Abstract
Förster (or fluorescence) resonance energy transfer (FRET) is a nonradiative energy transfer process between two fluorophores located in close proximity to each other. To date, a variety of biosensors based on the principle of FRET have been developed to monitor the activity of kinases, proteases, GTPases or lipid concentration in living cells. In addition, generation of biosensors that can monitor physical stresses such as mechanical power, heat, or electric/magnetic fields is also expected based on recent discoveries on the effects of these stressors on cell behavior. These biosensors can now be stably expressed in cells and mice by transposon technologies. In addition, two-photon excitation microscopy can be used to detect the activities or concentrations of bioactive molecules in vivo. In the future, more sophisticated techniques for image acquisition and quantitative analysis will be needed to obtain more precise FRET signals in spatiotemporal dimensions. Improvement of tissue/organ position fixation methods for mouse imaging is the first step toward effective image acquisition. Progress in the development of fluorescent proteins that can be excited with longer wavelength should be applied to FRET biosensors to obtain deeper structures. The development of computational programs that can separately quantify signals from single cells embedded in complicated three-dimensional environments is also expected. Along with the progress in these methodologies, two-photon excitation intravital FRET microscopy will be a powerful and valuable tool for the comprehensive understanding of biomedical phenomena.
Main Text
Since the discovery of green fluorescent protein (GFP) (1), scientists have been widely employing this gene-encoded fluorescent protein (FP) to investigate the spatiotemporal dynamics of molecules with subcellular resolution. One of the applications of FP engineering is the development of biosensors based on the principle of Förster (or fluorescence) resonance energy transfer (FRET) (2). FRET is a nonradiative energy transfer process between two fluorophores located in close proximity to each other, and its efficiency is strongly dependent on the distance between the fluorophores. More precisely, the energy transfer rate (kt) from a donor to an acceptor fluorophore is calculated in the following Förster’s equation;
where J is the overlap of donor emission and acceptor excitation spectra, κ2 is an orientation factor of transition moment (a factor determined by the relative orientation of the two fluorophores), n is a refractive index, R is the distance between the donor and acceptor fluorophores, and kf is the donor fluorescence emission speed. Therefore, FRET efficiency is determined by the distance and orientation of the two fluorophores. To measure FRET efficiency, there are roughly two methods; ratiometric analysis and lifetime analysis. Ratiometric analysis utilizes fluorescence intensity of an acceptor (e.g., yellow FP (YFP)) and a donor (e.g., cyan FP (CFP)) upon the donor excitation. Because acceptor fluorescence increases and donor fluorescence decreases when FRET occurs, the acceptor/donor intensity ratio positively correlates with FRET efficiency (3, 4). Lifetime analysis is based on the fact that donor fluorophore lifetime is shortened when energy is transferred to an acceptor fluorophore. Therefore, FRET efficiency can be quantified by detecting the lifetime change (5). Based on these analytical approaches, the FRET phenomenon has been utilized by creating two FP-fused target proteins and quantifying the FRET efficiency between them, which is used as an index of the “distance,” “orientation,” or, thus, “interaction” between the molecules. The specially designed molecules are known as FRET biosensors and can be classified into two categories, intermolecular and intramolecular (single-molecule) FRET biosensors (4). Because of their high signal/noise ratio and wider application in live imaging, and because their use does not require calculation of the corrective FRET efficiency, many laboratories have preferentially created single-molecule FRET biosensors that monitor specific molecules such as kinases (6, 7, 8, 9, 10), proteases (11, 12, 13), GTPases (14, 15, 16, 17) or lipids (18). Recently, a breakthrough was reported in the generation of transgenic mice expressing single-molecular FRET biosensors, in which the activities of numerous kinases, proteases, or GTPases are visualized in living tissues and organs (19, 20, 21). Despite the remarkable technical expansion of FRET microscopy, a couple of problems in image acquisition and data analysis have hindered precise signal quantification in intravital application. Further, emerging demands for quantitative visualization of physical stresses, such as mechanical power, heat, or electric/magnetic fields, have encouraged physicists and biologists to develop new FRET biosensors. In this issue of Perspective, we briefly summarize the history of the development and improvement of single-molecule FRET biosensors, delineate the problem and resolution in their application to intravital imaging, and discuss the future prospects for these tools and how they can contribute to the future understanding of biomedical phenomena in vivo.
