Abstract
The bacterial single-stranded DNA binding protein (SSB) acts as an organizer of DNA repair complexes. The radD gene was recently identified as having an unspecified role in repair of radiation damage and, more specifically, DNA double-strand breaks. Purified RadD protein displays a DNA-independent ATPase activity. However, ATP hydrolytic rates are stimulated by SSB through its C terminus. The RadD and SSB proteins also directly interact in vivo in a yeast two-hybrid assay and in vitro through ammonium sulfate co-precipitation. Therefore, it is likely that the repair function of RadD is mediated through interaction with SSB at the site of damage.
Keywords: ATPase, DNA repair, protein-DNA interaction, protein-protein interaction, radiation biology, SSB, helicase
Introduction
In Escherichia coli the single-stranded DNA-binding protein (SSB)2 functions as a molecular organizer of a variety of DNA replication and repair functions (1). Nearly a score of different proteins directly interact with the C-terminal eight amino acids of SSB (1–9), and newly discovered members of the SSB interactome are being described regularly. In some cases the activity of the interacting protein is stimulated by SSB. All of the proteins that interact with SSB to date have a role in some aspect of DNA metabolism. Therefore, an interaction with the C terminus of SSB provides an increasingly reliable indicator of DNA metabolic function.
The radD gene (formerly yejH) was recently implicated in the repair of DNA double-strand breaks after radiation or chemical damage (10, 11). The peptide sequence of the RadD protein includes all seven of the motifs associated with a superfamily 2 (SF2) helicase (10). Although there is little homology outside of these regions, the helicase motifs align well to other E. coli SF2 helicases including RecG and RecQ. RadD also contains a putative zinc finger motif in the C terminus that could aid in binding to DNA substrates. Both the ATPase and zinc binding domains are necessary for the rescue of DNA repair activities of radD deletion strains in vivo (10).
Here, we present the first biochemical characterization of the RadD protein. Although helicase activity was not observed, RadD hydrolyzes ATP in the absence of DNA. This activity is stimulated by the SSB protein, specifically the C-terminal amino acid residues of SSB. The RadD and SSB proteins interact in vivo and directly in vitro. The previous study and this work, taken together, suggest that SSB likely recruits and activates RadD in the cell during DNA repair.
Results
Rationale
Based on the sequence similarity of RadD to known DNA metabolism proteins and a recent study implicating RadD in the repair of double-strand DNA breaks (10), we natively purified the RadD protein and screened for likely activities and interactions. A number of assays for helicase activity, including a variety of both DNA and RNA substrates, produced no positive results and thus are not reported. Presented here are results that demonstrate DNA-independent ATPase activity for RadD as well as an interaction with the SSB C terminus.
RadD Protein Has a DNA-independent ATPase Activity
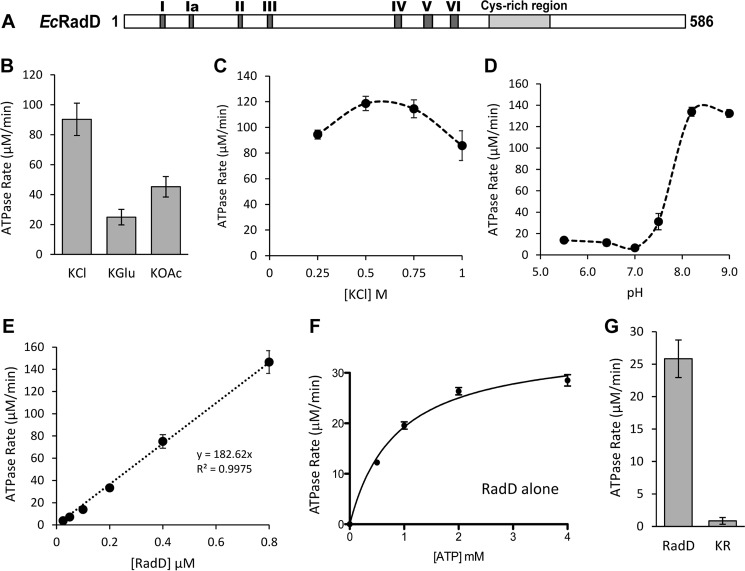
As a predicted SF2 helicase (10), RadD contains the classic Walker A and Walker B motifs (SF2 motifs I and II) that bind and hydrolyze nucleotide triphosphates (12). The relative location of the SF2 motifs is outlined in Fig. 1A. To test for ATP hydrolysis activity, RadD was incubated with ATP in a coupled spectrophotometric assay for ATP hydrolysis. As expected, RadD was able to hydrolyze ATP (Fig. 1). Further testing of buffer conditions revealed that RadD hydrolyzes ATP most efficiently in the presence of the chloride anion (Fig. 1B), peaking at a concentration of 0.5 m KCl (Fig. 1C) and at a pH above 7.5 (Fig. 1D). Surprisingly, RadD displayed a very high basal rate of hydrolysis, with an apparent kcat of up to 600 min−1 (in 0.5 m KCl, Fig. 1C), even in the absence of any DNA substrate. Conditions below 0.2 m salt were not tested, as the RadD protein spontaneously precipitates. For further study, potassium glutamate and pH 7.5 were chosen as the most physiologically relevant conditions. RadD hydrolysis activity increased linearly with increasing enzyme (Fig. 1E) and is, therefore, likely to act as a monomer. RadD failed to show any self-interaction in a yeast two-hybrid assay, further supporting this idea. Titration of the ATP substrate under physiologically relevant conditions resulted in a Km of 0.85 ± 0.11 μm and Vmax of 35.69 ± 1.48 μm min−1 (Fig. 1F, Table 1). A RadD variant containing a mutated Walker A motif (K37R) failed to show significant ATPase activity, as expected (Fig. 1G).
