Abstract
In eukaryotic cells, heme production is tightly controlled by heme itself through negative feedback-mediated regulation of nonspecific 5-aminolevulinate synthase (ALAS1), which is a rate-limiting enzyme for heme biosynthesis. However, the mechanism driving the heme-dependent degradation of the ALAS1 protein in mitochondria is largely unknown. In the current study, we provide evidence that the mitochondrial ATP-dependent protease ClpXP, which is a heteromultimer of CLPX and CLPP, is involved in the heme-dependent degradation of ALAS1 in mitochondria. We found that ALAS1 forms a complex with ClpXP in a heme-dependent manner and that siRNA-mediated suppression of either CLPX or CLPP expression induced ALAS1 accumulation in the HepG2 human hepatic cell line. We also found that a specific heme-binding motif on ALAS1, located at the N-terminal end of the mature protein, is required for the heme-dependent formation of this protein complex. Moreover, hemin-mediated oxidative modification of ALAS1 resulted in the recruitment of LONP1, another ATP-dependent protease in the mitochondrial matrix, into the ALAS1 protein complex. Notably, the heme-binding site in the N-terminal region of the mature ALAS1 protein is also necessary for the heme-dependent oxidation of ALAS1. These results suggest that ALAS1 undergoes a conformational change following the association of heme to the heme-binding motif on this protein. This change in the structure of ALAS1 may enhance the formation of complexes between ALAS1 and ATP-dependent proteases in the mitochondria, thereby accelerating the degradation of ALAS1 protein to maintain appropriate intracellular heme levels.
Keywords: ATP-dependent protease, heme, mass spectrometry (MS), mitochondria, post-translational modification (PTM), protein degradation, 5-aminolevulinate synthase
Introduction
Heme is an essential molecule to almost all organisms. Heme functions as a prosthetic group on several types of proteins, including cytochromes, catalases, hemoglobin, and myoglobin. Moreover, it has been reported that heme is also involved in numerous regulatory systems in mammals (1), including those that govern transcription (2), translation (3), microRNA processing (4), and the circadian rhythm (5). Excess quantities of heme or heme precursors result in the generation of reactive oxygen species, causing oxidative stress in cells (6). Thus, the production of heme and heme precursors must be tightly regulated. This regulation occurs through the precise control of the intracellular expression of nonspecific 5-aminolevulinatye synthases (ALAS-N or ALAS1). ALAS1 is the first and the rate-limiting enzyme of the heme biosynthetic pathway in mammalian cells, except for in erythroid cells, in which erythroid-specific 5-aminolevulinate synthase (ALAS-E or ALAS2) regulates the first step of heme biosynthesis (7). Although ALAS2 expression increases during erythroid differentiation, ALAS1 expression is suppressed by heme at the transcriptional, translational, and post-translational levels (8). ALAS1 and ALAS2 are encoded by independent genes (9); however, they both contain a conserved amino acid sequence called the heme regulatory motif (HRM),2 which is involved in heme-dependent inhibition of ALAS1 and ALAS2 translocation into the mitochondrial matrix (10). The HRM has also been referred to as the “CP motif” because it includes a core dipeptide motif composed of cysteine and proline residues (2). In humans, three CP motifs have been identified within the ALAS1 precursor protein. Two of these motifs (CP1 and CP2) are located within the presequence for mitochondrial translocation, and the remaining motif (CP3) is located within a region close to the N-terminal end of the mature ALAS1 protein (10). Interestingly, in addition to CP1 and CP2, CP3 has been reported to be involved in heme-dependent inhibition of the mitochondrial import of the ALAS1 protein, although CP3 is not located within the mitochondrial translocation presequence on the protein. Following mitochondrial import, this presequence is proteolytically removed, after which the mature ALAS1 protein catalyzes the condensation of glycine and succinyl-CoA to produce 5-aminolevulinic acid in the mitochondrial matrix (11). Although the mature ALAS1 protein has been suggested to retain CP3 within its N-terminal region (10), how CP3 affects the mature ALAS1 within the mitochondrial matrix remains unknown.
Several proteases and peptidases have been reported to play important roles in protein quality control within the mitochondrial matrix (12, 13). To date, two proteases, LONP1 (14) and ClpXP (15), have been reported to function as ATP-dependent proteases in the mitochondrial matrix of human cells. The human LONP1 protein acts as a homo-oligomeric hexamer (16), whereas ClpXP is a heteromultimer of two different proteins, CLPX and CLPP. CLPP consists of double heptameric rings, whereas CLPX is composed of two hexametric rings bound on each side of the CLPP ring (15). It has been suggested that CLPX recognizes and unfolds the tertiary structures of target proteins, after which CLPP degrades the unfolded target proteins (17). In fact, mammalian ClpXP has been reported to exhibit protease activity against model substrates of ClpXP, such as casein (15, 18); however, a specific substrate for mammalian ClpXP has not been identified. Moreover, Kardon et al. (18) recently reported that mammalian CLPX can associate with and activate ALAS by facilitating the insertion of pyridoxal 5-phosphate, a cofactor for ALAS, into the catalytic site of ALAS, whereas human CLPX does not act as a component of the ClpXP protease for ALAS. Conversely, it has been reported that LONP1 recognizes several different proteins, including mitochondrial aconitase (19) and COX4-1 (20). Moreover, Tian et al. (21) presented evidence that LONP1 was involved in heme-mediated proteolysis of ALAS1 in mitochondria, although the precise mechanism underlying the heme-dependent degradation of ALAS1 remains unclear.
In the present study, we aimed to identify proteins that associate with and regulate intracellular ALAS1 protein. Using immunoprecipitation followed by mass spectrometry analysis, we successfully identified several proteins. Interestingly, we found that ClpXP can form a protein complex with ALAS1. Additional experiments revealed that the formation of a complex between ALAS1 and ClpXP was inhibited by the suppression of endogenous heme biosynthesis and enhanced by the addition of hemin. Thus, we further examined the role of ClpXP in the regulation of heme biosynthesis in human cells.
Results
Several Different Proteins Co-immunoprecipitated with Human ALAS1
To identify proteins involved in the post-translational regulation of ALAS1, human ALAS1 was expressed as a FLAG-tagged protein in FT293 cells, designated FT293ALAS1F. The tagged protein, which was designated ALAS1F, was immunoprecipitated using anti-DDDDK-agarose (Medical and Biological Laboratories, Nagoya, Japan). Then components of the immunoprecipitated proteins were identified using nanoflow LC-MS as described under “Experimental Procedures.” FLAG-tagged firefly luciferase protein (designated LucF) was used as a control to determine what nonspecific proteins were pulled down during the immunoprecipitation. The proteins identified from the LucF immunoprecipitates were excluded as background from the list of proteins identified from the ALAS1F immunoprecipitates. As a result, we identified ∼60 different proteins capable of forming complexes with the ALAS1F protein specifically (Table 1). In addition to seven mitochondrial proteins (in boldface type in Table 1), several non-mitochondrial proteins, including cytosolic proteins, cytoskeletal proteins, the translation initiation protein complex, and RNA-binding proteins, were identified. To the best of our knowledge, the involvement of these proteins in the regulation of ALAS1 has not been reported previously. As such, the roles of these proteins in regulating ALAS1 remain unknown. Although we aim to elucidate the roles of all of the identified proteins in the regulation of ALAS1 in the near future, in the current work, we focused on the isolated mitochondrial matrix proteins because the increased number of total peptide spectrum matches identified by nanoflow LC-MS suggested a high correlation between mitochondrial matrix proteins and ALAS1 protein. Of the seven identified mitochondrial proteins, we chose to further analyze CLPX and CLPP, both of which function as ATP-dependent ClpXP proteases within the mitochondrial matrix, for their potential involvement in the degradation of the ALAS1 protein in the mitochondrial matrix.
TABLE 1.
Proteins that co-immunoprecipitated with FLAG-tagged ALAS1 protein
Coverage, percentage of the protein covered; Unique peptides, number of distinct peptides; PSMs, number of total peptide spectrum matches.
