SUMMARY
Dopamine neurons encode the difference between actual and predicted reward, or reward prediction error (RPE). Although many models have been proposed to account for this computation, it has been difficult to test these models experimentally. Here we established an awake electrophysiological recording system, combined with rabies virus and optogenetic cell-type identification, to characterize the firing patterns of monosynaptic inputs to dopamine neurons while mice performed classical conditioning tasks. We found that each variable required to compute RPE, including actual and predicted reward, was distributed in input neurons in multiple brain areas. Further, many input neurons across brain areas signaled combinations of these variables. These results demonstrate that even simple arithmetic computations such as RPE are not localized in specific brain areas but rather distributed across multiple nodes in a brain-wide network. Our systematic method to examine both activity and connectivity revealed unexpected redundancy for a simple computation in the brain.
eTOC blurb
Tian et al. combined rabies virus and photo-tagging to record from monosynaptic inputs to dopamine neurons. Information for reward prediction error computations is distributed and already mixed in input neurons, suggesting highly redundant and distributed computation for a simple arithmetic.
INTRODUCTION
One of the major goals in neuroscience is to understand how the brain performs computations. For some functions, the brain appears to perform simple arithmetic. For instance, multiplication-or division-like modulations of neural response – often called gain control – have been observed in a plethora of neural systems including attentional and “non-classical” modulations of visual responses (Carandini and Heeger, 2012; Williford and Maunsell, 2006) and the modulation of odor responses by overall input (Olsen et al., 2010; Uchida et al., 2013). In addition, various brain computations appear to rely on comparing two variables, requiring the brain to compute the difference between the two (Bell et al., 2008; Eshel et al., 2015; Johansen et al., 2010; Ohmae and Medina, 2015). However, how these computations are implemented in neural circuits remains elusive.
One potential strategy for these arithmetic computations is that one population of neurons encodes a variable X, and another population of neurons encodes another variable Y.
Downstream neurons combine inputs encoding these variables, and output the result of a simple arithmetic equation, such as X ÷ Y or X – Y. However, whether the above scenario is a plausible way to understand computations in complex neural circuits remains to be examined. First, the brain consists of neurons that are inter-connected in a highly complex manner. It is, therefore, unclear whether pure encoding of a single variable is a common or efficient way by which information is represented by neurons (Fusi et al., 2016), as it may be difficult to maintain a pure representation when receiving inputs from other neurons signaling other types of information. Second, a single neuron has to have the ability to perform precise computation, that is, its input-output function has to match a specific arithmetic. Previous modeling and experimental studies have shown that simple arithmetic computations may arise from a wealth of nonlinear mechanisms to transform synaptic inputs into output firing at the level of single neurons (Chance et al., 2002; Holt and Koch, 1997; Silver, 2010). However, it remains unknown whether these mechanisms underlie brain computations in a natural, behavioral context.
To address these questions, here we chose to study the midbrain dopamine system. It is thought that dopamine neurons in the ventral tegmental area (VTA) compute reward prediction error (RPE), that is, actual reward minus expected reward (or in temporal difference models, the value of the current state plus actual reward minus the value of the previous state) (Bayer and Glimcher, 2005; Cohen et al., 2012; Hart et al., 2014; Schultz et al., 1997), that is,
| (1) |
where Vactual and Vpredicted are the actual and predicted values of reward, respectively. The following experimental results supported this arithmetic operation. Dopamine neurons are activated by unpredicted reward. When reward is predicted by a preceding sensory cue, however, dopamine neurons’ response to reward is greatly reduced, while the reward-predictive cue starts to activate dopamine neurons (Mirenowicz and Schultz, 1994; Schultz et al., 1997). When a predicted reward is omitted, dopamine neurons reduce their firing below baseline at the time reward was expected. A recent study showed that the reduction of dopamine reward response by expectation occurs in a purely subtractive fashion, with some input from VTA GABA neurons (Eshel et al., 2015). Moreover, it was shown that single dopamine neurons in VTA compute RPEs in a very similar way (Eshel et al., 2016). Little is known, however, about the circuit mechanisms that underlie this computation.
Over the past twenty years, many electrophysiological recording experiments have been conducted with the goal of identifying specific brain areas conveying information about each variable in the above equations (Keiflin and Janak, 2015; Okada et al., 2009; Schultz, 2015). Based on these data, many models have been proposed to explain RPE computations (Brown et al., 1999; Daw et al., 2006; Doya, 2000; Houk et al., 1995; Joel et al., 2002; Kawato and Samejima, 2007; Keiflin and Janak, 2015; Kobayashi and Okada, 2007; Lee et al., 2012; Morita et al., 2012; O’Reilly et al., 2007; Schultz et al., 1997). These models typically assume that a particular brain area or a specific population of neurons within an area represents specific information such as Vactual or Vpredicted , and that these types of information are sent to dopamine neurons to compute RPEs. However, these hypotheses have rarely been tested experimentally. One obstacle in testing these models has been the inability to characterize both the activity of neurons and their precise synaptic connectivity (i.e., whether recorded neurons are presynaptic to dopamine neurons).
Several attempts have been made to examine neuronal activity with connectivity. Antidromic electrical stimulation has been used to identify neurons that project to a specific brain area (Hong and Hikosaka, 2014; Hong et al., 2011). More recently, optogenetic stimulation or viral infection at axon terminals has been used in a similar manner (Jennings and Stuber, 2014; Jennings et al., 2013; Nieh et al., 2015). However, these methods cannot distinguish cell types in a projection target. Importantly, ventral tegmental area (VTA) consists of different types of neurons, not only dopaminergic but also GABAergic and others (Nair-Roberts et al., 2008; Sesack and Grace, 2009). In the present study, we sought to characterize the activity of neurons presynaptic to dopamine neurons (input neurons). To record from input neurons directly, we combined rabies-virus-mediated trans-synaptic tracing and optogenetic-tagging during electrophysiological recording. Our results demonstrate that the information that dopamine neurons receive in order to compute RPE is not localized to specific brain areas and is not limited to pure signals of reward and expectation. Rather, information in monosynaptic inputs across multiple brain areas contained a spectrum of signals – including pure information and partial RPE – that dopamine neurons then combine into complete RPE. Our results challenge simple neural circuit models and point to the importance of understanding how neural circuits with complex connectivity can perform simple arithmetic.
RESULTS
Identification of monosynaptic inputs to midbrain dopamine neurons during awake electrophysiology
We aimed to record the activity of neurons that are presynaptic to dopamine neurons (“input neurons”). To achieve this, we used a rabies virus-based retrograde trans-synaptic tracing system (Wertz et al., 2015; Wickersham et al., 2007a). The rabies system with mouse genetics allows us to label only neurons that make synapses directly onto dopamine neurons (Watabe-Uchida et al., 2012). Using this system, the light-gated ion channel, channelrhodopsin-2 (ChR2) (Boyden et al., 2005; Osakada et al., 2011) was expressed in input neurons so that we can identify input neurons in behaving mice while we recorded their activity extracellularly (Cohen et al., 2012; Kvitsiani et al., 2013; Lima et al., 2009; Tian and Uchida, 2015).
Mice were trained in an odor-outcome association task where different odors signaled the probability of upcoming reward (90%, 50% or 0) or mildly aversive stimulus (air puff) (Cohen et al., 2012, 2015) (n = 81 mice) (Figure 1A). To minimize the period of rabies virus infection, we injected rabies virus (SADΔG-ChR2-mCherry (envA)) after animals acquired the association, and started recording neural activity soon after recovery from the surgery (Figure 1B). We first examined the mice’s behavior during the task. Rabies-infected mice showed anticipatory licking behavior, monotonically increasing with the probability of upcoming water, over the period of 5 to 15 days after rabies injection (Figure 1C). We also observed that over the course of retraining, our mice tended to show increasing anticipatory licking, suggesting either that the odor-outcome association became stronger or that mice became more motivated to perform the task during this period (Figure 1C).
Figure 1. Rabies-virus-infected putative dopamine neurons show aspects of reward prediction error signals.
(A) Probabilistic reward association task. Odor A and B predicted reward with different probabilities (90 and 50%, respectively). Odor C predicted no outcome. Odor D predicted air puff with 80% probability. (B) Experimental design and configuration of electrophysiological recording from rabies virus-infected VTA neurons. (C) Anticipatory licking behavior (1–2 s after odor onset) after rabies injection (mean ± SEM). The dash line separated the data into early and late days in D and E. For all days plotted, the strength of anticipatory licking follows odor A > odor B > odor C (p < 0.001, Wilcoxon signed rank test). (D) Average firing rates of rabies infected putative dopamine neurons during the early (5–9 days, top) and late (10–15 days, middle) periods after rabies virus injection. Putative dopamine neurons that were infected by AAV were shown in the bottom. Only rewarded trials are shown. (E) Quantifications of RPE signals (rabies group at 10–15 days after rabies injection versus AAV control). ***, p < 0.001 (Wilcoxon signed rank test against 0 spikes/s). ** p < 0.01; n.s., p >0.05 (Wilcoxon rank-sum test. Rabies versus AAV control). The edges of the boxes are the 25th and 75th percentiles, and the whiskers extend to the most extreme data points not considered outliers. Points are drawn as outliers if they are larger than Q3+1.5*(Q3-Q1). See also Figure S1.
