Abstract
Misfolded α-synuclein amyloid fibrils are the principal components of Lewy bodies and neurites, hallmarks of Parkinson’s disease (PD). Here we present a high-resolution structure of an α-synuclein fibril, in a form that induces robust pathology in primary neuronal culture, determined by solid-state NMR spectroscopy and validated by electron microscopy and X-ray fiber diffraction. Over 200 unique long-range distance restraints define a consensus structure with common amyloid features including parallel in-register β-sheets and hydrophobic core residues, but also substantial complexity, arising from diverse structural features: an intermolecular salt bridge, a glutamine ladder, close backbone interactions involving small residues, and several steric zippers stabilizing a novel, orthogonal Greek-key topology. These characteristics contribute to the robust propagation of this fibril form, as evidenced by structural similarity of early-onset PD mutants. The structure provides a framework for understanding the interactions of α-synuclein with other proteins and small molecules to diagnose and treat PD.
Introduction
Parkinson’s disease (PD) is pathologically characterized by Lewy bodies (LBs) and Lewy neurites (LNs)1, intracytoplasmic aggregates containing α-synuclein (α-syn) fibrils1. Exogenous α-syn fibrils seed LB- and LN-like inclusions in cell culture models2,3, and neuron-to-neuron α-syn transmission propagates PD-like pathology4. Inoculation of preformed α-syn fibrils into wild-type (WT) non-transgenic mice seeds aggregation of endogenous mouse α-syn and reproduces key features of the neurodegenerative cascade5. Additionally, recent rat model studies have established that α-syn fibril strains cause distinct synucleinopathies with differing toxicity profiles6.
Although secondary structures have been examined for several α-syn fibril forms by solid-state NMR (SSNMR) spectroscopy7–10, thus far there is no reported high-resolution three-dimensional (3D) structure. The α-syn monomer (14.5 kDa) is substantially larger than other amyloid peptides or proteins whose structures have been solved, such as HET-s (8.7 kDa)11, a β2-microglobulin fragment (2.5 kDa)12, amyloid-β(1–40) (Aβ(1–40), 4.3 kDa)13–15 and amyloid-β(1–42) (4.5 kDa)16. In addition to size, the highly repetitive secondary structure and residue type degeneracy in α-syn present major challenges for high-resolution structure determination. To address these challenges, we performed a comprehensive structural study of an α-syn fibril form previously reported by our group8,17,18. Utilizing extensive sample preparation (six isotopically labeled samples), data collection (68 multidimensional spectra), and computational analysis (interpretation of >7,500 cross peaks), we determined a single unique conformation in which the core residues are arranged in parallel, in-register β-sheets with a Greek-key topology. The structure was validated with measurements of fibril width, intermolecular stacking and β-sheet spacing by electron microscopy (EM) and X-ray fiber diffraction. These structural insights establish the basis for an improved understanding of α-syn fibril nucleation, propagation, and interactions with small molecules of potential utility for the diagnosis and treatment of PD.
Results
In vitro α-syn fibrils are pathogenic to neuronal cells
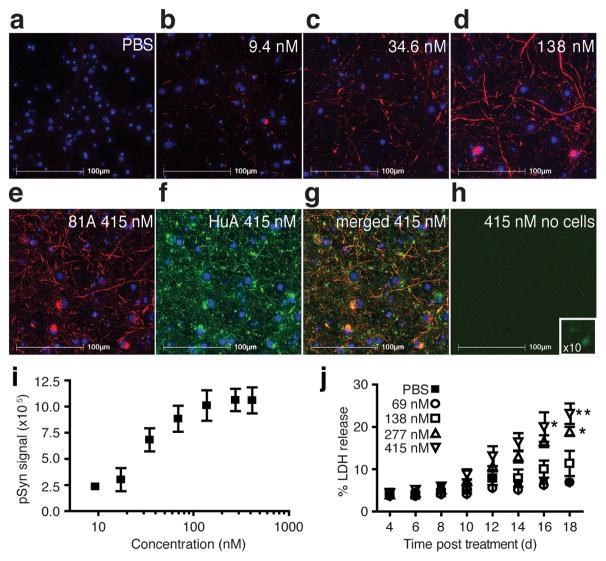
To determine if the α-syn fibrils used in this study for high-resolution 3D structural determination were pathophysiologically indistinguishable from those previously evaluated using cell-based models, we added fibrils to primary hippocampal neurons and showed the induction of insoluble, phosphorylated α-syn (pSyn) inclusions2,3 as indicated by immunostaining with 81A, an antibody specific for phosphorylated Ser129 (Fig. 1a–e). Under these conditions, untreated neurons show no pSyn signal. Importantly, exposure of the coverslips to a second α-syn antibody, HuA, also labeled the insoluble material identified by 81A (Fig. 1f–g) as well as the exogenously added material (Fig. 1h). A dose-dependent effect of fibril treatment on the development of misfolded, neuronal α-syn was confirmed by quantitation of the insoluble pSyn (Fig. 1i). Furthermore, treatment at doses >250 nM showed neuronal injury or death as determined by lactate dehydrogenase (LDH) release into the culture media (Fig. 1j). This result is consistent with our previously published data using fibril preparations in primary culture and WT mice3,5. To ensure that LDH release was not from residual endotoxin present from the protein expression in E. coli, we measured the endotoxin level in the fibrils and determined a range of 0.0094–0.0452 endotoxin units (EU) per treatment (0.0031–0.0151 EU per μg α-syn fibrils), a level below the threshold contributing to α-syn pathology and astrogliosis in α-syn toxicity assays19. Thus, the α-syn fibrils used in this study act as pathological seeds capable of initiating a disease-like cascade.
Figure 1. Effects of pre-formed α-synuclein (α-syn) fibril samples on neurons.
(a–e) Immunocytochemistry of fixed and extracted non-transgenic mouse primary hippocampal neurons labeled for phosphorylated α-syn (pSyn, red) with phosphorylated Ser129 specific (81A) antibody and the deoxyribonucleic acid binding dye 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI, blue) after treatment with (a) phosphate buffered saline (PBS), (b–e) α-syn fibrils in PBS with concentrations (b) 9.4 nM, (c) 34.6 nM, (d) 138 nM, and (e) 415 nM. (f) Immunocytochemical staining of the neurons in condition (e) for total α-syn with the polyclonal antibody, HuA (green), and (g) the co-localization of the total α-syn signal with pSyn (merged 81A and HuA, yellow). (h) Immunostaining of total α-syn (HuA, green) and pSyn (81A, red) staining of the fibrils on a coverslip in the absence of neurons. Inset is a 10X magnification. (i) Quantitation of insoluble, pSyn signal from coverslips treated with increasing doses of α-syn fibrils. Error bars, s.e.m (n = 3 coverslips). (j) Plot of lactate dehydrogenase (LDH) detected in the media of primary neuronal cultures over time for five doses of α-syn fibrils. Error bars, s.e.m. (n = 3 wells) *P=0.05; **P=0.01 by a two-way analysis of variance with Bonferroni correction.
