Abstract
Objective
Long noncoding RNAs (lncRNA) represent a growing class of noncoding genes with diverse cellular functions. We previously reported on SENCR, a lncRNA that appears to support the vascular smooth muscle cell (VSMC) contractile phenotype. However, information about the vascular smooth muscle cell (VSMC)-specific lncRNAs regulated by myocardin (MYOCD)/SRF, the master switch for VSMC differentiation, is virtually unknown.
Approach and Results
To define novel lncRNAs with functions related to VSMC differentiation, we performed RNA sequencing in human coronary artery SMCs (HCASMCs) that overexpress MYOCD. A number of novel lncRNAs showed altered expression with MYOCD overexpression and one, named MYOcardin-induced Smooth muscle Long non-coding RNA, Inducer of Differentiation (MYOSLID), was activated by MYOCD and selectively expressed in VSMCs. MYOSLID was a direct transcriptional target of both MYOCD/SRF and TGFβ/SMAD pathways. Functional studies revealed that MYOSLID promotes VSMC differentiation and inhibits VSMC proliferation. MYOSLID showed reduced expression in failed human arteriovenous fistula (AVF) samples compared with healthy veins. While MYOSLID did not affect gene expression of transcription factors such as SRF and MYOCD, its depletion in VSMCs disrupted actin stress fiber formation and blocked nuclear translocation of MYOCD-related transcription factor A (MKL1). Finally, loss of MYOSLID abrogated TGFβ1-induced SMAD2 phosphorylation.
Conclusion
We have demonstrated that MYOSLID, the first human VSMC-selective and SRF/CArG-dependent lncRNA, is a novel modulator in amplifying the VSMC differentiation program, likely through feed-forward actions of both MKL1 and TGFβ/SMAD pathways.
Keywords: Vascular smooth muscle, Differentiation, Myocardin, long noncoding RNA
Introduction
Mature vascular smooth muscle cells (VSMCs) are genetically wired to exhibit low proliferation and migration while carrying out functions related to contractility and the normal distribution of blood flow. However, VSMCs are also endowed with remarkable phenotypic plasticity and switch their contractile/differentiated phenotype to a synthetic/de-differentiated state in response to diverse environmental stimuli.1–3 A large body of work has documented that de-differentiated VSMCs contribute to the pathogenesis of vascular diseases such as atherosclerosis, restenosis, transplant arteriopathy, and hypertension.1, 2, 4 Work from the last decade has established that VSMC differentiation is regulated primarily by serum response factor (SRF), a widely expressed transcription factor that binds CArG boxes located in the regulatory region of most VSMC-specific genes, and its potent cofactor, Myocardin (MYOCD).2, 5, 6 The target genes of the CArG/SRF/MYOCD triad include a number of cyto-contractile, ion channel, signal transducer, and matrix-associated genes that collectively define the differentiated state of VSMC.7–9 In recent years, an increasing number of non-coding RNA genes have also been identified as MYOCD/SRF targets. For example, the bi-cistronic microRNA cluster miR143/145 is a direct target of SRF/MYOCD and promotes VSMC contractile gene expression.10, 11 The presence of these and other SRF-dependent microRNAs12, 13 in VSMC ensures phenotypic stability. However, given the pervasive transcription in the human genome,14 we surmise that other non-coding genes may play a role in the regulation of VSMC differentiation.
Long non-coding RNAs (lncRNAs) are defined as processed transcripts longer than 200 nucleotides that do not encode for proteins. Thus far, more than 100,000 lncRNA genes have been defined in the human genome, which far outnumber all protein-coding and microRNA genes combined, suggesting a dominant role of this class of genes in the mammalian genome.15 Indeed, accumulating evidence has demonstrated that lncRNAs function as important regulators in stem cell pluripotency,16 cellular differentiation,17, 18 cell cycle progression19 and human diseases.20 Unlike microRNAs, which exhibit well-defined actions in negatively regulating gene expression by targeting the 3’-UTR of transcripts, lncRNAs display diverse and unpredictable regulatory roles.18, 21, 22 In this context, although numerous lncRNAs have been intensively investigated in the stem cell and cancer fields, studies of lncRNA in vascular biology and disease are only beginning to be undertaken.20, 23
Lnc-Ang362 (aka HG-MIR222) was the first VSMC lncRNA discovered by RNA-seq analysis.24 This lncRNA was shown to be elevated in rat aortic SMCs following angiotensin II stimulation and it appears to regulate VSMC proliferation via controlling levels of miR221/222.24 The p53-induced LncRNA-p21 was found to have a role in suppressing the progression of atherosclerosis in the ApoE null mouse by repressing VSMC proliferation and inducing apoptosis.25 A smooth muscle and endothelial cell enriched lncRNA called SENCR was revealed to play a role in maintaining VSMC contractile phenotype and concomitantly inhibiting VSMC migration.17 Although lncRNAs tend to be more tissue-specific, none of the aforementioned lncRNAs exhibit a strict VSMC-specific expression profile. Moreover, whether there is interplay between these or other unknown lncRNAs and the regulatory axis of MYOCD-SRF is unknown.
We hypothesized there must exist inducible VSMC-specific lncRNAs comprising a new layer of molecular regulation of the VSMC contractile phenotype. To this end, we used RNA-seq in MYOCD overexpressing human coronary artery SMCs (HCASMCs) to identify new lncRNAs regulated in a CArG-SRF-MYOCD dependent manner. A number of lncRNAs were revealed to be induced or repressed by MYOCD. Here, we report on a new lncRNA we named, MYOcardin-induced Smooth muscle Long noncoding RNA, Inducer of Differentiation (MYOSLID). We demonstrated that MYOSLID is a VSMC-selective lncRNA, which is a direct transcriptional target of MYOCD/SRF and TGFβ/SMAD pathways. Most importantly, we defined a critical role of MYOSLID in amplifying the VSMC differentiation program, likely through feed-forward actions of MYOCD related transcription factor A (MKL1) and TGFβ-SMAD pathways.
Materials and Methods are available in the online-only Data Supplement.
Results
MYOSLID is a MYOCD-inducible and VSMC-selective natural antisense (NAT) lncRNA
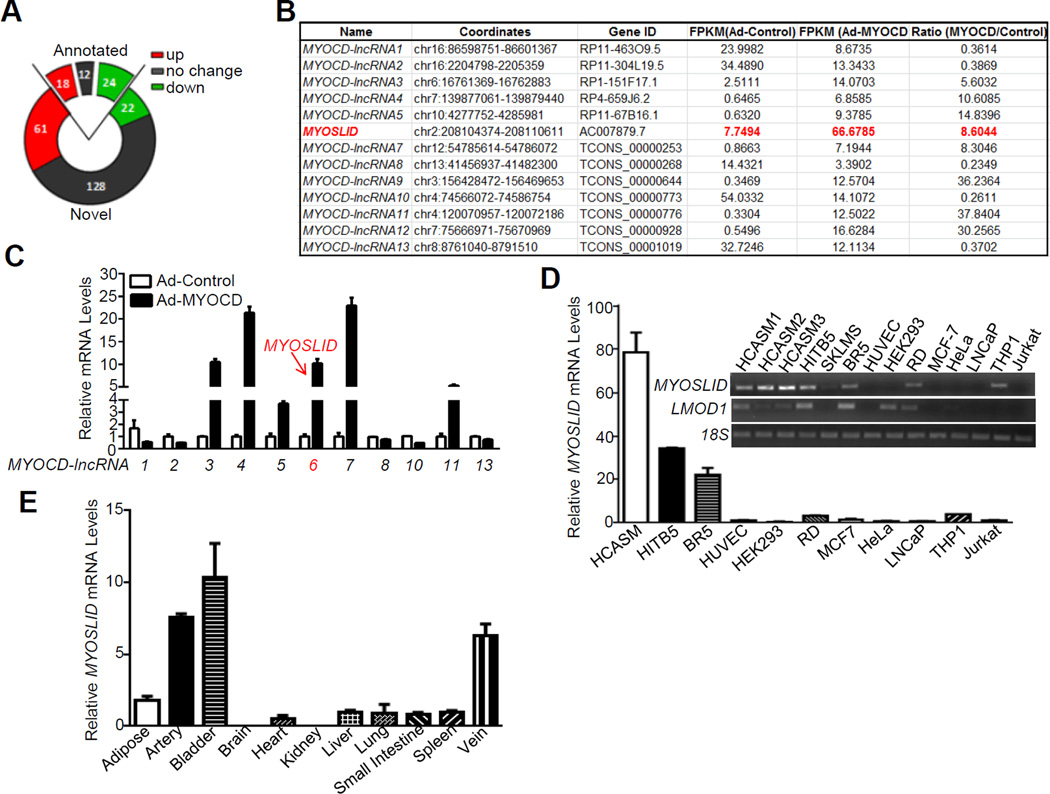
Cultured VSMCs exhibit reduced expression of MYOCD and various contractile genes with acquisition of the synthetic phenotype.6 To induce a VSMC contractile phenotype similar to that in the intact vessel wall, we transduced HCASMCs with adenovirus carrying MYOCD, a potent SRF cofactor for the VSMC differentiation program.26 We performed RNA-seq under these conditions to define novel lncRNAs associated with VSMC differentiation. As expected, RNA-seq revealed the induction of many MYOCD/SRF target genes such as MYH11, ACTA2 and CNN1 (data not shown). A total of 265 lncRNAs were found to be expressed in HCASMCs, among which, 54 were annotated lncRNAs in the UCSC Genome Browser and the remaining were novel lncRNAs. 137 lncRNAs were significantly regulated by MYOCD with 79 upregulated and 46 downregulated (FPKM fold change ≥2, Figure 1A). We selected 13 lncRNAs (8 upregulated and 5 downregulated), named here as MYOCD-lncRNAs, exhibiting the highest change in FPKM expression (Figure 1B) and validated each by qRT-PCR (Figure 1C). We examined the expression profile for all 13 validated MYOCD-lncRNAs in different human cell culture models and human tissues (data not shown). MYOSLID (previously AC007879.7), is enriched in three independent HCASMC isolates and the VSMC cell line, HITB5 (Figure 1D). MYOSLID is also selectively expressed in blood vessels (artery and vein) and bladder (Figure 1E), but not in other tissues with a paucity of SMC. The VSMC-selective expression profile is similar to LMOD1, a previously characterized VSMC-specific gene.8 While transcripts of all 13 MYOCD-lncRNAs are not conserved in mouse genome, they share high conservation with primates such as Rhesus monkey and Chimp, indicating that transcripts of these MYOCD-lncRNAs are unique to these animals (data not shown). These data identify MYOSLID as a novel MYOCD-induced and smooth muscle-selective lncRNA.
