Summary
Genome wide association studies (GWAS) have identified numerous loci that influence risk for psychiatric diseases. Genetically engineered mice are often used to characterize genes implicated by GWAS. These studies are based on the assumption that observed genotype-phenotype relationships will generalize to humans, implying that the results would at least generalize to other inbred mouse strains. Given current concerns about reproducibility we sought to directly test this assumption. We produced F1 crosses between male C57BL/6J mice heterozygous for null alleles of Cacna1c and Tcf7l2 and wild-type females from 30 inbred laboratory strains. We found extremely strong interactions with genetic background that sometimes supported diametrically opposing conclusions. These results do not negate the invaluable contributions of mouse genetics to biomedical science, but they do show that genotype-phenotype relationships cannot be reliably inferred by studying a single genetic background, and thus constitute a major challenge to the status quo.
Introduction
The mouse is the premiere mammalian model organism. Since the development of transgenic and knock out mice, thousands of publications have used mice to define relationships between genotypes and phenotypes (Brandon et al., 1995; Capecchi, 1989). The advent of nuclease-mediated genome editing strategies including CRISPR/Cas9 is expected to accelerate the use of mice for this purpose (Singh et al., 2014). Mutant alleles are typically studied on a single inbred strain background, reflecting the widely held reductionist world-view that seeks to examine a single genetic difference while holding all other genetic and environmental variables constant (Little et al., 2016). Moreover, most studies focus on relatively young animals and, until recently, most mutant alleles were studied in only one sex, typically males. As human genome wide association studies (GWAS) continue to implicate new loci in a wide range of common diseases, mutant mice are being used to examine which genes within these loci may influence disease liability and to examine the underlying molecular mechanisms. At the same time, the International Mouse Knockout Consortium (IKMC) and affiliated groups are currently generating and phenotyping mice with null alleles of every gene in the genome on a single genetic background (Bradley et al., 2012; Collins et al., 2007). Thus, a huge literature continues to accumulate that describes the phenotypes caused by mutant alleles, typically on a single inbred strain background.
Concerns about reproducibility have prompted recent discussions about failures to replicate results in biomedical science. Replicability is a paramount consideration when conducting phenotypic screening among mutant mice in single laboratories and is also a major issue for high-throughput efforts such as the IKMC. There are integrated resources that describe standard operating procedures for phenotyping as well as robust methodologies for data analysis that maximize power and reproducibility (Hrabě de Angelis et al., 2015; Karp et al., 2015; Kilkenny et al., 2010; Mallon et al., 2008). In addition, the National Institutes of Health has implemented a major policy change to encourage consideration of sex as a biological variable (Tannenbaum et al., 2016). Yet there is no standard in the field for establishing generalizability across inbred strains, despite an awareness among mouse geneticists that genetic background is another potential source of variability (Doetschman, 2009; Phillips et al., 1999; Sanford et al., 2001; Sibilia and Wagner, 1995; Threadgill et al., 1995). Examples of epistatic interactions among naturally occurring alleles were reported a century ago (Castle, 1919; Dexter, 1914).
We examined the phenotypic effects of two null alleles using F1 crosses between C57BL/6J and a panel of 30 different inbred strains. The two genes that we chose to study have been strongly implicated in multiple disorders by human GWAS. The first gene, CACNA1C, has been associated with bipolar disorder (Ferreira et al., 2008; Green et al., 2012; Sklar et al., 2008) and schizophrenia (Hamshere et al., 2012; Nyegaard et al., 2010; Ripke et al., 2011), as well as with cross-disorder risk for several other major psychiatric disorders (Smoller et al., 2013). Mice with a null allele for Cacna1c have been reported to display numerous behavioral phenotypes (Dao et al., 2010), which lends support to the human studies implicating CACNA1C in psychiatric diseases. The second gene we studied, TCF7L2, is among the most strongly associated and best replicated genetic risk factors for type 2 diabetes (Fuchsberger et al., 2016; Grant et al., 2006; Morris et al., 2012). In addition to its metabolic role, TCF7L2 has also been associated with schizophrenia and bipolar disorder (Alkelai et al., 2012; Hansen et al., 2011; Winham et al., 2013). Mice with a null allele for Tcf7l2 exhibit both physiological and behavioral phenotypes including improved glucose tolerance, altered fear learning, and anxiety (Savic et al., 2011a, 2011b). Here we evaluated the generalizability of Cacna1c +/- and Tcf7l2 +/-phenotypes using many different genetic backgrounds. Our results illustrate how the reproducibility and robustness suffer when only a single strain is considered.
