Abstract
Kinesin-1, -2, -5, and -7 generate processive hand-over-hand 8-nm steps to transport intracellular cargoes toward the microtubule plus end. This processive motility requires gating mechanisms to coordinate the mechanochemical cycles of the two motor heads to sustain the processive run. A key structural element believed to regulate the degree of processivity is the neck-linker, a short peptide of 12–18 residues, which connects the motor domain to its coiled-coil stalk. Although a shorter neck-linker has been correlated with longer run lengths, the structural data to support this hypothesis have been lacking. To test this hypothesis, seven kinesin structures were determined by x-ray crystallography. Each included the neck-linker motif, followed by helix α7 that constitutes the start of the coiled-coil stalk. In the majority of the structures, the neck-linker length differed from predictions because helix α7, which initiates the coiled-coil, started earlier in the sequence than predicted. A further examination of structures in the Protein Data Bank reveals that there is a great disparity between the predicted and observed starting residues. This suggests that an accurate prediction of the start of a coiled-coil is currently difficult to achieve. These results are significant because they now exclude simple comparisons between members of the kinesin superfamily and add a further layer of complexity when interpreting the results of mutagenesis or protein fusion. They also re-emphasize the need to consider factors beyond the kinesin neck-linker motif when attempting to understand how inter-head communication is tuned to achieve the degree of processivity required for cellular function.
Keywords: kinesin, microtubule, molecular motor, protein structure, x-ray crystallography, coiled-coil, kinesin neck-linker
Introduction
The coiled-coil was the first quaternary structural arrangement described and has been predicted to occur in ∼3% of all proteins (1, 2). This apparently simple motif shows considerable structural variation, but because of the characteristic distribution of hydrophobic and polar residues, it can be detected readily through sequence analysis (3–5). Coiled-coils play diverse functional roles, but in many cases, they serve as oligomerization domains. Under these circumstances, the exact length or starting point of the coiled-coil is structurally and functionally important. Unfortunately, this parameter is not well defined or uniformly predicted by any computational algorithm.
The precise starting point of the coiled-coil is particularly important in the kinesin and myosin family of molecular motors, where the dimerization module coordinates the activities of individual motor domains on separate polypeptide chains. Typically, in these motor families, there is a flexible section of the polypeptide that connects the motor to the dimerization module. The length of this connecting unit plays a critical role in processive molecular motors and is especially important for many members of the kinesin superfamily in which this domain is known as the neck-linker.
Kinesin motor proteins are classified into 15 different kinesin families, which share a structurally conserved kinesin motor domain (6–10). These families perform a diverse set of cellular functions, all of which involve moving along a microtubule track for cargo transport or modulating microtubule dynamics (11–17). Part of the classification is dictated by the location of their motor domains. N-terminally located motors comprise the majority of kinesin families. The exceptions are the kinesin-14A and -14B families that contain C-terminal motors and the kinesin-13 family in which the motor domain is located in the middle of the polypeptide (6).
This study is focused on N-terminal kinesins, which are composed of an N-terminal motor domain connected to a long α-helical region that dimerizes into a coiled-coil stalk that ends with a C-terminal cargo domain that may interact with other partner proteins or substrates. The N-terminal motor domain is responsible for ATP turnover coupled to force production. This group of motors is typically dimeric and shows processive movement along microtubules tracks. The ability of the N-terminal kinesin family to remain on the microtubule lattice is critical to their function.
Dimeric N-terminal kinesins employ an asymmetric hand-over-hand stepping motion to move processively along a microtubule as they hydrolyze ATP (Fig. 1) (18–23). A general outline of the hydrolytic cycle begins arbitrarily in an ATP waiting state, where the leading head without nucleotide is strongly bound to a microtubule, whereas the lagging head is bound to ADP, but only weakly associated with a microtubule (Fig. 1, E1) (18, 19). To proceed with stepping, the leading head binds ATP, and the dimer undergoes a structural transition transmitted through the neck-linker motif, a 12–18-amino acid flexible peptide that connects the motor domain to the coiled-coil stalk (Fig. 1, E2) (24, 25). This structural transition, designated neck-linker docking, shifts the lagging unbound head forward 16 nm to the next microtubule-binding site toward the microtubule plus end (Fig. 1, E3) (24, 26). Subsequently, ADP is released from this new leading head resulting in both heads bound to the microtubule (Fig. 1, E4 and E5) (27).
FIGURE 1.
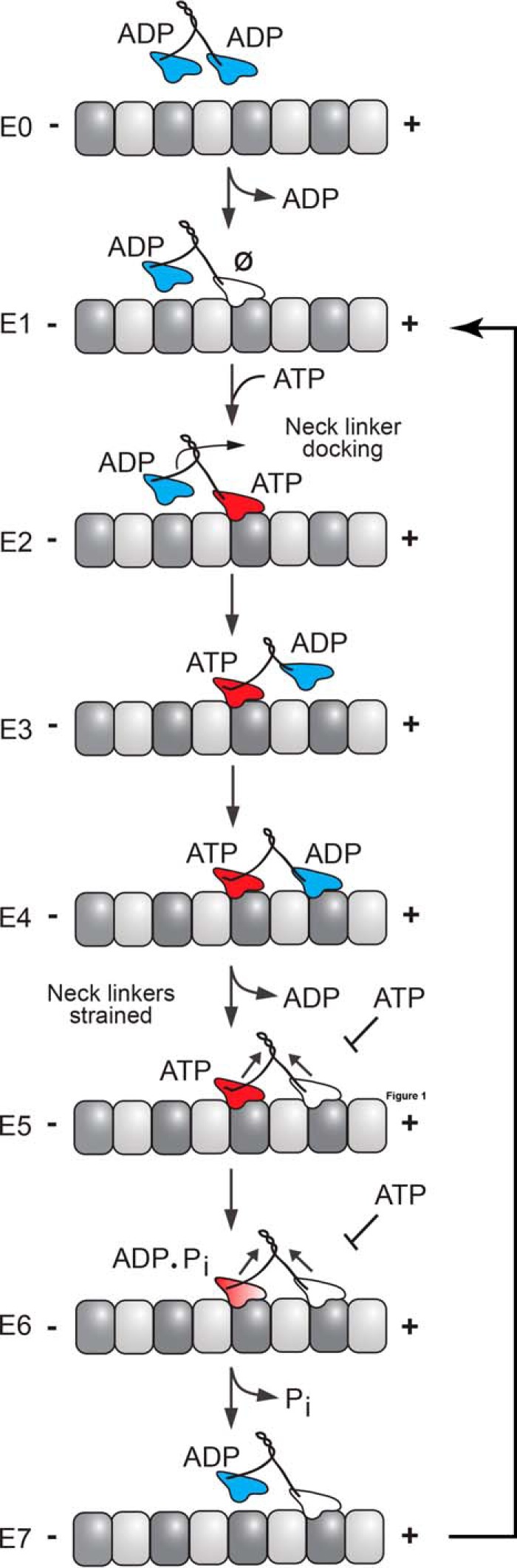
Schematic representation of the kinesin chemomechanical cycle that illustrates key transitions that influence processivity. A processive run is started by binding of either head followed by rapid ADP release to form the E1 intermediate where the leading head that forms the initial contact is microtubule-bound but nucleotide-free (Ø), whereas the trailing head is detached with ADP tightly bound. ATP binding to the leading head induces a series of structural transitions, including neck-linker docking that allows the trailing ADP-head to move 16 nm ahead to its new microtubule-binding site (E2–E4). ADP release from the new leading head (E4 and E5) results in the E5 two-head bound state, thereby generating intermolecular strain, which inhibits ATP binding at the now leading head. ATP hydrolysis at the trailing head followed by phosphate (Pi) release generates a microtubule weakly bound ADP state (E6 and E7). Detachment of the trailing head relieves the intermolecular strain (E7), and initiates the next motor cycle.
When both heads are bound to the microtubule, the neck-linker domains are oriented in opposite directions (28, 29). The neck-linker of the leading nucleotide-free head is oriented backward, inhibiting ATP binding and hydrolysis by the front head until the rear head detaches from the microtubule (Fig. 1, E6) (30). The neck-linker of the lagging ATP-bound head remains docked onto the catalytic core and directed forward. ATP hydrolysis on the microtubule-bound lagging head results in another structural rearrangement that leads to phosphate release. Thereby, the lagging head transitions into a weakly bound ADP state and detaches from the microtubule, thus starting the cycle anew (Fig. 1, E7).
During the stepping cycle, each motor domain must remain out-of-phase with the other. If both motor domains enter an unbound state at the same time, the processive run ends. The run length indicates the distance a motor steps along the microtubule before dissociation and gives a measure of processivity. Each kinesin family member has different average run lengths ranging from <0.2 μm, as in Eg5, to 2.1 μm, as in conventional kinesin-1, although this is construct-dependent for kinesin-1 and can range from 1.3 to 2.1 μm (25, 31, 32). Despite the structural conservation between different kinesin motors, there are clear kinetic differences between the families.
One domain hypothesized to contribute to processivity is the kinesin neck-linker, a small flexible peptide consisting of 12–18 amino acids (33–35). The neck-linker connects each motor domain to the coiled-coil stalk where the junction between these two entities has been assumed to occur at the same position in the sequence as that observed in kinesin-1 (36). The neck-linker itself undergoes a series of structural transitions as outlined in the kinesin mechanochemical cycle. It is hypothesized that longer neck-linkers increase the diffusional search area and therefore could slow down stepping, allowing time for the forward head to release from the microtubule (37). Recently, alterations in neck-linker length were shown to affect the kinetic cycle (38). Increasing neck-linker length resulted in increased rear head binding, and decreasing neck-linker length resulted in slower release of ADP from the unbound head. Both effects result in slowing the productive kinetic cycle (38). Because the neck-linker is involved in connecting the motor domain and coiled-coil stalk, changes in the length of the neck-linker are expected to alter communication between the two motor heads.
