Abstract
Although the elaborate combination of histone and non-histone protein complexes defines chromatin organization and hence regulates numerous nuclear processes, the role of chromatin organizing proteins remains unexplored at the organismal level. The highly abundant, multifunctional, chromatin-associated protein and transcriptional coactivator positive coactivator 4 (PC4/Sub1) is absolutely critical for life, because its absence leads to embryonic lethality. Here, we report results obtained with conditional PC4 knock-out (PC4f/f Nestin-Cre) mice where PC4 is knocked out specifically in the brain. Compared with the control (PC4+/+ Nestin-Cre) mice, PC4f/f Nestin-Cre mice are smaller with decreased nocturnal activity but are fertile and show no motor dysfunction. Neurons in different areas of the brains of these mice show sensitivity to hypoxia/anoxia, and decreased adult neurogenesis was observed in the dentate gyrus. Interestingly, PC4f/f Nestin-Cre mice exhibit a severe deficit in spatial memory extinction, whereas acquisition and long term retention were unaffected. Gene expression analysis of the dorsal hippocampus of PC4f/f Nestin-Cre mice revealed dysregulated expression of several neural function-associated genes, and PC4 was consistently found to localize on the promoters of these genes, indicating that PC4 regulates their expression. These observations indicate that non-histone chromatin-associated proteins like PC4 play a significant role in neuronal plasticity.
Keywords: chromatin structure, gene expression, hypoxia, neurogenesis, transgenic mice, Knockout, Spatial memory
Introduction
The complex eukaryotic genome is packaged in the nucleus as a dynamic DNA-protein structure, chromatin, by means of a multitude of factors. Among the various factors responsible for modulating chromatin architecture, non-histone chromatin-associated proteins such as heterochromatin protein 1 (HP1), poly(ADP-ribose) polymerase (PARP1), positive coactivator 4 (PC4/Sub1), and high mobility group (HMG) proteins play an important role in regulating the dynamicity of genome organization and thus the underlying gene function. Although the role of non-histone chromatin-associated proteins in the context of gene expression and chromatin organization has been explored, there are still very few reports of the specific functions of these proteins at the organismal level, other than those related to the Rett syndrome-associated MeCP2 protein (1–5).
Although initially reported as a potent transcriptional coactivator of activator-dependent RNA polymerase II-dependent transcription (6, 7), PC4/Sub1 has also been implicated in RNA polymerase III-driven transcription, DNA repair, and replication (8). We previously found that human transcriptional coactivator 4 (PC4) is a bona fide non-histone component of chromatin that is involved in chromatin organization through its ability to interact with histones (9). Silencing of PC4 leads to a global alteration of nuclear architecture and epigenetic landscape and overexpression of neural genes in non-neural cells. Interestingly, we also found that PC4 interacts with HP1α and recruits REST/CoREST to silence neural genes in these cells (10). However, if and how PC4 functions in neuronal cells and more generally in the brain is so far undetermined.
Considering the global role of PC4 in chromatin organization and the indications of its significant role in gene expression, we generated PC4 knock-out mice. Knocking out this highly abundant nuclear protein in the mouse germline causes embryonic lethality; however, knocking out PC4 specifically in the brain is not lethal, and these mice are fertile. Although there are no major brain-associated changes, these mice show several specific and discrete phenotypes including nocturnal hypoactivity, impaired adult neurogenesis, and the inability to extinguish spatial memory. Transcriptomic analysis of the dorsal hippocampus of the PC4f/f Nestin-Cre mice showed significant alteration in gene expression compared with the PC4+/+ Nestin-Cre control mice, and using chromatin immunoprecipitation analysis, PC4 was found to localize on some of these gene promoters. These data reveal new gene networks that could be regulated by PC4 and that are involved in neuronal activity and plasticity and therefore likely to play a role in memory processes. These results not only propose a role for PC4 at the organismal level for the first time but also indicate a connection between chromatin and hippocampus-dependent memory processes.
Results
Generation of a Brain-specific PC4 Knock-out Strain Due to the Lethality of Germline PC4 Knock-out
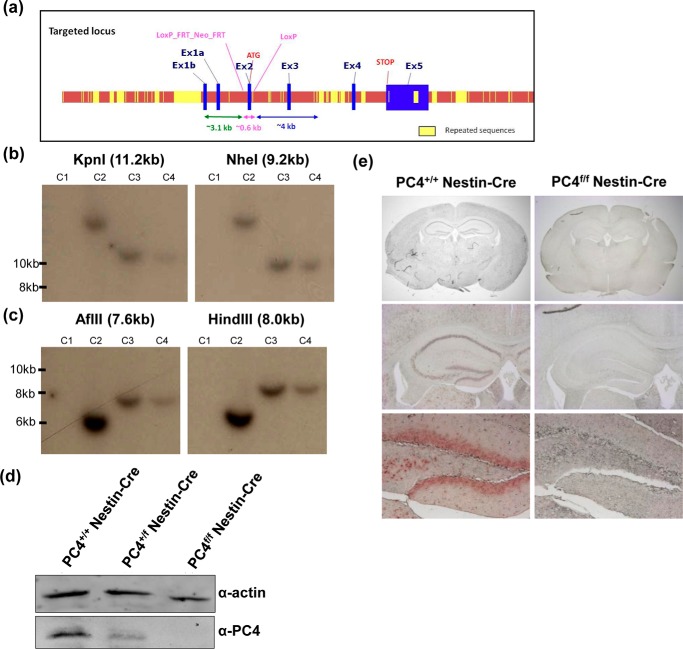
The mouse gene expression database shows that PC4 expression begins early during embryonic development in different tissues. The mouse PC4 gene consists of five exons and is located on chromosome 15. To knock out PC4 in mice, exon 2 was floxed (Fig. 1a). 5′ and 3′ arm validation of the targeted allele was performed by restriction digestion and Southern blot analysis (Fig. 1, b and c). Crosses between PC4f/f mice and mice expressing Cre in the germline (11) generated viable and fertile heterozygotes. However, further crossing of heterozygotes (PC4+/Δ) never resulted in the birth of homozygotes (PC4Δ/Δ); rather than the Mendelian ratio of 1:2:1, a 1:1 ratio was obtained (Table 1). Hence, complete knock-out of PC4 in the germline was lethal. The regulation of neural gene expression by PC4, and its role in the recruitment of the REST-CoREST complex to chromatin for gene silencing indicated a possible role for PC4 in the neural system (10). To investigate the role of PC4 in the neural system, the floxed mice were crossed with Nestin-Cre mice expressing Cre specifically in neural and glial progenitor cells beginning in the early stages of development (12). Surprisingly, despite the high expression of PC4 in many regions of the developing brain, knocking out PC4 in the brain in neuronal and glial precursors early in neural development did not lead to a loss of viability or very serious phenotypes up to 2 years of age (data not shown). PC4 knock-out in the brain was confirmed by Western blot analysis (Fig. 1d), and a dose-dependent decrease was observed in the heterozygous (PC4+/f Nestin-Cre) and homozygous knock-out (PC4f/f Nestin-Cre) mice. Total absence of PC4 in the PC4f/f Nestin-Cre mice was also observed in the immunohistological analysis of brain sections from these mice (Fig. 1e, upper panel), an absence also evidenced when focusing, for example, on the hippocampus (Fig. 1e, middle panel) and the dentate gyrus (Fig. 1e, lower panel). The PC4f/f Nestin-Cre mice were fertile but were smaller and lighter (Figs. 2, a and b). It was also observed that PC4f/f Nestin-Cre mice exhibited swift and aggressive movements compared with the PC4+/+ Nestin-Cre controls. However, on assessing the sleep-wake activity patterns of the mice using actigraphy, the overall nocturnal activity was significantly reduced compared with that of PC4+/+ Nestin-Cre mice (Fig. 2c). Nevertheless, the sleep-wake patterns remained unchanged, with PC4f/f Nestin-Cre mice being significantly more active at night than during the day. To assess whether the reduced activity was due to motor deficits, the motor functions of the PC4f/f Nestin-Cre mice were further analyzed with the rotarod test (13). Following 1 day of habituation, the mice were subjected to 3 trials each on 3 consecutive days. However, there were no evident motor defects observed in the PC4f/f Nestin-Cre mice, because these mice could stay on the rotating rod for similar time as the PC4+/+ Nestin-Cre mice (Fig. 2d). Hence, although PC4 knock-out in the germline was lethal, knocking out PC4 in neural tissue resulted in smaller mice showing nocturnal hypoactivity without any motor defects.
