Abstract
Zika virus (ZIKV) is unique among human-pathogenic flaviviruses by its association with congenital anomalies and trans-placental and sexual human-to-human transmission. Although the pathogenesis of ZIKV-associated neurological complications has been reported in recent studies, key questions on the pathogenesis of the other clinical manifestations, non-vector-borne transmission and potential animal reservoirs of ZIKV remain unanswered. We systematically characterized the differential cell line susceptibility of 18 human and 15 nonhuman cell lines to two ZIKV isolates (human and primate) and dengue virus type 2 (DENV-2). Productive ZIKV replication (⩾2 log increase in viral load, ZIKV nonstructural protein-1 (NS1) protein expression and cytopathic effects (CPE)) was found in the placental (JEG-3), neuronal (SF268), muscle (RD), retinal (ARPE19), pulmonary (Hep-2 and HFL), colonic (Caco-2),and hepatic (Huh-7) cell lines. These findings helped to explain the trans-placental transmission and other clinical manifestations of ZIKV. Notably, the prostatic (LNCaP), testicular (833KE) and renal (HEK) cell lines showed increased ZIKV load and/or NS1 protein expression without inducing CPE, suggesting their potential roles in sexual transmission with persistent viral replication at these anatomical sites. Comparatively, none of the placental and genital tract cell lines allowed efficient DENV-2 replication. Among the nonhuman cell lines, nonhuman primate (Vero and LLC-MK2), pig (PK-15), rabbit (RK-13), hamster (BHK21) and chicken (DF-1) cell lines supported productive ZIKV replication. These animal species may be important reservoirs and/or potential animal models for ZIKV. The findings in our study help to explain the viral shedding pattern, transmission and pathogenesis of the rapidly disseminating ZIKV, and are useful for optimizing laboratory diagnostics and studies on the pathogenesis and counter-measures of ZIKV.
Keywords: animal, cell line, flavivirus, placenta, transmission, tropism, virus, Zika
Introduction
Zika virus (ZIKV) is a mosquito-borne flavivirus that has been largely neglected for 60 years after its first isolation from the serum of a febrile sentinel rhesus macaque in the Zika Forest of Uganda in 1947.1 Since 2007, ZIKV has emerged to cause large epidemics of a dengue-like illness in the Pacific islands and Latin America, with imported cases also reported in other continents.2, 3 As of 14 July 2016, more than 60 countries/territories have reported continuing mosquito-borne transmission of ZIKV.4 Although most patients infected with ZIKV have an asymptomatic or a self-limiting acute febrile illness, some patients may develop severe neurological complications, such as Guillain–Barré syndrome, or fatal disseminated infections.2, 5, 6, 7 Moreover, recent clinico-epidemiological, laboratory and animal studies on pathogenesis have established the association between ZIKV infection and congenital anomalies, such as microcephaly, central nervous system malformations and ophthalmological abnormalities.8, 9, 10, 11, 12, 13
ZIKV research has so far been mainly focused on the pathogenesis of ZIKV-associated neurological complications. Questions on the pathogenesis of ZIKV's other clinical manifestations, its unique ability among flaviviruses to be transmitted trans-placentally and sexually, and its potential animal reservoirs remain mostly unanswered. Although in vivo phenomena may not be completely reproducible in the in vitro setting, cell line susceptibility studies are often useful for providing critical data for these knowledge gaps in the early phases of epidemics caused by emerging RNA viruses.14, 15 In this study, we correlated the differential cell line susceptibility, species tropism, viral replication efficiency and antigen expression patterns with the clinical and epidemiological characteristics of ZIKV, and compared them with those of the closely related dengue virus type 2 (DENV-2). Our findings provided novel implications on the disease pathogenesis, transmission and potential animal reservoirs that could be applied to optimize laboratory testing protocols and infection control strategies for this global health emergency.
Materials and Methods
VIRAL ISOLATES
To investigate on the differential cell line susceptibility to epidemic ZIKV and pre-epidemic ZIKV, we included a clinical isolate of ZIKV (Puerto Rico strain PRVABC59) obtained from a patient in the recent South American epidemic (ZIKV-PR; kindly provided by Brandy Russell and Barbara Johnson, Centers for Disease Control and Prevention, USA), and another pre-epidemic ZIKV strain isolated from a nonhuman primate in Uganda in 1947 (976 Uganda strain) (ZIKV-U; kindly provided by Tatjana Avšič Županc, University of Ljubljana, Slovenia, the European Virus Archive). An archived clinical isolate of DENV-2 was obtained from Department of Microbiology, The University of Hong Kong, for comparison with ZIKV, as the two viruses are closely related phylogenetically and have similar clinical manifestations. The ZIKV and DENV-2 isolates were amplified by three additional passages in Vero cells and C6/36 cells, respectively, to make working stocks of the viruses.
ETHICS STATEMENT
Institutional Review Board approval for use of the vial isolates was obtained and all samples were anonymized. The cell lines used in this study were obtained from sources listed in Table 1.
Table 1. Human and nonhuman cell lines used in the present study.
| Organism, anatomic site, cell line | Abbreviations | Source |
|---|---|---|
| Human | ||
| Placenta | ||
| Placental choriocarcinoma | JEG-3 | ATCC no. HTB-36 |
| Genitourinary tract | ||
| Fetal kidney | HEK | In-house |
| Cervical adenocarcinoma | HeLa | ATCC no. CCL-2.2 |
| Ovarian surface epithelium | HOSE6-3 | In-house |
| Metastatic prostatic adenocarcinoma | LNCaP | ATCC no. CRL-1740 |
| Testicular germ cell tumor | 833KE | Sigma-Aldrich, St Louis, MO, USA |
| Neuromuscular cells | ||
| Anaplastic astrocytoma (non-epithelial) | SF268 | Research Center for Emerging Viral Infections, Chang Gung University, Taiwan |
| Rhabdomyosarcoma | RD | ATCC no. CCL-136 |
| Retina | ||
| Retinal pigment epithelium | ARPE19 | ATCC no. CRL-2302 |
| Respiratory tract | ||
| Laryngeal epidermoid carcinoma | Hep-2 | ATCC no. CCL-23 |
| Lung adenocarcinoma | Calu-3 | ATCC no. HTB-55 |
| Embryonic lung fibroblasts | HFL | In-house |
| Gastrointestinal tract | ||
| Colorectal adenocarcinoma | Caco-2 | ATCC no. HTB-37 |
| Liver | ||
| Hepatocellular carcinoma | Huh-7 | JCRB cell bank of Okayama University, Japan |
| Immune cells | ||
| Peripheral blood monocytes from acute monocytic leukemia | THP-1 | ATCC no. TIB-202 |
| Monocytes from histiocytic lymphoma | U937 | ATCC no. CRL-1593.2 |
| B lymphocytes from Burkitt's lymphoma | Raji | ATCC no. CCL-86 |
| T lymphocytes | H9 | ATCC no. HTB-176 |
| Nonhuman | ||
| Mammals | ||
| African green monkey kidney | Vero | ATCC no. CCL-81 |
| Rhesus monkey kidney | LLC-MK2 | ATCC no. CCL-7 |
| Porcine kidney | PK-15 | ATCC no. CCL-33 |
| Madin–Darby canine kidney | MDCK | ATCC no. CCL-34 |
| Crandell Rees feline kidney | CRFK | ATCC no. CCL-94 |
| Bat brain | TP2 | In-house |
| Mouse subcutaneous areolar and adipose tissue | L929 | ATCC no. CCL-1 |
| Immortalized mouse microglia | BV2 | Cell Resource Center, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences/Peking Union Medical College |
| Primary mouse embryonic fibroblasts | 3T3 | ATCC no. CCL-92 |
| Rat retinal precursor cells | R28 | Kerafast, Inc., Boston, MA, USA |
| Rat kidney | RK3E | ATCC no. CRL-1895 |
| Rabbit kidney | RK-13 | ATCC no. CCL-37 |
| Baby hamster kidney | BHK21 | ATCC no. CCL-10 |
| Others | ||
| Chicken fibroblasts | DF-1 | ATCC no. CRL-12203 |
| Mosquito (Aedes albopictus) | C6/36 | ATCC no. CRL-1660 |
Abbreviation: American Tissue Culture Collection, ATCC.