FIGURE 1.
RadD is active in ATP hydrolysis. A, schematic of the primary sequence of RadD protein. The dark gray boxes show the sites of the conserved superfamily 2 helicase motifs, whereas the light gray box shows the position of the suspected zinc finger domain. B, RadD hydrolysis rates in the presence of the chloride (KCl), acetate (KOAc), or glutamate (K-Glu) anions. C, RadD hydrolysis activity with increasing concentrations of potassium chloride. D, RadD hydrolysis rates at different pH levels. MES buffer was used for pH < 7 and Tris acetate was used for pH ≥ 7. E, RadD hydrolysis rates at increasing concentrations of RadD. F, Michaelis-Menten curve for RadD hydrolysis in the presence of increasing ATP concentrations. G, ATP hydrolysis rates of wild type RadD protein versus the RadD K37R (KR) ATPase-deficient protein. Error bars indicate the S.D. of at least three experiments.
TABLE 1.
Kinetic parameters of RadD ATP hydrolysis
| Proteins | Km | Vmax |
|---|---|---|
| μm | μm min−1 | |
| RadD | 0.85 ± 0.11 | 35.69 ± 1.48 |
| RadD + SSB | 0.63 ± 0.09 | 64.56 ± 3.10 |
| RadD + SSB-C10 | 0.40 ± 0.043 | 80.21 ± 2.25 |
RadD and SSB Interact in Vivo and in Vitro
To find potential interaction partners of RadD, a tandem affinity purification assay was implemented (2, 13, 14). Dual-tagged RadD was expressed in E. coli, and the tags were used to purify RadD along with any associated proteins. The resulting complexes were analyzed by mass spectrometry.
Aside from RadD itself, the most prominent protein was SSB, the single-stranded DNA-binding protein, with 13 unique peptides and 61% sequence coverage (Table 2). Several other proteins pulled down in the complex were metabolic DNA binding proteins, including RNA polymerase (subunits β and β'), two topoisomerases, and the nucleoid-associating factors dps, H-NS, and SeqA (Table 2). As an SSB tetramer can interact with more than one protein at a time, some of the proteins detected may be co-associated with SSB. In particular, Top3 and RecQ are known to interact with SSB (1).
TABLE 2.
Potential protein interactors identified by mass spectrometry
| Gene | Description | MW | Unique peptides | Total spectra | Sequence coverage |
|---|---|---|---|---|---|
| ssb | Single-stranded DNA-binding protein | 18,974 | 13 | 1508 | 61% |
| radD | Predicted ATP-dependent DNA or RNA helicase | 66,414 | 44 | 1120 | 67% |
| tnaA | Tryptophanase/l-cysteine desulfhydrase | 52,775 | 16 | 136 | 46% |
| ruvA | Regulatory subunit of RuvABC resolvasome | 22,086 | 6 | 73 | 30% |
| topA | DNA topoisomerase I | 97,351 | 20 | 48 | 38% |
| topB | DNA topoisomerase III | 73,218 | 16 | 43 | 33% |
| dps | Nucleoid component, sequesters iron | 18,696 | 9 | 37 | 61% |
| rpoB | RNA polymerase, β subunit | 150,636 | 22 | 29 | 28% |
| rpoC | RNA polymerase, β prime subunit | 155,164 | 23 | 28 | 30% |
| seqA | Regulator of replication initiation | 20,316 | 6 | 26 | 54% |
| hns | DNA binding transcriptional dual regulator | 15,540 | 4 | 25 | 38% |
| ahpC | Alkyl hydroperoxide reductase, C22 subunit | 20,762 | 7 | 25 | 54% |
| recQ | ATP-dependent helicase | 68,365 | 12 | 25 | 29% |
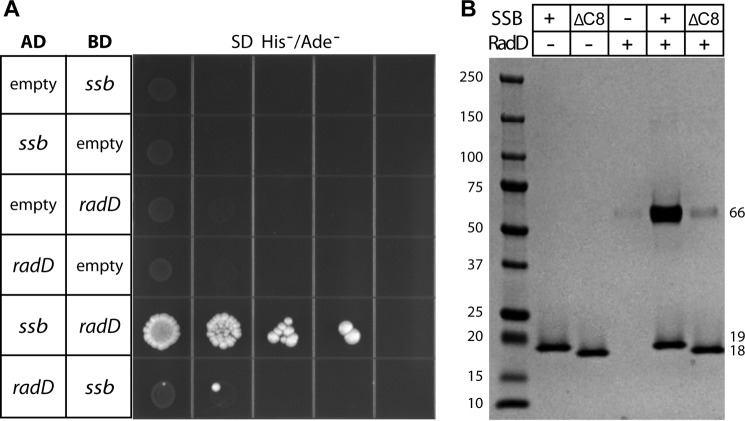
To provide support for RadD and SSB interacting in vivo, the two genes were cloned into vectors for yeast two-hybrid analysis. The DNA binding and activation domain fusions of each protein were then transformed pairwise into the reporter yeast strain. When grown on selective media, colonies appeared when the RadD and SSB vectors were both present in the yeast (Fig. 2A) but not when empty vectors were used. A much stronger signal was observed when ssb was fused to the activation domain and radD to the binding domain, although signal is also observable in the opposite orientation.