Figure 1.
Identification of lncRNAs regulated by MYOCD in human coronary arterial smooth muscle cells (HCASMCs). HCASMCs were transduced with adenovirus carrying MYOCD (Ad-MYOCD) or empty control adenovirus (Ad-Control) for 3 days and RNA samples were subjected to RNA-seq. A, Venn diagrams depicting the number of lncRNAs in HCASMCs from RNA-Seq analysis. Annotated refers to the lncRNAs annotated in GenBank; novel refers to those newly discovered lncRNAs based on the RNA-seq data. B, lncRNA targets (arbitrarily numbered as MYOCD-lncRNAs) most dramatically regulated by MYOCD from RNA-seq analysis. C, Quantitative RT-PCR (qRT-PCR) validation of the MYOCD-regulated lncRNAs listed in Figure 1B. qRT-PCR analysis of MYOSLID expression in the indicated cultured human cells (D) and human tissues (E). Relative MYOSLID expression was defined as fold increase to Jurkat cells and liver (set to 1) in (D) and (E), respectively. HITB5, human smooth muscle cell line; SKLMS, human uterine leiomyosarcoma cell line; BR5, human foreskin fibroblast; HUVEC, human umbilical vein endothelial cell; RD, human rhabdomyosarcoma cell line. Inset panel in (D) showing the semi-quantitative RT-PCR assessment of MYOSLID in cultured human cells. F, Genome browser track of MYOSLID gene locus, MYOLSID splicing variants and cis effect evaluation. Three adjacent lncRNAs (LOC101927865, AC007879.5 and AC007879.3) are transcribed antisense to MYOSLID (AC007879.7). Blue squares highlighted sequences denoting the 3 computer-predicted CArG boxes. Note serum response factor (SRF) ChIP-seq peak in HCASMCs flanking the putative −2 kb promoter region of MYOSLID and overlapping all three highlighted CArG boxes. G, Gene structure of MYOSLID splicing variants based on Rapid Amplification of cDNA end (RACE) analysis. Red highlighted numbers are the length of sequences derived from RACE analysis.
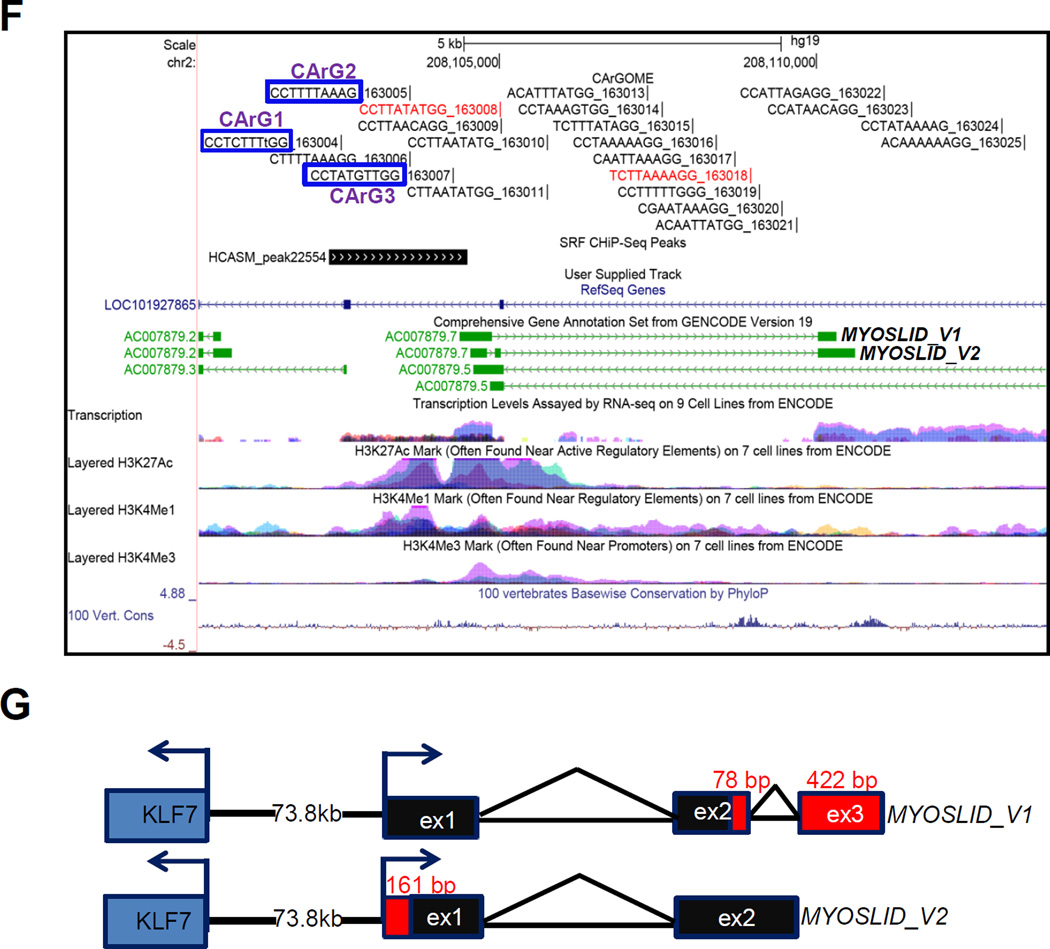
MYOSLID is located in a lncRNA-rich genomic region of chromosome 2 where the closest protein coding genes are 70 kb (KLF7) and 200 kb (CREB1) away from its 5’ end and 3’ end, respectively (Figure IA in the online-only Data Supplement). Two splice variants of MYOSLID were annotated in the UCSC Genome Browser. Interestingly, the entire MYOSLID locus falls within an opposing transcribed lncRNA (LOC101927865) with partially overlapping exonic sequence (Figure 1F). Thus, MYOSLID is best classified as a natural antisense (NAT) lncRNA.23 AC007879.3 is a lncRNA located in the proximal promoter region of MYOSLID. Another NAT lncRNA, AC007879.5, also partially overlaps with MYOSLID (Figure 1F). Expression levels of LOC101927865 and AC007879.5 are much lower than MYOSLID and there is no detectable level of AC007879.3 in HCASMCs (data not shown). Further, none of these 3 lncRNAs can be induced by MYOCD or TGFβ1 (Figure IB and IC in the online-only Data Supplement). The presence of H3K4Me1, H3K4Me3 and H3K27Ac histone marks within the −2 kb region of MYOSLID indicates this region likely harbors an active promoter for MYOSLID. Interestingly, sequence analysis shows 3 computer-predicted CArG boxes in the putative MYOSLID promoter. Further, ChIP-seq data of HCASMCs (manuscript in preparation) support the presence of SRF binding in the same region (Figure 1F). Collectively, these data define MYOSLID as a NAT lncRNA that may be a direct target of the MYOCD/SRF/CArG triad.
