Abstract
Pseudomonas aeruginosa acquires extracellular heme via the Phu (Pseudomonas heme uptake) and Has (heme assimilation system) systems. We have previously shown the catalytic actions of heme oxygenase (HemO) along with the cytoplasmic heme transport protein PhuS control heme flux into the cell. To further investigate the role of the PhuS-HemO couple in modulating heme uptake, we have characterized two HemO variants, one that is catalytically inactive (HemO H26A/K34A/K132A or HemOin) and one that has altered regioselectivity (HemO N19K/K34A/F117Y/K132A or HemOα), producing biliverdin IXα (BVIXα). HemOα similar to wild type was able to interact and acquire heme from holo-PhuS. In contrast, the HemOin variant did not interact with holo-PhuS and showed no enzymatic activity. Complementation of a hemO deletion strain with the hemOin or hemOα variants in combination with [13C]heme isotopic labeling experiments revealed that the absence of BVIXβ and BVIXδ leads to a decrease in extracellular levels of hemophore HasA. We propose BVIXβ and/or BVIXδ transcriptionally or post-transcriptionally regulates HasA. Thus, coupling the PhuS-dependent flux of heme through HemO to feedback regulation of the cell surface signaling system through HasA allows P. aeruginosa to rapidly respond to fluctuating extracellular heme levels independent of the iron status of the cell.
Keywords: bacterial genetics, heme, heme oxygenase, mass spectrometry (MS), Pseudomonas aeruginosa (P. aeruginosa), signaling, biliverdin
Introduction
Iron is an essential micronutrient for the survival and virulence of Gram-negative pathogens (1–4). Many pathogens, including the opportunistic pathogen Pseudomonas aeruginosa, encode systems that utilize the host heme-containing proteins as a source of iron (5, 6). P. aeruginosa encodes two heme uptake systems, the Pseudomonas heme uptake (phu) and the heme assimilation system (has). The phu system encodes a TonB-dependent outer membrane receptor (PhuR) that transports heme to the periplasm, where a soluble heme-binding protein (PhuT) interacts with an ATP-dependent permease (ABC transporter) (PhuUV), which then translocates heme to the cytoplasmic heme-binding protein (PhuS). In contrast the has operon encodes a soluble secreted hemophore (HasA) that scavenges heme and transfers it to the TonB-dependent outer membrane receptor (HasR) (7–9). In Serratia marcescens HasA and HasR additionally function in cell surface signaling by sensing heme and transmitting the signal through HasR to the extracytoplasmic σ function (ECF)3 system (HasS/HasI) (10). We have recently shown through a combination of bacterial genetics and [13C]heme metabolite studies that HasR functions primarily as a heme sensor, whereas PhuR is the major heme transporter (11).
Once translocated into the cell, heme is sequestered by the cytoplasmic binding protein PhuS and transferred to the BVIXβ/δ regioselective HemO. We have previously shown deletion of hemO shuts down the flux of extracellular heme into the cell despite up-regulation of the heme transport system (12). In contrast, deletion of phuS leads to inefficient heme utilization at low heme concentration, whereas at higher heme concentrations heme is degraded through both HemO and the bacterial phytochrome-associated BVIXα-selective BphO (13). The shunting of heme through both HemO and BphO is consistent with previous in vitro studies that have shown PhuS specifically interacts with HemO but not BphO (14). We concluded from these studies that PhuS and HemO are metabolically coupled to regulate heme flux into the cell.
To further dissect the role of the PhuS-HemO couple in driving and regulating extracellular heme flux, we have characterized two HemO variants, one that is catalytically inactive HemOin and the other that has altered regioselectivity HemOα, producing biliverdin IXα (BVIXα). We confirmed the previous observation that a catalytic HemO is required to drive extracellular heme uptake into the cell. Furthermore, we show the HemO-derived BVIXβ and BVIXδ metabolites act as positive feedback regulators of the extracellular hemophore, HasA. The studies herein suggest a mechanism that couples the regulation heme flux through PhuS-HemO to transcriptional and/or post-transcriptional regulation of the has operon that allows the cell to rapidly adapt to changes in extracellular heme levels.
Results
Characterization of the HemOin and HemOα Proteins
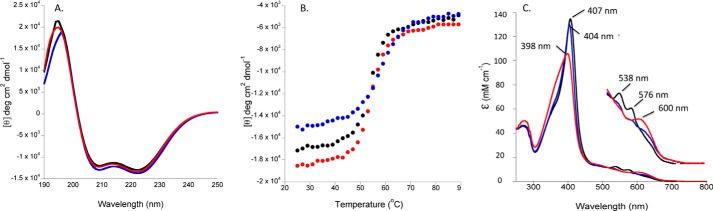
The crystal structure of the P. aeruginosa holo-HemO revealed the unique BVIXβ/δ regioselectivity was the result of a 90° in-plane rotation of the heme compared with the BVIXα-selective enzymes (15). To generate a BVIXα-selective HemO, we mutated the heme propionate coordinating Lys-34 and Lys-132 residues of HemO to Ala, and we introduced alternative propionate interactions through mutation of Asn-19 to Lys and Phe-117 to Tyr as seen in the BVIXα-selective Neisseriae meningitidis HemO (16). To generate the inactive enzyme HemOin, we removed the proximal His-26 in addition to the residues critical in anchoring the heme (Lys-34 and Lys-132). The HemO mutants expressed and purified as for the WT protein. The structural integrity of the purified HemO variants was confirmed by CD spectroscopy. The spectra of apo-HemOα and HemOin show proteins with α-helical content similar to that of the WT protein (Fig. 1A). Thermal denaturation curves show a single transition on unfolding with Tm values for the WT, HemOin, and HemOα within the range 54–56 °C (Fig. 1B). The similar Tm values indicate the intrinsic thermal stability of the mutant HemO proteins are not significantly different from the WT. The UV-visible absorption spectrum shows a Soret band at 407 nm for the WT and a slightly blue-shifted Soret band at 404 nm for the HemOα protein (Fig. 1C). As expected, the HemOin on addition of a molar equivalent of heme gave a spectrum with a peak at 398 nm typical of non-protein coordinated aqua heme (Fig. 1C).
FIGURE 1.
Spectroscopic characterization of the holo-HemOα and HemOin proteins. A, CD spectra of holo-HemOwt (black line), HemOα (blue line), and HemOin (red line). Spectra were recorded in 20 mm potassium phosphate (pH 7.8) at 25 °C with a final protein concentration of 20 μm. B, thermal denaturation curves θ at 222 nm for holo-HemOwt (black circles), HemOα (blue circles), and HemOin (red circles). Buffer and protein conditions as in A. C, absorbance spectra of 10 μm holo-HemOwt (black lines), HemOα (blue lines), and HemOin (red lines). Buffer conditions are as in A.
Holo-PhuS Interacts with HemOα but Not HemOin
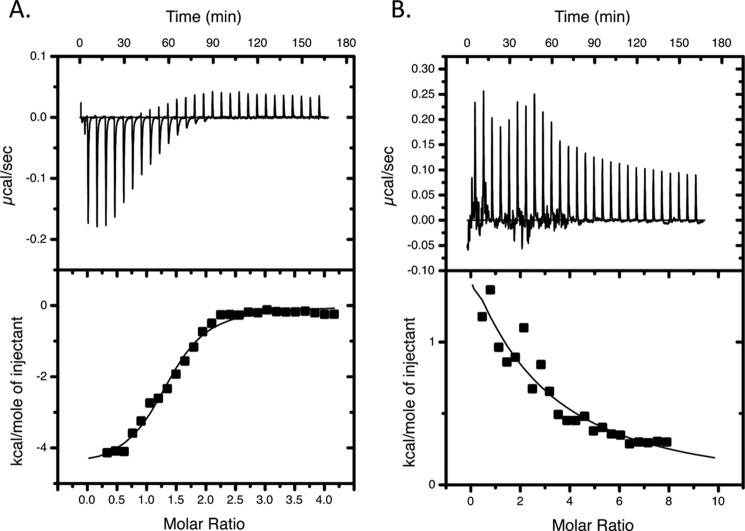
As reported previously, titration of apo-HemO with holo-PhuS gave a negative enthalpic signal with a 1:1 stoichiometry and an apparent Ka of 5.4 × 105 m−1 (Table 1) (14). Similarly, interaction of holo-PhuS with apo-HemOα gave a favorable binding enthalpy with a Ka value similar to that for the wild type HemO (Fig. 2A and Table 2). In contrast, the titration of holo-PhuS with HemOin is entropically driven and indicative of a nonspecific hydrophobic interaction (Fig. 2B). The inability of the HemOin variant to interact with holo-PhuS and transfer heme suggests the charged residues required for anchoring heme within the HemO active site may also be required for protein-protein interaction and transfer of heme from holo-PhuS.
