Abstract
Newborns are unable to mount antibody responses towards certain antigens. This has been related to the restricted repertoire of immunoglobulin (Ig) genes of their B cells. The mechanisms underlying the restricted fetal Ig gene repertoire are currently unresolved. We here addressed this with detailed molecular and cellular analysis of human precursor-B cells from fetal liver, fetal bone marrow (BM), and pediatric BM. In the absence of selection processes, fetal B-cell progenitors more frequently used proximal V, D and J genes in complete IGH gene rearrangements, despite normal Ig locus contraction. Fewer N-nucleotides were added in IGH gene rearrangements in the context of low TdT and XRCC4 expression. Moreover, fetal progenitor-B cells expressed lower levels of IL7Rα than their pediatric counterparts. Analysis of progenitor-B cells from IL7Rα-deficient patients revealed that TdT expression and N-nucleotides additions in Dh-Jh junctions were dependent on functional IL7Rα. Thus, IL7Rα affects TdT expression, and decreased expression of this receptor underlies at least in part the skewed Ig repertoire formation in fetal B-cell precursors. These new insights provide a better understanding of the formation of adaptive immunity in the developing fetus.
During the second trimester of human fetal development, B cells are generated in the liver and bone marrow (BM), providing the neonate with a diverse immunoglobulin (Ig) repertoire. Still, antibody responses towards certain antigens (e.g. tetanus or diphtheria toxoid) are impaired in neonates, and the ability to respond is only acquired with age1,2,3. Various processes can underlie this initial inability to mount such responses, including a “pre-mature” diversity of the Ig repertoire.
B-cell development has been extensively studied in human postnatal BM, where 5 distinct stages can be identified. In the early pro-B and pre-BI cell stages, D to J gene rearrangements and V to DJ gene rearrangements are induced by the lymphoid-specific recombination activating gene proteins 1 and 2 (Rag1 and Rag2) in the Ig heavy chain (IGH) locus4,5. Subsequently, the cells induce V to J gene rearrangements in their Ig light chain loci (IGK or IGL) as pre-B-II small cells and the complete Ig molecule is selected for functionality at the immature-B cell stage6,7. The Rag-induced double stranded DNA breaks activate cell cycle checkpoint protein ATM and the non-homologous end joining (NHEJ) pathway8,9,10, which enhances diversity in the junctional region that encodes the antigen-binding complementary determining region (CDR)3. A central player is the lymphoid-specific deoxynucleotidyl transferase protein (TdT) that randomly adds N-nucleotides to the junction11,12,13.
Several studies have addressed the Ig gene repertoire in human fetuses, and reported restricted repertoires due to shorter CDR3 regions in IGH. This restriction was the result of limited N-nucleotides in the junctions and more frequent usage of the relatively short Dh7-27, Jh2 and Jh3 genes1,2,14,15,16,17,18,19,20,21,22,23. Finally, the Dh-proximal Vh6-1 and Vh1-2 genes appear to be more frequently used in fetal B cells1,24,25,26,27.
Similar skewing of the IGH gene repertoire has been observed in mouse embryos14,15,18,19. Embryonic B-cell progenitors in the mouse liver have been shown to generate a separate B-cell lineage, and can develop independent of functional IL7R signaling28,29. Postnatal B-cell development in IL7R-deficient mice is completely blocked due to defects in proliferation and survival29,30. Furthermore, IL7R-deficient progenitor-B cells are impaired in the capacity to rearrange DJh-distal Vh genes31. In contrast to mouse, human B cells can develop in the absence of IL7R signaling32,33. However, the Ig gene repertoire in progenitor-B cells of IL7Rα-deficient patients has not been studied so far. Thus, it remains unclear what underlies the skewed Ig repertoire in fetal B cells and if IL-7R signaling is involved.
We here studied the nature of Ig gene repertoire formation during human fetal B cell development in B-cell progenitors, and the role of IL7Rα in this process. Through analysis of patients with genetic defects in IL7Rα, we were able to demonstrate a direct link between IL7Rα, TdT and N-nucleotide additions in IGH gene rearrangements.
Results
Skewed Ig repertoire generation in fetal B cell precursors
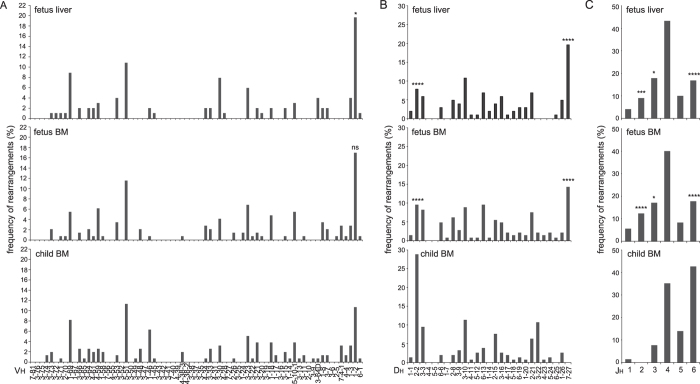
To study the Ig repertoire formation in fetal development, we purified B-cell subsets from 2nd trimester fetal BM and fetal liver, as well as from neonatal cord blood and pediatric BM. In line with previous observations1,16,17,20,21,24,25,27,34, mature B cells (CD19+CD20+CD10-IgM+IgD+) in fetal BM showed more frequent usage of Vh1-2, Dh7-27, Jh2 and Jh3 genes than pediatric B cells (Supplementary Fig. 1)23. This IGH gene usage was typical for early development, because the IGH repertoire in B cells from neonatal cord blood was more similar to pediatric than to fetal B cells (Supplementary Fig. 1). Moreover, CD19+CD34+CD10+ pre-BI cells derived from either fetal liver or fetal BM contained the same skewed IGH repertoire (Fig. 1A–C). These pre-BI cells initiate Vh to DJh gene rearrangements, but do not yet express cytoplasmic IgM and are not selected for functional IGH genes35. This was supported by analysis of only the out-of-frame and thus unproductive IGH gene rearrangements (Supplementary Fig. 2A–C). Approximately 1/3rd of the pre-BI cells carried in-frame and thereby potentially productive IGH gene rearrangements (31% in fetal liver, 29% in fetal BM and 33% in pediatric BM). Thus, fetal B-cell progenitors are generated with a skewed IGH gene repertoire, which is similar between fetal liver and fetal BM, and does not seem to be affected by selection processes.
Figure 1. Skewed Ig repertoire generation in fetal B cell precursors.
(A) IGHV, (B) IGHD, (C) IGHJ gene usage in pre-BI cells from fetal liver, fetal BM and pediatric BM. In-frame and out-of-frame rearrangements were included. 102 sequences were derived from fetal liver (3 donors), 148 from fetal BM (4 donors) and 160 from pediatric BM (5 donors). Statistics: χ2 test; *p < 0.05, ***p < 0.001, ****p < 0.0001.
In contrast to IGH, the IGK gene repertoire was not different between fetal and pediatric mature B cells nor small pre-BII cells (Supplementary Figs 3 and 4). Because the vast majority of IGK gene rearrangements are formed in small pre-B-II cells, the formation and selection processes for the IGK repertoire do not seem to differ between pediatric and fetal B-cell development.
