Abstract
The tail-anchored protein Fis1 is implicated as a passive tether in yeast mitochondrial fission. We probed the functional role of Fis1 Glu-78, whose elevated side chain pKa suggests participation in protein interactions. Fis1 binds partners Mdv1 or Dnm1 tightly, but mutation E78A weakens Fis1 interaction with Mdv1, alters mitochondrial morphology, and abolishes fission in a growth assay. In fis1Δ rescue experiments, Fis1-E78A causes a novel localization pattern in which Dnm1 uniformly coats the mitochondria. By contrast, Fis1-E78A at lower expression levels recruits Dnm1 into mitochondrial punctate structures but fails to support normal fission. Thus, Fis1 makes multiple interactions that support Dnm1 puncta formation and may be essential after this step, supporting a revised model for assembly of the mitochondrial fission machinery. The insights gained by mutating a residue with a perturbed pKa suggest that side chain pKa values inferred from routine NMR sample pH optimization could provide useful leads for functional investigations.
Keywords: confocal microscopy, mitochondria, nuclear magnetic resonance (NMR), protein assembly, protein-protein interaction, structural model, macromolecular machine, mitochondrial dynamics, pKa measurements, tetratricopeptide repeat (TPR) domains
Introduction
Mitochondrial fission is essential for organelle and cell viability (1), and altered fission contributes to a broad range of diseases, including neurodegeneration, diabetes, and cancer (2–6). Despite its significance, mitochondrial fission is not well understood beyond the identification of proteins involved. Only two proteins necessary for mitochondrial fission are conserved in all species containing mitochondria (7): the tail-anchored mitochondrial protein Fis1, which exposes a tetratricopeptide repeat (TPR)5 domain to the cytoplasm, and the large cytosolic GTPase Dnm1 (dynamin-like protein 1), which acts as a mechanoenzyme once assembled on the outer mitochondrial membrane. In budding yeast, Fis1 is uniformly distributed on the surface of the mitochondria, but Fis1-dependent Dnm1 assembly occurs at distinct, prescission foci on the mitochondria, suggesting that additional events determine a site of scission (8).
A prevalent model for the early steps in yeast mitochondrial fission holds that localizing Dnm1 from the cytoplasm to the passive anchor Fis1 requires the fungus-specific adaptor Mdv1 (7, 9, 10). Mdv1 then drives Dnm1 assembly into punctate foci on the mitochondrial surface with subsequent GTP hydrolysis and membrane scission. Complicating interpretation of these experiments is the presence of Caf4, an Mdv1-like protein that interacts with both Fis1 and Dnm1 but is not essential for fission (11, 12) and promotes mitochondrial fission much less effectively than Mdv1 (12). A passive role for Fis1 in this sequential model has been inferred from the ability of artificially tethered Mdv1 to partially support normal mitochondrial morphology in the absence of Fis1 (13). At odds with this model are the observations that Mdv1 arrives at mitochondrial foci after Dnm1 in time lapse experiments, not before (14). Also, in cells overexpressing Mdv1 domains, Dnm1 remains mitochondrial (15), suggesting that Fis1 might directly recruit Dnm1. This possibility is supported by a direct interaction between recombinant Fis1 and Dnm1, as demonstrated in biochemical experiments (16). Thus, although Fis1 is an essential conserved factor for fission, and Mdv1 is an essential factor in yeast, their exact roles remain unclear.
Point mutations that minimally perturb the balance of protein interactions necessary for proper mitochondrial fission might permit a clearer dissection of this machinery, but rationally identifying mutagenesis candidate sites is challenging even for small proteins such as Fis1 (15 kDa), which presents a large surface area that might mediate interactions (17–20). Here, we use a novel approach to identify target residues for mutational analysis. From NMR-based measurements of side chain ionization constants (pKa values), we identified Fis1 residues with pKa values perturbed toward neutral pH, which would be expected to lie at protein interfaces. Among the 16 Fis1 acidic residues that we assessed, Glu-78 has the most elevated apparent pKa. When Glu-78 is mutated to Ala, Fis1 no longer supports WT levels of mitochondrial fission. Fis1-E78A expression results in a novel Dnm1 localization pattern; instead of the normal discrete foci, Dnm1 instead has uniform mitochondrial localization, although at reduced Fis1-E78A expression levels some Dnm1 puncta are still detectable. Fis1-E78A lacks the WT Fis1 ability to bind Mdv1 but only moderately disrupts mitochondrial localization of Mdv1, suggesting that Fis1/Mdv1 interactions are not the only contacts necessary to drive native Mdv1/Dnm1 mitochondrial localization. Both Fis1 and Fis1-E78A bind directly to yeast-expressed Dnm1 in vitro, providing a mechanism for the observed localization. We propose a fission model in which Fis1 helps to directly recruit Dnm1 and actively cooperates with Mdv1 to drive Dnm1 assembly into prescission foci. Reassessing the roles of Fis1 and Mdv1 helps align yeast mitochondrial fission more closely with the mammalian process, which has Fis1 and Dnm1 (but not Mdv1) orthologs. More generally, these results suggest that identifying perturbed side chain pKa values can provide useful starting points for investigations into biological function.
Results
The pKa of Fis1 Glu-78 Is Significantly Perturbed
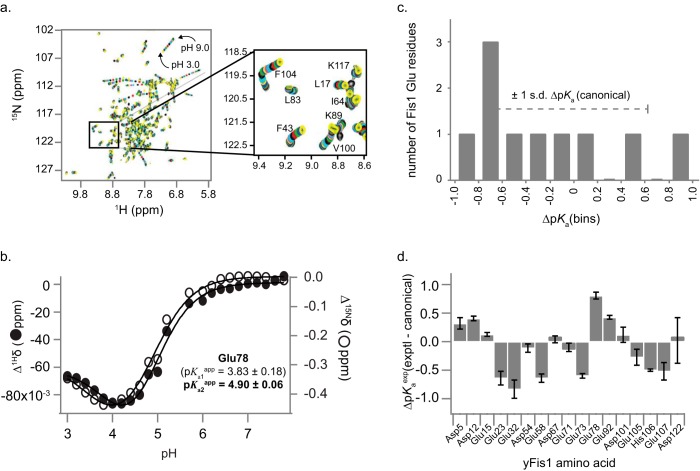
Protein interfaces are mediated by both polar and nonpolar amino acids, but the majority of buried surface area is nonpolar (21–23). Consequently, nonpolar residues probably surround most charged residues that will reside at a protein interface, even in the absence of the protein binding partner. The hydrophobic environment should perturb the pKa values of titratable residues toward neutral pH, allowing these residues to be identified. Direct side chain pKa measurements require side chain assignments (24), so we used the known sensitivity of amide backbone NMR chemical shifts to proximal titration events (25, 26) to infer apparent side chain pKa values in yeast Fis1. We fit Fis1 backbone amide proton and nitrogen NMR chemical shift changes as a function of pH (Fig. 1) to the Henderson-Hasselbalch equation, focusing on acidic residues because these were well described in the pH range covered by our titration (Fig. 1D, supplemental Results, and supplemental Fig. 1). Based on established criteria (27), we determined apparent pKa values for 6 of 7 Asp residues and 10 of 12 Glu residues (supplemental Table 1). Glu-78, with an apparent pKa of 4.9, is a full pH unit higher than the Fis1 Glu pKa average (3.9 ± 0.5) and is the most elevated pKa identified (>1σ; Fig. 1, C and D) relative to the average intrinsic pKa for acidic residues in proteins in the literature (supplemental Fig. 2).
FIGURE 1.
Fis1 amide chemical shift changes with pH. a, overlay of 26 1H-15N HSQC spectra recorded from pH 3.0 to 9.0 in increments of ∼0.2 pH units. Each spectrum is color-coded from pH 9.0 to pH 3.0 using a repeating sequence of black, red, green, blue, yellow, magenta, and cyan. b, backbone 1H and 15N chemical shift data for Glu-78 from a individually fit to either 1-, 2-, or 3-site Henderson-Hasselbalch equations to determine apparent pKa values. For Glu-78, the data were best fit to two pKa values, but only one of which (in bold) was deemed reliable (Supplemental Table 1). c, distribution of apparent pKa values of Fis1 Glu residues compared with the S.D. from canonical values. d, deviation of Fis1 apparent pKa values from “canonical” pKa values. The difference between experimentally determined pKa values from Fis1 NMR data from a compilation of canonical values of 3.38 (Asp), 4.09 (Glu), and 6.50 (His) is shown. Error bars, S.D.
The Fis1 Glu-78 Side Chain Is Essential for Fission
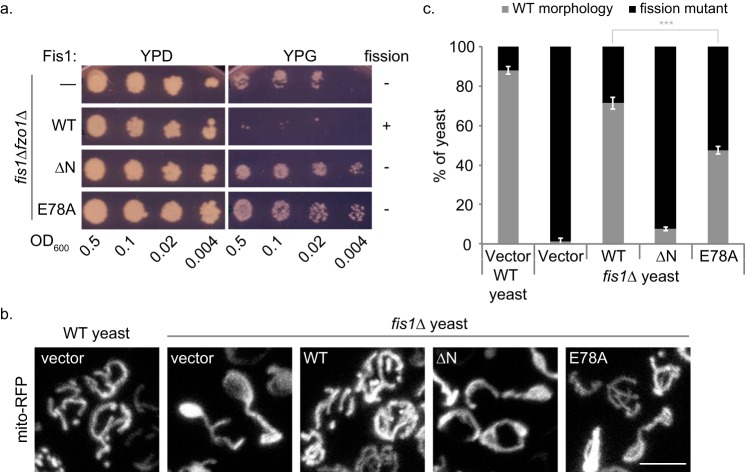
To determine whether Glu-78 is important for Fis1 activity, we used a yeast-based assay that relies on inhibition of mitochondrial fission for growth (8, 28). A strain of yeast lacking both Fis1 and the mitochondrial fusion protein Fzo1 (fis1Δfzo1Δ) is deficient for both fission and fusion; these cells contain mitochondria capable of respiration, which allows for growth on non-fermentable carbon sources, such as glycerol. When fission (but not fusion) is restored in these cells by a wild type copy of Fis1 on a plasmid, the mitochondria become fragmented, and the mitochondrial genome is lost (29, 30), rendering the cells incapable of growth on glycerol. We substituted Ala for Glu-78 and used this assay to assess whether Fis1-E78A could support fission. Strains of fis1Δfzo1Δ yeast were transformed with plasmids bearing either WT Fis1, Fis1-ΔN, or Fis1-E78A and replica-plated on fermentable (dextrose) or non-fermentable (glycerol) media. Yeast expressing WT Fis1 did not grow on glycerol, indicating that fission is restored, whereas yeast expressing Fis1-ΔN, a variant lacking the functionally important Fis1 arm (17, 31), did grow on glycerol, indicating that fission remained impaired (Fig. 2A). For Fis1-E78A, we observed a phenotype similar to Fis1-ΔN; cells expressing Fis1-E78A grew on glycerol, indicating that this single residue change renders Fis1 incapable of restoring fission in vivo (Fig. 2A).