We next examined whether infection of rabies virus affected neurons’ firing patterns. In this experiment, both AAV helper viruses and rabies virus were injected into VTA and recording was performed at the injection site (Figure 1B). We compared the firing patterns in reward trials of rabies-infected VTA neurons against control VTA neurons recorded in a different set of mice that were infected with AAV encoding ChR2 (Tian and Uchida, 2015). Using responses to light stimulation, we identified 76 rabies-infected VTA neurons from 12 animals. Among these neurons, rabies-infected, putative dopamine neurons showed aspects of RPE coding: (1) they were excited by reward-predictive cues with a monotonic increase in firing rate with increasing reward probability, (2) their reward responses were more strongly reduced with higher expectation, and (3) they showed inhibitory responses upon reward omission (Figures 1D and E, Figure S1). Overall, we noticed that the strength of responses was weaker in rabies-infected putative dopamine neurons compared to those in control mice (Figure 1E). This difference is unlikely to be due to the toxicity of rabies virus because we observed that dopamine neurons showed stronger responses to reward predictive cues in later days (10–15 days after rabies injection) compared to early days (5–9 days), contrary to the potential toxicity that would grow over longer infections (Figure 1D). These results show that recorded putative dopamine neurons exhibited relatively intact RPE coding, suggesting that inputs to dopamine neurons must be sufficient to generate these responses.
Diverse activities of monosynaptic inputs to dopamine neurons
Previous studies have mapped monosynaptic inputs to dopamine neurons using rabies virus and showed that input neurons are densely accumulated around the medial forebrain bundle (Menegas et al., 2015; Watabe-Uchida et al., 2012). We chose to record from those input-dense areas along this ventral stream that have most often been used in RPE models: the ventral and dorsal striatum (Aggarwal et al., 2012; Doya, 2008; Kawato and Samejima, 2007), the ventral pallidum (Kawato and Samejima, 2007; Keiflin and Janak, 2015; Tachibana and Hikosaka, 2012), lateral hypothalamus (Nakamura and Ono, 1986; Watabe-Uchida et al., 2012), subthalamic nucleus (Watabe-Uchida et al., 2012), rostromedial tegmental nucleus (RMTg) (Barrot et al., 2012), and pedunculopontine tegmental nucleus (PPTg) (Keiflin and Janak, 2015; Kobayashi and Okada, 2007; Pan and Hyland, 2005) (Figure S2, Table 1). We recorded from 1,931 neurons from 69 mice, among which 205 neurons were identified as being monosynaptic inputs of dopamine neurons based on their responses to light (see Experimental Procedures). We analyzed both identified input neurons and other neurons (hereafter “unidentified neurons”) separately.
Table 1.
Summary of animals and neurons
| Brain Area | mice | neuron | mean neuron per mouse |
Std neuron per mouse |
inputs | Mean inputs per mouse |
Std inputs per mouse |
inputs/ neurons |
|---|---|---|---|---|---|---|---|---|
| Ventral striatum |
19 | 360 | 18.9 | 10.6 | 35 | 1.8 | 1.9 | 10% |
| Dorsal striatum |
4 | 169 | 42.3 | 18.3 | 23 | 5.8 | 3.3 | 14% |
| Ventral pallidum |
11 | 198 | 18 | 12.4 | 18 | 1.6 | 1.4 | 9% |
| Subthalamic | 1 | 80 | 80 | 0 | 7 | 7 | 0 | 9% |
| Lateral hypothalamus |
15 | 586 | 39.1 | 21.6 | 46 | 3.1 | 3.0 | 8% |
| RMTg | 6 | 151 | 25.2 | 15.9 | 21 | 3.5 | 2.3 | 14% |
| PPTg | 9 | 177 | 19.7 | 10.4 | 55 | 6.1 | 4.5 | 31% |
| Central amygdala |
4 | 210 | 52.5 | 42.2 | 0 | 0 | 0 | 0% |
| Total | 69 | 1931 | 205 |
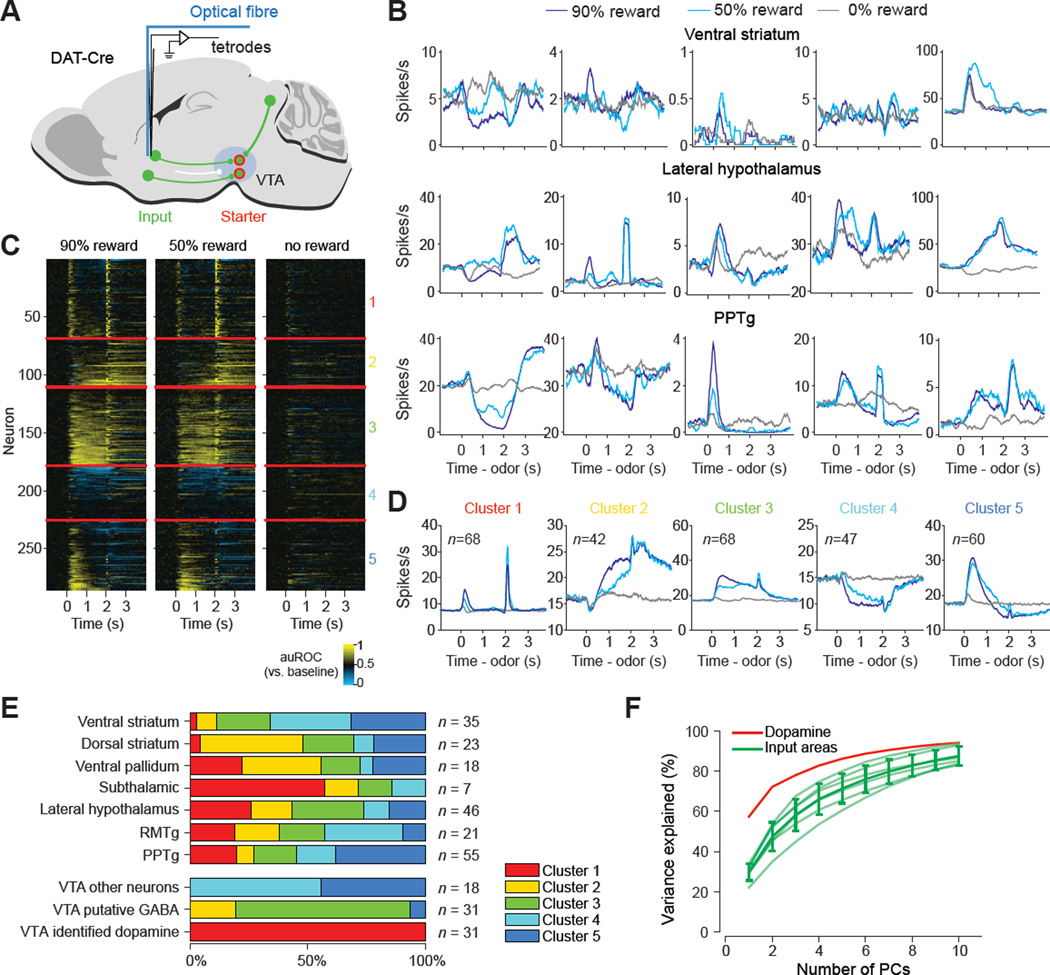
We first characterized each input neuron’s activity patterns in different trial types (Figure 2). We found that monosynaptic input neurons exhibited diverse firing patterns and complex temporal dynamics (Figure 2B and S3). In this and the following sections, we will examine the characteristics of activities of input neurons using different methods of analysis, focusing on reward trials. We first used k-means clustering to classify the activity profiles of individual neurons into five types based on a similarity metric (Person’s correlation coefficient) (Figure 2C, see Experimental Procedures). Cluster 1 showed phasic excitation to reward predictive cues and/or reward, similar to dopamine neurons (Figure 2D). Clusters 2 and 3 showed sustained excitation to reward predictive cues. Cluster 4 mainly showed sustained inhibition to reward predictive cues. Cluster 5 often showed biphasic activities, first excitation followed by inhibition, to reward predictive cues. Each input area contained several different clusters of neurons, whereas all dopamine neurons fell into Cluster 1, suggesting that in contrast to dopamine neurons, input neurons in each brain area are diverse in their response profiles (Figure 2E, see Experimental Procedures). Supporting this idea, when we extracted principal components (PCs) of normalized neuronal activities in dopamine neurons and each input area, the first PC accounted for 57% of firing of dopamine neurons, whereas the first PC explained only 22–34% of the activities in input neurons in each brain area (Figure 2F).
Figure 2. Each input area contains input neurons with diverse response profiles.
(A) Configuration of input neuron recordings. (B) Firing rates of randomly sampled individual identified input neurons from three input areas. (C) Temporal profiles of all input neurons and VTA neurons. Colors indicate an increase (yellow) or decrease (blue) from baseline, as quantified using a sliding–window auROC analysis (time bin: 100 ms, against baseline). Neurons were clustered into five clusters, separated by horizontal red lines. Within each cluster, neurons are ordered by their average responses during 2 s after odor onset. (D) Average firing rates of neurons from each cluster in (C). n, number of neurons. (E) The percentage of neurons that belong to each of the five clusters in individual input areas and VTA. VTA putative GABA and VTA other neurons are cluster 2 and 3 neurons respectively from the AAV control group (see Figure S1) (F) The percentage of variance explained by the first n principle components (PC) in identified inputs and dopamine neurons. PCs were computed for each individual input area and plotted as light green traces. The solid green trace is mean ± SD of traces from all input areas. Subthalamic nucleus has only 7 identified inputs and is excluded from the analysis.
See also Figure S2 and S3.