TABLE 1.
Thermodynamic and binding parameters for holo-PhuS to wild type and mutant HemO proteins
| HemO variant | n | Ka (× 105 m−1) | ΔH | −TΔS | ΔG |
|---|---|---|---|---|---|
| kcal mol−1 | kcal mol−1 | kcal mol−1 | |||
| HemOwt | 0.8 | 5.4 ± 0.5 | −5.9 ± 0.1 | −1.5 ± 0.8 | −7.4 ± 0.7 |
| HemOα | 1.3 | 9.7 ± 1.5 | −4.6 ± 0.1 | −3.1 ± 1.3 | −7.8 ± 1.2 |
| HemOin | 1.0 | 0.28 ± 0.02 | 9.0 ± 0.7 | −14.5 ± 1.5 | −5.5 ± 0.8 |
FIGURE 2.
ITC of holo-PhuS with HemOα (A) and HemOin (B) proteins. Titrations were performed in 40 mm sodium phosphate buffer (pH 8.0) at 298 K. Upper panel, time-dependent release of heat during the titration. Lower panel, peak integrals plotted as a function of the molar ratio of heme to protein. The data were fit to a single binding model with Origin software, supplied by Microcal.
TABLE 2.
Bacterial strains used in this study
| Relevant genotype or description | Refs. | |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1-λ-pir | pro thi hsdR_Tpr Smr; chromosome:: RP4-2 Tc:: Mu-Km::Tn7/λpir | 27 |
| BL21 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Stratagene |
| P. aeruginosa | ||
| PAO1 | Wild type | 28 |
| ΔhemO | Chromosomal hemO in-frame deletion in PAO1 | This study |
| ΔhemO::hemOwt | ΔhemO complemented by hemO wild type allele inserted at attB site of the PAO1 chromosome | This study |
| ΔhemO::hemOα | ΔhemO complemented by hemOα inserted at attB site of the PAO1 chromosome | This study |
| ΔhemO::hemOin | ΔhemO complemented by hemOTM inserted at attB site of the PAO1 chromosome | This study |
| Plasmids | ||
| pEX18Tc | TcR; allelic replacement vector | 15 |
| pFLP2 | ApR; source of Flp recombinase | 15 |
| pPS858 | ApR, GmR; Source of GmR conferring fragment flanked by FRT sites | 15 |
| pMRL2 | ApR; pET-11a derivative harboring a water-soluble domain of the rat liver cytochrome b5 | 22 |
| pET21a-HemO | ApR; wild-type hemO gene and promoter region | 20 |
| pET21a-HemOin | ApR; hemO H26F/K34A/K132A derived from site-directed mutagenesis on pET21a-HemO | This study |
| pBSP-HemOα | ApR; hemOα gene cloned into a derivative of pBBR1MCS3 under control of the araC-PBAD cassette | 17 |
| Mini-CTX1 | TcR; self-proficient integration vector | 16 |
| Mini-CTX1-hemOwt | TcR; PCR-amplified promoter and hemO wild type coding sequence cloned as a BamHI-HindIII fragment between same sites of mini-CTX1-PhemO | |
| Mini-CTX1-PhemO | TcR; PCR amplified hemO and promoter region cloned as a BamHI-HindIII fragment between same sites of mini-CTX1 | This study |
| Mini-CTX1-hemOα | TcR; PCR-amplified hemOa coding sequence cloned as a EcoRI-HindIII fragment between same sites of mini-CTX1-PhemO | This study |
| Mini-CTX1-hemOin | TcR; PCR-amplified hemOTM coding sequence cloned as a EcoRI-HindIII fragment between same sites of mini-CTX1-PhemO | This study |
Heme Utilization in the ΔhemO Complemented Strains
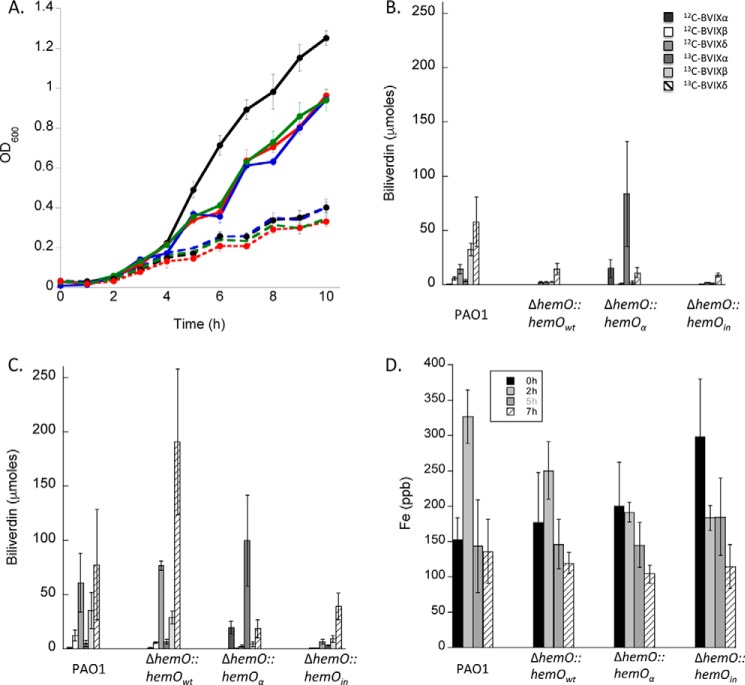
We have previously shown deletion of the hemO gene shuts down extracellular heme flux into the cell (12). To understand the potential role of HemO regioselectivity in regulating heme flux into the cell, we complemented the ΔhemO strain with the BVIXα regioselective hemOα and a catalytically inactive hemOin gene at the phage attachment (attB) site. For comparative purposes, we also cloned the hemOwt gene into the attB site. All of the strains grew at similar rates in iron-depleted M9 media (Fig. 3A). On addition of 0.5 μm heme, the strains grew at an increased rate, although the ΔhemO complemented strains lagged WT PAO1.
FIGURE 3.
Characterization of heme utilization in the ΔhemO complemented strains. A, growth curve of PAO1 and the ΔhemO complemented strains. Cells were grown in M9 minimal media PAO1 (black dashed line), ΔhemO::hemOwt (red dashed line), ΔhemO::hemOα (blue dashed line), ΔhemO::hemOin (green dashed line), or in the presence of 0.5 μm heme PAO1 (black solid line), ΔhemO::hemOwt (red solid line), ΔhemO::hemOα (blue solid line), ΔhemO::hemOin (green solid line). B, LC-MS/MS analysis of the BVIX isomers supplemented with 0.5 μm heme at 5 h. C, biliverdin isomer pattern at 7 h. Biliverdin values represent the standard deviation of three separate experiments. D, ICP-MS analysis of intracellular iron levels. Values for each strain and time point represent the average and standard deviation of n = 6.
To address the differences in growth rate between strains, we tested the ability of the complemented strains to utilize extracellular heme as an iron source under the same growth conditions. We have previously shown that supplementation with 0.5 μm [13C]heme as the sole source of iron is sufficient to detect BVIX metabolites and does not lead to Fur repression of the heme uptake systems within 3–5 h (11, 12). As reported previously, analysis of the BVIX metabolites in PAO1 at 5 h yields predominantly extracellular [13C]heme-derived BVIXβ and BVIXδ (Fig. 3B) (12). The peak in extracellular heme-derived [13C]BVIX is consistent with the increase in intracellular iron at 2 h as measured by ICP-MS (Fig. 3D). We expect to see a lag between peak intracellular iron and extracellular BVIX levels. At the 7-h time point, the ratio of [13C]heme to [12C]heme-derived BVIXβ and BVIXδ is close to 50% (Fig. 3C). The increase in 12C-derived BVIX metabolites is also consistent with the lower intracellular iron levels between 5–7 h as heme becomes limiting (Fig. 3D). In contrast, the ΔhemO::hemOwt strain showed a lag in extracellular [13C]BVIXβ and [13C]BVIXδ levels and as a consequence decreased intracellular iron levels compared with PAO1 (Fig. 3, B and D). Despite the fact the differences in mRNA and protein levels of the heme uptake proteins and HemO are not physiologically significant, the complemented strain is unable to utilize heme as efficiently as the WT PAO1 as shown by the lag in growth (Fig. 3A).
