Abstract
Methylmalonic aciduria (MMAuria), caused by deficiency of methylmalonyl-CoA mutase (MUT), usually presents in the newborn period with failure to thrive and metabolic crisis leading to coma or even death. Survivors remain at risk of metabolic decompensations and severe long term complications, notably renal failure and neurological impairment. We generated clinically relevant mouse models of MMAuria using a constitutive Mut knock-in (KI) allele based on the p.Met700Lys patient mutation, used homozygously (KI/KI) or combined with a knockout allele (KO/KI), to study biochemical and clinical MMAuria disease aspects. Transgenic Mutki/ki and Mutko/ki mice survive post-weaning, show failure to thrive, and show increased methylmalonic acid, propionylcarnitine, odd chain fatty acids, and sphingoid bases, a new potential biomarker of MMAuria. Consistent with genetic dosage, Mutko/ki mice have lower Mut activity, are smaller, and show higher metabolite levels than Mutki/ki mice. Further, Mutko/ki mice exhibit manifestations of kidney and brain damage, including increased plasma urea, impaired diuresis, elevated biomarkers, and changes in brain weight. On a high protein diet, mutant mice display disease exacerbation, including elevated blood ammonia, and catastrophic weight loss, which, in Mutki/ki mice, is rescued by hydroxocobalamin treatment. This study expands knowledge of MMAuria, introduces the discovery of new biomarkers, and constitutes the first in vivo proof of principle of cobalamin treatment in mut-type MMAuria.
Keywords: amino acid, ammonia, enzyme mutation, inborn error of metabolism, metabolic disease, mitochondrial disease, genotype-phenotype correlation, knock-in mouse model, methylmalonic aciduria, vitamin B12 metabolism
Introduction
Methylmalonyl-CoA mutase (MUT)2 is a homodimeric enzyme that catalyzes the reversible isomerization of l-methylmalonyl-CoA to succinyl-CoA by using vitamin B12 (cobalamin, Cbl) in the cofactor form (5′-deoxyadenosylcobalamin, AdoCbl). In this anaplerotic reaction, several metabolic pathways, including the breakdown of branched chain amino acids, odd chain fatty acids, and the side chain of cholesterol, converge on the propionate pathway. The vital importance of this enzyme is demonstrated by the severe inborn error of metabolism methylmalonic aciduria (MMAuria), which is caused by mutations in the MUT gene (mut-type MMAuria, OMIM no. 251000) or by defects of AdoCbl synthesis. Dysfunction of MUT leads to an accumulation of methylmalonic acid (MMA), 2-methylcitrate (2-MC), propionylcarnitine (C3), and other metabolites in body fluids and tissues (1, 2). Although the clinical symptoms observed in mut-type MMAuria patients are variable, they often present in the newborn period with ketoacidosis, lethargy, and failure to thrive, leading to coma or even death if untreated. In surviving patients, the chronic course involves organ-specific complications, the most common being neurological impairment and chronic kidney disease, manifesting as tubulointerstitial nephritis and renal tubular acidosis. End stage renal disease, occurring as early as the second decade of life in patients with severe MMAuria, is frequently seen (3–6). In addition to symptomatic care, there are a few rational treatment approaches, including reduction of protein intake, carnitine and Cbl supplementation to decrease the flux through the propionate pathway, and the formation of toxic metabolites (6). Although this treatment appears to partly improve metabolic control in some cases, most patients still suffer severe long term complications.
The first effort to generate a mouse model of mut-type MMAuria was made in 2003, when a Mut knockout (KO) model on C57BL/6 background resulted in neonatal lethality of all homozygous (KO/KO) pups (7). Later, Peters et al. (8) expressed varying copy numbers of the intact human MUT locus on the KO background and demonstrated a copy number-dependent rescue of the KO lethality. They also developed another transgenic model containing an introduced stop codon on the human MUT locus crossed to the KO background, resulting in a severe MMAuria phenotype (9). In 2009, Chandler et al. (10) generated a Mut-KO mouse on the modified ((C57BL/6X129Sv/Ev) × FVB/N) background, resulting in some KO mice surviving the neonatal period, although nearly all died within 25 days (11). Using this KO model, they applied adeno-associated virus-mediated gene therapy (11, 12) and stable transgenic Mut expression restricted to the liver (13) to provide a long term rescue of lethality. However, whereas these models have proven to be useful to study some aspects and rescue of disease, no model to date has been able to provide an accurate depiction of the patient situation. Neonatal or early lethality hamper long term follow-up studies and the understanding of chronic disease manifestation, whereas transgenic rescue of Mut does not resemble the pathologic situation, where a patient carries the same genetic defect in all cells.
To devise a model of MMAuria reflective of the patient situation using a known human mutation and amenable to the study of the long term disease course, we opted to knock-in (KI) a hypomorphic allele based on a patient missense mutation. From our previous in-depth MUT missense mutation characterization (14), we identified p.Met700Lys (c.2099T>A human cDNA; p.Met698Lys in mouse) as an ideal candidate for this allele because of the residual enzymatic activity and in vitro response to hydroxocobalamin (OHCbl) found in MUT protein carrying this mutation. p.Met700Lys has been detected heterozygously in four MMAuria patients in conjunction with severe mutations (15, 16), but at least three of these patients have had an intermediate disease course with a relatively later age of onset (between 14 days and 6 months) (16), supporting the potential of this mutation to cause moderate MMAuria. To assess the effect of genetic dosage on the phenotype and to generate a more severe disease model, the new KI mutation was combined with a KO allele (7). Here we present a thorough characterization of these newly generated Mutki/ki and Mutko/ki mouse models, which survive long term and at the same time recapitulate the key biochemical and clinical features of MMAuria.
Results
In Vitro Characterization of the p.Met698Lys Knock-in Allele
To characterize the selected KI mutation, we tested the biochemical properties of the human mutation p.Met700Lys (hs-p.Met700Lys) and the equivalent mutation in mouse (mm-p.Met698Lys). Separate constructs incorporating each allele were overexpressed in human fibroblasts carrying a homozygous null allele of MUT to determine enzyme activity and kinetics. In both, the residual activity was reduced to a few percent of wt (Table 1) with an almost 100-fold increased Km for AdoCbl (Table 1). These nearly identical biochemical results for the human and mouse equivalent mutations led us to conclude that the mm-p.Met698Lys mutation would be expected to produce an intermediate phenotype of MMAuria, similar to its hs-p.Met700Lys counterpart.
TABLE 1.
