Abstract
Optical Ca2+ indicators are powerful tools for investigating intracellular Ca2+ signals in living cells. Although a variety of Ca2+ indicators have been developed, deciphering the physiological functions and spatiotemporal dynamics of Ca2+ in intracellular organelles remains challenging. Genetically encoded Ca2+ indicators (GECIs) using fluorescent proteins are promising tools for organellar Ca2+ imaging, and much effort has been devoted to their development. In this review, we first discuss the key points of organellar Ca2+ imaging and summarize the requirements for optimal organellar Ca2+ indicators. Then, we highlight some of the recent advances in the engineering of fluorescent GECIs targeted to specific organelles. Finally, we discuss the limitations of currently available GECIs and the requirements for advancing the research on intraorganellar Ca2+ signaling.
Main Text
Ca2+ is an important second messenger that regulates numerous physiological cellular functions (1). The versatility of intracellular Ca2+ signaling relies on the precise control of spatiotemporal Ca2+ dynamics (2). Intracellular organelles play a pivotal role in generating Ca2+ signaling patterns by acting as sinks and sources of Ca2+. Furthermore, intraorganellar Ca2+ concentrations regulate the activities of enzymes residing in each organelle, and disturbances in organellar Ca2+ signaling are implicated in various pathophysiological conditions (3, 4). Thus, investigations into organellar Ca2+ dynamics are indispensable for advancing our understanding of the role of intracellular Ca2+ signals in cell physiology and pathophysiology. Optical Ca2+ indicators, which alter their spectral properties depending on the surrounding Ca2+ concentration, are useful tools for the quantitative analysis of organellar Ca2+ signals in living organisms.
Ideally, organellar Ca2+ indicators should fulfill the following requirements. First, they should localize specifically in the target organelle. Second, they should exhibit strong fluorescence, high signal/noise, and fast kinetics, sufficient to resolve the spatial and temporal dynamics of organellar Ca2+ signals. Third, they should possess affinity for Ca2+ suitable for detecting changes in Ca2+ concentration in the target organelle, because the range of intraorganellar Ca2+ concentrations can vary from nanomolar to submillimolar (see Fig. 1). Furthermore, it is desirable that they exhibit a monophasic Ca2+ titration curve, so that the fluorescence intensity changes are proportionally converted to Ca2+ concentration changes. Fourth, their signals should not be disturbed by organelle-specific environmental factors such as acidic or basic pH, because organelles have unique environments optimized for their functions. Fifth, a frequent requirement is noninterference with the measurement of other fluorescent molecules that may be used simultaneously. Last, the indicators should not be toxic to cells. In the light of these requirements, a wide variety of organellar Ca2+ indicators have been developed.
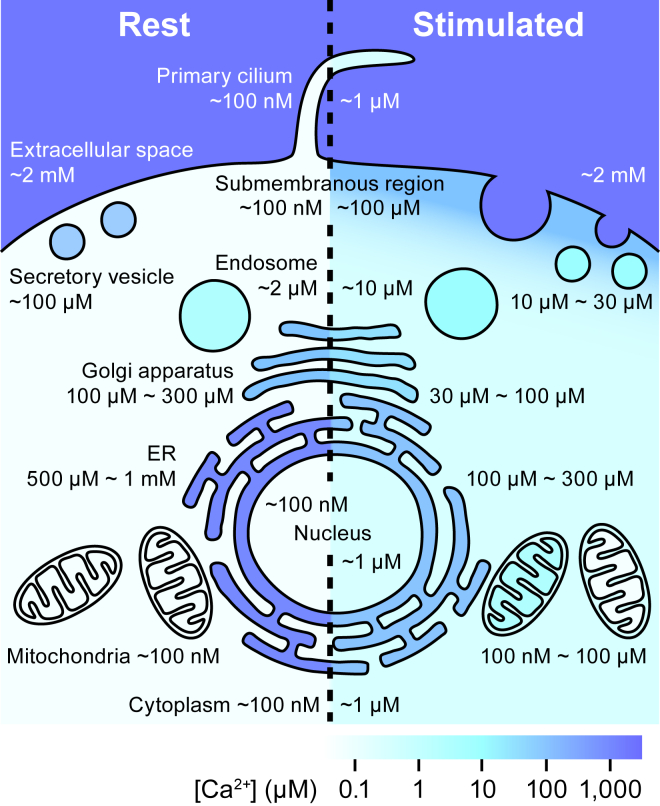
Figure 1.
Ca2+ concentrations in subcellular compartments. Ca2+ concentrations in the intracellular organelles and subcellular compartments in the resting state (left) and after stimulation (right). This represents a summary of the results obtained to date using the organellar genetically encoded Ca2+ indicators reviewed here. To see this figure in color, go online.
Small molecular fluorescent Ca2+ indicators have been used for Ca2+ imaging in several organelles. For example, furaptra and rhod-2 have been used for Ca2+ imaging in the sarco/endoplasmic reticulum (SR/ER) and mitochondria, respectively (5). These dyes are easily loaded into the intracellular space by using their acetoxymethyl esters, enabling imaging with high spatiotemporal resolution. However, these indicators face a drawback in that they cannot be selectively localized in the desired organelle; they are instead inevitably retained in the cytoplasm. Therefore, organellar Ca2+ imaging cannot be performed before the excess cytosolic indicator molecules are removed either by dialysis through a micropipette or by permeabilization of the plasma membrane. These procedures may preclude organellar Ca2+ imaging under physiological conditions. Furthermore, it is also difficult to use these dyes for long-term imaging, because they leak out of the organelles in a time-dependent manner.