Long-range distance restraints from solid-state NMR spectra
We then proceeded to prepare and validate isotopically labeled WT human α-syn fibril samples (Supplementary Fig. 1) for magic-angle spinning (MAS) SSNMR spectroscopy studies (Supplementary Fig. 2). Importantly, the fibril samples consistently reproduced the morphology observed in our prior reports8,17,18, enabling us to prepare several ~20 mg samples of identical form but with different 13C and 15N isotopic labeling patterns, customized in each case to enable collection of the crucial SSNMR data sets containing unique structural restraints (Table 1). Specifically, we utilized the uniformly-13C, 15N-labeled (U-13C,15N) α-syn fibril (sample A) to confirm and extend the resonance assignments using well-established pulse sequence methods20. We employed the α-syn fibril samples, which we had prepared with 1,3-13C-glycerol or 2-13C-glycerol (samples B and C) as the primary 13C sources in the growth medium21, to enhance spectral resolution and sensitivity; we identified many new long-range restraints in a series of 2D 13C-13C and 3D 15N-13C-13C spectra using dipolar-assisted rotational resonance (DARR) mixing22. We prepared a sample of U-13C, 15N α-syn monomer diluted in natural abundance α-syn prior to fibrillization (sample D), facilitating SSNMR measurements that detect unambiguously intramolecular 13C-13C correlations23 (Supplementary Fig. 2a). Finally, we generated mixtures of 13C-labeled α-syn monomer (from each of the glycerol sources) with 15N-labeled monomer (samples E and F), yielding isotopically mixed fibril samples that permitted the unambiguous determination of intermolecular restraints with 15N-13C transferred-echo double resonance TEDOR according to protocols24 (Supplementary Fig. 2b–e).
Table 1.
Summary of samples used to determine the structure of the α-syn fibril, the structural information each sample provided, and the number of restraints from these samples.
| α-Syn Fibril Sample | ID | Restraint Type | No. Restraints |
|---|---|---|---|
| Uniform-13C, 15N | A | Intraresidue | 807 |
| Interresidue | 562 | ||
| Medium-range | 32 | ||
| Long-range | 5 | ||
|
| |||
| 1,3-13C-Glycerol, 15N | B | Intraresidue | 1320 |
| Interresidue | 1449 | ||
| Medium-range | 595 | ||
| Long-range | 131 | ||
|
| |||
| 2-13C-Glycerol, 15N | C | Intraresidue | 765 |
| Interresidue | 1079 | ||
| Medium-range | 223 | ||
| Long-range | 164 | ||
|
| |||
| 25% U-13C, 15N, 75% natural abundance* | D | Intraresidue | 329 |
| Interresidue | 83 | ||
| Medium-range | 33 | ||
| Long-range Intramolecular | 5 | ||
|
| |||
| 50% 1,3-13C-Glycerol, 50% 15N† | E | Intermolecular Registry | 58 |
|
| |||
| 50% 2-13C-Glycerol, 50% 15N† | F | Intermolecular Registry | 64 |
| Total Assigned Peaks | 7704 | ||
|
|
|||
This isotope labeling scheme detects intramolecular interactions
This isotope labeling scheme detects intermolecular interactions
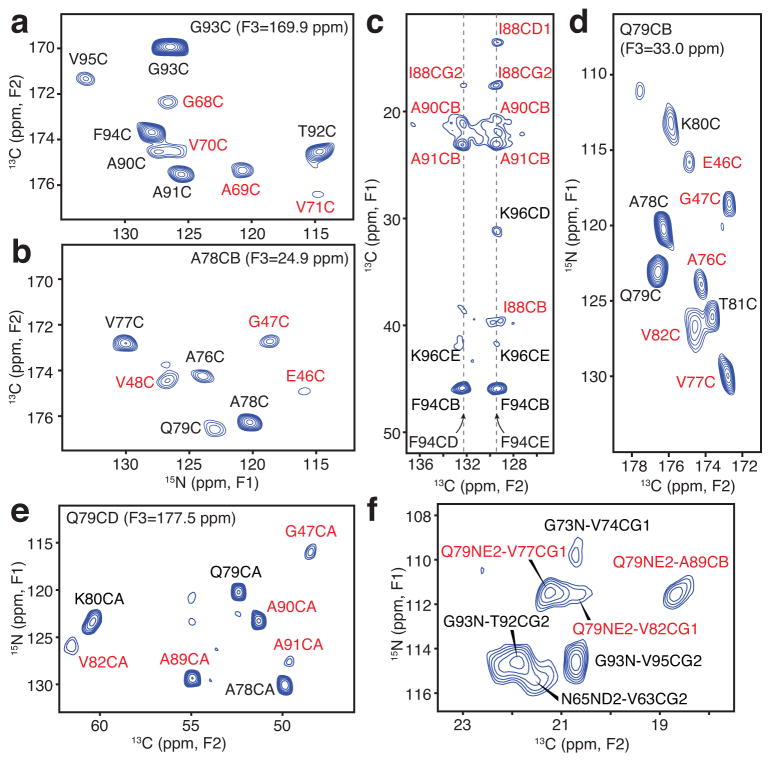
In total, we collected 52 2D and 16 3D SSNMR spectra (Supplementary Table 1), exhibiting 7,704 total assigned cross peaks, from which hundreds of long-range distance restraints were identified (Table 1). Most critically, we collected the 3D 15N-13C-13C and 2D 13C-13C spectra with samples B and C at a series of DARR 13C-13C mixing times from 50 ms to 500 ms, to enable the identification of 180 unambiguous and 80 ambiguous long-range correlations; the ambiguous cross peaks each exhibited less than six possible assignments within the guessing thresholds of ±0.2 ppm. We also assigned 30 unambiguous and five ambiguous long-range 15N-13C correlations in TEDOR experiments24. Taken together, many of the unambiguous correlations reported on the key structural features of the folded monomer within the fibril: residues G68 and A69 are packed close to the G93 carbonyl (Fig. 2a); G47 and V48 are close to the A78 β-carbon (Fig. 2b); the sidechains of I88, A91 and F94 exhibit a number of correlations defining a hydrophobic pocket (Fig. 2c); Q79 is positioned centrally in the core, with backbone interactions to E46, G47, A76, V77 and V82 (Fig. 2d), sidechain 13C-13C correlations with A89 and A90 (Fig. 2e), and a sidechain 15N exhibiting correlations to methyl groups of V77, V82 and A89 (Fig. 2f). Notably, many of these diagnostic correlations were clearly evident at high sensitivity even at short mixing times in the TEDOR (Supplementary Fig. 2b–e) and DARR (Supplementary Fig. 2f–k) data sets. We converted the cross peak intensities into internuclear distance restraints according to our prior studies24,25. In addition, TALOS-N analysis provided 90 backbone dihedral restraints26.
Figure 2. Long-range solid-state NMR structural restraints for an α-synuclein fibril.
(a–b) Two-dimensional (2D) 15N-13C planes from three-dimensional (3D) 15N-13CO-13CX spectrum with 500 ms dipolar-assisted rotational resonance (DARR) mixing22, collected with sample B (500 MHz 1H frequency, 11.111 kHz magic-angle spinning (MAS) rate, signal averaged 181.3 hours). (c) Aromatic-to-aliphatic region of 2D 13C-13C spectrum with 300 ms DARR mixing, collected with sample B (750 MHz 1H frequency, 12.5 kHz MAS rate, signal averaged 12.7 hours). (d) 2D plane from 15N-13CO-13CX spectrum with 500 ms DARR mixing collected with sample B (500 MHz 1H frequency, 11.111 kHz MAS rate, signal averaged 181.3 hours). (e) 2D plane from 15N-13CA-13CX spectrum with 500 ms DARR mixing collected with sample C (500 MHz 1H frequency, 11.111 kHz MAS rate, signal averaged 152.1 hours). (f) Region of 15N-13C spectrum for sample B with 6.4 ms transferred-echo double resonance (TEDOR) mixing24 (600 MHz 1H frequency, 10 kHz MAS rate, signal averaged 8.7 hours). Red labels indicate long-range correlations. distance restraints; black labels represent intraresidue and sequential correlations.