| Accession no. | Protein name | Gene name | Coverage | Unique peptides | PSMs |
|---|---|---|---|---|---|
| % | |||||
| P62857 | 40S ribosomal protein S28 | RPS28 | 23.19 | 1 | 58 |
| P08865 | 40S ribosomal protein SA | RPSA | 3.39 | 1 | 17 |
| P13196 | 5-Aminolevulinate synthase, nonspecific, mitochondrial | ALAS1 | 39.06 | 28 | 7612 |
| P18124 | 60S ribosomal protein L7 | RPL7 | 20.16 | 5 | 144 |
| P62424 | 60S ribosomal protein L7a | RPL7A | 9.77 | 2 | 334 |
| O15143 | Actin-related protein 2/3 complex subunit 1B | ARPC1B | 8.60 | 2 | 93 |
| P61158 | Actin-related protein 3 | ACTR3 | 4.07 | 1 | 25 |
| P68032 | Actin, α cardiac muscle 1 | ACTC1 | 19.89 | 1 | 1360 |
| P63261 | Actin, cytoplasmic 2 | ACTG1 | 28.80 | 1 | 1861 |
| Q53FZ2 | Acyl-coenzyme A synthetase ACSM3, mitochondrial | ACSM3 | 1.71 | 1 | 414 |
| P01011 | α-1-Antichymotrypsin | SERPINA3 | 8.75 | 2 | 227 |
| P35609 | α-Actinin-2 | ACTN2 | 6.04 | 1 | 29 |
| O76031 | ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial | CLPX | 28.12 | 18 | 2808 |
| Q16740 | ATP-dependent Clp protease proteolytic subunit, mitochondria | CLPP | 18.77 | 5 | 260 |
| Q562R1 | β-Actin-like protein 2 | ACTBL2 | 9.04 | 1 | 368 |
| P80723 | Brain acid-soluble protein 1 | BASP1 | 22.03 | 3 | 211 |
| O43852 | Calumenin | CALU | 21.27 | 5 | 263 |
| P31944 | Caspase-14 | CASP14 | 4.55 | 1 | 176 |
| P23528 | Cofilin-1 | CFL1 | 33.13 | 2 | 360 |
| Q9Y281 | Cofilin-2 | CFL2 | 41.57 | 3 | 303 |
| Q9Y6H1 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial | CHCHD2 | 15.23 | 2 | 116 |
| Q9ULV4 | Coronin-1C | CORO1C | 8.44 | 5 | 194 |
| P01040 | Cystatin-A | CSTA | 30.61 | 2 | 192 |
| O95793 | Double-stranded RNA-binding protein Staufen homolog 1 | STAU1 | 6.93 | 3 | 135 |
| Q16643 | Drebrin | DBN1 | 2.47 | 1 | 100 |
| P24534 | Elongation factor 1-β | EEF1B2 | 24.00 | 3 | 402 |
| P26641 | Elongation factor 1-γ | EEF1G | 2.06 | 1 | 25 |
| O94905 | Erlin-2 | ERLIN2 | 7.96 | 2 | 84 |
| O43324 | Eukaryotic translation elongation factor 1 ϵ-1 | EEF1E1 | 6.32 | 1 | 10 |
| P05198 | Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | 6.67 | 2 | 116 |
| O15371 | Eukaryotic translation initiation factor 3 subunit D | EIF3D | 5.11 | 2 | 33 |
| O75821 | Eukaryotic translation initiation factor 3 subunit G | EIF3G | 4.06 | 1 | 138 |
| Q9Y262 | Eukaryotic translation initiation factor 3 subunit L | EIF3L | 8.87 | 3 | 162 |
| P21333 | Filamin-A | FLNA | 3.25 | 6 | 383 |
| Q86SX6 | Glutaredoxin-related protein 5, mitochondrial | GLRX5 | 35.03 | 10 | 1501 |
| P28799 | Granulins | GRN | 2.70 | 1 | 130 |
| P22626 | Heterogeneous nuclear ribonucleoproteins A2/B1 | HNRNPA2B1 | 8.78 | 3 | 136 |
| O60814 | Histone H2B type 1-K | HIST1H2BK | 7.14 | 1 | 13 |
| A0M8Q6 | Ig λ-7 chain C region | IGLC7 | 14.15 | 1 | 125 |
| Q14974 | Importin subunit β-1 | KPNB1 | 1.71 | 1 | 48 |
| Q9Y6Y0 | Influenza virus NS1A-binding protein | IVNS1ABP | 3.58 | 2 | 139 |
| Q9NZI8 | Insulin-like growth factor 2 mRNA-binding protein 1 | IGF2BP1 | 8.32 | 3 | 45 |
| Q9UHB6 | LIM domain and actin-binding protein 1 | LIMA1 | 6.72 | 4 | 171 |
| P49006 | MARCKS-related protein | MARCKSL1 | 9.23 | 1 | 63 |
| O00264 | Membrane-associated progesterone receptor component 1 | PGRMC1 | 14.87 | 2 | 65 |
| Q99453 | Paired mesoderm homeobox protein 2B | PHOX2B | 4.46 | 1 | 20 |
| P30044 | Peroxiredoxin-5, mitochondrial | PRDX5 | 5.14 | 1 | 1 |
| Q13310 | Polyadenylate-binding protein 4 | PABPC4 | 8.85 | 3 | 251 |
| Q9Y2S7 | Polymerase δ-interacting protein 2 | POLDIP2 | 18.48 | 6 | 980 |
| Q92841 | Probable ATP-dependent RNA helicase DDX17 | DDX17 | 1.78 | 1 | 58 |
| Q15185 | Prostaglandin E synthase 3 | PTGES3 | 7.50 | 1 | 69 |
| Q9P258 | Protein RCC2 | RCC2 | 2.87 | 1 | 51 |
| Q14257 | Reticulocalbin-2 | RCN2 | 5.99 | 2 | 55 |
| P49458 | Signal recognition particle 9-kDa protein | SRP9 | 17.44 | 1 | 29 |
| O60749 | Sorting nexin-2 | SNX2 | 2.12 | 1 | 14 |
| Q9NYL9 | Tropomodulin-3 | TMOD3 | 30.97 | 9 | 1193 |
| Q13509 | Tubulin β-3 chain | TBB3 | 6.67 | 2 | 274 |
| Q8NFA0 | Ubiquitin carboxyl-terminal hydrolase 32 | USP32 | 0.69 | 1 | 2 |
| P62987 | Ubiquitin-60S ribosomal protein L40 | UBA52 | 33.59 | 2 | 116 |
| P16989 | Y-box-binding protein 3 | YBX3 | 13.44 | 2 | 152 |
| O15231 | Zinc finger protein 185 | ZNF185 | 2.32 | 1 | 6 |
The ALAS1 Protein Forms a Complex with ClpXP in a Heme-dependent Manner in the Mitochondria
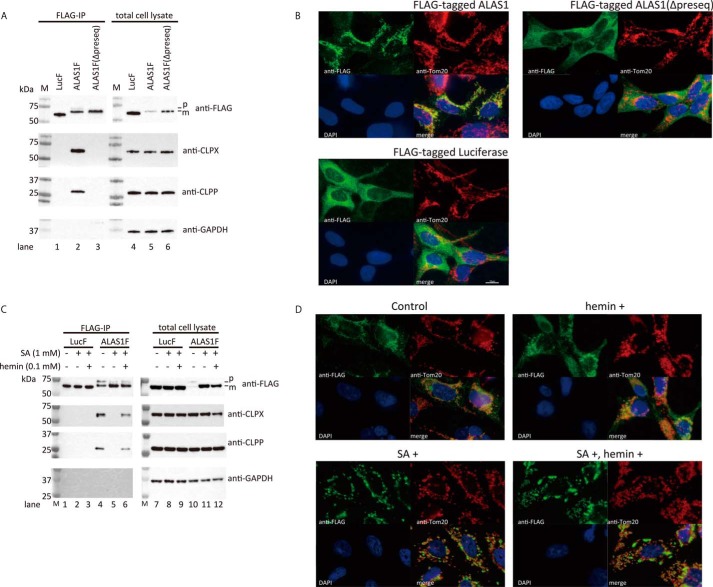
We first confirmed that a complex forms between ALAS1F and ClpXP using immunoprecipitation followed by Western blotting analysis. As shown in Fig. 1A, the co-immunoprecipitation of ALAS1F and ClpXP was reproducible (lane 2), whereas ClpXP did not co-immunoprecipitate with LucF (lane 1). Interestingly, ClpXP was not detected in the immunoprecipitates of the presequence-deleted ALAS1F protein (lane 3), which lacks a signal sequence for mitochondrial translocation. This result suggests that the formation of the ALAS1F-ClpXP complex occurs within mitochondria. The mitochondrial localization of ALAS1F, but not of LucF or ALAS1F(Δpreseq), was also verified by immunofluorescence (Fig. 1B).
FIGURE 1.
The formation of a complex between FLAG-tagged wild-type ALAS1 and endogenous ClpXP and the intracellular distribution of the FLAG-tagged protein. A, Western blotting analysis of immunoprecipitated FLAG-tagged ALAS1 protein. Luciferase (Luc), wild-type ALAS1 (ALAS1), or presequence-deleted ALAS1 (Δpreseq) was expressed in FT293 cells as FLAG-tagged protein and immunoprecipitated using anti-DDDDK-agarose. FLAG-tagged proteins, CLPX, and CLPP in the immunoprecipitates (left lanes, FLAG-IP) and in total cell lysates (right lanes) were detected by Western blotting analysis using specific antibodies. GAPDH was detected as an internal control. M, molecular size marker. B, immunofluorescence analysis of FT293 cells expressing FLAG-tagged proteins. Nucleus, mitochondria, and FLAG-tagged proteins were stained with DAPI (blue), anti-Tom20 (red), and anti-FLAG (green), respectively. C, intracellular heme levels are influenced by the formation of ALAS1 protein complexes. Shown is expression of FLAG-tagged luciferase (LucF) or ALAS1 (ALAS1F) proteins in FT293 cells following incubation with or without 1 mm SA for 24 h. The SA-treated cells were subsequently incubated with or without hemin for 30 min before harvest. In A and C, the loading volumes of the immunoprecipitates were adjusted according to the intensities of the FLAG-tagged proteins; therefore, each sample contains a similar quantity of FLAG-tagged protein. For the total cell lysate, 10 μg of protein was loaded into each lane. p or m, precursor or mature FLAG-tagged ALAS1 protein, respectively. D, immunofluorescence analysis of FT293 cells expressing FLAG-tagged ALAS1 protein after hemin and/or SA treatment. Nucleus, mitochondria, and FLAG-tagged ALAS1 protein were stained with DAPI (blue), anti-Tom20 (red), and anti-FLAG (green), respectively.
