Abstract
The signaling pathway downstream of stimulation of receptor activator of nuclear factor κB (RANK) by RANK ligand is crucial for osteoclastogenesis. RANK recruits TNF receptor-associated factor 6 (TRAF6) to TRAF6-binding sites (T6BSs) in the RANK cytoplasmic tail (RANKcyto) to trigger downstream osteoclastogenic signaling cascades. RANKcyto harbors an additional highly conserved domain (HCR) that also activates crucial signaling during RANK-mediated osteoclastogenesis. However, the functional cross-talk between T6BSs and the HCR in the RANK signaling complex remains unclear. To characterize the cross-talk between T6BSs and the HCR, we screened TRAF6-interacting proteins using a proteomics approach. We identified Vav3 as a novel TRAF6 binding partner and evaluated the functional importance of the TRAF6-Vav3 interaction in the RANK signaling complex. We demonstrated that the coiled-coil domain of TRAF6 interacts directly with the Dbl homology domain of Vav3 to form the RANK signaling complex independent of the TRAF6 ubiquitination pathway. TRAF6 is recruited to the RANKcyto mutant, which lacks T6BSs, via the Vav3 interaction; conversely, Vav3 is recruited to the RANKcyto mutant, which lacks the IVVY motif, via the TRAF6 interaction. Finally, we determined that the TRAF6-Vav3 interaction resulting from cross-talk between T6BSs and the IVVY motif in RANKcyto enhances downstream NF-κB, MAPK, and NFATc1 activation by further strengthening TRAF6 signaling, thereby inducing RANK-mediated osteoclastogenesis. Thus, Vav3 is a novel TRAF6 interaction partner that functions in the activation of cooperative signaling between T6BSs and the IVVY motif in the RANK signaling complex.
Keywords: mitogen-activated protein kinase (MAPK), NFAT transcription factor, osteoclast, osteoporosis, signal transduction
Introduction
The integrity of the skeletal system is maintained by the process of bone remodeling, in which old and damaged bone is continuously replaced with new bone through the balanced action of bone-resorbing osteoclasts (OCs)3 and bone-forming osteoblasts (1–3). As the only cells with bone-resorbing activity, OCs are clinically important in bone-related diseases; accelerated bone destruction by OCs results in pathological bone loss associated with rheumatoid arthritis, multiple myeloma, metastatic cancer, and osteoporosis, whereas impaired function of OCs leads to osteopetrotic disorders (2, 4).
OCs are multinucleated giant cells that are formed by the fusion of monocyte/macrophage lineage precursors through the process of OC differentiation or osteoclastogenesis (2). The signaling pathways of receptor activator of nuclear factor κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF) play key roles in osteoclastogenesis (2, 5). Mice lacking either RANK or RANKL exhibit severe osteopetrotic phenotypes, highlighting the importance of RANK-RANKL signaling as a crucial regulator of OC differentiation, activation, and survival (6, 7). RANK-RANKL signaling is initiated primarily by the recruitment of cytoplasmic TNF receptor-associated factors (TRAFs) that trigger the activation of signaling cascades downstream of adaptors/kinases, such as nuclear factor κB (NF-κB) essential modulator (NEMO), inhibitor of IκB kinases (IKKs), Akt kinase, and MAPKs, including p38, JNK, and ERK (1, 2). The downstream consequence of RANK-RANKL signaling is the activation of osteoclastogenic transcription factors, including NF-κB, activator protein 1 (AP-1), cAMP response element-binding protein (CREB), and nuclear factor of activated T cell c1 (NFATc1), which induce osteoclastogenic genes, such as Trap, Dcstamp, Atp6v0d2, and Oscar; these genes code for the proteins tartrate-resistant acid phosphatase (TRAP), dendritic cell-specific transmembrane protein (DC-STAMP), V-ATPase subunit d2 (Atp6v0d2), and OC-associated receptor (OSCAR), respectively (1–3).
TRAF6 is a TRAF adaptor molecule recruited to the RANK cytoplasmic tail (RANKcyto) signaling complexes and is critical for activating downstream targets. Similar to RANKL- and RANK-deficient mice, TRAF6-deficient mice exhibit severe osteopetrosis caused by a defect in OC activity (8). TRAF6 contains an N-terminal RING domain and zinc finger motifs, a central coiled-coil (CC) domain, and a conserved C-terminal TRAF domain. The RING domain has E3 ubiquitin (Ub) ligase activity in conjunction with the E2 Ub-conjugating enzyme Ubc13/Uev1A, which promotes Lys63-linked autoubiquitination (9). Lys63-linked TRAF6 autoubiquitination induced by RANKL stimulation leads to the recruitment and activation of downstream adaptors/kinases, including TGF-β-activated kinase 1 (TAK1), TAK1-binding proteins, NEMO, IKKs, and MAPKs (10, 11). Upon RANKL stimulation, the C-terminal TRAF domain of TRAF6 directly interacts with three TRAF6 binding sites (T6BS-I, -II, and -III), which are defined by a PXEXX[Ar/Ac] consensus sequence, where Ar is an aromatic residue, and Ac is an acidic residue in RANKcyto (2). Triple-mutant analyses have revealed a crucial role of the T6BSs in RANK-mediated osteoclastogenesis (12, 13). T6BS-I has the highest binding affinity for TRAF6, and T6BS-II and -III exhibit 10-fold lower binding affinity (12, 14). Despite different binding affinities for TRAF6, all T6BSs in RANKcyto have the potential to induce osteoclastogenesis via signaling (13). However, osteoclastogenesis is not completely disrupted in TRAF6-deficient mice (8, 15), suggesting that RANKcyto has additional motifs that are functionally independent of T6BS activity and that recruit specific adaptor/kinase molecules to promote osteoclastogenesis.
Studies of deletion mutations of RANKcyto have revealed that RANKcyto harbors a unique domain named the highly conserved domain in RANK (HCR, amino acids 487–546) that functions as a crucial regulatory motif in RANK-mediated osteoclastogenesis (16, 17). Upon RANKL stimulation, HCR recruits additional adaptor/kinase molecules, such as Vav3, Grb-2-associated binder 2 (Gab2), and phospholipase C γ2 (PLCγ2), to induce the late phase of RANK-mediated signaling (16, 18, 19). Fine deletion analyses of HCR further demonstrated that the IVVY motif (amino acids 535–538) in HCR plays a pivotal role in late phase signaling of RANK-mediated osteoclastogenesis by recruiting Vav3 to the RANK signaling complex (16, 18, 20, 21). Vav3 is a Rho-guanine nucleotide exchange factor belonging to the Vav family, which shares a common domain structure comprising a calponin homology domain, an acidic domain (AD), a Dbl homology (DH) domain, a pleckstrin homology (PH) domain, a cysteine-rich (C1) domain, a Src homology 2 domain (SH2), and a Src homology 3 (SH3) domain (22, 23). Interestingly, a direct link between TRAF6 and HCR-recruited molecules, such as Vav3, Gab2, and PLCγ2, in the RANK signaling complex has been proposed (18, 19). However, how T6BSs and HCR function in concert to regulate cooperative signaling networks between TRAF6 and HCR-recruited Vav3, Gab2, and PLCγ2 or unidentified additional adaptors/kinases during RANK-mediated osteoclastogenesis remains to be elucidated at the molecular level.
In this study, we attempted to identify novel TRAF6-interacting proteins using a proteomics approach to investigate the regulation of RANK-mediated osteoclastogenesis by TRAF6. We determined that TRAF6 interacts directly with Vav3 in the RANK signaling complex, thereby enhancing osteoclastogenesis. The DH domain of Vav3 is required for the interaction with the CC domain of TRAF6. TRAF6 recruits Vav3 and vice versa by interacting directly with the RANK signaling complex independent of its binding motifs in RANKcyto. Finally, we demonstrated that the TRAF6-Vav3 interaction in the RANK signaling complex enhances OC differentiation and function. Thus, we have discovered a new role of Vav3 as a novel TRAF6-interacting partner in RANK-mediated osteoclastogenesis.
Results
TRAF6 Interacts with Vav3
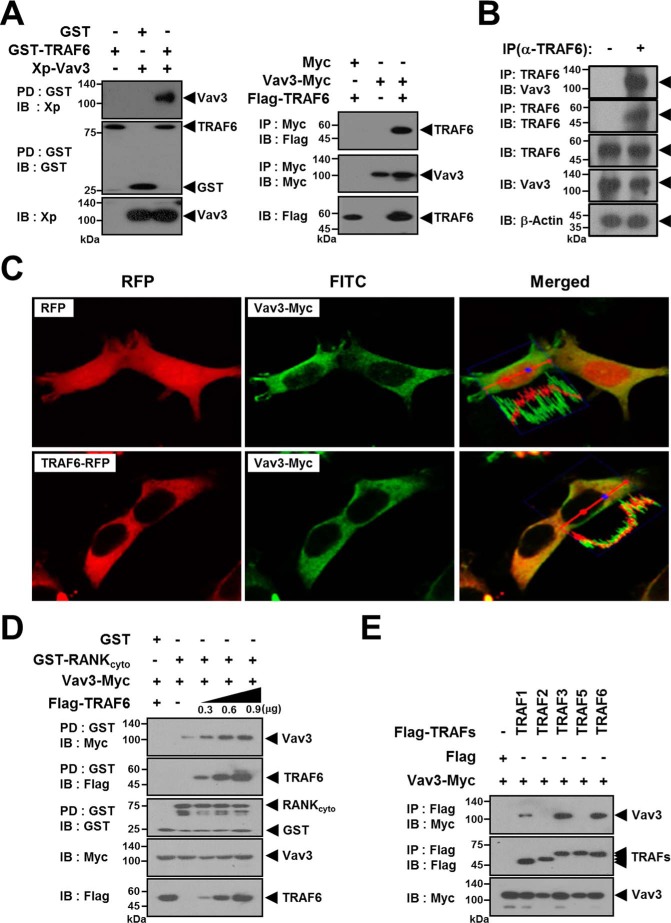
To identify novel protein partners that interact with TRAF6, lysates of FLAG-TRAF6-expressing 293T cells were immunoprecipitated with FLAG beads, and the TRAF6-bound protein band at ∼90 kDa was identified by nanoscale liquid chromatography coupled with tandem mass spectrometry (Table 1). The interaction of TRAF6 and Vav3 was confirmed by a GST bead pulldown (PD) assay and anti-Myc immunoprecipitation (IP) assay (Fig. 1A). To further confirm the TRAF6-Vav3 interaction at the endogenous protein level, we conducted an endogenous TRAF6 IP assay with an anti-TRAF6 antibody in bone marrow-derived macrophages (BMMs) and observed the interaction between endogenous TRAF6 and Vav3 (Fig. 1B). Consistent with these results, TRAF6 co-localized with Vav3 in subcellular localization analysis (Fig. 1C). Next, we examined the effect of the TRAF6-Vav3 interaction on the RANK signaling complex. The GST-PD assay with GST-RANKcyto revealed that Vav3 recruitment to GST-RANKcyto was increased by TRAF6 expression in a dose-dependent manner (Fig. 1D). Because RANK can recruit TRAFs 1, 2, 3, 5, and 6 (2), we also examined whether Vav3 interacts with other TRAF family members. We detected interaction bands for TRAFs 1, 3, and 6 but not TRAFs 2 or 5 in an anti-FLAG IP assay (Fig. 1E).
TABLE 1.