Development and improvement of single-molecule FRET biosensors
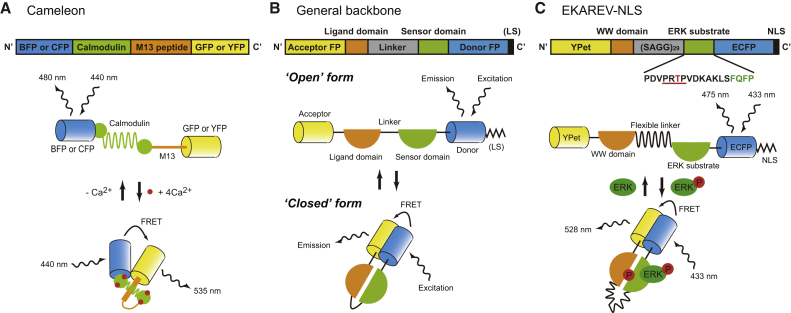
The first gene-encodable single-molecule FRET biosensor, Cameleon, was reported by Miyawaki et al. in 1997 (22) (Fig. 1 A). In this FRET biosensor, blue FP (BFP) or CFP, calmodulin, calmodulin-binding peptide M13, and GFP or YFP are tandemly fused in this order. The binding of Ca2+ to calmodulin induces a conformational change of the biosensor, resulting in a dramatic increase of FRET efficiency, which reflects the Ca2+ concentration within a cell. Based on the operating principle, BFP/CFP, GFP/YFP, calmodulin, and M13 are expediently called a donor FP, an acceptor FP, a sensor domain, and a ligand domain, respectively. Since the development of Cameleon, a variety of FRET biosensors based on a similar operating principle have been created and improved by many laboratories (Fig. 1 B). Due to the lack of a three-dimensional (3D) structure of a FRET biosensor, and to the contribution of multiple factors to FRET efficiency, such as the distance and orientation of the two FPs, it is complicated and time-consuming to develop and improve a single-molecule FRET biosensor. That is, the selection of appropriate sensor/ligand domains and FP pairs, the order of the domains in the biosensor, and the optimization of the linker length that connects each functional domain all must proceed by trial and error. Recently, Komatsu et al. reported the rational design and optimized backbone of a single-molecule FRET biosensor called the Eevee system; in this biosensor, the effect of the orientation-dependent FRET between donor and acceptor FPs is minimized by using a long flexible linker and reduced basal FRET efficiency when a biosensor presents an “open form” (Fig. 1 C). As a result, they successfully improved the dynamic range of some preexisting FRET biosensors by 1.2- to 1.5-fold (6).
Figure 1.
Schematic of single-molecule FRET biosensors. Schematics of constructions and operating principles are shown for Cameleon (A), the general backbone of single-molecule FRET biosensors (B), and EKAREV-NLS (C). Note that the order of donor/acceptor FPs and sensor/ligand domains from the N-terminus (N′) to the C-terminus (C′) is arbitrary, and optimization is required for each biosensor. LS, localization signal.
Generation of stable cell lines and transgenic mice expressing a FRET biosensor
Transient expression of a FRET biosensor is not the best method for a long-time observation such as intravital imaging, because the vast majority of the cells lose the expression of FRET biosensors after ∼5–10 cell divisions. The other problem with transient expression is that the expression levels of a biosensor greatly differ among cells. Therefore, one challenge in FRET imaging had been to establish cell lines stably expressing the biosensors, but all the conventional methods, including linearized DNA transfection or retrovirus/lentivirus-mediated gene transfer, have failed to maintain the CFP-YFP FRET biosensor structures when inserted into the host genome (23). This has been considered to be due to a homologous recombination between donor and acceptor FPs with high nucleotide sequential homology. In the case of linearized DNA transfection, the constructs are usually inserted into the host genome as a tandem concatamer, where a homologous recombination occurs between FPs located in proximity during cell divisions. In the case of virus-mediated gene transfer, a homologous recombination might happen during the reverse transcription process in the virus, which carries the construct as an RNA pair. Therefore, one solution to avoid this recombination is to use an FP pair originating from different species with less sequential homology, or to reorganize the codon of one FP to a different species (24). Indeed, by replacing CFP with teal FP, an FP derived from Clavularia sp. (coral) (25) with less homology to CFP, we established stable cell lines expressing FRET biosensors for small GTPases and phosphoinositides and observed their activity and concentration in organoids (26, 27, 28). Also, many laboratories have utilized a GFP-red-FP (RFP) pair, which originates from Aequorea victoria (jellyfish) and Discosoma sp. (sea anemone), respectively, for retrovirus/lentivirus-mediated generation of stable cell lines (29). Another method is to utilize a transposon-mediated gene transfer system, e.g., the Piggybac system, in which the biosensor construct is designed to be sandwiched between transposon-specific inverted terminal repeat sequences (30, 31). With this system, the construct between inverted terminal repeat sequences is randomly inserted into a host genome as a single structure by cotransfection with a DNA coding the specific transposase. Using this method, we have effectively obtained stable cell lines that express sufficient amounts of FRET biosensors (32, 33), and to date we have not experienced any problems related to the recombination or destruction of the constructs.
After the initial successes of transposon-mediated stable gene transfer, Kamioka et al. applied this technique to establish transgenic mice expressing FRET biosensors (19). In their study, they used cytoplasmic injection of Tol2 transposase together with the plasmid for the respective biosensors, and obtained neonatal pups with very high integration efficiency. The same group have also generated Cre-inducible FRET mice in which a biosensor cDNA is inserted behind a loxP-Keima-stop-loxP cassette to label all the cells with the RFP before the Cre recombinase action (34). When these mice are crossed with lineage-specific Cre mice, the cells with Cre recombinase begin to express FRET biosensors. Finally, another group succeeded in generating a transgenic mouse expressing SCAT3, a single-molecule FRET biosensor that reports Caspase-3 activity, by a conventional, linearized DNA injection method but using chicken HS4 insulator sequences to overcome silencing of the transgene (20).
Application to intravital imaging
The establishment of stable cell lines and mice with FRET biosensors has enabled us to investigate various biological and pathological events in vivo. For example, Mizuno et al. clarified the positive and negative roles of ERK and PKA, respectively, in neutrophil recruitment into the inflamed intestine (35). Johnsson et al. visualized Rac1 signaling during disease development in multiple organs using Rac1-FRET mice (21). Kumagai et al. revealed that breast cancer cells with low ERK activity possess cancer stem-cell-like properties by isolating the cells depending on the FRET efficiency from MMTV-Neu:Eisuke mice, a transgenic mouse line expressing the ERK FRET biosensor (36). And in this work, we focus on an example of intravital imaging with the EKAREV-NLS FRET biosensor, which successfully resolved an urgent question about drug resistance in cancer treatment.
