Abstract
Cell migration is orchestrated by dynamic interactions of microtubules with the plasma membrane cortex. How these interactions facilitate these dynamic processes is still being actively investigated. TIP150 is a newly characterized microtubule plus end tracking protein essential for mitosis and entosis (Ward, T., Wang, M., Liu, X., Wang, Z., Xia, P., Chu, Y., Wang, X., Liu, L., Jiang, K., Yu, H., Yan, M., Wang, J., Hill, D. L., Huang, Y., Zhu, T., and Yao, X. (2013) Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771–15785; Xia, P., Zhou, J., Song, X., Wu, B., Liu, X., Li, D., Zhang, S., Wang, Z., Yu, H., Ward, T., Zhang, J., Li, Y., Wang, X., Chen, Y., Guo, Z., and Yao, X. (2014) Aurora A orchestrates entosis by regulating a dynamic MCAK-TIP150 interaction. J. Mol. Cell Biol. 6, 240–254). Here we show that TIP150 links dynamic microtubules to steer cell migration by interacting with cortactin. Mechanistically, TIP150 binds to cortactin via its C-terminal tail. Interestingly, the C-terminal TIP150 proline-rich region (CT150) binds to the Src homology 3 domain of cortactin specifically, and such an interaction is negatively regulated by EGF-elicited tyrosine phosphorylation of cortactin. Importantly, suppression of TIP150 or overexpression of phospho-mimicking cortactin inhibits polarized cell migration. In addition, CT150 disrupts the biochemical interaction between TIP150 and cortactin in vitro, and perturbation of the TIP150-cortactin interaction in vivo using a membrane-permeable TAT-CT150 peptide results in an inhibition of directional cell migration. We reason that a dynamic TIP150-cortactin interaction orchestrates directional cell migration via coupling dynamic microtubule plus ends to the cortical cytoskeleton.
Keywords: cell invasion, cell migration, cell motility, cellular regulation, microtubule, microtubule-associated protein (MAP)
Introduction
Cell migration is an essential physiological function that is tightly regulated in numerous biological processes, including development, tissue remodeling, wound healing, and tumor metastasis (1, 2). These processes require coordination of dynamic reorganization between the actin filaments and microtubules.
Cortactin (CTN)6 is a multidomain-containing scaffold protein that consists of an N-terminal acidic domain, six and a half tandem repeats, an α helix, a proline-rich region, and an Src Homology (SH3) domain at its C terminus (3). CTN is an important scaffold for promoting the polymerization and rearrangement of the actin cytoskeleton (4). Functionally, CTN, distributed at protrusions and the leading edges, is necessary for cell migration by regulating intercellular adhesion and cell spreading (5). Importantly, CTN is phosphorylated at tyrosine residues in response to growth factor receptors, which regulates cell migration (6).
Microtubules, one of the three major components of the cytoskeleton, are required for maintaining the physical and plastic properties of migrating cells (7). The plus ends of microtubules extend into the cell peripheral regions, dynamically switching between growing and shortening phases (8). The cytoskeletal interactions controlling cell migration are mediated by mechanisms that promote dynamic regulation of microtubule-associated proteins (MAPs) (9). Previous studies suggest that MAP-dependent regulatory pathways control cell migration by stimulating actin polymerization at the leading edge of migrating cells and, through the modulation of dynamic cell adhesion properties, regulate both the stabilization and disassembly of focal adhesion complexes (10). Focal adhesion complexes are initiated outside of the cell by binding to the extracellular matrix, where they mechanically connect the extracellular matrix to the actin cytoskeleton and provide the anchor points required for cell migration (1, 10, 11). Migrating cells organize a dynamic extracellular matrix, activate proto-oncogene tyrosine-protein kinase Src based on extracellular matrix stiffness, and synergize various organelle dynamics (12). One group of MAPs regulates microtubule dynamics and interactions within cells via their localization at the plus ends of microtubules, which are called microtubule plus end tracking proteins (+TIPs) (13). +TIPs have been identified based on their actual localization at the growing ends of microtubules and their regulation by virtual interactions with the end-binding (EB) family of +TIP proteins.
Our early studies revealed that TIP150 is an EB1-based microtubule plus end tracking protein necessary for accurate microtubule-kinetochore attachment during cell division (14, 15). Our recent work showed that TIP150 is required for dynamic microtubule-plasma membrane interaction during entosis (16). Linkage of microtubule plus ends with the cell cortex is required for directional cell migration. However, the functionality and mechanistic role of TIP150 in cell migration remain unclear.
In this study, we identify that TIP150 directly associates with the actin-binding protein, CTN, via its C-terminal proline-rich region (CT150). Interestingly, the CT150 associates with the SH3 domain of CTN, and the interaction is negatively regulated by EGF-elicited tyrosine phosphorylation of CTN. Moreover, suppression of either TIP150 or CTN inhibited directional cell migration. Perturbation of the TIP150-CTN interaction using a membrane-permeable peptide, CT150, inhibited cell migration. Thus, our study suggested that the TIP150-CTN interaction orchestrates directional cell migration via linking dynamic microtubule plus ends to the cortical cytoskeleton.
Results
Identification of a Novel TIP150-CTN Complex in Interphase Cells
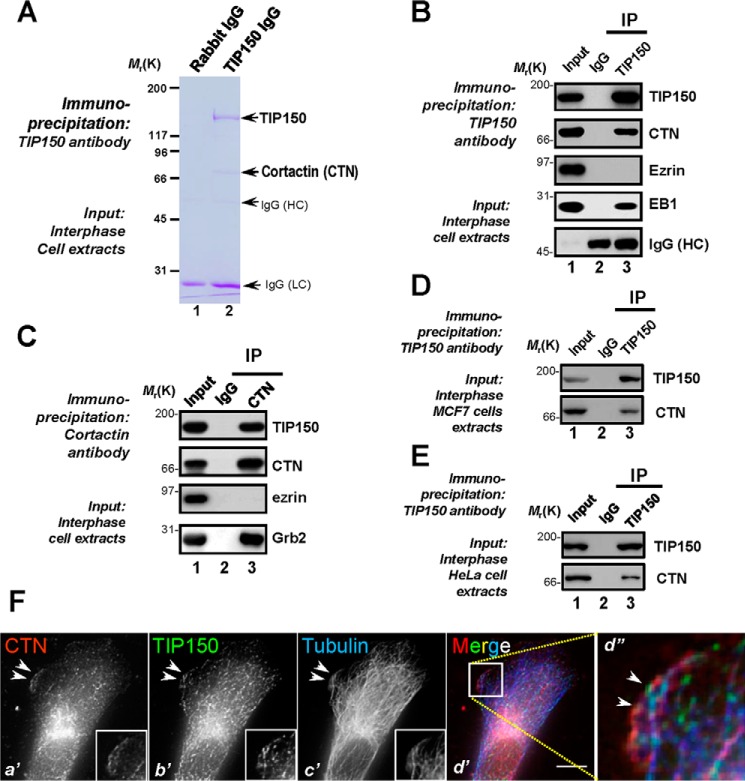
Our previous studies have revealed the functional importance of TIP150 in kinetochore and microtubule plus end dynamics during mitosis and entosis (14, 16). Because TIP150 protein levels remain unchanged during the cell cycle, we sought to examine the role of TIP150 in interphase cells. To study the molecular association of TIP150 with other microtubule end-binding proteins, we carried out an affinity isolation of a TIP150-containing protein complex followed by mass spectrometric identification of tryptic peptides derived from the individually sliced bands as described previously (17, 18). As shown in Fig. 1A, anti-TIP150 affinity beads covalently coupled to an anti-TIP150 antibody isolated a major polypeptide of 150 kDa in addition to three visible bands of polypeptides with approximate masses of 73, 55, and 25 kDa (lane 2). The 150-kDa polypeptide was absent from control IgG affinity matrix isolation (Fig. 1A, lane 1) and was confirmed as 150 kDa based on mass spectrometry and Western blotting analyses. Our mass spectrometric analyses identified the 73-kDa polypeptide as CTN, a characterized cortical actin-binding protein interacting with Grb2 in cell migration and cellular polarity establishment (19, 20). Western blotting analyses validated that the microtubule plus end protein EB1 also existed in the immunoprecipitates of TIP150, whereas the actin-binding protein ezrin was absent (Fig. 1B). Reciprocal immunoprecipitation using an anti-CTN antibody confirmed that TIP150 forms a cognate complex with CTN (Fig. 1C, lane 3, second panel). Ezrin was absent in the CTN immunoprecipitates (Fig. 1C, lane 3, third panel), suggesting that the CTN-TIP150 interaction is selective. As a positive control, Grb2 was retained by CTN immunoprecipitation (Fig. 1C, lane 3, fourth panel). Consistently, the TIP150-CTN interaction also occurs in MCF7 and HeLa cells (Fig. 1, D and E). Thus, we conclude that TIP150 forms a cognate complex with CTN in interphase cells.
FIGURE 1.