Results
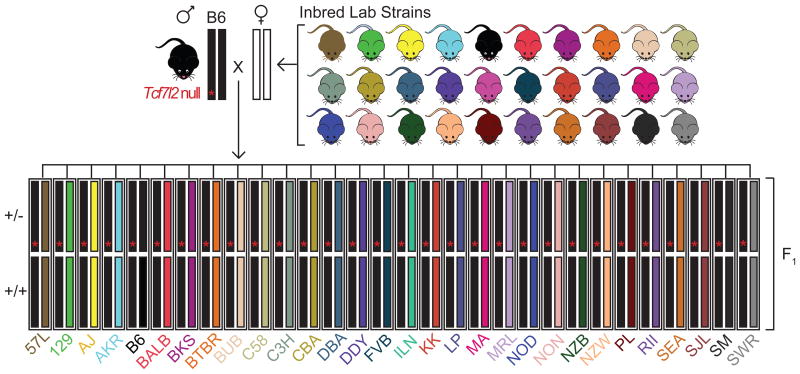
We generated a structured panel of heterozygous null and wild-type mice from 30 different F1 backgrounds by breeding +/- C57BL/6J males to +/+ females from 30 inbred strains (Figure 1). This produced a panel of +/+ and +/- littermates that were isogenic at all other loci. We used this breeding design to generate two cohorts of mice, one for the Cacna1c null allele and one for the Tcf7l2 null allele. Each cohort of mice was produced independently. Mice in the Cacna1c cohort (N=723) were tested for anxiety, methamphetamine sensitivity, depression-like behavior, and acoustic startle response. Mice in the Tcf7l2 cohort (N=630) were tested for several behavioral traits: anxiety, fear conditioning, and sensorimotor gating, as well as several metabolic traits: body weight, baseline blood glucose levels and fasted blood glucose levels. Thus, we obtained data for 15 phenotypes, 12 of which were behavioral (Table S3).
Figure 1.
Breeding design for placing null alleles on 30 different genetic backgrounds. Males heterozygous for the null allele mutation (red asterisk) on a C57BL/6J background were bred with females from 30 inbred laboratory mouse strains (represented by arbitrarily selected colors). This generated 30 distinct F1s that were genetically identical except at the targeted allele, where they were either +/- or +/+.
We estimated the variance in each phenotype explained by the factors of interest in our experimental design: genotype (+/+ or +/-), F1 genetic background (“strain”), sex and the two- and three-way interactions among these factors. The interaction between genotype and strain indicates the degree to which the null allele's effect depended on strain and is therefore a measure of generalizability of the null allele's phenotypic effects. Strikingly, the only Cacna1c +/- phenotypes that were generalizable across strain (i.e. for which there was not a significant genotype × strain interaction) were light avoidance in the light/dark box and acoustic startle response. The only Tcf7l2 +/- phenotypes that were generalizable were decreased body weight and decreased contextual fear learning (Table 1 and Table S3). The remaining phenotypes depended on the strain in which the null allele was expressed (Figure 2A-D and Table S3). The interactions affected phenotypes with highly penetrant, weak, and non-existent main effects of the null allele (Figure S1).
Table 1.
Genotype × strain interactions.
| Phenotype | Genetic background × null allele (ANOVA p-value) | Phenotypic variance explained |
|---|---|---|
| Cacna1c study | ||
| Percent time in the light box | 0.20‡ | 4.3% |
| Exploratory activity | 0.01* | 3.4% |
| Duration in center | 9.9e-08*** | 7.4% |
| Methamphetamine sensitivity | 9.5e-3** | 4.1% |
| Immobility | 7.4e-3** | 4.4% |
| Acoustic startle response | 0.08‡ | 1.6% |
| Tcf7l2 study | ||
| Body weight | 0.42 | 0.5% |
| Exploratory activity | 6.8e-06*** | 7.0% |
| Duration in center | 6.8e-07*** | 5.8% |
| Contextual fear learning | 0.23 | 3.5% |
| Cued fear learning | 0.08‡ | 4.2% |
| Acoustic startle response | 8.9e-08***‡ | 4.2% |
| Prepulse inhibition | 8.7e-04*** | 8.1% |
| Fasted blood glucose levels | 0.24 | 3.4% |
| Baseline blood glucose levels | 0.05 | 4.2% |
Figure 2.