Human and mouse mutant proteins show similar enzyme parameters when expressed in a mut0 patient cell line
Mean enzyme activities were measured in fibroblast homogenates after overexpression of the respective constructs. Mutant values were calculated in relation to their wt counterparts and normalized to the percentage of residual activity of the respective wt (set to 100%). Km values for AdoCbl were determined by measuring MUT activity with increasing concentrations of AdoCbl ranging from 0.0025 to 50 μm in the same homogenates as above. The displayed numbers represent mean values ± S.D.
| Human |
Mouse |
|||
|---|---|---|---|---|
| wt | p.M700K | wt | p.M698K | |
| Activity | 100.0 ± 22.4 | 3.46 ± 0.38 | 100.0 ± 32.1 | 6.04 ± 0.24 |
| Km | 2.59 ± 0.87 | 575.6 ± 46.0 | 8.70 ± 0.02 | 463.6 ± 37.3 |
Generation of KI Allele and Initial Characterization
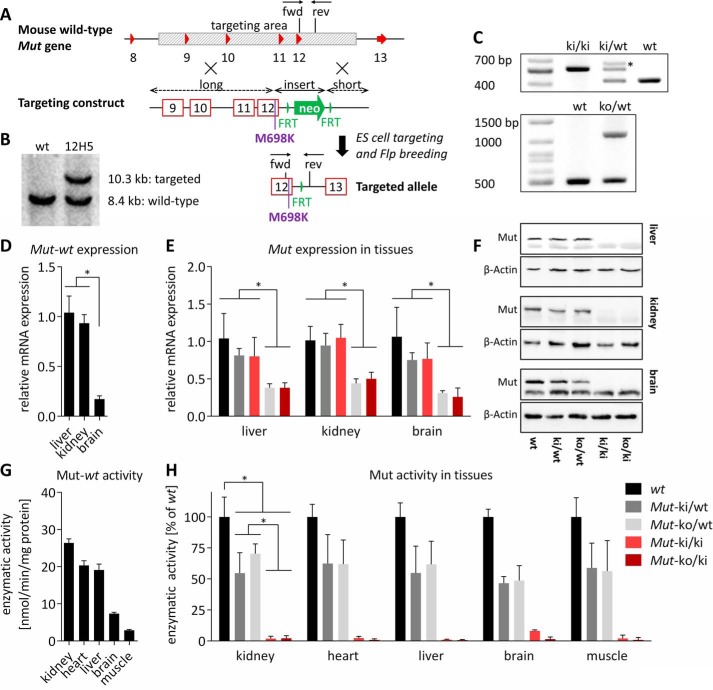
To generate the knock-in p.Met698Lys mutation, a targeting vector was designed to replace thymine with adenine at position c.2093 of Mut (based on NM_008650.3). The construct (Fig. 1A) was used for embryonic stem cell (C57BL/6-derived) targeting, and a positive clone was confirmed by Southern hybridization (Fig. 1B), which was used for blastocyst injection. Screening for positive germ line transmission was done by PCR genotyping (Fig. 1C, upper panel). Additionally, Mutko/ki mice were generated as a second disease model by crossing the previously established Mutko/wt mice (7) with our Mutki/ki mice and followed by PCR genotyping of the KO allele (Fig. 1C, lower panel).
FIGURE 1.
Generation of KI allele and initial characterization. A, based on the wt mouse Mut gene, a targeting construct was generated to replace the Mut gene region spanning from introns 8 to 12 (shaded in gray), consisting of long and short arms of homology, as well as an insert with the missense mutation (p.Met698Lys) and a neomycin cassette (neo, green arrow) flanked by FRT sites (vertical green triangles). Homologous recombination and Flp deletor breeding resulted in the final targeted KI allele with the mutation and one remaining FRT site. Exons are represented by red triangles or boxes. Arrows marked fwd and rev indicate forward and reverse genotyping primers, respectively. B, Southern blotting of targeted clone 12H5. C, upper panel, genotyping PCR of ear biopsy DNA for the KI allele with forward and reverse primers depicted in A. The asterisk indicates an additional heterodimer band as confirmed by Sanger sequencing. Lower panel, genotyping PCR of KO allele. D and E, Mut expression among wt tissues (error bars are S.E.; n = 4; *, p < 0.01) (D) and KO allele-dependent loss of Mut expression (bars are means normalized to wt in each tissue; error bars depict S.E.; n ≥ 4; *, p < 0.05) (E). F, Western blot analysis of Mut protein levels using β-actin as loading control. The lower band in the top panel of each organ represents an unspecific band. G and H, Mut activity varies by tissue (G) and by genotype (H) (bars are mean values and in each tissue normalized to the wt value; error bars are S.E.; n ≥ 4; *, p < 0.0001). Significance levels are the same in all five tissues (not depicted for clarity).
To determine the direct consequences of the missense change on Mut transcript, stability, and function, we investigated Mut expression and activity in various tissues. In wt mice, we found Mut expression to be lower in brain than in liver or kidney (Fig. 1D), similar to previously published results (17). The KI allele did not result in an appreciable decrease of Mut transcript levels in all tissues tested; however, mice with one KO allele had ∼50% of Mut transcript levels compared with wt (Fig. 1E), likely indicative of nonsense-mediated decay. At the protein level, the amount of the homozygous Mutki/ki protein was significantly reduced compared with controls, suggesting an unstable mutant protein, whereas the KO/KI Mut protein showed similar reduction (Fig. 1F). Mut activity from wt mice significantly varied among tissue types, with the highest activities in kidney, liver, and heart (Fig. 1G). Activities from KI/KI and KO/KI mice were greatly decreased in all of the tissues tested (Fig. 1H), comparable with the in vitro measurements (Table 1). Furthermore, a gene dosage-dependent decrease of enzyme activity was identified, with Mutko/ki activity slightly lower than Mutki/ki in most tissues (Fig. 1H). As expected, ∼50% of residual activity was maintained in the Mutki/wt and Mutko/wt animals, reflecting the activity of the remaining wt allele (Fig. 1H).
Gene Dosage-dependent Biochemical and Clinical Phenotype in Mutki/ki and Mutko/ki Mice
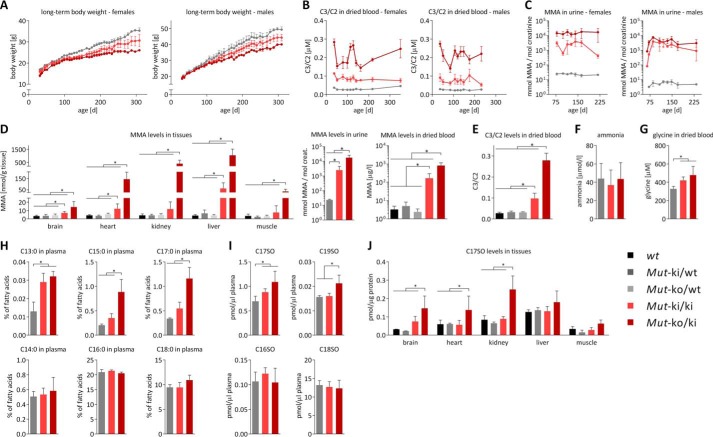
Mice homozygous for the targeted point mutation (Mutki/ki) were viable and, when crossed with heterozygous Mutki/wt mice, reproduced at the expected Mendelian ratio of ∼50% per genotype (51% Mutki/wt, 49% Mutki/ki, litters = 33). Breeding of Mutko/ki also resulted in viable animals and displayed the expected Mendelian distribution of the genotypes (51% Mutko/ki, 49% Mutki/wt, litters = 36). Mice of both genotypes survived over 1 year, allowing us to monitor long term development. At early stages of life, Mutki/ki and Mutko/ki mice were largely indistinguishable from their heterozygous littermates (Mutki/wt) and wt control mice. However, after the age of ∼100 days in females and ∼150 days in males (Fig. 2A), mice of both genotypes showed signs of significant growth retardation. The difference in body weight between littermate control (Mutki/wt) and Mutko/ki female mice was 30% after ∼1 year (Fig. 2A), whereas food intake remained constant (e.g. relative food intake at the age of 218 days in Mutki/wt was 0.079 grams of chow normalized to body weight in grams, S.D. ± 0.043 and in Mutko/ki 0.078 ± 0.048). In this cohort, C3 (normalized to acetylcarnitine (C2)) in blood and MMA in urine were constantly elevated over the entire time span investigated (Fig. 2, B and C). Survival was the same in all three groups (all mice still alive after 1 year except for one Mutki/ki and one Mutko/ki, both of which died during blood collection).
