Abstract
Primary cilia are highly specialized small antenna-like cellular protrusions that extend from the cell surface of many eukaryotic cell types. The protein content inside cilia and cytoplasm is very different, but details of the sorting process are not understood for most ciliary proteins. Recently, we have shown that prenylated proteins are sorted according to their affinity to the carrier protein PDE6δ and the ability of Arl3 but not Arl2 to release high affinity cargo inside the cilia (Fansa, E. K., Kösling, S. K., Zent, E., Wittinghofer, A., and Ismail, S. (2016) Nat. Commun. 7, 11366). Here we address the question whether a similar principle governs the transport of myristoylated cargo by the carrier proteins Unc119a and Unc119b. We thus analyzed the binding strength of N-terminal myristoylated cargo peptides (GNAT1, NPHP3, Cystin1, RP2, and Src) to Unc119a and Unc119b proteins. The affinity between myristoylated cargo and carrier protein, Unc119, varies between subnanomolar and micromolar. Peptides derived from ciliary localizing proteins (GNAT1, NPHP3, and Cystin1) bind with high affinity to Unc119 proteins, whereas a peptide derived from a non-ciliary localizing protein (Src) has low affinity. The peptide with intermediate affinity (RP2) is localized at the ciliary transition zone as a gate keeper. We show that the low affinity peptides are released by both Arl2·GppNHp and Arl3·GppNHp, whereas the high affinity peptides are exclusively released by only Arl3·GppNHp. Determination of the x-ray structure of myristoylated NPHP3 peptide in complex with Unc119a reveals the molecular details of high affinity binding and suggests the importance of the residues at the +2 and +3 positions relative to the myristoylated glycine for high and low affinities. The mutational analysis of swapping the residues at the +2 and +3 positions between high and low affinity peptides results in reversing their affinities for Unc119a and leads to a partial mislocalization of a low affinity mutant of NPHP3.
Keywords: cilia; lipid binding protein; protein myristoylation; small GTPase; sorting; cilia, Unc119 proteins, releasing factor, myristoylated proteins, Arl2/3
Introduction
The existence of cilia in higher organisms was discovered a century ago, but research to explore the functional importance of cilia has only recently intensified. Cilia are highly specialized small antenna-like cellular protrusions that extend from the cell surface of almost all eukaryotic cell types. Primary cilia as sensory organelles are important for many cellular functions. They work as control centers of developmental signaling pathways, such as Hedgehog, or the induction of left-right asymmetry (2–4). Ciliary dysfunction leads to a range of diseases like Meckel-Gruber syndrome, Joubert syndrome, Bardet-Biedl syndrome, nephronophthisis, and polycystic kidney disease, which are commonly identified as ciliopathies (5).
Although the ciliary membrane is the extension of plasma membrane, the lipid composition is different from that of the plasma membrane (6). The composition and concentrations of proteins inside this compartment are also very different from the entire cell (7). A membrane barrier between cilia and the rest of the cell formed by a septin ring has been postulated such that the entry of transmembrane and membrane-associated proteins is tightly regulated (8–10). This renders the ciliary membrane a very specialized compartment of the cell, which orchestrates proteins to achieve spatially controlled signaling pathways. However, the regulation of entry into and retention inside cilia and signals for such processes are still incompletely understood. It has been proposed that partition of proteins between cilium and cell body is directed by steric hindrance and/or cytoskeletal structures (11) or binding affinities between proteins and cytoplasmic elements (12).
We and others have previously shown that proteins with a C-terminal CaaX box motif, which are post-translationally modified with a farnesyl or geranylgeranyl moiety, are transported via the δ subunit of phosphodiesterase 6 (PDE6δ) (1, 13–15). PDE6δ is a structural homologue to RhoGDI. It forms a β-sandwich fold with a deep hydrophobic pocket, which binds the prenylated cysteine and the adjacent three amino acid residues, two of which determine the affinity of the interaction. We have shown recently that the sorting of cargo between cilia and the rest of the cell depends on the affinity between cargo and PDE6δ, such that cargo with high affinity is destined for cilia, whereas low affinity cargo stays in the rest of the cell or is no longer exclusively retained inside the cilium (1).
Unc119a (uncoordinated), also known as HRG4 (human retina gene 4) and Unc119b share 58% sequence identity and are homologous to PDE6δ. The C termini of Unc119a/b share the PDEδ-like β-sandwich domain, whereas they are considerably divergent in the N-terminal part. Unc119a has been shown to be localized at the basal body of the cilium, whereas the homologous Unc119b localizes to the transition zone and proximal cilium of RPE cells (16). Unc119a is also found in the eye and highly enriched in photoreceptor cells, in both rods and cones (17), supporting the notion that photoreceptors are considered as a specialized version of cilia (18–21). Unc119a has been shown to be expressed in eosinophils, T-cells, lung fibroblasts, adrenal glands, cerebellum, and kidney (22–25). Numerous studies have shown the involvement of Unc119a in the function of Src family kinases, Lyn, Fyn, Lck, and Hck, or as an inhibitor of Abl family tyrosine kinases, although the nature of such interactions had remained obscure (23–28).
N-terminal myristoylation of proteins facilitates their reversible membrane binding activity (29). The mechanism by which myristoylated proteins are recruited to the proper membrane of cellular organelles remains unclear. Unc119a/b have been shown to bind specifically to N-terminal myristoylated proteins (30), and biochemical studies have shown that Unc119a/b proteins are involved in binding and shuttling of N-myristoylated proteins (16, 30–32), suggesting that the early findings on the involvement of Unc119 in Src kinase function can be related to that observation. The determination of a structure between Unc119a and a lauroylated N-terminal peptide from the α subunit of transducin has shown that Unc119a forms a β-sandwich structure very similar to that of PDE6δ (32, 33). Unc119a forms a hydrophobic pocket that accommodates a lipid moiety and a number of N-terminal residues of the peptide. Thus, like PDE6δ, Unc119a/b work as a carrier of these post-transitionally modified membrane-associated proteins. These proteins can also be considered chaperones that shield the lipid from the solvent (16, 32–34).
Previous studies have revealed a number of myristoylated proteins that interact with Unc119 (16, 31, 32). Binding of Unc119a/b to their interacting partners has been analyzed either qualitatively via yeast two-hybrid screening and in vitro pull-down assays or quantitatively through isothermal titration calorimetry (32) and polarization measurements (31). Previous studies have measured the affinities of either Unc1119a or Unc119b with the N-terminal myristoylated peptides of GNAT1, NPHP3, and Src (16, 30–32). It has also been shown for Cystin1 and NPHP3 that the cargo is released from Unc119a only by Arl3·GTP and not Arl2·GTP (16, 31). Recently, we have shown that high affinity binding between farnesylated cargo INPP5E and PDE6δ leads to INPP5E recruitment to the ciliary membrane, and its release from PDE6δ by Arl3 exclusively in cilia, whereas low affinity binding between farnesylated Rheb and PDE6δ results in Rheb localization to the cytosol (1).
Here we have set out to perform a comparative more comprehensive analysis of the interaction between Unc119a/b and different myristoylated proteins, the interaction with Arl2 and Arl3, and finally the release of cargo by Arl2 and Arl3. The myristoylated proteins analyzed in this study are NPHP3, Cystin1, GNAT1, RP2, and Src. We find different affinities of Unc119a/b for Arl2 and Arl3 and for cargo peptides and differences in cargo release by Arl2/3. By structure-guided mutational analysis, we show how the difference in affinities can be manipulated between high and low affinity cargo. Our results suggest that similar to the transport of prenylated cargo into cilia, the import of myristoylated cargo might be determined by the cargo-carrier binding affinity and the cargo release specificity mediated by Arl2 and Arl3.