FIGURE 2.
RadD and SSB interact. A, yeast two-hybrid vectors containing ssb and radD are able to drive binding to the activation domain, leading to growth on His−/Ade− SD plates. Empty vectors do not support growth. AD = activation domain, BD = binding domain. B, purified SSB protein is able to pull down purified RadD during precipitation in 15% ammonium sulfate. Pelleted proteins were resuspended and run on a 4–15% acrylamide gel. SSB alone precipitates well, but RadD alone does not. SSB lacking the C terminus (ΔC8) is unable to co-precipitate RadD.
The yeast two-hybrid results indicate that RadD and SSB proteins interact in the context of a cell. To test for a direct interaction in vitro, the two proteins were overexpressed and natively purified to homogeneity. It was previously demonstrated that SSB precipitates at low concentrations of ammonium sulfate, and specific protein interactors will co-precipitate into the pellet (15). RadD is clearly enriched in the pellet when SSB is present (Fig. 2B). Furthermore, SSB protein lacking the C-terminal eight amino acids (SSBΔC8), which are critical for interaction of all previously described interacting proteins (1), failed to enrich RadD in the ammonium sulfate pellet (Fig. 2B).
SSB Stimulates the DNA-independent ATP Hydrolysis Activity of RadD
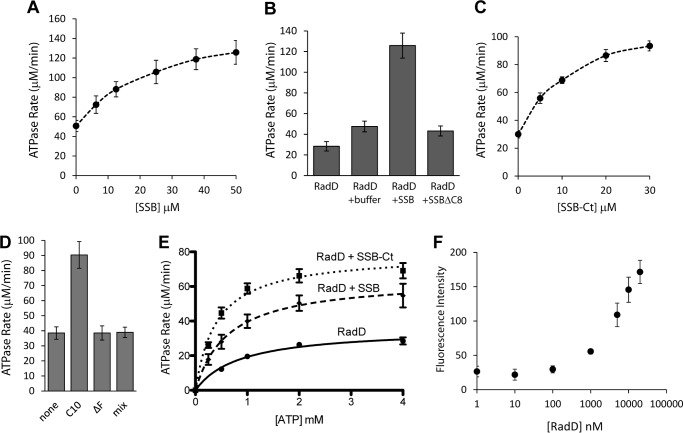
The demonstrable interaction of SSB with RadD led directly to tests for a functional interaction. Increasing concentrations of SSB were added to the RadD ATPase reaction, and a clear concentration-dependent stimulation effect was observed (Fig. 3A). The storage buffer used for SSB contains glycerol and sodium chloride, both of which can independently cause a slight increase in RadD ATPase rate. Therefore, all reactions involving SSB were adjusted to contain the same amount of glycerol and salt, leading to a slightly higher than expected hydrolysis rate even in the 0 μm SSB sample. An SSB variant lacking the C-terminal eight amino acids (SSBΔC8) shows the same level of stimulation as SSB storage buffer alone (Fig. 3B).
FIGURE 3.
SSB stimulates the ATP hydrolysis of RadD. A, rates of RadD ATP hydrolysis at varying concentrations of full-length SSB. B, rates of RadD ATP hydrolysis in the presence of RadD alone, compensating SSB storage buffer, SSB protein (50 μm), or SSBΔC8 protein (50 μm). C, rates of RadD ATP hydrolysis at varying concentrations of SSB tail peptide (SSB-Ct). D, rates of RadD ATP hydrolysis in the presence of buffer (none), wild type SSB tail peptide (C10), SSB tail peptide missing the final phenylalanine (ΔF), or scrambled tail sequence peptide (mix) at 30 μm. E, rates of RadD ATP hydrolysis at varying concentrations of ATP when RadD alone, full-length SSB (10 μm), or SSB tail peptide (10 μm) is present. F, fluorescence polarization of increasing concentrations of RadD with 0.5 nm fluorescein-labeled SSB tail peptide. Error bars indicate the S.D. of at least three experiments.
