Abstract
Cancer testis antigens (CTAs) are of clinical interest as biomarkers and present valuable targets for immunotherapy. To comprehensively characterize the CTA landscape of non–small-cell lung cancer (NSCLC), we compared RNAseq data from 199 NSCLC tissues to the normal transcriptome of 142 samples from 32 different normal organs. Of 232 CTAs currently annotated in the Caner Testis Database (CTdatabase), 96 were confirmed in NSCLC. To obtain an unbiased CTA profile of NSCLC, we applied stringent criteria on our RNAseq data set and defined 90 genes as CTAs, of which 55 genes were not annotated in the CTdatabase, thus representing potential new CTAs. Cluster analysis revealed that CTA expression is histology dependent and concurrent expression is common. IHC confirmed tissue-specific protein expression of selected new CTAs (TKTL1, TGIF2LX, VCX, and CXORF67). Furthermore, methylation was identified as a regulatory mechanism of CTA expression based on independent data from The Cancer Genome Atlas. The proposed prognostic impact of CTAs in lung cancer was not confirmed, neither in our RNAseq cohort nor in an independent meta-analysis of 1,117 NSCLC cases. In summary, we defined a set of 90 reliable CTAs, including information on protein expression, methylation, and survival association. The detailed RNAseq catalog can guide biomarker studies and efforts to identify targets for immunotherapeutic strategies.
A comprehensive analysis of cancer-testis antigens in non-small cell lung cancer based on RNAseq and complemented with immunohistochemistry is provided.
Introduction
The mutational landscape of non–small-cell lung cancer (NSCLC) has been extensively explored; this has led to the identification of cancer driver mutations, several of which have been successfully exploited as therapeutic targets in clinical practice (1–3). However, targetable genomic aberrations are only present in minor subgroups of patients and therapy response is only temporary (4–6). As a conceptual alternative, immunotherapeutic strategies have emerged, taking advantage of the patient’s own immune system to stimulate an antitumor response (7). As a proof of concept, so-called checkpoint inhibitors have been approved for the treatment of advanced NSCLC with impressive long-lasting tumor response (8). Along with antibody-based modalities, vaccination is also used as an immune modulatory approach (9–11). In principal, all cancer immunotherapies share the concept that the immune system can exclusively attack cancer cells but spare normal cells. Cancer-specific structures are thus an advantageous attribute for cancer cell recognition and elimination (12–14).
Cancer testis antigens (CTAs) have attracted the attention of cancer researchers as potential mediators of cancer cell recognition (15, 16). The members of this group are expressed in a wide range of cancers, including lung cancer, while in normal tissues their expression is restricted to immune privileged sites, such as testis and placenta. Indeed, CTAs have been shown to encode immunogenic proteins that can induce spontaneous humoral or cellular antitumor responses in cancer patients (17, 18). Because of the downregulation of the MHC and the production of immune-suppressive factors, autoimmunity in testis and placenta is prevented (19, 20). In order to systematically summarize the accumulating knowledge, the Ludwig Institute for Cancer Research established the Cancer Testis database (CTdatabase) (21), providing researchers with a detailed and, importantly, manually curated list of proposed CTAs, including splicing variants, immunogenic information, and the gene expression levels per cancer type, together with source of information.
NSCLC, along with melanoma and ovarian cancer, has been found to have the highest frequency of CTA expression of the different analyzed cancers. Until now the majority of CTA studies on NSCLC have focused on evaluating the expression of only a subset of CTAs (22–25). Therefore, the aim of this study was to explore the comprehensive landscape of CTAs in NSCLC. The basis for this analysis was RNA-sequencing (RNAseq) data from 199 NSCLC patients and 142 samples from 32 different normal human tissue types. We evaluated in a supervised approach the expression of all previously described CTAs (n = 232) annotated in the CTdatabase. This was followed by identification of 90 genes with substantial CTA features, defining the unbiased CTA profile of NSCLC. Since the majority of CTAs has only been described on mRNA levels, we complemented the transcriptomic information with protein profiling for a subset of CTAs, using antibodies to provide information on tissue distribution and subcellular localization in the in situ environment of NSCLC. Methylation status of each CTA supplemented the gene expression data to explain potential regulatory mechanisms of CTA expression. Finally, we explored the prognostic impact of the 90 NSCLC CTAs in our data set and in a meta-analysis of 1,117 patients from 7 independent data sets.
Results
The lung cancer transcriptome.
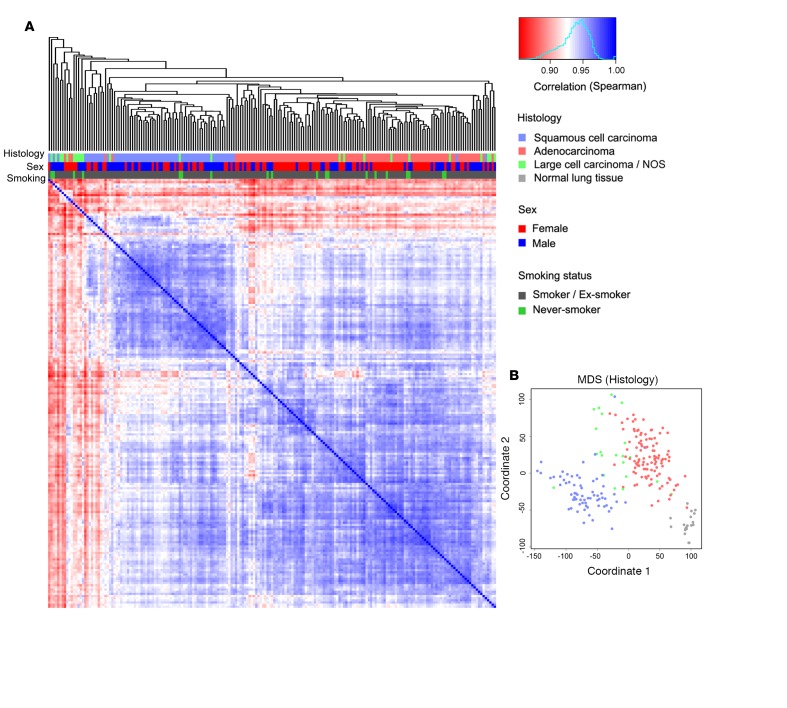
RNAseq analysis was performed on 199 fresh-frozen NSCLC samples (Table 1). mRNA expression of 20,293 putative protein-coding genes (Ensembl v.73) in the 199 NSCLC samples was used in unsupervised hierarchical cluster analysis, as well as multidimensional scaling, and revealed that cancer histology is by far the most dominant factor for gene expression differences and responsible for clustering. Based on these analyses, the two main groups containing either adenocarcinoma or squamous cell carcinomas were separated; other NSCLC subtypes were clustered within these two groups (Figure 1). This pattern is in accordance with previous results of NSCLC profiling using microarray technology (26, 27), indicating that the Uppsala lung cancer cohort is representative of NSCLC.
Table 1. Clinical characteristics of 199 NSCLC cases included in the RNAseq analysis.
Figure 1. The transcriptomic profile of non–small-cell lung cancer (NSCLC).
(A) Hierarchical cluster analysis demonstrating the pair-wise correlation between all 199 NSCLC cases based on mRNA expression of 20,293 putative protein-coding genes (Ensembl v.73). (B) Multidimensional scaling (MDS) plot with regard to histology showing the level of similarity of the 199 NSCLC cases analyzed as well as of 19 normal lung samples. NOS, not otherwise specified.
The mRNA expression of reported CTAs.