To determine the role of intracellular heme in the formation of a complex between ALAS1F and ClpXP, LucF- or ALAS1F-expressing cells were treated with succinylacetone (SA), which specifically inhibits heme biosynthesis, for 24 h. The cells were then further incubated with or without hemin for 30 min before being harvested. As shown in Fig. 1C, ALAS1F formed protein complexes with ClpXP (lane 4), whereas the suppression of endogenous heme synthesis by SA treatment prevented the ClpXP-ALAS1F complex formation (lane 5). Importantly, additional treatment with hemin restored the formation of ClpXP-ALAS1F complexes within 30 min (lane 6). Co-immunoprecipitation of LucF and ClpXP was not observed after SA or hemin treatment (lanes 1–3), although the expression levels of CLPX and CLPP in cell lysates were similar to those in cells expressing LucF (lanes 7–9) or ALAS1F (lanes 10–12). These results suggest that ALAS1F specifically forms complexes with ClpXP within the mitochondrial matrix in a heme-dependent manner. The immunofluorescence study revealed that the FLAG-tagged ALAS1 still localized in the mitochondria after treatment with SA and/or hemin, although SA treatment of these cells resulted in the enlargement of some mitochondria due to extensive accumulation of FLAG-tagged ALAS1 (Fig. 1D).
A Specific Heme-binding Motif on the ALAS1 Protein Is Involved in the Heme-dependent Association between ALAS1 and ClpXP
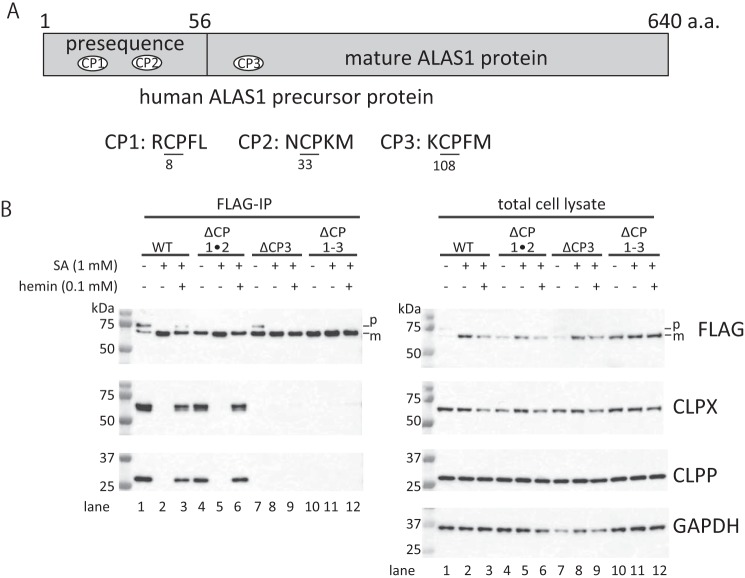
It has been reported that heme binds to ALAS1 via a conserved amino acid sequence, termed the HRM (10). This motif contains a cysteine-proline dipeptide motif (CP motif) as a core sequence (2). Human ALAS1 and ALAS2 proteins each contain three independent CP motifs, which are involved in regulating the mitochondrial translocation of ALAS1 and ALAS2 (10, 22). Two of these CP motifs (CP1 and CP2) are located within the presequences found on ALAS proteins, whereas the final CP motif (CP3) is located within the N-terminal regions of mature ALAS proteins (Fig. 2A). Interestingly, it has been suggested that CP3, along with CP1 and CP2, is also involved in regulating the mitochondrial translocation of ALAS1 (22). However, the role of CP3 with respect to mature ALAS1 proteins present in the mitochondria remains unclear. To determine the independent roles of CP1, CP2, and CP3 in the formation of the ALAS1-ClpXP complex, site-directed mutagenesis was utilized to substitute each cysteine residue within each CP motif to alanine, which inhibits the binding of heme to the motif (2). The resultant constructs expressed ALAS1F proteins in which both CP1 and CP2, only CP3, or all CP motifs were mutated; these constructs were designated ALAS1FΔCP1·2, ALAS1FΔCP3, and ALAS1FΔCP1–3, respectively. As shown in the left panel of Fig. 2B, CLPX and CLPP were detectable in the ALAS1F immunoprecipitates (lane 1), whereas incubation with SA led to the disappearance of CLPX and CLPP from the ALAS1F immunoprecipitate (lane 2). It should be noted that additional treatment with hemin restored the formation of complexes between ALAS1F and ClpXP within 30 min (lane 3). Similar results were observed when ALAS1FΔCP1·2 was immunoprecipitated (lanes 4–6). However, CLPX and CLPP proteins were barely detectable in the immunoprecipitates of the mutant ALAS1 proteins in which only CP3 (ALAS1FΔCP3; lanes 7–9) or all CP motifs (ALAS1FΔCP1–3; lanes 10–12) were mutated, whereas CLPX and CLPP expressed similarly in cells (lanes 1–12 in the right panel of Fig. 2B). These results strongly suggest that the CP3 motif on ALAS1 is involved in the formation of complexes between ALAS1 and ClpXP proteins.
FIGURE 2.
The CP3 motif in the mature ALAS1 protein is involved in the formation of complexes between ALAS1 and ClpXP. A, schematic presentation of the location of the CP motifs in the ALAS1 protein. The precursor of human ALAS1 consists of 640 amino acids. The presequence of ALAS1, which is located at the N-terminal end of the precursor protein (amino acids 1–56), contains two independent CP motifs (CP1 and CP2). A third CP motif (CP3) is located within the N-terminal region of the mature ALAS1 protein, which is the active form of ALAS1 in mitochondria. The amino acid sequences of each CP motif are shown, and cysteine and proline dipeptides are underlined. The numbers below each amino acid indicate the amino acid positions of the cysteine residues within each CP motif in the human ALAS1 precursor protein (GenBankTM number CAA39794). B, ClpXP protein quantities decreased in CP3-mutated ALAS1 immunoprecipitates. Wild-type ALAS1 (WT), CP1- and CP2-mutated ALAS1 (ΔCP1·2), CP3-mutated ALAS1 (ΔCP3), and ALAS1 with all CPs mutated (ΔCP1–3) were expressed in FT293 cells as FLAG-tagged proteins. The cells were treated with SA and/or hemin. Each FLAG-tagged protein was immunoprecipitated using anti-DDDDK-agarose. For the immunoprecipitated samples, the loading volume was adjusted according to the intensity of each FLAG-tagged protein; therefore, each sample contains a similar amount of FLAG-tagged protein. For the cell lysate, 10 μg of protein was loaded into each lane. p or m, precursor or mature FLAG-tagged ALAS1 protein, respectively.
Endogenous ALAS1 and ClpXP Form a Complex in Mitochondria in HepG2 Cells
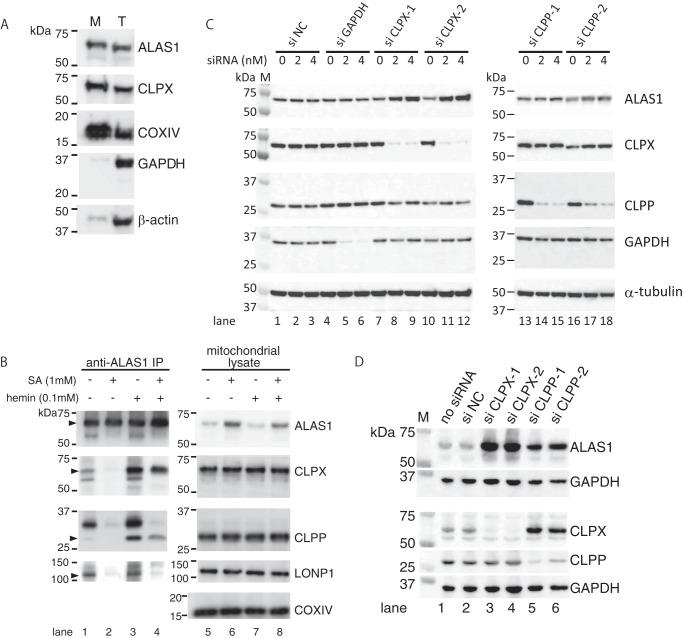
To confirm that endogenous ALAS1 and ClpXP form a complex in mitochondria in the HepG2 human hepatic cell line, we first prepared the mitochondria-rich fraction and confirmed the purity of the fraction. As shown in Fig. 3A, mitochondrial proteins (ALAS1, CLPX, and COXIV), cytosolic protein (GAPDH), and cytoskeletal protein (β-actin) were abundantly expressed in the total lysate, whereas GAPDH and β-actin proteins were marginally detected in the mitochondria-rich fraction, suggesting that the mitochondria were concentrated in this mitochondria-rich fraction. Thus, to identify proteins that associate with endogenous ALAS1 in the mitochondria, mitochondria-rich fraction prepared from HepG2 cells was subjected to immunoprecipitation of endogenous ALAS1 protein. Using an anti-human ALAS1 mouse monoclonal antibody for immunoprecipitation led to detectable levels of CLPX and CLPP proteins in ALAS1 immunoprecipitates (Fig. 3B, lane 1). However, treating the cells with SA decreased the quantity of CLPX and CLPP proteins in the immunoprecipitates (lane 2). Conversely, incubating the cells with hemin for 30 min resulted in increased quantities of CLPX and CLPP proteins in the immunoprecipitates (lane 3). Treatment of the cells with hemin increased the quantities of immunoprecipitated CLPX and CLPP proteins even after treatment with SA (lane 4). These results strongly suggest that ClpXP forms complexes with ALAS1 in mitochondria in HepG2 cells in a heme-dependent manner. Another ATP-dependent mitochondrial protease, LONP1, has been reported to be involved in the heme-dependent degradation of ALAS1 (21). We therefore also examined whether the immunoprecipitates of ALAS1 protein included LONP1. As shown in Fig. 3B, LONP1 was detected in the immunoprecipitates of ALAS1 without (lane 1) or with hemin treatment (lane 3) but was hardly detected after treatment with SA (lane 2). Interestingly, LONP1 was not clearly recovered in the immunoprecipitates after treatment with hemin for 30 min (lane 4), suggesting that the mode of complex formation between ALAS1 and LONP1 is different from that between ALAS1 and ClpXP.