VIRAL CULTURE
Viral culture was performed as we previously described with modifications.14 Briefly, 33 cell lines derived from different tissues or organs and host species were each inoculated with one multiplicity of infection of the ZIKV and DENV-2 isolates for 1 h (Table 1). Non-attached virus was removed by washing the cells twice in phosphate-buffered saline (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). The monolayer cells were maintained in minimum essential medium, Dulbecco's modified Eagle's medium (DMEM), DMEM/F12 or RPMI medium, with 1% fetal calf serum, depending on the type of cell line according to supplier's instructions (Gibco). The suspension cells were maintained in RPMI medium with 2% fetal calf serum (Gibco). All infected cell lines were incubated at 37 °C for five days, except C6/36 which was incubated at 28 °C. Cytopathic effects (CPE) were examined at one, three and five days post-virus inoculation (d.p.i) with inverted light microscopy.
RNA EXTRACTION AND QUANTITATIVE REVERSE-TRANSCRIPTION PCR
Total nucleic acid (TNA) was extracted from culture supernatants of the 33 cell lines infected by ZIKV or DENV-2 at 1, 3 and 5 d.p.i. using EZ1 Virus Mini Kit v2.0 (Qiagen, Hilden, Germany) as we previously described.16, 17 Briefly, 200 μL of supernatant were used for extraction and the TNA was eluted in 60 μL of AVE buffer. Real-time one-step quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was used for the detection of ZIKV and DENV-2 using QuantiNova Probe RT-PCR Kit (Qiagen) in a LightCycler 96 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland). Five microliters of purified TNA was amplified in a 20 μL-reaction containing 10 μL of 2 × QuantiNova Probe RT-PCR Master Mix, 0.2 μL QN Probe RT-mix, 0.8 μM forward primer, 0.8 μM reverse primer and 200 nM probe. ZIKV detection was performed as previously described, with slight modifications.18 Forward primer (5′-CGY TGC CCA ACA CAA GG-3′), reverse primer (5′-CCA CYA AYG TTC TTTT GCA BAC A-3′) and probe (5′-HEX-AGC CTA CCT TGA YAA GCA RTC AGA CAC TC-IABkFQ-3′) targeting the ZIKV envelope gene were used. For DENV-2 detection, forward primer (5′-GCA TAT TGA CGC TGG GAR AGA C-3′), reverse primer (5′-CGY TCT GTG CCT GGA WWG ATG-3′) and probe (5′-FAM-CAG AGA TCC TGC TGT C-MGB-NFQ-3′) targeting the DENV-2 3'-untranslated region were used. Reactions were incubated at 45 °C for 10 min, followed by 95 °C for 5 min, and then thermal cycled for 50 cycles (95 °C for 5 s, 55 °C for 30 s). A series of 10-fold dilutions equivalent to 1 × 102–1 × 106 copies/reaction mixture were prepared to generate calibration curves and run in parallel with the test samples.
CLONING AND PURIFICATION OF HIS6-TAGGED RECOMBINANT ZIKV AND DENV-2 NS1
Primers (5′-CAT ATG GAT GTG GGG TGC TCG GTG GAC T-3′ and 5′-CTC GAG TGC AGT CAC CAT TGA CCT TAC T-3′) were used to amplify the gene encoding the ZIKV–nonstructural protein 1 (NS1) protein by RT-PCR. The sequence encoding a 352-amino-acid fragment of ZIKV–NS1 protein was amplified and cloned into the NdeI and XolI sites of expression vector pETH in frame and downstream of the series of six histidine residues. The recombinant ZIKV–NS1 protein was expressed in Escherichia coli, denatured in 8 M urea, and purified with Ni-nitrilotriacetic acid affinity chromatography (Qiagen) according to the manufacturer's instructions. The solubilized protein was refolded by slowly dialyzing with refolding buffers. Expression of the recombinant ZIKV–NS1 protein was confirmed by western blot analysis using mouse anti-His monoclonal antibody (Sigma-Aldrich, St Louis, MO, USA). Preparation of DENV-2-NS1 protein was performed as previously described.19
PREPARATION OF SPECIFIC ANTIBODIES AGAINST ZIKV- AND DENV-2-NS1 PROTEINS
This was performed as we previously described with modifications.14 Briefly, 30 μg of purified recombinant ZIKV-NS1 protein was mixed with an equal volume of complete Freund's adjuvant (Sigma-Aldrich) and injected subcutaneously into BALB/c mice. Incomplete Freund's adjuvant (Sigma-Aldrich) was used in subsequent injections at 14-day intervals for three times. Serum samples were collected 14 days after the fourth injection. Preparation of specific antibodies against DENV-2-NS1 protein was performed as previously described.19
ANTIGEN DETECTION OF INFECTED CELL LINES BY IMMUNOFLUORESCENCE
This was performed as we previously described with modifications.14 Cell smears at 1, 3 and 5 d.p.i. were prepared and fixed in chilled acetone at −20 °C for 10 min. The fixed cells were incubated with mouse antiserum against the ZIKV- or DENV-2-NS1 protein, followed by FITC-rabbit anti-mouse IgG (Invitrogen, Carlsbad, CA, USA). Cells were then examined under a fluorescence microscope. Three representative microscopic fields were chosen and 100 cells were counted for characteristic cytoplasmic apple green fluorescence. The mean percentage of positive cells was rounded up to the nearest multiplicity of 10. Uninoculated cells were used as negative control.
STATISTICAL ANALYSIS
The Student's t-test was used to compare the mean viral load of the different cell lines at 1, 3 and 5 d.p.i. with the mean baseline viral load at 0 d.p.i. All calculations were based on log-transformed viral loads. P-value of <0.01 was considered as statistically significant. Computation was performed using the Predictive Analytics Software v18.0 for Windows.
