Abstract
Emerging evidence suggests that n-3 polyunsaturated fatty acids (PUFA) promote brown adipose tissue thermogenesis. However, the underlying mechanisms remain elusive. Here, we hypothesize that n-3 PUFA promotes brown adipogenesis by modulating miRNAs. To test this hypothesis, murine brown preadipocytes were induced to differentiate the fatty acids of palmitic, oleate, or eicosapentaenoic acid (EPA). The increases of brown-specific signature genes and oxygen consumption rate by EPA were concurrent with up-regulation of miR-30b and 378 but not by oleate or palmitic acid. Next, we hypothesize that free fatty acid receptor 4 (Ffar4), a functional receptor for n-3 PUFA, modulates miR-30b and 378. Treatment of Ffar4 agonist (GW9508) recapitulated the thermogenic activation of EPA by increasing oxygen consumption rate, brown-specific marker genes, and miR-30b and 378, which were abrogated in Ffar4-silenced cells. Intriguingly, addition of the miR-30b mimic was unable to restore EPA-induced Ucp1 expression in Ffar4-depleted cells, implicating that Ffar4 signaling activity is required for up-regulating the brown adipogenic program. Moreover, blockage of miR-30b or 378 by locked nucleic acid inhibitors significantly attenuated Ffar4 as well as brown-specific signature gene expression, suggesting the signaling interplay between Ffar4 and miR-30b/378. The association between miR-30b/378 and brown thermogenesis was also confirmed in fish oil-fed C57/BL6 mice. Interestingly, the Ffar4 agonism-mediated signaling axis of Ffar4-miR-30b/378-Ucp1 was linked with an elevation of cAMP in brown adipocytes, similar to cold-exposed or fish oil-fed brown fat. Taken together, our work identifies a novel function of Ffar4 in modulating brown adipogenesis partly through a mechanism involving cAMP activation and up-regulation of miR-30b and miR-378.
Keywords: adipose tissue metabolism, insulin resistance, microRNA (miRNA), obesity, polyunsaturated fatty acid (PUFA), uncoupling protein, free fatty acid receptor 4, brown adipogenesis, brown fat
Introduction
There are two specific types of fat with opposite functions, brown adipose tissue (BAT)3 and white adipose tissue (WAT). WAT stores energy in the form of triglyceride, whereas BAT dissipates energy in the form of heat via uncoupling protein 1 (UCP1) (1). Recent advances in our understanding of BAT biology in humans have provided a new insight into weight loss strategies by boosting BAT thermogenesis. Despite the surge of research for identifying cellular and molecular regulators of brown fat development (2, 3), the role of dietary constituents, particularly dietary fatty acids (FA), in thermogenic activation is poorly understood. Depending on the degree of desaturation and the n-6/n-3 ratio, FA differentially control adiposity, insulin sensitivity, immune response (4), and probably energy expenditure (5–7). Lately, it has been suggested that n-3 polyunsaturated FA (PUFA) stimulate beige/brown adipogenesis in primary murine adipogenic precursor cells (5) as well as C57BL/6 mice (6, 7). The latter animal study suggests that n-3 PUFA are associated with sympathetic activation and increased β3-adrenergic receptor (Adrb3) signaling (7). However, the underlying mechanistic details of how dietary n-3 PUFA up-regulate brown fat development and thermogenesis remain unknown.
MicroRNAs (miRNAs) play critical functions in brown adipocyte differentiation and maintenance. Deletion of adipose-specific dicer promotes white-like phenotype of brown precursor cells (8). Similarly, disruption of miRNA biogenesis by deleting fat-specific Dgcr8, a molecular anchor necessary for the recognition of pri-miRNA, results in defective BAT formation and severe cold intolerance (9). Recently, miR-193b/365 cluster was identified to be enriched in BAT and demonstrated as a key regulator to promote brown adipogenesis by targeting Runx1t1, a transcription factor that drives myocyte formation (10). In addition to suppression of myogenic lineage commitment, miR-196a promotes brown adipogenesis by inhibiting white adipocyte commitment (11). The brown transcriptional program could be corroborated by miR-378 that increases cAMP production via targeted degradation of phosphodiesterase 1B (Pde1b) (12) or by miR-30b that promotes degradation of Rip140, a transcriptional corepressor (13). Conversely, decreased levels of miR-133, miR-27a, miR-106b/93, and miR-155 have shown to enhance brown adipogenesis by preventing targeted degradation of transcription factors of Prdm16 (14), Pparγ (15), Pparα (16), and C/ebpβ (17), respectively (see Fig. 1E). However, it has not been investigated whether n-3 PUFA play a role in regulating the miRNA network for brown adipogenesis.
FIGURE 1.
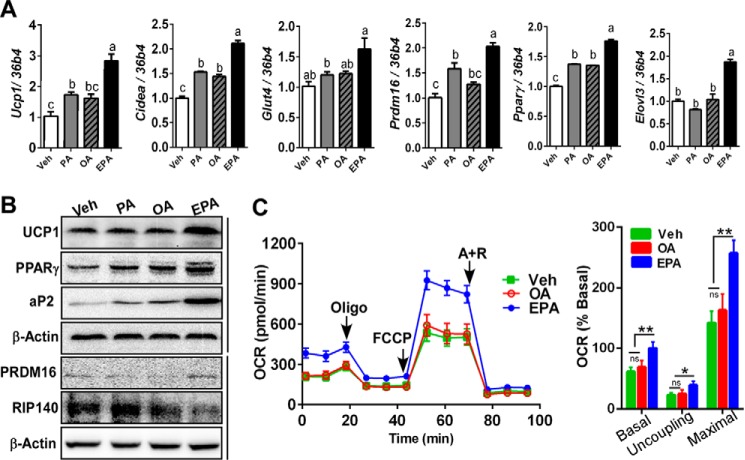
Dietary EPA promotes brown adipogenesis and OCR in primary murine brown progenitor cells. Brown progenitor cells were isolated from iBAT, and differentiation was induced for 7 days with either vehicle (Veh) (BSA) or 100 μm palmitate (PA), OA, or EPA. A, brown signature gene expression profile of Ucp1, Cidea, Glut4, Prdm16, Pparγ, and Elovl3 by qPCR. B, Western blotting analysis of Ucp1, Pparγ, aP2, Prdm16, and Rip140 and β-actin (loading control). C, OCR in brown adipocytes treated with vehicle (green), OA (red), and EPA (blue) determined by Seahorse extracellular analyzer. Arrow indicates the addition of respiratory inhibitors of oligomycin (Oligo), FCCP, and antimycin A plus rotenone (A+R). All data are presented as mean ± S.E. A, values not sharing a common letter differ significantly (p < 0.05) by one-way ANOVA (n = 5 per group). C, *, p < 0.05; **, p < 0.01 by one-way ANOVA with comparison to the vehicle control (n = 6–7 per treatment). n.s., not significant.
Supplementation of n-3 PUFA exerts multiple health benefits via activation of free fatty acid receptor 4 (Ffar4), previously known as G-protein-coupled receptor 120 (Gpr120) (18). Oh et al. (19) have demonstrated that Ffar4 senses n-3 PUFA triggering a broad spectrum of anti-inflammatory effects in macrophages. Furthermore, Ffar4 agonist has been shown to induce potent insulin-sensitizing and anti-diabetic effects against high fat (HF)-mediated obesity (20, 21). In addition, Ffar4-deficient mice develop hepatic steatosis and insulin resistance, implicating the crucial role of Ffar4 in maintaining energy balance. In humans, inhibition of FFAR4 signaling activity due to the genetic mutation of the FFAR4 gene (p.R2700H) was highly correlated with obesity risk (22). Although it is known that n-3 PUFA or pharmacological FFAR agonist modulate inflammation and glucose sensitivity in white adipocytes, little is known regarding the effects of FFAR activation in brown adipogenesis and BAT thermogenesis.
The objective of this study was to determine the underlying mechanism by which n-3 PUFA promote BAT thermogenesis. In this study, we hypothesized that n-3 PUFA stimulate brown adipogenesis by modulating the miRNA network, thereby promoting brown transcriptional activation. We also hypothesized that FFAR signaling activity will orchestrate activation of BAT thermogenesis by n-3 PUFA. To test these hypotheses, we investigated the signaling interplay that EPA coordinates Ffar4 and miRNAs in primary mouse brown preadipocytes. By using miRNA mimics and inhibitors, silencing of Ffar4, as well as complementary analyses of diet-fed animals, we report that Ffar4 plays a key role in promoting brown adipogenesis by EPA in part through miR-30b and 378.