The development of genetically encoded Ca2+ indicators (GECIs) is expected to lift the constraints associated with small molecular Ca2+ indicators, because GECIs can be selectively expressed and retained in organelles by fusing organelle-specific targeting sequences to the indicator molecule. The first organelle-targeted GECI was engineered using aequorin, a Ca2+-sensitive bioluminescent protein (LP), which emits light via enzymatic reaction in the presence of both Ca2+ and the cofactor coelenterazine (6, 7). Aequorin derivatives have been successfully used for detecting Ca2+ dynamics with a high signal/noise in several organelles including the ER, mitochondria, nucleus, Golgi apparatus, plasma membrane, peroxisomes, and secretory vesicles (8). Because excitation illumination is not required for their observation, their measurements are free from autofluorescence or phototoxicity. However, their low photon emission rate makes it difficult to obtain subcellular resolution. In addition, the irreversible nature of the luminescence limits the duration of measurements.
GECIs based on fluorescent proteins (FPs) have opened up a new era in organellar Ca2+ imaging. FP-based GECIs have several advantages over the LP-based indicators; they are bright enough to visualize Ca2+ dynamics with subcellular resolution, and their reaction is reversible and does not require any cofactors. Thus, fluorescent GECIs have properties suitable for quantitative measurements of organellar Ca2+ signals. In general, FP-based GECIs are categorized into two classes: dual-FP type and single-FP type. The dual-FP type GECIs, which comprise two FPs linked by a Ca2+-responsive element, are founded on the principle of Förster resonance energy transfer (FRET), a process whereby energy liberated from the donor FP is transferred to the neighboring acceptor FP. The efficiency of FRET depends on the donor-acceptor distance and orientation as well as the spectral overlap between the donor emission and the acceptor excitation. Ca2+ binding to the Ca2+-responsive element causes a conformational change in the indicator, which alters the FRET efficiency between the two FPs. This results in a change in fluorescence intensity of both donor and acceptor FPs, enabling ratiometric measurements. The first and most widely used FRET-type GECI was cameleon, which consists of cyan and yellow FPs as the donor and acceptor, respectively, linked by calmodulin (CaM) and the M13 peptide from the myosin light-chain kinase (9). Derivative indicators of cameleon have been used for Ca2+ imaging in a variety of organelles including the ER, mitochondrial inner matrix, mitochondrial outer membrane, Golgi apparatus, nucleus, secretory granules, plasma membrane, and peroxisomes (9, 10, 11, 12, 13, 14, 15, 16, 17, 18).
The single-FP-type GECIs are made up of circularly permutated FP and a Ca2+-responsive element. Ca2+-dependent conformational change of the indicators alters the protonation state of the fluorophores of the FPs, resulting in a change in fluorescent intensity. The advantages of single-FP-type GECIs compared with the FRET types are that they generally have a higher dynamic range and use a relatively narrower range of excitation and emission wavelengths, which makes it easy to perform simultaneous imaging with other fluorescent molecules. Although most of these indicators allow only intensiometric Ca2+ measurements because they contain only one FP, certain single-FP indicators can be used for ratiometric imaging (19, 20, 21, 22). The single-FP GECIs have been used for Ca2+ imaging in various organelles including the ER, ER membrane, mitochondrial inner matrix, mitochondrial outer membrane, Golgi apparatus, endosomes, nucleus, plasma membrane, and primary cilia (11, 19, 21, 23, 24, 25, 26, 27, 28, 29, 30).
As described above, a large variety of organellar Ca2+ indicators have been developed. In the following sections, we review the design strategies and properties of fluorescent GECIs for each target organelle. A list of organellar GECIs discussed in this review is provided in Table 1, along with their respective characteristics. Intraorganellar Ca2+ concentrations obtained with the organellar GECIs reviewed here are summarized in Fig. 1.
Table 1.
Properties of Fluorescent GECIs for Organelles
| Locus | Name | Typea | Tag | Kd for Ca2+ | Dynamic range | Hill coefficient | pKa (Ca2+ ±) | Ca2+-responsive elements | Fluorophore | Excitation peak (nm) | Emission peak (nm) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ER | YC3er | FRET | Calreticulin, KDEL | 4.4 μM | — | 0.76 | — | CaM, M13 | ECFP, EYFP | 433 | 475, 527 | (9) |
| YC4er | FRET | Calreticulin, KDEL | 0.083, 700 μM | — | 1.5, 0.87 | — | CaM, M13 | ECFP, EYFP | 433 | 475, 527 | (9) | |