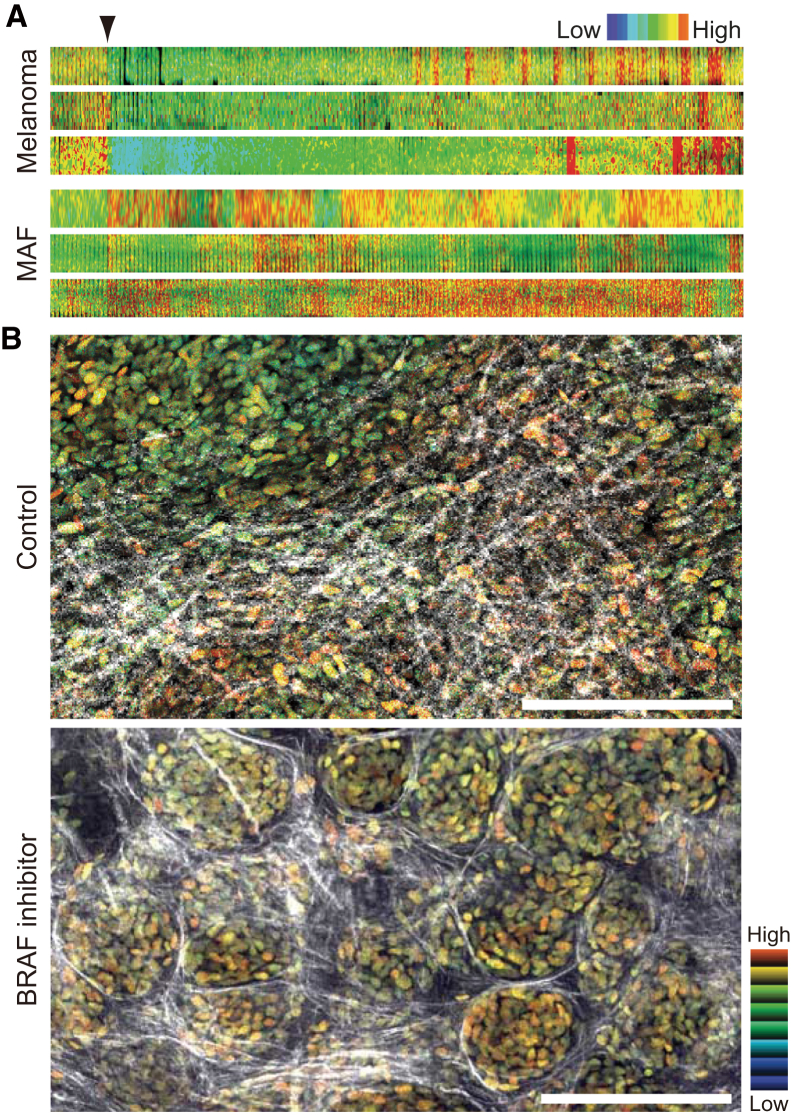
BRAF mutant melanomas dramatically respond to selective BRAF inhibitors. However, there is significant variability in the magnitude of the initial response (37, 38), and most of the cases develop robust drug resistance after six months of treatment (39), despite the fact that no genetically resistant subclones have been detected before treatment, even in tumors that show modest responses. Further, different genetic resistance mechanisms can develop in the same patient (40), often at multiple metastatic sites (41). It is therefore suspected that certain extrinsic mechanisms underlie and determine the initial response, following the adaptation and acquisition of genetically robust drug resistance. We utilized an EKAREV-NLS biosensor under two-photon excitation intravital FRET microscopy and demonstrated that melanoma cells initially responded to the drug, but rapid reactivation of ERK/MAPK was observed in areas of high stromal density (42). This effect was linked to the “paradoxical” activation of melanoma-associated fibroblasts (MAFs) (Fig. 2 A) and to the promotion of matrix production/remodeling, which led to the reactivation of ERK/MAPK through cell-adhesion-mediated signals in melanoma cells (Fig. 2 B).
Figure 2.
