Abstract
The nuclear protein IκBζ, comprising the N-terminal trans-activation domain and the C-terminal ankyrin repeat (ANK) domain composed of seven ANK motifs, activates transcription of a subset of nuclear factor-κB (NF-κB)-dependent innate immune genes such as Lcn2 encoding the antibacterial protein lipocalin-2. Lcn2 activation requires formation of a complex containing IκBζ and NF-κB p50, a transcription factor that harbors the DNA-binding Rel homology region but lacks a trans-activation domain, on the promoter with the canonical NF-κB-binding site (κB site) and its downstream cytosine-rich element. Here we show that IκBζ productively interacts with p50 via Asp-451 in the N terminus of ANK1, a residue that is evolutionarily conserved among IκBζ and the related nuclear IκB proteins Bcl-3 and IκBNS. Threonine substitution for Asp-451 abrogates direct association with the κB-site-binding protein p50, complex formation with the Lcn2 promoter DNA, and activation of Lcn2 transcription. The basic residues Lys-717 and Lys-719 in the C-terminal region of ANK7 contribute to IκBζ binding to the Lcn2 promoter, probably via interaction with the cytosine-rich element required for Lcn2 activation; glutamate substitution for both lysines results in a loss of transcriptionally active complex formation without affecting direct contact of IκBζ with p50. Both termini of the ANK domain in Bcl-3 and IκBNS function in a manner similar to that of IκBζ to interact with promoter DNA, indicating a common mechanism in which the nuclear IκBs form a regulatory complex with NF-κB and promoter DNA via the invariant aspartate in ANK1 and the conserved basic residues in ANK7.
Keywords: gene expression, NF-κB transcription factor, protein motif, protein-DNA interaction, protein-protein interaction, Bcl-3, IκBζ, IκBNS, ankyrin repeat, lipocalin-2
Introduction
Nuclear factor-κB (NF-κB)2 plays central roles in host defense and inflammation as a homo- or heterodimer of NF-κB/Rel family proteins by controlling the expression of genes for pro-inflammatory cytokines, chemokines, and antibacterial proteins (1–4). The mammalian NF-κB family is composed of five structurally related polypeptides: p50, p52, p105 (the precursor of p50), p100 (the precursor of p52), p65 (also known as RelA), RelB, and c-Rel. They share the Rel homology region, which mediates dimerization, nuclear translocation, binding to specific DNA sequences known as NF-κB-binding elements (κB sites), and association with one of the IκB family proteins (1–4). Among the members of the family, p65, RelB, and c-Rel have an ability to activate transcription by themselves via the C-terminal trans-activation domain, which is absent in the smaller p50 and p52 proteins. In resting cells, NF-κB dimers are retained in the cytoplasm by associating with a member of the prototypical/cytoplasmic IκB proteins including IκBα, IκBβ, and IκBϵ (1–4). Cell activation with appropriate stimuli such as bacterial LPS leads to phosphorylation-induced degradation of cytoplasmic IκBs and resultant liberation of NF-κB dimers (1, 5, 6). The released NF-κB dimers subsequently translocate to the nucleus and thus induce the expression of primary response genes via binding to κB sites on their promoter/enhancer regions (1–4).
The primarily induced gene products include the atypical/nuclear IκB proteins Bcl-3 (7, 8), IκBζ (also known as MAIL or INAP) (9–11), and IκBNS (12), which are not expressed in resting cells. In contrast to the cytoplasmic IκBs, which preferentially associate with NF-κB dimers that possess at least one p65 or c-Rel subunit such as the p50-p65 heterodimer (13), the nuclear IκBs prefer the p50 (or p52) homodimer as a partner to directly regulate the activation of secondary response genes for appropriate host defense responses (14–19). Thus, a part of the secondary response genes is controlled by NF-κB-dependent induction of nuclear IκBs and their subsequent association with NF-κB. For instance, the secondarily induced genes Lcn2 (encoding the antibacterial protein lipocalin-2) and Ptx3 (encoding the antibacterial protein pentraxin 3) are activated by the nuclear IκB protein IκBζ (20–24), which serves as a key regulator in the immune system (16, 18, 25, 26). On the other hand, Bcl-3 participates in transcriptional control of the chemokine-encoding genes Ccl2 for monocyte chemoattractant protein-1 (MCP-1) and Cxcl10 for interferon-γ-induced protein 10 (IP-10) (27), and IκBNS regulates transcription of Il6 (encoding the pro-inflammatory cytokine interleukin-6) and Il12b (encoding interleukin-12 subunit p40) (28, 29).
The IκB proteins are characterized by the presence of the ankyrin repeat domain (ARD). The ARD of the cytoplasmic or nuclear IκBs contains six or seven ankyrin repeat (ANK) motifs, respectively. The ANK is an evolutionarily conserved protein motif of about 33 amino acid residues that forms an L-shaped structure comprising a β hairpin and two antiparallel α helices. Consecutive ANK motifs generally stack together to serve as an underlying architecture of a modular specific protein-interacting interface (30–32). The ARD of IκBs mediates the association with NF-κB dimers via direct binding to the NF-κB Rel homology region (23, 33, 34). Although the ARD of nuclear IκBs displays a high sequence similarity to that of cytoplasmic IκBs except for the additional seventh motif ANK7 (10, 12, 35, 36), the preference for NF-κB dimer species differs between the nuclear and cytoplasmic IκBs, as described above (7–9, 12, 16, 37). Furthermore, there exists a difference in interaction with κB DNA. Although IκBα promotes the dissociation of a p65-containing dimer from the promoter DNA (38), the nuclear IκBs are generally assumed to interact indirectly with the κB site via binding to NF-κB p50 or p52 (7, 8, 23, 33, 34). In addition, we have recently shown that IκBζ interaction with the Lcn2 promoter also requires a region downstream of the κB site (23). However, the molecular mechanism underlying assembly of the nuclear IκB-containing regulatory complex has not been well elucidated.
In the present study, we show that IκBζ, containing the N-terminal trans-activation domain and the C-terminal ARD, forms a transcriptionally active complex with the Lcn2 promoter via both Asp-451-mediated association with the κB-site-binding protein p50/p52 and Lys-717/Lys-719-dependent interaction with the downstream extra κB site on the Lcn2 promoter. Asp-451, an invariant residue among the nuclear IκBs, is present in the N terminus of ANK1; Lys-717 and Lys-719 exist in the region C-terminal to the second α helix of ANK7, and the corresponding sites are also occupied by basic residues in Bcl-3 and IκBNS. Both termini of the ARD in these proteins serve in a manner similar to that of IκBζ, indicative of a common mechanism by which nuclear IκBs form a p50/p52-containing regulatory complex on target gene promoters.
Results
The Invariant Aspartate in ANK1 of IκBζ and Other Nuclear IκBs Is Crucial for Interaction with NF-κB
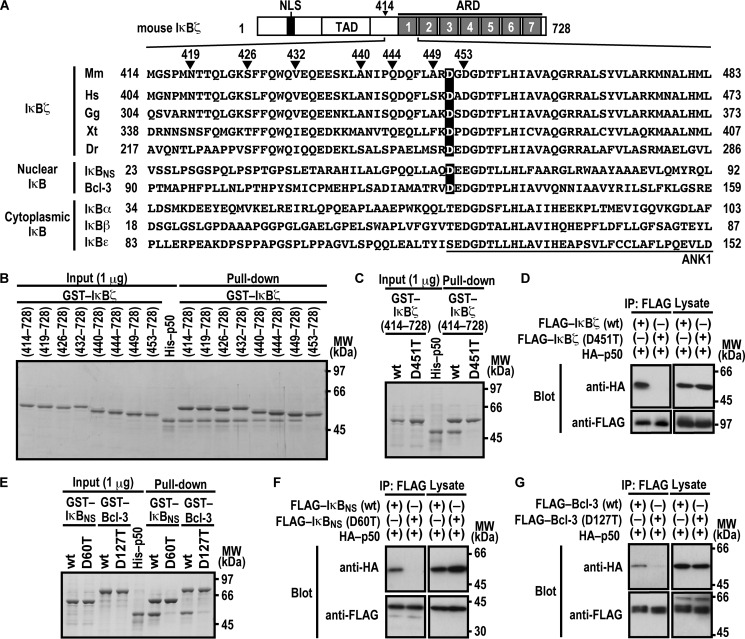
It is known that the ARD of the nuclear IκB proteins IκBζ, IκBNS, and Bcl-3 (Fig. 1A) is responsible for binding to NF-κB p50 (33, 34). To determine the N-terminal boundary of the IκBζ region required for interaction with p50, we expressed and purified a series of N-terminally truncated IκBζ as GST-fused proteins and tested their ability to bind to p50. As shown in Fig. 1B, IκBζ-(449–728) fully interacted with p50, as did IκBζ-(414–728). On the other hand, IκBζ-(453–728) failed to bind to p50 (Fig. 1B), suggesting a role for amino acid residues 449–452. Among the four residues, Asp-451 in ANK1 is the only one that is completely conserved during evolution (Fig. 1A). Intriguingly, the aspartate also exists in the other nuclear IκB proteins IκBNS and Bcl-3 (Asp-60 and Asp-127, respectively) but is replaced by threonine or serine in cytoplasmic IκB members (Fig. 1A). Consistent with the conservation, the replacement of Asp-451 by threonine impaired IκBζ binding to p50 in a GST pulldown assay (Fig. 1C). The D451T substitution in IκBζ also resulted in a loss of its co-immunoprecipitation with p50 when FLAG-tagged full-length IκBζ and HA-p50 were expressed in HEK293T cells (Fig. 1D). Thus, the invariant residue Asp-451 in ANK1 plays a crucial role in IκBζ interaction with p50.
FIGURE 1.
Asp-451 in ANK1 of IκBζ and the corresponding residues of IκBNS and Bcl-3 are involved in interaction with NF-κB p50. A, domain organization of mouse IκBζ and comparison of the amino acid sequences of ANK1 and its N-terminally flanking region. Mouse IκBζ of 728 amino acids contains the nuclear localization signal (NLS), the trans-activation domain (TAD), and the ARD comprised of seven ANK motifs. The amino acid sequences of ANK1 and its N-terminally flanking region of IκBζ from various species and those of other mouse IκB proteins are aligned. Mm, Mus musculus; Hs, Homo sapiens; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio. B, the IκBζ N-terminal boundary required for interaction with NF-κB p50. GST-fused IκBζ with the indicated truncation was incubated with His-p50 and pulled down with glutathione-Sepharose-4B beads, followed by SDS-PAGE analysis with CBB staining. MW, molecular weight. C and E, the role for IκBζ Asp-451, IκBNS Asp-60, and Bcl-3 Asp-127 in interaction with NF-κB p50 in vitro. GST-fused IκBζ (C), IκBNS (E), or Bcl-3 (E) with or without the indicated amino acid substitution was incubated with His-p50, followed by analysis as in B. D, F, and G, the role of IκBζ Asp-451, IκBNS Asp-60, and Bcl-3 Asp-127 in interaction with NF-κB p50 in vivo. FLAG-tagged IκBζ (D), IκBNS (F), or Bcl-3 (G) with or without the indicated amino acid substitution was co-expressed with HA-p50 in HEK293T cells, and proteins in the cell lysate were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblot analysis with the indicated antibody (Blot). Positions for marker proteins are indicated in kilodaltons.