List of proteins identified as TRAF6-interacting candidates
| Protein name | Protein symbol | Gene ID | Protein function |
|---|---|---|---|
| ATP-dependent RNA helicase DDX3X | DDX3X | 1654 | Translation regulation |
| ATP-dependent RNA helicase DDX17 | DDX17 | 10521 | Translation regulation |
| Carboxypeptidase N subunit 2 | CPN2 | 1370 | Plasma protein |
| Cysteinyl-tRNA synthetase | CARS | 833 | tRNA regulation |
| Elongation factor 1-α1 | EEF1A-1 | 1915 | Translation regulation |
| FUS RNA-binding protein | FUS | 2521 | RNA splicing |
| Keratin 1 | KRT1 | 3848 | Keratin formation |
| Keratin 2 | KRT2 | 3849 | Keratin formation |
| Keratin 3 | KRT3 | 3850 | Keratin formation |
| Keratin 5 | KRT5 | 3852 | Keratin formation |
| Keratin 9 | KRT9 | 3857 | Keratin formation |
| Keratin 10 | KRT10 | 3858 | Keratin formation |
| Keratin 12 | KRT12 | 3859 | Keratin formation |
| Neurofibromin 1 | NF1 | 4763 | Neuronal differentiation |
| SH3 and PX domain-containing protein 2A | SH3PXD2A | 9644 | Invadopodia formation |
| ST3 β-galactoside α2,3-sialyltransferase 1 | ST3GAL1 | 6482 | Protein sialylation |
| Ubiquitin | UBC | 7316 | Signal transduction |
| Vav3 | VAV3 | 10451 | Cytoskeleton formation |
| Zinc finger protein 326 | ZNF326 | 284695 | Unknown |
| Zw10 kinetochore protein | ZW10 | 9183 | Cell division |
FIGURE 1.
TRAF6 interacts with Vav3. A, TRAF6 interacts with Vav3 in vitro. Left panels, 293T cells were transfected with GST-TRAF6 (1.0 μg) with or without Xp-Vav3 (1.0 μg). The cell lysates were normalized for total protein content. GST-TRAF6 was pulled down (PD) using GST beads, and the PD GST-TRAF6 was visualized via anti-GST (B-14) immunoblotting (IB, middle panel). Vav3 bound to the TRAF6 was visualized via anti-Xp IB (top panel). The level of Vav3 expression was detected via anti-Xp IB (bottom panel). GST alone was used as a control. Right panels, Vav3-Myc was immunoprecipitated (IP) using anti-Myc beads. TRAF6 bound to Vav3 was visualized via anti-FLAG (M2) IB (top panel). B, identification of the endogenous TRAF6-Vav3 interaction. Endogenous TRAF6 was IP with an anti-(α)-TRAF6 (D-10) antibody. The cell lysates were normalized for total protein content. Protein bound to TRAF6 was visualized via anti-Vav3 IB. β-Actin was used as a loading control. C, subcellular localization of TRAF6 and Vav3. 293T cells were transfected with Vav3-Myc and TRAF6-RFP. The transfected cells were immunostained with an anti-Myc (9E10) antibody and a FITC-labeled secondary antibody, and the subcellular localization of TRAF6 (red) and Vav3 (green) was visualized by confocal microscopy (magnification, ×800). D, TRAF6 interacts with Vav3 in RANKcyto. 293T cells were transfected with GST-RANKcyto (0.8 μg) and Vav3-Myc (0.4 μg) with or without FLAG-TRAF6 (0.3–0.9 μg). The cell lysates were normalized for total protein content. Proteins bound to GST-RANKcyto were visualized via anti-Myc IB (top panel) or anti-FLAG IB (second panel). E, identification of Vav3 interactions with other TRAF members recruited to RANKcyto. 293T cells were co-transfected with FLAG-TRAFs (1.0 μg) and Vav3-Myc (1.0 μg). The cell lysates were normalized for total protein content. Vav3 bound to TRAF was visualized via anti-Myc IB (top panel). All experiments were performed at least three times with similar results.
The DH Domain of Vav3 Physically Interacts with the CC Domain of TRAF6
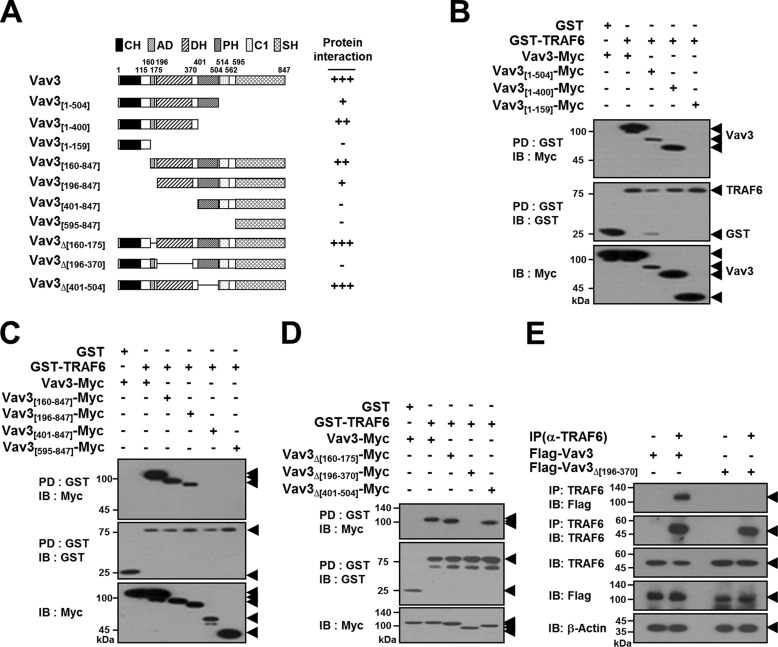
To identify the specific domain of Vav3 that interacts with TRAF6, we constructed expression plasmids for Vav3 deletion mutants lacking the N- and C-terminal regions (Fig. 2A). The GST-TRAF6 PD assay of the C-terminal deletion mutants of Vav3 revealed that TRAF6 bound to the Vav3[1–504] and Vav3[1–400] mutants but not to Vav3[1–159], indicating that the region between residues 160 and 847 in Vav3 is involved in the TRAF6 interaction (Fig. 2B). Among the N-terminal deletion mutants of Vav3, TRAF6 bound to the Vav3[160–847] and Vav3[196–847] mutants but not to Vav3[401–847] or Vav3[595–847], indicating that the region between residues 1 and 400 in Vav3 is required for the TRAF6 interaction (Fig. 2C). Collectively, these results indicate that the region between residues 160 and 400, including the AD and DH domains of Vav3, may be involved in the TRAF6 interaction. To further determine the precise domain of Vav3 that interacts with TRAF6, we constructed internal deletion mutants of Vav3 (Fig. 2A). In the GST-PD assay, TRAF6 interacted with the Vav3Δ[160–175] and Vav3Δ[401–504] mutants but not Vav3Δ[196-370] (Fig. 2D). To further confirm the specific domain of Vav3 interacting with TRAF6 in BMMs, we conducted an endogenous TRAF6 IP assay with an anti-TRAF6 antibody in BMMs, and we observed the specific interaction between the DH domain of Vav3 and endogenous TRAF6 (Fig. 2E). As summarized in Fig. 2A, these results indicate that the DH domain of Vav3 containing residues 196–370 is required for the TRAF6 interaction.
FIGURE 2.
TRAF6 interacts with the DH domain of Vav3. A, schematic diagram of Vav3 mutants. The CH domain, AD, DH domain, PH domain, C1 domain, and SH2/3 domain are indicated. The internal deletion is shown using lines. The number of amino acid residues is shown. The relative interaction of Vav3 mutants with TRAF6 is summarized. B, analysis of C-terminal deletion mutants of Vav3 interacting with TRAF6. 293T cells were co-transfected with GST-TRAF6 (1.0 μg) and Vav3-Myc (1.0 μg) or its mutants (1.0 μg). The cell lysates were normalized for total protein content. GST-TRAF6 was pulled down (PD) using GST beads, and the PD GST-TRAF6 was visualized by anti-GST (B-14) immunoblotting (IB, middle panel). Vav3 bound to TRAF6 was visualized via anti-Myc (9E10) IB (top panel). The level of Vav3 expression was detected by anti-Myc IB (bottom panel). C, analysis of N-terminal deletion mutants of Vav3 interacting with TRAF6. D, analysis of internal deletion mutants of Vav3 interacting with TRAF6. E, identification of the domain of Vav3 interacting with the endogenous TRAF6. BMMs cultured with M-CSF for 2 days were infected by retroviruses harboring Vav3 expression and puromycin-resistant cassettes. Vav3-expressing BMMs were further cultured with M-CSF and puromycin for 2 days. The cell lysates were normalized for total protein content. Endogenous TRAF6 was immunoprecipitated with an anti-(α)-TRAF6 (D-10) antibody. Protein bound to TRAF6 was visualized via anti-FLAG (M2) immunoblotting. β-Actin was used as a loading control. All experiments were performed at least three times with similar results.
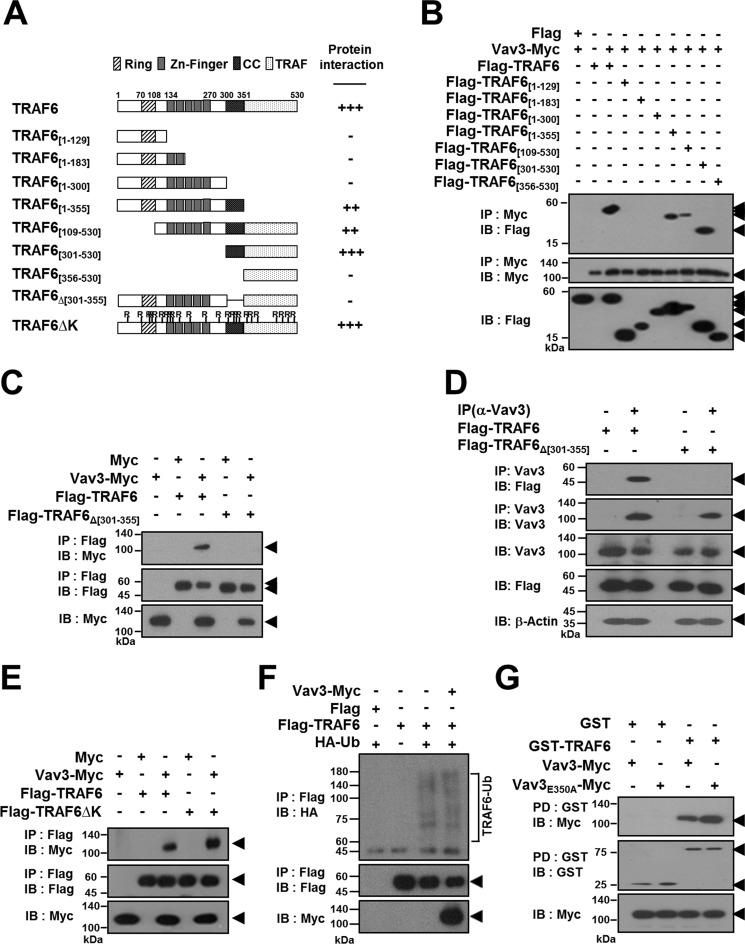
To identify the specific domain in TRAF6 that interacts with Vav3, we generated a series of TRAF6 deletion mutants lacking the N-terminal, C-terminal, or internal regions (Fig. 3A). In an anti-Myc IP assay, Vav3 interacted only with TRAF6 [1–355], TRAF6[109–530], and TRAF6[301–530], indicating that the region between residues 301 and 355 containing the CC domain of TRAF6 is required for the Vav3 interaction (Fig. 3B). Moreover, in the internal deletion mutant assay, we did not observe any protein-protein interaction of the TRAF6Δ[301–355] mutant, which harbors an internal deletion of the CC domain of TRAF6 (Fig. 3C). To further confirm the specific domain of TRAF6 interacting with Vav3 in BMMs, we conducted an endogenous Vav3 IP assay with an anti-Vav3 antibody in BMMs, and we observed the specific interaction between the CC domain of TRAF6 and endogenous Vav3 (Fig. 3D). As summarized in Fig. 3A, these results indicate that residues 301–355 of TRAF6 containing the CC domain are required for the Vav3 interaction.
FIGURE 3.