To further quantify the diversity of response profiles, we computed the correlation coefficient of response profiles in all trial types for pairs of neurons. This analysis showed that all the dopamine neurons showed high correlation of activities to one another (indicated by a dense yellow square at the top left in Figure 3A), whereas input neurons in each brain area exhibited more complicated correlation patterns, often grouped as indicated by our clustering analysis (diagonal patches in Figure 3A, see Figure 3B for examples). When we compared the correlation coefficient of pairs of neurons within each area, dopamine neurons tended to have higher correlations than input neurons in each area (Figure 3C). These analyses show that the diversity of response profiles is dramatically reduced as information converges from the presynaptic neurons to postsynaptic dopamine neurons.
Figure 3. Correlation analysis of dopamine neurons and their inputs.
(A) Correlation matrix of firing patterns of dopamine neurons and their input neurons. The similarity of response profiles between pairs of neurons was quantified based on the Pearson’s correlation coefficient. The red lines separate neurons of each brain area. The purple ticks mark the boundaries of different clusters within each area (see Figure 2). Within each area, neurons are ordered in the same way as in Figure 2C. (B) Example firing profiles of dopamine neurons and PPTg neurons (n = 7 each) in 90% reward trials. Note that dopamine neurons’ firing profiles are more similar to one another than PPTg neurons. (C) Histogram of pairwise correlations for all pairs of neurons in each input area and dopamine neurons. The red line indicates the mean pairwise correlation for each area. The grayscale indicates the probability density.
Distributed and mixed information in monosynaptic inputs to dopamine neurons
We next examined what information each monosynaptic input neuron conveyed to dopamine neurons with respect to the RPE computation (Figures 4 and 5, Table S1). To compute RPEs in this task, dopamine neurons need to take into account at least two types of information: the value of predicted reward, which was provided by odor cues, and the value of actual reward, which was signaled by the delivery of water. The probabilistic reward task that we used (Figure 1A) allowed us to operationally classify the types of information that each neuron conveyed related to RPE computation. Because the amount of reward was held constant among 90% and 50% reward trials, those neurons that conveyed pure reward information should show the same response regardless of the level of expectation. Conversely, those neurons that conveyed reward expectation should change their activity depending on the probability of reward.
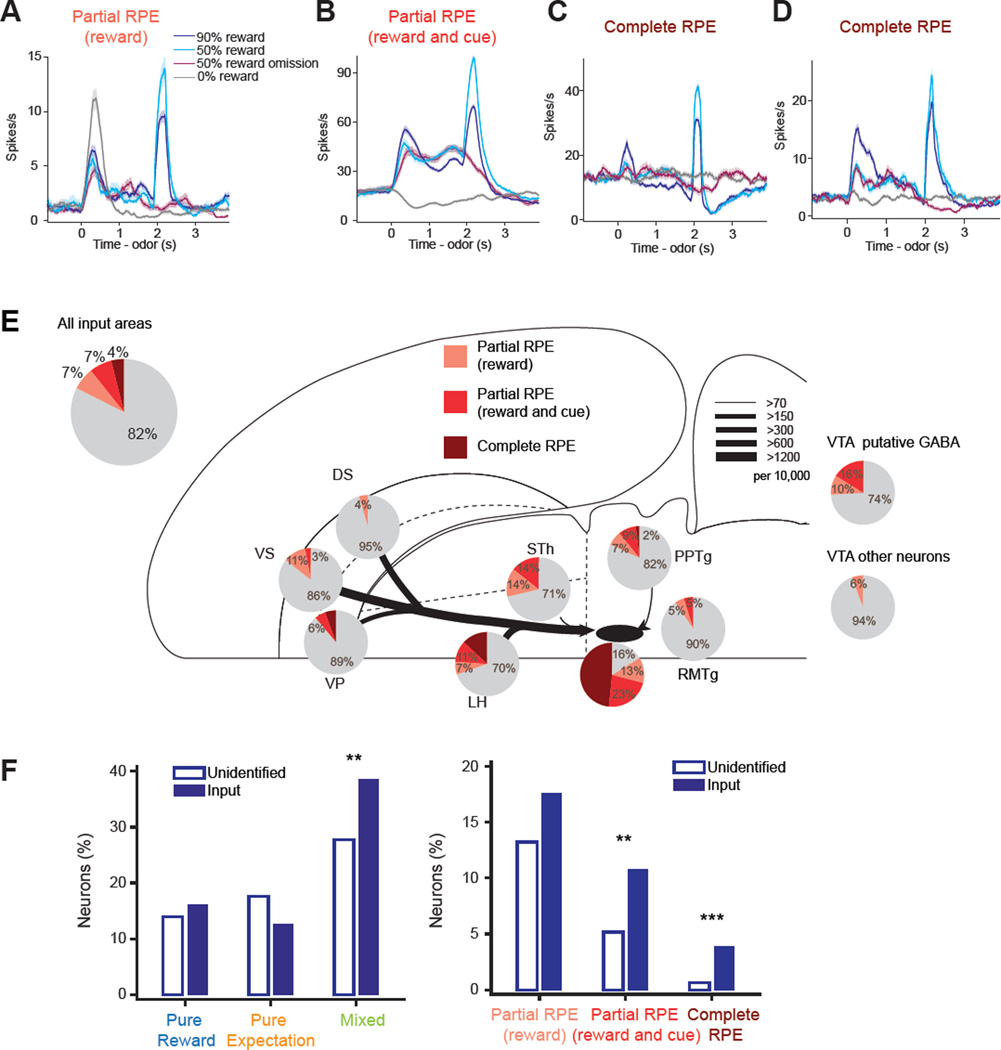
Figure 4. Mixed coding of reward and expectation signals in input neurons.
(A–D) Firing rates (mean ± SEM) of example neurons that were categorized as different response types. Neurons are from dorsal striatum (A), lateral hypothalamus (B), RMTg (C) and ventral striatum (D). (E) Flatmap summary (Swanson, 2000) of the percentage of input neurons and control VTA neurons that were classified into each response type. See Experimental Procedures and Figure S4 for criteria. VTA putative GABA and VTA other neurons are cluster 2 and 3 neurons respectively from the AAV control group (see Figure S1). The thickness of each line indicates the number of input neurons in each area (per 10,000 total inputs in the entire brain) as defined at the top right corner (Watabe-Uchida et al., 2012). DS, dorsal striatum; VS, ventral striatum; VP, ventral pallidum; LH, lateral hypothalamus; STh, subthalamic nucleus; DA, dopamine neurons. See also Figure S4.
Figure 5. Input neurons encode reward prediction error signals.
(A–D) Firing rates (mean ± SEM) of example neurons that were categorized as encoding different parts of RPE signals. Neurons are from RMTg (A, B), PPTg (C) and ventral pallidum (D). (E) The percentage of input neurons and control VTA neurons that showed RPE signals. See Figure S6 for criteria. The conventions are the same as Figure 4E. (F) The percentage of neurons in each response type for identified input neurons and other (unidentified) neurons. Pure reward type includes neurons with cue responses. Mixed type of responses as well as RPE types are more concentrated in input neurons. * p < 0.05; ** p < 0.01; *** p < 0.001; no labels, p > 0.05 (Chi-squared test, p-values are after Bonferroni correction for multiple comparisons) See also Figure S5 and S6.
We examined whether there were neurons that conveyed actual reward values. Although previous studies found that local GABA neurons in VTA may signal reward expectation to dopamine neurons (Cohen et al., 2012; Eshel et al., 2015), these studies did not find neurons that signal actual reward in VTA. Some of the areas that we recorded are good candidates for the source of excitatory inputs for reward. For example, the lateral hypothalamus and PPTg are the main sources of the few glutamatergic inputs onto dopamine neurons, whereas many other input areas of dopamine neurons are dominated by GABA neurons (Geisler and Zahm, 2005; Geisler et al., 2007; Watabe-Uchida et al., 2012). Indeed, previous recording studies found reward-responsive neurons in these areas (Nakamura and Ono, 1986; Okada et al., 2009). We found some input neurons that appeared to convey the value of actual reward, as predicted by previous models (Figures 4A and 4B). However, we noticed that many other neurons were modulated by reward in more complex ways (Figure 4D). To further quantify this observation, we first identified reward-responsive neurons based on statistically significant differences in firing among 50% reward trials, comparing when the reward was actually delivered to when it was not (p < 0.05, Wilcoxon rank-sum test; 0–500 ms from reward onset timing, see Figure S4). Although 54.6% (112 of 205) of input neurons were classified as reward responsive, a large portion (79 of 112) of these reward-responsive neurons was also modulated by the predicted value of reward (p < 0.05, Wilcoxon rank-sum test, 90% reward vs 50% reward, 0–500 ms from reward onset) (e.g. Figure 4D; Figure 4E, green “mixed”). Reward responsive neurons that did not show a significant modulation by predicted value of reward can be classified as “pure reward” neurons, the type of neuron assumed in many models (e.g. Figure 4A and B; Figure 4E, light blue and blue). However, “pure reward” neurons often showed responses in an earlier time window, responding to odor cues (e.g. Figure 4B). Removing those neurons responding to odor cues (see Figure S4), only 4.9% of input neurons (10 out of 205) signaled actual reward in a pure fashion (e.g. Figure 4A; Figure 4E, blue). These “pure reward” neurons were not localized to a specific brain area such as the lateral hypothalamus or PPTg. We found slightly more pure reward neurons in the ventral pallidum, which also contains glutamatergic projections to VTA (Geisler et al., 2007).