KLF7 is the nearest protein coding gene located 73.8 kb upstream of MYOSLID and transcribed in an antisense orientation to MYOSLID (Figure IA in the online-only Data Supplement). Genomic analysis and ChIP-seq for SRF binding in HCASMCs revealed 3 putative CArG boxes in the proximal promoter region of KLF7 (data not shown). KLF7 is also strongly induced by MYOCD in HCASMCs (Figure ID in the online-only Data Supplement). Depletion of MYOSLID by two different dicer substrate siRNAs (D-siRNAs) in HCASMCs did not change mRNA levels of KLF7 and two other neighboring lncRNAs (Figure IE and IF in the online-only Data Supplement), suggesting no cis or trans effect of MYOSLID on the expression of these neighboring genes. Thus, the induction of KLF7 by MYOCD is probably through its own CArG-containing promoter.
We performed RACE to define the full-length sequence of both MYOSLID variants. We extended more than 500 bp at the 3’ end, identifying a small portion of exon 2 (78 bp) and an additional new exon in MYOSLID_V1, exon 3 (422 bp) (Figure 1G). We were unable to extend the 5’ end of MYOSLID_V1, indicating a complete 5’ end of MYOSLID_V1 as annotated in GenBank. However, we extended the original exon 1 of MYOSLID_V2 to an additional 161 bp and confirmed that exon 1 of MYOSLID_V2 was identical to that of MYOSLID_V1 by sequencing (Figure 1G). Finally, we PCR amplified both variants and confirmed the entire 1,305 bp of the MYOSLID_V1 sequence based on the original annotation and the RACE results. However, sequencing results of MYOSLID_V2 revealed a mis-annotated small intron 1 with the original annotation which is actually included in exon 1 of MYOSLID_V2 (Figure IIA and IIB in the online-only Data Supplement). Further, using multiple variant-specific primer pairs, we confirmed that both variants were induced by MYOCD (Figure IIA in the online-only Data Supplement). No coding potential of MYOSLID was predicted by PhyloCSF27 (Figure IIIA in the online-only Data Supplement). The longest open reading frame of MYOSLID_V1 predicted by ExPASy (http://web.expasy.org/translate/) is 76 amino acids. However, no conserved protein domain has been found with any of the predicted open reading frames in MYOSLID. Quick coupled in vitro transcription/translation assays did not reveal production of peptides/proteins encoded by the open reading frames predicted by ExPASy, supportive of MYOSLID having no protein-coding potential (Figure IIIB in the online-only Data Supplement). We selected the longer MYOSLID_V1 transcript for overexpression studies below.
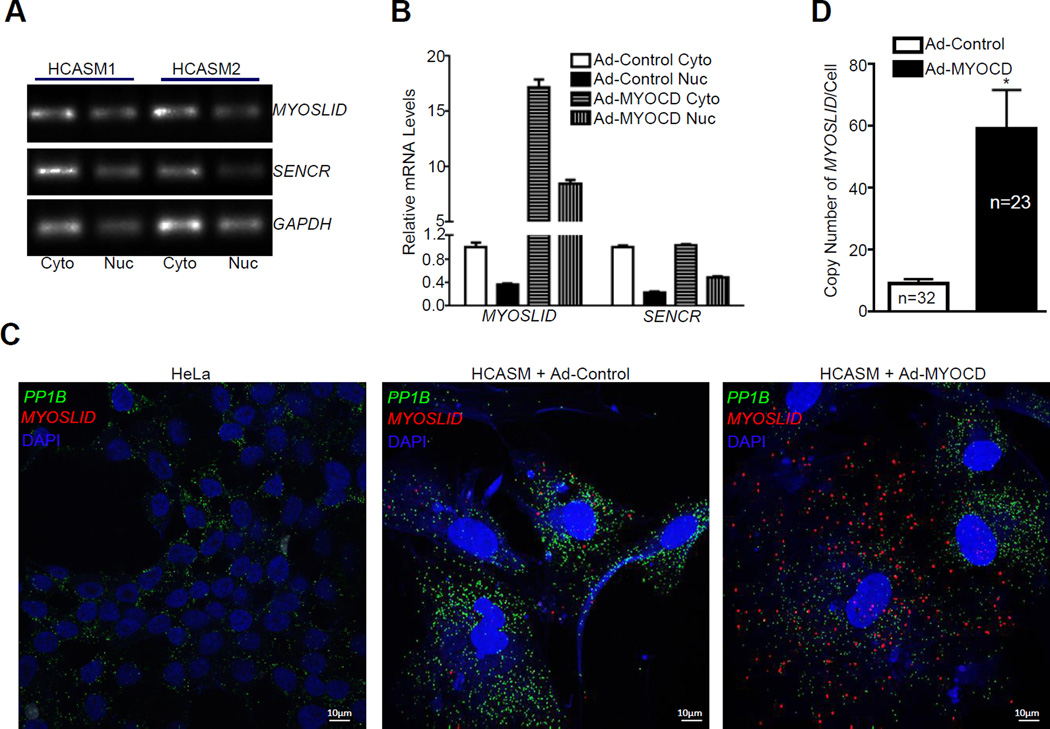
MYOSLID is a cytoplasmic lncRNA
Insight into the function of lncRNAs can be predicted by their localization in a cell.23 Conventional RT-PCR analysis of the fractionated RNA from cytosolic and nuclear compartments in HCASMCs revealed that MYOSLID transcripts were predominately localized in the cytosol, a pattern similar to SENCR, a known cytosolic lncRNA,17 and the mRNA of the protein coding gene, GAPDH (Figure 2A). MYOCD was able to induce both cytosolic and nuclear MYOSLID while it failed to promote the expression of SENCR (Figure 2B). Consistent with the qRT-PCR data, RNA-FISH documented the majority of MYOSLID transcripts in the cytosol with very few transcripts seen in the nucleus. In contrast to the abundant signal of PP1B mRNA, the basal level of MYOSLID was much lower, consistent with the low abundance of most lncRNAs compared with protein coding genes.19 In agreement with the qRT-PCR results, when HCASMCs were treated with Ad-MYOCD, both cytosolic and nuclear MYOSLID signals were increased (Figure 2C). No signal was detected by RNA-FISH in HeLa cells under baseline conditions (Figure 2C), consistent with the qRT-PCR data shown in Figure 1D. Quantitative analysis of the signal from RNA-FISH revealed 10 or 60 copies per single cell under basal versus MYOCD-induced conditions in HCASMCs, respectively (Figure 2D). Taken together, MYOSLID is a cytosolic enriched RNA induced by MYOCD. The cytoplasmic localization of MYOSLID is consistent with the finding that MYOSLID has no detectable cis-acting effect on the expression of neighboring genes (Figure IE and IF in the online-only Data Supplement).
Figure 2.
Cellular localization of MYOSLID in HCASMCs. A, Semi-quantitative RT-PCR analysis of MYOSLID and the indicated cytoplasmic positive control RNAs in cytosolic versus nuclear RNA fractions from two separate HCASMC isolates. Cyto, Cytosol; Nuc, Nucleus. B, qRT-PCR of MYOSLID and SENCR in the cytosolic (Cyto) and nuclear (Nuc) RNA fractions from HCASMCs transduced with either Ad-MYOCD or Ad-Control for 3 days. Results were representative of ≥3 separate experiments. C, RNA fluorescence in situ hybridization (RNA-FISH) analysis of the cellular localization of MYOSLID (red) and a positive control cytoplasmic RNA, PP1B (green) in HeLa cells under baseline conditions and HCASMCs transduced with either Ad-MYOCD or Ad-Control for 3 days. DAPI (Blue) reveals the nucleus compartment. D, Quantitative analysis of the average copy number of MYOSLID per single cell from the indicated number of cells randomly counted from HCASMCs transduced with either Ad-MYOCD or Ad-Control for 3 days.