A novel Greek-key topology
To determine a unique 3D structure consistent with all NMR data, we implemented simulated annealing calculations on a set of ten monomers of α-syn within XPLOR-NIH27. We specifically configured the distance potential to consider the explicit contributions from intramolecular as well as intermolecular distances from each of the ten monomers, for the restraints that were measured on undiluted samples. This approach avoids the bias of assuming that individual pairs of nuclei give rise to the observed correlations, and resulted in a single backbone fold consistent with the available experimental data.
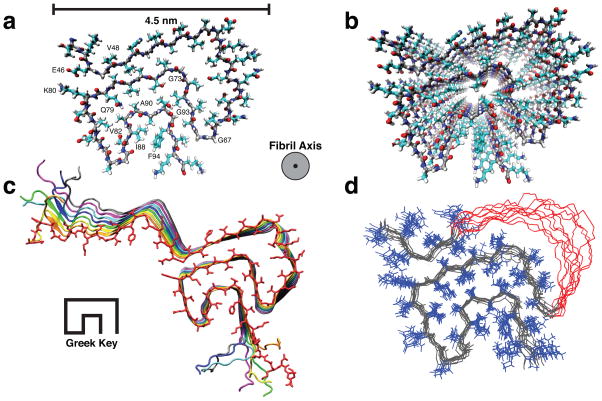
The resulting structure adopts a β-serpentine arrangement with a Greek-key β-sheet topology (Fig. 3a–d). In detail, the fold here exhibits hydrogen bonds in register along the fibril axis, which is orthogonal to the hydrogen bond geometry in a standard Greek-key motif28 (Fig. 3c and Supplementary Fig. 3a,b). The innermost β-sheet of the core includes residues 71–82, which are necessary for fibril formation29. Sidechains in the core are tightly packed (Supplementary Fig. 3c–f) and well-defined in the ensemble (Fig. 3d). Although the stagger of individual sidechains is not uniquely determined13, the lowest energy structures (e.g., Fig. 3a) exhibit short distances between E46 and K80 (Supplementary Fig. 3c,g,h) and among I88, A91 and F94 (Supplementary Fig. 3f,i,j) of neighboring molecules. In two instances, compact residues facilitate an especially close backbone-backbone interaction: (1) A69-G93 bridges the distal loops of the Greek key, and (2) G47-A78 adopts a geometry in which a stable intermolecular salt bridge can be formed between E46 and K80. Hydrophobic sidechain packing among I88, A91 and F94 establish the innermost portion of the Greek key (Supplementary Fig. 3f,i,j). V77, V82, A89, and A90 comprise a hydrophobic pocket within which the Q79 sidechain forms a glutamine ladder along the fibril axis (Supplementary Fig. 3k,l). The turns consist mostly of glycine and alanine residues (G67-G68-A69, G73, G84-A85-G86, A89-A90-A91). Residues 55–62 are disordered, consistent with the incomplete resonance assignments and slightly reduced order parameters in this region as measured by Comellas et al.8. The fibril core (residues 46–54 and 63–96) exhibits a 1.5 Å backbone root-mean-squared deviation (RMSD) and 2.0 Å heavy atom RMSD (Table 2). The structure is consistent with all observed NMR restraints (Supplementary Fig. 4a) and torsion angles for restrained residues (Supplementary Fig. 4b,c).
Figure 3. Three-dimensional structure of an α-synuclein fibril.
(a) View of a central monomer from residues 44 to 96 looking down the fibril axis showing the Greek-key motif of the fibril core. (b) A view of the stacked monomers showing the sidechain alignment between each monomer down the fibril axis (c) Residues 25 to 105 of 8 monomers showing the β-sheet alignment of each monomer in the fibril and the Greek-key topology of the core. (d) Overlay of the 10 lowest energy structures showing agreement of sidechain positions within the core corresponding to an RMSD of 2.0 Å for all heavy atoms for residues 46 to 54 and 63 to 96. Residues 51–57 are shown in red with side chains removed for clarity.
Table 2.
NMR and refinement statistics for protein structures
| α-syn fibril | |
|---|---|
| NMR distance and dihedral constraints per monomer | |
| Distance restraints | |
| Total | 405 |
| Inter-residue | |
| Medium range (1 < |i−j| < 5) | 90 |
| Long-range (|i−j| ≥ 5) | 205 |
| Intermolecular | 110 |
| Total dihedral angle restraints | |
| Phi | 45 |
| Psi | 45 |
| Structure statistics from lowest 32 structures | |
| Violations (mean and s.d.) | |
| Distance restraints ≥ 5 Å (Å) | 0.58 ± 0.05 Å |
| Dihedral angle restraints ≥ 5° (°) | 8.6 ± 3.1° |
| Max. dihedral angle violation (°) | 18.44° |
| Max. distance restraint violation (Å) | 0.64 Å |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.00 ± 0.00 Å |
| Bond angles (°) | 0.45 ± 0.01° |
| Impropers (°) | 0.27 ± 0.02° |
| Average pairwise r.m.s.d.** (Å) | |
| Heavy | 2.04 Å |
| Backbone | 1.48 Å |
Pairwise r.m.s.d. was calculated among 10 refined structures for structured residues 46 to 54 and 63 to 96. Residues 55 to 62 are disordered.
Structural validation
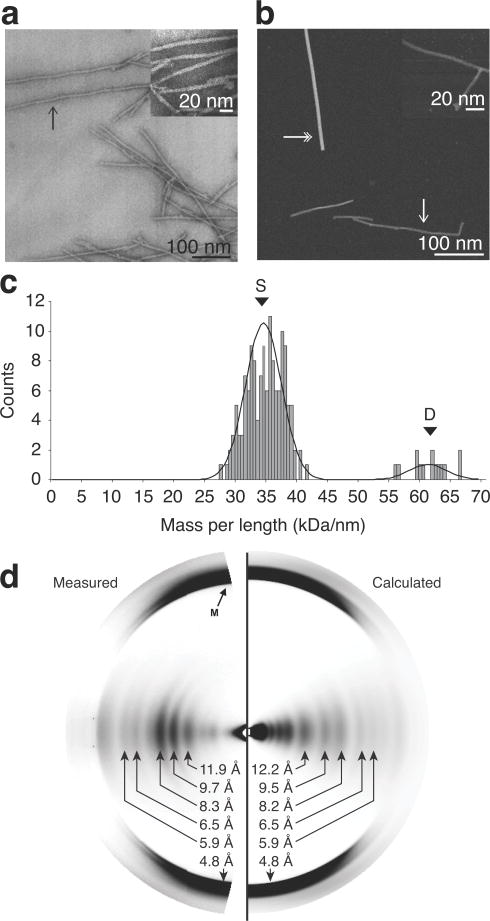
Low-resolution structural characterization by bright-field negatively stained transmission electron microscopy (TEM) (Fig. 4a) of single, isolated fibrils demonstrated features of highly homogeneous, microscopically ordered fibrils with a width of 4.6 ± 0.4 nm. This observation was consistent with our prior results8 and other α-syn fibril studies30–32. Further, we applied scanning transmission electron microscopy (STEM) measurements (Fig. 4b) to obtain a mass-per-length (MPL) ratio33 of 34.5 ± 3.0 kDa/nm (Fig. 4c), supporting an in-register parallel β-sheet structure with one monomer per β-sheet spacing. The high precision of MPL ratio data, combined with narrow SSNMR linewidths (~0.2 ppm) and only one set of resonance assignments, indicated that the fibril samples were highly homogenous and adopted a single core conformation. To test whether the fibrils used in the STEM measurements were in the same conformation as those used in the SSNMR study, we prepared a sample of fibrils using the same buffer and sonication conditions as used for the EM sample preparation and confirmed that the chemical shifts were identical to those throughout the rest of this study (Supplementary Fig. 5a,b).