Application of a FRET biosensor to 3D live imaging. (A) Representative kymographs of ERK activity in melanoma cells and MAFs. Cells stably expressing EKAREV-NLS were embedded in 3D gels, treated with a BRAF inhibitor at the indicated time point (black arrowhead), and imaged for 13 h in total. Shown are pseudo-colored ratiometric FRET images depicted with an intensity-modulated display (IMD) mode of 8-ratio 32-intensity. High ERK activities are depicted in red (warm) hues and low activities in blue (cold) hues. (B) WM266.4 human melanoma cells stably expressing EKAREV-NLS were subcutaneously injected into nude mice and treated with DMSO (control) or PLX4720 (BRAF inhibitor) for 13 and 11 days, respectively. Shown are ratiometric FRET images merged with signals of second harmonic generation (white). Please note that cells treated with BRAF inhibitors still exhibit high levels of ERK activities within reorganized nest-like structures consisting of thick collagen fibers. Scale, 200 μm. (This image was modified from Hirata et al. (42))
Sophisticated image acquisition and signal quantification in vivo
Artifacts caused by nonuniform environments
One of the biggest concerns in intravital FRET imaging is artifacts caused by the nonuniform environments the signals travel through. Because the transmission property of light with longer wavelength (e.g., YFP) is higher than that of light with shorter wavelength (e.g., CFP), the apparent FRET efficiency calculated by a conventional ratiometric method becomes higher when the emission light is coming from deeper or going through denser tissues. Another problem is autofluorescence; various cell types emit different levels and wavelengths of endogenous fluorescence, which directly affects the total intensities (43, 44). Therefore, we must verify that the obtained FRET ratio (FRET/CFP) actually reflects the net FRET efficiency. One of the easiest ways to accomplish this is acceptor photobleaching (45). If the fluorescence from the donor FP were found to increase upon bleaching of the acceptor FP, we could conclude that the fluorescent signal from the acceptor FP contains FRET. Another method would be to couple ratiometric FRET analysis with fluorescence lifetime imaging microscopy (FLIM). The FLIM system most compatible with intravital imaging is two-photon time-correlated single photon counting (5). Because the lifetime of an FP is not affected by its intensity (and thus is not affected by the transmission property of FPs or the light scattering in nonuniform environments), but is shortened by FRET-induced energy loss, the lifetime of the donor FP parallels FRET efficiency with more accuracy than the ratiometric method. Nonetheless, at present, conventional two-channel imaging for ratiometric analysis surpasses FLIM, especially in terms of the feasibility of 3D time-lapse aquisition. Generally, it takes longer to acquire a single image for FLIM-FRET analysis (>20 s/section) than for ratiometric analysis (<5 s/section) (5). Therefore, FLIM-FRET analysis has a limitation in 3D time-lapse imaging, and for now, we consider that ratiometric FRET analysis is practicable and can provide wider application for intravital “4D” FRET microscopy, the results of which should ideally be confirmed by single-scan FLIM-FRET. Finally, we should mention one other method for determining the net FRET efficiency: imaging of a negative control biosensor. A representative example is our previously described preparation of the nonphosphorylated mutant EKAREV-TA-NLS, which was made by substituting threonine in the sensor domain to alanine (42). As expected, the FRET efficiency of the mutant biosensor was not increased upon growth factor stimulation, and the heterogeneous FRET efficiency distribution was not detected in vivo in the same range as in the wild-type biosensor (42). In some cases, however, there seem to be problems in generating negative control biosensors. For example, in the case of the Eevee-Akt FRET biosensor, the negative control biosensor Eevee-Akt-TA also responds to the stimulants to some extent (6). This is due to the translocation of the biosensor (i.e., from cytosol to the plasma membrane) upon stimulation, because the biosensor carries the AktPH domain at its N-terminus, which binds to phosphatidylinositol (3, 4, 5) trisphosphates in the plasma membrane. This change of localization may cause bystander FRET (or intermolecular FRET) at the plasma membrane, which contributes to the apparent increase in the FRET ratio after stimulation. One of the most effective ways to generate a FRET biosensor with high sensitivity is to attach a localization signal where the molecule of interest is activated (e.g., the CAAX domain for Ras biosensor), because it can reduce the biosensor fraction, which is not contributing to FRET upon stimulation (6, 16). However, like Eevee-Akt, there is sometimes a trade-off between the increase of the sensitivity and incidence of pseudo-FRET artifacts. Therefore, we need to examine the best construction in each case and optimize the difference of FRET gain between a biosensor and its negative control biosensor.
Tissue/organ position fixation during live imaging
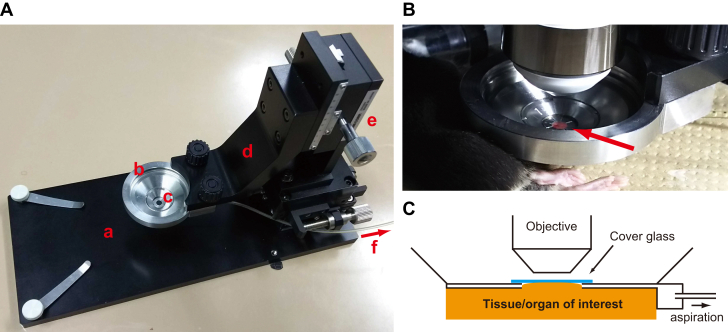
Another practical problem in intravital imaging is tissue/organ fixation during live imaging. Because the FRET ratio is calculated pixel by pixel in each image, sample movement or drift caused by breathing or heartbeat can raise critical errors in FRET microscopy. Recently, we developed an aspiration fixation system that can be applied to mouse intravital imaging of various organs (Fig. 3). In this system, a tissue or organ is appressed by negative pressure (Fig. 3 A E) beneath a glass attached to the bottom of a dish chamber (Fig. 3 A, B and C). The region of interest can then be imaged with an upright microscope through a chamber filled with water or an appropriate solution (Fig. 3, B and C). With this system, the stomach, liver, intestine, rectum, pancreas, spleen, lymph nodes, testis, muscle, and skin of anesthetized mice have been successfully imaged (35, 46) and unpublished data). Another group has also reported a simple aspiration fixation system for lung imaging (47). Therefore, although they are not suitable for longitudinal applications (as discussed below), we predict that aspiration fixation systems will become the method of choice for tissue stabilization in intravital imaging.
Figure 3.
Aspiration fixation system for mouse intravital imaging. (A) Overview of the aspiration fixation system, which consists of a stage (a), a chamber dish (b), a cover glass (c), an arm for x-y adjustment (d), a height adjustment device (e), and an aspiration tube (f), which is connected to a pump. (B) Intravital intestinal imaging of an anesthetized mouse. The red arrow indicates the mouse intestine appressed beneath the coverglass by negative pressure. (C) Schematic of the aspiration fixation system.