Given that the overall size of the kinesin motor domain is similar across all families and that they bind to microtubules in a similar manner, it is surprising that the predicted length of the neck-linker between families shows considerable variation even though within each family the neck-linker/α7 sequences are almost completely conserved (36). This is especially puzzling because a multitude of studies show increasing the neck-linker for a given kinesin by even one residue results in decreases in run length and processivity (25, 31, 34, 39, 40). This conundrum is particularly evident in members of the kinesin-2 family, KIF3AB and KIF3AC, which are unique in forming a heterodimeric motor. KIF3AB and KIF3AC further interact with an adaptor protein to bind a variety of cargoes for intraflagellar and neuronal transport (11, 15, 41–45). KIF3AC in particular has been implicated in neuronal repair (11). Both KIF3AB and KIF3AC are highly processive motors (46). Yet their neck-linker is predicted to be three residues longer than conventional kinesin, which would suggest that these motors should not be so processive (36). KIF3AB and KIF3AC have identical neck-linker lengths and nearly identical neck-linker sequences, differing only at Thr-380 in KIF3C, which is an alanine in KIF3A and KIF3B. Both motors are processive; KIF3AB and KIF3AC have run lengths of 1.6 and 1.2 μm, respectively (37, 46). However, the kinetic parameters vary significantly between the two motors (46–48). Furthermore, homodimeric species KIF3AA, KIF3BB, and KIF3CC also exhibit vastly different processivity parameters. KIF3AA and KIF3BB are highly processive, but KIF3CC travels at only 7.5 nm/s with an average run length of just 0.6 μm, which suggests that the neck-linker is not the only determinant of processivity (46). These observations prompted an investigation of the true length of the neck-linker in the best studied classes of N-terminal kinesin families.
The original estimates of the neck-linker length made the assumption that the coiled-coil would begin on a hydrophobic residue that lie in either the a or d position of the coiled-coil heptad repeat (36). However, the predictions of the first residue to adopt a helical conformation in any coiled-coil are ambiguous, even though the body of a coiled-coil is well indicated by current software (3, 4). Prediction software recognizes the heptad repeat in a coiled-coil domain. However, because the beginning and end of sequences may not follow that pattern, there is insufficient information to make accurate predictions of the start or of the end of a native coiled-coil. Given the considerable variation in the amino acid sequence of neck-linkers and associated coiled-coils, this raises the question whether the true length of the neck-linker has been accurately identified across the kinesin superfamily with the original assumption described above.
There is only one structure for a dimeric N-terminal kinesin, rat kinesin-1 (3KIN) because dimeric kinesins are difficult to crystallize (14). The 3KIN structure provided the first picture of the true α7 start and neck-linker length in the context of a dimeric motor. Most kinesin motor structures are monomeric, allowing the neck-linker to adopt varying conformations that may not reflect that experienced by the dimeric motor in vivo. For example, in the structure of the kinesin-2 KIF3B (3B6U) the neck-linker includes a cis-proline; therefore, this structure is unlikely to reflect the native neck-linker conformation. This study is directed toward providing an experimental foundation for determining the start site for the coiled-coil and by inference the length of the neck-linker. We determined the structures of the neck-linker and α7 helix from the following four different kinesin families: kinesin-1, -2, -5, and -7. Kinesin-1 is the canonical N-terminal processive kinesin motor. Kinesin-2 family members, KIF3A and KIF3C, are involved in long range transport and neuronal repair (11, 15). Kinesin-5 family member, Eg5, is unique within this subset as it is a bipolar tetrameric kinesin whose role is to cross-link microtubules during cell division (12, 13, 49). Kinesin-7 family member CENP-E is responsible for transporting misaligned chromosomes during congression in mitosis (16). In addition, we compared structures in the PDB2 that include a native start to their coiled-coil with the predictions generated from either MARCOIL or COILS-28 (3, 4). Overall, we find that structures of proteins are necessary to determine the true start site of the coiled-coil rather than relying on prediction software alone.
Results
Crystal Structures of Kinesin-1, -2, -5, and -7 Neck-linker α7 Helix Proteins
All neck-linker structures were homodimeric and solved to a resolution of 2.3 Å or higher allowing for accurate determination of secondary structure transitions. The extent of the ordered structure in each construct is given in Table 1. Several structures had multiple monomers in the asymmetric unit. Individual monomers were similar as shown in Table 1. For each neck-linker structure, the start of the coiled-coil helix was determined using the Dictionary of Secondary Structure of Proteins algorithm (50). In each structure, varying lengths of the neck-linker were ordered; thus we focused on the initiation point of the α7 helix to determine neck-linker length.
TABLE 1.
Average pairwise root means square differences between independent chains within each structural determination
First ordered residue is listed for the longest monomer and may not be ordered in other monomers in the asymmetric unit. r.m.s.d. is root mean square deviation. NA is not applicable.
| Construct | Monomers in asymmetric unit | Average r.m.s.d. | First ordered residue |
|---|---|---|---|
| Å | |||
| CENP-E | 6 | 0.862 | Asn-336a |
| Eg5 | 4 | 0.450 | Asn-336a |
| KIF3A-kinesin-1 | 8 | 0.739 | Asn-352 (KIF3A)a |
| KIF3C | 2 | 0.578 | Asn-374a |
| KIF3A | 1 | NA | Asp-354 |
| Kinesin-1 | 2 | 0.529 | Asn-340 |
| Kinesin-1 + DAL | 1 | NA | Asp of DAL (Ala-345-2) |
a In these structures two residues that remain from the recombinant tobacco etch virus cleavage site were also observed.
All of the structures were determined as fusions with the C-terminal dimerization domain of EB1. Previous studies have shown that inclusion of globular folding domains considerably increases the ability to express and crystallize sections of coiled-coil proteins and that they do not perturb the structure more than one heptad from the point of fusion (51, 52).
The crystal structure of the kinesin-1 neck-linker (Fig. 2) shows that the start of α7 and the end of the kinesin-1 neck-linker observed here is identical to that seen in the structure of the kinesin-1 dimer (PDB code 3KIN) (14). An overlay of the kinesin-1 neck-linker structure and dimeric kinesin-1 is shown in Fig. 3. Neck-linker residues Asn-340–Thr-344 were ordered in the crystal structure and adopt a random coil conformation, and the α7 helix begins at Ala-345 as expected. The consistency of the kinesin-1 neck-linker structure with predictions and previous work supports the assertion that the structures from other families likely represent the solution state of the junction between their neck-linker and α7.
FIGURE 2.
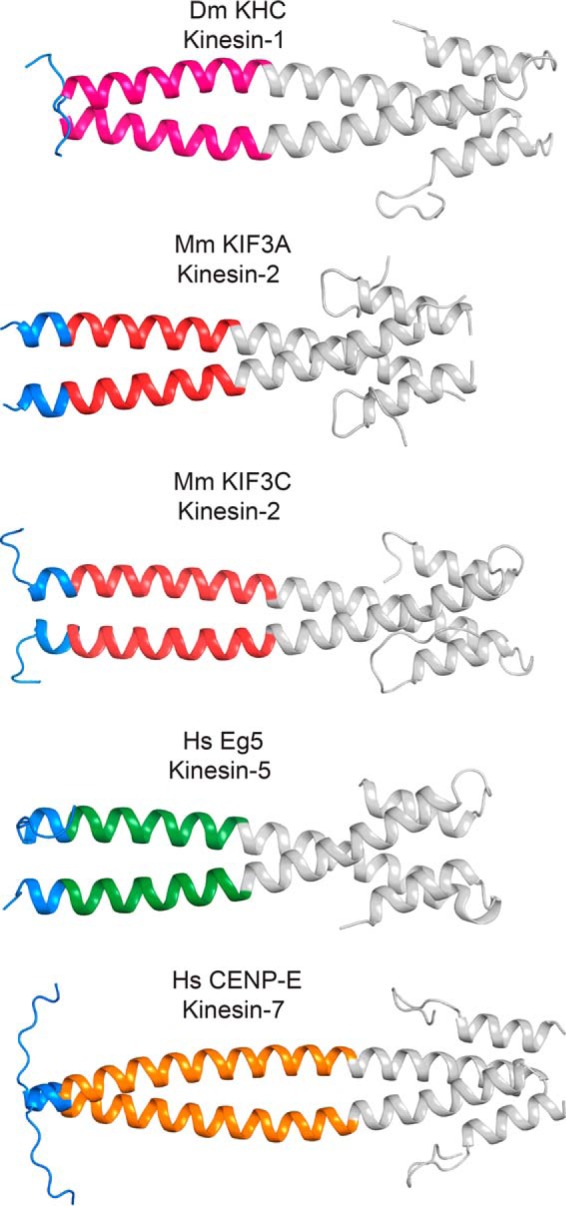
Crystal structures of kinesin-1, -2, -5, and -7 reveal unique class-specific neck-linker α7 neck-coil domains. The neck-linker motif predicted to occur based on the earlier studies of kinesin-1 is colored blue. Helix α7, as observed in kinesin-1, is colored according to the kinesin family to which it belongs. EB1, a coiled-coil fusion protein, is colored gray. There are varied amounts of ordered neck-linker motifs. These structures show that the true start of helix α7 is variable across the kinesin superfamily. Table 4 provides the protein sequence of each fusion protein and their coiled-coil registry. Figs. 2–6 were prepared in part with PyMOL.
FIGURE 3.
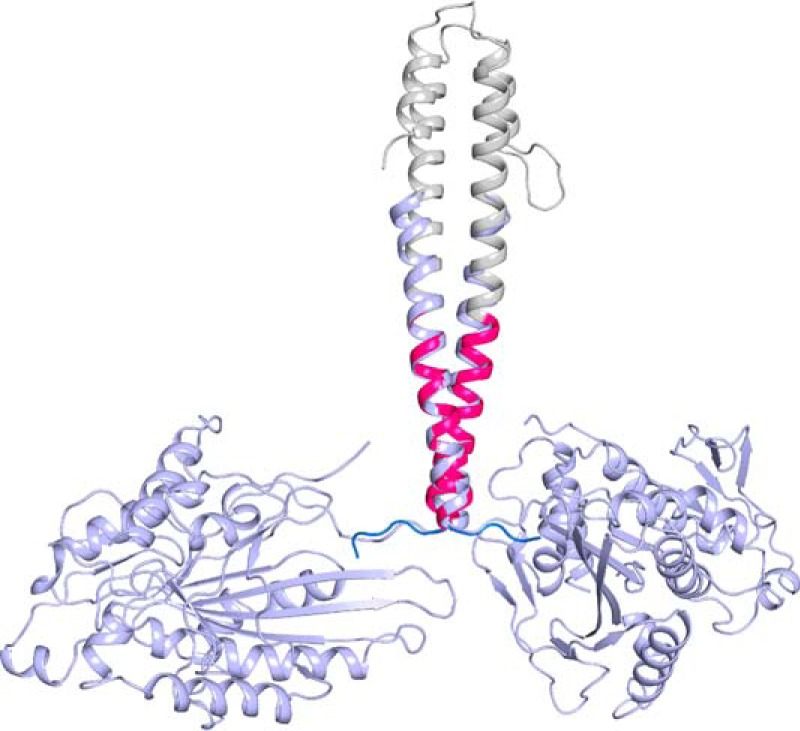
Overlay of the kinesin-1 neck-linker structure and dimeric kinesin-1 crystal structure (PDB code 3KIN) (14). The kinesin-1 neck-linker is colored as in Fig. 2. Dimeric kinesin-1 is shown in light blue.