Results
EPA Promotes Brown Adipogenesis by Altering miRNA Network
To investigate the impact of different dietary FA on brown adipogenesis, murine primary brown adipogenic precursor cells from iBAT were treated with either vehicle (BSA) or 100 μm PA, OA, or EPA during brown adipogenesis. There was a significant increase of brown signature gene expression of Ucp-1, Cidea, Prdm16, PPARγ, Glut4, and Elovl3 in EPA-treated culture compared with PA, OA, or vehicle (BSA) (Fig. 1A). In parallel with the mRNA profile, the EPA treatment increased Ucp1, Pparγ, Prdm16, and aP2 protein expression (Fig. 1B). To determine the impacts of EPA treatment on mitochondrial energetics, oxygen consumption rates (OCR) were determined. EPA treatment significantly increased the basal, uncoupling, and maximal respiration compared with vehicle or OA treatment (Fig. 1C).
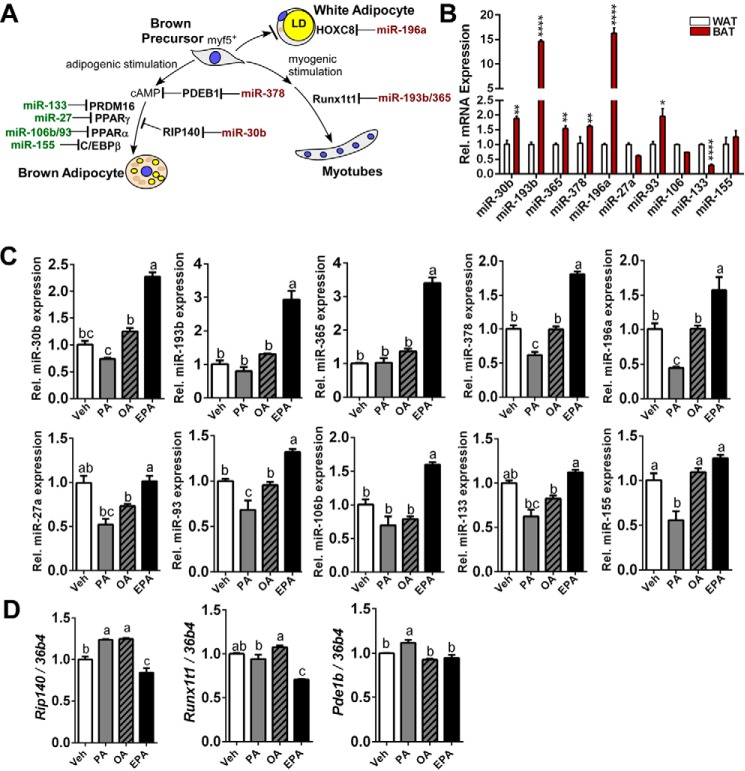
Next, we examined whether EPA-induced up-regulation of brown adipogenesis is associated with modification of miRNA expression. Based on literature reviews, we selected 10 miRNAs that alter the brown adipogenic program either by suppressing or enhancing brown adipogenesis (Fig. 2A). To conform the changes of miRNAs in BAT, miRNA levels in inguinal WAT and iBAT were compared; the expression levels of miR-30b, miR-193b, miR-365, miR-378, and miR-196a were significantly higher in BAT than WAT, whereas those of miR-27a and miR-133 were higher in WAT. There was no significant difference in miR-106 and miR-133 expression between groups (Fig. 2B). The brown differentiation with EPA, but not OA or PA, significantly increased miR-193/365 (3.4/2.9-fold), miR-378 (1.7-fold), miR-196a (1.5-fold), miR-30b (2.3-fold), and miR-106/93 (1.3/1.6-fold) compared with vehicle (BSA). In contrast, EPA did not significantly alter miRNA expression levels of miR-27a, miR-93, miR-133, and miR-155 (Fig. 2C). It is also interesting to note that OA had no significant impacts on the selected miRNAs for brown adipogenesis, whereas PA decreased the overall miRNA levels except for miR-193b/365 cluster and miR-106 (Fig. 2C). Consistently, mRNA expressions of Rip140 and Runx1t1, the proposed target of miR-30b and miR-193b/365 respectively, were significantly reduced compared with vehicle control. It is also notable that EPA treatment is associated with decreased expression of RIP140 (Fig. 1B). Despite a significant increase of miR-378, its proposed target of Pdeb1b (12) did not differ from the vehicle control (Fig. 2D). In summary, EPA promotes brown adipogenesis and uncoupled respiration, which was associated with modulation of miRNAs.
FIGURE 2.
Dietary EPA modulates miRNA network for brown adipogenesis. A, schematic diagram depicting the miRNA network that regulates brown adipogenesis. B, comparison of 10 representative miRNA expressions between subcutaneous WAT and interscapular BAT. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test. C, changes of miRNA expression of miR-30b, miR-193b, miR-365, miR-378, miR-196a, miR-27a, miR-93, miR-106b, miR-133, and miR-155. D, mRNA expression of Rip140, Runxt1, and Pde1b by qPCR. All data are presented as mean ± S.E. B, *, p < 0.05; **, p < 0.01; ***, p < 0.001 by Student's t test (from n = 5 individual animal per depot). C and D, values not sharing a common letter differ significantly (p < 0.05) by one-way ANOVA (n = 4 per group). Veh, vehicle.
FFAR4 Agonist Recapitulates Increase of Brown Adipogenesis and miRNA Network for Brown Fat Development
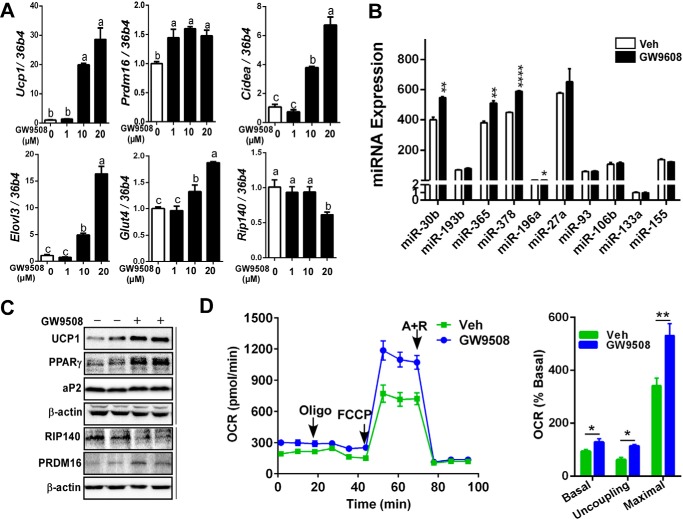
It is well established that FFAR4 (also known as GPR120) recognizes long or medium chain of FA, including docosahexaenoic acid and EPA, and exerts a plethora of health benefits (23–25). Based on these backgrounds, we asked whether FFAR4 signaling is also involved in EPA-mediated brown adipogenesis and miRNA modulation. To address this question, an increasing dose (0, 1, 10, and 20 μm) of GW9508, a FFAR4 agonist, was added to the differentiating brown progenitor cells. Prdm16 expression, a master transcription factor for brown fat, was significantly increased even with 1 μm GW9508 treatment. In addition, Ucp1, Cidea, Elov3, and Glut4 mRNA levels were increased in a dose-dependent manner, whereas Rip140 mRNA expression was reduced stepwise with GW9508 (Fig. 3A). In parallel with the changes of miRNA network by EPA (Fig. 2C), expression levels of miR-30b, miR-365, miR-378, and miR-196a were significantly elevated in GW9508-treated brown adipocytes (Fig. 3B). Similar to EPA treatment, GW9508 treatment increased Ucp1, Pparγ, aP2, and Prdm16 protein expression, whereas decreased RIP140 protein levels compared with vehicle control (Fig. 3C). Next, brown adipocytes grown with or without GW9508 were used to determine OCR. FFAR4 agonist GW9508 significantly increased basal, uncoupling, and maximal respiration rate similar to EPA treatment (Fig. 3D). Collectively, these data suggest that FFAR4 signaling participates in up-regulation of brown thermogenic program probably by modulating miRNAs.
FIGURE 3.