Results
Binding of N-terminal Myristoylated Peptides to Unc119a and Unc119b
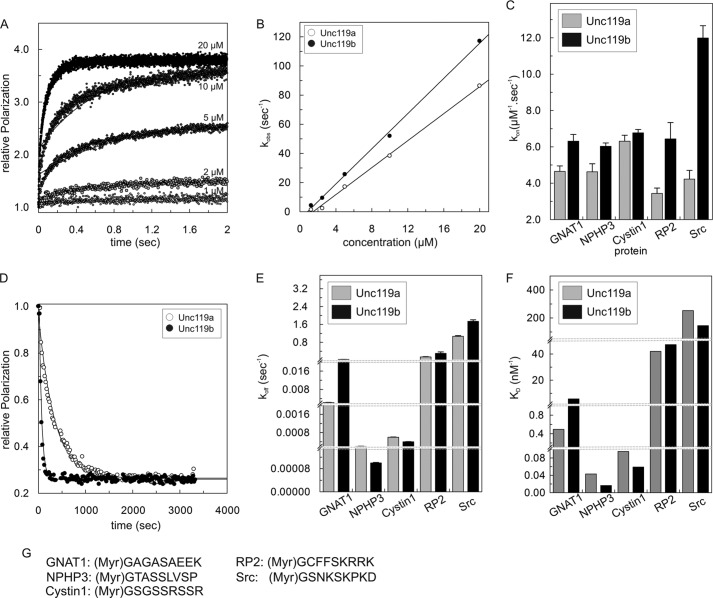
Using fluorescence polarization, we measured the binding affinity of both Unc119a and Unc119b to N-terminal myristoylated peptides derived from transducin-α (GNAT1), NPHP3, Cystin1, RP2, and Src, which were labeled with fluorescein at the C terminus. Preliminary experiments had suggested that some of the affinities were too high to be reliably measured by equilibrium methods (data not shown; also see below). Thus, we used kinetic measurements via stopped flow instead to determine association and dissociation rate constants and obtain KD (Fig. 1). Fig. 1A shows the association of 1 μm fluorescein-labeled N-terminal myristoylated GNAT1 peptide with increasing concentrations of Unc119a. The association of the myristoylated GNAT1 peptide with Unc119a (0.2–20 μm) leads to the increase in fluorescence polarization. Plotting of the observed rate constants (kobs) against the concentration of Unc119a resulted in the determination of the association rate constant (kon) of the reaction (Fig. 1B). The association rate constant (kon) for Unc119a is 4.6 μm−1 s−1 and is very similar to Unc119b (6.3 μm−1 s−1) measured under the same conditions (Fig. 1B). Association rate measurements were also measured for the N-terminal myristoylated peptides from NPHP3, Cystin1, RP2, and Src under similar experimental conditions, and the association rate constants (kon) obtained are shown as a bar diagram in Fig. 1C and numerically in Table 1. These values are very similar and range from 3.4 to 12 μm−1 s−1. Association of myristoylated cargo is generally slightly faster for Unc119b than for Unc119a.
FIGURE 1.
Interaction of Unc119a/b with N-terminal myristoylated peptides. A, kinetics of association between fluorescently labeled GNAT1 peptide (1 μm) and different concentrations (1–20 μm) of Unc119a. Rates were measured as change in fluorescence polarization using a stopped flow instrument. Reactions were carried out at 25 °C in buffer A. Observed rate constants (kobs) of associations were obtained by single exponential fitting of individual curves. B, the observed association rate constants kobs for interaction of Unc119a and Unc119b with N-terminal myristoylated cargo peptide (GNAT1) obtained as in A were plotted against the concentration of Unc119a and Unc119b. The association rate (kon) is represented by the slope. The kon values for all N-terminal myristoylated cargo peptides (GNAT1, NPHP3, Cystin1, RP2, and Src) are summarized in a bar diagram (C) and in Table 1. D, dissociation of cargo was measured at 25 °C in buffer A by monitoring the decrease of fluorescence polarization after incubating a complex of fluorescein labeled peptide with Unc119 (1 μm) with a 100-fold excess (100 μm) of unlabeled peptide. Dissociation rate constants (koff) were obtained by single exponential fitting of the data. All koff values are summarized in a bar diagram (E) and in Table 1. F, equilibrium dissociation constants (KD) were calculated as ratios, koff/kon, and the values are plotted as a bar diagram and summarized in Table 1. G, the sequences of N-terminal myristoylated peptides used in this study. For labeled peptides, fluorescein fluorophore was attached at the C terminus of N-myristoylated peptides. All bar graphs show the average of 5–7 measurements for the experiments performed by stopped flow instruments and an average of three measurements for the experiments performed by the Fluoromax instrument. Error bars, S.D., for (C) n = 5–7 and for (E) n = 3.
TABLE 1.
N terminus-myristoylated peptide binding properties for Unc119 proteins
| Protein | GNAT1 | NPHP3 | Cystin1 | RP2 | Src |
|---|---|---|---|---|---|
| Association constants (kon) of Unc119a/b for N terminus-myristoylated peptides (s−1 μm−1) | |||||
| Unc119a | 4.64 | 4.26 | 6.31 | 3.44 | 4.23 |
| Unc119b | 6.32 | 6.04 | 6.77 | 6.44 | 11.99 |
| Dissociation rates (koff) by 100× non-labeled peptide (s−1) | |||||
| Unc119a | 0.0023 | 0.0002 | 0.0006 | 0.145 | 1.07 |
| Unc119b | 0.0364 | 0.0001 | 0.0004 | 0.304 | 1.74 |
| Equilibrium dissociation constants (KD) of Unc119a/b for N-terminal myristoylated peptides (nm) | |||||
| Unc119a | 0.49 | 0.043 | 0.095 | 42.1 | 252.96 |
| Unc119b | 5.76 | 0.02 | 0.059 | 47.2 | 145.12 |
| Dissociation rates (koff) by 10× Arl3 in the presence of 100× non-labeled peptide (s−1) | |||||
| Unc119a | 2.8 | 1.72 | 5.2 | 0.61 | 2.73 |
| Unc119b | 5.6 | 2.5 | 11.3 | 0.52 | 2.82 |
| Dissociation rates (koff) by 10× Arl2 in the presence of 100× non-labeled peptide (s−1) | |||||
| Unc119a | 0.045 | 0.018 | 0.002 | 0.56 | 2.83 |
| Unc119b | 0.658 | 0.032 | 0.001 | 0.38 | 3.99 |
The dissociation rate constants were determined by incubating a complex of C-terminal fluorescein labeled N-terminal myristoylated peptides with Unc119a/b in the presence of a 100-fold excess of unlabeled peptides, as shown for the GNAT1 peptide (Fig. 1D). In contrast to kon, the koff values vary considerably among the different peptides over a 900-fold range from 0.0002 to 1.74 s−1 (Fig. 1E and Table 1). NPHP3 and Cystin1 show the slowest release from Unc119a with 0.002 and 0.006 s−1, whereas Src peptide has the fastest off rate close to 1 s−1. The rate for RP2 is intermediate between very slow and very fast rates. The dissociation rate constants of Unc119a and -b for NPHP3, Cystin1, RP2, and Src are rather similar, but for GNAT1, there is a >10-fold difference between Unc119a and Unc119b, with koff values of 0.0023 and 0.035 s−1, respectively.
The equilibrium dissociation constants (KD) for the complexes between Unc119a/b and myristoylated peptides were calculated as the ratio of koff/kon (Fig. 1F and Table 1). The data revealed three different categories of affinity: (i) the very tight binding with subnanomolar affinities for Cystin1 and NPHP3 and low nanomolar affinity for GNAT1, (ii) the intermediate binding affinity in the two-digit nanomolar range for RP2, and (iii) the low affinity in the submicromolar range for Src (145–252 nm) (Fig. 1F and Table 1). The affinities of Unc119a and Unc119b for myristoylated peptides are similar except for GNAT1, which shows >10-fold higher affinity for Unc119a as compared with Unc119b, exclusively due to the different dissociation rates. The affinities determined here by kinetics are different from those determined earlier by an equilibrium method (31). As indicated above, equilibrium binding assays are not quite suitable for such high affinities (picomolar or sub-nanomolar) because such affinity measurements by titration result in straight lines that look like active site titrations and only give upper limits for the KD.
Binding Affinity of Unc119a and Unc119b to Arl Proteins
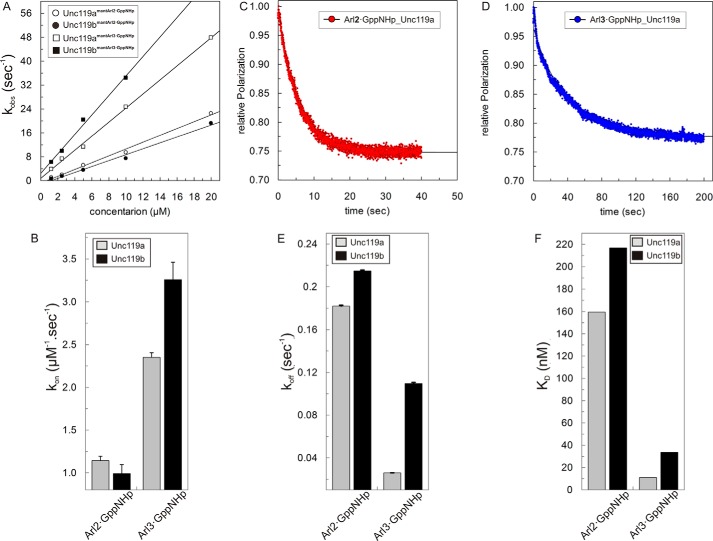
We have shown previously that Unc119a and PDE6δ bind to Arl2/3 with high affinity (1, 35, 36). We showed that the binding affinities of Unc119a and PDE6δ are 20–30-fold higher for Arl3 as compared with Arl2 (1, 36) because of the additional contribution by the N-terminal helix of Arl3 to the interaction. To verify whether the same holds true for Unc119b and to compare with Unc119a, we measured the kinetics of interaction using full-length Arl2/3 labeled with mant-GppNHp3 (fluorescent, non-hydrolyzable analog of GTP). The association rates with increasing concentrations of Unc119a and -b were measured with fluorescence polarization using a stopped flow instrument (Fig. 2). Association rate constants for Arl2/3·GppNHp were obtained as described above by plotting the observed rate constants of association against the concentration of Unc119a/b (Fig. 2A). The results are shown as a bar diagram in Fig. 2B and in numerical form in Table 2. The association rate constants (kon) are on the order of 1–3 μm−1 s−1. The association rates are >3-fold higher for Arl3 than for Arl2 in the case of Unc119b, whereas it is only 2-fold in the case of Unc119a. Dissociation rates were measured by incubating the fluorescent complex of Unc119a/b and Arl2/3·mant-GppNHp with a 100-fold excess of unlabeled Arl2/3·GppNHp, as shown for Arl2 and Unc119a/b (Fig. 2, C and D). The dissociation rate constants (koff) are shown as a bar diagram in Fig. 2E and numerically in Table 2. The dissociation rate constants (koff) are slower for Arl3 than Arl2 for both Unc119a and Unc119b. The equilibrium dissociation constants for Unc119a/b toward full-length Arl2·GppNHp and Arl3·GppNHp were calculated as the ratio of koff/kon (Fig. 2F and Table 2). The kinetic data suggested the binding affinities of Arl3 toward both Unc119a and -b are higher as compared with Arl2. Whereas, the 14-fold difference in affinity between Arl3 and Arl2 toward Unc119a is mediated only by the dissociation rates, the difference for Unc119b is due to a combination of different association and dissociation rates.