The hemO::hemOα strain was able to utilize heme as efficiently as PAO1 as shown by the comparable levels of [13C]BVIXα to [13C]BVIXβ and -BVIXδ at 5 h (Fig. 3B). However, despite the ability to utilize [13C]heme, the intracellular iron levels did not increase over the first 2 h as observed for WT PAO1 (Fig. 3D).
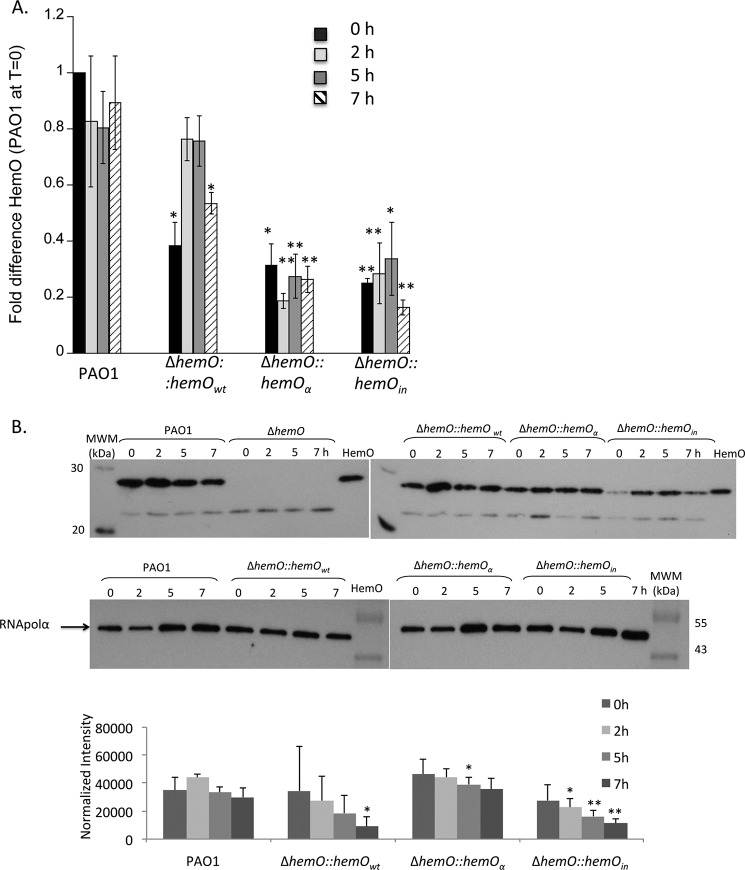
In contrast, the ΔhemO::hemOin strain is unable to acquire extracellular [13C]heme in the absence of a catalytically active HemO as confirmed by the lack of [13C]BVIX metabolites and a progressive decrease in intracellular iron levels over time (Fig. 3, B and D). These results as observed previously for the ΔhemO knock-out confirm that the lack of HemO activity cannot be compensated for by the alternative BVIXα-selective heme oxygenase, BphO (12). At the 7-h time point, background levels of BVIX are detected in the extracellular media, a result of nonspecific heme degradation and release of iron by redox-active molecules (Fig. 3C). These redox-active molecules include pyocyanin and phenazines that are secreted in higher amounts in the ΔhemO and ΔhemO::hemOin mutant strains (data not shown). Interestingly, the inability to utilize heme and the lack of [13C]BVIXδ and -BVIXβ is accompanied by a down-regulation in both hemOin mRNA and protein expression (Fig. 4B).
FIGURE 4.
HemO mRNA levels in PAO1 and the ΔhemO complemented strains. A, mRNA isolated at 0, 2, 5, and 7 h. mRNA values represent the mean from three biological experiments each performed in triplicate and normalized to PAO1 at 0 h. Error bars represent the standard deviation from three independent experiments performed in triplicate. The indicated p values were normalized to mRNA levels of PAO1 at the same time point, where *, p < 0.05; **, p < 0.005. B, representative Western blot of HemO. Total protein (25 μg) was loaded in each well. RNA polymerase α was used as an equal loading control. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. The indicated p values were normalized to PAO1 at the same time point where *, p < 0.05, and **, p < 0.005.
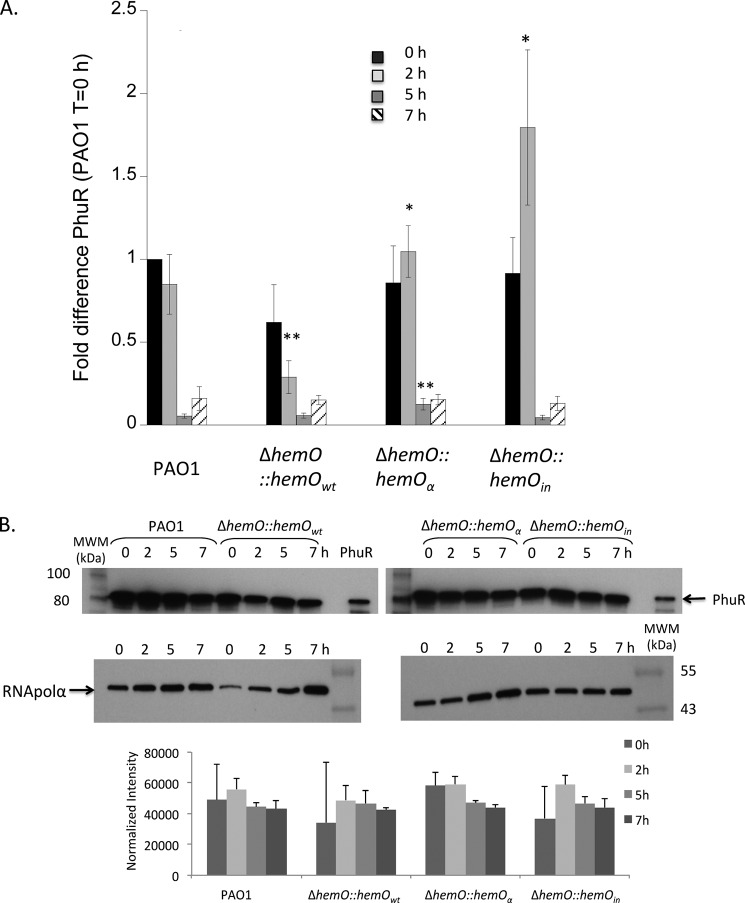
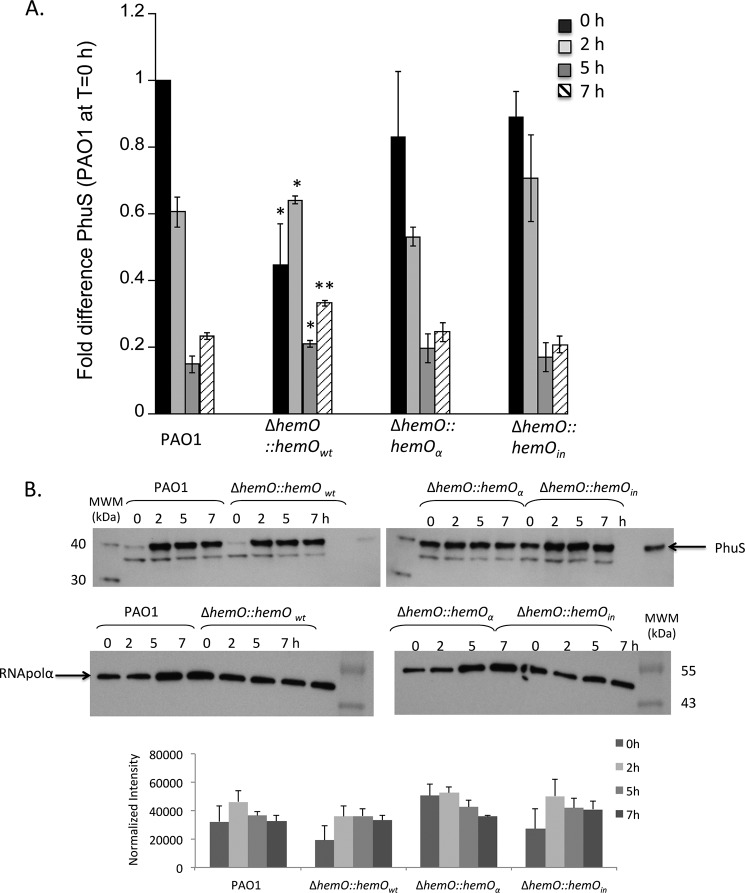
We have previously shown PhuR is the major heme transporter (11), and PhuS is required for regulation of heme flux through HemO (12). Therefore, to confirm that the decreased efficiency in heme utilization by the hemO::hemOwt and hemO::hemOα strains was not due to changes in the expression levels of PhuR and PhuS, we analyzed the mRNA and protein levels in each of the strains. As shown in Fig. 5, the difference in PhuR mRNA levels was not physiologically significant as the protein expression was similar in all strains (Fig. 5B). Although the protein expression levels of PhuS show variation between strains following initial iron starvation, on addition of 0.5 μm heme the mRNA and protein expression profiles between 2 and 7 h are similar (Fig. 6). Therefore, the lag in growth and decrease in the ability to efficiently utilize heme in the hemO::hemOwt and hemO::hemOα strains is not due to decreased expression of the major heme transporter PhuR or heme flux through PhuS. It is unclear at this time why complementation with hemOwt at the phage site does not restore heme utilization efficiency to that of PAO1. However, as will be outlined below, there exists an underlying link between BVIXδ and BVIXβ metabolite levels and the regulation of the hemophore HasA. The consequence of decreased BVIXδ and/or BVIXβ leads to a decrease in HasA and the cells' ability to sense extracellular heme.