The CTdatabase (http://www.cta.lncc.br) is a systematic data repository for CTAs, currently including 276 genes designated as CTAs, with curated information about gene and protein expression in normal and cancer tissues (21). All 276 genes were initially included, but 44 of these genes were excluded from further analysis; 31 genes were not mappable to the Ensembl database, 8 genes lack protein-coding transcripts, 1 gene was not expressed in any normal or NSCLC samples, and 4 genes were double annotated in the CTdatabase. In summary, 232 unique CTAs were used in all further analyses of previously reported CTAs. To determine the number of previously reported CTAs expressed in NSCLC and to validate their restricted expression pattern in testis/placenta, RNAseq data from 199 NSCLC tissues and 142 normal tissues from 32 different human organs sites were analyzed.
A CTA was designated as “confirmed CTA” when the expression in testis or placenta and at least one of the NSCLC samples was 5 times higher than the expression in any other normal tissue type. Based on these criteria, 96 of the 232 genes (41%) were classified as “confirmed,” i.e., having validated testis/placenta-specific expression and also being expressed in NSCLC (Figure 2 and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.86837DS1). Of the 96 confirmed CTAs, only 24 genes were previously described as CTAs in the CTdatabase on both the mRNA and protein level in lung cancer, whereas 44 genes had only been described in NSCLC on mRNA levels. Finally, 28 genes had previously not been reported in NSCLC/lung cancer, neither on the mRNA level nor on the protein level, but only in other cancer types.
Figure 2. The expression of reported cancer testis antigens (CTAs) in non–small-cell lung cancer (NSCLC).
Reported CTAs in the CTdatabase (n = 232) were analyzed in 199 NSCLC cases and 142 normal tissues from 32 different organs. Based on the mRNA expression for each CTA in NSCLC and normal tissues, these 232 genes were grouped in either confirmed CTAs (green shades) or not confirmed CTAs and subdivided in testis/placenta-specific genes without expression in NSCLC (pink) or CTAs with expression in somatic tissues (blue shades). From each of these 6 categories the mRNA expression of a representative CTA is shown. The y axes show the mRNA levels as fragments per kilobase of transcript per million mapped reads (FPKM) values, and the x axes represent each analyzed case: NSCLC samples (red), normal tissues except testis (black), and testis (gray).
Further analysis of the 232 CTAs revealed that 59 genes (25%) were defined as testis/placenta-specific genes, i.e., expressed in testis or placenta and not expressed in other somatic tissue, but demonstrated no expression in our NSCLC cohort (Supplemental Table 2), thus not representing relevantly expressed CTAs in NSCLC. Unexpectedly, 77 of 232 genes (33%) were also expressed in other normal tissues than testis and placenta (Supplemental Table 3). Of these 77 genes, 34 genes have at least one testis-specific transcript; therefore, we cannot exclude that these transcripts are expressed as isoform-specific CTAs (Supplemental Table 4). We suggest that these 77 nonconfirmed CTAs should be reevaluated and potentially removed as CTA candidates in the CTdatabase.
The protein expression of reported CTAs in NSCLC.
Of 232 genes, 68 genes were reported to be expressed in NSCLC according to the CTdatabase. Of these, 24 were described on both the mRNA and protein level, whereas the remaining 44 CTAs were only defined based on the mRNA level (Figure 2). Using the image database of Human Protein Atlas (HPA) (28), we confirmed protein expression of 8 CTAs in NSCLC (MAGEC2, MAGEB6, PAGE2, PAGE5, PAGE2B, CT45A2, SAGE1, and MAGEA8; Figure 3A). Furthermore, the 59 genes that only showed testis/placenta-specific expression, but no expression in NSCLC, were evaluated. Indeed, testis-specific protein expression in normal tissues was confirmed in the image database of the HPA for 14 genes (AKAP3, PRM2, CRISP2, ODF2, AKAP4, FATE1, LDHC, SSX2, ACRBP, CALR3, HORMAD1, LUZP4, PRM1, and DKKL). The protein expression for 6 of these genes is shown in Figure 3B, in which expression is demonstrated in both early (e.g., LUZP4) and in later stages (e.g., DKKL1) of spermatogenesis.
Figure 3. Immunohistochemical staining of confirmed cancer testis antigens (CTAs) and not confirmed CTAs in non–small-cell lung cancer (NSCLC) and normal human tissues.
(A) Tissue sections of normal testis (MAGEC2, MAGEB6, PAGE2/5/2B, CT45A2, and SAGE1) and placenta (MAGEA8) as well as adenocarcinoma and squamous cell carcinoma were stained with antibodies targeting 8 confirmed CTAs. (B) Tissue sections showing normal testis, adenocarcinoma, and squamous cell carcinoma stained with antibodies targeting 6 not confirmed CTAs, encoded by testis-specific genes without expression in NSCLC. Scale bar: 100 μm.
The evaluation of the 77 nonconfirmed CTAs (mRNA expression was not restricted to testis/placenta in our data set) demonstrated protein expression in somatic tissue for 18 of these genes, thus confirming the uncertain annotation as CTAs in the CTdatabase (HEMGN, SPA17, SPAG6, SPAG8, ANKRD45, KDM5B, CTAG1A, CT45A6, CT45A3, CT45A1, CT45A5, SPACA3, SPEF2, GAGE12G, GAGE12F, GAGE12I, PRSS50, and PBK), of which 6 examples are shown in Figure 4. Several of these uncertain CTAs were also expressed in the epithelial cells of the fallopian tube (SPA17, SPAG6, SPAG8, and ANKRD45) and are related to the motility apparatus of cilia and flagella.
Figure 4. Immunohistochemical staining of not confirmed cancer testis antigens (CTAs) that are expressed in somatic tissues.
Tissue sections showing normal testis, fallopian tube, skin, lung, and bone marrow stained with antibodies targeting not confirmed CTAs with gene expression in somatic tissues.
Identification of new CTAs in NSCLC.
To complement the information of the CTdatabase and to provide a comprehensive CTA profile in NSCLC, we extended our analysis. To identify CTAs in an unbiased manner and with robust expression frequencies, we applied the following two criteria to our transcriptomic data: (a) a gene should have at least 5 times higher expression in NSCLC than any normal tissue (excluding testis and placenta) to be considered specifically expressed in cancer tissues and (b) the gene must be expressed in at least 2% of either adenocarcinomas or squamous cell carcinomas. Based on these criteria, a list of 96 genes was generated (Supplemental Table 5), of which 35 genes were already annotated as CTAs in the CTdatabase. Consequently, the remaining 61 genes were putative new CTAs.
The list of these 61 candidate CTAs was manually curated utilizing the independent resource of transcriptomic data from post-mortem tissues called the Genotype-Tissue Expression Project (GTEx), which contains mRNA expression data from 29 solid organ sites and 11 brain subregions (29). Of our 61 candidates, 6 genes (FGF3, TCHHL1, GABRR1, KRT85, TFPI2, and HMGB3) were excluded, because they showed mRNA expression in normal tissues other than testis and placenta in this independent set of samples. Additionally, 7 of the genes were expressed in nerve tissue and different regions of the brain. Since these tissue compartments are often also considered to be immune privileged (30), we marked these genes as candidate CTAs with neural-related expression. The testis/placenta-specific expression of the remaining 55 genes was confirmed in all analyzed normal tissue specimens in GTEx. Interestingly, 9 of these genes were located on the X chromosome (XAGE1A, CT83, MAGEB16, CXorf67, VCX, TGIF2LX, USP26, H2BFM, and TKTL1), a typical feature for CTAs (16). As expected, we observed a histology-dependent expression of the new CTAs, with many genes, such as C12orf54, TSPY10, LIN28B, CXorf67, TDRD12, LETM2, and DPEP3, exclusively expressed in squamous carcinoma patients and others, such as PAGE5, SKOR2, XAGE1A, TUBA3C, and STK31, exclusively or preferentially expressed in adenocarcinoma patients. Altogether, we defined 90 NSCLC CTAs, of them 35 previously described and 55 novel CTAs (Supplemental Table 11).