Short Dh genes and fewer N-nucleotides in fetal B cells
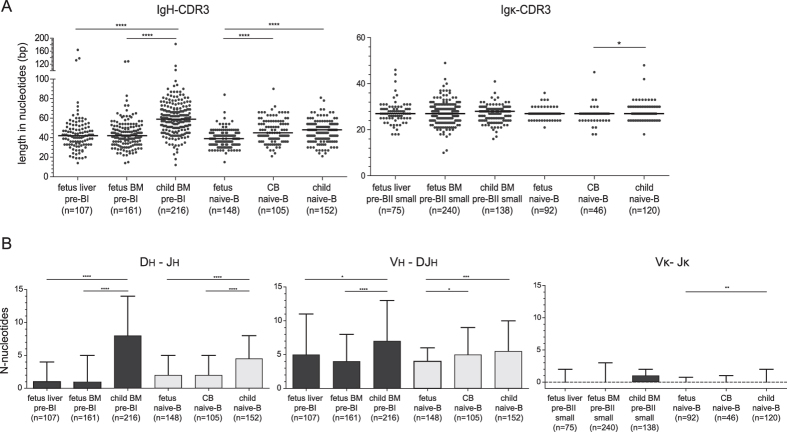
In addition to the skewed V, D and J gene usage, IGH gene rearrangements in fetal naive mature B cells carried shorter IgH-CDR3 than pediatric B cells (Fig. 2A)1,2,16,17,20,21,36. This was mostly the result of fewer N-nucleotide additions in both the Dh-Jh and Vh-DJh junctions (Fig. 2B), as well as high usage of the short Dh7-27 gene (Supplementary Fig. 1). The IgH-CDR3 of B cells from neonatal cord blood were intermediate in size between fetal and pediatric B cells with slightly more N-nucleotides in the Vh-DJh junction than fetal B cells. Both in fetal and in pediatric tissue, the IgH-CDR3 regions in pre-BI cells were larger than in naive mature B cells. Still, fetal pre-BI carried shorter IgH-CDR3 with fewer N-nucleotides than their pediatric counterparts (Fig. 2B, Supplementary Fig. 2D,E, Supplementary Table 1).
Figure 2. Fetal B cells have short IgH-CDR3 regions and few N-nucleotides.
(A) CDR3 length of complete IGH and IGK gene rearrangements. Grey dots represent unique clones, black lines median values. (B) The median numbers of N-nucleotides in Dh-Jh, Vh-DJh and Vκ-Jκ junctions with inter-quartile range. Numbers of analyzed sequences in brackets. In-frame and out-of-frame rearrangements were included in the analysis of precursor-B cells, whereas only in-frame rearrangements for naive mature-B cells. Statistics: Mann-Whitney U test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In addition to junctional region processing, the IgH-CDR3 size is affected by the Dh gene usage as these differ greatly in size. Indeed, the Dh7-27 gene that was abundantly found in fetal B cells is the shortest of all D genes. In addition, the large Dh genes were less frequently observed in fetal than in pediatric B cells (Supplementary Fig. 5a,b). Still, as previously suggested16, N-nucleotide additions seem dependent on Dh gene usage. The incomplete Dh-Jh rearrangements involving the short Dh7-27 carried more N-nucleotides than rearrangements involving any other Dh gene families (Supplementary Fig. 5c). Finally, the shorter IgH-CDR3 in naive mature B cells than in pre-BI cells (Fig. 2) was associated with selection against long Dh genes. Thus, the IgH-CDR3 size seems tightly regulated through N-nucleotide additions and Dh gene usage, which differ between fetal and pediatric B-cell progenitors.
In contrast to IGH, the IGK locus does not contain D genes, and Vκ-Jκ gene rearrangements hardly contain any N-nucleotide, thus rendering small CDR3 regions. Indeed, the Igκ-CDR3 regions of the fetal, neonatal and pediatric B-cell subsets were much smaller than IgH-CDR3 and did not show overt differences between any of the subsets (Fig. 2A, Supplementary Table 2). Although the pediatric naive mature B cells tended to have more N-nucleotide additions (Fig. 2B), there was no indication of a different Igκ-CDR3 repertoire in fetus.
B cells in BCP-ALL of presumed fetal origin show fetal Ig repertoire characteristics
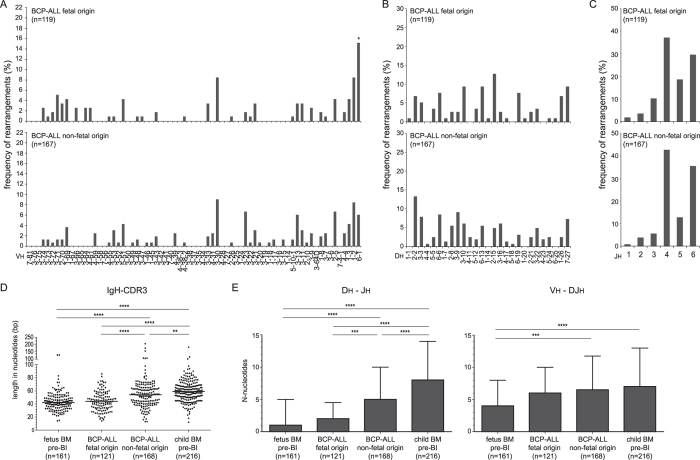
Our data indicate that normal B-cell precursors from fetal tissue differ from children. To study whether these cellular and molecular differences were retained in a disease model, we analyzed the IGH gene repertoire in BCP-ALL samples. BCP-ALL with MLL rearrangements or with TEL-AML1 fusion genes can originate in utero37,38,39,40. We compared the IGH gene repertoire of these presumed fetal BCP-ALL subgroups and with the rest, i.e. subgroups containing E2A-PBX translocations, BCR-ABL translocations, hyperdiploidy or none of these abnormalities. The average age in years at diagnosis was 0.65 ± 0.68 for MLL rearranged BCP-ALL and 4.85 ± 2.95 for TEL-AML1-positive BCP-ALL, while the non-fetal origin group aged 6.39 ± 4.18.
The BCP-ALL of presumed fetal origin indeed showed increased usage of the Dh-proximal Vh6-1 gene, similar to fetal B cells (Fig. 3A). In contrast, the usage of Dh and Jh genes did not differ between the BCP-ALL patients of presumed fetal and non-fetal origin (Fig. 3B,C). Still, BCP-ALL of presumed fetal origin showed a trend to less frequently use long Dh genes than the other BCP-ALL (Supplementary Figs 5a and 6a), and the IgH-CDR3 length was shorter in BCP-ALL of presumed fetal origin. The latter was due to significantly fewer N-nucleotides in Dh-Jh junctions (Fig. 3D,E, Supplementary Table 1), and equally apparent in BCP-ALL with MLL-rearrangements and with TEL-AML1 translocations (Supplementary Fig. 6b,c).
Figure 3. BCP-ALL of presumed fetal origin show fetal Ig repertoire characteristics.
(A) IGHV, (B) IGHD, (C) IGHJ gene usage in IGH gene rearrangments. (D) IgH-CDR3 length. Grey dots represent unique clones, black lines median values. (E) Median numbers of N-nucleotides in Dh-Jh and Vh-DJh junctions with inter-quartile range. In-frame and out-of-frame rearrangements were included. The numbers of sequences are indicated between brackets. Statistics: χ2 (panels A/B/C), or Mann-Whitney (panels D/E); *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Concluding, these observations in BCP-ALL confirm and extend our results that the IGH gene repertoire differences between fetus and child are the result of altered V(D)J recombination processes in early progenitors rather than Ig repertoire selection processes in mature B cells.
