Abstract
Single-molecule microscopy and stopped-flow kinetics assays were carried out to understand the microtubule polymerase activity of kinesin-5 (Eg5). Four lines of evidence argue that the motor primarily resides in a two-heads-bound (2HB) state. First, upon microtubule binding, dimeric Eg5 releases both bound ADPs. Second, microtubule dissociation in saturating ADP is 20-fold slower for the dimer than for the monomer. Third, ATP-triggered mant-ADP release is 5-fold faster than the stepping rate. Fourth, ATP binding is relatively fast when the motor is locked in a 2HB state. Shortening the neck-linker does not facilitate rear-head detachment, suggesting a minimal role for rear-head-gating. This 2HB state may enable Eg5 to stabilize incoming tubulin at the growing microtubule plus-end. The finding that slowly hydrolyzable ATP analogs trigger slower nucleotide release than ATP suggests that ATP hydrolysis in the bound head precedes stepping by the tethered head, leading to a mechanochemical cycle in which processivity is determined by the race between unbinding of the bound head and attachment of the tethered head.
Keywords: cytoskeleton, kinesin, microtubule, mitotic spindle, molecular motor, single-molecule biophysics
Introduction
In dividing cells, tetrameric kinesin-5 (Eg5)2 motors slide apart antiparallel microtubules and thereby generate outward pushing forces to stabilize the mitotic spindle (1–3). Due to its crucial mitotic function, Eg5 has been a target for development of anti-cancer drugs, and there are a number of established Eg5 inhibitors (1). In addition to its ability to generate outward pushing forces, Eg5 has been shown to pause at microtubule plus-ends and enhance microtubule polymerization by accelerating growth and slowing catastrophe (3–5). By allowing sustained force generation at the end of microtubule tracks, this end binding property may maximize the ability of tetrameric Eg5 to push apart antiparallel microtubules. The polymerase activity potentially enhances the antiparallel sliding activity by generating more track for the motor to walk along, and it may be important for balancing microtubule depolymerase activities at poles that lead to poleward microtubule flux (6, 7). Despite considerable study, the specific features of the kinesin-5 chemomechanical cycle that underlie the end binding and polymerase activity of this motor are not understood.
Eg5 is minimally processive in vitro, a feature that has been accounted for by the fact that these motors normally operate in large teams (2, 3). Shortening the 18-residue Eg5 neck linker domain to the 14-residue length of kinesin-1 resulted in Eg5 having similar run lengths to kinesin-1 (8), suggesting that the shorter wild-type run lengths result from lack of interhead coordination rather than the specific biochemistry inherent to the Eg5 core catalytic domain. However, beyond the neck linker there are important differences between Eg5 and transport motors such as kinesin-1. Eg5 is roughly 10-fold slower than kinesin-1, although how this speed relates to the motor's processivity and force-generating ability is unclear. From CryoEM reconstructions, the Eg5 neck linker domain adopts a rearward-facing orientation in the ADP state, in contrast to the more disordered kinesin-1 (9, 10), although recent structural kinetics data identifies discrete neck linker positions in both motor classes (11). Eg5 has an extended Loop 5, which has been shown to affect nucleotide binding and serve as a latch for structural coupling within the Switch I domain (12, 13) and as a binding target of several potential drugs (14–16). Finally, it has been proposed that upon initial collision with a microtubule there is an ∼1-s structural transition before the motor begins walking (17); however, the relationship of this possible initial pause to the observed plus-end pausing (5) is unclear.
Interestingly, despite its apparent lack of processivity, Eg5 is able to generate and withstand relatively large mechanical loads. For instance, a 4-piconewton assisting load that reduced kinesin-1 run lengths by roughly 10-fold only reduced kinesin-5 run length by a factor of 2 (18, 19), and in mixed motor gliding assays kinesin-5 motors were able to efficiently slow the transport of microtubules driven by faster transport motors (20). This ability to resist loads is consistent with the proposed role of kinesin-5 as a “brake” that stabilizes microtubule bundles in axons; motor inhibition leads to faster axonal outgrowth and longer branches in culture (7). Maintaining spindle integrity also requires sustained force generation, so this property also aligns with the motor's mitotic function.
By observing dynamic microtubules in the presence of an engineered Eg5 dimer, we recently showed that Eg5 doubles the microtubule growth rate and reduces catastrophe by a factor of three in vitro (5). The motor also pauses for ∼7 s at the plus-end of both taxol-stabilized and dynamic microtubules, a property that is assumed to relate to its polymerase activity.
The goal of the present study was to identify features of the Eg5 chemomechanical cycle that underlie its end binding and polymerase activity. Our approach was to measure the microtubule and nucleotide binding kinetics of dimeric and monomeric Eg5 on the microtubule lattice and to integrate these rates into a model for the dimeric Eg5 hydrolysis cycle. The results support a model in which the motor resides primarily in a 2HB state. The kinesin-5 chemomechanical cycle developed here highlights unique features of kinesin-5 mechanochemistry that may underlie its polymerase activity.
Results
The goal of this study was to uncover specific features of the kinesin-5 chemomechanical cycle that underlie its ability to pause at microtubule plus-ends and enhance microtubule polymerization (5). Achieving this goal requires first characterizing the chemomechanical cycle of kinesin-5 as it steps along the microtubule lattice, and this characterization in turn requires understanding family-specific properties of the individual heads. Therefore, we began our study by investigating the ATPase and microtubule binding properties of a monomeric kinesin-5 construct, Eg5M, which consists of the first 367 amino acids of Xenopus Eg5 and contains the head and entire 18-residue neck linker domain. This construct is similar to the human Eg5367 used by other groups, except Eg5M contains seven more amino acids in the neck linker (21, 22) that are not present in human Eg5367. Work was then extended to an Eg5 dimer construct, consisting of the Xenopus Eg5 head and neck linker up to residue 367, dimerized through the coiled-coil (residues 345–405) of Drosophila kinesin-1. This dimeric construct was described previously (8) and was shown to be a good model for a dimeric form of tetrameric kinesin-5. Throughout the text Eg5M refers specifically to the monomeric species, and Eg5 refers to this dimeric species. All experiments were carried out in BRB80 buffer at 23 °C.
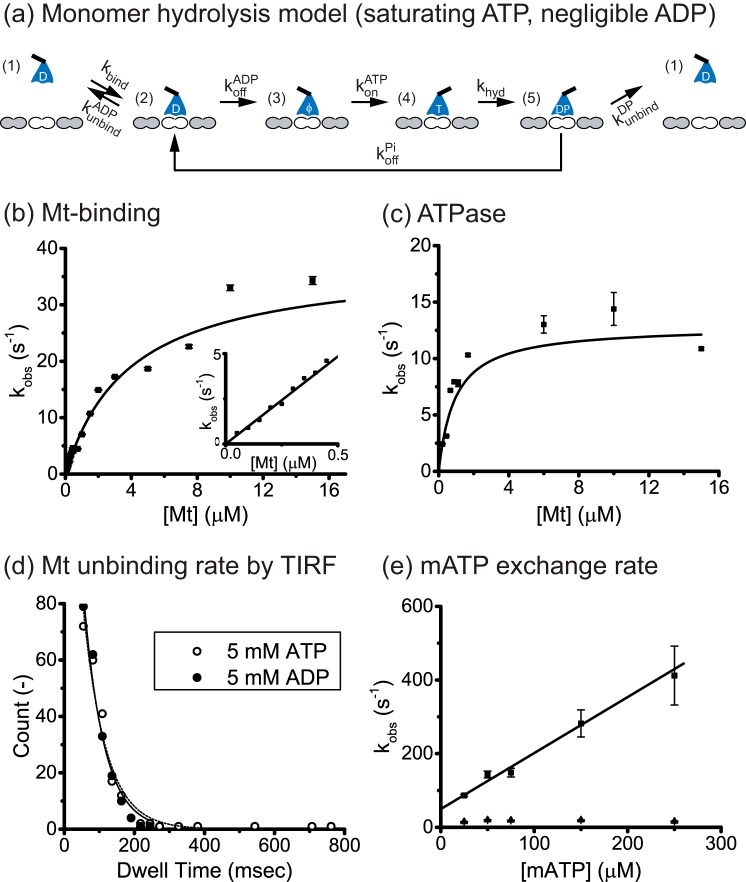
To interpret our Eg5M experiments, we used the monomer hydrolysis cycle shown in Fig. 1a (21–23). The cycle starts from free motors in solution with a bound ADP (state 1) that encounter the microtubule in the ADP state (state 2) and then release ADP to generate a strongly bound apo state (state 3) (24). ATP then binds, generating another strongly bound state (state 4) and is hydrolyzed to ADP-Pi (state 5) (25). If the motor detaches in the ADP-Pi state, then each Mt encounter the motor will hydrolyze only 1 ATP molecule. Alternatively, if the motor releases its phosphate and returns to the weak binding ADP-state (state 2), this is defined as a “futile cycle”; the number of futile ATP hydrolysis cycles per Mt encounter is determined by the race between the Mt-unbinding rate (state 2 to state 1) and the ADP off-rate (state 2 to state 3).