FIGURE 2.
Fis1-E78A is unable to restore fission to WT levels in vivo. a, fission/fusion-deficient yeast cells (fis1Δfzo1Δ) were transformed with either empty vector, Fis1 WT, or variants (ΔN or E78A) and were replica-plated on agar containing dextrose (YPD, fermentable) or glycerol (YPG, non-fermentable). Lack of growth on YPG indicates that fission has been restored due to the excessive mitochondrial fragmentation and the ensuing loss of mitochondrial DNA. Nonfunctional Fis1 mutants (ΔN or E78A) allow growth on YPG. b, WT or fis1Δ yeast transformed with mito-RFP and either empty vector, 1×HA-Fis1 WT, or variants (ΔN or E78A) were grown to mid-log phase and plated on an agarose bed for imaging. Representative confocal image projections are shown. Scale bar, 5 μm. c, mitochondrial morphology phenotypes from b were categorized into either WT or fission mutant. Using ImageJ, at least 100 cells were counted from six confocal image projections. Data represent the average ± S.D. (error bars) from three independent experiments. ***, p < 0.001 (supplemental Table 2).
We determined how the E78A substitution affects mitochondrial morphology using fis1Δ yeast expressing Fis1 variants and mitochondrially targeted red fluorescent protein (mito-RFP) (Fig. 2, B and C, and supplemental Videos 1–5). In accordance with prior findings, ∼90% of wild type yeast cells show WT mitochondrial morphology (a continuous, interconnected reticulum with >2 termini), whereas >95% of fis1Δ yeast show defects in mitochondrial fission (netted, collapsed mitochondria or a mitochondrial network with <3 termini) (32). Expression of Fis1 in fis1Δ yeast restores WT morphology to ∼80% of the level observed for wild type yeast. Consistent with earlier findings (17, 33), Fis1-ΔN restores WT morphology to only 10% of the wild type level. A mixed morphology phenotype is seen for fis1Δ yeast expressing Fis1-E78A, with only half of cells exhibiting WT morphology. As controls, we also examined mitochondrial morphology for three acidic residues (Asp-12, Glu-15, and Glu-92) with weakly elevated pKa values (<1σ and ΔpKa of 0.41, 0.13, and 0.43, respectively). Substitution of these residues with alanine restored fission to WT levels in vivo (supplemental Fig. 3). We conclude that the Glu-78 side chain is important for promoting mitochondrial fission by Fis1 but that the E78A phenotype is less severe than Fis1 deletion or truncation.
TPR folds usually support protein-protein interactions (34), so Fis1 Glu-78 may help mediate interactions with other fission factors. Fis1 has been reported to interact with both the fission adaptor Mdv1 (15, 16, 18, 35) and the fission mechanoenzyme Dnm1 (16), although the significance of the Fis1/Dnm1 interaction is not as firmly established as the Fis1/Mdv1 interaction (36, 37). After determining that the E78A substitution has little effect on Fis1 structure by NMR, its folding by chemical denaturation, its propensity to form dimers (38), or its ability to bind membranes (16) (supplemental Fig. 4), we tested the hypothesis that the fission defect caused by the E78A substitution (Fig. 2) could result from weakened binding between Fis1 and either of these partners using pairwise biochemical pull-down assays with purified proteins and co-immunoprecipitation (co-IP) assays with total yeast extracts.
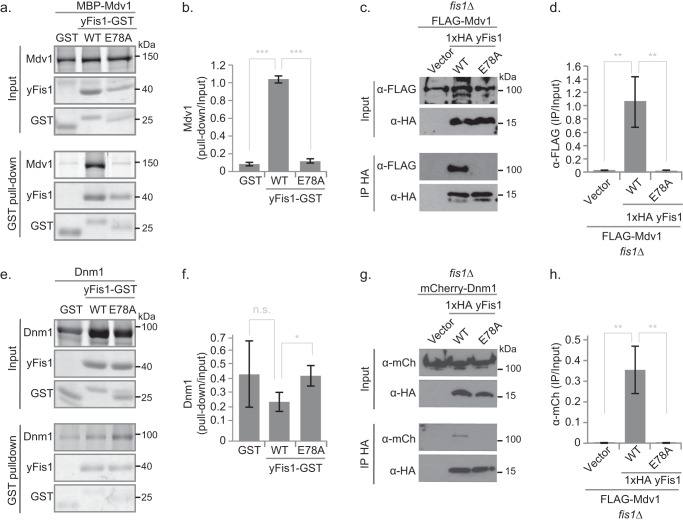
Glu-78 Is Critical for Direct Fis1/Mdv1 Binding and Fis1/Mdv1 Co-IP from Cell Extracts
Direct interactions between Fis1 and Mdv1 are central to every model for yeast fission proposed to date (7, 39, 40). In GST pull-down experiments, the Fis1 cytosolic domain tagged with GST (Fis1ΔTM-GST) bound well to yeast-expressed MBP-Mdv1 (Fig. 3, A and B), demonstrating a direct physical interaction between Fis1 and full-length Mdv1 that is consistent with previous co-IP, yeast two-hybrid, and GST pull-down studies (15, 16, 33, 35). This robust pull-down was almost completely lost for Fis1-E78A (Fig. 3, A and B), showing that direct Fis1/Mdv1 binding in vitro depends strongly on the Glu-78 side chain. In anti-HA co-IP experiments of fis1Δ yeast ectopically expressing HA-Fis1 and FLAG-Mdv1, the E78A substitution also significantly decreased the amount of Mdv1 that bound to Fis1 (Fig. 3, C and D). Loss of a hydrogen bond between Fis1 Glu-78 and the backbone of Mdv1 Leu-148, as seen in a Fis1/Mdv1 peptide co-crystal (18) (supplemental Fig. 5), is the likely physical basis for this decreased interaction. The E78A mutation also disrupted Fis1 co-IP of the Mdv1-like protein Caf4 (supplemental Fig. 6), which structural data (18) suggest is probably due to disrupting a similar hydrogen bonding interaction (supplemental Fig. 5).
FIGURE 3.
Fis1-E78A impairs Mdv1, but not Dnm1, binding and abrogates co-immunoprecipitation with both. a and e, representative images from GST pull-down experiments between purified Fis1ΔTM-GST (WT or E78A) and either MBP-Mdv1 (a) or Dnm1 (e). ImageQuant analysis from three independent experiments (average ± S.D. (error bars)) is shown for MBP-Mdv1 (b) or Dnm1 (f). c and g, representative images from anti-HA co-IP experiments using extracts from fis1Δ yeast expressing 1×HA Fis1 (WT or E78A) and either FLAG-Mdv1 (c) or Dnm1-mCherry (g). ImageJ64 analysis from three independent experiments (average ± S.D.) is shown for FLAG-Mdv1 (d) or Dnm1-mCherry (h). Statistical analysis is represented as follows. n.s., non-significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (supplemental Table 2).
Fis1 Directly Binds Dnm1 but E78A Does Not Disrupt This Interaction
Contacts between Fis1 and Dnm1 have been invoked in some models of yeast fission (16, 41, 42), but strong direct interactions have been shown only for bacterially expressed Fis1ΔNΔTM-GST and bacterially expressed MBP-Dnm1 (weak interactions are seen for Fis1ΔTM-GST) (16). Here we show that bacterially expressed Fis1ΔTM-GST consistently pulls down yeast-expressed untagged Dnm1 and that this interaction is unaffected by the Fis1 mutation E78A (Fig. 3). Direct Fis1/Dnm1 binding is at odds with the current, sequential model for yeast mitochondrial fission, but the specificity of this interaction was previously demonstrated by the ability of substitutions at some positions (Arg-77, Tyr-82, Ile-85, and Lys-89) but not others (Gln-21, Ile-24, Gln-28, Gln-40, Asn-44, and Gln-112) to abolish Fis1/Dnm1 pull-down (16). Because the Fis1/Dnm1 pull-down was not diminished by the E78A substitution (Fig. 3, E and F), we conclude that direct Fis1/Dnm1 contacts do not involve Glu-78.
A Weak Fis1/Dnm1 Co-IP Depends on Fis1 Glu-78
Co-IP experiments using fis1Δ yeast ectopically expressing HA-Fis1 and endogenously expressing tagged Dnm1-mCherry reveal a faint but reproducible Fis1/Dnm1 interaction (Fig. 3G) consistent with previous observations (43). Although E78A did not alter direct Fis1/Dnm1 pull-down (Fig. 3E), the Fis1/Dnm1 co-IP signal was surprisingly abolished for the E78A variant (Fig. 3, G and H). We infer that the loss of Fis1/Dnm1 co-IP signal results from a loss of interactions with other protein(s) involved in this Fis1/Dnm1 complex. Candidates include Mdv1, which is thought to act as an adaptor between Fis1 and Dnm1 (7, 11, 33, 43), and Caf4, which also binds Dnm1 and Fis1 (11, 12), both of which have reduced binding to Fis1-E78A (Fig. 3 and supplemental Fig. 6). We propose that Mdv1 (or Caf4) forms a modestly stable ternary complex with Dnm1-mCherry and WT Fis1 that is destabilized by the E78A variant.
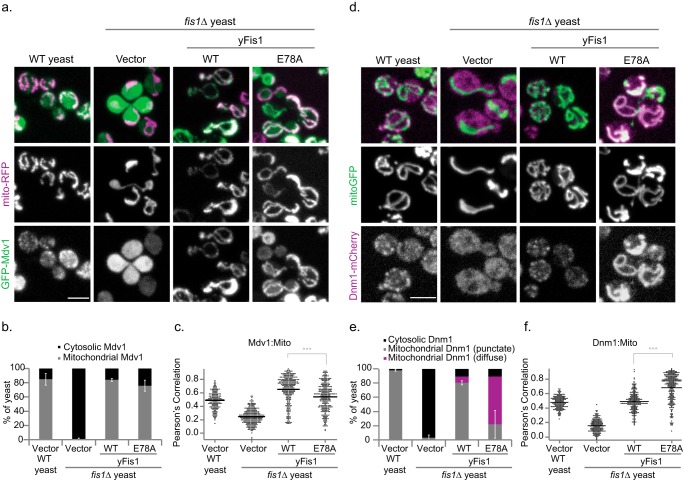
E78A Modestly Affects Mdv1 Mitochondrial Localization
We next determined how Fis1-E78A affects the localization of Mdv1 and Dnm1, which normally co-localize on the mitochondrial surface in distinct punctate structures, or foci, that mark future scission sites (15, 35, 44, 45). As in previous findings (17, 33), the absence of Fis1 ablated Mdv1 mitochondrial localization; 85% of wild type cells show punctate mitochondrial Mdv1, whereas 99% of fis1Δ cells show cytoplasmic Mdv1 (Fig. 4, A and B). Expression of WT Fis1 from a plasmid restored mitochondrial Mdv1 localization in 84% of fis1Δ cells but with a more uniform, less punctate distribution (17) that is reflected in an elevated Pearson's correlation (Fig. 4B). Expression of Fis1-E78A rescued mitochondrial Mdv1 localization in just 76% of cells with a significant downward shift in the Pearson's correlation values (Fig. 4, A and B). Thus, the mutation E78A only modestly altered Fis1-dependent Mdv1 mitochondrial localization despite dramatically decreasing the Fis1/Mdv1 pull-down and co-IP interactions (Fig. 3, A and C).