FIGURE 5.
PhuR mRNA and protein levels in PAO1 and the ΔhemO complemented strains. A, mRNA isolated at 0, 2, 5, and 7 h. mRNA values represent the standard deviation from three independent experiments performed in triplicate and normalized to PAO1 at 0 h. The indicated p values were normalized to mRNA levels of PAO1 at the same time point, where *, p < 0.05, and **, p < 0.005. B, representative Western blot of PhuR. Total protein (25 μg) was loaded in each well. RNA polymerase α was used as an equal loading control. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. The indicated p values were normalized to PAO1 at the same time point where *, p < 0.05, and **, p < 0.005.
FIGURE 6.
PhuS mRNA and protein levels in PAO1 and the ΔhemO complemented strains. A, mRNA isolated at 0, 2, 5, and 7 h. mRNA values represent the standard deviation from three independent experiments performed in triplicate and normalized to PAO1 at 0 h. The indicated p values were normalized to mRNA levels of PAO1 at the same time point, where *, p < 0.05, and **, p < 0.005. B, representative Western blot of PhuS. Total protein (25 μg) was loaded in each well. RNA polymerase α was used as an equal loading control. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. The indicated p values were normalized to PAO1 at the same time point where *, p < 0.05, and **, p < 0.005.
Heme and BVIX-dependent Regulation of hasA and hasR
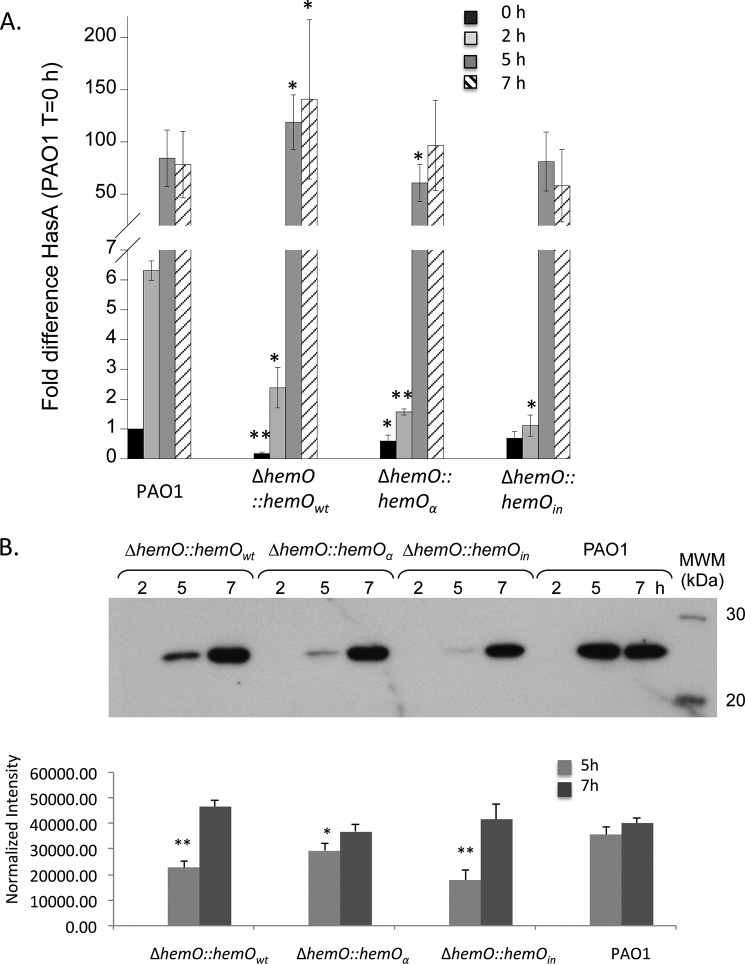
The P. aeruginosa-encoded has ECF σ-factor signaling cascade like that of S. marcescens is thought to autoregulate the hasA and hasR genes in response to heme (10). Consistent with activation of the ECF signaling cascade by the heme-loaded HasA, the hasA and hasR mRNA levels show a time-dependent increase on addition of heme to the media (Figs. 7A and 8A). In PAO1, the steady state mRNA levels of HasA and HasR peak at 5 h with an ∼100- and ∼10-fold increase, respectively. As the cells are washed and resuspended in fresh media prior to the addition of [13C]heme, there is a lag before the appearance of HasA in the extracellular medium (Fig. 7B). Protein expression of HasA peaks at 5 h consistent with the mRNA profile (Fig. 7A) and [13C]BVIXδ and -BVIXβ levels (Fig. 3B). A similar protein expression profile is observed for the complemented ΔhemO::hemOwt strain, although there is an initial lag in hasA mRNA levels and protein expression compared with PAO1 (Fig. 7). The lag in up-regulation of hasA coincides with the reduced extracellular [13C]BVIXβ and [13C]BVIXδ metabolites and lower intracellular iron compared with the parent PAO1 strain (Fig. 3, B and D). The decreased levels of extracellular HasA as a function of lower BVIXβ and BVIXδ metabolites were confirmed in the hemO::hemOin strain where HasA protein levels are significantly reduced over the first 5 h (Fig. 7B).
FIGURE 7.
HasA mRNA and protein levels in PAO1 and the ΔhemO complemented strains. A, mRNA isolated at 0, 2, 5, and 7 h. mRNA values represent the standard deviation from three independent experiments performed in triplicate and normalized to PAO1 at 0 h. The indicated p values were normalized to mRNA levels of PAO1 at the same time point, where *, p < 0.05, and **, p < 0.005. B, Western blotting analysis of HasA in the extracellular medium. Supernatant (5–10 μl) adjusted for differences in A600 was loaded in each well. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. The indicated p values were normalized to PAO1 at the same time point where *, p < 0.05, and **, p < 0.005.
FIGURE 8.