In all cases where the SSB interaction has been characterized, the SSB-interacting proteins bind SSB through its C-terminal eight amino acid residues (1). To further determine if this region was sufficient for the ATPase stimulation observed in Fig. 3, a peptide containing the last 10 amino acids of SSB (WMDFDDDIPF) was added to the RadD ATPase reactions. Indeed, this SSB tail peptide alone was sufficient to stimulate RadD ATP hydrolytic reaction in a concentration-dependent manner (Fig. 3C), reaching saturation at a lower concentration than full-length SSB (30 μm versus 50 μm). Peptides with the same amino acid composition but in random order (WDFMDDPFID) or with the penultimate phenylalanine (critical for most protein-SSB interactions (1)) missing (WMDFDDDIP) were unable to stimulate RadD ATPase activity even at the highest concentration used (30 μm, Fig. 3D).
The addition of full-length SSB (10 μm) led to a clear decrease in Km (26%) and increase in Vmax (1.8-fold) of ATP hydrolysis by RadD (Fig. 3E, Table 1). The addition of SSB C-terminal peptide shifted Km and Vmax more dramatically than full-length protein, roughly halving Km and doubling Vmax (Fig. 3E, Table 1).
The concentration of SSB or SSB tail peptide needed to induce stimulation of RadD activity was far in excess of the concentration of RadD (30–50 μm versus 200 nm), suggesting a fairly weak interaction. To determine the dissociation constant (KD) between the two proteins, fluorescence polarization was used with a labeled SSB tail peptide and increasing concentrations of RadD. However, the apparent dissociation constant exceeded the concentration of our preparation of RadD, and we could only determine that it is >10 μm (Fig. 3F), which is consistent with our ATPase results. Other proteins shown to interact with SSB are also typically in the low micromolar range (χψ subunits of polymerase III, KD = 2.7 μm; RecQ, KD = 6.4 μm; PriA, KD = 2.4 μm; Exo I, Ka = 7.1 μm; uracil DNA glycosylase, Ka = 5.9 μm; Refs. 16–20, respectively).
RadD Interacts with ssDNA, but ssDNA Does Not Stimulate Hydrolysis
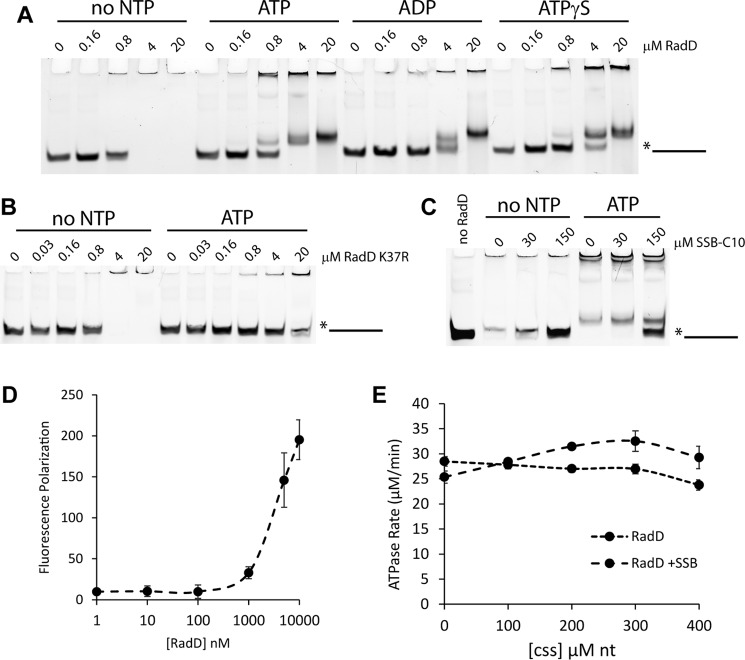
As RadD is a predicted helicase that interacts with SSB, its ability to interact with DNA was evaluated. When incubated with a single-stranded oligonucleotide (poly-dT100), RadD forms a stable higher order complex in the presence or absence of nucleotide cofactor (Fig. 4A). More of the larger complex that does not enter the gel (super shifted) was formed in the absence of nucleotide. A shifted complex (within the gel but above substrate level) formed more readily in the presence of ATP versus ATPγS or ADP, but the differences are slight. No strong interaction was seen with other DNA or RNA substrates. In contrast to wild type RadD protein, the RadD K37R mutant, which is not able to bind ATP, did not show the formation of the smaller, in-gel complex with the addition of ATP, as expected (Fig. 4B).
FIGURE 4.