Similarly, we categorized “pure expectation” neurons that encoded predicted reward value monotonically and were not reward-responsive (e.g. Figure 4C; Figure 4E, orange; see Figure S4 for criteria). In addition to VTA GABA neurons (Cohen et al., 2012; Eshel et al., 2015), the striatum and ventral pallidum have been proposed to send reward expectation signals to dopamine neurons (Kawato and Samejima, 2007). However, we found that relatively few neurons (26 of 205 neurons) signaled reward expectation in a pure fashion. In fact, many putative GABA neurons in VTA were also reward responsive, in addition to encoding expectation (Cohen et al., 2012; Eshel et al., 2015). Similar to reward neurons, expectation neurons were also distributed in several brain areas. The striatum and ventral pallidum had slightly more expectation neurons, although these areas also had a comparable number of mixed-coding neurons.
In total, these results demonstrate two characteristics of input neurons of dopamine neurons. First, each brain area that we tested contained input neurons encoding different types of information. In other words, information about the predicted and actual reward values was conveyed by input neurons spread across multiple, distributed brain areas. Second, these components of information were already mixed in most of the single input neurons we recorded.
Among the neurons with mixed coding of predicted and actual reward, we found that some (17.6%, 36 out of 205) input neurons showed an important aspect of RPE coding (Figure 5): their reward responses were suppressed more by 90% reward expectation than by 50% reward expectation (Figure 5E, pink bar; see Experimental Procedures and Figure S5 for criteria). This type of expectation-dependent suppression of reward response was not localized to a specific area, such as RMTg, which has been proposed to send RPE to dopamine neurons (Hong et al., 2011), but were found in input neurons in all areas that we examined (Figure 5E). However, RPE coding in these neurons was not as complete as it was in dopamine neurons. In addition to expectation-dependent reductions of reward responses, dopamine neurons respond to reward-predictive cues and reward omission, which are additional hallmarks of RPE signals. Only 3.9% of input neurons showed all three features of RPE signals, but many other neurons showed partial RPE signaling (Figure 5E). Because we applied multiple criteria to identify RPE-related signals, the numbers reported above are expected to be conservative. Indeed, the false discovery rates (FDRs), estimated by applying the same criteria to trial-shuffled data in each neuron, were much lower (FDRs: 0.7% for reward RPE type, and 0.01% for complete RPEs, see Experimental Procedures). These results demonstrate that a significant fraction of monosynaptic input neurons already convey aspects of RPE signals, although the quality of RPE signals is not as robust and mature as in dopamine neurons.
Our identification of input neurons relied on optogenetic activation. It should be noted that laser stimulation could indirectly excite neurons postsynaptic to ChR2-expressing neurons. To distinguish direct versus indirect activation, we used high-frequency light stimulation (20 and 50 Hz) because synaptic transmission tends to fail at high frequency. To further eliminate contamination with indirect stimulation, we repeated the above analysis using only neurons that responded to laser with short latency (< 6 ms, 118 out of 205 neurons). The data still supported our assertion that the plurality of input neurons encoded mixed information (Figure S6) and this is unlikely due to the contamination of neurons indirectly activated by ChR2. Next, we examined whether the results differed in early versus late days after rabies injection. Mixed coding was a dominant feature regardless of the recording period (Figure S6). Furthermore, mixed coding and partial RPE were observed more in later periods after rabies injection (25% and 42% of mixed coding neurons from 5–9 and 10–16 days after injection, respectively), the time course of which is potentially related to the development of RPE in putative dopamine neurons (Figure 1).
So far, our analyses have focused on neurons identified as monosynaptic inputs of dopamine neurons. Next, we compared these results with unidentified neurons. Task-responsive neurons were slightly more often observed in inputs (83.4%, n = 205 neurons) than in unidentified neurons in the same areas (74.5%, n = 1516 neurons) (p = 0.0036, Chi-squared test). We also found that during the reward period (0–500 ms after reward onset), mixed coding neurons and RPE neurons were more concentrated in inputs than in unidentified neurons (mixed, p = 0.0013; complete RPE, p < 10−5; Chi-squared test), whereas pure signals of reward or expectation were observed at similar frequencies (pure reward, p = 0.39; pure expectation, p = 0.076; Chi-squared test) (Figure 5F). These results show that the identified input neurons differed significantly from unidentified neurons. In other words, labeling monosynaptic inputs was considerably more revealing than randomly sampling neurons in known input areas.
Contribution of negative and positive responses in input neurons to dopamine responses to rewarding or aversive stimuli
To further understand which brain areas are important for generating the phasic responses of dopamine neurons upon reward-predictive cues and reward, we broke down the responses into positive and negative coding types (Figure 6) with “positive” referring to neurons that showed a monotonic increase in firing rate response with increasing reward probabilities.
Figure 6. Positive and negative responses in inputs.
(A) Firing rates (mean ± SEM) of example input neurons (top) and the percentage of neurons (bottom) encoding reward positively and negatively during CS (0–500 ms after odor onset) in identified inputs. Filled bars indicate the percentage of neurons, that monotonically encode reward probability faster than median latency of dopamine neurons (see Table S2, Experimental Procedures). (B) The percentage of neurons encoding reward value positively or negatively during late delay (1500–2000 ms after odor onset) in identified inputs. (C) Firing rates (mean ± SEM) of example neurons (top) and the percentage of neurons (bottom) encoding air puff positively and negatively during CS (0–500 ms after odor onset) in identified inputs. Filled bars indicate the percentage of neurons that distinguish air puff CS from nothing CS faster than median latency of dopamine neurons (see Table S2, Experimental Procedures). (D) The percentage of neurons encoding air puff positively or negatively during late delay (1500–2000 ms after odor onset) in identified inputs. See also Figure S7.
We first examined the number of neurons whose firing rates were modulated monotonically by reward probability during the CS (conditioned stimulus, cue) period (0–500 ms after CS onset). We found that neurons whose firing was modulated monotonically by reward probabilities were found in all the brain areas that we examined (Figure 6A). The responses of most of these input neurons were positive, whereas negative reward coding was rare except in RMTg (Figure 6A). Note that this percentage would be even higher if we could limit the analysis to RMTg core, which was shown to have mainly negative reward coding in a previous study (Jhou et al., 2009a). Next, we compared onset latency of reward coding in inputs and dopamine neurons. We examined neurons which showed shorter latency than the median latency of dopamine neurons. Inputs that met the two criteria – those that showed monotonic reward coding and short latency – were all positive reward coding (Figure 6A, Table S2). Considering that dopamine neurons encode predictive reward positively, these results suggest that input neurons which are able to initiate reward-value-coding responses during the CS in dopamine neurons were distributed among excitatory neurons in multiple brain areas, including the ventral pallidum, lateral hypothalamus and PPTg.
On the other hand, during the late delay period before reward (1.5 – 2 s after CS onset), both positive and negative reward coding were distributed in multiple brain areas (Figure 6B). Monotonic negative reward coding was observed in 34.6% of input neurons in PPTg, suggesting a unique function of PPTg in signaling reward expectation, potentially via its excitatory projections (Figure 6B).
Next, we examined responses to a mildly aversive stimulus (Figure 6C–D). In our previous studies, dopamine neurons showed a mixture of excitation, inhibition or no response upon air puffs or air puff-predicting cue (Cohen et al., 2012; Tian and Uchida, 2015). These results were consistent with electrophysiology studies in monkeys, which showed a continuous array of responses ranging from net inhibition to net excitation (Bromberg-Martin et al., 2010; Fiorillo et al., 2013), which were interpreted as “value”- or “salience”-coding, respectively (Matsumoto and Hikosaka, 2009). In input neurons, we observed a much smaller number of neurons that signal aversive outcomes during CS periods than neurons signaling upcoming reward. Similar to the reward CS, these neurons were distributed across brain areas. During US (unconditioned stimulus, outcome) periods, representations of reward and aversive stimuli were observed in input neurons in multiple brain areas and were dominated by excitatory responses, except in the RMTg, which showed relatively frequent inhibitory responses to reward (Figures S7A–B). Among input neurons that encoded both reward and aversive stimuli, most input neurons were excited by both (Figure S7C–D). In short, excitatory inputs in multiple brain areas have the potential to provide “salience” signals to dopamine neurons, whereas the “value” representation in dopamine neurons may form out of separate signals of reward and aversion, the former from excitatory inputs and the latter from inhibitory inputs.
In summary, most inputs were activated by reward predictive cues or rewards, suggesting an important role of excitatory inputs in generating phasic responses of dopamine neurons. On the other hand, signals of aversive stimuli may be conveyed by both inhibitory and excitatory inputs, resulting in “value” and “salience”- coding in dopamine neurons, respectively.