Enrichment of early B-cell precursors in fetal liver and BM
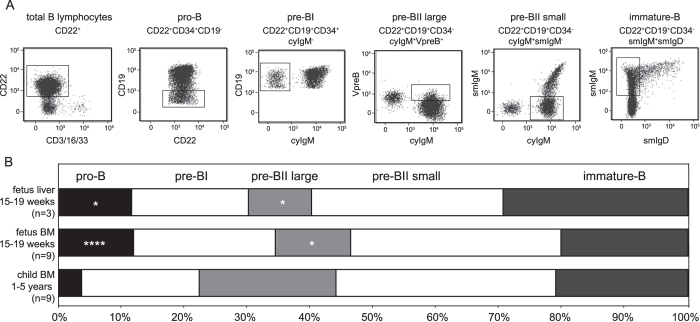
To study the nature of the skewed IGH gene repertoire generation in prenatal development, we first analyzed the precursor-B cells in fetal liver and BM in more detail by flow cytometry. The relative distributions of the five major precursor-B-cell subsets in these fetal tissues were determined using a panel of membrane and intracellular markers and compared with pediatric BM (Fig. 4A)41,42. The frequencies of the pro-B, pre-BII large and pre-BII small within the total CD22+ B-cell precursors were similar between fetal liver and fetal BM. Fetal livers contained on average less pre-BI and more immature-B cells, but these differences were not significant. Both fetal tissues contained more CD34+ precursors than pediatric BM (Fig. 4B). This was mostly the result of increased pro-B cell frequencies (~12% vs 4%), and was to the expense of reduced pre-BII large frequencies (~10% vs 20%).
Figure 4. Relative increase of early precursor-B cells in fetus.
(A) Gating strategy to define 5 precursor B-cell subsets according to previous studies35,42. (B) Relative distributions of the 5 precursor-B-cell subsets in fetal liver, fetal BM and pediatric BM. Statistics: Mann-Whitney U test; *p < 0.05, ****p < 0.0001.
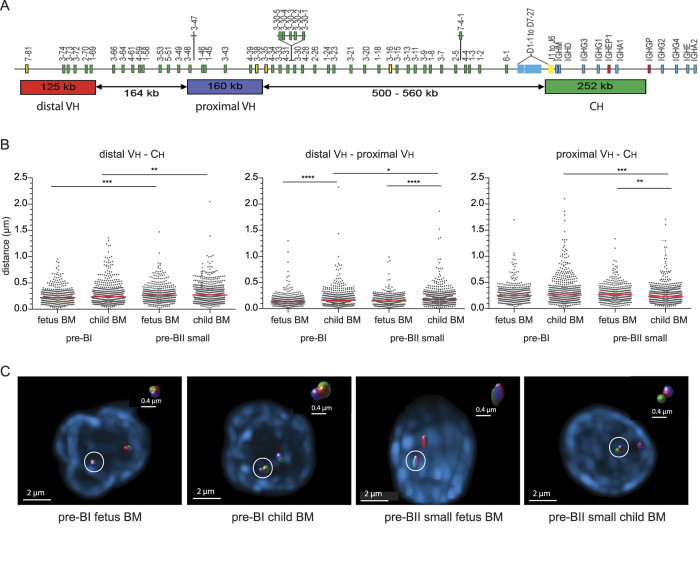
IGH locus contraction in fetal precursor-B cells
Previous studies have shown large-scale contraction of the IGH locus in mouse pro-B cells and the corresponding human pre-BI cells that facilitate rearrangements involving Dh-distal Vh genes43,44,45,46,47. Thus, the skewed Ig repertoire formation and developmental shift observed in fetus at pro-B/pre-BI stage could potentially result from inefficiently formed IGH gene rearrangements due to aberrant IGH locus topology. To study whether the IGH topology provided optimal conditions for V(D)J recombination, we measured spatial distances between distal Vh, proximal Vh, and Ch regions with 3D DNA FISH in precursor-B cells of fetal and pediatric BM (Fig. 5A). In line with our previous studies44,46, spatial distances between all three DNA regions were short in pediatric pre-BI cells and slightly, yet significantly, larger in pediatric pre-BII small cells (Fig. 5B,C). IGH locus contraction did not seem defective in fetal pre-BI cells, as all spatial distances between Ch and both Vh regions were similarly small and distances between both Vh regions were even smaller than in pediatric pre-BI cells. IGH locus contraction is therefore likely to facilitate rearrangement of both proximal and distal Vh gene rearrangements similarly between fetal and pediatric B-cell precursors and it does not provide further insight into the relative abundance of Vh1-2 in the fetal IGH gene repertoire.
Figure 5. IGH locus contraction in fetal and pediatric B-cell precursors.
(A) The human IGH locus with details of BAC clones used as 3D FISH probes44,46. (B) Spatial distances separating distal Vh, proximal Vh, and Ch regions in precursor-B cells from fetal and pediatric BM. Grey dots represent individual alleles, red bars median values. Per group, >200 alleles from 2 donors were analyzed. Statistics: Mann–Whitney U test; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) Representative images of stained IGH loci.
Mechanisms causing formation of skewed Ig repertoire in fetuses
To delve deeper into the mechanism responsible for generation of the different Ig repertoires between fetal and pediatric early B-cell progenitors, we performed genome-wide gene expression profiling of fetal and childhood B-cell progenitors (Supplementary Fig. 7A). The analysis was performed in fetal pro-B and pre-BI cells, which were compared to previously published childhood subsets44, and was focused on genes important for B-cell development and V(D)J recombination to provide insights into the molecular environment in which Ig gene rearrangements are formed.
The expression of RAG1 and RAG2 was similar between fetus and child for both pro-B and pre-BI cells. The genes encoding key B-cell transcription factors E2A, EBF1 and PAX5 were also highly expressed in fetal progenitors, as was FOXO1, which regulates RAG gene expression48,49. These data are in line with the diverse IGH gene repertoire in fetus. Furthermore, the genes encoding pre-BCR signaling components were normally expressed and did not seem rate-limiting for differentiation into pre-BII cells (Table 1).
Table 1. Gene expression profiling of fetal and pediatric precursor-B cells.