We next investigated the role of the corresponding aspartate residue in other nuclear IκB proteins (Asp-60 in IκBNS and Asp-127 in Bcl-3). Threonine substitution for Asp-60 in IκBNS led to an impaired interaction with p50 both in a GST pulldown assay using purified proteins (Fig. 1E) and in a co-immunoprecipitation assay using proteins expressed in HEK293T cells (Fig. 1F). Similarly, compared with wild-type Bcl-3, a mutant protein with threonine substitution for Asp-127 interacted with p50 much less efficiently both in vitro (Fig. 1E) and in vivo (Fig. 1G). These findings highlight a conservative role for the invariant aspartate of ANK1 in direct interaction of nuclear IκBs with p50.
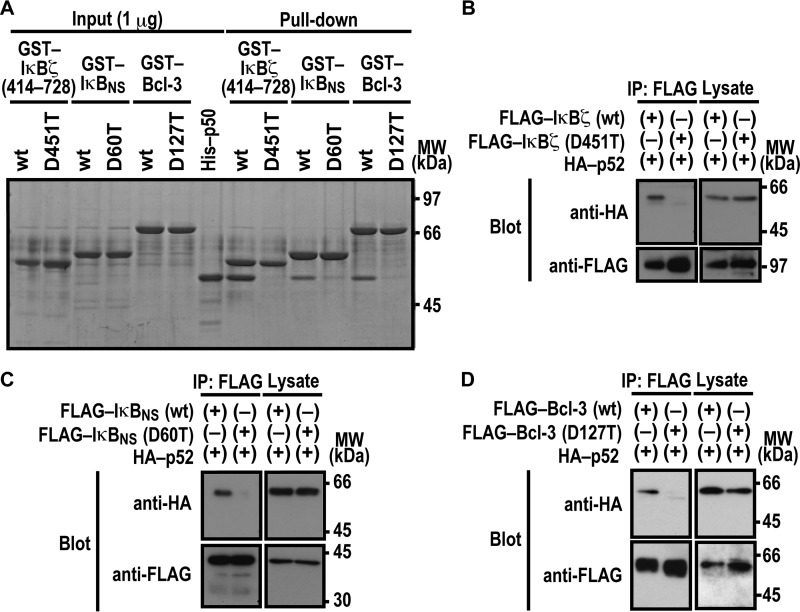
Nuclear IκB proteins are also known to form a complex with NF-κB p52, a protein homologous to p50 (7, 12, 37). As expected from the homology, in vitro complex formation with p52 was impaired by threonine substitution for the invariant aspartate in ANK1 of nuclear IκBs: Asp-451 in IκBζ, Asp-60 in IκBNS, and Asp-127 in Bcl-3 (Fig. 2A). The critical role for the aspartates was confirmed by co-precipitation of p52 with IκBζ (Fig. 2B), IκBNS (Fig. 2C), and Bcl-3 (Fig. 2D) when ectopically expressed in HEK293T cells. Thus, p52 likely interacts with nuclear IκB proteins in a manner similar to the way p50 does.
FIGURE 2.
Asp-451 in ANK1 of IκBζ and the corresponding residue of IκBNS and Bcl-3 are involved in interaction with NF-κB p52. A, the role for IκBζ Asp-451, IκBNS Asp-60, and Bcl-3 Asp-127 in interaction with NF-κB p50 in vitro. GST-fused IκBζ, IκBNS, and Bcl-3 with or without the indicated amino acid substitution were incubated with His-p52-(1–341), and pulled down with glutathione-Sepharose-4B beads, followed by SDS-PAGE analysis with CBB staining. MW, molecular weight. B–D, the role of IκBζ Asp-451, IκBNS Asp-60, and Bcl-3 Asp-127 in interaction with NF-κB p50 in vivo. FLAG-tagged IκBζ (B), IκBNS (C), or Bcl-3 (D) with or without the indicated amino acid substitution was co-expressed with HA-p52 in HEK293T cells, and proteins in the cell lysate were immunoprecipitated (IP) with the anti-FLAG antibody, followed by immunoblot analysis with the indicated antibody (Blot). Positions for marker proteins are indicated in kilodaltons.
Asp-451 of IκBζ Is Involved in Transcriptional Activation
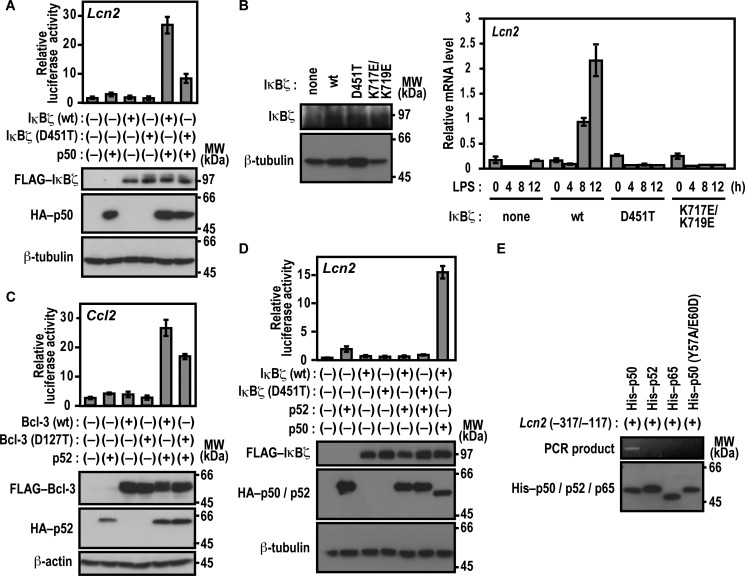
As we have shown previously (23), expression of IκBζ along with p50 results in transcriptional activation of the promoter of the lipocalin-2-encoding gene Lcn2 in p50-/IκBζ-deficient mouse embryonic fibroblasts (MEFs) (Nfkb1−/−;Nfkbiz−/− MEFs) (Fig. 3A). To examine the role for Asp-451 of IκBζ in p50-dependent Lcn2 activation, we used p50-/IκBζ-deficient MEFs in which a luciferase reporter is regulated by the Lcn2 promoter (23). As shown in Fig. 3A, IκBζ (D451T), defective in associating with p50, activated the Lcn2 promoter much less effectively than the wild-type protein even in the presence of p50. We next tested the role for Asp-451 of ANK1 in IκBζ-mediated activation of the endogenous Lcn2 gene. As shown in Fig. 3B, exogenous expression of wild-type IκBζ in IκBζ-deficient bone marrow-derived macrophages (BMMs) resulted in time-dependent activation of the endogenous Lcn2 gene in response to LPS. On the other hand, IκBζ (D451T), a mutant protein defective in direct interaction with NF-κB p50 (Fig. 1), failed to activate the endogenous Lcn2 gene, although IκBζ (D451T) was expressed at a level similar to that of the wild-type protein (Fig. 3B). Thus, the interaction between IκBζ and p50 appears to be involved in transcriptional activation of Lcn2.
FIGURE 3.
Asp-451 of IκBζ and Asp-127 of Bcl-3 participate in transcriptional activation. A and D, the role of IκBζ Asp-451 in Lcn2 activation. p50-/IκBζ-deficient MEFs were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of Lcn2 (−1031/+54), the internal control plasmid pRL-TK, pcDNA3 for expression of FLAG-IκBζ (WT) or FLAG-IκBζ (D451T), and HA-p50 (A) or HA-p52 (D). Luciferase activities were determined as described under “Experimental Procedures.” Each graph represents the mean ± S.D. obtained from three independent transfections. Cell lysates were analyzed by immunoblot with anti-FLAG, anti-HA, or anti-β-tubulin antibody. MW, molecular weight. B, IκBζ-mediated activation of the endogenous Lcn2 gene. IκBζ-deficient BMMs were retrovirally transduced for expression of WT IκBζ or a mutant protein with the D451T or K717E/K719E substitution. Proteins in the cell lysate were analyzed by immunoblot with the anti-IκBζ or anti-β-tubulin antibody (left panel). The transduced BMMs were stimulated for the indicated time with LPS, and the relative amounts of mRNA transcribed from the endogenous Lcn2 gene were estimated by quantitative real-time RT-PCR as described under “Experimental Procedures” (right panel). Each graph represents the mean ± S.D. in triplicate determinations. C, the role of Bcl-3 Asp-127 in Ccl2 activation. RAW264.7 cells were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of Ccl2 (−2777/+76), the internal control plasmid pRL-TK, and pcDNA3 for expression of FLAG-Bcl-3 (WT) or FLAG-Bcl-3 (D127T) and HA-p52. Luciferase activities were determined as described under “Experimental Procedures.” Each graph represents the mean ± S.D. obtained from three independent transfections. Cell lysates were analyzed by immunoblot with anti-FLAG, anti-HA, or anti-β-actin antibody. E, association of p50 with the Lcn2 gene promoter. His-tagged p50 (WT), p52 (WT), p65 (WT), or p50 (Y57A/E60D) was incubated with the Lcn2 gene promoter (−317/−117). After the protein-DNA complex was pulled down with COSMOGEL® His-Accept, the co-precipitated DNA was amplified by PCR, and the product was analyzed by agarose gel electrophoresis. The precipitated proteins were subjected to immunoblot analysis with anti-His antibody. Positions for marker proteins are indicated in kilodaltons.
We also investigated the function of Bcl-3 Asp-127, a residue that corresponds to Asp-451 in IκBζ and is crucial for binding to p50 and p52 (Figs. 1 and 2), in gene activation. As shown in Fig. 3C, Bcl-3 (D127T) activated a reporter of the Ccl2 promoter in LPS-stimulated RAW264.7 cells that ectopically expressed p52 but to a significantly lesser extent than the wild-type protein. Thus, Bcl-3 binding to p52 plays a role in Ccl2 activation. On the other hand, IκBζ did not activate Lcn2 in combination with p52 under conditions where p50 fully supported IκBζ (Fig. 3D). This raises the possibility that p52 is incapable of directly interacting with the Lcn2 promoter DNA, although p52 binds to IκBζ as p50 does (Fig. 2B). To address this question, we analyzed the formation of the protein-promoter complex using a DNA-binding assay in which DNA bound to a tagged protein is pulled down with tag affinity beads and amplified by PCR (for details, see “Experimental Procedures”). As shown in Fig. 3E, the Lcn2 promoter efficiently interacted with wild-type p50 but not with p50 (Y57A/E60D), a mutant protein defective in binding to the κB site (23), or p65. In contrast to p50, wild-type p52 failed to directly bind to the Lcn2 promoter (Fig. 3E). Thus, IκBζ appears to activate Lcn2 by specifically interacting with p50.