Results
HUMAN CELL LINES
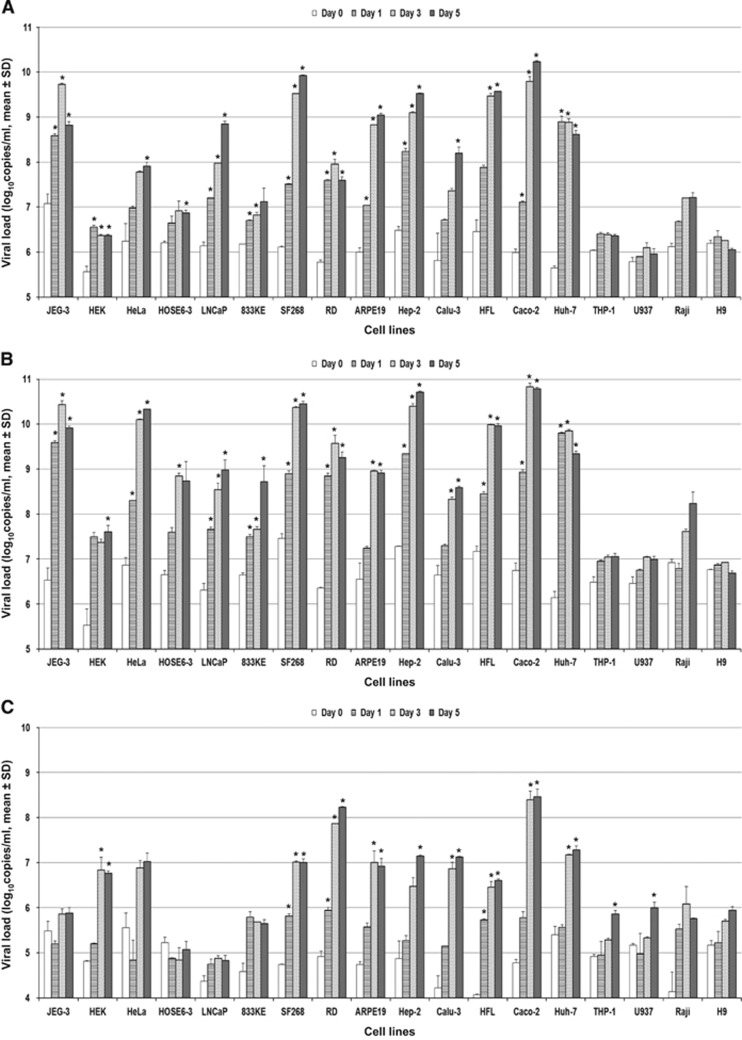
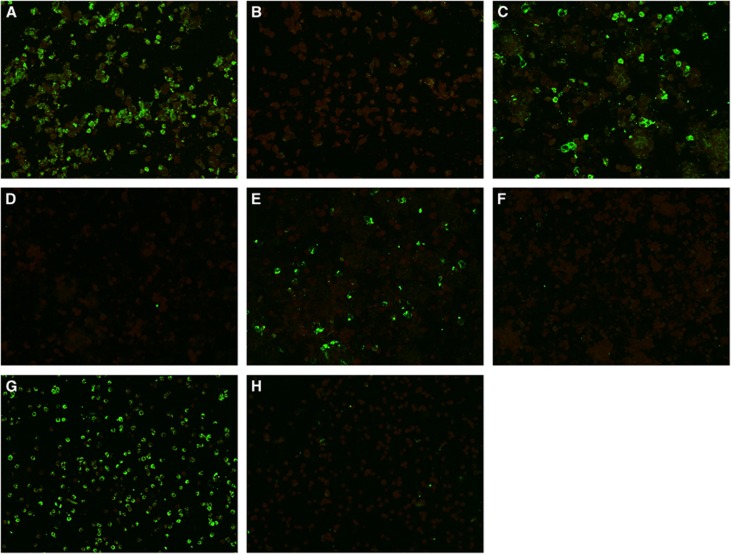
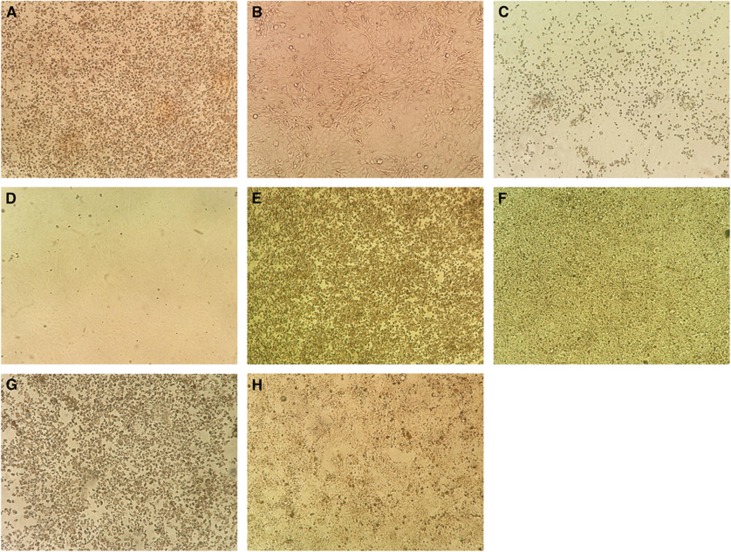
Eighteen human cell lines were tested (Table 1). ZIKV-PR and ZIKV-U showed similar tropism as defined by viral load, immunofluorescent antigen staining and CPE. A ⩾2 log increase in mean viral load (P<0.01) within 5 d.p.i. was observed in 14/18 cell lines infected with ZIKV-PR or ZIKV-U (Figures 1A and 1B). These included the placental (JEG-3), genitourinary (HEK, HeLa, HOSE6-3, LNCaP and 833KE), neuromuscular (SF268 and RD), retinal (ARPE19), respiratory (Hep2, Calu-3 and HFL), intestinal (Caco-2) and hepatic (Huh-7) cell lines. There was a slight increase in mean viral load of about 0.5–1 lgcopies/mL in the monocyte (THP-1) and B lymphocyte (Raji) cell lines (P>0.01), but none of the immune cell lines achieved a ⩾2 log increase in mean viral load within 5 d.p.i. The highest mean viral loads (⩾10 lgcopies/mL) were observed in the placental (JEG-3), cervical (HeLa), neuronal (SF268), upper respiratory tract (Hep-2) and intestinal (Caco-2) cell lines. Twelve of these 14 cell lines (JEG-3, HEK, HeLa, LNCaP, SF268, RD, ARPE19, Hep-2, Calu-3, HFL, Caco-2 and Huh-7) showed obvious ZIKV-NS1 protein expression by IF in addition to a high viral load (Table 2). The peak ZIKV-NS1 protein expression in these cell lines occurred on 3–5 d.p.i. ZIKV-NS1 protein expression was most prominent (⩾50% of infected cells) in the placental (JEG-3), neuronal (SF268), muscle (RD), retinal (APRE19), lower respiratory tract (HFL) and hepatic (Huh-7) cell lines (Figure 2). Although ovarian (HOSE6-3) and testicular (833KE) cells showed high mean viral loads in qRT-PCR, ZIKV-NS1 protein expression was consistently found in only ⩽5% of infected cells. CPE were observed in 8 of these 14 cell lines, namely, JEG-3, SF268, RD, ARPE19, Hep-2, HFL, Caco-2 and Huh-7 (Table 3). CPE were most prominent (>50% involvement) in the placental (JEG-3), neuronal (SF268), muscle (RD), intestinal (Caco-2) and hepatic (Huh-7) cell lines (Figure 3). The gradual onset of CPE in most of these cell lines at 3 d.p.i. correlated with the steady increase of viral load and ZIKV-NS1 protein expression at 3–5 d.p.i.
Figure 1.