| YC3.3er | FRET | Calreticulin, KDEL | — | — | — | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (11) | |
| D1ER | FRET | Calreticulin, KDEL | 0.081, 60 μM | — | 1.18, 1.67 | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (12) | |
| split-YC7.3er | FRET | Calreticulin, KDEL | 130 μM | — | 1.4 | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (35) | |
| D4ER | FRET | Calreticulin, KDEL | 195 μM | — | — | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (36) | |
| mTFP1-YC3.3 | FRET | Calreticulin, KDEL | — | — | — | — | CaM, M13 | mTFP1, Citrine | 462 | 492, 529 | (89) | |
| D1ERCmR2 | FRET | Calreticulin, KDEL | 200 μM | — | 0.5 | — | CaM, M13 | Clover, mRuby2 | 490 | 510, 560 | (37) | |
| T1ER | FRET | Calreticulin, KDEL | — | — | — | — | CaM, M13 | mTurquoise, Citrine | 434 | 474, 529 | (90) | |
| apoK1-er | FRET | Calreticulin, KDEL | 124 μM | — | 1.2 | — | kringle | ECFP, EYFP | 433 | 475, 527 | (34) | |
| G-CEPIA1er | Intensio | Immunoglobulin Vh, KDEL | 672 μM | 4.7 | 1.95 | 8.0/8.7 | CaM, M13 | cpEGFP | 497 | 511 | (24) | |
| R-CEPIA1er | Intensio | Immunoglobulin Vh, KDEL | 565 μM | 8.8 | 1.7 | 6.5, 9.0/8.9 | CaM, M13 | cpmApple | 562 | 584 | (24) | |
| GEM-CEPIA1er | Ratio | Immunoglobulin Vh, KDEL | 558 μM | 21.7 | 1.37 | 6.5, 10.6/6.1 | CaM, M13 | cpEGFP | 391 | 462, 510 | (24) | |
| ER-LAR-GECO1 | Intensio | Calreticulin, KDEL | 24 μM | 10 | 1.3 | 5.4, 8.8/8.6 | CaM, M13 | cpmApple | 561 | 589 | (25) | |
| GCaMPer(10.19) | Intensio | Calreticulin, KDEL | 400 μM | 14 | 1.9 | 7.2/8.0 | CaM, M13 | cpEGFP | 496 | 513 | (32) | |
| Ca-G1-ER | Ratio | Calreticulin, KDEL | 800 μM | 1.8 | 1 | 7.45 | Graft Ca2+ binding site |
EGFP | 398, 490 | 510 | (33) | |
| CatchER | Intensio | Calreticulin, KDEL | 180 μM | 2.3 | 0.94 | 6.91/7.59 | Create Ca2+ binding site |
EGFP | 398, 490 | 510 | (23) | |
| erGAP1 | Ratio | Calreticulin, KDEL | 12 μM | 2.7 | 1 | — | Aequorin | GFP, Aequorin | 403, 470 | 510 | (21) | |
| ER membrane | GCaMP6f-T/J | Intensio | Triadin or junctin 1 | 375 nM | 51.8 | 2.27 | 6.34/8.77 | CaM, M13 | cpEGFP | 497 | 515 | (29) |
| Golgi apparatus | GT-YC3.3 | FRET | N81 of galactosyltransferase type II | 1.5 μM | — | — | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (11) |
| Go-D1cpv | FRET | N69 of sialyl-transferase I | — | — | — | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (42) | |
| medialGo-D1cpv | FRET | N32 of N-acetylglucosaminyl transferase | 27.4 μM | — | — | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (43) | |
| goGAP1 | Ratio | N81 of galactosyltransferase type II | 12 μM | — | 1 | — | Aequorin | GFP variant, Aequorin | 403, 470 | 510 | (21) | |
| Secretory granules | D1-SG | FRET | tPA | 0.081, 60 μM | — | — | — | CaM, M13 | ECFP, Citrine | 433 | 475, 529 | (17) |
| Endosomes | TiVAMP-GEM-GECO1 | Ratio | C61 of TiVAMP | 340 nM | 110 | 2.94 | 6.16 | CaM, M13 | cpEGFP | 390 | 455, 510 | (27) |
| Mitochondria | YC2mt | FRET | 1× COX VIII | 1.24 μM | — | 0.79 | — | CaM, M13 | EGFP, EYFP | 433 | 475, 527 | (10) |
| YC2.1mt | FRET | 1× COX VIII | — | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (10) | |
| YC3.1mt | FRET | 1× COX VIII | 3.98 μM | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (10) | |
| YC4.1mt | FRET | 1× COX VIII | 0.105, 104 μM | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (10) | |
| 2mt8YC2 | FRET | 2× COX VIII | 1.24 μM | — | — | — | CaM, M13 | EGFP, EYFP | 433 | 475, 527 | (13) | |
| 2mt8YC2.1 | FRET | 2× COX VIII | 1.24 μM | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (13) | |
| 2mt8YC2.12 | FRET | 2× COX VIII | 1.24 μM | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (13) | |
| 2mt8YC2.3 | FRET | 2× COX VIII | 1.24 μM | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (13) | |
| 2mtD1cpv | FRET | 2× COX VIII | — | — | — | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (16) | |
| 2mtD2cpv | FRET | 2× COX VIII | 0.097, 7.67 μM | 5.3 | 1.34, 0.77 | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (91) | |
| 2mtD3cpv | FRET | 2× COX VIII | 760 nM | 5.1 | 0.74 | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (91) | |
| 2mtD4cpv | FRET | 2× COX VIII | 49.68 μM | 3.8 | 1.35 | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (91) | |
| 4mtD1cpv | FRET | 4× COX VIII | — | — | — | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (92) | |
| 4mtD3cpv | FRET | 4× COX VIII | 760 nM | 5.1 | 0.74 | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (14) | |
| 4mtD1GO-Cam | FRET | 4× COX VIII | 1.53 μM | — | — | — | CaM, M13 | cpEGFP, mKOκ | 477 | 510, 560 | (93) | |
| GCaMP2-mt | Intensio | 1× COX VIII | 124 nMb | — | 1.6b | — | CaM, M13 | cpEGFP | 491 | 516 | (94) | |