FIGURE 4.
Expression of Fis1-E78A partially inhibits Mdv1 mitochondrial localization and induces novel uniform distribution of Dnm1. a and c, representative confocal image projections of WT or fis1Δ yeast expressing either empty vector, 1×HA-Fis1 WT or E78A, and mito-RFP/GFP-Mdv1 (a) or mito-GFP/Dnm1-mCherry (c). Mdv1 localization was recorded as either cytosolic or mitochondrial from at least 50 cells. The column on the right indicates the percentage of yeast containing mitochondrial Mdv1 and represents the average ± S.D. (error bars) from three independent experiments. Scale bar, 5 μm. b and d, Pearson's correlation co-localization index was determined from all cellular data from the three independent experiments for GFP-Mdv1/mito-RFP (b) or Dnm1-mCherry/mito-GFP (d). Data are displayed as a scatter dot plot using Igor Pro, and the mean value is indicated with the black line. e, Dnm1 localization was recorded as either cytosolic or mitochondrial from at least 50 cells. Data represent the average ± S.D. from three independent experiments. f, for Dnm1, mitochondrial localization was further assessed for either punctate or uniform Dnm1 localization. ***, p < 0.001 (supplemental Table 2).
Fis1 Glu-78 Induces Uniform Mitochondrial Dnm1 Localization
In WT yeast expressing endogenous levels of Dnm1-mCherry, Dnm1 localized to mitochondria in distinct punctate structures, but in fis1Δ yeast, these puncta were lost, and most Dnm1 remained cytoplasmic (Fig. 4C), showing that Fis1 is critical to Dnm1 mitochondrial recruitment in vivo, as reported previously (41). Expression of either Fis1 or Fis1-E78A restored mitochondrial localization of Dnm1 in 90% of yeast (Fig. 4, C and D). However, in most yeast expressing Fis1-E78A, Dnm1-mCherry was uniformly distributed on the mitochondrial surface, and only in a minority of cells did it form occasional puncta (Fig. 4, C and D). This localization pattern has not previously been reported for Dnm1, but it resembles the reported distributions of Fis1 in wild type yeast (8) or of Mdv1 in dnm1Δ yeast (44, 46, 47). The uniform Dnm1 distribution induced by Fis1-E78A significantly increased its co-localization with mitochondria (Pearson's correlation; Fig. 4, D–F). We infer that Fis1-E78A populates a state in the mitochondrial fission scheme in which Dnm1 is at the mitochondrial surface but is not assembled into the higher order oligomers that give rise to visible puncta.
Mdv1 (or Caf4) Is Necessary for Dnm1 Uniform Mitochondrial Localization
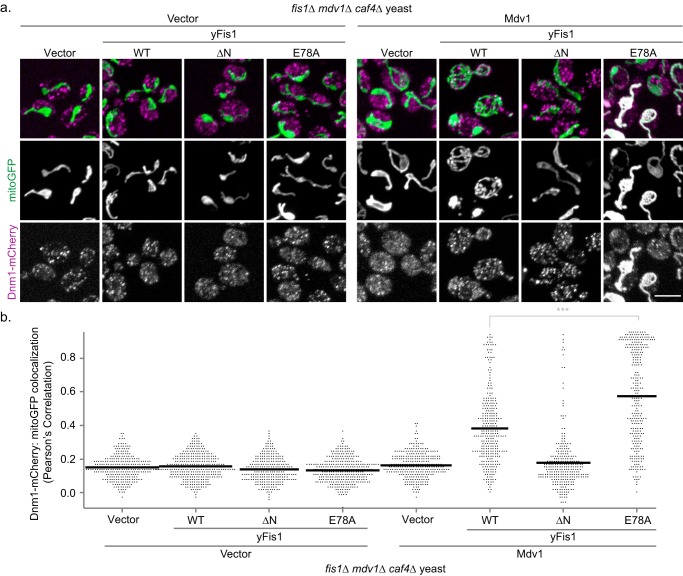
To determine which fission partners are necessary for Dnm1 uniform distribution on mitochondria, we examined Dnm1-mCherry localization in yeast lacking Fis1, Mdv1, and Caf4 (fis1Δmdv1Δcaf4Δ; Fig. 5A, left). In these yeast, the Dnm1-mCherry signal correlated poorly with mitochondria (Pearson 0.18) and appeared largely as punctate cytoplasmic structures (Fig. 5, A and B). These data are consistent with earlier findings that Dnm1 is primarily cytoplasmic in the absence of other fission proteins (11, 15, 35). Expression of Fis1 alone (Fig. 5A, left, second row) or Mdv1 alone (Fig. 5A, right, top row) from plasmids did not alter Dnm1 localization, consistent with earlier findings (8, 11, 15, 35, 44). Thus, the physical interaction detected between Fis1 and Dnm1 is not sufficient to direct Dnm1 mitochondrial recruitment in the absence of Mdv1. Expression of both Mdv1 and Fis1 lacking its N-terminal domain (Fis1-ΔN) did not alter Dnm1 localization, but expression of both Mdv1 and WT Fis1 drove Dnm1 to co-localize with mitochondria, where it was found predominantly in punctate structures (Fig. 5, A and B). Expression of both Mdv1 and Fis1-E78A also drove Dnm1 to co-localize with mitochondria, but in this case, Dnm1 uniformly coated the entire mitochondrial surface, although small puncta were seen in some cells. Similar localization occurred upon expression of Fis1-E78A with the Mdv1 homolog Caf4 (supplemental Fig. 7), although the proportion of cells with prominent uniform Dnm1 localization was much lower. We conclude that the uniform distribution of Dnm1 induced by Fis1-E78A requires the participation of Mdv1. Because Fis1-E78A localizes Dnm1 (and Mdv1) to mitochondria despite failing to pull down Mdv1, we conclude that strong direct interactions between Fis1 and Mdv1 are not necessary for mitochondrial localization of wild type Mdv1/Dnm1, although such interactions are important for proper Dnm1 puncta formation and fission. Surprisingly, Mdv1 itself is critical for Dnm1 mitochondrial localization even when strong Fis1/Mdv1 interactions are abolished. We therefore propose that Fis1 and Mdv1 cooperate to help assemble Dnm1 into productive foci on mitochondria. The dependence of the Dnm1 localization pattern, uniform or punctate, on a single Fis1 residue supports a more active role for Fis1 in yeast fission than as merely a passive anchor.
FIGURE 5.
Mdv1 is required for uniform Dnm1 distribution on mitochondria upon Fis1-E78A expression. a, representative confocal images from fis1Δ mdv1Δ caf4Δ cells expressing Dnm1-mCherry, mito-GFP, either empty vector (a, left) or 3×FLAG-Mdv1 (a, right), and either empty vector, 1×HA-Fis1 WT, ΔN, or E78A. The yeast were first grown overnight in medium containing 5× methionine and 2% glucose (to suppress Mdv1 and yFis1 expression, respectively) and then back-diluted into medium containing 1× methionine and 2% galactose and grown for 3 h to induce Mdv1 and yFis1 expression, respectively. Scale bar, 5 μm. b, Pearson's correlation co-localization index between Dnm1-mCherry and mito-GFP was determined from all cellular data from the three independent experiments from a. Data are displayed as a scatter dot plot using Igor Pro, and the mean value is indicated with the black line. ***, p < 0.001 (supplemental Table 2).
Fis1 Expression Levels Can Control Dnm1 Puncta Formation
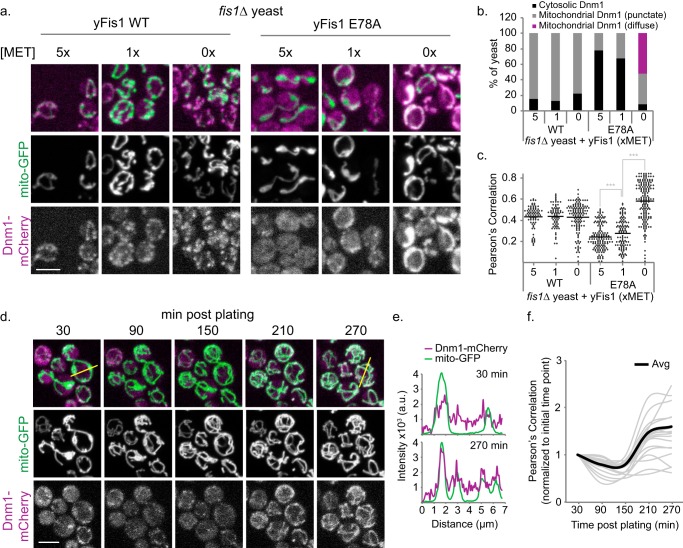
If pairwise interactions among all three fission partners contribute to assembly of Dnm1 foci, variations in Fis1 expression levels might affect the ternary complexes formed in vivo. Accordingly, we altered HA-Fis1 expression levels under the control of the repressive methionine promoter (MET25) (48) and measured Dnm1-mCherry localization in fis1Δ yeast. Overnight fis1Δ cultures grown under repressive conditions (5× methionine) were shifted to medium containing methionine at concentrations that allowed low, medium, or high levels of Fis1 expression (methionine = 5×, 1×, or 0×, respectively) for 3 h. Undeterred by varying expression levels, WT Fis1 was able to recruit Dnm1-mCherry to punctate mitochondrial structures in a majority of yeast (∼80%) (Fig. 6, A–C). By contrast, Fis1-E78A exhibited a greater sensitivity than wild type and recruited Dnm1-mCherry to punctate mitochondrial structures in an expression level-dependent manner (5×, 22%; 1×, 33%; 0×, 40%). These mitochondrial Dnm1 puncta appeared non-functional, given the presence of collapsed mitochondrial networks (Fig. 6, A–C). Only at high Fis1-E78A expression levels (0× methionine) was the diffuse population of Dnm1 evident, with >50% of the yeast containing Dnm1 in a uniform mitochondrial localization (Fig. 6, A–C). This uniform distribution is similar to that in Figs. 4 and 5, which utilized 1× methionine for the entire experiments, and ultimately suggests that elevated levels of Fis1-E78A lead to accumulation of Dnm1-mCherry over time. To test this idea, we took sequential images of yeast expressing Dnm1-mCherry and mito-GFP as they shifted from low to high Fis1-E78A expression (Fig. 6D). Over the course of 4.5 h, Dnm1 accumulated on the mitochondria and transitioned from a punctate to a uniform distribution (Fig. 6, E and F). Decreasing puncta with increasing levels of Fis1-E78A is inconsistent with Fis1 being a passive anchor that binds a single partner at 1:1 stoichiometry. We conclude that at low levels, Fis1-E78A can drive Dnm1 into the assemblies that we detect as puncta, but when present in excess, Fis1-E78A sequesters Mdv1 and/or Dnm1 in a manner that precludes mature puncta formation.