The CC domain of TRAF6 interacts with Vav3. A, schematic diagram of TRAF6 mutants. The RING finger domain (Ring), zinc finger domain (Zn-Finger), coiled-coil (CC) structure, and TRAF domain are indicated. The internal deletion is shown using lines. The number of amino acid residues is shown. The relative interaction of TRAF6 mutants with Vav3 is summarized. B, mapping the interaction domain of TRAF6 with Vav3 by deletion mutation. 293T cells were co-transfected with Vav3-Myc (1.0 μg) and FLAG-TRAF6 (1.0 μg) or its mutants (1.0 μg). The cell lysates were normalized for total protein content. Vav3-Myc was immunoprecipitated (IP) using anti-Myc beads, and the IP Vav3 was visualized by anti-Myc (9E10) immunoblotting (IB, middle panel). TRAF6 bound to Vav3 was visualized via anti-FLAG (M2) IB (top panel). The expression level of TRAF6 mutants was detected via anti-FLAG IB (bottom panel). C, interaction of the CC domain of TRAF6 with Vav3. D, identification of the domain of TRAF6 interacting with the endogenous Vav3. BMMs cultured with M-CSF for 2 days were infected by a retroviruses harboring TRAF6 expression and puromycin-resistant cassettes. TRAF6-expressing BMMs were further cultured with M-CSF and puromycin for 2 days. The cell lysates were normalized for total protein content. Endogenous Vav3 was IP with an anti-(α)-Vav3 (K-19) antibody. Protein bound to Vav3 was visualized via anti-FLAG (M2) IB. β-Actin was used as a loading control. E, interaction of the Lys-deficient TRAF6 mutant with Vav3. F, the effect of Vav3 interaction on the TRAF6 E3 Ub ligase activity. 293T cells were transfected with FLAG-TRAF6 (0.5 μg) and HA-Ub (0.5 μg) with or without Vav3-Myc (0.5 μg). FLAG-TRAF6 was IP using FLAG beads, and TRAF6 ubiquitination (TRAF6-Ub) was visualized via anti-HA IB. Vector alone (FLAG) was used as a negative control. G, interaction of the Vav3E350A mutant with TRAF6. 293T cells were co-transfected with GST-TRAF6 (1.0 μg) and Vav3-Myc (1.0 μg) or Vav3E350A-Myc (1.0 μg). The cell lysates were normalized for total protein content. GST-TRAF6 was pulled down (PD) using GST beads, and the PD GST-TRAF6 was visualized via anti-GST (B-14) IB (middle panel). Vav3 bound to TRAF6 was visualized via anti-Myc IB (top panel). The level of Vav3 expression was detected via anti-Myc IB (bottom panel). All experiments were performed at least three times with similar results.
The CC domain of TRAF6 is essential for autoubiquitination and NF-κB activation (24, 25). Thus, we next examined whether the TRAF6-Vav3 interaction is involved in a Ub-dependent pathway. The binding assay results for Lys-deficient TRAF6 (TRAF6ΔK) demonstrated that the TRAF6-Vav3 interaction is not dependent on TRAF6 ubiquitination (Fig. 3E). Moreover, the E3 Ub ligase activity of TRAF6 was not affected by Vav3 expression (Fig. 3F). Thus, we conclude that the TRAF6-Vav3 interaction is not dependent on the TRAF6 ubiquitination pathway.
Because earlier studies suggested that Vav3 is recruited to RANKcyto (18, 20), we next attempted to identify the specific domain of Vav3 that is recruited to RANKcyto. However, no specific domain of Vav3 interacted with the RANKcyto in the GST-RANKcyto PD assay, indicating that the entire protein or a broad range of Vav3 domains is required for the interaction with RANKcyto (data not shown). TRAF6 interacts directly with consensus T6BSs (PXEXX[Ar/Ac]) in RANKcyto through its C-terminal TRAF domain (2). We identified one similar sequence with a consensus T6BS at residues 348–353 in the DH domain of Vav3. To assess the involvement of this putative T6BS sequence in the TRAF6-Vav3 interaction, we constructed a Vav3E350A point mutant (mutation of the Glu residue at position 350 in Vav3 to Ala). The Vav3-TRAF6 interaction was not affected by the Vav3E350A point mutation in the GST-PD assay (Fig. 3G). Moreover, the TRAF6[356–530] mutant harboring the C-terminal TRAF domain that binds to T6BSs in RANKcyto did not interact with Vav3 (Fig. 3B). Taken together, these results indicate that the DH domain of Vav3 physically interacts with the CC domain of TRAF6 and that the interaction domain between TRAF6 and Vav3 does not overlap with the RANKcyto-recruited domains of the proteins.
Vav3 Enhances Osteoclastogenesis through the Activation of TRAF6 Signaling
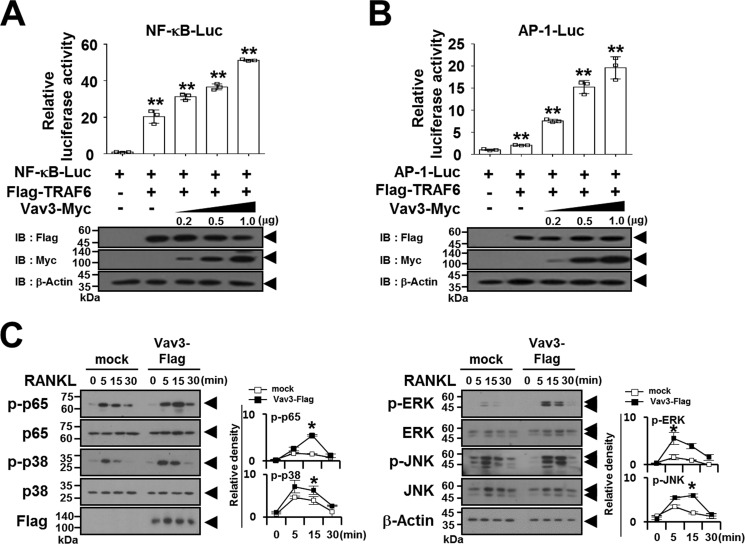
Because TRAF6 functions as a crucial adaptor molecule in the RANK signaling complex that activates downstream targets of NF-κB and MAPKs (2), we determined whether the TRAF6-Vav3 interaction enhances the RANK-mediated signaling pathway. In a reporter assay, we observed that the transcription activities of TRAF6-induced NF-κB and AP-1 were significantly enhanced by Vav3 expression in a dose-dependent manner (Fig. 4, A and B). Consistent with these results, upon RANKL stimulation, the phosphorylation of NF-κB p65 and MAPKs (p38, ERK, and JNK) was induced by Vav3 expression (Fig. 4C).
FIGURE 4.
Vav3 expression activates the TRAF6 downstream signaling pathway. A, the effect of Vav3 expression on TRAF6-induced NF-κB activation. 293T cells were transfected with FLAG-TRAF6 (0.15 μg), Vav3-Myc (0.2–1.0 μg), and reporters. At 24 h post-transfection, the cells were analyzed using luciferase assays. The cell lysates were normalized for total protein content. The level of Vav3 or TRAF6 expression was detected via anti-Myc (9E10) immunoblotting (IB) or anti-FLAG (M2) IB. β-Actin was used as a loading control. **, p < 0.01. B, the effect of Vav3 expression on TRAF6-induced AP-1 activation. C, the effect of Vav3 expression on RANKL-induced signaling pathways. Puromycin-selected Vav3-expressing BMMs were cultured for 2 h under serum-free conditions. The BMMs were stimulated with RANKL (100 ng/ml) for the indicated times and subsequently analyzed via IB with antibodies that recognized phosphorylated and total NF-κB p65 (Ser536), p38 (Thr180/Tyr182), ERK (Thr202/Thr204), and JNK (Thr183/Tyr185). The cell lysates were normalized for total protein content. The level of Vav3 expression was detected via anti-FLAG IB. The relative level of phosphorylated forms was calculated after normalization to total protein input (right). *, p < 0.05. All experiments were performed at least three times with similar results.
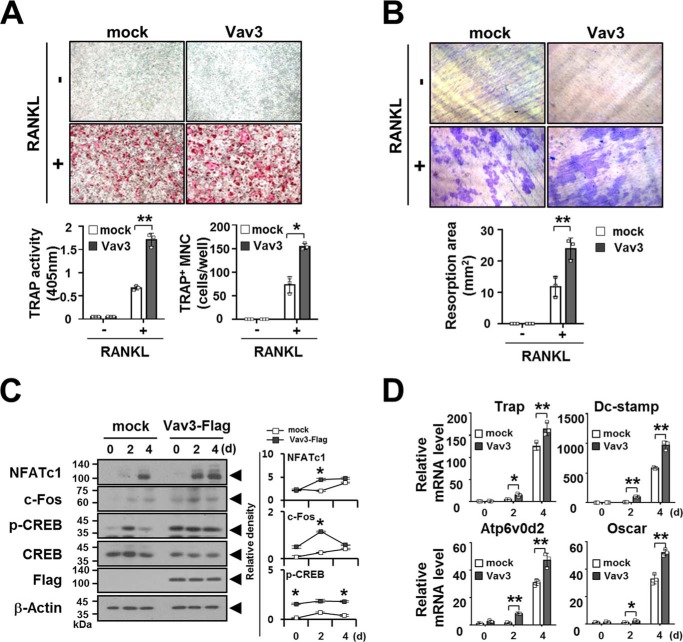
The activation of NF-κB and MAPKs is a key signaling pathway in osteoclastogenesis (2, 3). Thus, we next examined whether Vav3 expression is involved in osteoclastogenesis using a retroviral gene transfer system. Vav3-transduced BMMs were selected with puromycin, and puromycin-resistant BMMs were differentiated into OCs with M-CSF and RANKL for 4 days. TRAP activity and the number of TRAP-positive multinucleated OCs (TRAP+ MNCs) in Vav3-expressed OCs were increased significantly (1.7 ± 0.1 and 154.7 ± 6.7, respectively) compared with the mock control (0.67 ± 0.05 and 73.3 ± 17.4, respectively) (Fig. 5A). Similar results were obtained in a bone resorption assay, with a 2.0-fold increase in the resorption area after Vav3 expression (24.0 ± 3.4) compared with the mock control (11.9 ± 3.2) (Fig. 5B). Consistent with these results, Vav3 expression enhanced the expression or activation of the osteoclastogenic transcription factors NFATc1, c-Fos, and CREB (Fig. 5C). The activation of the transcription factors NFATc1, c-Fos, and CREB is crucial for the expression of OC marker genes (2, 3). Thus, we also examined the expression of OC differentiation markers. The gene expression of Trap, Dcstamp, Atp6v0d2, and Oscar was significantly increased by Vav3 expression (164.3 ± 14.4, 968.4 ± 67.9, 47.1 ± 5.1, and 52.1 ± 2.3, respectively) compared with the mock control (125.2 ± 8.8, 584.8 ± 11.3, 30.8 ± 2.2, and 32.7 ± 3.6, respectively) at day 4 (Fig. 5D).
FIGURE 5.
Vav3 expression enhances osteoclastogenesis. A, the effect of Vav3 expression on OC differentiation. BMMs cultured with M-CSF (150 ng/ml) for 2 days were transduced with retroviral supernatants harboring Vav3 expression and puromycin-resistant cassettes and were further cultured with puromycin and M-CSF (150 ng/ml) for 2 days. Puromycin-selected Vav3-expressing BMMs were differentiated into OCs with RANKL (100 ng/ml) and M-CSF (50 ng/ml) for 4 days. The OCs were photographed (top panel, original magnification, ×50) after TRAP staining. The OCs were analyzed using the TRAP solution assay (bottom left panel). The number of TRAP+ MNCs (at least three nuclei) was counted (bottom right panel). mock, empty vector control. *, p < 0.05; **, p < 0.01. B, the effect of Vav3 expression on resorption pit formation. Resorption pits were visualized (top panel, original magnification, ×50). The summarized data from the resorption pit assays are shown in the bottom panel. C, the effect of Vav3 expression on the activation of osteoclastogenic factors. OCs were differentiated from BMMs with RANKL (100 ng/ml) and M-CSF (50 ng/ml) for the indicated times and subsequently analyzed via immunoblotting (IB) with antibodies that recognized phosphorylated or total NFATc1 (7A6), c-Fos (D-1), and CREB (Ser133). The cell lysates were normalized for total protein content. The level of Vav3 expression was detected via anti-FLAG IB. The relative protein level was calculated after normalization to β-actin protein input (right). The relative level of phosphorylated CREB was calculated after normalization to total CREB protein input (right). β-Actin was used as the loading control. D, the effect of Vav3 on the expression of osteoclastogenic markers. Total RNA was prepared from OCs and subjected to real time PCR analysis. The data were normalized to β-actin. All experiments were performed at least three times with similar results.