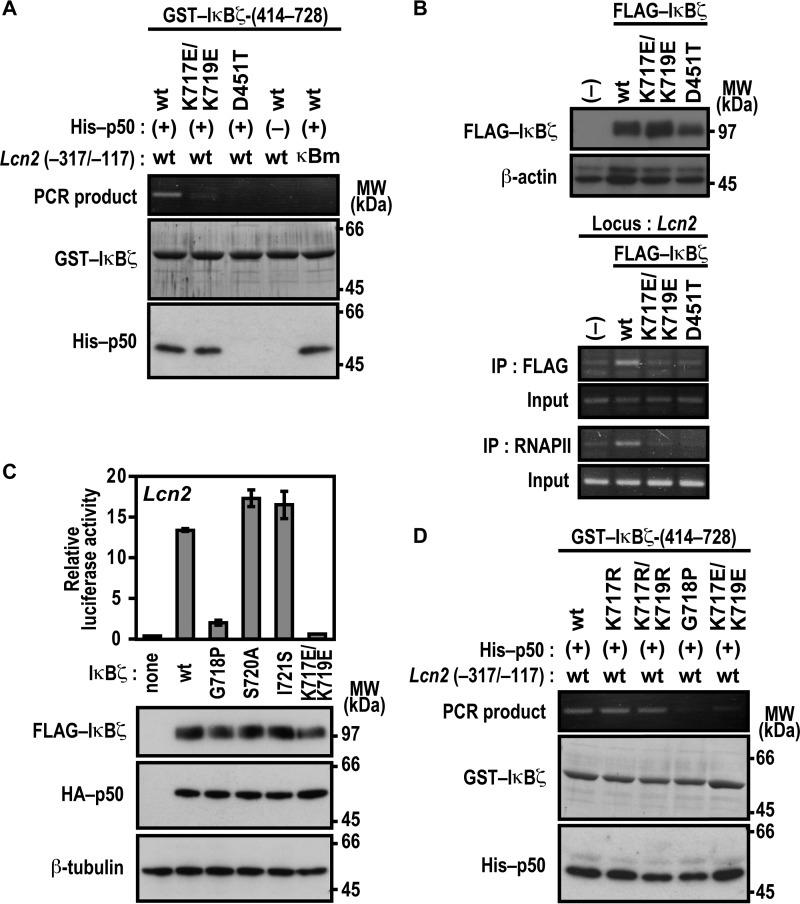
Lys-717 and Lys-719 in ANK7 of IκBζ Participate in Lcn2 Activation
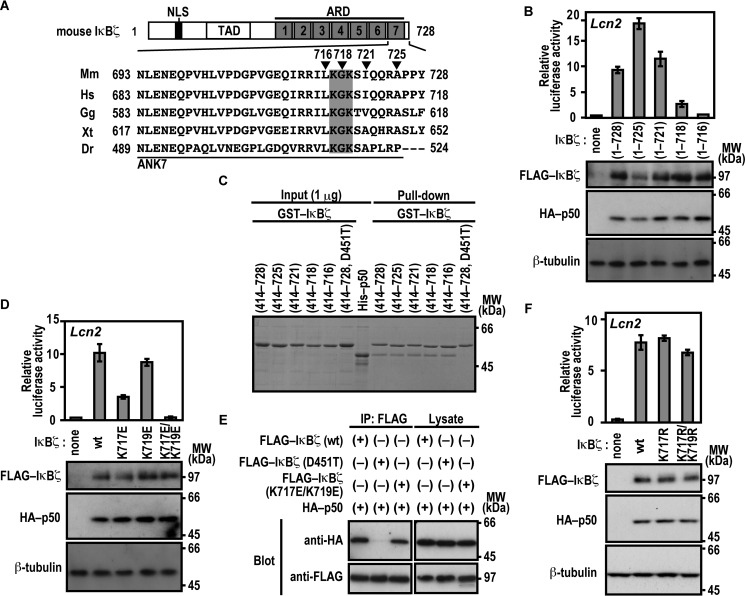
We next studied the role for the C-terminal region of the IκBζ ARD, which comprises seven ANK repeats (Fig. 4A). For this purpose, we expressed a series of C-terminally truncated IκBζ proteins in Nfkb1−/−;Nfkbiz−/− MEFs to test their ability to activate the Lcn2 promoter. As shown in Fig. 4B, IκBζ-(1–721) was as active as full-length IκBζ of 728 amino acids. By contrast, Lcn2 was activated much more weakly by IκBζ-(1–718) and only marginally by IκBζ-(1–716) (Fig. 4B), indicating a crucial role for the C-terminal region of IκBζ ANK7 (amino acids 717–721) in Lcn2 activation. On the other hand, this region was dispensable for direct contact of IκBζ with p50, as estimated by the GST pulldown assay (Fig. 4C). The dispensability is in contrast with the involvement of IκBζ ANK1 in direct interaction with p50 (Fig. 1) as well as in Lcn2 activation (Fig. 3). The IκBζ C-terminal region crucial for Lcn2 activation contains the basic residues Lys-717 and Lys-719, both of which are evolutionarily well conserved (Fig. 4A). The Lcn2 promoter was activated by a mutant IκBζ carrying substitution of the acidic residue glutamate for Lys-717, but to a lesser extent than the wild-type protein (Fig. 4D), and double glutamate substitution for Lys-717 and Lys-719 led to an almost complete loss of Lcn2 activation (Fig. 4D) without affecting the ability to directly interact with p50 (Fig. 4E). Furthermore, LPS-induced activation of the endogenous Lcn2 gene was not observed in BMMs expressing a mutant IκBζ with the K717E/K719E substitution (Fig. 3B). In contrast, simultaneous replacement of Lys-717 and Lys-719 by the other basic residue arginine hardly affected Lcn2 activation (Fig. 4F). These observations imply a possible role for the positive charge at amino acid positions 717 and 719 in IκBζ ANK7.
FIGURE 4.
Lys-717 and Lys-719 in ANK7 of IκBζ are crucial for activation of the Lcn2 gene. A, amino acid sequence alignment of ANK7 in IκBζ from various species. TAD, trans-activation domain; Mm, Mus musculus; Hs, Homo sapiens; Gg, Gallus gallus; Xt, Xenopus tropicalis, Dr, Danio rerio. B, the IκBζ C-terminal boundary required for Lcn2 activation. p50-/IκBζ-deficient MEFs were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of Lcn2 (−1031/+54); the internal control plasmid pRL-TK, and pcDNA3 for expression of FLAG-IκBζ with the indicated truncation and HA-p50. Luciferase activities were determined as described under “Experimental Procedures.” Each graph represents the mean ± S.D. obtained from three independent transfections. Cell lysates were analyzed by immunoblot with anti-FLAG, anti-HA, or anti-β-tubulin antibody. MW, molecular weight. C, the IκBζ C-terminal boundary required for interaction with NF-κB p50. GST-fused IκBζ with the indicated truncation was incubated with His-p50 and pulled down with glutathione-Sepharose-4B beads, followed by SDS-PAGE analysis with CBB staining. D, the role of Lys-717 and Lys-719 of IκBζ ANK7 in Lcn2 activation. The ability of FLAG-IκBζ with the indicated amino acid substitution was analyzed as in B. E, the role of Lys-717 and Lys-719 of IκBζ ANK7 in interaction with NF-κB p50 in vivo. FLAG-tagged IκBζ with the K717E/K719E or D451T substitution was co-expressed with HA-p50 in HEK293T cells, and proteins in the cell lysate were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblot analysis with the indicated antibody (Blot). F, the effect of arginine substitution for Lys-717 and Lys-719 on IκBζ-mediated Lcn2 activation. The ability of FLAG-IκBζ with the indicated amino acid substitution was analyzed as in B. Positions for marker proteins are indicated in kilodaltons.
Lys-717 and Lys-719 in ANK7 of IκBζ Participate in Complex Formation with the Lcn2 Promoter and Not via p50
It should be noted that the basic residues lysine and arginine are both capable of not only forming a salt bridge with a phosphate group of DNA but also of directly interacting with a base of DNA, especially guanine (39, 40). Furthermore, we have shown recently that a cytosine-rich region downstream of the κB site is involved in formation of the active three-species complex IκBζ-p50-DNA (23), suggesting that the extra-κB site makes a direct contact with p50, IκBζ, or both. It seems thus possible that the arginine-replaceable residues Lys-717 and Lys-719 in ANK7 participate in IκBζ interaction with the Lcn2 promoter DNA. To test this possibility, we analyzed the in vitro formation of the IκBζ-p50-DNA complex using the method used in Fig. 3E. Under conditions where the Lcn2 promoter fragment was fully co-precipitated with wild-type IκBζ, the co-precipitation was not caused by IκBζ (D451T), defective in direct binding to p50 (Fig. 5A). The D451T substitution also disrupted IκBζ interaction with the endogenous promoter of Lcn2, as shown by ChIP analysis using cells that stably expressed the wild-type or mutant protein (Fig. 5B). Further analysis with the anti-RNA polymerase II antibody revealed that the impairment of IκBζ-p50 association resulted in loss of the in vivo formation of a transcriptionally active complex on the Lcn2 promoter (Fig. 5B). The requirement of IκBζ-p50 association for interaction with the promoter is in good agreement with the following observation. Both p50 protein and the p50-binding site on the promoter (the κB site) were required for IκBζ interaction with DNA (Fig. 5A). Thus, Asp-451 indirectly participates in complex formation with the Lcn2 promoter via direct binding to p50. On the other hand, Lys-717 and Lys-719 in IκBζ may directly interact with target DNA because the K717E/K719E substitution abolished not only the recruitment to the Lcn2 endogenous promoter for subsequent active complex formation (Fig. 5B) but also the in vitro interaction with the target DNA without affecting binding to p50 (Fig. 5A). As indicated from the finding that arginine can replace Lys-717 and Lys-719 in Lcn2 activation (Fig. 4F), the replacement did not affect active complex formation with the Lcn2 promoter (Fig. 5D). These findings indicate that IκBζ interacts with the Lcn2 promoter via the arginine-replaceable residues Lys-717 and Lys-719 in a manner independent of direct binding to p50.
FIGURE 5.
Lys-717 and Lys-719 in ANK7 of IκBζ are crucial for association with the Lcn2 promoter DNA. A and D, GST-fused IκBζ-(414–728) or a mutant IκBζ carrying the indicated substitution were incubated with His-p50 in the presence of WT Lcn2 (−317/−117) or a mutated κB site (κBm)-carrying Lcn2 (−317/−117). After the protein-DNA complex was pulled down with glutathione-Sepharose-4B beads, the co-precipitated DNA was amplified by PCR, and the product was analyzed by agarose gel electrophoresis. The precipitated proteins were subjected to SDS-PAGE, followed by staining with CBB or immunoblot with anti-His antibody. MW, molecular weight. B, formaldehyde-fixed chromatin was prepared from RAW264.7 cells stably expressing FLAG-IκBζ (WT), FLAG-IκBζ (K717E/K719E), or FLAG-IκBζ (D451T) (top panel) and subjected to ChIP assay using anti-FLAG (M2) mouse monoclonal antibody or anti-RNA polymerase II antibody (bottom panel). Precipitated DNA was analyzed by PCR using primers corresponding to the Lcn2 locus. The results are representative of experiments from at least three independent experiments. IP, immunoprecipitation. C, the role of Gly-718, Ser-720, and Ile-721 in Lcn2 activation. p50-/IκBζ-deficient MEFs were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of Lcn2 (−1031/+54), the internal control plasmid pRL-TK, and pcDNA3 for expression of FLAG-IκBζ with the indicated amino acid substitution and HA-p50. Luciferase activities were determined as described under “Experimental Procedures.” Each graph represents the mean ± S.D. obtained from three independent transfections. Cell lysates were analyzed by immunoblot with anti-FLAG, anti-HA, or anti-β-tubulin antibody. Positions for marker proteins are indicated in kilodaltons.