FIGURE 1.
Eg5 monomer kinetics. a, monomer hydrolysis model. Monomeric motors in solution (state 1) bind reversibly to the Mt (state 2), and release their bound ADP to generate a strongly bound state (state 3). In sufficient ATP and negligible ADP, ATP binds (state 4) and is hydrolyzed (state 5). If the motor detaches in the ADP-Pi state, 1 ATP is hydrolyzed per Mt encounter. Alternatively, if the ADP-Pi state is strongly bound and/or Pi release is fast, then hydrolysis and detachment are not tightly coupled and futile hydrolysis cycles may occur (return to state 2). The number of futile cycle is determined in state 2 by the race between the ADP off-rate, koffADP, and the Mt-unbinding rate, kunbindADP. b, microtubule-stimulated mADP release. Eg5M incubated in mADP was flushed against varying Mt concentrations with 2 mm ATP added to prevent mADP rebinding. Transients (average of n = 5–7) were fit with a single exponential, and observed rates were plotted as a function of Mt concentration. Fitting to a hyperbola generated a maximal mADP off-rate of koffmADP = 37.8 ± 5.3 s−1 and half-max Mt concentration of K0.5Mt = 3.9 ± 0.7 μm Mt. Inset, blow up of the linear range of the curve with fit of effective bimolecular on-rate, kbiADP = 9.7 ± 0.2 μm−1 s−1. c, Mt-stimulated ATPase activity of Eg5M. From the hyperbolic fit (n = 4–5 determinations per point), kcat = 12.9 ± 1.9 s−1 and K0.5Mt = 0.93 ± 0.25 μm Mt. The calculated kbiATPase = kcat/K0.5Mt = 13.9 ± 4.3 μm−1 s−1, results in kbiratio = kbiATPase/kbiADP = 1.4 ± 0.4 ATP molecules hydrolyzed per Mt encounter. d, Mt-unbinding rates of QD655-conjugated Eg5M in saturating ATP (open circle and dashed curve) or ADP (solid circle and curve). Single-molecule dwell-time distributions were measured from movies taken at 40 frames/s, and binned data (excluding first 25-ms wide bin) were fit by single exponentials. Calculated monomer unbinding rates were kunbindATP = 15.6 ± 1.5 s−1 and kunbindADP = 17.0 ± 2.0 s−1. e, mATP exchange rate of the Eg5M-Mt complex. Fluorescence transients (average of n = 16–18) were fit with a bi-exponential and two rate constants plotted at varying mATP concentrations. Fitting the fast phase by linear regression yielded a slope corresponding to the on-rate of mATP to the motor-Mt complex, konmATP = 1.5 ± 0.2 μm−1 s−1. The slow phase had a rate in the range of 15 - 20 s−1, which is similar to the ATP turnover rate and was interpreted as hydrolysis after mATP binding rather than rapid unbinding. Both phases had similar amplitudes, and the amplitudes did not vary with mATP concentration.
Kinesin-5 Monomer Does Not Display Chemical Processivity
To quantify the Mt unbinding and nucleotide exchange rates of the monomer, the first question we addressed is whether the ATPase cycle is tightly coupled to detachment from the microtubule. In tightly coupled kinesin dimers like kinesin-1, each head hydrolyzes one ATP per attachment/detachment cycle (26); however, in some monomer species the hydrolysis and detachment cycles are uncoupled, resulting in futile hydrolysis cycles and multiple ATP hydrolyzed per encounter with the microtubule (27). The “chemical processivity,” defined as the number ATP molecules hydrolyzed per Mt encounter, is described by the parameter kbiratio, which is a ratio of the calculated bimolecular collision rate from the ATPase (kbiATPase) divided by the measured bimolecular collision rate from mADP off-rate experiments (kbiADP). To determine kbiADP, motors with bound mADP were flushed against varying Mt concentrations (Fig. 1b). Microtubule binding triggers mADP release, which results in a drop in fluorescence, and excess unlabeled ADP included with the Mt prevents mADP rebinding. At high Mt concentrations, mADP release (state 2 to state 3) is rate-limiting, and the plateau denotes the mADP off-rate of the bound head (koffmADP = 37.8 ± 5.3 s−1). At limiting Mt concentrations (Fig. 1b, inset), Mt binding by the motor is rate-limiting, and the linear dependence reveals a bimolecular on-rate, kbiADP = 9.7 ± 0.2 μm−1 s−1, which is consistent with previous monomer studies (21–23).
To calculate kbiATPase, the Eg5M ATPase activity was quantified to determine the maximal hydrolysis rate and apparent microtubule affinity. The maximal hydrolysis rate of the monomer, kcat = 12.9 ± 1.9 s−1, was similar to the dimeric stepping rate (8.5 ± 1.6 s−1; Fig. 2c), suggesting that the monomer is a good model for the processive dimer. The apparent Mt affinity in the ATPase, K0.5Mt = 0.93 ± 0.25 μm Mt, yielded an apparent bimolecular Mt binding rate kbiATPase = kcat/K0.5Mt = 13.9 ± 4.3 μm−1 s−1 for Eg5M. The final kbiratio = kbiATPase/kbiADP = 1.4 ± 0.4 indicates that only 1 ATP molecule is hydrolyzed per Mt encounter, consistent with previous kinesin-5 monomer studies (21).
FIGURE 2.
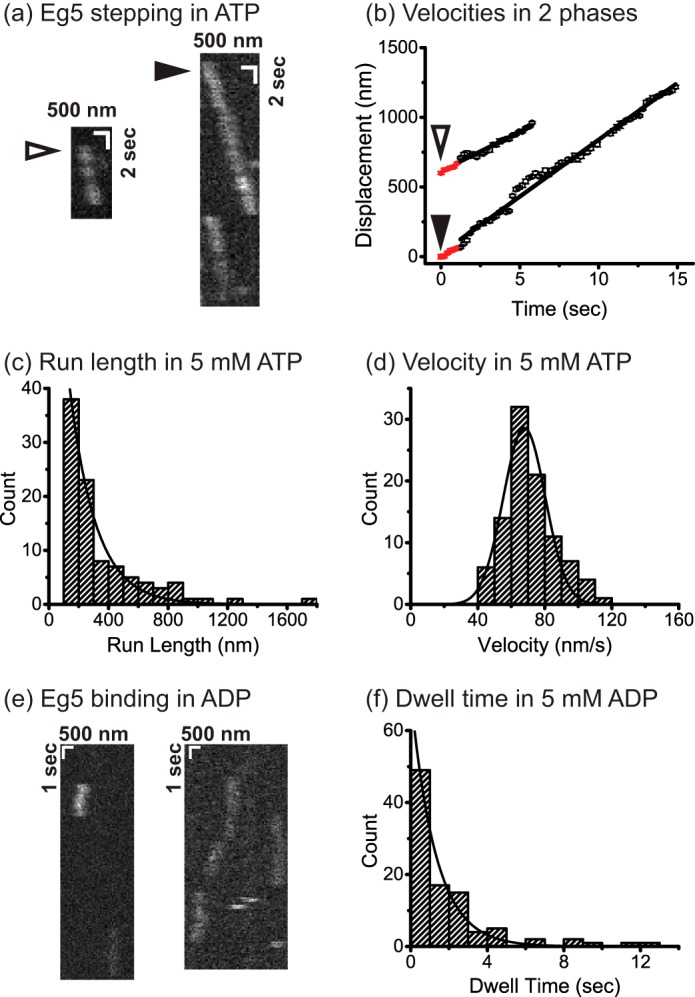
Single-molecule motility of dimeric Eg5 in 5 mm ATP (a–d) and 5 mm ADP (e–f). a, high resolution tracking of QD655-labeled Eg5 stepping at 5 mm ATP. Kymographs with open and closed triangles indicate the two traces analyzed in initial velocity determination (b). Each trace was separated into two segments, including the initial 1-s velocity (60 ± 9 nm/s, 65 ± 7 nm/s, mean ± S.D.) and the steady-state velocity (56 ± 3 nm/s, 81.5 ± 1 nm/s, mean ± S.D.). No initial pausing was observed. Run length of 177 ± 13 nm (c) and velocity of 68 ± 13 nm/sec (mean ± S.D.) (d) were determined by fitting cumulative density function to run lengths above 100 nm (n = 96). e, kymograph of Eg5 binding at 5 mm ADP. f, dwell time of 1.3 ± 0.1 s (Mt-unbinding rate: 0.77 ± 0.06 s−1) was measured (n = 97), which is significantly slower than monomer Mt-unbinding rate in Fig. 1d.