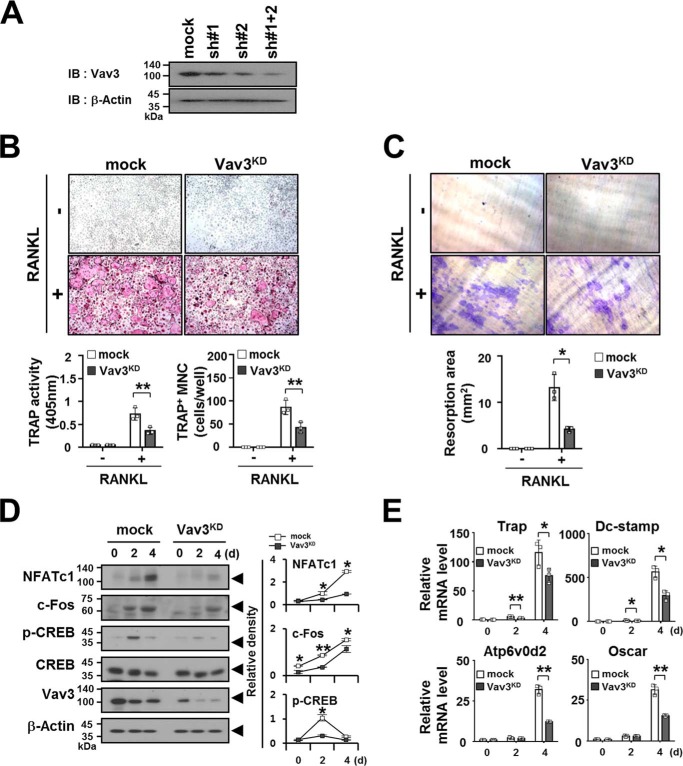
Next, we investigated whether Vav3 knockdown (Vav3KD) is involved in osteoclastogenesis. As a preliminary experiment, BMMs transduced by retroviral supernatant harboring Vav3 shRNA expression cassettes were selected with puromycin and further tested for the expression of Vav3. Vav3 expression was reduced significantly by the combination of shRNAs 1 and 2 (sh1+2) compared with shRNA 1 (sh 1) or shRNA 2 (sh 2) alone (Fig. 6A). To examine the effects of Vav3KD, puromycin-resistant BMMs transduced by sh1+2 were differentiated into OCs with M-CSF and RANKL for 4 days. TRAP activity and the number of TRAP+ MNCs in Vav3KD OCs were reduced significantly (0.36 ± 0.08 and 42.7 ± 11.6, respectively) compared with the mock control (0.73 ± 0.14 and 86.3 ± 15.5, respectively) (Fig. 6B). Similar results were obtained in a bone resorption assay, with a 3.1-fold decrease in the resorption area after Vav3KD (13.2 ± 1.8) compared with the mock control (41.3 ± 8.8) (Fig. 6C). Consistent with these results, Vav3KD reduced the expression or activation of the osteoclastogenic transcription factors (Fig. 6D) and markers (Fig. 6E). Collectively, these data indicate that Vav3 can induce the signaling pathway downstream of TRAF6, thereby enhancing osteoclastogenesis.
FIGURE 6.
Knockdown of Vav3 expression in BMMs by shRNAs resulted in reduced osteoclastogenesis. A, analysis of Vav3 knockdown (Vav3KD) effects via immunoblot analysis. Puromycin-selected BMMs transduced with retroviral supernatants harboring Vav3KD cassettes expressing Vav3 shRNA 1 (sh#1) and Vav3 shRNA 2 (sh#2) separately or in combination (sh#1+2) were cultured with M-CSF (150 ng/ml) for 2 days. The cell lysates were normalized for total protein content. Vav3KD was visualized via anti-Vav3 (K-19) immunoblotting (IB). β-Actin was used as a loading control. mock, empty vector control. B, the effect of Vav3KD on OC differentiation. BMMs were infected by Vav3KD-retroviral supernatants harboring shRNAs 1 and 2, and the Vav3KD-BMMs were further cultured with puromycin and M-CSF (150 ng/ml) for 2 days. Puromycin-selected Vav3KD-BMMs were differentiated into OCs with RANKL (100 ng/ml) and M-CSF (50 ng/ml) for 4 days. The OCs were photographed (top panel, original magnification, ×50) after TRAP staining. The OCs were analyzed using the TRAP solution assay (bottom left panel). The number of TRAP+ MNCs (at least three nuclei) was counted (bottom right panel). mock, empty vector control. **, p < 0.01. C, the effect of Vav3KD on resorption pit formation. Resorption pits were visualized (top panel, original magnification, ×50). The summarized data from the resorption pit assays are shown in the bottom panel. *, p < 0.05. D, the effect of Vav3KD expression on the activation of osteoclastogenic factors. OCs were differentiated from Vav3KD-BMMs with RANKL (100 ng/ml) and M-CSF (50 ng/ml) for the indicated times and subsequently analyzed via IB with antibodies that recognized phosphorylated or total NFATc1 (7A6), c-Fos (D-1), and CREB (Ser133). The cell lysates were normalized for total protein content. The level of Vav3 expression was detected via anti-Vav3 IB. The relative protein level was calculated after normalization to β-actin protein input (right panel). The relative level of phosphorylated CREB was calculated after normalization to total CREB protein input (right panel). β-Actin was used as the loading control. E, the effect of Vav3KD on the expression of osteoclastogenic markers. Total RNA was prepared from Vav3KD-OCs and subjected to real time PCR analysis. The data were normalized to β-actin. All experiments were performed at least three times with similar results.
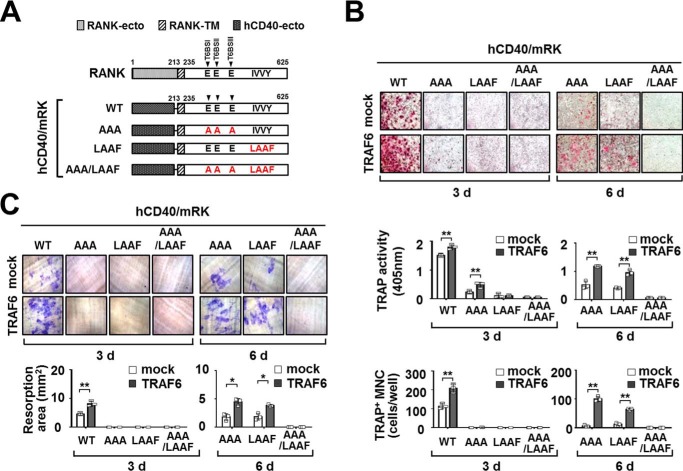
Physical Interaction of TRAF6 and Vav3 Forms a Receptor Complex with RANKcyto Lacking Either T6BSs or the IVVY Motif
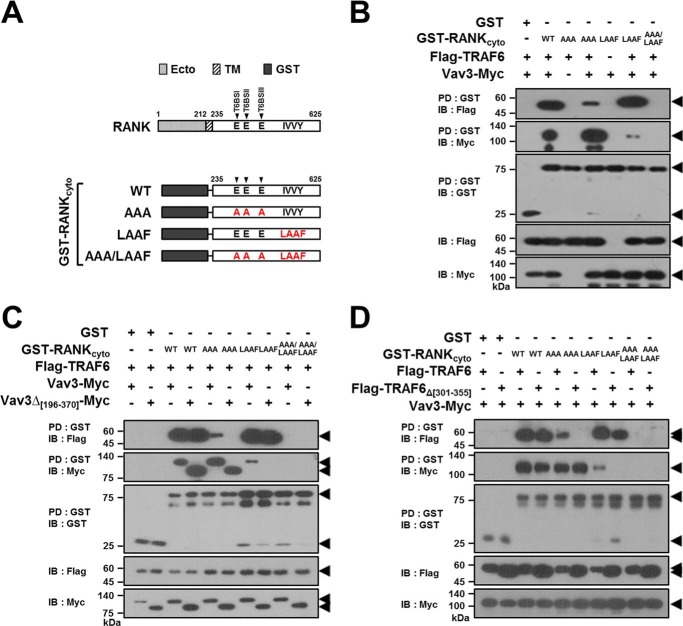
RANK has distinct cytoplasmic T6BSs, and an IVVY motif that can recruit TRAF6 and Vav3, respectively, to form the RANK signaling complex during osteoclastogenesis (12, 14, 18, 20). Thus, to confirm that TRAF6-Vav3 interaction can lead to RANK signaling complex formation independent of the binding motifs in RANKcyto, we constructed GST-RANKcyto mutants lacking either T6BSs or the IVVY motif (Fig. 7A). In a GST-RANKcyto-PD assay, TRAF6 was clearly recruited to the GST-RANKcyto-AAA mutant lacking T6BSs by Vav3, although a large fraction of the TRAF6 was not bound to this mutant (Fig. 7B, fourth lane). Similarly, a small fraction of Vav3 was recruited to the GST-RANKcyto-LAAF mutant lacking the IVVY motif by TRAF6 (Fig. 7B, sixth lane). However, neither TRAF6 nor Vav3 binding was detected for GST-RANKcyto-AAA/LAAF, which lacks both T6BSs and the IVVY motif (Fig. 7B, seventh lane). To further confirm the formation of the RANK signaling complex by direct TRAF6-Vav3 interaction, we conducted a GST-RANKcyto-PD assay with Vav3Δ[196–370], which lacks the DH domain. In a GST-PD assay with GST-RANKcyto-AAA or GST-RANKcyto-LAAF, the formation of the RANKcyto-TRAF6-Vav3 ternary complex via the direct TRAF6-Vav3 interaction was not detected for Vav3Δ[196–370] (Fig. 7C, sixth and eighth lanes). We obtained similar results in a GST-RANKcyto-PD assay with TRAF6Δ[301–355], which lacks the CC domain (Fig. 7D, sixth and eighth lanes).
FIGURE 7.
TRAF6-Vav3 interaction forms a receptor complex with RANKcyto lacking either T6BSs or the IVVY motif. A, schematic diagram of GST-fused RANKcyto mutants. The extracellular domain (Ecto) and transmembrane domain (TM) in RANK are indicated. The numbers shown above each domain diagram denote the amino acid numbers. The Ala mutations of the Glu residues for the T6BS consensus sequences (PXEXX[Ar/Ac]) and the LAAF mutation for the IVVY motif are indicated in RANKcyto. The arrow indicates the position of T6BSs. B, signaling complex formation by the TRAF6-Vav3 interaction in RANKcyto lacking either T6BSs or the IVVY motif. 293T cells were transfected with GST-RANKcyto (1.0 μg), Vav3-Myc (1.0 μg), and FLAG-TRAF6 (1.0 μg). The cell lysates were normalized for total protein content. GST-RANKcyto was pulled down (PD) using GST beads, and the PD GST-RANKcyto was visualized via anti-GST (B-14) immunoblotting (IB, third panel). The proteins bound to the GST-RANKcyto were visualized via anti-FLAG (M2) immunoblotting (top panel) or anti-Myc (9E10) IB (second panel). The level of TRAF6 or Vav3 expression was detected via anti-FLAG immunoblotting (fourth panel) or anti-Myc IB (bottom panel). C, the effect of the deletion of the DH domain of Vav3 on signaling complex formation by the RANKcyto lacking either T6BSs or the IVVY motif. D, the effect of the deletion of the CC domain of TRAF6 on signaling complex formation by the RANKcyto lacking either T6BSs or the IVVY motif. All experiments were performed at least three times with similar results.