Ffar4 agonist recapitulates EPA-mediated brown thermogenesis. Brown progenitor cells were isolated from iBAT and induced differentiation for 7 days with an increasing dose (0, 1, 10, and 20 μm) of GW9508, a FFAR4 agonist. A, brown signature gene expression profile of Ucp1, Prdm16, Cidea, Elovl3, Glut4, and Rip140 by qPCR. B, changes of miRNA expression of miR-30b, miR-193b, miR-365, miR-378, miR-196a, miR-27a, miR-93, miR-106b, miR-133, and miR-155. C, representative Western blotting analysis of Ucp1, Pparγ, aP2, and β-actin. D, OCR in brown adipocytes grown with vehicle (Veh) (green) or 20 μm GW9508 (blue) determined by Seahorse extracellular analyzer. Arrow indicates the addition of respiratory inhibitors of oligomycin (Oligo), FCCP, and antimycin A plus rotenone (A + R). All data are presented as mean ± S.E. A, values not sharing a common letter differ significantly (p < 0.05) by one-way ANOVA (n = 5 per group). B (n = 5); D (n = 6), *, p < 0.05; **, p < 0.01; ****, p < 0.0001 by Student's t test.
miR-30b Regulates Brown Adipogenesis in FFAR4-dependent Mechanism
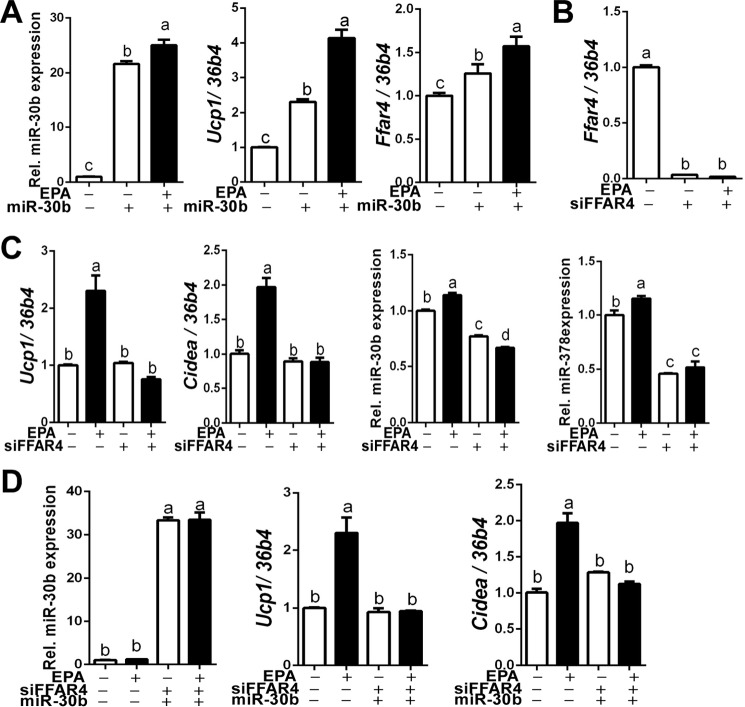
It has been recently identified that miR-30b participates in brown thermogenesis by targeting Rip140, a nuclear receptor corepressor (13). Among the 10 miRNAs that are relevant to brown adipogenesis, miR-30b seems to be a viable candidate responsible for EPA and FFAR4 agonist's effect. To test this notion, brown preadipocytes were transfected with either miR-30b mimics or non-targeting control and then the cells were differentiated to brown adipocytes in the presence or absence of EPA incubation. The transfection of miR-30b mimics increased intracellular miR-30b levels ∼20-fold. miR-30b expression level was significantly higher in cultures that received EPA + miR-30b mimics than those with miR-30b alone, confirming that endogenous miR-30b was produced in response to EPA treatment (Fig. 4A, left). Consistent with the literature (13), exogenous miR-30b increased Ucp1 gene expression. Intriguingly, EPA treatment with miR-30b exhibited an additive increase in Ucp1 mRNA expression (Fig. 4A, middle). Unexpectedly, Ffar4 expression was increased by exogenous miR-30b, which was further increased by EPA addition (Fig. 4A, right) suggesting a synergistic effect of exogenous and endogenous miR-30b for UCP1 up-regulation. It also suggest that miR-30b may regulate Ffar4 as well as Ucp1 expression.
FIGURE 4.
Ffar4 expression is required for EPA-mediated brown gene expression. A, miR-30b, Ucp1, and Ffar4 expression in EPA-treated brown adipocytes with or without exogenous miR-30b mimics. B, transfection efficiency of siFFAR4. C, Ffar4-depleted brown preadipocytes were differentiated with or without EPA and measured mRNA levels of Ucp1, Cidea, miR-30b, and miR-378 expression by qPCR. D, FFAR4-depleted brown preadipocytes were differentiated with or without EPA along with exogenous miR-30b mimic. All data are presented as mean ± S.E., n = 6/group. Values not sharing a common letter differ significantly (p < 0.05) by one-way ANOVA.
Subsequently, we attempted to determine the impact of FFAR4 signaling activity on miR-30b function to augment brown adipogenesis. For this purpose, we depleted FFAR4 by transfecting siRNA. siRNA transfection effectively decreased FFAR4 mRNA levels (∼98%) quantified by qPCR analysis, which was not reversed by EPA incubation (Fig. 4B). The silencing of Ffar4 completely dampened the EPA-mediated elevation of Ucp1 and Cidea mRNA expression as well as miR-30b and miR-378 expression (Fig. 4C), suggesting that Ffar4 signaling might be required for miRNA modulation. To further determine whether miR-30b plays an independent role, exogenous miR-30b mimics were added to the FFAR4-silenced preadipocytes. Despite effective transfection of miR-30b mimics, miR-30b failed to trigger up-regulation of Ucp1 and Cidea in the absence of FFAR4 (Fig. 4D). Collectively, these results suggest the following: 1) Ffar4 and miR-30b have mutual interaction, and 2) Ffar4 signaling is required for miR-30b production as well as its function to up-regulate brown-specific marker gene expression.
Inhibition of miR-30b Attenuates Brown Adipogenesis
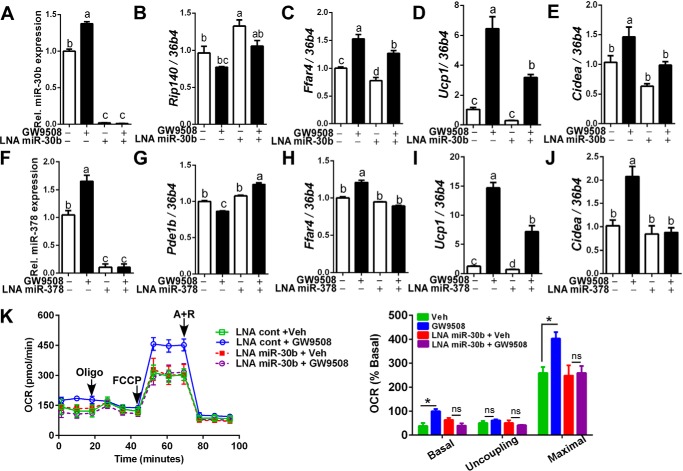
To further determine the degree in which miR-30b contributes to the Ffar4-mediated brown adipogenesis, miR-30b was sequestered by transfecting with antisense-LNA (locked nucleic acid) miR-30b inhibitor. Transfection of LNA miR-30b inhibitor completely blunted miR-30b production regardless of GW9508 incubation (Fig. 5A). Accordingly, the decrease of Rip140 gene expression by GW9508 was reversed by LNA miR-30b inhibitor (Fig. 5B). Consistent with miR-30b up-regulation of Ffar4 (Fig. 4A), sequestration of miR-30b by LNA miR30b inhibitor resulted in a significant decrease of Ffar4 gene expression in both basal and GW9508-treated cultures (Fig. 5C). It is important to note that inhibition of miR-30b attenuated GW9508-induced Ucp1 expression by ∼50% (Fig. 5D) and completely abolished the increase of Cidea mRNA expression by GW9508 (Fig. 5E). Similarly, miR-378 was sequestered by antisense-LNA miR-378 inhibitor. The transfection with LNA miR-378 inhibitor effectively blocked the miR-378 expression (Fig. 5F). LNA miR-378 transfection resulted in an increase of Pde1b in the presence of GW9508 (Fig. 5G, 2nd and 4th lanes) and abolished GW9508-mediated elevation of Ffar4 (Fig. 5H), Ucp1 (Fig. 5I), and Cidea (Fig. 5J). These results suggest that miR-378 and miR-30b may play a complementary role in up-regulating brown adipogenesis. It is noteworthy that the inhibition of miR-30b production by antisense-LNA miR-30b significantly dampened the GW9508-mediated elevation of OCR similar to the vehicle control (Fig. 5K). In summary, these data suggest the following: 1) miR-30b and miR-378 are not only downstream targets of Ffar4 signaling, but they also reversely affect Ffar4 expression, and 2) blockage of miR-30b or miR-378 significantly attenuates Ffar4-induced up-regulation of the brown adipogenic program.
FIGURE 5.
Inhibition of miR-30b or miR-378 attenuates Ffar4 agonist-mediated brown-specific gene activation. Brown preadipocytes were transfected with LNA miR-30b inhibitor (LNA miR-30b) (A–E) or LNA miR-378 (LAN miR-378) (F–J) before stimulation of brown adipogenesis with or without GW9508. A, miR-30b after transfection with LNA miR-30b. B–E, mRNA expression of Rip140 (B), Ffar4 (C), UCP1 (D), and Cidea (E). F, miR-378 after transfection with LNA miR-378. G–J, mRNA expression of Pdeb1(G), Ffar4 (H), UCP1 (I), and Cidea (J). K, OCR by extracellular analyzer in brown adipocytes pretreated with either non-targeting LNA (square) in combination with vehicle (Veh) (green) or GW9508 (blue), or LNA miR-30b (circle) in combination of vehicle (red) or GW9508 (violet). Arrow indicates the addition of respiratory inhibitors of oligomycin (Oligo), FCCP, and antimycin A plus rotenone (A + R). All data are presented as mean ± S.E., n = 6/group. Values not sharing a common letter differ significantly (p < 0.05) by one-way ANOVA. *, p < 0.05; ns, not significant by Student's t test .