FIGURE 6.
Increased E78A expression results in Dnm1 accumulation on mitochondria. a, representative confocal image projections from fis1Δ yeast expressing Dnm1-mCherry, mito-GFP, and either 1×HA-Fis1 WT or E78A that were grown overnight in medium containing 5× methionine; back-diluted into 5×, 1×, or 0× methionine medium (to induce Fis1 expression); and grown to mid-log phase. b, using the confocal image projections from a, Dnm1 localization was recorded as either cytosolic, mitochondrial with punctate Dnm1, or mitochondrial with circumscribing Dnm1 from at least 85 cells. c, Pearson's correlation co-localization index was determined for all of the cells from the experiment conducted for a. d, Fis1Δ yeast expressing Dnm1-mCherry, mito-GFP and 1×HA-Fis1-E78A were grown overnight in medium containing 5× methionine. The yeast were then back-diluted in medium containing 0× methionine, grown to mid-log phase, and plated on an agarose bed containing 0× methionine. A field of yeast containing existing Dnm1 puncta was imaged every hour for 5 h, assaying for the accumulation of Dnm1. e, line scan plots of Dnm1-mCherry (purple) and mito-GFP (green) from the yellow lines indicated on the 30- or 270-min merged image. f, Pearson's correlation was determined for each yeast cell in the image from d and tracked through the subsequent time points. Individual data are displayed in the gray line plots, whereas the black line represents the average from the 19 yeast cells imaged for the experiment. Scale bar, 5 μm. Note the initial time points were normalized to 1. ***, p < 0.001 (supplemental Table 2).
Discussion
Yeast mitochondrial fission has long been known to depend on Fis1 as a membrane-tethered anchor for recruiting Mdv1 and Dnm1 (8, 44, 46, 47), but the idea that Fis1 acts only as an anchor without further contribution to the mechanism of membrane scission has gained favor with the discovery of other natural anchors (49–51) and with the demonstration that artificially tethering the C-terminal portion of Mdv1 to mitochondria can partially rescue morphology in yeast cells that lack Fis1, Mdv1, and Caf4 (13). A nominal role for Fis1 in Dnm1 assembly and hydrolysis was shown with recombinant proteins (36), although these studies involved Fis1 lacking its transmembrane domain that when present would dramatically increase the effective concentration at the membrane. Our demonstration that the single Fis1 substitution E78A strongly alters Dnm1 localization and assembly despite only modestly altering Mdv1 localization is inconsistent with models in which a passive Fis1 serves merely to anchor Mdv1, which is then entirely responsible for recruiting Dnm1 and regulating its assembly. Instead, Fis1, and particularly Glu-78, must participate in Dnm1 mitochondrial recruitment and assembly in vivo.
In addition to prompting a reassessment of the Fis1 role in yeast fission, our findings confirm the importance of Mdv1. The mutation E78A largely abrogates Fis1 pull-down of Mdv1 and the Fis1/Mdv1 co-IP, but Mdv1 is nevertheless partially localized to mitochondria by Fis1-E78A in qualitatively the same way as by WT Fis1 (Fig. 4). This phenotype is consistent with mutations in either Fis1 or Mdv1 that drastically reduce the Fis1/Mdv1 co-IP yet retain significant mitochondrial localization of Mdv1 (19). Note that such localization is opposite to what would be expected if Mdv1 were an obligate adaptor, in which case failure of Mdv1 to bind tethered Fis1 should cause both Mdv1 and Dnm1 to remain cytosolic.
Residual Mdv1 localization by Fis1-E78A may be due to structural or dynamic changes in the Fis1 arm known to be important for Mdv1 interactions (28). However, this possibility is eliminated by our NMR chemical shift and line width analyses (supplemental Fig. 4) that show little or no changes between WT and E78A. Residual Mdv1 localization more likely occurs from weakened but not obliterated Fis1/Mdv1 interactions via interaction sites not disrupted by the E78A mutation. Crystallographic data with a fragment of Mdv1 in complex with Fis1 show stabilizing interactions at three sites, A, B, and C (supplemental Fig. 8) (18). Glu-78 lies at the site B interface, leaving the other sites with the capacity to bind, but probably at a much weaker affinity (given the robust decline in both pull-down and co-IP experiments). In addition to this reduced affinity, we infer that Fis1-E78A may also recruit Mdv1 to mitochondria in vivo indirectly through contacts with Dnm1 (Fig. 4). Dnm1 is known to bind tightly to Mdv1 (15, 36, 44), and Fis1 residues implicated in Dnm1 binding (16) (Arg-77, Tyr-82, Ile-85, and Lys-89) lie on helix 4 adjacent to Glu-78 and a Fis1/Mdv1 contact point (site B). Furthermore, mutation at residue Ile-85 of Fis1 disrupts pull-down between Fis1/Dnm1, but not Fis1/Mdv1, and leads to cytoplasmic localization of both Mdv1 and Dnm1 (16) (data not shown). Thus, we speculate that Dnm1 binding to Fis1 could facilitate Fis1 binding to Mdv1. Direct Fis1/Dnm1 contacts have been reported previously (16), albeit not for untagged Dnm1 as described here. Although we identify robust direct Fis1/Dnm1 interactions in vitro (Fig. 3), neither Fis1 nor Fis1-E78A significantly recruit Dnm1 to mitochondria in the absence of Mdv1 (Fig. 5). Because Mdv1 is required for Dnm1 mitochondrial recruitment even by Fis1-E78A, which shows little direct binding to Mdv1 (Fig. 3), we infer that Mdv1 regulates the Fis1/Dnm1 interaction. The reciprocal nature of the proposed interactions between these three proteins suggests that a ternary complex may be formed via mutually stabilizing interactions. Consistent with this idea are biochemical pull-down data showing that Fis1-GST was able to pull down both Dnm1 and a truncated construct of Mdv1 incapable of supporting Dnm1 interaction on its own (16). In these experiments, increasing concentrations of Mdv1 decreased, but did not eliminate, the Fis1-Dnm1 interaction, suggesting that a ternary complex is formed.
The capacity of Fis1-E78A to support some degree of Dnm1 puncta formation (Fig. 6) shows that strong Fis1/Mdv1 interactions are not absolutely required for assembly of Dnm1 on mitochondria and suggests that E78A hinders fission by affecting some step after initial puncta formation. We hypothesize that Fis1-E78A can be incorporated into near native complexes because the large-to-small substitution causes no steric clashes, only a loss of favorable enthalpic contacts. The profound effects on Dnm1 localization and negative impact on mitochondrial fission that result from the removal of four non-hydrogen atoms show that native functional interactions among Fis1, Mdv1, and Dnm1 are delicately poised. Through mass action, elevating Fis1-E78A expression can cause the Fis1 variant to weakly (if at all) bind Mdv1 and suppress Dnm1 higher order assembly and the maturation of Dnm1 puncta but not mitochondrial recruitment in vivo. As a TPR-containing protein, Fis1 probably makes multivalent interactions with its fission partners. We hypothesize that WT Fis1 can function over a wide range of expression levels (15) because its affinities for Mdv1 and Dnm1 are balanced in a way that supports assembly of productive complexes. High expression levels of Fis1-E78A, which has lost tight binding to Mdv1, probably result in a sequestration of some fission partners away from others, similar to how overexpressing Dnm1, Mdv1, and Mdv1 domains and point mutants have been shown to affect morphology through sequestering of fission partners (8, 15, 33, 35, 43, 44, 47).
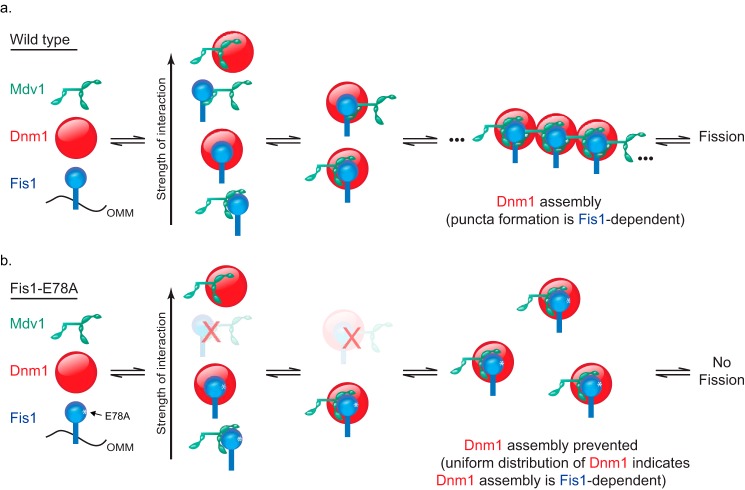
Previous models for yeast fission have cast Fis1 as a passive anchor and Mdv1 as an obligate adaptor that follow a sequential binding scheme; Fis1 recruits Mdv1, and then Mdv1 recruits and regulates the assembly of Dnm1 (supplemental Fig. 9). The data presented here show that a single Fis1 point mutation can abolish fission despite the presence of native Mdv1 and Dnm1 and can alter the localization of Dnm1 in ways that are inconsistent with a sequential binding model in which Fis1 is merely an anchor. We propose a mutual model in which Fis1, Mdv1, and Dnm1 cooperate through pairwise interactions among all three partners to assemble into puncta at the mitochondrial surface (Fig. 7). This model is supported by data herein and previous observations of binary interactions between Fis1-Dnm1 (16), Fis1-Mdv1/NTE domain (18, 19, 32, 33, 35), Fis1-Mdv1/WD domain (15), and Mdv1-Dnm1 (11, 14, 15, 35–37, 44) as well as the aforementioned pull-down data consistent with ternary interactions between all three proteins (16). These interactions are exquisitely balanced for fission machinery assembly (and subsequent fission) as indicated by the sensitivity of Dnm1 distribution to Fis1-E78A expression levels (Fig. 6). Because low expression of Fis1-E78A supports Dnm1 puncta formation (Fig. 6) but not WT morphology, we hypothesize that the E78A substitution has an effect after Dnm1 puncta formation that is due to weakened Fis1/Mdv1 interactions within assemblies. Further evidence for a set of mutually balanced interactions are data from a yeast suppressor screen in which conditional mutations in Fis1 are suppressed by mutations in the Mdv1 coiled coil domain, which is not the primary Mdv1 domain supporting interactions with Fis1 (19, 33).