Null allele phenotypes on 30 different genetic backgrounds. (A) Locomotor response to methamphetamine in Cacna1c +/- and +/+ mice. On days 1 and 2 a saline injection was given before a 30 min exposure to an open field. On day 3 methamphetamine (2 mg/kg i.p.) was given prior to the same open field test. The difference in locomotor activity on day 3 compared to activity on day 2 is shown. (B) Exploratory activity during a 30 min open field test in Tcf7l2 +/- and +/+ mice. (C) Baseline blood glucose levels in Tcf7l2 +/- and +/+ mice. (D) Startle response to a 120-dB acoustic stimulus in Tcf7l2 +/- and +/+ mice. Bars show the mean phenotype +/- s.e.m.. F1s are labeled according to the maternal strain, and are ordered along the x-axis according to the difference between +/- and +/+ mice. See also Table S3. Null allele effects within strains were evaluated using two-tailed t-tests. _p<0.1, *p<0.05, **p<0.01, ***p<0.001. –p<0.1, *p<0.05, **p<0.01, ***p<0.001.
Phenotypic data from different F1s frequently supported dramatically different conclusions about the effect of the null alleles. For example, Cacna1c genotype affected methamphetamine sensitivity in approximately half of the F1s, while the remaining F1 +/- mice were not significantly different from +/+ mice (Figure 2A). This spectrum of vulnerability was also observed for Tcf7l2 (e.g. Figure 2B, C). However, the impact of genetic background was not always merely a matter of degree; there were four instances in which directionally opposite effects of the same allele occurred in different genetic backgrounds (Table 1 and Table S3). For example, a genotype × strain interaction precluded a main effect of the Tcf7l2 allele on startle response (Figure 2D). Post-hoc tests demonstrated that for three of the F1s (DBA, BKS, ILN) Tcf7l2 haploinsufficiency decreased acoustic startle response, whereas the opposite was true for two other F1s (AJ, NZB).
Finally, we considered the role of sex as a biological variable. Three of the traits showed modest but significant interactions between genotype and sex (startle, body weight and prepulse inhibition). For body weight, there was a significant three-way interaction between genotype, strain and sex (Table S3).
Discussion
Single strains are often used to establish the presence, absence, and severity of phenotypes in genetically engineered mice. The conclusions from such experiments inform our understanding of genetic and physiological systems, yet these conclusions are predicated on the assumption that they generalize to humans, which presupposes that they would at least generalize to other inbred mouse strains. Here we evaluated the phenotypic effects of two null alleles on a panel of F1 mice derived from commonly used inbred mouse strains. Our breeding design produced heterozygous null littermates that were genetically identical to their respective wild-type controls at all loci besides the targeted allele. The majority of null phenotypes observed in Cacna1c +/- and Tcf7l2 +/- mice were not generalizable: phenotypic responses often varied from strongly affected to unaffected in different genetic backgrounds, and in several cases there were directionally opposite effects of the same allele. Overall, the prevalence and the strength of null allele interactions with genetic background were even stronger than null allele interactions with sex. This study illustrates that the choice of genetic background can have a dramatic effect on the null allele phenotype, challenging the reductionist idea that mutant alleles have a specific phenotype that can be readily determined using a single strain.