RadD interacts with DNA. A, Electrophoretic mobility shift of 5-fold dilution series of RadD incubated with poly-dT100 oligonucleotide and no nucleotide, ATP, ADP, or ATPγS (adenosine 5′-O-(thiotriphosphate); 5 mm). B, electrophoretic mobility shift of 5-fold dilution series of RadD K37R incubated with poly-dT100 oligonucleotide with or without ATP (5 mm). C, electrophoretic mobility shift of RadD incubated with poly-dT100 oligonucleotide with increasing concentrations of SSB tail peptide with or without ATP (5 mm). D, fluorescence polarization assay of increasing concentrations of RadD incubated with fluorescent poly-dT100 oligonucleotide. E, RadD ATP hydrolysis rates in the presence of various concentrations of M13mp18 circular single-stranded DNA (css) in the presence or absence of full-length SSB (10 μm).
Surprisingly, the addition of SSB C-terminal peptide at relative levels similar to those seen to stimulate RadD ATPase activity (10–30 μm SSB-Ct versus 200 nm RadD in ATPase; 30–150 μm SSB-Ct versus 2 μm RadD in this EMSA) clearly abrogated the single-stranded DNA binding (Fig. 4C), suggesting that RadD has two distinct modes of action: DNA binding versus ATPase activity. This is consistent with SSB-Ct stimulating RadD ATPase activity in the absence of DNA (Fig. 3).
To confirm that RadD interacts with single-stranded DNA in solution and is not simply the product of aggregation in the gel wells, a fluorescence polarization assay was used. In this assay, bound molecules tumble more slowly in solution. As with the SSB-Ct interaction (Fig. 3F), RadD displays an interaction with poly-dT100 when RadD >1 μm but does not reach saturation (Fig. 4D).
Despite this stable interaction, circular single-stranded DNA (M13mp18) does not stimulate the ATP hydrolysis rate of RadD (Fig. 4E). In the presence of both SSB and RadD, single-stranded DNA appears to confer a slight stimulation (Fig. 4E), with the maximal rate seen at approximately two 65-nt DNA binding sites per SSB tetramer (∼300 μm nucleotides). For this assay, RadD levels were lowered to 100 nm to increase the relative ratio of SSB and DNA to RadD; thus the overall rates appear lower.
Discussion
Many proteins involved in DNA replication, repair, replication restart, recombination, and other DNA metabolic functions interact with the C terminus of SSB (1–9). These interactions put SSB at the nexus of DNA replication and repair coordination. The present work adds RadD to the list of proteins coordinated by SSB. Because there are currently no proteins known to interact with SSB that do not use DNA as a substrate in some fashion, RadD presumably also plays a role in DNA metabolism. Previous genetic studies (10, 21) combined with our biochemical data suggest a role for RadD in protein clearing.
This work presents the first biochemical characterization of the RadD protein. We show for the first time that RadD is an active ATP-hydrolyzing enzyme, with a DNA-independent hydrolysis rate (kcat) of up to 600 min−1. The rate of hydrolysis increases with the addition of SSB, or SSB tail peptide, in a concentration-dependent manner. Interestingly, this is the first reported instance of protein activity being stimulated by the tail peptide alone. This suggests that binding of the SSB C terminus to RadD causes a conformational shift that increases the turnover rate of RadD. Both the Km and Vmax of RadD ATP hydrolysis are strongly affected by this interaction (Km is halved, and Vmax is doubled), supporting this idea. This stimulation is somewhat lower when full-length SSB is used, indicating that tail access is somewhat limited by the additional domains of the protein.
The conformational shift of RadD induced by interaction with SSB also appears to abrogate its DNA binding activity, suggesting that DNA binding and ATPase activity are two separate modes of RadD action. RadD may bind to exposed single-stranded DNA and await a neighboring bound SSB to stimulate its activity, and/or protein clearing activity by RadD may reveal a section of single-stranded DNA, which RadD would bind and then cease its motor activity (Fig. 5C). A small decrease in RadD ATP hydrolysis is seen with an increasing concentration of single-stranded DNA (Fig. 4E), consistent with this idea. The addition of single-stranded DNA stimulated the ATPase activity of RadD only when SSB was present, in which case it is likely that SSB binds the DNA. It would appear that the SSB-DNA complex provides further stimulation than SSB alone, but this difference was quite small and may not be physiologically relevant.
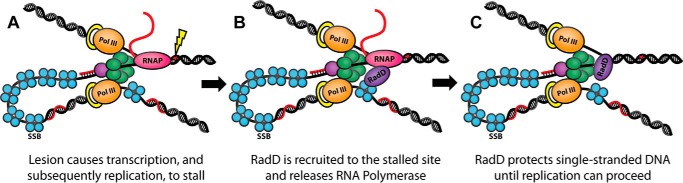
FIGURE 5.