Kinesin-5 Monomer Likely Detaches in the ADP-Pi State
To characterize the microtubule unbinding properties of the monomer, single-molecule dwell times were measured by total internal reflection fluorescence microscopy using a C-terminal biotinylated Eg5 monomer conjugated with a quantum dot (QD655; Molecular Probes) (Fig. 1d). If the monomeric motor carries out 2 or more hydrolysis cycles during each Mt encounter, then its Mt unbinding rate in saturating ATP should be at least 2-fold slower than ATP hydrolysis rate. Instead, the Mt unbinding rate of Eg5M in ATP (kunbindATP = 15.6 ± 1.5 s−1; Fig. 1d) matched the kcat from the ATPase assay (12.9 ± 1.9 s−1; Fig. 1c), providing independent evidence that the monomer hydrolyzes one ATP per attachment/detachment cycle.
The next question was whether the monomer detaches from the microtubule in the ADP-Pi state or the ADP state. In 5 mm ADP, the unbinding rate was similar to that in ATP, kunbindADP = 17.0 ± 2.0 s−1 (Fig. 1d). Although the most obvious conclusion is that the motor normally detaches in the ADP state, a closer analysis argues against this. If Pi release is fast and the motor returns to state 2 (Fig. 1a), then the motor could either unbind from this state or release its ADP and initiate another hydrolysis cycle. Because koffADP (37.8 s−1; Fig. 1b) is twice as fast as kunbind (17.0 s−1, Fig. 1d), the motor would most frequently release its ADP and ATP binding would follow, resulting in multiple ATP hydrolyzed per encounter. Hence, based on the chemical processivity near 1, we conclude that monomeric Eg5M normally detaches from the ADP-Pi state. If the Mt unbinding rate in the ADP-Pi state kunbindDP is not faster than the unbinding rate in the ADP state, which is supported by a body of work (9, 21, 22), then it follows that the rate-limiting step for the monomer is unbinding in the ADP-Pi state (state 5 in Fig. 1a).
To complete the analysis of the monomer hydrolysis cycle, we measured the mATP exchange rate when the motor is bound to the Mt. By mixing motor-Mt complexes with varying mATP, we observed a nucleotide on-rate (konmATP = 1.5 ± 0.2 μm−1 s−1; Fig. 1e), in agreement with previous reports (22, 23, 28).
Kinesin-5 Dimer Does Not Pause at the Start of a Processive Run
Previous work using a dimeric human kinesin-5 construct, Eg5513, dimerized through the proximal coiled-coil of the native motor rather than the kinesin-1 dimerization domain used here, concluded that upon its first interaction with the microtubule there is a slow (∼1 s−1) conformational change before the motor begins stepping (17). Because of the complications that this lag phase introduces into data interpretation, we directly tested whether such a pause occurs in the hydrolysis cycle of our Eg5 dimer. Single molecule fluorescence measurements were carried out by total internal reflection fluorescence using QD655-conjugated dimeric Eg5 motors (representative kymographs are shown in Fig. 2a). To test for initial pausing, point-spread function fitting (29) was used to determine motor position over time, and velocities during the first second were compared with velocity at later times. As seen in Fig. 2b, the initial phase velocities (60 ± 9 nm/s and 65 ± 7 nm/s, mean ± S.D.) were similar to the later velocities (56 ± 3 nm/s and 81.5 ± 1 nm/s, mean ± S.D.), and no visible pauses were apparent. Therefore, we conclude that in our Eg5 dimeric motors there is no ∼1 s−1 conformational change, and the motors begin stepping immediately upon first collision with the microtubule.
To quantify mean velocity and run length, we analyzed 96 traces using point-spread function fitting (29) with 100-ms temporal resolution and 20-nm spatial precision. These QD-labeled motors had a similar run length (177 ± 13 nm, n = 96; Fig. 2c) and velocity (68 ± 13 nm/s, mean ± S.D.; Fig. 2d) to GFP-labeled constructs analyzed previously (5, 8).
Kinesin-5 Dimer Has a Slow Mt Unbinding Rate in ADP
Having measured a 59-ms mean Mt binding duration for monomeric Eg5M in ADP (17 s−1 unbinding rate; Fig. 1d), we next measured the Mt unbinding rate of Eg5 dimer in ADP. If both heads of kinesin-5 interact with the microtubule, then the off-rate of kinesin-5 dimer in ADP might be expected to be slower than for the monomer. Using QD655-conjugated dimeric Eg5 in 1 mm ADP (Fig. 2e), we found that the motor binds to the lattice with a mean dwell time of 1.3 ± 0.1 s (off-rate of 0.77 ± 0.06 s−1; Fig. 2f), nearly 20-fold longer than the monomer. This result suggests that when one head is bound to the Mt in the ADP state, the second (tethered) head is able to interact with the microtubule and enhance the microtubule affinity of the motor.
Kinesin-5 Resides in a 2HB State in the Absence of ATP
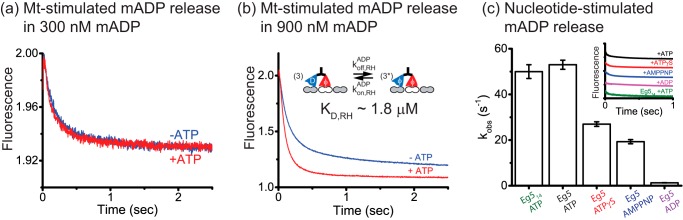
To further investigate coordination between the two heads of Eg5, we measured Mt-stimulated mADP release in the presence and absence of excess ATP. In kinesin-1, Mt binding triggers ADP release by one (bound) head, whereas the second (tethered) retains its bound ADP until ATP binding triggers a step (30). At low Eg5 concentrations, where the free mADP remains low, flushing motors against microtubules resulted in identical fluorescence amplitudes in the presence and absence of ATP, indicating that microtubule binding by Eg5 triggers release of both mADP (Fig. 3a). At higher mADP concentrations, Mt binding resulted in a smaller fluorescence fall in the absence of added ATP, indicating that the motor retained a portion of bound mADP (Fig. 3b). The difference in amplitudes is consistent with the second (tethered) head retaining 33% of its bound mADP, corresponding to a KDmADP of 1.8 μm mADP. This tethered-head mADP affinity is an order-of-magnitude stronger than that of the bound head (estimated 20∼40 μm mADP; Fig. 1b). Based on previous work on kinesin-1 (18), we interpret these data to represent a motor with both heads bound to the microtubule with the leading head having an undocked neck linker and low mADP affinity and the trailing head having a forward-facing neck linker and higher mADP affinity. The higher mADP affinity of the trailing head could be a result of the trailing head transiently unbinding and rebinding to the microtubule or it could represent a microtubule-bound state with a forward-facing neck linker that traps the nucleotide in the binding pocket (“lockdown model”) (31).
FIGURE 3.
Eg5 resides in a 2HB state during processive stepping. a, microtubule stimulated mADP release in the presence (red) or absence (blue) of ATP. In contrast to kinesin-1 half-site mADP release (30), Eg5 releases both bound mADP at limiting mADP concentrations. b, half-site mADP release is partially rescued by adding 0.9 μm free mADP. The fall in fluorescence in the absence of ATP was 83% that of the total, corresponding to 66% mADP release in the tethered head, consistent with a tethered head KDmADP of 1.8 μm. c, nucleotide-stimulated half-site mADP release. Complexes consisting of 0.35 μm Eg5, 8 μm Mt, and 2 μm free mADP were flushed against 2 mm ATP (black), 2 mm ATPγS (red), 2 mm AMPPNP (blue), and 20 μm ADP (magenta). All solutions contained 2 μm free mADP to maintain free concentration. The averaged transients (n = 17–18, inset) were fit by bi-exponentials (mono-exponential for ADP), yielding rate constants kmaxATP/HS = 53 ± 2 s−1, kmaxATPγS/HS = 27 ± 1 s−1, kmaxAMPPNP/HS = 19.3 ± 0.9 s−1, and kTH,offADP = 1.29 ± 0.07 s−1, respectively. ATP-stimulated half-site release rate of shorter neck linker construct Eg514 (n = 11) was kmaxATP/HS = 50 ± 3 s−1 (green). HS, half-site experiment.
Notably, when motors bound to microtubules in the presence of ATP (Fig. 3b, +ATP trace), there was no evidence for slow (∼1 s) isomerization preceding release of the second nucleotide. This is consistent with the motility data in Fig. 2, a and b, but differs from results of others (17).