Figure 4. Validation of α-synuclein (α-syn) fibril structure by microscopy and fiber diffraction.
(a) Bright-field, negatively stained transmission electron microscopy. (b) Dark-field unstained scanning transmission electron microscopy (STEM). Single-headed arrows indicate examples of individual fibrils. Double-headed arrows indicate tobacco mosaic virus (TMV) rods, an internal control for mass-per-length (MPL) ratio determination. Insets are of higher magnification of the fibril samples. (c) Histogram of the distributions of the STEM MPL measurement of unstained, freeze-dried, α-syn fibril. The peak labeled S indicates the mass of a single fibril and the peak labeled D indicates the mass of a double fibril. (d) Experimental and calculated fiber diffraction pattern from α-syn fibrils. Black arrow labeled M indicates the cross-β meridonal diffraction near 4.8 Å resolution.
X-ray fiber diffraction patterns (Fig. 4d) from our fibril form exhibited the archetypal amyloid meridional diffraction at 4.8 Å indicating a cross-β structure. Strong intensity on the equator at about 10 Å resolution is typical of diffraction patterns from amyloids in which β-sheets are stacked together. For our α-syn fibril, the intensity was divided into three strong peaks, whose center of mass corresponds to an average β-sheet separation of 9.1 Å. The 0.02 Å−1 spacing of the three peaks indicates an average diameter of the diffracting unit of ~50 Å. The width of the structured fibril core from Figure 3 is 4.5 nm, and the average β-sheet separation is ~8.9 Å, in excellent agreement with the STEM and fiber diffraction results. A diffraction pattern (Fig. 4d) calculated from the SSNMR structure (Fig. 3) agreed well with the observed pattern at resolutions between 13 Å and 2.5 Å, including the three peaks near 10 Å (Fig. 4d and Supplementary Fig. 5c). Agreement was not as good at lower resolution, near the center of Fig. 4d, but this result is not unexpected, since this part of the pattern was heavily influenced by the disordered terminal segments of the protein, whose structure is not known. The correlation coefficient for the intensities between 0.075 and 0.25 Å−1 (~13 and 4 Å resolution) was 0.77. This is strong support for the SSNMR structure, bearing in mind that a substantial part of the structure, including not only disordered parts but parts expected to have an intermediate degree of order, is uncertain. For comparison, the correlation coefficient is 0.85 between the pattern predicted from the SSNMR structure of the fungal prion HET-s11 and the observed fiber diffraction pattern equator34. Correlation coefficients between computed and experimental patterns for samples of HET-s under reduced humidity are also very close to the correlation coefficient that we observed for α-syn.
Discussion
The α-syn fibril structure exhibits many of the stabilizing features of common amyloid folds11,14,16,35, including parallel in-register β-sheet hydrogen bonding, an intermolecular salt bridge (E46-K80), steric zippers involving the hydrophobic sidechains (such as V49, V77 and V82), a glutamine ladder (Q79) along the fibril axis, and hydrophobic packing of a methyl-bearing and aromatic residues (I88, A91 and F94). Nevertheless, the α-syn fibril structure here is considerably more complex than the earliest predicted β-serpentine models of α-syn fibrils32 and recent computational models36. Most recently, a structural model for the core of α-syn fibrils was proposed by Rodriguez et al.37, on the basis of micro-electron diffraction structures of short peptides simulating the core of α-syn fibrils; this model contains a steric zipper as is commonly found in amyloid fibril structures, but it is comprised of mostly extended β-sheets. In this case, the peptides used lack the residues that most predominately contribute to the folding of the Greek key, including E46, Q79, K80, I88, A91 and F94. Indeed, structural complexity has been identified in general as an important factor in the stability of self-propagating amyloid fibrils38. For α-syn in particular, we note that the especially compact Greek-key topology is facilitated by a large fraction of small, flexible amino acid residues (glycine, alanine or serine) in the core of the fibril. Not only within the KTKEGV repeats, but also in the intervening residues, these sequence motifs enable α-syn to adapt from an intrinsically unstructured protein in solution to a well ordered hairpin of tightly pitched helices in complex with micelles39 or broken helices on lipid vesicles40, to the particular arrangement in the fibril core as observed here. Specifically, more than 40% structured residues in the α-syn fibril core (18 out of 43) are glycine, alanine or serine. We therefore attribute the novel yet highly specific ordering of the fibril core in part to the prevalence of small residues. Thus, the long β-sheets identified in our prior study8 are not fully extended, but adopt a more compact arrangement due to this high prevalence of residues with small sidechains that can maintain dihedral angles characteristic of β-sheet residues while relieving the energetic strain of extended parallel β-sheets, which are rarely longer than seven amino acid residues41.
α-Syn fibrils isolated from the substantia nigra of PD patient brains have a diameter of ~5 nm for single untwisted fibrils42, consistent with the value of 4.6 nm in our structure and further supported by our EM and fiber diffraction data (Fig. 4). The spacing of the three distinct strong peaks on the equator in our fiber diffraction pattern (Fig. 4d and Supplementary Fig. 5c) corresponds to a single, narrow fibril unit. Bousset et al. identified cylindrical fibrils that are substantially wider (13±2 nm)10, which could be attributable to the lateral association of two to three individual protofilaments, as observed in amyloid-β13,35. Other α-syn fibril forms examined by SSNMR exhibit different secondary structure elements from the form studied here, most notably near residues K45, V74 and V82 which in our form are in β-sheets, while in other forms are in turn or loop regions10,43. Based on these observations, it is likely that the additional fibril forms adopt tertiary core structures that differ from the one in this work, accounting for the observed SSNMR, EM, and fiber diffraction data10.
Several missense mutations of α-syn, including A30P, E46K, H50Q, G51D and A53T, have been implicated in autosomal-dominant PD inheritance44–48. Previous work by some of us presented the SSNMR chemical shift assignments of three early-onset PD mutants—A30P (Ref. 49) and E46K and A53T (Ref. 50)—and fibrils grown in the presence of phospholipids51. In the cases of A30P and A53T, we observed that the chemical shifts and secondary structures were nearly identical to those of this form with only small perturbations to sites proximal to the mutation49,50 (Supplementary Fig. 6). In the presence of phospholipids, we again noted that the chemical shifts of the core residues were largely unchanged, with differences localized to the N-terminal residues that are known to directly interact with lipids51. The lack of perturbations in the chemical shifts revealed that the structure of these different fibril samples must be very similar. A previous SSNMR study of fibrils formed from mouse α-syn52 also identified a secondary structure that is highly similar to the form examined in the present study. With the E46K mutant, however, we noticed large chemical shift perturbations for many sites throughout the core of the fibril, suggesting a substantial change in fibril structure50, which is understood readily in the context of our 3D structure, which demonstrates that residues E46 and K80 form an intermolecular salt bridge that is not compatible with the E46K mutant.