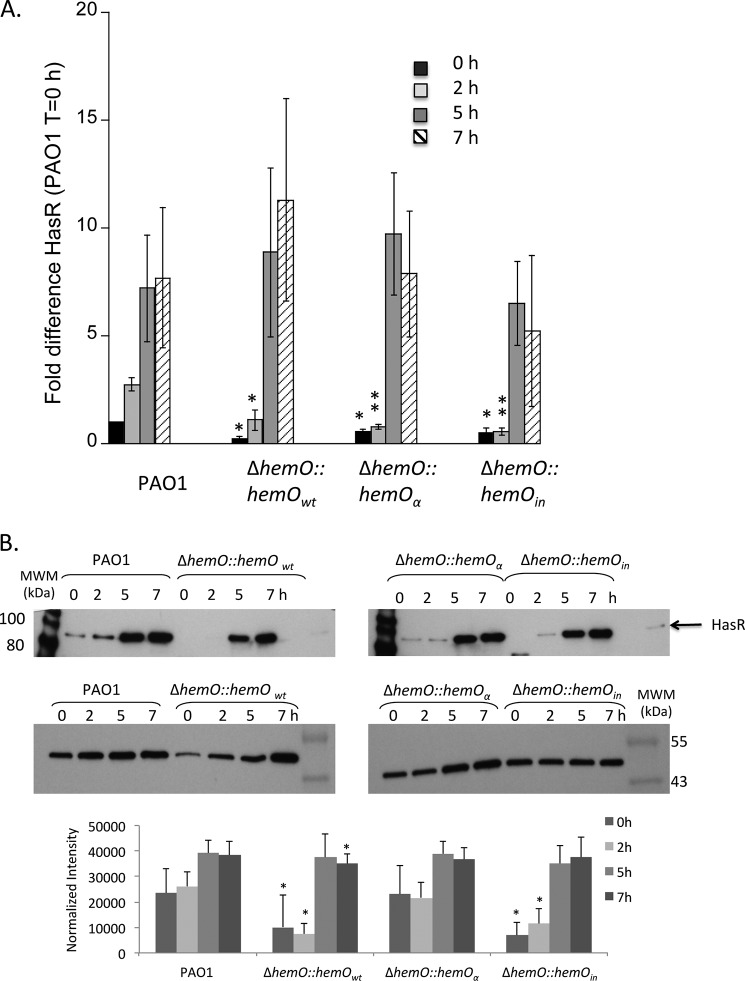
HasR mRNA and protein levels in PAO1 and the ΔhemO complemented strains. A, mRNA isolated at 0, 2, 5, and 7 h. mRNA values represent the standard deviation from three independent experiments performed in triplicate and normalized to PAO1 at 0 h. The indicated p values were normalized to mRNA levels of PAO1 at the same time point, where *, p < 0.05, and **, p < 0.005. B, representative Western blot of HasR. Total protein (25 μg) was loaded in each well. RNA polymerase α was used as an equal loading control. Normalized density (n = 3) was performed on Western blots for three separate biological replicates. The indicated p values were normalized to PAO1 at the same time point where *, p < 0.05, and **, p < 0.005.
Although the hemO:hemOα strain can utilize heme as efficiently as the parent PAO1 strain as judged by the similar overall BVIX levels in first 5 h (Fig. 3B), the extracellular HasA protein levels are significantly lower than in PAO1 (Fig. 7B). The lag in HasA protein levels in strains lacking a BVIXβ/δ regioselective HemO (ΔhemO::hemOin and hemO::hemOα) suggests one or both isomers may play a role in the regulation of hasA. Interestingly, the hasR steady state mRNA and protein levels in the complemented strains lag PAO1, presumably a result of the reduced levels of HasA dampening down activation of the σ-factor HasI (Fig. 8). The similar levels of HasA in all strains at the 7-h time point is due to derepression of Fur on iron limitation as extracellular heme becomes limiting (Fig. 3D). We conclude that hasA independent of hasR is regulated by heme flux through the BVIXβ and BVIXδ regioselective HemO.
Discussion
Based on our previous findings that PhuS acts as the regulator of heme flux through HemO, we hypothesized the alternative BVIXβ and BVIXδ regioselectivity may play a role in regulating extracellular heme uptake (12, 14). However, complementation of the ΔhemO strain at the phage integration locus with the hemOwt gene did not fully recover wild type levels of heme metabolites. When grown in low iron, the steady state hemO mRNA levels of the ΔhemO::hemOwt strain lag those of PAO1 (Fig. 3B). It is unclear at this time why complementation of the hemO gene at the attB site does not restore heme utilization efficiency to that of PAO1. It is possible that there are post-transcriptional regulatory elements within the hemO locus that are not within the currently defined promoter region. Regardless of the reason, complementation of the ΔhemO strain with the hemOwt gene at the attB site ultimately leads to reduced BVIXβ and BVIXδ and points to a potential metabolite-driven regulation of hasA. This was further supported on complementation of the PAO1 ΔhemO strain with the catalytically inactive HemOin enzyme where the lack of BVIX metabolites correlated with reduced HasA protein levels. The BVIXα-selective hemO::hemOα strain also shows a significant decrease in HasA expression compared with PAO1. Therefore, regulation of hasA is not due to the overall level of BVIX metabolites but is dependent specifically on the levels of HemO-derived BVIXβ and/or BVIXδ isomers.
As for S. marcescens, the extracellular heme is proposed to activate the Has system through a cell surface signaling cascade. In the presence of heme, holo-HasA interacts with the HasR receptor and activates the cell surface signaling cascade releasing the HasI σ-factor that then activates hasA and hasR expression (Figs. 7A and 8A). However, we noted the induction of hasA mRNA is almost 10-fold greater than that of hasR suggesting a separate and distinct mechanism of regulation. Previous studies by Vasil and co-workers (6) have shown that under conditions of low iron hasA and hasR are co-transcribed from the hasR promoter. They further confirmed constitutive but weak transcription of hasA from an independent promoter within the intergenic region of hasR and hasA. However, these studies only addressed the effects of iron and not extracellular heme. In this study, we have confirmed heme-dependent transcriptional activation of the has operon, and based on current and previous studies, we propose a revised model for heme and BVIX-dependent regulation of heme uptake.
In this revised model under low iron conditions, hasR and hasA are co-transcribed from the hasR promoter. In addition, hasA is transcribed from the previously identified weak promoter within the intergenic region (6). We have recently confirmed by 5′-rapid amplification of cDNA ends a hasA transcript originating from within the hasRA intergenic region (data not shown). This accounts for the differences in mRNA levels we observe in low iron or on addition of heme to the media. On encountering heme in the extracellular environment, the constitutively produced HasA activates the cell surface signaling system further up-regulating transcription of hasRA. We propose heme taken up by either the Has or Phu systems is degraded to BVIXβ and BVIXδ, which function as post-transcriptional regulators of hasA. Thus, heme-dependent transcriptional activation of the cell surface signaling system, in combination with post-transcriptional regulation of the HasA signal by BVIXβ and/or BVIXδ, allows the cell to rapidly respond to fluctuating extracellular heme levels. Although we presently do not understand the mechanism by which BVIXβ and/or BVIXδ post-transcriptionally regulate hasA, we propose this may be via a BVIX-specific ribosomal binding protein, an sRNA-dependent mechanism, or alternatively a BVIX-dependent riboswitch. Interestingly, within the intergenic region between hasR and hasA there is a previously predicted Rho-independent terminator (6) that may function as a regulatory element.
Furthermore, post-transcriptional BVIXβ/δ-dependent regulation of HasA provides a feedback loop coupling heme sensing by the Has system to the metabolic flux of heme through the PhuS-HemO couple. As shown previously, heme flux into the cell via the Has or Phu systems is driven by the catalytic activity of HemO and regulated by PhuS (12). PhuS acts as a “stop valve” controlling heme flux through its specific interaction with HemO. Therefore, if heme flux is down-regulated due to iron-dependent suppression of HemO, the subsequent drop in BVIXβ and BVIXδ levels rapidly down-regulates heme sensing via HasA, irrespective of external heme levels. Conversely, if external levels of heme are extremely low, the decreased flux of heme through HemO maintains a constitutive low level expression of HasA. To further address the mechanism by which BVIXβ and BVIXδ regulate heme uptake and utilization, we are constructing strains where the hemOα and hemOin genes are integrated into the original hemO locus. This will allow transcriptional and translational fusion assays to be performed in the hemOα and hemOin background without the complication of reduced BVIX levels.
Experimental Procedures
Bacterial Strains
All bacterial strains and plasmids used in this study are listed in Tables 2 and 3, respectively. Escherichia coli strains were routinely grown in Luria Bertani (LB) broth (American Bioanalytical) or on LB agar plates, and P. aeruginosa strains were freshly streaked and maintained on Pseudomonas isolation agar (BD Biosciences). All strains were stored frozen at −80 °C in LB broth with 20% glycerol. For BVIX metabolism, gene expression, and Western blotting analysis, a single isolated colony was inoculated into 10 ml of LB broth and grown overnight at 37 °C. The bacteria were then harvested and washed in 10 ml of M9 minimal medium (Nalgene). Cells were harvested by centrifugation, and the bacterial pellet was resuspended in 10 ml of M9 medium as the initial inoculum into 50 ml of fresh M9 medium at a starting A600 of 0.04. Cultures were grown at 37 °C with shaking at 210 rpm for 3 h before supplementation with [13C]heme at a final concentration of 0.5 μm (time point 0) and incubated for 2, 5, or 7 h. When required, antibiotics were used at the following concentrations (μg/ml): ampicillin, 100; tetracycline (Tc), 10 (for E. coli) and 150 (for P. aeruginosa); gentamicin (Gm), 200; carbenicillin (Cb), 500.