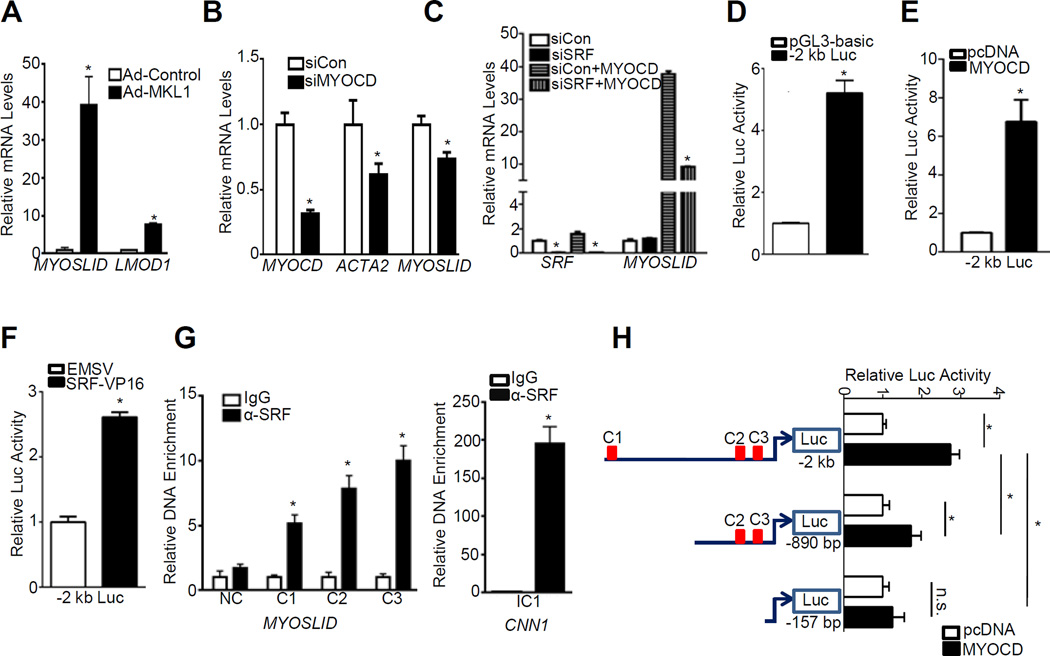
MYOSLID is a direct transcriptional target of MYOCD/SRF
Data above suggest MYOSLID could be a direct target of SRF/MYOCD. To test this hypothesis, we first evaluated if MYOSLID is induced by MKL1, a MYOCD related transcription factor family member whose function on VSMC differentiation is also associated with SRF/CArG28. Similar to MYOCD, overexpression of MKL1 caused a 40-fold increase of MYOSLID expression in HCASMCs (Figure 3A). Importantly, knockdown of MYOCD by siRNA resulted in significant downregulation of MYOSLID (Figure 3B). Surprisingly, depletion of endogenous SRF by siRNA had little effect on MYOSLID expression under basal conditions. However, SRF knockdown attenuated the induction of MYOSLID by MYOCD, indicating that MYOCD induction of MYOSLID expression requires normal SRF levels (Figure 3C).
Figure 3.
MYOCD/SRF-mediated transcriptional regulation of MYOSLID. A, qRT-PCR of the indicated genes in HCASMCs transduced with adenovirus carrying MKL1 (Ad-MKL1) or Ad-Control for 3 days. B, qRT-PCR analysis of the indicated genes in HCASMCs transfected with siRNA to MYOCD (siMYOCD) or negative control siRNA (siCon) for 3 days. C, HCASMCs were transfected with siRNA to SRF (siSRF) for 24 hrs, after which cells were transduced with Ad-MYOCD or Ad-Control for 48 hrs before RNA extraction for qRT-PCR analysis of the indicated genes. D, The −2 kb MYOSLID luciferase reporter (−2 kb luc) was transfected in SKLMS cells and baseline promoter activity was normalized to the control pGL3 basic reporter (set to 1). SKLMS were transfected with the −2 kb luc in the presence of either control pcDNA vector or MYOCD expression plasmid (E), EMSV control vector or SRF-VP16 (F) for 36 hrs. Luciferase activity is normalized to the internal control reporter Renilla. MYOCD or SRF-dependent activation of MYOSLID promoter was defined as fold increase to their respective vector control group (set to 1). Representative data from multiple independent experiments (n≥3) were expressed as the average of triplicates. G, Chromatin Immunoprecipitation (ChIP) assay was carried out with growing HCASMCs for assessment of binding of SRF to each individual putative CArG box denoted as C1, C2 and C3. Amplified DNA signal was normalized to the respective input control. Relative enrichment of CArG box containing fragment was expressed as fold increase to IgG control (set to 1). Primers to exon 3 of MYOSLID_V1 without any predicted CArG box (NC) and primers to CArG1 in intron1 (IC1) of CNN1 were used as negative and positive control, respectively. Representative data were shown from 3 independent experiments. H, Luciferase assay was done as described in (E) for the indicated luciferase reporters. Values are the mean ± SD and all data were representative of ≥3 separate experiments.
To further delineate the transcriptional regulation of MYOSLID, we PCR cloned the −2 kb promoter of MYOSLID into a luciferase reporter plasmid. This −2 kb MYOSLID luciferase reporter showed significant promoter activity compared with control vector in SKLMS, a human uterine leiomyosarcoma cell line which expresses similar markers of the contractile state as VSMCs (Figure 3D). Both MYOCD and SRF activated the −2 kb promoter in SKLMS (Figure 3E and 3F) and HEK293 cells (data not shown), providing evidence for CArG-dependent transactivation. ChIP assay with SRF antibody in HCASMCs revealed significant enrichment of the DNA flanking each of the predicted CArG boxes. In contrast, no obvious enrichment was seen in a negative control (NC) region of exon 3 where no predicted CArG box resides (Figure 3G, left panel), indicating the specificity of the binding between SRF-CArG. SRF binding to CArG boxes near MYOSLID was less than that seen in the intronic CArG1 (IC1) of CNN1 likely because of the greater affinity of SRF for the consensus CArG in CNN1 versus non-consensus CArGs near MYOSLID (Figure 3G, right panel).29
To ascertain if specific CArG boxes are critical for MYOCD transactivation, we constructed two truncated luciferase reporters and compared their response to MYOCD with the original −2 kb reporter. The −2 kb reporter encompassing 3 putative CArG boxes displayed the highest response to MYOCD. The truncated −890 bp reporter harboring CArG2/CArG3 showed a significantly reduced response to MYOCD. The −157 bp reporter lacking all three predicted CArG boxes was barely activated by MYOCD (Figure 3H). Collectively, these data support MYOSLID as a new direct transcriptional target of the MYOCD/SRF/CArG triad.
MYOSLID is an activator of the VSMC contractile phenotype
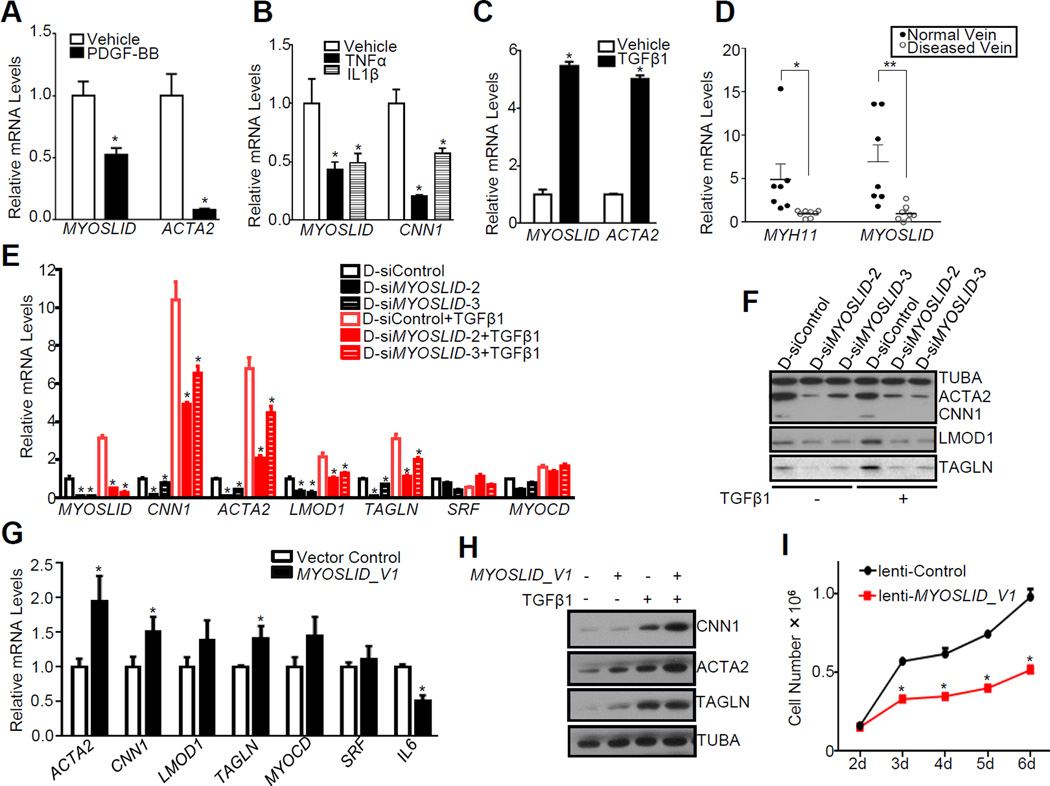
Having demonstrated MYOSLID as a direct transcriptional target of MYOCD/SRF, we sought to explore the association of MYOSLID with VSMC differentiation. Similar to ACTA2 or CNN1, MYOSLID was significantly downregulated with PDGF, TNFα and IL1β treatment (Figure 4A and 4B). On the other hand, MYOSLID was induced upon stimulation with TGFβ1, a potent activator of human VSMC differentiation30 (Figure 4C). To determine if MYOSLID is associated with vascular disease, we assessed MYOSLID levels by qRT-PCR in discarded and failed human arteriovenous fistula (AVF) samples from patients with end stage renal disease. The hyperplastic and stenotic response associated with AVF failure is thought to involve SMC de-differentiation.31 Similar to MYH11, the gold standard marker of the contractile VSMC phenotype, levels of MYOSLID were reduced in failed AVF specimens compared with normal control vein samples obtained at the time of AVF creation (Figure 4D). While no significant difference of SRF mRNA was seen between the normal and diseased AVF samples, MYOCD mRNA was sharply decreased in diseased vessels (Figure IV in the online-only Data Supplement), a result consistent with published data showing mRNA levels of MYOCD are decreased in vascular diseases.26 These data suggest that MYOSLID is a new member of the SRF-dependent program of human VSMC differentiation and, like other differentiation marker genes, is reduced under pathological conditions promoting VSMC de-differentiation.