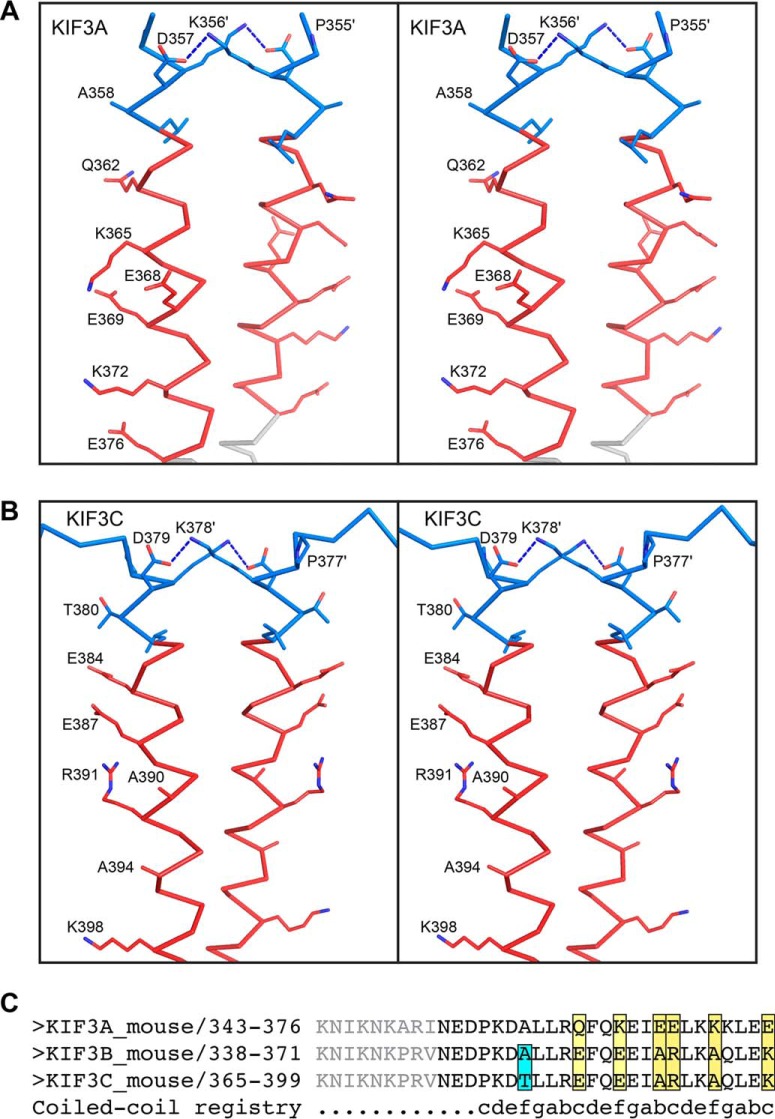
For the remaining neck-linker structures, the length of the neck-linker and the α7 helix deduced from prediction algorithms do not agree with the experimental structural data. Although the neck-linker itself is flexible as evidenced by docking, it is unlikely that the start residue for the α7 helix changes. Studies using high resolution atomic force microscopy and computational modeling have shown that there is no local conformational unwinding or “breathing” at helix α7 (53–55). Thus, the α7 start residues determined here are most likely the same as in the full dimeric motor. The kinesin-2 family members, KIF3A and KIF3C (Fig. 2), have significantly shorter neck-linkers and longer α7 helices than earlier models based on the kinesin-1 family. Although predictions suggested that the α7 helix would begin at Leu-360 in KIF3A and at Leu-382 in KIF3C, in the crystal structures α7 starts five residues earlier at Pro-355 and Pro-377 in KIF3A and KIF3C, respectively (Fig. 4). The discrepancy on the start of α7 leads to a shortening of the neck-linker from 17 residues to 12. This appears to be in contrast with the neck-linker observed in monomeric KIF3B structure (PDB code 3B6U), which has a length of 16 residues. However, the KIF3B structure (PDB code 3B6U) is monomeric where it is unlikely that there were enough residues to form a stable α-helix, leading to the discrepancy in the neck-linker lengths. Although the native kinesin-2s are heterodimeric, the crystallized constructs are homodimeric. It is not anticipated that the length of the neck-linker or structures of the coiled-coils will differ between the homodimer as compared with the heterodimer, as both KIF3A and KIF3C resulted in a neck-linker of 12 residues. Additionally, the buried residues in the first heptad of the coiled-coil are similar in both KIF3A and KIF3C, and thus it should not affect neck-linker length. Furthermore, the native coiled-coil is stabilized by a hydrogen bond between a lysine and aspartate in both KIF3A and KIF3C; thus this interaction should also be present in the heterodimer (Fig. 4). There are sequence differences in α7 between KIF3A and KIF3C, but these occur in positions that are solvent-exposed and do not interact with the adjacent α-helix (Fig. 4) and hence are not expected to greatly influence the structure of the heterodimer. The structure of the kinesin-2 neck-linkers in Fig. 2 show that the neck-linker is shorter than that of kinesin-1, which has a 14-residue neck-linker and the longest run length of all families tested (39).
FIGURE 4.
Selected side-chain interactions in and the sequence differences between KIF3A and KIF3C neck-linkers shown in stereo. KIF3A and KIF3C are colored as in Fig. 2, with side chains shown as sticks and colored by element. In both KIF3A (A) and KIF3C (B), there is a hydrogen bonding interaction between a lysine and an aspartate that stabilizes part of the coiled-coil. The sequences of the neck-linker and α7 are also shown (C) where the residues depicted in gray were not included in the constructs but represent the full-length linker. Interestingly, none of the differences in sequence between KIF3A and KIF3C are predicted to influence formation of a heterodimer.
The Eg5 neck-linker crystal structure (Fig. 2) shows differences in length as well. The α7 helix begins at Lys-371 and not Ile-375 as predicted, resulting in a neck-linker that was only 14 residues long. As in Eg5, the CENP-E neck-linker was predicted to be 18 residues long. The coiled-coil of CENP-E begins at Asp-341 rather than Leu-345, thus shortening the neck-linker to 14 residues (Fig. 2). These crystal structures show that both the CENP-E and Eg5 neck-linkers are the same length as that of kinesin-1. A summary of the predicted neck-linker lengths and actual neck-linker lengths along with the average run length is listed in Table 2.
TABLE 2.
Predicted and observed kinesin neck-linker lengths together with the average run length
| Neck-linker | Predicted length (36) | Observed length | Average run length | Refs. |
|---|---|---|---|---|
| μm | ||||
| DmKinesin-1 (kinesin-1) | 14 | 14 | 2.1 ± 0.1 | 39 |
| DmKinesin-1 + DAL | 17 | 15 | 0.39 ± 0.02 | 39 |
| MmKIF3AB (kinesin-2) | 17 | 12 | 1.23 ± 0.09 | 46 |
| MmKIF3AC (kinesin-2) | 17 | 12 | 1.62 ± 0.11 | 46 |
| HsEg5 (kinesin-5) (dimer) | 18 | 14 | 0.67 ± 0.07 | 32 |
| KIF3A-kinesin-1 (hybrid) | 17 | 15 | 0.7 ± 0.03 | 39 |
| HsCENP-E (kinesin-7) | 18 | 14 | 2.7 ± 0.2 | 80 |
Crystal Structures of the Kinesin-1 Extended Neck-linker and KIF3A-Kinesin-1 Hybrid
The coiled-coil stalk of the kinesin-1 motor has often been fused to the motor domain of other kinesin family members for single molecule studies where this served as a template for understanding the effect of length differences in the neck-linker seen across the entire kinesin superfamily (25, 39, 56–59). This hybrid was used in part to ensure that kinetic differences were derived from the differences in the neck-linker domain and not due to the coiled-coil stalk or other charged regions (39). Additionally, the hybrid constructs were easily expressed in Escherichia coli, rather than baculovirus (39). To determine the molecular consequences of these engineered hybrids and how they might affect the interpretation of changes in kinetic or motile behavior, structural studies were performed on a KIF3A-kinesin-1 hybrid and a kinesin-1 in which three-residues (kinesin-1 + DAL) were inserted (Fig. 5). This extension has been previously used to examine processivity changes in the kinesin-1 motor (25, 39).
FIGURE 5.
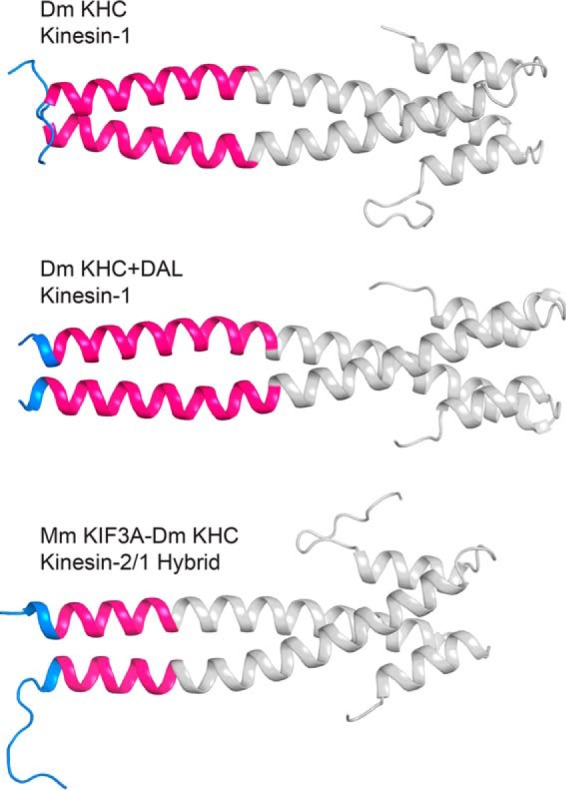
Crystal structures of native D. melanogaster kinesin-1 neck-linker protein, kinesin-1 + DAL, and the hybrid of the M. musculus KIF3A neck-linker with the kinesin-1 helix α7 coil. All are fused to EB1 dimerization domain (gray). Each helix α7 domain, as predicted based on the earlier studies of kinesin-1, is colored hot pink with the neck-linker peptide colored blue.