Hydrolysis Accelerates Tether-head Attachment
To determine the sequence of nucleotide hydrolysis and neck linker docking during the forward step, we carried out nucleotide-stimulated half-site mADP release assays with different nucleotide analogs. Motors were preincubated with microtubules in 2 μm mADP, which is sufficiently high to occupy the tethered head but not sufficient to occupy the bound head. Flushing against saturating ATP-triggered mADP release at 53 ± 2 s−1 (Fig. 3c, Eg5 + ATP). Shortening the neck linker domain from 18 to 14 residues, which increases motor processivity (8), had no effect on this rate (Fig. 3c, Eg514 + ATP). Flushing against slow or non-hydrolyzing ATP analogs led to 2-fold slower mADP release (Eg5 + ATPγS: 27 ± 1 s−1; Eg5 + AMPPNP: 19.3 ± 0.9 s−1; Fig. 3c). These slower rates suggest that tethered head attachment can occur either before or after ATP hydrolysis, but the kinetically favored pathway is hydrolysis preceding tethered head attachment, as described for kinesin-1 and kinesin-2 (32–34). Importantly, the ATP-stimulated half-site release rate is 6 times faster than the motor stepping rate (8.5 ± 1.6 s−1; Fig. 2c), meaning that ATP binding, ATP hydrolysis, tethered head attachment, and ADP release are all too fast to be rate-limiting. Therefore, the data support a model in which trailing-head detachment is the rate-limiting step.
Flushing against ADP triggered mADP release at 1.29 ± 0.07 s−1 (Fig. 3c, Eg5 + ADP), which is faster than dimer unbinding rate of 0.77 s−1 in 1 mm ADP (Fig. 2f). Because KDmADP of the tethered head was estimated to be 1.8 μm (Fig. 3b), we interpret this 1.29 s−1 as the mADP off-rate of the tethered head, which leads to an estimate of 0.7 μm−1 s−1 for the tethered head mADP on-rate.
ATP Binds to the Front Head in the 2HB State
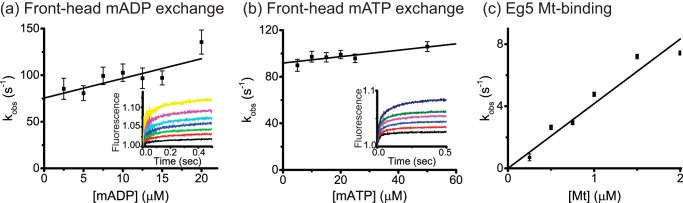
To determine the sequence of ATP binding and trailing head detachment, we investigated nucleotide exchange in the 2HB state. We generated a 2HB species by mixing motors with microtubules and AMPPNP, as described previously (33), and flushed this complex against varying concentrations of mADP (Fig. 4a) or mATP (Fig. 4b). The observed rate constants from the fast phase represent reversible binding of mant nucleotide and thus were fit to the function, kobs = kon[mAxP] + koff. The fits yielded front-head ADP exchange rates of kFH,onADP = 2.1 ± 0.25 μm−1s−1 and kFH,offADP = 75 ± 8 s−1 and front-head ATP exchange rates of kFH,onATP = 0.28 ± 0.08 μm−1s−1 and kFH,offATP = 92 ± 2 s−1. The on-rate for mATP binding to the front head in the 2HB species is 5-fold slower than the corresponding on-rate to the microtubule-bound monomer (Fig. 1e), providing evidence that rearward strain and/or orientation of the neck linker slows nucleotide binding to the front head (front-head gating; Ref. 35). However, at physiological ATP concentrations the time taken for ATP binding (4 ms at 1 mm ATP) should be negligible with respect to the total cycle time (118 ms based on the 8.5 s−1 stepping rate in Fig. 2c). Finally, we measured an on-rate for Mt binding of kbind = 4.2 ± 0.2 μm−1s−1 (Fig. 4c), which is roughly half that of the monomer (Fig. 1b).
FIGURE 4.
Eg5 displays front-head-gating but not rear-head-gating. a, front-head mADP exchange. 0.4 μm motor was incubated with 8 μm Mt and 100 μm AMPPNP for 10 min to generate a 2HB species. The complexes were then flushed against varying mADP concentrations, and the rise in fluorescence due to mADP binding averaged for n = 16–18 traces for each mADP concentration (see the inset for averaged fluorescence traces). For each condition n = 16–18 transients were averaged and fit with a bi-exponential, and the fast phases were fit to the equation kobs = kon,FHmADP[mADP] + koff,FHmADP. The mADP exchange rates were kon,FHmADP = 2.1 ± 0.8 μm−1 s−1 and koff,FHmADP = 75 ± 8 s−1. b, front-head mATP exchange. Identical procedures were used, and the final rates were kon,FHmATP = 0.28 ± 0.08 μm−1 s−1 and koff,FHmATP = 92 ± 2 s−1. Inset, the averaged transients (n = 16–18). c, bimolecular on-rate for Mt binding. 0.2 μm motor with 0.5 μm mADP were flushed against varying Mt concentrations containing 2 mm ADP, yielding a Mt binding on-rate constant for dimeric kinesin-5 of kbind = 4.2 ± 0.2 μm−1 s−1.
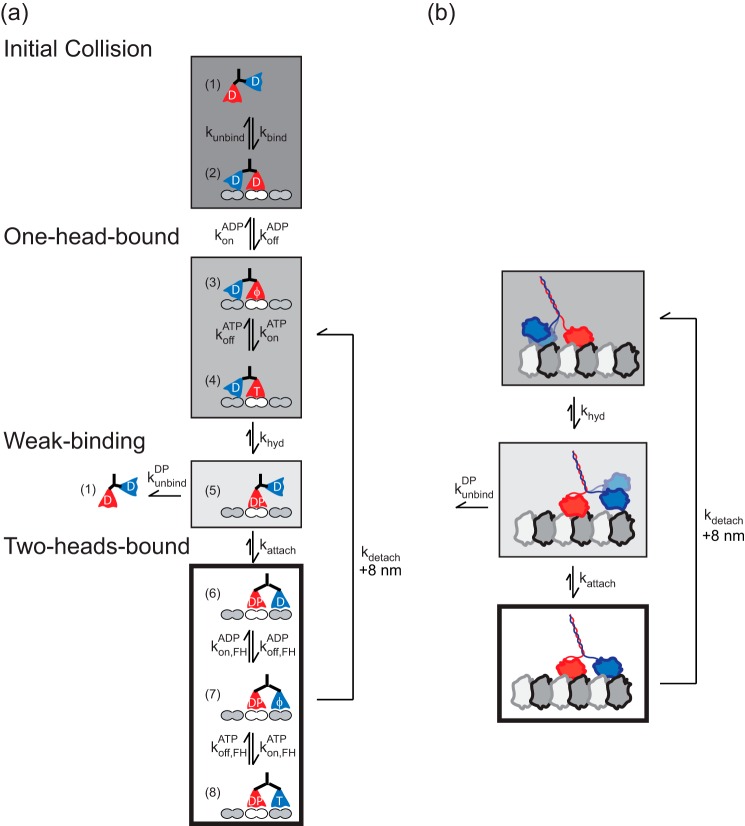
The results were integrated into a kinetic model for the kinesin-5 chemomechanical cycle at saturating ATP concentrations (Fig. 5, a and b). Measured and calculated rate constants are provided in Table 1.
FIGURE 5.
Conceptual model of the kinesin-5 chemomechanical cycle. a, detailed model for dimeric kinesin-5 stepping. Free motors in solution (state 1) bind to the Mt (state 2), which triggers ADP release (state 3). ATP binding (state 4) is followed by ATP hydrolysis (state 5) and tethered-head attachment (state 6). In this 2HB state, the motor releases the ADP in its front-head (state 7) and allows ATP binding (state 8). The 2HB motor detaches its trailing head to complete a step (states 6–8 returns to states 3–4). In this model the processivity is determined by the race between tethered-head attachment (state 5 to state 6) and Mt unbinding (state 5 to state 2). b, simplified model that lumps the cycle into a strong binding one-head-bound state (dark gray), a weakly bound one-head-bound state (light gray), and a strongly bound two-head bound state (white). Processivity is determined by the race occurring at the weak binding state between tethered head attachment and bound head dissociation. The results presented here indicate that the motor spends most of its time in the 2HB strong binding state. Measured and calculated rate constants are given in Table 1, and details are described in Table 1 footnotes.
TABLE 1.