Differential human cell line susceptibility to (A) ZIKV-PR, (B) ZIKV-U and (C) DENV-2, as defined by viral load, on days 1, 3 and 5 post-ZIKV inoculation. All experiments were done in triplicate. The mean viral loads on days 1, 3 and 5 were compared with the mean baseline viral load on day 0 (1 h post-ZIKV inoculation). All calculations were based on log-transformed viral loads. *P-value of <0.01.
Table 2. Differential cell line susceptibility to ZIKV-PR, ZIKV-U and DENV-2, as defined by immunofluorescent antigen staining on days 1, 3 and 5 after infection.
| Cell lines |
Immunofluorescencea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
ZIKV-PR |
ZIKV-U |
DENV-2 |
|||||||
| Day 1 | Day 3 | Day 5 | Day 1 | Day 3 | Day 5 | Day 1 | Day 3 | Day 5 | |
| Human | |||||||||
| Placenta | |||||||||
| JEG-3 (placenta) | 50 | 60 | 80 | 60 | 70 | 80 | N | N | N |
| Genitourinary tract | |||||||||
| HEK (kidney) | <1 | 1 | 5 | 10 | 20 | 20 | 1 | 20 | 20 |
| HeLa (cervix) | N | <1 | 1 | 5 | 20 | 30 | 10 | 30 | 30 |
| HOSE6-3 (endometrium) | 1 | 1 | 1 | 1 | 5 | 5 | N | N | N |
| LNCaP (prostate) | 1 | 5 | 40 | 1 | 20 | 30 | N | N | N |
| 833KE (testis) | N | 1 | 1 | N | N | 5 | N | N | N |
| Neuromuscular cells | |||||||||
| SF268 (neuron) | 5 | 20 | 40 | 30 | 70 | 80 | 40 | 40 | 30 |
| RD (muscle) | 1 | 10 | 20 | 30 | 70 | 50 | 50 | 90 | 80 |
| Retina | |||||||||
| ARPE19 (retina) | 5 | 60 | 50 | 20 | 60 | 60 | 10 | 30 | 30 |
| Respiratory tract | |||||||||
| Hep-2 (larynx) | 1 | 5 | 10 | 10 | 30 | 30 | 10 | 20 | 50 |
| Calu-3 (lung) | 1 | 5 | 10 | 1 | 5 | 10 | 10 | 20 | 10 |
| HFL (lung) | 5 | 40 | 60 | 10 | 60 | 60 | 40 | 40 | 40 |
| Gastrointestinal tract | |||||||||
| Caco-2 (colon) | 10 | 40 | 40 | 30 | 40 | 30 | 5 | 50 | 70 |
| Liver | |||||||||
| Huh-7 (liver) | 20 | 60 | 100 | 50 | 80 | 100 | 50 | 80 | 80 |
| Immune cells | |||||||||
| THP-1 (monocyte) | <1 | <1 | <1 | N | 1 | 1 | N | N | N |
| U937 (monocyte) | N | N | N | N | N | N | N | N | N |
| Raji (B lymphocyte) | N | N | <1 | <1 | <1 | 5 | N | 1 | 40 |
| H9 (T lymphocyte) | N | N | N | N | <1 | N | N | N | <1 |
| Nonhuman | |||||||||
| Mammals | |||||||||
| Vero (monkey) | 40 | 50 | 70 | 70 | 80 | 90 | 50 | 90 | 100 |
| LLC-MK2 (monkey) | 1 | 20 | 30 | 20 | 90 | 90 | 40 | 80 | 70 |
| PK-15 (pig) | 10 | 40 | 40 | 30 | 70 | 70 | 20 | 60 | 70 |
| MDCK (dog) | 1 | 1 | 5 | 5 | 10 | 10 | N | N | 5 |
| CRFK (cat) | N | 1 | 1 | N | 5 | 5 | N | N | 5 |
| TP2 (bat) | N | N | N | N | 5 | 10 | 20 | 30 | 30 |
| L929 (mouse) | N | N | N | 5 | 5 | 1 | 1 | 1 | 1 |
| BV2 (mouse) | N | N | N | N | N | N | 5 | 5 | N |
| 3T3 (mouse) | 1 | 5 | 10 | 5 | 20 | 10 | <1 | <1 | <1 |
| R28 (rat) | N | N | N | N | N | N | <1 | <1 | <1 |
| RK3E (rat) | 5 | 5 | 5 | 10 | 10 | 10 | N | <1 | <1 |
| RK-13 (rabbit) | 20 | 30 | 20 | 40 | 40 | 30 | N | N | N |
| BHK21 (hamster) | 10 | 70 | 80 | 30 | 50 | 50 | 50 | 50 | 50 |
| Others | |||||||||
| DF-1 (chicken) | 1 | 30 | 70 | 40 | 70 | 70 | N | <1 | <1 |
| C6/36 (mosquito) | 10 | 40 | 90 | 10 | 60 | 90 | N | 30 | 40 |
N is defined as negative. The numerals denote the mean percentage of positive cells from three representative microscopic fields, rounded up to the nearest multiplicity of 10.
Figure 2.
ZIKV nonstructural protein 1 expression by immunofluorescence in cell lines representing the potential trans-placental, sexual and vector-borne transmission routes, and/or genitourinary tissues with persistent viral replication of ZIKV. Expression of ZIKV nonstructural protein 1 as intense apple green cytoplasmic fluorescence in different cell lines stained by monospecific polyclonal serum from BALB/c mice immunized with His6-tagged recombinant ZIKV nonstructural protein 1 at day 5 post-ZIKV inoculation (original magnification × 200). (A) Infected placental (JEG-3) cells; (B) uninfected JEG-3 control; (C) infected prostatic (LNCaP) cells; (D) uninfected LNCaP control; (E) infected renal (HEK) cells; (F) uninfected HEK control; (G) infected mosquito (C6/36) cells; (H) uninfected C6/36 control.
Table 3. Differential cell line susceptibility to ZIKV-PR, ZIKV-U and DENV-2, as defined by cytopathic effect (CPE) on days 1, 3 and 5 after infection.