The kinesin-1 + DAL structure shows the effect of adding three additional residues to the kinesin-1 neck-linker. Previous studies have added these three residues, DAL, as an extension to mimic kinesin-2, as the three final residues of its neck-linker are DAL (DTL in KIF3C) (25, 39). The addition of these residues to the end of the kinesin-1 neck-linker should result in a 17-residue neck-linker, as in kinesin-2, rather than the native 14-residue neck-linker. Interestingly, in our structure, even though three residues (DAL) were added to the putative end of the neck-linker, two of the three residues become a part of the α7 helix and thus only lengthens the neck-linker by one residue.
The KIF3A-kinesin-1 neck-linker construct fuses the KIF3A neck-linker domain to the α7 helix of kinesin-1 (25, 39). In the native KIF3A structure determined here, the neck-linker was 12 residues long; however, in the hybrid, it lengthens to 14–15 amino acids (Figs. 5 and 6). There is variation in the start of the coiled-coil due to slight differences in crystal packing. However, the length is clearly different from that of the native kinesin-2 neck-linker. These results indicate that studies of the kinetic and motile properties of kinesins should be performed in the context of the native neck-linker and coiled-coil.
FIGURE 6.
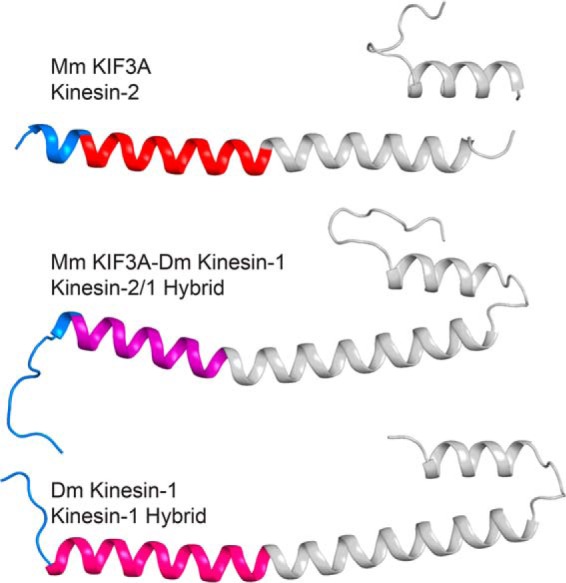
Comparison of KIF3A, the KIF3A-kinesin-1 hybrid protein, and kinesin-1 structures. Predicted neck-linkers are shown in blue and the EB1 domain in gray. KIF3A is at the top with its helix α7 colored red, KIF3A-kinesin-1 hybrid protein in the middle with its helix α7 in purple, and kinesin-1 at the bottom with its helix α7 in hot pink. Note the variability in neck-linker length based on the start of helix α7.
Temperature Factor Trends at the Neck-linker/α7 Junction
As noted earlier, atomic force microscopy measurements suggest that the start of α7 in kinesin-1 is particularly stable. It is not known whether this phenomenon holds true for all N-terminal kinesins, but is an important consideration when assessing the length of the neck-linker. The structures give a clear indication of the start of α7 but do not necessarily give an assessment of stability. In principle, examination of the temperature factors across the neck-linker/α7 junction could provide some insight as to whether the longer helices are less stable. As shown in Fig. 7, there is a general trend that the residues at the N terminus of the neck-linker have a higher temperature factor than the coiled-coil, but there is a continuum leading into the α-helix. This is often observed for the N termini of proteins or linkers between domains. This analysis, though fraught with reservations because temperature factors are susceptible to modification by crystal packing, radiation decay, and lattice disorder, suggests that the residues that increase the length of α7 are no less stable than those that make up the canonical helix in kinesin-1. Interestingly, the same trends in B-values are also seen at the N termini of other native coiled-coils as discussed later (Fig. 8).
FIGURE 7.
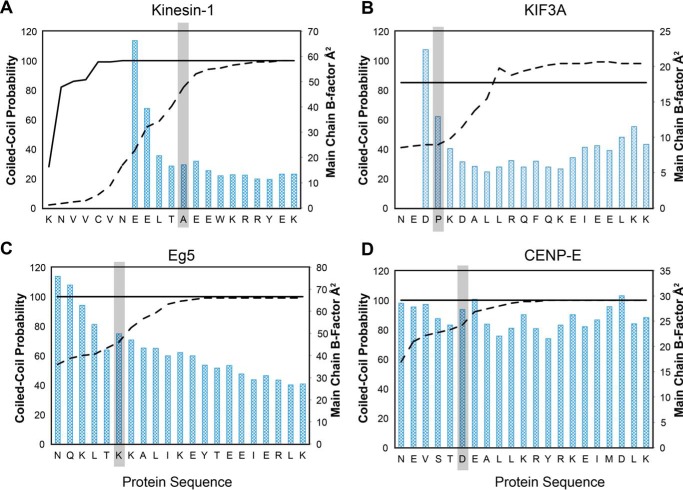
Predicted coiled-coil probability and temperature factors versus sequence for the class-specific kinesins. A, kinesin-1; B, KIF3A; C, Eg5; and D, CENP-E. The COILS-28 probabilities are shown as a solid line with MARCOIL probabilities shown with a dashed line. The start residue of the helix α7 coil as determined in the x-ray crystal structure is highlighted in gray. The predicted starting residues of the coiled-coil for KIF3A, Eg5, and CENP-E do not agree with the observed coiled-coil in the x-ray structures.
FIGURE 8.
Coiled-coil probability and temperature factors versus sequence for selected PDB files. COILS-28 probabilities shown as a solid line and MARCOIL probabilities shown as a dashed line. The start of the coiled-coil is highlighted in gray. All sequences begin on a g residue. PDB codes included are (A) 2FXM (63), (B) 1GD2 (64), (C) 3HNW, and (D) 1UII (79).
Coiled-coil Predictions Do Not Accurately Reflect the Start Residues of Coiled-coils
As noted earlier, the current prediction algorithms provide a robust estimate of the existence of a coiled-coil, although the exact start of the structural motif is ambiguous. A robust prediction for a residue in a coiled-coil will often be close to 1.0, and a value greater than 0.5 is commonly considered to indicate a coiled-coil (60). The question is as follows. What value for the probability should be accepted as a reliable indication of the first residue? To gain insight into this area of uncertainty, the structures of the kinesin neck-linkers and α7 were compared with the calculated probabilities for two algorithms. The register of the coiled-coil and prediction for the start site were determined with COILS, a Position-specific Scoring Matrix model, and MARCOIL, a Hidden Markov Model (3, 4). Both COILS and MARCOIL gave similar registers for the body of the coiled-coils, However, neither program accurately predicted the observed start sites for the α7 helix (Fig. 7, Table 3). In the calculations of the probabilities, the 28-residue window in COILS was used as it gives the lowest false-positive rate of the three options (14, 21, or 28 residue windows).
TABLE 3.
Predicted coiled-coil start sites, actual start sites, and the corresponding probabilities of the residue being in a coiled-coil according to MARCOIL and COILS using the 28-residue window
| Kinesin | Observed start residuea | MARCOIL | COILS-28 | Predicted start residueb | MARCOIL | COILS-28 |
|---|---|---|---|---|---|---|
| Kinesin-1 | Ala-345 | 0.815 | 1.000 | Ala-345 | 0.815 | 1.000 |
| KIF3A | Pro-355 | 0.432 | 0.846 | Leu-360 | 0.853 | 0.846 |
| KIF3C | Pro-377 | 0.086 | 0.056 | Leu-382 | 0.578 | 0.056 |
| Eg5 | Lys-371 | 0.694 | 1.000 | Ile-375 | 0.945 | 1.000 |
| CENP-E | Asp-341 | 0.828 | 1.000 | Leu-345 | 0.978 | 1.000 |
| KIF3A-kinesin-1 (hybrid) | Ala-358 (A345-2)c | 0.416 | 1.000 | Kinesin-1-Ala-345 | 0.728 | 1.000 |
| Leu-359 (A345-1)c | 0.546 | 1.000 |
a Observed start residues are determined using the first residue in a helical conformation in the crystal structures of the neck-linker protein defined as helix as determined by Dictionary of Secondary Structure of Proteins.
b Predicted start residue as determined in Ref. 36. This start was assumed in earlier kinetic studies of kinesins to determine the influence of the neck-linker length.
c The KIF3A neck-linker residue where the coiled coil starts is shown first, followed by the corresponding position in kinesin-1 relative to the kinesin-1 α7 start.
Several coiled-coil prediction algorithms were recently reviewed to check for both the accuracy in prediction of coiled-coil and also the accuracy of the oligomeric state of the coiled-coil (60). This study found the CCHMM_PROF algorithm to give the best indicator of coiled-coil; however, it does not yield a registry prediction for the coiled-coil. Thus, it was not used in this study (61). Multicoil2 also performed well, but its results were consistent with that of MARCOIL and COILS-28 (62). In general, for the kinesin coiled-coil domains, MARCOIL predicted coiled-coil start sites more conservatively than COILS-28. MARCOIL probabilities were nearly always lower than the COILS-28 prediction, except for the coiled-coil of KIF3C (60). Both programs poorly predicted the KIF3C coiled-coil. MARCOIL yielded a probability of 0.09 for the propensity of Pro-377 to form a coiled-coil, and COILS predicted a probability of 0.06 for the 28-residue window.
For kinesin-1, where the structure was previously known, the algorithms differ in the coiled-coil probabilities. Ala-345 is the α7 helix start. MARCOIL gives a conservative probability of 0.82, although COILS reaches a probability of 1, five residues earlier in the sequence where there is no coiled-coil. COILS tended to over-predict the neck-linker coiled-coil, reaching high probabilities earlier in the sequence. MARCOIL is a better estimator, but its predictions for the coiled-coil start sites ranged from 0.09 to 0.83. Neither program yielded reliable predictions for the start site of coiled-coils. The variation between predictive approaches creates a dilemma for deciding the coiled-coil start site based on bioinformatics approaches. Indeed, structural results reveal a fundamental weakness in the prediction algorithms because they are unable to categorically indicate the first residue that will adopt the α-helical conformation.