The protein expression of new CTAs.
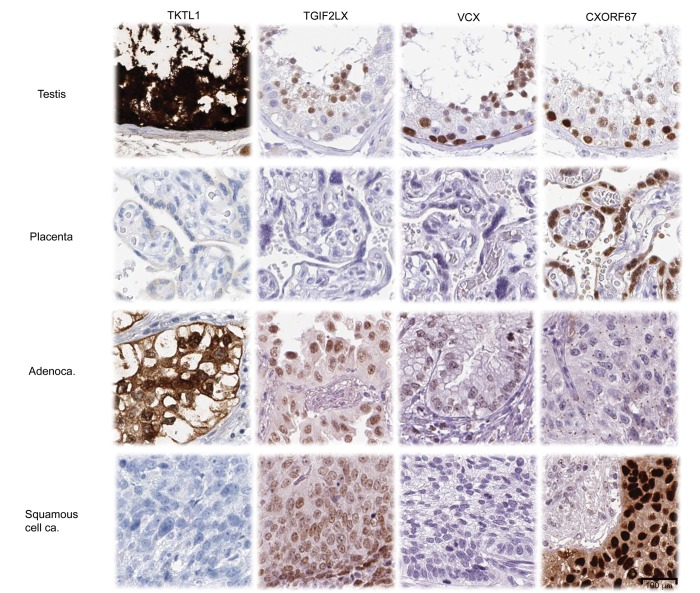
We utilized the HPA database to evaluate protein expression in a subset of the novel CTAs. Four antibodies directed to TKTL1, TGIF2LX, VCX, and CXorf67 showed, in accordance to mRNA expression, protein expression in testis or placenta, but no staining in all other normal tissues. These antibodies were applied to stain a tissue microarray (TMA) of 35 NSCLC specimens, confirming protein expression in at least one NSCLC sample (Figure 5). Transketolase-like protein 1 (TKTL1) is an enzyme involved in the nonoxidative pentose-phosphate pathway that is reported to be overexpressed in several human cancers, including lung cancer (31), but it has not previously been reported to be a CTA. In our analysis, TKTL1 was expressed in both histologic subtypes of NSCLC and showed cytoplasmic and nuclear positivity in the IHC analysis in 2.9% of NSCLC cases. TGF-β–induced transcription factor 2-like protein (TGIF2LX) is a poorly characterized protein with a putative transcriptional role in testis. TGIF2LX expression has previously only been reported in prostate cancers among human cancers (32). In our analysis, TGIF2LX was expressed in both histological subtypes of NSCLC and showed dominant nuclear staining in 5.7% of NSCLC cases. Variable charge X-linked protein 1 (VCX) is involved in spermatogenesis and has recently been suggested to be a CTA in NSCLC (33). We confirmed nuclear staining in the IHC analysis in 62.9% of NSCLC cases. The uncharacterized protein CXorf67 was hitherto not described on the protein level. In our data set, CXorf67 showed strong nuclear expression in the squamous histology (11.4%).
Figure 5. Immunohistochemical staining of new cancer testis antigens (CTAs) in testis, placenta, and non–small-cell lung cancer (NSCLC) tissues.
Tissue sections of normal testis or placenta as well as adenocarcinoma and squamous cell carcinoma were stained with antibodies towards 4 new CTAs.
The methylation status of CTAs in NSCLC.
The expression of CTAs is often considered to be regulated by gene promoter hypomethylation. Therefore, we analyzed the relationship between DNA methylation and gene expression in an independent NSCLC data set from The Cancer Genome Atlas (TCGA), including in total 845 NSCLC and 74 normal lung samples. The methylation status for 65 of the 90 NSCLC CTAs (72%) could be retrieved from these data sets. In adenocarcinoma 31.8% of the CTA genes and in squamous cell carcinoma 47.0% of the CTA genes were hypomethylated (Supplemental Table 6). This was significantly greater compared with the hypomethylation status of all other genes (non-CTAs). In adenocarcinoma 7.6% of all non-CTAs were defined as hypomethylated (P < 0.01), whereas 12.6% of all non-CTAs were hypomethylated in squamous cell cancer (P < 0.01).
Furthermore, we analyzed the correlation between CTA methylation and gene expression to evaluate whether CTA expression was associated with hypomethylation in NSCLC tissue. A relevant negative correlation (r < –0.4) between methylation and mRNA levels was demonstrated for 12 CTAs (19%) in adenocarcinoma and 15 CTAs in squamous cell cancer (23%). This was significantly more than expected compared with the number of genes with a coefficient of < –0.4 when all other non-CTA genes were analyzed (P < 0.01 for both comparisons; Supplemental Figure 1).
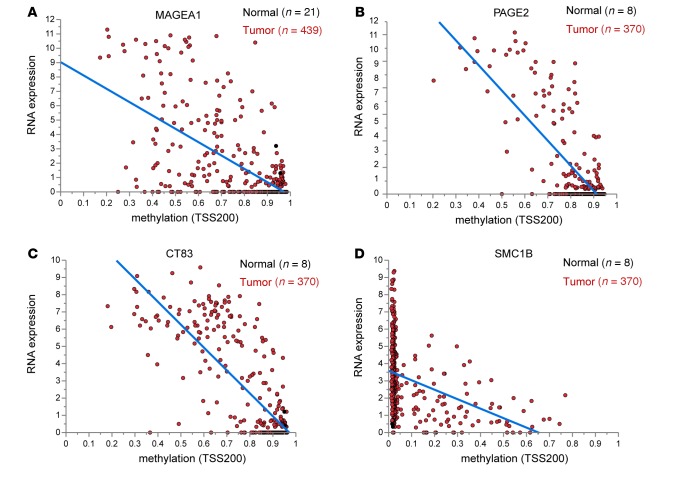
Most often the promoter region TSS200 (200 bp upstream of the transcription start site) or the first exon was involved. Relevant associations were observed for established CTAs and X chromosomal CTAs (e.g., MAGEA1 and PAGE2; Figure 6, A and B) as well as new and non–X chromosomal CTAs (e.g., CT83) and SMC1B; Figure 6, C and D).
Figure 6. Correlation of DNA methylation with gene expression.
Matched methylation and gene expression data from The Cancer Genome Atlas were plotted to illustrate the correlation for selected cancer testis antigens (CTAs): (A) MAGEA1, (B) PAGE2, (C) CT83, and (D) SMC1B. The x axes give the β value of the region of the CTA 200 bp upstream of the transcription start site (TSS200) as a degree of methylation. The y axes give the gene expression as log2 transformed 1+ reads per kilobase of transcript per million mapped reads values of RNAseq.
That methylation is a possible mechanism of regulation in a subset of CTAs was confirmed by the analysis of gene expression data of human bronchial epithelial cell lines (NHBE) and human small airway epithelial cells treated with the demethylating agents 5-aza-2′-deoxycytidine (AZA) and trichostatin A. AZA and trichostatin A–associated regulation of CTA gene expression in at least one of both cell lines was observed for 23 of 68 evaluable CTAs (Supplemental Table 6).
When evaluating the effect of AZA-based demethylation in NSCLC cell lines, an even higher fraction of CTAs demonstrated an increased expression (47 genes in at least one of the adenocarcinoma cell lines and 34 genes in at least one of the squamous cell carcinoma cell lines of 63 evaluable CTAs).
Taken together, the in silico results based on independent NSCLC cases indicated that demethylation plays an important role in the regulation of CTA expression in NSCLC.
Coordinately expressed genes and cluster analysis.