FIGURE 7.
Mutual model for yeast mitochondrial fission inferred from current and previous observations in which Mdv1 facilitates Dnm1 assembly in a Fis1-dependent manner. a, WT Fis1 is anchored to the mitochondrial outer membrane (OMM), where it makes transient interactions with Dnm1 (Fig. 3) that are stabilized in vivo by Mdv1 (Figs. 4 and 5). This leads to Dnm1 assembly into prescission complexes visualized by light microscopy as puncta (Figs. 4 and 5). b, the Fis1-E78A substitution prevents one of the stable Fis1-Mdv1 interactions from forming (Fig. 3), preventing Dnm1 assembly (Figs. 4–6). In this model, Fis1-Mdv1 interactions occur through either the NTE (15, 16, 18, 19, 32, 35) or WD domain (15) with the Fis1-WD interaction too weak to be detected in pull-down experiments. Note that each protein is reported to undergo homotypic interactions (19, 32, 35, 38, 65) that are not represented in this model but would aid in building the lattice architecture.
A more active role for Fis1 is concordant with early reports of dual activity for Fis1 (35), which identified fis1-L80P as a mutant allele functional for Dnm1 assembly into discrete puncta on mitochondria but incompetent in mitochondrial fission. Similar to E78A, L80P impaired Mdv1 interactions, indicating the importance of Fis1-Mdv1 contacts in membrane scission. Contrary to E78A, L80P was subsequently found to enhance dimerization of Fis1ΔTM (38), although the impact of Fis1 dimerization on Dnm1 and Mdv1 localization was not reported. Our current model neither invokes nor excludes Fis1 dimerization, but we note that the differential localization of Dnm1 at low and high Fis1-E78A expression levels could be explained by Fis1 dimers acting in Mdv1/Dnm1 recruitment or assembly.
Our findings may help resolve discrepancies between the roles of yeast and mammalian Fis1 in fission (7). In sequential models, Mdv1 acts as an obligate adaptor between Fis1 and Dnm1, but no mammalian Mdv1 ortholog has been identified, suggesting that mammalian and yeast fission mechanisms are divergent. This view is supported by a potentially divergent role for Fis1 in mitophagy (9, 52, 53) and the presence of non-fungal tail anchors (Mff and Mid-49/51) capable of mammalian Dnm1 (Drp1) recruitment independent of Fis1 (13, 40, 49, 51, 54, 55). However, Fis1 and Dnm1 (Drp1 in mammals) are the only two proteins conserved for mitochondrial fission in all species, and mutating conserved residues in yeast Fis1 disrupts Dnm1 pull-downs, suggesting conserved Fis1/Dnm1 interactions in fission (16). A mutual model for assembly in which Mdv1 is not an obligate adaptor, but rather stabilizes or directs Fis1/Dnm1 interaction, can accommodate these observations and may indicate that Fis1 activity in mammalian and yeast fission are less divergent than currently thought.
Interrogating the intricate balance of Fis1-directed protein interactions necessary for proper mitochondrial fission was possible because we identified Fis1 Glu-78 as a mutagenesis candidate on the basis of a perturbed pKa value. Ionizable residues can specify distance- and angle-dependent interactions that are more stringent than the hydrophobic effect (56). Such residues may often have perturbed pKa values (57, 58) because protein-protein interfaces are predominantly hydrophobic (21–23), which favors the neutral species even in the absence of a binding partner. Consistent with this, Fis1 Glu-78 is situated in a hydrophobic environment in structures of both the Fis1 monomer (17, 20) and complexes with Mdv1 or Caf4 (18, 19). We propose that identifying perturbed side chain pKa values could provide useful starting points for investigations into biological function. Acidic residues at interfaces are particularly attractive candidates for mutagenesis studies because they offer the option of a large-to-small substitution that can preserve the global geometry of contacts but decrease binding such as we find here for Fis1-E78A.
Experimental Procedures
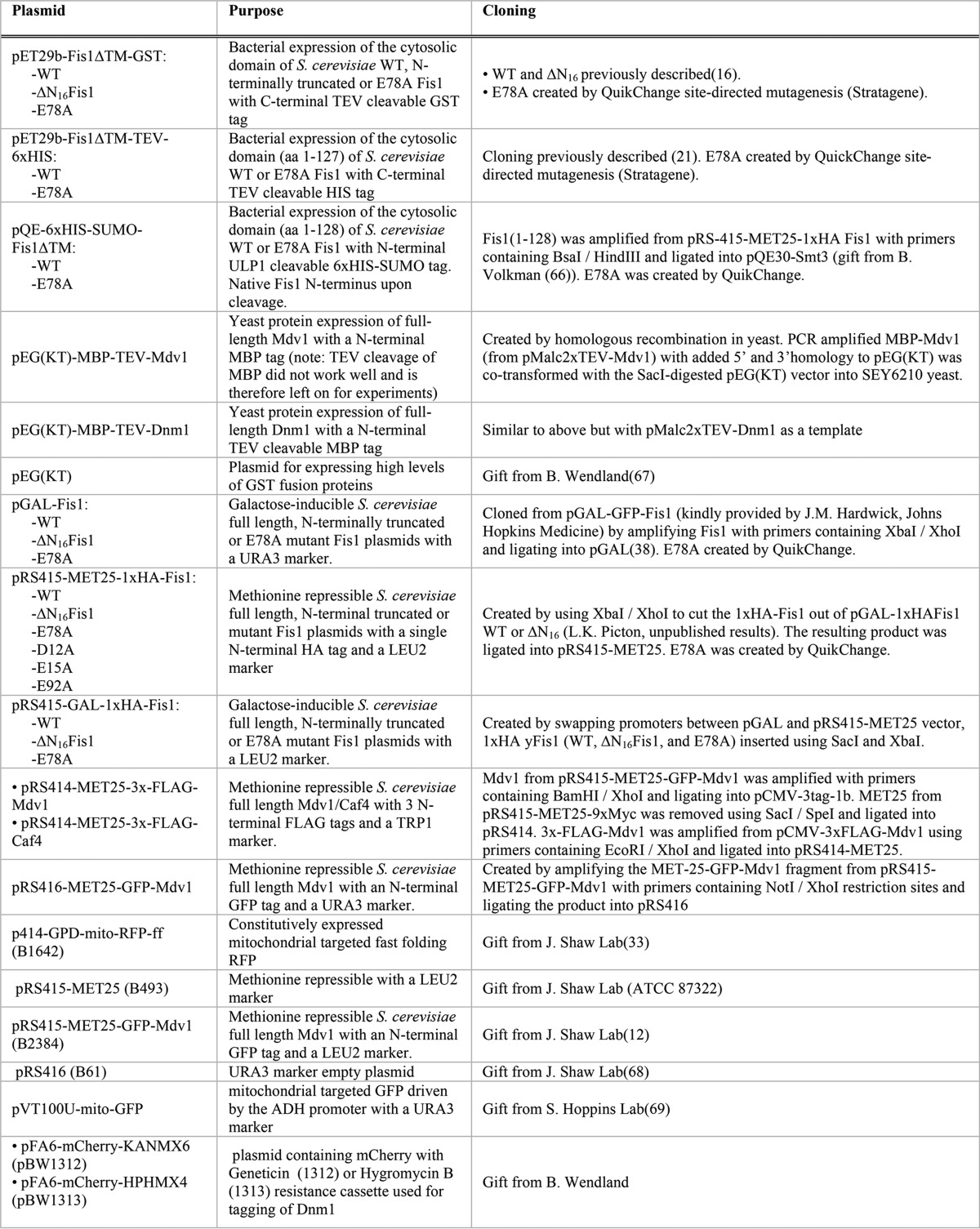
All plasmids used in this work are reported in Table 1. All yeast strains are reported in Table 2.
TABLE 1.
Plasmids used in this study
TABLE 2.
Yeast strains used in this study
Protein Expression and Purification
Bacterial Expression
Recombinant Saccharomyces cerevisiae Fis1 cytosolic domain, pET29b-Fis1ΔTM(1–127)-TEV-GST, was expressed in Escherichia coli Rosetta cells (Novagen) as described previously (38). For isotope labeling, transformed cells were grown at 37 °C in Luria broth containing kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml) to an A600 of 0.7–0.8, harvested by centrifugation, and transferred to M9 minimal medium containing 1 g/liter 15NH4Cl. Protein expression was induced by the addition of 0.4 mm isopropyl β-d-1-thiogalactopyranoside at 18 °C. After 18 h, cells were harvested by centrifugation and resuspended in column buffer (25 mm Tris·HCl, pH 7.4, 50 mm NaCl, 30 mm imidazole) containing protease inhibitors (Roche Applied Science). Cells were lysed with an EmulsiFlex C3 homogenizer (Avestin) at 15,000 p.s.i., followed by the addition of DNase (1 μg/ml) and MgCl2 (10 mm) and centrifugation to remove cell debris. The supernatant was purified by affinity chromatography with nickel-Sepharose 6 Fast Flow resin (5-ml column, GE Healthcare) using a 100-ml linear elution gradient of buffer containing 500 mm imidazole. Fractions containing 15N-labeled Fis1ΔTM-His were pooled and further purified by size exclusion chromatography (Superdex 75, GE Healthcare). Sample purity was evaluated by Coomassie-stained SDS-PAGE and was typically >95%. Some amount of free GST contaminant of varying length is routinely observed in the Fis1ΔTM-GST purification. Free GST for use as a negative control in pull-down experiments was expressed in Rosetta E. coli cells using the pGEX-4T-2 plasmid (Sigma-Aldrich) and purified as described for Fis1ΔTM-GST. This construct has a molecular weight slightly different from that of the GST contaminant in Fis1ΔTM-GST purifications due to the linker present in the fusion construct.