FIGURE 2.
Clinical and biochemical phenotype of Mutki/ki and Mutko/ki mice (genotypes see legend, data in panels D–J are derived from female animals). A, monitoring of body weight over time. Decreased body weight in female Mutki/ki mice is significant compared with Mutki/wt from day 162, and in Mutko/ki significant compared with Mutki/wt from day 114. For both, p < 0.05. B, C3 levels in dried blood spots normalized to C2. C, MMA levels measured by LC-MS/MS in urine collected overnight in metabolic cages. In A–C points represent mean values, error bars depict S.E. and n = 5/group. D, MMA levels in tissues (left panel), urine (middle panel), and dried blood (right panel), determined by tandem mass spectrometry. E, C3 levels normalized to C2 in dried blood. F, ammonia levels measured in whole blood. G, glycine concentration determined in dried blood by tandem mass spectrometry. H, fatty acid levels determined in plasma and expressed as percentage of total fatty acids. I, plasma levels of sphingoid bases (expressed as pmol/μl plasma). J, 17-carbon chain sphingoid base levels in different tissues expressed as pmol/μg protein. D–J, bars represent mean values from 35 days old mice (error bars depict S.D.; n ≥ 4; *, p < 0.05).
To gain further insight into the consequences of the clearly reduced Mut enzymatic activity, we performed a cross-sectional analysis of hallmark metabolites of MMAuria (18) at 35 days of age. MMA was found elevated in a genetic dosage-dependent manner in tissues, urine and dried blood, with the highest levels found in Mutko/ki mice (Fig. 2D). Although Mutki/ki animals showed considerably lower levels, they were still significantly elevated compared with controls (Fig. 2D). C3 levels (normalized to C2) showed the same trend (Fig. 2E). Mutki/wt and Mutko/wt did not show any elevations, suggesting that one functional wt allele is sufficient to cope with the demands of the propionate pathway (Fig. 2, D and E). Ammonia levels, which are elevated during metabolic crisis in patients, were normal (Fig. 2F), suggesting that these animals did not suffer from acute crises on normal chow. Similar to patients (19–21), both Mutko/ki and Mutki/ki showed elevated glycine levels (Fig. 2G), whereas other amino acid levels were mostly unchanged.
In addition to classic metabolites, we performed a comprehensive analysis of fatty acids and sphingoid bases. We found increased levels of odd chain fatty acids, including the 17-carbon chain length fatty acid, which has recently been proposed as a biomarker of MMAuria (22, 23), with normal levels of even chain fatty acids (Fig. 2H). Further, we show here for the first time an elevation of odd chain length sphingoid bases in plasma (Fig. 2I) and tissues (Fig. 2J), possibly as a consequence of the increased odd chain fatty acids, whereas levels of even chain sphingoid bases remained normal (Fig. 2I). Specific fatty acid and sphingoid base levels are freely available upon request.
Initial Characterization of the Renal and Neurological Phenotypes
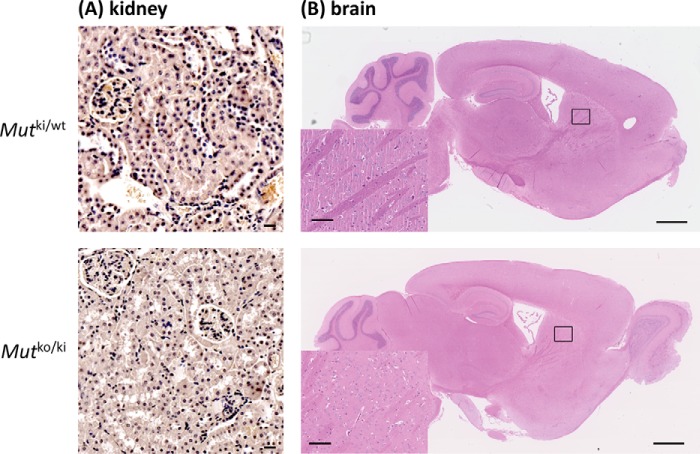
We investigated whether Mutko/ki mice suffer from similar long term complications as patients with MMAuria,e.g. renal dysfunction. When normalized to body weight, Mutko/ki mice consistently produced less urine during adulthood than littermate controls (Mutki/wt) (Fig. 3A), although water intake was not significantly different between the two groups (Fig. 3B). Consistent with renal dysfunction, these mice also had increased plasma urea throughout the time course measured (Fig. 3C), which has been observed in the context of a decreased glomerular filtration rate (24). Renal tubular dysfunction in these animals was evidenced by disturbed excretion of several electrolytes in urine (Fig. 3D). Kidney damage was further supported by increased levels of the biomarker lipocalin-2 (Lcn2) in kidney tissue (Fig. 3E). Of note, light microscopy examination of hematoxylin-eosin stained sections of kidneys from Mutki/wt and Mutko/ki mice (female, 1 year old) did not reveal any structural abnormalities (Fig. 4A).
FIGURE 3.
Renal and neurological phenotype. A, urine production measured in metabolic cages overnight and expressed as microliters of urine per hour normalized to body weight. B, overnight water intake measured in grams of water normalized to body weight. C, urea in plasma expressed in mg per deciliter. D, electrolytes measured in urine and normalized to grams of creatinine. E, Western blot of Mut and Lcn2 in kidney at two different time points. F, brain weight normalized to body weight at three different time points. G, expression of Lcn2 in brain tissue on mRNA level normalized to β-actin and on protein level (β-actin as loading control). In A–D, F, and G, points and bars represent mean values (error bars depict S.E.; *, p < 0.05).
FIGURE 4.
Histological analysis by hematoxylin-eosin staining of kidney (A) and brain (B) sections from Mutki/wt and Mutko/ki female mice at the age of 1 year. Scale bars are 10 μm in the kidney pictures, 1 mm on brain overview pictures, and 100 μm in the insets of the basal ganglia.
Neurological dysfunction is the other major long term complication of MMAuria (25, 26), often manifesting as movement disorder because of lesions in the basal ganglia, and in acute metabolic crisis brain edema may be observed. In the Mutko/ki mice we observed an increase in brain weight at the age of 1 month as well as 1 year (Fig. 3F), which may be suggestive of brain edema. Although brain histology was normal in the basal ganglia (Fig. 4B), we also identified an up-regulation in Lcn2 at the mRNA and protein level (Fig. 3G). Together, the initial characterization of both the brain and kidney, in combination with the metabolites already identified in these organs, suggest an initial dysfunction in each, which has not yet manifested at the gross level.