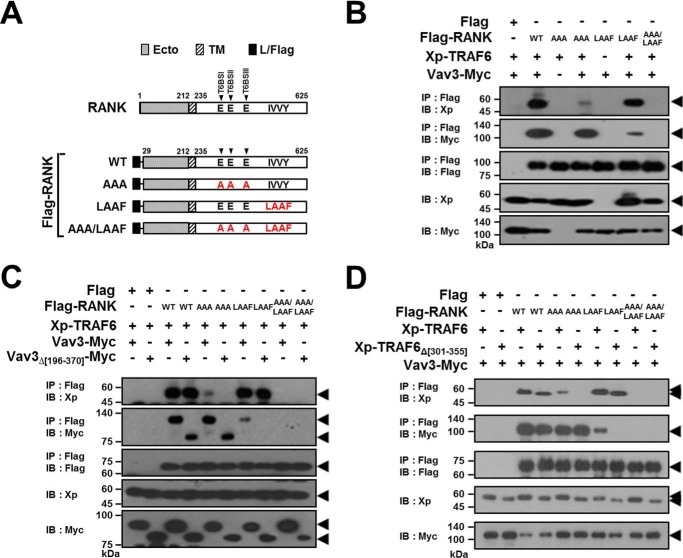
To further confirm the formation of the receptor complex by the TRAF6-Vav3 interaction, we generated FLAG-tagged full-length RANK (FLAG-RANK) mutants that lacked either T6BSs or the IVVY motif (Fig. 8A). Consistent with the data shown in Fig. 7B, the formation of the receptor complex via the direct TRAF6-Vav3 interaction was detected in an anti-FLAG IP assay with RANK mutants lacking either T6BSs or the IVVY motif (Fig. 8B, fourth and sixth lanes), whereas the ternary receptor complex was not observed in a RANK-AAA/LAAF mutant that lacked both T6BSs and the IVVY motif (Fig. 8B, seventh lane). Moreover, consistent with the data shown in Fig. 7 (C and D), we did not detect RANK-TRAF6-Vav3 receptor complex formation for Vav3Δ[196–370] (Fig. 8C, sixth and eighth lanes) or TRAF6Δ[301–355] (Fig. 8D, sixth and eighth lanes) in an anti-FLAG IP assay with FLAG-RANK-AAA or FLAG-RANK-LAAF. These results indicate that TRAF6 is recruited to RANKcyto mutants that lack their own T6BSs through the Vav3 interaction and that Vav3 is also recruited to a RANKcyto mutant lacking the IVVY motif through the TRAF6 interaction. Taken together, we conclude that TRAF6 recruits Vav3 to the RANK signaling complex and vice versa via a direct protein-protein interaction that is independent of the binding motifs in RANKcyto.
FIGURE 8.
TRAF6-Vav3 interaction forms a signaling complex with RANK lacking either T6BSs or the IVVY motif. A, schematic diagram of FLAG-tagged RANK mutants. The extracellular domain (Ecto) and transmembrane domain (TM) in RANK are indicated. The numbers shown above each domain diagram denote the amino acid numbers. The leader sequence of the full-length RANK was replaced with the leader sequence from preprotrypsin with a FLAG tag (L/FLAG). The Ala mutations of the Glu residues for the T6BS consensus sequences (PXEXX[Ar/Ac]) and the LAAF mutation for the IVVY motif are indicated in RANKcyto. The arrows indicate the positions of the T6BSs. B, signaling complex formation by the TRAF6-Vav3 interaction in FLAG-RANK lacking either T6BSs or the IVVY motif. 293T cells were transfected with FLAG-RANK (1.0 μg), Vav3-Myc (1.0 μg), and Xp-TRAF6 (1.0 μg). The cell lysates were normalized for total protein content. FLAG-RANK was immunoprecipitated (IP) using anti-FLAG beads, and the IP FLAG-RANK was visualized via anti-FLAG (M2) immunoblotting (IB, third panel). The proteins bound to the FLAG-RANK were visualized via anti-Xp IB (top panel) or anti-Myc (9E10) IB (second panel). The level of TRAF6 or Vav3 expression was detected via anti-Xp IB (fourth panel) or anti-Myc IB (bottom panel). C, the effect of the DH domain of Vav3 on signaling complex formation by FLAG-RANK lacking either T6BSs or the IVVY motif. D, the effect of the CC domain of TRAF6 on signaling complex formation by FLAG-RANK lacking either T6BSs or the IVVY motif. All experiments were performed at least three times with similar results.
The TRAF6-Vav3 Interaction Enhances Osteoclastogenesis
To address the role of the TRAF6-Vav3 direct interaction in osteoclastogenesis, we determined whether the TRAF6-Vav3 interaction in the RANK signaling complex induces OC differentiation and function. First, we generated retroviral hCD40/mRK chimeric receptor mutants that lacked either T6BSs or the IVVY motif to examine the effect of the TRAF6-Vav3 interaction under conditions of binding motif deficiency (Fig. 9A). BMMs transduced with a retroviral hCD40/mRK mutant and TRAF6 were differentiated into OCs by treatment with an anti-human CD40 monoclonal antibody to exclude the possibility that endogenous RANKL signaling affects osteoclastogenesis. TRAF6-expressing hCD40/mRK-WT BMMs treated with anti-human CD40 stimulation for 3 days exhibited increased (1.8 ± 0.08 and 209.3 ± 23.5, respectively) osteoclastogenesis compared with the mock control (1.5 ± 0.05 and 114.3 ± 14.6, respectively), as indicated by both TRAP activity and TRAP staining (Fig. 9B, left panel). Similar to previous reports that both T6BSs and the IVVY motif are crucial for osteoclastogenesis (12, 13, 16, 17), OC formation was not detected in hCD40/mRK mutants that lacked either T6BSs (hCD40/mRK-AAA) or the IVVY motif (hCD40/mRK-LAAF) after 3-day culture (Fig. 9B, left panel). However, when the OC culture was extended to day 6, we observed enhanced OC formation (1.0 ± 0.08 to 1.2 ± 0.04 and 64.3 ± 2.9 to 100.3 ± 8.7, respectively) in hCD40/mRK mutants that lacked either T6BSs or the IVVY motif, as indicated by TRAF6 expression, compared with the mock control (0.4 ± 0.05 to 0.5 ± 0.1 and 6.3 ± 3.2 to 12.3 ± 4.5, respectively), although the total number of OCs was relatively low compared with hCD40/mRK-WT in 3-day culture (Fig. 9B, right panel). However, even at the end of this 6-day extended culture, OC formation was not detected in an hCD40/mRK-AAA/LAAF mutant that lacked both T6BSs and the IVVY motif (Fig. 9B, right panel). Moreover, we also observed that the resorption area formed by OCs was increased (3.9 ± 0.2 to 4.5 ± 0.6) by TRAF6 expression in hCD40/mRK mutants that lacked either T6BSs or the IVVY motif in this 6-day extended OC culture compared with the mock control (1.8 ± 0.6 to 1.9 ± 0.6) (Fig. 9C, right panel).
FIGURE 9.
TRAF6 expression enhances osteoclastogenesis through a chimeric hCD40-RANKcyto receptor lacking either T6BSs or the IVVY motif. A, schematic diagram of chimeric hCD40-RANKcyto receptor mutants. The extracellular domain (RANK-ecto) and transmembrane domain (RANK-TM) in RANK are indicated. The numbers shown above each domain diagram denote the amino acid numbers. RANK-ecto of full-length RANK was replaced with the extracellular domain of human CD40 (hCD40-ecto). The Ala mutations of the Glu residues in the T6BS consensus sequences (PXEXX[Ar/Ac]) and the LAAF mutation for the IVVY motif are indicated in RANKcyto. The arrow indicates the position of T6BSs. B, the effect of TRAF6 expression on OC differentiation via a chimeric hCD40-RANKcyto receptor lacking either T6BSs or the IVVY motif. Puromycin-selected BMMs expressing hCD40-RANKcyto with or without TRAF6 were differentiated into OCs with M-CSF (50 ng/ml) and the anti-human CD40 monoclonal antibody G28–5 (100 ng/ml) for 3 or 6 days. The OCs were photographed (top panel, original magnification, ×50) after TRAP staining. The OCs were analyzed using a TRAP solution assay (middle panel). The number of TRAP+ MNCs (at least three nuclei) was counted (bottom panel). mock, empty vector control. *, p < 0.05; **, p < 0.01. C, the effect of TRAF6 expression on resorption pit formation via a chimeric hCD40-RANKcyto receptor lacking either T6BSs or the IVVY motif. The resorption pits were visualized (top panel, original magnification, ×50). The summarized data from the resorption pit assays are shown in the bottom panel. All experiments were performed at least three times with similar results.
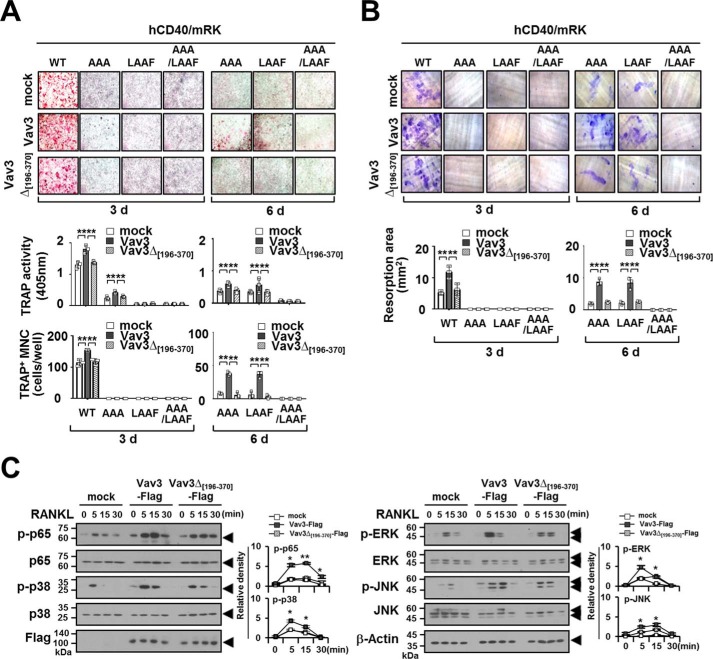
To further confirm the effect of the TRAF6-Vav3 interaction on osteoclastogenesis, we conducted an OC differentiation assay with BMMs transduced with retroviral hCD40/mRK mutants and Vav3. Consistent with the data obtained after RANKL stimulation (shown in Fig. 5A), Vav3-expressing hCD40/mRK-WT BMMs exhibited increased (1.8 ± 0.1 and 154.0 ± 3.0, respectively) OC differentiation in terms of both TRAP activity and TRAP staining after anti-human CD40 stimulation for 3 days compared with the mock control (1.3 ± 0.1 and 114.7 ± 10.5, respectively). However, similar results were not obtained for a Vav3Δ[196–370] mutant (1.4 ± 0.04 and 118.0 ± 9.6, respectively) that lacked the DH domain (Fig. 10A, left panel). In a 3-day culture, OC formation was also not detected in hCD40/mRK mutants that lacked either T6BSs (hCD40/mRK-AAA) or the IVVY motif (hCD40/mRK-LAAF) (Fig. 10A, left panel). However, when the OC culture was extended to day 6, OC formation by either the hCD40/mRK-AAA or hCD40/mRK-LAAF mutant was enhanced (0.6 ± 0.2 to 0.7 ± 0.07 and 36.3 ± 4.6 to 37.7 ± 3.1, respectively) by Vav3 expression but not by the Vav3Δ[196–370] mutant (0.3 ± 0.06 to 0.5 ± 0.06 and 2.7 ± 3.1 to 5.3 ± 4.2, respectively) (Fig. 10A, right panel). Consistent with these results, the resorption area was increased (8.4 ± 1.7 ∼ 8.7 ± 1.0) by Vav3 expression in hCD40/mRK mutants that lacked either T6BSs or the IVVY motif but not by the Vav3Δ[196–370] mutant (2.4 ± 0.2 ∼ 2.6 ± 0.1) in a 6-day extended OC culture (Fig. 10B, right panel). However, OC differentiation and bone resorption were not detected in the hCD40/mRK-AAA/LAAF mutant that lacked both T6BSs and the IVVY motif (Fig. 10, A and B, right panels). Finally, we analyzed whether the TRAF6-Vav3 interaction enhances RANK signaling. Upon RANKL stimulation, phosphorylation of NF-κB p65, p38, ERK, and JNK was significantly reduced after expression of the Vav3Δ[196–370] mutant compared with Vav3 (Fig. 10C). Taken together, these data indicate that the direct TRAF6-Vav3 interaction in the RANK signaling complex enhances OC differentiation and bone-resorbing function.