Rate constants for the kinesin-5 chemomechanical cycle
| State | Rate constant | Description | Source | Experiment | Units |
|---|---|---|---|---|---|
| 1-2 | kbind | Bound-head Mt on-rate | Fig. 4c | 4.2 ± 0.2 | μm−1 s−1 |
| 2-1 | kunbind | Bound-head Mt off-rate | Fig. 2f | 0.77 ± 0.06 | s−1 |
| 2-3 | koffADP | Bound-head ADP off-rate | Fig. 1b | 37.8 ± 5.3 | s−1 |
| 3-4 | konATP | Bound-head ATP on-rate | Fig. 1e | 1.5 ± 0.2 | μm−1 s−1 |
| 4-5 | khyd | Hydrolysis rate | Deriveda | 250 (79, 414)b | s−1 |
| 5-1 | kunbindDP | Mt off-rate at ADP-Pi state | Deriveda | 21.3 ± 4.5 | s−1 |
| 5-6 | kattach | Front-head attachment rate | Deriveda | 476 (79, 1073)b | s−1 |
| 6-7 | koff,FHADP | Front-head ADP off-rate | Fig. 4a | 75 ± 8 | s−1 |
| 7-6 | kon,FHADP | Front-head ADP on-rate | Fig. 4a | 2.1 ± 0.8 | μm−1 s−1 |
| 6-2 | kdetach | Trailing-head detachment rate | Derivedc | 10.1 ± 0.3 | s−1 |
| 7-3 | kdetach | ||||
| 8-4 | kdetach | ||||
| 7-8 | kon,FHATP | Front-head ATP on-rate | Fig. 4b | 0.28 ± 0.08 | μm−1 s−1 |
| 8-7 | koff,FHATP | Front-head ATP off-rate | Fig. 4b | 92 ± 2 | s−1 |
a In Fig. 1, c and d, 1/kcat − 1/ koffADP = 1/khyd + 1/ kunbindDP = 51 ± 10 ms. In Figs. 3c and 4a, 1/kmaxATP/HS − 1/kFH,offADP = 1/khyd + 1/kattach = 5.6 ± 1.6 ms. In Fig. 2c, processivity was constrained by kattach/kunbindDP = 22. These three equations give the solution set of the three unknowns. HS, half-site experiment.
b 95% confidence interval with truncated gaussian model. Lower bound: experimental constraints in a; upper bound: one-tailed, α = 5%.
Eg5 Stepping Cycle Has One Rate-limiting Step
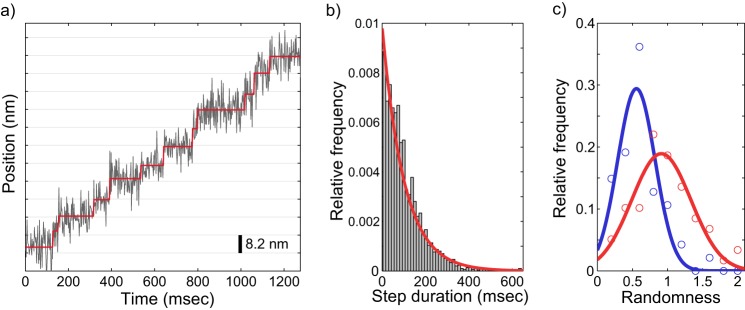
To provide an independent estimate for the number of rate-limiting steps in the Eg5 stepping cycle, we carried out high resolution tracking of gold nanoparticle-labeled Eg5 motors stepping along immobilized microtubules. Single motors with a 40-nm gold nanoparticle attached to the C terminus were tracked with 2-ms temporal resolution and 1–2 nm localization precision, which was sufficient to detect 8.2-nm steps (Fig. 6a), and to quantify the duration between successive steps. The resulting distribution of step durations was well estimated by an exponential distribution with the time constant fixed to the population mean of 101.1 ms (Fig. 6b), consistent with Eg5 having one rate-limiting step under no load.
FIGURE 6.
Stepping kinetics of kinesin-5 is consistent with one rate-limiting step per ATPase cycle. a, example raw trace for kinesin-5 stepping at saturating ATP concentrations (2 mm). A 40-nm gold nanoparticle was attached to the C terminus using biotin-streptavidin and imaged under total internal reflection dark field microscopy at 500 frames per second (see “Experimental Procedures”). Output of a model-free step-finding algorithm is overlaid in red, showing clear measurement of 8.2-nm steps. b, distribution of step durations (n = 2478 steps from 59 traces) as called by the step-finding algorithm. The data are well described by an exponential distribution, P(t) = λe−λt. Fixing λ at the measured population mean (101.1 ms) and performing regression (red line) returned R2 = 0.96. c, distribution of measured randomness values for kinesin-1 (blue, n = 47 traces) and kinesin-5 (red, n = 59 traces). Gaussian fits to the distributions returned a center value of 0.56 for kinesin-1 and 0.91 for kinesin-5.
It was shown previously that at minimal loads, the randomness parameter (see “Experimental Procedures”) for kinesin-1 was 0.5, consistent with the hydrolysis cycle having two rate-limiting steps (36). In agreement with this previous work, we found a randomness of 0.56 for kinesin-1 (Fig. 6c). In contrast, we found a randomness of 0.91 for Eg5 (Fig. 6c). Previous work measured a randomness near 0.5 for Eg5 under external load (37), but this is the first measurement of Eg5 randomness under zero load, matching the conditions of our biochemical experiments. Simulations of our Eg5 kinetic model, in which rear-head detachment is the sole rate-limiting step, generated a value of 0.80–0.90 for the randomness parameter, in good agreement with the experimental determination. The match between the experimental data and simulations provides further evidence that the Eg5 stepping cycle has only 1 rate-limiting step, in support of the biochemical data.
Discussion
In contrast to transport kinesins that rapidly dissociate upon reaching the microtubule plus-end (38), kinesin-5 remain bound to microtubule plus-ends for more than 7 s before dissociating (5). More importantly, these end-bound motors increase the microtubule growth rate and suppress the catastrophe frequency, meaning that in addition to generating outward-directed forces to maintain spindle integrity, kinesin-5 motors help to build the mitotic spindle. The goal of the present study was to uncover aspects of the kinesin-5 chemomechanical cycle that contribute to this end binding and polymerase activity. We previously put forward three potential mechanisms that could explain the plus-end binding activity of kinesin-5 (5). First, motors may dissociate slowly from the microtubule in the ADP state when unable to take a forward step. Second, ATP hydrolysis may require stepping by the tethered head, such that end-bound motors wait in an ATP-bound state. Third, due to specific features of its hydrolysis cycle, kinesin-5 motors may spend a large fraction of their hydrolysis cycle in a 2HB state, which results in long-lived plus-end binding and stabilization of incoming tubulin. Below, we describe data that rule out the first two hypotheses and support the third hypothesis. We then present a model for the kinesin-5 hydrolysis cycle that diverges in some aspects from previous models and explains unique features of kinesin-5 mechanochemistry that may underlie its polymerase activity.
We found that monomeric Eg5M hydrolyzes one ATP per microtubule encounter and that the monomer turnover rate matches the stepping rate of the dimer, consistent with previous work (39). Furthermore, in single-molecule assays, monomeric motors dissociated at similar rates of ∼15 s−1 in both ATP and ADP, consistent with previous work (22). Thus, to the extent that a monomer is equivalent to an end-bound dimer in which the tethered head is unable to take a forward step, there is no unique feature of Eg5 monomers that explains end binding by the dimer. Specifically, both monomers and dimers in ADP dissociate much more rapidly than end-bound dimers. This result rules out the first hypothesis that motors dissociate slowly from the microtubule in the ADP state when unable to take a forward step.
The second hypothesis, that tethered head attachment is a prerequisite for ATP hydrolysis, is also ruled out by the experiments. The 15 s−1 monomer hydrolysis rate demonstrates that hydrolysis does not need a second head. Also, in the dimer, ATP triggers half-site mADP release at a faster rate than does AMPPNP or ATPγS, consistent with hydrolysis accelerating tethered head attachment rather than tethered head attachment triggering hydrolysis (32, 36). Instead, the results support our third hypothesis, namely that kinesin-5 motors spend a large fraction of their hydrolysis cycle in a 2HB state.
Our results are consistent with a kinesin-5 mechanochemical cycle (Fig. 5), that has a number of specific features that diverge from previous kinesin-5 work. For clarity, we separate the initial microtubule interaction (states 1→2) from the subsequent states (3→8) that make up the normal hydrolysis cycle. The measured Mt binding and ADP release rates are similar to rates measured previously (21), but the tethered head in State 3 had a relatively low ADP affinity of 1.8 μm. We found previously for kinesin-1 that extending the neck linker domain from wild-type 14 residues to 18 residues (which matches the native neck linker of kinesin-5; Ref. 40) results in the rate of tethered head ADP exchange increasing from 0.4 s−1 to 1.3 s−1 (18). The measured kinesin-5 tethered head ADP off-rate of 1.3 s−1 (based on mADP exchange with ADP in Fig. 3c) matches this rate and suggests that the faster exchange relative to wild-type kinesin-1 results from the longer 18-residue neck linker of kinesin-5. We propose that the trailing head in State 2 and 3 is actually bound to the microtubule with its neck linker in a forward orientation due to tension from the second head. For kinesin-1, evidence that the tethered head interacts with the microtubule comes from the faster rate of reversal of ATP hydrolysis in the tethered head compared with a free motor in solution (31). Evidence that a forward-facing neck-linker domain traps the nucleotide come from the finding that “lock-down” of the neck linker by binding of the tail domain or by cross-linking traps ADP in the binding pocket even in the presence of microtubules (41).
Further evidence that the tethered head is bound to the microtubule comes from the difference in monomer and dimer unbinding rates. In saturating ADP, which induces a weak binding state, the monomer dissociates at 17 s−1 (Fig. 1d), whereas the dimer dissociates nearly 20-fold slower at 0.77 s−1 (Fig. 2f), suggesting that both heads must unbind for the dimer to dissociate from the microtubule.