Fish Oil Supplementation Increases Brown Thermogenesis and Modifies miRNA Expression in Brown Fat
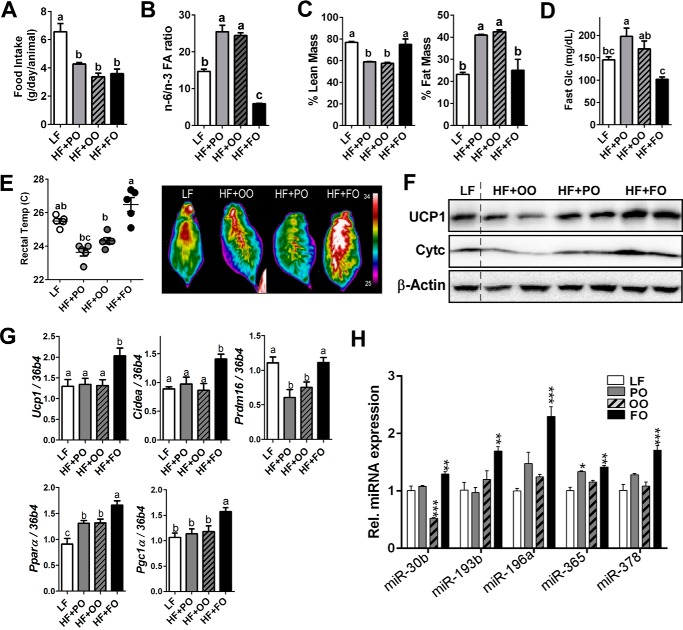
To confirm our results in vivo, C57BL/6 mice were fed with either a LF diet (10% of total calorie from fat) or an iso-caloric HF diet (50% of total calorie from fat) enriched with palm oil (HF + PO), olive oil (HF + OO), or fish oil (HF + FO) for 12 weeks. There was no significant difference in food intake among the HF-fed diet groups (Fig. 6A). The n-6/n-3 FA ratio was significantly higher in HF + OO- and HF + PA-fed mice but lower in HF + FO-fed mice than the LF control, implying that n-3 PUFA are accumulated in adipose tissue (Fig. 6B). In accordance with other studies of n-3 PUFA supplementation (26), the FO-fed mice were resistant to HF-mediated increase of fat mass and decrease of lean mass by DEXA measurement (Fig. 6C). The HF + FO-fed mice exhibited substantially lower fasting glucose levels than HF + PO- or HF + OO-fed mice (Fig. 6D). Importantly, when the mice were exposed to cold (8 °C) acutely (for 45 min), HF + FO-fed mice were able to maintain a core body temperature higher than HF + PO or HF + OO. Furthermore, heat release was significantly higher in HF + FO-fed than HF + PO- or HF + OO-fed mice (Fig. 6E). The increases of UCP1 protein and cytochrome c (Fig. 6F) and brown signature gene profiles of Ucp-1, Cidea, Prdm16, PPARγ, and PPARα (Fig. 6G) were confirmed in iBAT from HF + FO-fed mice compared with HF + PO- or HF + OO-fed mice. These results are consistent with recent publications showing thermogenic activation with FO supplementation (6, 7). Subsequently, we analyzed the five candidate miRNAs that were up-regulated with EPA treatment in vitro; all five miRNAs, miR-30b, miR-193b, miR-365, miR-196a, and miR-378, were significantly up-regulated in iBAT from mice fed a HF + FO diet compared with HF + OO- or HF + PO diet-fed mice (Fig. 6H).
FIGURE 6.
Fish oil supplementation promotes brown thermogenesis. C57BL/6 mice were fed with LF or an iso-caloric HF diet containing 15% of palm oil (HF + PO), olive oil (HF + OO), and fish oil (HF + FO) for 12 weeks (n = 8). A, food intake per day. B, n-6/n-3 ratio after 12 weeks of diet. C, lean and fat mass by DEXA. D, fasting glucose levels. E, core body temperature measured by the rectal thermometer after acute cold exposure (8 °C) for 45 min (left) and representative thermography captured by IR camera (right). F, Western blotting analysis of Ucp1, cytochrome c (Cyt c), and β-actin. G, gene expression of Ucp1, Cidea, Prdm16, Pparγ, and Pgc1α by qPCR of iBAT (n = 6/group), H, miRNA expression of miR-30b, miR-193b, miR-196a, miR-365, and miR-378 (n = 6/group). All data are presented as mean ± S.E. A and G, values not sharing a common letter differ significantly (p < 0.05) by one-way ANOVA. H, *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA with comparison to LF control (n = 6–7 per treatment).
Given that miR-30b and 378 are downstream targets of cAMP (12, 13), we questioned whether cAMP signaling is associated with FFAR4 activation in brown adipocytes. The cold exposure (5–8 °C) for 2 weeks (samples from our previous studies (27)) significantly increased Ucp1 and Ffar4 expression along with the increases of miR-30b and miR-378 in iBAT (Fig. 7A). Moreover, FO supplementation showed an additional increase of Ffar4 as well as miR-30b and 378 expression (Fig. 7B). Next, we raised a question whether FFAR4 activation per se triggers elevation of intracellular cAMP in brown adipocytes linking the modulation of miR-30b and 378. There was a significant increase of cAMP levels in HF + FO-fed iBAT compared with iBAT from mice fed a HF + OO. In fact, there was small but a significant decrease of cAMP levels in HF + PO-fed mice compared with HF + OO control (Fig. 7C). More surprisingly, FFAR4 activation by GW9508 treatment significantly increased cAMP levels in cultures of brown adipocytes without exogenous supply of catecholamine or Adrb3 activation (Fig. 7D). These results suggest that FFAR4 agonism may promote autonomous brown differentiation program by elevating cAMP levels and by subsequent activation of functional miRNA network for brown-specific gene transcription.
FIGURE 7.
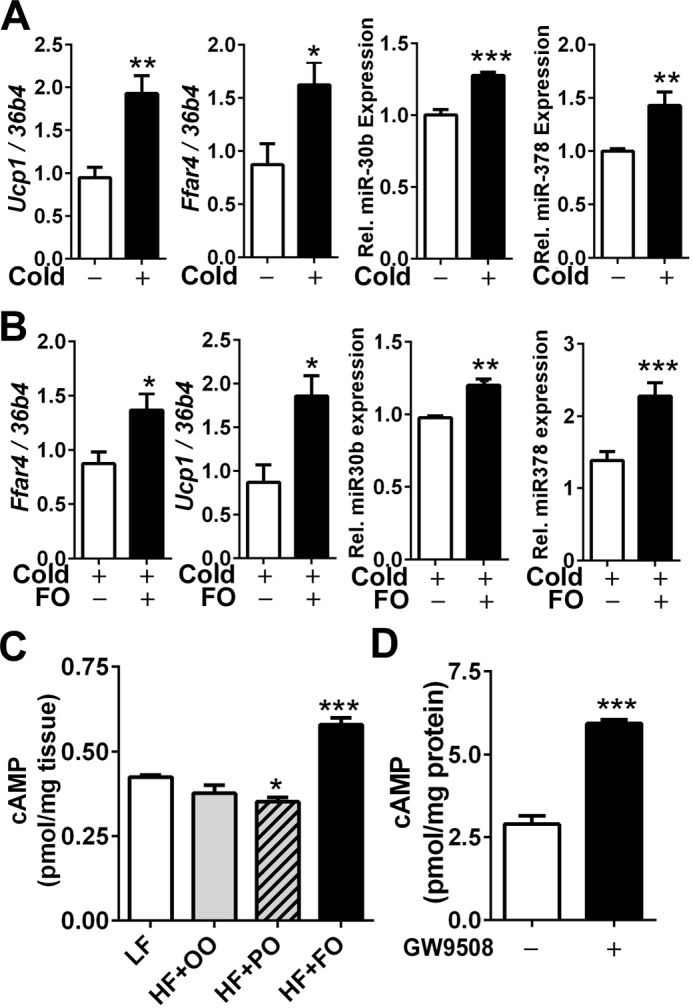
FFAR4 agonism elevates intracellular cAMP levels. A and B, qPCR analysis of gene expression levels of Ucp1 and Ffar4 and miRNA 30b and 378 of iBAT before and after cold exposure (8 °C) for 2 weeks (A) or cold exposure with or without FO supplementation (B). C, cAMP levels in iBAT after supplementation with LF or an iso-caloric HF diet containing 15% of palm oil (HF + PO), olive oil (HF + OO), and fish oil (HF + FO), for 12 weeks (n = 6). D, cAMP levels in brown adipocytes grown in the presence (+) and absence (−) of 20 μm GW9508 (n = 4). C, *, p < 0.05; ***, p < 0.001 by one-way ANOVA with comparison to LF control (n = 6 per treatment). A, B, and D, *, p < 0.05; **, p < 0.01; ***, p < 0.0001 by Student's t test.