Model of the role of RadD in response to DNA damage. A, a DNA lesion causes the RNA polymerase (RNAP) to stall, which in turn causes the replication fork to stall. B, SSB recruits RadD to the site of the stalled replisome. C, RadD removes RNA polymerase and stabilizes the revealed single-stranded DNA.
Given the results of our biochemical assays, the in vivo role of RadD is likely in stabilizing DNA intermediates and/or clearing protein blocks ahead of replication or repair processes rather than direct DNA remodeling. Previous genetic data demonstrated a role for RadD in responding to DNA damaging agents (10). The ATPase activity of RadD was necessary for this effect, with an ATPase-dead RadD displaying a dominant negative effect in response to UV. This role also showed overlap with the functions of RadA, a RecA paralogue involved in homologous recombination (22, 23), and RecG, a superfamily 2 helicase involved in stabilizing recombination intermediates and preventing origin-independent replication (24, 25).
The stable DNA binding of RadD suggests that RadD may also be involved in stabilizing DNA intermediates formed by lesions along DNA. As the ATP hydrolysis activity of RadD is DNA-independent and has not been linked to any DNA remodeling, it is also possible that RadD is involved in protein clearing during DNA replication or repair. Indeed, there is evidence in Vibrio cholerae that RadD (YejH) clears RNA polymerase from UV-induced DNA lesions (21), and an interaction with both RpoB and RpoC was seen in our TAP tag assay (Table 2). SSB would assist in this process by binding to the single-stranded DNA exposed by stalled replication forks, after encountering either a DNA lesion or a stalled RNA polymerase, and recruiting and activating RadD to help prevent DNA loss and clear the blockage (Fig. 5). Although Mfd normally performs this function, it may be overwhelmed in the presence of extensive damage. Further experiments will be necessary to clarify the role of RadD in DNA metabolism.
Experimental Procedures
Tandem Affinity Purification
The open reading frame of E. coli radD was amplified by PCR and subcloned in-frame into cloning vector pCN70 to produce an expression vector encoding RadD with an N-terminal dual affinity “TAP” tag (includes protein A and calmodulin peptide binding domains separated by a tobacco etch virus protease cleavage site). E. coli K12 strain MG1655 (DE3) transformed with pTAP-RadD was grown at 37 °C in 4 liters of LB medium supplemented with 50 μg/ml ampicillin to midlog phase (A600 of ∼0.5), induced by the addition of 1 μm isopropyl 1-thio-β-d-galactopyranoside and grown for an additional 3 h. Cells were harvested by centrifugation, suspended in 50 ml of Nonidet P-40 buffer (6 mm dibasic sodium phosphate, 4 mm monobasic sodium phosphate, 150 mm NaCl, 2 mm EDTA, 50 mm NaF, 4 mg/liter leupeptin, 0.1 mm sodium vanadate, 19.5 mg/liter benzamidine, 8.7 mg/liter phenylmethylsulfonyl fluoride (PMSF), 1% Nonidet P-40 substitute), and lysed by sonication. Tandem affinity purification was performed as described previously (2), except the TCA precipitation pellet was resuspended in buffer, digested with trypsin, and subjected to MALDI-TOF mass spectrometry for identification of peptides (University of Wisconsin Mass Spectrometry facility).
RadD Purification
The open reading frame of E. coli radD was amplified by PCR and subcloned in-frame into NdeI/BamHI-digested pET21a resulting in plasmid pEAW724. Changes were made for better codon usage in codons 4, 5, and 584. QuikChange site-directed mutagenesis (Agilent Technologies) of pEAW724 using primers consisting of the radD gene bases 98–125, and their complement, were used to create the radD K37R mutant. The AAA bases coding for the lysine at amino acid 37 were changed to CGT to code for an arginine. The resulting plasmid, designated pEAW755, was directly sequenced to confirm the presence of the radD K37R mutant.
The Rosetta (DE3) strain was transformed with the pEAW724, and cells were grown in Luria-Bertani medium containing 100 μg/ml ampicillin at 37 °C to an A600 of ∼0.5. RadD expression was induced by the addition of 0.8 mm isopropyl 1-thio-β-d-galactopyranoside, and cells were grown for an additional 4 h at 37 °C. Cells were pelleted and resuspended in sucrose solution (25% (w/v) sucrose, 250 mm Tris chloride (pH 7.7), 7 mm EDTA, 1 μm pepstatin, 1 μm leupeptin, 1 μm E-64) and lysed by sonication and the addition of 1.6 g/liter of lysozyme. Insoluble material was removed by centrifugation. RadD was precipitated by the addition of solid (NH4)2SO4. The (NH4)2SO4 pellet was resuspended in 20 mm Tris chloride (pH 7.7), 1 mm EDTA, 10% glycerol (R-buffer) containing 800 mm (NH4)2SO4 and applied to a butyl-Sepharose column. RadD was eluted using a linear gradient from 1 m to 0 mm (NH4)2SO4 in R-buffer. RadD containing fractions were pooled, dialyzed into P buffer (20 mm phosphate (pH 7.0), 1 mm EDTA, and 10% glycerol) and loaded onto a ceramic hydroxyapatite column. RadD came off late in the wash. Fractions containing RadD were dialyzed into R-buffer + 200 mm NaCl and loaded onto a source-15Q column. RadD flowed through but a peak containing nuclease came off in gradient before being run over a size exclusion (S300) column. The resulting RadD was dialyzed into R-buffer +200 mm NaCl, aliquoted, and flash-frozen in liquid nitrogen before being stored at −80 °C. The purified protein was >95% pure by gel and free of any detectable nuclease activity. The purification of RadD K37R was identical, except that plasmid pEAW755 was used for expression. The concentration was determined utilizing the extinction coefficient 5.59 × 104 m−1cm−1 for both proteins.