FIGURE 1.
Strategy for knocking out PC4. a, strategy for creation of the knock-out mice. The second exon of the PC4 gene was floxed. b, representative images of 5′ arm validation of the targeted allele. The restriction enzymes KpnI and NheI were used, where digest products of 11.2 and 9.2 kb are expected with the targeted allele, and no product is expected in wild type. The clone numbers are indicated above as C1–C4; C3 and C4 are positive clones. The two panels are from different lanes of the same autoradiogram. c, 3′ arm validation of the targeted allele. The restriction enzymes AflII and HindIII digests were used, and products of 7.6 and 8 kb were expected with the targeted allele, and no product is expected in the wild type. Of the 372 clones tested, 5 clones were positive, of which C5 was used further for generation of the floxed mice. d, Western blotting of whole brain tissue lysates to examine PC4 levels in wild type mice expressing Cre (PC4+/+ Nestin-Cre) (control), heterozygous knock-out mice (PC4+/f Nestin-Cre), and homozygous knock-out mice (PC4f/f Nestin-Cre). β-Actin served as a loading control. e, immunohistochemistry showing the absence of PC4 protein in the brain of the PC4f/f Nestin-Cre mice; PC4+/+ Nestin-Cre mice served as controls. The hippocampus and dentate gyrus are magnified to show the absence of staining in the lower panels.
TABLE 1.
Offspring ratio of PC4 +/Δ crossings (Cre expressed in germline)
| Genotype | Expected | Observed at birth | Number of total progeny from multiple crosses |
|---|---|---|---|
| % | % | ||
| PC4+/+ | 25 | 54 | 15 |
| PC4+/Δ | 50 | 46 | 13 |
| PC4Δ/Δ | 25 | 0 | 0 |
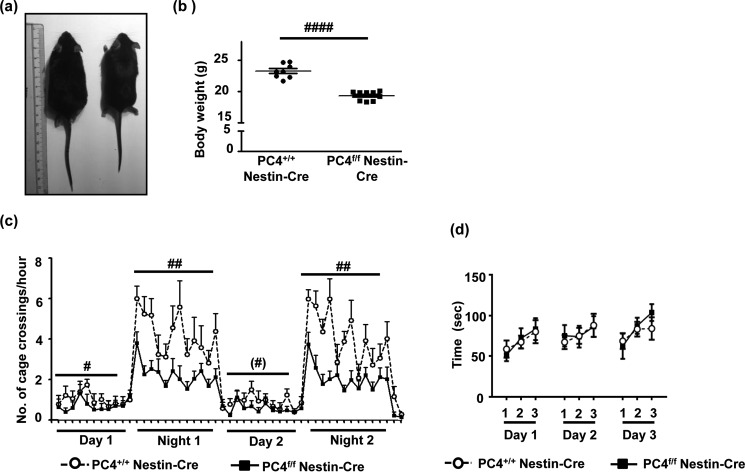
FIGURE 2.
Knocking out PC4 leads to hypoactivity with no motor dysfunction. a, comparison of overall size of PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice. The PC4f/f Nestin-Cre mice were visibly smaller in size that the PC4+/+ Nestin-Cre mice. b, average body weight of adult male mice in grams (n = 8; unpaired t test, p < 0. 0001). c, activity graph with the number of cage crossings/h plotted for 2 days and 2 nights. Statistical analyses revealed significant general hypoactivity for the PC4f/f Nestin-Cre mice compared with the PC4+/+ Nestin-Cre mice (night 1: F(1, 17) = 13.13, p = 0. 002; night 2: F(1,17) = 14. 99, p = 0. 0012; day 1: F(1,17) = 6.36, p = 0.021; day 2: F(1,17) = 3.95, p = 0.062). However, the PC4f/f Nestin-Cre mice followed the day-night cycle comparable with the PC4+/+ Nestin-Cre mice (day 1 versus night 1 t(9) = 8.65, p = 0.000012 and day 2 versus night 2 t(9) = 7.06, p = 0.000005). d, rotarod test showing the average time (s) before which the mice fell off the rotarod. No effect of genotype was observed (F(1,14) = 0.15, p = 0.70).
Knocking Out PC4 in the Brain Induces Morphological Changes and Ischemic Sensitivity in Neurons
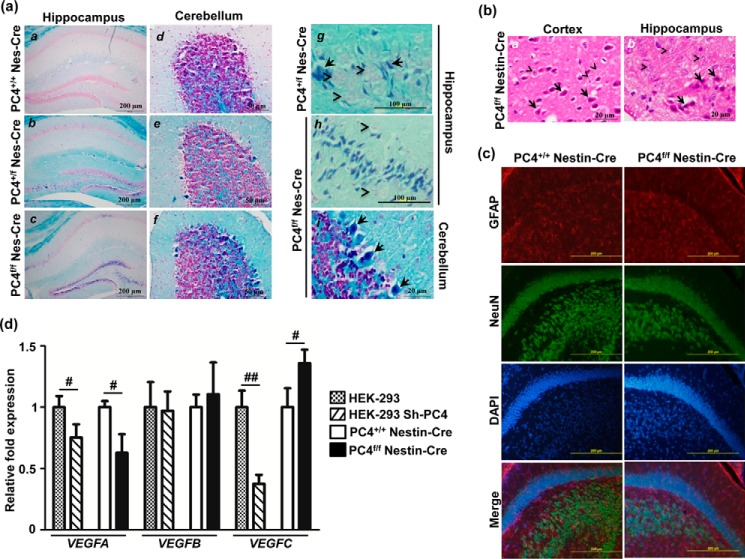
The brain was analyzed at the morphological level using specific staining procedures. Although the main structural domains of the brain appeared normal in the PC4f/f Nestin-Cre mice (Fig. 1e), staining of myelinated fibers with Luxol fast blue followed by nuclear red counter-staining revealed striking differences in various parts of the brain. In many structures, darkly stained neurons were observed in PC4f/fNestin-Cre mice, whereas they were uncommon in stained sections from the PC4+/+ Nestin-Cre mice brains (Fig. 3a). These neurons have a shrunken appearance and an increased affinity for stains, which is indicative of neuronal vulnerability resulting from an excitotoxic mechanism secondary to acute anoxia or hypoglycemia, especially when observed in clinical and experimental neuropathology (14). A closer examination of sections stained with hematoxylin/eosin (Fig. 3b) showed that the cytoplasm was markedly eosinophilic, with the nucleus appearing shrunken and darkly stained with condensed chromatin forming clumps, similar to what is observed in ischemic neurons (14). Among the brain structures, abnormal neurons were numerous in the hippocampus, in the inner layer of the dentate gyrus (Fig. 3a, panels b and c), and CA1 and CA3 neurons, especially in the homozygotes (Fig. 3a, panel c). Purkinje cells of the cerebellum were also often affected (Fig. 3a, panels e, f, and i). However, no obvious defects/changes were observed in the density/distribution of neurons and/or glia (Fig. 3c). Upon observing sections from heterozygotes, fewer darkly stained cells were observed, showing that a decrease in levels of PC4 also led to hypoxic conditions, although they were definitely much fewer in number than the knock-out mice (Fig. 3a, panels b, e, and g).