FIGURE 10.
Vav3 expression enhances osteoclastogenesis via a chimeric hCD40-RANKcyto receptor lacking either T6BSs or the IVVY motif. A, the effect of Vav3 or DH domain-deficient Vav3 expression on OC differentiation via a chimeric hCD40-RANKcyto receptor lacking either T6BSs or the IVVY motif. Puromycin-selected BMMs expressing hCD40-RANKcyto with Vav3 or DH domain-deficient Vav3 were differentiated into OCs with M-CSF (50 ng/ml) and the anti-human CD40 monoclonal antibody G28–5 (100 ng/ml) for 3 or 6 days. The OCs were photographed (top panel, original magnification, ×50) after TRAP staining. The OCs were analyzed using a TRAP solution assay (middle panel). The number of TRAP+ MNCs (at least three nuclei) was counted (bottom panel). mock, empty vector control. **, p < 0.01. B, the effect of Vav3 or DH domain-deficient Vav3 expression on resorption pit formation via a chimeric hCD40-RANKcyto receptor lacking either T6BSs or the IVVY motif. Resorption pits were visualized (top panel, original magnification, ×50). The summarized data from the resorption pit assays are shown in the bottom panel. C, the effect of Vav3 or DH domain-deficient Vav3 expression on RANKL-induced signaling pathways. Puromycin-selected BMMs expressing Vav3 or DH domain-deficient Vav3 were cultured for 2 h under serum-free conditions. The BMMs were stimulated with RANKL (100 ng/ml) for the indicated times and subsequently analyzed via immunoblotting (IB) with antibodies that recognized phosphorylated and total NF-κB p65 (Ser536), p38 (Thr180/Tyr182), ERK (Thr202/Thr204), and JNK (Thr183/Tyr185). The cell lysates were normalized for total protein content. The level of Vav3 expression was detected via anti-FLAG (M2) IB. The relative level of phosphorylated forms was calculated after normalization to total protein input (right). β-Actin was used as a loading control. The relative level of phosphorylated forms was calculated after normalization to total protein input (right). *, p < 0.05. All experiments were performed at least three times with similar results.
Discussion
The crucial role of T6BSs in RANK-mediated osteoclastogenesis through the recruitment of TRAF6 is well documented (2, 12, 13). An additional HCR motif in RANKcyto was also recently demonstrated to play a pivotal role in RANK-mediated osteoclastogenesis (16, 17). However, the functional cross-talk between T6BSs and HCR through the recruitment of specific adaptors in the RANK signaling complex remains to be elucidated. Thus, in this study, we screened Vav3 as a novel TRAF6 binding partner and identified the functional importance of the TRAF6-Vav3 interaction in the RANK signaling complex. We demonstrated that the interaction between the CC domain in TRAF6 and the DH domain in Vav3 synergistically activates RANK-mediated NF-κB and MAPK signaling, as well as NFATc1 induction during osteoclastogenesis. The CC domain of TRAF6 is important for TRAF6 autoubiquitination and downstream NF-κB activation via binding to Ubc13/Uev1A or NEMO (24, 25). Moreover, Lys63-linked TRAF6 autoubiquitination induced by RANKL stimulation is associated with the activation of downstream adaptors/kinases, including TAK1, TAK1-binding protein 1/2, NEMO, IKKs, and MAPKs, in RANK-mediated signaling (10, 11). Thus, it was important to examine whether the TRAF6-Vav3 interaction is involved in the TRAF6 ubiquitination pathway. However, the TRAF6-Vav3 interaction was not dependent on TRAF6 ubiquitination (Fig. 3E), and the E3 Ub ligase activity of TRAF6 was not affected by Vav3 expression (Fig. 3F). Vav3-deficient BMMs have been consistently reported to behave normally in terms of both RANKL-stimulated NF-κB activation and M-CSF-stimulated ERK activation, but mice lacking Vav3 display an osteopetrotic phenotype caused by defective OC function (26). Thus, it is possible that Vav3 per se is not directly involved in RANK-mediated signaling but may be required for the regulation process in RANK-mediated osteoclastogenesis. Interestingly, it has been proposed that Vav3 is involved in both OC fusion and actin ring formation via recruitment to the HCR motif during osteoclastogenesis (20). Hence, considering our results and those of previous studies, it is reasonable to presume that the direct TRAF6-Vav3 interaction strengthens the TRAF6 signaling complex via cross-talk between T6BSs and the HCR motif in RANKcyto, thereby further enhancing TRAF6-induced downstream NF-κB and MAPK signaling in osteoclastogenesis.
Previous studies of RANKcyto deletion mutants demonstrated that the IVVY motif in the HCR plays a crucial role in RANK-mediated osteoclastogenesis by inducing NFATc1 (16, 17, 21). Blocking peptides that harbor the IVVY motif attenuate RANK-mediated osteoclastogenesis, possibly by inhibiting the recruitment of Vav3 to the RANK signaling complex (18, 20). Moreover, Vav3 can bind to the IVVY motif in vitro (18, 20). Thus, Vav3 is a strong candidate for an adaptor molecule that binds directly to the IVVY motif. However, the IVVY motif per se is not directly involved in NF-κB and MAPK activation (16, 17, 20), and treatment with the IVVY peptide alone is not sufficient for inhibiting OC differentiation (20). Moreover, a requirement for an additional adaptor molecule in the Vav3-IVVY motif interaction has been proposed by Kim et al. (20) because Vav3 does not directly interact with RANKcyto in a yeast two-hybrid system. Thus, the direct binding of Vav3 to the IVVY motif remains unclear. Further studies are required to elucidate the Vav3-IVVY motif interaction.
Interestingly, Vav3 and Gab2 share an IVVY binding motif to form the RANK signaling complex, indicating that Gab2 may be involved in the binding of Vav3 to the IVVY motif, although Gab2 binding requires additional binding residues in the HCR of the RANKcyto (16, 18). Moreover, Gab2 can interact with TRAF6 and recruits PLCγ2 to the RANK signaling complex (16, 27). RANK-mediated NF-κB, MAPK, and NFATc1 activation are reduced in Gab2-deficient OCs (19). Mice lacking Gab2 also exhibit an osteopetrotic phenotype, with decreased bone resorption caused by partially impaired osteoclastogenesis (19). Thus, Gab2 functions as a crucial molecular scaffold in the RANK signaling complex (19, 27). However, whether Gab2 associates directly with RANKcyto and how Gab2 function is linked to TRAF6 or Vav3 in the RANK signaling complex remain unclear. Choi et al. (28) recently identified early estrogen-induced gene 1 (EEIG1) as an another HCR-binding adaptor molecule. EEIG1 also shares an IVVY binding motif to form the RANK signaling complex with Gab2 and PLCγ2, although Vav3 recruitment to this signaling complex has not been defined. Interestingly, EEIG1 is involved in RANK-mediated PLCγ2 activation and NFATc1 induction but not NF-κB or MAPK activation (28). Thus, the activation of RANK signaling by EEIG1 can be distinguished from TRAF6, Gab2, or Vav3 signaling (19, 28). These previous studies and our current observations emphasize the need for further investigations of the functional relevance of the adaptor molecules TRAF6, Gab2, PLCγ2, EEIG1, and Vav3 for the formation of the RANK signaling complex via cross-talk between T6BSs and the HCR.
Upon RANKL stimulation, RANK can recruit TRAF1, 2, 3, and 5, in addition to TRAF6, through three TRAF-binding motifs (motif 1 (PFQEP; amino acids 369–373), motif 2 (PVQEET; amino acids 559–564), and motif 3 (PVQEQG; amino acids 604–609)) in RANKcyto (29, 30). Under inflammatory conditions, these TRAF-binding motifs play important roles in TNF/IL-1-mediated osteoclastogenesis supported by RANKL signaling (31–33). Interestingly, the IVVY motif is also crucial for TNF/IL-1-mediated osteoclastogenesis (21, 34). Furthermore, Jules et al. (35) recently demonstrated that TRAF-binding motifs cooperate with the IVVY motif in TNF/IL-1-mediated osteoclastogenesis. Motif 1 is capable of NF-κB and MAPK activation, and motifs 2 and 3 are responsible for NFATc1 induction via cooperation with the IVVY motif (35). Although each motif contributes to TNF/IL-1-mediated osteoclastogenesis to a different extent, motifs 2 and 3 are more potent than motif 1 in promoting TNF/IL-1-mediated osteoclastogenesis (35). Interestingly, motifs 2 and 3 are crucial for the recruitment of both TRAF2 and TRAF5 (16, 29, 30). Thus, it is possible that TRAF2 and TRAF5, among the RANK-recruited TRAF members, are primarily involved in the cooperation between TRAF-binding motifs and the IVVY motif to promote TNF/IL-1-mediated osteoclastogenesis. In this study, we demonstrated that Vav3 interacts with TRAF1, 3, and 6 but not TRAF2 and 5 (Fig. 1E). Hence, we presume that the functional cross-talk that occurs between T6BSs and the IVVY motif through the direct TRAF6-Vav3 interaction is dispensable for the cooperation between the TRAF-binding motifs and the IVVY motif in TNF/IL-1-mediated osteoclastogenesis.
In conclusion, in this study, we demonstrated that T6BSs cooperate with the IVVY motif to enhance RANK-mediated osteoclastogenesis via the TRAF6-Vav3 interaction in the RANK signaling complex. We demonstrated that the CC domain of TRAF6 interacts directly with the DH domain of Vav3 to form the RANK signaling complex independent of the TRAF6 ubiquitination pathway. Via cross-talk between T6BSs and the IVVY motif in RANKcyto, the TRAF6-Vav3 interaction enhances downstream TRAF6 signaling and NFATc1 induction by further strengthening the TRAF6 signaling complex, thereby inducing osteoclastogenesis.
Experimental Procedures
Antibodies, Reagents, Cell Lines, and Mice
Specific antibodies were purchased from the following commercial sources: anti-FLAG epitope, anti-Myc epitope, and anti-β-actin from Sigma-Aldrich; anti-Xpress (Xp) epitope from Invitrogen; anti-GST, anti-c-Fos, anti-NFATc1, anti-Vav3, and anti-TRAF6 from Santa Cruz Biotechnology (Santa Cruz, CA); anti-CREB, anti-phospho-CREB, anti-p65, anti-phospho-p65, anti-ERK, anti-phospho-ERK, anti-p38, anti-phospho-p38, anti-JNK, and anti-phospho-JNK from Cell Signaling Technology (Danvers, MA); and anti-human CD40 monoclonal antibody G28–5 was obtained from Biolegend (San Diego, CA). Glutathione-Sepharose 4B (GST beads), anti-Myc affinity gels (Myc beads), and anti-FLAG affinity gels (FLAG beads) were purchased from Sigma-Aldrich. Protein G beads were purchased from Amersham Biosciences-Pharmacia Biotech. Recombinant human soluble RANKL and human M-CSF were described previously (36, 37). 293T and PlatE cells were cultured in DMEM (Welgene, Daegue, Korea) supplemented with 10% FBS (Invitrogen) and an antibiotic/antimycotic solution (Welgene) at 37 °C in a 5% CO2 atmosphere. C57BL6/J mice were purchased from Daehan Biolink (Umsung, Korea) and maintained under pathogen-free conditions. All animal work was approved (approval CNU-00584) by the Animal Experiment Ethics Committee of Chungnam National University.