Yeast Protein Expression
Recombinant full-length S. cerevisiae Mdv1 and Dnm1 constructs, pEG(KT)-MBP-TEV-Mdv1 and pEG(KT)-MBP-TEV-Dnm1, were transformed into the protease-deficient yeast strain DDY1810 using the LiAc method. Transformed DDY1810 yeast were grown in a 50-ml starter culture of complete synthetic medium lacking uracil with 2% dextrose (CSM+D−U) at 30 °C with shaking to an A600 of ∼2. 25 ml were then used to inoculate 1 liter of CSM lacking uracil and leucine with 2% raffinose (CSM+R−UL) and 40 mg/liter tryptophan. The cells were grown at 26 °C with shaking to an A600 of ∼2 before protein expression was induced with 2% galactose supplemented with 20 g/liter bacto-peptone and 10 g/liter yeast extract. Cells were harvested by centrifugation after 16–18 h of induction. Washed cells were resuspended in a minimal amount of sterile water with 24 μg/ml 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride and stored at −80 °C for later use. For purification, cells were thawed on ice in column buffer (50 mm phosphate, pH 7.4, 500 mm NaCl, 5 mm MgCl2, 1 mm DTT) containing protease inhibitors (Roche Applied Science). Cells were lysed with five passes through an EmulsiFlex C3 homogenizer (Avestin) at 20,000 p.s.i. DNase was added to the lysate to 2 μg/ml, and cell debris was removed by centrifugation. Protein was isolated from the supernatant by amylose affinity chromatography using 20 mm maltose in the elution buffer. Sample purity was assessed by Coomassie-stained SDS-PAGE and was typically 80%. For MBP-Dnm1 (but not MBP-Mdv1, due to technical difficulties), the MBP tag was removed by TEV cleavage and subsequent size exclusion chromatography.
NMR Chemical Shift Perturbation by pH Titration
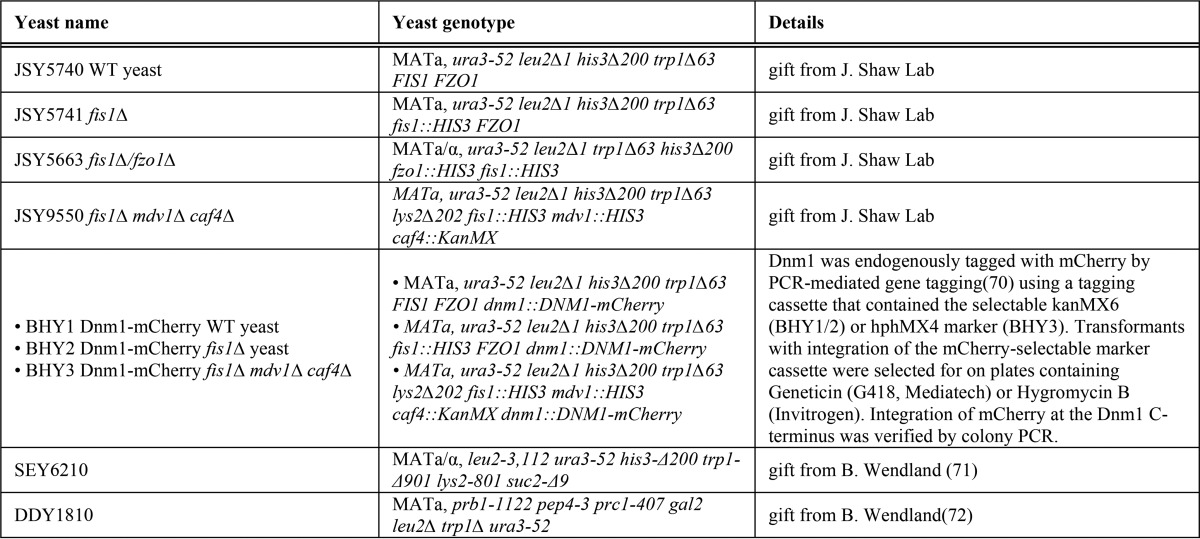
Samples of 250 μm uniformly 15N-labeled Fis1ΔTM were used for NMR experiments to determine pH-dependent chemical shift changes. The samples also contained 50 mm NaCl, 10 mm EDTA, 1 mm DTT, and 10% 2H2O. One sample starting at pH 5.7 was incrementally titrated to pH 9.0 by the addition of NaOH, whereas another sample was incrementally acidified to pH 3.0 by the addition of HCl. A total of 26 two-dimensional 1H-15N heteronuclear single-quantum correlation (HSQC) experiments were collected every ∼0.2 pH units on a 600-MHz Bruker AVANCE I equipped with TXI cryoprobe at 25 °C using WATERGATE for solvent suppression (59). HSQC spectra were collected at a rate of eight scans per increment, 1280 (t2) × 256 (t1) points with acquisition times of 128 ms (1H) and 78 ms (15N). NMR data processing was conducted with NMRPipe (60) and zero-filled twice to give a digital resolution of 3.9 and 6.4 Hz/point for 1H and 15N, respectively. Spectra were subsequently analyzed with NMRView (61) using a Titration.tcl script (kindly provided by Dr. Kevin Gardner, CUNY Advanced Science Research Center) to track peaks through the titration. The pH 7.4 spectrum was referenced to an external standard (2,2-dimethyl-2-silapentanesulfonic acid). The pH-dependent changes in amide backbone proton (Δ1H) and nitrogen (Δ15N) chemical shifts were determined from the differences in peak centroids relative to the pH 7.4 reference spectrum, plotted as a function of pH, and fit to the Henderson-Hasselbalch equation to determine apparent pKa values. Single chemical shift data sets were fit to the above equations, and the appropriate model for each data set was determined by χ2 difference tests. In addition to intra-residue ionizations, some backbone amides report on ionizations from non-neighboring, but proximal, side chains that are within 10 Å (see below) (supplemental Table 1).
The Henderson-Hasselbalch equations are as follows,
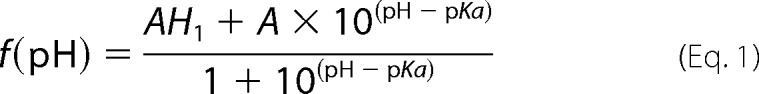 |
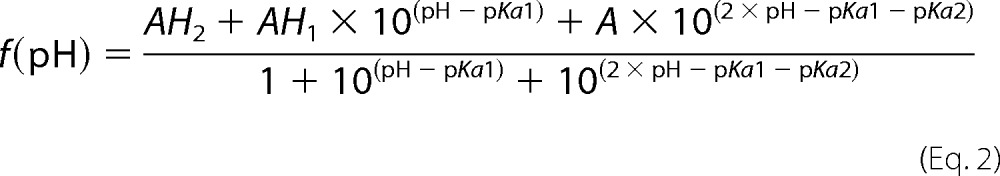 |
 |
where AHn is baseline for the protonated species (where n is the number of protons = 1, 2, or 3 (Equations 1–3, respectively)) and A is the baseline for the deprotonated species. Errors reported for the fit include uncertainties in the NMR chemical shift differences (0.009 and 0.01 ppm for 1H and 15N, respectively) estimated from the variance in the chemical shifts that exhibited little pH dependence from pH 3 to 9. Note that these uncertainties do not reflect the larger uncertainty in pH determination of the sample, which we estimate is 0.1 pH units.
Criteria similar to those proposed by Nielsen and co-workers (27) are applied, and only those pKa values that meet all three conditions are “reliable.” 1) Magnitude criteria (Δδ ≥ cut-off, where cut-off is 0.060 ppm for 1H and 0.30 ppm for 15N) ensure that the magnitude of the chemical shift change is large enough to provide a good fit. 2) The uniqueness criterion (pKa1app − pKa2app ≥ 0.2) is for multiple pKa fits and ensures that one is fitting distinct titration events. 3) Baseline criteria (pKaapp − pHmin ≥ 0.25; pHmax − pKaapp ≥ 0.25) ensure that reasonable pre- or post-transition baselines are present for fitting. In the absence of good baselines, an upper or lower limit to the pKa can be determined.
Glycerol Growth Assay
To assess the functionality of Fis1-E78A in vivo, fis1ΔfzoΔ yeast (derived from JSY5663, a gift from Dr. Janet Shaw, University of Utah) were transformed with plasmid encoding Fis1, Fis1-E78A, or Fis1 lacking 16 N-terminal residues (ΔNFis1) under control of the galactose promoter (pGAL-Fis1 (38)) using standard techniques. Overnight cultures were grown in CSM+D−U at 30 °C. As a control, fis1ΔfzoΔ cells lacking plasmid were grown in CSM+D−U. Serial dilutions of A600 0.5, 0.1, 0.02, and 0.004 were made from these cultures and spotted onto YP medium containing either 2% glucose (YPD) or 3% glycerol (YPG). Plates were incubated at 30 °C for 5 days and scored for growth on YPG (growth indicates non-functional fission activity).
Biochemical Pull-down Assay
Purified MBP-Mdv1 and Dnm1 were labeled with AlexaFluor Maleimide (AFM)-488 (Invitrogen) per manufacturer's protocol for easier detection in pull-down assays. 1 μm Fis1ΔTM-GST was incubated with 2 μm AFM-labeled MBP-Mdv1 or 4 μm Dnm1 in pull-down buffer (50 mm sodium phosphate, pH 7.4, 150 mm NaCl, and 10 mm DTT with 5 mm MgCl2 added for experiments with Dnm1) for 2 h at room temperature with rotating. 5 μl of washed and equilibrated magnetic GST bead slurry (Promega) was added, and the samples were incubated for 30 min, rotating at room temperature. Beads were washed 5 times with 500 μl of pull-down buffer, and the bound proteins were released by incubating in 24 μl of pull-down buffer containing 50 mm glutathione, pH 8.2. After 30 min rotating at room temperature, the elution was removed and mixed with SDS loading dye for analysis by SDS-PAGE. Gels were scanned on a Typhoon 9410 variable mode imager (GE Healthcare) with an excitation of 488 nm and a 520BP40 filter to detect AFM-labeled MBP-Mdv1 or Dnm1 and then subsequently stained with Coomassie Blue Silver. ImageQuant TL (GE Healthcare) was used to quantitate protein bands.
Fis1ΔTM Dimerization by Refolding
Samples of 25 μm Fis1ΔTM-His WT and E78A were used to assess the effect of E78A on Fis1 dimerization. To unfold the protein, samples were dialyzed overnight at 4 °C against 6 m guanidine hydrochloride (GdnHCl) buffered with either 100 mm phosphate (for pH 7.0) or 100 mm acetate (for pH 4.5) and containing 50 mm NaCl and 10 mm DTT. Samples were then transferred to buffer without GdnHCl for refolding and dialyzed extensively to remove the denaturant. Dimer formation was analyzed by size exclusion chromatography with detection of intrinsic tryptophan fluorescence emission at 350 nm (excitation at 280 nm).
Fis1ΔTM Membrane Binding
Synthetic lipids were obtained from Avanti Polar Lipids (Alabaster, AL). For sedimentation assays, Br4DSPC:DOPG vesicles were prepared containing 60% Br4DSPC, which allows for sedimentation at low centrifugal forces, and 40% DOPG. Vesicles also contained 0.25% Rh-DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)) to aid visualization of lipid pellets. Lipids were measured from chloroform stocks in the correct ratio, mixed for at least 30 min, and dried in a thin film under a stream of nitrogen. Dried films were lyophilized for 16 h to remove excess chloroform and then resuspended in deionized water to make a 13.1 mm lipid stock. Lipid solutions were freeze-thawed 11 times in a dry ice/ethanol bath and 37 °C water bath and extruded 11 times through a 100-nm nucleopore track etch membrane (Whatman) using an Avanti syringe extruder apparatus (Avanti Polar Lipids) to create unilamellar vesicles.