FIGURE 2.
Interaction of Unc119a/b with Arl2/3. A, observed association rate constants between 0.5 μm mant-GppNHp-loaded Arl2 and Arl3 and different concentrations (1–20 μm) of Unc119a and Unc119b measured with fluorescence polarization using a stopped flow instrument as described in the legend to Fig. 1A at 25 °C in buffer A. The association rate constants (kon) of Unc119a (open circles/squares) and Unc119b (closed circles/squares) for Arl2·GppNHp (open/closed circles) and Arl3·GppNHp (open/closed squares) binding were calculated from the slope of the linear regression of the kobs values plotted against the concentration of Unc119a and Unc119b proteins. All kon values are plotted as a bar diagram (B) and appear in Table 2. C and D, the dissociations of full-length Arl2·GppNHp and Arl3·GppNHp from a complex with Unc119a/b (0.5 μm) at 25 °C in buffer A were measured as decreases of fluorescence polarization after the addition of a 100-fold excess (50 μm) of unlabeled Arl2/3·GppNHp. E, observed dissociation rate constants (koff) were obtained by single exponential fitting of the data. All koff values are plotted as a bar diagram and summarized in Table 2. F, equilibrium dissociation constants (KD) were calculated as ratios of koff/kon, and the values here are plotted as a bar diagram and appear in Table 2. All bar graphs show the average of 5–7 measurements for the experiments performed by stopped flow instruments and an average of three measurements for the experiments performed by the Fluoromax instrument. Error bars, S.D., n = 5–7.
TABLE 2.
Arl2 and Arl3 binding properties for Unc119 proteins
| Unc119a | Unc119b | |
|---|---|---|
| Association constants (kon) (s−1 μm−1) | ||
| Arl2·GppNHp | 1.14 | 0.99 |
| Arl3·GppNHp | 2.35 | 3.26 |
| Dissociation rates (koff) (s−1) | ||
| Arl2·GppNHp | 0.18 | 0.21 |
| Arl3·GppNHp | 0.026 | 0.11 |
| Equilibrium dissociation constants (KD) (nm) | ||
| Arl2·GppNHp | 159.36 | 216.82 |
| Arl3·GppNHp | 11.02 | 33.61 |
Release of Myristoylated Cargo by Arl2/3 Proteins
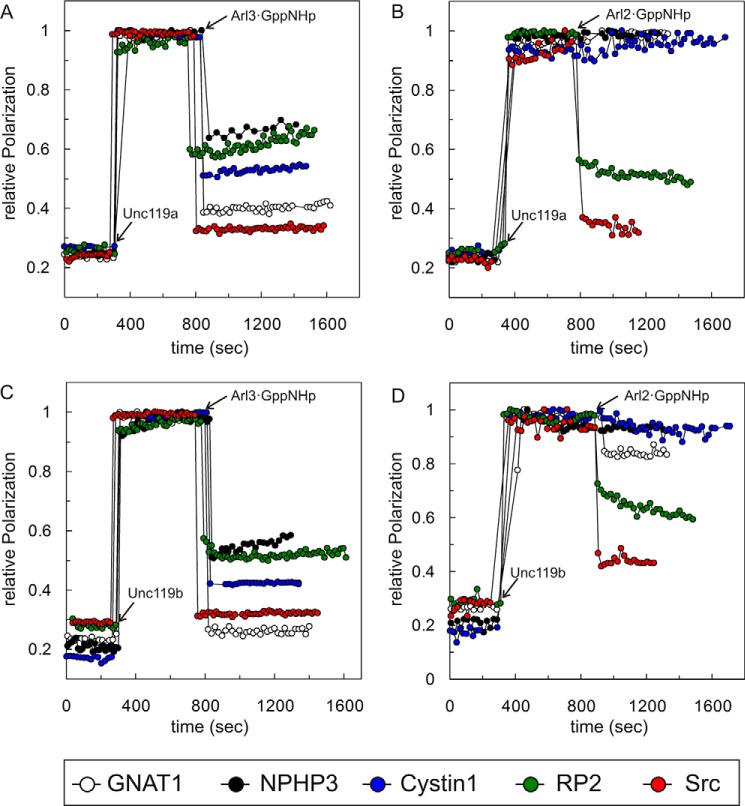
The Arf-like small GTP-binding proteins Arl2 and Arl3 act as displacement factors for lipid-modified proteins bound to the GDI-like solubilizing factor PDE6δ as well as to Unc119a/b (15, 31, 33, 34). Previously, it has been shown that Arl3, but not Arl2, allosterically regulates the release of ciliary proteins from Unc119a/b (16, 31). We thus analyzed the specificity of Arl2- and Arl3-mediated release of the high and low affinity peptides analyzed above for both Unc119a and Unc119b (Fig. 3). Arl2·GppNHp or Arl3·GppNHp was added to the preformed complex of fluorescent peptides from GNAT1, NPHP3, Cystin1, RP2, and Src with Unc119a and Unc119b. The release of peptide from the complex is scored as a decrease in fluorescence polarization. Under the conditions used, Arl3·GppNHp was able to disrupt the complex of Unc119a with all five peptides (Fig. 3, A and C). In contrast, Arl2·GppNHp was only able to disrupt the complex of Unc119a with the low or intermediate affinity RP2 or Src peptides, whereas the high affinity GNAT1, Cystin1, and NPHP3 peptides were fully resistant to release by Arl2 under the conditions used (Fig. 3, B and D). The results were consistent with the previous studies, which used GNAT1 and NPHP3 bound to Unc119a (16, 31). When comparing Unc119a and -b, similar results were obtained for the specificity of Arl2 and Arl3 (Fig. 3, A and B versus C and D). These results indicate that the high affinity cargos GNAT1/NPHP3/Cystin1 are only released by Arl3·GppNHp, whereas Arl2·GppNHp can release only the low affinity cargo RP2 and Src. Myristoylated GNAT1 was partially released by Arl2·GppNHp when bound to Unc119b but not Unc119a, possibly due to the 10-fold affinity difference observed above (Fig. 3D).
FIGURE 3.
Cargo release from Unc119a/b by Arl2 and Arl3 proteins. The cargo release by Arl proteins was observed by fluorescence polarization at 25 °C in buffer A. 0.2 μm Unc119a (A and B) or Unc119b (C and D) was added to a solution of 0.1 μm fluorescein-labeled N-terminal myristoylated GNAT1 (open circles), NPHP3 (black, closed circles), Cystin1 (blue, closed circles), RP2 (green, closed circles), and Src (red, closed circles) peptides followed by the addition of a 10-fold excess (2 μm) full-length Arl3·GppNHp (A and C) or Arl2·GppNHp (B and D). The addition time points are indicated by arrows. All experiments were repeated three times independently.