FIGURE 3.
PC4f/f Nestin-Cre mice show indications of hypoxia in the brain. a, Luxol fast blue staining of paraffin sections shows numerous hyperchromatic dark blue cells throughout the brain of PC4f/f Nestin-Cre mice, and fewer in the brain of PC4+/f Nestin-Cre mice, particularly in the hippocampus (panels a–c) and cerebellum (panels d–f). High magnification images of PC4+/f Nestin-Cre (panel g) and PC4f/f Nestin-Cre (panel h and i) mice are shown in the right column. Normal neurons (pink, arrowheads) can be observed. Dark blue-stained neurons (arrows) are few in the PC4+/f Nestin-Cre mice and numerous in PC4f/f Nestin-Cre mice in the dentate gyrus and CA3 of the hippocampus (panels b, c, g, and h), and in Purkinje cells from the cerebellum (panels e, f, and i). Scale bars, 200 μm in a–c; 50 μm in d–f; 100 μm in g and h; and 20 μm in i. b, hematoxylin/eosin staining showing eosinophilic neurons (arrows) compared with normal neurons (arrowheads) in the cortex and hippocampus of PC4f/f Nestin-Cre brains. Scale bars, 20 μm. c, immunofluorescence analysis of neuronal and glial markers, NeuN and GFAP, respectively, counterstained with DAPI; in PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice, hippocampal sections did not show any obvious change. Scale bars, 200 μm. d, quantitative RT-PCR analysis for indicators of hypoxia. Levels of VEGF isoforms in HEK-293 cells in the presence of normal and down-regulated levels of PC4 and in dorsal hippocampus of PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice were compared. mRNA levels were normalized to GAPDH levels and are represented as fold changes, with the value obtained with the control expressing normal PC4 levels set at 1. Significant changes of VEGFA and VEGFC were observed with very minimal effects on VEGFB (n = 3). #, p < 0.05; ##, p < 0.01. Student's t test p values for dorsal hippocampus were as follows: VEGFA, 0.0160; VEGFB, 0.5497; and VEGFC, 0.0312. Student's t test p values for stable PC4 knockdown HEK-293 cells were as follows: VEGFA, 0.0378; VEGFB, 0.8515; and VEGFC, 0.0022.
To further investigate whether the absence of PC4 in the brain results in hypoxia, the expression of downstream target genes of hypoxia-inducible factor-1α (HIF-1α),4 such as VEGF, were analyzed. In addition, the VEGFC isoform has recently been suggested to be up-regulated and serve as an adaptive response to ischemic and oxidative stress in the hippocampus, where it can have neurotrophic and neuroprotective effects (15–17). HIF-1α levels were not analyzed because HIF-1α has been shown in multiple earlier studies to be regulated at the level of protein stability in hypoxic conditions, rather than of gene expression (18, 19). Expression of the three VEGF isoforms was assessed in PC4f/f Nestin-Cre mice, as well as in stable PC4 knockdown HEK-293 cells (Fig. 3d). We observed an increase in the levels of VEGFC, and there was a significant reduction in the levels of VEGFA in the dorsal hippocampus of the knock-out mice, whereas VEGFB levels remained unchanged (Fig. 3d). However, VEGFC levels were not increased in PC4 knockdown HEK-293 cells (Fig. 3d), indicating a specificity for VEGF regulation in vivo that is likely to result from the ischemic conditions observed in the absence of PC4, especially in the brain where oxygen tension is critical.
Knocking Out PC4 in the Brain Affects the Number of Newly Generated Neurons in the Adult
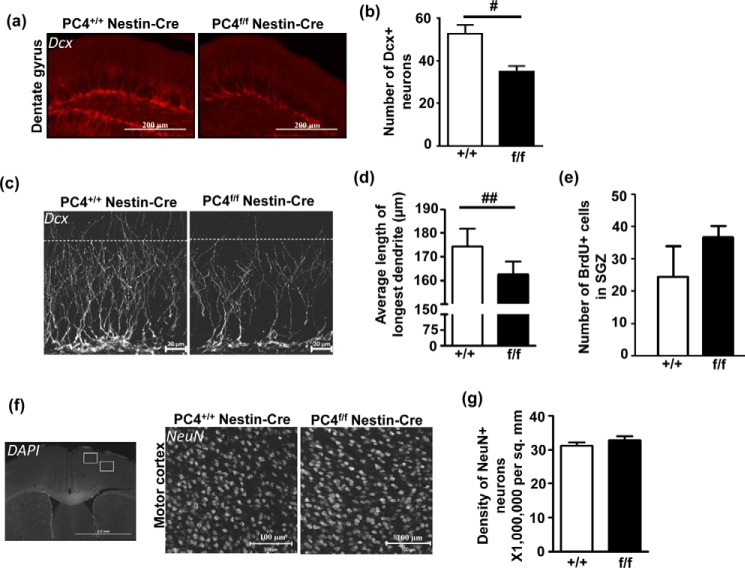
The subgranular zone (SGZ) of the dentate gyrus from the hippocampus is one of the primary sites for neurogenesis in the mammalian adult brain (20). Because darkly stained cells were located in the inner layer of the dentate gyrus, we investigated possible changes in adult neurogenesis by immunostaining for DCX (doublecortin), a microtubule-associated protein found exclusively in developing and immature neurons (21) (Fig. 4a). We found that the number of doublecortin-positive neurons was significantly reduced in the PC4f/f Nestin-Cre mice (Fig. 4b). In addition, we observed that the length of the dendrites from the newly generated neurons present in the middle upper part of the hippocampus showed a small but significant decrease in the PC4f/f Nestin-Cre compared with control mice (Fig. 4, c and d). This decreased number of DCX-positive cells could result from a reduced generation of new neurons because of the PC4 knock-out or from decreased cell survival (or increased death) because of the large number of hypoxic cells observed in the inner layer of the dentate gyrus (Fig. 3, b and c). Mice from both genotypes were injected with BrdU and killed 1 h later to measure the exact number of rapidly proliferating BrdU-positive cells produced at that time (22) (Fig. 4e). However, the number of BrdU+ cells was not significantly different between the genotypes, indicating that PC4 does not have an effect on the proliferation step of adult neurogenesis. Lastly, although no major alteration in brain structures was observed (e.g. hippocampal formation; Figs. 1e and 2a), we also verified whether knocking out PC4 could have impacted embryonic neurogenesis. The density of cortical neurons in the motor cortical region of adult mice ultimately reflects the number of viable cortical progenitors produced during development. Such counting did not reveal a significant difference in the density of NeuN-positive mature neurons in this region (Fig. 4, f and g), suggesting that there was no major impairment of embryonic neurodevelopment. Thus, the decrease in number of newly generated doublecortin-positive neurons does not seem to result from a direct effect of PC4 on proliferation but rather from a secondary effect of PC4 knock-out, such as increased hypoxic conditions.
FIGURE 4.