Improvement of biosensors for two-photon intravital imaging
To date, CFP-YFP and derived mutants are preferentially used as an FP pair for single-molecule FRET biosensors because of their large spectral overlap integral (i.e., large overlap between the CFP emission and YFP excitation spectrum profile), which strongly affects the FRET efficiency (45). Recently, CFP mutants with a single, long lifetime and increased brightness and photostability (e.g., mTurquoise or mCerulean3) have been developed that are suitable for donor FPs in FRET biosensors, with great potential for in vivo application (48, 49). However, there is a paradoxical concern about the application of this pair to two-photon excitation intravital FRET microscopy, mainly due to the proximity of their excitation profiles at relatively short wavelength. Because the two-photon excitation wavelength for CFP (usually 820–840 nm) also directly excites YFP, and because YFP is much brighter than CFP, the effect of direct excitation sometimes becomes nonnegligible. As a consequence, two-photon excitation FRET images sometimes exhibit a positive correlation between the YFP intensity and FRET ratio, whereas single-photon excitation or epifluorescent FRET images exhibit a negative correlation, as mentioned above. This problem can be circumvented by certain conventional approaches, such as using a biosensor with high sensitivity and dynamic range (6), choosing a two-photon laser that possesses a steep profile around the wavelength of use, or excluding from quantification the cells that express extremely high/low fluorescent levels of the biosensor (42). However, there are also possible avenues toward optimization of the biosensors for two-photon intravital imaging. One idea is to use a nonradiative mutant FP as an acceptor, such as sREACh, which has already been utilized in two-photon excitation FLIM-FRET (50). Another approach is to change the FP pair to an FP pair with a large gap between the excitation wavelength. For example, RFP is not excited by the two-photon excitation wavelength for GFP (usually 850–880 nm); thus, we can easily overcome the problem of direct excitation by using an RFP-GFP pair or their mutants. Although the FRET efficiency between GFP and RFP is generally lower than that between CFP and YFP, Lam et al. reported that a GFP mutant, Clover, and an RFP mutant, Ruby, are the optimal FP pair for two-photon-excitation FRET microscopy (29). Moreover, FPs with a longer emission/excitation wavelength profile are more ideal for intravital imaging because of their advantage for deep tissue penetration. Therefore, the establishment of FRET biosensors with near-infrared FPs, such as iRFP713 and its mutants (51), in parallel with the development of a multiphoton laser with longer wavelength, are also anticipated in the next generation of intravital FRET microscopy.
Development of algorithms for signal quantification from a single cell
Another eagerly awaited event in intravital FRET microscopy is the development of a method for quantifying the signals obtained from a complex 3D environment. Although qualitatively precise intravital FRET images can be obtained with the latest FRET biosensors and leading microscopy techniques, precise quantification of the FRET signals at the single-cell level is still challenging. In the case of biosensors localized in the nucleus, some image analysis software packages (e.g., Metamorph), or the new algorithm recently reported by Chittajallu et al., have already enabled semiautomatic quantification in thousands of cells individually (52). However, in the case of membrane-localized biosensors (e.g., RaichuEV-Rac1) in crowded cells with complicated morphologies (e.g., glioma cells in brain tissue), there is no automated program to separate individual cells. Furthermore, although detecting signaling events in a cell in motion would reveal the molecular mechanisms for biological reactions in vivo, 3D tracking methodologies for FRET quantification are not yet established. Recently, Kikuta et al. successfully traced time-dependent morphological changes in osteoclasts in vivo (53), and it might be possible to extend this method to semiautomated FRET quantification. Regarding intravital FLIM-FRET analysis, spectral phasor analysis with Global for Images SimFCS 4 will effectively remove signals from autofluorescence, which makes the analysis more reliable (54), and FLIMfit software provides an excellent platform with efficient use of computer processor and memory resources, which enables us to quickly analyze large FLIM data sets (55). There is no doubt that further development and improvement of the algorithms, programs, and software for automated 4D FRET analysis will accelerate our understanding of the signal transduction in vivo.
Visualization of physical stresses in vivo
One of the issues that we should tackle in the next decade is the effect of physical stresses, such as mechanical power, heat or local temperature, pH, redox state, oxygen concentration, and electric/magnetic fields, some of which are already being visualized with FRET biosensors (56, 57). In addition, there is much evidence that physical stress directly affects the biological behavior of cells. For example, mechanical pressures from flanking cells accelerate the cell-cycle progression in epithelial cells (58), and mechanotransduction derived from substrate rigidity can affect stem cell differentiation (59). Another report has shown that local metabolic heat production affects the electrochemical gradient in mitochondria (60). And although it is quite challenging to visualize physical stresses in vivo, we believe that FRET biosensors hold great potential for this purpose.