Discussion
Dietary FA are not only major nutrients but also key signaling molecules that regulate endocrine function, immunological responses, and energy metabolism (4). Emerging evidence suggests that dietary n-3 PUFA also trigger brown/beige fat activation (5, 7). It has been demonstrated that miRNAs play a crucial role in regulating brown adipogenic program (28–30). However, it has not been investigated whether n-3 PUFA participate in regulating miRNA for brown/beige fat activation. The objective of this work was to determine the impacts of n-3 PUFA on classical brown adipogenesis by examining the miRNA network. Here, we demonstrated that the increase of brown adipogenesis by EPA was associated with the increase of several miRNAs that are involved in functional cluster to stimulate transcriptional activation of brown adipogenesis (Figs. 1 and 2). The increase of these miRNAs was also confirmed in fish oil-fed mice (Fig. 6). We have also demonstrated that activation of Ffar4, n-3 PUFA receptor, is sufficient to simulate up-regulation of brown adipogenesis (Fig. 3), and these effects were abolished in the absence of Ffar4 expression (Fig. 4). By taking advantage of mimics and LNA inhibitors, we identified that elevation of miR-30b was dependent on Ffar4 signaling, and associated with EPA-mediated brown adipogenesis (Figs. 4 and 5). Our results also suggest that intracellular cAMP would be a potential link between Ffar4 activation and brown-specific miRNA modulation (Fig. 7). Based on these results, we propose a model in which Ffar4 activation by dietary n-3 PUFA provides an additional layer of transcriptional activation for brown adipogenesis in part through miR-30b and 378 (Fig. 8). To our knowledge, our work is the first study reporting that Ffar4 functions to mediate n-3 PUFA-promoted brown adipogenic program via modulation of miRNAs.
FIGURE 8.
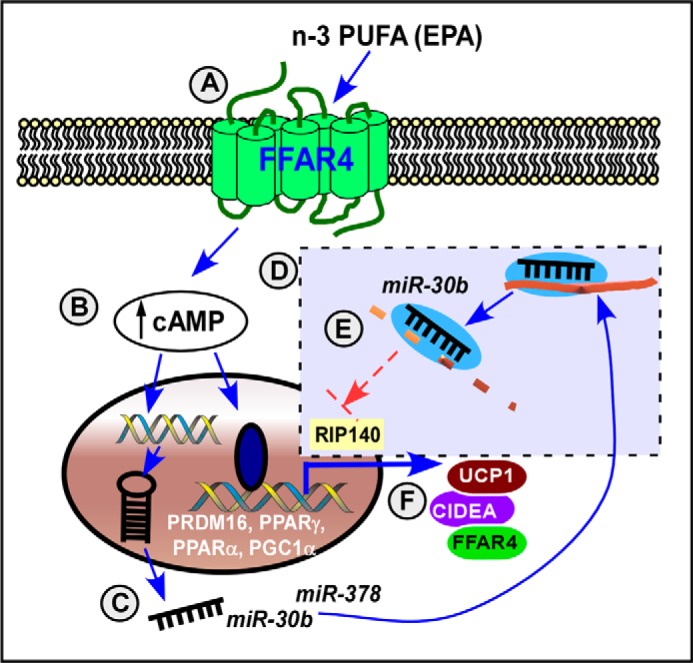
Working model that n-3 PUFA promotes brown adipogenesis through FFAR4. n-3 PUFA such as EPA are sensed by membrane receptor FFAR4 (A), which triggers a rise of intracellular cAMP levels (B) and biogenesis of miRNAs, including miR-30b and miR-378 (C). Mature miRNA targets to their complementary targets forming the miRNA (miR)-RISC complex (dotted box) (D). For example, miR-30b attenuates its target gene expression of Rip140, a nuclear receptor corepressor, thereby promoting transcriptional activity of brown adipogenesis, including Prdm16, Pparγ, Pparα, and Pgc1α (E). As a consequence, n-3 PUFA up-regulate brown-specific transcriptional program, including UCP1 and CIDEA as well as FFAR4 (F). In summary, Ffar4 plays a significant role in mediating dietary n-3 PUFA-induced brown adipogenesis partly through miR-30b; and miR-378-dependent mechanisms.
miRNAs are single-stranded noncoding RNA composed of 19–22 nucleotides. An individual miRNA can modulate hundreds of genes by regulating mRNA availability or translational activity depending on the degree of complementarity with its targets (31–33). No more than two dozen miRNAs have been identified so far to up-regulate or down-regulate brown adipogenesis (34). As there was a tendency to increase the overall miRNA formation by EPA (Fig. 2C), we first speculated whether EPA facilitated miRNA biogenesis or RNA-induced silencing complex (RISC) formation. However, there was no difference in Drosha, Dicer, and Ago2 expression (data not shown), and thus we excluded the possibility that EPA alters early steps for miRNA biogenesis or RISC formation. There is increasing literature support that the benefits of n-3 PUFA consumption are mediated by miRNA regulation; Recchiuti et al. (35) demonstrated that the anti-inflammatory role of resolvin, n-3 PUFA-derived eicosanoids, is associated with specific miRNA target genes involved in resolution of acute inflammation (e.g. miR-21, miR-146b, miR-208a, miR-203, miR-142, miR-302d, and miR-219); Antal et al. (36) showed that miR-146 and miR-181a are induced by docosahexaenoic acid in glioma cells increasing sensitivity of radiation therapy. More recently, Ortega et al. (37) suggested that dietary PUFA consumption is linked with changes in plasma circulatory miRNA profiles. Given the broad spectrum of health benefits by n-3 PUFA consumption from cognitive function, cancer, inflammation, and dyslipidemia to insulin sensitivity, miRNA regulation could provide a plausible explanation on how n-3 PUFA alter hundreds of genes. In our study, we have demonstrated that EPA or fish oil treatments were associated with an increase of positive regulatory miRNA for browning as a functional cluster (miR-196a, miR-193b/365, miR-378, and miR-30b), but no significant changes were found in a group of miRNAs negatively regulating brown adipogenesis (miR-133, miR-27a, miR-106b/93, and miR-155) (Figs. 2C and 6H). It is also important that FFAR4 agonism by GW9508 has mirrored the miRNA expression patterns in brown adipocytes (Fig. 3B), strongly suggesting that Ffar4 signaling may directly regulate miRNA synthesis in brown adipocytes. As far as we are concerned, it is the first report to demonstrate that n-3 PUFA-induced BAT activation is linked with up-regulation of miRNA in an Ffar4-dependent mechanism.
Among the set of miRNAs elevated by EPA treatment, we have focused on miR-30b and miR378. As we used the primary brown precursor cells (brown preadipocytes) isolated from iBAT, we ruled out the possibility that miRNAs regulating lineage switch or brown commitment could play a major role, i.e. miR-193b/365 (myoblast → brown precursor) (10) and miR-196a (white → brown precursor) (11). Between the two miRNAs tested, miR-30b and miR-378, miR-30b was our first candidate for further investigation. This selection was based upon the results that Rip140, the proposed target of miR-30b in brown adipocytes, was decreased in response to an increase of miR-30b in both EPA- and GW9508-treated brown adipocytes (Figs. 2C and 3B), whereas the Pde1b expression level, miR-378 proposed target, was inconsistent with increased miR-387 expression. In addition, the decreased expression of Rip140 could induce a broad spectrum of changes in brown-specific genes (38) by liberating restrictions on the nuclear receptors of Pparγ, Pparα, C/ebpβ, and Prdm16 (39, 40). Moreover, Rip140 plays an important role in anti-inflammation (41), which is well aligned with the immunomodulatory role of n-3 PUFA. However, we cannot completely exclude the possibility that miR-378 may contribute to EPA-mediated BAT activation. Based on study by Kim et al. (7), fish oil supplementation is linked with sympathetic activation and Adrb3 signaling in brown and inguinal white adipocytes. This implies the activation of cAMP production and its potential role of miR-378, specifically in brown fat (12). To support this notion, sequestration of miR-378 by the LNA inhibitor triggered a partial reduction of Ucp1 expression (Fig. 5). It is worth noting that the silencing of Ffar4 abolished the EPA-induced increase in miRNA expression of both miR-30b and miR-378 (Fig. 4C). Intriguingly, the exogenous miR-30b mimic failed to rescue UCP1 up-regulation in Ffar-silenced cells (Fig. 4D). These data implicate that Ffar4 signaling event must precede the production of miRNA functional cluster for up-regulation of brown thermogenesis. Based on our current results, it is also speculated that once miRNAs are formed via Ffar4 signaling, individual miRNAs may exert separate impacts on their target genes, leading to augmented brown adipogenesis as the sum of collaborative activities of each miRNA. In addition, considering that FFAR4 agonism induces cAMP responses (Fig. 7, C and D) and miRNA inhibitors only partly inhibit the Ffar4-mediated Ucp1 expression, we do not exclude the possibility that FFAR4 agonism also promotes UCP1 transcription by microRNA-independent but cAMP-dependent mechanisms.