Absence of PC4 results in altered neurogenesis. a, a typical example of the immunofluorescence analysis of Dcx (doublecortin) in cryosections. Scale bars, 200 μm. b, the number of Dcx-positive neurons in the dentate gyrus was counted in several sections per animal (n = 3; p < 0.05), and a significant decrease was observed in the number of Dcx-positive neurons in PC4f/f Nestin-Cre mice. c, typical magnified images of Dcx-stained sections showing the reduced number of neurons. The dotted line indicates a length of 100 μm from the SGZ, and most dendrites were restricted to this length in the PC4f/f Nestin-Cre mice, whereas visibly more dendrites were longer than 100 μm in the PC4+/+ Nestin-Cre mice. Scale bars, 20 μm. d, measuring the length of the longest Dcx-positive dendrite in the hippocampus showed that this length was significantly lesser in the PC4f/f Nestin-Cre mice compared with the PC4+/+ Nestin-Cre mice (PC4+/+ Nestin-Cre: 174.4 μm versus PC4f/f Nestin-Cre: 162.4 μm; n = 3; p < 0.01). e, effect of PC4 knock-out on proliferation in the SGZ after BrdU injection (100 mg/kg). Control and PC4 knock-out mice were injected with BrdU 1 h before they were sacrificed. The numbers of BrdU-positive cells in the SGZ were not statistically different between the two genotypes (n = 2–3; 4 sections/animal). f, immunofluorescence analysis of NeuN (right panel), counterstained with DAPI (left panel) in cryosections of PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice showing density of neurons in the cortical regions (white boxes depict M1 and M2 motor cortical regions). Representative images showing NeuN-positive cells in the motor M1 cortical region are shown for both genotypes (right panel). Scale bars, 100 μm. g, the number of NeuN-positive cells in the motor cortical region was counted, and the density of neurons was seen to be similar in the PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice (n = 3; 4–5 sections/animal).
Absence of PC4 in the Brain Does Not Impact Spatial Memory Formation but Prevents Its Extinction
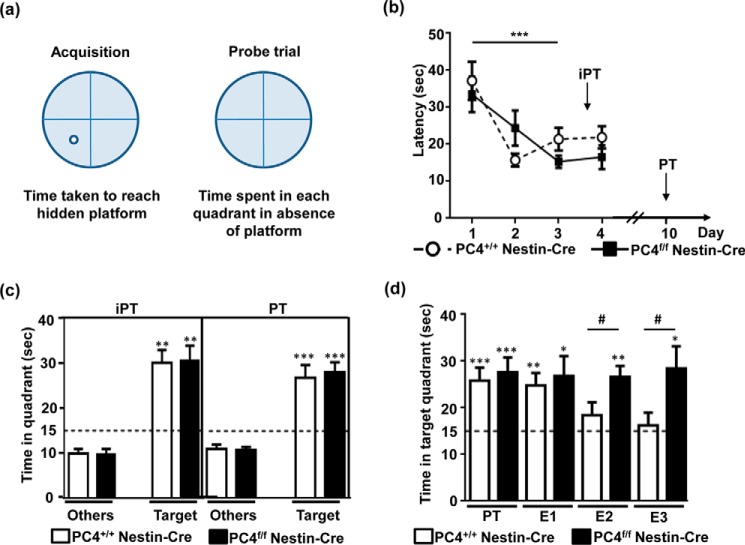
The hippocampus plays a major role in memory formation, and newly generated hippocampal neurons have been implicated in recent and long term spatial memory formation (23). We thus tested spatial memory in PC4f/f Nestin-Cre mice, using the Morris water maze (Fig. 5a). This test consists of a circular pool divided into 4 virtual quadrants surrounded by spatial cues, where an escape platform is hidden below the surface of opaque water. The mice were trained for 4 consecutive days with four trials/day to learn the platform location. We observed that the acquisition was comparable in PC4f/f Nestin-Cre and PC4+/+ Nestin-Cre mice, with both requiring less time to reach the platform over the days. On the third day, both strains reached a plateau, showing that the PC4f/f Nestin-Cre mice did not display any learning defects (Fig. 5b).
FIGURE 5.
Altered memory extinction in the absence of PC4 in the brain. a, scheme depicting the Morris water maze experiment. b, the two mice strains (n = 9/group) were trained using a Morris water maze for 4 days. Latencies to reach the platform are represented in seconds. Acquisition in the Morris water maze was comparable between PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice, with both strains learning significantly during the 4 days (days effect: F(3,48) = 10.273, p < 0.001; genotype effect: F(1,16) = 0.341, p = 0.56). c, Morris water maze probe trials show that memory retention was comparable between PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice 24 h after the 3rd day of acquisition in an intermediate probe trial (iPT) and 6 days after a supplemental 4th day of acquisition (PT). The time spent in the target quadrant (Target) is represented and compared with the average time spent in the three other quadrants (Others). The chance level (15 s in one of the four quadrants in a span of 1 min) is represented by a dashed line. The time spent in the target quadrant was significantly longer than 15 s (intermediate PT, p < 0.01; PT, p < 0.001) and longer than the time spent in the other quadrants (p < 0.001). d, extinction tests were performed at 2-h intervals after the PT. Whereas the PC4+/+ Nestin-Cre mice showed a decrease in the time spent in the target quadrant, no decrease was observed in the PC4f/f Nestin-Cre mice. A significant difference between PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice was observed for extinction trial 2 (#, p < 0.05) and extinction trial 3 (#, p < 0.01). For PC4f/f Nestin-Cre mice, all extinction values were significantly greater than 15 s (*, p < 0.05; **, p < 0.01).
Memory retention was tested in two different probe trials (PT), in which the mice were left in the water for 60 s in the absence of the platform, and the time spent in each of the four virtual quadrants of the pool was quantified. We first tested memory retention in a probe trial on day 4 (i.e. 24 h after the third day of acquisition) to assess recent memory (intermediate PT or intermediate probe trial). This was followed by additional training to avoid extinction. We then tested the mice in another PT, which was performed on Day 10 (6 days after the last day of training to assess memory persistence). In both probe trials, the PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice spent a comparable amount of time (∼30 s) in the target quadrant (where the platform was located during training), which was significantly higher than chance (15 s; Fig. 5c). This suggests that the acquisition, consolidation, and retention of recent memory, as well as memory persistence, were not affected in the PC4f/f Nestin-Cre mice.
The PC4f/f Nestin-Cre mice were then subjected to several trials of extinction. To this end, both groups of mice were tested repeatedly (at 2-h intervals) as was done for the probe trial to assess extinction processes. Although the PC4+/+ Nestin-Cre mice realized that the platform was no longer in place, they started swimming more randomly in the pool (spending about 15 s in each quadrant) as early as in extinction trial 2 (Fig. 5d). However, the most striking observation was that the PC4f/f Nestin-Cre mice were unable to extinguish their memory (Fig. 5d) and still spent a significantly longer time in the target quadrant, even after the third extinction trial, showing that the PC4f/f Nestin-Cre mice were indeed strongly deficient in memory extinction.