TABLE 3.
Oligonucleotides used in this study
F is forward; R is reverse; qPCR is quantitative PCR.
| Name | Sequence and restriction site (enzyme) |
|---|---|
| RT-qPCR primers and probes | |
| qPCR-phuS probe | 5′-/56-FAM/CTT TCG GCC GCC GCT TCG A/3BQH_1/-3′ |
| qPCR-phuS F | 5′-TGC CGA CGA ACA CCA TGA-3′ |
| qPCR-phuS R | 5′-TGG CGA CCT GGC GAA A-3′ |
| qPCR-hemO probe | 5′-/56-FAM/TTC GTC GCC GCC CAG TAC CTC TTC CAG CAT/3BHQ_1/-3′ |
| qPCR-hemO F | 5′-TGG TGA AGA GCA AGG AAC CCT TC-3′ |
| qPCR-hemO R | 5′-TTC GTT GCG ATA AAG CGG CTC CA-3′ |
| qPCR-phuR probe | 5′/56-FAM/TAC GCG CAG ACC CAC CGC AA/3BQH_1/-3′ |
| qPCR-phuR F | 5′-ACT GCC CAA CGA CTT CTT CAG-3′ |
| qPCR-phuR R | 5′-TTA CGA TGT CCG GAT CGA CGT A-3′ |
| qPCR-hasR probe | 5′-/FAM/CTG GCC TAC GGG CAG CTC TCC TA/3BHQ_1/-3′ |
| qPCR-hasR F | 5′-CGT GGC GTC GAG TAC CAG-3′ |
| qPCR-hasR R | 5′-GGT CTT CGA ACA GAA GTC GTT G-3′ |
| qPCR-hasA probe | 5′-/56-FAM/TCG ACC CGA GCC TGT/3BHQ_1/-3′ |
| qPCR-hasA F | 5′-ATC GAC GCG CTG CTG AA-3′ |
| qPCR-hasA R | 5′-TGG TCG AAG GTG GAG TTG ATC-3′ |
| qPCR-bphO probe | 5′-/56-FAM/CCC GGC AGA TCG ACA GCC CC/3BHQ_1/-3′ |
| qPCR-bphO F | 5′-GCG CTG GCA GGA GTT TCT C-3′ |
| qPCR-bphO R | 5′-ATC GAC GAA ACG AAA GGA ATG T-3′ |
| qPCR-oprF probe | 5′-/56-FAM/CGG TGA GTA CCA TGA CGT TCG TGG C/3BHQ_1/-3′ |
| qPCR-oprF F | 5′-CAG CTG GAC GTG AAG TTC GA-3′ |
| qPCR-oprF R | 5′-GAA GTC AGC CAG GTT CTT GAT GT-3′ |
| Gene deletion and complementation | |
| HemO-A | 5′-GCG AGC TCC GGG TTT CGA TGC ACA GGC G-3′ (SacI) |
| HemO-B | 5′-GCG GTA CCG GTC AGC AGG TTG AGG CGT TG-3′ (KpnI) |
| HemO-C | 5′-GCG GTAC CGG CGC CAG CGA TGC CTT CAA T-3′ (KpnI) |
| HemO-D | 5′-GCG CTG CAG TAC CTG CGG TGA CAT GTT TT-3′ (PstI) |
| E-HemO | 5′-GCG AAT TCC CCT ATG GAT ACC CTG GCC CC-3′ (EcoRI) |
| H-HemOrev | 5′-GCA AGC TTC CCC TCA GGC GAA GGT ACG CTC-3′ (HindIII) |
| PhemO | 5′-GCG GAT CCT TTC GCT CAG GGC ATG CTC C-3′ (BamHI) |
| PhemOrev | 5′-GGG AAT TCC TTT CGA GGG ACG GAA CGC A-3′ (EcoRI) |
| Pser-up | 5′-CGA GTG GTT TAA GGC AAC GGT CTT GA-3′ |
| Pser-down | 5′-AGT TCG GCC TGG TGG AGC AAC TCG-3′ |
| Site-directed mutagenesis | |
| HemO-K132A F | 5′-CTG CGT TCC TGT TCA AGG CAG CCG CC-3′ |
| HemO-K132A R | 5′-AGC GCG GCG GCT GCC TTG AAC AGG AA-3′ |
| HemO-K34A | 5′-CCT GGA GAG CCT GGT GGC GAG CAA GG-3′ |
| HemO-K34A R | 5′-GAA GGG TTC CTT GCT CGC CAC CAG GC-3′ |
| HemO-H26F F | 5′-GAC CAA CGA GCC GTT CCA GCG CCT GG-3′ |
| HemO-H26F R | 5′-CTC CAG GCG CTG GAA CGG CTC GTT GG-3′ |
Construction of the Unmarked PAO1 ΔhemO and Complemented Strains
The in-frame P. aeruginosa hemO mutant was constructed using two primer pairs, HemO-A/HemO-B and HemO-C/HemO-D (Table 3), designed to amplify the amino and carboxyl termini of the gene from PAO1 genomic DNA. The gene sequence was confirmed by DNA sequencing (Eurofins MWG Operon). Following SacI-PstI digestion, the fragments were cloned into the counter-selective suicide plasmid pEX18Tc (17). The resulting pEX18TcΔhemO was transformed into E. coli S17-1-λpir, and a 0.8-kb KpnI fragment containing the FRT-Gm-FRT resistance cassette from pPS858 was inserted into the KpnI site of pEX18TcΔhemO. The resulting pEX18Tc-ΔhemO-GmFRT plasmid was transferred from E. coli S17-1-λpir into P. aeruginosa PAO1 by conjugation. Clones in which a double event of homologous recombination resulted in the chromosomal integration of the ΔhemO::GmFRT allele were selected on PIA plates containing Gm and 5% sucrose. To obtain the unmarked ΔhemO-deleted strain, the pFLP2 plasmid was mobilized into the PAO1 ΔhemO::GmFRT strain. This plasmid encoding the Flippase recombinase (Flp) promotes recombination between FRT sequences flanking the Gm cassette. Isolated colonies were selected on PIA plates containing Cb, and loss of the hemO gene was confirmed by screening for GmS. Plasmid pFLP2 was then cured by streaking CbR GmS colonies on PIA plates supplemented with 5% sucrose. Loss of pFLP2 was tested selecting for sucrose resistance and CbS. Southern blotting hybridization, PCR, and sequencing analyses were used to verify allelic exchange of the parental gene and to ensure that the construct was non-polar (data not shown).
Complemented strains ΔhemO::hemOwt, ΔhemO::hemOα, and ΔhemO::hemOin were constructed by site-specific integration into the attB locus in the ΔhemO mutant strain (18). Briefly, a 0.4-kb fragment encoding the hemO promoter region was PCR-amplified with primers PhemO/PhemOrev and cloned into the BamHI-EcoRI sites of mini-CTX1 plasmid to obtain the construct mini-CTX1-PhemOwt. The hemOα and hemOin coding sequences were PCR-amplified with primer pair E-HemO/H-HemOrev using pBSP-HemOα (19) and pET21a-HemOin (this study) as template, respectively. After EcoRI-HindIII digestion, the 0.6-kb fragments were cloned into the mini-CTX1-PhemO, and the final constructs, mini-CTX1-hemOα and mini-CTX1-hemOin, were confirmed by DNA sequencing. Recombinant plasmids were transferred from E. coli S17-1-λpir into PAO1 ΔhemO by conjugation. Screening for TcR yielded derivatives containing the plasmid integrated into the CTX phage attB site of the P. aeruginosa chromosome. Excision of the unwanted plasmid DNA sequences was achieved by Flp recombinase as described previously (17, 18). To verify the correct integration of complemented sequences at the ΔhemO phage attachment (attB) site, this region was PCR-amplified from chromosomal DNA and sequenced by employing primers Pser-up and Pser-down (18).