RPE can be robustly reconstructed by a combination of monosynaptic inputs
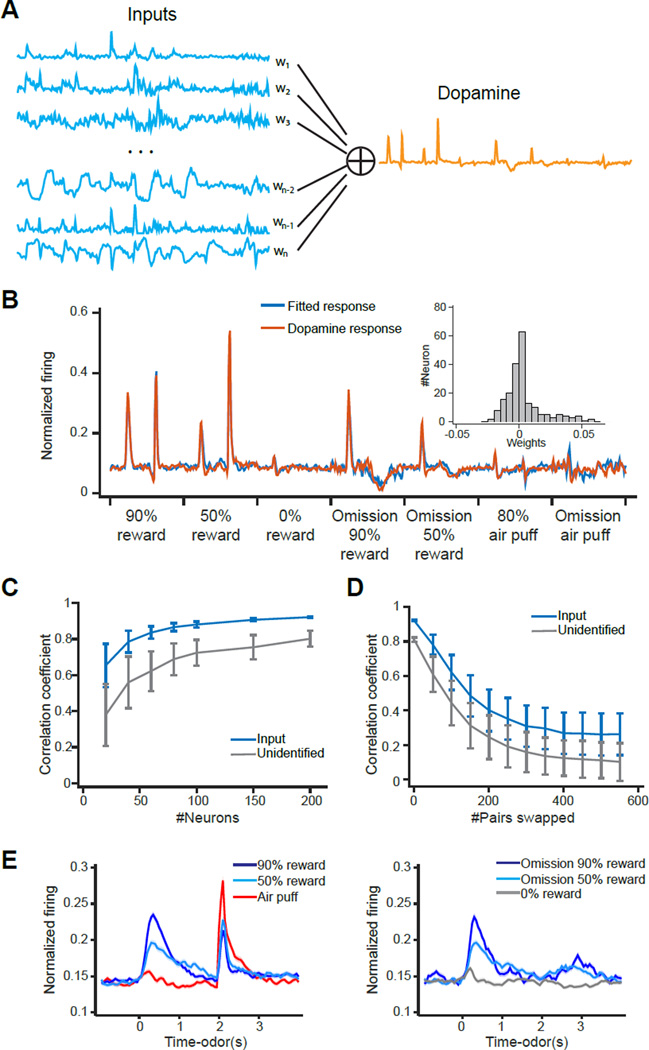
We next explored the nature of input data using a simple linear model that tries to reconstruct mean dopamine activities by taking a weighted sum of the firing patterns of input neurons (Figure 7A). Since the actual strength of connection of each input neuron is unknown (which may differ depending on synaptic strength, number and location of synapses etc.), the weights of each input were assigned to best fit the dopamine activities. Assuming that all the inputs in the striatum are inhibitory and all the inputs in the subthalamic nucleus are excitatory, the weights of the striatum and subthalamic nucleus were constrained to negative and positive, respectively (see Experimental Procedures). Using the activity patterns of all of our input neurons, we first confirmed that a linear combination of inputs can provide a good fit of dopamine activities (cross-validated r = 0.92, Pearson’ correlation coefficient, Figure 7B). Even if one entire area is removed, we obtained a good fit of dopamine firing patterns (r ≥0.89 for removal of any single input area). These results support a highly redundant distribution of information across multiple brain areas.
Figure 7. A linear model can reliably extract RPE signals from inputs.
(A) In a regularized linear model, dopamine neurons’ responses in the task are modeled as the weighted sum of all input neurons. See Experimental Procedures for details of the model. (B) The linear model fitted dopamine activity (fitted response) and the actual dopamine activity (dopamine response). Inset, distribution of fitted weights. (C) Comparison of quality of fitting using randomly sampled neurons from identified input neurons and other (unidentified) neurons in model fitting (mean ± SD). (D) Comparison of quality of fitting when shuffling the fitted weights using identified input neurons and other neurons (mean ± SD, 500 simulations). To shuffle the data, the weights of two randomly chosen neurons are swapped. “#Pairs swapped” indicates how many times this procedure is repeated for each simulation. (E) Mean ± SEM (100 simulations) of the fitted activity when the fitted weights of input neurons are completely shuffled.
The above results are not necessarily surprising because a complex “waveform” can be reconstructed from a combination of time-varying functions as long as these functions have enough complexity. Yet, we sought to use this method to quantify the nature of inputs. First, we examined whether RPE-related signals are more readily reconstructed from input neurons than from unidentified neurons in the same areas. We found that for a given number of randomly picked neurons, dopamine activities were more readily reconstructed from input neurons than from unidentified neurons (Figure 7C). We also performed the same analysis only using task-responsive neurons, and acquired very similar results (data not shown), excluding the possibility that the above results are merely due to the enrichment of task-related neurons in inputs. Further, we examined how much our linear reconstruction depends on the exact weights. In the above, we obtained the weights that best fit the dopamine activities. We also examined how much the fit degrades after increasing the number of neurons whose weights were randomly swapped with one another. After shuffling the weights, we computed the similarity between the weighted sum of the activity of input neurons and the activity of dopamine neurons using a Pearson’s correlation coefficient. We found that the similarity gradually decreased (Figure 7D). Even after complete random shuffling, the sum of input activities resembled RPE signals of dopamine neurons (Figure 7E); it showed monotonic reward value coding at the CS and decreased reward responses with increasing expectation. For comparison, we next performed the same analysis using unidentified neurons. We observed that the similarity between the reconstructed and actual responses decreased more dramatically after shuffling, compared to the analysis using input neurons. These results suggest that the observed activity patterns of input neurons can provide not only a sufficient but also a robust substrate for constructing RPE-related activities of dopamine neurons in the sense that precise tuning of weights (i.e. precise connectivity) is not required. The comparison with unidentified neurons further suggests that the activity profiles of input neurons are “pre-formatted” such that the downstream neurons can more easily generate RPE signals. On the other hand, we also noticed several differences from activities of dopamine neurons; e.g., CS responses were more sustained, reward omission responses were not reconstructed, and air puff responses were bigger (Figure 7E), suggesting that some aspects of dopamine signaling may require additional mechanisms.
DISCUSSION
The activity of dopamine neurons can be approximated by simple arithmetic equations (Bayer and Glimcher, 2005; Cohen et al., 2012; Eshel et al., 2015, 2016; Hart et al., 2014; Schultz et al., 1997). It is, therefore, tempting to suggest simple models in which distinct populations of neurons that are presynaptic to dopamine neurons represent different terms in these equations, and these inputs are combined at the level of dopamine neurons. As a result, previous experimental work has focused on finding neurons encoding these types of information in relevant brain areas (Cohen et al., 2012; Eshel et al., 2015; Keiflin and Janak, 2015; Okada et al., 2009; Schultz, 2015). However, recording experiments typically find that these “pure” neurons are intermingled with other neurons with diverse firing patterns conveying different types of information. Thus, without confirming the assumed synaptic connectivity, it is unclear whether these models are valid. Here, for the first time, we addressed this issue by directly recording the activity of neurons that are presynaptic to dopamine neurons, in multiple brain areas. In stark contrast to the simple models proposed previously, we observed that information about reward and expectation are distributed among multiple brain areas, and are already mixed in many individual input neurons. Our data suggest that the RPE computation is not a one-step process combining pure information about reward and expectation in dopamine neurons. Nor do dopamine neurons receive complete RPE from a specific brain area. Instead, the prevalence of input neurons encoding both pure and mixed information, partial and complete RPE signals appears to be a sign of redundant computations distributed in a complex neuronal network, which ultimately converge onto dopamine neurons to construct a more complete RPE signal.
Distributed and multi-step computation of RPEs
Midbrain dopamine neurons receive monosynaptic inputs from many brain areas (Geisler and Zahm, 2005; Menegas et al., 2015; Watabe-Uchida et al., 2012). This is unlikely due to the potential diversity of dopamine neurons, because subpopulations of dopamine neurons projecting to specific targets nonetheless receives inputs from many brain areas (Beier et al., 2015; Lerner et al., 2015; Menegas et al., 2015). Which brain areas among these are important for the RPE computation? Why do so many brain areas project to dopamine neurons? One possibility is that in simple tasks such as the one used in the present study, only some brain areas ‘come online’ to provide relevant information. In contrast to this prediction, 83.4% of input neurons (p < 0.05, ANOVA, see Experimental Procedures for details) in 7 different brain areas were modulated by reward-predictive cues or reward. We further found that both critical components of RPE computations – information about predicted and actual reward – were distributed in input neurons in these brain areas.
Our data also showed that the two types of information are often mixed in single input neurons. Very few neurons represented information about expected reward and actual reward in a pure fashion. Why is the information mixed? First, anatomical constraints may make coding of pure information unrealistic. The brain is a dynamical system consisting of a large number of neurons that are interconnected in a highly complex manner within and across brain areas. Indeed, all the areas that we recorded in this study are highly connected with one another, either directly or indirectly (Geisler and Zahm, 2005; Heimer et al., 1991; Jhou et al., 2009b). Only by removing these connections will it be possible to understand the original information which each brain area might receive from upstream sources. This is a common challenge for study of neuronal computation.
Second, it has been proposed that mixed coding – single neurons representing multiple types of information in a mixed fashion – may garner computational and representational advantages (Fusi et al., 2016). For instance, mixing information in a non-linear way may expand the space of information coding, facilitating the readout of downstream neurons (Fusi et al., 2016; Rigotti et al., 2013). This line of work suggests that the brain exploits these properties to actively mix information at the single neuron level, and mixed coding is the normal way by which the brain represents information, rather than the exception. In this framework, it can be seen that particular information and computations are embedded in a complex pattern of activity distributed in a network of many neurons. At a glance, one might think that subtraction may not require such complex representations. Yet, mixed coding might provide a robust substrate for RPE computations in more complex task conditions.
Third, mixing information can be seen as a reflection of multi-step RPE computations. It has been reported that RPE signals can be found in multiple brain areas such as prefrontal cortex, the striatum, GPi, lateral habenula and RMTg (Asaad and Eskandar, 2011; Bromberg-Martin and Hikosaka, 2011; Hong and Hikosaka, 2008; Matsumoto and Hikosaka, 2007, 2008; Oyama et al., 2015). This is potentially because RPE signals are useful for a variety of behaviors outside of reinforcement learning (Den Ouden et al., 2012; Schultz and Dickinson, 2000). In other words, these RPE-related activities might have been computed to subserve different brain functions, yet at the same time, became a good source of inputs for dopamine neurons.