The null alleles we evaluated had been phenotyped previously using single genetic backgrounds, allowing us to compare our results to those previous studies. The importance of procedural and environmental differences complicate direct comparisons of our phenotypic data to previous results (Crabbe et al., 1999)(Wahlsten et al., 2003)(Chesler et al., 2002)(Sorge et al., 2014). However, in the case of Cacna1c, the one assay that was performed and analyzed with very similar procedures replicated a sex-specific decrease of the startle response in C57BL/6J mice (Dao et al., 2010) (Table S3). For Tcf7l2, the null allele was previously evaluated in mice from a CD-1 outbred background in our laboratory (Savic et al., 2011a). We identified five phenotypes from that study that could be compared to our current results, three of which exhibited strong interactions with genetic background that precluded cross-study comparisons of single phenotypic effects. The other two were different from what was reported in CD-1 mice, and for one of them, contextual fear learning, the direction of the effect was directionally opposite to that previously reported. These results further illustrate how genetic background can dramatically alter experimental conclusions.
The statistical power to detect effects of the null alleles was limited by sample size in our study design. Certain F1s that appeared sensitive or insensitive to one of the mutant alleles may represent type 1 or 2 errors; accordingly, the number of significant post-hoc tests was reduced when we performed permutations of the phenotypic data (data not shown). However, correction for multiple testing would not have been appropriate because our goal was to compare the experimental conclusions that would have been reached if only a single genetic background was considered. Our study design could have been extended to include reciprocal crosses or even a full diallele cross; however, our design was sufficient to demonstrate the importance of genetic background. Our use of F1 mice most likely reduced interactions when compared to what would be seen across fully inbred strains. Had we introduced the same mutation using nuclease-mediated genome editing onto multiple pure inbred lines rather than F1s, we might have seen stronger interactions with genetic background, but these could have been due to either off-target mutations that would have differed from strain to strain or true interactions with genetic background.
This study was not designed to address the prevalence of epistatic interactions involving CACNA1C and TCF7L2 in humans; indeed, we examined null alleles, which are presumably more severe than the corresponding human risk alleles. Furthermore, it is possible that the unique population history of inbred mouse strains contributes to the strength of the observed interactions, for example by amplifying the frequency of alleles that are rare in wild mouse populations. It is not clear whether interactions with genetic background would have been as prevalent had we used wild rather than laboratory mice. It has been argued that evaluation of mutant phenotypes in single strains of F1 hybrid mice, such as those we used here, is a preferable strategy to using single inbred strains because F1s capture greater genetic diversity and are less likely to show anomalous phenotypic effects (Silva et al., 1997). However, our data suggest that using F1 animals does not circumvent the confounding effects of genetic background.
What can be done to remedy this problem? The strong effects of genetic background represent both a blessing and a curse. While they complicate the evaluation of mutant alleles, they also create new experimental opportunities. It is now feasible to create mutations in several strains using nuclease-mediated genome editing or, when mutant phenotypes of interest are dominant, by using an F1 breeding scheme similar to the one presented here. Differences in susceptibility across mouse strains allow the identification of gene-gene interactions (Nadeau, 2001), an approach that has been used successfully in the past (Dietrich, 1993; Hamilton and Yu, 2012; Heydemann et al., 2009; Hide et al., 2002; Pinto et al., 2013; Rozmahel et al., 1996). There are major opportunities to leverage the genetic diversity among inbred mouse strains to reveal functional biological networks that underlie disease processes, especially as the engineering of mutant alleles becomes increasingly efficient. Doing so will be key to unraveling the genetic basis for disease-relevant traits as well as developing new therapeutic avenues to intervene in the associated pathophysiology. We must broaden our focus beyond single strains to realize this potential.
Experimental Procedures
Null alleles
Cacna1c +/- mice were originally developed by Deltagen (San Mateo, CA) and were obtained from the Jackson Laboratory (Strain 005783). The line was backcrossed to C57BL/6J for at least five generations prior to arrival at our facility. We backcrossed the line for three additional generations. We examined residual heterozygosity in the backcrossed Cacna1c +/- animals using the Mega Mouse Universal Genotyping Array (MegaMUGA) (http://csbio.unc.edu/ccstatus). The founders used to breed the F1 panel were 99.8% identical to C57BL/6J based on 73,178 informative SNPs.
Tcf7l2 +/- mice on a C57BL/6J background were generated using a zinc finger nuclease construct obtained from Dr. Marcelo Nobrega (University of Chicago). We obtained a mutant line with a 10 bp frameshift-inducing deletion. The founder male was backcrossed to C57BL/6J for one generation before generating F1 crosses. This same construct was previously used to generate Tcf7l2 +/- mice on a CD-1 outbred background (Savic et al., 2011b).