Diet-induced Metabolic Decompensation
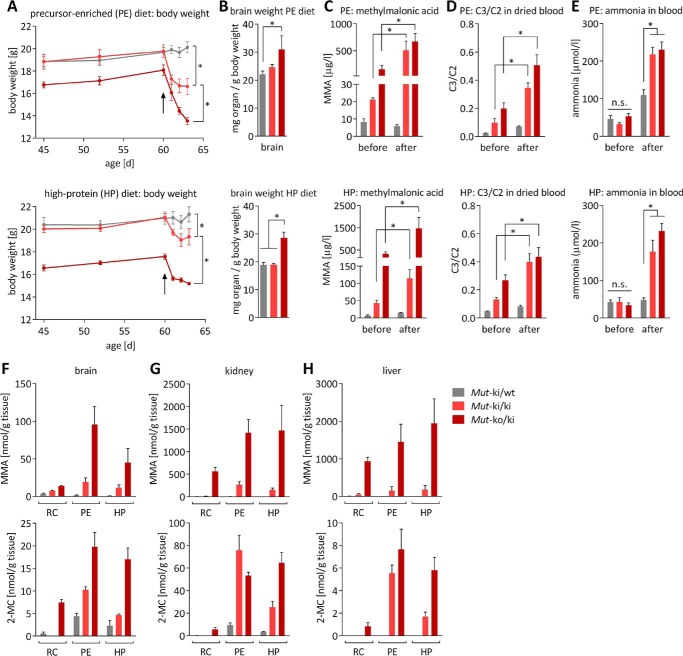
An increased throughput of the propionate pathway in patients with MMAuria,e.g. caused by a catabolic state or an augmented protein intake, often results in metabolic decompensation (crisis). To replicate this state in our MMAuria mouse models and exacerbate their clinical phenotype, we used dietary challenge. Mice were fed a high protein (HP) diet, as used previously (13), or a precursor-enriched (PE) diet comprised of increased levels of precursor amino acids of propionate pathway metabolites, i.e. isoleucine, valine, and threonine. Upon initiation of both diets at the age of 2 months, both Mutko/ki and Mutki/ki mice experienced rapid weight loss, which was so severe that the study had to be terminated after 3 days (Fig. 5A). The effect exerted by the PE diet appeared to be stronger than the HP diet, as shown by the more pronounced weight loss (Fig. 5A), likely reflecting the 2-fold higher isoleucine, valine, and threonine content in the PE diet (70, 84, and 53 g/kg compared with 35, 42, and 27 g/kg, respectively). Brain weight (normalized to body weight) was increased in Mutko/ki mice under both diets, which was not the case on reference chow (RC: isoleucine, 10 g/kg; valine, 12 g/kg; threonine, 7.6 g/kg) at this age (Fig. 5B). Investigation of the plasma before and after the diet change revealed, from both diets, further elevated levels of MMA, C3, and for the first time in these mice, increased ammonia levels in Mutki/ki and Mutko/ki mice (Fig. 5, C–E), consistent with the induction of metabolic crisis. Further investigation of MMA and 2-MC concentrations in the brain, kidney, and liver demonstrated a gene dosage-dependent increase of both metabolites in mice on either diet, compared with mice on the control diet (RC) (Fig. 5, F–H). Indeed, although these metabolites were already elevated in Mutko/ki and Mutki/ki mice on RC (e.g. brain levels of MMA in Mutko/ki were 4.4 times higher than in Mutki/wt; Fig. 5F), they were further increased up to 53-fold when the diet was changed to HP or PE (Fig. 5F).
FIGURE 5.
Modified diet leads to disease acceleration. A, start of both modified diets at 60 days of age is indicated by the black arrows. Upper panel, PE; lower panel, HP. Monitoring of body weight showed significant differences among the three groups on day 63 for both diets. B, weight of brain normalized to body weight after diet study. C, MMA in dried blood. D, C3 (normalized to C2) in dried blood. E, whole blood ammonia before and after diet modification. F–H, MMA and 2-MC levels in homogenates of brain (F), kidney (G), and liver (H) tissue on RC, HP, and PE as determined by GC-MS. For all panels, from five animals for each genotype, bars and points represent means; error bars are S.E. For A–E, *, p < 0.01; n.s., not significant; statistics were not calculated for F–H.
Cobalamin Treatment Partly Rescues Diet-induced Metabolic Crisis
To attempt to mitigate the effects exerted by the HP diet, we treated the mice with daily i.p. injections of OHCbl (0.3 μg/g) for a week before and continuously during the dietary challenge. These injections resulted in nearly doubled levels of plasma Cbl (Fig. 6A), indicating efficient uptake of the vitamin into the blood stream. Mutki/ki mice appeared to be partially protected by this treatment, as, in contrast to mice on the HP diet alone (Fig. 5A), OHCbl-treated Mutki/ki mice did not show significant weight loss (Fig. 6B). Further, elevations in C3 and ammonia were markedly delayed, although they increased considerably, along with MMA, by the study's end (Fig. 6, C–E). Mutko/ki mice showed a less obvious but slight protective effect. Their weight loss was delayed compared with their untreated counterparts (compare Fig. 5A, HP, and Fig. 6B), and they did not have the significantly increased brain weight (Fig. 6F) seen previously (Fig. 5B). However, they did show immediate and striking increases in metabolite levels (Fig. 6, C–E) suggesting they remained metabolically compromised.
FIGURE 6.
Treatment of HP diet-induced phenotype by OHCbl. A, cobalamin levels in plasma of untreated mice and OHCbl-treated mice (*, p < 0.0001). B, time course of body weight. The black arrow indicates start of HP diet on day 60. Differences of body weight on day 64 were significant between Mutko/ki and the other two genotypes (Mutki/ki and wt), which did not differ significantly from each other. C–E, C3 (normalized to C2) (C), MMA levels in dried blood (D), and whole blood ammonia levels (E) were measured on the day the diet was started (day 60) and on days 62 and 64 following diet commencement. F, weights of brain whole organs in sacrificed animals at the end of the study. In B–E duration of HP diet was from days 60 to 64 and OHCbl treatment from days 53 to 64 as indicated by capped horizontal lines. Points and bars represent mean values; error bars in all panels depict S.E.
Discussion
Novel Mouse Models Recapitulate Clinical and Biochemical Phenotype of MMAuria
In this study, we aimed to generate novel mouse models of MMAuria that survive through adulthood but show clear biochemical and clinical signs of disease. To achieve these models, we knocked in a missense mutation to the mouse Mut gene. This constitutive KI allele causes Mut deficiency, which is further aggravated by combination with a KO allele. The most striking phenotypic sign in our mice was growth retardation, which is likely a correlate of failure to thrive described in human patients (3, 6). This lack of weight gain is not readily explained by reduced energy intake, because we identified no difference in overnight food consumption between the mutant mice and controls, suggesting that other, possibly subcellular, processes are affected. Previous studies point toward inhibited tricarboxylic acid cycle enzymes and interference with oxidative phosphorylation in MMAuria (27–29). It will be important to assess these potential mechanisms in future studies.