Furthermore, proline substitution for the evolutionarily well conserved residue Gly-718, which locates between Lys-717 and Lys-719 in IκBζ ANK7 (Fig. 4A), resulted in a loss of both interaction with the Lcn2 promoter (Fig. 5A) and Lcn2 activation (Fig. 5C). The observation suggests that correct orientation of Lys-717 and Lys-719 toward the target DNA may be required for association of IκBζ to the Lcn2 promoter. On the other hand, neither S720A nor I721S substitution affected Lcn2 activation (Fig. 5C).
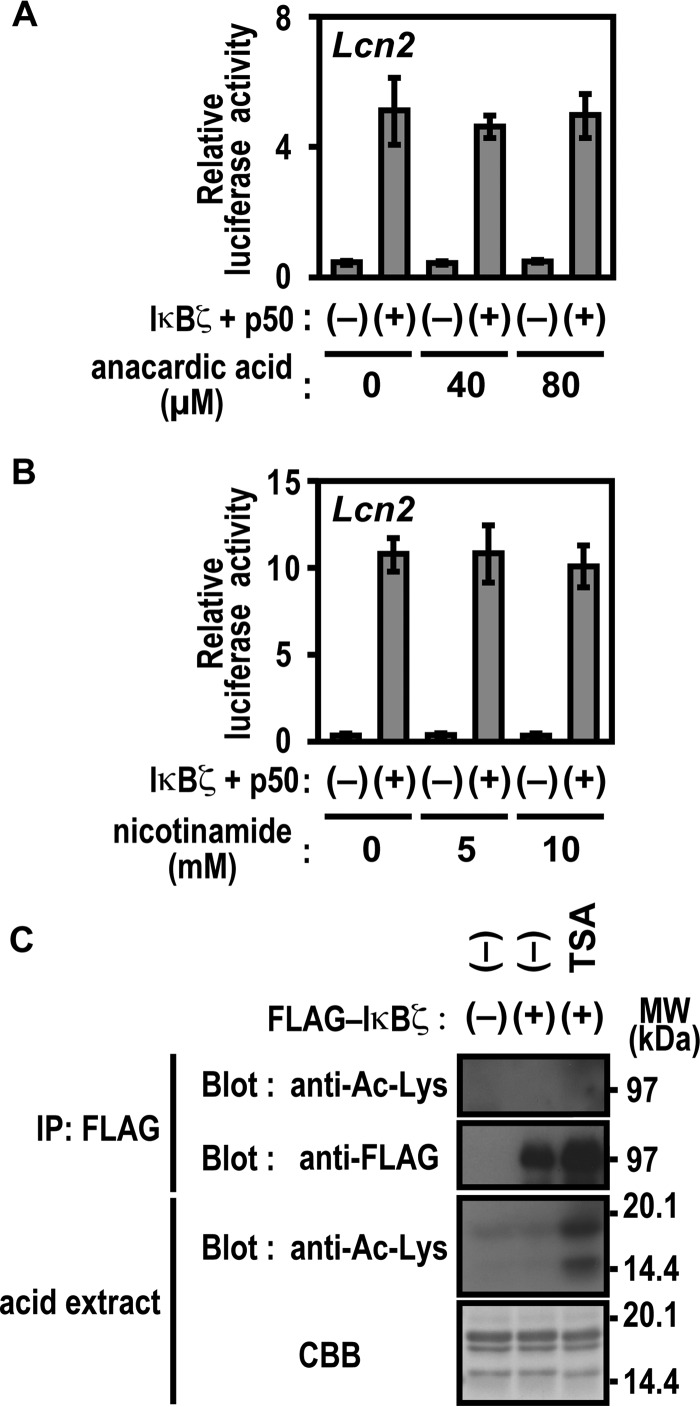
Acetylation of IκBζ Does Not Seem to Be Involved in Lcn2 Activation
The significance of Lys-717 and Lys-719 of IκBζ in Lcn2 activation may suggest the involvement of posttranslational modification of these lysine residues, such as acetylation. To test this possibility, we treated cells with the potent histone acetyltransferase inhibitor anacardic acid (41, 42) and nicotinamide, an agent that inhibits the NAD+-dependent class III (sirtuin) family of histone deacetylases (HDACs) (43, 44). Lcn2 activation by IκBζ remained unaffected by these inhibitors (Fig. 6, A and B). Under conditions where histone acetylation was significantly enhanced by the presence of trichostatin A (TSA), an inhibitor of class I/II HDAC (43, 44), lysine residues in IκBζ were not acetylated (Fig. 6C). Furthermore, a mobility shift of IκBζ on an SDS-PAGE gel, which was expected to occur by posttranslational modification, was not induced by TSA (Fig. 6C). These observations suggest that acetylation of Lys-717 or Lys-719 of IκBζ does not participate in Lcn2 activation, which also appears to be supported by the finding that neither gene activation nor promoter association are prevented by replacement of the lysines with arginine, a residue that does not undergo acetylation (Figs. 4F and 5D).
FIGURE 6.
Protein acetylation is not involved in IκBζ-mediated Lcn2 activation. A and B, effect of anacardic acid (A) and nicotinamide (B) on IκBζ-mediated Lcn2 activation. p50-/IκBζ-deficient MEFs were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of Lcn2 (−1031/+54), the internal control plasmid pRL-TK, and the pcDNA3 vector or pcDNA3 for expression of FLAG-IκBζ and HA-p50. Cells were incubated for 12 h with the indicated concentrations of the histone acetyl transferase inhibitor anacardic acid (A) or the HDAC inhibitor nicotinamide (B), and luciferase activities were determined as described under “Experimental Procedures.” Results are means ± S.D. for three independent transfections. C, effect of the HDAC inhibitor TSA on IκBζ. HEK293T cells expressing FLAG-IκBζ were incubated for 6 h with 2 μm TSA. The cell lysates were applied to immunoprecipitation (IP) with anti-FLAG antibody. Precipitated FLAG-IκBζ as well as acid extracted histones in the nuclear extracts were analyzed by immunoblot with the indicated antibodies or by staining with CBB. Positions for marker proteins are indicated in kilodaltons.
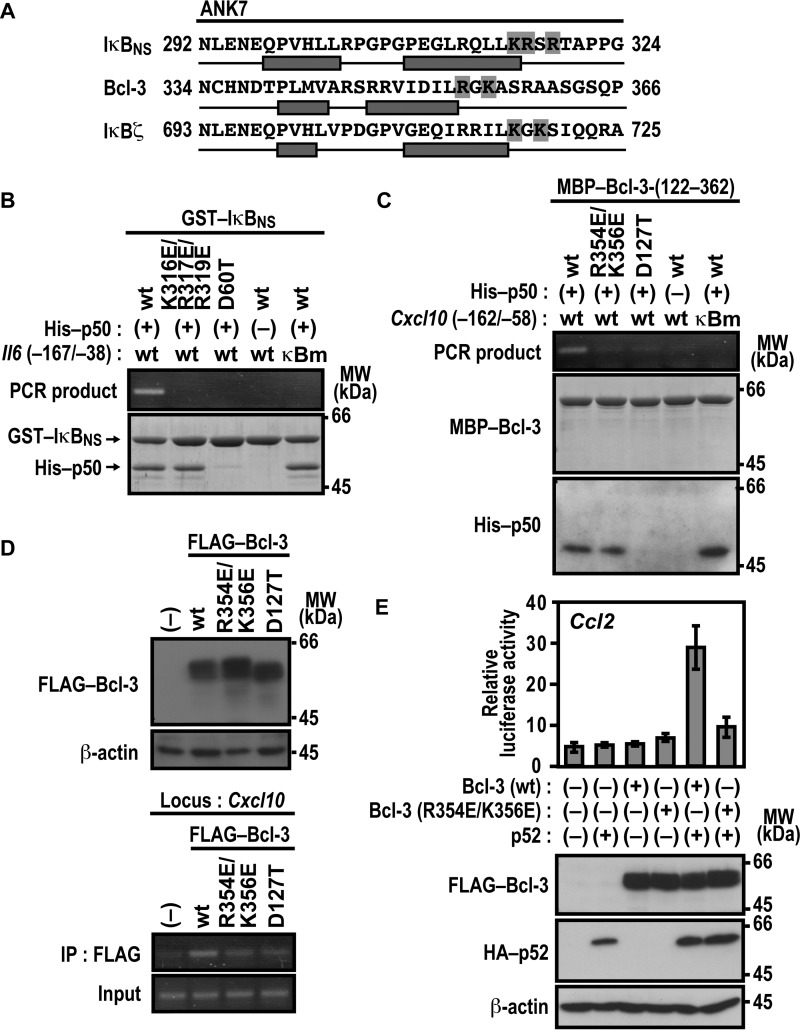
Basic Residues in ANK7 of IκBNS and Bcl-3 Are Involved in Association with Target DNA
Lys-717 and Lys-719 of IκBζ are predicted to follow the second α helix in ANK7, and basic residues also exist at the corresponding sites in ANK7 of IκBNS and Bcl-3 (Fig. 7A). As shown in Fig. 7B, association of IκBNS with the promoter of the IκBNS-regulated gene Il6 was prevented by simultaneous glutamate substitution for Lys-316, Arg-317, and Arg-319 in ANK7, although the substitution did not affect IκBNS binding to p50. Similarly, glutamate substitution for Arg-354 and Lys-356 in ANK7 of Bcl-3 resulted in a loss of complex formation with the promoter of the Bcl-3-dependent gene Cxcl10 without affecting Bcl-3 interaction with p50 (Fig. 7C), and Bcl-3 (R354E/K356E) also failed to associate with the endogenous promoter of Cxcl10, as indicated by ChIP analysis (Fig. 7D). Thus, in IκBNS and Bcl-3, basic residues that follow the second α helix in ANK7 appear to be involved in recognition of their target gene promoters. Furthermore, the R354E/K356E substitution in Bcl-3 resulted in loss of the p52-dependent Ccl2 activation in LPS-stimulated RAW264.7 cells (Fig. 7E), confirming the significance of the basic residues in ANK7. The DNA-binding assays also revealed that the invariant aspartate residues in ANK1 of IκBNS (Asp-60) (Fig. 7B) and Bcl-3 (Asp-127) (Fig. 7, C and D) participate in association with their target gene promoters via direct binding to p50, similar to the corresponding residue of IκBζ (Asp-451) (Fig. 5A).
FIGURE 7.