| Pro-B |
Pre-BI |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean fetus | Mean child | Fold change | p-value | Mean fetus | Mean child | Fold change | p-value | ||
| transcription factors / B-cell commitment | E2A (TCF3) | 226.09 | 154.98 | 1.5 | 0.02 | 259.49 | 209.27 | 1.2 | 0.02 |
| ID2 | 121.79 | 43.51 | 2.8 | 0.23 | 195.40 | 80.31 | 2.4 | 0.03 | |
| EBF1 | 389.42 | 876.11 | −2.3 | 0.33 | 1182.40 | 1090.52 | 1.1 | 0.12 | |
| PAX5 | 257.97 | 189.37 | 1.4 | 0.52 | 618.04 | 456.95 | 1.4 | 0.02 | |
| HEB (TCF12) | 87.07 | 224.12 | −2.6 | 0.07 | 204.00 | 279.67 | −1.4 | 0.01 | |
| PU.1 (Spi1) | 90.22 | 57.57 | 1.6 | <0.01 | 91.75 | 72.83 | 1.3 | 0.06 | |
| Foxo1 | 182.02 | 167.64 | 1.1 | 0.56 | 231.92 | 224.62 | 1 | 0.58 | |
| IRF4 | 99.81 | 81.96 | 1.2 | 0.45 | 140.36 | 135.16 | 1 | 0.87 | |
| IRF8 | 106.55 | 48.62 | 2.2 | 0.03 | 113.48 | 74.69 | 1.5 | 0.02 | |
| IKZF1 (Ikaros) | 86.01 | 151.17 | −1.8 | 0.05 | 201.46 | 172.66 | 1.2 | 0.25 | |
| IKZF2 (Helios) | 59.77 | 65.55 | −1.1 | 0.77 | 248.02 | 277.30 | −1.1 | 0.66 | |
| IKZF3 (Aiolos) | 21.92 | 17.95 | 1.2 | 0.43 | 58.31 | 76.44 | −1.3 | 0.32 | |
| V(D)J/DNA repair | RAG1 | 103.62 | 121.84 | −1.2 | 0.47 | 165.91 | 150.29 | 1.1 | 0.39 |
| RAG2 | 113.90 | 209.38 | −1.8 | 0.43 | 227.32 | 309.86 | −1.4 | 0.05 | |
| XRCC5 (Ku80) | 186.08 | 554.95 | −2 | 0.15 | 606.18 | 725.66 | −1.2 | 0.04 | |
| XRCC6 (Ku70) | 44.06 | 39.20 | 1.1 | 0.50 | 48.62 | 31.89 | 1.5 | 0.09 | |
| DNA-PKcs (PRKDC) | 60.68 | 196.41 | −3.2 | 0.01 | 149.73 | 290.76 | −1.9 | 0.02 | |
| Artemis (DCLRE1C) | 41.31 | 71.56 | −1.7 | 0.02 | 87.42 | 121.32 | −1.4 | 0.03 | |
| XRCC4 | 31.30 | 80.04 | −2.6 | 0.049 | 79.37 | 124.96 | −1.6 | 0.049 | |
| XLF (NHEJ1) | 51.80 | 49.12 | −1.1 | 0.26 | 54.44 | 49.08 | 1.1 | 0.14 | |
| LIG4 | 243.02 | 300.93 | −1.2 | 0.84 | 746.53 | 472.78 | 1.6 | 0.14 | |
| TdT (DNTT) | 399.84 | 1693.51 | −4.2 | 0.17 | 1111.20 | 1656.59 | −1.5 | 0.02 | |
| pol λ | 47.36 | 31.85 | 1.5 | 0.15 | 33.68 | 31.84 | 1.1 | 0.52 | |
| pol μ | 85.47 | 55.48 | 1.5 | 0.02 | 74.14 | 58.40 | 1.3 | 0.05 | |
| ATM | 48.57 | 166.38 | −3.4 | 0.04 | 106.49 | 199.37 | −1.9 | 0.01 | |
| MRE11A | 41.67 | 169.07 | −4.1 | 0.02 | 96.98 | 212.86 | −2.2 | <0.01 | |
| NBS (NBN) | 43.04 | 174.16 | −4.1 | 0.02 | 85.99 | 178.98 | −2.1 | 0.02 | |
| Rad50 | 96.36 | 290.60 | −3 | 0.15 | 264.57 | 326.31 | −1.2 | 0.049 | |
| pre-BCR signaling | CD22 | 83.09 | 64.89 | 1.3 | 0.18 | 545.90 | 295.23 | 1.9 | <0.01 |
| CD19 | 117.52 | 68.81 | 1.7 | 0.01 | 138.91 | 82.83 | 1.7 | <0.01 | |
| CD34 | 210.88 | 295.12 | −1.4 | 0.42 | 329.68 | 268.36 | 1.2 | 0.48 | |
| CD10 | 217.69 | 969.99 | −4.5 | 0.15 | 880.24 | 1275.02 | −1.5 | 0.03 | |
| CD79A | 150.68 | 139.54 | 1.1 | 0.86 | 268.79 | 143.01 | 1.9 | <0.01 | |
| CD79B | 183.05 | 103.54 | 1.8 | 0.27 | 228.40 | 158.76 | 1.4 | 0.05 | |
| VPREB1 | 297.75 | 501.40 | −1.7 | 0.49 | 778.90 | 644.68 | 1.2 | 0.18 | |
| λ14.1 (IGLL5) | 38.86 | 29.63 | 1.3 | 0.10 | 36.88 | 35.34 | 1 | 0.24 | |
| BTK | 106.92 | 142.97 | −1.3 | 0.50 | 193.35 | 183.15 | 1.1 | 0.64 | |
| SYK | 106.83 | 170.55 | −1.6 | 0.23 | 208.60 | 219.74 | −1.1 | 0.14 | |
| BLNK | 284.80 | 812.93 | −2.9 | 0.20 | 696.87 | 672.31 | 1 | 0.68 | |
| IGHG1 | 89.03 | 65.57 | 1.4 | <0.01 | 110.02 | 82.20 | 1.3 | 0.01 | |
| IGKC | 137.41 | 54.08 | 2.5 | 0.09 | 313.73 | 192.59 | 1.6 | 0.10 | |
| IL2RG | 100.12 | 238.93 | −2.4 | 0.05 | 174.79 | 155.79 | 1.1 | 0.28 | |
| proliferation/survival | Ki−67 (MKI67) | 182.64 | 305.69 | −1.7 | 0.39 | 689.30 | 833.39 | −1.2 | 0.24 |
| IL7Rα | 44.38 | 171.98 | −3.9 | 0.04 | 169.35 | 150.13 | 1.1 | 0.75 | |
| LEF1 | 258.38 | 355.16 | −1.4 | 0.7 | 893.34 | 1002.85 | −1.1 | 0.03 | |
| STAT5A | 80.84 | 67.61 | 1.2 | 0.23 | 89.54 | 77.34 | 1.2 | 0.06 | |
| STAT5B | 66.58 | 92.58 | −1.4 | 0.09 | 85.84 | 90.19 | −1.1 | 0.43 | |
| JAK1 | 170.76 | 303.35 | −1.8 | 0.35 | 419.79 | 366.42 | 1.2 | 0.01 | |
| JAK2 | 38.05 | 81.16 | −2.1 | <0.01 | 39.67 | 76.44 | −1.9 | 0.07 | |
| JAK3 | 61.46 | 42.30 | 1.5 | 0.06 | 60.12 | 48.21 | 1.3 | 0.02 | |
| TYK2 | 77.17 | 56.01 | 1.4 | 0.049 | 75.25 | 64.44 | 1.2 | 0.64 | |
| FLT3 | 34.20 | 279.61 | −8.2 | 0.01 | 64.21 | 157.15 | −2.5 | 0.30 | |
| c-KIT | 35.86 | 73.68 | −2.1 | 0.03 | 31.01 | 32.66 | −1.1 | 0.02 | |
Expression levels are shown as mean signal values. Differences in gene expression levels between fetus and child which showed p < 0.05 and fold change >1.5 are marked in bold. Statistical significance was calculated with one-way ANOVA test. Gene expression profiling was performed on pro-B and pre-BI cells purified from 3 fetal BM and 4 pediatric BM donors.