| Cell lines |
CPE, gradea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
ZIKV-PR |
ZIKV-U |
DENV-2 |
|||||||
| Day 1 | Day 3 | Day 5 | Day 1 | Day 3 | Day 5 | Day 1 | Day 3 | Day 5 | |
| Human | |||||||||
| Placenta | |||||||||
| JEG-3 (placenta) | N | 1+ | 4+ | 1+ | 4+ | 4+ | N | N | N |
| Genitourinary tract | |||||||||
| HEK (kidney) | N | N | N | N | N | N | N | N | N |
| HeLa (cervix) | N | N | N | N | 1+ | 1+ | N | N | N |
| HOSE6-3 (endometrium) | N | N | N | N | N | N | N | N | N |
| LNCaP (prostate) | N | N | N | N | N | N | N | N | N |
| 833KE (testis) | N | N | N | N | N | N | N | N | N |
| Neuromuscular cells | |||||||||
| SF268 (neuron) | N | 2+ | 2+ | N | 2+ | 3+ | N | 1+ | 2+ |
| RD (muscle) | N | 1+ | 2+ | N | 2+ | 4+ | N | 4+ | 4+ |
| Retina | |||||||||
| ARPE19 (retina) | N | 1+ | 1+ | N | 1+ | 2+ | N | 1+ | 2+ |
| Respiratory tract | |||||||||
| Hep-2 (larynx) | N | N | N | N | 1+ | 1+ | N | N | 1+ |
| Calu-3 (lung) | N | N | N | N | N | N | N | N | 1+ |
| HFL (lung) | N | 1+ | 1+ | N | 2+ | 3+ | N | 2+ | 2+ |
| Gastrointestinal tract | |||||||||
| Caco-2 (colon) | N | N | 3+ | N | 3+ | 4+ | N | N | 1+ |
| Liver | |||||||||
| Huh-7 (liver) | N | 3+ | 4+ | 1+ | 4+ | 4+ | N | 2+ | 4+ |
| Immune cells | |||||||||
| THP-1 (monocyte) | N | N | N | N | N | N | N | N | N |
| U937 (monocyte) | N | N | N | N | N | N | N | N | N |
| Raji (B lymphocyte) | N | N | N | N | N | N | N | 2+ | 4+ |
| H9 (T lymphocyte) | N | N | N | N | N | N | N | N | 1+ |
| Nonhuman | |||||||||
| Mammals | |||||||||
| Vero (monkey) | N | 3+ | 4+ | N | 4+ | 4+ | N | 1+ | 1+ |
| LLC-MK2 (monkey) | N | 1+ | 2+ | N | 2+ | 4+ | N | 4+ | 4+ |
| PK-15 (pig) | N | 2+ | 2+ | N | 2+ | 3+ | N | 1+ | 2+ |
| MDCK (dog) | N | N | 1+ | N | 1+ | 1+ | N | N | 1+ |
| CRFK (cat) | N | N | N | N | N | N | N | 1+ | 1+ |
| TP2 (bat) | N | N | N | N | N | N | N | 1+ | 2+ |
| L929 (mouse) | N | N | N | N | N | N | N | N | N |
| BV2 (mouse) | N | N | N | N | N | N | N | N | N |
| 3T3 (mouse) | N | N | N | N | N | N | N | N | N |
| R28 (rat) | N | N | N | N | N | N | N | 1+ | 1+ |
| RK3E (rat) | N | N | N | N | N | N | N | N | N |
| RK-13 (rabbit) | N | N | 1+ | N | 1+ | 1+ | N | N | N |
| BHK21 (hamster) | N | 1+ | 2+ | N | 4+ | 4+ | 1+ | 4+ | 4+ |
| Others | |||||||||
| DF-1 (chicken) | N | N | 1+ | N | 4+ | 4+ | N | N | N |
| C6/36 (mosquito) | N | N | N | N | N | N | N | N | N |
N is defined as negative, 1+ is defined as 1–25% involvement, 2+ is defined as >25% to 50% involvement, 3+ is defined as >50% to 75% involvement and 4+ is defined as >75% involvement.
Figure 3.
Cytopathic effects in cell lines representing the major organs with clinical manifestations in ZIKV infection. Cytopathic effects consisting of cell rounding, detachment and degeneration were observed in different cell lines at day 5 post-ZIKV inoculation under inverted microscopy (original magnification × 100). (A) Infected neuronal (SF268) cells; (B) uninfected SF268 control; (C) infected retinal (ARPE19) cells; (D) uninfected ARPE19 control; (E) infected muscle (RD) cells; (F) uninfected RD3 control; (G) infected hepatic (Huh-7) cells; (H) uninfected Huh-7 control.
In contrast, DENV-2 showed a much narrower range of human tissue tropism, with ⩾2 log increase in the mean viral load (P<0.01) in only 9/18 cell lines (HEK, SF268, RD, ARPE19, Hep-2, Calu-3, HFL, Caco-2 and Huh-7; Figure 1C). Notably, unlike ZIKV, the DENV-2 load did not significantly increase by ⩾2 lgcopies/mL in any of the genital tract and placental cell lines, with only mildly increased viral load (P>0.01) and DENV-2-NS1 protein expression in the HeLa cell line. Most of these nine cell lines with increased viral loads demonstrated DENV-2-NS1 protein expression (Table 2) and CPE (Table 3). The CPE were generally delayed in onset (5 d.p.i.) and limited (⩽50%), except in the RD and Huh-7 cell lines.
NONHUMAN CELL LINES
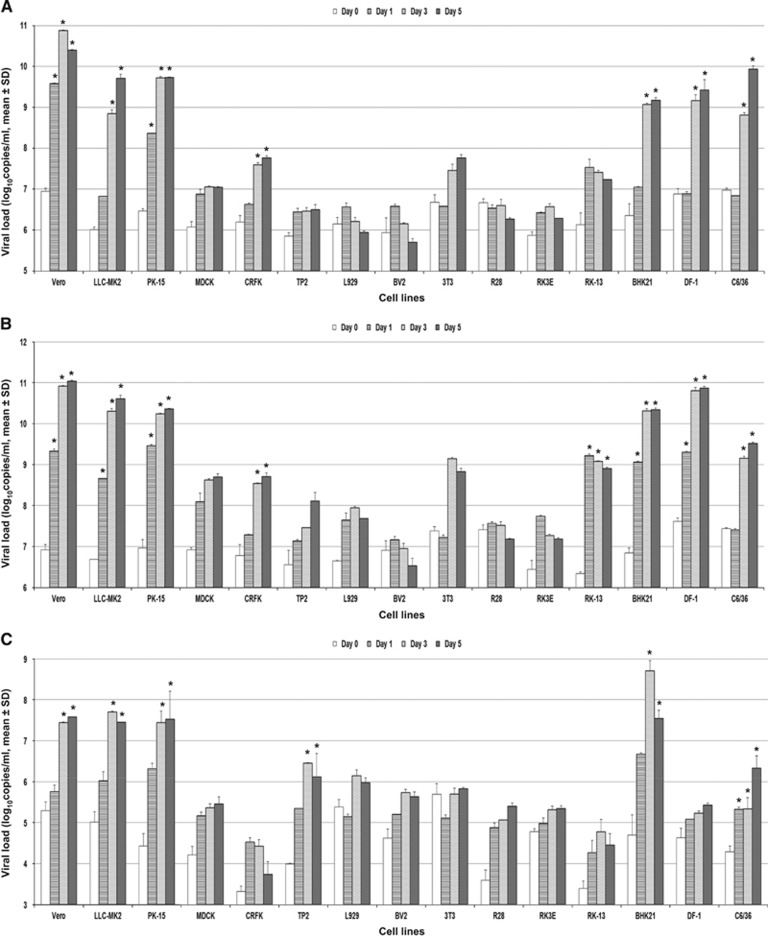
Fifteen nonhuman cell lines were tested (Table 1). ZIKV-PR and ZIKV-U again showed similar tropism in these cell lines. A ⩾2 log increase in mean viral load (P<0.01) was observed within 5 d.p.i. in 8/15 cell lines (Figures 4A and 4B). These included the nonhuman primate (Vero and LLC-MK2), pig (PK-15), cat (CRFK), rabbit (RK-13), hamster (BHK21), chicken (DF-1) and mosquito (C6/36) cell lines. None of the mouse, rat and bat cell lines showed a ⩾2 log increase in viral load within 5 d.p.i. The highest mean viral loads (⩾10 lgcopies/mL) were observed in the nonhuman primate (Vero and LLC-MK2), pig (PK-15), hamster (BHK21) and chicken (DF-1) cell lines. All of these eight cell lines showed ZIKV-NS1 protein expression by IF, usually starting on 3 d.p.i. and peaked on 5 d.p.i. (Table 2). ZIKV-NS1 protein expression was most prominent (⩾50% of infected cells) in the nonhuman primate (Vero and LLC-MK2), pig (PK-15), hamster (BHK21), chicken (DF-1) and mosquito (C6/36) cell lines (Figure 2). Some of the mouse, rat and bat cell lines also showed ZIKV-NS1 protein expression, but these expressions were much less prominent than that in the other cell lines. CPE were observed in six of these eight cell lines, including the nonhuman primate (Vero and LLC-MK2), pig (PK-15), rabbit (RK-13), hamster (BHK21) and chicken (DF-1) cell lines, and were most prominent (>50% involvement) in the nonhuman primate, pig, hamster and chicken cell lines (Table 3). CPE were not observed in the mosquito (C6/36) cell line within 5 d.p.i.