We envision that the α-syn fibril structure presented here will enable the systematic development of improved ligands for diagnosis of PD and development of biomarker assays. This idea has been demonstrated in the case of Aβ fibrils implicated in Alzheimer’s disease (AD), where seeding monomeric Aβ with material from AD patients’ brains can produce different strains with structural changes that are associated with different disease progression patterns14. Such high-resolution Aβ fibril structures have enabled structure-based discovery of small molecule ligands that bind longitudinally53. In the case of α-syn fibrils, phenothiazine derivatives have been identified as lead compounds for positron emission tomography imaging54. In the structure, we see exposed surfaces on β-sheets surrounding the central non-amyloid component of the core, including residues 46–67 and 82–86, that may be potential binding regions for such ligands, although a majority of the residues in the non-amyloid component are inaccessible, consistent with proteinase-K digestion studies of α-syn fibrils55. Prior studies have established that two other strains of α-syn fibrils possess different secondary structures, levels of toxicity, and propagation properties,6 which may be due to different exposed regions in their fibril structures10. The detailed 3D structures of these forms therefore will be highly beneficial for comparison in the future. Meanwhile, the structure reported here establishes a basis for understanding of the structural differences among α-syn fibril strains, for improving the mechanistic understanding of templated recruitment and propagation in α-synucleinopathies, and for developing imaging agents and PD drug candidates.
Methods
α-Synuclein fibril sample preparation for solid-state NMR studies
Samples of α-synuclein (α-syn) protein used in this study were expressed and purified as described previously17. The expression protocol of Marley et al.56 was modified for sparse labeling using a glycerol carbon source57 as follows: A pre-culture of E. coli BL21(DE3)/pET28a-AS was prepared in Studier medium M (Ref. 58) containing 2 g/L ammonium chloride, 2 g/L glycerol, 1 mL/L BME vitamins (Sigma-Aldrich no. B6891), and kanamycin. The culture (200 mL/2-L baffled flask) grew with shaking (250 rpm) at 37 °C to a cell density of near saturation (absorbance at 600 nm (A600) of 2.2–2.7). Cells were then collected by centrifugation at 25 °C and resuspended in half the original culture volume of fresh medium M containing 2 g/L 15N-ammonium chloride, and either 4 g/L 1,3-13C-glycerol and 1 g/L natural abundance sodium carbonate or 4 g/L 2-13C-glycerol and 1 g/L sodium 13C-carbonate. The sodium carbonate was added to prevent isotopic dilution due to reversible carboxylase activity57. The resuspended culture (100 mL/1-L flask) was shaken for 30 min at 25 °C, and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside. Cells were harvested at 13 h. The cell density (A600) at induction was in the range 5–6 and at harvest in the range 7–9. Protein was purified using hydrophobic interaction chromatography and size exclusion chromatography17 with a yield of 130–150 mg α-syn per L culture medium. Isotopically-labeled (1,3-13C and 2-13C) glycerol, 15N-ammonium chloride and sodium 13C-carbonate were purchased from Cambridge Isotope Laboratories, Andover, MA. The monomer solution was concentrated to a solution of 15 mg/mL in sodium phosphate buffer (50 mM, pH 7.4) with 0.1 mM ethylenediaminetetraacetic acid (EDTA) and 0.02% sodium azide (w/v), and was fibrillized at 37 °C with shaking at 200 rpm for 3 weeks, after which the fibrils were pelleted, washed with deionized water, dried under nitrogen gas overnight and packed into 3.2 mm rotors as described by Comellas et al.8.
Solid-state NMR spectroscopy and data processing
Magic-angle spinning (MAS) SSNMR experiments were conducted at 11.7 T (500 MHz 1H frequency), 14.1 T (600 MHz), and 17.6 T (750 MHz) on Agilent Technologies, Inc. (Santa Clara, CA) VNMRS (500 and 750 MHz) and Varian Inc. (formerly of Palo Alto, CA and Fort Collins, CO) InfinityPlus (600 MHz) spectrometers using 3.2 mm Balun probes (Varian). Spinning was controlled with a Varian MAS controller to 11,111 ± 3 Hz (500 MHz 1H frequency), 10,000 kHz or 13,333 kHz (600 MHz 1H frequency), and 12,500 Hz (750 MHz 1H frequency). Typical pulse widths were ~2–2.5 μs for 1H, ~3 μs for 13C, and ~5 μs for 15N. We used 75–80 kHz small phase incremental alternation (SPINAL) decoupling during evolution and acquisition optimized according to Ref. 59. Experiments were performed at a variable-temperature (VT) setting of 10 °C, which is calibrated by ethylene glycol to a sample temperature of 12–17 °C. Chemical shifts were externally referenced using adamantane with the downfield peak set to 40.48 ppm60. Chemical shifts for each sample were confirmed to agree with those previously reported8 using 2D 13C-13C, 3D 15N-13C-13C, 13C-15N-13C and 13C-13C-13C correlation experiments.
Spectra were processed with NMRPipe61 with back linear prediction with a polynomial baseline correction to the first dimension. Lorentzian-to-Gaussian apodization, phase-shifted cosine bells, and zero filling were applied in each dimension prior to Fourier transformation and phasing. Spectra were analyzed in Sparky (https://www.cgl.ucsf.edu/home/sparky/; Goddard, T.D. & Kneller, D.G. Sparky 3. University of California, San Francisco).
XPLOR-NIH structure calculations
XPLOR-NIH version 2.33.4 was used for structure calculations. Ten extended monomers of full-length α-syn were aligned along the z-axis with a 20 Å separation from one monomer to the next. Since we observed a single chemical shift for each residue in the core region (residues 30 to 95), these residues were restrained in each monomer using non-crystallographic symmetry restraints implemented using PosDiffPot term in XPLOR-NIH. NOE restraints from samples where it was not possible to determine whether a correlation was inter- or intramolecular were included in a separate potential by setting nMono to 10 within the NOE potential; this approach avoids the bias of assuming that individual pairs of nuclei give rise to the observed correlations. Restraints that were explicitly intermolecular or intramolecular were included in NOE potentials with nMono set to one with a copy of each restraint for each monomer. Because the distance restraints were applied to all subunits, translational symmetry was thus implicitly applied. The initial annealing calculation of 256 structures started at 5,000 K with a high temperature dynamics run for 10 ps or 5,000 steps, whichever comes first, beginning with a 0.001 ps timestep that self adjusts depending on energy conservation from one step to the next. After the high-temperature calculation, the temperature was reduced to 20 K in steps of 20K. At each temperature step, a trajectory was run 0.4 ps or 500 steps, whichever came first, with an initial 0.001 ps timestep. A gyration volume term62 was applied to residues 46–94 of all subunits.
The empirical hydrogen bond database63 and a statistical torsion angle potential64 were utilized in addition to terms for bond lengths, bond angles and improper angles. After the initial structure calculation phase, each structure was refined using slow annealing from 3000 K to 20 K in 4 K steps. Force constants for the restraint terms were ramped as follows: ambiguous distance restraints were ramped from 0.01 to 40 kcal/mol/Å2 in the initial phase and from 40 to 50 kcal/mol/Å2 in the refinement phase; unambiguous distance restraints were ramped from 0.01 to 20 kcal/mol/Å2 in the initial phase and from 20 to 25 kcal/mol/Å2 in the refinement phase; dihedral restraints were ramped from 10 to 150 kcal/mol/rad2 in the initial phase and from 150 to 300 kcal/mol/rad2 during refinement; the gyration volume was scaled from 10−3 to 0.1 during the initial phase and from 0.1 to 5.0 during refinement. From the resulting 256 structures we took the best 32 structures based on their relative energies for further analysis. In each of the structures the backbone and relative sidechain orientations were identical, but because the XPLOR-NIH calculation did not explicitly specify whether a restraint was intermolecular or intramolecular, there was a differing degree of structurally distinct intermolecular sidechain packing (the intrafibril quaternary structural features). Similar quaternary contacts have been observed in previous studies with Aβ (Ref. 13). For instance, the data support an intermolecular sidechain interaction between K80 and E46, but we could not uniquely determine whether the stagger of those sidechains would be +1 or −1. We observed this same phenomenon with the I88-A91-F94 interactions, with three different populations of intermolecular sidechain orientations, one with moderate sidechain interactions with a + orientation, as well as the +1 and −1 sidechain stagger. The structure presented in the main text corresponds to the most highly populated state in the XPLOR-NIH calculations, with a moderate + stagger for the aromatic sidechain of F94 and a +1 stagger for K80. The resulting structure had 77.2% of residues in the most favored Ramachandran space, 18.1% in allowed regions, 3.1% in generous regions, and 1.6% in disallowed regions. All residues with disallowed Ramachandran conformations were in disordered regions or unrestrained termini, which is consistent with the MolProbity65 clash score (4.76) and overall score (1.95) showing that the structure is of high quality except in these few disallowed Ramachandran conformations.