A previous study showed that neurons signaling negative RPE in RMTg project to SNc (Hong et al., 2011). By using transsynaptic optogenetic tagging, we confirmed and extended these results, revealing for the first time that these RPE-coding neurons indeed provide monosynaptic inputs to dopamine neurons, and that RPE-coding neurons exist in all 7 brain areas we recorded, not only in RMTg. Furthermore, the RPE signals we discovered in various input neurons tended to be incomplete; for example, many did not respond to reward-predicting cues or to reward omissions. Thus, it is not the case that RMTg provides dopamine neurons with a complete RPE signal; rather, our results imply that the calculation of complete RPE mostly happens at the level of the dopamine neuron.
By comparing input neurons to other neurons in the same brain area, we found that input neurons were more enriched for mixed signals and RPE signals. Furthermore, our simulation showed that reconstruction of RPE signals was easier using the activity of input neurons compared to that of unidentified neurons. The enrichment of mixed coding in input neurons thus appears to be a reflection of multiple steps toward RPE computation. In other words, dopamine neurons need not combine pure signals of reward and expectation, but can rather refine signals that already exist in partial form. In parallel, some input neurons do send pure information of predicted and actual reward values. This might further reinforce the fidelity of RPE computations. Taken together, RPEs appear to form in a multi-layered, distributed network that eventually converges onto dopamine neurons.
Comparison to past models
Although dopamine neurons receive a significant number of glutamatergic projections, the major projections to dopamine neurons are GABAergic (Tepper and Lee, 2007). Because of this anatomical observation, many models proposed that phasic excitation of dopamine neurons is caused by disinhibition, i.e., a release of inhibition from GABA neurons onto dopamine neurons (Gale and Perkel, 2010; Lobb et al., 2010, 2011). More specifically, the basal ganglia have been proposed to provide a disinhibition circuit. Indeed, dopamine neurons receive large numbers of monosynaptic inputs from both the striatum and pallidum (Watabe-Uchida et al., 2012). In the present study, however, we observed that input neurons in most areas, including the ventral and dorsal striatum and ventral pallidum, were excited by positive values (e.g. reward-predictive cues). In our dataset of the striatum or ventral pallidum (n = 58 and 18 input neurons, respectively), we found no neuron that was inhibited in a monotonically reward value-dependent manner by reward-predictive CS with short latency, whereas we observed significant numbers of inputs that were excited with short latency. These results suggest that the phasic excitation of dopamine neurons is likely caused by excitatory inputs rather than disinhibitions, although we cannot exclude the possibility that a small fraction of neurons in these brain areas or other brain areas from which we did not record may provide a disinhibition circuit.
If GABA neurons in inputs may not provide phasic excitation of dopamine neurons, what is the function of the huge amounts of inhibitory inputs? In previous studies, we found that GABA neurons in VTA show activities which correlate with expected reward values (Cohen et al., 2012). Furthermore, inhibition of these neurons increased phasic responses to rewards in dopamine neurons (Eshel et al., 2015). These observations suggest that VTA GABA neurons are a source of inhibitory inputs that suppress dopamine reward responses depending on reward expectation. In the present study, we observed similar activities in input neurons in each area (Cluster 2 and 3 in Figure 2C, E, monotonic value coding in Figure 6B), especially in the ventral and dorsal striatum and ventral pallidum during the US period (“pure expectation” in Figure 4E). These results suggest redundancy of reward expectation signals in inputs. Yet, because of the vicinity of local inputs and weak electrical connections from the striatum to dopamine neurons (Bocklisch et al., 2013; Chuhma et al., 2011; Xia et al., 2011), it is possible that local GABA neurons have a larger influence on dopamine neurons than more distant inhibitory inputs. It will be important to examine the difference of synaptic weights and functions between local and long distance inputs.
In addition to providing expectation signals, inhibitory inputs may have other functions. We noticed three related characteristics in the activity of input neurons. First, input neurons tended to show more sustained activities than dopamine neurons, although a relatively small fraction of input neurons showed transient phasic activities. It is possible that inhibitory inputs may contribute to make the dopamine activities more phasic than the sum of excitatory inputs, akin to taking the derivative of otherwise sustained inputs. Note that taking the derivative is an important component of temporal difference error calculation (Schultz et al., 1997; Uchida, 2014). Second, many input neurons showed stronger excitation at air puff than dopamine neurons. Because inputs in the striatum, pallidum and RMTg as well as GABA neurons in VTA also showed excitation to air puff, these inputs may function to cancel out excitatory inputs that do not encode values. Thus, the air puff response of dopamine neurons may depend on a balance of incoming excitatory and inhibitory inputs. Third, some inputs in RMTg neurons had strong positive responses to reward omission, consistent with previous findings (Hong et al., 2011; Jhou et al., 2009a). RMTg receives projections from the lateral hebenula (Hb), which also encodes negative RPE. Interestingly, we previously showed that lesioning the whole Hb complex did not remove RPE in dopamine neurons, except that the response to reward omission in dopamine neurons was greatly reduced (Tian and Uchida, 2015). Whereas other information for RPE computation is redundant in multiple brain areas, the Hb-RMTg system may have a unique role in conveying signals of reward omission to dopamine neurons.
Technical considerations
In our previous recordings and the present study, we have focused on dopamine neurons in the lateral VTA. In this location, most dopamine neurons showed a uniform response function for RPE computations (Eshel et al., 2016). However, increasing studies indicate the diversity of midbrain dopamine neurons. For example, putative dopamine neurons in the lateral SNc in monkeys were excited by aversive stimuli (“motivational salience” or “absolute value”) whereas putative dopamine neurons in VTA and medial SNc were inhibited (“value”) (Matsumoto and Hikosaka, 2009). Other studies also suggested that responses to aversive stimuli in dopamine neurons are variable depending on their projection targets (Lammel et al., 2014; Lerner et al., 2015). There is also diversity of dopamine in the nucleus accumbens (parts of rodent ventral striatum) between the medial shell and core regions, anatomically (Beier et al., 2015) and physiologically (Brown et al., 2011; Saddoris et al., 2015). To address how different inputs contribute to the diversity of dopamine responses, it is important to distinguish different subsets of dopamine neurons which project to different brain areas. By incorporating newly developed rabies methods with retrograde infection of helper CAV or AAV (Beier et al., 2015; Menegas et al., 2015) into the present recording system, it will be possible to record from inputs to a subset of dopamine neurons which project to a specific brain area. We also have to remember that modification of the signal at the level of dopamine axon terminals may play an important role in the variation of dopamine signaling in different brain areas.
The possibility of recording from axons during extra-cellular recording experiments was indicated in previous studies (Barry, 2015). In the present study, the data do not support significant contamination from dopamine axon recording: dopamine-like activity (“type 1” neuron, or RPE-coding neurons) was observed more often in dopamine fiber-poor areas rather than dopamine projections-heavy areas such as the striatum. Further, RPE coding in input neurons was not as complete as RPE coding in dopamine neurons.
A previous study showed that cortical pyramidal neurons infected with SADΔG virus showed electrophysiological properties similar to uninfected neurons 5–12 days after injection, and the survival was roughly constant until 16 days, although the survival of neurons dropped substantially 18 days after injection (Wickersham et al., 2007b). We thus restricted our recording time window to 5–16 days after injection of the rabies virus. This meant that we had a maximum of only 12 days of recording per animal. This was the biggest obstacle in the present study because it was very difficult to achieve optimal behavior (Figure 1C) and precise targeting of recording electrodes in this short time window. Achievement of higher yields is necessary to cover more areas such as the central amygdala and to conduct powerful statistical analyses. A recent study found that a different line of rabies virus or helper virus may lengthen the survival of infected neurons and/or have more efficient trans-synaptic spreads (Kim et al., 2016; Reardon et al., 2016). Improving the methodology using an improved version of rabies virus such as this will aid greatly in advancing our ability to study neural computations.
Conclusions
By recording from monosynaptic inputs to dopamine neurons in behaving mice, we found that the computation of reward prediction error does not arise from simply combining pure inputs. Instead, our data show that components of information required for RPE computation are distributed and mixed in monosynaptic inputs from multiple brain areas. It is remarkable that, despite this complexity at the level of inputs, once these inputs are combined, the activity of dopamine neurons becomes homogeneous and largely follows simple arithmetic equations (Bayer and Glimcher, 2005; Eshel et al., 2015, 2016; Schultz et al., 1997). These results, at a first glance, might appear to indicate that dopamine neurons have the ability to weigh different inputs precisely to compute RPEs. Unfortunately, our methods did not allow us to measure the precise weights of each of these input neurons, but our analysis also indicated that precise tuning of synaptic weights is not as important as one might imagine. We propose that the mixed and distributed representation of information in the brain network may help dopamine neurons to robustly combine inputs to converge on a single solution.
EXPERIMENTAL PROCEDURES
Viral injection and electrophysiology
99 adult male mice heterozygous for Cre recombinase under the control of the DAT gene (B6.SJL-Slc6a3tm1.1(cre) Bkmn/J, The Jackson Laboratory, RRID: MGI_3689434) (Bäckman et al., 2006), were used. Mice underwent injection of helper viruses and then injection of SADΔG-ChR2-mCherry(envA) into VTA.Recording techniques were similar to those described in previous studies (Cohen et al., 2012; Tian and Uchida, 2015). On the day 16 after injection of rabies virus, recording locations were marked by making electrolytic lesions. All procedures were approved by the Harvard University Institutional Animal Care and Use Committee.