Further clinical signs could be observed when the disease was accelerated by means of a high protein challenge. The success of this approach was demonstrated by the substantial elevation of metabolites and immediate loss of body weight in both diets (HP and PE) for both mouse genotypes (Mutko/ki and Mutki/ki), suggesting that the propionate pathway was unable to process the additional metabolic load contributed by the modified diets. As indicated by the significantly elevated ammonia levels, this induced situation is consistent with acute metabolic crisis, where aggressive treatment is required to prevent encephalopathy when ammonia levels rise above a threshold of 200 μmol/liter (30).
Along with these general organism-wide symptoms, we were able to identify organ-specific disturbances in the kidney and the brain. Under normal diet conditions, adult Mutko/ki mice showed manifestations of kidney dysfunction, as evidenced by increased plasma urea, impaired diuresis, and changes in the urinary excretion of electrolytes. These changes mirror the organ manifestations of MMAuria patients, in whom chronic kidney disease occurs in almost half of all cases in an age-dependent manner (3–6). In our mice, these changes were accompanied by increased renal levels of Lcn2, a protein that has been suggested to be an early biomarker of chronic kidney damage (31), including in MMAuria (13). Further, we found increased brain weight in the Mutko/ki mice at 1 month and 1 year of age, possibly indicative of cytotoxic edema as a sign of neurotoxicity. Interestingly, we also found increased Lcn2 mRNA and protein levels in the brain of these mice. In contrast to its potential role as a kidney specific marker, up-regulation of Lcn2 in the brain has been suggested to be a biomarker in multiple sclerosis (32) and was found to be up-regulated in animal models of brain-specific diseases (33, 34). Hence, the specific role of Lcn2 as an organ-specific biomarker remains unclear; however, the finding of up-regulation in affected tissues suggests at least a general role in dysfunctional cells.
Biochemically, the phenotype of human patients was accurately emulated, as illustrated by the types of metabolites that were increased and the extent of their elevation. For example, MMA levels in the urine were detected in a range comparable with human mut− patients (35), whereas glycine, thought to be increased by inhibition of the intramitochondrial glycine cleavage enzyme caused by accumulated organic acids or their CoA esters (36), was found at high levels in our mice, similar to the description in the first MMAuria patients (37). Further, we found elevated levels of odd chain fatty acids, most likely as a consequence of increased C3, which is used as a primer in fatty acid synthesis (6, 23, 38, 39). In addition, we present for the first time elevations of odd-numbered chain sphingoid bases in plasma and tissue. Their elevation may be explained by the disturbed fatty acid metabolism, i.e. serine palmitoyltransferase, a key enzyme in sphingoid base synthesis, may more often utilize 17- or 19-carbon fatty acid CoAs as substrate, instead of the normally preferred 14- or 18-chain length fatty acids. The pathophysiological significance of this newly discovered phenomenon remains unknown.
Overall, these novel models of MMAuria recapitulate many of the biochemical and clinical hallmarks of MMAuria in patients and thus are excellent tools to study the pathogenesis of disease. Application of a long term dietary challenge with less potent diets will potentially allow monitoring of chronic disease progression, leading to more pronounced impacts on brain and kidney tissue.
Gene Dose Dependence Allows Titration of Disease Severity
Patients carrying the p.Met700Lys mutation show an intermediate phenotype with relatively late onset and residual enzyme and pathway activity in fibroblast cell homogenates (15, 16, 40). Based on these observations, the measurable residual enzyme activity in vitro, and the missense type of the variant used for the generation of the KI allele, we expected that the KI allele would result in a milder phenotype than the KO allele, which is in fact a truncating mutation. Indeed, a gene dose-dependent effect was corroborated by all experimental data: Mutko/ki mice displayed higher concentrations of MMA, 2-MC, and C3, more pronounced growth retardation, and a stronger response to dietary challenge than homozygous Mutki/ki animals. Therefore, the gene dose effect detected in our study is analogous to the phenotypic differences observed in patients. This circumstance is a true benefit of this study because the novel animal models allow us to modulate the phenotypic severity of MMAuria and to systematically assess milder and more severe forms of MMAuria.
Response to Cobalamin Treatment
It is recommended to evaluate every mut-type MMAuria patient for responsiveness to cobalamin treatment (6, 35). However, no systematic study has thus far investigated the effectiveness of cofactor treatment in mut-type MMAuria. If effective, the cofactor would be expected to ameliorate disease by supporting the enzyme activity and/or stability, similar to the cofactor response displayed by phenylalanine hydroxylase (41). The human mutant p.Met700Lys is an excellent candidate to test cobalamin responsiveness, because patient fibroblasts compound heterozygous for this and a premature stop mutation showed 4.5-fold times increased incorporation of [14C]propionate into acid precipitable material when supplemented with OHCbl (40), and biochemically this mutation was found to affect the Km of MUT for AdoCbl (14). Indeed, in our HP diet study, Mutki/ki mice appear to have been at least partly protected by pre-administration of OHCbl. These mice did not show the immediate weight loss associated with the initiation of the HP diet and showed delayed accumulation of metabolites. The reduced degree of protection for Mutko/ki mice suggests that with the level of Cbl achieved by these injections, both mutant alleles are required to be potentially Cbl-responsive. An even better rescue may be facilitated by the application of higher Cbl doses in these mice, potentially reaching levels suggested for treatment in human patients (35). Nevertheless, our results set the stage for further exploration of cofactor response, a concept that may be applicable to many other missense mutations, since approximately one-quarter of all mut-type mutations show an in vitro response to cobalamin (3).
Experimental Procedures
In Vitro Cell Culture and Enzyme Activity Assay
Constructs of human (hs) MUT wild-type and mouse (mm) Mut wild-type and the mutants hs-p.Met700Lys and mm-p.Met698Lys were made by site-directed mutagenesis in the pTracer-CMV2 vector (Thermo Fisher Scientific) using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. Constructs were transiently transfected into immortalized fibroblasts carrying the homozygous MUT mutation p.Gln30* by electroporation. Cell lysates were obtained by from frozen, pelleted cells by sonication in 5 mm potassium phosphate buffer (pH 7.4) and enzymatic activity and Km determined as previously described (14).
Mouse Tissue Preparation
Liver, kidney, brain, heart, and muscle as well as blood samples were removed from euthanized mice and stored at −80 °C. Lysates of homogenized (TissueLyserII; Qiagen) tissues were resuspended in 300–600 μl of a lysis buffer dependent on the subsequent usage of the sample, either buffer A (Triton X-100 0.5% (v/v), 10 mm HEPES at pH 7.4, 2 mm DTT, 1 tablet of Complete Protease Inhibitor Mixture Tablets; Roche) for Western blot and activity assay or buffer B (250 mm sucrose, 50 mm KCl, 5 mm MgCl2, 20 mm Tris base, adjusted to pH 7.4) for determination of MMA and 2-MC levels. The homogenate was then centrifuged at 14,000 rpm for 10 min at 4 °C, and the supernatant was used for MUT activity assay (14), Western blotting, and quantitative RT-PCR (see below).