The second feature of the hydrolysis cycle is that in States 4→6, ATP hydrolysis precedes attachment of the tethered head to the next binding site. Evidence for this sequence in kinesin-1 came from the finding that slowing hydrolysis using ATPγS does not enhance motor run length and that under assisting loads added Pi enhances the run length (32, 36). For kinesin-5, we found that nucleotide-triggered mADP release from the tethered head is faster in ATP than in ATPγS or AMPPNP (Fig. 3c), consistent with hydrolysis triggering complete neck linker docking and tethered head binding. Previous work suggested that ATP hydrolysis is the rate-limiting step in the kinesin-5 hydrolysis cycle (42), but the fast rate of ATP-triggered mADP release (53 ± 2 s−1; Fig. 3c) argues against this. The finding that the monomer dissociation rate is similar in ATP and ADP (Fig. 1d) also argues against ATP hydrolysis being rate-limiting, as follows. If the monomer unbinding rate in the ADP-Pi state is equal to or slower than the unbinding rate in ADP, then the similar unbinding rates in ADP and ATP mean that hydrolysis must be significantly faster than the 17 s−1 unbinding rate. Thus, an important feature in the chemomechanical cycle in Fig. 4c is that termination of a run occurs from the ADP-Pi-bound state 5 and that run length is determined by a race between kunbindDP and kattach at state 5.
The third important feature of the hydrolysis cycle is that ATP binds in a 2HB state (state 6→8). The key piece of evidence is the nucleotide exchange kinetics when motors are locked in a 2HB state with AMPPNP (Fig. 4, a and b). This analog is thought to generate a 2HB state with AMPPNP trapped in the trailing head and an empty front head (33, 35, 43), and the rapid binding kinetics demonstrate that nucleotide readily binds to the front head. The front-head mATP on-rate in Fig. 4b was 5-fold slower than the corresponding on-rate for monomeric Eg5M (Fig. 1e), providing evidence for front-head gating (44). However, the front-head on-rate of 0.3 μm−1s−1 means that at a concentration of 1 mm, ATP binding will only take 3 ms, which is very fast relative to the roughly 100-ms cycle time. Thus, under physiological conditions ATP binds when the motor is in a 2HB state.
The final defining feature of the chemomechanical cycle in Fig. 5 is that trailing head detachment (state 8→4) is the rate-limiting step in the cycle at saturating ATP. Evidence comes from comparing the rate ATP-triggered half-site release (53 s−1 in Fig. 3c) to the overall stepping rate (9.5 s−1 in Fig. 2d). In the hydrolysis cycle of Fig. 5, the half-site release experiment begins in state 3 and terminates in state 7 when ADP is released. The remaining steps in the cycle are the ATP binding transition 7→8, which we show to be fast as described above, and the trailing head detachment rate (state 8→4). Thus, it follows that trailing head detachment (estimated at 10 s−1 by comparison of half-site and stepping rates) is the slowest transition in the cycle and the motor will remain in a 2HB state for most of the cycle. As trailing head detachment at 10 s−1 is near the monomer unbinding rate of 15 s−1 (Fig. 1d), there is no evidence of rear-head gating, meaning that interhead strain does not appear to accelerate trailing head detachment. A recent optical trapping study using kinesin-1 with extended neck linkers suggested that the interhead tension present in wild-type kinesin is relieved when the neck linker is extended from 14 to 15 or more residues (18), in agreement with this lack of rear-head gating of kinesin-5 (which has an 18 residues neck linker). Based on the tethered head exchange results (ADP in Fig. 3c), nucleotide release from the trailing head in this 2HB state 8 is expected to be slow (1.3 s−1) due to the forward neck linker orientation that traps nucleotide in the pocket. One possibility is that Pi release is the rate-limiting transition that determines detachment; alternatively, Pi release could be coupled with or immediately follow detachment (11).
One hallmark of kinesin-5 motility is its minimal processivity, which has been functionally accounted for by the fact that the motors work intracellularly in teams (19, 37, 39, 45). We previously showed that shortening the Eg5 neck linker to match the 14 residues of kinesin-1 results in ∼1-μm run lengths similar to wild-type kinesin-1 (8). The present work is consistent with that result; in the framework developed in Fig. 5, run length is determined simply by the race between kunbindDP and kattach at state 5. Based on the 15 s−1 monomer unbinding rate (Fig. 1d), we estimate kunbindDP (5→1 transition in Fig. 5) at 15 s−1, putting kattach at ∼300 s−1 to match the observed unloaded run lengths measured here. Shortening the neck linker to 14 residues, which increases the run length by roughly a factor of four (8), is hypothesized in our model to enhance kattach to >1000 s−1. The finding that the ATP-stimulated mADP release for Eg514 matches the rate of Eg5 (half-site data in Fig. 3c) is consistent with this prediction, although it does not provide stringent constraints.
Another distinctive feature of kinesin-5 processivity is its relative insensitivity to load. Whereas a 4-piconewton-assisting load decreases the kinesin-1 run length by roughly 10-fold (18), a similar assisting load diminishes the kinesin-5 run length by only a factor of 2 (19). This property was also observed in mixed motor gliding assays; kinesin-5 motors were able to efficiently slow the transport of microtubules driven by faster transport motors (20). This ability to resist loads is consistent with the proposed role of kinesin-5 as a brake that stabilizes microtubule bundles in axons; motor inhibition leads to faster axonal outgrowth and longer branches in culture (7). Maintaining spindle integrity requires sustained force generation, so this property also aligns with the motor's mitotic function. The finding that kinesin-5 spends the majority of its hydrolysis cycle in a 2HB state provides a mechanism to explain its ability to resist loads.
Recent work using time-resolved kinetics suggested that in monomeric Eg5 there is a weak conformational coordination between switch-1 closure and neck linker docking (11). Interestingly, we found that monomeric Eg5 has a tightly coupled chemomechanical cycle (kbiratio = 1), whereas monomeric kinesin-1 does not (kbiratio = 3–7; Ref. 27). The Eg5 kbiratio could be explained by the race between the opening of the switch-1 cleft and motor unbinding as follows. After ATP hydrolysis, Eg5 opens its switch-1 cleft on a time scale of 100 ms (10 s−1) to allow nucleotide release (11). We measured a detachment rate of 15 s−1 for monomeric Eg5, which makes a second ATP turnover event unlikely to occur.
The finding that kinesin-5 is predominantly in a 2HB state provides a general mechanism to account for the motors' observed microtubule polymerase activity; the 2HB state bridges adjacent tubulin subunits on a single protofilament and thereby enhances microtubule stability. Family-specific features of kinesin-5 that may contribute to this behavior include a rearward-facing orientation of the neck linker in the apo or ADP states (9) and the 18-residue neck linker domain, which could provide added interhead flexibility.
A full accounting of the cellular function of kinesin-5 requires accounting for all of the structural and regulatory elements present in the wild-type motor. The construct employed here, a dimer in which the heads are connected through a stable kinesin-1 coiled-coil domain, provides an excellent model system for identifying features of interhead coordination. However, the wild-type motor is a tetramer; the tail domains reside near the opposite head domains and may bind microtubules and regulate the motor activity (3). There is also evidence of communication through the tetrameric coiled-coil domain meaning that mechanical activity of one pair of heads may regulate the behavior of the motors at the opposite end of the molecule (46). Additionally, this model provides open questions on the ionic strength-dependent directional switching of yeast kinesin Cin8 (47, 48) and the microtubule depolymerizing properties of yeast kinesin-5 Kip1p (49, 50). Finally, phosphorylation may regulate motor activity (51). Hence, with the current core hydrolysis cycle, a framework is in place to understand these other modulating factors that determine behavior of the wild-type tetramer in cells.
Experimental Procedures
Protein Constructs, Purification, and Quantification
All kinesin-5 dimeric constructs consisted of the head and neck linker domain of Xenopus laevis Eg5 (residues 1–367) fused to the coiled-coil (residues 345–559) of Drosophila KHC, as previously described (8). The dimeric motor Eg5 and Eg514 denote the neck linker lengths of wild-type 18 residues and truncated 14 residues, respectively. Monomeric Eg5 (Eg5M) includes the entire catalytic core and entire 18 residue neck linker (residues 1–367) fused to a hexa-His tag or to an AviTag (GGGLNDIFEAQKIEWHE) followed by a hexa-His tag. Details of the GFP fusion, biotinylation procedures, bacterial expression, and protein purification and storage were described previously (33, 36, 52). Active motor concentration was quantified by mADP chase-off assays (33), and all motor concentrations described correspond to the active motor concentration. Active motor concentrations were typically 30–40% that of total motor concentrations quantified by SDS-PAGE using Coomassie Blue staining. Stock concentrations of dimeric motors were 1–30 μm, and monomeric motors were 30–100 μm. Experiments were carried out in BRB80 buffer (80 mm PIPES, 1 mm EGTA, 1 mm MgCl2, pH 6.8) at 23 °C.