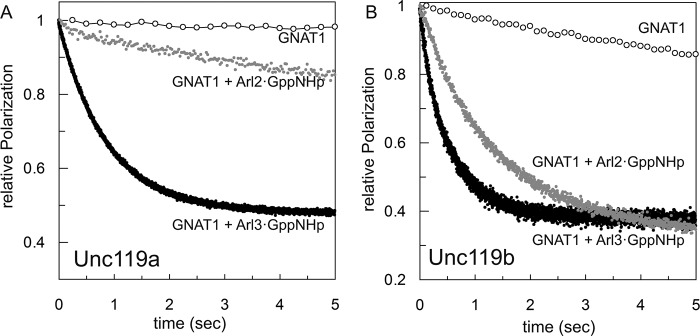
Because these data were obtained under equilibrium conditions and are somewhat difficult to compare quantitatively, we determined the Arl2/3·GppNHp cargo release acceleration from Unc119a/b in the presence of excess unlabeled peptides. The effects of Arl3·GppNHp and Arl2·GppNHp on the dissociation rates were determined in the presence of a 100-fold excess of unlabeled peptides to silence the back-reaction and were measured as a decrease in polarization in a stopped flow instrument. The data for the release of GNAT1 peptide from either Unc119a or -b are shown in Fig. 4, A and B, respectively, and the dissociation rates for release of GNAT1, NHP3, Cystin1, RP2, and Src are summarized in Table 1. The presence of Arl3 increases the off rates for high affinity peptides GNAT1 (2.8 s−1), NPHP3 (1.72 s−1), and Cystin1 (5.2 s−1) 1217-, 8600-, and 8667-fold, respectively, for Unc119a such that these rates are now very similar, on the order of 2–5 s−1. There is a similar pattern of release from the Unc119b complex by Arl3; the only significant difference is the >2-fold faster release for Cystin1, which amounts to an almost 19,000-fold acceleration of the dissociation. The release rates for the intermediate affinity RP2 peptide (0.61 s−1) and low affinity Src peptide (2.73 s−1) are only marginally increased by Arl3·GppNHp, by 4.3- and 2.5-fold for RP2 and Src, respectively. The presence of Arl2·GppNHp has a much smaller effect on the release rates for high affinity peptides, which are increased 19-, 90-, and 3-fold for GNAT1, NPHP3, and Cystin1 peptides, respectively. There is no major difference between Unc119a and Unc119b complexes, except for GNAT1. The latter has a weaker affinity to Unc119b and is also much more effectively released, such that the dissociation in the presence of Arl2·GppNHp is 0.66 s−1. The allosteric effect on the release of intermediate (RP2) and low (Src) affinity peptide is, however, very similar for both Arl3 and Arl2. In comparison, the dissociation rates of the high affinity peptides GNAT1, NPHP3, and Cystin1 are increased 10–20-fold by Arl2 but up to 1000–10,000-fold by Arl3. However, for intermediate RP2 or low affinity Src peptides, the dissociation rates are similarly increased 2–4-fold by both Arl2 and Arl3.
FIGURE 4.
Dissociation rates of cargo from Unc119a/b in the presence and absence of Arl proteins. The dissociation rates of cargo release in the presence and absence of Arl proteins were measured by a stopped flow instrument in the fluorescence polarization mode at 25 °C in buffer A. 1 μm preformed complex of Unc119a (A) or Unc119b (B) with fluorescein-labeled N-terminal myristoylated GNAT1 peptide was mixed with a 100-fold excess of unlabeled N-terminal myristoylated GNAT1 peptide in the presence or absence of a 10-fold excess of Arl2/3·GppNHp proteins as indicated. The kinetics of dissociation was monitored, and dissociation rate constants were calculated using first order rate equations. All bar graphs show the average of 5–7 measurements.
Crystallization and Structural Analysis of Unc119a-NPHP3 Complex
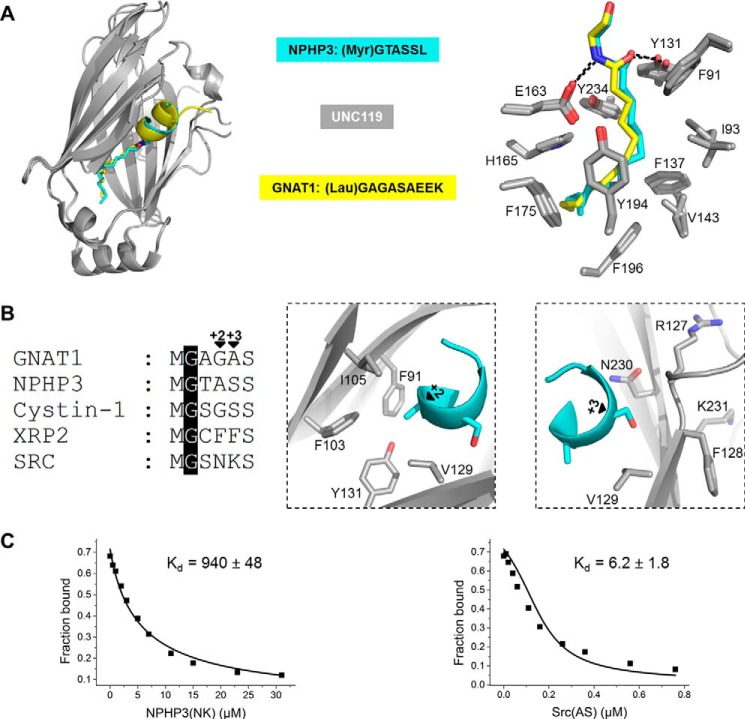
To gain molecular insights and to understand the nature of the binding affinity difference between low and high affinity myristoylated peptides toward Unc119a/b, we aimed to solve the crystal structure of peptides in complex with Unc119a or Unc119b. Using full-length Unc119a/b and myristoylated GNAT1, NPHP3, RP2, and Src peptides of 10-residue length, we did not obtain any suitable crystals. The sequence alignment of Unc119a and Unc119b reveals that they share 58% sequence identity. The N-terminal 50 residues of Unc119b are the most variable region. Because the first 57 residues of Unc119a do not have any effect on peptide binding (16, 33), we deleted these residues of Unc119a. However, the complex of the truncated Unc119a with peptides did not crystallize either. Finally, we used myristoylated peptides of 6-residue length and in addition added limited amounts of both trypsin and chymotrypsin to the crystallization setup. With this strategy, we were able to obtain crystals and solve the crystal structure of myristoylated NPHP3 peptide in complex with Unc119a at 2.1 Å resolution (data collection and refinement statistics summarized in Table 3). Trypsin and chymotrypsin protease were used for in situ proteolysis. The flexible loop of Unc119a, which is located at the entry of the myristoylated peptide, is cleaved by trypsin and chymotrypsin and apparently facilitates crystal packing. Superimposition of the NPHP3 peptide-Unc119a complex with the previously solved structure of Unc119a in complex with lauroylated GNAT1 peptide (PDB code 3RBQ) shows that the β-sandwich fold of Unc119a in both complexes superimposes well with a root mean square deviation of 0.9 Å (Fig. 5A, left). The myristoyl moiety of NPHP3 inserts into the hydrophobic cavity of Unc119a in a very similar pattern as the lauroyl moiety of GNAT1. The two additional carbon atoms of the myristoyl moiety are suited inside the hydrophobic cavity by a higher level of ramification and slightly deeper insertion. Nevertheless, both moieties maintain an identical interaction pattern with the surrounding hydrophobic residues of Unc119a (Fig. 5A, left).
TABLE 3.
Data collection and refinement statistics (molecular replacement)
Numbers in parentheses represent the highest resolution bin.
| Unc119a(58–240)·Myr-NPHP3 peptide (myrGTASSL) | |
|---|---|
| Data collection | |
| Space group | I41 |
| Cell dimensions | |
| a, b, c (Å) | 82.27, 82.27, 105.94 |
| α, β, γ (degrees) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 28.53–2.10 |
| Rsym or Rmerge | 12.3 (74.6) |
| I/σI | 13.47 (3.38) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 11.3 (13.6) |
| Refinement | |
| Resolution (Å) | 2.10 |
| No. of reflections | 22,466 |
| Rwork/Rfree | 22.11/27.82 |
| No. of atoms | |
| Protein | 2425 |
| Ligand/ion | 118 |
| Water | 116 |
| B-factors | |
| Protein | 31.85 |
| Ligand/ion | 37.58 |
| Water | 41.47 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.018 |
| Bond angles (degrees) | 1.925 |
| PDB code | 5L7K |
FIGURE 5.
Structural analysis of a complex of Unc119a with myr-NPHP3. A, superimposition of the crystal structures of Unc119a·myr-NPHP3 and Unc119a·lau-GNAT1 (PDB code 3RBQ) with the lauroyl group in yellow and myristoyl in blue (left). The right panel shows the Unc119a residues interacting with myristoylated NPHP3 and lauroylated GNAT1. B, sequence alignment of the N-terminal part of myristoylated proteins involved in this study (left). Residues of Unc119a around the +2 (middle) and +3 (right) positions of the myr-NPHP3 are shown. C, titration of a complex between 100 nm fluorescein-labeled RP2 peptide and 200 nm Unc119a with increasing concentration of NPHP3(NK) (left) and Src(AS) (right) mutant peptides leads to a decrease in fluorescence polarization. Titration data were fitted with a competition model.
The myristoyl-glycine ester bond makes hydrophilic interactions with Tyr-131 and Glu-163 (Fig. 5A, right). As in the structure with the GNAT1 peptide, the first residues of NPHP3 also form helical turns. The interaction of the carbon chain with the pocket involves the hydrophobic residues Ile-93, Val-143, Phe-175, Tyr-194, Tyr-134, Phe-91, Phe-137, and Phe-196. Fig. 5B shows that the alanine and serine side chain of the NPHP3 peptide at positions +2 and +3 after the myristoylated glycine (the 0 position) are situated in a tightly structured environment. Comparing the equivalent +2 and +3 residues from the other peptides analyzed here, we observe that the high affinity Unc119 binders (GNAT1, NPHP3, and Cystin1) have small size side chain residues, such as alanine, serine, or glycine (Fig. 5B). In contrast, the low or intermediate affinity Unc119 binders (Src and RP2) possess residues with bigger side chains, such as lysine, phenylalanine, or glutamine. We thus hypothesized that the residues with bigger side chains at the +2 and +3 positions would create steric clashes with Unc119a, resulting in lower binding affinities, as found for Src and RP2.