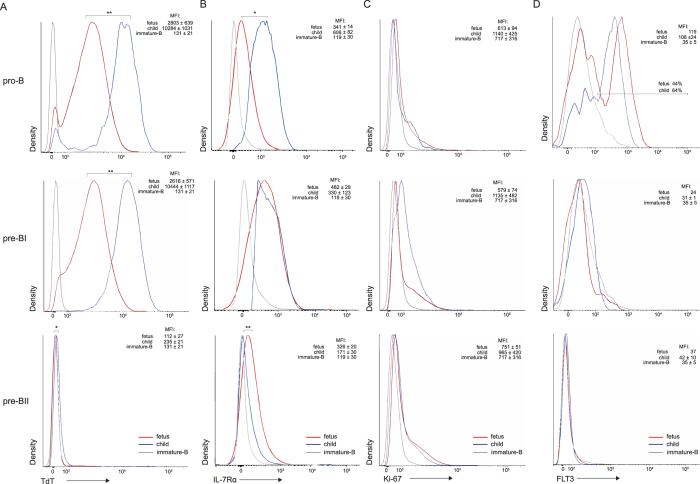
However, the components of the DNA damage response pathways were downregulated in fetuses as compared to children. Among these was ATM (Table 1 and Supplementary Fig. 7b), which has a critical role in cell cycle arrest and recruitment of DNA repair factors to the broken ends8,9,50,51. Furthermore, the gene encoding XRCC4 that is part of the NHEJ pathway required for repair of Rag-induced DNA breaks10, was downregulated in fetal cells (Table 1 and Supplementary Fig. 7b). XRCC4 forms a protein complex with TdT, the enzyme responsible for N-nucleotide additions, and it was shown that XRCC4 promotes N-nucleotides addition by TdT52. Importantly, we also observed decreased expression of DNTT, which encodes TdT (Table 1). Additional intracellular TdT staining by flow cytometry revealed that these decreased transcripts resulted in decreased protein levels in both pro-B and pre-BI in fetuses (Fig. 6A).
Figure 6. Protein expression levels in B-cell progenitors.
(A) TdT, (B) IL-7Rα, (C) Ki-67, (D) FLT. Graphs represent merged data from 3–5 fetal BM (aged 15–19 weeks) and 3–6 pediatric BM samples (aged 7 years) as determined by flow cytometry analysis. Immature-B cells of pediatric BM were included as a biological negative control. In each plot, the indicated median fluorescence intensities (MFI) are shown as the averages of the individual donors with standard error of mean (SEM). Statistics were performed with an unpaired t-test on MFI values from individual donors; *p < 0.05, **p < 0.01.
In addition to DNA repair, the components of signaling pathways which increase accessibility of Ig loci for rearrangements were downregulated in fetus. IL-7R signaling affects IGH gene rearrangements and inhibits IGK gene rearrangements46,53,54. Fetal pro-B cells showed low expression of IL7RA transcripts and membrane proteins (Table 1 and Fig. 6B). Moreover, transcripts encoding the IL-7R signaling molecule JAK2 were reduced in pro-B cells, whereas transcripts of ID2 and germline IGK that are normally suppressed by IL-7R signaling55,56,57 were upregulated (Table 1). The decreased IL-7Rα expression did not seem to affect proliferation because transcripts nor nuclear Ki-67 protein were different between fetuses and children (Table 1 and Fig. 6C). Fms-related tyrosine kinase-3 (FLT3) was also downregulated in fetal pro-B cells (Table 1 and Fig. 6D). This is the receptor for FLT3L that has synergistic effects with IL-7 on survival and proliferation of B-cell progenitors58.
Together, fetal B-cell progenitors differ from their pediatric counterparts in expression levels of NHEJ components and TdT, as well as surface receptors IL-7R and FLT3.
TdT expression and N-nucleotide additions in IL7Rα-deficient patients
Ingenuity pathway analysis suggests that IL-7R and FLT3 signaling interact with the NHEJ pathway (Fig. 7A, Supplementary Fig. 8), and that these proteins regulate expression of TdT and N-nucleotide insertions in postnatal B-cell precursors. To confirm this relation, we tested the effects of lack of IL7R signaling: B-cell precursors from IL7Rα-deficient children. If IL7R signaling is important for TdT expression and N-nucleotide additions in postnatal B-cell precursors, IL7Rα-deficient B-cells should be defective in both. CD79A+CD19- BM-derived pro-B cells of IL7Rα-deficient children showed lower nuclear TdT protein levels than healthy children (Fig. 7B). Out-of-frame IGH gene rearrangements in B cells of these patients contained fewer N-nucleotides in the Dh-Jh junctions (Fig. 7C). Since these Dh-Jh junctions were generated in pro-B cells35, and were not subjected to selection processes, this confirms that functional IL-7R expression affects IGH junction processing.
Figure 7. IL7Rα-regulated expression of TdT and IGH junctional processing.
(A) Functional network of molecules interacting with IL-7R, FLT3, ATM, XRCC4 and TdT (DNTT) in pro-B cells. Upregulated (red) and downregulated (green) transcripts in fetus vs child. (B) TdT protein expression in cyCD79A+CD19- BM derived pro-B cells of IL7Rα-deficient children (aged 3-9 months) and pediatric controls (aged 7 years). Histograms include merged data from 3 donors for each group. In each plot, the indicated median fluorescence intensities (MFI) are shown as the averages of the individual donors with standard error of mean (SEM). Statistics were performed with an unpaired t-test on MFI values from individual donors; ***p < 0.001. (C) N-nucleotide additions in B cells from IL7RA-deficient and healthy children. The median numbers of N-nucleotides in Dh-Jh junctions from out-of-frame IGH gene rearrangements with inter-quartile range. Statistics: Mann-Whitney U test; **p < 0.01, ****p < 0.0001.
Discussion
In the present study, we showed that during fetal development a diverse Ig repertoire is formed. Still, this repertoire is skewed with regards to combinatorial diversity. IgH-CDR3 regions were shorter due to fewer N-nucleotide addition, and this was directly associated with reduced TdT and IL7Rα expression. Indeed, TdT expression and N-nucleotide additions were affected in B cells of patients with genetic defects in IL7RA. Thus, we here provide the first link between IL7Rα with IGH gene rearrangements in man, as well as a role for IL7Rα in the differences between fetal and childhood Ig gene repertoires.
The fetal Ig repertoire has been a subject of study for the past 3 decades1,2,16,17,20,21,24,25,26,27,34. Reported differences with postnatal B cells include V, D, and J gene usage, as well as N-nucleotide additions1,16,17,20,34. Still, not all findings were consistent, and several studies reported different findings1,24,34. We acknowledged the apparent difficulties in the assessment of the fetal Ig repertoire, and took a two-step approach to tackle these. First, we assessed the repertoire in single-sorted naive mature B cells, thereby avoiding potentially preferential PCR-based amplification of V genes. Consequently, we obtained an unbiased view of the naive B cell-repertoire23. Secondly, we amplified IGH and IGK gene rearrangements from early precursors that had not yet been selected for in-frame rearrangements35. This approach allowed us to study which of the characteristics in the mature repertoire were the result of altered V(D)J recombination processes rather than selection. Thus, we confirmed that the fetal naive mature Ig repertoire contained more Vh1-2, Dh7-27, Jh2 and Jh3 genes than pediatric B cells1,16,17,20. The high frequencies of Dh7-27 and Jh2 and Jh3 could in part be the result of absence of secondary rearrangements involving upstream Dh and downstream Jh genes. However, other than Dh7-27 usage, we did not see trends for rearrangements between upstream Dh and downstream Jh genes (not shown). Despite the high Vh1-2 usage, the Dh-proximal Vh genes were in general not more frequently used in the human fetal repertoire24,25,26, in contrast to the mouse14,15,18,19. Importantly, we now established that these changes in the IGH gene repertoire were primarily the result of altered V(D)J recombination, and not due to changes in Ig repertoire selection.