Vesicle sedimentation assays were performed as described (62). Briefly, 100-μl samples of 5 μm protein buffered with 20 mm phosphate, 20 mm acetate at the desired pH were prepared with and without vesicles and incubated for 6 h at 25 °C with rotation. Samples were then subjected to three 30-min centrifugation steps at 1500, 6000, and 18,000 × g to pellet vesicles. The top 80 μl of supernatant were diluted into SDS loading dye for analysis. The next 20 μl were discarded, and the pellets were resuspended in 100 μl of buffer and diluted into SDS loading dye. Supernatant and pellet fractions were analyzed by SDS-PAGE and stained with Coomassie G-250 Blue Silver stain. Protein bands were quantitated using ImageQuant TL (GE Healthcare). The fraction of lipid-bound protein was calculated as volumepellet/(volumepellet + volumesupernatant).
Chemical Denaturation of Fis1-E78A
To assess the structural integrity of the E78A substitution, chemical denaturation experiments were performed by monitoring the intrinsic fluorescence of the two tryptophan WT and E78A constructs, as a function of increasing GdnHCl concentration at 25 °C using a SLM-48000 spectrofluorometer (SLM-AMINCO, Urbana, IL). Samples were prepared by mixing 5 μm protein stock solutions (0 m and 6 m GdnHCl) buffered with 25 mm Tris-HCl (pH 7.4), 150 mm NaCl, 10 mm EDTA, and 1 mm DTT. Excitation was at 295 nm, and emission spectra were collected from 310 to 480 nm at a scan rate of 2 nm/s with slit widths of 2 and 4 nm for excitation and emission, respectively. The Trp emission at 330 nm was plotted as a function of GdnHCl concentration and fit to a two-state model using the method of Santoro and Bolen (63) with the fit equation, f(x) = {(yf + mfx) + (yu + mux) × exp[−(ΔG + mgx)/RT]}/{1 + exp[−(ΔG + mgx)/RT]} to determine the midpoint of the unfolding transition, Cm.
Co-IP Assay
Co-IP assays between Fis1 and Mdv1, Fis1 and Dnm1, or Fis1 and Caf4 were performed on fis1Δ yeast expressing 1×HA-Fis1 (pRS415-MET25–1×HA-Fis1) and either 3×-FLAG-Mdv1 (pRS414-MET25–3×-FLAG-Mdv1), genomic Dnm1-mCherry or 3×-FLAG-Caf4 (pRS414-MET25–3×-FLAG-Caf4), respectively. All co-IP assays were adapted from earlier protocols (33). An overnight starter culture of the transformed yeast was back-diluted to 0.3 A600 in complete synthetic medium lacking leucine and tryptophan with 2% dextrose and 130 μm methionine (CSM+D−LT+1×MET; Mdv1 and Caf4) or just lacking leucine (CSM+D−L+1×MET; Dnm1), grown to mid-log phase at 30 °C (∼3 h of shaking at 250 rpm), and collected by centrifugation (50 A600 units). The cells were washed in water, resuspended in Buffer Z (0.1 mm Tris-SO4, pH 9.4, 10 mm DTT) at 0.5 g cells/ml, and incubated at 30 °C for 10 min. The cells were collected, washed in Buffer W (20 mm potassium phosphate, pH 7.4, 1.2 m sorbitol), resuspended at 0.15 g cells/ml Buffer W containing Zymolyase 100T (5 mg/g of cells; Seikagaku Biobusiness, Tokyo, Japan), and incubated at 37 °C. After 45 min, 2.5 mm DSP (Thermo Fisher Scientific) was added, and the cells were incubated for an additional 30 min at 37 °C. The cells were quenched with 50 mm glycine and washed two times in Buffer W containing 50 mm glycine. The spheroplasted yeast were then chilled on ice, lysed in co-IP buffer (1% Triton X-100, 150 mm NaCl, 30 mm HEPES-KOH, pH 7.4, 50 mm glycine, and 1:200 yeast protease inhibitor mixture (Sigma)), and incubated on ice for 30 min. The cell debris was cleared by centrifugation, and the supernatant (42 A600 equivalents) was combined with magnetic HA beads (Thermo Fisher Scientific), rotating overnight at 4 °C, while the input fraction (5 A600 equivalents) was boiled in sample buffer. The magnetic beads were collected and washed five times in co-IP buffer and boiled in 2× sample buffer to release bound proteins. The supernatant was separated from the magnetic beads, β-mercaptoethanol was added to reverse the cross-linking, and the sample was boiled again. 14 A600 equivalents of the co-IP reaction and 2.4 A600 equivalents of the input reaction were resolved on an SDS-polyacrylamide gel followed by Western blotting for HA (Roche Applied Sciences), FLAG (Sigma-Aldrich), or mCherry (Abcam).
Confocal Microscopy
Mitochondrial Morphology Assay
JSY5740 WT and JSY5741 fis1Δ yeast were co-transformed using the LiAc method with p414GPD-mito-RFP-ff and either pRS415-MET25 empty vector, pRS415-MET25–1×HA-yFis1 WT, or E78A and plated on complete synthetic media lacking both leucine and tryptophan and supplemented with 2% dextrose and 650 μm methionine (CSM+D−LT+5×MET). An overnight starter culture of the transformed yeast was back-diluted to 0.3 A600 in CSM+D−LT+1×MET, grown to mid-log phase at 30 °C (∼3 h of shaking at 250 rpm), and collected by centrifugation. The yeast were then washed in CSM+D−LT+1×MET, resuspended in a small volume of CSM+D−LT+1×MET, plated on an agarose imaging bed, and covered by a 22 × 22-mm No. 1.5 coverglass (Ted Pella, Inc.). This agarose bed was created according to Ref. 64. Briefly, a 2% low melt agarose (Thermo Fisher Scientific) solution containing CSM+D−LT+1×MET was added to a 3-well depression slide (VWR) and was leveled off by covering with an additional slide. The yeast were imaged for mito-RFP, and the resulting confocal projections were assessed for WT or fission mutant morphology according to published procedures (32).
Mdv1 Localization
JSY5740 WT and JSY5741 fis1Δ yeast were co-transformed using the LiAc method with p414GPD-mito-RFP-ff and pRS416-MET25-GFP-Mdv1 and plated on CSM+D−UT and incubated at 30 °C. The resulting yeast were then transformed with either pRS415-MET25 empty vector, pRS415-MET25–1×HA-yFis1 WT, or E78A and plated on CSM+D−ULT+5×MET. The yeast were then grown overnight, back-diluted to 0.3 A600 in CSM+D−ULT+1×MET, and ultimately grown to mid-log phase at 30 °C (∼3 h of shaking at 250 rpm) and processed for GFP fixation. Following the fixation, cells were imaged for mito-RFP and GFP-Mdv1.
Dnm1 Localization
BHY1 Dnm1-mCherry WT and BHY2 Dnm1-mCherry fis1Δ yeast were transformed using the LiAc method with pVT100U-mito-GFP, plated on CSM+D−U, and incubated at 30 °C. The resulting yeast were then transformed with either pRS415-MET25 empty vector, pRS415-MET25–1×HA-Fis1 WT, or E78A, plated on CSM+D−UL+5×MET, and incubated at 30 °C. The yeast were then grown overnight, back-diluted to 0.3 A600 in CSM+D−UL+1×MET, and ultimately grown to mid-log phase at 30 °C and processed for GFP fixation. Following the fixation, cells were imaged for mito-GFP and Dnm1-mCherry. For the time course experiment, the cells were plated on an agarose bed similar to the morphology assay above, and a single image window was maintained and revisited every hour for 5 h total.
BHY3 Dnm1-mCherry fis1Δ mdv1Δ caf4Δ yeast were transformed using the LiAc method with pVT100U-mito-GFP and either pRS414-MET25-vector, pRS414-MET25–3×FLAG-Mdv1, or pRS414-MET25–3×-FLAG-Caf4; plated on CSM+D−UT+5×MET; and incubated at 30 °C. The resulting yeast were then transformed with either pRS415-GAL empty vector, pRS415-GAL-1×HA-Fis1 WT, ΔN16, or E78A; plated on CSM+D−ULT+5×MET; and incubated at 30 °C. The yeast were then grown overnight in CSM+D-ULT+5×MET, back-diluted to 0.3 A600 in CSM+GAL-ULT+1×MET (GAL; 2% galactose), and ultimately grown to mid-log phase at 30 °C and processed for GFP fixation. Following the fixation, cells were imaged for mito-GFP and Dnm1-mCherry.
GFP Fixation
Mid-log phase yeast were collected by centrifugation; resuspended in 4% paraformaldehyde, 3.4% sucrose; and incubated for 15 min at room temperature. Cells were then washed one time in 500 μl of 100 mm KPO4, 1.2 m sorbitol and resuspended in a small volume of 100 mm KPO4, 1.2 m sorbitol. Cells were then stored at 4 °C.
Colocalization
Confocal image projections for the localization experiments were imported into Volocity. Individual yeast were selected and measured for the Pearson's correlation co-localization index value. Data are displayed as a scatter dot plot using Igor Pro, and the mean value is indicated with a black line.
Confocal Microscope
All images were collected using a Nikon D-eclipse C1 confocal microscope and a ×100/1.45 numerical aperture Plan Apo oil objective. Images were acquired at zoom 3 with z-sections of 0.5 μm, and projections were generated using ImageJ. Images were pseudocolored to purple and green using Adobe Photoshop. All scale bars in the images represent 5 μm. For the time course experiment, a single image window was maintained and revisited every hour for 5 h total.
Author Contributions
M. K. R. and R. B. H. conceived the project and with K. R. M. analyzed the pKa data. M. K. R. performed the NMR experiments, recombinant protein experiments, and in vivo fission growth assays and also created the Dnm1-mCherry yeast strains. M. C. H. created the strains for and performed the confocal imaging and the coimmunoprecipitation experiments and also analyzed the confocal images. A. E. P. and K. R. M. created scripts to query protein and pKa databases and analyzed these data. J. M. E. collected and analyzed E78A NMR data. M. K. R., M. C. H., K. R. M., J. M. E., and R. B. H. prepared the figures. The manuscript was written by M. K. R., M. C. H., K. R. M., and R. B. H.
Supplementary Material
Acknowledgments
We thank Amber Bakkum for the pQE30 yFis1(1–128) plasmid, Drs. J. Corbett and A. Marchese for the use of their laboratory's confocal microscopes, Dr. J. Shaw for generously sharing yeast strains and plasmids, Dr. B. Wendland for mentorship and shared resources, Dr. B. Smith for helpful advice, and Dr. B. Volkman for shared resources and the pQE30 plasmid.