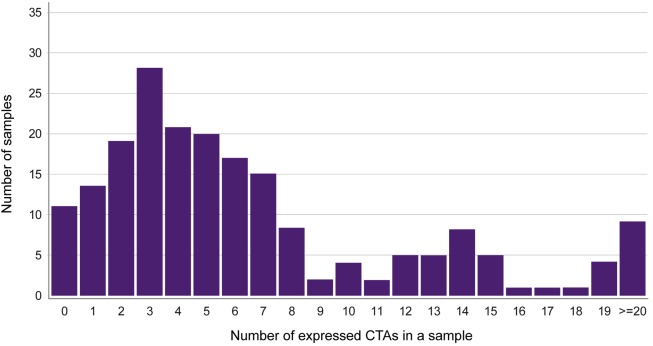
In our data set, 90 CTAs were identified (35 previously described CTAs and 55 new CTAs) and their expression patterns were analyzed. Most of the CTAs were coexpressed; only 5.5% of the NSCLC cases did not express any CTA; 64.3% expressed more than 3 CTAs, 22.6% expressed more than 10 CTAs, and 4.5% expressed more than 20 CTAs (Figure 7). Hierarchical clustering and network analysis were performed to illustrate CTA expression patterns among the 199 NSCLC cases (Figure 8). The hierarchical cluster analysis also illustrated that CTA expression is histology dependent. Clustering stratified cancer cases in histological groups, for example, with MAGE family members in squamous cell cancers and the XAGE family members in adenocarcinomas. However, there were several exceptions, with single cases scattered within the main groups. Many of CTAs were coexpressed, i.e., one gene expressed together with one gene or several other genes. As expected, this was observed for the known CTA families, like SAGE, MAGE, and XAGE.
Figure 7. Coexpression analysis of cancer testis antigens (CTAs).
Coexpression of the number of CTAs expressed in a non–small-cell lung cancer tumor sample (x axis) and the number of tumor samples (y axis).
Figure 8. Hierarchical cluster analysis of cancer testis antigens (CTAs) in non–small-cell lung cancer (NSCLC).
The 199 NSCLC cases were clustered based on the expression of the 90 CTAs identified in our data set. The genes previously identified as CTAs from the CTdatabase are marked in gray, the newly identified genes are marked in green, and the newly identified genes that are also expressed in neuronal tissue according to the Genotype-Tissue Expression are marked in yellow. X chromosome–bound genes are marked in brown, and non–X chromosomal genes are in white.
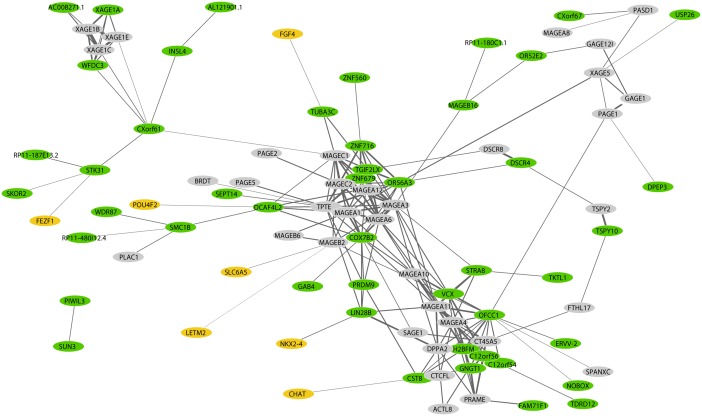
The relationships are also illustrated in the coexpression network, with three main clusters of XAGE, MAGE, and a mixed cluster with uncertain family affiliation (Figure 9). The newly identified CTAs often group together with known CTAs. For example, COX7B2 and ZNF679 are grouped with the MAGE family. Additionally, seemingly unrelated CTAs are concurrently expressed, such as XAGE1E and WFDC3. Noteworthily, some genes did not show strong correlations, including the novel CTAs with brain expression.
Figure 9. Network analysis of cancer testis antigens (CTAs) in non–small-cell lung cancer (NSCLC).
A network analysis illustrating the coexpression pattern of the 90 identified CTAs. The genes previously identified as CTAs from the CTdatabase are marked in gray, the newly identified genes are marked in green, and the newly identified genes that are also expressed in neuronal tissue according to the Genotype-Tissue Expression are marked in yellow.
Association of NSCLC CTAs with survival.
Several studies suggest that CTA expression is associated with poor prognosis in NSCLC (23, 24, 34). Therefore, we explored the prognostic impact of CTA gene expression in our RNAseq cohort of 199 NSCLC patients. The Cox regression model revealed no significant association with survival for any of the 90 NSCLC CTAs, neither in the univariate nor in the multivariate analysis, including the dichotomized clinicopathological parameters of age (≤70 and >70 years of age), performance status (0 and 1–2 according to the WHO score), and pathological tumor-node-metastasis stage (stage I; stage II–IV) (Supplemental Table 7). This was also true when the adenocarcinoma and squamous cell cancer subtypes were analyzed separately.
Since adjustment for multiple testing is maybe too vigorous to detect minor influence in a single cohort, we applied a meta-analysis approach, including 1,117 patients from 7 studies. In total, 68 of the 90 CTAs were presented with at least one probe set on the Affymetrix HG U133 Plus 2.0 Array. After adjustment for multiple testing again, none of the CTAs showed any significant association with survival in the meta-analysis. However, 16 genes (15 different probe sets) were significant without stringent adjustment for multiple testing (Supplemental Table 8). The proportion (15 of 98) of these potential prognostic probe sets is not higher than expected compared with the proportion of significant probe sets in the rest of all Affymetrix HG U133 Plus 2.0 probe sets (9,866 of 54,577 probe sets, Fisher test, P = 0.80). The meta-analysis for adenocarcinoma and squamous cell cancer separately demonstrated similar results, without evidence for a particular prognostic value of NSCLC CTAs. Thus, the hypothesis that CTA expression is associated with poor prognosis is doubtful.
Discussion
Currently, this study provides the most comprehensive mapping of the CTA landscape in NSCLC based on transcriptomic, methylation, and protein data. The basis for this analysis was RNAseq data from 199 NSCLC cases together with 142 cases representing normal tissue from 32 different organ sites. Initially, we evaluated the expression of all CTAs annotated in the CTdatabase (21). We confirmed the expression of 96 established CTAs in NSCLC. However, the CTdatabase does not appear to include the whole spectrum of genes with cancer- and testis-specific expression in NSCLC. Therefore, we took advantage of our data set to search for potential new CTAs and identified 90 genes with specific expression in testis/placenta that were also expressed in at least 2% of adenocarcinoma or squamous cell cancer cases. The majority (n = 55) were previously not described in the CTdatabase, of them 46 genes that have not been previously discussed, even in the context of lung cancer. To demonstrate, that the results based on mRNA expression can be translated to the protein level, we provided in situ protein data for CTA expression in testis and in NSCLC tissue, adding another dimension of evidence. We confirmed that one regulatory mechanism of CTA activation is gene demethylation, implicated in the regulation of approximately one-third of the CTAs. In contrast, another common assumption, that CTA expression is associated with unfavorable prognosis, was not confirmed in our data set or in an independent meta-analysis.
To our knowledge, only one previous study evaluated CTA expression in NSCLC in a comprehensive fashion (35), applying another strategy to identify CTAs. In the first step, Rousseaux et al. used expressed sequence tags and microarray data sets to identify 506 testis- and placenta-restricted genes. These genes were further evaluated in microarray data of 1,776 different tumor tissues and finally in a cohort of 293 NSCLC patients. The authors identified 100 genes to be expressed in lung cancer in at least 1% of the patients. As expected, the list demonstrates an overlap with our CTA list (Supplemental Table 9). However, we identified 48 additional new CTAs, and 84 of their CTAs were not confirmed in our data set. This discrepancy can be explained by different selection criteria. For example, testis/placenta specificity was defined as <3 times standard deviation above the mean of all other tissues in the Rousseaux study, whereas in our study the specificity was defined as 5 times higher expression in one sample than the highest expression value of any other tissue. Clearly, the choice of the cut-off has a strong effect on the number of identified CTA candidates. However, based on our previous studies describing organ-specific gene expression (28, 36, 37), we believe that this study provides CTA candidates with high confidence.