The first FRET mechanosensor was reported by Meng et al. in 2008. In that report, they generated a tension-sensing module (called stFRET) in which the FPs Cerulean (donor FP) and Venus (acceptor FP) were connected by the coiled-coil spring of a helix protein (61). By inserting this module into a target molecule, they visualized the tension loaded on the molecule as the distance between the FPs (and thus as the FRET efficiency). Two years later, Grashoff et al. modified the linker to a 40-amino-acid-long elastic peptide derived from the spider-silk flagelliform protein, and by inserting this new tension-sensing module (named TSMod) into vinculin, a member of the focal adhesion complex, they successfully visualized the tension force at the focal adhesion complex with piconewton sensitivity (62). After their report was published, many laboratories applied TSMod to various molecules to visualize mechanical stress in living cells (63, 64). In two representative examples, a tension-sensor targeting β-spectrin was applied to intravital imaging of Caenorhabditis elegans (65), and a tension sensor targeting E-cadherin was applied to Drosophila ovary imaging (66). However, application to higher-order animals such as mice has not yet succeeded. Since these FRET biosensors are designed to sense the structural changes in proteins that organize the actin cytoskeleton for cell-cell or cell-substrate interactions, other molecules such as N-cadherin, α-catenin, integrins, paxillin, talin, and p130Cas (67, 68) can be utilized for biosensors with better sensitivity and dynamic range. Another candidate may be transient receptor potential (TRP) channel proteins, which respond to a variety of stimuli, including mechanical or chemical stimuli, temperature, or osmolarity (69). After selecting the most appropriate protein, it is necessary to optimize the FPs for deep tissue imaging, as discussed above. Regarding heat or electric/magnetic fields, there has been no report of FRET biosensors to date, but in addition to the possibility of TRP channel proteins, Kiyonaka et al. developed a GFP dimerization-based thermosensor by utilizing TlpA, a heat-sensitive protein derived from Salmonella (60).
Future perspective of intravital FRET microscopy
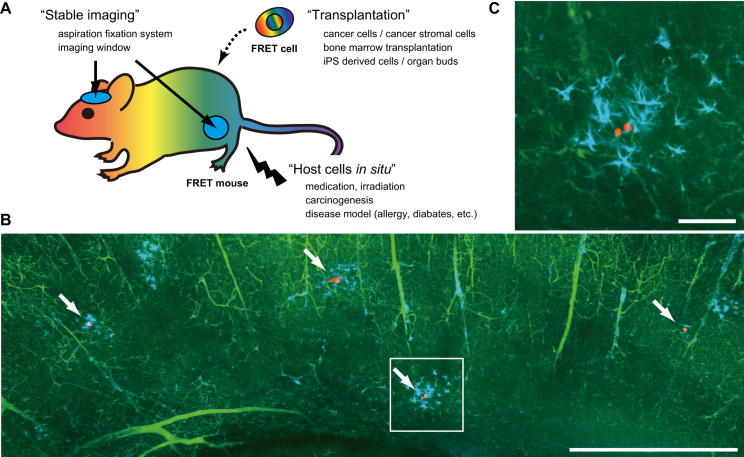
We are now able to observe moving or proliferating cells in the short term (hours to days). One of the challenges in the near future will be to observe such cells for longer durations (months to years), which would contribute to our understanding of various biomedical phenomena, such as cancers, the immune system, disease-specific alterations in metabolism, and tissue regeneration. To this end, we consider that cell transplantation, host-cell visualization in situ, and stable imaging methods will be the key technical factors (Fig. 4 A). Cell transplantation techniques will continue to be a major part of intravital FRET microscopy. These includes cancer cells, cancer stromal cells, bone marrow cells or iPS derived cells, all of which can express FRET biosensors. In the future, a series of FRET mice will be generated that can be irradiated, treated with carcinogens, crossed with genetically modified mice, and/or used to investigate cell- or tissue-specific toxic effects of newly developed drugs. Cell transplantation into these FRET mice will also provide insight into how the cells in the microenvironment regulate the signaling pathways. For example, organ-bud transplantation into FRET mice might clarify specific signaling pathways in host organs that contribute to effective engraftment and regeneration (70). In addition, for stable imaging of these exciting phenomena, aspiration fixation systems will play an effective role in intravital microscopy (Fig. 4 A). Also of note, as mentioned above, long-duration (months to years) microscopy is a next challenge in intravital FRET microscopy. In this context, “cancer dormancy” is certainly among the most mysterious biomedical phenomena in the human body, and we expect that intravital FRET microscopy will provide critical information to solve this mystery. Several mouse models that exhibit cancer dormancy have already been established in the lung (71, 72, 73) and brain (Fig. 4, B and C); thus, by introducing FRET biosensors into the cancer cells, we can visualize the variance of specific signals with spatiotemporal resolution. It should also be useful to utilize “imaging window techniques” to follow the dormant cells over time (42, 74). In addition, we can visualize the signals in stromal cells that shape the microenvironment for dormant cancer cells by using FRET mice. Further improvement of the existing biosensors and development of new stress biosensors will synergistically advance our understanding of how cancer dormancy is regulated and disrupted in the body.
Figure 4.
Cancer dormancy in brain tissue. (A) A schematic illustration of mouse intravital FRET microscopy in the future. (B and C) Cancer cells were injected into the left ventricle of a nude mouse and the brain was excised after 32 days. The brain was made transparent by the tissue-clearing CUBIC method (75), stained with first/second antibodies, and imaged under a two-photon excitation microscope. The white rectangle in (B) is magnified in (C). Red, cancer cells; green, blood vessels; cyan, activated astrocytes. Scale, (B) 1 mm; (C) 100 μm.
In conclusion, single-molecule FRET biosensors have a broad range of applications, and intravital FRET microscopy holds great advantages in that it can be used to visualize intravital signals in situ. Over the long term, by consolidating the knowledge and skills that we have obtained through the development and application of FRET biosensors, it might be possible to create innovative tools for clinical application, such as novel contrast agents for magnetic resonance imaging or photoacoustic tomography/spectroscopy.
Author Contributions
E.H. and E.K. wrote the article.
Acknowledgments
Support was received in the form of a Grant-in-Aid for Scientific Research on Innovative Areas (26112719) from the Japan Society for the Promotion of Science (JSPS).