PC4 Regulates Gene Expression Pattern in the Hippocampus through Direct and Indirect Mechanisms
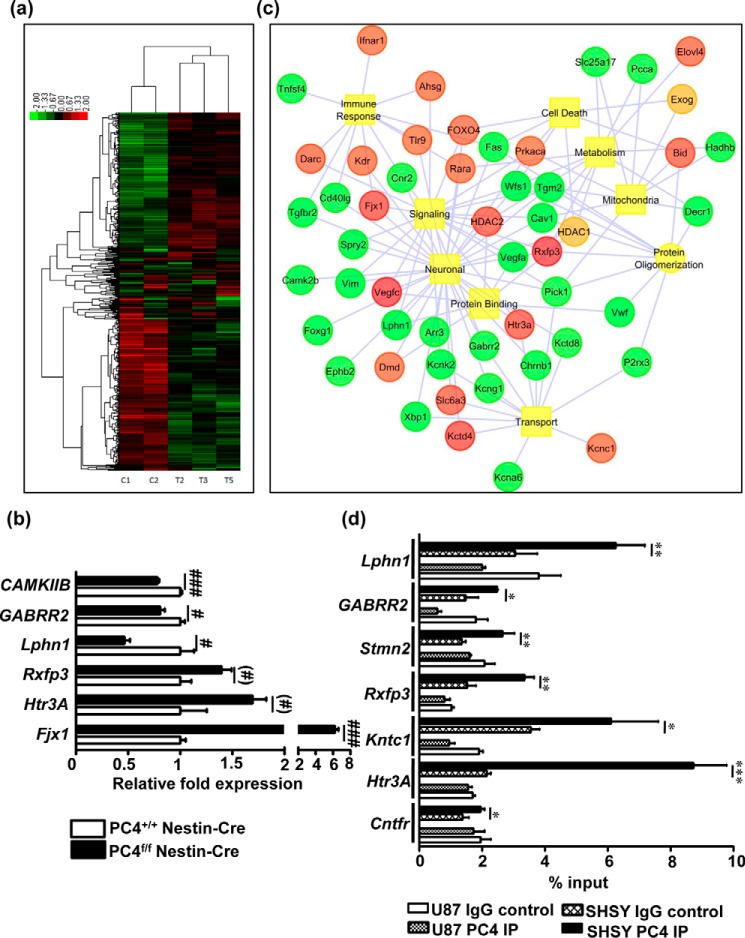
To gain further insight into the molecular basis of these observations, a comparative gene expression analysis was performed with the dorsal hippocampi isolated from the PC4f/f Nestin-Cre mice and the PC4+/+ Nestin-Cre mice (Fig. 6a). Although PC4 is a global chromatin organizer, a specific subset of 377 genes was found to be significantly down-regulated, and 347 were significantly up-regulated (p < 0. 05). A few genes that were highly dysregulated in the microarray data were chosen for validation (Fig. 6b). Some of these genes are known to be involved in neural function, either generally (such as the serotonin receptor 3A (Htr3a) and the rho2 subunit of the GABA C receptor (Gabrr2)) or specifically (such as the calcium calmodulin kinase 2b (Camk2b), which was recently shown to remove unwanted synapses together with Arc (24), and the rodent four-jointed ortholog (Fjx1), which inhibits dendritic extension (25) (Fig. 6b)). The expression of these genes followed the pattern observed in the microarray analysis.
FIGURE 6.
PC4 directly regulates gene expression patterns in the mouse hippocampus. a, heat map of the gene expression analysis performed using RNA extracted from the dorsal hippocampus of PC4+/+ Nestin-Cre and PC4f/f Nestin-Cre mice. b, quantitative RT-PCR validation of a few relevant genes that were up-regulated or down-regulated upon PC4 knock-out (n = 3/group). mRNA levels were normalized to GAPDH levels and are represented as fold inductions, with the value obtained with the PC4+/+ Nestin-Cre mice set at 1 (t test; #, p < 0.05; ##, p < 0.01; ###, p < 0.001). c, network map of the genes dysregulated upon PC4 knock-out classified according to GO category. d, ChIP performed with U87 gliobastoma and SHSY-5Y neuroblastoma cells. Specific promoters of genes that were highly dysregulated in the gene expression analysis were evaluated by quantitative PCR. Quantification of immunoprecipitated DNA was represented relative to the input DNA (n = 3; *, p < 0.05; **, p < 0.01, ***, p < 0.001).
Interestingly, many potassium channel-related members were significantly altered, suggesting possible damage to neuronal excitability and/or neuronal/glial interactions (Fig. 6b and Table 2). Likewise, a series of mitochondrial NADH deshydrogenase enzymes were found to be dysregulated, suggesting potential perturbation of oxidative phosphorylation (Table 2). We further performed functional analyses using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt, GSAT) (p < 0.05, FC > 1.10). Strikingly, phenotype analyses highlighted abnormal synaptic plasticity in the down-regulated genes (Table 3) and hippocampus-related morphological abnormalities in the up-regulated genes (Table 3). Wikipathways and analysis by Kyoto Encyclopedia of Genes and Genomes also highlighted a significant down-regulation of plasticity-related pathways, including the MAPK pathways, pathways related to plasticity for drug addiction, and oxidative phosphorylation pathways, whereas the VEGF and apoptosis signaling pathways were significantly up-regulated (Table 3). A network map connecting the gene ontology class with the relevant dysregulated genes was generated (Fig. 6c). This network mapped a significant set of molecules involved in multiple pathways, including neuronal signaling, immune response, transport, mitochondria, protein binding and oligomerization, metabolism, and cell death.
TABLE 2.
List of genes in the microarray analyses
List of genes related to potassium channels and mitochondrial NADH deshydrogenase enzymes, that were found significantly up- or down-regulated (p < 0.05) in the microarray analyses. Gene ID, symbol, and absolute fold change are given.
| Gene ID | Symbol | Absolute fold change | p value | Regulated |
|---|---|---|---|---|
| Potassium channel-related genes | ||||
| 16506 | Kcnd1 | 1.14 | 0.03 | Up |
| 67516 | Kctd4 | 1.41 | 0.01 | Up |
| 16494 | Kcna6 | 1.12 | 0.03 | Down |
| 241794 | Kcng1 | 1.1 | 0.02 | Down |
| 16520 | Kcnj4 | 1.37 | 0.04 | Down |
| 16526 | Kcnk2 | 1.22 | 0.01 | Down |
| 243043 | Kctd8 | 1.11 | 0.04 | Down |
| Mitochondria-related genes | ||||
| 66091 | Ndufa3 | 1.13 | 0.04 | Up |
| 70316 | Ndufab1 | 1.12 | 0.04 | Up |
| 68342 | Ndufb10 | 1.21 | 0.05 | Down |
| 67264 | Ndufb8 | 1.1 | 0.01 | Down |
| 66218 | Ndufb9 | 1.27 | 0.04 | Down |
TABLE 3.
Functional analyses with WebGestalt
Functional analyses using WEB-based GEne SeT AnaLysis Toolkit (WebGestalt, GSAT) performed with either down- or up-regulated gene lists, showing significant regulation (Adj_p value<0.05, represented as −log10) for Phenotype analysis (A) and Wikipathways/KEEG pathways (B).

To examine the role of PC4 in the expression of these genes, a chromatin immunoprecipitation was performed using U87 glioblastoma and SHSY-5Y neuroblastoma cells. A significant enrichment in the binding of PC4 was seen on all the selected gene promoters in SHSY-5Y neuroblastoma cells, but not in U87 cells (Fig. 6d). Among these genes, although some genes showed down-regulation (Cntfr, Lphn1, and Gabrr2) in the gene expression analysis, some were up-regulated (Htr3a, Rxfp3, Stmn2, and Kntc1) upon PC4 knock-out.
Discussion
Gene expression is regulated by chromatin organization in the eukaryotic nucleus, and non-histone chromatin-associated proteins act to fine-tune chromatin states, among other factors. Our knowledge of the role of PC4/Sub1, a highly and ubiquitously expressed chromatin-associated multifunctional protein, in organ-specific development is rather limited; thus, this study is the first in which a conditional knock-out of PC4 in the brain has been generated, because a complete PC4 knock-out in the germline was lethal. Here, we show that specific PC4 knock-out in the brain leads to abnormal phenotypes related to neuronal morphology, sensitivity, plasticity, and memory. Indeed, compared with the PC4+/+ Nestin-Cre mice, the PC4f/f Nestin-Cre mice showed no evident brain anatomical abnormalities but exhibited alteration in sensitivity to hypoxia/anoxia, decreased number of newly generated neurons in the hippocampus, and strong deficits in spatial memory extinction. A dysregulation of a series of genes implicated in different neural processes was also observed, some of which were under the direct regulation of PC4.
The mouse gene expression database shows that PC4 expression begins early during embryonic development in different tissues. We have observed that a complete knock-out of PC4 leads to embryonic lethality in mice, whereas heterozygotes with just one functional copy of the gene were normal and fertile, showing that although mice can survive haploinsufficiency, complete absence of PC4 leads to death in utero. Young adult PC4f/f Nestin-Cre mice were smaller in size and lighter compared with the PC4+/+ Nestin-Cre mice; this phenomenon is observed often in transgenic mice (26). Although motricity was unaffected, the PC4f/f Nestin-Cre mice showed significantly less nocturnal activity despite exhibiting fast, darting movements.