Mutational Analysis of +2 and +3 Positions
To verify our model, we generated two 10-residue mutant peptides, where the residues in the +2 and +3 positions were swapped between high (NPHP3) and low (Src) affinity binders, creating NPHP3(NK) and Src(AS) peptides. The affinities of these swapped peptides toward Unc119a were measured by titrating increasing amounts of unlabeled NPHP3(NK) (Myr-GTNKSLVSP) and Src(AS) (Myr-GSASSKPKD) peptides into a preformed complex of a fluorescent RP2 peptide (0.1 μm) with Unc119a (0.2 μm) and monitoring the displacement of the fluorescent peptide by the unlabeled ones, scored as the decrease of fluorescence polarization. The data were analyzed with a competition model equation derived from the law of mass action as described before (1, 37, 38), using Origin software. The affinity of NPHP3(NK) to Unc119a decreased to a KD of 940 ± 48 nm, whereas the affinity of Src(AS) increased to a KD value of 6.2 ± 1.8 nm (Fig. 5C). These results clearly show that increasing the size of the amino acid side chains at the positions +2 and +3 decreases the binding affinity with Unc119a, and vice versa. Taken together, the residues at the +2 and +3 positions relative to the myristoylated glycine seem to determine the binding affinity between Unc119a and cargo.
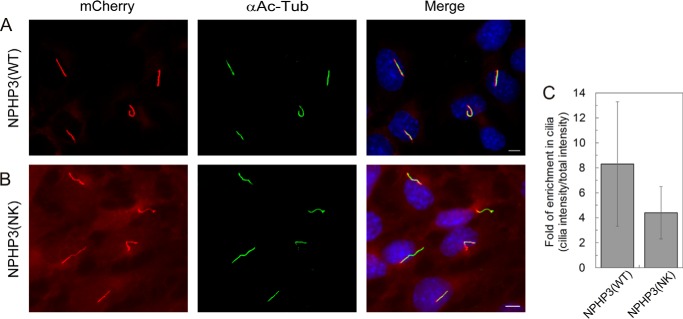
To test whether the reduced affinity of NPHP3(NK) affects its ciliary localization, we used a fragment of NPHP3 (residues 1–203) that is very similar to one described previously for its consistent ciliary localization (16). Constructs containing wild type NPHP3 and mutant NPHP3(NK) were stably transfected into IMCD3 cells, and the ciliary localization of the proteins was measured by quantifying and comparing the fluorescence inside and outside the cilium in ciliated cells. Fig. 6 shows that NPHP3(WT) is highly enriched in cilia compared with the cell body (Fig. 6A), whereas considerable amounts of the NPHP3(NK) mutant are localized to the rest of the cell (Fig. 6B). Quantification of the fluorescence intensity of mCherry-tagged protein was done using CellProfiler. The mean fluorescence intensity ratio between cilia and whole cell shows that the wild type protein has an 8.3-fold enrichment inside the cilia. The NPHP3(NK) mutant loses its almost exclusive ciliary localization and is more distributed over the entire cell, with only a 4.4-fold ciliary enrichment (Fig. 6C).
FIGURE 6.
Localization of NPHP3(WT)-mCherry and NPHP3(NK)-mCherry in IMCD3 cells. A, mCherry fluorescence (LAP-tagged) (red) shows stably expressed NPHP3(WT)-mCherry almost exclusively localizing to primary cilia, which are immunostained with an antibody against acetylated tubulin (green). B, NPHP3(NK)-mCherry localizes both to primary cilia and to the rest of the cell. White bar, 5 μm. C, ciliary enrichment of NPHP3(WT)-mCherry was compared with that of the NPHP3(NK) mutant. The bar graph shows the ratios of the mCherry fluorescence intensity in cilia relative to the total mCherry intensity outside the cilium. Ratios indicate the enrichment of mCherry-tagged NPHP3 inside the cilia. Data analysis of 40 ciliated cells each stably expressing wild type or mutant was accomplished using CellProfiler. Error bars, S.D., n = 40 (p < 0.05; Student's t test).
Discussion
Nephrocystin-3 (NPHP3) is a ciliopathy protein and localizes to primary cilia (39, 40). It has been shown to be a myristoylation-dependent binding partner of Unc119, and this myristoylation occurs at the conserved glycine at position 2 (16). Cystin1 is another cilia-associated protein (41) that has been shown to interact with Unc119 protein via myristoyl binding (16), and its N-terminal myristoylation is required for its proper ciliary membrane localization (42). Myristoylated transducin-α (GNAT1) also interacts with Unc119 protein (31, 32) and is localized in the outer segment of photoreceptor cells, a specialized form of primary cilia (43). Retinitis pigmentosa 2 (RP2) is the Arl3-specific GTPase-activating protein (GAP) (44) that is mutated in X-linked retinitis pigmentosa (45, 46) and has been shown to be localized to the basal body or periciliary region (47, 48). Src kinases Lyn, Hck, and Src itself, non-ciliary proteins, are known to be myristoylated (34, 49), and their biology has been shown to be dependent on Unc119a/b (24–26, 28, 34, 50).
Our experiments reveal that ciliary cargo proteins NPHP3, Cystin1, and GNAT1 bind to Unc119a and Unc119b with picomolar to low nanomolar affinity, whereas the non-ciliary cargo Src has a submicromolar affinity. RP2, which has been shown to localize at the ciliary base, binds with an affinity in the 2-digit nanomolar range. As is typical for such binary interactions, the difference in affinity is mostly dictated by the dissociation rates, which differ between Unc119a and -b by a similar factor, whereas the association rates are very similar. There is no significant difference between Unc119a and -b for the binding affinities of myristoylated peptides except for GNAT1. Its interaction with Unc119a is 10-fold tighter and is determined by a 10-fold difference in the off-rate. Unc119a was originally found as a retina-specific gene named HRG4. It is highly expressed in photoreceptor cells, a specialized form of cilia, and the protein is reported to be localized in the inner segments and photoreceptor synapses (51). Our quantitative and comparative study for Unc119a/b binding to myristoylated cargo suggests that the binding affinity between cargo and Unc119a/b seems to be very important for the Unc119a/b-mediated sorting into the ciliary compartment. This is a scenario similar to what we have recently demonstrated for the sorting of prenylated cargo, such as INPP5E, into cilia (1). The cell biology experiments show the mislocalization of the mutant NPHP3 construct, which is no longer highly enriched in cilia but is now visible over the whole cell. We propose that this mislocalization results from its reduced affinity to Unc119 protein. The fact that the localization cannot be completely reversed by reducing the affinity argues for an additional retention signal operating inside cilia, something that has also been proposed for the localization of prenylated proteins (1).
Although Arl2 and Arl3 share several GTP-dependent interacting partners like the GDI-like solubilizing factors Unc119a, Unc119b, and PDE6δ, in addition to BART and BARTL1 (52), their cellular functions are distinct. Our data show that high affinity myristoylated cargoes (GNAT1, NPHP3, and Cystin1) bound to Unc119a/b are specifically released only by Arl3, whereas lower affinity cargo is released by both Arl2 and Arl3. One exception is GNAT1, which can be partially released from its complex with Unc119b but not with Unc119a by Arl2, possibly due to the 10-fold lower affinity of GNAT1 to Unc119b as compared with Unc119a. We have previously shown that the Arl3-specific GEF Arl13B (53) is exclusively localized inside cilia, whereas RP2, the Arl3-specific GAP (44), localizes to the base of cilia and the periciliary region (47, 48, 52), thus making cilia an Arl3·GTP-enriched compartment. This suggests that Arl3 can release NPHP3 and Cystin1 cargo only inside the cilia and GNAT1 in the outer segment of photoreceptors, whereas Arl2 is capable of releasing intermediate affinity (RP2) and low affinity cargo (Src) outside cilia. Our data support the notion that (i) Unc119a/b proteins regulate the trafficking of myristoylated transducin α subunit (GNAT1), NPHP3, and Cystin1 into the cilia, and (ii) Arl2/3 proteins regulate the sorting of myristoylated cargo into the cilia. The fact that the affinities of Unc119 for its ciliary myristoylated cargo are higher than for Arl3 suggests a very high concentration of activated Arl3 inside the cilium and/or that, in addition to the release mechanism by Arl3, a retention signal for cargo proteins must be in operation to drive the equilibrium toward full release. Such retention could be achieved by the interaction with the ciliary membrane or other ciliary interacting partners.