The skewed IGH gene repertoire in fetus did not seem to result from changes in chromatin organization as the IGH locus in fetal B-cell precursors was contracted to a similar extent as in children. Moreover, critical transcription factors for this process, such as E2A and Pax5 were highly expressed. The 3D spatial organization could thus support the utilization of diverse Vh genes in fetal B-cell precursors independent of genomic proximity to the Dh-Jh-Ch region14,15,18,19. It is possible that the Vh1-2, Dh7-27, Jh2 and Jh3 genes were more efficiently recombined due to their optimal RSS sequences (www.imgt.org). However, RAG gene expression was equally high in both fetal and pediatric B-cell progenitors, making it difficult to understand how these could be rate-limiting in fetus. Alternatively, the Vh1-2 gene promoter activity could be affected by B-cell intrinsic or extrinsic factors. A likely candidate pathway would be IL-7R signaling that is known to affect IGH locus activity46,59. We were unable to study the production of IL-7 in fetal or pediatric BM, but we did find evidence for reduced IL-7Rα expression and lower IL-7R signaling in fetal B-cell progenitors, indicating that specific changes in Vh gene usage could be the result of altered susceptibility to IL-7. Furthermore, we observed increased usage of the Vh1-2 gene in out-of-frame rearrangements from IL-7Rα-deficient patients (not shown). This would suggest a similar role for IL-7R signaling in human as in mouse B-cell precursors, as the latter were found to be impaired in the capacity to rearrange DJh-distal Vh genes when IL7R was defective31.
We found additional evidence for differences in the generation of IGH gene rearrangements in fetal precursors through analysis of BCP-ALL. ALL cases with MLL rearrangements or TEL-AML1 translocations can arise already during fetal development. Although not all cases will actually have a prenatal origin, our comparison of these cases with other BCP-ALL subsets confirmed previously observed paucity of N and P nucleotides in IGH junctions in ALL cells in neonatal blood spots60 and low TdT transcript levels in MLL-rearranged BCP-ALL61. Furthermore, we report similar changes in the Vh, Dh and Jh gene usage as in normal fetal B-cell precursors. Considering the distinct gene expression repertoires between fetal and postnatal B-cell precursors, our observations in BCP-ALL indicate that those of in utero origin will be derived from distinct precursors than other ALL. Thus, it is possible that the differences between BCP-ALL subsets will not only be determined by their genetic abnormalities, but also by their origin from fetal or postnatal precursor-B cells.
Through analysis of precursor-B cells, we could defined the cause of the reported reduction in N-nucleotides in IGH gene rearrangements1,17,20,22,34. Fetal precursor B cells expressed low levels of TdT, the enzyme that inserts these during NHEJ, as well as XRCC4, the NHEJ factor that directly affects TdT function52. Moreover, we could link the reduced TdT expression to the low expression of IL7Rα, as our analysis of precursor B cells from IL7Rα-deficient patients revealed low TdT expression and reduced N-nucleotide additions in D-Jh junctions. This is the first evidence for a role of IL7Rα in human IGH gene rearrangements. In mouse models, precursor B cells in postnatal BM completely depend on IL7Rα signaling, and IL7R-deficient cells show defects in IGH gene rearrangements, proliferation and survival29,30,31. Following the identification of similar roles for human and mouse IL7Rα in preventing premature IGK gene rearrangements46,62, it now appears that the function of IL7Rα in human and mouse B-cell precursors is more similar than has been previously appreciated.
Despite complete absence of a functional IL-7R, IL7Rα-deficient patients showed more N-nucleotide additions in IGH than fetal progenitor-B cells that still expressed low levels of IL-7Rα. It is unlikely that this is due to separate effects of IL-7 and TSLP, as IL7Rα is a crucial component for the receptors of both cytokines63,64. However, FLT3L can support the development of B cells in absence of IL7Rα28, and acts in synergy with IL-7R to expand the hematopoietic progenitors58. The combined reduction in expression of both the receptor for FLT3L, FLT3, and IL7Ra in human fetal B-cell precursors might have a stronger impact on TdT and XRCC4 function than in IL7Rα-deficient patients.
A potential explanation can be derived from studies in TdT deficient mice65. In a competitive setting with TdT-proficient B-cell precursors, TdT-deficient B cells developed quicker and demonstrated an advantage in populating peripheral lymphoid tissues. Through low TdT expression, the developing fetus would thus be able to provide a fast wave of B cells. Decreased ATM expression likely further enhanced expansion of early progenitor-B cells in fetus with less strict processing of IGH junctions. Considering the low levels of TdT in fetal thymus66,67,68,69, and the low numbers of N-nucleotides in TRB junctions22, it is likely that similar mechanisms apply for T cells. In addition to relatively fast generation, the shorter CDR3 regions could equip the early waves of B cells with a relevant autoreactive repertoire to assist in clearance of apoptotic cells36. How the IL7Rα expression levels are regulated to mediate between a fast and skewed versus a more random repertoire remains to be determined. A likely mechanism would be the amount of FLT3L produced by stromal cells, as FLT3 signaling is shown to enhance IL7Rα expression levels70.
Taken together, we here showed that the skewed IGH gene repertoire of fetal B cells is the result of altered V(D)J recombination in progenitor B cells rather than repertoire selection. Specifically, IGH gene rearrangements contained fewer N-nucleotide additions due to low XRCC4 and TdT expression in fetal pro-B and pre-BI cells. Since fetal B-cell progenitors expressed low levels of IL7Rα, and TdT expression was found to be directly linked to functional IL7Rα, we here provide the first evidence for a role of IL7Rα in human IGH gene repertoire formation. These new insights into precursor-B cell development could contribute to a better understanding of the formation of adaptive immunity in the developing fetus.
Methods
Tissue sampling and flow cytometry
The use of human fetal tissue (3 fetal liver and 9 fetal BM donors) and pediatric tissue (12 pediatric BM donors), and cord blood (4 donors) was approved by the Medical Ethical Committees of the Erasmus MC and Eastern Norway, and was contingent on informed consent in accordance with the Declaration of Helsinki and the Dutch Fetal tissue act. Fetal tissues were obtained from elective abortions, and informed consent was obtained after the decision to abort was finalized. Inclusion in the study did not affect the treatment regime in any way, and did not involve any form of payment. Gestational ages of the fetuses ranged between 15 and 19 weeks, and no abnormalities were present that could potentially affect the immune-repertoire. PBMC and BM B cells from 3 IL7Rα-deficient patients aged 3-9 months were obtained from left-over material of diagnostic work-up.
All methods were carried out in accordance with guidelines and regulations of the Immunology Department, Erasmus MC.
Flow cytometric immunophenotyping and purification of B-cell subsets from pediatric BM aspirates, fetal limb BM suspensions and fetal liver homogenates were performed as described previously (see Supplementary Methods)35,44.