We believe that our study extends this previous study with more detailed information based on RNAseq, protein, methylation, and survival data. Another study (33) applied a more focused approach and utilized the BioGPS database to identify testis-specific genes located on the X chromosome and finally suggested 4 new CTAs (BEX1, NXF3, TCEAL3, and VCX2). In our data set, BEX1, NXF3, and TCEAL3 demonstrate clear expression in somatic tissues and may not represent true CTAs. Only VCX2 showed high testis specificity and is expressed in two adenocarcinoma patients and one squamous carcinoma patient but is not included in our catalog because of the low frequency of expression (<2%).
The identified 90 genes were independently confirmed as true CTAs by manual curation using the GTEx database (29). This validation step lead to the exclusion of 6 genes, mainly due to the inclusion of additional tissue types in GTEx compared with our collection. An additional potential source of discrepancies could be tissues affected by inflammation, leading to a change of tissue composition and consequently the expression profile. Surprisingly, we identified and excluded only one gene (TFPI2) due to effects likely related to inflammation (38, 39). Furthermore, the coexpression analyses demonstrate that newly identified CTAs largely group together with established CTAs, indicating a similar regulatory mechanism and coordinated expression, which is a known feature of CTAs (22, 40, 41). This finding was also supported by the methylation analysis of TCGA data. Indeed, hypomethylation-associated CTA expression was observed for established X chromosomal CTAs and novel non–X chromosomal CTAs in around one-third of our NSCLC CTA list. This is in line with previous studies suggesting that epigenetic regulation is one major regulatory mechanism of CTA activation (35, 42, 43).
Recent evidence suggests that CTAs expression is not a result of uncontrolled cancerous dedifferentiation but that the activation of CTA implies oncogenic function (44). As promising immunotherapeutic targets, the cancer-specific identification of CTAs is of obvious clinical interest. Early strategies for CTA identification were mainly based on immunological methods, like T cell epitope cloning and antibody screening (45). Later strategies used differential gene expression analysis to compare pooled mRNA of normal and cancer tissue by differential display or microarrays (18, 35, 46). Subsequently, the group of genes reported to possess CTA features was considerably expanded, and we show evidence that some of the existing CTAs should most likely be omitted from the database.
It is important to point out that the cancer testis-specific gene expression pattern does not automatically translate genes into immunogenic antigens that are either directly presented on the cancer cell surface or processed as MHC-bound peptides. The fragmented knowledge about immune-inducing mechanisms may also present the largest hindrance to implementing vaccination strategies in cancer therapy (11). This is best exemplified by the results of the recent randomized phase III trial in NSCLC patients receiving adjuvant MAGE-A3 vaccine (47). Although the primary selection of the CTA seems of natural importance, numerous other factors, like type of adjuvant, vaccination schedule, and the patient’s HLA also play major roles in the successful induction of an anticancer response (48–51). These results support that more work is needed to understand the molecular mechanisms that induce anticancer immune response. Strategies to enhance the effect of cancer vaccines include combination with the recently developed checkpoint inhibitor, showing promising results in preclinical and clinical studies (52).
Our study extends previous data on several levels. First, earlier studies applied complementary techniques, including quantitative real-time PCR or oligonucleotide arrays, while we provide global genome-wide quantitative data based on next-generation sequencing. Second, several previous studies have been predominantly based on gene expression in cell lines that are difficult to extrapolate to human cancer tissue (53, 54). Our study is based on a large, representative NSCLC patient cohort, resulting in mRNA levels and frequencies from the in situ environment of cancer. Furthermore, the analysis of the expression of CTAs in normal tissues was here performed with samples from the same biobank; RNA preparation was performed under the same conditions and technical analysis was performed on the same platform at the same facility. Furthermore, the characterization of CTAs has previously mostly been based only on mRNA analysis. Here, we supplement the RNA data with protein images from human NSCLC tissue and testis, and, based on methylation data, we suggest regulatory mechanisms for each single CTA. Thus, we believe that our publically available data can contribute to the ambitious efforts of the CTdatabase by extending the existing catalog of CTAs and by curating current information.
As an unexpected finding, we were unable to confirm the previous assumptions that CTAs are valuable biomarkers for cancer prognostication and CTA expression indicates higher malignant potential (24, 35, 55). Instead, we demonstrated, based on our data set and 7 independent data sets, that CTAs did not possess more prognostic information than other genes. This unexpected result stresses the need for appropriate statistics and independent validation data sets in the evaluation process of prognostic markers (56–58).
In summary, our study provides RNAseq expression profiles of the whole CTA repertoire in NSCLC, including an analysis of previously proposed CTAs, as well as the identification of novel CTAs. The detailed catalog can guide biomarker studies and also help researchers to select candidates for focused experimental or clinical exploration.
Methods
Study cohort and patient characteristics.
Patient material used for transcript profiling (RNAseq) consisted of fresh-frozen tumor tissue from 199 patients diagnosed with NSCLC and surgically treated from 2006 to 2010 at the Uppsala University Hospital (Table 1). The original cases were reevaluated by two lung pathologists (H. Brunnström and P. Micke) in accordance to the WHO classification from 2004 (59) and the new proposed adenocarcinoma classification (60). Thirty-five of the 199 NSCLC patients analyzed with RNAseq were used to construct a TMA for the immunohistochemical analysis (Supplemental Table 10).
Transcript profiling (RNAseq).
Freshly frozen tumor tissues were embedded in Optimal Cutting Temperature compound and stored at –80°C. For RNA extraction, the tissue was cut in sections (10 μM) using a cryostat (Leica). One section was H&E stained and used to decide whether the tumor cell content was sufficient to be included in the analysis. Subsequently, 5 sections (10 μm) were cut and used for RNA extraction. The 5 sections were transferred to RLT buffer using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Afterwards one additional section was H&E stained for eventual tumor cell content correction. Only tissues with more than 10% tumor content were included in the analysis. Extracted RNA samples were analyzed by the Agilent 2100 Bioanalyzer system (Agilent Biotechnologies) with the RNA 6000 Nano Labchip Kit. For the following mRNA sample preparation for sequencing, the vast majority (188 of 199) of the samples were of high-quality RNA, with a RNA integrity number (RIN) ≥7.5. In addition, 11 samples had RIN values ranging from 2.6 to 7.4. Since these samples passed the internal quality control test and did not show any deviation from the other samples in the multidimensional scaling analysis, they were included in the further analysis. Samples were prepared for sequencing using the Illumina TruSeq RNA Sample Prep Kit v2, using polyA selection. The sequencing was performed multiplexed with 5 samples per lane on Illumina HiSeq2500 machines (Illumina) using the standard Illumina RNAseq protocol with a read length of 2 × 100 bases. The raw data has been uploaded together with clinical information on GEO, with the accession number GSE81089 (http://www.ncbi.nlm.nih.gov/geo/).
RNAseq data analysis.