Basic residues in ANK7 of IκBNS and Bcl-3 participate in association with their target gene promoter. A, amino acid sequence alignment of ANK7 in the mouse nuclear IκB proteins IκBζ, Bcl-3, and IκBNS. The two antiparallel α helices in ANK7 of Bcl-3 were determined in crystal structures of Bcl-3 (36), whereas those of IκBζ and IκBNS were predicted from their primary sequences by using the program PSIPRED (45, 46). Residues boxed in gray are the basic residues that follow the second α helix of ANK7. B, complex formation of IκBNS with p50 and the Il6 promoter. GST-fused WT IκBNS or a mutant IκBNS with the K316E/R317E/R319E or D60T substitution was incubated with or without His-p50 in the presence of WT Il6 (−167/−38) or a mutated κB site (κBm)-carrying Il6 (−167/−38). After the protein-DNA complex was pulled down with glutathione-Sepharose-4B beads, the co-precipitated DNA was amplified by PCR, and the product was analyzed by agarose gel electrophoresis. The precipitated proteins were subjected to SDS-PAGE, followed by staining with CBB. MW, molecular weight. C, complex formation of Bcl-3 with p50 and the Cxcl10 promoter. MBP-fused WT Bcl-3-(122–362) or a mutant protein with the R354E/K356E or D127T substitution was incubated with or without His-p50 in the presence of Cxcl10 (−162/−58) or a mutated κB site (κBm)-carrying Cxcl10 (−162/−58). After the protein-DNA complex was pulled down with amylose resins, the co-precipitated DNA was analyzed as in B. The precipitated proteins were subjected to SDS-PAGE, followed by staining with CBB or immunoblot with the anti-His antibody. D, formaldehyde-fixed chromatin was prepared from RAW264.7 cells stably expressing FLAG-Bcl-3 (WT), FLAG-Bcl-3 (R354E/K356E), or FLAG-Bcl-3 (D127T) (top panel) and subjected to ChIP assay using anti-FLAG (M2) mouse monoclonal antibody (bottom panel). Precipitated DNA was analyzed by PCR using primers corresponding to the Cxcl10 locus. The results are representative of experiments from at least three independent experiments. IP, immunoprecipitation. E, the role of Arg-354 and Lys-356 of Bcl-3 ANK7 in Ccl2 activation. RAW264.7 cells were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of Ccl2 (−2777/+76), the internal control plasmid pRL-TK, and pcDNA3 for expression of FLAG-Bcl-3 (WT) or FLAG-Bcl-3 (R354E/K356E) and HA-p52. Luciferase activities were determined as described under “Experimental Procedures.” Each graph represents the mean ± S.D. obtained from three independent transfections. Cell lysates were analyzed by immunoblot with anti-FLAG, anti-HA, or anti-β-actin antibody. Positions for marker proteins are indicated in kilodaltons.
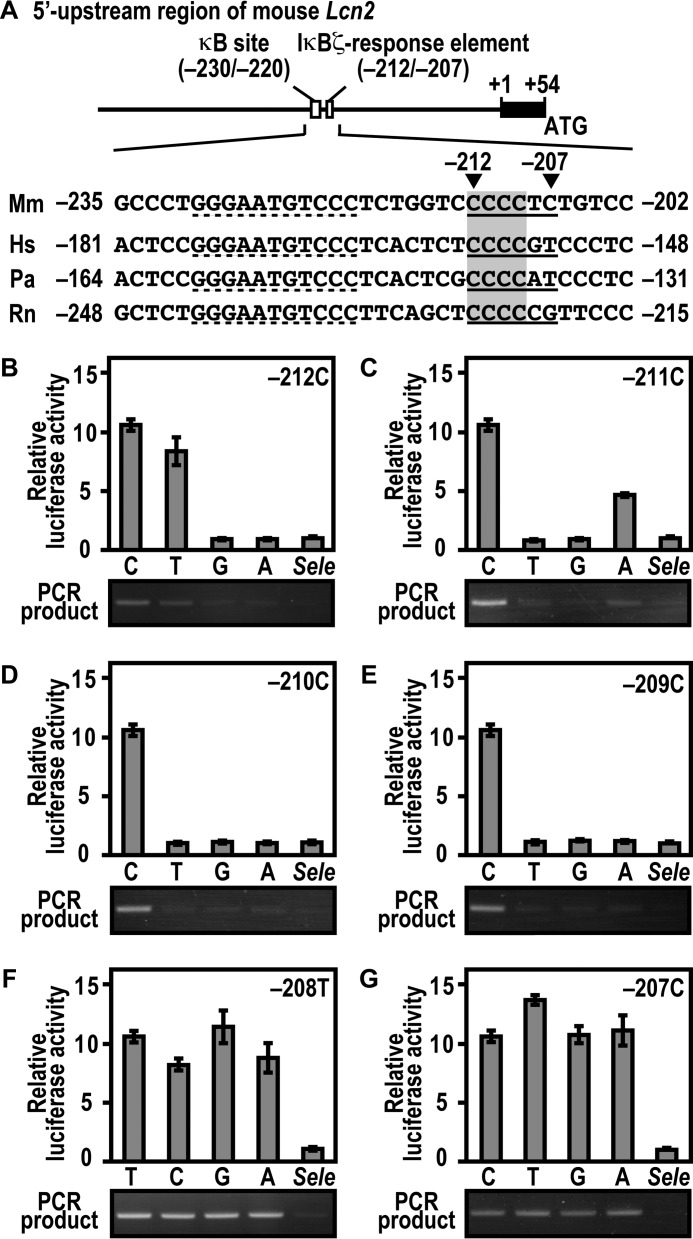
Activation of Lcn2 Involves Promoter Association with IκBζ via an Extra-κB Site in a Sequence-specific Manner
Activation of the mouse Lcn2 gene requires promoter association with the IκBζ-p50 complex via both the κB site (5′-GGGAATGTCCC-3′ at positions −230 to −220 relative to the transcription start site) and its downstream region of 5′-CCCCTC-3′ at positions −212 to −207 (23) (see Fig. 8A). To elucidate base specificity in the downstream sequence, we constructed a series of mutant Lcn2 promoters and tested their ability. Substitution of either guanine or adenine for cytosine at position −212 led to a loss of both interaction with IκBζ and IκBζ-mediated Lcn2 activation (Fig. 8B). On the other hand, they were only marginally impaired by thymine replacement at the corresponding position (Fig. 8B). Adenine but not thymine or guanine partially replaced cytosine at position −211 (Fig. 8C). At positions −210 (Fig. 8D) and −209 (Fig. 8E), cytosine was strictly required for both IκBζ binding to the Lcn2 promoter and IκBζ-mediated Lcn2 activation. By contrast, any of four bases fully functioned at positions −208 (Fig. 8F) and −207 (Fig. 8G). In the sequence CCCCTC at positions −212 to −207 of the mouse Lcn2 promoter, the preference for cytosine at positions −212 to −209 but not at position −208 or −207 (Fig. 8, B–G) appears to be consistent with the conservation of the first four cytosines but not the last two bases in the corresponding region from other mammals (Fig. 8A). These mutational analyses indicate that the IκBζ-responsive element (positions −212 to −209) requires the sequence 5′-YCCC-3′ (Y is pyrimidine). Taken together with the present findings, IκBζ activates the endogenous Lcn2 gene both via Asp-451-dependent direct interaction with p50 and Lys-717/Lys-719-involved association with the sequence 5′-YCCC-3′ downstream of the κB site.
FIGURE 8.
Interaction of IκBζ with the Lcn2 promoter, and its activation requires the 5′-YCCC-3′ sequence downstream of the κB site. A, the 5′ upstream promoter region of the mouse Lcn2 gene. The κB site and its downstream IκBζ-response element in the region are underlined. The nucleotide sequences of the corresponding region from various species are aligned. Mm, Mus musculus; Hs, Homo sapiens; Pa, Papio anubis; Rn, Rattus norvegicus. B–G, base preference of the extra-κB site in Lcn2 activation (top panels) and in formation of the three-species complex containing IκBζ, p50, and promoter DNA (bottom panels). For estimation of Lcn2 activation, p50-/IκBζ-deficient MEFs were transfected with the following plasmids: the luciferase reporter plasmid pGL3-Basic containing the upstream region of wild-type Lcn2 (−500/+50); a mutant Lcn2 with the indicated base replacement, or the IκBζ-independent gene Sele (−445/+105); the internal control plasmid pRL-TK; and pcDNA3 for expression of FLAG-IκBζ and HA-p50. Luciferase activities were determined as described under “Experimental Procedures.” Each graph represents the mean ± S.D. obtained from three independent transfections. For estimation of protein-DNA complex formation, GST-IκBζ was incubated with His-p50 in the presence of the DNA fragment of wild-type Lcn2 (−500/+50), a mutant Lcn2 with the indicated base replacement, or Sele (−445/+105). After the complex was pulled down with glutathione-Sepharose-4B beads, the co-precipitated DNA was amplified by PCR, and the product was analyzed by agarose gel electrophoresis.
Discussion
The nuclear IκB proteins IκBζ, Bcl-3, and IκBNS are thought to regulate NF-κB-dependent transcription by directly interacting with a p50 or p52 homodimer that binds to the κB site on target genes. However, the mechanism for formation of the regulatory complex has not been fully elucidated. In the present study, we show that IκBζ, comprising the N-terminal trans-activation domain and the C-terminal ARD composed of seven ANK motifs, forms a transcriptionally active complex on its target gene Lcn2 both via Asp-451-mediated binding to p50 and via Lys-717/Lys-719-dependent interaction with the extra-κB site of the Lcn2 promoter. Asp-451 is present in the N-terminal region of ANK1, whereas the basic residues Lys-717 and Lys-719 exist in the C-terminal region of ANK7. We also demonstrate similar roles for both termini of the ARD in Bcl-3 and IκBNS, proposing a model for a common mechanism by which nuclear IκBs form a p50/p52-containing complex on target gene promoters.
Asp-451 in IκBζ ANK1 is strictly conserved during evolution (Fig. 1). Replacement of Asp-451 by threonine abrogates both association with a homodimer of the NF-κB subunit p50 (Fig. 1) and activation of Lcn2 via formation of the IκBζ-p50-DNA complex on the promoter (Figs. 3 and 5). The aspartate residue is also conserved among the nuclear IκBs, including Bcl-3 and IκBNS; however, it is replaced by threonine or serine in cytoplasmic IκBs such as IκBα and IκBβ (Fig. 1), which associate with a p50/p52 homodimer much less efficiently (13). The conservation among the nuclear IκBs is consistent with the present finding that the corresponding aspartate residues (Asp-127 in Bcl-3 and Asp-60 in IκBNS) are also crucial for interaction with a p50 or p52 homodimer (Figs. 1 and 2). It should be noted that Asp-451 in IκBζ is one of the very few residues that are completely conserved among nuclear IκBs but replaced in cytoplasmic IκBs. On the other hand, the presence of aspartate at this position by itself does not seem to be sufficient because a mutant IκBα carrying aspartate substitution for Thr-71 at the corresponding position of ANK1 as well as the wild-type protein fails to bind to a p50 homodimer (data not shown).