At the structural level, though there were no gross changes in the brain structures of the PC4f/f Nestin-Cre mice compared with the PC4+/+ Nestin-Cre mice, certain cells in the PC4f/f Nestin-Cre mice brain showed strong affinity for Luxol fast blue/nuclear red. Further, hematoxylin/eosin staining supported the notion that such cells are usually neurons that show increased vulnerability to acute anoxia/hypoxia or hypoglycemia (14). It is noteworthy that the levels of VEGFC, which has neurotrophic and neuroprotective effects, were increased significantly, probably as an adaptive response to overcome the effects of hypoxia (17). Regarding the function of the hippocampus, we noticed inhibition of adult neurogenesis with a decreased number of newly generated doublecortin-positive neurons (Fig. 4, a–d) in the SGZ, a region mostly represented by hypoxic cells in the PC4 knock-out mice (Fig. 3, a–c). Overall, the proliferation of progenitors was not impaired in PC4 knock-out mice, despite the hypoxic conditions in the hippocampus. Interestingly, it has been recently shown that low oxygen environment supports the early precursors, whereas surrounding higher oxygen levels can be toxic to those cells that start becoming neurons (27). Precursors of new neurons have to undergo oxidative damage when becoming mature neurons, leaving the SGZ, and only a proportion of them ultimately reach this differentiated state (27). Thus, it is likely that new neurons from PC4 knock-out mice may be more sensitive to this oxidative damage during maturation, despite induction of the VEGF pathway. Additionally, VEGFA, which is secreted by stem cells to maintain the stem cell niche (28, 29), was significantly reduced. This reduction may also account for the decrease in neurogenesis observed. Along this line, many isoforms of mitochondrial NADH dehydrogenase enzymes and oxidative phosphorylation pathway were dysregulated, suggesting a possible defect in the respiratory chain, energy production, and/or intracellular calcium homeostasis regulation, which could ultimately lead to mitochondrial damage and cell death. Lastly, several genes related to apoptosis and inflammation were altered in our microarray analyses (Table 3). These alterations indicate a possibility of higher rate of progenitor death in the dentate gyrus under oxidative conditions. Taken together, these data suggest that PC4 may play an important role in the regulation of hypoxia/anoxia and cell death.
When exploring hippocampus-dependent memory functions, no impairments were found in the processes of learning, consolidation or retention of recent, and persistent spatial memory, because the PC4f/f Nestin-Cre mice were able to learn and remember the location of the hidden platform even 6 days after training. However, the memory extinction of the PC4f/f Nestin-Cre mice was severely impaired. Even upon repeated introduction into the pool, these mice spent a considerable period of time in the target quadrant despite the absence of the platform, whereas the PC4+/+ Nestin-Cre mice eventually spent an approximately average time (15 s) in each quadrant. Brain regions underlying memory extinction have mostly been studied in the fear memory paradigm and have established that the prefrontal cortex is required for consolidation of extinction (30, 31). Fear extinction requires long term synaptic changes in the infralimbic prefrontal cortex that are mediated by a variety of mechanisms, including NMDA receptor-dependent plasticity (32, 33). In addition, a few reports have described the role of factors involved in extinction processes, such as CAMKIIα (34–36). GSAT analysis revealed abnormal synaptic plasticity as a significant phenotype, and genes belonging to the CAMKII family, NMDA receptors, and MAPK kinase pathway were all significantly down-regulated in our microarray study (Fig. 6, b and c, and Table 3). VEGFA, which was significantly down-regulated in the absence of PC4 (Fig. 3d), has also been shown to have an effect on neuronal plasticity and memory. Of note, the serotonin receptor, Htr3A, which was up-regulated and showed a strong enrichment of PC4 binding on its promoter (Fig. 6, b–d), has been recently associated with fear memory extinction (37, 38). Furthermore, the hippocampus is among the primary areas of the brain that are particularly susceptible to injury because of inadequate availability of oxygen/glucose. The defect in the memory extinction ability of the PC4f/f Nestin-Cre mice might also be a reflection of the sensitivity of the hippocampus to such conditions.
Finally, we believe that no major impairment occurred during embryonic neurodevelopment in the PC4f/f Nestin-Cre mice, because we found that the density of cortical neurons in motor regions (neurons originating from the same number of progenitors produced during development) was similar between the control and knock-out mice (Fig. 4, f and g). We cannot exclude, however, that improper network connections have been established, which could ultimately alter the extinction network and/or process. Of note, Four-jointed ortholog Fjx1 levels were significantly increased in the PC4f/f Nestin-Cre mouse brain compared with the PC4+/+Nestin-Cre mouse brain (Fig. 6b). Because Fjx1 is known to be a regulator of dendrite branching and extension in the hippocampus (25), this abnormal increase may result in altered or defective network connections. However, the phenotype observed in the PC4f/f Nestin-Cre mice could be due to changes during development or due to changes in the adult (or both), which only the generation of conditional adult PC4 knock-out mouse will be able to conclusively decipher.
Lastly, the presence of PC4 on all the tested gene promoters in SHSY-5Y cells indicates a strong and specific involvement of PC4 in regulating gene expression in neuronal cells. The presence of PC4 on both down-regulated and up-regulated gene promoters indicates the possible association of PC4 with different protein complexes to mediate gene transcription or repression, because PC4 is known to be a transcriptional coactivator and also associates with heterochromatin proteins (10) to compact chromatin acting as a chromatin organizer.
In conclusion, this work highlights for the first time the importance of non-histone chromatin proteins in brain function and behavior, by revealing that the abundant nuclear protein PC4 plays a significant role in response to hypoxia, memory extinction, and adult neurogenesis, by regulating a small subset of genes associated with brain plasticity in an organismal context. In addition, this study also gives a gene expression profile of a mouse strain defective in memory extinction, the molecular mechanisms of which are still not clearly understood. Because memory extinction plays an important role in replacing original memories with more relevant ones after new experiences, especially in conditions like anxiety and spatial changes, the genes and proteins described in this study could contribute to a further understanding of the phenomenon.
Experimental Procedures
Mouse Maintenance and Generation of Transgenic Mice
C57BL6 mice were maintained in compliance with international laws and guidelines under a 12-h light/dark cycle with controlled temperature (22 ± 2 °C) and humidity. Transgenic mice with a floxed exon of PC4 were generated by the Mouse Clinical Institute (Strasbourg, France). For the germline knock-out, PC4f/f mice were crossed with mice that express Cre in the germline (11). In subsequent crosses, heterozygotes were crossed to obtain homozygous PC4−/− mice lacking PC4 because of a deletion in the germline. Similarly, the generation of the conditional brain-specific knock-out was performed using Nestin-Cre (FT) mice, in which Cre is expressed specifically in neuronal and glial precursors beginning in early developmental stages (12). 10–12-week-old adult male mice were used for all the behavioral tests after blinding the experimenter to the genotype.
Western Blotting, Immunohistology, and Immunofluorescence
Western blotting, immunohistology, and immunofluorescence analyses were performed using routine procedures as described in Ref. 39. For the BrdU labeling, a single dose of BrdU (100 mg/kg) was injected in 2-month-old mice for 1 h before sacrifice. An average of 35 cryosections (40 mm thick) were made in the dorsal hippocampus between Bregma −1.06 and −2.46, and sections were then processed for immunostaining.
Actigraphy
The mice were placed individually in transparent cages and adapted to the shelves of the testing device. Two infrared light beams passing through each cage were targeted on two photocells that were placed 2.5 cm above the cage floor level and 28 cm apart. The number of cage crossings was recorded by a computer over 2 day-night cycles.