FIGURE 3.
Endogenous ALAS1 forms a complex with ClpXP in a heme-dependent manner in mitochondria, and the specific suppression of CLPX or CLPP results in the accumulation of ALAS1 proteins within HepG2 cells. A, the purity of the mitochondria in the mitochondria-rich fraction was determined by detecting mitochondria-specific proteins (ALAS1, CLPX, and COXIV), cytosolic protein (GAPDH), or cytoskeleton protein (β-actin) using Western blotting analysis. B, HepG2 cells were incubated with (lanes 2, 4, 6, and 8) or without (lanes 1, 3, 5, and 7) SA for 24 h. Some cell samples were subsequently treated with hemin for 30 min (lanes 3, 4, 7, and 8). Endogenous ALAS1 protein was immunoprecipitated using an anti-ALAS1 monoclonal antibody, and the loading volume of each eluate was adjusted according to the intensity of each immunoprecipitated ALAS1 protein; therefore, each sample contained a similar amount of ALAS1 protein (lanes 1–4). For total cell lysate, 10 μg of protein was loaded into each lane (lanes 5–8). C, specific siRNA-mediated reduction in CLPX expression causes accumulation of ALAS1 proteins in HepG2 cells. The transfected siRNAs and their concentrations are indicated at the top of the panel. Total cell lysate was prepared for each cell sample, and 10 μg of protein was loaded into each lane. D, prolonged suppression of CLPP or CLPX induced the accumulation of endogenous ALAS1 protein in HepG2 cells. The cells were harvested 10 days after the initial transfection of each siRNA. The details of this procedure are described under “Experimental Procedures.” A total of 10 μg of protein was loaded into each lane.
Next, we used siRNA to suppress CLPX or CLPP expression to examine the roles of these proteins in regulating ALAS1 expression. As shown in Fig. 3C, the introduction of siCLPX-1 (lanes 7–9) and siCLPX-2 (lanes 10–12), both of which effectively suppressed CLPX expression, increased ALAS1 expression in HepG2 cells. Conversely, the introduction of the siNegative control (lanes 1–3) or siGAPDH (lanes 4–6), a positive control for siRNA transfection, did not influence ALAS1 expression. Surprisingly, transient suppression of CLPP using siCLPP1 (lanes 13–15) or siCLPP2 (lanes 16–18) had only a marginal effect on ALAS1 expression at 48 h after transfection. Therefore, we prolonged the suppression of CLPP expression by performing sequential transfection of siRNA 4 days after the initial transfection. Ten days after the initial transfection of siCLPP to suppress CLPP expression, endogenous ALAS1 expression was found to increase in HepG2 cells (Fig. 3D, lanes 5 and 6) compared with that found in cells transfected with the siNegative control (Fig. 3D, lane 2). The prolonged suppression of CLPX more effectively induced the accumulation of ALAS1 protein in HepG2 cells (lanes 3 and 4). These results suggest that a complex forms between ClpXP and ALAS1 proteins in HepG2 cells and that ClpXP is involved in the degradation of ALAS1 protein in these cells.
CLPX Is Essential for Heme-dependent Degradation of Endogenous ALAS1 Protein in FT293 Cells
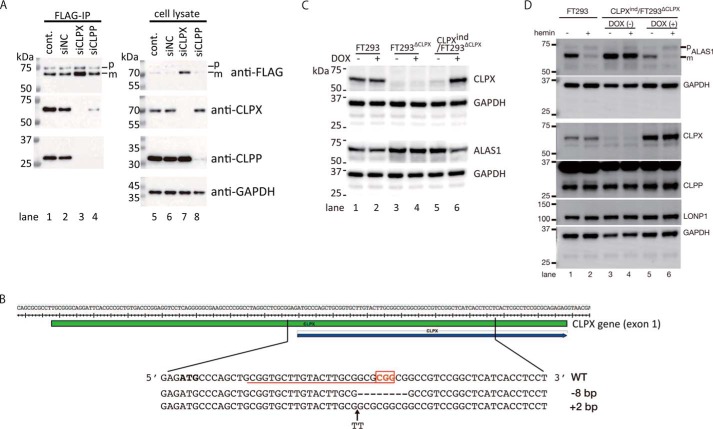
To determine which protein, CLPX or CLPP, is closely associated with ALAS1, we immunoprecipitated FLAG-tagged ALAS1 (ALAS1F) after the introduction of siRNA against CLPX or CLPP in FT293ALAS1F cells. As shown in Fig. 4A, CLPX-specific or CLPP-specific siRNA successfully decreased the expression of CLPX and CLPP, respectively (lanes 7 and 8), in FT293ALAS1F cells. Both CLPX and CLPP were detected in the immunoprecipitates of ALAS1F from FT293ALAS1F cells treated without (lane 1) or with siNC (lane 2). CLPX was also detected in the immunoprecipitates from siCLPP-treated FT293ALAS1F cells (lane 4), whereas CLPP was not detectable in the immunoprecipitates from siCLPX-treated FT293ALAS1F cells (lane 3). These results suggest that CLPP requires CLPX to form a complex with ALAS1, whereas CLPX can associate with ALAS1 without CLPP. We next further examined the involvement of CLPX in the regulation of ALAS1. We established CLPX knock-out FT293 cells using the CRSPR/Cas9 system (designated FT293ΔCLPX). Then a CLPX expression vector (pcDNA5/FRT/TO-CLPX) was introduced as described previously to establish CLPXind/FT293ΔCLPX, in which the expression of CLPX is inducible in a doxycycline-dependent manner. Fig. 4B presents the target sequence for Cas9 nuclease and the genotype of the FT293ΔCLPX cells we established. As shown in Fig. 4C, CLPX was not detected in FT293ΔCLPX in the absence or presence of doxycycline (lane 3 or 4, respectively). Although CLPX was highly expressed after incubation with doxycycline (lane 6), it was marginally detected in CLPXind/FT293ΔCLPX cells even in the absence of doxycycline (lane 5). Although the reason for the basal expression of CLPX in CLPXind/FT293ΔCLPX cells is unclear, it is possible that the FBS used to culture CLPXind/FT293ΔCLPX cells contained tetracycline, as indicated in the manual for the Flp-In T-Rex system (Invitrogen). Using these cells, we examined the expression level of endogenous ALAS1 protein in the presence or absence of CLPX. As shown in Fig. 4C, deletion of CLPX resulted in an increase in the expression of the ALAS1 protein (lanes 3 and 4) compared with that in FT293 cells (lanes 1 and 2). ALAS1 expression in CLPXind/FT293ΔCLPX cells was also increased without doxycycline (lane 5), whereas it was decreased after the induction of CLPX protein expression (lane 6). Using these cells, we have further examined the effect of hemin treatment on the expression level of ALAS1 (Fig. 4D). Interestingly, hemin treatment resulted in a decrease in ALAS1 protein in FT293 cells (lane 2, compared with lane 1) or CLPX-induced CLPXind/FT293ΔCLPX cells (lane 6, compared with lane 5); ALAS1 protein levels were unchanged after treatment with hemin in uninduced CLPXind/FT293ΔCLPX cells (lanes 3 and 4). These results strongly suggested that the expression of CLPX is essential for heme-dependent degradation of ALAS1 protein.
FIGURE 4.
CLPX is essential for heme-dependent degradation of ALAS1. A, along with the induction of ALAS1F, siNegative control (siNC), siCLPX-1 (siCLPX), or siCLPP-1 (siCLPP) RNA or no siRNA (cont.) was introduced into FT293ALAS1F cells. Then ALAS1F protein was immunoprecipitated as described under “Experimental Procedures.” After immunoprecipitation, equal volumes of immunoprecipitates (FLAG-IP) or equal amounts of total cell lysate (10 μg/lane; cell lysate) were loaded for the detection of each protein using SDS-PAGE followed by immunoblotting analysis. FLAG-tagged ALAS1 protein, CLPX, or CLPP was detected in immunoprecipitates and cell lysate. GAPDH was also detected in the total cell lysate as a loading control. B, target sequence of Cas9 in the human CLPX gene and the DNA sequence of the flanking region of the CLPX gene in FT293ΔCLPX. The red underline and open red box of the reference sequence (WT) indicate the target sequence of Cas9 and protospacer adjacent motif (PAM), respectively. C, expression of CLPX and ALAS1 after incubation with or without doxycycline. GAPDH was detected as a loading control. D, effect of CLPX expression on the decrease in ALAS1 after hemin treatment. The influence of hemin treatment was determined in FT293 cells (lanes 1 and 2) or CLPXind/FT293ΔCLPX without (lanes 3 and 4) or with (lanes 5 and 6) induction of CLPX expression by doxycycline. p or m, precursor or mature FLAG-tagged ALAS1 protein, respectively.