Supplementary Material
Highlights.
Electrophysiological recording from monosynaptic inputs of dopamine neurons
Rabies virus tracing was combined with optogenetic tagging in awake recording
Information required to compute reward prediction errors (RPEs) is distributed
Mixed representations of variables and partially computed RPEs in input neurons
Acknowledgments
We thank N. Eshel for comments, C. Dulac for sharing resources, V. Rao for technical assistance, M. Shoji for training assistance and M. Nagashima for histology assistance. This work was supported by Dr. Mortier and Theresa Sackler Foundation (J.T.); National Institute of Mental Health R01MH095953, R01MH101207 (N.U.); NIH MH063912 (E.M.C.); Japan Society for the Promotion of Science, Japan Science and Technology Agency (F.O.); and Bial Foundation 389/14 (D.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
JYC and MWU initiated the project. FO and EMC established SADΔG-ChR2-mCherry. JT and MWU designed experiments and collected data. RH trained mice. JT, DK, CKM and MWU analyzed data. JT, NU and MWU wrote the paper.
REFERENCE
- Aggarwal M, Hyland BI, Wickens JR. Neural control of dopamine neurotransmission: implications for reinforcement learning. European Journal of Neuroscience. 2012;35:1115–1123. doi: 10.1111/j.1460-9568.2012.08055.x. [DOI] [PubMed] [Google Scholar]
- Asaad WF, Eskandar EN. Encoding of Both Positive and Negative Reward Prediction Errors by Neurons of the Primate Lateral Prefrontal Cortex and Caudate Nucleus. J. Neurosci. 2011;31:17772–17787. doi: 10.1523/JNEUROSCI.3793-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, Tomac AC. Characterization of a mouse strain expressing Cre recombinase from the 3' untranslated region of the dopamine transporter locus. Genesis. 2006;44:383–390. doi: 10.1002/dvg.20228. [DOI] [PubMed] [Google Scholar]
- Barrot M, Sesack SR, Georges F, Pistis M, Hong S, Jhou TC. Braking Dopamine Systems: A New GABA Master Structure for Mesolimbic and Nigrostriatal Functions. J. Neurosci. 2012;32:14094–14101. doi: 10.1523/JNEUROSCI.3370-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry JM. Axonal activity in vivo: technical considerations and implications for the exploration of neural circuits in freely moving animals. Front Neurosci. 2015;9 doi: 10.3389/fnins.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer HM, Glimcher PW. Midbrain dopamine neurons encode a quantitative reward prediction error signal. Neuron. 2005;47:129–141. doi: 10.1016/j.neuron.2005.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, Luo L. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Han V, Sawtell NB. Cerebellum-like structures and their implications for cerebellar function. Annu. Rev. Neurosci. 2008;31:1–24. doi: 10.1146/annurev.neuro.30.051606.094225. [DOI] [PubMed] [Google Scholar]
- Bocklisch C, Pascoli V, Wong JCY, House DRC, Yvon C, de Roo M, Tan KR, Luscher C. Cocaine Disinhibits Dopamine Neurons by Potentiation of GABA Transmission in the Ventral Tegmental Area. Science. 2013;341:1521–1525. doi: 10.1126/science.1237059. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Bromberg-Martin ES, Hikosaka O. Lateral habenula neurons signal errors in the prediction of reward information. Nat Neurosci. 2011;14:1209–1216. doi: 10.1038/nn.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg-Martin ES, Matsumoto M, Hikosaka O. Dopamine in motivational control: rewarding, aversive, and alerting. Neuron. 2010;68:815–834. doi: 10.1016/j.neuron.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HD, McCutcheon JE, Cone JJ, Ragozzino ME, Roitman MF. Primary food reward and reward-predictive stimuli evoke different patterns of phasic dopamine signaling throughout the striatum. European Journal of Neuroscience. 2011;34:1997–2006. doi: 10.1111/j.1460-9568.2011.07914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Bullock D, Grossberg S. How the Basal Ganglia Use Parallel Excitatory and Inhibitory Learning Pathways to Selectively Respond to Unexpected Rewarding Cues. J. Neurosci. 1999;19:10502–10511. doi: 10.1523/JNEUROSCI.19-23-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carandini M, Heeger DJ. Normalization as a canonical neural computation. Nat. Rev. Neurosci. 2012;13:51–62. doi: 10.1038/nrn3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance FS, Abbott LF, Reyes AD. Gain modulation from background synaptic input. Neuron. 2002;35:773–782. doi: 10.1016/s0896-6273(02)00820-6. [DOI] [PubMed] [Google Scholar]
- Chuhma N, Tanaka KF, Hen R, Rayport S. Functional Connectome of the Striatal Medium Spiny Neuron. J. Neurosci. 2011;31:1183–1192. doi: 10.1523/JNEUROSCI.3833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JY, Haesler S, Vong L, Lowell BB, Uchida N. Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature. 2012;482:85–88. doi: 10.1038/nature10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JY, Amoroso MW, Uchida N. Serotonergic neurons signal reward and punishment on multiple timescales. eLife Sciences. 2015;4:e06346. doi: 10.7554/eLife.06346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw ND, Courville AC, Touretzky DS. Representation and Timing in Theories of the Dopamine System. Neural Computation. 2006;18:1637–1677. doi: 10.1162/neco.2006.18.7.1637. [DOI] [PubMed] [Google Scholar]
- Doya K. Complementary roles of basal ganglia and cerebellum in learning and motor control. Current Opinion in Neurobiology. 2000;10:732–739. doi: 10.1016/s0959-4388(00)00153-7. [DOI] [PubMed] [Google Scholar]
- Doya K. Modulators of decision making. Nat Neurosci. 2008;11:410–416. doi: 10.1038/nn2077. [DOI] [PubMed] [Google Scholar]
- Eshel N, Bukwich M, Rao V, Hemmelder V, Tian J, Uchida N. Arithmetic and local circuitry underlying dopamine prediction errors. Nature. 2015;525:243–246. doi: 10.1038/nature14855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel N, Tian J, Bukwich M, Uchida N. Dopamine neurons share common response function for reward prediction error. Nat. Neurosci. 2016;19:479–486. doi: 10.1038/nn.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo CD, Yun SR, Song MR. Diversity and homogeneity in responses of midbrain dopamine neurons. Journal of Neuroscience. 2013;33:4693–4709. doi: 10.1523/JNEUROSCI.3886-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi S, Miller EK, Rigotti M. Why neurons mix: high dimensionality for higher cognition. Current Opinion in Neurobiology. 2016;37:66–74. doi: 10.1016/j.conb.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Gale SD, Perkel DJ. A Basal Ganglia Pathway Drives Selective Auditory Responses in Songbird Dopaminergic Neurons via Disinhibition. J. Neurosci. 2010;30:1027–1037. doi: 10.1523/JNEUROSCI.3585-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Zahm DS. Afferents of the ventral tegmental area in the rat-anatomical substratum for integrative functions. J. Comp. Neurol. 2005;490:270–294. doi: 10.1002/cne.20668. [DOI] [PubMed] [Google Scholar]
- Geisler S, Derst C, Veh RW, Zahm DS. Glutamatergic afferents of the ventral tegmental area in the rat. J. Neurosci. 2007;27:5730–5743. doi: 10.1523/JNEUROSCI.0012-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AS, Rutledge RB, Glimcher PW, Phillips PEM. Phasic dopamine release in the rat nucleus accumbens symmetrically encodes a reward prediction error term. J. Neurosci. 2014;34:698–704. doi: 10.1523/JNEUROSCI.2489-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimer L, Zahm DS, Churchill L, Kalivas PW, Wohltmann C. Specificity in the projection patterns of accumbal core and shell in the rat. Neuroscience. 1991;41:89–125. doi: 10.1016/0306-4522(91)90202-y. [DOI] [PubMed] [Google Scholar]
- Holt GR, Koch C. Shunting inhibition does not have a divisive effect on firing rates. Neural Comput. 1997;9:1001–1013. doi: 10.1162/neco.1997.9.5.1001. [DOI] [PubMed] [Google Scholar]
- Hong S, Hikosaka O. The globus pallidus sends reward-related signals to the lateral habenula. Neuron. 2008;60:720–729. doi: 10.1016/j.neuron.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Hikosaka O. Pedunculopontine tegmental nucleus neurons provide reward, sensorimotor, and alerting signals to midbrain dopamine neurons. Neuroscience. 2014;282:139–155. doi: 10.1016/j.neuroscience.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Jhou TC, Smith M, Saleem KS, Hikosaka O. Negative reward signals from the lateral habenula to dopamine neurons are mediated by rostromedial tegmental nucleus in primates. J. Neurosci. 2011;31:11457–11471. doi: 10.1523/JNEUROSCI.1384-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk JC, Adams JL, Barto AG. A model of how the basal ganglia generate and use neural signals that predict reinforcement. Models of Information Processing in the Basal Ganglia. 1995:249–270. [Google Scholar]