Editor: Katharina Gaus.
Contributor Information
Eishu Hirata, Email: ehirata@kanazawa-med.ac.jp.
Etsuko Kiyokawa, Email: kiyokawa@kanazawa-med.ac.jp.
References
- 1.Shimomura O., Johnson F.H., Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Förster T. Energy migration and fluorescence. 1946. J. Biomed. Opt. 2012;17:011002. doi: 10.1117/1.JBO.17.1.011002. [DOI] [PubMed] [Google Scholar]
- 3.Jares-Erijman E.A., Jovin T.M. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 4.Aoki K., Kamioka Y., Matsuda M. Fluorescence resonance energy transfer imaging of cell signaling from in vitro to in vivo: basis of biosensor construction, live imaging, and image processing. Dev. Growth Differ. 2013;55:515–522. doi: 10.1111/dgd.12039. [DOI] [PubMed] [Google Scholar]
- 5.Becker W. Fluorescence lifetime imaging—techniques and applications. J. Microsc. 2012;247:119–136. doi: 10.1111/j.1365-2818.2012.03618.x. [DOI] [PubMed] [Google Scholar]
- 6.Komatsu N., Aoki K., Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terai K., Matsuda M. Ras binding opens c-Raf to expose the docking site for mitogen-activated protein kinase kinase. EMBO Rep. 2005;6:251–255. doi: 10.1038/sj.embor.7400349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terai K., Matsuda M. The amino-terminal B-Raf-specific region mediates calcium-dependent homo- and hetero-dimerization of Raf. EMBO J. 2006;25:3556–3564. doi: 10.1038/sj.emboj.7601241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen M.D., Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem. Biophys. Res. Commun. 2006;348:716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 10.Harvey C.D., Ehrhardt A.G., Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. USA. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouyang M., Lu S., Wang Y. Visualization of polarized membrane type 1 matrix metalloproteinase activity in live cells by fluorescence resonance energy transfer imaging. J. Biol. Chem. 2008;283:17740–17748. doi: 10.1074/jbc.M709872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Zhang Z., Luo Q. Detection of MMP activity in living cells by a genetically encoded surface-displayed FRET sensor. Biochim. Biophys. Acta. 2007;1773:400–407. doi: 10.1016/j.bbamcr.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Joseph J., Seervi M., Retnabai S.T. High throughput ratio imaging to profile caspase activity: potential application in multiparameter high content apoptosis analysis and drug screening. PLoS One. 2011;6:e20114. doi: 10.1371/journal.pone.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyokawa E., Aoki K., Matsuda M. Spatiotemporal regulation of small GTPases as revealed by probes based on the principle of Förster Resonance Energy Transfer (FRET): Implications for signaling and pharmacology. Annu. Rev. Pharmacol. Toxicol. 2011;51:337–358. doi: 10.1146/annurev-pharmtox-010510-100234. [DOI] [PubMed] [Google Scholar]
- 15.Kitano M., Nakaya M., Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 16.Mochizuki N., Yamashita S., Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 17.Kaláb P., Pralle A., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 18.Sato M., Ueda Y., Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 2003;5:1016–1022. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- 19.Kamioka Y., Sumiyama K., Matsuda M. Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct. Funct. 2012;37:65–73. doi: 10.1247/csf.11045. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi Y., Shinotsuka N., Miura M. Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. J. Cell Biol. 2011;195:1047–1060. doi: 10.1083/jcb.201104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnsson A.K., Dai Y., Welch H.C. The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Reports. 2014;6:1153–1164. doi: 10.1016/j.celrep.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyawaki A., Llopis J., Tsien R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 23.Aoki K., Komatsu N., Matsuda M. Stable expression of FRET biosensors: a new light in cancer research. Cancer Sci. 2012;103:614–619. doi: 10.1111/j.1349-7006.2011.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsubara A.T., Matsuda M., Aoki K. Quantitative analysis of recombination between YFP and CFP genes of FRET biosensors introduced by lentiviral or retroviral gene transfer. Sci. Rep. 2015;5:13283. doi: 10.1038/srep13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ai H.W., Henderson J.N., Campbell R.E. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirata E., Yukinaga H., Matsuda M. In vivo fluorescence resonance energy transfer imaging reveals differential activation of Rho-family GTPases in glioblastoma cell invasion. J. Cell Sci. 2012;125:858–868. doi: 10.1242/jcs.089995. [DOI] [PubMed] [Google Scholar]
- 27.Yagi S., Matsuda M., Kiyokawa E. Chimaerin suppresses Rac1 activation at the apical membrane to maintain the cyst structure. PLoS One. 2012;7:e52258. doi: 10.1371/journal.pone.0052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagi S., Matsuda M., Kiyokawa E. Suppression of Rac1 activity at the apical membrane of MDCK cells is essential for cyst structure maintenance. EMBO Rep. 2012;13:237–243. doi: 10.1038/embor.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam A.J., St-Pierre F., Lin M.Z. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivics Z., Li M.A., Izsvák Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodard L.E., Wilson M.H. piggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 2015;33:525–533. doi: 10.1016/j.tibtech.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aoki K., Kumagai Y., Matsuda M. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell. 2013;52:529–540. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 33.Yukinaga H., Shionyu C., Matsuda M. Fluctuation of Rac1 activity is associated with the phenotypic and transcriptional heterogeneity of glioma cells. J. Cell Sci. 2014;127:1805–1815. doi: 10.1242/jcs.139733. [DOI] [PubMed] [Google Scholar]