Mutation of the CP3 Motif on the ALAS1 Protein Extends the ALAS1 Protein Half-life in Mitochondria
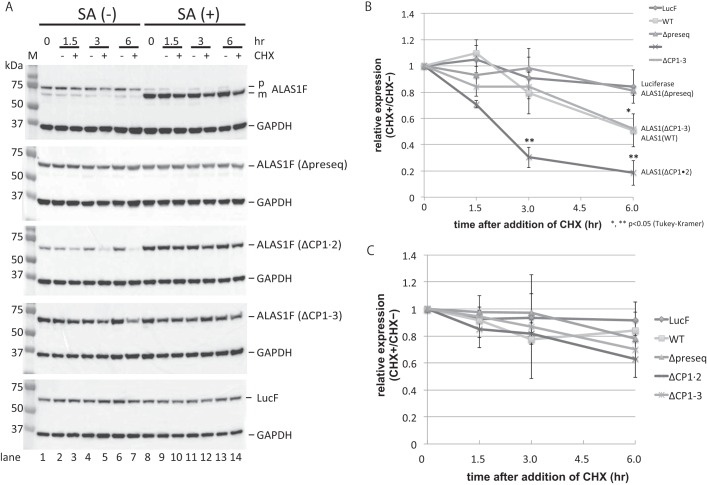
Because specific knockdown of CLPX or CLPP expression resulted in the accumulation of ALAS1 protein in HepG2 cells, we hypothesized that ClpXP is involved in the degradation of ALAS1 proteins within mitochondria. To test this hypothesis, we investigated the turnover rates of ALAS1F proteins in which the presequence (Δpreseq), CP1 and CP2 (CP1·2) motifs, or all three CP motifs (CP1–3) were mutated, and we compared these rates with that of WT ALAS1F protein. Each expression vector was introduced into FT293 cells, which allowed transcription to be induced by the addition of doxycycline. At 24 h after inducing the expression of each protein, translation was inhibited using cycloheximide (CHX). ALAS1F protein expression was examined 1.5, 3, and 6 h after the addition of CHX. A representative result is shown in Fig. 5A. The intensity produced by the mature protein in each lane was measured for statistical analysis. The means and S.D. of three independent experiments are shown in Fig. 5, B (without SA) and C (with SA). As shown in Fig. 5B, the relative expression levels of ALAS1FΔCP1·2 at 3 and 6 h after the addition of CHX are significantly lower than those of the other proteins (**, p < 0.05). The relative expression levels of WT ALAS1 and ALAS1FΔCP1–3 at 6 h after the initiation of CHX treatment were lower than those of luciferase or ALAS1(Δpreseq) (*, p < 0.05). As shown in Fig. 5B, the expected half-lives of WT ALAS1 and ALAS1FΔCP1–3 ALAS1 are each ∼6 h, whereas the half-life of ALAS1FΔCP1·2 is ∼2 h. These results suggest that the presence of CP3 in the mature ALAS1 protein is related to the shorter half-life of mature ALAS1. Although we expected WT ALAS1 to have a similar half-life to that of ALAS1FΔCP1·2, the half-life was actually longer than that of ALAS1FΔCP1·2. This discrepancy might be caused by the accumulation of precursor WT proteins in the cells, which might partially compensate for degraded mature proteins by translocating into mitochondria. Indeed, the accumulation of precursor wild-type ALAS1 protein was detected in cells not treated with SA (Fig. 5A, top, SA (−)), whereas there was virtually no accumulation of ALAS1FΔCP1·2 or ALAS1FΔCP1–3, both of which contain mutated CP motifs within their presequences. Thus, to determine the functional role of the CP3 motif on ALAS1F with respect to the half-life of the mature protein in mitochondria, it might be suitable to compare the half-life of ALAS1FΔCP1·2 with that of ALAS1FΔCP1–3. Conversely, following treatment with SA, WT ALAS1, ALAS1FΔCP1·2, and ALAS1FΔCP1–3 ALAS1F all became very stable, and no differences were found in the relative expression levels within each sample at any time point (Fig. 5C). These results suggest that the presence of the CP3 motif on ALAS1 correlates to the short half-life of this protein in mitochondria, although this effect is masked when endogenous heme biosynthesis is suppressed by treatment with SA.
FIGURE 5.
Determination of ALAS1 protein turnover. A, FLAG-tagged proteins, including luciferase (LucF), wild-type ALAS1 (WT), presequence-deleted ALAS1 (Δpreseq), ALAS1 protein with mutations in its presequence CP motifs (ΔCP1·2), and ALAS1 protein with mutations in all of its CP motifs (ΔCP1–3), were expressed in FT293 cells. Total cell lysates were prepared at 1.5, 3, and 6 h after the addition of cycloheximide into the culture medium. The intensities of the signals produced by the FLAG-tagged proteins were normalized against that produced by GAPDH. All signals were measured using ImageQuant TL software, and expression levels relative to that of cells not treated with cycloheximide were calculated. Each experiments was repeated at least three times, and one representative result is shown per panel. A total of 10 μg of protein was loaded into each lane. B and C, expression levels of FLAG-tagged proteins at each time point relative to those at 0 h were used to determine the half-life of each protein. In C, cells were treated with succinylacetone for 24 h before and during incubation with cycloheximide. The average of three independent experiments was used to prepare the graph, and the error bars indicate S.D. p or m, precursor or mature FLAG-tagged ALAS1 protein, respectively.
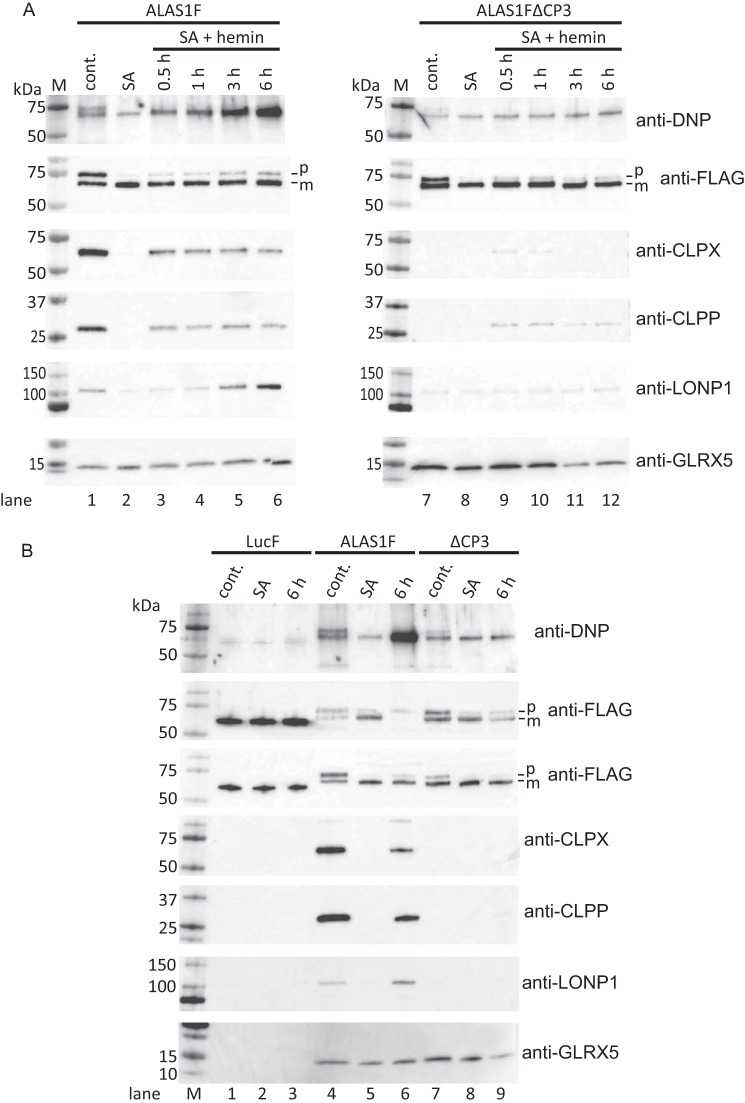
The CP3 Motif Is Involved in the Oxidation of ALAS1 Protein and the Recruitment of LONP1 into the ALAS1 Protein Complex
Because it has been reported that the binding of free heme to the CP motif on iron regulatory protein 2 (IRP2) leads to the oxidation of the IRP2 protein (23), we attempted to determine whether the CP3 motif on ALAS1 is involved in heme-mediated oxidation of ALAS1. To accomplish this, we incubated ALAS1F-expressing cells in which endogenous heme production was suppressed by SA with hemin for several hours before immunoprecipitation. Following this, we examined the carbonylation of immunoprecipitated ALAS1 because carbonylation is the most common oxidative modification of a protein. As shown in Fig. 6A (top left), the immunoprecipitated ALAS1F proteins contained carbonylation, and their molecular sizes correspond to precursor and mature ALAS1F proteins (lane 1). Furthermore, treating the cells with SA slightly inhibited the oxidation of the protein (lane 2), whereas incubating the cells with hemin after the SA treatment enhanced the accumulation of carbonylated proteins, the molecular size of which corresponded to mature ALAS1 protein. This effect lasted for up to 6 h (lanes 3–6). We also attempted to determine whether immunoprecipitated ALAS1FΔCP3 proteins showed an accumulation of carbonylated proteins because the CP3 motif is present in the mature ALAS1F protein. We found that carbonylated proteins were only faintly detectable in ALAS1FΔCP3 immunoprecipitates, even after a 6-h incubation with hemin (Fig. 6A, top right, lanes 7–12). These results suggest that the presence of the CP3 motif on ALAS1 and the longer exposure of cells to hemin are both related to the oxidation of the ALAS1 protein. Interestingly, CLPX and CLPP proteins were barely detectable in ALAS1FΔCP3 immunoprecipitates, even after a 6-h-long hemin treatment (Fig. 6A, middle panels, lane 12). These results suggest that the CP3 motif on ALAS1 is involved in the heme-dependent oxidative modification of the ALAS1 protein and in the formation of a complex between ClpXP and ALAS1. However, it is unclear whether ClpXP is involved in the degradation of oxidized ALAS1 because the quantities of CLPX and CLPP proteins in the ALAS1F immunoprecipitates did not change during the incubation with hemin (Fig. 6A, middle panels, lanes 3–6). Previously, it was reported that another ATP-dependent mitochondrial matrix protease, LONP1, is involved in the heme-dependent degradation of ALAS1 (21) and in the removal of oxidized mitochondrial proteins (19, 24). We thus examined whether ALAS1F immunoprecipitates contained LONP1, although LC-MS analysis of immunoprecipitated ALAS1F proteins did not identify LONP1 protein. As shown in the second panels from the bottom in Fig. 6A, LONP1 protein was detected in WT ALAS1 immunoprecipitates (lane 1). Furthermore, the quantity of LONP1 protein in ALAS1F immunoprecipitates increased after the cells were incubated with hemin (lanes 3–6). It should be noted that the presence of LONP1 in the ALAS1F immunoprecipitates is also related to the presence of the CP3 motif on ALAS1 because LONP1 was barely detectable in ALAS1FΔCP3 immunoprecipitates, even after incubation with hemin (lanes 7–12). We also assessed whether ALAS1F immunoprecipitates contained GLRX5, which was identified in these immunoprecipitates via LC-MS analysis. As shown in the bottom panels of Fig. 6A, GLRX5 was detected in ALAS1F and ALAS1FΔCP3 immunoprecipitates, and treatment of cells with SA and/or hemin did not influence the formation of complexes between these proteins. These results suggest that intracellular heme levels and the presence of the CP3 motif on ALAS1 do not affect the formation of complexes between GLRX5 and ALAS1 (lanes 1–12). Several of the above samples as well as immunoprecipitates of LucF were selected and loaded onto the same acrylamide gel for comparison against each other (Fig. 6B). The results clearly demonstrated differences between ALAS1F (lanes 4–6) and ALAS1FΔCP3 (lanes 7–9) with respect to heme-dependent oxidative modifications and the formation of complexes with CLPX, CLPP, and LONP1. In contrast, no heme-dependent carbonylation or complex formation was detected in the LucF immunoprecipitates (lanes 1–3). Again, only GLRX5 was similarly detected in the immunoprecipitates of ALAS1F and ALAS1FΔCP3 (bottom). This finding was true regardless of whether the cells were treated with SA or hemin before immunoprecipitation (Fig. 6B, bottom, lanes 4–9).