CTN is a novel TIP150-interacting partner at leading edges of migrating cells. A, TIP150 forms a novel complex with CTN in interphase cells. Aliquots of MDA-MB-231 cells were collected by centrifugation and extracted with Triton X-100-containing buffer as described under “Materials and Methods.” Clarified cell lysates were incubated with Sepharose bead affinity matrix covalently coupled with TIP150 antibody and control rabbit IgG, as described under “Materials and Methods.” The beads were washed successively with PBS before elution with 0.2 m glycine (pH 2.3). All samples were separated by SDS-PAGE. HC, heavy chain; LC, light chain. B, TIP150 immunoprecipitates from MDA-MB-231 cells in interphase were immunoblotted with antibodies against TIP150, CTN, ezrin, and EB1. Note that TIP150 immunoprecipitation (IP) brought down CTN and EB1 but not ezrin. C, CTN immunoprecipitates from MDA-MB-231 cells in interphase were immunoblotted with antibodies against TIP150, CTN, ezrin, and Grb2. Note that CTN immunoprecipitation brought down CTN and Grb2 but not ezrin. D, TIP150 immunoprecipitates from MCF7 cells in interphase were immunoblotted with antibodies against TIP150 and CTN. Note that TIP150 immunoprecipitation brought down CTN. E, immunoprecipitation of the endogenous TIP150-CTN protein complex in HeLa cells using TIP150 antibody or preimmune IgG. F, MDA-MB-231 cells were labeled with antibodies against CTN (red), TIP150 (green), and tubulin (blue). Scale bar = 10 μm. Note that the membrane ruffle-like localization of TIP150 is superimposed onto that of CTN, as shown in the magnified montage (d”).
The cortical cytoskeleton constitutes an important subcellular structure that determines cell shape and orchestrates cellular dynamics. CTN is an actin-binding protein located at the cortical cytoskeleton for integration of diverse signaling pathways. The identification of CTN in the TIP150 protein complex prompted us to examine the distribution profiles of TIP150 and CTN in interphase cells. To validate the hypothesis, we carried out immunofluorescence microscopic analyses of MDA-MB-231 cells triple-stained for CTN (red), TIP150 (green), and microtubules (blue). As shown in Fig. 1F, b', green, TIP150 distributes in the cytoplasm of interphase MDA-MB-231 cells and appears as comet-like structures, reminiscent of microtubule plus ends. Careful examination revealed a brief localization of TIP150 around plasma membrane regions reminiscent of membrane ruffle structures (Fig. 1F, b', arrowheads). Surprisingly, this membrane ruffle-like localization of TIP150 is partially superimposed onto that of CTN signal (Fig. 1F, red), as shown in the magnified montage of triple color merged images (Fig. 1F, d”, arrowheads). Thus, we conclude that TIP150 interacts and co-distributes with CTN near the plasma membrane in interphase cells.
Characterization of the Physical Interaction between TIP150 and CTN
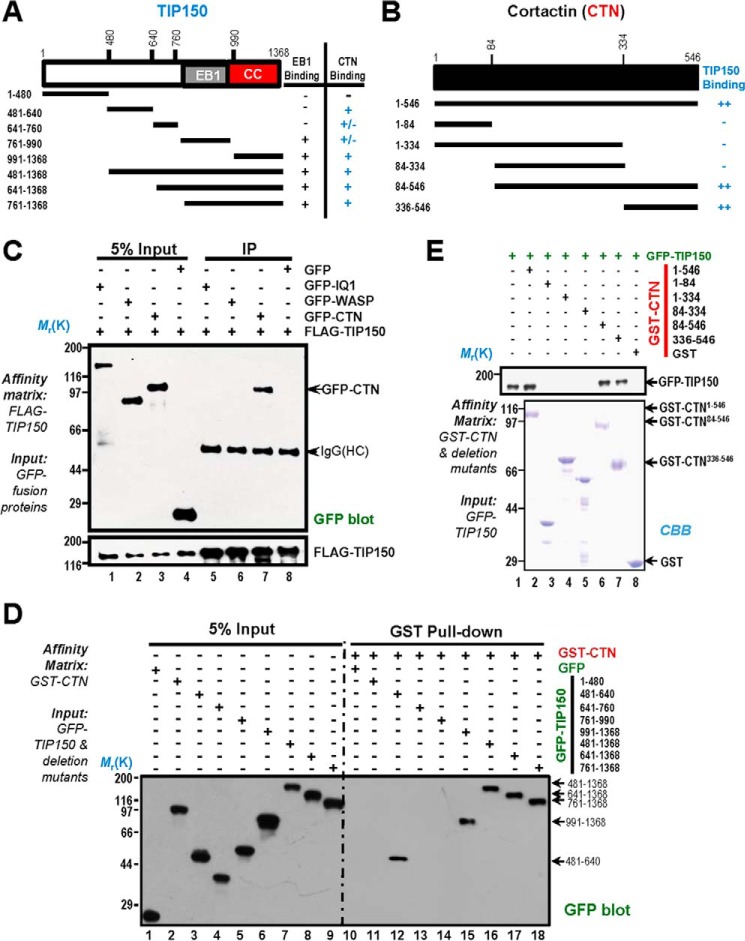
CTN is a signaling scaffold molecule containing several structural modules that facilitate the orchestration of protein-protein interactions underlying the signaling cascade during cell polarity establishment and cell migration. On the other hand, TIP150 also contains structural determinants essential for interacting with EB1 (14). We hypothesized that TIP150 physically interacts with CTN and that the binding interface and perhaps structural determinants could be uncovered by biochemical characterization. To test this hypothesis, we designed a series of deletion mutants of TIP150 and CTN according to their structural features (Fig. 2, A and Fig. B) and expressed those proteins tagged with GFP in HEK293T cells.
FIGURE 2.
Biochemical characterization reveals a physical link between the TIP150 and CTN. A, schematic of TIP150 truncation mutants. Residue numbers at domain boundaries are indicated. CC, coiled coil. B, schematic of CTN truncation mutants. Residue numbers at domain boundaries are indicated. C, confirmation of the TIP150-CTN interaction. MDA-MB-231 cells transfected with FLAG-TIP150 and GFP-IQGAP, N-WASP, and CTN were lysed and incubated with anti-FLAG M2 affinity matrix. Immunoprecipitates were resolved by SDS-PAGE and detected by immunoblotting with anti-GFP antibody (top) and anti-FLAG antibody (bottom). IP, immunoprecipitation; HC, heavy chain. D, the TIP150 C terminus binds to the CTN in vitro. Purified GST-CTN fusion proteins were used to isolate GFP-TIP150 deletion mutants from HEK293T cell lysates and fractionated by anti-GFP blotting analysis. E, GST-tagged CTN deletion mutants were purified on glutathione beads and used as an affinity matrix for absorbing a full-length GFP-TIP150 protein from HEK293T cells. GST protein-bound agarose beads were used as a negative control. Anti-TIP150 immunoblotting analyses confirmed that the C-terminal of CTN (amino acids 336–546) is responsible for TIP150-binding (top panel, lane 7). CBB, Coomassie Brilliant Blue.
To first determine the specificity of TIP150-CTN interaction, aliquots of HEK293T cells were transiently transfected to express FLAG-TIP150 with GFP-tagged IQGAP1 (GFP-IQ1), N-WASP (GFP-WASP), CTN (GFP-CTN), and GFP alone. Anti-FLAG immunoprecipitation using M2 affinity matrix followed by anti-GFP blotting analyses confirmed that FLAG-TIP150 forms a cognate complex with GFP-CTN (Fig. 2C, lane 7, top panel). Neither IQGAP1 nor N-WASP was present in the FLAG-TIP150 immunoprecipitates (Fig. 2C, lanes 5 and 6, top panel), confirming that the CTN-TIP150 interaction is selective.
To further evaluate the physical interaction and pinpoint the regions of TIP150 involved in the interaction, we carried out a pulldown assay using GST-CTN as an affinity matrix to absorb the GFP-TIP150 and its deletion mutants from HEK293T cell lysates. As shown in Fig. 2D, right panel, lanes 15–18, Western blotting analyses with a monoclonal antibody of GFP demonstrated that the GFP fusion proteins containing the C-terminal TIP150, in addition to full-length GFP-TIP150, were readily retained on GST-CTN matrix. The N-terminal domain also exhibited weak binding activity toward CTN (Fig. 2D, right panel, lane 12). To pinpoint the precise region of CTN involved in TIP150 binding, we expressed recombinant GST-CTN and its deletion mutants in bacteria. Purified GST-CTN proteins (full-length, N terminus, and C terminus) were used as affinity matrices to absorb the GFP-TIP150 from HEK293T cell lysates. As shown in Fig. 2E, top panel, lanes 6–7, Western blotting analysis with an anti-GFP antibody indicated that the TIP150 binds to the C-terminal domain of CTN. The Coomassie Brilliant Blue-stained gel showed the quality of purified GST-CTN proteins (Fig. 2E, bottom panel).