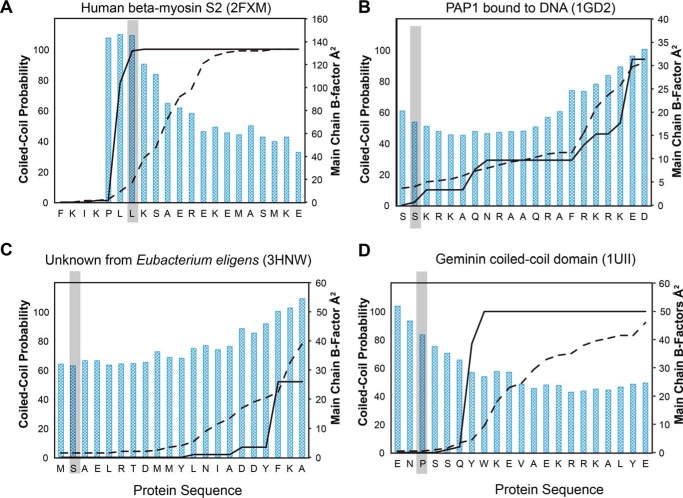
To see whether this problem of predicting the coiled-coil start sites in kinesins is a general phenomenon, structures of coiled-coil proteins in the PDB that contain a native transition from random-coil to parallel coiled-coil were also examined (Fig. 8 and Table 4). Although there are a large number of structures of dimeric coiled-coils in the Protein Data Bank, there are only about a dozen that contain the native start sequence. Most structures in the PDB represent fragments of a larger protein or are fused to the canonical coiled-coil found in GCN4. Interestingly, the performance of the algorithms on this restricted set was similar to that observed for the kinesin neck-linker proteins.
TABLE 4.
COILS-28 and MARCOIL prediction for structures of coiled-coil containing proteins that contain a native start sequence
All sequences are in the same register as listed in the top chart. The start residues for each coiled-coil as determined by Dictionary of Secondary Structure of Proteins and the PDB structure are shaded and underlined. Probabilities (×100) as predicted by COILS-28 and MARCOIL are shown successively below each amino acid sequence. References for each PDB if available are included below the PDB code.
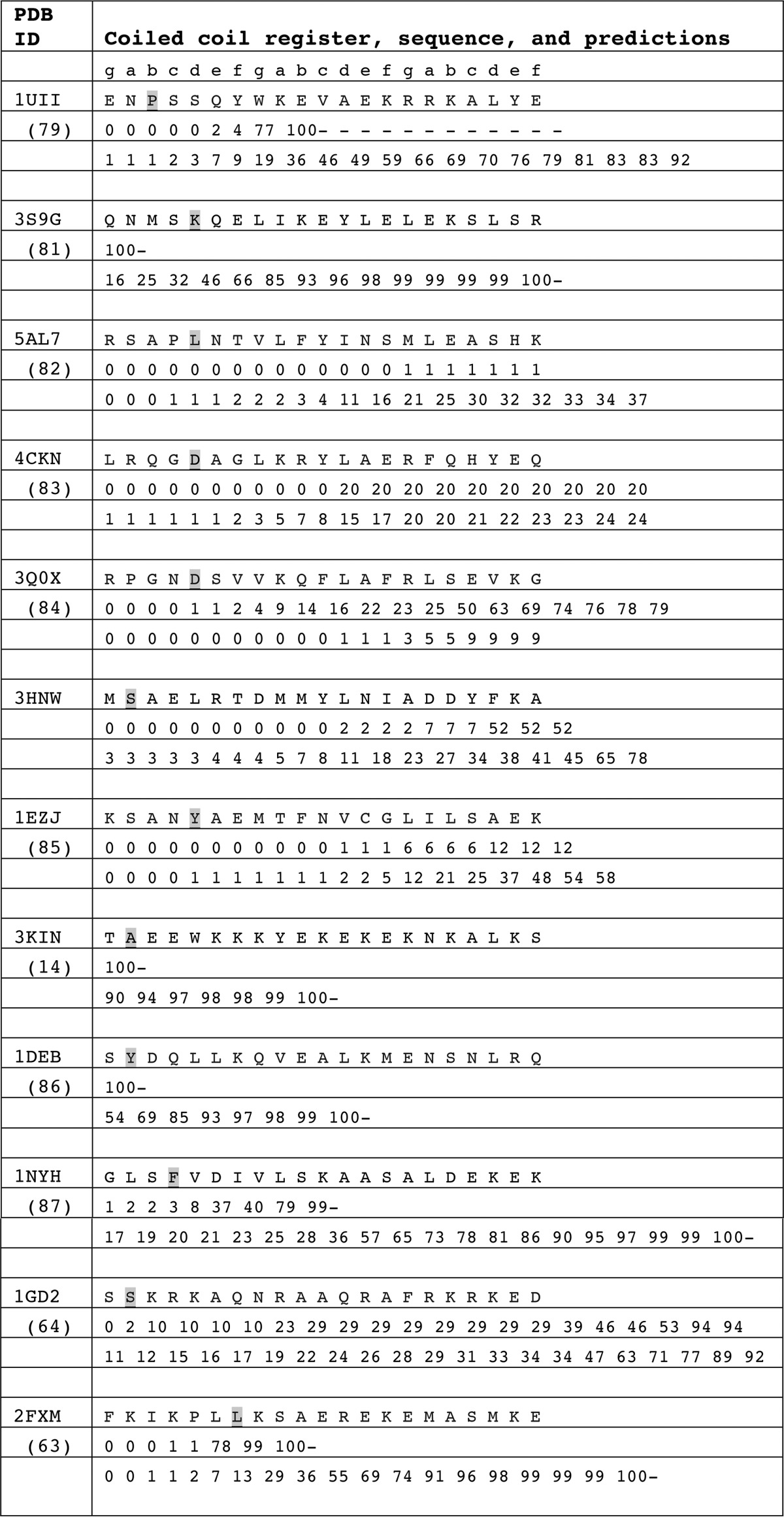
In almost every case, the algorithms miss the start site of the coiled-coil. As with the neck-linker proteins, COILS-28 over-predicts the propensity for coiled-coil, and MARCOIL is much more conservative. Neither algorithm accurately predicted the coiled-coil start, often only reaching a reasonable probability of coiled-coil formation until 10 or more residues after the structurally observed start site.
A notable exception is 2FXM, the N-terminal region of the S2 fragment of cardiac β-myosin II (63). COILS-28 reaches a probability of 99% for the start of the coiled-coil to begin at the site corresponding with the coiled-coil start in the structure. MARCOIL is close, but still under-predicts the probability of a coiled-coil in that region.
In 1GD2, a structure of the bZIP transcription factor, both COILS and MARCOIL under-predict the possibility of a coiled-coil and do not reach an ∼90% probability until 17 residues later in the sequence, corresponding to ∼2.5 heptads of coiled-coil missed (64). Results are similar for 3HNW, a coiled-coil protein from Eubacterium eligens with unknown function. Neither COILS nor MARCOIL reaches a reasonable coiled-coil prediction until more than three heptads into the coiled-coil domain. Overall, it is clear that algorithms can predict the presence and the register of a coiled-coil but do not provide an accurate guide to the start of the helical structure.
Discussion
In this study, the crystal structures of seven kinesin neck-linker α7 helices were determined from four different kinesin families representing N-terminal kinesin motors. In these structures, there are differences in the length of the neck-linker and the start of the coiled-coil stalk as compared with previous predictions. These differences are most likely the result of inaccurate assumptions in the prediction of coiled-coil start sites. The coiled-coil algorithms, COILS-28 and MARCOIL, were further investigated with respect to start site accuracy for other non-kinesin proteins and were shown to be unable to predict the start sites for the majority of coiled-coil-containing structures.
In the kinesin neck-linker structures, the coiled-coil start site was 4–5 residues earlier in the sequence than was predicted. The original assignment of neck-linker length by Hariharan and Hancock in 2009 (36) was based upon the assumption that the α7 helix would begin on an a or d residue in the coiled-coil registry, and this was true for the kinesin-1 motors. However, the choice of this residue registry was arbitrary and as shown by the results in this paper does not universally apply. KIF3A and KIF3C both begin on a c position, CENP-E and Eg5 begin on d positions, and the KIF3A-kinesin-1 hybrid begins on an f position. The addition of the DAL to the kinesin-1 neck-linker changes the start site from an a position to an f position. Of the non-kinesin proteins also examined, the most common start residue was the d position consistent with the assumption of a d position beginning the coiled-coil. The varying start sites show that most positions in the heptad are utilized for the start site of a coiled-coil domain.
Numerous studies have shown differences in the neck-linker length can drastically alter the single molecular parameters (25, 31, 37, 39, 65). However, our study suggests that rather than a direct association of shorter neck-linkers leading to greater processivity, it may be that neck-linker length is tuned to its motor and relative changes in length can increase or decrease the processivity.
Many studies have inserted residues in or deleted residues from the presumptive end point of the neck-linker where it was assumed that the additional residues would add to the neck-linker and have no effect on the coiled-coil (25, 39). As shown in this study, inserted residues can be incorporated directly into the α7 helix, rather than increasing the length of the neck-linker. Even when there are added residues at the appropriate end of the neck-linker, as in kinesin-1, the residues may be incorporated into the coiled-coil domain, rather than extending the neck-linker. It is possible that the added residues may disrupt coiled-coil formation because the coiled-coil motif depends on specific residues at each position to fulfill the canonical knobs-into-holes packing. Thus, the disruption of coiled-coil formation, rather than direct altering of neck-linker length, could be leading to artificial lengthening of the neck-linker. The changes in coiled-coil formation may also underlie the changes in processivity seen in other studies (25, 57, 58).
Furthermore, we have shown that fusing the α7 helix from kinesin-1 to kinesin-2 results in changes in the true neck-linker length. This accounts for the observed difference in kinetic and motile activities between synthetic and native fusions. In a study where the KIF3A motor domain and neck-linker were fused to the kinesin-1 α7 helix, the average run length was 0.7 μm, although a separate study showed the run length was nearly 50% longer at 1 μm when using the native coiled-coil (39, 46, 65). In contrast, the speed of the KIF3A-kinesin-1 construct, 480 nm/s, was nearly twice as fast as the native, 240 nm/s, showing that there are clear alterations in kinetic properties when proteins are fused to different coiled-coil domains (39, 46).