SSB Purification
SSB was purified as previously (26). The concentration was determined utilizing the extinction coefficient 2.38 × 104 m−1cm−1.
Ammonium Sulfate Co-precipitation
Co-precipitation experiments were performed as described previously (15), and pellet fractions were suspended in 30 μl of loading buffer before SDS-PAGE on 4–15% polyacrylamide gradient gels (Bio-Rad).
Yeast Two-hybrid Assays
The open reading frame of E. coli radD was amplified by PCR and subcloned in-frame into pGAD-C3 and pGBD-ADC3 (27). This created an N-terminal fusion with either the GAL4 activation or binding domain. Plasmids containing the open reading frame of ssb with the GAL4 activation or binding domain were a gift from Aimee Marceau. The plasmids were designated pGAD-protein name or pGBD-protein name. The vectors were transformed into pJK69-4A (27) in various combinations to test for interaction with RadD. Cultures of pJK69-4A were grown in YPD (yeast extract/peptone/dextrose) at 30 °C with shaking. One ml of overnight night culture was transferred to an Eppendorf tube and centrifuged for 1 min at 5000 × g. The supernatant was removed, and 125 μl of solution A (80% polyethylene glycol 3350, 20% 1 m lithium acetate), 5 μl of carrier DNA (herring testes DNA), and 3 μl of each vector was added to each tube. The cells were heat-shocked at 42 °C for 1 h, with vortexing every 10 min. The cells were spun at 5000 × g for 1 min, and the supernatant was removed. The cell pellets were resuspended in 200 μl of 2% glucose and plated on Leu−/Trp− SD medium. The transformed cells were grown at 30 °C for several days. Transformants were then selected and grown overnight in Leu−/Trp− SD liquid medium. Cells were centrifuged, the supernatant was removed, and the cells were resuspended in an equal volume of 2% glucose and then serially diluted (1:10). The dilutions were spot plated on His−/Ade− SD plates and grown at 30 °C for several days. A positive protein-protein interaction was assessed by growth of the experimental transformants and no growth of the negative controls (empty plasmids).
ATP Hydrolysis Assay
A coupled spectrophotometric enzyme assay (28) was used to measure the ATPase activities of RadD. The assays were carried out in a Varian Cary 300 dual beam spectrophotometer equipped with a temperature controller and a 12-position cell changer. The reactions were carried out at 37 °C in 25 mm Tris acetate (80% cation), 1 mm DTT, 200 mm potassium glutamate, 10 mm magnesium acetate, 20% (w/v) glycerol, an ATP regeneration system (10 units/ml pyruvate kinase, 2.2 mm phosphoenolpyruvate), a coupling system (3 mm NADH and 10 units/ml lactate dehydrogenase), and purified RadD (200 nm, unless otherwise noted).
Electrophoretic Mobility Shift Assay
Increasing concentrations of RadD protein or equivalent volumes of storage buffer were incubated with 50 nm fluorescein-labeled poly-dT100 oligonucleotide in a buffer of 25 mm Tris acetate (pH 7.5), 10 mm magnesium acetate, 200 mm potassium glutamate, 1 mm dithiothreitol, and 5 mm nucleotide (as appropriate) for 30 min at 37 °C. Ficoll was added to each sample before running on a TBE (Tris/boric acid/EDTA) acrylamide gel for 1–2 h at 100V.
Fluorescence Polarization
Increasing concentrations of RadD protein or equivalent volumes of storage buffer were incubated with 0.5 nm fluorescein amidite-labeled SSB C-terminal polypeptide or 10 nm fluorescein-labeled poly-dT100 oligonucleotide in a buffer of 25 mm Tris acetate (pH 7.5), 10 mm magnesium acetate, 200 mm potassium glutamate, 0.1 μg/ml bovine serum albumin for 30 min at room temperature. Fluorescence was determined using a Beacon instrument.
Author Contributions
S. H. C. conducted most of the experiments, analyzed the results, and wrote most of the paper. R. T. B purified the RadD protein, performed preliminary ATP hydrolysis tests, and performed the ammonium sulfate precipitation. M. M. C. assisted in conceiving the experiments, analyzing results, and revising the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank James L. Keck for reagents and thoughtful discussions and Elizabeth A. Wood for assistance in cloning.