ATPase Assays
ATPase was carried out by enzyme-coupled assay with 50 nm active monomeric motor, and hydrolysis rates were estimated by steady-state absorbance decrease at 340 nm as previously described (33). The parameter kbiATPase was calculated by dividing the maximal ATP hydrolysis rate by the Mt concentrations at half-maximal ATPase, as carried out previously for kinesin-1 dimer (26) and monomer (27).
Single-molecule Fluorescence
Single-molecule fluorescence of GFP-labeled or QD655-labeled (Thermo-Fisher Q10121MP) motors was carried out using a Nikon TE-2000 total internal reflection fluorescence microscope, as previously described (33). Microtubules were polymerized by mixing a 1:20 ratio of Cy5-labeled and unlabeled calf brain tubulin. A monoclonal anti-β-tubulin antibody (Sigma T7816) or kinesin-1 rigor mutant (36) was used to immobilize microtubules followed by blocking with 0.5 mg/ml casein. To image vivid QD655-labeled motor, β-mercaptoethanol was not used in an oxygen scavenger system due to quenching, and the imaging time was kept below 60 s to prevent photo-damage. Dwell time data were analyzed with the MultipleKymograph plug-in in ImageJ (imagej.nih.gov) using specific macros to ensure the fluorescence events were aligned with the microtubule tracks.
To quantify initial pausing events, run lengths, and velocities, single-molecule measurements on QD655-labeled motors were analyzed by point-spread fitting using FIESTA software (29). The initial velocity was determined by linear fit of displacement during the first second, and the remainder of the trace was fit to obtain the steady-state velocity. Student's t tests were used to determine the occurrence of the “initial pause” and the difference between the initial velocity and the steady-state velocity. For run length and velocity measurements, analyses included only run lengths >100 nm and velocities between 40 and 120 nm/s to rule out pausing and diffusive events.
High Speed Single-molecule Tracking by Total Internal Reflection Dark Field Microscopy
High speed single-molecule tracking experiments were carried out using a custom-built total internal reflection dark field microscope utilizing micro-mirrors (53). The microscope was built around a Mad City Labs RM21 base (54) using micromirrors both to provide illumination and to pick off the totally internally reflected beam. Objective-type total-internal reflection was achieved using an Olympus 1.49 NA objective, and illumination was provided by a Coherent Sapphire-LP 532-nm laser. Images were recorded using a Basler Ace acA2000–165 μm CMOS camera. Kinesin-1 and -5 motors were prepared with a biotinylated C-terminal Avitag as previously (36) and mixed with 40-nm neutravidin-coated gold nanoparticles (Nanopartz) at the lowest molar ratio of total motor concentration (including non-biotinylated and inactive) to gold that produced landing events (16:1 for kinesin-1 and 4:1 for kinesin-5). Microtubules were attached to the glass substrate using full-length rigor kinesin-1 mutant, as previously described (36).
High resolution position versus time traces were obtained from 500 frame/s movies by fitting the point spread function with a two-dimensional Gaussian using FIESTA software (29). x-y trajectories were rotated to minimize the standard deviation in the x direction, resulting in alignment of the y axis with the microtubule axis. Step finding was performed on the y-displacement versus time traces using the tDetector algorithm (55), which returned step sizes and durations. Randomness measurements were calculated using the formula (56, 57),
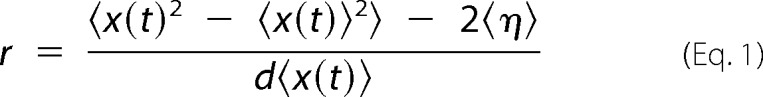 |
All data analysis was done in MATLAB.
Stopped-flow Setup
Stopped-flow measurements using mant-nucleotides were carried out on an Applied Physics SX20 spectrofluorometer using 356-nm excitation with a HQ480SP emission filter. Note that the mant moiety does not change nucleotide affinity for kinesin-5, unlike the 20-fold difference measured for kinesin-2 (33) and 2-fold difference for kinesin-1 (36). All nucleotides contained equimolar Mg2+.
konMt and kbiADP Experiments
Experiments were carried out by mixing 0.2 μm mADP-incubated motors against solutions containing 2 mm ADP and varying Mt concentrations. Data were fit to a bi-exponential, and the observed rate of the fast phase, kobs was fit to,
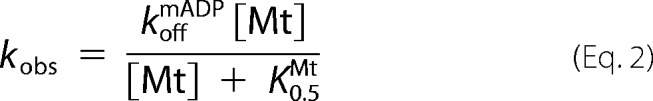 |
where koffmADP and K0.5Mt correspond to the mADP off-rate and the apparent Mt affinity, respectively. At low Mt concentration, the rates can be fit by linear regression, where the slope corresponds to kbiADP, the apparent bimolecular Mt on-rate (26).
Nucleotide-stimulated Half-site Release Assays
Motors (0.3–0.4 μm) were incubated with microtubules and 2 μm mADP to generate a species with no nucleotide in the bound head and mADP in the tethered head (30). To quantify the nucleotide-induced tethered-head stepping rate, this complex was flushed against a solution containing 2 mm ATP, 2 mm ATPγS, or 2 mm AMPPNP with additional 2 μm mADP to maintain a constant mADP concentration. To measure the intrinsic tethered-head mADP release rate, the complex was flushed against a solution containing 20 μm ADP to prevent mADP rebinding to the tethered head; this lower concentration ensures that the bound head has minimal dissociation events. Signals were fit by a bi-exponential for ATP, ATPγS, and AMPPNP and a single exponential for ADP. Control experiments confirmed that 1 mm nucleotide was sufficient to achieve maximal half-site mADP release rate.
Front-head Nucleotide Exchange Kinetics
Motors at 1–2 μm were mixed with 40 μm Mt and 500 μm AMPPNP to generate a 2HB motor. After 10 min of incubation, complexes were diluted 5-fold with BRB80 buffer to maximize the species with AMPPNP in the bound head and no nucleotide in the front head. These complexes were then flushed against varying mant-nucleotide concentrations to estimate the nucleotide on- and off-rates, as determined previously (33).
Simulation of Single-molecule Traces
Stochastic simulations were carried out using the parameter set obtained from single-molecule and ensemble measurements, as previously described (33). After recapitulating the stepping traces, the randomness was quantified using both temporal duration (58) and spatial fluctuation (56) to check its robustness. The code can be found in the supplemental Randomness.txt file.
Data Analysis
All curve fitting for stopped-flow data were carried out using regression with instrumental weighting function in OriginPro 9.1 software. Data are presented as the means ± S.E. of fit (S.E.) unless otherwise noted.
Author Contributions
G.-Y. C. and W. O. H. designed the study and wrote the paper. K. J. M. acquired and analyzed the high resolution tracking data. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We gratefully acknowledge David Arginteanu for assistance in protein purification, Steve Rosenfeld for thoughtful discussions, and Yalei Chen for previous experimental work on the Eg5 polymerase mechanism. A portion of the bacterial culture work was carried out at the Penn State Huck Institutes for Life Sciences Fermentation Facility with the assistance of Mark Signs.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM076476 (to W. O. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as a Paper of the Week.

This article contains a supplemental file.
- Eg5
- kinesin-5
- Eg5M
- monomeric Eg5 construct
- AMPPNP
- adenosine 5′-[(β,γ)-imido] triphosphate
- mant-
- 2′-(or-3′)-O-(N-methylanthraniloyl) group
- mATP
- mant-ATP
- mADP
- mant-ADP
- Mt
- microtubule
- FH
- front head
- 2HB
- two-heads bound.