Housing of Mice and Generation of Knock-in Allele
Animal experiments were performed in accordance with policies of the Veterinary Office of the State of Zurich and Swiss law on animal protection. Animal studies were approved by the Cantonal Veterinary Office Zurich under license number 202/2014. Animals were kept in single-ventilated cages and under controlled humidity and temperature (21–23 °C). Whenever possible, littermate controls were used to compare experimental groups.
The generation of mice carrying the Mut-p.Met698Lys mutation was performed by Polygene (Rümlang, Switzerland) using embryonic stem cell targeting. To generate Mutko/ki mice, female Mutko/wt (7) were crossed to Mutki/ki males. Mouse genotyping was performed on genomic DNA from ear punch biopsies using the primers 5′-GTGGGTGTCAGCACACTTG-3′ (forward) and 5′-CGTATGACTGGGATGCCT-3′ (reverse) for the ki allele and 5′-ACAACTCCTTGTGTAGGTC-3′ (forward) and 5′-CCTTTAGGATGTCATTCTG-3′ (reverse) for the ko allele.
Long Term Studies and Metabolic Cage Studies
Long term monitoring of mice entailed weekly weight measurements and regular blood collections, as well as urine collections. The animals were single caged overnight to collect urine and measure individual chow and water intake. Urine was collected in the morning, the sediment was removed, and supernatant was frozen at −80 °C.
Quantitative Real Time PCR Analysis and Western Blot
Total RNA (QIAmp RNA blood mini kit; Qiagen) was extracted from frozen mouse tissue lysates and analyzed for expression using specific probes for Mut (Mm00485312_m1) and Lcn2 (Mm01324470_m1) via the TaqMan Gene Expression Master Mix on a 7900HT Fast Real-Time PCR System, with normalization to β-actin (Mm00607939_s1) (all Thermo Fisher Scientific). Experiments were performed in triplicate. For Western blotting analysis, antibodies against MUT (ab67869, 1:500; Abcam), Lcn2 (ab63929, 1:1000; Abcam), and β-actin (A2228; Sigma-Aldrich) were detected by HRP-labeled goat anti-mouse (sc-2302; Santa Cruz) or goat anti-rabbit (sc-2301; Santa Cruz) IgG at a dilution of 1:5000. Bands were quantified by normalization to internal β-actin control bands.
Metabolite Measurements
MMA in mice urine was analyzed by liquid chromatography mass spectrometry (LC-MS/MS) on a Thermo Scientific UltiMate 3000 rapid separation LC coupled to an AB Sciex 5500 TripleQuad mass spectrometer using a commercial kit (Recipe ClinMass® advanced). For determination of the concentrations of amino acids, acylcarnitines (C2 and C3), as well as MMA in dried blood spots, blood was collected from tail vein onto a filter card and dried. Punches from filter cards were analyzed by tandem mass spectrometry similar to Ref. 42. Total fatty acid profiles from plasma/serum were determined by direct methylation and gas chromatography, as described (43). Sphingoid bases were analyzed in tissue homogenates, as published (44). Ammonia levels in whole blood were measured by the PocketChem blood ammonia meter (PA-4140; Arkray). Electrolytes in urine were measured by a timed end point method (SYNCHRON system kit; Beckman Coulter). For urea determination in plasma the SYNCHRON CX3 Delta System was used (kit no. 443350; Beckman Coulter).
Diet and Cobalamin Therapy
We applied a HP (60% protein, TD.140830; Harlan) and PE (700% isoleucine, valine and threonine compared with reference, TD.140829; Harlan) diet to 60-day-old mice. For the PE diet, leucine (19 g/kg, 119%) was enriched because its uptake might compete with the uptake of the other amino acids, which are increased in the diet, and cystine was increased (3.5 g/kg, 700%) to elevate the overall sulfur content. For cobalamin rescue, mice were injected with 0.3 μg hydroxocobalamin (Streuli Pharma AG) i.p. 7 days before and throughout diet treatment.
Author Contributions
P. F. designed the research together with M. R. B., D. S. F., and O. D. and performed most of the experiments. Experimental work was also carried out by A. S., M. M., N. N., and A. Z. New reagents or analytic tools were contributed by D. M., C.-D. L., J. H., L. S., R. F., H. L. P., T. H., B. T., S. K., and O. D. The data were analyzed by P. F., M. R. B., D. S. F., O. D., P. B., and T. H. The manuscript was written by P. F., M. R. B., D. S. F., and O. D.
Acknowledgments
We thank Adam Guenzel for discussion and Elisabeth Rushing and the Institute of Neuropathology, University Hospital Zurich for brain histology, and Museer Lone for analysis of sphingoid bases. We thank Dr. Hendrica Belge and Huguette Debaix for help with phenotyping the kidney.
This work was supported by the Rare Disease Initiative Zurich (radiz), a clinical research priority program for rare diseases of the University of Zurich; the Swiss National Science Foundation Grant 31003A_138521; the Wolferman-Nägeli Foundation; the European Community's Seventh Framework Programme FP7/2007–2013 Grant 305608 (EURenOmics) (to O. D.); the Swiss National Science Foundation Project Grant 310030-146490 (to O. D.); and a Swiss National Science Foundation MD-PhD fellowship (Swiss National Science Foundation Grant 323530_145248; to P. F.). The authors declare that they have no conflicts of interest with the contents of this article.
- MUT
- methylmalonyl-CoA mutase
- MMA
- methylmalonic acid
- MMAuria
- methylmalonic aciduria
- KI
- knock-in
- Cbl
- cobalamin
- AdoCbl
- 5′-deoxyadenosylcobalamin
- 2-MC
- 2-methylcitrate
- C3
- propionylcarnitine
- OHCbl
- hydroxocobalamin
- C2
- acetylcarnitine
- Lcn2
- lipocalin-2
- HP
- high protein
- PE
- precursor-enriched
- RC
- reference chow
- KO
- knockout.