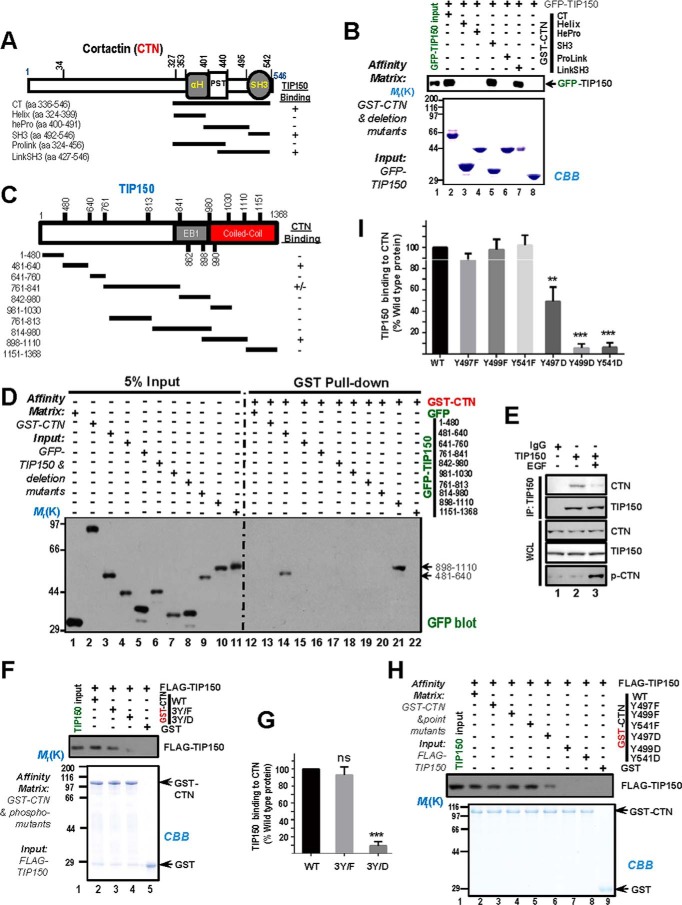
To define the exact domain(s) essential for their physical interaction, we carried out an additional GST pulldown assay using fine deletion mutants of CTN as affinity matrices to absorb GFP-TIP150 from HEK293T cell lysates. Immunoblotting with an anti-GFP antibody confirmed that GFP-TIP150 was retained on GST-CTN, indicating a direct interaction between the CTN SH3 domain and the TIP150 C-terminal (amino acids 898–1110) in vitro (Fig. 3B, lane 5, and D, lane 21). The EB1-binding domain alone exhibited no detectable binding activity toward CTN protein (Fig. 3D, lane 17). Thus, we conclude that TIP150 directly interacts with the CTN SH3 domain through its C-terminal coiled-coil domain.
FIGURE 3.
Phosphorylation of CTN in its SH3 domain regulates the CTN-TIP150 interaction. A, schematic of deletion mutants of CTN C-terminal. +, positive; −, negative. aa, amino acids. B, purified GST-CTN and its binding domains (CT, C-terminal; Helix, helical; hePro, helical proline-rich; SH3; ProLink, proline-rich; and LinkSH3) fragments were used to absorb GFP-TIP150 from the HEK293T cell lysates. GST-CTN and its bound materials were resolved by SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining and immunoblotted with an anti-GFP antibody. C, schematic of TIP150 deletion mutants. +, positive; −, negative. D, purified GST-CTN was used as an affinity matrix to isolate proline-rich mutants of GFP-TIP150 from the lysates of HEK293T cells. Western blotting was performed as described in B. E, serum-starved MDA-MB-231 cells were treated with or without EGF. Endogenous TIP150 was immunoprecipitated, and precipitated endogenous CTN was detected by Western blotting. IP, immunoprecipitation. WCL, whole cell lysate. F, phospho-mutants of purified GST-CTN at tyrosine sites 421, 466, and 482 were used as affinity matrices to isolate FLAG-TIP150 from the lysates of HEK293T cells. Western blotting was performed as described previously. G, the intensity of bands as shown in F was quantified and normalized to that of CTNWT. Data represent mean ± S.E. from three independent experiments. ***, p < 0.001; ns, not significant. H, phospho-mutants of purified GST-CTN were used to absorb FLAG-TIP150 from the HEK293T cell lysates. After washing, proteins bound to agarose beads were analyzed by Coomassie Brilliant Blue staining and Western blotting using an anti-FLAG antibody. I, the intensity of bands as shown in H was quantified and normalized to that of CTNWT. Data represent mean ± S.E. from three independent experiments. **, p < 0.01; ***, p < 0.001.
Phosphorylation of CTN at Its SH3 Domain Regulates CTN-TIP150 Interaction
Phosphorylation of CTN regulates its activity, including its ability to promote actin polymerization and cell migration (21). In particular, CTN is phosphorylated on tyrosines 421, 466, and 482 in response to EGF stimulation (22). Cell motility could be inhibited by truncating the N-terminal F-actin binding domains of CTN or by blocking tyrosine phosphorylation mutation sites (Y421F/Y466F/Y482F, non-phosphorylatable) (23). Next we tested whether EGF stimulation modulates TIP150-CTN interaction in cells. MDA-MB-231 cells were stimulated with EGF (100 ng/ml) for 5 min after serum starvation of 8 h. The cell lysates were then incubated with TIP150 affinity matrix beads. The results showed that the TIP150-CTN association in MDA-MB-231 cells was reduced by EGF stimulation (Fig. 3E). Interestingly, mutation of CTN phosphorylation sites (Y421E/Y466E/Y482E, phosphomimetic mutants) have been shown to reduce the ability to interact with focal adhesion kinase and promote cell migration (23). We sought to investigate whether mutations at these phosphorylation sites had any effect on TIP150 interaction. To determine whether the phospho-mimicking mutants modulate the TIP150 interaction with CTN, we used anti-GFP antibody to probe GFP-TIP150 bound to the affinity beads preloaded with GST-CTN and a triple site mutation: GST-CTNY421F/Y466F/Y482F (Fig. 3F, 3Y/F) and GST-CTNY421D/Y466D/Y482D (Fig. 3F, 3Y/D). Quantitatively, the relative binding efficiency of TIP150 was significantly less, p < 0.001, when tyrosine was changed to aspartic acid, mimicking the phosphorylated form of CTN (Fig. 3G), suggesting that reversible phosphorylation may regulate the dynamic interaction between TIP150 and CTN.
The C-terminal region of CTN contains a series of tyrosine residues: 421, 466, 482, 497, 499, and 541, which are conserved between mouse and human (24). To further evaluate the effect of tyrosine phosphorylation of CTN on the TIP150-CTN interaction, we carried out another pulldown assay using GST-CTN phosphorylation mutants as affinity matrices. As shown in Fig. 3H, FLAG-TIP150 from HEK293T cell extracts directly bound to GST-CTNY497F, GST-CTNY499F, and GST-CTNY541F but not to GST-CTNY497D, GST-CTNY499D, or GST-CTNY541D (lanes 3–5 and lanes 6–8, respectively). We compared the relative binding efficiency of TIP150 to wild-type CTN and its phosphorylation-mimicking forms and found that TIP150 exhibited no apparent difference in binding to wild-type CTN and non-phosphorylatable mutants. However, the binding of TIP150 to phospho-mimicking CTN was impaired (Fig. 3H, lanes 6–8). Quantitative analyses showed that GST-CTNY497D binding of TIP150 was reduced by half. GST-CTNY499D and GST-CTNY541D virtually lost their binding to TIP150 (Fig. 3I), suggesting that these sites are major residues essential for TIP150-CTN interactions.