Figure 4.
Differential nonhuman cell line susceptibility to (A) ZIKV-PR, (B) ZIKV-U and (C) DENV-2, as defined by viral load, on days 1, 3 and 5 post-ZIKV inoculation. All experiments were done in triplicate. The mean viral loads on days 1, 3 and 5 were compared with the mean baseline viral load on day 0 (1 h post-ZIKV inoculation). All calculations were based on log-transformed viral loads. *P-value of <0.01.
A narrower range of nonhuman cell lines were susceptible to infection by DENV-2 than ZIKV. A ⩾2 log increase in mean viral load (P<0.01) within 5 d.p.i. was observed in only 6/15 cell lines, including the nonhuman primate (Vero and LLC-MK2), pig (PK-15), bat (TP2), hamster (BHK21) and mosquito (C6/36) cell lines (Figure 4C). All of these six cell lines demonstrated DENV-2-NS1 protein expression by IF (Table 2). CPE were observed in five of these six cell lines (all except C6/36), but was usually not prominent (⩽50%) except in the LLC-MK2 and BHK21 cell lines (Table 3).
Discussion
Although ZIKV infection has been described as a dengue-like illness in the past, recent evidence suggests that ZIKV is unique among known human-pathogenic flaviviruses. ZIKV is the first flavivirus known to be associated with congenital malformations, and sexual and vertical transmissions in human.1, 8, 9, 20, 21, 22 The pathogenesis of these unusual clinical manifestations and transmission routes is incompletely understood. Moreover, a comprehensive surveillance for potential animal reservoirs of ZIKV, which are likely important in facilitating the spread of the virus after 60 years of quiescence, has not been reported. A virus' ability to grow in cells of different host species might provide insight into its ability to cross interspecies barriers. We therefore conducted this systematic study to characterize the tropism of the emerging ZIKV in a broad range of human and nonhuman cell lines derived from different tissues or organs and host species. The findings in our study may have important clinical and epidemiological implications for this rapidly expanding viral epidemic.
Productive ZIKV replication as defined by ⩾2 log increase in mean viral load, ZIKV-NS1 protein expression by IF, and presence of CPE was observed within 5 d.p.i. in 8/18 human cell lines, which represented cells of placental, neuronal, muscle, retinal, pulmonary, intestinal and hepatic origin. This broad tissue tropism is likely related to the wide distribution of the candidate receptors of ZIKV, including AXL, TYRO3 and DC-SIGN, in different organs.23 The placental (JEG-3) and neuronal (SF268) cell line tropism corroborated with the congenital and neurological manifestations of ZIKV infection. These manifestations include congenital microcephaly and other central nervous system malformations in infected fetuses, and meningoencephalitis, myelitis, and post-infectious Guillain–Barré syndrome in adults.1, 5, 8, 24 ZIKV RNA, full-length viral genome and/or viral particles could be detected in the brain tissue of fetuses with congenital microcephaly.8, 25 Microscopically, flavivirus-like virions were observed together with extensive inflammation, astrocyte and microglial activation, and injury in the cortex and the lateral corticospinal tracts in the fetal brain tissue.8, 25 Moreover, ZIKV could impair growth and deplete neural progenitor cells in human neurospheres and cerebral organoids.26, 27
Consistent with ZIKV's tropism for the human rhabdomyosarcoma (RD) cell line, myalgia and arthralgia with periarticular edema are common features in symptomatic ZIKV infection. Fetuses with congenital ZIKV infection may develop arthrogryposis.28 Myositis and myocarditis were observed in suckling mice inoculated with ZIKV intracerebrally, and viral RNA could be detected in the muscle and heart of type I interferon receptor-deficient mice infected with ZIKV.29, 30
Viral tropism for the retinal (ARPE19) cell line corroborates with the ophthalmological complications of congenital ZIKV infection, including chorioretinal atrophy and pigmentary maculopathy.10, 13 Abdominal symptoms including diarrhea, vomiting, abdominal pain and jaundice with hepatic dysfunction have been reported in patients with ZIKV infection, and are in agreement with the susceptibility of the intestinal (Caco-2) and hepatic (Huh-7) cell lines to ZIKV.1, 6, 7, 31 Respiratory distress may occur in severe ZIKV infection, and this correlates with ZIKV's tropism for respiratory (Hep-2, Calu-3 and HFL) cell lines.6, 7 Similarly, the clinical features of DENV infection, including encephalopathy/encephalitis (SF268), myalgia and rhabdomyolysis (RD), retinitis (ARPE19), respiratory symptoms (Hep-2 and HFL), diarrhea (Caco-2) and hepatic dysfunction (Huh-7), also correlated with the cell line susceptibility to DENV-2 in our study.32, 33
Although neurological manifestations may also be seen in infections caused by other flaviviruses, such as DENV, yellow fever virus, West Nile virus (WNV) and Japanese encephalitis virus (JEV), none of these flaviviruses have been associated with congenital microcephaly and central nervous system malformations. These unique clinical features of ZIKV infection require the virus to be able to infect neural progenitor cells of the fetus in utero during early gestation. A striking difference between the cell line susceptibilities of ZIKV and DENV-2 observed in our study was ZIKV's ability to productively replicate in placental cells (JEG-3). JEG-3 cells infected with ZIKV produced high viral loads (~10 lgcopies/mL), abundant ZIKV-NS1 protein expression and prominent CPE. This finding helped to explain the unique ability of ZIKV to infect the fetus via the placenta and cause congenital anomalies, particularly when the infection occurred during the first trimester.34 Although a previous study suggested that type III interferons produced by human placental trophoblasts could confer protection against ZIKV infection, it is notable that the study utilized primary human trophoblasts of full-term instead of early gestation placenta.35 Indeed, our in vitro finding concurred with the recent reports of detectable viral particles and RNA in trophoblasts of the maternal and fetal placenta of ZIKV-infected type I interferon signaling-deficient mice and permissibility of placental macrophages and cytotrophoblasts to productive ZIKV infection.12, 36
Besides trans-placental transmission, our findings also provided insights to explain the pathogenesis of other non-vector-borne human transmission routes of ZIKV. Probable sexual transmission of ZIKV has been reported.20, 21, 22 ZIKV-infected male patients might develop hematospermia and perineal pain compatible with prostatitis, and might have detectable viral RNA and/or infectious viral particles in their semen.20, 21, 22 In contrast to DENV-2, which did not efficiently replicate in any of the genital tract cell lines, the mean ZIKV load significantly increased in the prostatic (LNCaP) and testicular (833KE) cell lines without inducing CPE. This suggested that the prostate and testes might be important sites of ZIKV replication that led to prolonged virus shedding in the semen of infected men.20, 21, 37 Interestingly, abundant ZIKV-NS1 protein expression was detected in ZIKV-infected LNCaP cells, but not 833KE cells. This finding might imply that ZIKV could replicate more efficiently in the prostate than the testes. Recent studies only reported high viral loads in the testes of ZIKV-infected mice deficient in interferon signaling and/or receptor, but did not report the viral loads in their prostates.30, 38 Further studies should be conducted to investigate the prostate's role as a replication site of ZIKV. The female genital tract cell lines, including HeLa and HOSE6-3, were also susceptible to ZIKV, supporting the hypothesis that non-immune women might acquire the infection through sexual intercourse with their ZIKV-infected male partners.