- 34.Goto A., Nakahara I., Funabiki K. Circuit-dependent striatal PKA and ERK signaling underlies rapid behavioral shift in mating reaction of male mice. Proc. Natl. Acad. Sci. USA. 2015;112:6718–6723. doi: 10.1073/pnas.1507121112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno R., Kamioka Y., Matsuda M. In vivo imaging reveals PKA regulation of ERK activity during neutrophil recruitment to inflamed intestines. J. Exp. Med. 2014;211:1123–1136. doi: 10.1084/jem.20132112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumagai Y., Naoki H., Matsuda M. Heterogeneity in ERK activity as visualized by in vivo FRET imaging of mammary tumor cells developed in MMTV-Neu mice. Oncogene. 2015;34:1051–1057. doi: 10.1038/onc.2014.28. [DOI] [PubMed] [Google Scholar]
- 37.Chapman P.B., Hauschild A., McArthur G.A., BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sosman J.A., Kim K.B., Ribas A. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flaherty K.T., Puzanov I., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Allen E.M., Wagle N., Schadendorf D., Dermatologic Cooperative Oncology Group of Germany (DeCOG) The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi H., Hugo W., Lo R.S. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirata E., Girotti M.R., Sahai E. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell. 2015;27:574–588. doi: 10.1016/j.ccell.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monici M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol. Annu. Rev. 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 44.Zipfel W.R., Williams R.M., Webb W.W. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl. Acad. Sci. USA. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day R.N., Davidson M.W. Fluorescent proteins for FRET microscopy: monitoring protein interactions in living cells. BioEssays. 2012;34:341–350. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiratsuka T., Fujita Y., Matsuda M. Intercellular propagation of extracellular signal-regulated kinase activation revealed by in vivo imaging of mouse skin. eLife. 2015;4:e05178. doi: 10.7554/eLife.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Looney M.R., Thornton E.E., Krummel M.F. Stabilized imaging of immune surveillance in the mouse lung. Nat. Methods. 2011;8:91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goedhart J., van Weeren L., Gadella T.W., Jr. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods. 2010;7:137–139. doi: 10.1038/nmeth.1415. [DOI] [PubMed] [Google Scholar]
- 49.Markwardt M.L., Kremers G.J., Rizzo M.A. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakoshi H., Lee S.J., Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shcherbakova D.M., Verkhusha V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chittajallu D.R., Florian S., Mitchison T.J. In vivo cell-cycle profiling in xenograft tumors by quantitative intravital microscopy. Nat. Methods. 2015;12:577–585. doi: 10.1038/nmeth.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikuta J., Wada Y., Ishii M. Dynamic visualization of RANKL and Th17-mediated osteoclast function. J. Clin. Invest. 2013;123:866–873. doi: 10.1172/JCI65054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Digman M.A., Caiolfa V.R., Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warren S.C., Margineanu A., French P.M. Rapid global fitting of large fluorescence lifetime imaging microscopy datasets. PLoS One. 2013;8:e70687. doi: 10.1371/journal.pone.0070687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newman R.H., Fosbrink M.D., Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hochreiter B., Garcia A.P., Schmid J.A. Fluorescent proteins as genetically encoded FRET biosensors in life sciences. Sensors (Basel) 2015;15:26281–26314. doi: 10.3390/s151026281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benham-Pyle B.W., Pruitt B.L., Nelson W.J. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engler A.J., Sen S., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 60.Kiyonaka S., Kajimoto T., Mori Y. Genetically encoded fluorescent thermosensors visualize subcellular thermoregulation in living cells. Nat. Methods. 2013;10:1232–1238. doi: 10.1038/nmeth.2690. [DOI] [PubMed] [Google Scholar]
- 61.Meng F., Suchyna T.M., Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275:3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grashoff C., Hoffman B.D., Schwartz M.A. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gayrard C., Borghi N. FRET-based molecular tension microscopy. Methods. 2015;94:33–42. doi: 10.1016/j.ymeth.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 64.Jurchenko C., Salaita K.S. Lighting up the force: investigating mechanisms of mechanotransduction using fluorescent tension probes. Mol. Cell. Biol. 2015;35:2570–2582. doi: 10.1128/MCB.00195-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krieg M., Dunn A.R., Goodman M.B. Mechanical control of the sense of touch by β-spectrin. Nat. Cell Biol. 2014;16:224–233. doi: 10.1038/ncb2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai D., Chen S.C., Montell D.J. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sawada Y., Tamada M., Sheetz M.P. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cost A.L., Ringer P., Grashoff C. How to measure molecular forces in cells: a guide to evaluating genetically-encoded FRET-based tension sensors. Cell. Mol. Bioeng. 2015;8:96–105. doi: 10.1007/s12195-014-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smani T., Shapovalov G., Rosado J.A. Functional and physiopathological implications of TRP channels. Biochim. Biophys. Acta. 2015;1853:1772–1782. doi: 10.1016/j.bbamcr.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 70.Takebe T., Sekine K., Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 71.El Touny L.H., Vieira A., Green J.E. Combined SFK/MEK inhibition prevents metastatic outgrowth of dormant tumor cells. J. Clin. Invest. 2014;124:156–168. doi: 10.1172/JCI70259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao H., Chakraborty G., Giancotti F.G. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghajar C.M., Peinado H., Bissell M.J. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kienast Y., von Baumgarten L., Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 75.Susaki E.A., Tainaka K., Ueda H.R. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]