Unc119a has been shown to localize to the centrosome at the ciliary base, whereas Unc119b localizes to the transition zone and proximal cilium of RPE cells (16). Unc119a and Unc119b proteins share a 58% identity. The sequence comparison of Unc119a and -b shows that the N terminus (residues 1–55) is the most variable region and is not involved in the binding to myristoylated proteins. However, the N terminus might be important and responsible for the different localization of the orthologues. The crystal structure revealed that Unc119a forms a hydrophobic pocket to accommodate the myristoyl moiety that is formed by the residues Phe-91, Ile-93, Tyr-131, Phe-137, Val-143, Glu-163, His-165, Phe-175, Tyr-194, Phe-196, and Tyr-234 of Unc119a (Fig. 5A). The sequence comparison between Unc119a and -b shows that these residues are conserved in Unc119b (Fig. 7), suggesting a similar mode of interaction between Unc119a/b and myristoylated cargo.
FIGURE 7.
Sequence comparison of Unc119 isoforms. Sequence alignment of Unc119 isoforms Unc119a and Unc119b was generated by using ClustalW program. The residues involved in the interaction of Unc119a and the myristoyl moiety are marked by an asterisk and are conserved between Unc119a and -b.
Experimental Procedures
Plasmids and Proteins
Constructs of full-length human Unc119a and Unc119b were cloned into the pGEX-4T3 and pET28a vectors containing an N-terminal GST and His6 tag, respectively. The construct of Unc119a(58–240) in pGEX-2T we received as a gift from the laboratory of Prof. Baehr and was later recloned in pET28 vector. The C-terminal His6 tag full-length constructs of Arl2 and Arl3 in pET20b vector were already available in the laboratory. All proteins were expressed as GST or His6 fusion proteins from Escherichia coli BL21(DE3) CodonPlusRIL, isolated in a first step by affinity chromatography on a glutathione-Sepharose and Talon column respectively, and purified after proteolytic cleavage of GST in a second step by size exclusion chromatography (Superdex S75), as described before (35, 44, 54). After purification, both Arl2 and Arl3 full-length proteins were bound to GDP, as detected via HPLC. The exchange for the GTP analog GppNHp and to mant fluorophore-labeled mant-GppNHp was done as described before (35, 54). The amount of protein-bound nucleotide was analyzed and quantified by C18 reversed-phase column with HPLC.
Plasmids for Cell Culture
Plasmids for transfection of IMCD3 Flp-In cells were created using Gateway cloning technology (Life Technologies), following the manufacturer's recommendations. Full-length human NPHP3 was amplified by PCR from a human Wi38 cDNA library, using the following primers: 5′-GAGAGCTAGCGCCGGTCCGATGGGGACCGCCTCGTCGCTCG-3′ (forward) and 5′-GAGTGTCGACTTACCTTTGTCCTTGCTGAAGG-3′ (reverse). It was located to a modified pACEBac1 vector (ATG Biosynthetics) by restriction enzyme cloning and used as a template for the Gateway entry clone. The entry clone was obtained by PCR using the primers 5′-ATGGGGACCGCCTCGTCGCTCGTG-3′ (forward) and 5′-CCTTTGTCCTTGCTGAAGGAAAAC-3′ (reverse), and the PCR fragment was integrated into the pCR8/GW/TOPO vector (Life Technologies) and located to a pG-LAP5 destination vector (Addgene) (55) by LR recombination. The pG-LAP5 vector originally encoded a LAP tag (S-peptide-PreScission site-GFP) C-terminal to the gene of interest where the region encoding GFP was exchanged against mCherry by restriction enzyme cloning. The C-terminal truncated construct NPHP3(1–203) (NPHP3(WT)) was generated using NPHP3-pG-LAP5-mCherry as a template and the following primers: 5′-ATGGGGACCGCCTCGTCGCTCGTG-3′ (forward) and 5′-CTGAGCCTGTAGCCTCTGAAGTTTGC-3′ (reverse). Mutant NPHP3(1–203) A4N/S5K (NPHP3(NK)) was created using NPHP3(1–203) (WT)-pG-LAP5-mCherry as template and the following mutagenesis primer: 5′-CCGAATTCGCCCTTATGGGGACCAACAAGTCGCTCGTGAGCCCCGCGG-3′ (forward).
Peptides
The N-terminal myristoylated peptides GNAT1 (Myr-GAGASAEEK) and NPHP3 (Myr-GTASSLVSP), unlabeled and labeled with fluorescein at the C terminus, were obtained from AltaBioscience. C-terminal fluorescein-labeled and -unlabeled N-terminal myristoylated Cystin1 (Myr-GSGSSRSSR), NPHP3(NK) (Myr-GTNKSLVSP), and Src(AS) (Myr-GSASSKPKD) were obtained by CambridgePeptides. The C-terminal fluorescein-labeled and unlabeled N-terminal myristoylated RP2 (Myr-GCFFSKRRK) and Src peptides (Myr-GSNKSKPKD) were prepared as described below.
For crystallization, 6-amino acid length N-terminal myristoylated peptides of GNAT1 (Myr-GAGASA), NPHP3 (Myr-GTASSL), Cystin1 (Myr-GSGSSR), RP2 (Myr-GCFFSK), and Src (Myr-GSNKSK) were obtained from Cambridge Peptides.
Synthesis of the Peptides
Solid phase peptide synthesis was carried out on a 0.1-mmol scale using a CEM-Liberty peptide synthesizer equipped with a CEM-Discover microwave. Washing steps between coupling and deprotection were carried out in DMF and DCM using 1 ml of solvent per 100 mg of resin. The Fmoc protecting group was removed with a solution of piperidine in DMF (20%, v/v), 1 min at 30 °C (intensity = 40 watts), and 5 min at 70 °C (intensity = 40 watts). All amino acid couplings were performed in DMF and repeated twice. Typically, Fmoc-Xaa-OH (4 eq, 0.2 m), HCTU (4 eq), and DIPEA (8 eq) were reacted for 10 min at 80 °C (intensity = 20 watts). Upon the completion of the automated synthetic cycles, the resin was washed with DCM (5 ml × 5), DMF (5 ml × 5), and DCM (5 ml × 5). The C-terminal allyl group was removed with a mixture of Pd(PPh3)4 (20 mol %), PhSiH3 (14 eq) in dry THF (3 ml for 0.1 mmol of peptide on the resin) under an argon atmosphere for 12 h to deprotect them. Upon the completion of the reaction, the resin was filtered under vacuum and washed with dry THF (5 ml × 5), DCM (5 ml × 5), DMF (5 ml × 5), and DCM (5 ml × 5). Fmoc-(PEG3)-NH2 (4 eq with regard to the resin loading) was dissolved in dry DMF (0.2 m). [O-(6-chloro-1H-benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate] (HCTU) (4 eq) and diisopropylethylamine (DIPEA) (8 eq) were subsequently added, and the resulting mixture was shaken for 5 min. The peptide-containing resin was shaken with the reaction mixture for 4 h at the ambient temperature. The resin was filtered under vacuum and washed with DCM (5 ml × 5), DMF (5 ml × 5), and DCM (5 ml × 5). The coupling and the washing step were repeated twice. The Fmoc group was removed by shaking the resin with piperidine (20% in DMF, 5 ml) for 40 min at the ambient temperature twice. The resin was washed with DMF (5 ml × 5). Fluorescein isothiocyanate (5 eq) and DIPEA (5 eq) were dissolved in dry DMF (0.2 m), and the resin was shaken with the mixture for 4 h at the ambient temperature. The resin was washed with DCM (5 ml × 5), DMF (5 ml × 5) and DCM (5 ml × 5). The coupling and the washing steps were repeated twice. The peptides were fully deprotected and cleaved from the resin with 5 ml of the mixture of DCM/TFA/TES (50:25:25). The resin was shaken with the deprotection mixture for 2 h and filtered into a round bottom flask. The resin was washed with DCM (5 ml × 3), DCM/MeOH (1:1, 5 ml × 3), and MeOH (5 ml × 3). The combined liquids were diluted with toluene (10 ml) and concentrated under reduced pressure. The resulting slurry was diluted again in toluene (10 ml) and co-evaporated again. The co-evaporation was repeated three times. The resulting crude products were purified by preparative HPLC using a C4 column and characterized by HRMS. The purity of the peptides exceeded 95%.
Fluorescence Measurements
All fluorescence polarization measurements were performed in buffer containing 30 mm HEPES, pH 7.5, 5 mm MgCl2, 100 mm NaCl, and 3 mm DTE (buffer A) at 20 °C. The kinetic measurements were performed with a stopped flow instrument (Applied Photophysics, Leatherhead, UK) in the polarization mode, and fluorescence polarization experiments were performed with a Fluoromax-2 spectrophotometer instrument (Horiba Jobin Yvon, Munich, Germany). The excitation wavelengths were 366 nm for mant and 490 nm for fluorescein fluorophore, whereas the emission wavelengths used for mant and fluorescein fluorophore were 450 and 520 nm, respectively. Emission in stopped flow was detected through a cut-off filter (Schott glass) of 420 and 500 nm for mant and fluorescein, respectively. Data were analyzed using GraFit version 5.0 (Erithracus Software).