This work was supported by National Institutes of Health Grant R01 GM067180. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Results, Tables 1 and 2, Figs. 1–9, and Videos 1–5.
- TPR
- tetratricopeptide repeat
- Br4DSPC
- 1,2-dibromostearoyl-sn-glycero-3-phosphocholine
- DOPG
- dioleoylphosphatidylglycerol
- IP
- immunoprecipitation
- HSQC
- heteronuclear single-quantum correlation
- GdnHCl
- guanidine hydrochloride
- TEV
- tobacco etch virus
- MBP
- maltose-binding protein.
References
- 1. Scheffler I. E. (2008) Mitochondria, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 2. Archer S. L. (2013) Mitochondrial dynamics: mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 369, 2236–2251 [DOI] [PubMed] [Google Scholar]
- 3. DuBoff B., Feany M., and Götz J. (2013) Why size matters: balancing mitochondrial dynamics in Alzheimer's disease. Trends Neurosci. 36, 325–335 [DOI] [PubMed] [Google Scholar]
- 4. Chan D. C. (2007) Mitochondrial dynamics in disease. N. Engl. J. Med. 356, 1707–1709 [DOI] [PubMed] [Google Scholar]
- 5. Nunnari J., and Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shenouda S. M., Widlansky M. E., Chen K., Xu G., Holbrook M., Tabit C. E., Hamburg N. M., Frame A. A., Caiano T. L., Kluge M. A., Duess M.-A., Levit A., Kim B., Hartman M.-L., Joseph L., et al. (2011) Altered mitochondrial dynamics contributes to endothelial dysfunction in diabetes mellitus. Circulation 124, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bui H. T., and Shaw J. M. (2013) Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr. Biol. 23, R891–R899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mozdy A. D., McCaffery J. M., and Shaw J. M. (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Onoue K., Jofuku A., Ban-Ishihara R., Ishihara T., Maeda M., Koshiba T., Itoh T., Fukuda M., Otera H., Oka T., Takano H., Mizushima N., Mihara K., and Ishihara N. (2013) Fis1 acts as a mitochondrial recruitment factor for TBC1D15 that is involved in regulation of mitochondrial morphology. J. Cell Sci. 126, 176–185 [DOI] [PubMed] [Google Scholar]
- 10. Friedman J. R., and Nunnari J. (2014) Mitochondrial form and function. Nature 505, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffin E. E., Graumann J., and Chan D. C. (2005) The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170, 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo Q., Koirala S., Perkins E. M., McCaffery J. M., and Shaw J. M. (2012) The mitochondrial fission adaptors Caf4 and Mdv1 are not functionally equivalent. PLoS One 7, e53523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koirala S., Guo Q., Kalia R., Bui H. T., Eckert D. M., Frost A., and Shaw J. M. (2013) Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. U.S.A. 110, E1342–E1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J. E., and Nunnari J. (2006) Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281, 2177–2183 [DOI] [PubMed] [Google Scholar]
- 15. Cerveny K. L., and Jensen R. E. (2003) The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 14, 4126–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wells R. C., Picton L. K., Williams S. C., Tan F. J., and Hill R. B. (2007) Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J. Biol. Chem. 282, 33769–33775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki M., Neutzner A., Tjandra N., and Youle R. (2005) Novel structure of the N terminus in yeast Fis1 correlates with a specialized function in mitochondrial fission. J. Biol. Chem. 280, 21444–21452 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y., and Chan D. (2007) Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. U.S.A. 104, 18526–18530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Chan N. C., Ngo H. B., Gristick H., and Chan D. C. (2012) Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (Mdv1) coiled coil. J. Biol. Chem. 287, 9855–9861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tooley J. E., Khangulov V., Lees J. P., Schlessman J. L., Bewley M. C., Heroux A., Bosch J., and Hill R. B. (2011) The 1.75 Å resolution structure of fission protein Fis1 from Saccharomyces cerevisiae reveals elusive interactions of the autoinhibitory domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Young L., Jernigan R. L., and Covell D. G. (1994) A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 3, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo Conte L., Chothia C., and Janin J. (1999) The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 23. Chakrabarti P., and Janin J. (2002) Dissecting protein-protein recognition sites. Proteins 47, 334–343 [DOI] [PubMed] [Google Scholar]
- 24. Castañeda C. A., Fitch C. A., Majumdar A., Khangulov V., Schlessman J. L., and García-Moreno B. E. (2009) Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins 77, 570–588 [DOI] [PubMed] [Google Scholar]
- 25. Schaller W., and Robertson A. (1995) pH, ionic strength, and temperature dependences of ionization equilibria for the carboxyl groups in turkey ovomucoid third domain. Biochemistry 34, 4714–4723 [DOI] [PubMed] [Google Scholar]
- 26. Wilkens S. J., Xia B., Weinhold F., Markley J. L., and Westler W. M. (1998) NMR investigations of Clostridium pasteurianum rubredoxin: origin of hyperfine 1H, 2H, 13C, and 15N NMR chemical shifts in iron-sulfur proteins as determined by comparison of experimental data with hybrid density functional calculations. J. Am. Chem. Soc. 120, 4806–4814 [Google Scholar]
- 27. Farrell D., Miranda E. S., Webb H., Georgi N., Crowley P. B., McIntosh L. P., and Nielsen J. E. (2010) Titration_DB: storage and analysis of NMR-monitored protein pH titration curves. Proteins 78, 843–857 [DOI] [PubMed] [Google Scholar]
- 28. Sesaki H., and Jensen R. E. (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hermann G. J., and Shaw J. M. (1998) Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14, 265–303 [DOI] [PubMed] [Google Scholar]
- 30. Rapaport D., Brunner M., Neupert W., and Westermann B. (1998) Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150–20155 [DOI] [PubMed] [Google Scholar]
- 31. Picton L. K., Casares S., Monahan A. C., Majumdar A., and Hill R. B. (2009) Evidence for conformational heterogeneity of fission protein Fis1 from Saccharomyces cerevisiae. Biochemistry 48, 6598–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koirala S., Bui H. T., Schubert H. L., Eckert D. M., Hill C. P., Kay M. S., and Shaw J. M. (2010) Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J. Cell Biol. 191, 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karren M. A., Coonrod E. M., Anderson T. K., and Shaw J. M. (2005) The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 171, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blatch G. L., and Lässle M. (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 35. Tieu Q., Okreglak V., Naylor K., and Nunnari J. (2002) The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lackner L. L., Horner J. S., and Nunnari J. (2009) Mechanistic analysis of a dynamin effector. Science 325, 874–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bui H. T., Karren M. A., Bhar D., and Shaw J. M. (2012) A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment. J. Cell Biol. 199, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lees J. P., Manlandro C. M., Picton L. K., Tan A. Z., Casares S., Flanagan J. M., Fleming K. G., and Hill R. B. (2012) A designed point mutant in Fis1 disrupts dimerization and mitochondrial fission. J. Mol. Biol. 423, 143–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lackner L. L., and Nunnari J. M. (2009) The molecular mechanism and cellular functions of mitochondrial division. Biochim. Biophys. Acta 1792, 1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Losón O. C., Song Z., Chen H., and Chan D. C. (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shaw J. M., and Nunnari J. (2002) Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12, 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cerveny K. L., Tamura Y., Zhang Z., Jensen R. E., and Sesaki H. (2007) Regulation of mitochondrial fusion and division. Trends Cell Biol. 17, 563–569 [DOI] [PubMed] [Google Scholar]
- 43. Bhar D., Karren M. A., Babst M., and Shaw J. M. (2006) Dimeric Dnm1-G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 281, 17312–17320 [DOI] [PubMed] [Google Scholar]
- 44. Tieu Q., and Nunnari J. (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Legesse-Miller A., Massol R. H., and Kirchhausen T. (2003) Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 14, 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fekkes P., Shepard K. A., and Yaffe M. P. (2000) Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 151, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cerveny K. L., McCaffery J. M., and Jensen R. E. (2001) Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell 12, 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mumberg D., Müller R., and Funk M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gandre-Babbe S., and van der Bliek A. M. (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Palmer C. S., Osellame L. D., Laine D., Koutsopoulos O. S., Frazier A. E., and Ryan M. T. (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., and Nistér M. (2011) Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30, 2762–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seaman M. N. J., Harbour M. E., Tattersall D., Read E., and Bright N. (2009) Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J. Cell Sci. 122, 2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamano K., Fogel A. I., Wang C., van der Bliek A. M., and Youle R. J. (2014) Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife 3, e01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Otera H., and Mihara K. (2011) Discovery of the membrane receptor for mitochondrial fission GTPase Drp1. Small GTPases 2, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Palmer C. S., Elgass K. D., Parton R. G., Osellame L. D., Stojanovski D., and Ryan M. T. (2013) Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 288, 27584–27593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hill R. B., Hong J. K., and DeGrado W. F. (2000) Hydrogen bonded cluster can specify the native state of a protein. J. Am. Chem. Soc. 122, 746–747 [Google Scholar]
- 57. Norel R., Sheinerman F., Petrey D., and Honig B. (2001) Electrostatic contributions to protein-protein interactions: fast energetic filters for docking and their physical basis. Protein Sci. 10, 2147–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sheinerman F. B., Norel R., and Honig B. (2000) Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 10, 153–159 [DOI] [PubMed] [Google Scholar]
- 59. Piotto M., Saudek V., and Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 60. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 61. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 62. Wells R. C., and Hill R. B. (2011) The cytosolic domain of Fis1 binds and reversibly clusters lipid vesicles. PLoS One 6, e21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Santoro M. M., and Bolen D. W. (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 64. Rines D. R., Thomann D., Dorn J. F., Goodwin P., and Sorger P. K. (2011) Live cell imaging of yeast. Cold Spring Harbor Protoc. 10.1101/pdb.top065482 [DOI] [PubMed] [Google Scholar]
- 65. Ingerman E., Perkins E. M., Marino M., Mears J. A., McCaffery J. M., Hinshaw J. E., and Nunnari J. (2005) Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Waltner J. K., Peterson F. C., Lytle B. L., and Volkman B. F. (2005) Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Sci. 14, 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pierce B. D., and Wendland B. (2009) Sequence of the yeast protein expression plasmid pEG(KT). Yeast 26, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., and Hieter P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]
- 69. Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., and Westermann B. (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 71. Shaw J. D., Hama H., Sohrabi F., DeWald D. B., and Wendland B. (2003) PtdIns(3,5)P2 is required for delivery of endocytic cargo into the multivesicular body. Traffic 4, 479–490 [DOI] [PubMed] [Google Scholar]
- 72. Whitworth K., Bradford M. K., Camara N., and Wendland B. (2014) Targeted disruption of an EH-domain protein endocytic complex, Pan1-End3. Traffic 15, 43–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.