| CEPIA2mt | Intensio | 2× COX VIII | 160 nMb | 1.7b | — | — | CaM, M13 | cpEGFP | 487 | 508 | (24) | |
| CEPIA3mt | Intensio | 2× COX VIII | 11 μMb | 1.6b | — | — | CaM, M13 | cpEGFP | 487 | 508 | (24) | |
| CEPIA4mt | Intensio | 2× COX VIII | 56 μMb | 1.5b | — | — | CaM, M13 | cpEGFP | 487 | 508 | (24) | |
| mitGC3 | Intensio | 1× COX VIII | 542 nM | 12 | 2.73 | 6.6/8.73 | CaM, M13 | cpEGFP | 496 | 513 | (95) | |
| mito-GCaMP5G | Intensio | 1× COX VIII | 460 nM | 45.4 | 2.46 | 6.96/9.14 | CaM, M13 | cpEGFP | 497 | 515 | (96) | |
| mito-GCaMP6s | Intensio | 1× COX VIII | 144 nM | 63.2 | 2.9 | 6.20/9.77 | CaM, M13 | cpEGFP | 497 | 515 | (96) | |
| 2mtGCaMP6m | Intensio | 2× COX VIII | 167 nM | 38.1 | 2.96 | 6.90/8.68 | CaM, M13 | cpEGFP | 497 | 515 | (46) | |
| mitochondrial R-GECO1 | Intensio | 2× COX VIII | 482 nM | 16 | 2.06 | 6.59/8.9 | CaM, M13 | cpmApple | 561 | 589 | (97) | |
| mito-LAR-GECO1.2 | Intensio | 1× COX VIII | 12 μM | 8.7 | 1.4 | 5.8, 8.9/9.0 | CaM, M13 | cpmApple | 557 | 584 | (25) | |
| mito-GEM-GECO1 | Ratio | 2× COX VIII | 340 nM | 110 | 2.94 | 6.16 | CaM, M13 | cpEGFP | 390 | 455, 511 | (20) | |
| mtCamgaroo-2 | Intensio | 1× COX VIII | 5.3 μM | 7 | 1.24 | — | CaM | cpEYFP variant | 490 | 514 | (11) | |
| 2mt8CG2 | Intensio | 2× COX VIII | 5.3 μM | 7 | 1.24 | — | CaM | cpEYFP variant | 490 | 514 | (13) | |
| Ratiometric-Pericam-mt | Ratio | 1× COX IV | 1.7 μM | — | 1.1 | — | CaM, M13 | cpEYFP | 415, 494 | 517 | (19) | |
| mt8PR | FRET | 1× COX VIII | 1.7 μM | — | 1.1 | — | CaM, M13 | cpEYFP | 415, 494 | 517 | (13) | |
| 2mt8PR | FRET | 2× COX VIII | 1.7 μM | — | 1.1 | — | CaM, M13 | cpEYFP | 415, 494 | 517 | (13) | |
| RP3.1mt | Ratio | 1× COX IV | — | — | — | — | CaM, M13 | cpEYFP | 410, 480 | 510 | (98) | |
| mt-inverse pericam 2 | Intensio | 1× COX VIII | 80 nMb | — | 1.4b | — | CaM, M13 | cpEYFP | 503 | 515 | (35) | |
| Mitycam-E67Q | Intensio | 1× COX VIII | 255 nMb | — | — | — | CaM, M13 | cpEYFP | 498 | 515 | (99) | |
| Mitycam-E31Q | Intensio | 1× COX VIII | 1.8 μMb | — | — | — | CaM, M13 | cpEYFP | 498 | 515 | (48) | |
| mito-Case12 | Intensio | 1× COX VIII | 1 μM | 12 | — | 7.2 | CaM, M13 | cpEGFP | 491 | 516 | (100) | |
| mitGAP | Ratio | 1× COX VIII | 200 nM | — | 1 | — | Aequorin | GFP variant, Aequorin | 403, 470 | 510 | (21) | |
| Mitochondrial outer membrane | N33D1cpv | FRET | N33 of TOM20 | — | — | — | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (16) |
| OMM-pcm-ER | Ratio | N30 of mAKAP1 | 460 nM | — | — | — | CaM, M13 | cpEYFP | 415, 494 | 517 | (26) | |
| OMM-pcmD2-ER | Ratio | N30 of mAKAP1 | 3.14 μM | — | — | — | CaM, M13 | cpEYFP | 415, 494 | 517 | (26) | |
| Nucleus | Cameleon-2nu | FRET | NLS | 1.24 μM | — | — | — | CaM, M13 | EBFP, EGFP | 380 | 460, 507 | (9) |
| H2BD1cpv | FRET | Histone 2B | — | — | — | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (16) | |
| CaYang1NLS | FRET | NLS | 1.04, 66.8 μM | — | — | — | CaM, M13 | mTFP1, mCitrine | 462 | 492, 529 | (57) | |
| NLS-GCaMP2 | Intensio | NLS | 146 nM | — | 3.8 | — | CaM, M13 | cpEGFP | 487 | 508 | (56) | |
| NLS-R-GECO1 | Intensio | NLS | 482 nM | 16 | 2.06 | 6.59/8.9 | CaM, M13 | cpmApple | 561 | 589 | (20) | |
| Ratiometric-pericam-nu | Ratio | NLS | 1.7 μM | — | 1.1 | — | CaM, M13 | cpEYFP | 415, 494 | 517 | (19) | |
| nucGAP | Ratio | Nucleoplasmin | 200 nM | — | 1 | — | Aequorin | GFP variant, Aequorin | 403, 470 | 510 | (21) | |
| Peroxisomes | D3cpv-KVK-SKL | FRET | KVK-SKL | 1 μM | — | — | — | CaM, M13 | ECFP, cpVenus | 433 | 475, 528 | (15) |
| Plasma membrane | P2X2-cam | FRET | P2X2 | 3.98 μM | — | — | — | CaM, M13 | EGFP, EYFP variant | 433 | 475, 527 | (10) |
| Cav2.2-TN-XL | FRET | Cav2.2 | 2.5 μM | 1.7 | — | — | Troponin C | CFP, Citrine | 432 | 475, 527 | (66) | |
| Orai-G-GECO1 | Intensio | Orai1 | 749 nM | 25 | 2.97 | 7.57/10.05 | CaM, M13 | cpEGFP | 496 | 512 | (67) | |
| Orai-G-GECO1.2 | Intensio | Orai1 | 1.15 μM | 23 | 2.11 | 7.24/10.44 | CaM, M13 | cpEGFP | 498 | 513 | (67) | |
| Primary cilia | 5HT6-G-GECO1.0 | Intensio | 5-HT6 receptor | 749 nM | 25 | 2.97 | 7.57/10.05 | CaM, M13 | cpEGFP | 496 | 512 | (28) |
| SMO–GCaMP3 | Intensio | Smoothened | 542 nM | 12 | 2.73 | 6.6/8.73 | CaM, M13 | cpEGFP | 496 | 513 | (70) | |
| CTS-G-CaMP3 | Intensio | C-tail of fibrocystin | 542 nM | 12 | 2.73 | 6.6/8.73 | CaM, M13 | cpEGFP | 496 | 513 | (72) | |
| Arl13b-GCaMP5 | Intensio | Arl13b | 447 nM | 45.4 | 2.46 | 6.58/8.61 | CaM, M13 | cpEGFP | 496 | 513 | (71) | |
| Arl13b-GCaMP6 | Intensio | Arl13b | 144 nM | 63.2 | 2.9 | 6.20/9.77 | CaM, M13 | cpEGFP | 496 | 513 | (71) |
AKAP1, A-kinase anchor protein 1; COX, cytochrome c oxidase; NLS, nuclear localization sequences; TOM20, translocase of outer mitochondrial membranes 20 kDA; tPA, tissue plasminogen activator.