Crystallization and Structure Determination
The myristoylated N-terminal peptide of NPHP3 (Myr-GTASSL) was dissolved in 100% DMSO to make 50 mm stock solution. 500 μm solution of Unc119a(58–240) was mixed with N-terminal myristoylated NPHP3 peptide at a 1:1 molar ratio in a buffer containing 25 mm Tris-HCl (pH 7.5), 150 mm NaCl and 3 mm DTE. In situ proteolysis was applied prior to the screening at 20 °C by the addition of both proteases, trypsin and chymotrypsin (at a 1:1000 (w/w) ratio each), as described before (56). The crystals appeared in several conditions containing ammonium sulfate. The final crystallization condition that was optimized was 1.75 m (NH4)2SO4, 0.1 m CAPS (pH 10.0), and 0.2 m Li2SO4. Cryoprotectant solution containing the mother liquor in addition to 20% (v/v) glycerol was used for flash freezing the crystals. X-ray diffraction data were collected at the X10SA beamline of the Suisse Light Source, Villigen. Data were processed by the XDS program. Molecular replacement was carried out using Molrep from the CCP4 suite and Unc119a from the Unc119a-N-terminal lauroylated transducin-α-mimicking peptide complex (PDB code 3RBQ) used as a search model. The model was further built by WinCoot, and the refinement was done with REFMAC5. Refinement and data collection statistics are summarized in Table 3.
Cell Culture and Generation of Stable Cell Lines
Mouse renal epithelial cells from the inner medullary collecting duct (IMCD3) were cultured at 37 °C and 5% CO2 in DMEM/F-12, HEPES complemented with 10% fetal bovine serum and 2 mm l-Glutamine (Life Technologies). The genome of the IMCD3 cells contained a stably integrated FRT cassette (IMCD3 Flp-In, a kind gift from M. V. Nachury; Flp-In cell line technology by Life Technologies), which enabled the generation of stable cell lines as described previously (55, 57). Briefly, IMCD3 Flp-In cells were seeded in 6-well plates at a density of 100,000 cells/well. On the next day, the cells reached a confluence of 40–60% and were cotransfected with the following two vectors: (i) the modified pG-LAP5-mCherry vector containing the gene of interest, a flippase recognition target site, and a hygromycin resistance gene and, (ii) the pOG44 vector (Life Technologies) encoding the FLP recombinase. Transfection was accomplished using Lipofectamine 2000 (Life Technologies). Beginning 2 days after transfection, cells were selected with 100–200 mg/ml hygromycin in the complemented culture medium. Expression of the respective mCherry-tagged proteins was verified by immunoblotting with an antibody against mCherry (1:2000; MPI-CBG antibody facility).
Immunostaining and Microscopy
IMCD3 Flp-In cells stably expressing the respective mCherry-tagged protein were plated on poly-l-lysine-coated coverslips in 6-well plates. Each well contained 100,000 cells. On the following day, cilia were induced by serum starvation for 48 h. After washing in PBS, cells were fixed with 4% formaldehyde in cytoskeletal buffer (2.75 m NaCl, 100 mm KCl, 25 mm Na2HPO4, 8 mm KH2PO4, 40 mm MgCl2, 40 mm EGTA, 100 mm PIPES, 100 mm glucose, pH 6.0) for 20 min. Cells were washed twice in PBS and permeabilized with 0.3% Triton X-100 in cytoskeletal buffer for 10 min. After rinsing in 0.1% Tween 20 in PBS, cells were incubated in 10% FBS in PBS for 30 min for blocking. To immunostain primary cilia, mouse 6-11B-1 anti-acetylated tubulin antibody (1:5000; Sigma T6793) in 10% FBS in PBS was incubated overnight at 4 °C. After washing four times with 0.1% Tween 20 in PBS, Alexa Fluor 488 anti-mouse secondary antibody (1:800; Life Technologies A-11001) was added for 45 min at room temperature. Coverslips were rinsed three times in 0.1% Tween 20 in PBS and once in PBS. Nuclei were stained with DAPI (Serva) for 1 min, diluted 1:10,000 in PBS. Cells were washed three times in PBS, and the coverslips were fixed on glass slides using Mowiol (Merck). Images were taken using an Olympus IX81 microscope with a CCD camera and a 60× oil immersion objective with a numerical aperture of 1.35 using identical settings for each image to ensure comparability.
Author Contributions
M. J., E. K. F., and A. W. designed the experiments and wrote the manuscript. M. J., E. K. F., S. K. K., and T. M. performed the experiments. H. W. critically reviewed the manuscript. All authors read and approved the manuscript.
Acknowledgments
We thank C. Koerner for cloning the Unc119b construct and J. A. Seidel, M. Lokaj, and K. Gotthardt for helping in generating the plasmids used for the generation of stable cell lines. We are also very thankful to Prof. Wolfgang Baehr and Prof. M. V. Nachury for providing the Unc119a plasmid and IMCD3 Flp-In system, respectively.
This work was supported by European Research Council (ERC) Grant 268782 and Sonderforschungsbereich-DFG Grant SFB 642. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5L7K) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- mant-GppNHp
- 2′/3′-O-(N-methyl-anthraniloyl)-guanosine-5′-[(β,γ)-imido]triphosphate, triethylammonium salt
- GppNHp
- 5′-guanylyl imidodiphosphate
- PDB
- Protein Data Bank
- GAP
- GTPase-activating protein
- DMF
- N,N-dimethylformamide
- DCM
- dichloromethane
- DIPEA
- diisopropylethylamine
- HCTU
- [O-(6-chloro-1H-benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium hexafluorophosphate].
References
- 1. Fansa E. K., Kösling S. K., Zent E., Wittinghofer A., and Ismail S. (2016) PDE6delta-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat. Commun. 7, 11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Veland I. R., Awan A., Pedersen L. B., Yoder B. K., and Christensen S. T. (2009) Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol. 111, p39–p53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., and Reiter J. F. (2008) Kif3a constrains β-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat. Cell Biol. 10, 70–76 [DOI] [PubMed] [Google Scholar]
- 4. Berbari N. F., O'Connor A. K., Haycraft C. J., and Yoder B. K. (2009) The primary cilium as a complex signaling center. Curr. Biol. 19, R526–R535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waters A. M., and Beales P. L. (2011) Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. 26, 1039–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tyler K. M., Fridberg A., Toriello K. M., Olson C. L., Cieslak J. A., Hazlett T. L., and Engman D. M. (2009) Flagellar membrane localization via association with lipid rafts. J. Cell Sci. 122, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nachury M. V. (2014) How do cilia organize signalling cascades? Philos. Trans. R. Soc. Lond. B Biol. Sci. 10.1098/rstb.2013.0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., Spiliotis E. T., and Nelson W. J. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caudron F., and Barral Y. (2009) Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell 16, 493–506 [DOI] [PubMed] [Google Scholar]
- 10. Kim S. K., Shindo A., Park T. J., Oh E. C., Ghosh S., Gray R. S., Lewis R. A., Johnson C. A., Attie-Bittach T., Katsanis N., and Wallingford J. B. (2010) Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science 329, 1337–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Najafi M., Maza N. A., and Calvert P. D. (2012) Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc. Natl. Acad. Sci. U.S.A. 109, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Najafi M., and Calvert P. D. (2012) Transport and localization of signaling proteins in ciliated cells. Vision Res. 75, 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomas S., Wright K. J., Le Corre S., Micalizzi A., Romani M., Abhyankar A., Saada J., Perrault I., Amiel J., Litzler J., Filhol E., Elkhartoufi N., Kwong M., Casanova J. L., Boddaert N., et al. (2014) A homozygous PDE6D mutation in Joubert syndrome impairs targeting of farnesylated INPP5E protein to the primary cilium. Hum. Mutat. 35, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chandra A., Grecco H. E., Pisupati V., Perera D., Cassidy L., Skoulidis F., Ismail S. A., Hedberg C., Hanzal-Bayer M., Venkitaraman A. R., Wittinghofer A., and Bastiaens P. I. (2012) The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 14, 148–158 [DOI] [PubMed] [Google Scholar]
- 15. Zhang H., Constantine R., Frederick J. M., and Baehr W. (2012) The prenyl-binding protein PrBP/δ: a chaperone participating in intracellular trafficking. Vision Res. 75, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wright K. J., Baye L. M., Olivier-Mason A., Mukhopadhyay S., Sang L., Kwong M., Wang W., Pretorius P. R., Sheffield V. C., Sengupta P., Slusarski D. C., and Jackson P. K. (2011) An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 25, 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kobayashi A., Kubota S., Mori N., McLaren M. J., and Inana G. (2003) Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 534, 26–32 [DOI] [PubMed] [Google Scholar]
- 18. Allen R. A. (1965) Isolated cilia in inner retinal neurons and in retinal pigment epithelium. J. Ultrastruct. Res. 12, 730–747 [DOI] [PubMed] [Google Scholar]