References
- 1. Goulet A., and Moores C. (2013) New insights into the mechanism of force generation by kinesin-5 molecular motors. Int. Rev. Cell Mol. Biol. 304, 419–466 [DOI] [PubMed] [Google Scholar]
- 2. Shimamoto Y., Forth S., and Kapoor T. M. (2015) Measuring pushing and braking forces generated by ensembles of kinesin-5 cross-linking two articles measuring pushing and braking forces generated by ensembles of kinesin-5 cross-linking two microtubules. Dev. Cell 34, 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kapitein L. C., Peterman E. J., Kwok B. H., Kim J. H., Kapoor T. M., and Schmidt C. F. (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it cross-links. Nature 435, 114–118 [DOI] [PubMed] [Google Scholar]
- 4. Fridman V., Gerson-Gurwitz A., Shapira O., Movshovich N., Lakämper S., Schmidt C. F., and Gheber L. (2013) Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 126, 4147–4159 [DOI] [PubMed] [Google Scholar]
- 5. Chen Y., and Hancock W. O. (2015) Kinesin-5 is a microtubule polymerase. Nat. Commun. 6, 8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nadar V. C., Ketschek A., Myers K. A., Gallo G., and Baas P. W. (2008) Kinesin-5 is essential for growth-cone turning. Curr. Biol. 18, 1972–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Myers K. A., and Baas P. W. (2007) Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J. Cell Biol. 178, 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shastry S., and Hancock W. O. (2011) Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U.S.A. 108, 16253–16258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goulet A., Major J., Jun Y., Gross S. P., Rosenfeld S. S., and Moores C. A. (2014) Comprehensive structural model of the mechanochemical cycle of a mitotic motor highlights molecular adaptations in the kinesin family. Proc. Natl. Acad. Sci. U.S.A. 111, 1837–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sindelar C. V., and Downing K. H. (2010) An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. U.S.A. 107, 4111–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muretta J. M., Jun Y., Gross S. P., Major J., Thomas D. D., and Rosenfeld S. S. (2015) The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc. Natl. Acad. Sci. U.S.A. 112, E6606–E6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Behnke-Parks W. M., Vendome J., Honig B., Maliga Z., Moores C., and Rosenfeld S. S. (2011) Loop L5 acts as a conformational latch in the mitotic kinesin Eg5. J. Biol. Chem. 286, 5242–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muretta J. M., Behnke-Parks W. M., Major J., Petersen K. J., Goulet A., Moores C. A., Thomas D. D., and Rosenfeld S. S. (2013) Loop L5 assumes three distinct orientations during the ATPase cycle of the mitotic kinesin Eg5: a transient and time-resolved fluorescence study. J. Biol. Chem. 288, 34839–34849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kapoor T. M., Mayer T. U., Coughlin M. L., and Mitchison T. J. (2000) Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150, 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Talapatra S. K., Schüttelkopf A. W., and Kozielski F. (2012) The structure of the ternary Eg5-ADP-ispinesib complex. Acta Crystallogr. D. Biol. Crystallogr. 68, 1311–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skoufias D. A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R. H., Hackney D., and Kozielski F. (2006) S-trityl-l-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281, 17559–17569 [DOI] [PubMed] [Google Scholar]
- 17. Krzysiak T. C., Grabe M., and Gilbert S. P. (2008) Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J. Biol. Chem. 283, 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Andreasson J. O. L., Milic B., Chen G.-Y., Guydosh N. R., Hancock W. O., and Block S. M. (2015) Examining kinesin processivity within a general gating framework. eLife 4, e07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valentine M. T., and Block S. M. (2009) Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys. J. 97, 1671–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arpağ G., Shastry S., Hancock W. O., and Tüzel E. (2014) Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys. J. 107, 1896–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cochran J. C., Sontag C. A., Maliga Z., Kapoor T. M., Correia J. J., and Gilbert S. P. (2004) Mechanistic analysis of the mitotic kinesin Eg5. J. Biol. Chem. 279, 38861–38870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenfeld S. S., Xing J., Jefferson G. M., and King P. H. (2005) Docking and rolling, a model of how the mitotic motor Eg5 works. J. Biol. Chem. 280, 35684–35695 [DOI] [PubMed] [Google Scholar]
- 23. Cochran J. C., Krzysiak T. C., and Gilbert S. P. (2006) Pathway of ATP hydrolysis by monomeric kinesin Eg5. Biochemistry 45, 12334–12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hackney D. D. (1988) Kinesin ATPase: rate-limiting ADP release. Proc. Natl. Acad. Sci. U.S.A. 85, 6314–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hackney D. D., Malik A. S., and Wright K. W. (1989) Nucleotide-free kinesin hydrolyzes ATP with burst kinetics. J. Biol. Chem. 264, 15943–15948 [PubMed] [Google Scholar]
- 26. Hackney D. D. (1995) Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature 377, 448–450 [DOI] [PubMed] [Google Scholar]
- 27. Jiang W., Stock M. F., Li X., and Hackney D. D. (1997) Influence of the kinesin neck domain on dimerization and ATPase kinetics. J. Biol. Chem. 272, 7626–7632 [DOI] [PubMed] [Google Scholar]
- 28. Cochran J. C., and Gilbert S. P. (2005) ATPase mechanism of Eg5 in the absence of microtubules: Insight into microtubule activation and allosteric inhibition by monastrol. Biochemistry 44, 16633–16648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruhnow F., Zwicker D., and Diez S. (2011) Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 100, 2820–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hackney D. D. (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 91, 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hackney D. D. (2005) The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc. Natl. Acad. Sci. U.S.A. 102, 18338–18343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Milic B., Andreasson J. O., Hancock W. O., and Block S. M. (2014) Kinesin processivity is gated by phosphate release. Proc. Natl. Acad. Sci. U.S.A. 111, 14136–14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen G.-Y., Arginteanu D. F., and Hancock W. O. (2015) Processivity of the kinesin-2 KIF3A results from rear-head gating and not front-head gating. J. Biol. Chem. 290, 10274–10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma Y. Z., and Taylor E. W. (1997) Interacting head mechanism of microtubule-kinesin ATPase. J. Biol. Chem. 272, 724–730 [DOI] [PubMed] [Google Scholar]
- 35. Rosenfeld S. S., Fordyce P. M., Jefferson G. M., King P. H., and Block S. M. (2003) Stepping and stretching. How kinesin uses internal strain to walk processively. J. Biol. Chem. 278, 18550–18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mickolajczyk K. J., Deffenbaugh N. C., Arroyo J. O., Andrecka J., Kukura P., and Hancock W. O. (2015) Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. U.S.A. 112, E7186–E7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valentine M. T., Fordyce P. M., Krzysiak T. C., Gilbert S. P., and Block S. M. (2006) Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 8, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varga V., Leduc C., Bormuth V., Diez S., and Howard J. (2009) Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174–1183 [DOI] [PubMed] [Google Scholar]
- 39. Kaseda K., Crevel I., Hirose K., and Cross R. A. (2008) Single-headed mode of kinesin-5. EMBO Rep. 9, 761–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hariharan V., and Hancock W. O. (2009) Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol. Bioeng. 2, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaan H. Y., Hackney D. D., and Kozielski F. (2011) The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333, 883–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krzysiak T. C., and Gilbert S. P. (2006) Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J. Biol. Chem. 281, 39444–39454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guydosh N. R., and Block S. M. (2006) Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc. Natl. Acad. Sci. U.S.A. 103, 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Block S. M. (2007) Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 92, 2986–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Valentine M. T., and Gilbert S. P. (2007) To step or not to step? How biochemistry and mechanics influence processivity in kinesin and Eg5. Curr. Opin. Cell Biol. 19, 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scholey J. E., Nithianantham S., Scholey J. M., and Al-Bassam J. (2014) Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers. Elife 3, e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gerson-Gurwitz A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D. R., Schmidt C. F., and Gheber L. (2011) Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength, and microtubule geometry. EMBO J. 30, 4942–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roostalu J., Hentrich C., Bieling P., Telley I. A., Schiebel E., and Surrey T. (2011) Directional switching of the kinesin Cin8 through motor coupling. Science 332, 94–99 [DOI] [PubMed] [Google Scholar]
- 49. Gardner M. K., Bouck D. C., Paliulis L. V., Meehl J. B., O'Toole E. T., Haase J., Soubry A., Joglekar A. P., Winey M., Salmon E. D., Bloom K., and Odde D. J. (2008) Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 135, 894–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McCoy K. M., Tubman E. S., Claas A., Tank D., Clancy S. A., O'Toole E. T., Berman J., and Odde D. J. (2015) Physical limits on kinesin-5 mediated chromosome congression in the smallest mitotic spindles. Mol. Biol. Cell 26, 3999–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Avunie-Masala R., Movshovich N., Nissenkorn Y., Gerson-Gurwitz A., Fridman V., Kõivomägi M., Loog M., Hoyt M. A., Zaritsky A., and Gheber L. (2011) Phospho-regulation of kinesin-5 during anaphase spindle elongation. J. Cell Sci. 124, 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shastry S., and Hancock W. O. (2010) Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 20, 939–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lebel P., Basu A., Oberstrass F. C., Tretter E. M., and Bryant Z. (2014) Gold rotor bead tracking for high-speed measurements of DNA twist, torque, and extension. Nat. Methods 11, 456–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Larson J., Kirk M., Drier E. A., O'Brien W., MacKay J. F., Friedman L. J., and Hoskins A. A. (2014) Design and construction of a multiwavelength, micromirror total internal reflectance fluorescence microscope. Nat. Protoc. 9, 2317–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen Y., Deffenbaugh N. C., Anderson C. T., and Hancock W. O. (2014) Molecular counting by photobleaching in protein complexes with many subunits: best practices and application to the cellulose synthesis complex. Mol. Biol. Cell 25, 3630–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Svoboda K., Mitra P. P., and Block S. M. (1994) Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc. Natl. Acad. Sci. U.S.A. 91, 11782–11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Verbrugge S., van den Wildenberg S. M., and Peterman E. J. (2009) Novel ways to determine kinesin-1's run length and randomness using fluorescence microscopy. Biophys. J. 97, 2287–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schnitzer M. J., and Block S. M. (1995) Statistical kinetics of processive enzymes. Cold Spring Harb. Symp. Quant. Biol. 60, 793–802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.