The raw sequencing data were mapped to the human reference genome (GRCh37) and the Ensembl version 73 gene annotation using TopHat version 2.0.8b (61, 62). Gene fragments per kilobase of transcript per million mapped reads values were calculated from the generated alignments using Cufflinks version 2.1.1. Raw read counts were calculated using featureCounts from the Subread package version 1.4.0-p1 (63). RNAseq data for 32 normal tissues from 123 individuals altogether was previously reported and analyzed using the same methodological pipeline as that for the NSCLC data. These samples included testis (n = 7), thyroid gland (n = 4), placenta (n = 4), cerebral cortex (n = 3), liver (n = 3), gallbladder (n = 3), pancreas (n = 2), salivary gland (n = 3), esophagus (n = 3), stomach (n = 3), duodenum (n = 2), small intestine (n = 4), appendix (n = 3), colon (n = 7), rectum (n = 4), kidney (n = 4), urinary bladder (n = 2), prostate (n = 4), endometrium (n = 5), fallopian tube (n = 5), ovary (n = 3), adipose tissue (n = 5), skin (n = 3), bone marrow (n = 4), lymph node (n = 5), tonsil (n = 5), spleen (n = 4), adrenal gland (n = 3), lung (n = 5), heart muscle (n = 4), skeletal muscle (n = 5), and smooth muscle (n = 2). In addition to the tumor tissue samples from NSCLC, 19 paired normal lung tissues were also sampled and analyzed, yielding 142 individual normal tissue samples altogether.
Differential expression analysis was performed on read counts from featureCounts using DESeq with Benjamini-Hochberg correction using false discovery rate (FDR) of P values. Potential CTAs were determined by requiring that at least 2% of adenocarcinoma or squamous cell carcinoma samples, i.e., two adenocarcinoma samples or squamous cell carcinoma sample, expressed the gene at least 5 times higher than any normal tissue sample, excluding testis and placenta.
TMA production and IHC.
Representative formalin-fixed, paraffin-embedded (FFPE) material from donor blocks was punched (1 mm in diameter) using a manual tissue arrayer (MTA-1, Beecher Instruments) and placed in a recipient block, generating a TMA block containing 35 NSCLC cases represented in duplicates. Four-μm sections of the TMA blocks were cut using a microtome (HM 355S, Microm), mounted on adhesive slides (SuperFrost Plus, Thermo Scientific), and baked for 45 minutes at 60°C (64). Deparaffinization and hydration was performed in xylene and graded alcohols to distilled water prior to the IHC staining. Endogenous peroxidase was blocked using 0.3% hydrogen peroxide in 95% ethanol for 5 minutes. For antigen retrieval, a pressure boiler (Decloaking Chamber, Biocare Medical) was used, and the slides were boiled for 4 minutes at 125°C in citrate buffer, pH 6 (Lab Vision). Automated IHC was performed as previously described using an Autostainer 480 instrument (Thermo Fisher Scientific) on TMAs consisting of FFPE material from 35 of the NSCLC patients analyzed with RNAseq (Supplemental Table 10). Primary antibodies diluted in UltraAb Diluent (Lab Vision) and the secondary reagent UltraVision LP HRP polymer (Lab Vision) were incubated for 30 minutes each at room temperature. Following the washing steps, the slides were developed for 10 minutes at room temperature, adding diaminobenzidine (Lab Vision) as chromogen and thereafter counterstaining with Mayer’s hematoxylin (Histolab) and mounting with Pertex (Histolab). The IHC-stained slides were scanned at ×20 magnification using an Aperio ScanScope XT Slide Scanner (Aperio Technologies) to obtain high-resolution digital images.
Antibodies.
Antibodies were selected based on images and data available from the HPA database (http://www.proteinatlas.org/). The HPA database contains gene expression data corresponding to 46 normal tissue types, of which 32 tissue types also have been analyzed using RNAseq. Antibodies were only chosen for which the staining pattern in normal tissue was in accordance with corresponding RNAseq values of the same tissue, i.e., selected antibodies had revealed testis/placenta specificity and were in concordance with literature regarding subcellular localization. Antibodies against novel CTAs were required to show weak positivity in more than 1% of cancer cells in at least one of the 35 NSCLC specimens included in the TMA (Supplemental Table 10). The IDs of all antibodies used in the IHC analysis are summarized here: MAGEC2 (HPA062230), MAGEB6 (HPA041853), PAGE2/5/2B (HPA052619), CT45A2 (HPA046872), SAGE1 (HPA003208), MAGEA8 (HPA003998), ACRBP (HPA039082), CALR3 (AF2927, R&D Systems), HORMAD1 (HPA037850), LUZP4 (HPA046436), PRM1 (HPA055150), DKKL1 (HPA047174), HEMGN (HPA019572), SPA17 (HPA037568), SPAG6 (HPA038440), SPAG8 (HPA068012), ANKRD45 (HPA031657), KDM5B (AMAb90860, Atlas Antibodies), TKTL-1 (T001, R-Biopharm AG), TGIF2LX (HPA034543), VCX (HPA049357), and CXORF67 (HPA061280). All antibodies are from the HPA project, unless otherwise stated. For detailed information, see the HPA database (http://www.proteinatlas.org/) (28).
Methylation status.
To determine the association of gene expression of each CTA to the corresponding methylation status we utilized TCGA data. Expression data from RNAseq and methylation data from the Illumina 450K methylation array were obtained from TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). For comparison of methylation status between tumor and normal lung, methylation data of 475 adenocarcinomas and 32 normal lung tissues as well as 370 squamous cell carcinoma and 42 normal lungs were evaluated with probes for the TSS200. If there were no probes for TSS200 available, the average of probes mapping the first exon was used. If these probes were also absent, we used the average of probes covering TSS1500 (1,500 bp upstream of the transcription start site). Gene expression was considered to be associated with methylation if the Spearman correlation coefficient was less than –0.4, demonstrating inverse correlation between methylation status and gene expression. To analyze the effect of DNA methylation in nonmalignant lung cell lines, we used publically available gene expression data sets in which the human bronchial epithelial cells and human small airway epithelial cells were treated with AZA (GSE18454) (65). Mean β values of each probe were calculated in both tumor and normal lungs, and Δβ values (normal – tumor) > 0.1 were defined as hypomethylated. For correlation analyses, 439 adenocarcinomas and 370 squamous cell carcinomas with matched RNAseq and methylation data were used. The Spearman correlation coefficient was calculated using the β value of each probe and is shown as log2 transformed 1+ reads per kilobase of transcript per million mapped reads values (Figure 6). In accordance with the algorithm presented by Jiao et al. (66) for each gene, the average β value of probes for the TSS200 was calculated.
Survival analysis and meta-analysis of public data sets.
A Cox proportional hazards model, as described by Klein and Moeschberger (67) and implemented in the R package “survival” (68), was used to determine the association between CTA mRNA expression and overall survival. Genes having a total sum of less than 10 raw counts across all patients were excluded from this analysis. After normalization using DESeq (69), the mRNA expression data were logarithmized and standardized as proposed by Zwiener et al. (70) before fitting a Cox proportional hazards model to the 90 CTAs. Multiple testing adjustments of significance levels were performed using the FDR with Benjamini-Hochberg correction of P values (71). To validate significant survival associations in independent patient cohorts, the R package “meta” (72) was applied to perform a meta-analysis across 7 publicly available NSCLC data sets with Affymetrix HG U133 Plus 2.0 Array expression data and corresponding information on overall survival (in total 1,117 patients): GSE29013 (73); GSE30219 (35); GSE31210 (74); GSE19188 (75); GSE3141 (76); GSE50081 (77); and GSE37745 (27). All data sets were downloaded from the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/). The raw data were normalized using frozen robust multiarray analysis (78), apart from GSE3141, for which only MAS-normalized data were available. Normal (nontumoral) samples and small cell carcinomas were removed. All data sets were checked for duplicates so that patients across all data sets were independent. In GSE37745, two different values of clinical data for one patient were both removed.