It is possible that off target mutations in the Tcf7l2 line or residual heterozygosity in the Cacna1c line may have had phenotypic effects that were falsely attributed as phenotypes of the null allele. However, our use of littermate controls ensures that these could not have been the source of the interactions between the null allele and genetic background. Additional details about the mouse lines and genotyping the null alleles are found in Supplemental Experimental Procedures.
Generation of F1 offspring and phenotyping
All animal procedures were approved by the University of Chicago Institutional Animal Care and Use Committee. Mice were housed in a single, pathogen-free barrier facility. Lights were on a 12h on/12h off cycle with lights on at 0600h. Mice were housed in standard polycarbonate cages with corn cob bedding and ad libitum access to water and laboratory chow. Water (filtered by reverse osmosis) was available in each cage. Breeders received Envigo (formerly Harlan) 2919 19% protein chow. F1 offspring received Envigo 2918 18% protein chow after weaning. The length of time required to produce and phenotype all F1 mice was 8 months for the Tcf7l2 cohort and 6 months for the Cacna1c cohort. The housing room, caging systems, diet, water source, and husbandry practices were held constant throughout each cohort.
The 30 classical laboratory mouse strains we selected are priority strains in large community genotyping and phenotyping efforts (Table S1) (Grubb et al., 2014). Females from each strain were obtained from the Jackson Laboratory at 6-8 weeks of age and acclimated for one week before being placed in harems with a heterozygous (+/-) C57BL/6J male. Once females were visibly pregnant they were singly housed. F1 offspring were weaned at 21-24 days of age. They were housed in same-sex cages containing at least two and no more than five littermates of the same genetic background. In the event that there was only one male or female littermate, the animal was excluded from the study. Randomization of genotype and sex occurred as a result of our breeding scheme as wild type vs. heterozygous and male vs. female littermates were produced at equal ratios. Litters were combined into testing groups containing up to 48 mice. Females from all 30 strains were bred on a rotating basis such that the production of litters, and the composition of each test group of F1 mice, was randomized with respect to strain.
All mice within the same cohort (e.g. Cacna1c or Tcf7l2) had the same behavioral testing schedule (Table S2). All testing was conducted between 0900h - 1600h. The experimenter conducting each assay was held constant. Experiments began after 7-11 weeks of age and progressed from relatively least stressful to more stressful with at least four days between tests. Before each test, mice were acclimated to the test room for at least 30 min in their home cages. They were then placed in a clean holding cage to await the start of testing. Each animal was assigned an arbitrary 5 digit identification number that obscured their strain and genotype from the experimenter. Animals were placed in the appropriate testing apparatus by the experimenter, and were then directly returned to their home cages after completion of the test. Testing equipment was cleaned with 10% isopropanol between animals. All behavior was monitored and scored by automated software systems, removing the possibility of human bias. Factors specific to the testing environment including the particular test box in which the animal was placed and the time of day (morning or afternoon) were recorded as potential covariates. Details of each phenotypic test are given in the Supplemental Experimental Procedures.
Statistics
The R Statistical environment was used to fit a linear regression model for each phenotype. When appropriate, we transformed some phenotypes onto the logarithmic (base 10) scale, and transformed others using the logit function, logit10(x) = log10(x/(1-x)) to ensure that the distribution of residuals met the assumption of normality. We modeled each phenotype as a linear combination of covariates including sex, body weight, coat color, and other experimental factors relevant to each phenotypic analysis. Covariates explaining less than 2% of the variance were not used in the linear regression. Outlying data points with a residual more than two standard deviations away from the mean of each strain were removed. We confirmed that the residuals of our linear models had empirical quantiles that closely matched the expected quantiles under the normal distribution. Analysis of variance was conducted to evaluate main effects and interactions using the anova.lm function in R (see Supplemental Information). Because we were interested in the effect of the null alleles in each individual F1, we performed t-tests comparing +/+ to +/- within each F1. We did not apply a multiple testing correction (e.g. Bonferroni) because we wanted to illustrate the different conclusions that would have been reached had only one genetic background been considered, which is the common practice when evaluating the phenotypic consequences of a mutant allele. The raw phenotype data and code used to complete the analyses are available at https://github.com/pcarbo.