Primary neuronal cultures and fibril treatment
Primary neuronal cultures were prepared from E15–E17 embryos of CD1 mice (Charles River). All procedures were performed according to the US National Institutes of Health Guide for the Care and Use of Experimental Animals and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Dissociated hippocampal neurons were plated onto poly-D-lysine coated 13 mm coverslips in a 24 well plate at 100,000 cells/coverslip and allowed to mature for 10 days. Pre-formed fibril (PFF) treatment was performed at 10 days in vitro, whereby α-syn PFFs were diluted to 2.3 μM or 0.23 μM (33.3 μg/mL or 3.3 μg/mL) in Dulbecco’s phosphate-buffered saline (PBS), calcium and magnesium-free, and sonicated for 5 min on the high setting with a Diagenode Biorupter™ (30 s on, 30 s off, 10 °C bath temp). In total, 11 samples of each dilution were independently prepared. Neurons were then treated with PBS or sonicated PFFs to give final α-syn concentrations of 0, 9.4, 17.3, 34.6, 69.2, 138, 277, and 415 nM. Three coverslips were treated at each concentration. Beginning at 4 days post treatment, 200 μL of the media was carefully removed from each well and transferred to a 96 well plate and frozen for later cytotoxicity testing. Fresh, pre-warmed neuronal media (200 μL) was then added back to each well. Treated neurons were harvested for immunocytochemistry at 18 days post treatment (28DIV). Endotoxin levels were measured for the fibrils in this study using the PierceTM LAL Chromogenic Endotoxin Quantitation Kit per the manufactures instructions and found to be between 0.0094–0.0452 endotoxin unit (EU) per treatment (0.0031–0.0151 EU per μg α-syn).
Immunocytochemistry
Neurons were fixed and permeabilized with pre-warmed 4% paraformaldehyde (PFA) in PBS containing 4% sucrose and 1% Triton X-100 for 15 min to remove soluble proteins. After blocking with 3% bovine serum albumin and 3% fetal bovine serium in PBS for at least 1 hr at room temperature, neurons were incubated with 81A (pSer129 α-syn, ab184674, Abcam®) and HuA (total synuclein, doi: 10.1083/jcb.200403061) overnight at 4 °C followed by staining with appropriate Alexa fluor 594 or 488-conjugated secondary antibodies (Life Technologies, Thermo Fisher Scientific Inc., Waltham, MA, USA, A-11032 and A-11034, 1:1000) for 2 h at room temperature. Coverslips were then mounted on to glass slides with eBioscience Fluoromount G™ with deoxyribonucleic acid binding dye 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) and scanned on a Perkin Elmer Lamina™ scanner. Quantification was performed using Indica Labs HALO™ software ((area occupied × average intensity)/DAPI count) and data is reported as an average of three coverslips. Graphing (Figure 1i) was performed with Graphpad Prism 4™, Error bar, S.E.M.
Cytotoxicity assay
Toxicity was assessed using media sampled from neurons every other day beginning at 4 days post-treatment through 18 days post-treatment using the Pierce™ LDH Cytotoxicity assay kit per the manufacturer’s instructions. Data was recorded using a Molecular Devices Spectramax™ plate reader. Graphing (Figure 1C) was performed using Graphpad Prism 4 and statistical significance was determined with the same software using a two-way analysis of variance with Bonferroni correction. * = p < 0.05, ** = p < 0.01, Error bar, s.e.m.
Preparation of the fibril samples for electron microscopy
After ultracentrifugation, the pellet was resuspended in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.4, 0.02 % sodium azide and 0.1 mM EDTA) and ultracentrifuged. This cycle was repeated several times and the final solution was sonicated (600 volts, 5 amps) three times using 1 min pulses.
Transmission electron microscopy (TEM)
Negatively stained α-syn samples for TEM were prepared by resuspending the fibrils in 20 mM HEPES buffer (pH 7.4, 0.02% sodium azide and 0.1 mM EDTA); Karnovsky’s fixative was added and a drop was placed over Parafilm. A formvar carbon coated grid (300 mesh) was positioned face down on the drop for 15 min to adsorb the fibrils. The grid was then placed for 2.5 min in a drop of 0.2% ammonium molybdate (w/v) for negative staining, wicked in a drop of water and allowed to dry. Electron micrographs were recorded in a Hitachi H600 Transmission Electron Microscope, operating at 75 kV.
Scanning Transmission Electron Microscopy (STEM)
α-Syn samples for STEM were prepared using the wet film approach with freeze-dried specimens. Titanium grids (2.3 mm) were coated with thick holey carbon film that supported a thin carbon film. The thin carbon film was prepared by ultra-high vacuum evaporation onto a freshly cleaved crystal of rock salt and floated on a dish of clean water to avoid non-specific absorption of material. Grids were then positioned face down on the floating thin carbon films and picked so that the thin carbon film retained a droplet of water. This water was exchanged by washing with injection buffer containing 2 μL of tobacco mosaic virus (TMV) as quantitative internal control at a concentration of 100 μg/mL, incubated for 1 min and washed four times. Three different grids were prepared by adding 2 μL of α-syn fibrils at the concentrations of 120 μg/mL, 300 μg/mL and 1200 μg/mL in 20 mM HEPES buffer, respectively, incubated for 1 min and washed ten times with the buffer and water. After the final wash, the grid was pinched between two pieces of paper filters and immediately plunged into liquid nitrogen slush. The grids were then transferred under liquid nitrogen to an ion-pumped freeze dryer with an efficient cold trap, freeze-dried overnight by gradually warming to −80°C and transferred under vacuum to the STEM. Dark-field micrographs were recorded on a custom-built STEM, operating at 40 kV, with a probe focused at 0.25 nm and a sample temperature of −150° C.
Mass-per-length measurement and analysis of STEM images
Dark-field STEM micrographs with 512 × 512 points and a dwell of 30 μs per pixel were analyzed using the PCMass software available from the Brookhaven STEM resource (ftp://ftp.stem.bnl.gov/pub/PCMass32)66. The mass-per-length (MPL) measurements were conducted using boxes of 30 nm length and the appropriate widths from the #21 (TMV) and #55 (amyloid) fitting functions, respectively, from PCMass31. The fitting function was used for alignment and quality control only and had no effect on measured MPL. The resulting data were normalized to the known MPL of TMV (131.4 kDa/nm). Histograms of the α-syn fibril MPL measurement were calculated with 1-kDa/nm bins and the main peak was fitted to a Gaussian distribution using the Marquardt-Levenberg algorithm, as implemented in Sigma Plot (Systat Software; https://systatsoftware.com/products/sigmaplot/). STEM images were also used to measure the variation of the fibril widths in steps of 5-nm along the fibrils using the PCMass software66. At each step, the fibril density was determined by averaging each point with the 5 nm length of fibril in each direction to increase the signal-to noise. As previously demonstrated for HET-s (Ref. 67), fibrils composed of multiple single fibrils wound around each other vary systematically in projected diameter along their length, while for double fibrils consisting of two singlet fibrils, the projected diameters varies from a single fibril diameter to twice that measurement.