Single-FP-type GECIs are classified into two groups: intensiometric (Intensio) and ratiometric (Ratio) indicators.
Measured at pH 8.0.
Sarco/endoplasmic reticulum
The SR/ER accumulates Ca2+ in its luminal space and acts as a major source of Ca2+ for generating intracellular Ca2+ signals. Indeed, Ca2+ release from the SR/ER increases cytosolic Ca2+ concentrations both in excitable and nonexcitable cells, and it also regulates a variety of cell functions, such as contraction, fertilization, development, vesicular secretion, and synaptic plasticity (1). Furthermore, the Ca2+ level within the ER is an important factor in ER function because ER-residing chaperones require Ca2+ for their activities (31). In contrast, Ca2+ depletion of the ER triggers ER stress through the accumulation of misfolded proteins, causing apoptosis (3). Consequently, much effort has been devoted to the development of GECIs for visualizing ER Ca2+ dynamics.
To achieve selective localization of GECIs in the lumen of the ER, an ER-targeting sequence and the ER retention signal KDEL were attached to the N- and C-terminus of the GECIs, respectively. A high value of apparent dissociation constant (Kd) is a critical factor for ER GECIs because free Ca2+ concentrations in the ER lumen may reach the submillimolar range, which is ∼10,000 times higher than that in the cytoplasm (Fig. 1). Introduction of mutations in the Ca2+-responsive element of the cytosolic GECIs has been used to increase the Kd value, but such alterations often have an adverse effect in decreasing the dynamic range of these indicators. Several strategies have been employed to optimize these contradictory effects, such as substituting multiple amino acids in the Ca2+-binding sites, using Ca2+-responsive elements derived from endogenous ER-residing Ca2+-binding proteins, and designing artificial Ca2+-binding sites (12, 23, 24, 25, 32, 33, 34).
The first fluorescent ER GECIs to be developed were the cameleon-type indicators, YC3er and YC4er (9). To lower the Ca2+ affinity of their parent GECI, YC2, YC3er, and YC4er contain point mutations in the Ca2+-binding sites of CaM within the indicator molecule. YC3er has a Kd of 4.4 μM, which is too low to faithfully report the entire range of ER Ca2+ dynamics. YC4er has a biphasic Ca2+ titration curve with Kd values of 0.083 and 700 μM, the higher of which is sufficient to detect ER Ca2+ dynamics, but with a limited dynamic range. YC4er was subsequently improved to generate D1ER by redesigning the binding interface between CaM and M13 to minimize nonspecific interactions with endogenous CaM (12), although it also has a biphasic Ca2+ titration curve (Kd = 0.81 and 60 μM). Further improvements have led to the generation of low-affinity variants such as D4ER (Kd = 195 μM) and split-YC7.3er (Kd = 130 μM), and a red-shifted variant named D1ERCmR2, by substituting CFP and YFP with Clover and mRuby FPs, respectively (35, 36, 37). In parallel with these improvements of the cameleon-type GECIs, a unique FRET-type ER GECI named “apoK1er” has been developed (34). Instead of CaM and M13, apoK1er employs a kringle domain as the Ca2+-responsive element, which is derived from apolipoprotein A and shifts from a linear form to a compact structure via the Ca2+-dependent activities of ER-resident chaperones. This approach has the advantage that Ca2+ buffering within the ER is not affected by the expression of the Ca2+ indicator because apoK1er itself does not bind Ca2+. However, it also has the disadvantage that the indicator is sensitive not only to Ca2+, but also to the redox state of the ER.
In addition to these FRET-type ER GECIs, several types of single-FP ER GECIs have been developed. The first report of such indicators is Ca-G1-ER, which was created by grafting a Ca2+-binding motif from CaM into fluorophore-sensitive locations in EGFP (33). Ca-G1-ER has a low sensitivity toward Ca2+ with a Kd of ∼600–800 μM. Ca2+ binding to Ca-G1-ER results in an increase in fluorescence with excitation at 398 nm and a decrease with excitation at 490 nm. The ratio of the fluorescence intensity at these two excitation wavelengths increases by 80% upon Ca2+ binding. Later, the same research group developed CatchER, which also has a Ca2+-binding site introduced near the fluorophore in EGFP. The Ca2+-binding site was created by introducing point mutations instead of CaM grafting (23). CatchER has a Kd of 180 μM, fast kinetics, and a dynamic range value of 230%.
A recent entry to this category is a family of CEPIAs (24), generated on the basis of single-FP-based cytosolic GECIs (GECOs) (20). CEPIA indicators were generated by introducing point mutations in the Ca2+-binding domains of CaM of GECOs, and have low Ca2+ affinities (Kd = 500–700 μM) favorable for detecting ER Ca2+ dynamics. CEPIAs have three unique features. First, CEPIAs have three color variants, G-CEPIA1er (green), R-CEPIA1er (red), and GEM-CEPIA1er (blue and green), utilizing the color palette of GECOs. Second, they have high signal/noise and large dynamic ranges (470–2100%), enabling visualization of ER Ca2+ dynamics with subcellular resolution such that wavelike Ca2+ release from the ER could be visualized for the first time (Fig. 2) (24). Third, they allow the simultaneous use of other indicators, enabling multiparametric measurements such as simultaneous Ca2+ imaging in three subcellular compartments, i.e., the ER, mitochondria, and cytosol (Fig. 3) (24). Owing to these characteristics, CEPIAs have been successfully used to shed new light on ER Ca2+ dynamics, such as the importance of Ca2+ diffusion within the ER (Ca2+ tunneling) for the replenishment of ER Ca2+ after synaptic inputs in cerebellar Purkinje cells (38), and abnormal ER Ca2+ handling associated with disease-related mutations of the ryanodine receptor Ca2+ release channel (39). Other groups also reported ER GECIs based on GECO or its analog GCaMP3. ER-LAR-GECO1 was generated by combining the strategies of random and site-directed mutagenesis of the CaM-M13 interaction sites in R-GECO1, resulting in a Kd of 24 μM and a dynamic range of 1000% (25). GCaMPer (10.19) was created by introducing multiple mutations in the Ca2+-binding sites of GCaMP3, yielding a Kd of 400 μM and a dynamic range of 1400% (32).