Rotarod Test
Each mouse was placed on a rotating rod, and the speed of the rod could be increased. On day 1 of the test, the mice were allowed to habituate to the task, and the speed of the rod was kept constant at 4 rpm. Beginning on day 2, the speed of the rod was increased from 4 to 40 rpm. The mouse had to keep up with the increasing speed until it could no longer stay on the rod. The latency before the fall was measured over 3 consecutive days, with three trials on each day. The last day of training is considered as the test day, and the statistical analyses were performed on this day. Following the test, the rod was cleaned well before introducing the next mouse.
Morris Water Maze
A Morris water maze test was performed as described in Ref. 39. The acquisition was performed over 4 days. two probe trials, the first on day 4 before the start of the 4th day training sessions and the second on day 10, were performed. Three extinction trials were performed on day 10 at 2-h intervals. Acquisition was analyzed statistically using two-way analysis of variance with “day” and “genotype” as variables, and probe trial performance was analyzed using t test.
RNA Isolation, Microarray, Data Analysis, and Quantitative RT-PCR Analysis
RNA isolation, microarray, data analysis, and quantitative RT-PCR analysis were performed as described in Ref. 40. The primers used are as follows: CAMK2B, forward, 5′-CGTTTCACCGACGAGTACCAG-3′, and reverse, 5′-GCGTACAATGTTGGAATGCTTC-3′; GABRR2, forward, 5′-ATGCCTTATTTGATGAGACTCGC-3′, and reverse, 5′-CCACACCTACAGGGATGGC-3′; Lphn1, forward, 5′-ACCCTTTCCAGATGGAGAATGTG-3′, and reverse, 5′-GGGACACAGTCGTACTGCAC-3′; Rxfp3, forward, 5′-TCCTCATCAGCGCGGTTTAC-3′, and reverse, 5′-CAGTGCCAGGTTAGTGACAAAG-3′; Fjx1, forward, 5′-ATCCTCTTCGATTACCTGACGG-3′, and reverse, 5′-TTGTCCAGAAAGACCAACGCC-3′; Htr3A, forward, 5′-CCTGGCTAACTACAAGAAGGGG-3′, and reverse, 5′-TGCAGAAACTCATCAGTCCAGTA-3′; VEGFA, forward, 5′AGGGCAGAATCATCACGAAGTGGTG-3′, and reverse, 5′-GTGGGCACACACTCCAGGCCC-3′; VEGFB, forward, 5′-CCTGACGATGGCCTGGAGTGTG-3′, and reverse, 5′-CCTGGGGCTGTCTGGCTTCAC-3′; and VEGFC, forward, 5′-GGTGTGTATAGATGTGGGGAAGG-3′, and reverse, 5′-CGGCAGGAAGTGTGATTGG-3′.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed using SHSY-5Y and U87 cells from ATCC, using the protocol described in Ref. 41. The primers used are as follows: GABRR2, forward, 5′-GTCTGCTGAGGTCCAAAGGATC-3′, and reverse, 5′-ACCAGGTCAGAGAGGGGCATG-3′; Lphn1, forward, 5′-AGTCGCCGCCCGGGACTGGCC-3′, and reverse, 5′-CCTTATACTTCACTGGAAAATCC-3′; Rxfp3, forward, 5′-CTGCTGAAACTCTGGAGAGG-3′, and reverse, 5′-GGCTCTCACCAGTTACGGTG-3′; Cntfr, forward, 5′-GTCCTAAGGTGTCCTTGGACC-3′, and reverse, 5′-GGCTCCTTTTAGGATGGAGGC-3′; Htr3A, forward, 5′-ACCCCTTGGAAAGATGCTGG-3′, and reverse, 5′-CAACCGGCCAGTTAGACAGTA-3′; Stmn2, forward, 5′-GACGCCTAAGTCCAGCTGTGC-3′, and reverse, 5′-TTGAGAAGTCCTCACTATATG-3′; and Kntc1, forward, 5′-CAGTGAGGCGAAAGGAACGTAG-3′, and reverse, 5′-CGGCTCCCAGAACCTTGTTCG-3′.
Author Contributions
T. K. K. designed and initiated the project. A. S., H. D., S. K., P. B., and T. K. K. were involved in designing the knock-out strategy. A. S., H. D., and N. M. performed genotyping analyses and initial characterization of the knock-out. A. S., L. B.-D., H. D., and B. C. carried out the sectioning. H. D., F. M., B. L., and A.-L. B. performed the immunostaining and analysis. A.-L. B., J.-C. C., P. B., and T. K. K. designed the behavioral experiments, which were performed by A. S., S. C., and R. C. A.-L. B., J.-C. C., and R. C. analyzed the behavioral experiments. A. S. performed the gene expression and chromatin immunoprecipitation analyses. All authors approved the final version of the manuscript.
Acknowledgments
We thank the Plateau de Biologie Expérimentale de la Souris (PBES, UMS3444/US8) for help in mice breeding. We are grateful to Madavan Vasudevan (Bionivid Technologies Pvt. Ltd., Bangalore, India) for fruitful discussions on analyzing the gene expression data. We thank B. S. Suma, Confocal Facility, JNCASR for technical help.
This work was supported by the Charpak fellowship from the French Embassy in India, Region Rhône-Alpes COOPERA 2012 Grant 12 004948 01, the Department of Science and Technology, Government of India through a Sir J. C. Bose Fellowship (to T. K. K.), Council of Scientific and Industrial Research Senior Research Fellow funding (to A. S.), Department of Biotechnology Grant BT/01/CEIB/10/III/01 (to T. K. K.), and CNRS-International Programs for Scientific Cooperation Grant 6449 (to P. B.). Work performed in the laboratory of A.-L. B. was supported by CNRS, Université de Strasbourg, Grant ANR-12-MALZ-0002-01, IFCPAR Grant 4803-3, and funds from the Alsace Alzheimer 67 Association. The authors declare that they have no conflicts of interest with the contents of this article.
- HIF
- hypoxia-inducible factor
- SGZ
- subgranular zone
- PT
- probe trial.