- Jennings JH, Stuber GD. Tools for resolving functional activity and connectivity within intact neural circuits. Curr. Biol. 2014;24:R41–R50. doi: 10.1016/j.cub.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Sparta DR, Stamatakis AM, Ung RL, Pleil KE, Kash TL, Stuber GD. Distinct extended amygdala circuits for divergent motivational states. Nature. 2013;496:224–228. doi: 10.1038/nature12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhou TC, Fields HL, Baxter MG, Saper CB, Holland PC. The rostromedial tegmental nucleus (RMTg), a GABAergic afferent to midbrain dopamine neurons, encodes aversive stimuli and inhibits motor responses. Neuron. 2009a;61:786–800. doi: 10.1016/j.neuron.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhou TC, Geisler S, Marinelli M, Degarmo BA, Zahm DS. The mesopontine rostromedial tegmental nucleus: A structure targeted by the lateral habenula that projects to the ventral tegmental area of Tsai and substantia nigra compacta. J Comp Neurol. 2009b;513:566–596. doi: 10.1002/cne.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel D, Niv Y, Ruppin E. Actor-critic models of the basal ganglia: new anatomical and computational perspectives. Neural Netw. 2002;15:535–547. doi: 10.1016/s0893-6080(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Johansen JP, Tarpley JW, LeDoux JE, Blair HT. Neural substrates for expectation-modulated fear learning in the amygdala and periaqueductal gray. Nat. Neurosci. 2010;13:979–986. doi: 10.1038/nn.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawato M, Samejima K. Efficient reinforcement learning: computational theories, neuroscience and robotics. Curr Opin Neurobiol. 2007;17:205–212. doi: 10.1016/j.conb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Keiflin R, Janak PH. Dopamine Prediction Errors in Reward Learning and Addiction: From Theory to Neural Circuitry. Neuron. 2015;88:247–263. doi: 10.1016/j.neuron.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Jacobs MW, Ito-Cole T, Callaway EM. Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Reports. 2016;15:692–699. doi: 10.1016/j.celrep.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Okada K-I. Reward Prediction Error Computation in the Pedunculopontine Tegmental Nucleus Neurons. Annals of the New York Academy of Sciences. 2007;1104:310–323. doi: 10.1196/annals.1390.003. [DOI] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Malenka RC. Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology. 2014;76(Part B):351–359. doi: 10.1016/j.neuropharm.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Seo H, Jung MW. Neural basis of reinforcement learning and decision making. Annu. Rev. Neurosci. 2012;35:287–308. doi: 10.1146/annurev-neuro-062111-150512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, et al. Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell. 2015;162:635–647. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb CJ, Wilson CJ, Paladini CA. A Dynamic Role for GABA Receptors on the Firing Pattern of Midbrain Dopaminergic Neurons. J Neurophysiol. 2010;104:403–413. doi: 10.1152/jn.00204.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb CJ, Wilson CJ, Paladini CA. High-frequency, short-latency disinhibition bursting of midbrain dopaminergic neurons. Journal of Neurophysiology. 2011;105:2501–2511. doi: 10.1152/jn.01076.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hikosaka 0. Lateral habenula as a source of negative reward signals in dopamine neurons. Nature. 2007;447:1111–1115. doi: 10.1038/nature05860. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Hikosaka 0. Representation of negative motivational value in the primate lateral habenula. Nat Neurosci. 2008;12:77–84. doi: 10.1038/nn.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hikosaka 0. Two types of dopamine neuron distinctly convey positive and negative motivational signals. Nature. 2009;459:837–U4. doi: 10.1038/nature08028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Venkataraju KU, Osten P, Uchida N, Watabe-Uchida M. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife Sciences. 2015:e10032. doi: 10.7554/eLife.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirenowicz J, Schultz W. Importance of unpredictability for reward responses in primate dopamine neurons. J. Neurophysiol. 1994;72:1024–1027. doi: 10.1152/jn.1994.72.2.1024. [DOI] [PubMed] [Google Scholar]
- Morita K, Morishima M, Sakai K, Kawaguchi Y. Reinforcement learning: computing the temporal difference of values via distinct corticostriatal pathways. Trends Neurosci. 2012;35:457–467. doi: 10.1016/j.tins.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Nair-Roberts RG, Chatelain-Badie SD, Benson E, White-Cooper H, Bolam JP, Ungless MA. Stereological estimates of dopaminergic, GABAergic and glutamatergic neurons in the ventral tegmental area, substantia nigra and retrorubral field in the rat. Neuroscience. 2008;152:1024–1031. doi: 10.1016/j.neuroscience.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Ono T. Lateral hypothalamus neuron involvement in integration of natural and artificial rewards and cue signals. J Neurophysiol. 1986;55:163–181. doi: 10.1152/jn.1986.55.1.163. [DOI] [PubMed] [Google Scholar]
- Ohmae S, Medina JF. Climbing fibers encode a temporal-difference prediction error during cerebellar learning in mice. Nat. Neurosci. 2015;18:1798–1803. doi: 10.1038/nn.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Toyama K, Inoue Y, Isa T, Kobayashi Y. Different pedunculopontine tegmental neurons signal predicted and actual task rewards. J. Neurosci. 2009;29:4858–4870. doi: 10.1523/JNEUROSCI.4415-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bhandawat V, Wilson RI. Divisive normalization in olfactory population codes. Neuron. 2010;66:287–299. doi: 10.1016/j.neuron.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly RC, Frank MJ, Hazy TE, Watz B. PVLV: the primary value and learned value Pavlovian learning algorithm. Behav. Neurosci. 2007;121:31–49. doi: 10.1037/0735-7044.121.1.31. [DOI] [PubMed] [Google Scholar]
- Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;77:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Ouden HE, Kok P, De Lange FP. How prediction errors shape perception, attention, and motivation. Front. Psychol. 2012;3:10–3389. doi: 10.3389/fpsyg.2012.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama K, Tateyama Y, Hernadi I, Tobler PN, Iijima T, Tsutsui K-I. Discrete coding of stimulus value, reward expectation, and reward prediction error in the dorsal striatum. Journal of Neurophysiology. 2015;114:2600–2615. doi: 10.1152/jn.00097.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W-X, Hyland BI. Pedunculopontine tegmental nucleus controls conditioned responses of midbrain dopamine neurons in behaving rats. Journal of Neuroscience. 2005;25:4725–4732. doi: 10.1523/JNEUROSCI.0277-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, Losonczy A. Rabies Virus CVS-N2cAG Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron. 2016;89:711–724. doi: 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigotti M, Barak O, Warden MR, Wang X-J, Daw ND, Miller EK, Fusi S. The importance of mixed selectivity in complex cognitive tasks. Nature. 2013;497:585–590. doi: 10.1038/nature12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddoris MP, Cacciapaglia F, Wightman RM, Carelli RM. Differential Dopamine Release Dynamics in the Nucleus Accumbens Core and Shell Reveal Complementary Signals for Error Prediction and Incentive Motivation. J. Neurosci. 2015;35:11572–11582. doi: 10.1523/JNEUROSCI.2344-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W. Neuronal Reward and Decision Signals: From Theories to Data. Physiological Reviews. 2015;95:853–951. doi: 10.1152/physrev.00023.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W, Dickinson A. Neuronal Coding of Prediction Errors. Annual Review of Neuroscience. 2000;23:473–500. doi: 10.1146/annurev.neuro.23.1.473. [DOI] [PubMed] [Google Scholar]
- Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Science. 1997;275:1593–1599. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- Sesack SR, Grace AA. Cortico-Basal Ganglia Reward Network: Microcircuitry. Neuropsychopharmacology. 2009;35:27–47. doi: 10.1038/npp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver RA. Neuronal arithmetic. Nat. Rev. Neurosci. 2010;11:474–489. doi: 10.1038/nrn2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain Res. 2000;886:113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- Tachibana Y, Hikosaka O. The primate ventral pallidum encodes expected reward value and regulates motor action. Neuron. 2012;76:826–837. doi: 10.1016/j.neuron.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper JM, Lee CR. GABAergic control of substantia nigra dopaminergic neurons. In: EDA, James JPB, Tepper M, editors. Progress in Brain Research. Elsevier; 2007. pp. 189–208. [DOI] [PubMed] [Google Scholar]
- Tian J, Uchida N. Habenula Lesions Reveal that Multiple Mechanisms Underlie Dopamine Prediction Errors. Neuron. 2015;87:1304–1316. doi: 10.1016/j.neuron.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N. Bilingual neurons release glutamate and GABA. Nat Neurosci. 2014;17:1432–1434. doi: 10.1038/nn.3840. [DOI] [PubMed] [Google Scholar]
- Uchida N, Eshel N, Watabe-Uchida M. Division of labor for division: inhibitory interneurons with different spatial landscapes in the olfactory system. Neuron. 2013;80:1106–1109. doi: 10.1016/j.neuron.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wertz A, Trenholm S, Yonehara K, Hillier D, Raics Z, Leinweber M, Szalay G, Ghanem A, Keller G, Rózsa B, et al. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science. 2015;349:70–74. doi: 10.1126/science.aab1687. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann K-K, Young JAT, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007a;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann K-K, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Meth. 2007b;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williford T, Maunsell JHR. Effects of spatial attention on contrast response functions in macaque area V4. J. Neurophysiol. 2006;96:40–54. doi: 10.1152/jn.01207.2005. [DOI] [PubMed] [Google Scholar]
- Xia Y, Driscoll JR, Wilbrecht L, Margolis EB, Fields HL, Hjelmstad GO. Nucleus Accumbens Medium Spiny Neurons Target Non-Dopaminergic Neurons in the Ventral Tegmental Area. J. Neurosci. 2011;31:7811–7816. doi: 10.1523/JNEUROSCI.1504-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.