Our study also shows that although the coiled-coil prediction algorithms, COILS and MARCOIL, are able to predict both the occurrence of a coiled-coil accurately and the registry, both algorithms struggle to identify the start site of a coiled-coil. The probabilities given by both MARCOIL and COILS cannot be relied upon to yield the correct initiation point because there is no a priori way to decide which probability should be chosen to indicate the absolute start of the helical conformation. Thus, when evaluating sequences through bioinformatics, it should be understood that there is considerable ambiguity in where the coiled-coil starts. This might not be important for many proteins that include coiled-coils, but it is critical when the junction between globular domains and coiled-coils plays a role in function, as seen in motor proteins.
Kinesin neck-linker domains are clearly important for processivity, but it is not as simple as a short neck-linker leading to increased processivity. Further studies must consider the length of the neck-linkers carefully and ensure that when mutations or substitutions are made for kinetic studies the constructs are altered in the way that was intended.
Experimental Procedures
Construct Preparation
Mus musculus KIF3A cDNA was a generous gift from William O. Hancock (Pennsylvania State University, University Park, PA). M. musculus KIF3C cDNA was synthesized by Open Biosystems (GE Healthcare, Lafayette, CO). Homo sapiens Eg5 was obtained from an expression plasmid containing Eg5(1–513) (66). H. sapiens CENP-E was obtained from an expression plasmid containing CENP-E(1–407) (67). Drosophila melanogaster kinesin-1 neck-linker was synthesized by Integrated DNA Technologies (Coralville, IA). Kinesin-1 neck-linker mutagenesis was accomplished through a QuikChange-like protocol to introduce gatgcgctg for the +DAL mutation and to mutate cysteine 338 to alanine.
Neck-linker proteins were cloned using a modified pET-31 vector (Novagen) containing an N-terminal 8-histidine tag linked to the protein via a tobacco etch virus protease site and a C-terminal EB1 protein used as a coiled-coil fusion. EB1 is a coiled-coil protein with a C-terminal globular domain used to improve crystallization and expression of coiled-coil-containing proteins (46, 51, 68, 69). Great care was taken to maintain the coiled-coil registration across the fusion boundary and to avoid conflicts between structurally adjacent residues (68). A complete description of all constructs is included in Table 5. Cloning was accomplished using a protocol similar to the QuikChange method (Agilent) as described previously (68). Briefly, the QuikChange method allows genes to be inserted into vectors via linear amplification using PfuUltra II Fusion HF polymerase (Agilent), avoiding the introduction of cloning artifacts and resulting in faster preparation of constructs (70, 71). The sequences of the constructs and coiled-coil registry are detailed in Table 5.
TABLE 5.
Neck-linker construct sequences and coiled-coil registry
The neck-linker and α7 helices are shown in black text; His8 tags are shown in purple; the recombinant tobacco etch virus protease recognition site and linker are shown in red, and EB1 is depicted in blue.

a The native kinesin-1 neck-linker includes a cysteine at residue 338. Initial structural studies of this construct yielded a structure that included a spurious cross-link between adjacent chains. This problem was solved by mutating this residue (underlined) to an alanine to avoid this crystallization artifact.
Neck-linker proteins were expressed in an E. coli BL21-CodonPlus (DE3)-RIL strain (Agilent). For natively expressed neck-linker proteins, 6 liters of Lysogeny Broth culture were inoculated with an overnight culture formed from a single colony and allowed to grow to an A600 between 0.6 and 1.0 at 37 °C. Upon reaching the appropriate A600, the cells were chilled to 16 °C, induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside, and grown at 16 °C for 18 h before harvesting via centrifugation. For production of selenomethionine-derived proteins, 6 liters of M9 media were inoculated with 50 ml per 500 ml of overnight culture. Cells were grown to an A600 between 0.6 and 1.0 at 37 °C. Upon reaching the appropriate A600, the cells were chilled to 16 °C, and 5 ml of an amino acid mixture (100 mg of lysine, 100 mg of threonine, 100 mg of phenylalanine, 50 mg of leucine, 50 mg of isoleucine, 50 mg of valine, 50 mg of selenomethionine per 30 ml of mixture) was added. Cells were grown with shaking at 16 °C for 30 min before induction with 1 mm isopropyl β-d-1-thiogalactopyranoside. Cells were grown for 24 h before harvesting via centrifugation.
Protein Purification
All purification steps occurred at 4 °C. Ten g of cell paste were lysed via sonication in 100 ml of lysis buffer (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 0.1 mm EGTA, 0.2 mm tris(2-carboxyethyl)phosphine (TCEP), 30 mm imidazole) with 1 mm phenylmethylsulfonyl fluoride, 50 nm leupeptin (Peptide International), 70 nm E-65 (Peptide International), 2 nm aprotinin (ProSpec), and 2 μm 4-(2-aminoethyl)benzenesulfonyl fluoride (Gold BioTechnology). The lysate was clarified via centrifugation at 125,000 × g for 30 min at 4 °C. The supernatant was loaded onto a 7-ml nickel-nitrilotriacetic acid (Qiagen) column at 1 ml/min and washed with 20 column volumes of lysis buffer. Protein was eluted in 4 column volumes of elution buffer (lysis buffer with 200 mm imidazole). The octa-histidine tag was cleaved using a 1:100 molar ratio of a recombinant tobacco etch virus protease and was dialyzed against lysis buffer without imidazole (72). After overnight digestion at 4 °C, the octa-histidine tag was removed via a second 7-ml nickel-nitrilotriacetic acid column. Protein was loaded at 1 ml/min and washed with 3 column volumes of buffer (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 0.1 mm EGTA, 0.2 mm TCEP, 30 mm imidazole), followed by 3 column volumes of lysis buffer. The protein was concentrated using an Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-10 membrane (Millipore). The concentrated protein was dialyzed against 10 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.1 mm EGTA, 0.2 mm TCEP and flash-frozen in 30-μl droplets in liquid nitrogen and stored at −80 °C prior to crystallization.
Crystallization
Kinesin-1 neck-linker crystals were grown at 20 °C via vapor diffusion from a 1:1 mixture of 15 mg/ml protein solution and 20% (w/v) methoxy polyethylene glycol (MEPEG) 5000, 100 mm Li2SO4, 100 mm MES/acetate, pH 5.5, 10 mm γ-caprolactone. Kinesin-1 crystals appeared spontaneously after 1–2 days. Crystals were layered plates reaching maximal dimensions of 200 × 200 × 25 μm. Crystals were flash-frozen in liquid nitrogen in synthetic mother liquor (20% (w/v) MEPEG 5000, 100 mm Li2SO4, 100 mm MES/acetate, pH 5.5, 10 mm γ-caprolactone) supplemented with 10% ethylene glycol.
Kinesin-1 + DAL crystals were grown at 20 °C via vapor diffusion from a 1:1 mixture of 15 mg/ml protein solution and 100 mm sodium acetate, pH 5.0, 24% (w/v) polyethylene glycol (PEG) 400, 80 mm MgCl2, 3 mm ZnSO4. Drops were streak-seeded after 24 h, and cube-like crystals grew to a maximum size of 100 × 100 ×100 μm after 1 week. Crystals were flash-frozen in liquid nitrogen with synthetic mother liquor (100 mm sodium acetate, pH 5.0, 24% (w/v) polyethylene glycol (PEG) 400, 80 mm MgCl2, 3 mm ZnSO4) supplemented with 8% (w/v) ethylene glycol.
KIF3A crystals were grown via vapor diffusion from a 1:1 mixture of 15 mg/ml protein solution at 20 °C in 24% (w/v) 2-methyl-2,4-pentanediol, 10% (w/v) PEG 4000, 100 mm CaCl2, 100 mm MOPS, pH 7.0, 1 mm CdCl2. Drops were streak-seeded after 24 h, and rod-shaped crystals reached a maximum size of 200 × 20 × 20 μm within 2 days and were flash-frozen for data collection directly from the drop.
KIF3A-kinesin-1 hybrid (KIF3A-KHC) crystals were grown at 20 °C via vapor diffusion from a 1:1 mixture of 15 mg/ml protein solution in 32% (w/v) polyethylene glycol dimethyl ether 500, 30 mm MgCl2, 10 mm diethylenetriamine, 100 mm sodium 3-[4-(2-hydroxyethyl)-1-piperazinyl]propanesulfonic acid, pH 8.5. Drops were streak-seeded after 24 h, and rod-shaped crystals reached a maximum size of 350 × 20 × 20 μm within 2 days and were flash-frozen in liquid nitrogen for data collection directly from the drop.
KIF3C crystals were grown via vapor diffusion from a 1:1 mixture of 15 mg/ml protein solution at 20 °C in 30% (w/v) pentaerythritol ethoxylate (PEE) 797, 1.5% (w/v) ethylene glycol monoethyl ether, 400 mm MgCl2, 100 mm BTP, pH 9.0. Drops were streak-seeded after 24 h, and rod-shaped crystals reached a maximum size of 200 × 30 × 30 μm. For data collection, crystals were flash-frozen in liquid nitrogen directly from the drop.
Eg5 crystals were grown at 4 °C via vapor diffusion from a 1:1 mixture of 15 mg/ml protein solution in 24% (w/v) PEE 797, 100 mm CaCl2, 100 mm MOPS, pH 7.0, 0.1% (w/v) octyl glucoside. Crystals were streak-seeded after 24 h and cube-shaped crystals reached maximum dimensions of 100 × 100 × 100 μm after 3 days. Crystals were flash-frozen in liquid nitrogen in synthetic mother liquor (24% (w/v) PEE797, 100 mm CaCl2, 100 mm MOPS, pH 7.0, 0.1% (w/v) octylglucoside) supplemented with 16% (w/v) sucrose.
The CENP-E selenomethionine crystals were grown via vapor diffusion from a 1:1 mixture of 10 mg/ml protein solution at 20 °C in 18% (w/v) MEPEG 2000, 175 mm Li2SO4, 100 mm MES, pH 6.0, 1 mm CdCl2. Drops were streak-seeded after 24 h, and hexagonal crystals reached maximum dimensions of 100 × 100 × 100 μm after 5 days. CENP-E selenomethionine crystals were flash-frozen in liquid nitrogen for data collection in synthetic mother liquor (20 °C in 18% (w/v) MEPEG 2000, 175 mm Li2SO4, 100 mm MES, pH 6.0, 1 mm CdCl2) supplemented with 15% (w/v) ethylene glycol.