References
- 1. Singh P. B. (2010) HP1 proteins: what is the essential interaction? Genetika 46, 1424–1429 [PubMed] [Google Scholar]
- 2. Hock R., Furusawa T., Ueda T., and Bustin M. (2007) HMG chromosomal proteins in development and disease. Trends Cell Biol. 17, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Z. Q., Auer B., Stingl L., Berghammer H., Haidacher D., Schweiger M., and Wagner E. F. (1995) Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9, 509–520 [DOI] [PubMed] [Google Scholar]
- 4. Ménissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Amé J. C., Dierich A., LeMeur M., Sabatier L., Chambon P., and de Murcia G. (2003) Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22, 2255–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guy J., Cheval H., Selfridge J., and Bird A. (2011) The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 27, 631–652 [DOI] [PubMed] [Google Scholar]
- 6. Ge H., and Roeder R. G. (1994) Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78, 513–523 [DOI] [PubMed] [Google Scholar]
- 7. Kretzschmar M., Kaiser K., Lottspeich F., and Meisterernst M. (1994) A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 78, 525–534 [DOI] [PubMed] [Google Scholar]
- 8. Conesa C., and Acker J. (2010) Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol. 7, 287–290 [DOI] [PubMed] [Google Scholar]
- 9. Das C., Hizume K., Batta K., Kumar B. R., Gadad S. S., Ganguly S., Lorain S., Verreault A., Sadhale P. P., Takeyasu K., and Kundu T. K. (2006) Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol. Cell Biol. 26, 8303–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Das C., Gadad S. S., and Kundu T. K. (2010) Human positive coactivator 4 controls heterochromatinization and silencing of neural gene expression by interacting with REST/NRSF and CoREST. J. Mol. Biol. 397, 1–12 [DOI] [PubMed] [Google Scholar]
- 11. Dubois N. C., Hofmann D., Kaloulis K., Bishop J. M., and Trumpp A. (2006) Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis 44, 355–360 [DOI] [PubMed] [Google Scholar]
- 12. Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R., and Schütz G. (1999) Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 [DOI] [PubMed] [Google Scholar]
- 13. Brooks S. P., and Dunnett S. B. (2009) Tests to assess motor phenotype in mice: a user's guide. Nat. Rev. Neurosci. 10, 519–529 [DOI] [PubMed] [Google Scholar]
- 14. Auer R. N., and Sutherland G. R. (2002) Hypoxia and related conditions. In Greenfield's Neuropathology (Graham D. I., and Lantos P. L., eds.) 7th Ed., Arnold, London [Google Scholar]
- 15. Bhuiyan M. I., Kim J. C., Hwang S. N., Lee M. Y., and Kim S. Y. (2015) Ischemic tolerance is associated with VEGF-C and VEGFR-3 signaling in the mouse hippocampus. Neuroscience 290, 90–102 [DOI] [PubMed] [Google Scholar]
- 16. Le Bras B., Barallobre M. J., Homman-Ludiye J., Ny A., Wyns S., Tammela T., Haiko P., Karkkainen M. J., Yuan L., Muriel M. P., Chatzopoulou E., Bréant C., Zalc B., Carmeliet P., Alitalo K., et al. (2006) VEGF-C is a trophic factor for neural progenitors in the vertebrate embryonic brain. Nat. Neurosci. 9, 340–348 [DOI] [PubMed] [Google Scholar]
- 17. Wang J. F., Zhang X., and Groopman J. E. (2004) Activation of vascular endothelial growth factor receptor-3 and its downstream signaling promote cell survival under oxidative stress. J. Biol. Chem. 279, 27088–27097 [DOI] [PubMed] [Google Scholar]
- 18. Conde E., Alegre L., Blanco-Sánchez I., Sáenz-Morales D., Aguado-Fraile E., Ponte B., Ramos E., Sáiz A., Jiménez C., Ordoñez A., López-Cabrera M., del Peso L., de Landázuri M. O., Liaño F., Selgas R., et al. (2012) Hypoxia inducible factor 1-α (HIF-1α) is induced during reperfusion after renal ischemia and is critical for proximal tubule cell survival. PLoS One 7, e33258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Powell J. D., Elshtein R., Forest D. J., and Palladino M. A. (2002) Stimulation of hypoxia-inducible factor-1α (HIF-1α) protein in the adult rat testis following ischemic injury occurs without an increase in HIF-1α messenger RNA expression. Biol. Reprod. 67, 995–1002 [DOI] [PubMed] [Google Scholar]
- 20. Ming G. L., and Song H. (2011) Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown J. P., Couillard-Després S., Cooper-Kuhn C. M., Winkler J., Aigner L., and Kuhn H. G. (2003) Transient expression of doublecortin during adult neurogenesis. J. Comp. Neurol. 467, 1–10 [DOI] [PubMed] [Google Scholar]
- 22. Belvindrah R., Rougon G., and Chazal G. (2002) Increased neurogenesis in adult mCD24-deficient mice. J. Neurosci. 22, 3594–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodman T., Trouche S., Massou I., Verret L., Zerwas M., Roullet P., and Rampon C. (2010) Young hippocampal neurons are critical for recent and remote spatial memory in adult mice. Neuroscience 171, 769–778 [DOI] [PubMed] [Google Scholar]
- 24. Okuno H., Akashi K., Ishii Y., Yagishita-Kyo N., Suzuki K., Nonaka M., Kawashima T., Fujii H., Takemoto-Kimura S., Abe M., Natsume R., Chowdhury S., Sakimura K., Worley P. F., and Bito H. (2012) Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell 149, 886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Probst B., Rock R., Gessler M., Vortkamp A., and Püschel A. W. (2007) The rodent Four-jointed ortholog Fjx1 regulates dendrite extension. Dev. Biol. 312, 461–470 [DOI] [PubMed] [Google Scholar]
- 26. Reed D. R., Lawler M. P., and Tordoff M. G. (2008) Reduced body weight is a common effect of gene knockout in mice. BMC Genet. 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chatzi C., Schnell E., and Westbrook G. L. (2015) Localized hypoxia within the subgranular zone determines the early survival of newborn hippocampal granule cells. Elife 4, e08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jin K., Zhu Y., Sun Y., Mao X. O., Xie L., and Greenberg D. A. (2002) Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 99, 11946–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirby E. D., Kuwahara A. A., Messer R. L., and Wyss-Coray T. (2015) Adult hippocampal neural stem and progenitor cells regulate the neurogenic niche by secreting VEGF. Proc. Natl. Acad. Sci. U.S.A. 112, 4128–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgan M. A., Romanski L. M., and LeDoux J. E. (1993) Extinction of emotional learning: contribution of medial prefrontal cortex. Neurosci. Lett. 163, 109–113 [DOI] [PubMed] [Google Scholar]
- 31. Marek R., Strobel C., Bredy T. W., and Sah P. (2013) The amygdala and medial prefrontal cortex: partners in the fear circuit. J. Physiol. 591, 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burgos-Robles A., Vidal-Gonzalez I., Santini E., and Quirk G. J. (2007) Consolidation of fear extinction requires NMDA receptor-dependent bursting in the ventromedial prefrontal cortex. Neuron 53, 871–880 [DOI] [PubMed] [Google Scholar]
- 33. Milad M. R., and Quirk G. J. (2012) Fear extinction as a model for translational neuroscience: ten years of progress. Annu. Rev. Psychol. 63, 129–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimura R., Silva A. J., and Ohno M. (2008) Autophosphorylation of αCaMKII is differentially involved in new learning and unlearning mechanisms of memory extinction. Learn. Mem. 15, 837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pang K. C., Jiao X., Sinha S., Beck K. D., and Servatius R. J. (2011) Damage of GABAergic neurons in the medial septum impairs spatial working memory and extinction of active avoidance: effects on proactive interference. Hippocampus 21, 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cestari V., Rossi-Arnaud C., Saraulli D., and Costanzi M. (2014) The MAP(K) of fear: from memory consolidation to memory extinction. Brain Res. Bull. 105, 8–16 [DOI] [PubMed] [Google Scholar]
- 37. Kondo M., Nakamura Y., Ishida Y., Yamada T., and Shimada S. (2014) The 5-HT3A receptor is essential for fear extinction. Learn. Mem. 21, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park S. M., and Williams C. L. (2012) Contribution of serotonin type 3 receptors in the successful extinction of cued or contextual fear conditioned responses: interactions with GABAergic signaling. Rev. Neurosci. 23, 555–569 [DOI] [PubMed] [Google Scholar]
- 39. Chatterjee S., Mizar P., Cassel R., Neidl R., Selvi B. R., Mohankrishna D. V., Vedamurthy B. M., Schneider A., Bousiges O., Mathis C., Cassel J. C., Eswaramoorthy M., Kundu T. K., and Boutillier A. L. (2013) A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J. Neurosci. 33, 10698–10712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Selvi B. R., Swaminathan A., Maheshwari U., Nagabhushana A., Mishra R. K., and Kundu T. K. (2015) CARM1 regulates astroglial lineage through transcriptional regulation of Nanog and posttranscriptional regulation by miR92a. Mol. Biol. Cell 26, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carey M. F., Peterson C. L., and Smale S. T. (2009) Chromatin immunoprecipitation (ChIP). Cold Spring Harb. Protoc. 2009, pdb.prot5279 [DOI] [PubMed] [Google Scholar]