We showed that the renal (HEK) and respiratory tract (Hep-2, Calu-3, HFL) cell lines were also susceptible to ZIKV. This helped to explain the high viral loads in the urine and nasopharyngeal swabs of ZIKV-infected patients.39, 40, 41 The finding of high viral load and abundant ZIKV-NS1 protein expression without inducing CPE in HEK cells is particularly interesting, as it suggests that the kidney might be another site of persistent ZIKV replication. Similarly, DENV-2 infection also caused increased mean viral load and DENV-2-NS1 protein expression without inducing CPE in HEK cells, and viral RNA might be detected in urine of dengue patients.42 An important implication of this finding is that kidney transplant donors in ZIKV-affected regions should be tested for the infection before organ donation, as virus shedding in urine might be prolonged.39, 40, 41 The intestinal (Caco-2) cell line was also highly susceptible to ZIKV, which corroborated with the high viral load detected in the intestines of ZIKV-infected mice.43 Serial testing of fecal samples of ZIKV-infected patients should be performed to determine whether feces represent another possible transmission route of ZIKV. It would also be interesting to investigate ZIKV's ability to productively replicate in salivary gland cells, as high viral load could also be detected in the saliva of ZIKV-infected patients.41, 44
The differential cell line susceptibility of our nonhuman cell lines to ZIKV may provide insights into the virus' potential animal reservoirs and alternative animal models. The finding of high viral loads and abundant ZIKV-NS1 protein expression without induction of CPE within 5 d.p.i. in the Aedes albopictus (C6/36) cell line was quite expected, and corroborated with the high non-fatal infection, dissemination and transmission rates of ZIKV in infected A. albopictus.45 A ⩾2 log increase in mean viral load with ZIKV-NS1 protein expression and CPE were observed in the nonhuman primate (Vero and LLC-MK2), pig (PK-15), rabbit (RK-13), hamster (BHK21) and chicken (DF-1) cell lines. It is well known that the transmission of flaviviruses may involve sylvatic cycles between the mosquito vectors and susceptible wild animals, and urban cycles between the vectors and domestic animals.46, 47 Nonhuman primates are known wild animal reservoirs of ZIKV and DENV.1, 46 Our findings of efficient ZIKV replication in pig, rabbit, hamster and chicken cell lines suggest that these mammalian or avian species might have a role in the transmission of ZIKV. Similar to domestic pigs and wild birds for JEV, and domestic and wild birds for WNV, these species may serve as the animal reservoirs or amplifying hosts of ZIKV.46 Moreover, vector-free JEV transmission due to persistence of virus in tonsils and virus shedding in oronasal secretions of pigs has recently been reported.48 Alternatively, these animal species may be susceptible hosts with less importance in the transmission cycle, which is analogous to horses for JEV and many vertebrate species for WNV.46 Systematic sampling of domestic animals in ZIKV-affected regions would help to confirm these findings. Pigs, rabbits and hamsters should be evaluated as animal models for ZIKV infection, as interferon signaling- or receptor-deficient mice are not routinely available in most laboratories. Pig and/or hamster models have been previously reported for DENV and WNV infection.49, 50
Our findings are useful for optimizing laboratory diagnostics and studies on the pathogenesis and counter-measures of ZIKV. Some of the laboratories in resource-limited areas affected by ZIKV may not have the capability to routinely perform molecular diagnostic tests. In addition to the few cell lines previously reported, our study has identified a broad range of cell lines that are suitable for growing ZIKV as an alternative diagnostic tool in clinical virology laboratories.51, 52, 53, 54 These cell lines may also be used in pathogenesis studies conducted by laboratories without the technical expertise to handle brain organoids, and to evaluate antiviral treatment and vaccines in vitro prior to animal experiments.
Acknowledgments
We thank Tony Schountz (Colorado State University, Fort Collins, CO, USA), Kwok-Fai So (The University of Hong Kong, Hong Kong Special Administrative Region, China), Stephen Yuen-Wing Shiu (The University of Hong Kong, Hong Kong Special Administrative Region, China) and Annie Nga-Yin Cheung (The University of Hong Kong, Hong Kong Special Administrative Region, China) for facilitation of the study. This work was partly supported by the donations of Larry Chi-Kin Yung, and Hui Hoy and Chow Sin Lan Charity Fund Limited; and funding from the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases of the Department of Health, Hong Kong Special Administrative Region; the Seed Funding Programme for Basic Research, the University Research Committee, The University of Hong Kong; the Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, the Ministry of Education of China; and the European Virus Archive goes Global (EVAg) project that has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement NO 653316.