Although little is known about the tertiary structure of nuclear IκB·NF-κB complexes, crystal structures of the cytoplasmic IκB proteins IκBα and IκBβ complexed with a p65-p50 heterodimer and a p65 homodimer, respectively, have been solved (45–47). If IκBζ interacts with NF-κB subunits in a manner similar to the cytoplasmic IκBs, then it seems likely that Asp-451 in IκBζ is positioned toward the C-terminally localized nuclear localization signal (NLS) in p50, a region that is required not only for nuclear localization of p50 but also for direct interaction of p50 with IκBζ (23, 48). Because of the dual role of the p50 NLS-containing region, the requirement of direct p50-IκBζ interaction for gene activation has not been established, although an NLS-truncated p50 protein did not activate IκBζ-dependent transcription, and thus IκBζ was assumed to interact with the promoter DNA via association with p50, which directly binds to the κB site (23, 48). The conclusion appears to be strongly supported by the present findings that the D451T substitution in IκBζ abrogates interaction with p50 (Fig. 1), complex formation on both endogenous and exogenous Lcn2 promoter (Fig. 5), and activation of Lcn2 transcription (Fig. 3).
In addition to direct association with the κB-site-binding protein p50, IκBζ activates Lcn2 transcription by interacting with the extra-κB site of the Lcn2 promoter in which the basic residues Lys-717 and Lys-719 in the C-terminal region of IκBζ ANK7 likely play a major role (Figs. 4 and 5). The association does not appear to be mediated via direct binding to p50 because the K717E/K719E substitution in IκBζ leads to a loss of both promoter association and Lcn2 activation without affecting direct contact of IκBζ with p50 (Figs. 4 and 5). In addition, the function of the lysine residues does not seem to require their modification, such as acetylation (Fig. 6), which is supported by the finding that arginine, a residue insusceptible to acetylation, fully serves in place of them (Figs. 4 and 5).
The extra-κB site required for IκBζ-dependent Lcn2 activation localizes seven bases downstream of the κB site in the Lcn2 promoter (23) and strictly requires the sequence 5′-YCCC-3′ as an IκBζ-responsive element, as shown in this study (Fig. 8). In both promoter association and Lcn2 activation, Lys-717 and Lys-719 in IκBζ are fully replaced by arginine residues (Figs. 4 and 5). It is known that lysine and arginine, but not glutamate, are often involved in direct interaction with DNA, which can be mediated not only via nonspecific interaction with the phosphate moiety of DNA but also via recognition of a DNA base, especially a guanine (39, 40). In this context, it should be noted that the IκBζ-responsive element (5′-YCCC-3′) of the Lcn2 promoter is abundant in the Lys/Arg-recognizing base guanine in the antisense strand. The presence of multiple guanines in the element may be in agreement with the present conclusion that Lys-717 and Lys-719 each contribute to element recognition because single glutamate substitution for either lysine residue results in only a partial loss of the activity of IκBζ (Fig. 4). Correct orientation of the side chains of Lys-717 and Lys-719 toward target DNA also seems to be important for element recognition, as indicated by the finding that replacement of the flexible residue Gly-718, which is the intervening amino acid between the lysines and is strictly conserved during evolution, by the inflexible residue proline abrogates both binding to the Lcn2 promoter and transcription of Lcn2 (Fig. 5).
The NF-κB family proteins serve as a homo- or heterodimer to bind to a κB DNA response element in the promoters of distinct inducible genes, thereby playing their respective roles in gene regulation (1–4). The transcriptional specificity of NF-κB dimers is generally thought to be coded within the κB site sequences (27, 49). Indeed, in the case of the Lcn2 promoter, its κB site effectively interacts with the NF-κB p50 homodimer but not with the p52 homodimer (Fig. 3), although both homodimers are capable of binding to IκBζ (Fig. 2). As a result, in contrast to p50, p52 fails to induce IκBζ-dependent activation of the Lcn2 gene (Fig. 3). In addition to the κB site sequence itself, the ability of NF-κB dimers to function on a particular promoter is also considered to be determined by extra-κB sites recognized via co-activator proteins (16, 50). However, such a co-activator was not previously identified. This study demonstrates that IκBζ, a co-activator of p50, activates the Lcn2 gene not only via binding to p50 but also by associating with the IκBζ-responsive element (5′-YCCC-3′) downstream of the κB site, providing evidence that an extra-κB site and its interacting co-activator do determine the transcriptional specificity of NF-κB dimers.
Bcl-3 and IκBNS are the putative second and third examples that determine the transcriptional specificity of NF-κB dimers via association with extra-κB sites. These nuclear IκB proteins, capable of binding to a p50 and p52 homodimer (Figs. 1 and 2), have conserved basic residues that follow the second α helix in the C-terminal region of ANK7; the residues appear to be involved in association of Bcl-3 and IκBNS with promoters of their target genes (Fig. 7). The crucial role for Arg-354 and Lys-356 of Bcl-3 ANK7 in promoter binding and gene activation agrees well with a model in which the C-terminal region of Bcl-3 ARD in complex with the p50 homodimer is placed near the promoter DNA (36). Future studies should aim to elucidate a Bcl-3- or IκBNS-interacting cis element out of κB sites in Bcl-3- or IκBNS-dependent gene promoters, respectively.
Combined with the present observation that the aspartate residue in the N terminus of the ARD of Bcl-3 and IκBNS as well as that of IκBζ plays a crucial role in direct interaction with a p50 or p52 homodimer (Figs. 1 and 2), we propose a common mechanism for nuclear-IκB-mediated regulation of NF-κB-dependent genes. The nuclear IκB proteins with seven ANK motifs (IκBζ, Bcl-3, and IκBNS) form a p50/p52-containing regulatory complex on promoter DNA via the following two interactions: direct association with the κB-site-binding protein p50/p52 via the invariant aspartate residue in the N-terminal region of ANK1 and interaction with an extra-κB site via recognition by the conserved lysine/arginine residues that locate C-terminally to the second α helix of ANK7.
Experimental Procedures
Cells, Antibodies, and Reagents
MEFs doubly deficient in NF-κB p50 and IκBζ (Nfkb1−/−;Nfkbiz−/− MEFs) were prepared as described previously (23). Mouse BMMs were obtained as described previously (22, 24). All animals were housed and maintained in a specific pathogen-free animal facility at Kyushu University. All experiments were performed in strict accordance with the guidelines for proper conduct of animal experiments (Science Council of Japan). The experimental protocol was approved by the Animal Care and Use Committee of Kyushu University (permit numbers: A24-042 and A26-102). All efforts were made to minimize the numbers of animals and their suffering. MEFs, BMMs, HEK293T cells, and mouse macrophage-like RAW 264.7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C under 5% CO2.
Anti-FLAG (M2) (catalog no. F3165) and anti-β-tubulin (TUB 2.1, catalog no. T4026) mouse monoclonal antibodies and anti-FLAG rabbit polyclonal antibodies (catalog no. F7425) were purchased from Sigma-Aldrich, an anti-HA rat monoclonal antibody (3F10) (catalog no. 11 867 431 001) from Roche Applied Science, an anti-His5 monoclonal antibody (catalog no. 34660) from Qiagen, an anti-β-actin mouse monoclonal antibody (catalog no. sc-47778) and anti-RNA polymerase II rabbit polyclonal antibody (catalog no. sc-899) from Santa Cruz Biotechnology, and an anti-acetylated lysine rabbit polyclonal antibody (catalog no. 9441) from Cell Signaling Technology. Anti-IκBζ rabbit polyclonal antibodies were raised against IκBζ-(1–100) and prepared as described previously (51). TSA and anacardic acid were purchased from Calbiochem and nicotinamide from Sigma-Aldrich
Plasmid Construction
The mouse cDNAs encoding full-length IκBζ (amino acid residues 1–728), IκBζ-(414–728), p50 of 366 amino acid residues, and p65 of 325 amino acids were prepared as described previously (9, 33). The cDNA for mouse full-length IκBNS (amino acid residues 1–327) were prepared by RT-PCR using mRNA from mouse BMMs stimulated with LPS (List Biological Laboratories). The cDNAs for mouse full-length Bcl-3 (amino acid residues 1–448) and p52 of 415 amino acid residues were prepared by RT-PCR using mRNA from LPS-stimulated RAW 264.7 cells. The cDNA fragments encoding Bcl-3-(122–362) and p52-(1–341) for bacterial expression were prepared by PCR using their respective full-length cDNAs. Mutations leading to the indicated amino acid substitutions or truncations were introduced by PCR-mediated site-directed mutagenesis. The cDNA fragments were ligated to the following expression vectors: pGEX-6P-2 (GE Healthcare) and pMALc2 (New England Biolabs) for bacterial expression of proteins fused to GST and maltose-binding protein (MBP), respectively; pRSFDuet-1 (Novagen) modified for expression of hexahistidine (His)-tagged proteins in Escherichia coli (52); pcDNA3 (Invitrogen) for expression of FLAG- or HA-tagged proteins in mammalian cells; and pBABE-puro (Cell Biolabs) for retroviral transduction in BMMs. The 5′ upstream region (−1031/+54 or −500/+50) of the lipocalin-2-encoding gene Lcn2 or the 5′ upstream region (−445/+105) of the E-selectin-encoding gene Sele was ligated to the luciferase reporter plasmid pGL3-Basic (Promega) as described previously (23). The 5′ upstream regions of the following genes were amplified by PCR using mouse genomic DNA as a template and subcloned into pGL3-Basic: the IL-6-encoding gene Il6 (−1217/+50), the IP-10-encoding gene Cxcl10 (−500/+50), and the MCP-1-encoding gene Ccl2 (−2777/+76). Mutations in the κB site of the following gene promoters were introduced by PCR-mediated site-directed mutagenesis: Lcn2 (−230/−220), 5′-TGGGAATGTCCCT-3′ to 5′-TAATAATGTTAAT-3′ (23, 24); Il6 (−73/−64), 5′-TGGGATTTTCCCA-3′ to 5′-TAATATTTTTAAA-3′; and Cxcl10 (−113/−103), 5′-AGGGGACTTCCCT-3′ to 5′-ATAATACTTTAAT-3′ (note that mutated nucleotides are underlined). All of the constructs were sequenced for confirmation of their identity.
GST Pulldown Assay
GST- and His-tagged proteins were purified as described previously (52, 53) and incubated for 20 min at 4 °C in 500 μl of buffer A (137 mm NaCl, 2.7 mm KCl, 0.5% Triton X-100, 1 mm DTT, 8.1 mm Na2HPO4, and 1.5 mm KH2PO4 (pH 7.4)). A slurry of glutathione-Sepharose 4B beads (GE Healthcare) was subsequently added, followed by further incubation for 40 min at 4 °C. After washing three times with the buffer above, the proteins were eluted from the beads with 20 mm glutathione in 150 mm NaCl, 2 mm DTT, and 100 mm Tris-HCl (pH 8.8). The eluate was subjected to SDS-PAGE, followed by staining with Coomassie Brilliant Blue (CBB).