Ig gene rearrangements and IGH locus contraction
Complete IGH and IGK gene rearrangements were amplified from DNA of bulk-sorted pediatric and fetal pre-BI and pre-BII small cells35,71, pre-BI cells of IL-7Rα-deficient patients and from umbilical cord blood mononuclear cells (Supplementary Table 3), cloned into the pGEM-T Easy vector (Promega Benelux BV, Leiden, The Netherlands) and sequenced. From RNA of single-cell sorted naive mature-B cells, cDNA was prepared and gene rearrangements were amplified in nested PCR reactions72. IGH gene rearrangements in BCP-ALL samples were determined as part of the diagnostic work-up73. Sequences were analyzed with ImMunoGeneTics (IMGT) information system (http://imgt.cines.fr/)74. All sequence data have been deposited in GenBank (accession numbers KP771015 - KP771662).
IGH locus contraction was studied with 3D DNA FISH as described previously (details in Supplementary Methods)44,46,47.
Gene expression profiling
Microarray analysis was performed on 3 biological replicates of pro-B and pre-BI subsets that were bulk-sorted from fetal BM, and compared with previously described pro-B and pre-BI subsets from pediatric donors (details provided in Expanded View; ArrayExpress accession number E-MTAB-1422)44. Gene networks and canonical pathways representing key genes were identified using Ingenuity Pathways Analysis (www.ingenuity.com) following criteria of fold change >1.5 and p < 0.05 in differences in gene expression. Gene expression was confirmed with specific RQ-PCR assays (see Supplementary Methods).
Statistics
Statistical significance was calculated by a non-parametric Mann-Whitney U test, an unpaired t-test, χ2 test or one-way ANOVA as indicated in the Figure legends. P values < 0.05 were considered as significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant.
Additional Information
How to cite this article: Rother, M. B. et al. Decreased IL7Rα and TdT expression underlie the skewed immunoglobulin repertoire of human B-cell precursors from fetal origin. Sci. Rep. 6, 33924; doi: 10.1038/srep33924 (2016).
Supplementary Material
Acknowledgments
We thank Drs W.A. van Cappellen and G. Kremers (Optical Imaging Center, Erasmus MC) for support with confocal microscopy, Mr. E. Oole (Biomics, Erasmus MC) for support with gene expression profiling, Mr. S.J.W. Bartol and Mrs H. Bouallouch-Charif for their help with high-speed cell sorting, Dr. E. van Roon (Dept. Pediatrics, Erasmus MC) for help with processing fetal tissue, and Ms. P. Hoogeveen for support with retrospective IGH gene rearrangement analysis in BCP-ALL. This study was performed in the department of Immunology as part of the Molecular Medicine Postgraduate School of the Erasmus MC, and was supported by ZonMW Veni grant 916.116.090 to MCvZ and ZonMW Vidi grant 91712323 to MvdB.
Footnotes
Author Contributions Conceived and designed the experiments: M.B.R. and M.C.v.Z. Performed the experiments: M.B.R., K.J., F.S.v.d.B., R.K. and W.F.J.v.I. Analyzed the data: M.B.R., K.J., F.S.v.d.B., R.K. and O.K.O. Wrote the paper: M.B.R., M.C.v.Z. Reviewed and commented on the manuscript: V.H.J.v.d.V., T.C., M.v.d.B. and J.J.M.v.D.
References
- Schroeder H. W. Jr. et al. Developmental regulation of the human antibody repertoire. Ann N Y Acad Sci 764, 242–260 (1995). [DOI] [PubMed] [Google Scholar]
- Schroeder H. W. Jr, Zhang L. & Philips J. B. 3rd. Slow, programmed maturation of the immunoglobulin HCDR3 repertoire during the third trimester of fetal life. Blood 98, 2745–2751 (2001). [DOI] [PubMed] [Google Scholar]
- Stein K. E. Thymus-independent and thymus-dependent responses to polysaccharide antigens. J Infect Dis 165 Suppl 1, S49–52 (1992). [DOI] [PubMed] [Google Scholar]
- Alt F. W. et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J 3, 1209–1219 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghia P. et al. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med 184, 2217–2229 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossen C., Mansson R. & Murre C. Chromatin Topology and the Regulation of Antigen Receptor Assembly. Annu Rev Immunol 30, 337–356 (2012). [DOI] [PubMed] [Google Scholar]
- Hendriks R. W. & Middendorp S. The pre-BCR checkpoint as a cell-autonomous proliferation switch. Trends Immunol 25, 249–256 (2004). [DOI] [PubMed] [Google Scholar]
- Matsuoka S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007). [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3, 155–168 (2003). [DOI] [PubMed] [Google Scholar]
- van Gent D. C. & van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene 26, 7731–7740 (2007). [DOI] [PubMed] [Google Scholar]
- Alt F. W. & Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci USA 79, 4118–4122 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C. L., Gilfillan S., Thai T. H. & Kearney J. F. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev 175, 150–157 (2000). [PubMed] [Google Scholar]
- Desiderio S. V. et al. Insertion of N Regions into Heavy-Chain Genes Is Correlated with Expression of Terminal Deoxytransferase in B-Cells. Nature 311, 752–755 (1984). [DOI] [PubMed] [Google Scholar]
- Jeong H. D. & Teale J. M. Comparison of the Fetal and Adult Functional B-Cell Repertoires by Analysis of Vh Gene Family Expression. J Exp Med 168, 589–603 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter R. M., Kearney J. F., Chang S. P. & Hood L. E. Developmentally Controlled Expression of Immunoglobulin-Vh Genes. Science 227, 1597–1601 (1985). [DOI] [PubMed] [Google Scholar]
- Shiokawa S. et al. IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol 162, 6060–6070 (1999). [PubMed] [Google Scholar]
- Souto-Carneiro M. M., Sims G. P., Girschik H., Lee J. & Lipsky P. E. Developmental changes in the human heavy chain CDR3. J Immunol 175, 7425–7436 (2005). [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D. et al. Preferential Utilization of the Most Jh-Proximal Vh Gene Segments in Pre-B-Cell Lines. Nature 311, 727–733 (1984). [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Malynn B. A. & Alt F. W. Developmentally Regulated and Strain-Specific Expression of Murine Vh Gene Families. J Exp Med 168, 417–435 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlin M., Schelonka R. L., Bauer K. & Schroeder H. W. Jr. Regulation and chance in the ontogeny of B and T cell antigen receptor repertoires. Immunol Res 26, 265–278 (2002). [DOI] [PubMed] [Google Scholar]
- Schroeder H. W., Ippolito G. C. & Shiokawa S. Regulation of the antibody repertoire through control of HCDR3 diversity. Vaccine 16, 1383–1390 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi E. et al. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci Transl Med 7, 276ra225 (2015). [DOI] [PubMed] [Google Scholar]
- Rother M. B. et al. The Human Thymus Is Enriched for Autoreactive B Cells. J Immunol 197, 441–448 (2016). [DOI] [PubMed] [Google Scholar]
- Pascual V., Verkruyse L., Casey M. L. & Capra J. D. Analysis of Ig H chain gene segment utilization in human fetal liver. Revisiting the “proximal utilization hypothesis”. J Immunol 151, 4164–4172 (1993). [PubMed] [Google Scholar]
- Raaphorst F. M., Langlois van den Bergh R., Waaijer J. L., Vossen J. M. & van Tol M. J. Expression of the human immunoglobulin heavy chain VH6 gene element by fetal B lymphocytes. Scand J Immunol 46, 292–297 (1997). [DOI] [PubMed] [Google Scholar]
- Schroeder H. W. & Wang J. Y. Preferential Utilization of Conserved Immunoglobulin Heavy-Chain Variable Gene Segments during Human Fetal Life. Proc Natl Acad Sci USA 87, 6146–6150 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H. W. Jr., Hillson J. L. & Perlmutter R. M. Early restriction of the human antibody repertoire. Science 238, 791–793 (1987). [DOI] [PubMed] [Google Scholar]
- Vosshenrich C. A., Cumano A., Muller W., Di Santo J. P. & Vieira P. Thymic stromal-derived lymphopoietin distinguishes fetal from adult B cell development. Nat Immunol 4, 773–779 (2003). [DOI] [PubMed] [Google Scholar]