Supplementary Material
Acknowledgments
The authors thank Dr. Marcelo Nobrega for the Tcf7l2 targeting construct and Dr. Bruce Hamilton for providing scholarly input on a draft of the manuscript.
Footnotes
Author contributions: AP and LS conceived and designed the experiments; LS, KE, KK and CB performed the experiments; PC and LS analyzed the data; LS, AP, and PC wrote and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkelai A, Greenbaum L, Lupoli S, Kohn Y, Sarner-Kanyas K, Ben-Asher E, Lancet D, Macciardi F, Lerer B. Association of the type 2 diabetes mellitus susceptibility gene, TCF7L2, with schizophrenia in an Arab-Israeli family sample. PLoS One. 2012;7:e29228. doi: 10.1371/journal.pone.0029228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Bürger A, Bult CJ, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon EP, Idzerda RL, McKnight GS. Targeting the mouse genome: a compendium of knockouts (part I) Curr Biol. 1995;5:625–634. doi: 10.1016/s0960-9822(95)00152-7. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- Castle WE. Piebald Rats and the Theory of Genes. Proc Natl Acad Sci U S A. 1919;5:126–130. doi: 10.1073/pnas.5.4.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS. Influences of laboratory environment on behavior. Nat Neurosci. 2002;5:1101–1102. doi: 10.1038/nn1102-1101. [DOI] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- Dao DT, Mahon PB, Cai X, Kovacsics CE, Blackwell RA, Arad M, Shi J, Zandi PP, O'Donnell P, Knowles JA, et al. Mood disorder susceptibility gene CACNA1C modifies mood-related behaviors in mice and interacts with sex to influence behavior in mice and diagnosis in humans. Biol Psychiatry. 2010;68:801–810. doi: 10.1016/j.biopsych.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J. The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am Nat 1914 [Google Scholar]
- Dietrich W. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- Doetschman T. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol Biol. 2009;530:423–433. doi: 10.1007/978-1-59745-471-1_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MAR, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SFA, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E, Grozeva D, Kirov G, Holmans P, Moran JL, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–D834. doi: 10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS Genet. 2012;8:e1002644. doi: 10.1371/journal.pgen.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere ML, Walters JTR, Smith R, Richards AL, Green E, Grozeva D, Jones I, Forty L, Jones L, Gordon-Smith K, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T, Ingason A, Djurovic S, Melle I, Fenger M, Gustafsson O, Jakobsen KD, Rasmussen HB, Tosato S, Rietschel M, et al. At-risk variant in TCF7L2 for type II diabetes increases risk of schizophrenia. Biol Psychiatry. 2011;70:59–63. doi: 10.1016/j.biopsych.2011.01.031. [DOI] [PubMed] [Google Scholar]
- Heydemann A, Ceco E, Lim JE, Hadhazy M, Ryder P, Moran JL, Beier DR, Palmer AA, McNally EM. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J Clin Invest. 2009;119:3703–3712. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hide T, Hatakeyama J, Kimura-Yoshida C, Tian E, Takeda N, Ushio Y, Shiroishi T, Aizawa S, Matsuo I. Genetic modifiers of otocephalic phenotypes in Otx2 heterozygous mutant mice. Development. 2002;129:4347–4357. doi: 10.1242/dev.129.18.4347. [DOI] [PubMed] [Google Scholar]