Immunoprecipitation Analysis
HEK293T cells were transfected with the indicated expression plasmids using X-tremeGENE HP DNA Transfection Reagent (Roche Applied Science) and cultured for 24 or 48 h. Cells were lysed by sonication at 4 °C in 500 μl of lysis buffer (137 mm NaCl, 2.7 mm KCl, 1% Nonidet P-40, 8.1 mm Na2HPO4, and 1.5 mm KH2PO4 (pH 7.4)) supplemented with CompleteTM Protease Inhibitor Cocktail (Roche Applied Science). Proteins in the cell lysate were immunoprecipitated using anti-FLAG antibody (M2) and protein G-Sepharose (GE Healthcare). The precipitants were analyzed by immunoblot with the anti-FLAG rabbit polyclonal antibody or the anti-HA (3F10) rat monoclonal antibody. The blots were developed using ECL-Prime (GE Healthcare) for visualization of the antibodies.
Detection of Acetylated Proteins
FLAG-IκBζ expressed in HEK293T cells was incubated for 6 h with 2 μm TSA and immunoprecipitated from the cell lysates as described above, with the exception that 2 μm TSA and 20 mm nicotinamide were used during immunoprecipitation to prevent deacetylation. The precipitated proteins were applied to immunoblot analysis with anti-acetylated lysine rabbit polyclonal antibody. Histones were prepared from HEK293T cells by acid extraction as described by Nightingale et al. (54). For detection of acetylated histones, the acid extracts were subsequently analyzed by immunoblot with the anti-acetylated lysine rabbit polyclonal antibody.
Luciferase Reporter Assay
Nfkb1−/−;Nfkbiz−/− MEFs or RAW264.7 cells were transfected with the luciferase reporter plasmid pGL3-Basic containing the indicated promoter, the internal control plasmid pRL-TK (Promega), and the indicated plasmids for protein expression using X-tremeGENE HP DNA Transfection Reagent. The luciferase activities were determined by the Dual-Luciferase® reporter assay system (Promega). For estimation of protein levels, cell lysates were analyzed by immunoblot with anti-FLAG (M2), anti-HA, anti-β-tubulin, or anti-β-actin antibody.
Analysis of Protein-DNA Interaction
Analysis of protein-DNA interaction was performed as described previously (23). GST-IκBζ, GST-IκBNS, or MBP-Bcl-3 was incubated with His-p50 for 20 min at 4 °C in 500 μl of buffer A containing salmon sperm DNA (0.1 mg/ml), or His-p50, His-p52, or His-p65 alone was incubated for 20 min at 4 °C in 500 μl of buffer B (150 mm NaCl, 5% glycerol, 1 mm DTT, 0.5% Triton X-100, and 25 mm Tris-HCl (pH 8.0)) containing salmon sperm DNA (0.1 mg/ml). A slurry of glutathione-Sepharose 4B, Amylose Resin (New England Biolabs), or COSMOGEL® His-Accept (Nacalai Tesque) was added in a GST, MBP, or His pulldown assay, respectively. The indicated DNA fragment (0.1 pmol) was subsequently added to the mixture, followed by further incubation for 40 min at 4 °C. After washing with buffer A for a GST or MBP pulldown assay or with buffer B containing 25 mm imidazole for a His pulldown assay, the protein-DNA complex was eluted from glutathione-Sepharose 4B with an elution buffer (20 mm glutathione, 150 mm NaCl, 2 mm DTT, and 100 mm Tris-HCl (pH 8.8)), from amylose resin with buffer A containing 20 mm maltose, or from COSMOGEL® His-Accept with buffer B containing 1 m imidazole. The eluted protein was applied to SDS-PAGE, followed by CBB staining or immunoblot analysis with the anti-His5 monoclonal antibody. The DNA in the eluted complex was analyzed by PCR using the following primer pairs: 5′-TACAGGGTTATGGGAGTGGAC-3′ and 5′-TCTGTTGAAATACTTGGCAAGAT-3′ for detection of the Lcn2 promoter region; 5′-CCATGGAAGACGCCAAAAACA-3′ and 5′-CATATCGTTTCATAGCTTCTGC-3′ for the 263-bp region of the pGL3-Basic vector (23); 5′-CTTAATAAGGTTTCCAATCAGCC-3′ and 5′-GTCTCATCTTTATTAGGAGTCAAC-3′ for the Il6 promoter region; and 5′-TCCAAGTTCATGGGTCACAA-3′ and 5′-TGATTGGCTGACTTTGGAGA-3′ for the Cxcl10 promoter region.
Activation of the Endogenous Lcn2 Gene by IκBζ
Retroviral expression of wild-type IκBζ or a mutant protein with the D451T or K717E/K719E substitution was performed according to the method of He et al. (55). Briefly, HEK293T cells were transfected with the Moloney Murine Leukemia virus-ΨE helper plasmid and pBABE-puro-3×FLAG-IκBζ, and the culture supernatant containing the retrovirus was collected. IκBζ-deficient BMMs were infected with the retrovirus and cultured for 2 days. The cells were treated for 4, 8, or 12 h with or without LPS (100 ng/ml). Expression of IκBζ was detected by immunoblot with the anti-IκBζ antibody. The mRNA product of the endogenous Lcn2 gene was estimated by quantitative real-time RT-PCR as described previously (56). Briefly, total RNAs were extracted using TRIsure (BIOLINE) according to the instructions of the manufacturer, and 1 μg of the RNA was reverse-transcribed by ReverTra Ace® reverse transcriptase (TOYOBO) using an oligo(dT) primer, followed by real-time PCR using SYBR® premix Ex TaqTM (Takara Bio) on the Roter-Gene 6200 system (Corbett). The primer pairs used were 5′-AAGGAGCTGTCCCCTGAACT-3′ and 5′-GGTGGGGACAGAGAAGATGA-3′ for Lcn2 and 5′-AAGCGAAACTGGCGGAAAC-3′ and 5′-TAACCGATGTTGGGCATCAG-3′ for the control gene Rpl32.
ChIP Analysis
RAW264.7 cells expressing IκBζ or Bcl-3 under the control of the sheep metallothionein Ia promoter (−600/+72) were prepared as described previously (22). The stable transformants were incubated in the presence of 50 μm ZnSO4 and LPS (100 ng/ml), and ChIP analysis was performed according to a method described previously (24, 56). Cells were fixed for 10 min at 25 °C with 1% formaldehyde and washed with ice-cold phosphate-buffered saline. After sonication, a chromatin-containing solution was precleared with Protein G-Sepharose 4 Fast Flow (GE Healthcare), followed by incubation overnight at 4 °C with the indicated antibody. Antigen-antibody complexes on the resin were washed sequentially with a low-salt wash buffer (150 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% Triton X-100, and 20 mm Tris-HCl (pH 8.0)), a high-salt wash buffer (500 mm NaCl, 2 mm EDTA, 0.1% SDS, 1% Triton X-100, and 20 mm Tris-HCl (pH 8.0)); a LiCl wash buffer (250 mm LiCl, 1 mm EDTA, 1% sodium deoxycholate, 1% Nonidet-P40, and 10 mm Tris-HCl (pH 8.0)), and TE (1 mm EDTA and 10 mm Tris-HCl (pH 8.0)). DNA-protein complexes were eluted with elution buffer containing 1% SDS and 100 mm sodium bicarbonate. After cross-links were reversed by overnight incubation at 65 °C, Proteinase K (0.1 mg/ml) was added and incubated for 3 h at 45 °C. Purified DNA was subjected to PCR to detect the following regions using specific primers as follows: the Lcn2 promoter region containing the CCCCTC element (−212/−207), 5′-CCCCTCTGTCCCCTGCAGC-3′, and 5′-TCTGTTGAAATACTTGGCAAGAT-3′ and the Cxcl10 promoter region, 5′-TCCAAGTTCATGGGTCACAA-3′ and 5′-GGGAAGTCCCCTGTAAACCGA-3′.
Secondary Structure Prediction
The secondary structure of the ANK7 in IκBζ and IκBNS was predicted using the server-side program PSIPRED (57, 58).
Author Contributions
A. K., S. Y., and H. S. designed the study. A. K. performed the experiments. A. K., S. Y., and H. S. analyzed the data and wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Namiko Kubo (Kyushu University) and Yohko Kage (Kyushu University) for technical assistance, Shoko Miura (Kyushu University) for secretarial assistance, and the technical staff from the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences, Fukuoka, Japan.
This work was supported in part by Grant-in-Aid for Scientific Research on Innovative Areas “Oxygen Biology: a New Criterion for Integrated Understanding of Life” (no. 26111009). The authors declare that they have no conflicts of interest with the contents of this article.
- NF-κB
- nuclear factor-κB
- ARD
- ankyrin repeat domain
- ANK
- ankyrin repeat
- MEF
- mouse embryonic fibroblast
- BMM
- bone marrow-derived macrophage
- HDAC
- histone deacetylase
- TSA
- trichostatin A
- MBP
- maltose-binding protein
- CBB
- Coomassie brilliant blue.