In terms of signaling mediator, we propose that cAMP seems to be the second messenger coordinating Ffar4 to transcriptional regulation of brown fat (Fig. 8). It is based on our following observations: 1) cold treatment triggers Ffar4 in iBAT (Fig. 7A), which was consistent with the studies by Rosell et al. (42); 2) FO supplementation showed an additional up-regulation Ffar4; and 3) Ffar4 agonism is associated with a rise of cAMP levels in brown adipocyte in vitro as well as in iBAT in vivo (Fig. 7, C and D). These data strongly suggest the positive association between Ffar4 activation and cAMP levels. Considering that Ffar4 (Gpr120) is a family of G protein-coupled receptors with Gαq subunits, the primary signaling mediator is intracellular calcium ([Ca2+]i) (43). Although it could be controversial, Moran et al. (44) demonstrated that augmented insulin secretion by FFAR4 agonism in pancreatic β-cell is accompanied by significant elevation of cAMP as well as [Ca2+]i. Our results showed that cAMP levels were significantly higher in fish oil-fed iBAT (Fig. 7). It could be due to an activation of sympathetic nervous system and Adrb3 pathway by n-3 PUFA, as proposed by Kim et al. (7). Therefore, it was surprising when we were able to determine the rise of cAMP levels in EPA-treated brown adipocytes, an isolated brown adipocyte system. Although we could not completely exclude the possibility that Ffar4 signaling may be involved in catecholamine production within brown adipocytes (45), we speculate that either Ffar4 signaling contributes to a delay of cAMP degradation or a rise of cAMP via miRNA-mediated mechanism. We are currently investigating whether Ffar4 agonism is linked with PKA activation or [Ca2+]i signaling for cAMP regulation.
It is well accepted that the n-6/n-3 ratio is a biomarker used for prediction of chronic disease risk, including cardiovascular diseases and diabetes. Our animal experiment showed that fish oil supplementation decreased the n-6/n-3 ratio by ∼5-fold compared with palm oil or olive oil supplementation. However, our in vitro experiment revealed that Ffar4 signaling activity is necessary and sufficient to simulate EPA-mediated brown thermogenesis (Figs. 3 and 4). It is uncertain whether Ffar4 signaling is more crucial than n-6/n-3 ratio itself. Interestingly, Li et al. showed that the anti-inflammatory function of n-3 PUFA is dependent on Ffar4 signaling. They used transgenic fat-1 mice, an engineered mice to endogenously synthesize n-3 PUFA from n-6 PUFA by overexpressing n-3 desaturase fat-1, thereby the n-6/n-3 ratio remains low (46). When FFAR4 is deleted in Fat-1 mice (FFAR4−/−/fat-1), the n-3 PUFA-mediated anti-inflammation effects in the vasculature disappeared despite the similar levels in the n-6/n-3 ratio (47). These results suggest that Ffar4 signaling activity, rather than cellular loading of n-3 PUFA, may mediate health benefits. Consistent with this concept, treatment of selective Ffar4 agonist effectively reproduced n-3 PUFA-mediated anti-inflammatory and insulin-sensitizing effects without n-3 PUFA consumption. Until now, little information is available regarding the browning of Ffar4 knock-out animals, although it has been demonstrated that Ffar4 deletion precipitates obesity and insulin resistance (21). We are currently investigating whether Fat-1 mice alter the miRNA network that we observed in Fig. 2 without any supplementation or FFAR4 agonist treatment.
In this study, we mainly focused on investigating the effects of EPA on brown adipogenesis using brown progenitor cells, but we did not specifically address the role of EPA on the beigeing of white adipocytes. It has been suggested that EPA (or fish oil) could trigger brown-like phenotypes in adipocyte progenitor cells isolated from rodent subcutaneous fat in vitro (5) and primary human adipose-derived stem cells (48). These results are inconsistent with our preliminary study that GW9508 treatment in human adipose-derived stem cells increased the adipogenesis but decreased UCP1 expression without any significant changes in oxygen consumption rate (data not shown). In addition, GW9508 treatment decreased the bone morphogenetic protein 7 (BMP7)-mediated UCP1 expression in primary human white adipocytes (data not shown). Further studies are warranted to reconcile this controversy by measuring metabolic activities in the presence or absence of ADRB3 stimulation rather than UCP1 gene expression. There is a study showing that depletion of Ffar4 impairs white adipocyte differentiation (49). Therefore, it is plausible that white adipose tissue browning by n-3 PUFA might be a separate mechanism from augmented brown adipogenesis of classical brown fat. As white adipose tissue browning (or beige fat formation) is more critically regulated by the immunological status of WAT (50), we envision that the decrease of lipotoxicity and modulation of immune responses in white adipose tissue (e.g. macrophage M2 polarization) by n-3 PUFA facilitate white adipocyte browning, whereas FFAR4 signaling accompanying the miRNA network activates classical brown fat development upon n-3 PUFA supplementation.
In summary, here we report that the activation of FFAR4, a functional receptor for sensing n-3 PUFA, potentiates transcriptional activation of brown adipogenesis through modulation of miRNAs, including miR-30b and miR-378. We also propose that elevation of cAMP is a likely downstream pathway coordinating Ffar4 signaling to the transcriptional activation of brown adipogenic program. Although further investigations are necessary to validate these results in human classical brown adipocytes or pluripotent stem cells, our study provides novel insights into the development of dietary intervention strategies to increase BAT development. In contrast to the temporal and reversible Ucp1 activation of beige fat, classical brown fat constitutively activates thermogenesis through Ucp1. Given this clinical relevance of BAT to energy metabolism, this study will provide evidence for therapeutic application to boost BAT via activation of Ffar4/miRNA signaling axis.
Experimental Procedures
Chemicals
Fetal bovine serum (FBS) was purchased from Gibco. Rosiglitazone (BRL) was purchased from Cayman Chemical. Insulin, 3-isobutyl-1-methylxanthine, and trifluoride (BF3)/methanol reagent were purchased from Sigma. Palmitic acid, oleic acid, EPA, and fatty acid standard were purchased from NuChek Prep (Elysian, MN). GW9508, an FFAR4 agonist, was purchased from Tocris Bioscience (Bristol, UK).
Animals
All protocols and procedures were approved by the Institutional Animal Care and Use Committee at the University of Nebraska, Lincoln. C57BL/6 male mice were purchased at 6 weeks of age from The Jackson Laboratory. After 2 weeks of acclimation, the mice were randomly assigned into four diet groups (n = 8 per group) receiving either a low fat (LF; 10% calories from fat) diet or an iso-caloric high fat (HF; 50% calories from fat) diet that was enriched with different FA for 12 weeks. The AIN-93G rodent formulation was modified for fat composition containing 15% of fat (w/w) from either palm oil (HF + PO), olive oil (HF + OO), or fish oil (HF + FO). All diets contained the same amount of soy bean oil (5% w/w) as a source for essential FA. Gas chromatography (GC) was used to analyze the fatty acid profile as described under “n-6/n-3 Ratio.” At the time of necropsy, blood, subcutaneous (inguinal) WAT, and iBAT were collected, snap-frozen in liquid nitrogen, and kept at −80 °C until analysis.
Brown Progenitor Cell Isolation and Differentiation
The 6–7-week-old C57BL/6 mice were purchased from The Jackson Laboratory. iBAT were used for brown progenitor cell isolation. Excised iBAT were immediately washed in PBS, minced into small pieces (1–2 mm), and digested with collagenase (1 mg/ml) in a Krebs-Ringer bicarbonate buffer in a shaking water bath (37 °C). After 1 h of digestion, stromal vascular cells were collected by centrifugation, and the resulting pellet was filtered through a 100-μm cell strainer. After centrifugation, the obtained stromal vascular fraction was seeded into a 12-well plate and cultured in a complete medium (DMEM plus 10% FBS) until confluence. Adipocyte differentiation was induced by adding the differentiation mixture, which contained 20 nm insulin, 1 nm triiodothyronine, 0.5 mm isobutylmethylxanthine, 5 μm dexamethasone, 0.125 mm indomethacin, and 0.5 μm rosiglitazone brl 49653 for 2 days. Then, a fresh medium containing 20 nm insulin and 1 nm triiodothyronine were added to cells along with either 100 μm BSA-complexed FA, i.e. PA, OA, EPA, or 20 μm GW9508 for additional 5 days.
Rectal Temperature and Thermogenesis
To measure the core body temperature, a digital thermometer (TC Thermocouple Meter) was used in combination with a copper thermocouple rectal probe (RET-3) (Kent Scientific Corp). The probe was properly positioned into the anal ducts of the adult mice to record temperature. An average of three readings obtained from the same mouse was calculated. For the detection of thermal release, an infrared (IR) camera (A655sc, FLIR Systems) was used to acquire images of the body surface temperature of each mouse. The images were displayed using the rainbow high contrast color palette in the FLIR Research IR program using a temperature linear display between 25 and 34 °C.
