potential involvement of Wnt and Notch signalling. Gut. 2010;59:247–257. doi: 10.1136/gut.2009.188367. [DOI] [PubMed] [Google Scholar]
- 27.Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 29.Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sackett SD, Fulmer JT, Friedman JR, Kaestner KH. Foxl1-Cre BAC transgenic mice: a new tool for gene ablation in the gastrointestinal mesenchyme. Genesis. 2007;45:518–522. doi: 10.1002/dvg.20315. [DOI] [PubMed] [Google Scholar]
- 31.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin S, Upadhyay N, Greenbaum LE, Kaestner KH. Ablation of Foxl1-Cre-Labeled Hepatic Progenitor Cells and Their Descendants Impairs Recovery of Mice From Liver Injury. Gastroenterology. 2015;148:192–202. doi: 10.1053/j.gastro.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malhi H, Guicciardi ME, Gores GJ. Hepatocyte death: a clear and present danger. Physiological reviews. 2010;90:1165–1194. doi: 10.1152/physrev.00061.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jors S, Jeliazkova P, Ringelhan M, Thalhammer J, Durl S, Ferrer J, Sander M, et al. Lineage fate of ductular reactions in liver injury and carcinogenesis. J Clin Invest. 2015;125:2445–2457. doi: 10.1172/JCI78585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu X, Espanol-Suner R, Mederacke I, Affo S, Manco R, Sempoux C, Lemaigre FP, et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J Clin Invest. 2015;125:3891–3903. doi: 10.1172/JCI77995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H, Zhang XF, Zhou HJ, Xue YH, Dong QZ, Ye QH, Qin LX. Expression and prognostic significance of osteopontin and caspase-3 in hepatocellular carcinoma patients after curative resection. Cancer Sci. 2010;101:1314–1319. doi: 10.1111/j.1349-7006.2010.01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wee A. Diagnostic utility of immunohistochemistry in hepatocellular carcinoma, its variants and their mimics. Appl Immunohistochem Mol Morphol. 2006;14:266–272. doi: 10.1097/00129039-200609000-00003. [DOI] [PubMed] [Google Scholar]
- 41.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Español-Suñer R, Carpentier R, Van Hul N, Legry V, Achouri Y, Cordi S, Jacquemin P, et al. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143:1564–1575. e1567. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 43.Strazzabosco M, Fabris L, Albano E. Osteopontin: a new player in regulating hepatic ductular reaction and hepatic progenitor cell responses during chronic liver injury. Gut. 2014;63:1693–1694. doi: 10.1136/gutjnl-2014-307712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhlmann WD, Peschke P. Hepatic progenitor cells, stem cells, and AFP expression in models of liver injury. Int J Exp Pathol. 2006;87:343–359. doi: 10.1111/j.1365-2613.2006.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holczbauer A, Factor VM, Andersen JB, Marquardt JU, Kleiner DE, Raggi C, Kitade M, et al. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology. 2013;145:221–231. doi: 10.1053/j.gastro.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villanueva A, Alsinet C, Yanger K, Hoshida Y, Zong Y, Toffanin S, Rodriguez-Carunchio L, et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143:1660–1669. e1667. doi: 10.1053/j.gastro.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braeuning A, Heubach Y, Knorpp T, Kowalik MA, Templin M, Columbano A, Schwarz M. Gender-specific interplay of signaling through beta-catenin and CAR in the regulation of xenobiotic-induced hepatocyte proliferation. Toxicol Sci. 2011;123:113–122. doi: 10.1093/toxsci/kfr166. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Meng Z, Wang X, Zeng S, Huang W. The nuclear receptor CAR modulates alcohol-induced liver injury. Lab Invest. 2011;91:1136–1145. doi: 10.1038/labinvest.2011.68. [DOI] [PubMed] [Google Scholar]
- 49.Takizawa D, Kakizaki S, Horiguchi N, Yamazaki Y, Tojima H, Mori M. Constitutive active/androstane receptor promotes hepatocarcinogenesis in a mouse model of non-alcoholic steatohepatitis. Carcinogenesis. 2011;32:576–583. doi: 10.1093/carcin/bgq277. [DOI] [PubMed] [Google Scholar]
- 50.Cao L, Fan X, Jing W, Liang Y, Chen R, Liu Y, Zhu M, et al. Osteopontin promotes a cancer stem cell-like phenotype in hepatocellular carcinoma cells via an integrin-NF-kappaB-HIF-1alpha pathway. Oncotarget. 2015;6:6627–6640. doi: 10.18632/oncotarget.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colombo F, Baldan F, Mazzucchelli S, Martin-Padura I, Marighetti P, Cattaneo A, Foglieni B, et al. Evidence of distinct tumour-propagating cell populations with different properties in primary human hepatocellular carcinoma. PloS one. 2011;6:e21369. doi: 10.1371/journal.pone.0021369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Font-Burgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A, Taniguchi K, et al. Hybrid Periportal Hepatocytes Regenerate the Injured Liver without Giving Rise to Cancer. Cell. 2015;162:766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Wong PP, Sjeklocha L, Steer CJ, Sahin MB. Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture. Hepatology. 2012;55:563–574. doi: 10.1002/hep.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell stem cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.