Figure 4.
MYOSLID is an activator of the VSMC contractile phenotype. HCASMCs (A, C) or cultured Venous SMCs (B) were serum starved for 24 hrs, cells were then treated with the indicated growth factors for another 24 hrs before RNA isolation for qRT-PCR analysis of the indicated genes. Results were representative of 3 independent experiments. D, qRT-PCR of the relative expression of MYOSLID and MYH11 in diseased arteriovenous fistula vein (failed revision AVF samples) versus normal replacement vein samples. AVF samples were derived from unidentified discarded segments from patients with chronic kidney disease undergoing surgical revision of failed AVFs at Albany Medical College. E, HCASMCs transfected with two separate D-siMYOSLIDs (25 nM) or the same amount of D-siControl for 24 hrs and then serum starved for overnight, were subsequently treated with TGFβ1 (4 ng/ml) or vehicle control for another 24 hrs before RNA extraction for qRT-PCR analysis of the indicated genes. Values are the mean ± SD and all data were representative of ≥3 separate experiments. F, A representative Western blot of MYOSLID knockdown mediated by two separate D-siMYOSLIDs on the levels of the indicated proteins in basal or TGFβ1-treated HCASMCs as described above. Similar results were obtained in two additional HCASMC isolates (data not shown). Data were the representative of ≥3 separate experiments. G, HCASMCs were transfected with either MYOSLID_V1 expression plasmid or pcDNA vector control using electroporation machine (Nucleofector 2b, Lonza) for 72 hrs before total RNA extraction for qRT-PCR assessment of the indicated genes. Results are the mean ± SD from 3 independent experiments. H, Western blot for the effect of MYOSLID_V1 overexpression (described as above) on the indicated proteins. Results are representative of 3 independent experiments. I, HCASMCs were transduced with lentivirus carrying MYOSLID_V1 (lenti-MYOSLID_V1) or same amount of empty control lentivirus (lenti-Control) and cells were counted at the indicated days after transduction. Values are the mean ± SD and data were representative of three separate experiments.
The association of MYOSLID expression with the differentiated VSMC phenotype prompted us to investigate if MYOSLID confers a regulatory role in VSMC differentiation. Two D-siRNAs targeting different regions of the MYOSLID transcript achieved more than 80% knockdown efficiency in HCASMCs (Figure 4E). Depletion of MYOSLID resulted in significant decreases in mRNA levels of VSMC contractile genes under both basal and TGFβ1-induced conditions; no consistent effect was seen for SRF or MYOCD (Figure 4E). Western blotting validated the downregulation of VSMC contractile proteins upon knockdown of MYOSLID in HCASMCs (Figure 4F). We also demonstrated downregulation of the contractile proteins with a third D-siRNA to MYOSLID (data not shown). Conversely, overexpression of MYOSLID_V1 in HCASMCs caused a modest but significant increase in the mRNA levels of most VSMC genes such as ACTA2, CNN1 and TAGLN. This change was not applied to every gene as IL6 mRNA was reduced when MYOSLID_V1 was overexpressed (Figure 4G), suggesting the effect of overexpression of MYOSLID_V1 is specific to VSMC contractile genes. Western blot confirmed that protein levels of CNN1 and ACTA2 were also induced when MYOSLID_V1 was overexpressed (Figure 4H). Overexpression of MYOSLID_V1 increased TAGLN protein in basal conditions but not in TGFβ1-induced conditions, which is likely due to the fact that the strong induction of TAGLN protein exerted by TGFβ1 masks the effect of MYOSLID overexpression. Because differentiated VSMCs display less cell proliferation, we tested whether overexpression of MYOSLID_V1 could suppress growth and migration of HCASMCs. Indeed, lentiviral transduction of MYOSLID_V1 in serum-stimulated HCASMCs attenuated cell proliferation (Figure 4I) and migration (Figure VA in the online-only Data Supplement) compared with the negative control viral transduced cells. Conversely, depletion of MYOSLID promoted migration of HCASMs upon serum stimulation (Figure VB in the online-only Data Supplement). Taken together, these data demonstrate that MYOSLID is a novel positive regulator of the VSMC differentiation program.
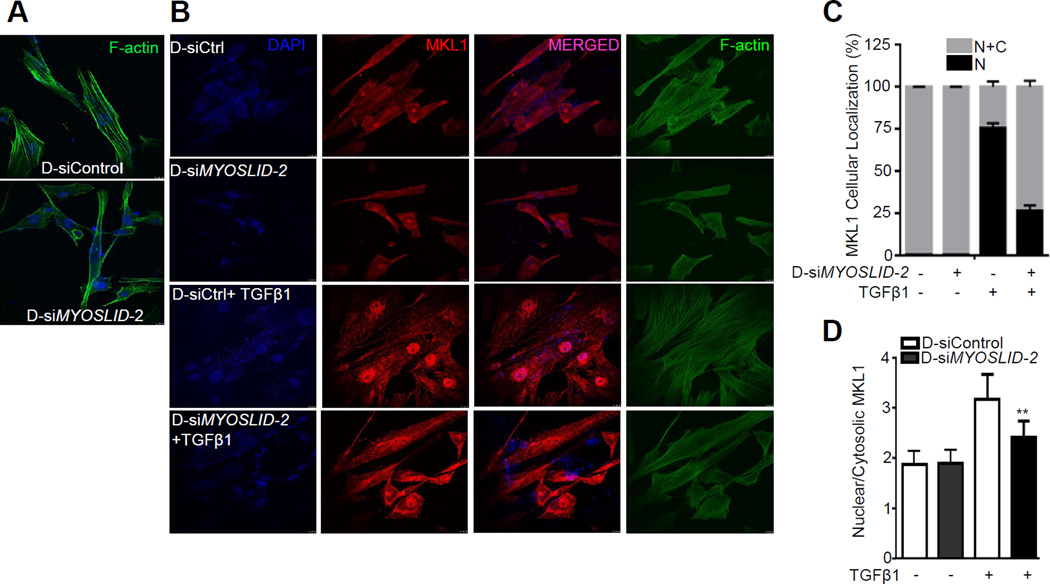
Depletion of MYOSLID disrupts stress fiber assembly and inhibits TGFβ1-induced MKL1 nuclear translocation
Depletion of MYOSLID resulted in an obvious disruption of stress fiber assembly visualized by F-actin staining in HCASMCs (Figure 5A). It has been established that MKL1 cellular shuttling and downstream SRF-dependent gene expression are tightly regulated by actin dynamics.32, 33 Accordingly, we considered the possibility that MYOSLID might affect MKL1 nucleocytoplasmic shuttling, and thus transcriptional activation of VSMC contractile genes, through effects of MYOSLID on the actin cytoskeleton. To test this hypothesis, MYOSLID expression was depleted with siRNA in HCASMCs followed by TGFβ1 treatment to stimulate nuclear translocation of MKL1.34 F-actin assembly was disrupted upon MYOSLID knockdown in both vehicle and TGFβ1 treated groups (Figure 5B). Further, forced expression of MYOSLID_V1 rescued F-actin formation in HCASMCs when SRF was depleted although no obvious differences were seen under the basal conditions (Figure VI in the online-only Data Supplement). Under unstimulated conditions, MKL1 was distributed in both cytosolic and nuclear fractions of control D-siRNA and D-siMYOSLID treated HCASMCs. TGFβ1 promoted a striking nuclear translocation of MKL1 in the control D-siRNA treated cells; however, this nuclear shuttling was attenuated upon knockdown of MYOSLID (Figure 5B). Quantitative analysis demonstrated about 75% of control D-siRNA treated cells had exclusively nuclear MKL1, and a small portion (25%) of cells showed both cytosolic and nuclear distribution of MKL1 after TGFβ1 treatment (Figure 5C). Conversely, the majority of cells (~75%) from the D-siMYOSLID treated group failed to undergo TGFβ1-induced MKL1 nuclear translocation, as MKL1 was retained in both cytosolic and nuclear compartments with only 25% of the cells exhibiting nuclear MKL1 (Figure 5C). D-siMYOSLID treated cells displayed a significantly lower ratio of nuclear to cytosolic MKL1 content compared with the control D-siRNA treated cells (Figure 5D), providing further evidence that depletion of MYOSLID attenuated MKL1 nuclear shuttling. Taken together, these data suggest MYOSLID promotes F-actin assembly leading to MKL1 nuclear translocation and the subsequent transcriptional activation of downstream VSMC contractile genes.