References
- 1. Smale S. T. (2010) Selective transcription in response to an inflammatory stimulus. Cell 140, 833–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Dea E., and Hoffmann A. (2010) The regulatory logic of the NF-κB signaling system. Cold Spring Harb. Perspect. Biol. 2, a000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghosh G., Wang V.-Y., Huang D.-B., and Fusco A. (2012) NF-κB regulation: lessons from structures. Immunol. Rev. 246, 36–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayden M. S., and Ghosh S. (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kawai T., and Akira S. (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 6. Natoli G., Ghisletti S., and Barozzi I. (2011) The genomic landscapes of inflammation. Genes Dev. 25, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bours V., Franzoso G., Azarenko V., Park S., Kanno T., Brown K., and Siebenlist U. (1993) The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell 72, 729–739 [DOI] [PubMed] [Google Scholar]
- 8. Fujita T., Nolan G. P., Liou H.-C., Scott M. L., and Baltimore D. (1993) The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev. 7, 1354–1363 [DOI] [PubMed] [Google Scholar]
- 9. Yamazaki S., Muta T., and Takeshige K. (2001) A novel IκB protein, IκB-ζ, induced by proinflammatory stimuli, negatively regulates nuclear factor-κB in the nuclei. J. Biol. Chem. 276, 27657–27662 [DOI] [PubMed] [Google Scholar]
- 10. Kitamura H., Kanehira K., Okita K., Morimatsu M., and Saito M. (2000) MAIL, a novel nuclear IκB protein that potentiates LPS-induced IL-6 production. FEBS Lett. 485, 53–56 [DOI] [PubMed] [Google Scholar]
- 11. Haruta H., Kato A., and Todokoro K. (2001) Isolation of a novel interleukin-1-inducible nuclear protein bearing ankyrin-repeat motifs. J. Biol. Chem. 276, 12485–12488 [DOI] [PubMed] [Google Scholar]
- 12. Fiorini E., Schmitz I., Marissen W. E., Osborn S. L., Touma M., Sasada T., Reche P. A., Tibaldi E. V., Hussey R. E., Kruisbeek A. M., Reinherz E. L., and Clayton L. K. (2002) Peptide-induced negative selection of thymocytes activates transcription of an NF-κB inhibitor. Mol. Cell 9, 637–648 [DOI] [PubMed] [Google Scholar]
- 13. Huxford T., and Ghosh G. (2009) A structural guide to proteins of the NF-κB signaling module. Cold Spring Harb. Perspect. Biol. 1, a000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz E. M., Krimpenfort P., Berns A., and Verma I. M. (1997) Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev. 11, 187–197 [DOI] [PubMed] [Google Scholar]
- 15. Franzoso G., Carlson L., Xing L., Poljak L., Shores E. W., Brown K. D., Leonardi A., Tran T., Boyce B. F., and Siebenlist U. (1997) Requirement for NF-κB in osteoclast and B-cell development. Genes Dev. 11, 3482–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., Saitoh T., Yamaoka S., Yamamoto N., Yamamoto S., Muta T., et al. (2004) Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430, 218–222 [DOI] [PubMed] [Google Scholar]
- 17. Touma M., Antonini V., Kumar M., Osborn S. L., Bobenchik A. M., Keskin D. B., Connolly J. E., Grusby M. J., Reinherz E. L., and Clayton L. K. (2007) Functional role for IκBNS in T cell cytokine regulation as revealed by targeted gene disruption. J. Immunol. 179, 1681–1692 [DOI] [PubMed] [Google Scholar]
- 18. Okamoto K., Iwai Y., Oh-Hora M., Yamamoto M., Morio T., Aoki K., Ohya K., Jetten A. M., Akira S., Muta T., and Takayanagi H. (2010) IκBζ regulates TH17 development by cooperating with ROR nuclear receptors. Nature 464, 1381–1385 [DOI] [PubMed] [Google Scholar]
- 19. Schuster M., Glauben R., Plaza-Sirvent C., Schreiber L., Annemann M., Floess S., Kühl A. A., Clayton L. K., Sparwasser T., Schulze-Osthoff K., Pfeffer K., Huehn J., Siegmund B., and Schmitz I. (2012) IκBNS protein mediates regulatory T cell development via induction of the Foxp3 transcription factor. Immunity 37, 998–1008 [DOI] [PubMed] [Google Scholar]
- 20. Cowland J. B., Muta T., and Borregaard N. (2006) IL-1β-specific up-regulation of neutrophil gelatinase-associated lipocalin is controlled by IκB-ζ. J. Immunol. 176, 5559–5566 [DOI] [PubMed] [Google Scholar]
- 21. Kayama H., Ramirez-Carrozzi V. R., Yamamoto M., Mizutani T., Kuwata H., Iba H., Matsumoto M., Honda K., Smale S. T., and Takeda K. (2008) Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J. Biol. Chem. 283, 12468–12477 [DOI] [PubMed] [Google Scholar]
- 22. Yamazaki S., Matsuo S., Muta T., Yamamoto M., Akira S., and Takeshige K. (2008) Gene-specific requirement of a nuclear protein, IκB-ζ, for promoter association of inflammatory transcription regulators. J. Biol. Chem. 283, 32404–32411 [DOI] [PubMed] [Google Scholar]
- 23. Kohda A., Yamazaki S., and Sumimoto H. (2014) DNA element downstream of the κB site in the Lcn2 promoter is required for transcriptional activation by IκBζ and NF-κB p50. Genes Cells 19, 620–628 [DOI] [PubMed] [Google Scholar]
- 24. Yamazaki S., Akira S., and Sumimoto H. (2015) Glucocorticoid augments lipopolysaccharide-induced activation of the IκBζ-dependent genes encoding the anti-microbial glycoproteins lipocalin 2 and pentraxin 3. J. Biochem. 157, 399–410 [DOI] [PubMed] [Google Scholar]
- 25. Okuma A., Hoshino K., Ohba T., Fukushi S., Aiba S., Akira S., Ono M., Kaisho T., and Muta T. (2013) Enhanced apoptosis by disruption of the STAT3-IκB-ζ signaling pathway in epithelial cells induces Sjögren's syndrome-like autoimmune disease. Immunity 38, 450–460 [DOI] [PubMed] [Google Scholar]
- 26. Johansen C., Mose M., Ommen P., Bertelsen T., Vinter H., Hailfinger S., Lorscheid S., Schulze-Osthoff K., and Iversen L. (2015) IκBζ is a key driver in the development of psoriasis. Proc. Natl. Acad. Sci. U.S.A. 112, E5825–E5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang V. Y.-F., Huang W., Asagiri M., Spann N., Hoffmann A., Glass C., and Ghosh G. (2012) The transcriptional specificity of NF-κB dimers is coded within the κB DNA response elements. Cell Rep. 2, 824–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirotani T., Lee P. Y., Kuwata H., Yamamoto M., Matsumoto M., Kawase I., Akira S., and Takeda K. (2005) The nuclear IκB protein IκBNS selectively inhibits lipopolysaccharide-induced IL-6 production in macrophages of the colonic lamina propria. J. Immunol. 174, 3650–3657 [DOI] [PubMed] [Google Scholar]
- 29. Kuwata H., Matsumoto M., Atarashi K., Morishita H., Hirotani T., Koga R., and Takeda K. (2006) IκBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity 24, 41–51 [DOI] [PubMed] [Google Scholar]
- 30. Sedgwick S. G., and Smerdon S. J. (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316 [DOI] [PubMed] [Google Scholar]
- 31. Mosavi L. K., Cammett T. J., Desrosiers D. C., and Peng Z. Y. (2004) The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J., Mahajan A., Tsai M.-D. (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45, 15168–15178 [DOI] [PubMed] [Google Scholar]
- 33. Trinh D. V., Zhu N., Farhang G., Kim B. J., and Huxford T. (2008) The nuclear IκB protein IκBζ specifically binds NF-κB p50 homodimers and forms a ternary complex on κB DNA. J. Mol. Biol. 379, 122–135 [DOI] [PubMed] [Google Scholar]
- 34. Hinz M., Arslan SÇ., and Scheidereit C. (2012) It takes two to tango: IκBs, the multifunctional partners of NF-κB. Immunol. Rev. 246, 59–76 [DOI] [PubMed] [Google Scholar]
- 35. Hatada E. N., Nieters A., Wulczyn F. G., Naumann M., Meyer R., Nucifora G., McKeithan T. W., and Scheidereit C. (1992) The ankyrin repeat domains of the NF-κB precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-κB DNA binding. Proc. Natl. Acad. Sci. U.S.A. 89, 2489–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Michel F., Soler-Lopez M., Petosa C., Cramer P., Siebenlist U., and Müller C. W. (2001) Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IκB protein family. EMBO J. 20, 6180–6190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nogai H., Wenzel S. S., Hailfinger S., Grau M., Kaergel E., Seitz V., Wollert-Wulf B., Pfeifer M., Wolf A., Frick M., Dietze K., Madle H., Tzankov A., Hummel M., Dörken B., et al. (2013) IκB-ζ controls the constitutive NF-κB target gene network and survival of ABC DLBCL. Blood 122, 2242–2250 [DOI] [PubMed] [Google Scholar]
- 38. Bergqvist S., Alverdi V., Mengel B., Hoffmann A., Ghosh G., and Komives E. A. (2009) Kinetic enhancement of NF-κB·DNA dissociation by IκBα. Proc. Natl. Acad. Sci. U.S.A. 106, 19328–19333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kono H., and Sarai A. (1999) Structure-based prediction of DNA target sites by regulatory proteins. Proteins 35, 114–131 [PubMed] [Google Scholar]
- 40. Luscombe N. M., Laskowski R. A., and Thornton J. M. (2001) Amino acid-base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 29, 2860–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y., Jiang X., Chen S., and Price B. D. (2006) Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 580, 4353–4356 [DOI] [PubMed] [Google Scholar]
- 42. Sung B., Pandey M. K., Ahn K. S., Yi T., Chaturvedi M. M., Liu M., and Aggarwal B. B. (2008) Anacardic acid (6-nonadecyl salicylic acid), an inhibitor of histone acetyltransferase, suppresses expression of nuclear factor-κB-regulated gene products involved in cell survival, proliferation, invasion, and inflammation through inhibition of the inhibitory subunit of nuclear factor-κBα kinase, leading to potentiation of apoptosis. Blood 111, 4880–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., and Guarente L. (2004) Mammalian SIRT1 represses forkhead transcription factors. Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 44. Luo J., Li M., Tang Y., Laszkowska M., Roeder R. G., and Gu W. (2004) Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 101, 2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jacobs M. D., and Harrison S. C. (1998) Structure of an IκBα/NF-κB complex. Cell 95, 749–758 [DOI] [PubMed] [Google Scholar]
- 46. Huxford T., Huang D. B., Malek S., and Ghosh G. (1998) The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell 95, 759–770 [DOI] [PubMed] [Google Scholar]
- 47. Malek S., Huang D.-B., Huxford T., Ghosh S., and Ghosh G. (2003) X-ray crystal structure of an IκBβ·NF-κB p65 homodimer complex. J. Biol. Chem. 278, 23094–23100 [DOI] [PubMed] [Google Scholar]
- 48. Ryzhakov G., Teixeira A., Saliba D., Blazek K., Muta T., Ragoussis J., and Udalova I. A. (2013) Cross-species analysis reveals evolving and conserved features of the nuclear factor κB (NF-κB) proteins. J. Biol. Chem. 288, 11546–11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siggers T., Chang A. B., Teixeira A., Wong D., Williams K. J., Ahmed B., Ragoussis J., Udalova I. A., Smale S. T., and Bulyk M. L. (2012) Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 13, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoffmann A., Leung T. H., and Baltimore D. (2003) Genetic analysis of NF-κB/Rel transcription factors defines functional specifics. EMBO J. 22, 5530–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamazaki S., Muta T., Matsuo S., and Takeshige K. (2005) Stimulus-specific induction of a novel NF-κB regulator, IκB-ζ, via Toll/interleukin-1 receptor is mediated by mRNA stabilization. J. Biol. Chem. 280, 1678–1687 [DOI] [PubMed] [Google Scholar]
- 52. Yuzawa S., Kamakura S., Iwakiri Y., Hayase J., and Sumimoto H. (2011) Structural basis for interaction between the conserved cell polarity proteins Inscuteable and Leu-Gly-Asn repeat-enriched protein (LGN). Proc. Natl. Acad. Sci. U.S.A. 108, 19210–19215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamakura S., Nomura M., Hayase J., Iwakiri Y., Nishikimi A., Takayanagi R., Fukui Y., and Sumimoto H. (2013) The cell polarity protein mInsc regulates neutrophil chemotaxis via a noncanonical G protein signaling pathway. Dev. Cell 26, 292–302 [DOI] [PubMed] [Google Scholar]
- 54. Nightingale K. P., Gendreizig S., White D. A., Bradbury C., Hollfelder F., and Turner B. M. (2007) Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 282, 4408–4416 [DOI] [PubMed] [Google Scholar]
- 55. He J. Q., Zarnegar B., Oganesyan G., Saha S. K., Yamazaki S., Doyle S. E., Dempsey P. W., and Cheng G. (2006) Rescue of TRAF3-null mice by p100 NF-κB deficiency. J. Exp. Med. 203, 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamazaki S., and Takeshige K. (2008) Protein synthesis inhibitors enhance the expression of mRNAs for early inducible inflammatory genes via mRNA stabilization. Biochim. Biophys. Acta 1779, 108–114 [DOI] [PubMed] [Google Scholar]
- 57. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 58. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 [DOI] [PMC free article] [PubMed] [Google Scholar]