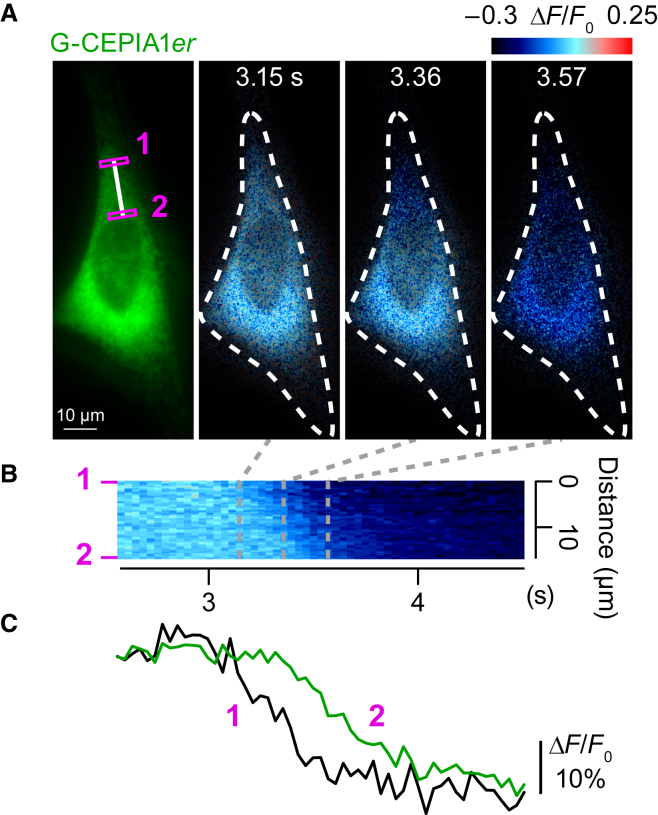
Figure 2.
High spatiotemporal resolution of ER Ca2+ release using CEPIA. (A) Time-lapse images of a wavelike decrease in the ER Ca2+ concentration in a HeLa cell visualized with G-CEPIA1er. Perfusion of 10 μM histamine is started at 0 s. (B) Time-course of ER Ca2+ dynamics along the white line indicated in (A). (C) Comparison of ER Ca2+ dynamics in the two regions of interest indicated in (A). The fluorescence intensity is normalized by the initial intensity. These results show that Ca2+ release from the ER initiates at the tips of cells, and then it propagates to the perinuclear region. (Black line) Region 1; (green line) region 2. This figure is adapted from Suzuki et al. (24). To see this figure in color, go online.
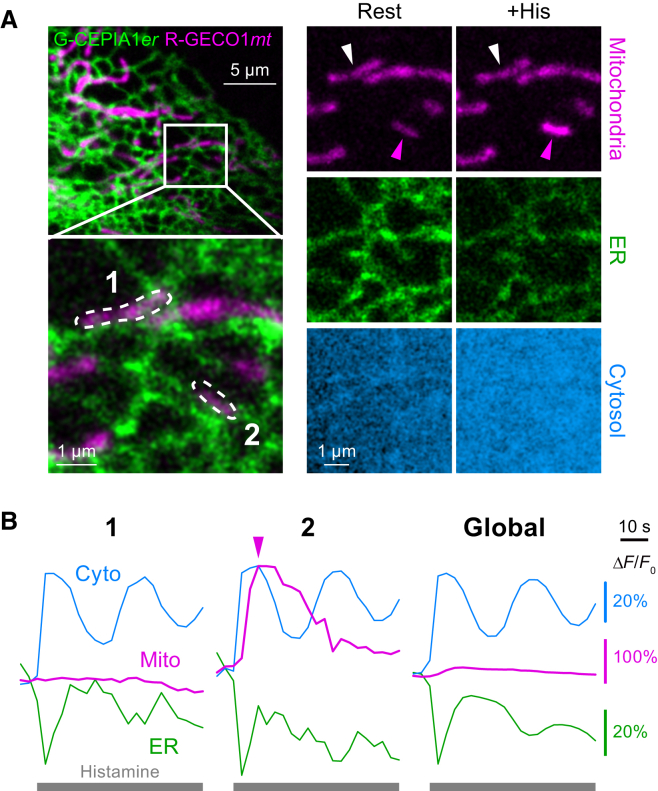
Figure 3.
Simultaneous imaging of Ca2+ signal in three subcellular compartments. (A) Ca2+ response of a HeLa cell in the resting state (upper) and after 10 μM histamine application (lower) in the mitochondria (left), ER (middle), and cytosol (right) visualized with R-GECO1mt, G-CEPIA1er, and GEM-GECO1, respectively. (B) Time-courses of Ca2+ signal in the mitochondria (magenta), ER (green), and cytosol (blue) within or surrounding the two regions of interest indicated in (A). These results show that there is a considerable intracellular inhomogeneity in mitochondrial Ca2+ signals after agonist-induced Ca2+ release from the ER, while homogeneous responses are observed in the cytosol and the ER. This figure is adapted from Suzuki et al. (24). To see this figure in color, go online.