Figure 5.
Loss of MYOSLID disrupts stress fiber assembly and inhibits TGFβ1-induced MKL1 nuclear translocation. A, HCASMCs were transfected with the indicated D-siRNA to MYOSLID (D-siMYOSLID-2) or same amount of negative control D-siRNA (D-siControl) for 72 hrs followed by phalloidin staining. B, Growing HCASMCs were transfected with D-siMYOSLID-2 or D-siControl for 24 hrs, and starved with serum free medium overnight. Cells were then stimulated with TGFβ1 (4 ng/ml) for at least 24 hrs for phalloidin and MKL1 immunofluorescent staining. Quantitative analysis of MKL1 cellular localization and the relative intensity of MKL1 signal (Nuclear/Cytosolic) in each group were shown in (C) and (D), respectively. N+C (Nuclear+Cytosolic); N (Nuclear). >100 cells from 3 independent studies were analyzed.
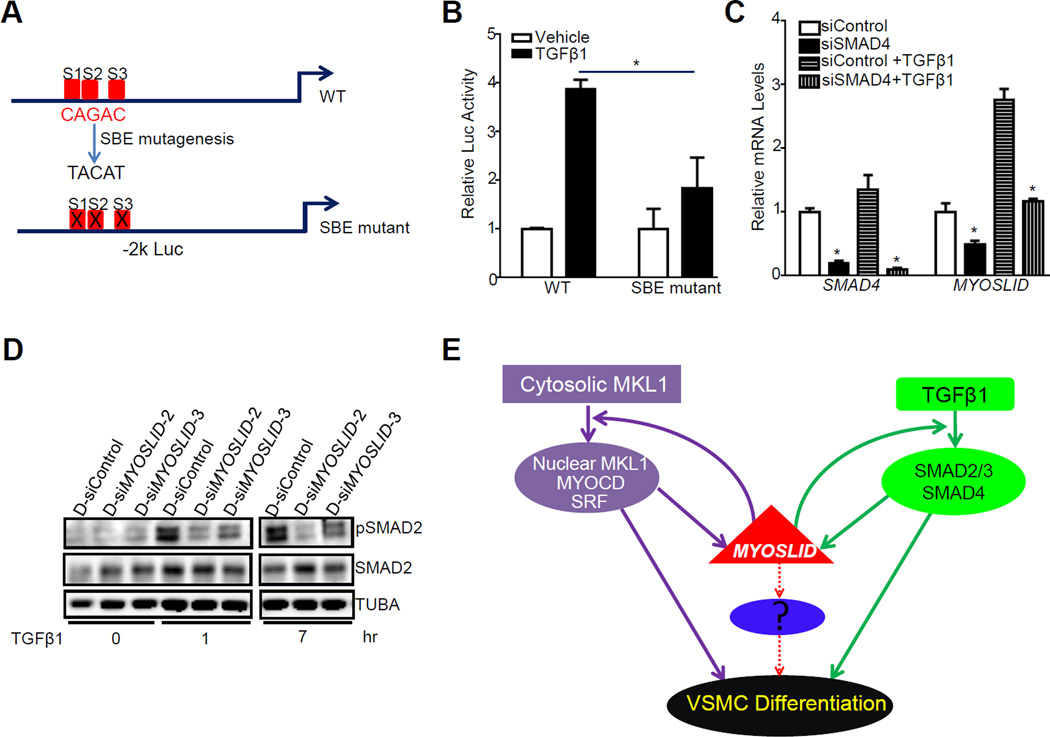
A positive feedback loop between TGFβ1/SMAD pathway and MYOSLID
Like most VSMC-specific genes, MYOSLID is also induced by TGFβ1 (Figure 4C). Sequence analysis of the −2 kb MYOSLID promoter indicated 3 putative SMAD-binding elements (SBEs) (Figure 6A), suggesting MYOSLID could be a direct transcriptional target of the classical TGFβ1/SMAD pathway as seen with the miR-143/145 locus.30 As expected, luciferase assay revealed that TGFβ1 induced a 4-fold increase in MYOSLID promoter activity whereas this activation was significantly attenuated when all 3 SBEs were mutated (Figure 6B), suggesting TGFβ1 activates MYOSLID promoter via one or more of these 3 SBEs. Consistent with the luciferase data, knockdown of endogenous SMAD4, the common mediator SMAD (co-SMAD), reduced the basal level of MYOSLID and abolished TGFβ1-induced upregulation of MYOSLID (Figure 6C). Based on these data, we conclude that MYOSLID is a direct transcriptional target of the classical TGFβ1/SMAD pathway.
Figure 6.
A positive feedback loop between TGFβ1/SMAD and MYOSLID and a working model of MYOSLID in VSMCs. A, Schematic for the 3 computer-predicted SMAD binding elements (SBEs, denoted as S1, S2 and S3, respectively) within the −2 kb MYOSLID promoter and the mutagenesis strategy of the SBEs. B, The −2 kb MYOSLID wildtype (WT) luciferase reporter and its SBE mutant (with all three SBEs mutated) were transfected in 10T1/2 cells. Cells were starved overnight and then stimulated by TGFβ1 (5 ng/ml) for 24 hrs before luciferase activity assessment. The TGFβ1 activation was defined as fold increase to the vehicle treated control group (set to 1). Representative data from multiple independent experiments (n≥3) were expressed as the average of triplicates. C, HCASMCs were transfected with siRNA to SMAD4 (siSMAD4) for 24 hrs, cells were then starved overnight followed by treatment with TGFβ1 (4 ng/ml) for another 24 hrs before RNA extraction for qRT-PCR analysis of the indicated genes. D, HCASMCs were transfected with the indicated D-siMYOSLID for 24 hrs, cells were then starved overnight followed by the treatment of TGFβ1 (4 ng/ml) for the indicated time. Western blot was done for the analysis of the indicated proteins. Data were representative of three separate experiments. E, Schematic model illustrating the regulation and the functional roles of MYOSLID in VSMCs: Two parallel pathways, MKL1/SRF and TGFβ1/SMAD, are critical to transcriptionally regulate MYOSLID in VSMCs; MYOSLID works with both pathways in a feedforward fashion to amplify VSMC differentiation. Solid lines represent the pathways suggested by the experimental evidence reported here and the hypothetical unknown pathway is indicated with dotted lines.
We next sought to evaluate the effect of loss of MYOSLID on SMAD2 activation, a key process for TGFβ1/SMAD in transactivating VSMC gene expression.35 Depletion of MYOSLID significantly decreased TGFβ1-induced SMAD2 phosphorylation at both 1 hr and 7 hrs after TGFβ1 treatment in HCASMCs (Figure 6D). These data indicate a positive feedback loop between TGFβ1/SMAD and MYOSLID in regulating VMSC contractile gene expression.
Discussion
The human genome undergoes pervasive transcription, thus debunking the traditional view of widespread “junk DNA”.36 LncRNAs, the byproduct of much transcription in the genome, are emerging as critical regulators of virtually every aspect of VSMC biology.23 Here, we report MYOSLID as the first CArG-SRF-MYOCD dependent lncRNA. MYOSLID appears to amplify the VSMC contractile program through an effect on two parallel pathways (Figure 6E). Loss of MYOSLID disrupts F-actin assembly, a key process governing MKL1 nuclear shuttling, which is necessary for MKL1 to transactivate VSMC contractile genes. Accordingly, loss of MYOSLID blocks TGFβ1-induced MKL1 nuclear translocation, implicating a novel mechanism through which MYOSLID may function to amplify VSMC contractile gene expression. Further, TGFβ1/SMAD, a critical regulatory pathway for VSMC development and differentiation,35, 37 directly transactivates MYOSLID. Reduced expression of MYOSLID attenuates TGFβ1-induced SMAD2 phosphorylation, indicating a positive feedback loop between TGFβ1/SMAD and MYOSLID in regulating VSMC differentiation. Since decreased MYOSLID is seen in the human failed AVF samples where VSMC differentiation is compromised, our results imply that loss of MYOSLID may play an important role in the vascular remodeling underlying the pathogenesis of vascular diseases. We surmise such loss in expression of MYOSLID is due to attenuated expression of MYOCD.