References
- Chan JF, Choi GK, Yip CC, Cheng VC, Yuen KY. Zika fever and congenital Zika syndrome: an unexpected emerging arboviral disease. J Infect 2016; 72: 507–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MR, Chen TH, Hancock WT et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 2009; 360: 2536–2543. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chan JF, Tee KM et al. Comparative genomic analysis of pre-epidemic and epidemic Zika virus strains for virological factors potentially associated with the rapidly expanding epidemic. Emerg Microbes Infect 2016; 5: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health OrganizationEmergencies: Zika situation report. WHO: Geneva. 2016. Available at http://who.int/emergencies/zika-virus/situation-report/14-july-2016/en/ (accessed on 14 July 2016). [Google Scholar]
- Brasil P, Sequeira PC, Freitas AD et al. Guillain-Barre syndrome associated with Zika virus infection. Lancet 2016; 387: 1482. [DOI] [PubMed] [Google Scholar]
- Arzuza-Ortega L, Polo A, Pérez-Tatis G et al. Fatal sickle cell disease and Zika virus infection in girl from Colombia. Emerg Infect Dis 2016; 22: 925–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento-Ospina A, Vasquez-Serna H, Jimenez-Canizales CE, Villamil-Gomez WE, Rodriguez-Morales AJ. Zika virus associated deaths in Colombia. Lancet Infect Dis 2016; S1473-3099: 30006–30008. [DOI] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N et al. Zika virus associated with microcephaly. N Engl J Med 2016; 374: 951–958. [DOI] [PubMed] [Google Scholar]
- Brasil P, Pereira Jr JP, Raja Gabaglia C et al. Zika virus infection in pregnant women in Rio de Janeiro - preliminary report. N Engl J Med 2016. [DOI] [PMC free article] [PubMed]
- Ventura CV, Maia M, Travassos SB et al. Risk factors associated with the ophthalmoscopic findings identified in infants with presumed Zika virus congenital infection. JAMA Ophthalmol 2016; 134: 912–918. [DOI] [PubMed] [Google Scholar]
- Wu KY, Zuo GL, Li XF et al. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res 2016; 26: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, Cao B, Govero J et al. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 2016; 165: 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula Freitas B, de Oliveira Dias JR, Prazeres J et al. Ocular findings in infants with microcephaly associated with presumed Zika virus congenital infection in Salvador, Brazil. JAMA Ophthalmol 2016. e-pub ahead of print 9 February 2016 doi:10.1001/jamaophthalmol.2016.0267. [DOI] [PMC free article] [PubMed]
- Chan JF, Chan KH, Choi GK et al. Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J Infect Dis 2013; 207: 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li IW, Chan KH, To KW et al. Differential susceptibility of different cell lines to swine-origin influenza A H1N1, seasonal human influenza A H1N1, and avian influenza A H5N1 viruses. J Clin Virol 2009; 46: 325–330. [DOI] [PubMed] [Google Scholar]
- Chan JF, Choi GK, Tsang AK et al. Development and evaluation of novel real-time reverse transcription-PCR assays with locked nucleic acid probes targeting leader sequences of human-pathogenic coronaviruses. J Clin Microbiol 2015; 53: 2722–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CC, Lau SK, Lo JY et al. Genetic characterization of EV71 isolates from 2004 to 2010 reveals predominance and persistent circulation of the newly proposed genotype D and recent emergence of a distinct lineage of subgenotype C2 in Hong Kong. Virol J 2013; 10: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Kosoy OL, Laven JJ et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 2008; 14: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu LW, Di B, Wen K et al. Development of an antigen capture immunoassay based on monoclonal antibodies specific for dengue virus serotype 2 nonstructural protein 1 for early and rapid identification of dengue virus serotype 2 infections. Clin Vaccine Immunol 2009; 16: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy BD, Kobylinski KC, Chilson Foy JL et al. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 2011; 17: 880–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D, Roche C, Robin E et al. Potential sexual transmission of Zika virus. Emerg Infect Dis 2015; 21: 359–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ortenzio E, Matheron S, de Lamballerie X et al. Evidence of sexual transmission of Zika virus. N Engl J Med 2016; 374: 2195–2198. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Di Lullo E et al. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell 2016; 18: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteaux G, Maquart M, Bedet A et al. Zika virus associated with meningoencephalitis. N Engl J Med 2016; 374: 1595–1596. [DOI] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM et al. Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med 2016; 374: 2142–2151. [DOI] [PubMed] [Google Scholar]
- Dang J, Tiwari SK, Lichinchi G et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 2016; 19: 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016; 352: 816–818. [DOI] [PubMed] [Google Scholar]
- Schuler-Faccini L, Ribeiro EM, Feitosa IM et al. Possible association between Zika virus infection and microcephaly - Brazil, 2015. MMWR Morb Mortal Wkly Rep 2016; 65: 59–62. [DOI] [PubMed] [Google Scholar]
- Weinbren MP, Williams MC. Zika virus: further isolations in the Zika area, and some studies on the strains isolated. Trans R Soc Trop Med Hyg 1958; 52: 263–268. [DOI] [PubMed] [Google Scholar]
- Rossi SL, Tesh RB, Azar SR et al. Characterization of a novel murine model to study Zika virus. Am J Trop Med Hyg 2016; 94: 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller-Luque P, Dominguez-Gil Gonzalez M, Alvarez-Manzanares J et al. First case of imported Zika virus infection in Spain. Enferm Infecc Microbiol Clin 2016; 34: 243–246. [DOI] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 1998; 11: 480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carod-Artal FJ, Wichmann O, Farrar J, Gascon J. Neurological complications of dengue virus infection. Lancet Neurol 2013; 12: 906–919. [DOI] [PubMed] [Google Scholar]
- Cauchemez S, Besnard M, Bompard P et al. Association between Zika virus and microcephaly in French Polynesia, 2013-15: a retrospective study. Lancet 2016; 387: 2125–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A, Lennemann NJ, Ouyang Y et al. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 2016; 19: 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quicke KM, Bowen JR, Johnson EL et al. Zika virus infects human placental macrophages. Cell Host Microbe 2016; 20: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson B, Hearn P, Afrough B et al. Detection of Zika virus in Semen. Emerg Infect Dis 2016; 22: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Govero J, Smith AM et al. A mouse model of Zika virus pathogenesis. Cell Host Microbe 2016; 19: 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis 2015; 21: 84–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos Rde M, Cirne-Santos C, Meira GL et al. Prolonged detection of Zika virus RNA in urine samples during the ongoing Zika virus epidemic in Brazil. J Clin Virol 2016; 77: 69–70. [DOI] [PubMed] [Google Scholar]
- Bingham AM, Cone M, Mock V et al. Comparison of test results for Zika virus RNA in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease – Florida, 2016. MMWR Morb Mortal Wkly Rep 2016; 65: 475–478. [DOI] [PubMed] [Google Scholar]
- Poloni TR, Dornas FP, Dos Santos NNJ et al. High prevalence of clinically unsuspected dengue disease among children in Ribeirao Preto City, Brazil. J Med Virol 2016; 88: 1711–1719. [DOI] [PubMed] [Google Scholar]
- Aliota MT, Caine EA, Walker EC et al. Characterization of lethal Zika virus infection in AG129 mice. PLoS Negl Trop Dis 2016; 10: e0004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D, Roche C, Nhan TX et al. Detection of Zika virus in saliva. J Clin Virol 2016; 68: 53–55. [DOI] [PubMed] [Google Scholar]
- Wong PS, Li MZ, Chong CS, Ng LC, Tan CH. Aedes (Stegomyia) albopictus (Skuse): a potential vector of Zika virus in Singapore. PLoS Negl Trop Dis 2013; 7: e2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean AG, Baker ML, Stewart CR et al. Studying immunity to zoonotic diseases in the natural host - keeping it real. Nat Rev Immunol 2013; 13: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF, To KK, Chen H, Yuen KY. Cross-species transmission and emergence of novel viruses from birds. Curr Opin Virol 2015; 10: 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin ME, Garcia-Nicolas O, Brechbuhl D et al. Vector-free transmission and persistence of Japanese encephalitis virus in pigs. Nat Commun 2016; 7: 10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetti MC, Durbin A, Harris E et al. Report of an NIAID workshop on dengue animal models. Vaccine 2010; 28: 4229–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Sasaki M, Okumura M, Kim E, Sawa H. Flavivirus encephalitis: pathological aspects of mouse and other animal models. Vet Pathol 2010; 47: 806–818. [DOI] [PubMed] [Google Scholar]
- Frumence E, Roche M, Krejbich-Trotot P et al. The South Pacific epidemic strain of Zika virus replicates efficiently in human epithelial A549 cells leading to IFN-β production and apoptosis induction. Virology 2016; 493: 217–226. [DOI] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 2016; 18: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R, Dejarnac O, Wichit S et al. Biology of Zika virus infection in human skin cells. J Virol 2015; 89: 8880–8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Liu S, Zhang Q et al. Isolation and characterization of Zika virus imported to China using C6/36 mosquito cells. Virol Sin 2016; 31: 176–179. [DOI] [PMC free article] [PubMed] [Google Scholar]