FIGURE 6.
Hemin treatment induced oxidation of the ALAS1 protein. A, cells expressing FLAG-tagged ALAS1 (ALAS1F) or ALAS1ΔCP3 (ALAS1FΔCP3) were treated with succinylacetone for 24 h. Following this, the cells were treated with hemin for the indicated times before immunoprecipitation. The loading volume for each immunoprecipitated sample was adjusted according to the intensity of the FLAG-tagged protein so that similar quantities of FLAG-tagged protein were loaded into each lane. One membrane was used to detect carbonylated protein (top), and another membrane was used to detect co-immunoprecipitated proteins (ALAS1F, CLPX, CLPP, LONP1, and GLRX5; bottom five panels) because treating the membrane with dinitrophenylhydrazine (DNPH) to detect carbonylated proteins modified the reactivity of some proteins toward their specific antibodies (data not shown). B, untreated (cont.), SA-treated and hemin-treated (6 h) samples of immunoprecipitated FLAG-tagged Luciferase (LucF), wild-type ALAS1 (ALAS1F), and CP3-mutated ALAS1 (ΔCP3) were selected for comparison. p or m, precursor or mature FLAG-tagged ALAS1 protein, respectively.
Discussion
In the present study, using immunoprecipitation followed by MS analysis, we identified several proteins capable of forming complexes with ALAS1. The presence of CLPX, CLPP, or GLRX5 proteins in the immunoprecipitates of FLAG-tagged ALAS1 proteins was also confirmed using Western blotting analysis. We further demonstrated that hemin treatment stimulates the formation of complexes between ALAS1 and ClpXP in human hepatic cells. Moreover, siRNA-mediated suppression of CLPX or CLPP resulted in the accumulation of ALAS1 proteins in mitochondria, suggesting that ClpXP is involved in the degradation of the ALAS1 protein within mitochondria. Furthermore, using CLPXind/FT293ΔCLPX cells, we successfully demonstrated that the presence of CLPX is essential for heme-mediated degradation of endogenous ALAS1 protein. Interestingly, the HRM on the mature ALAS1 protein (CP3), which has previously been reported to be involved in heme-dependent suppression of the mitochondrial translocation of the ALAS1 precursor protein, was found to play a crucial role in the formation of complexes between ALAS1 and ClpXP and in the heme-dependent degradation of ALAS1. Taken together, our results demonstrate that ClpXP is involved in negative feedback-mediated regulation of heme biosynthesis via the heme-dependent degradation of ALAS1.
Recently, Kardon et al. (18) reported that Mcx1, a yeast homolog of CLPX, stimulates heme biosynthesis in yeast by enhancing the insertion of co-enzymes into the Hem1 protein. These authors also demonstrated that human CLPX can activate ALAS2 both in vitro and in vivo, whereas recombinant ClpXP, which degrades the typical ClpXP substrate, is unable to digest recombinant human apoALAS2 proteins in vitro. The referenced work indicates a novel function for mammalian CLPX as an activator of ALAS2 enzymatic activity in vertebrates. However, it is also important to note that recombinant ClpXP, which consists of mouse CLPX and human CLPP, is able to digest the general ClpXP substrate “casein,” confirming that mammalian ClpXP is an active protease. Indeed, other groups have previously reported on the proteolytic ability of human ClpXP using casein as a substrate (15, 25), although the specific substrate used by human ClpXP remains unclear. Thus, based on our results, we hypothesize that ClpXP can recognize and modify ALAS1 for degradation under special conditions, such as in the presence of heme, as in our assay conditions. It is still unclear whether ClpXP directly digests ALAS1, which can be determined in vitro using a combination of recombinant CLPX, CLPP, and ALAS1 proteins. Such experiments are ongoing projects in our laboratory, and we are actively attempting to express and purify these recombinant proteins to create an in vitro assay system. Although a procedure for purifying recombinant active human ClpXP protein has already been reported (25), a method for purifying recombinant human ALAS1 protein has not yet been established. Indeed, human ALAS1 could not be purified using the same methods that we used to purify recombinant ALAS2 (26, 27) because the recombinant ALAS1 protein degraded during purification (data not shown). Thus, we are currently attempting to modify our method for the purification of recombinant ALAS1 for the purpose of creating an in vitro assay system.
In our initial experiment using LC/MS, we did not identify LONP1 in ALAS1F immunoprecipitates (Table 1), although Tian et al. (21) reported that heme-mediated breakdown of ALAS1 protein is dependent on the presence of LONP1. However, our Western blotting analysis revealed the presence of LONP1 in ALAS1 precipitates from HepG2 cells (Fig. 3B) and in ALAS1F immunoprecipitates from FT293 cells (Fig. 6A), and LONP1 levels were enhanced following hemin treatment (Fig. 6A). Thus, it is possible that LONP1 and ClpXP cooperatively regulate ALAS1 degradation within the mitochondria of mammalian cells. Interestingly, the sequential exposure of cells to SA and hemin restored the formation of complexes between ClpXP and ALAS1F within 30 min after the initiation of the hemin treatment. LONP1 became evident in these immunoprecipitates several hours after the initiation of the hemin treatment (Fig. 6A). Furthermore, increases in LONP1 quantities in the ALAS1F immunoprecipitates appeared to be related to the accumulation of oxidized ALAS1 in the immunoprecipitates, suggesting that heme-dependent oxidation of ALAS1 might be required for LONP1-dependent degradation. Conversely, ClpXP recognizes heme-bound ALAS1 as an earlier response to intracellular increases in free heme, mediating the modification of ALAS1 for degradation by LONP1. It should be noted that the CP3 motif on mature ALAS1 plays an important role in the formation of complexes between ALAS1 and ClpXP and in the oxidation of the ALAS1 protein. These results suggest that the binding of heme to the CP3 motif triggers the recognition of ALAS1 by ClpXP and the oxidative modification of ALAS1.
Our study revealed that both of the examined ATP-dependent proteases might be involved in the heme-dependent degradation of the ALAS1 protein within the mitochondrial matrix. This degradation occurs as a part of a negative feedback mechanism for heme biosynthesis and might be initiated by the direct binding of heme to the CP3 motif on ALAS1. In addition to the above-discussed proteins, several other proteins were identified as possible candidates for the formation of complexes with ALAS1. However, it is still unclear whether these proteins form complexes independently or form one large complex together. The answer to this question should be better elucidated in future work performed by our group, in which we intend to separate immunoprecipitates using isoelectric focusing or glycerol gradient centrifugation before LC/MS analysis.