Structure Determination
The x-ray diffraction data for all neck-linker structures were collected at the SBC 19-ID beam line at the Advanced Photon Source (Argonne, IL). The datasets were integrated and scaled with the program HKL2000 (73). The kinesin-1, KIF3A-kinesin1 hybrid, KIF3A, KIF3C, and Eg5 neck-linker structures were solved via molecular replacement using Phaser with PDB structure 1YIB (74, 75). The CENP-E neck-linker structure was solved independently using selenomethionine-containing crystals, where single anomalous diffraction data were processed using Phaser (75). After the initial solutions were obtained, structures were refined by iterative cycles of manual model building in Coot and refinement with phenix.refine (76, 77). Data collection and refinement statistics for all structural determinations are given in Table 6. Secondary structure assignment was calculated with the Dictionary of Secondary Structure of Proteins Algorithm (50). Structural overlays were done using Superpose (78).
TABLE 6.
Crystallographic data collection and refinement statistics
RMS means root mean square; SeMet is selenomethionine.
| Kinesin-1 Neck-linker | KIF3A Neck-linkera | KIF3C Neck-linkera | Eg5 Neck-linker | CENP-E Neck-linker (SeMet) a | Kinesin-1+DAL Neck-linkera | KIF3A-Kinesin-1 hybrid Neck-linker | |
|---|---|---|---|---|---|---|---|
| Wavelength | 0.97934 | 0.97924 | 0.97924 | 0.97918 | 0.97929 | 0.97934 | 0.97929 |
| Resolution rangeb | 28.26–2.0 (2.072–2.0)a | 27.18–1.67 (1.73–1.67) | 35.99–1.57 (1.62–1.57) | 29.75–2.0 (2.09–2.0) | 45.78–2.1 (2.18–2.1) | 46.34–2.24 (2.28–2.24) | 42.62–2.1 (2.175–2.1) |
| Space group | P 1 21 1 | C 2 2 21 | P 21 21 21 | P 1 21 1 | P 32 2 1 | I 41 2 2 | P 21 21 21 |
| Unit cell dimensions (Å,°) | 46.97 37.78 52.71 β = 115.00 | 45.81 80.53 36.83 | 72.07 71.97 43.14 | 34.86 59.51 72.67 β = 100.53 | 100.65 100.65 107.65 | 40.0 40.0 185.45 | 53.12 80.20 155.37 |
| Total reflections | 59,704 | 71,250 | 222,535 | 80,554 | 1,138,524 | 48,657 | 52,4708 |
| Unique reflections | 11,394 (1046) | 8218 (805) | 30,981 (2513) | 19,128 (1641) | 37,185 (3647) | 3537 (153) | 39,515 (3835) |
| Multiplicity | 4.8 (3.6) | 8.6 (5.9) | 7.1 (3.0) | 4.1 (4.1) | 16.6 (8.7) | 12.5 (4.9) | 13.2 (7.6) |
| Completeness (%) | 0.99 | 1 | 0.96 | 0.97 | 1 | 0.90 | 1 |
| Mean I/σ(I) | 16.6 (2.1) | 50.1 (12.4) | 21.7 (1.6) | 25.9 (3.8) | 34.8 (12.7) | 37.5 (4.6) | 31.1 (3.2) |
| Wilson B-factor(Å2) | 18.88 | 12.06 | 13.98 | 25.03 | 20.45 | 28.41 | 21.94 |
| R-merge | 0.083 (0.589) | 0.050 (0.295) | 0.073 (0.595) | 0.048 (0.414) | 0.114 (0.360) | 0.058 (0.522) | 0.089 (0.571) |
| R-meas | 0.094 (0.687) | 0.051 (0.319) | 0.075 (0.707) | 0.056 (0.474) | 0.112 (0.364) | 0.061 (0.567) | 0.092 (0.613) |
| Reflections used in refinement | 11,391 (1046) | 8218 (805) | 30,979 (2513) | 19,127 (1642) | 37,176 (3649) | 3535 (153) | 39,505 (3835) |
| Reflections used for R-free | 570 (58) | 412 (41) | 1552 (127) | 962 (90) | 1846 (179) | 176 (8) | 1975 (192) |
| R-work | 0.208 (0.241) | 0.192 (0.196) | 0.179 (0.251) | 0.221 (0.271) | 0.235 (0.263) | 0.198 (0.273) | 0.190 (0.238) |
| R-free | 0.250 (0.284) | 0.228 (0.204) | 0.204 (0.265) | 0.252 (0.307) | 0.270 (0.299) | 0.236 (0.212) | 0.228 (0.283) |
| No. of chains | 2 | 1 | 2 | 4 | 6 | 1 | 8 |
| No. of non-hydrogen atoms | 1395 | 637 | 1618 | 2516 | 4893 | 605 | 4833 |
| Macromolecules | 1244 | 566 | 1343 | 2255 | 4400 | 567 | 4426 |
| Ligands | 1 | 1 | 3 | 5 | |||
| Protein residues | 145 | 67 | 159 | 269 | 531 | 65 | 519 |
| RMS(bonds) | 0.005 | 0.006 | 0.005 | 0.001 | 0.003 | 0.002 | 0.002 |
| RMS(angles) | 0.7 | 0.72 | 0.71 | 0.29 | 0.48 | 0.34 | 0.38 |
| Ramachandran favored (%) | 99 | 97 | 99 | 98 | 96 | 100 | 99 |
| Ramachandran allowed (%) | 0.7 | 1.6 | 0.62 | 1.5 | 2.5 | 0 | 1.2 |
| Ramachandran outliers (%) | 0 | 1.6 | 0 | 0 | 2 | 0 | 0.2 |
| Rotamer outliers (%) | 0 | 1.6 | 0.67 | 0.82 | 1.4 | 0 | 1.9 |
| Clashscore | 8.09 | 5.28 | 1.49 | 1.98 | 3.52 | 1.76 | 2.64 |
| Average B-factor Å2 | 28.0 | 19.8 | 22.3 | 43.5 | 33.9 | 49.9 | 42.5 |
| Macromolecules | 26.7 | 18.5 | 19.8 | 43.6 | 33.9 | 49.2 | 42.3 |
| Ligands | 18.7 | 40.3 | 158.3 | 119.3 | |||
| Solvent | 38.1 | 30.3 | 34.4 | 43.1 | 33.2 | 52.5 | 44.7 |
| No. of TLS groups | 4 | 2 | 7 | 10 | 17 | 2 | 18 |
| PDB codes | 5JVU | 5JX1 | 5JVM | 5JV3 | 5JVP | 5JVS | 5JVR |
a Friedel pairs were averaged when calculating reflection statistics.
b Data in parentheses represent the highest resolution shell.
Author Contributions
R. K. P., L. G. P., S. P. G., and I. R. designed the research. R. K. P. and L. G. P. performed the research, and R. K. P., S. P. G., and I. R. analyzed the data and wrote the manuscript.
Acknowledgments
Use of the Structural Biology ID19 and BM19 beamlines, Argonne National Laboratory Advanced Photon Source, was supported by the United States Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38. We also thank Dr. Alessandro Senes (University of Wisconsin-Madison) for enthusiastic and helpful discussions.
This work was supported by National Institutes of Health Grant R37-GM054141 (to S. P. G.) and in part by National Science Foundation Graduate Research Fellowship Program DGE-0718123 (to R. K. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PDB
- Protein Data Bank
- MEPEG
- methoxy polyethylene glycol
- PEE
- pentaerythritol ethoxylate
- PEG
- polyethylene glycol
- TCEP
- tris(2-carboxyethyl)phosphine
- BTP
- 1,3-bis[tris(hydroxymethyl)methylamino]propane.