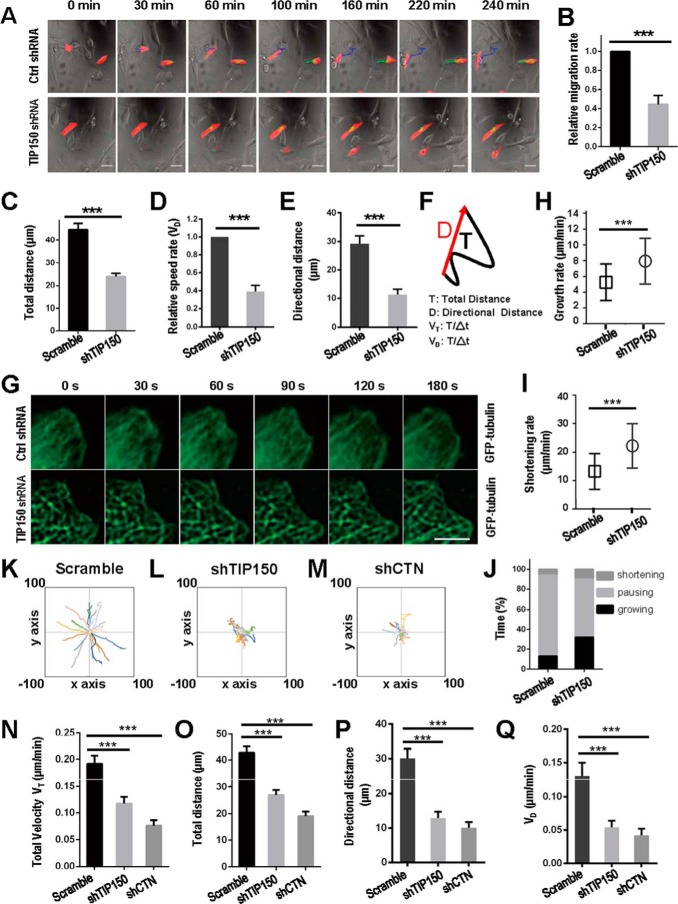
TIP150 Participates in Directional Cell Migration
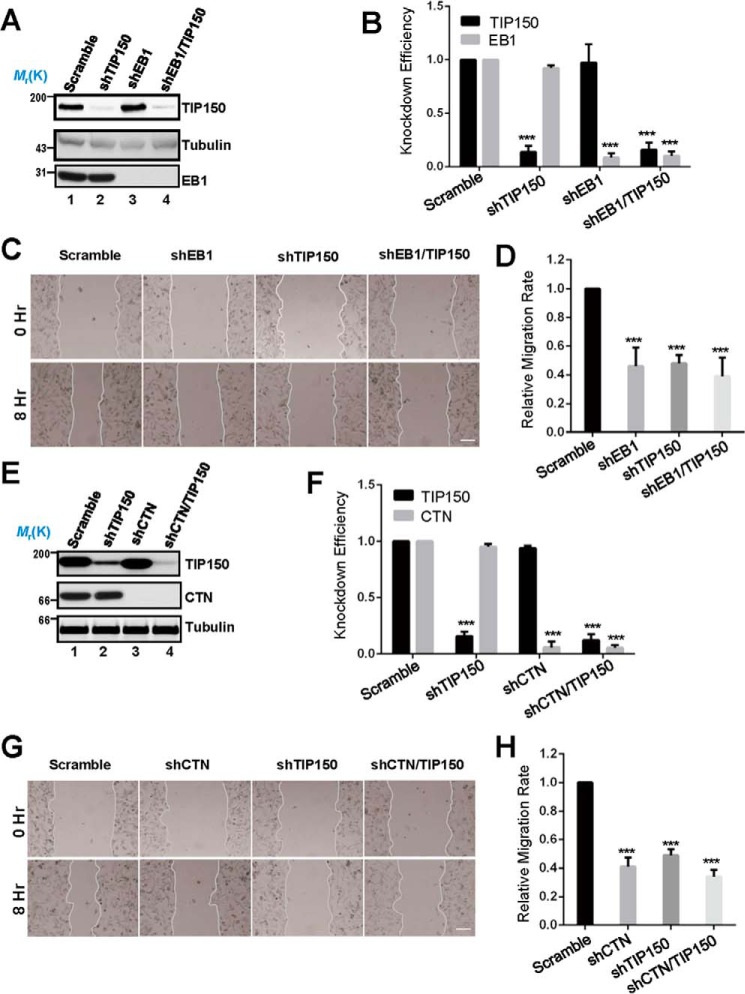
Several microtubule plus end tracking proteins have been reported to modulate cell migration by different mechanisms (7, 13). Our immunofluorescence microscopic analyses show that TIP150 co-distributes with CTN near the plasma membrane in interphase cells (Fig. 1F). To examine the possible function of endogenous TIP150 and TIP150-CTN interactions in cell migration, we sought to suppress TIP150 and CTN in MDA-MB-231 cells using the indicated shRNAs. Because EB1 is involved in cell migration (25), we included EB1 as a positive control. Western blotting analysis revealed that TIP150 was efficiently depleted by specific shRNAs but not by scrambled sequences, whereas the protein levels of tubulin and EB1 were unaffected (Fig. 4A, lane 2). Our quantitative analyses showed knockdown efficiencies of 86% and 91% for TIP150 and EB1, respectively (Fig. 4B).
FIGURE 4.
TIP150 and CTN are essential for EGF-elicited cell migration. A, characterization of the knockdown efficiency of TIP150 shRNA and EB1 shRNA. The protein levels of TIP150 and EB1 were significantly decreased in cells transfected with corresponding shRNAs. B, quantitative analysis of the knockdown efficiency of TIP150 shRNA in A. The data represent mean ± S.E. from three independent experiments. ***, p < 0.001. C, in MDA-MB-231 cells, suppression of either TIP150 or EB1 perturbed directional migration. The cells were treated with the indicated shRNAs for 72 h and were then scratched, followed by visualization with phase-contrast microscopy at the indicated time points. Scale bar = 100 μm. D, relative migration rates in C were calculated and graphed. Statistical significance was evaluated by Student's t test. Data are presented as mean ± S.E. from three independent experiments. ***, p < 0.001. E, knockdown efficiency of CTN shRNA. MDA-MB-231 cells were transfected with the indicated shRNA for 72 h and then subjected to immunoblotting. TIP150 shRNA was used as a control. F, quantitative analysis of the knockdown efficiency of CTN shRNA in E. Data represent mean ± S.E. from three independent experiments. ***, p < 0.001. G, in MDA-MB-231 cells, suppression of CTN or TIP150 caused defects in directional migration. The cells were treated with the indicated shRNAs for 72 h and were then scratched, followed by visualization with phase-contrast microscopy at the indicated time points. Scale bar = 100 μm. H, quantitative analysis of the relative migration velocities of cells toward the opposite side in G. Data are presented as mean ± S.E. from three independent experiments. ***, p < 0.001.
To probe whether TIP150 is involved in cell migration, we used wound healing assays to judge whether suppression of TIP150 alters the cell dynamics. Specifically, aliquots of MDA-MB-231 cells were transfected with TIP150 shRNAs, followed by starvation and a linear scratch generated by a sterile pipette tip as described previously (26). As shown in Fig. 4C, suppression of TIP150 protein inhibited cell migration, as determined by the wound healing assay. Our quantitative analyses showed that suppression of TIP150 alone, EB1 alone, or combined suppression of EB1 and TIP150 proteins reduced the relative migration rate to 46.2%, 47.9%, and 39.4%, respectively, compared with that of the scrambled shRNA (Fig. 4D). These results demonstrate that TIP150 functions in cell migration.
After demonstrating the involvement of TIP150 in cell migration, we next examined whether CTN is involved in cell migration. As shown in Fig. 4, E and F, Western blotting analyses revealed that CTN was efficiently depleted by specific shRNAs but not by the scramble sequences, whereas the protein levels of tubulin were unaffected (Fig. 4E, lanes 3 and 4). As shown in Fig. 4G, suppression of CTN protein inhibited cell migration, as determined by the wound healing assay. Our quantitative analyses show that suppression of CTN alone, TIP150 alone, or combined suppression of CTN and TIP150 reduced the relative migration rate to 41.3%, 49.2%, and 34.1%, respectively, compared with that of the scrambled shRNA (Fig. 4H). These results suggest that endogenous CTN and TIP150 are regulators responsible for the EGF-stimulated cell migration.
The TIP150-CTN Interaction Is Essential for Cell Migration
To further validate our results at the single-cell level, MDA-MB-231 cells transfected with mCherry-tagged control shRNA or TIP150 shRNA were starved and then stimulated by serum to migrate. Suppression of TIP150 induced a measurable defect in cell migration (Fig. 5A, bottom panels). To quantify these results, the migration speeds of cells transfected with control shRNA or TIP150 shRNA were calculated. The quantitative analysis showed that TIP150 knockdown reduced the velocity of MDA-MB-231 cells by roughly 55% of that observed in control cells (Fig. 5B). We next examined the migration distance in the absence of TIP150. As shown in Fig. 5C, the total distance of travel from initial to final point upon stimulation with EGF was statistically different, whereas shRNA-TIP150-transfected cells migrated a shorter distance compared with the non-transfected group. Quantitative analyses showed that the distance of cell migration in the TIP150-depleted cells was half of that in control cells (Fig. 5C, p < 0.001). Our quantitative analyses also show that shRNA-TIP150 significantly reduced the directional velocity of cell migration (Fig. 5, D–F). Thus, we conclude that TIP150 is required for directionally persistent cell migration in MDA-MB-231 cells.
FIGURE 5.
TIP150 and CTN are essential for directional cell movement. A, MDA-MB-231 cells transfected with control or TIP150 shRNA (red) were treated as described under “Materials and Methods,” and images were collected at 10-min intervals. Ctrl, control. B, migration rates of TIP150-supppressed MDA-MB-231 cells were measured, and statistical significance was determined using Student's t test. ***, p < 0.001. C, quantitative analysis measuring total migration distance of control or TIP150 shRNA-treated groups. Data represent mean ± S.E. of 65 cells collected from three independent experiments. Statistical significance was determined by Student's t test. ***, p < 0.001. D, quantitative analysis of relative cell speed. Data represent mean ± S.E. n = 65, respectively. ***, p < 0.001 by t test. E, quantitative analysis of directional distance. Data represent mean ± S.E. n = 65. ***, p < 0.001 by t test. F, visual demonstration of path length. The total distance between starting and ending points (T) and the actual trajectory (D) are indicated. G, TIP150 is required for microtubule plus end stabilization in the region of the cell cortex. Time-lapse images show the microtubule plus end dynamics in TIP150 knockdown cells. MDA-MD-231 cells were co-transfected with GFP-tubulin and control or TIP150 shRNAs for 72 h. Live-cell images were collected at 10-s intervals. Scale bar = 5 μm. H and I, statistical analysis of microtubule growth rate and shortening rate at the cell cortex in F. Data are presented as mean ± S.D. and derived from ∼100 microtubules in 25 cells. ***, p < 0.001 by t test. J, the percentages of growing, pausing, and shortening times microtubules displayed at the cell cortex in F. Data are derived from ∼100 microtubules in 25 cells. K–M, migration tracks of the indicated groups are presented from at least 20 cells. Data were collected from three independent experiments. N, quantitative analysis of total velocity in K–M. Data represent mean ± S.E. from three independent experiments. Statistical significance was determined by Student's t test. ***, p < 0.001. O, quantitative analysis of measured total distance in K–M. ***, p < 0.001 by t test. P, quantitative analysis of directional distance in K–M. Data represent mean ± S.E. from three independent experiments. Statistical significance was determined by Student's t test. ***, p < 0.001. Q, quantitative analysis of measured speed rates in K–M. ***, p < 0.001 by t test.