This work was supported in whole or part by the National Institutes of Health Grant GM032335 (to M. M. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SSB
- single-stranded DNA-binding protein
- SF2
- superfamily 2.
References
- 1. Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., and Keck J. L. (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Page A. N., George N. P., Marceau A. H., Cox M. M., and Keck J. L. (2011) Structure and biochemical activities of Escherichia coli MgsA. J. Biol. Chem. 286, 12075–12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu D., Myers A. R., George N. P., and Keck J. L. (2011) Mechanism of exonuclease I stimulation by the single-stranded DNA-binding protein. Nucleic Acids Res. 39, 6536–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ryzhikov M., Koroleva O., Postnov D., Tran A., and Korolev S. (2011) Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 39, 6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antony E., Weiland E., Yuan Q., Manhart C. M., Nguyen B., Kozlov A. G., McHenry C. S., and Lohman T. M. (2013) Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 425, 4802–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wessel S. R., Marceau A. H., Massoni S. C., Zhou R., Ha T., Sandler S. J., and Keck J. L. (2013) PriC-mediated DNA replication restart requires PriC complex formation with the single-stranded DNA-binding protein. J. Biol. Chem. 288, 17569–17578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furukohri A., Nishikawa Y., Akiyama M. T., and Maki H. (2012) Interaction between Escherichia coli DNA polymerase IV and single-stranded DNA-binding protein is required for DNA synthesis on SSB-coated DNA. Nucleic Acids Res. 40, 6039–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aramaki T., Abe Y., Furutani K., Katayama T., and Ueda T. (2015) Basic and aromatic residues in the C-terminal domain of PriC are involved in ssDNA and SSB binding. J. Biochem. 157, 529–537 [DOI] [PubMed] [Google Scholar]
- 9. Petzold C., Marceau A. H., Miller K. H., Marqusee S., and Keck J. L. (2015) Interaction with single-stranded DNA-binding protein stimulates Escherichia coli ribonuclease HI enzymatic activity. J. Biol. Chem. 290, 14626–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen S. H., Byrne R. T., Wood E. A., and Cox M. M. (2015) Escherichia coli radD (yejH) gene: a novel function involved in radiation resistance and double-strand break repair. Mol. Microbiol. 95, 754–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byrne R. T., Chen S. H., Wood E. A., Cabot E. L., and Cox M. M. (2014) Escherichia coli genes and pathways involved in surviving extreme exposure to ionizing radiation. J. Bacteriol. 196, 3534–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker J. E., Saraste M., Runswick M. J., and Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., and Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 14. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., and Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 15. Marceau A. H. (2012) Ammonium sulfate co-precipitation of SSB and interacting proteins. Methods Mol. Biol. 922, 151–153 [DOI] [PubMed] [Google Scholar]
- 16. Glover B. P., and McHenry C. S. (1998) The χψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem. 273, 23476–23484 [DOI] [PubMed] [Google Scholar]
- 17. Shereda R. D., Bernstein D. A., and Keck J. L. (2007) A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 282, 19247–19258 [DOI] [PubMed] [Google Scholar]
- 18. Cadman C. J., and McGlynn P. (2004) PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 32, 6378–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu D., Windsor M. A., Gellman S. H., and Keck J. L. (2009) Peptide inhibitors identify roles for SSB C-terminal residues in SSB/exonuclease I complex formation. Biochemistry 48, 6764–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Handa P., Acharya N., and Varshney U. (2001) Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil DNA glycosylases. J. Biol. Chem. 276, 16992–16997 [DOI] [PubMed] [Google Scholar]
- 21. Baharoglu Z., Babosan A., and Mazel D. (2014) Identification of genes involved in low aminoglycoside-induced SOS response in Vibrio cholerae: a role for transcription stalling and Mfd helicase. Nucleic Acids Res. 42, 2366–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beam C. E., Saveson C. J., and Lovett S. T. (2002) Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184, 6836–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cooper D. L., and Lovett S. T. (2016) Recombinational branch migration by the RadA/Sms paralog of RecA in Escherichia coli. Elife 5, e10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mawer J. S., and Leach D. R. (2014) Branch migration prevents DNA loss during double-strand break repair. PLoS Genet. 10, e1004485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rudolph C. J., Upton A. L., Stockum A., Nieduszynski C. A., and Lloyd R. G. (2013) Avoiding chromosome pathology when replication forks collide. Nature 500, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lohman T. M., and Overman L. B. (1985) Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes: modulation by NaCl concentration. J. Biol. Chem. 260, 3594–3603 [PubMed] [Google Scholar]
- 27. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morrical S. W., Lee J., and Cox M. M. (1986) Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single-stranded DNA. Biochemistry 25, 1482–1494 [DOI] [PubMed] [Google Scholar]