References
- 1. Crick F. H. (1953) The packing of α-helices: simple coiled-coils. Acta Crystallogr. 6, 689–697 [Google Scholar]
- 2. Rackham O. J., Madera M., Armstrong C. T., Vincent T. L., Woolfson D. N., and Gough J. (2010) The evolution and structure prediction of coiled coils across all genomes. J. Mol. Biol. 403, 480–493 [DOI] [PubMed] [Google Scholar]
- 3. Delorenzi M., and Speed T. (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 [DOI] [PubMed] [Google Scholar]
- 4. Lupas A., Van Dyke M., and Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 5. Parry D. A., Fraser R. D., and Squire J. M. (2008) Fifty years of coiled-coils and α-helical bundles: a close relationship between sequence and structure. J. Struct. Biol. 163, 258–269 [DOI] [PubMed] [Google Scholar]
- 6. Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 7. Lawrence C. J., Dawe R. K., Christie K. R., Cleveland D. W., Dawson S. C., Endow S. A., Goldstein L. S., Goodson H. V., Hirokawa N., Howard J., Malmberg R. L., McIntosh J. R., Miki H., Mitchison T. J., Okada Y., et al. (2004) A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Endow S. A., Kull F. J., and Liu H. (2010) Kinesins at a glance. J. Cell Sci. 123, 3420–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kull F. J., Sablin E. P., Lau R., Fletterick R. J., and Vale R. D. (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marx A., Müller J., and Mandelkow E. (2005) The structure of microtubule motor proteins. Adv. Protein Chem. 71, 299–344 [DOI] [PubMed] [Google Scholar]
- 11. Gumy L. F., Chew D. J., Tortosa E., Katrukha E. A., Kapitein L. C., Tolkovsky A. M., Hoogenraad C. C., and Fawcett J. W. (2013) The kinesin-2 family member KIF3C regulates microtubule dynamics and is required for axon growth and regeneration. J. Neurosci. 33, 11329–11345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kashina A. S., Rogers G. C., and Scholey J. M. (1997) The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta 1357, 257–271 [DOI] [PubMed] [Google Scholar]
- 13. Kashina A. S., Baskin R. J., Cole D. G., Wedaman K. P., Saxton W. M., and Scholey J. M. (1996) A bipolar kinesin. Nature 379, 270–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kozielski F., Sack S., Marx A., Thormählen M., Schönbrunn E., Biou V., Thompson A., Mandelkow E. M., and Mandelkow E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91, 985–994 [DOI] [PubMed] [Google Scholar]
- 15. Muresan V., Abramson T., Lyass A., Winter D., Porro E., Hong F., Chamberlin N. L., and Schnapp B. J. (1998) KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol. Biol. Cell 9, 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaar B. T., Chan G. K., Maddox P., Salmon E. D., and Yen T. J. (1997) CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vale R. D., Reese T. S., and Sheetz M. P. (1985) Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hackney D. D. (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 91, 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilbert S. P., Webb M. R., Brune M., and Johnson K. A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature 373, 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schnitzer M. J., and Block S. M. (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388, 386–390 [DOI] [PubMed] [Google Scholar]
- 21. Asbury C. L., Fehr A. N., and Block S. M. (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302, 2130–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kaseda K., Higuchi H., and Hirose K. (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 5, 1079–1082 [DOI] [PubMed] [Google Scholar]
- 23. Yildiz A., Tomishige M., Vale R. D., and Selvin P. R. (2004) Kinesin walks hand-over-hand. Science 303, 676–678 [DOI] [PubMed] [Google Scholar]
- 24. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., and Vale R. D. (1999) A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 25. Shastry S., and Hancock W. O. (2011) Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U. S. A. 108, 16253–16258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muretta J. M., Jun Y., Gross S. P., Major J., Thomas D. D., and Rosenfeld S. S. (2015) The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc. Natl. Acad. Sci. U. S. A. 112, E6606–E6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mickolajczyk K. J., Deffenbaugh N. C., Arroyo J. O., Andrecka J., Kukura P., and Hancock W. O. (2015) Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. U. S. A. 112, E7186–E7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoenger A., Sack S., Thormählen M., Marx A., Müller J., Gross H., and Mandelkow E. (1998) Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: comparison with x-ray structure and implications for motility. J. Cell Biol. 141, 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skiniotis G., Surrey T., Altmann S., Gross H., Song Y. H., Mandelkow E., and Hoenger A. (2003) Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 22, 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dogan M. Y., Can S., Cleary F. B., Purde V., and Yildiz A. (2015) Kinesin's front head is gated by the backward orientation of its neck linker. Cell Rep. 10, 1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yildiz A., Tomishige M., Gennerich A., and Vale R. D. (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134, 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valentine M. T., Fordyce P. M., Krzysiak T. C., Gilbert S. P., and Block S. M. (2006) Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 8, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Romberg L., Pierce D. W., and Vale R. D. (1998) Role of the kinesin neck region in processive microtubule-based motility. J. Cell Biol. 140, 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thorn K. S., Ubersax J. A., and Vale R. D. (2000) Engineering the processive run length of the kinesin motor. J. Cell Biol. 151, 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomishige M., and Vale R. D. (2000) Controlling kinesin by reversible disulfide cross-linking: identifying the motility-producing conformational change. J. Cell Biol. 151, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hariharan V., and Hancock W. O. (2009) Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell Mol. Bioeng. 2, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andreasson J. O., Milic B., Chen G.-Y., Guydosh N. R., Hancock W. O., and Block S. M. (2015) Examining kinesin processivity within a general gating framework. Elife 4, e07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Isojima H., Iino R., Niitani Y., Noji H., and Tomishige M. (2016) Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. 12, 290–297 [DOI] [PubMed] [Google Scholar]
- 39. Shastry S., and Hancock W. O. (2010) Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 20, 939–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Düselder A., Thiede C., Schmidt C. F., and Lakämper S. (2012) Neck-linker length dependence of processive kinesin-5 motility. J. Mol. Biol. 423, 159–168 [DOI] [PubMed] [Google Scholar]
- 41. Aizawa H., Sekine Y., Takemura R., Zhang Z., Nangaku M., and Hirokawa N. (1992) Kinesin family in murine central nervous system. J. Cell Biol. 119, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kondo S., Sato-Yoshitake R., Noda Y., Aizawa H., Nakata T., Matsuura Y., and Hirokawa N. (1994) KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 125, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1995) KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130, 1387–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., and Rosenbaum J. L. (1998) Chlamydomonas kinesin-II–dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carpenter B. S., Barry R. L., Verhey K. J., and Allen B. L. (2015) The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J. Cell Sci. 128, 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guzik-Lendrum S., Rank K. C., Bensel B. M., Taylor K. C., Rayment I., and Gilbert S. P. (2015) Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules. Biophys. J. 109, 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Albracht C. D., Rank K. C., Obrzut S., Rayment I., and Gilbert S. P. (2014) Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 289, 27836–27848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang P., Rayment I., and Gilbert S. P. (2016) Fast or slow, either head can start the processive run of kinesin-2 KIF3AC. J. Biol. Chem. 291, 4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scholey J. E., Nithianantham S., Scholey J. M., and Al-Bassam J. (2014) Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers. eLife 3, e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kabsch W., and Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 51. Frye J., Klenchin V. A., and Rayment I. (2010) Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition. Biochemistry 49, 4908–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Korkmaz E. N., Taylor K. C., Andreas M. P., Ajay G., Heinze N. T., Cui Q., and Rayment I. (2016) A composite approach towards a complete model of the myosin rod. Proteins 84, 172–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hyeon C., and Onuchic J. N. (2007) Mechanical control of the directional stepping dynamics of the kinesin motor. Proc. Natl. Acad. Sci. U. S. A. 104, 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hyeon C., and Onuchic J. N. (2007) Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc. Natl. Acad. Sci. U.S.A. 104, 2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bornschlögl T., Woehlke G., and Rief M. (2009) Single molecule mechanics of the kinesin neck. Proc. Natl. Acad. Sci. U. S. A. 106, 6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Arpağ G., Shastry S., Hancock W. O., and Tüzel E. (2014) Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys. J. 107, 1896–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen G.-Y., Arginteanu D. F., and Hancock W. O. (2015) Processivity of the kinesin-2 KIF3A results from rear head gating and not front head gating. J. Biol. Chem. 290, 10274–10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen Y., and Hancock W. O. (2015) Kinesin-5 is a microtubule polymerase. Nat. Commun. 6, 8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoeprich G. J., Thompson A. R., McVicker D. P., Hancock W. O., and Berger C. L. (2014) Kinesin's neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 106, 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li C., Ching Han Chang C., Nagel J., Porebski B. T., Hayashida M., Akutsu T., Song J., and Buckle A. M. (2016) Critical evaluation of in silico methods for prediction of coiled-coil domains in proteins. Brief. Bioinform. 17, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bartoli L., Fariselli P., Krogh A., and Casadio R. (2009) CCHMM_PROF: a HMM-based coiled-coil predictor with evolutionary information. Bioinformatics 25, 2757–2763 [DOI] [PubMed] [Google Scholar]
- 62. Trigg J., Gutwin K., Keating A. E., and Berger B. (2011) Multicoil2: Predicting coiled coils and their oligomerization states from sequence in the twilight zone. PLoS ONE 6, e23519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Blankenfeldt W., Thomä N. H., Wray J. S., Gautel M., and Schlichting I. (2006) Crystal structures of human cardiac β-myosin II S2-Δ provide insight into the functional role of the S2 subfragment. Proc. Natl. Acad. Sci. U. S. A. 103, 17713–17717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fujii Y., Shimizu T., Toda T., Yanagida M., and Hakoshima T. (2000) Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 7, 889–893 [DOI] [PubMed] [Google Scholar]
- 65. Andreasson J. O., Shastry S., Hancock W. O., and Block S. M. (2015) The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 25, 1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krzysiak T. C., Wendt T., Sproul L. R., Tittmann P., Gross H., Gilbert S. P., and Hoenger A. (2006) A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 25, 2263–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sardar H. S., Luczak V. G., Lopez M. M., Lister B. C., and Gilbert S. P. (2010) Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Curr. Biol. 20, 1648–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klenchin V. A., Frye J. J., Jones M. H., Winey M., and Rayment I. (2011) Structure function analysis of the C-terminal domain of CNM67, a core component of the Saccharomyces cerevisiae spindle pole body. J. Biol. Chem. 286, 18240–18250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Taylor K. C., Buvoli M., Korkmaz E. N., Buvoli A., Zheng Y., Heinze N. T., Cui Q., Leinwand L. A., and Rayment I. (2015) Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E3806–E3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen G. J., Qiu N., Karrer C., Caspers P., and Page M. G. (2000) Restriction site-free insertion of PCR products directionally into vectors. BioTechniques 28, 498–500 [DOI] [PubMed] [Google Scholar]
- 71. van den Ent F., and Löwe J. (2006) RF cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67–74 [DOI] [PubMed] [Google Scholar]
- 72. Blommel P. G., and Fox B. G. (2007) A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 55, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 74. Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A., and Vale R. D. (2005) Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 168, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maiti R., Van Domselaar G. H., Zhang H., and Wishart D. S. (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 32, W590–W594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Saxena S., Yuan P., Dhar S. K., Senga T., Takeda D., Robinson H., Kornbluth S., Swaminathan K., and Dutta A. (2004) A dimerized coiled-coil domain and an adjoining part of geminin interact with two sites on Cdt1 for replication inhibition. Mol. Cell 15, 245–258 [DOI] [PubMed] [Google Scholar]
- 80. Gudimchuk N., Vitre B., Kim Y., Kiyatkin A., Cleveland D. W., Ataullakhanov F. I., and Grishchuk E. L. (2013) Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bigalke J. M., Dames S. A., Blankenfeldt W., Grzesiek S., and Geyer M. (2011) Structure and dynamics of a stabilized coiled-coil domain in the P-TEFb regulator Hexim1. J. Mol. Biol. 414, 639–653 [DOI] [PubMed] [Google Scholar]
- 82. Cottee M. A., Muschalik N., Johnson S., Leveson J., Raff J. W., and Lea S. M. (2015) The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. eLife 4, e07236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. van Breugel M., Wilcken R., McLaughlin S. H., Rutherford T. J., and Johnson C. M. (2014) Structure of the SAS-6 cartwheel hub from Leishmania major. eLife 3, e01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M. C., Flückiger I., Gönczy P., and Steinmetz M. O. (2011) Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tarbouriech N., Curran J., Ruigrok R. W., and Burmeister W. P. (2000) Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7, 777–781 [DOI] [PubMed] [Google Scholar]
- 86. Day C. L., and Alber T. (2000) Crystal structure of the amino-terminal coiled-coil domain of the APC tumor suppressor. J. Mol. Biol. 301, 147–156 [DOI] [PubMed] [Google Scholar]
- 87. Chang J. F., Hall B. E., Tanny J. C., Moazed D., Filman D., and Ellenberger T. (2003) Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure 11, 637–649 [DOI] [PubMed] [Google Scholar]