Plasmids
FLAG epitope-tagged eukaryotic expression plasmids for TRAF6 (FLAG-TRAF6) and lysine-deficient TRAF6 (FLAG-TRAF6ΔK; all Lys residues in TRAF6 mutated to Arg) were described previously (38). The GST-fused (GST-TRAF6), red fluorescent protein-fused (RFP-TRAF6), and Xp-tagged TRAF6 (Xp-TRAF6) and the FLAG-tagged TRAF6 deletion mutants were constructed by PCR amplification and insertion in pEBG (39), pDsRed-Monomer-N1 (BD Biosciences Clontech, Mountain View, CA), pcDNA3.1-His (Invitrogen), pFLAG-CMV2 (Sigma-Aldrich), and pMX-puro-FLAG (37), respectively. The FLAG-tagged TRAF members were described previously (29). The Myc epitope-tagged eukaryotic expression plasmid for Vav3 (Vav3-Myc) was described previously (40). The deletion mutants for Xp-tagged (Xp-Vav3), Myc-tagged Vav3 (Vav3-Myc), and FLAG-tagged (Vav3-FLAG) were constructed by PCR amplification and insertion in pcDNA3.1-His, pEF4/Myc-His (Invitrogen), and pMXs-puro, respectively. The FLAG-tagged RANK (FLAG-RANK) expression plasmid was described previously (13). The GST-fused RANKcyto (GST-RANKcyto) expression plasmid was described previously (20). To generate T6BS-deficient mutants, all consensus Glu residues in the T6BSs were mutated to Ala by PCR amplification as previously described (39), and the mutated fragments were subcloned into pFLAG-CMV1 (FLAG-RANK-AAA) or pEBG (GST-RANKcyto-AAA). The retroviral expression plasmids for the chimeric receptor of human CD40 and murine RANKcyto (hCD40/mRK) and its T6BS-deficient mutant (hCD40/mRK-AAA) were described previously (12). To generate the IVVY motif mutants, the LAAF mutation was introduced into the IVVY motif in the RANKcyto by PCR amplification, and the fragments of the LAAF mutant were subcloned into pFLAG-CMV1 (FLAG-RANK-LAAF), pEBG (GST-RANKcyto-LAAF), or pMXs-puro (hCD40/mRK-LAAF). The expression plasmids carrying a double mutation of both T6BS and the IVVY motif were constructed by PCR amplification and insertion in pFLAG-CMV1 (FLAG-RANK-AAA/LAAF), pEBG (GST-RANKcyto-AAA/LAAF), and pMXs-puro (hCD40/mRK-AAA/LAAF), respectively. The plasmids p(κB)3-NF-κB-Luc (NF-κB-Luc), pAP-1-luciferase (AP-1-Luc), and pcDNA3.0HisLacZ (Invitrogen) were described previously (38, 39). The expression plasmid of HA-tagged ubiquitin (HA-Ub) was described previously (38, 39). For the Vav3 knockdown (Vav3KD) plasmid DNA, a shRNA targeting the murine Vav3 gene was designed as previously described (39) and subcloned into pSuper-retro-puro (Oligoengine, Seattle, WA). The primers used for Vav3KD vector construction were as follows: Vav3 shRNA 1 (sh1), 5′-GAT CCC CCA CTC TGG ACA AGC ATA CAT TCA AGA GAT GTA TGC TTG TTC CAG AGT TTT TTA-3′ and 5′-AGC TTA AAA ACA CTC TGG ACA AGC ATA CAT CTC TTG AAT GTA TGC TTG TTC CAG AGT GGG-3′; and Vav3 shRNA 2 (sh2), 5′-GAT CCC CAA GGG CCA TTC AAA CCT TCA AGA GAG GTT TGA ATG GCC CTT GTT TTT TTA-3′ and 5′-AGC TTA AAA CAA GGG CCA TTC AAA CCT CTC TTG AAG GTT TGA ATG GCC CTT GTT GGG-3′.
Osteoclastogenesis and OC Analysis
Murine BMMs were prepared as previously described (37, 41). In brief, bone marrow cells isolated from the femurs of 8-week-old C57BL/6 male mice were differentiated into BMMs by culturing them in α-minimum essential medium containing 10% FBS and M-CSF (150 ng/ml) for 2 days. To infect the BMMs with a retrovirus carrying a foreign gene (Vav3 or TRAF6) and puromycin-resistant expression cassettes, Vav3- or TRAF6-retroviral expression vectors were transfected into the retroviral packaging cell line PlatE using TurboFect transfection reagent (Fermentas). The BMMs were infected by retroviral supernatants harboring Vav3 or TRAF6 expression and puromycin-resistant cassettes with Polybrene (8 μg/ml) for 6 h, and the infected BMMs were then selected with puromycin (2 μg/ml) and M-CSF (150 ng/ml) for 2 days. Then BMMs were analyzed via flow cytometry with a FITC-conjugated anti-CD11b antibody (eBioscience, San Diego, CA). The puromycin-resistant BMMs (CD11b+ cells with a purity of >99.0%; 1.5 × 104 cells/well) in 96-well plates were differentiated into OCs in the presence of M-CSF (50 ng/ml) and RANKL (100 ng/ml) or the anti-human CD40 monoclonal antibody G28–5 (100 ng/ml) for 3–6 days. To analyze OCs, the cells were fixed in 10% formalin and an ethanol/acetone (50:50 v/v) mixture. To measure TRAP activity, the cells were incubated with p-nitrophenyl phosphate substrate (Sigma-Aldrich) solution for 30 min, and the solution was then mixed with 1 n NaOH. The TRAP solution activity of the collected supernatant was measured at 405 nm using a microplate reader (Bio-Rad). For TRAP staining, the TRAP solution supernatant was discarded from the cells, and the cells were incubated in TRAP staining solution containing naphthol AS phosphate (Sigma-Aldrich) and fast red violet (Sigma-Aldrich) for 20–30 min at room temperature. Finally, the morphology of the TRAP-stained OCs was photographed under a microscope. TRAP+ MNCs with more than three nuclei were considered multinucleated OCs. Analysis of the RANKL-induced signaling pathway was performed as previously described (37, 41). For the bone resorbing activity of OCs, retroviral-transduced BMMs (1.5 × 104 cells/well) were cultured with M-CSF (50 ng/ml) and RANKL (100 ng/ml) or the anti-human CD40 monoclonal antibody G28–5 (100 ng/ml) on dentin slices for 3–6 days, as previously described (37, 41). The cells were removed with 10% sodium hypochlorite solution to permit observation of the resorption pit. The resorption pit was stained with 1% toluidine blue and photographed under a microscope, and the resorbing activity was evaluated by measuring the area of the resorption pit. To analyze osteoclastogenic gene expression, real time PCR was performed as previously described (37, 41). The primers used for real time PCR amplification were as follows: TRAP (sense), 5′-AAA TCA CTC TTC AAG ACC AG-3′; TRAP (antisense), 5′-TTA TTG AAC AGC AGT GAC AG-3′; DC-STAMP (sense), 5′-TCC TCC ATG AAC AAA CAG TTC CAA-3′; DC-STAMP (antisense), 5′-AGA CGT GGT TTA GGA ATG CAG CTC-3′; Atp6v0d2 (sense), 5′-TTC AGT TGC TAT CCA GGA CTC GGA-3′; Atp6v0d2 (antisense), 5′-GCA TGT CAT GTA GGT GAG AAA TGT GCT CA-3′; OSCAR (sense), 5′-TCT GCC CCC TAT GTG CTA TCA-3′; and OSCAR (antisense), 5′-AGG AGC CAG AAC CTT CGA AAC-3′. β-Actin expression served as an internal control.
Protein-Protein Interaction and IP Assay
For mass spectrometry analysis, 293T cells (1 × 106 cells/ml) in 100-mm culture plates were transfected with FLAG-TRAF6 (20 μg) using TurboFect transfection reagent (Fermentas, Hanover, MD). At 24 h post-transfection, the transfected cells were lysed in lysis buffer (250 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 0.5% Triton X-100, and 5% glycerol). After centrifugation, the total protein concentrations of the supernatants were determined using BCA protein assay kits (Pierce). An equal amount of each supernatant (normalized for total protein content) was precipitated with FLAG beads (50% slurry) for 16 h at 4 °C. The beads were washed three times with lysis buffer and then suspended in loading buffer for 10% SDS-PAGE. The gel was stained with Coomassie Blue, and the selected bands were analyzed by nanoscale liquid chromatography coupled with tandem mass spectrometry by the proteomics core facility at the University of Pennsylvania School of Medicine. Protein-protein interaction and IP assays were performed as described previously (39). In brief, 293T cells (3.5 × 105 cells/ml) in 6-well plates were co-transfected with epitope-tagged TRAF6 (1.0 μg) and Vav3 (1.0 μg) using TurboFect (Fermentas). At 24 h post-transfection, the transfected cells were lysed in lysis buffer, and the lysate (normalized for total protein content) was precipitated with GST beads, Myc beads, or FLAG beads (50% slurry) for 16 h at 4 °C. The beads were washed three times with lysis buffer. The samples were separated by 10% SDS-PAGE, transferred to PVDF membranes, and immunoblotted with antibodies. For the endogenous protein-protein interaction assay, endogenous TRAF6 in murine BMM lysates (normalized for total protein content) was immunoprecipitated with an anti-TRAF6 antibody and protein G beads (50% slurry) overnight at 4 °C, followed by immunoblotting analysis with antibodies.
Luciferase Reporter Assay
Luciferase assays were performed as previously described (39). In brief, 293T cells were plated in 24-well plates at a density of 1 × 105 cells/well. After 12 h, the cells were co-transfected with luciferase reporter (0.5 μg), FLAG-TRAF6 (0.15 μg), Vav3-Myc (0.2–1 μg), and pcDNA3.0HisLacZ (0.15 μg) in triplicate using TurboFect transfection reagent. At 24 h post-transfection, the cells were lysed in reporter lysis buffer (Promega, Madison, WI). The cell lysates were normalized for total protein content, and 50-μg aliquots were analyzed by a luciferase assay. Luciferase activity was measured using a luciferase reporter assay kit (Promega) and normalized to β-galactosidase activity using a β-galactosidase assay kit (Applied Biosystems, Bedford, MA), according to the manufacturer's instructions.
Subcellular Localization
293T cells (1.0 × 104 cells/ml) were plated on sterile glass coverslips in 6-well plates. After 12 h of culture, the cells were co-transfected with RFP-TRAF6 (1.5 μg) and Vav3-Myc (1.5 μg). At 24 h post-transfection, the cells were stained with an anti-Myc primary antibody and a FITC-conjugated anti-rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Finally, the fluorescent signals were photographed under an Axiovert 135M laser scanning confocal microscope (Carl Zeiss).
Ubiquitination Assay
The TRAF6 ubiquitination assay was performed as previously described (39). In brief, the expression plasmids for FLAG-TRAF6 (0.5 μg), HA-Ub (0.5 μg), and Vav3-Myc (0.5 μg) were transfected into 293T cells seeded (3.5 × 105 cells/ml) in 6-well plates. At 24 h post-transfection, the cells were lysed. The cell lysates (normalized for total protein content) were subsequently subjected to IP and immunoblotting analysis with antibodies.
Statistical Analysis
All experiments were performed at least three times. The data represent the means ± standard deviations (n = 3 or n = 4/group). Student's t test was used to determine the significance of the differences between the experimental samples.
Author Contributions
J. Y., H. Y., and B. S. conducted research and wrote the manuscript; Y. K., E.-S. P., S. C., J. Y., D. S. A., and S. K. performed plasmid construction, cell differentiation, and gene expression analysis; J.-I. I., M. C. W., and Y. C. contributed to the experimental design and data analysis; M. T. provided dentin slices and technical support; and J. R. supervised the project and wrote the manuscript.