Meta-analysis was performed with random effects models based on the parameter estimates of log hazard ratios of the univariate Cox survival models and their standard errors. Inverse variance weighting was used to combine the single estimates into a pooled estimate. Significance of the overall effect was assessed by the P value of the random effects model. All analyses were performed using R version 3.2.1.
Data availability.
The RNAseq data of normal tissue are available at http://www.proteinatlas.org/about/publicationdata The raw sequencing data for normal tissue are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2836/). The raw sequencing data for the NSCLC samples and additional normal lung cancer samples were deposited at http://www.ncbi.nlm.nih.gov/geo/, with the accession number GSE81089
Statistics.
DESeq with Benjamini-Hochberg correction (FDR) of P values was used for differential expression analysis. A P value less than 0.05 was considered significant.
To determine the methylation status of CTAs in NSCLC (Supplemental Table 6) and to determine the correlation between CTA methylation and gene expression in NSCLC (Supplemental Figure 1 and Supplemental Table 6), Fisher’s exact test was used. A P value less than 0.01 was considered significant.
To test the association of NSCLC CTAs with survival in the Cox proportional hazards model, the P values of the Wald test were used, and a P value of 0.05 was considered significant.
Study approval.
The study was approved by the Uppsala Ethical Review Board (reference 2012/532) and did not require written informed consent from each patient.
Author contributions
DD collected data and performed IHC, data analysis, and manuscript preparation. BMH performed bioinformatics, data analysis, and manuscript preparation. MH performed data analysis and manuscript preparation. JSMM performed compilation of the cohort, sample annotation, data collection, and sample preparation. LLF performed sample annotation and sample preparation. LF performed bioinformatics and data analysis. HB performed sample annotation. CL performed IHC and manuscript preparation. KM performed data analysis and manuscript preparation. JR performed data analysis and manuscript preparation. SE performed sample annotation. ES performed sample annotation. HK performed compilation of the cohort and sample annotation. EB performed compilation of the cohort and sample annotation. KE performed compilation of the cohort, data interpretation, and manuscript preparation. JGH performed data interpretation and manuscript preparation. ML performed compilation of the cohort and sample annotation. AS performed data analysis and manuscript preparation. JB performed compilation of the cohort, sample annotation, and data interpretation. FP performed data collection, data analysis, IHC, and data interpretation. PM and MU performed study design, data collection, data analysis, data interpretation, and manuscript preparation.
Supplementary Material
Acknowledgments
We acknowledge the HPA team; support from Science for Life Laboratory, the National Genomics Infrastructure (NGI); and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure. We thank Clinical Pathology at the Uppsala University Hospital and Simin Tahmasebpoor for assistance with tissue samples and for assistance with sample preparations. This work was supported by the Swedish Cancer Society Cancerfonden (2010/871, 2012/738 to P.M and 2012/598 to F.P), Lions Cancer Foundation Uppsala (to P.M) the Knut and Alice Wallenberg Foundation (2008.0143 to F.P and M.U) and the Erik, Karin and Gösta Selanders Foundation, Sweden (to P.M).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(10):e86837. doi:10.1172/jci.insight.86837.
Contributor Information
Dijana Djureinovic, Email: dijana.djureinovic@igp.uu.se.
Masafumi Horie, Email: mhorie25@gmail.com.
Linnea La Fleur, Email: linnea.la_fleur@igp.uu.se.
Linn Fagerberg, Email: linn.fagerberg@scilifelab.se.
Hans Brunnström, Email: hans.brunnstrom@med.lu.se.
Cecilia Lindskog, Email: cecilia.lindskog@igp.uu.se.
Katrin Madjar, Email: madjar@statistik.tu-dortmund.de.
Jörg Rahnenführer, Email: rahnenfuehrer@statistik.tu-dortmund.de.
Simon Ekman, Email: simon.ekman@igp.uu.se.
Elisabeth Ståhle, Email: Elisabeth.Stahle@akademiska.se.
Hirsh Koyi, Email: hirsh.koyi@regiongavleborg.se.
Eva Brandén, Email: eva.branden@regiongavleborg.se.
Karolina Edlund, Email: edlund@ifado.de.
Mats Lambe, Email: Mats.Lambe@ki.se.
Akira Saito, Email: akirasaito0314@gmail.com.
Johan Botling, Email: johan.botling@igp.uu.se.
Fredrik Pontén, Email: fredrik.ponten@igp.uu.se.
Patrick Micke, Email: patrick.micke@igp.uu.se.
References
- 1.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical Lung Cancer Genome Project (CLCGP), Network Genomic Medicine (NGM) A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5(209):e86837. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurent-Puig P, Lievre A, Blons H. Mutations and response to epidermal growth factor receptor inhibitors. Clin Cancer Res. 2009;15(4):1133–1139. doi: 10.1158/1078-0432.CCR-08-0905. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest. 2015;125(9):3335–3337. doi: 10.1172/JCI83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg SA, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17(13):4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125(9):3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anagnostou VK, Brahmer JR. Cancer immunotherapy: a future paradigm shift in the treatment of non-small cell lung cancer. Clin Cancer Res. 2015;21(5):976–984. doi: 10.1158/1078-0432.CCR-14-1187. [DOI] [PubMed] [Google Scholar]
- 10.Madureira P, de Mello RA, de Vasconcelos A, Zhang Y. Immunotherapy for lung cancer: for whom the bell tolls? Tumour Biol. 2015;36(3):1411–1422. doi: 10.1007/s13277-015-3285-6. [DOI] [PubMed] [Google Scholar]
- 11.Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH. Therapeutic cancer vaccines. J Clin Invest. 2015;125(9):3401–3412. doi: 10.1172/JCI80009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen CJ, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015;125(10):3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 15.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100(11):2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 17.Knuth A, Danowski B, Oettgen HF, Old LJ. T-cell-mediated cytotoxicity against autologous malignant melanoma: analysis with interleukin 2-dependent T-cell cultures. Proc Natl Acad Sci U S A. 1984;81(11):3511–3515. doi: 10.1073/pnas.81.11.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin U, et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A. 1995;92(25):11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt JS. Stranger in a strange land. Immunol Rev. 2006;213:36–47. doi: 10.1111/j.1600-065X.2006.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janitz M, et al. Analysis of mRNA for class I HLA on human gametogenic cells. Mol Reprod Dev. 1994;38(2):231–237. doi: 10.1002/mrd.1080380215. [DOI] [PubMed] [Google Scholar]
- 21.Almeida LG, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37(Database issue):D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grunwald C, et al. Expression of multiple epigenetically regulated cancer/germline genes in nonsmall cell lung cancer. Int J Cancer. 2006;118(10):2522–2528. doi: 10.1002/ijc.21669. [DOI] [PubMed] [Google Scholar]
- 23.Gure AO, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11(22):8055–8062. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 24.Shigematsu Y, et al. Clinical significance of cancer/testis antigens expression in patients with non-small cell lung cancer. Lung Cancer. 2010;68(1):105–110. doi: 10.1016/j.lungcan.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Tajima K, et al. Expression of cancer/testis (CT) antigens in lung cancer. Lung Cancer. 2003;42(1):23–33. doi: 10.1016/S0169-5002(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botling J, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19(1):194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 28.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):e86837. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carson MJ, Doose JM, Melchior B, Schmid CD, Ploix CC. CNS immune privilege: hiding in plain sight. Immunol Rev. 2006;213:48–65. doi: 10.1111/j.1600-065X.2006.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritz P, Coy JF, Mürdter TE, Ott G, Alscher MD, Friedel G. TKTL-1 expression in lung cancer. Pathol Res Pract. 2012;208(4):203–209. doi: 10.1016/j.prp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Ousati Ashtiani Z, et al. Association of TGIFLX/Y mRNA expression with prostate cancer. Med Oncol. 2009;26(1):73–77. doi: 10.1007/s12032-008-9086-7. [DOI] [PubMed] [Google Scholar]