X-ray fiber diffraction
Fibers were prepared68 by suspending a 5–10 μl drop of fibril suspension between two glass rods approximately 1.5 mm apart. The fibers were allowed to dry for several days in a closed chamber under high humidity (nominally 100% in equilibrium with water). Diffraction data were collected at beamline 4-2 at the Stanford Synchrotron Radiation Laboratory. Fibers were dusted with calcite and specimen-to-detector distances were determined from the 012 calcite diffraction ring at 3.8547 Å resolution69. Diffraction data were transformed into reciprocal space70 and background subtracted using the program WCEN71. Equatorial intensity plots were produced from processed patterns using WCEN by integrating over a 20° angle centered on the equator. Plots from each diffraction pattern were linearly scaled using a coefficient determined by least-squares fitting of the continuous equatorial diffraction data between 0.075 and 0.25 Å−1 (~13−4 Å).
Diffraction patterns were calculated using DISORDER, a program that calculates Fourier-Bessel transforms72 and simulates disorientation73. DISORDER and its associated documentation are available on request. B-factors of 4000 Å2 were used for disordered regions (residues 1–29 and 114–140) to simulate large disorder. B-factors of 30 Å2 were used for the ordered core (residues 42–98). B-factors of 100 Å2 were used for the remaining residues, to simulate an intermediate degree of order.
Supplementary Material
Acknowledgments
This study was supported by the US National Institutes of Health (NIH) (R01-GM073770 to C.M.R., P50-NS053488 to V.M-Y.L. and P01-AG002132 to G.S.) and utilized solid-state NMR instrumentation procured with support of S10-RR025037 (to C.M.R.) from the US NIH National Center for Research Resources (NCRR). M.D.T., A.J.N. and A.M.B. were members of the US NIH Molecular Biophysics Training Grant at the University of Illinois at Urbana-Champaign (T32-GM008276) and D.J.C. is supported by T32-AG000255. J.M.C. was supported by a US National Science Foundation Graduate Research Fellowship. C.D.S. is supported by the Intramural Research Program of the Center for Information Technology at NIH. The authors thank J. Wall and B. Lin (Brookhaven National Laboratory) for STEM MPL sample preparations and data collection. The Brookhaven National Laboratory STEM was a US NIH-supported Resource Center (P41-EB2181), with additional support provided by the US Department of Energy (DOE), Office of Biological and Environmental Research. TEM images were collected at the Frederick Seitz Materials Research Laboratory Central Facilities, University of Illinois, which are partially supported by the US DOE under grants DE-FG02-07ER46453 and DE-FG02-07ER46471. The Voyager-DE STR MALDI TOF mass spectrometer was purchased in part with a grant from the US NIH NCRR (S10-RR011966). The Stanford Synchrotron Radiation Lightsource (SSRL) is a national user facility operated by Stanford University on behalf of the US DOE, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the US DOE and the US NIH. The authors thank M. Tang (College of Staten Island, New York, NY) for helpful discussions.
Footnotes
Author Contributions
M.D.T. analyzed and collected SSNMR data, performed structure calculations, analyzed structural features and was the primary author on the manuscript. G.C analyzed and collected SSNMR data, performed MPL data analysis and provided input on the manuscript. A.J.N. analyzed and collected SSNMR data, performed initial structure calculations and provided input on the manuscript. D.J.C. performed the immunofluorescence, biochemical and biophysical assays and helped prepare the manuscript. D.A.B. prepared isotopically labeled samples and provided input on the manuscript. K.D.K. aided in sample preparation and manuscript preparation. J.M.C. created scripts for data conversion, analysis of final structures, and provided input on the manuscript. J.K.K. contributed to the sample preparation methods. A.M.B contributed to the mass spectrometry and solution NMR data and analysis. A.K. and W.W. prepared fiber diffraction samples and collected and analyzed fiber diffraction data. G.S. analyzed fiber diffraction data and aided in manuscript preparation. C.D.S. supported the development of structure calculations in XPLOR-NIH. V.M.Y.L., J.M.G. and C.M.R. were the primary investigators that designed the experiments for this study and aided in manuscript preparation, data collection, and interpretation.
Accession Codes
Coordinates have been deposited in the Protein Data Bank under accession number 2N0A and chemical shifts have been deposited to the Biological Magnetic Resonance Bank under entry 25518.
References
- 1.Spillantini MG, et al. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.Luk KC, et al. Exogenous α-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc Natl Acad Sci USA. 2009;106:20051–20056. doi: 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volpicelli-Daley LA, et al. Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron. 2011;72:57–71. doi: 10.1016/j.neuron.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desplats P, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of α-synuclein. Proc Natl Acad Sci USA. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luk KC, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peelaerts W, et al. α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature. 2015;522:340–344. doi: 10.1038/nature14547. [DOI] [PubMed] [Google Scholar]
- 7.Heise H, et al. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comellas G, et al. Structured regions of α-synuclein fibrils include the early-onset Parkinson’s disease mutation sites. J Mol Biol. 2011;411:881–895. doi: 10.1016/j.jmb.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gath J, et al. Solid-state NMR sequential assignments of α-synuclein. Biomol NMR Assign. 2012;6:51–55. doi: 10.1007/s12104-011-9324-3. [DOI] [PubMed] [Google Scholar]
- 10.Bousset L, et al. Structural and functional characterization of two α-synuclein strains. Nat Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasmer C, et al. Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 12.Iwata K, et al. 3D structure of amyloid protofilaments β2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci USA. 2006;103:18119–18124. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu JX, et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schütz AK, et al. Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, et al. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloepper KD, Woods WS, Winter KA, George JM, Rienstra CM. Preparation of α-synuclein fibrils for solid-state NMR: Expression, purification, and incubation of wild-type and mutant forms. Protein Expres Purif. 2006;48:112–117. doi: 10.1016/j.pep.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Kloepper KD, Hartman KL, Ladror DT, Rienstra CM. Solid-state NMR spectroscopy reveals that water is nonessential to the core structure of α-synuclein fibrils. J Phys Chem B. 2007;111:13353–13356. doi: 10.1021/jp077036z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford NJ, et al. Studies of lipopolysaccharide effects on the induction of α-synuclein pathology by exogenous fibrils in transgenic mice. Mol Neurodegener. 2015;10:32. doi: 10.1186/s13024-015-0029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comellas G, Rienstra CM. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annu Rev Biophys. 2013;42:515–536. doi: 10.1146/annurev-biophys-083012-130356. [DOI] [PubMed] [Google Scholar]
- 21.Castellani F, et al. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 22.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 23.Lange A, Luca S, Baldus M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J Am Chem Soc. 2002;124:9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- 24.Nieuwkoop AJ, Wylie BJ, Franks WT, Shah GJ, Rienstra CM. Atomic resolution protein structure determination by three-dimensional transferred echo double resonance solid-state nuclear magnetic resonance spectroscopy. J Chem Phys. 2009;131:095101. doi: 10.1063/1.3211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franks WT, et al. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc Natl Acad Sci USA. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor–NIH for NMR molecular structure determination. Prog Nucl Magn Reson Sp. 2006;48:47–62. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson EG, Thornton JM. The Greek key motif: extraction, classification and analysis. Protein Eng. 1993;6:233–245. doi: 10.1093/protein/6.3.233. [DOI] [PubMed] [Google Scholar]