Experimental Procedures
Reagents
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich, Wako Pure Chemical Industries (Osaka, Japan), or Nacalai Tesque (Kyoto, Japan). Complete EDTA-free protease inhibitor tablets were purchased from Roche Diagnostics GmbH (Mannheim, Germany). Anti-DDDDK-agarose and DDDDK peptides for the purification of FLAG-tagged proteins were purchased from Medical and Biological Laboratories Co., Ltd. (Nagoya, Japan).
cDNA Cloning and Site-directed Mutagenesis
Human ALAS1 cDNA (GenBankTM number NM_000688) encoding an ALAS1 precursor protein (GenBankTM number CAA39794) was amplified by PCR with the following primers: 5′-CTCAGCGCAGTCTTTCCACAGG-3′ and 5′-GTCGACGCTAGCCTGAGCAGATACCAACTTG-3′. The amplified products were cloned into a pGEM-T Easy Vector (Promega Corp., Madison, WI). The resultant plasmid was digested with the SalI restriction enzyme to isolate ALAS1 cDNA, and the isolated fragment was then used to replace ALAS2 cDNA in a pGEM-AET vector (28). The resultant pGEM-ALAS1F vector contained ALAS1 cDNA that encoded an ALAS1 precursor protein with a FLAG tag at its C-terminal end. Using a PrimerStar Max site-directed mutagenesis kit (Takara Bio, Shiga, Japan), the mutations c.22_23TG>GC, c.97_98TG>GC, and c.322_323TG>GC were introduced into the pGEM-ALAS1F plasmid, respectively, resulting in p.C8A, p.C33A, and p.C108A amino acid substitutions within the ALAS1 protein (Takara Bio). Because p.Cys-8, p.Cys-33, and p.Cys-108 correspond to conserved cysteine residues within CP1, CP2, and CP3, respectively, each mutation was expected to inhibit heme binding to these CP motifs within the HRM (10). To prepare cDNA encoding the mature ALAS1 protein, which lacks a mitochondrial targeting signal, the pGEM-ALAS1F plasmid was subjected to amplification by PCR using the following primers: 5′-GCGGCCGCGATGGAACAGATCAAAGAAACCCCTC-3′ and 5′-GTCGACGCTAGCCTGAGCAGATACCAACTTG-3′. Amplified products were cloned into a pGEM-T Easy Vector, the plasmid was digested with NotI and SalI, and the isolated fragments were used to replace ALAS2 cDNA within a pGEM-AET plasmid. The resultant plasmid, pGEM-ΔpreseqALAS1F, contained cDNA encoding the mature ALAS1 protein with a FLAG tag at its C-terminal end. The above-described plasmids were digested with NotI, and each cDNA construct was cloned into the NotI site of a pcDNA5/FRT/TO vector (Invitrogen). The resultant plasmids pFRT-ALAS1F, pFRT-ALAS1Δpreseq, pFRT-ALAS1ΔCP1·2, pFRT-ALAS1ΔCP3, and pFRT-ALAS1ΔCP1–3 were used to express FLAG-tagged, wild-type, presequence-deleted ALAS1 proteins containing mutations in both HRM1 and HRM2, in HRM3 alone, and in all three HRMs, respectively. Each plasmid was then co-transfected with a pOG44 vector in Flp-In T-REx 293 cells (FT293 cells, Invitrogen) to establish stable transformants. These transformants individually expressed each FLAG-tagged ALAS1 protein construct (ALAS1F) in a tetracycline/doxycycline-inducible manner. The conditions used to select the transformants and the establishment of cells expressing FLAG-tagged luciferase (LucF) were performed as described previously (27).
Cell Culture
The culture conditions used to grow the FT293 cells were as described previously (27). HepG2 cells were purchased from the European Collection of Cell Cultures (ECACC), a public cell culture collection maintained in the United Kingdom. The cells were maintained in Eagle's minimum essential medium supplemented with 10% FBS, 2 mm glutamine, 1% non-essential amino acids, 50 units/ml penicillin, and 50 μg/ml streptomycin.
Immunoprecipitation and Western Blotting Analysis
Unless otherwise noted, the preparation, incubation, and centrifugation of the collected samples were performed at 4 °C. The cells were lysed in lysis buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 1% Triton X-100, 10% glycerol, complete EDTA-free protease inhibitor mix, 1 mm EDTA, 1 mm NaF, and 0.4 mm Na3VO4) by pipetting vigorously. The samples were then incubated on ice for 10 min and centrifuged at 21,000 × g for 15 min. Supernatant protein concentrations were determined using Pierce 660nm protein assay reagent (Thermo Fisher Scientific) with bovine serum albumin as a standard. An aliquot of total cell lysate was subjected to Western blotting analysis. The majority of the samples were incubated with anti-DDDDK-agarose beads in tubes appropriately sized for immunoprecipitation, which was performed by rotating the tubes for 90 min at 4 °C. Then the beads were washed once with lysis buffer, followed by three sequential washes with wash buffer (20 mm HEPES, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, and 10% glycerol). FLAG-tagged proteins were eluted with elution buffer (0.1 mg/ml DDDDK peptides in wash buffer). For immunoprecipitation of endogenous ALAS1 protein, cell lysates were incubated with a mouse monoclonal anti-Alas1 antibody (ab54758, Abcam, Cambridge, UK) overnight at 4 °C, and the proteins were then purified using μMACS Protein G MicroBeads and a μMACS column (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to the manufacturer's instructions. For immunoprecipitation of endogenous ALAS1 in HepG2 cells, the mitochondria-rich fraction was prepared using the Qproteome mitochondria purification kit (Qiagen GmbH Germany) before immunoprecipitation. For Western blotting analysis, samples were mixed with 6× SDS-PAGE sample buffer with reducing reagent (Nacalai Tesque) and were boiled for 10 min before being loaded onto a TGX acrylamide gel (Bio-Rad). The samples were electrophoresed according to the manufacturer's instructions and then electrically transferred onto PVDF membranes. For the detection of specific proteins, these membranes were blocked with 5% skim milk in Tris-buffered saline containing 0.05% Tween 20 (TBS-T) for 1 h. The blocked membranes were incubated with diluted primary antibody for 1 h at room temperature and then washed with TBS-T three times for 5 min each. The membranes were then incubated with an HRP-conjugated secondary antibody for 1 h at room temperature and washed with TBS-T three times for 10 min each at room temperature. Signals were detected using Clarity ECL Western substrate (Bio-Rad) or Immobilon Western chemiluminescent HRP substrate (Millipore Corp., Billerica, MA) and visualized using an ImageQuant LAS500 image analyzer (GE Healthcare, Uppsala, Sweden). For the detection of FLAG-tagged proteins and GAPDH protein, an HRP-conjugated primary antibody was used. Signal intensity was measured using ImageQuant TL software (GE Healthcare). A protein carbonyl Western blot detection kit (Shima Laboratories, Tokyo, Japan) was used for the detection of oxidized proteins according to the manufacturer's instructions. An anti-FLAG, HRP-conjugated M2 monoclonal antibody (A8592); anti-GAPDH, HRP-conjugated monoclonal antibody (G9295); anti-LONP1 antibody (HPA002192); and anti-GLRX5 (HPA042465) antibody were purchased from Sigma-Aldrich. Anti-CLPX (ab168338), anti-CLPP (ab124822), and anti-ALAS1 (ab54758, ab154860) antibodies were purchased from Abcam.
Nano-HPLC/MS/MS Analysis and Protein Sequence Database Searching
Immunoprecipitated FLAG-tagged ALAS1 protein was digested with trypsin and dried as described previously (29). The dried peptide extract (20 μg) was dissolved in 80 μl of sample solution (5% acetonitrile and 0.1% TFA). Each sample (1.25 μg/5 μl) was injected into an EasynLC-1000 system (Thermo Fisher Scientific), which was connected to an EASY-Spray column (25-cm length × C18 ODS 75 μm, Thermo Fisher Scientific). Peptides were eluted with a 180-min gradient of 4–25% solvent B (0.1% formic acid in acetonitrile, v/v) in solvent A (0.1% formic acid in water, v/v) at a flow rate of 300–400 nl/min. Peptides were then ionized and analyzed using a Fusion mass spectrometer (Thermo Fisher Scientific) coupled to a nanospray source. High-resolution full scan MS spectra (from m/z 380 to 1800) were acquired in the Orbitrap with a resolution of 140,000 at m/z 400 and lock mass enabled (m/z at 445.12003 and 391.28429), followed by MS/MS fragmentation of the 10 most intense ions in the linear ion trap with collisionally activated dissociation energy of 35%. The exclusion duration for the data-dependent scan was 0 s, and the isolation window was set at 10.0 m/z.
The MS/MS data were analyzed by sequence alignment with variable and static modifications using Mascot and Sequest algorithms. We utilized the UniProt protein database to search each tryptic peptide sequence. The specific parameters used to search the protein sequence database included oxidation of methionine, deamination of asparagine or glutamine, acetylation of N-terminal of peptide, and pyroglutamation as variable modifications and carbamidomethylation as a static modification. Other parameters used in our data analysis included two allowed missing cleavages, a mass error of 10 ppm for precursor ions, and 0.02 Da for fragment ions. Charge states of +2 to +4 were considered for parent ions. If more than one spectrum was assigned to a peptide, only the spectrum with the highest Mascot score was selected for manual analysis. All peptides with Mascot scores >20 were manually examined using rules described previously (30).
Immunofluorescence
Cells grown on coverslips were fixed with 4% formaldehyde in PBS for 10 min at room temperature and permeabilized in blocking solution (2% skim milk, 0.2% BSA, and 0.2% Triton X-100 in PBS) for 10 min. The fixed cells were incubated with an anti-FLAG M2 mouse monoclonal antibody (1 μg/ml; F3165, Sigma-Aldrich) and an anti-Tom20 rabbit polyclonal antibody (1:250; sc-11415, Santa Cruz Biotechnology, Inc., Dallas, TX) in 2% BSA/PBS for 30 min at 37 °C, followed by incubation with Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 588-conjugated anti-rabbit IgG antibodies (1:1000 each; Thermo Fisher Scientific), respectively. The coverslips were washed with PBS and sealed with ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific). The slides were imaged using a fluorescence microscope fitted with a ×60 objective lens (BZ-9000, Keyence Corp., Osaka, Japan).