As microtubule plus end dynamics in the cell cortex region are involved in cell morphogenesis, polarity, and migration, we focused on the role of TIP150 on microtubule plus end dynamics in this region. As shown Fig. 5F, TIP150 knockdown increased the unstable microtubule ends in the cell cortex region. In TIP150-depleted cells, the microtubule plus end growth rate and shrinking rate increased by 42.6% and 67.6%, respectively, relative to that in control cells (Fig. 5, H and I). At the cortex of TIP150 knockdown cells, microtubule plus ends displayed a shorter time in pausing but a longer time in growing and shortening (Fig. 5J). The data suggest that TIP150 is likely responsible for microtubule stabilization in the cortex region.
The biochemical characterization of the TIP150-CTN physical interaction prompted us to investigate whether TIP150 cooperates with CTN in linking microtubule plus ends to the plasma membrane cortex. To this end, we first examined whether CTN alters TIP150-dependent microtubule plus end tracking in vitro using a total internal reflection fluorescence microscopy (TIRFM)-based assay reported previously (15, 20). Analyses of kymographs showed that wild-type TIP150 tracked the growing ends of microtubules in the presence of EB1. Addition of CTN did not alter the TIP150-dependent microtubule plus end tracking.7 Statistical analyses indicated that CTN did not modulate the TIP150-dependent microtubule dynamics. We next sought to examine whether TIP150 alters CTN function to modulate actin dynamics using the pyrene-actin assay reported previously (23). CTN promotes actin filament elongation. However, addition of TIP150 did not alter CTN-dependent actin filament elongation.7 Thus, these experiments show that TIP150 does not alter CTN function to modulate actin dynamics and that CTN does not modulate TIP150-dependent microtubule dynamics.
Next we examined the respective roles of TIP150 and CTN in cell migration. As shown in Fig. 5, K–M, suppression of CTN perturbed the trajectory in a similar pattern as seen in TIP150-depleted cells (Fig. 5L). Quantitative analysis showed that TIP150 knockdown reduced the velocity of MDA-MB-231 cells to roughly 60.9% and that CTN suppression reduced the velocity of MDA-MB-231 cells to roughly 39.6% compared with that of cells transfected with scramble shRNA (Fig. 5N). Further analyses of cell migration distances demonstrated that suppression of CTN resulted in a dramatic reduction of persistent migration (Fig. 5O). In addition, our quantitative analyses also showed that suppression of TIP150 or CTN significantly impaired the directional velocity of cell migration (Fig. 5, P and Q, p < 0.001). Thus, both CTN and TIP150 are essential for EGF-elicited cell migration in MDA-MB-231 cells.
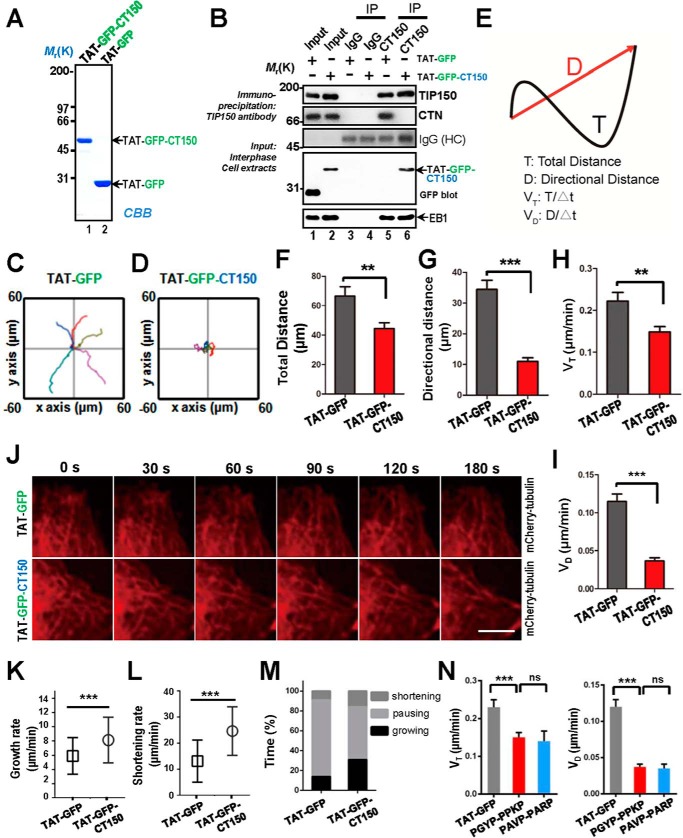
To assess the precise function of TIP150-CTN interaction in EGF-elicited cell migration, we designed a competitive peptide that would specifically perturb TIP150 binding to CTN. Consequently, we engineered a membrane-permeable peptide containing the PXXP motif of TIP150, which is involved in binding to CTN. This was achieved by introducing an 11-amino acid peptide derived from the TAT protein transduction motif into a fusion protein containing amino acids 898–1110 of TIP150, named TAT-GFP-CT150, that was involved in the binding interface between TIP150 and CTN (Fig. 3D). The recombinant protein was histidine-tagged and purified to homogeneity using nickel affinity beads (Fig. 6A). As predicted, the recombinant TAT-GFP-CT150 peptide disrupted the TIP150-CTN association in vivo, as judged by anti-TIP150 immunoprecipitation (Fig. 6B, lane 6). As a negative control, TAT-GFP did not interfere with the interaction of TIP150-CTN (Fig. 6B, lane 5). MDA-MB-231 cells were treated with TAT-GFP or TAT-GFP-CT150 peptide for 30 min before imaging cell migration elicited by EGF; real-time imaging of the cells began 5 min after EGF addition. The migration tracks illustrated that treatment of cells with the TAT-GFP-CT150 peptide perturbed EGF-elicited cell migration relative to cells treated with the TAT-GFP control (Fig. 6, C and D). Our quantitative analyses show that the TAT-GFP-CT150 peptide significantly reduced the directional velocity of cell migration (Fig. 6, E–I). In addition, TAT-GFP-CT150-treated cells increased the unstable microtubule ends in the cell cortex region (Fig. 6J), suggesting that the TIP150-CTN interaction promotes microtubule dynamics. In TAT-GFP-CT150-treated cells, the microtubule growth rate and shortening rate increased by 37.8% and 60.2%, respectively, relative to that in control cells (Fig. 6, K and L). At the cortex of TAT-GFP-CT150-treated cells, microtubule plus ends displayed a shorter time in pausing but a longer time in growing and shortening (Fig. 6M). The data suggest that TIP150 may be responsible for microtubule stabilization in the cortex region.
FIGURE 6.
Perturbation of the TIP150-CTN interaction inhibited cell migration. A, Coomassie Brilliant Blue (CBB) staining of SDS-PAGE gel showed the quality and quantities of the purified TAT-GFP-His6-tagged proteins. Bacteria expressing TAT-GFP-peptide (TAT-GFP-CT150) and TAT-GFP-His6 (TAT-GFP) were purified with nickel-nitrilotriacetic acid affinity chromatography and desalted into L-15. B, TIP150 immunoprecipitates from MDA-MB-231 cells incubated with TAT-GFP or TAT-GFP-CT150 peptide in interphase were immunoblotted with TIP150, CTN, EB1, and GFP antibodies. Nonspecific IgG-coupled beads were used as a control. Note that the EB1-TIP150 interaction was not altered by addition of the TAT-GFP-CT150 peptide but that the endogenous TIP150-CTN interaction was perturbed by addition of the TAT-GFP-CT150 peptide. IP, immunoprecipitation; HC, heavy chain. C and D, MDA-MB-231 cells were treated with the TAT-GFP or TAT-GFP-CT150 peptide for 30 min. The 5-h tracks of five randomly picked cells are presented for each group. E, visual demonstration of path length. The total distance between starting and ending points (T) and the actual trajectory (D) are indicated. F and G, quantitative analyses of total distance and directional distance. Data represent mean ± S.E. n = 35 and 29, respectively. **, p < 0.01; ***, p < 0.001. H and I, quantitative analyses of cell speed and velocity. Data represent mean ± S.E. n = 35 and 29, respectively. **, p < 0.01, ***, p < 0.001. J, the TIP150-CTN interaction is required for microtubule plus end stabilization in the region of the cell cortex. Time-lapse images show the microtubule plus end dynamics in mCherry-tubulin-expressing cells, which were treated with the TAT-GFP or TAT-GFP-CT150 for 30 min, and then live-cell images were collected at 10-s intervals. TAT-GFP-CT150-treated cells show microtubule retraction and bending. Scale bar = 5 μm. K and L, statistical analysis of microtubule growth rate and shortening rate at the cell cortex in J. Data are presented as mean ± S.D. and derived from approximately 100 microtubule bundles from 25 cells. ***, p < 0.001 by t test. M, the percentages of growing, pausing, and shortening times microtubules displayed at the cell cortex in F. Data are derived from ∼100 microtubule bundles from 25 cells. N, MDA-MB-231 cells were treated with the TAT-GFP-CT150 (PGYP-PPKP) or TAT-GFP-CT150 mutant (PAVP-PARP) for 30 min Presented are quantitative analyses of total distance and directional distance for 5 h. Data represent mean ± S.E. n = 36. ns, non-significant. Also presented are quantitative analyses of cell speed and velocity. Data represent mean ± S.E. n = 36.