DEXA
Body composition for the mice was measured by using a dual energy x-ray absorptiometry (DEXA). The mice were anesthetized with CO2 and scanned using a Lunar PIXImus2 densitometer (software version 2.1; Lunar Corp). All of the mice were weighed before the DEXA measurements. Immediately after the DEXA analysis, blood was collected, and the mice were killed by cervical dislocation.
n-6/n-3 Ratio
The fatty acid profiles of the fat samples were determined by GC (51). Briefly, ∼0.1g of tissue was homogenized, and total lipids were extracted. They were then subjected to fatty acid methylation by 14% BF3/methanol reagent at 100 °C for 1 h. An Agilent Technologies HP-88 column (100 m × 0.25 mm × 0.2 μm film thickness) was used. The fatty acid peaks were identified by comparing their relative retention times with the commercial mixed FA standard (NuChek PreP), and the area percentages for all resolved peaks were analyzed using the GC ChemStation software.
OCR by Seahorse
To determine the mitochondrial respiration activities, the O2 concentration in the cells was measured using XF24 extracellular flux analyzer (Seahorse). Briefly, mouse brown primary cells were seeded in gelatin-coated seahorse microplates (24-well) until confluence was reached followed by brown differentiation as we described above. The mitochondrial basal respiration was assessed in untreated cells. The cells were then treated with oligomycin (1 μm) to measure the ATP turnover. The maximum respiratory capacity was assessed by addition of carbonyl cyanide 4-trifluoromethoxy phenylhydrazone (FCCP, 0.3 μm), a chemical uncoupler of electron transport and oxidative phosphorylation. The mitochondrial respiration was blocked by antimycin A (1 μm) plus rotenone (1 μm). The OCR was calculated by plotting the O2 tension of the medium in the microenvironment above the cells as a function of time (pmol of O2/min).
qPCR of mRNA and microRNA
Total RNA was extracted using TRIzol® reagent (Invitrogen) from homogenized tissues or cells. RNA was purified using DNase (5 PRIME). One μg of RNA was converted into cDNA (iScript, Bio-Rad). Relative gene expression was determined based on the 2−ΔΔCT method with normalization of the raw data to 36b4 (primer sequences are available upon request). For miRNA analysis, miRNA was converted to cDNA using the miScript reverse transcription kit (Qiagen) according to the manufacturer's instructions. miRNA expression was measured by using the commercial miScript Universal Primer, with the miScript primer assay kit (Qiagen). Primers of miR-16, -27a, -30b, -93, -106b, -133a, -155, -193b, -196, -365, and -378 and RNU6-2 were purchased from Qiagen. Values were normalized to RNU6-2 (U6 small nuclear) and also confirmed with miR-16.
Transfection of miRNA Mimic, Inhibitors, and Silencing FFAR4
For transfection of miR-30b mimic, brown precursor cells were first seeded in a 12-well plate at 60–70% confluence. The next day, transfection was performed for 2 days with 10 nm miRCURY locked nucleic acid (LNATM) miR-30b mimic (miR-30b mimic, Exiqon471216-001) or non-targeting negative control (Exiqon479902-001). To deplete FFAR4 expression, 100 nm ON TARGET plus SMARTpool siRNAs (Mouse FFAR4) or Non-Targeting siRNA Pool (Dharmacon) were transfected into cells using DharmaFECT-1 (GE Dharmacon) according to the manufacturer's instructions. After 48 h of post-transfection, adipocyte differentiation was initiated by adding the differentiation mixture as we described previously. miRCURY LNA Power inhibitors of miR-30b (4102487-101, Exiqon), miR-378 (4100837-101, Exiqon), or respective negative control (199006-001, Exiqon) were transfected according to the manufacturer's instructions. The final oligonucleotide concentrations were 10 nm for LNA miR-30b inhibitor and 100 nm for LNA miR-378a inhibitor. After 48 h of post-transfection, the cells were induced to differentiation.
Western Blotting Analysis
Total cell extracts were prepared as described previously (27). Proteins were fractionated using 4–15% precast polyacrylamide gel (Bio-Rad) and transferred to PVDF membranes with a semi-dry transfer unit (Hoefer TE77X). Antibody targeting UCP1 (14670), PPARγ (2444), and β-actin (4967) was purchased from Cell Signaling Technology. aP2 (sc-271529) and PRDM16 (sc-55697) were purchased from Santa Cruz Biotechnology. Blots were visualized with FluorChemTME imaging system (Protein Simple).
cAMP Analysis
To measure cAMP levels, immunoassay was conducted by using mouse/rat cAMP parameter assay kit (R&D System) according to the manufacturer's protocol. cAMP levels were normalized with either total protein or milligram of tissue used.
Statistical Analysis
All data are presented as means of ± S.E. Independent samples were statistically analyzed using one-way ANOVA followed by Tukey's multiple comparison tests for comparisons between two groups or comparison to the control. All statistical analyses were conducted by Graph Pad Prism 6 (Version 6.02).
Author Contributions
S. C. and J. K. conceived and coordinated the study and wrote the paper. J. K. and M. O. performed experiments and analyzed the data. S. K. N. assisted miRNA assays and critically reviewed the manuscript. T. C. and A. E. conducted GC analysis to determine FA profiles. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Sunwoo Ha for reviewing the manuscript.
This work was supported by National Institutes of Health Grant 1P20GM104320 (Project 5) (to S. C.) and in part by Nebraska Tobacco Settlement Biomedical Research Enhancement Funds. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- BAT
- brown adipose tissue
- WAT
- white adipose tissue
- PA
- palmitic acid
- OA
- oleate
- EPA
- eicosapentaenoic acid
- OCR
- oxygen consumption rate
- LNA
- locked nucleic acid
- PPAR
- peroxisome proliferator-activated receptor
- FA
- fatty acid
- ANOVA
- analysis of variance
- FCCP
- carbonyl cyanide 4-trifluoromethoxyphenylhydrazone
- qPCR
- quantitative PCR
- FFAR
- free fatty acid receptor
- HF
- high fat
- LF
- low fat
- PO
- palm oil
- OO
- olive oil
- FO
- fish oil
- BF3
- boron trifluoride
- [Ca2+]i
- intracellular calcium
- iBAT
- interscapular brown adipose tissue
- miRNA
- microRNA
- DEXA
- dexamethasone
- RISC
- RNA-induced silencing complex.
References
- 1. Giralt M., and Villarroya F. (2013) White, brown, beige/brite: different adipose cells for different functions? Endocrinology 154, 2992–3000 [DOI] [PubMed] [Google Scholar]
- 2. Harms M., and Seale P. (2013) Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 [DOI] [PubMed] [Google Scholar]
- 3. Seale P., and Lazar M. A. (2009) Brown fat in humans: turning up the heat on obesity. Diabetes 58, 1482–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakamura M. T., Yudell B. E., and Loor J. J. (2014) Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 53, 124–144 [DOI] [PubMed] [Google Scholar]
- 5. Zhao M., and Chen X. (2014) Eicosapentaenoic acid promotes thermogenic and fatty acid storage capacity in mouse subcutaneous adipocytes. Biochem. Biophys. Res. Commun. 450, 1446–1451 [DOI] [PubMed] [Google Scholar]
- 6. Bargut T. C., Silva-e-Silva A. C., Souza-Mello V., Mandarim-de-Lacerda C. A., and Aguila M. B. (2016) Mice fed fish oil diet and up-regulation of brown adipose tissue thermogenic markers. Eur. J. Nutr. 55, 159–169 [DOI] [PubMed] [Google Scholar]
- 7. Kim M., Goto T., Yu R., Uchida K., Tominaga M., Kano Y., Takahashi N., and Kawada T. (2015) Fish oil intake induces UCP1 upregulation in brown and white adipose tissue via the sympathetic nervous system. Sci. Rep. 5, 18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mori M. A., Thomou T., Boucher J., Lee K. Y., Lallukka S., Kim J. K., Torriani M., Yki-Järvinen H., Grinspoon S. K., Cypess A. M., and Kahn C. R. (2014) Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J. Clin. Invest. 124, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim H. J., Cho H., Alexander R., Patterson H. C., Gu M., Lo K. A., Xu D., Goh V. J., Nguyen L. N., Chai X., Huang C. X., Kovalik J. P., Ghosh S., Trajkovski M., Silver D. L., et al. (2014) MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes 63, 4045–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun L., Xie H., Mori M. A., Alexander R., Yuan B., Hattangadi S. M., Liu Q., Kahn C. R., and Lodish H. F. (2011) Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 13, 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mori M., Nakagami H., Rodriguez-Araujo G., Nimura K., and Kaneda Y. (2012) Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLos Biol. 10, e1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan D., Mao C., Quattrochi B., Friedline R. H., Zhu L. J., Jung D. Y., Kim J. K., Lewis B., and Wang Y. X. (2014) MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 5, 4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu F., Wang M., Xiao T., Yin B., He L., Meng W., Dong M., and Liu F. (2015) miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes 64, 2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trajkovski M., Ahmed K., Esau C. C., and Stoffel M. (2012) MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 14, 1330–1335 [DOI] [PubMed] [Google Scholar]