- Kikuchi K. & Kondo M. Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc Natl Acad Sci USA 103, 17852–17857 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz C. A., Harmon I. R., O’Neil J. J., Burchill M. A. & Farrar M. A. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol 172, 4770–4778 (2004). [DOI] [PubMed] [Google Scholar]
- Corcoran A. E., Riddell A., Krooshoop D. & Venkitaraman A. R. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature 391, 904–907 (1998). [DOI] [PubMed] [Google Scholar]
- Noguchi M. et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73, 147–157 (1993). [PubMed] [Google Scholar]
- Puel A., Ziegler S. F., Buckley R. H. & Leonard W. J. Defective IL7R expression in T(−)B(+)NK(+) severe combined immunodeficiency. Nat Genet 20, 394–397 (1998). [DOI] [PubMed] [Google Scholar]
- Kolar G. R., Yokota T., Rossi M. I., Nath S. K. & Capra J. D. Human fetal, cord blood, and adult lymphocyte progenitors have similar potential for generating B cells with a diverse immunoglobulin repertoire. Blood 104, 2981–2987 (2004). [DOI] [PubMed] [Google Scholar]
- van Zelm M. C. et al. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol 175, 5912–5922 (2005). [DOI] [PubMed] [Google Scholar]
- Meffre E. & Salmon J. E. Autoantibody selection and production in early human life. J Clin Invest 117, 598–601 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno C., Montes R., Catalina P., Rodriguez R. & Menendez P. Insights into the cellular origin and etiology of the infant pro-B acute lymphoblastic leukemia with MLL-AF4 rearrangement. Leukemia 25, 400–410 (2011). [DOI] [PubMed] [Google Scholar]
- Ford A. M. et al. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc Natl Acad Sci USA 95, 4584–4588 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemels J. L. et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 354, 1499–1503 (1999). [DOI] [PubMed] [Google Scholar]
- Wasserman R. et al. Predominance of Fetal Type-Dj(H) Joining in Young-Children with B-Precursor Lymphoblastic-Leukemia as Evidence for an Inutero Transforming Event. J Exp Med 176, 1577–1581 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordzij J. G. et al. Composition of precursor B-cell compartment in bone marrow from patients with X-linked agammaglobulinemia compared with healthy children. Pediatr Res 51, 159–168 (2002). [DOI] [PubMed] [Google Scholar]
- Noordzij J. G. et al. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG-deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood 100, 2145–2152 (2002). [PubMed] [Google Scholar]
- Fuxa M. et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev 18, 411–422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. et al. Increased ID2 Levels in Adult Precursor B Cells as Compared with Children Is Associated with Impaired Ig Locus Contraction and Decreased Bone Marrow Output. J Immunol 191, 1210–1219 (2013). [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S. et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell 133, 265–279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodland S. E. et al. IL-7R expression and IL-7 signaling confer a distinct phenotype on developing human B-lineage cells. Blood 118, 2116–2127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh C. E., Jhunjhunwala S., Riblet R. & Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev 19, 322–327 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler H. S. et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol 9, 1388–1398 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. C. et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11, 635–U109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. & Jackson S. P. Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547–551 (2002). [DOI] [PubMed] [Google Scholar]
- Zhou B. B. & Elledge S. J. The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 (2000). [DOI] [PubMed] [Google Scholar]
- Boubakour-Azzouz I., Bertrand P., Claes A., Lopez B. S. & Rougeon F. Terminal deoxynucleotidyl transferase requires KU80 and XRCC4 to promote N-addition at non-V(D)J chromosomal breaks in non-lymphoid cells. Nucleic Acids Res 40, 8381–8391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M. et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol 12, 1212–1220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfe S. A. & Paige C. J. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin Immunol 24, 198–208 (2012). [DOI] [PubMed] [Google Scholar]
- Kee B. L. E. and ID proteins branch out. Nat Rev Immunol 9, 175–184 (2009). [DOI] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol 6, 1079–1086 (2005). [DOI] [PubMed] [Google Scholar]
- Bain G. et al. Both E12 and E47 allow commitment to the B cell lineage. Immunity 6, 145–154 (1997). [DOI] [PubMed] [Google Scholar]
- Ahsberg J. et al. Interleukin-7-induced Stat-5 acts in synergy with Flt-3 signaling to stimulate expansion of hematopoietic progenitor cells. J Biol Chem 285, 36275–36284 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D. & Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J 20, 6394–6403 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. et al. Detection of clonotypic IGH and TCR rearrangements in the neonatal blood spots of infants and children with B-cell precursor acute lymphoblastic leukemia. Blood 96, 264–268 (2000). [PubMed] [Google Scholar]
- Armstrong S. A. et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 30, 41–47 (2002). [DOI] [PubMed] [Google Scholar]
- Malin S. et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 11, 171–179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A. et al. Cloning of a receptor subunit required for signaling by thymic stromal lymphopoietin. Nat Immunol 1, 59–64 (2000). [DOI] [PubMed] [Google Scholar]
- Park L. S. et al. Cloning of the murine thymic stromal lymphopoietin (TSLP) receptor: Formation of a functional heteromeric complex requires interleukin 7 receptor. J Exp Med 192, 659–670 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelonka R. L. et al. Absence of N addition facilitates B cell development, but impairs immune responses. Immunogenetics 63, 599–609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J. F. & Schroeder H. W. Developmental Regulation of D-Beta Reading Frame and Junctional Diversity in T-Cell Receptor-Beta Transcripts from Human Thymus. J Immunol 148, 1230–1239 (1992). [PubMed] [Google Scholar]
- Bodger M. P., Janossy G., Bollum F. J., Burford G. D. & Hoffbrand A. V. The Ontogeny of Terminal Deoxynucleotidyl Transferase Positive Cells in the Human-Fetus. Blood 61, 1125–1131 (1983). [PubMed] [Google Scholar]
- Bonati A., Casoli C., Starcich B. & Buscaglia M. Terminal deoxynucleotidyl transferase (TdT) in human foetuses. An immunofluorescent and biochemical study. Scand J Haematol 31, 447–453 (1983). [DOI] [PubMed] [Google Scholar]
- Deibel M. R. Jr. et al. Expression of terminal deoxynucleotidyl transferase in human thymus during ontogeny and development. J Immunol 131, 195–200 (1983). [PubMed] [Google Scholar]
- Li L. X., Goetz C. A., Katerndahl C. D., Sakaguchi N. & Farrar M. A. A Flt3- and Ras-dependent pathway primes B cell development by inducing a state of IL-7 responsiveness. J Immunol 184, 1728–1736 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J. J. et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17, 2257–2317 (2003). [DOI] [PubMed] [Google Scholar]
- Tiller T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden V. H. & van Dongen J. J. MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol Biol 538, 115–150 (2009). [DOI] [PubMed] [Google Scholar]
- Alamyar E., Duroux P., Lefranc M. P. & Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol 882, 569–604 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.