To ensure that the observed cell crawling phenotype is not simply due to an imbalance in SH3-binding tyrosine kinase activity caused by an abundance of a PXXP-containing peptide, we mutated the PGYP-PPKP sequence of the TAT-GFP-CT150 peptide into PAVP-PARP. As shown Fig. 6N, PGYP-PPKP containing the TAT-GFP-CT150 peptide did not reduce the cell speed and directional velocity of cell migration. Thus, the TIP150-CTN interaction, mediated by the PGYP-PPKP motif, is essential for directional cell migration. We conclude that the TIP150-CTN interaction provides a regulatory linkage between the microtubule plus ends and cell cortex to steer cell migration.
Discussion
The highly regulated process of cell migration is critical for morphogenesis, tissue homeostasis, and tumor metastasis. Microtubule plus end tracking proteins constitute a complex structural platform that orchestrates the molecular events involved in regulating microtubule dynamics required for cell migration (13, 27). Directional cell movement is orchestrated by extracellular cue gradients; however, the molecular mechanism of microtubule dynamics regulated by unique +TIPs to mediate directional cell movement has remained elusive. TIP150 is a novel microtubule plus end tracking protein involved in various cellular processes (14–16). Previous studies have shown that many of the +TIPs accumulating at the cell cortex are involved in regulating cell migration through different pathways (13). In this study, we explored the physiological function of TIP150 and its association with cytoskeletal proteins that regulate cell migration. Through a series of biochemical, immunofluorescence, and cell migration assays, we demonstrated that TIP150 acts as a linkage protein mediating microtubule-to-actin interaction with the actin nucleation-promoting factor CTN, a protein that functionally regulates cell migration. TIP150 is localized along the cell cortex in MDA-MB-231 cells, indicating its involvement in the stabilization of microtubule dynamics along the cell cortex and, thus, facilitation of directional cell migration. We found that TIP150 interacts with the SH3-binding domain of CTN in a similar fashion as determined for other proline-rich binding proteins (28–30). The most stringent binding activity was located at the C-terminal coiled-coil region rich with proline residues (amino acids 898–1110). Suppression of TIP150 by shRNA-mediated knockdown was demonstrated to attenuate the directional migration elicited by EGF. In addition, phosphomimetic mutants of CTN tyrosine were shown to reduce binding activity to TIP150, suggesting that reversible phosphorylation of CTN upon stimulation of EGF is a dynamic mechanism regulating TIP150 interaction with CTN in EGF-elicited migrating cells.
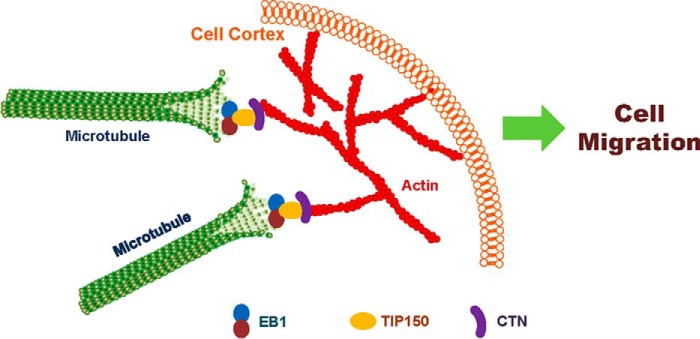
TIP150 was previously shown to be an EB1-binding protein tracking along microtubule plus ends (14). However, the function of TIP150 in cell migration and the interaction of TIP150 with other cytoskeletal components have not been examined. It was long speculated that the most likely candidate to assist microtubule guidance along the cell cortex is the actin cytoskeleton (7). Linkage of +TIPs with the cell cortex is required for microtubule polarization and plasticity, which are the characteristics of migrating cells. Our study of the TIP150-CTN interaction characterized here provides a novel insight into further delineation of how cortical molecules bind and stabilize the plus end of microtubules during cell migration (illustrated in Fig. 7).
FIGURE 7.
Proposed working model accounting for the TIP150-CTN interaction in directional cell migration. TIP150 functions as a microtubule plus end stabilizer that interacts with CTN to promote microtubule-actin cortex interaction. EGF stimulation promotes phosphorylation of CTN, which accelerates a dynamic TIP150-CTN interaction at the leading edges of migrating cells for directional movement. Thus, the TIP150-CTN complex serves as a link to orchestrate directional cell migration via coupling dynamic microtubule plus ends with the cell cortex.
Polarized dynamics and organization of the cytoskeleton are required for directional cell migration. Microtubules are organized by the centrosome, where many are anchored by the minus end and extended into the periphery by the plus end. These pioneering microtubules fortify the interactions with actin. It has been shown that microtubules could be captured and guided by stiff actin bundles regulating microtubule-actin interactions along the cell cortex (31). The leading edge lamellipodium protrusion relies on continuous polymerization of actin into a dense, cross-linked meshwork of filaments that moves away from the cell edge when polymerized in a process called retrograde flow (32). CTN is an actin-binding protein and was initially identified as an Src substrate (33). CTN is phosphorylated by the tyrosine kinase Src in growth factor signaling, transformation, and pathogen invasion (22, 34–36). Several studies have suggested that CTN and phosphorylated CTN dynamically regulate lamellipodial protrusion, cell spreading, intercellular adhesion, and cell motility. In addition, phosphorylation of CTN may enhance its ability to act as an adaptor or negatively regulate its function. Moreover, phosphorylation of CTN tyrosine is regulated by growing microtubules (28, 30, 34). This study suggests that EGF-elicited dissociation of the TIP150-CTN complex is needed to accelerate the recycling of TIP150 and CTN to promote leading edge membrane expansion and persistent microtubule growth, respectively. Thus, it would be of interest in the future to elucidate the structural basis of the TIP150 interaction with CTN. Elucidation of the structural basis of this interaction will enable us to consolidate the TIP150-CTN interaction and regulation into a mechanistic model by which the EGF-elicited tyrosine phosphorylation regulates the localization of microtubule plus ends to the cortical actin network.
Recently, we developed a new method for optically imaging intracellular protein interactions at a nanometer scale of spatial resolution in live cells using photoactivatable complementary fluorescent (PACF) proteins (37) based on photoactivation of complementary fluorescent proteins. Upon maturation, sparse subsets of PACF molecules were activated, localized, and then bleached (38). The aggregate position information from all PACF subsets was then assembled into a nanoscale map that allows imaging of a single-molecule copy of specific target protein-protein interactions in space and time in fixed and live cells. Using PACF, we obtained the precise localization of dynamic microtubule plus end hub protein EB1 dimers and their distinct distributions at the leading edges and cell bodies of migrating cells (37). Given the fact that TIP150 functions as dimers (15), in the future it would be of great interest to employ PACF-based analyses to probe how TIP150 dimers are precisely located to CTN in leading and trailing edges of migrating cells and whether alteration of the PXXP motif in a single TIP150 molecule of a dimer is sufficient to alter the TIP150-CTN linkage in cell migration at the single-molecule level. In addition, we can employ PACF to study the assembly and disassembly of the TIP150-CTN complex on a single microtubule and quantify how EGF-stimulated cell migration dynamics are correlated to spatial and temporal “turnover” of the TIP150-CTN complex.
In summary, we present evidence that TIP150 directly interacts with CTN. Our results also demonstrate that the TIP150-CTN interaction promotes microtubule dynamics and orchestrates EGF-elicited directional cell migration. However, more sophisticated mechanisms need to be delineated regarding how the reversible phosphorylation/dephosphorylation cycle regulates the aforementioned interaction dynamics during cell migration. Furthermore, it would be exciting and challenging to visualize and compare the single microtubule dynamics and corresponding cortex during cell migration.
Materials and Methods
Cell Culture
HEK293T cells obtained from the ATCC (Manassas, VA) were maintained as subconfluent monolayers in advanced DMEM (Gibco) with 10% FBS (Hyclone), 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco) at 37 °C with 8% CO2. MDA-MB-231 cells (ATCC) were maintained in L-15 medium (Gibco) containing 10% FBS at 37 °C. MDA-MB-231 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the protocol of the manufacturer.
Plasmids
Human TIP150 cDNA was obtained from the Kazuza DNA Research Institute (Chiba, Japan) as described previously (14, 15). Briefly, to generate GFP-tagged TIP150, PCR-amplified cDNA was cloned into the pEGFP-C2 vector (Clontech, Mountain View, CA) with EcoRI and SalI digestion. TIP150 was then subcloned into the following vectors: p3×FLAG (Sigma) and pET-28a (Novagen). All TIP150 deletion mutants were created by PCR and confirmed by DNA sequencing. Murine CTN cDNA constructs (full-length CTN (amino acids 1–546)), N-terminal/fragment(s), C-terminal/fragment(s), Helix, hePro, SH3, ProLink, and LinkSH3 were fused with GST. The murine CTN non-phosphorylatable mutant (Y421F/Y466F/Y482F), phospho-mimicking mutant (Y421D/Y466D/Y482D), non-phosphorylatable mutant (Y497F/Y499F/Y541F), and phospho-mimicking mutant (Y497D/Y499D/Y541D) were cloned into pGEX-6p-1 (GE Healthcare). GFP-tagged IQGAP1, N-WASP, and CTN full-length and/or truncations were generated by inserting PCR-amplified cDNAs into pEGFP-C2. All plasmid constructs were sequenced for verification.