- 29.Giasson BI, Murray IV, Trojanowski JQ, Lee VM. A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J Biol Chem. 2001;276:2380–2386. doi: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- 30.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant α-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 31.Khurana R, et al. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys J. 2003;85:1135–1144. doi: 10.1016/S0006-3495(03)74550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilar M, et al. The fold of α-synuclein fibrils. Proc Natl Acad Sci USA. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baxa U, et al. Architecture of Ure2p prion filaments: the N-terminal domains form a central core fiber. J Biol Chem. 2003;278:43717–43727. doi: 10.1074/jbc.M306004200. [DOI] [PubMed] [Google Scholar]
- 34.Wan W, Stubbs G. Fiber diffraction of the prion-forming domain HET-s(218–289) shows dehydration-induced deformation of a complex amyloid structure. Biochemistry. 2014;53:2366–2370. doi: 10.1021/bi5002807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petkova AT, et al. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atsmon-Raz Y, Miller Y. A proposed atomic structure of the self-assembly of the non-amyloid-β-component of human α-synuclein as derived by computational tools. J Phys Chem B. 2015;119:10005–15. doi: 10.1021/acs.jpcb.5b03760. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez JA, et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan W, Stubbs G. Fungal prion HET-s as a model for structural complexity and self-propagation in prions. Proc Natl Acad Sci USA. 2014;111:5201–5206. doi: 10.1073/pnas.1322933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human α-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 40.Lokappa SB, Ulmer TS. α-Synuclein populates both elongated and broken helix states on small unilamellar vesicles. J Biol Chem. 2011;286:21450–21457. doi: 10.1074/jbc.M111.224055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penel S, Morrison RG, Dobson PD, Mortishire-Smith RJ, Doig AJ. Length preferences and periodicity in β-strands. Antiparallel edge β-sheets are more likely to finish in non-hydrogen bonded rings. Protein Eng. 2003;16:957–961. doi: 10.1093/protein/gzg147. [DOI] [PubMed] [Google Scholar]
- 42.Crowther RA, Daniel SE, Goedert M. Characterisation of isolated α-synuclein filaments from substantia nigra of Parkinson’s disease brain. Neurosci Lett. 2000;292:128–130. doi: 10.1016/s0304-3940(00)01440-3. [DOI] [PubMed] [Google Scholar]
- 43.Gath J, et al. Unlike twins: an NMR comparison of two α-synuclein polymorphs featuring different toxicity. PLoS One. 2014;9:e90659. doi: 10.1371/journal.pone.0090659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polymeropoulos MH, et al. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 45.Kruger R, et al. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 46.Zarranz JJ, et al. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 47.Appel-Cresswell S, et al. α-Synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov Disord. 2013;28:811–813. doi: 10.1002/mds.25421. [DOI] [PubMed] [Google Scholar]
- 48.Lesage S, et al. G51D α-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann Neurol. 2013;73:459–71. doi: 10.1002/ana.23894. [DOI] [PubMed] [Google Scholar]
- 49.Lemkau LR, et al. Mutant protein A30P α-synuclein adopts wild-type fibril structure, despite slower fibrillation kinetics. J Biol Chem. 2012;287:11526–11532. doi: 10.1074/jbc.M111.306902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemkau LR, et al. Site-specific perturbations of α-synuclein fibril structure by the Parkinson’s disease associated mutations A53T and E46K. PLoS One. 2013;8:e49750. doi: 10.1371/journal.pone.0049750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Comellas G, Lemkau LR, Zhou DH, George JM, Rienstra CM. Structural intermediates during α-synuclein fibrillogenesis on phospholipid vesicles. J Am Chem Soc. 2012;134:5090–5099. doi: 10.1021/ja209019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lv G, et al. Structural comparison of mouse and human α-synuclein amyloid fibrils by solid-state NMR. J Mol Biol. 2012;420:99–111. doi: 10.1016/j.jmb.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 53.Jiang L, et al. Structure-based discovery of fiber-binding compounds that reduce the cytotoxicity of amyloid beta. eLife. 2013;2:e00857. doi: 10.7554/eLife.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bagchi DP, et al. Binding of the radioligand SIL23 to α-synuclein fibrils in Parkinson disease brain tissue establishes feasibility and screening approaches for developing a Parkinson disease imaging agent. PLoS One. 2013;8:e55031. doi: 10.1371/journal.pone.0055031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo JL, et al. Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell. 2013;154:103–117. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 57.Lemaster DM, Kushlan DM. Dynamical mapping of E. coli thioredoxin via 13C NMR relaxation analysis. J Am Chem Soc. 1996;118:9255–9264. [Google Scholar]
- 58.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expres Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 59.Comellas G, Lopez JJ, Nieuwkoop AJ, Lemkau LR, Rienstra CM. Straighforward, effective calibration of SPINAL-64 decoupling results in the enhancement of sensitivity and resolution of biomolecular solid-state NMR. J Magn Reson. 2011;209:131–135. doi: 10.1016/j.jmr.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morcombe CR, Zilm KW. Chemical shift referencing in MAS solid state NMR. J Magn Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- 61.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 62.Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J Am Chem Soc. 1999;121:2337–2338. [Google Scholar]
- 63.Grishaev A, Bax A. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J Am Chem Soc. 2004;126:7281–7292. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 64.Bermejo GA, Clore GM, Schwieters CD. Smooth statistical torsion angle potential derived from a large conformational database via adaptive kernel density estimation improves the quality of NMR protein structures. Protein Sci. 2012;21:1824–1836. doi: 10.1002/pro.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wall J, Simon M. Scanning transmission electron microscopy of DNA-protein complexes. Methods Mol Biol. 2001;148:589–601. doi: 10.1385/1-59259-208-2:589. [DOI] [PubMed] [Google Scholar]
- 67.Sen A, et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- 68.McDonald M, Kendall A, Tanaka M, Weissman JS, Stubbs G. Enclosed chambers for humidity control and sample containment in fiber diffraction. J Appl Crystallogr. 2008;41:206–209. [Google Scholar]
- 69.Effenberger H, Mereiter K, Zemann J. Crystal structure refinements of magnesite, calcite, rhodochrosite, siderite, smithonite, and dolomite, with discussion of some aspects of the stereochemistry of calcite type carbonates. Z für Kristallogr. 1981;156:233–243. [Google Scholar]
- 70.Fraser RDB, Macrae TP, Miller A, Rowlands RJ. Digital processing of fibre diffraction patterns. J Appl Crystallogr. 1976;9:81–94. [Google Scholar]
- 71.Bian W, Wang H, McCullough I, Stubbs G. WCEN: a computer program for initial processing of fiber diffraction patterns. J Appl Crystallogr. 2006;39:752–756. [Google Scholar]
- 72.Chandrasekaran R, Stubbs G. Fibre diffraction. In: Arnold E, Himmel DM, Rossman MG, editors. International Tables for Crystallography, Volume F: Crystallography of Biological Macromolecules. 2. Wiley; Chichester, UK: 2012. pp. 444–450. [Google Scholar]
- 73.Holmes KC, Leigh JB. The effect of disorientation on the intensity distribution of non-crystalline fibres. I Theory. Acta Crystallogr Sect A. 1974;30:635–638. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.