MYOCD is necessary and sufficient for driving a VSMC contractile phenotype through the induction of VSMC specific contractile genes and regulatory microRNAs.26 Since the vast majority of transcripts from the genome are lncRNAs, with more than 100,000 to date,15 we predicted there must be MYOCD regulated lncRNAs that augment or attenuate the VSMC contractile gene program. Indeed, RNA-seq in HCASMC revealed over 100 lncRNAs that are significantly regulated by MYOCD. Interestingly, careful analysis of 13 human MYOCD-lncRNAs revealed that all transcripts are only conserved in primates such as Rhesus monkey and Chimp, which is in agreement with the poorly conserved characteristic of most lncRNAs.38 Although MYOSLID lacks sequence conservation in rodents, it will be interesting to see if it possesses a conserved secondary structure, which might confer functional conservation in rodents. These human and primate-specific MYOCD-lncRNAs may render unique biological features to SMCs directly related to the pathogenesis of vascular diseases in humans. Therefore, elucidation of the regulation and functionality of MYOCD-lncRNAs will provide novel insights into the biology of MYOCD-dependent processes in VSMCs with high translational significance.
Thus far, several lncRNAs have been reported to differentially modulate VSMC proliferation, apoptosis, migration and differentiation17, 25, 39 and, among these, ANRIL and LincRNA-p21 have been suggested to regulate the progression of atherosclerosis.25, 39 However, these lncRNAs display a broad spectrum expression profile, which could be a potential obstacle for their clinical application. MYOSLID, on the other hand, is a VSMC-selective positive regulator of the VSMC differentiation program via promoting contractile gene expression and suppressing VSMC proliferation and migration. This result is consistent with data showing MYOSLID is induced by MYOCD and reduced in failed human AVF samples wherein the contractile VSMCs phenotypically switch to the synthetic state. We confirmed the knockdown effect of MYOSLID on contractile gene expression with multiple siRNAs in different HCASMC isolates, indicating MYOSLID is necessary for a fully executed VSMC contractile gene program. Of note, MYOSLID does not affect the gene expression of MYOCD, the molecular switch for the VSMC differentiation program6 (Figure 4E). Further, there was no consistent effect of MYOSLID on SRF or MKL1 (Figure 4E and Figure VIIA in the online-only Data Supplement), indicating the effect of MYOSLID on the downstream contractile gene expression is not attributed to altered gene expression of these regulators of the SMC contractile phenotype. Interestingly, overexpression of MYOSLID in non-SMC systems (such as HEK293 cells and BR5 fibroblast cells) was unable to induce contractile gene expression (data not shown), indicating the effect of MYOSLID on VSMC contractile gene expression is cell context-dependent and may only be restricted to VSMC. Thus, we do not consider MYOSLID as a master regulator of VSMC differentiation in the sense of MYOCD or the recent chromatin remodeling factor TET2.40 Given that MYOSLID is a VSMC-selective lncRNA that apparently amplifies the VSMC differentiation program, it could hold potential as a therapeutic target for treating vascular diseases.
MYOSLID is a direct transcriptional target of both MYOCD/SRF and TGFβ1/SMAD pathways, similar to miR143/145.30 Moreover, depletion of SMAD4 totally abolished TGFβ1-induced MYOSLID expression, indicating that the induction of MYOSLID by TGFβ1 is solely through the classical SMAD pathway, which is probably due to the transient induction of MYOCD by TGFβ1 in HCASMCs.30 Of note, 3 functionally active CArG boxes in the proximal promoter region of MYOSLID are not conserved in mouse, in keeping with the absence of a true ortholog of MYOSLID in mouse. By contrast, SENCR, a vascular cell-enriched lncRNA that also positively regulates the VSMC differentiation program,17 is a MYOCD/SRF-independent lncRNA despite the presence of several conserved CArG boxes in the proximal promoter (unpublished data). This underscores the fact that not every predicted CArG box is functional, especially those deviating from the consensus sequence whose SRF binding affinity is normally weak. There are thousands of conserved CArG boxes in the human genome41 and >100,000 lncRNAs annotated in the human genome.15 A big challenge therefore will be to identify additional SRF/CArG-dependent and human specific lncRNAs from the vast sea of lncRNAs through the integration of high throughput sequencing experiments, comprehensive bioinformatics analysis of CArG boxes and SRF ChIP-seq studies.
A notable paradigm to emerge from this study is the putative positive feedback loop between MYOSLID and TGFβ/SMAD, implying a novel mechanism in the regulation of MYOSLID on VSMC differentiation. The classical SMAD-dependent pathway, which necessitates the phosphorylation of SMAD transcription factors for its subsequent nuclear translocation, DNA-binding and ultimate transcriptional activation, has been demonstrated as an important pathway in vascular development and VSMC differentiation.35, 42 We found that depletion of MYOSLID decreases SMAD2 phosphorylation, indicating that MYOSLID is critical in the activation of SMAD and subsequent VSMC transcription. Inspired by the recent publications showing cytosolic lncRNAs influence the phosphorylation of signal proteins or transcription factors via direct physical association,21, 22 we attempted to detect the physical interaction between MYOSLID and SMAD2. However, both in vitro RNA pulldown and in vivo RNA precipitation assays have thus far failed to reveal such a physical interaction (data not shown). This may be attributed to one or more of the following facts: first, the interaction may be transient or weak and therefore difficult to establish with the assays employed; second, the effect of MYOSLID on SMAD2 phosphorylation may be an indirect phenomenon, involving unknown interactive partners. Thus, the mechanism through which MYOSLID influences SMAD2 phosphorylation needs to be further investigated with more sophisticated tools in RNA biology.
Actin dynamics are intimately linked to nuclear gene transcription, primarily through MKL1 nucleocytoplasmic shuttling.32, 33 In the static state, MKL1 binds G-actin and thus is retained in the cytoplasm. Upon G-actin polymerization, MKL1 is released and translocates to the nucleus where it binds SRF over CArG-dependent target genes.32, 33 We found depletion of MYOSLID can disrupt F-actin formation and block TGFβ1-induced MKL1 nuclear translocation. Further, forced expression of MYOSLID can rescue F-actin formation in VSMCs when SRF is depleted. We hypothesize that MYOSLID might impact the signal pathway(s) which is essential for F-actin assembly. F-actin formation is governed mainly by the upstream regulators of Rho family GTPases.32 Therefore, it will be important to examine whether MYOSLID can directly interact with Rho family GTPases and impact their activity.
The full-fledged VSMC contractile phenotype is orchestrated by a closely interactive network comprised of transcription factors, microRNAs and signal transducers.13, 23, 43 The emergence of lncRNAs in VSMCs renders this network even more comprehensive. However, given that MYOSLID is primarily localized in the cytosol, it is unlikely that MYOSLID directly interferes with the transactivation of SRF/MYOCD by physically interacting with them or effecting chromatin remodeling. Further, although a pathway involving a competing endogenous (“sponge”) RNA titrating a microRNA of VSMC differentiation (such as miR1) from its target transcript (such as MYOCD), is an attractive mechanism for MYOSLID to execute its positive role in governing VSMC differentiation, analysis of data from the LNCipedia database did not reveal any potential seed sequences corresponding to known microRNAs within the MYOSLID transcript (unpublished). In line with this, MYOSLID has little to no effect on MYOCD expression. We recently reported that SENCR is a vascular cell-enriched lncRNA that stabilizes the VSMC contractile phenotype.17 Though both MYOSLID and SENCR appear to act as positive regulators of VSMC contractile phenotype, depletion of MYOSLID has no effect on SENCR expression (Figure VIIB in the online-only Data Supplement). Therefore, the effect of MYOSLID on VSMC differentiation does not appear to be associated with SENCR expression. Knockdown of SENCR can decrease MYOCD gene expression, suggesting the regulation of SENCR on VSMC differentiation is likely through MYOCD expression. However, this effect was not seen with MYOSLID (Figure 4E). Altogether, we surmise that MYOSLID and SENCR may utilize their own distinct mechanisms in augmenting the VSMC contractile phenotype.
In summary, we report an array of lncRNAs regulated by MYOCD which we refer to as MYOCD-lncRNAs. We demonstrated that one of these MYOCD-lncRNAs, MYOSLID, is VSMC-selective, SRF/CArG-dependent, and an amplifier of the VSMC differentiation program, likely through effects on MKL1 nuclear translocation and activation of the TGFβ/SMAD pathway. These studies, together with our previous findings with SENCR, imply that VSMC lncRNAs constitute a new layer of molecular regulation of the differentiated VSMC phenotype.
Supplementary Material
Highlights.