To generate TAT-GFP-CT150-His fused proteins to perturb the TIP150-CTN interaction, an 11-amino acid TAT sequence followed by GFP and a His tag was inserted into the pET-22b vector. TAT-GFP-His fused proteins were expressed and purified as described previously (15).
siRNA and/or shRNA Transfection
The siRNA sequence used for silencing TIP150 was 5′-CAUACAGCAAAGUGCGAGA-3′, described previously (14) (synthesized by DharmaconResearch, Inc., Boulder, CO), or SMARTpool (L-022219-00, Thermo Fisher Scientific). EB1 siRNA (5′-AAGUGAAAUUCCAAGCUAAGC-3′) (39) and CTN siRNA (5′-CAAGCTTCGAGAGAATGTCTT-3′) were synthesized by Qiagen. As a control, a duplex-composing scrambled sequence was used (40, 41). The 21-mer oligonucleotide RNA duplexes were synthesized from Dharmacon Research, Inc. The shRNAs were constructed using the same targeting sequences as their siRNAs or as described previously (39). The vector was used as a control. All siRNAs and shRNAs were transfected into cells using Lipofectamine 2000 for 72 h, and the knockdown efficiency was confirmed by Western blotting and/or immunofluorescence.
Pulldown and Immunoprecipitation Assay
Pulldown assays were carried out as described previously (42). Briefly, the GST-tagged proteins were purified from bacteria by glutathione-agarose chromatography. GST fusion protein-bound Sepharose beads were incubated with HEK293T cell lysates ectopically expressing GFP- or FLAG-tagged proteins or their deletion mutants, or with purified His-tagged proteins expressed in bacterial cells. After extensive washing, bound proteins were boiled in sample buffer, split into two aliquots, and separated by SDS-PAGE gradient gels (5–15%) manually prepared for Western blotting analysis. All pulldown assays were conducted at least three times.
FLAG-TIP150 and pEGFP-C2 vector or GFP-tagged proteins were co-transfected and expressed in HEK293T cells. The FLAG-tagged proteins were precipitated with anti-FLAG-M2 beads (Sigma). Beads were washed three times with lysis buffer, and proteins were separated on SDS-PAGE for transferring onto a nitrocellulose membrane for Western blotting using GFP antibody.
Antibodies and Reagents
Mouse serum against TIP150 was generated using HIS-TIP150-P (amino acids 801–980) from bacteria according to a standard protocol. Rabbit antibody against TIP150 was developed by YenZym (San Francisco, CA) as described previously (14). The following antibodies were obtained from commercial sources: anti-EB1 mouse antibody (BD Biosciences), anti-CTN rabbit antibody (Sigma), phospho-CTN (Tyr-421) antibody (Cell Signaling Technology), anti-GFP antibody (BD Biosciences), anti-FLAG antibody (M2, Sigma), and anti-tubulin antibody (DM1A, Sigma). FITC-conjugated secondary antibodies (Pierce) and rhodamine phalloidin (Invitrogen) were also obtained commercially.
Immunofluorescence and Single Cell Migration
Cells were seeded onto sterile, acid-treated, 12-mm coverslips in 24-well plates (Corning Glass Works) for transfection or drug treatment (43). Cells were fixed with cold methanol for 2 min at −20 °C. After washing three times with PBST (0.05% Tween 20 in PBS), cells were blocked with 1% bovine serum albumin (Sigma) for 45 min at room temperature. Cells were subsequently incubated with the indicated primary antibodies in a humidified chamber for 1 h at room temperature, followed by secondary antibodies for 1 h at room temperature. DNA was stained with DAPI (Sigma). Images were acquired with a DeltaVision wide-field deconvolution microscope (Applied Precision Inc.) as described previously (44). For single cell migration, MDA-MB-231 cells were cultured in a glass-bottom culture dish (MatTek). During imaging, cells were maintained in CO2-independent medium (Gibco) containing 10% FBS and 2 mm glutamine in a sealed chamber at 37 °C. Images of living cells were taken with a DeltaVision microscopy system at 1 frame/10 min. Images were prepared for publication using Photoshop (Adobe).
Scratch Assay
For the wound healing assay, confluent MDA-MB-231 cells transfected with the indicated shRNAs were scratched with a 10-μl pipette tip and then stimulated by EGF (100 ng/ml) or 20% serum at 37 °C for the indicated times. Images were taken with a ×10 objective of an inverted microscope (Axiovert 200) coupled with an AxioCam-HS digital camera (Carl Zeiss). The relative healing velocities were measured using ImageJ software (National Institutes of Health).
Total Internal Reflection Fluorescence Microscopic Analyses
The microtubule plus end tracking experiment was performed as described recently (20). The GTP analogue guanylyl-(α,β)-methylene-diphosphonate (GMPCPP) microtubule seeds were prepared by polymerizing 30 μm tubulin (at a bovine tubulin/rhodamine-tubulin/biotin-tubulin ratio of 30:2:1) in the presence of 1 mm GTP analogue guanylyl-(α,β)-methylene-diphosphonate (GMPCPP) (Jena Bioscience) at 37 °C for 40 min. The seeds were then centrifuged and resuspended in BRB80 buffer (80 mm K-PIPES (pH 6.8), 1 mm MgCl2, and 1 mm EGTA). These seeds were sheared with a 25-gauge needle before they were used to generate short seeds.
Flow chambers were prepared as described previously (20) and coated with 10% monoclonal anti-biotin antibody (Sigma), followed by blocking with 5% Pluronic F-127 (Sigma). After a brief wash, sheared microtubule seeds (125 nm) were added into the chamber. Tubulin polymerization mixture (30 μm tubulin in total containing 1:30 rhodamine-labeled bovine tubulin in BRB80, 50 mm KCl, 5 mm DTT, 1.25 mm magnesium GTP, 0.25 mg/ml κ-casein, 0.15% methylcellulose (Sigma), an oxygen-scavenging system, and +TIPs) was introduced into the chamber to initiate polymerization. Unless stated otherwise, the final concentrations of +TIPs were 250 nm EB1 and 250 nm TIP150 or TIP150 plus CTN proteins. The temperature was kept at 25 °C. Images were collected with a superresolution microscope configured on an ELYRA system (Carl Zeiss). The laser intensities were kept at a low level to avoid photobleaching. For +TIPs tracking assays, 1 frame/s was taken. Plus end tracking of GFP-SKAP and its SXIP mutant was analyzed using kymographs in ZEN software (Carl Zeiss).
Pyrene-labeled Actin Assembly
To examine whether TIP150 alters actin assembly dynamics directly or indirectly via CTN, we adopted the pyrene-actin assembly assay reported previously (23). In short, pyrene-labeled actin was obtained from Cytoskeleton Inc. (catalog no. AP05-A) and used at a concentration of 5 μm. Because some polymerization of actin occurs during the freezing of samples, thawed actin was spun at 312,000 × g for 40 min before initiating assembly. Polymerization was initiated at the following final conditions: 5 mm Tris (pH 7.5), 0.5 mm ATP, 2 mm MgCl2, 10 mm KCl, and 0.2 mm DTT. Fluorescence was monitored continuously (excitation wavelength, 355 nm; emission wavelength, 407 nm) in the presence or absence of TIP150 and TIP150 plus CTN. Purified yeast cofilin was used as a control as reported previously (23).
Data Analyses
All fluorescence intensity measurements and statistical analyses were carried out using ImageJ software (National Institutes of Health) and GraphPad Prism. To determine the significant differences between means, unpaired Student's t test assuming unequal variance was performed; differences were considered significant when p < 0.05.
Author Contributions
X. D., and X. L. conceived the project. G. A., J. Z., H. W., P. X., Z. W., F. M., and S. Z. designed and performed most biochemical experiments. J. Z., Y. L., J. J., X. Z., W. W., K. T., J. Q., F. O. A., P. H., and X. D. designed and performed cell biological characterization. G. A., J. Z., S. Z., and K. T. performed the in vitro reconstitution experiment and data analyses. G. A., J. Z., D. L. H., P. H., X. L., X. D., and X. Y. wrote the manuscript, and all authors read and approved the manuscript.
Acknowledgments
We thank the members of our groups for insightful discussions during the course of this study and William Yao for editorial work.
This work was supported by Chinese 973 Project Grants 2012CB917204, 2013CB911203, and 2012CB713704; Chinese Academy of Science Grant KSCX2-YW-H-10; Anhui Province Key Project Grant 08040102005; Natural Science Foundation of China Grants 31320103904, 31430054, 91313303, 31501130, and 31501095; MOE Innovative Team Grant IRT13038, Fundamental Research Funds for the Central Universities Grant WK2070000066; Chinese Academy of Sciences Center of Excellence Grant 2015HSC-UE010; and National Institutes of Health Grants DK56292 and CA146133. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
J. Zhou and X. Ding, unpublished observations.
- CTN
- cortactin
- SH3
- Src homology 3
- MAP
- microtubule-associated protein
- +TIP
- plus end tracking protein
- EB
- end-binding
- PACF
- photoactivatable complementary fluorescent
- N-WASP
- neuronal Wiskott-Aldrich syndrome protein.