- 15. Sun L., and Trajkovski M. (2014) MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism 63, 272–282 [DOI] [PubMed] [Google Scholar]
- 16. Wu Y., Zuo J., Zhang Y., Xie Y., Hu F., Chen L., Liu B., and Liu F. (2013) Identification of miR-106b-93 as a negative regulator of brown adipocyte differentiation. Biochem. Biophys. Res. Commun. 438, 575–580 [DOI] [PubMed] [Google Scholar]
- 17. Chen Y., Siegel F., Kipschull S., Haas B., Fröhlich H., Meister G., and Pfeifer A. (2013) miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 4, 1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oh D. Y., and Lagakos W. S. (2011) The role of G-protein-coupled receptors in mediating the effect of fatty acids on inflammation and insulin sensitivity. Curr. Opin. Clin. Nutr. Metab. Care 14, 322–327 [DOI] [PubMed] [Google Scholar]
- 19. Oh D. Y., Talukdar S., Bae E. J., Imamura T., Morinaga H., Fan W., Li P., Lu W. J., Watkins S. M., and Olefsky J. M. (2010) GPR120 is an ω-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L., Wang Y., and Xu M. (2014) Acid hydrolysis of crude tannins from infructescence of Platycarya strobilacea Sieb. et Zucc to produce ellagic acid. Nat. Prod. Res. 28, 1637–1640 [DOI] [PubMed] [Google Scholar]
- 21. Oh da Y., Walenta E., Akiyama T. E., Lagakos W. S., Lackey D., Pessentheiner A. R., Sasik R., Hah N., Chi T. J., Cox J. M., Powels M. A., Di Salvo J., Sinz C., Watkins S. M., Armando A. M., et al. (2014) A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nat. Med. 20, 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ichimura A., Hirasawa A., Poulain-Godefroy O., Bonnefond A., Hara T., Yengo L., Kimura I., Leloire A., Liu N., Iida K., Choquet H., Besnard P., Lecoeur C., Vivequin S., Ayukawa K., et al. (2012) Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature 483, 350–354 [DOI] [PubMed] [Google Scholar]
- 23. Talukdar S., Olefsky J. M., and Osborn O. (2011) Targeting GPR120 and other fatty acid-sensing GPCRs ameliorates insulin resistance and inflammatory diseases. Trends Pharmacol. Sci. 32, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ulven T., and Christiansen E. (2015) Dietary fatty acids and their potential for controlling metabolic diseases through activation of FFA4/GPR120. Annu. Rev. Nutr. 35, 239–263 [DOI] [PubMed] [Google Scholar]
- 25. Moniri N. H. (2016) Free-fatty acid receptor-4 (GPR120): Cellular and molecular function and its role in metabolic disorders. Biochem. Pharmacol. 110, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martínez-Fernández L., Laiglesia L. M., Huerta A. E., Martínez J. A., and Moreno-Aliaga M. J. (2015) ω-3 fatty acids and adipose tissue function in obesity and metabolic syndrome. Prostaglandins Other Lipid Mediat. 121, 24–41 [DOI] [PubMed] [Google Scholar]
- 27. Okla M., Wang W., Kang I., Pashaj A., Carr T., and Chung S. (2015) Activation of Toll-like receptor 4 (TLR4) attenuates adaptive thermogenesis via endoplasmic reticulum stress. J. Biol. Chem. 290, 26476–26490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karbiener M., and Scheideler M. (2014) MicroRNA functions in Brite/Brown fat–novel perspectives towards anti-obesity strategies. Comput. Struct. Biotechnol. J. 11, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou J. Y., and Li L. (2014) MicroRNAs are key regulators of brown adipogenesis. Biochim. Biophys. Acta 1841, 1590–1595 [DOI] [PubMed] [Google Scholar]
- 30. Trajkovski M., and Lodish H. (2013) MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol. Metab. 24, 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hwang H. W., and Mendell J. T. (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94, 776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fernández-Hernando C., Suárez Y., Rayner K. J., and Moore K. J. (2011) MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 22, 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chakraborty C., Doss C. G., Bandyopadhyay S., and Agoramoorthy G. (2014) Influence of miRNA in insulin signaling pathway and insulin resistance: micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip. Rev. RNA 5, 697–712 [DOI] [PubMed] [Google Scholar]
- 34. Xu S., Chen P., and Sun L. (2015) Regulatory networks of non-coding RNAs in brown/beige adipogenesis. Biosci. Rep. 35, e00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Recchiuti A., Krishnamoorthy S., Fredman G., Chiang N., and Serhan C. N. (2011) MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 25, 544–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Antal O., Hackler L. Jr, Shen J., Mán I., Hideghéty K., Kitajka K., and Puskás L. G. (2014) Combination of unsaturated fatty acids and ionizing radiation on human glioma cells: cellular, biochemical and gene expression analysis. Lipids Health Dis. 13, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ortega F. J., Cardona-Alvarado M. I., Mercader J. M., Moreno-Navarrete J. M., Moreno M., Sabater M., Fuentes-Batllevell N., Ramírez-Chávez E., Ricart W., Molina-Torres J., Pérez-Luque E. L., and Fernández-Real J. M. (2015) Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 26, 1095–1101 [DOI] [PubMed] [Google Scholar]
- 38. Rosell M., Jones M. C., and Parker M. G. (2011) Role of nuclear receptor corepressor RIP140 in metabolic syndrome. Biochim. Biophys. Acta 1812, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leonardsson G., Steel J. H., Christian M., Pocock V., Milligan S., Bell J., So P. W., Medina-Gomez G., Vidal-Puig A., White R., and Parker M. G. (2004) Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. U.S.A. 101, 8437–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seale P. (2010) Transcriptional control of brown adipocyte development and thermogenesis. Int. J. Obes. 34, Suppl. 1, 17–22 [DOI] [PubMed] [Google Scholar]
- 41. Liu P. S., Lin Y. W., Lee B., McCrady-Spitzer S. K., Levine J. A., and Wei L. N. (2014) Reducing RIP140 expression in macrophage alters ATM infiltration, facilitates white adipose tissue browning, and prevents high-fat diet-induced insulin resistance. Diabetes 63, 4021–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosell M., Kaforou M., Frontini A., Okolo A., Chan Y. W., Nikolopoulou E., Millership S., Fenech M. E., MacIntyre D., Turner J. O., Moore J. D., Blackburn E., Gullick W. J., Cinti S., Montana G., et al. (2014) Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am. J. Physiol. Endocrinol. Metab. 306, E945–E964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blad C. C., Tang C., and Offermanns S. (2012) G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat. Rev. Drug Discov. 11, 603–619 [DOI] [PubMed] [Google Scholar]
- 44. Moran B. M., Abdel-Wahab Y. H., Flatt P. R., and McKillop A. M. (2014) Evaluation of the insulin-releasing and glucose-lowering effects of GPR120 activation in pancreatic beta-cells. Diabetes Obes. Metab. 16, 1128–1139 [DOI] [PubMed] [Google Scholar]
- 45. Vargovic P., Ukropec J., Laukova M., Cleary S., Manz B., Pacak K., and Kvetnansky R. (2011) Adipocytes as a new source of catecholamine production. FEBS Lett. 585, 2279–2284 [DOI] [PubMed] [Google Scholar]
- 46. Kang J. X., Wang J., Wu L., and Kang Z. B. (2004) Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature 427, 504. [DOI] [PubMed] [Google Scholar]
- 47. Li X., Ballantyne L. L., Che X., Mewburn J. D., Kang J. X., Barkley R. M., Murphy R. C., Yu Y., and Funk C. D. (2015) Endogenously generated ω-3 fatty acids attenuate vascular inflammation and neointimal hyperplasia by interaction with free fatty acid receptor 4 in mice. J. Am. Heart. Assoc. 4, e001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fleckenstein-Elsen M., Dinnies D., Jelenik T., Roden M., Romacho T., and Eckel J. (2016) Eicosapentaenoic acid and arachidonic acid differentially regulate adipogenesis, acquisition of a brite phenotype and mitochondrial function in primary human adipocytes. Mol. Nutr. Food Res. 10.1002/mnfr.201500892, in press [DOI] [PubMed] [Google Scholar]
- 49. Gotoh C., Hong Y. H., Iga T., Hishikawa D., Suzuki Y., Song S. H., Choi K. C., Adachi T., Hirasawa A., Tsujimoto G., Sasaki S., and Roh S. G. (2007) The regulation of adipogenesis through GPR120. Biochem. Biophys. Res. Commun. 354, 591–597 [DOI] [PubMed] [Google Scholar]
- 50. Lee M. W., Odegaard J. I., Mukundan L., Qiu Y., Molofsky A. B., Nussbaum J. C., Yun K., Locksley R. M., and Chawla A. (2015) Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 160, 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Metcalfe L., Schmitz A., and Pelka J. (1966) Rapid preparation of fatty acid esters from lipids for gas chromatographic analysis. Anal. Chem. 38, 514–515 [Google Scholar]