- 33.Taguchi A, et al. A search for novel cancer/testis antigens in lung cancer identifies VCX/Y genes, expanding the repertoire of potential immunotherapeutic targets. Cancer Res. 2014;74(17):4694–4705. doi: 10.1158/0008-5472.CAN-13-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melloni G, et al. Prognostic significance of cancer-testis gene expression in resected non-small cell lung cancer patients. Oncol Rep. 2004;12(1):145–151. [PubMed] [Google Scholar]
- 35.Rousseaux S, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5(186):e86837. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djureinovic D, et al. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod. 2014;20(6):476–488. doi: 10.1093/molehr/gau018. [DOI] [PubMed] [Google Scholar]
- 37.Lindskog C, et al. The lung-specific proteome defined by integration of transcriptomics and antibody-based profiling. FASEB J. 2014;28(12):5184–5196. doi: 10.1096/fj.14-254862. [DOI] [PubMed] [Google Scholar]
- 38.Pou J, et al. Tissue factor pathway inhibitor 2 is induced by thrombin in human macrophages. Biochim Biophys Acta. 2011;1813(6):1254–1260. doi: 10.1016/j.bbamcr.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt J, Weijdegård B, Mikkelsen AL, Lindenberg S, Nilsson L, Brännström M. Differential expression of inflammation-related genes in the ovarian stroma and granulosa cells of PCOS women. Mol Hum Reprod. 2014;20(1):49–58. doi: 10.1093/molehr/gat051. [DOI] [PubMed] [Google Scholar]
- 40.Karanikas V, et al. Co-expression patterns of tumor-associated antigen genes by non-small cell lung carcinomas: implications for immunotherapy. Cancer Biol Ther. 2008;7(3):345–352. doi: 10.4161/cbt.7.3.5424. [DOI] [PubMed] [Google Scholar]
- 41.Lin C, et al. Cancer/testis antigen CSAGE is concurrently expressed with MAGE in chondrosarcoma. Gene. 2002;285(1–2):269–278. doi: 10.1016/s0378-1119(02)00395-5. [DOI] [PubMed] [Google Scholar]
- 42.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A. 1996;93(14):7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim R, Kulkarni P, Hannenhalli S. Derepression of cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation. BMC Cancer. 2013;13:e86837. doi: 10.1186/1471-2407-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gjerstorff MF, Andersen MH, Ditzel HJ. Oncogenic cancer/testis antigens: prime candidates for immunotherapy. Oncotarget. 2015;6(18):15772–15787. doi: 10.18632/oncotarget.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Bruggen P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 46.Scanlan MJ, et al. Identification of cancer/testis genes by database mining and mRNA expression analysis. Int J Cancer. 2002;98(4):485–492. doi: 10.1002/ijc.10276. [DOI] [PubMed] [Google Scholar]
- 47.Vansteenkiste JF, et al. MAGRIT, a double-blind, randomized, placebo-controlled phase iii study to assess the efficacy of the recmage-A3 + AS15 cancer immunotherapeutic as adjuvant therapy in patients with resected mage-A3-positive non-small cell lung cancer (NSCLC) Ann Oncol. 2014;25(suppl 4):e86837. doi: 10.1016/S1470-2045(16)00099-1. [DOI] [PubMed] [Google Scholar]
- 48.Dutoit V, et al. Multiepitope CD8(+) T cell response to a NY-ESO-1 peptide vaccine results in imprecise tumor targeting. J Clin Invest. 2002;110(12):1813–1822. doi: 10.1172/JCI16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kissick HT, Sanda MG. The role of active vaccination in cancer immunotherapy: lessons from clinical trials. Curr Opin Immunol. 2015;35:15–22. doi: 10.1016/j.coi.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Nardelli-Haefliger D, Dudda JC, Romero P. Vaccination route matters for mucosal tumors. Sci Transl Med. 2013;5(172):e86837. doi: 10.1126/scitranslmed.3005638. [DOI] [PubMed] [Google Scholar]
- 51.Sasada T, Yamada A, Noguchi M, Itoh K. Personalized peptide vaccine for treatment of advanced cancer. Curr Med Chem. 2014;21(21):2332–2345. doi: 10.2174/0929867321666140205132936. [DOI] [PubMed] [Google Scholar]
- 52.Morse MA, Lyerly HK. Checkpoint blockade in combination with cancer vaccines. Vaccine. 2015;33(51):7377–7385. doi: 10.1016/j.vaccine.2015.10.057. [DOI] [PubMed] [Google Scholar]
- 53.Chen YT, et al. Identification of cancer/testis-antigen genes by massively parallel signature sequencing. Proc Natl Acad Sci U S A. 2005;102(22):7940–7945. doi: 10.1073/pnas.0502583102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugita M, et al. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62(14):3971–3979. [PubMed] [Google Scholar]
- 55.John T, et al. The role of cancer-testis antigens as predictive and prognostic markers in non-small cell lung cancer. PLoS ONE. 2013;8(7):e86837. doi: 10.1371/journal.pone.0067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindskog C, Edlund K, Mattsson JS, Micke P. Immunohistochemistry-based prognostic biomarkers in NSCLC: novel findings on the road to clinical use? Expert Rev Mol Diagn. 2015;15(4):471–490. doi: 10.1586/14737159.2015.1002772. [DOI] [PubMed] [Google Scholar]
- 57.Simon RM1, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102(7):464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Travis W, Brambilla E, Muller-Hermelink HK, Harris CC. Pathology And Genetics: Tumors Of The Lung, Pleura, Thymus And Heart. Vol. 1. Lyon, France: IARC; 2004. [Google Scholar]
- 60.Travis WD, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8(5):381–385. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 61.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40(Database issue):D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 64.Kampf C, Olsson I, Ryberg U, Sjostedt E, Ponten F. Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J Vis Exp. 2012;63 doi: 10.3791/3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glazer CA, et al. Integrative discovery of epigenetically derepressed cancer testis antigens in NSCLC. PLoS ONE. 2009;4(12):e86837. doi: 10.1371/journal.pone.0008189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiao Y, Widschwendter M, Teschendorff AE. A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics. 2014;30(16):2360–2366. doi: 10.1093/bioinformatics/btu316. [DOI] [PubMed] [Google Scholar]
- 67.Klein JP, Moeschberger ML. New York, New York, USA: Springer; 2003. Survival Analysis: Techniques For Censored And Truncated Data. 2nd ed. [Google Scholar]
- 68. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, New York, USA: Springer; 2000. [Google Scholar]
- 69.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):e86837. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zwiener I, Frisch B, Binder H. Transforming RNA-Seq data to improve the performance of prognostic gene signatures. PLoS ONE. 2014;9(1):e86837. doi: 10.1371/journal.pone.0085150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benjamini Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 72. Schwarzer G. Meta: general package for meta-analysis. Version: 4.3-0. 2015. [Google Scholar]
- 73.Xie Y, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res. 2011;17(17):5705–5714. doi: 10.1158/1078-0432.CCR-11-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okayama H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 75.Hou J, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5(4):e86837. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 77.Der SD, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9(1):59–64. doi: 10.1097/JTO.0000000000000042. [DOI] [PubMed] [Google Scholar]
- 78.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11(2):242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNAseq data of normal tissue are available at http://www.proteinatlas.org/about/publicationdata The raw sequencing data for normal tissue are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2836/). The raw sequencing data for the NSCLC samples and additional normal lung cancer samples were deposited at http://www.ncbi.nlm.nih.gov/geo/, with the accession number GSE81089