Site-directed Mutagenesis of the HemO
Site-directed mutagenesis was performed by PCR utilizing the QuikChange II mutagenesis kit and the previously constructed pET21a HemO expression plasmid (20). Primers used to generate HemO H26A/K34A/K132A (HemOin) and HemO N19K/K34A/F117Y/K132A (HemOα) are listed in Table 3. Mutations were verified by DNA sequencing (Eurofins MWG Operon).
Purification and Characterization of HemO Wild Type and Mutant Proteins
Proteins were expressed and purified as described previously (20, 21). The holo-HemO complexes and turnover experiments were performed as described previously (22). The integrity of the secondary structure was determined by circular dichroism (CD) spectroscopy recorded on a Jasco J-810 spectropolarimeter. All samples were recorded in 20 mm potassium phosphate (pH 7.4) at 25 °C from 19 to 260 nm at a scan rate of 20 nm/min, with each spectrum representing 10 accumulations. Data were acquired at 0.2-mm resolution and 1.0-cm bandwidth. The mean residue ellipticity (degrees cm2 dmol−1) was calculated using CDPRO software as recommended by Jasco. Thermal denaturation studies were performed over a temperature range of 20–90 °C at 222 nm.
HemO-PhuS Protein-Protein Interaction as Measured by Isothermal Titration Calorimetry
Titrations were performed at 25 °C using a MicroCal MCS titration calorimeter. All protein solutions were in 40 mm sodium phosphate buffer (pH 8.0). Samples were degassed before use and injections carried out at 5-min intervals. The heat of dilution of the protein was measured by injecting into the buffer alone. The value obtained was subtracted from the heat of reaction to give the effective heat of binding. Holo-PhuS was expressed and purified as described previously (23). For all titration experiments, the concentration of holo-PhuS protein ranged from 20 to 30 μm, and the titrated HemO variant was set to 10–20 times this value. The ITC data were analyzed using the Origin software package provided by MicroCal. A nonlinear least squares algorithm (minimization of χ2) was used to fit the heat flow per injection to an equation corresponding to an equilibrium binding model, which provides best fit values for the stoichiometry (nITC), change in enthalpy (ΔHITC), and binding constant (KITC). KD values and Gibbs free energy were calculated according to Kd = 1/Ka and ΔG = −RT lnKa. The thermodynamic relationship ΔG = ΔH − TΔS was used to find the entropic contribution to binding. The data were averaged from three independent ITC experiments.
Preparation and Isolation of [12C]Heme and [13C]Heme
Unlabeled δ-[12C]aminolevulinic acid (ALA) was purchased from Frontier Scientific. Labeled δ-[4-13C]ALA was purchased from Cambridge Isotope Laboratories. [12C]Heme or [13C]heme was prepared by a slight modification of the method described by Rivera and Walker (24) and as reported previously (12, 19). Briefly, mitochondrial rat cytochrome b5 was expressed in E. coli BL21 (DE3) cells in M9 minimal media with the following supplements: 2 mm MgSO4, 100 μm CaCl2, 150 nm (NH4)6Mo7O24, 40 μm FeSO4 (acidified with 1 n HCl), 17 μm EDTA, 3 μm CuSO4, 2 μm Co(NO3)2, 7.6 μm ZnSO4, 9.4 μm Na2B4O7·10H2O, and 1 mm thiamine. Following lysis and centrifugation, the cellular lysate was loaded onto a Q-Sepharose anion exchange column (2.5 × 10 cm) equilibrated in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 50 mm NaCl. The column was washed with the same buffer followed by an additional wash with 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 125 mm NaCl (5 column volumes). Cytochrome b5 was eluted in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 250 mm NaCl and dialyzed overnight in 50 mm Tris-HCl (pH 7.4), 5 mm EDTA, 50 mm NaCl. Heme extraction from cytochrome b5 was performed by the acid-butanone method as described previously (25). Heme concentrations were determined by the pyridine hemochrome assay (26). Heme stocks were prepared immediately prior to use by dissolving in 0.1 n NaOH and buffered to pH 7.4 with 1 m Tris-HCl, and the final concentration was determined by pyridine hemochrome (pH 7.4). As reported previously δ-[4-13C]ALA labeling shifts the heme mass by eight yielding distinct BVIX isomer fragmentation patterns shifted from the [12C]BVIX fragment ions by mass ranges of +2 to +4 (11, 12, 17).
Extraction of BVIX Isomers from P. aeruginosa Supernatants
Analysis of the BVIX isomers was performed by slight modification of the previously reported method (12, 19). PAO1 WT and mutant strains supplemented with 0.5 μm [13C]heme were allowed to grow for 2, 5, and 7 h at 37 °C in 250-ml baffled flasks with shaking. Culture supernatants following pelleting of the cells were filtered through a 0.2-μm PES syringe filter, and 1 nm BVIXα dimethyl ester as an internal standard was added to each culture. The supernatant was acidified to pH ∼3 with 10% trifluoroacetic acid (TFA). BVIX isomers were purified over a C18 Sep-Pak column (Waters) as follows. The column was previously equilibrated with 2 ml of acetonitrile, 2 ml of methanol, 2 ml of water, and 2 ml of 10% methanol in 0.1% TFA (v/v). The supernatant was loaded on the column and washed with 4 ml of 0.1% (v/v) TFA, 4 ml of acetonitrile, 0.1% TFA (v/v 20:80), 2 ml of methanol, 0.1% TFA (50:50), and 450 μl of methanol. BVIX isomers were eluted with additional 600 μl of methanol, air-dried, and stored at −80 °C prior to LC-MS/MS analysis.
LC-MS/MS Analysis of P. aeruginosa BVIX Isomers
For LC-MS/MS analysis, samples were resuspended in 10 μl of DMSO and diluted to 100 μl with 90 μl of H2O/acetonitrile (50:50). BVIX isomers were separated over an Ascentis RP-amide 2.7-μm (C18) HPLC column (10 cm × 2.1 mm) with a flow rate of 0.4 ml/min and analyzed on a Waters TQD triple quadrupole mass spectrometer with ACQUITY H-Class UPLC. Fragmentation patterns of the parent BVIX ions derived from [12C]heme or [13C]heme and the BVIXα dimethyl ester standard were analyzed using multiple reaction monitoring. The source temperature was set to 150 °C, the capillary voltage to 3.6 kV, and the cone voltage to 75 V. The collision energy was set to 43 for the internal standard, 34 V for [12C]BVIXα and [13C]BVIX isomers, 30 V for [12C]BVIXβ, and 36 V for [12C]BVIXδ, respectively. Standard curves for quantification were prepared with known concentrations of each of the BVIX isomers. The lower and higher limits of quantification for all isomers were 0.001 and 150 μm, respectively.
ICP-MS Analysis of Intracellular Iron Content
Aliquots (2 ml) were removed from cultures grown as described for the heme uptake studies at 2, 5, and 7 h, pelleted, and washed in fresh M9 media. The pellets were dissolved in trace-metal-free ultrapure 20% HNO3 and boiled overnight at 100 °C. Samples were further diluted with ultrapure water to a final concentration of 2% HNO3 and subjected to ICP-MS on Agilent 7700 ICP-MS (Agilent Technologies). ICP-MS runs were calibrated with high purity iron standard solution (Sigma), and raw ICP-MS data (ppb) were corrected for drift using values for scandium and germanium (CPI International) as internal standards and added to samples during processing. Corrected values were then normalized to culture density as determined by the absorbance at 600 nm. Experimental values and reported standard deviation were the average of six biological replicates.
Expression Studies
Total RNA was purified from a 1-ml aliquot of 0-, 2-, 5-, and 7-h liquid cultures. RNA stabilization was conducted on-site using 250 μl of RNALater Solution (Ambion), and samples were stored at −80 °C until sample preparation. Total RNA was isolated from each cell pellet using the RNeasy mini spin columns according to the manufacturer's directions (Qiagen). Total RNA was treated with RNase-free DNase I (New England Biolabs) for 3 h at 37 °C to remove contaminating chromosomal DNA and precipitated with 0.1× volume 3 m sodium acetate (pH 5.2) and 2× volume 100% (v/v) ethanol. RNA quantity and quality were assessed by UV absorption at 260 nm in a NanoDrop 2000c spectrophotometer (Thermo Scientific) and adjusted to 50 ng/μl. cDNA was generated using the ImProm-IITM reverse transcription system (Promega) from a total of 50 ng of RNA and 0.5 μg random primers. cDNA was analyzed using the StepOnePlus real time PCR system (Applied Biosystems) and TaqMan gene expression master mix (Applied Biosystems). The relative gene expression was calculated using the ΔΔCt method, and the cycle threshold (Ct) values at the 0-, 2-, 5-, and 7-h time point were normalized to the constitutively expressed oprF gene. mRNA values represent the standard deviation of three independent experiments performed in triplicate. All primers and probes used for quantitative PCR studies are listed in Table 2.