Knockdown of Endogenous Proteins
Gene-specific siRNAs (Silencer Select validated siRNA) were purchased from Thermo Fisher Scientific. Silencer Select GAPDH siRNA was used as a positive control for knockdown, and Silencer Select negative control no. 1 siRNA was used as a negative control. Each siRNA was introduced into HepG2 cells using Lipofectamine RNAiMax reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. Forty-eight hours after transfection, the cells were harvested and lysed, and the total cell lysates were subjected to Western blotting analysis. For prolonged suppression of the CLPX or CLPP protein, a second siRNA transfection was performed 4 days after the initial transfection, and the cells were cultured for an additional 7 days before harvest. The culture medium for the siRNA-transfected cells was replaced with fresh medium at least every 4 days. Total cell lysate was prepared using lysis buffer as described above.
Establishment of CLPX-knock-out FT293 Cells and Doxycycline-inducible Expression of CLPX
The pSpCas9(BB)-2A-Puro(PX459) expression vector was a gift from Feng Zhang (Addgene plasmid 48139), and the knock-out of the CLPX gene was constructed as described with slight modifications (31). The sequences of the oligonucleotides, which included a guiding RNA sequence, were 5′-CACCGCGGTGCTTGTACTTGCGGCG-3′ and AAACCGCCGCAAGTACAAGCACCGC-3′. After annealing, these oligonucleotides were inserted into the BbsI site of pSpCas9(BB)-2A-Puro(PX459). The resultant plasmid, PX459-CLPX, was used to transfect FT293 cells using Lipofectamine 2000 (Invitrogen). After cloning transfected cells, several clones were analyzed for the CLPX gene mutation using the Guide-it mutation detection kit (Clontech, Mountain View, CA). Positive clones were further subjected to Western blotting analysis to detect the CLPX protein. The selected CLPX negative clone was designated FT293ΔCLPX. The CLPX locus targeted by Cas9 in FT293ΔCLPX cells was amplified by PCR and subcloned into T-Vector pMD20 (Takara Bio) for sequencing. To construct the human wild-type CLPX protein expression vector, the CLPX cDNA carrying a silent mutation at Ala11 was inserted into the BamHI and NotI restriction sites of the pcDNA5/FRT/TO vector. The resulting expression vector, pcDNA5/FRT/TO-CLPX, was co-transfected with the pOG44 vector into FT293ΔCLPX cells to obtain stable transformants. After cloning the transformants as described (27), each clone was examined for the doxycycline-inducible expression of CLPX and zeocin sensitivity. The resultant clone was designated CLPXind/FT293ΔCLPX. For the induction of CLPX, cells were treated with 1 μg/ml doxycycline for 8 days and then further incubated with hemin for 6 h before harvesting cells.
Author Contributions
Y. Kubota, Y. Katoh, and K. F. designed the study, performed the experiments, and wrote the manuscript. K. N., R. Y., and K. K. performed the experiments and analyzed the data.
Acknowledgments
We thank Dr. Kazuhiko Igarashi for valuable discussion and Miyuki Ohno for excellent support in performing experiments.
This work was supported in part by Grant-in-aid for Scientific Research (C) 23590353 from the Japan Society for the Promotion of Science and by a Health Labor Sciences Research Grant from the Ministry of Health Labor and Welfare in Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- HRM
- heme regulatory motif
- SA
- succinylacetone
- CHX
- cycloheximide.
References
- 1. Furuyama K., Kaneko K., and Vargas P. D. (2007) Heme as a magnificent molecule with multiple missions: heme determines its own fate and governs cellular homeostasis. Tohoku J. Exp. Med. 213, 1–16 [DOI] [PubMed] [Google Scholar]
- 2. Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., Fujita H., and Igarashi K. (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen J. J. (2007) Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood 109, 2693–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faller M., Matsunaga M., Yin S., Loo J. A., and Guo F. (2007) Heme is involved in microRNA processing. Nat. Struct. Mol. Biol. 14, 23–29 [DOI] [PubMed] [Google Scholar]
- 5. Kaasik K., and Lee C. C. (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430, 467–471 [DOI] [PubMed] [Google Scholar]
- 6. Ryter S. W., and Tyrrell R. M. (2000) The heme synthesis and degradation pathways: role in oxidant sensitivity: heme oxygenase has both pro- and antioxidant properties. Free Radic. Biol. Med. 28, 289–309 [DOI] [PubMed] [Google Scholar]
- 7. Riddle R. D., Yamamoto M., and Engel J. D. (1989) Expression of δ-aminolevulinate synthase in avian cells: separate genes encode erythroid-specific and nonspecific isozymes. Proc. Natl. Acad. Sci. U.S.A. 86, 792–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Furuyama K., and Yamamoto M. (2013) Differential regulation of 5-aminolevulinate synthase isozymes in vertebrates in Handbook of Porphyrin Science, Vol. 27 (Ferreira G. C., Kadish K. M., Smith K. M., and Guilard R. eds) pp. 1–39, World Scientific, Hackensack, NJ [Google Scholar]
- 9. Bishop D. F., Henderson A. S., and Astrin K. H. (1990) Human δ-aminolevulinate synthase: assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics 7, 207–214 [DOI] [PubMed] [Google Scholar]
- 10. Lathrop J. T., and Timko M. P. (1993) Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 259, 522–525 [DOI] [PubMed] [Google Scholar]
- 11. Yamauchi K., Hayashi N., and Kikuchi G. (1980) Cell-free synthesis of rat liver δ-aminolevulinate synthase and possible occurrence of processing of the enzyme protein in the course of its translocation from the cytosol into the mitochondrial matrix. FEBS Lett. 115, 15–18 [DOI] [PubMed] [Google Scholar]
- 12. Koppen M., and Langer T. (2007) Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 42, 221–242 [DOI] [PubMed] [Google Scholar]
- 13. Baker B. M., and Haynes C. M. (2011) Mitochondrial protein quality control during biogenesis and aging. Trends Biochem. Sci. 36, 254–261 [DOI] [PubMed] [Google Scholar]
- 14. Wang N., Gottesman S., Willingham M. C., Gottesman M. M., and Maurizi M. R. (1993) A human mitochondrial ATP-dependent protease that is highly homologous to bacterial Lon protease. Proc. Natl. Acad. Sci. U.S.A. 90, 11247–11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang S. G., Ortega J., Singh S. K., Wang N., Huang N. N., Steven A. C., and Maurizi M. R. (2002) Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J. Biol. Chem. 277, 21095–21102 [DOI] [PubMed] [Google Scholar]
- 16. García-Nafría J., Ondrovicová G., Blagova E., Levdikov V. M., Bauer J. A., Suzuki C. K., Kutejová E., Wilkinson A. J., and Wilson K. S. (2010) Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 19, 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baker T. A., and Sauer R. T. (2012) ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kardon J. R., Yien Y. Y., Huston N. C., Branco D. S., Hildick-Smith G. J., Rhee K. Y., Paw B. H., and Baker T. A. (2015) Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 161, 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bota D. A., and Davies K. J. (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4, 674–680 [DOI] [PubMed] [Google Scholar]
- 20. Fukuda R., Zhang H., Kim J. W., Shimoda L., Dang C. V., and Semenza G. L. (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 [DOI] [PubMed] [Google Scholar]
- 21. Tian Q., Li T., Hou W., Zheng J., Schrum L. W., and Bonkovsky H. L. (2011) Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J. Biol. Chem. 286, 26424–26430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Munakata H., Sun J. Y., Yoshida K., Nakatani T., Honda E., Hayakawa S., Furuyama K., and Hayashi N. (2004) Role of the heme regulatory motif in the heme-mediated inhibition of mitochondrial import of 5-aminolevulinate synthase. J. Biochem. 136, 233–238 [DOI] [PubMed] [Google Scholar]
- 23. Ishikawa H., Kato M., Hori H., Ishimori K., Kirisako T., Tokunaga F., and Iwai K. (2005) Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol. Cell 19, 171–181 [DOI] [PubMed] [Google Scholar]
- 24. Nyström T. (2005) Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 24, 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowth B. R., Kirstein-Miles J., Saiyed T., Brötz-Oesterhelt H., Morimoto R. I., Truscott K. N., and Dougan D. A. (2012) Substrate recognition and processing by a Walker B mutant of the human mitochondrial AAA+ protein CLPX. J. Struct. Biol. 179, 193–201 [DOI] [PubMed] [Google Scholar]
- 26. Furuyama K., Fujita H., Nagai T., Yomogida K., Munakata H., Kondo M., Kimura A., Kuramoto A., Hayashi N., and Yamamoto M. (1997) Pyridoxine refractory X-linked sideroblastic anemia caused by a point mutation in the erythroid 5-aminolevulinate synthase gene. Blood 90, 822–830 [PubMed] [Google Scholar]
- 27. Kadirvel S., Furuyama K., Harigae H., Kaneko K., Tamai Y., Ishida Y., and Shibahara S. (2012) The carboxyl-terminal region of erythroid-specific 5-aminolevulinate synthase acts as an intrinsic modifier for its catalytic activity and protein stability. Exp. Hematol. 40, 477–486.e1 [DOI] [PubMed] [Google Scholar]
- 28. Furuyama K., and Sassa S. (2000) Interaction between succinyl CoA synthetase and the heme-biosynthetic enzyme ALAS-E is disrupted in sideroblastic anemia. J. Clin. Invest. 105, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh Y., Ikura T., Hoshikawa Y., Tashiro S., Ito T., Ohta M., Kera Y., Noda T., and Igarashi K. (2011) Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol. Cell 41, 554–566 [DOI] [PubMed] [Google Scholar]
- 30. Chen Y., Kwon S. W., Kim S. C., and Zhao Y. (2005) Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 31. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]