References
- 1. Kölker S., Schwab M., Hörster F., Sauer S., Hinz A., Wolf N. I., Mayatepek E., Hoffmann G. F., Smeitink J. A., and Okun J. G. (2003) Methylmalonic acid, a biochemical hallmark of methylmalonic acidurias but no inhibitor of mitochondrial respiratory chain. J. Biol. Chem. 278, 47388–47393 [DOI] [PubMed] [Google Scholar]
- 2. Schwab M. A., Sauer S. W., Okun J. G., Nijtmans L. G., Rodenburg R. J., van den Heuvel L. P., Dröse S., Brandt U., Hoffmann G. F., Ter Laak H., Kölker S., and Smeitink J. A. (2006) Secondary mitochondrial dysfunction in propionic aciduria: a pathogenic role for endogenous mitochondrial toxins. Biochem. J. 398, 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hörster F., Baumgartner M. R., Viardot C., Suormala T., Burgard P., Fowler B., Hoffmann G. F., Garbade S. F., Kölker S., and Baumgartner E. R. (2007) Long-term outcome in methylmalonic acidurias is influenced by the underlying defect (mut0, mut-, cblA, cblB). Pediat. Res. 62, 225–230 [DOI] [PubMed] [Google Scholar]
- 4. Hörster F., Garbade S. F., Zwickler T., Aydin H. I., Bodamer O. A., Burlina A. B., Das A. M., De Klerk J. B., Dionisi-Vici C., Geb S., Gökcay G., Guffon N., Maier E. M., Morava E., Walter J. H., et al. (2009) Prediction of outcome in isolated methylmalonic acidurias: combined use of clinical and biochemical parameters. J. Inherit. Metab. Dis. 32, 630–639 [DOI] [PubMed] [Google Scholar]
- 5. Cosson M. A., Benoist J. F., Touati G., Déchaux M., Royer N., Grandin L., Jais J. P., Boddaert N., Barbier V., Desguerre I., Campeau P. M., Rabier D., Valayannopoulos V., Niaudet P., and de Lonlay P. (2009) Long-term outcome in methylmalonic aciduria: a series of 30 French patients. Mol. Genet. Metab. 97, 172–178 [DOI] [PubMed] [Google Scholar]
- 6. Baumgartner M. R., Hörster F., Dionisi-Vici C., Haliloglu G., Karall D., Chapman K. A., Huemer M., Hochuli M., Assoun M., Ballhausen D., Burlina A., Fowler B., Grünert S. C., Grünewald S., Honzik T., et al. (2014) Proposed guidelines for the diagnosis and management of methylmalonic and propionic acidemia. Orphanet J. Rare Dis. 9, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peters H., Nefedov M., Sarsero J., Pitt J., Fowler K. J., Gazeas S., Kahler S. G., and Ioannou P. A. (2003) A knock-out mouse model for methylmalonic aciduria resulting in neonatal lethality. J. Biol. Chem. 278, 52909–52913 [DOI] [PubMed] [Google Scholar]
- 8. Peters H. L., Pitt J. J., Wood L. R., Hamilton N. J., Sarsero J. P., and Buck N. E. (2012) Mouse models for methylmalonic aciduria. PLoS One 7, e40609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buck N. E., Dashnow H., Pitt J. J., Wood L. R., and Peters H. L. (2012) Development of transgenic mice containing an introduced stop codon on the human methylmalonyl-CoA mutase locus. PLoS One 7, e44974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chandler R. J., Zerfas P. M., Shanske S., Sloan J., Hoffmann V., DiMauro S., and Venditti C. P. (2009) Mitochondrial dysfunction in mut methylmalonic acidemia. FASEB J. 23, 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chandler R. J., and Venditti C. P. (2010) Long-term rescue of a lethal murine model of methylmalonic acidemia using adeno-associated viral gene therapy. Mol. Ther. 18, 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chandler R. J., and Venditti C. P. (2008) Adenovirus-mediated gene delivery rescues a neonatal lethal murine model of mut(0) methylmalonic acidemia. Hum. Gene Ther. 19, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manoli I., Sysol J. R., Li L., Houillier P., Garone C., Wang C., Zerfas P. M., Cusmano-Ozog K., Young S., Trivedi N. S., Cheng J., Sloan J. L., Chandler R. J., Abu-Asab M., Tsokos M., et al. (2013) Targeting proximal tubule mitochondrial dysfunction attenuates the renal disease of methylmalonic acidemia. Proc. Natl. Acad. Sci. U.S.A. 110, 13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forny P., Froese D. S., Suormala T., Yue W. W., and Baumgartner M. R. (2014) Functional characterization and categorization of missense mutations that cause methylmalonyl-CoA mutase (MUT) deficiency. Hum. Mutat. 35, 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Acquaviva C., Benoist J. F., Pereira S., Callebaut I., Koskas T., Porquet D., and Elion J. (2005) Molecular basis of methylmalonyl-CoA mutase apoenzyme defect in 40 European patients affected by mut(o) and mut- forms of methylmalonic acidemia: identification of 29 novel mutations in the MUT gene. Hum. Mutat. 25, 167–176 [DOI] [PubMed] [Google Scholar]
- 16. Forny P., Schnellmann A.-S., Buerer C., Lutz S., Fowler B., Froese D. S., and Baumgartner M. R. (2016) Molecular genetic characterization of 151 mut-type methylmalonic aciduria patients and identification of 41 novel mutations in MUT. Hum. Mutat. 37, 745–754 [DOI] [PubMed] [Google Scholar]
- 17. Ballhausen D., Mittaz L., Boulat O., Bonafé L., and Braissant O. (2009) Evidence for catabolic pathway of propionate metabolism in CNS: expression pattern of methylmalonyl-CoA mutase and propionyl-CoA carboxylase α-subunit in developing and adult rat brain. Neuroscience 164, 578–587 [DOI] [PubMed] [Google Scholar]
- 18. Kölker S., and Okun J. G. (2005) Methylmalonic acid: an endogenous toxin? Cell. Mol. Life Sci. 62, 621–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perry T. L., Urquhart N., and Hansen S. (1977) Studies of the glycine cleavage enzyme system in brain from infants with glycine encephalopathy. Pediat. Res. 11, 1192–1197 [DOI] [PubMed] [Google Scholar]
- 20. Matsui S. M., Mahoney M. J., and Rosenberg L. E. (1983) The natural history of the inherited methylmalonic acidemias. N. Engl. J. Med. 308, 857–861 [DOI] [PubMed] [Google Scholar]
- 21. Kölker S., Garcia-Cazorla A., Valayannopoulos V., Lund A. M., Burlina A. B., Sykut-Cegielska J., Wijburg F. A., Teles E. L., Zeman J., Dionisi-Vici C., Barić I., Karall D., Augoustides-Savvopoulou P., Aksglaede L., Arnoux J. B., et al. (2015) The phenotypic spectrum of organic acidurias and urea cycle disorders: Part 1: the initial presentation. J. Inherit. Metab. Dis. 38, 1041–1057 [DOI] [PubMed] [Google Scholar]