Another entry to the ER GECIs is erGAP1, which is a derivative of GAP, a fusion protein of aequorin and GFP (21). Although aequorin is a Ca2+-sensitive bioluminescent protein, GAP is a fluorescent indicator and does not require the cofactor coelenterazine. The unique feature of GAP is that it has dual excitation peaks at ∼403 and ∼470 nm, and a single emission peak at ∼510 nm, enabling ratiometric measurements. The ratio of the fluorescence intensity at the two excitation wavelengths increases by 170% upon Ca2+ binding. Although it has not been fully understood how GAP detects changes in Ca2+ concentration, it is assumed that the aequorin molecule functions as a Ca2+-responsive element inducing a conformational change in the adjacent GFP. The Ca2+ affinity of GAP was decreased from a Kd of 200 nM to that of 12 μM by substituting amino acids in the Ca2+-binding sites of aequorin to generate erGAP1.
In addition to the GECIs for intra-ER Ca2+ imaging, another type of GECI was developed to visualize Ca2+ signals in proximity of the ER membrane. To detect Ca2+ release events via the ryanodine receptor Ca2+ release channels, GCaMP6f, a GECI for cytosolic Ca2+ measurement, was fused with triadin 1 or junctin, which are membrane proteins colocalizing with ryanodine receptor in the junctional regions of the SR in cardiac myocytes. GCaMP6f-T/J were successfully used to visualize Ca2+ nanosparks, which are ∼50 times smaller in volume than conventional Ca2+ sparks (29).
Acidic organelles
Similar to the ER, acidic organelles such as the Golgi apparatus (pH ≈ 6.0–6.7), endosomes (pH ≈ 5.5–6.5), and secretory granules (pH ≈ 5.5) engage in intracellular Ca2+ signaling by acting as Ca2+ sources (40, 41). Ca2+ is also thought to be important in the functioning of these acidic organelles, but it remains challenging to visualize Ca2+ signals in these organelles with fluorescent indicators because of their acidic environment, which often reduces the brightness of fluorescent indicators.
The first fluorescent GECI specifically developed for acidic organelles was GT-YC3.3 for the Golgi apparatus (11). GT-YC3.3 is a cameleon-type FRET indicator, in which YFP is replaced by a less pH-sensitive EYFP variant, Citrine. GT-YC3.3 was targeted to the Golgi by incorporating the N-terminal sequence of type II galactosyltransferase, and was used to successfully detect Ca2+ release from the Golgi upon depletion of the luminal Ca2+ with ionomycin, a Ca2+ ionophore, although its affinity to Ca2+ (Kd = 1.5 μM) appears rather high for Golgi Ca2+ concentrations. Later, cameleon-type Golgi GECIs with lower Ca2+ affinities were developed: Go-D1cpv and medial Go-D1cpv localized in the trans and medial Golgi through incorporation of the N-terminal sequence of sialyl-transferase I and 1,6 n-acetyl glucosaminyl transferase, respectively (42, 43). These indicators showed that Ca2+ uptake into the Golgi depends on both secretory pathway Ca2+ ATPase1 and sarco/endoplasmic reticulum Ca2+ ATPase, but their activities are differently regulated between the trans and medial Golgi. In addition, these indicators suggest that there is a gradient in luminal Ca2+ concentration among the ER, cis-Golgi, and trans-Golgi (Fig. 1). Recently, a single-FP-type Golgi GECI, goGAP1, was developed by modifying erGAP1 to utilize its low sensitivity to pH (21).
Fluorescent GECIs for visualizing Ca2+ signals in the lumen of secretory granules (SGs) and endosomes have also been developed. D1-SG is based on the cameleon-type GECI D1 attached to tissue plasminogen activator, which localizes in the SGs (17). Measurements using D1-SG suggest that a high concentration of Ca2+ (∼69 μM) is accumulated in the SGs, and that the Orai1 channel on the SG membranes is important for Ca2+ release from the SGs. For Ca2+ imaging in the endosomes, TiVAMP-GEM-GECO1 was developed by fusing single-FP-type ratiometric indicator GEM-GECO1 with tetanus-insensitive vesicle-associated membrane protein (TiVAMP), a soluble NSF attachment protein receptor protein localized in the endosomes (27). TiVAMP-GEM-GECO1 shows that the luminal Ca2+ concentration of the endosomes is maintained at ∼2 μM (Fig. 1), and that endosomes take up Ca2+ and act as a Ca2+ buffer in response to extracellular glucose stimulation in β-cells.
Mitochondria
Mitochondria, which are primarily involved in oxidative metabolism and cell survival, participate in intracellular Ca2+ signaling both as modulators and as sensors (4). In response to cytosolic Ca2+ elevation via Ca2+ release from the ER or to Ca2+ influx from the extracellular space, mitochondria take up Ca2+ into the matrix and buffer cytosolic Ca2+ elevation to prevent intracellular Ca2+ overload. Mitochondrial Ca2+ is also important for controlling the mitochondrial metabolism itself, because the activities of mitochondrial enzymes for ATP production rely on the mitochondrial Ca2+ concentration (44). Abnormal mitochondrial Ca2+ dynamics are involved in cell death or autophagic degradation of the mitochondria, which are implicated in several neurodegenerative diseases (45). Furthermore, loss-of-function mutations in MICU1, one of the regulator proteins of the mitochondrial Ca2+ uniporter, cause brain and muscle disorders in humans (46), even though mice lacking mitochondrial Ca2+ uniporter exhibit no severe phenotypes (47). Thus, mitochondrial Ca2+ is a critical parameter for cell function, and much attention has been paid to the development of mitochondrial fluorescent GECIs.