References
- 1. Chen J., Yao Y., Gong C., Yu F., Su S., Chen J., Liu B., Deng H., Wang F., Lin L., Yao H., Su F., Anderson K. S., Liu Q., Ewen M. E., et al. (2011) CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 19, 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richardson B. E., and Lehmann R. (2010) Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat. Rev. Mol. Cell Biol. 11, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosen-Binker L. I., and Kapus A. (2006) Cortactin: the gray eminence of the cytoskeleton. Physiology 21, 352–361 [DOI] [PubMed] [Google Scholar]
- 4. Murphy D. A., and Courtneidge S. A. (2011) The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MacGrath S. M., and Koleske A. J. (2012) Cortactin in cell migration and cancer at a glance. J. Cell Sci. 125, 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lua B. L., and Low B. C. (2005) Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 579, 577–585 [DOI] [PubMed] [Google Scholar]
- 7. Watanabe T., Noritake J., and Kaibuchi K. (2005) Regulation of microtubules in cell migration. Trends Cell Biol. 15, 76–83 [DOI] [PubMed] [Google Scholar]
- 8. Mitchison T., and Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237–242 [DOI] [PubMed] [Google Scholar]
- 9. Kaverina I., and Straube A. (2011) Regulation of cell migration by dynamic microtubules. Semin. Cell Dev. Biol. 22, 968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stehbens S., and Wittmann T. (2012) Targeting and transport: how microtubules control focal adhesion dynamics. J. Cell Biol. 198, 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stehbens S. J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T. (2014) CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calvo F., Ege N., Grande-Garcia A., Hooper S., Jenkins R. P., Chaudhry S. I., Harrington K., Williamson P., Moeendarbary E., Charras G., and Sahai E. (2013) Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akhmanova A., and Steinmetz M. O. (2008) Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 14. Jiang K., Wang J., Liu J., Ward T., Wordeman L., Davidson A., Wang F., and Yao X. (2009) TIP150 interacts with and targets MCAK at the microtubule plus ends. EMBO Rep. 10, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ward T., Wang M., Liu X., Wang Z., Xia P., Chu Y., Wang X., Liu L., Jiang K., Yu H., Yan M., Wang J., Hill D. L., Huang Y., Zhu T., and Yao X. (2013) Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 288, 15771–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xia P., Zhou J., Song X., Wu B., Liu X., Li D., Zhang S., Wang Z., Yu H., Ward T., Zhang J., Li Y., Wang X., Chen Y., Guo Z., and Yao X. (2014) Aurora A orchestrates entosis by regulating a dynamic MCAK-TIP150 interaction. J. Mol. Cell Biol. 6, 240–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang Z., Miao Y., Ding X., Deng H., Liu S., Wang F., Zhou R., Watson C., Fu C., Hu Q., Lillard J. W. Jr., Powell M., Chen Y., Forte J. G., and Yao X. (2006) Proteomic identification and functional characterization of a novel ARF6 GTPase-activating protein, ACAP4. Mol. Cell. Proteomics 5, 1437–1449 [DOI] [PubMed] [Google Scholar]
- 18. Huang Y., Wang W., Yao P., Wang X., Liu X., Zhuang X., Yan F., Zhou J., Du J., Ward T., Zou H., Zhang J., Fang G., Ding X., Dou Z., and Yao X. (2012) CENP-E kinesin interacts with SKAP protein to orchestrate accurate chromosome segregation in mitosis. J. Biol. Chem. 287, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crostella L., Lidder S., Williams R., and Skouteris G. G. (2001) Hepatocyte growth factor/scatter factor-induces phosphorylation of cortactin in A431 cells in a Src kinase-independent manner. Oncogene 20, 3735–3745 [DOI] [PubMed] [Google Scholar]
- 20. Zhou R., Guo Z., Watson C., Chen E., Kong R., Wang W., and Yao X. (2003) Polarized distribution of IQGAP proteins in gastric parietal cells and their roles in regulated epithelial cell secretion. Mol. Biol. Cell 14, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang C., Liu J., Haudenschild C. C., and Zhan X. (1998) The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J. Biol. Chem. 273, 25770–25776 [DOI] [PubMed] [Google Scholar]
- 22. Head J. A., Jiang D., Li M., Zorn L. J., Schaefer E. M., Parsons J. T., and Weed S. A. (2003) Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell 14, 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang W., Liu Y., and Liao K. (2011) Tyrosine phosphorylation of cortactin by the FAK-Src complex at focal adhesions regulates cell motility. BMC Cell Biol. 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang W., Chen L., Ding Y., Jin J., and Liao K. (2008) Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J. Cell Sci. 121, 1334–1343 [DOI] [PubMed] [Google Scholar]
- 25. Wen Y., Eng C. H., Schmoranzer J., Cabrera-Poch N., Morris E. J., Chen M., Wallar B. J., Alberts A. S., and Gundersen G. G. (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6, 820–830 [DOI] [PubMed] [Google Scholar]
- 26. Zhang L., Shao H., Zhu T., Xia P., Wang Z., Liu L., Yan M., Hill D. L., Fang G., Chen Z., Wang D., and Yao X. (2013) DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration. Sci. Rep. 3, 1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galjart N. (2010) Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20, R528–537 [DOI] [PubMed] [Google Scholar]
- 28. Lynch D. K., Winata S. C., Lyons R. J., Hughes W. E., Lehrbach G. M., Wasinger V., Corthals G., Cordwell S., and Daly R. J. (2003) A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J. Biol. Chem. 278, 21805–21813 [DOI] [PubMed] [Google Scholar]
- 29. Garcia J. G., Verin A. D., Schaphorst K., Siddiqui R., Patterson C. E., Csortos C., and Natarajan V. (1999) Regulation of endothelial cell myosin light chain kinase by Rho, cortactin, and p60(src). Am. J. Physiol. 276, L989–998 [DOI] [PubMed] [Google Scholar]
- 30. Biosse Duplan M., Zalli D., Stephens S., Zenger S., Neff L., Oelkers J. M., Lai F. P., Horne W., Rottner K., and Baron R. (2014) Microtubule dynamic instability controls podosome patterning in osteoclasts through EB1, cortactin, and Src. Mol. Cell. Biol. 34, 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Preciado López M., Huber F., Grigoriev I., Steinmetz M. O., Akhmanova A., Koenderink G. H., and Dogterom M. (2014) Actin-microtubule coordination at growing microtubule ends. Nat. Commun. 5, 4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Small J. V., Stradal T., Vignal E., and Rottner K. (2002) The lamellipodium: where motility begins. Trends Cell Biol. 12, 112–120 [DOI] [PubMed] [Google Scholar]
- 33. Wu H., Reynolds A. B., Kanner S. B., Vines R. R., and Parsons J. T. (1991) Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol. Cell. Biol. 11, 5113–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tehrani S., Tomasevic N., Weed S., Sakowicz R., and Cooper J. A. (2007) Src phosphorylation of cortactin enhances actin assembly. Proc. Natl. Acad. Sci. U.S.A. 104, 11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agerer F., Lux S., Michel A., Rohde M., Ohlsen K., and Hauck C. R. (2005) Cellular invasion by Staphylococcus aureus reveals a functional link between focal adhesion kinase and cortactin in integrin-mediated internalisation. J. Cell Sci. 118, 2189–2200 [DOI] [PubMed] [Google Scholar]
- 36. Bowden E. T., Onikoyi E., Slack R., Myoui A., Yoneda T., Yamada K. M., and Mueller S. C. (2006) Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp. Cell Res. 312, 1240–1253 [DOI] [PubMed] [Google Scholar]
- 37. Davoli T., Xu A. W., Mengwasser K. E., Sack L. M., Yoon J. C., Park P. J., and Elledge S. J. (2013) Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., and Hess H. F. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 [DOI] [PubMed] [Google Scholar]
- 39. Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., and Yao X. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16564–16569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang H., Hu X., Ding X., Dou Z., Yang Z., Shaw A. W., Teng M., Cleveland D. W., Goldberg M. L., Niu L., and Yao X. (2004) Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 279, 54590–54598 [DOI] [PubMed] [Google Scholar]
- 41. Lou Y., Yao J., Zereshki A., Dou Z., Ahmed K., Wang H., Hu J., Wang Y., and Yao X. (2004) NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J. Biol. Chem. 279, 20049–20057 [DOI] [PubMed] [Google Scholar]
- 42. Wang X., Zhuang X., Cao D., Chu Y., Yao P., Liu W., Liu L., Adams G., Fang G., Dou Z., Ding X., Huang Y., Wang D., and Yao X. (2012) Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 287, 39380–39390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yao X., Abrieu A., Zheng Y., Sullivan K. F., and Cleveland D. W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 44. Mo F., Zhuang X., Liu X., Yao P. Y., Qin B., Su Z., Zang J., Wang Z., Zhang J., Dou Z., Tian C., Teng M., Niu L., Hill D. L., Fang G., Ding X., Fu C., and Yao X. (2016) Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat. Chem. Biol. 12, 226–232 [DOI] [PMC free article] [PubMed] [Google Scholar]