- 22. Malvagia S., Haynes C. A., Grisotto L., Ombrone D., Funghini S., Moretti E., McGreevy K. S., Biggeri A., Guerrini R., Yahyaoui R., Garg U., Seeterlin M., Chace D., De Jesus V. R., and la Marca G. (2015) Heptadecanoylcarnitine (C17) a novel candidate biomarker for newborn screening of propionic and methylmalonic acidemias. Clin. Chim. Acta 450, 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wendel U., Baumgartner R., van der Meer S. B., and Spaapen L. J. (1991) Accumulation of odd-numbered long-chain fatty acids in fetuses and neonates with inherited disorders of propionate metabolism. Pediat. Res. 29, 403–405 [DOI] [PubMed] [Google Scholar]
- 24. Schwartz G. J., Muñoz A., Schneider M. F., Mak R. H., Kaskel F., Warady B. A., and Furth S. L. (2009) New equations to estimate GFR in children with CKD. J. Am. Soc. Nephrol. 20, 629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicolaides P., Leonard J., and Surtees R. (1998) Neurological outcome of methylmalonic acidaemia. Arch. Dis. Child. 78, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kölker S., Burgard P., Sauer S. W., and Okun J. G. (2013) Current concepts in organic acidurias: understanding intra- and extracerebral disease manifestation. J. Inherit. Metab. Dis. 36, 635–644 [DOI] [PubMed] [Google Scholar]
- 27. Morath M. A., Okun J. G., Müller I. B., Sauer S. W., Hörster F., Hoffmann G. F., and Kölker S. (2008) Neurodegeneration and chronic renal failure in methylmalonic aciduria: a pathophysiological approach. J. Inherit. Metab. Dis. 31, 35–43 [DOI] [PubMed] [Google Scholar]
- 28. de Keyzer Y., Valayannopoulos V., Benoist J. F., Batteux F., Lacaille F., Hubert L., Chrétien D., Chadefeaux-Vekemans B., Niaudet P., Touati G., Munnich A., and de Lonlay P. (2009) Multiple OXPHOS deficiency in the liver, kidney, heart, and skeletal muscle of patients with methylmalonic aciduria and propionic aciduria. Pediat. Res. 66, 91–95 [DOI] [PubMed] [Google Scholar]
- 29. Okun J. G., Hörster F., Farkas L. M., Feyh P., Hinz A., Sauer S., Hoffmann G. F., Unsicker K., Mayatepek E., and Kölker S. (2002) Neurodegeneration in methylmalonic aciduria involves inhibition of complex II and the tricarboxylic acid cycle, and synergistically acting excitotoxicity. J. Biol. Chem. 277, 14674–14680 [DOI] [PubMed] [Google Scholar]
- 30. Häberle J., Boddaert N., Burlina A., Chakrapani A., Dixon M., Huemer M., Karall D., Martinelli D., Crespo P. S., Santer R., Servais A., Valayannopoulos V., Lindner M., Rubio V., and Dionisi-Vici C. (2012) Suggested guidelines for the diagnosis and management of urea cycle disorders. Orphanet J. Rare Dis. 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malyszko J., Bachorzewska-Gajewska H., Sitniewska E., Malyszko J. S., Poniatowski B., and Dobrzycki S. (2008) Serum neutrophil gelatinase-associated lipocalin as a marker of renal function in non-diabetic patients with stage 2–4 chronic kidney disease. Renal Failure 30, 625–628 [DOI] [PubMed] [Google Scholar]
- 32. Berard J. L., Zarruk J. G., Arbour N., Prat A., Yong V. W., Jacques F. H., Akira S., and David S. (2012) Lipocalin 2 is a novel immune mediator of experimental autoimmune encephalomyelitis pathogenesis and is modulated in multiple sclerosis. Glia 60, 1145–1159 [DOI] [PubMed] [Google Scholar]
- 33. Ferreira A. C., Dá Mesquita S., Sousa J. C., Correia-Neves M., Sousa N., Palha J. A., and Marques F. (2015) From the periphery to the brain: Lipocalin-2, a friend or foe? Prog. Neurobiol. 131, 120–136 [DOI] [PubMed] [Google Scholar]
- 34. Naudé P. J., Nyakas C., Eiden L. E., Ait-Ali D., van der Heide R., Engelborghs S., Luiten P. G., De Deyn P. P., den Boer J. A., and Eisel U. L. (2012) Lipocalin 2: novel component of proinflammatory signaling in Alzheimer's disease. FASEB J. 26, 2811–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fowler B., Leonard J. V., and Baumgartner M. R. (2008) Causes of and diagnostic approach to methylmalonic acidurias. J. Inherit. Metab. Dis. 31, 350–360 [DOI] [PubMed] [Google Scholar]
- 36. Hillman R. E., and Otto E. F. (1974) Inhibition of glycine-serine interconversion in cultured human fibroblasts by products of isoleucine catabolism. Pediat. Res. 8, 941–945 [DOI] [PubMed] [Google Scholar]
- 37. Oberholzer V. G., Levin B., Burgess E. A., and Young W. F. (1967) Methylmalonic aciduria. An inborn error of metabolism leading to chronic metabolic acidosis. Arch. Dis. Child. 42, 492–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coker M., de Klerk J. B., Poll-The B. T., Huijmans J. G., and Duran M. (1996) Plasma total odd-chain fatty acids in the monitoring of disorders of propionate, methylmalonate and biotin metabolism. J. Inherit. Metab. Dis. 19, 743–751 [DOI] [PubMed] [Google Scholar]
- 39. Merinero B., Pérez B., Pérez-Cerdá C., Rincón A., Desviat L. R., Martínez M. A., Sala P. R., García M. J., Aldamiz-Echevarría L., Campos J., Cornejo V., Del Toro M., Mahfoud A., Martínez-Pardo M., Parini R., et al. (2008) Methylmalonic acidaemia: examination of genotype and biochemical data in 32 patients belonging to mut, cblA or cblB complementation group. J. Inherit. Metab. Dis. 31, 55–66 [DOI] [PubMed] [Google Scholar]
- 40. Lempp T. J., Suormala T., Siegenthaler R., Baumgartner E. R., Fowler B., Steinmann B., and Baumgartner M. R. (2007) Mutation and biochemical analysis of 19 probands with mut0 and 13 with mut- methylmalonic aciduria: identification of seven novel mutations. Mol. Genet. Metab. 90, 284–290 [DOI] [PubMed] [Google Scholar]
- 41. Ziesch B., Weigel J., Thiele A., Mütze U., Rohde C., Ceglarek U., Thiery J., Kiess W., and Beblo S. (2012) Tetrahydrobiopterin (BH4) in PKU: effect on dietary treatment, metabolic control, and quality of life. J. Inherit. Metab. Dis. 35, 983–992 [DOI] [PubMed] [Google Scholar]
- 42. Turgeon C. T., Magera M. J., Cuthbert C. D., Loken P. R., Gavrilov D. K., Tortorelli S., Raymond K. M., Oglesbee D., Rinaldo P., and Matern D. (2010) Determination of total homocysteine, methylmalonic acid, and 2-methylcitric acid in dried blood spots by tandem mass spectrometry. Clin. Chem. 56, 1686–1695 [DOI] [PubMed] [Google Scholar]
- 43. Scheja L., Toedter K., Mohr R., Niederfellner G., Michael M. D., Meissner A., Schoettler A., Pospisil H., Beisiegel U., and Heeren J. (2008) Liver TAG transiently decreases while PL n-3 and n-6 fatty acids are persistently elevated in insulin resistant mice. Lipids 43, 1039–1051 [DOI] [PubMed] [Google Scholar]
- 44. Othman A., Saely C. H., Muendlein A., Vonbank A., Drexel H., von Eckardstein A., and Hornemann T. (2015) Plasma C20-sphingolipids predict cardiovascular events independently from conventional cardiovascular risk factors in patients undergoing coronary angiography. Atherosclerosis 240, 216–221 [DOI] [PubMed] [Google Scholar]