Long noncoding RNAs (lncRNAs) represent an expansive class of genes in the mammalian genome and are emerging as important regulatory molecules in many aspects of cell biology and disease. We report here an array of MYOCD regulated lncRNAs (MYOCD-lncRNAs) and demonstrated one of these MYOCD-lncRNAs, MYOSLID, as the first VSMC-selective and SRF/CArG-dependent lncRNA. Importantly, we provided evidence showing that MYOSLID can amplify human VSMC differentiation, likely through effects on MKL1 nuclear translocation and activation of the TGFβ/SMAD pathway. Levels of MYOSLID are closely correlated with VSMC differentiation and reduced in human failed arteriovenous fistula samples where VSMC differentiation is compromised, indicating MYOSLID may play an important role in the vascular remodeling underlying the pathogenesis of vascular diseases. Our studies thus provided novel insights into the regulation of VSMC phenotypic plasticity and will potentially open new avenues for the development of novel therapeutic and preventive strategies in combating vascular disease.
Acknowledgments
We thank the University of Rochester Genomics Research Center for performing the RNA-seq experiments. We thank the following surgeons at Albany Medical Center for providing the precious human samples throughout the studies: David Conti, MD, AMC Surgery Group - Transplantation Department; Reynold Lopez-Soler, MD and PhD, AMC Surgery Group - Transplantation Department; Nikolaos Chandolias, MD, AMC Surgery Group - Transplantation Department and Paul Kreienberg, MD, the Vascular Group, PLLC, Albany.
Sources of Funding
This work is supported by American Heart Association Scientist Development Grant 10SDG3670036, National Institutes of Health R01HL122686, Paul Teschan Research Fund# 2015_04 and Albany Medical College faculty startup funding to X.L; R01HL117907 to J.M.M; R01HL49426 to H.A.S.
Nonstandard Abbreviations and Acronyms
- HCASMCs
human coronary artery smooth muscle cell(s)
- lncRNA
long noncoding RNA
- MYOCD
myocardin
- SRF
serum response factor
- RT-PCR
reverse transcription polymerase chain reaction
- MYOSLID
MYOcardin-induced Smooth muscle Long non-coding RNA, Inducer of Differentiation
- VSMC
vascular smooth muscle cell
Footnotes
Disclosures
None.
References
- 1.Thyberg J. Phenotypic modulation of smooth muscle cells during formation of neointimal thickenings following vascular injury. Histology and histopathology. 1998;13:871–891. doi: 10.14670/HH-13.871. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological reviews. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Herring BP, Hoggatt AM, Burlak C, Offermanns S. Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vascular cell. 2014;6:21. doi: 10.1186/2045-824X-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauletto P, Sarzani R, Rappelli A, Chiavegato A, Pessina AC, Sartore S. Differentiation and growth of vascular smooth muscle cells in experimental hypertension. American journal of hypertension. 1994;7:661–674. doi: 10.1093/ajh/7.7.661. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: A component of a molecular switch for smooth muscle differentiation. Journal of molecular and cellular cardiology. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 7.Long X, Tharp DL, Georger MA, Slivano OJ, Lee MY, Wamhoff BR, Bowles DK, Miano JM. The smooth muscle cell-restricted kcnmb1 ion channel subunit is a direct transcriptional target of serum response factor and myocardin. J Biol Chem. 2009;284:33671–33682. doi: 10.1074/jbc.M109.050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanda V, Miano JM. Leiomodin 1, a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. J Biol Chem. 2012;287:2459–2467. doi: 10.1074/jbc.M111.302224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsson E, McLean SE, Mecham RP, Lindahl P, Nelander S. Do two mutually exclusive gene modules define the phenotypic diversity of mammalian smooth muscle? Molecular genetics and genomics : MGG. 2008;280:127–137. doi: 10.1007/s00438-008-0349-y. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. Microrna-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circulation research. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. Mir-145 and mir-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidersbach A, Saxby C, Carver-Moore K, Huang Y, Ang YS, de Jong PJ, Ivey KN, Srivastava D. Microrna-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. eLife. 2013;2:e01323. doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albinsson S, Sessa WC. Can micrornas control vascular smooth muscle phenotypic modulation and the response to injury? Physiological genomics. 2011;43:529–533. doi: 10.1152/physiolgenomics.00146.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding rnas. PLoS genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, Chen R. Noncode 2016: An informative and valuable data source of long non-coding rnas. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, Head SR, Duester G, Dong PD, Rana TM. An evolutionarily conserved long noncoding rna tuna controls pluripotency and neural lineage commitment. Molecular cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding rna. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding rna transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun M, Gadad SS, Kim DS, Kraus WL. Discovery, annotation, and functional analysis of long noncoding rnas controlling cell-cycle gene expression and proliferation in breast cancer cells. Molecular cell. 2015;59:698–711. doi: 10.1016/j.molcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida S, Dimmeler S. Long noncoding rnas in cardiovascular diseases. Circulation research. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 21.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic nf-kappab interacting long noncoding rna blocks ikappab phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The stat3-binding long noncoding rna lnc-dc controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 23.Miano JM, Long X. The short and long of noncoding sequences in the control of vascular cell phenotypes. Cellular and molecular life sciences : CMLS. 2015;72:3457–3488. doi: 10.1007/s00018-015-1936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R. Novel long noncoding rnas are regulated by angiotensin ii in vascular smooth muscle cells. Circulation research. 2013;113:266–278. doi: 10.1161/CIRCRESAHA.112.300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C. Lincrna-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miano JM. Myocardin in biology and disease. J Biomed Res. 2015;29:3–19. doi: 10.7555/JBR.29.20140151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin MF, Jungreis I, Kellis M. Phylocsf: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics (Oxford, England) 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Fang H, Zhou J, Herring BP. A novel role of brg1 in the regulation of srf/mrtfa-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282:25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 29.Long X, Slivano OJ, Cowan SL, Georger MA, Lee TH, Miano JM. Smooth muscle calponin: An unconventional carg-dependent gene that antagonizes neointimal formation. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:2172–2180. doi: 10.1161/ATVBAHA.111.232785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long X, Miano JM. Transforming growth factor-beta1 (tgf-beta1) utilizes distinct pathways for the transcriptional activation of microrna 143/145 in human coronary artery smooth muscle cells. J Biol Chem. 2011;286:30119–30129. doi: 10.1074/jbc.M111.258814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang M, Wang Y, Liang A, Mitch WE, Roy-Chaudhury P, Han G, Cheng J. Migration of smooth muscle cells from the arterial anastomosis of arteriovenous fistulas requires notch activation to form neointima. Kidney international. 2015;88:490–502. doi: 10.1038/ki.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nature reviews. Molecular cell biology. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control srf activity by regulation of its coactivator mal. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 34.Trembley MA, Velasquez LS, de Mesy Bentley KL, Small EM. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development (Cambridge, England) 2015;142:21–30. doi: 10.1242/dev.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Y, Yang X, Friesel RE, Vary CP, Liaw L. Mechanisms of tgf-beta-induced differentiation in human vascular smooth muscle cells. Journal of vascular research. 2011;48:485–494. doi: 10.1159/000327776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno S. So much "junk" DNA in our genome. Brookhaven symposia in biology. 1972;23:366–370. [PubMed] [Google Scholar]
- 37.Chen S, Lechleider RJ. Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circulation research. 2004;94:1195–1202. doi: 10.1161/01.RES.0000126897.41658.81. [DOI] [PubMed] [Google Scholar]
- 38.Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding rnas and the evolution of gene expression. PLoS genetics. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, Yamamoto K, Onishi M, Sugimoto K, Katsuya T, Awata N, Ikebe K, Gondo Y, Oike Y, Ohishi M, Rakugi H. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of anril and cdkn2a/b. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Liu R, Jin Y, Tang WH, Qin L, Zhang X, Tellides G, Hwa J, Yu J, Martin KA. Ten-eleven translocation-2 (tet2) is a master regulator of smooth muscle cell plasticity. Circulation. 2013;128:2047–2057. doi: 10.1161/CIRCULATIONAHA.113.002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson CC, Zhou Q, Long X, Miano JM. Identifying functional single nucleotide polymorphisms in the human cargome. Physiological genomics. 2011;43:1038–1048. doi: 10.1152/physiolgenomics.00098.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao X, Debenedittis P, Sun Y, Chen J, Yuan K, Jiao K, Chen Y. Vascular smooth muscle cell smad4 gene is important for mouse vascular development. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:2171–2177. doi: 10.1161/ATVBAHA.112.253872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, Urs S, Boucher J, Bernaiche T, Venkatesh D, Spicer DB, Vary CP, Liaw L. Notch and transforming growth factor-beta (tgfbeta) signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. J Biol Chem. 2010;285:17556–17563. doi: 10.1074/jbc.M109.076414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.