- 19. Christensen S. T., Pedersen L. B., Schneider L., and Satir P. (2007) Sensory cilia and integration of signal transduction in human health and disease. Traffic 8, 97–109 [DOI] [PubMed] [Google Scholar]
- 20. Singla V., and Reiter J. F. (2006) The primary cilium as the cell's antenna: signaling at a sensory organelle. Science 313, 629–633 [DOI] [PubMed] [Google Scholar]
- 21. Wolfrum U., and Schmitt A. (2000) Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil. Cytoskeleton 46, 95–107 [DOI] [PubMed] [Google Scholar]
- 22. Swanson D. A., Chang J. T., Campochiaro P. A., Zack D. J., and Valle D. (1998) Mammalian orthologs of C. elegans unc-119 highly expressed in photoreceptors. Invest. Ophthalmol. Vis. Sci. 39, 2085–2094 [PubMed] [Google Scholar]
- 23. Vepachedu R., Karim Z., Patel O., Goplen N., and Alam R. (2009) Unc119 protects from Shigella infection by inhibiting the Abl family kinases. PLoS One 4, e5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cen O., Gorska M. M., Stafford S. J., Sur S., and Alam R. (2003) Identification of UNC119 as a novel activator of SRC-type tyrosine kinases. J. Biol. Chem. 278, 8837–8845 [DOI] [PubMed] [Google Scholar]
- 25. Gorska M. M., Stafford S. J., Cen O., Sur S., and Alam R. (2004) Unc119, a novel activator of Lck/Fyn, is essential for T cell activation. J. Exp. Med. 199, 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gorska M. M., Liang Q., Karim Z., and Alam R. (2009) Uncoordinated 119 protein controls trafficking of Lck via the Rab11 endosome and is critical for immunological synapse formation. J. Immunol. 183, 1675–1684 [DOI] [PubMed] [Google Scholar]
- 27. Gorska M. M., Cen O., Liang Q., Stafford S. J., and Alam R. (2006) Differential regulation of interleukin 5-stimulated signaling pathways by dynamin. J. Biol. Chem. 281, 14429–14439 [DOI] [PubMed] [Google Scholar]
- 28. Lee Y., Chung S., Baek I. K., Lee T. H., Paik S. Y., and Lee J. (2013) UNC119a bridges the transmission of Fyn signals to Rab11, leading to the completion of cytokinesis. Cell Cycle 12, 1303–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Resh M. D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16 [DOI] [PubMed] [Google Scholar]
- 30. Mejuch T., van Hattum H., Triola G., Jaiswal M., and Waldmann H. (2015) Specificity of lipoprotein chaperones for the characteristic lipidated structural motifs of their cognate lipoproteins. Chembiochem 16, 2460–2465 [DOI] [PubMed] [Google Scholar]
- 31. Ismail S. A., Chen Y. X., Miertzschke M., Vetter I. R., Koerner C., and Wittinghofer A. (2012) Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J. 31, 4085–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang H., Constantine R., Vorobiev S., Chen Y., Seetharaman J., Huang Y. J., Xiao R., Montelione G. T., Gerstner C. D., Davis M. W., Inana G., Whitby F. G., Jorgensen E. M., Hill C. P., Tong L., and Baehr W. (2011) UNC119 is required for G protein trafficking in sensory neurons. Nat. Neurosci. 14, 874–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ismail S. A., Chen Y. X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P. I., and Wittinghofer A. (2011) Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7, 942–949 [DOI] [PubMed] [Google Scholar]
- 34. Constantine R., Zhang H., Gerstner C. D., Frederick J. M., and Baehr W. (2012) Uncoordinated (UNC)119: coordinating the trafficking of myristoylated proteins. Vision Res. 75, 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Veltel S., Kravchenko A., Ismail S., and Wittinghofer A. (2008) Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 582, 2501–2507 [DOI] [PubMed] [Google Scholar]
- 36. Kapoor S., Fansa E. K., Möbitz S., Ismail S. A., Winter R., Wittinghofer A., and Weise K. (2015) Effect of the N-terminal helix and nucleotide loading on the membrane and effector binding of Arl2/3. Biophys. J. 109, 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roehrl M. H., Wang J. Y., and Wagner G. (2004) A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry 43, 16056–16066 [DOI] [PubMed] [Google Scholar]
- 38. Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M., Hahn S. A., Triola G., Wittinghofer A., Bastiaens P. I., and Waldmann H. (2013) Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497, 638–642 [DOI] [PubMed] [Google Scholar]
- 39. Olbrich H., Fliegauf M., Hoefele J., Kispert A., Otto E., Volz A., Wolf M. T., Sasmaz G., Trauer U., Reinhardt R., Sudbrak R., Antignac C., Gretz N., Walz G., Schermer B., et al. (2003) Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat. Genet. 34, 455–459 [DOI] [PubMed] [Google Scholar]
- 40. Shiba D., Manning D. K., Koga H., Beier D. R., and Yokoyama T. (2010) Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton 67, 112–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hou X., Mrug M., Yoder B. K., Lefkowitz E. J., Kremmidiotis G., D'Eustachio P., Beier D. R., and Guay-Woodford L. M. (2002) Cystin, a novel cilia-associated protein, is disrupted in the cpk mouse model of polycystic kidney disease. J. Clin. Invest. 109, 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tao B., Bu S., Yang Z., Siroky B., Kappes J. C., Kispert A., and Guay-Woodford L. M. (2009) Cystin localizes to primary cilia via membrane microdomains and a targeting motif. J. Am. Soc. Nephrol. 20, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lerea C. L., Bunt-Milam A. H., and Hurley J. B. (1989) α-Transducin is present in blue-, green-, and red-sensitive cone photoreceptors in the human retina. Neuron 3, 367–376 [DOI] [PubMed] [Google Scholar]
- 44. Veltel S., Gasper R., Eisenacher E., and Wittinghofer A. (2008) The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat. Struct. Mol. Biol. 15, 373–380 [DOI] [PubMed] [Google Scholar]
- 45. Schwahn U., Lenzner S., Dong J., Feil S., Hinzmann B., van Duijnhoven G., Kirschner R., Hemberger M., Bergen A. A., Rosenberg T., Pinckers A. J., Fundele R., Rosenthal A., Cremers F. P., Ropers H. H., and Berger W. (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 19, 327–332 [DOI] [PubMed] [Google Scholar]
- 46. Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., and Zuker C. S. (2004) Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117, 527–539 [DOI] [PubMed] [Google Scholar]
- 47. Evans R. J., Schwarz N., Nagel-Wolfrum K., Wolfrum U., Hardcastle A. J., and Cheetham M. E. (2010) The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum. Mol. Genet. 19, 1358–1367 [DOI] [PubMed] [Google Scholar]
- 48. Hurd T., Zhou W., Jenkins P., Liu C. J., Swaroop A., Khanna H., Martens J., Hildebrandt F., and Margolis B. (2010) The retinitis pigmentosa protein RP2 interacts with polycystin 2 and regulates cilia-mediated vertebrate development. Hum. Mol. Genet. 19, 4330–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silverman L., Sudol M., and Resh M. D. (1993) Members of the src family of nonreceptor tyrosine kinases share a common mechanism for membrane binding. Cell Growth Differ. 4, 475–482 [PubMed] [Google Scholar]
- 50. Gorska M. M., and Alam R. (2012) Consequences of a mutation in the UNC119 gene for T cell function in idiopathic CD4 lymphopenia. Curr. Allergy Asthma Rep. 12, 396–401 [DOI] [PubMed] [Google Scholar]
- 51. Higashide T., McLaren M. J., and Inana G. (1998) Localization of HRG4, a photoreceptor protein homologous to Unc-119, in ribbon synapse. Invest. Ophthalmol. Vis. Sci. 39, 690–698 [PubMed] [Google Scholar]
- 52. Lokaj M., Kösling S. K., Koerner C., Lange S. M., van Beersum S. E., van Reeuwijk J., Roepman R., Horn N., Ueffing M., Boldt K., and Wittinghofer A. (2015) The interaction of CCDC104/BARTL1 with Arl3 and implications for ciliary function. Structure 23, 2122–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gotthardt K., Lokaj M., Koerner C., Falk N., Giessl A., and Wittinghofer A. (2015) A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. Elife 4, 10.7554/eLife.11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jaiswal M., Dubey B. N., Koessmeier K. T., Gremer L., and Ahmadian M. R. (2012) Biochemical assays to characterize Rho GTPases. Methods Mol. Biol. 827, 37–58 [DOI] [PubMed] [Google Scholar]
- 55. Torres J. Z., Miller J. J., and Jackson P. K. (2009) High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9, 2888–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dong A., Xu X., Edwards A. M., Midwest Center for Structural Genomics, Structural Genomics Consortium, Chang C., Chruszcz M., Cuff M., Cymborowski M., Di Leo R., Egorova O., Evdokimova E., Filippova E., Gu J., Guthrie J., et al. (2007) In situ proteolysis for protein crystallization and structure determination. Nat. Methods 4, 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sang L., Miller J. J., Corbit K. C., Giles R. H., Brauer M. J., Otto E. A., Baye L. M., Wen X., Scales S. J., Kwong M., Huntzicker E. G., Sfakianos M. K., Sandoval W., Bazan J. F., Kulkarni P., et al. (2011) Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]