SDS-PAGE and Western Blotting Analysis
1-ml aliquots of PAO1 WT and mutant cultures were harvested at the 0-, 2-, 5-, and 7-h time points. Cell pellets were resuspended in 200 μl per A600 of 1.0 in Bugbuster (Novagen). Cells were incubated at room temperature for 30 min with occasional agitation to ensure complete cell lysis. Total protein concentrations were determined using the Bio-Rad RCDC assay. 25 μg of total protein in 2× SDS-loading buffer was run on 10% SDS-PAGE for PhuR or HasR detection, and 12% SDS-PAGE for PhuS, HemO, and HasA detection. Proteins were transferred by electrophoresis to a PVDF membrane (Bio-Rad). Membranes were blocked with blocking buffer (5% w/v skim milk in Tris-buffered saline (TBS) with 0.2% v/v Tween 20), washed, and probed with 1:750 dilution of anti-PhuR, anti-HasR, anti-HasA, anti-PhuS, or anti-HemO primary antibodies in hybridization buffer (1% w/v skim milk in TBS with 0.2% v/v Tween 20). Antibodies were obtained from Covance Custom Antibodies and generated from purified proteins supplied by our laboratory. Antibody specificity was checked against the respective purified proteins (10 ng), and all experimental Western blots were run with molecular weight markers as standards. RNA polymerase α was probed as the loading control with the E. coli RNA polymerase primary antibody (BioLegend) at a dilution of 1:1000. Membranes were rinsed three times in TBS with 0.2% (v/v) Tween 20 and probed with goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (KPL) at a dilution of 1:10,000 in hybridization buffer. Proteins were visualized by chemiluminescent detection using the SuperSignal chemiluminescence kit (Pierce) and hyperfilm ECL (Amersham Biosciences). The normalized density represents the relative abundance of each protein compared with RNA polymerase α as the loading control for n = 3 independent biological replicates. Densitometry analysis was performed on an AlphaImager HP System using the manufacturer's supplied AlphaView software.
Author Contributions
A. W. and S. M. conceived and designed the experiments. S. M., B. J. G., and H. R. C. performed the experiments. S. M., B. J. G., H. R. C., and A. W. analyzed the data. A. W. and S. M. wrote the paper.
Acknowledgments
All mass spectrometry was performed at the University of Maryland, School of Pharmacy, Mass Spectrometry Center. We thank Maureen Kane (Director) and Jace Jones (Associate Director) for helpful discussions regarding LC-MS method development and Heather Neu and Sarah Michel for the running of ICP-MS samples.
This work was supported by National Institutes of Health Grant AI102883 (to A. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ECF
- extracytoplasmic σ function
- HemO
- heme oxygenase
- phu
- Pseudomonas heme uptake
- has
- heme assimilation system
- HemOin
- HemO H26A/K34A/K132A
- HemOα
- HemO N19K/K34A/F117Y/K132A
- Cb
- carbenicillin
- Gm
- gentamicin
- Tc
- tetracycline
- BVIX
- biliverdin IX
- ITC
- isothermal titration calorimetry
- ICP-MS
- inductively coupled plasma-mass spectrometry
- δ-ALA
- δ-aminolevulinic acid.
References
- 1. Wilks A., and Barker K. D. (2011) in Handbook of Porphyrin Science (Kadish K. M., Smith K. M., and Guilard R., eds) 1st Ed., pp. 357–398, World Scientific, Singapore [Google Scholar]
- 2. Wilks A., and Burkhard K. A. (2007) Heme and virulence: how bacterial pathogens regulate, transport and utilize heme. Nat. Prod. Rep. 24, 511–522 [DOI] [PubMed] [Google Scholar]
- 3. Contreras H., Chim N., Credali A., and Goulding C. W. (2014) Heme uptake in bacterial pathogens. Curr. Opin. Chem. Biol. 19, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cornelis P. (2010) Iron uptake and metabolism in Pseudomonads. Appl. Microbiol. Biotechnol. 86, 1637–1645 [DOI] [PubMed] [Google Scholar]
- 5. Cornelis P., and Dingemans J. (2013) Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell Infect. Microbiol. 3, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ochsner U. A., Johnson Z., and Vasil M. L. (2000) Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146, 185–198 [DOI] [PubMed] [Google Scholar]
- 7. Létoffé S., Delepelaire P., and Wandersman C. (1996) Protein secretion in Gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 15, 5804–5811 [PMC free article] [PubMed] [Google Scholar]
- 8. Létoffé S., Delepelaire P., and Wandersman C. (2004) Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J. Bacteriol. 186, 4067–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cescau S., Cwerman H., Létoffé S., Delepelaire P., Wandersman C., and Biville F. (2007) Heme acquisition by hemophores. Biometals 20, 603–613 [DOI] [PubMed] [Google Scholar]
- 10. Biville F., Cwerman H., Létoffé S., Rossi M. S., Drouet V., Ghigo J. M., and Wandersman C. (2004) Haemophore-mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) σ factor involved in haem acquisition. Mol. Microbiol. 53, 1267–1277 [DOI] [PubMed] [Google Scholar]
- 11. Smith A. D., and Wilks A. (2015) Differential contributions of the outer membrane receptors PhuR and HasR to heme acquisition in Pseudomonas aeruginosa. J. Biol. Chem. 290, 7756–7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Neill M. J., and Wilks A. (2013) The P. aeruginosa heme binding protein PhuS is a heme oxygenase titratable regulator of heme uptake. ACS Chem. Biol. 8, 1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wegele R., Tasler R., Zeng Y., Rivera M., and Frankenberg-Dinkel N. (2004) The heme oxygenase(s)–Phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 279, 45791–45802 [DOI] [PubMed] [Google Scholar]
- 14. O'Neill M. J., Bhakta M. N., Fleming K. G., and Wilks A. (2012) Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO). Proc. Natl. Acad. Sci. U.S.A. 109, 5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedman J., Lad L., Li H., Wilks A., and Poulos T. L. (2004) Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry 43, 5239–5245 [DOI] [PubMed] [Google Scholar]
- 16. Schuller D. J., Zhu W., Stojiljkovic I., Wilks A., and Poulos T. L. (2001) Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry 40, 11552–11558 [DOI] [PubMed] [Google Scholar]
- 17. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., and Schweizer H. P. (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 [DOI] [PubMed] [Google Scholar]
- 18. Hoang T. T., Kutchma A. J., Becher A., and Schweizer H. P. (2000) Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 [DOI] [PubMed] [Google Scholar]
- 19. Barker K. D., Barkovits K., and Wilks A. (2012) Metabolic flux of extracellular heme uptake in Pseudomonas aeruginosa is driven by the iron-regulated heme oxygenase (HemO). J. Biol. Chem. 287, 18342–18350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ratliff M., Zhu W., Deshmukh R., Wilks A., and Stojiljkovic I. (2001) Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu W., Wilks A., and Stojiljkovic I. (2000) Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J. Bacteriol. 182, 6783–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilks A., and Schmitt M. P. (1998) Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273, 837–841 [DOI] [PubMed] [Google Scholar]
- 23. Lansky I. B., Lukat-Rodgers G. S., Block D., Rodgers K. R., Ratliff M., and Wilks A. (2006) The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase. J. Biol. Chem. 281, 13652–13662 [DOI] [PubMed] [Google Scholar]
- 24. Rivera M., and Walker F. A. (1995) Biosynthetic preparation of isotopically labeled heme. Anal. Biochem. 230, 295–302 [DOI] [PubMed] [Google Scholar]
- 25. Teale F. W. (1959) Cleavage of the haem-protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543. [DOI] [PubMed] [Google Scholar]
- 26. Fuhrop J. H., and Smith K. M. (eds) (1975) Porphyrins and Metalloporphyrins, pp. 804–807, Elsevier, Amsterdam [Google Scholar]
- 27. de Lorenzo V., and Timmis K. N. (1994) Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235, 386–405 [DOI] [PubMed] [Google Scholar]
- 28. Holloway B. W. (1955) Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13, 572–581 [DOI] [PubMed] [Google Scholar]