- Hrabě de Angelis M, Nicholson G, Selloum M, White JK, Morgan H, Ramirez-Solis R, Sorg T, Wells S, Fuchs H, Fray M, et al. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat Genet. 2015;47:969–978. doi: 10.1038/ng.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Meehan TF, Morgan H, Mason JC, Blake A, Kurbatova N, Smedley D, Jacobsen J, Mott RF, Iyer V, et al. Applying the ARRIVE Guidelines to an In Vivo Database. PLoS Biol. 2015;13:e1002151. doi: 10.1371/journal.pbio.1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TJ, Colegrave N, Viney M, Lazarou L, Abolins S, Abolins S, Pocock M, Hafalla J, Riley E, Viney M, et al. Caging and Uncaging Genetics. PLOS Biol. 2016;14:e1002525. doi: 10.1371/journal.pbio.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 2008;36:D715–D718. doi: 10.1093/nar/gkm728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- Nyegaard M, Demontis D, Foldager L, Hedemand A, Flint TJ, Sørensen KM, Andersen PS, Nordentoft M, Werge T, Pedersen CB, et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry. 2010;15:119–121. doi: 10.1038/mp.2009.69. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Hen R, Crabbe JC. Complications associated with genetic background effects in research using knockout mice. Psychopharmacology (Berl) 1999;147:5–7. doi: 10.1007/s002130051128. [DOI] [PubMed] [Google Scholar]
- Pinto RM, Dragileva E, Kirby A, Lloret A, Lopez E, St Claire J, Panigrahi GB, Hou C, Holloway K, Gillis T, et al. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington's disease mice: genome-wide and candidate approaches. PLoS Genet. 2013;9:e1003930. doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin D, Duan J, Ophoff RA. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozmahel R, Wilschanski M, Matin A, Plyte S, Oliver M, Auerbach W, Moore A, Forstner J, Durie P, Nadeau J, et al. Modulation of disease severity in cystic fibrosis transmembrane conductance regulator deficient mice by a secondary genetic factor. Nat Genet. 1996;12:280–287. doi: 10.1038/ng0396-280. [DOI] [PubMed] [Google Scholar]
- Sanford LP, Kallapur S, Ormsby I, Doetschman T. Influence of genetic background on knockout mouse phenotypes. Methods Mol Biol. 2001;158:217–225. doi: 10.1385/1-59259-220-1:217. [DOI] [PubMed] [Google Scholar]
- Savic D, Distler MG, Sokoloff G, Shanahan NA, Dulawa SC, Palmer AA, Nobrega MA. Modulation ofTcf7l2 expression alters behavior in mice. PLoS One. 2011a;6:e26897. doi: 10.1371/journal.pone.0026897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic D, Ye H, Aneas I, Park SY, Bell GI, Nobrega MA. Alterations in TCF7L2 expression define its role as a key regulator of glucose metabolism. Genome Res. 2011b;21:1417–1425. doi: 10.1101/gr.123745.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Simpson EM, Takahashi JS, Lipp HP, Nakanishi S, Wehner JM, Giese KP, Tully T, Abel T, Chapman PF, et al. Mutant mice and neuroscience: recommendations concerning genetic background. Banbury Conference on genetic background in mice. Neuron. 1997:755–759. doi: 10.1016/s0896-6273(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. A Mouse Geneticist's Practical Guide to CRISPR Applications. Genetics. 2014;199:1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MAR, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Ripke S, Lee PH, Neale BM, Nurnberger J, Santangelo S, Sullivan PF, Perlis RH, Purcell SM, Fanous A. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge RE, Martin LJ, Isbester KA, Sotocinal SG, Rosen S, Tuttle AH, Wieskopf JS, Acland EL, Dokova A, Kadoura B, et al. Olfactory exposure to males, including men, causes stress and related analgesia in rodents. Nat Methods. 2014;11:629–632. doi: 10.1038/nmeth.2935. [DOI] [PubMed] [Google Scholar]
- Tannenbaum C, Schwarz JM, Clayton JA, de Vries GJ, Sullivan C. Evaluating sex as a biological variable in preclinical research: the devil in the details. Biol Sex Differ. 2016;7:13. doi: 10.1186/s13293-016-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Metten P, Phillips TJ, Boehm SL, Burkhart-Kasch S, Dorow J, Doerksen S, Downing C, Fogarty J, Rodd-Henricks K, et al. Different data from different labs: lessons from studies of gene-environment interaction. J Neurobiol. 2003;54:283–311. doi: 10.1002/neu.10173. [DOI] [PubMed] [Google Scholar]
- Winham SJ, Cuellar-Barboza AB, Oliveros A, McElroy SL, Crow S, Colby C, Choi DS, Chauhan M, Frye M, Biernacka JM. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol Psychiatry. 2013;19:1010–1016. doi: 10.1038/mp.2013.159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.