Acknowledgment
We thank J. M. Hwang for technical help.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea, by Ministry of Education Grant NRF-2014R1A1A2057884, by the research fund of Chungnam National University, by Ministry of National Defense Foundation Grant ADD14-01-06-06, and National Institutes of Health Grants AR069546 and AR067726 (to Y. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- OC
- osteoclast
- RANK
- receptor activator of nuclear factor κB
- RANKL
- RANK ligand
- TRAF
- TNF receptor-associated factor
- T6BS
- TRAF6-binding site
- HCR
- highly conserved domain in RANK
- M-CSF
- macrophage colony-stimulating factor
- NEMO
- NF-κB essential modulator
- IKK
- IκB kinase
- AP-1
- activator protein 1
- CREB
- cAMP-response element-binding protein
- NFATc1
- nuclear factor of activated T cell c1
- TRAP
- tartrate-resistant acid phosphatase
- DC-STAMP
- dendritic cell-specific transmembrane protein
- Atp6v0d2
- V-ATPase subunit d2
- OSCAR
- OC-associated receptor
- CC
- coiled-coil
- Ub
- ubiquitin
- TAK1
- TGF-β-activated kinase 1
- Gab2
- Grb-2-associated binder 2
- PLC
- phospholipase C
- AD
- acidic domain
- DH
- Dbl homology
- C1
- cysteine-rich
- SH
- Src homology
- PD
- pulldown
- IP
- immunoprecipitation
- BMM
- bone marrow-derived macrophage
- MNC
- multinucleated OC
- EEIG1
- estrogen-induced gene 1
- Xp
- Xpress
- IB
- immunoblotting.
References
- 1. Rho J., Takami M., and Choi Y. (2004) Osteoimmunology: interactions of the immune and skeletal systems. Mol. Cells 17, 1–9 [PubMed] [Google Scholar]
- 2. Walsh M. C., Kim N., Kadono Y., Rho J., Lee S. Y., Lorenzo J., and Choi Y. (2006) Osteoimmunology: interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 24, 33–63 [DOI] [PubMed] [Google Scholar]
- 3. Takayanagi H. (2007) Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7, 292–304 [DOI] [PubMed] [Google Scholar]
- 4. Amarasekara D. S., Yu J., and Rho J. (2015) Bone loss triggered by the cytokine network in inflammatory autoimmune diseases. J. Immunol. Res. 2015, 832127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rauner M., Sipos W., and Pietschmann P. (2007) Osteoimmunology. Int. Arch. Allergy Immunol. 143, 31–48 [DOI] [PubMed] [Google Scholar]
- 6. Dougall W. C., Glaccum M., Charrier K., Rohrbach K., Brasel K., De Smedt T., Daro E., Smith J., Tometsko M. E., Maliszewski C. R., Armstrong A., Shen V., Bain S., Cosman D., Anderson D., et al. (1999) RANK is essential for osteoclast and lymph node development. Genes Dev. 13, 2412–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J., Sarosi I., Yan X.-Q., Morony S., Capparelli C., Tan H.-L., McCabe S., Elliott R., Scully S., Van G., Kaufman S., Juan S.-C., Sun Y., Tarpley J., Martin L., et al. (2000) RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc. Natl. Acad. Sci. U.S.A. 97, 1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lomaga M. A., Yeh W. C., Sarosi I., Duncan G. S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., van der Heiden A., Itie A., Wakeham A., Khoo W., Sasaki T., et al. (1999) TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lamothe B., Webster W. K., Gopinathan A., Besse A., Campos A. D., and Darnay B. G. (2007) TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochem. Biophys. Res. Commun. 359, 1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., and Chen Z. J. (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell 15, 535–548 [DOI] [PubMed] [Google Scholar]
- 11. Mizukami J., Takaesu G., Akatsuka H., Sakurai H., Ninomiya-Tsuji J., Matsumoto K., and Sakurai N. (2002) Receptor activator of NF-κB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol. Cell. Biol. 22, 992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gohda J., Akiyama T., Koga T., Takayanagi H., Tanaka S., and Inoue J. (2005) RANK-mediated amplification of TRAF6 signaling leads to NFATc1 induction during osteoclastogenesis. EMBO J. 24, 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kadono Y., Okada F., Perchonock C., Jang H. D., Lee S. Y., Kim N., and Choi Y. (2005) Strength of TRAF6 signalling determines osteoclastogenesis. EMBO Rep 6, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye H., Arron J. R., Lamothe B., Cirilli M., Kobayashi T., Shevde N. K., Segal D., Dzivenu O. K., Vologodskaia M., Yim M., Du K., Singh S., Pike J. W., Darnay B. G., Choi Y., et al. (2002) Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418, 443–447 [DOI] [PubMed] [Google Scholar]
- 15. Naito A., Azuma S., Tanaka S., Miyazaki T., Takaki S., Takatsu K., Nakao K., Nakamura K., Katsuki M., Yamamoto T., and Inoue J. (1999) Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells 4, 353–362 [DOI] [PubMed] [Google Scholar]
- 16. Taguchi Y., Gohda J., Koga T., Takayanagi H., and Inoue J. (2009) A unique domain in RANK is required for Gab2 and PLCgamma2 binding to establish osteoclastogenic signals. Genes Cells 14, 1331–1345 [DOI] [PubMed] [Google Scholar]
- 17. Xu D., Wang S., Liu W., Liu J., and Feng X. (2006) A novel receptor activator of NF-κB (RANK) cytoplasmic motif plays an essential role in osteoclastogenesis by committing macrophages to the osteoclast lineage. J. Biol. Chem. 281, 4678–4690 [DOI] [PubMed] [Google Scholar]
- 18. Taguchi Y., Kiga Y., Gohda J., and Inoue J. (2012) Identification and characterization of anti-osteoclastogenic peptides derived from the cytoplasmic tail of receptor activator of nuclear factor κB. J Bone Miner Metab 30, 543–553 [DOI] [PubMed] [Google Scholar]
- 19. Wada T., Nakashima T., Oliveira-dos-Santos A. J., Gasser J., Hara H., Schett G., and Penninger J. M. (2005) The molecular scaffold Gab2 is a crucial component of RANK signaling and osteoclastogenesis. Nat. Med. 11, 394–399 [DOI] [PubMed] [Google Scholar]
- 20. Kim H., Choi H. K., Shin J. H., Kim K. H., Huh J. Y., Lee S. A., Ko C. Y., Kim H. S., Shin H. I., Lee H. J., Jeong D., Kim N., Choi Y., and Lee S. Y. (2009) Selective inhibition of RANK blocks osteoclast maturation and function and prevents bone loss in mice. J. Clin. Invest. 119, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jules J., Shi Z., Liu J., Xu D., Wang S., and Feng X. (2010) Receptor activator of NF-κB (RANK) cytoplasmic IVVY535–538 motif plays an essential role in tumor necrosis factor-α (TNF)-mediated osteoclastogenesis. J. Biol. Chem. 285, 37427–37435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tybulewicz V. L. (2005) Vav-family proteins in T-cell signalling. Curr Opin Immunol 17, 267–274 [DOI] [PubMed] [Google Scholar]
- 23. Hornstein I., Alcover A., and Katzav S. (2004) Vav proteins, masters of the world of cytoskeleton organization. Cell Signal 16, 1–11 [DOI] [PubMed] [Google Scholar]
- 24. Yang K., Zhu J., Sun S., Tang Y., Zhang B., Diao L., and Wang C. (2004) The coiled-coil domain of TRAF6 is essential for its auto-ubiquitination. Biochem. Biophys. Res. Commun. 324, 432–439 [DOI] [PubMed] [Google Scholar]
- 25. Gautheron J., Pescatore A., Fusco F., Esposito E., Yamaoka S., Agou F., Ursini M. V., and Courtois G. (2010) Identification of a new NEMO/TRAF6 interface affected in incontinentia pigmenti pathology. Hum Mol Genet 19, 3138–3149 [DOI] [PubMed] [Google Scholar]
- 26. Faccio R., Teitelbaum S. L., Fujikawa K., Chappel J., Zallone A., Tybulewicz V. L., Ross F. P., and Swat W. (2005) Vav3 regulates osteoclast function and bone mass. Nat. Med. 11, 284–290 [DOI] [PubMed] [Google Scholar]
- 27. Mao D., Epple H., Uthgenannt B., Novack D. V., and Faccio R. (2006) PLCgamma2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J. Clin. Invest. 116, 2869–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi H. K., Kang H. R., Jung E., Kim T. E., Lin J. J., and Lee S. Y. (2013) Early estrogen-induced gene 1, a novel RANK signaling component, is essential for osteoclastogenesis. Cell Res 23, 524–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wong B. R., Josien R., Lee S. Y., Vologodskaia M., Steinman R. M., and Choi Y. (1998) The TRAF family of signal transducers mediates NF-κB activation by the TRANCE receptor. J. Biol. Chem. 273, 28355–28359 [DOI] [PubMed] [Google Scholar]
- 30. Galibert L., Tometsko M. E., Anderson D. M., Cosman D., and Dougall W. C. (1998) The involvement of multiple tumor necrosis factor receptor (TNFR)-associated factors in the signaling mechanisms of receptor activator of NF-κB, a member of the TNFR superfamily. J. Biol. Chem. 273, 34120–34127 [DOI] [PubMed] [Google Scholar]
- 31. Wei S., Kitaura H., Zhou P., Ross F. P., and Teitelbaum S. L. (2005) IL-1 mediates TNF-induced osteoclastogenesis. J. Clin. Invest. 115, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lam J., Takeshita S., Barker J. E., Kanagawa O., Ross F. P., and Teitelbaum S. L. (2000) TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Invest. 106, 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma T., Miyanishi K., Suen A., Epstein N. J., Tomita T., Smith R. L., and Goodman S. B. (2004) Human interleukin-1-induced murine osteoclastogenesis is dependent on RANKL, but independent of TNF-alpha. Cytokine 26, 138–144 [DOI] [PubMed] [Google Scholar]
- 34. Jules J., Zhang P., Ashley J. W., Wei S., Shi Z., Liu J., Michalek S. M., and Feng X. (2012) Molecular basis of requirement of receptor activator of nuclear factor κB signaling for interleukin 1-mediated osteoclastogenesis. J. Biol. Chem. 287, 15728–15738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jules J., Wang S., Shi Z., Liu J., Wei S., and Feng X. (2015) The IVVY motif and tumor necrosis factor receptor-associated factor (TRAF) sites in the cytoplasmic domain of the receptor activator of nuclear factor κB (RANK) cooperate to induce osteoclastogenesis. J. Biol. Chem. 290, 23738–23750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee S. H., Rho J., Jeong D., Sul J. Y., Kim T., Kim N., Kang J. S., Miyamoto T., Suda T., Lee S. K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., and Choi Y. (2006) v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 37. Shin B., Yu J., Park E. S., Choi S., Yu J., Hwang J. M., Yun H., Chung Y. H., Hong K. S., Choi J. S., Takami M., and Rho J. (2014) Secretion of a truncated osteopetrosis-associated transmembrane protein 1 (OSTM1) mutant inhibits osteoclastogenesis through down-regulation of the B lymphocyte-induced maturation protein 1 (BLIMP1)-nuclear factor of activated T cells c1 (NFATc1) axis. J. Biol. Chem. 289, 35868–35881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walsh M. C., Kim G. K., Maurizio P. L., Molnar E. E., and Choi Y. (2008) TRAF6 autoubiquitination-independent activation of the NFκB and MAPK pathways in response to IL-1 and RANKL. PLoS One 3, e4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park E. S., Choi S., Shin B., Yu J., Yu J., Hwang J. M., Yun H., Chung Y. H., Choi J. S., Choi Y., and Rho J. (2015) Tumor necrosis factor (TNF) receptor-associated factor (TRAF)-interacting protein (TRIP) negatively regulates the TRAF2 ubiquitin-dependent pathway by suppressing the TRAF2-sphingosine 1-phosphate (S1P) interaction. J. Biol. Chem. 290, 9660–9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Charvet C., Canonigo A. J., Billadeau D. D., and Altman A. (2005) Membrane localization and function of Vav3 in T cells depend on its association with the adapter SLP-76. J. Biol. Chem. 280, 15289–15299 [DOI] [PubMed] [Google Scholar]
- 41. Yu J., Choi S., Park E. S., Shin B., Yu J., Lee S. H., Takami M., Kang J. S., Meong H., and Rho J. (2012) d- chiro-inositol negatively regulates the formation of multinucleated osteoclasts by down-regulating NFATc1. J. Clin. Immunol. 32, 1360–1371 [DOI] [PubMed] [Google Scholar]