Selective localization of GECIs in mitochondria can be achieved by attaching a mitochondrial-targeting sequence (MTS) derived from human cytochrome c oxidase subunit VIII or cytochrome c oxidase subunit IV, to N-terminus of the indicators (10, 19). It has been reported that tandem repeats of the MTS enhance the specificity of the localization of these indicators (13). Because the resting Ca2+ level in mitochondria is assumed to be <100 nM, which is almost equal to that in the cytoplasm (Fig. 1), a number of cytosolic GECIs fused with the MTS have been successfully used to detect mitochondrial Ca2+ dynamics. However, Ca2+ affinity is an important parameter even for mitochondrial GECIs because mitochondrial Ca2+ concentration can reach up to ∼100 μM after certain types of stimulation (4) (Fig. 1). To this end, mitochondrial GECIs possessing low Ca2+ affinity have been generated by modifying their Ca2+-responsive element (10, 14, 16, 24, 25, 48).
In addition to these factors, pH sensitivity is another important factor for mitochondrial GECIs. pH in the mitochondrial matrix is estimated at ∼8.0 (40), but direct visualization of spatiotemporal pH dynamics using a genetically encoded mitochondrial pH indicator revealed two unique features of mitochondrial matrix pH: (1) an acidification during cytosolic Ca2+ elevation by the activation of Ca2+/H+ transporters and (2) a spontaneous and transient alkalization, named pH flash (49, 50). These mitochondrial pH changes have the potential to cause artifactual fluctuations in the fluorescent intensity of pH-sensitive GECIs. This problem may be alleviated by using mitochondrial GECIs with high pH stability (14, 21).
Apart from the mitochondrial matrix, GECIs for visualizing Ca2+ dynamics in the proximal region surrounding the mitochondrial outer membrane (OMM) have also been created (16, 26). Mitochondria form close contacts with the ER, and this ER-mitochondrial junction is presumed to be important for local regulation of mitochondrial function. In fact, GECIs targeted to the mitochondrial matrix have revealed that considerable heterogeneity is observed in mitochondrial Ca2+ signals in response to global elevation of cytosolic Ca2+ levels via the activation of inositol 1,4,5-trisphosphate receptors (19, 24, 51). OMM-attached GECIs have been developed by fusion with the OMM-targeting sequence from A-kinase anchor protein 1 or translocase of outer mitochondrial membrane, 20 kDa. These GECIs have shown that the surface of the mitochondria may be exposed to higher Ca2+ concentrations than that of the bulk cytosol in response to Ca2+ release from the ER (16, 26).
Nucleus
Ca2+ is one of the key molecules regulating gene transcription. Cytosolic Ca2+ elevation activates Ca2+-sensitive kinases or phosphatases in the cytoplasm and induces translocation of transcription factors from the cytoplasm to the nucleus (52). A Ca2+ increase in the nucleus also directly enhances gene transcription by deactivating Ca2+-dependent repressors (53). Thus, nuclear Ca2+ dynamics has gathered much attention. Because Ca2+ concentrations in the nucleus are assumed to be similar to that in the cytosol (Fig. 1), nuclear GECIs can be generated by attaching nuclear localization sequences to cytosolic GECIs. Actually, a variety of fluorescent indicators including FRET-type and single-FP-type GECIs have been used to visualize nuclear Ca2+ signals, showing that the nuclear Ca2+ signal is mostly synchronized with the cytosolic Ca2+ signal (9, 16, 19, 21, 54, 55, 56, 57).
Peroxisomes
Peroxisomes are multifunctional organelles regulating the oxidation of fatty acids, biosynthesis of lipids, and reduction of reactive oxygen species (58). Similar to other organelles, it is likely that peroxisomes play several roles in intracellular Ca2+ handling or that peroxisomal Ca2+ regulates enzyme activities. However, little information is currently available on the dynamics and function of Ca2+ in peroxisomes. A cameleon-based FRET GECI for peroxisomes was recently developed, named D3cpv-KVK-SKL (15). It is targeted to the peroxisomes by inserting a three-amino-acid positively charged sequence, Lys-Val-Lys (KVK), and a canonical peroxisomal-targeting signal sequence, Ser-Lys-Leu (SKL), at the C-terminus. Measurements using D3cpv-KVK-SKL show that peroxisomal Ca2+ increases in response to cytosolic Ca2+ increases, suggesting that peroxisomes function as a Ca2+ buffer. Further research is required to clarify the regulatory mechanisms and biological roles of peroxisomal Ca2+ dynamics.
Subplasmamembrane
In subplasmamembranous domains, Ca2+ signals regulate a variety of biological processes, such as vesicular secretion and ion channel modulation (59). Ca2+ influx from the extracellular space via Ca2+ channels elevates the local Ca2+ concentration up to 100 μM beneath the plasma membrane, resulting in the generation of a Ca2+ concentration gradient between the vicinity of the plasma membrane and the bulk of the cytosol (60) (Fig. 1). This suggests that Ca2+ signals in the subplasmamembranous regions can be separately regulated from those in the cytosol. Therefore, the subplasmamembranous domains have been one of the major targets of GECIs, and several types of GECIs have been localized to the plasma membrane by fusion with plasma membrane-anchoring motifs, such as myristoylation, palmitoylation, or farnesylation tags (18, 61), or with plasma membrane-localized transporters (62, 63). GEGIs fused with synaptic proteins, such as synaptosome-associated protein of 25 kDa (SNAP25), synaptophysin, and synaptobrevin, have been also used to detect microdomain Ca2+ signals in the synapse (18, 64).
Recently, optical recordings of channel activities on the plasma membrane have been reported, whereby GECIs were localized within close proximity of Ca2+ channels by a direct fusion with the channel proteins. P2X2-cam is a purinergic receptor fused with a FRET-type GECI, YC3.1, which successfully detected transmitter-gated Ca2+ currents both in vitro and in vivo (65). Another group successfully detected nanodomain Ca2+ signals in the surrounding region of CaV2.2, a voltage-dependent Ca2+ channel, by fusion with a FRET-typed GECI, TN-XL (66). A single-FP-typed GECI, G-GECO, was also used by attaching it to Orai1, a Ca2+ release-activated Ca2+ channel on the plasma membrane, and results from studies with G-GECO revealed that Ca2+ influx through Orai1 makes several characteristic patterns, such as rapid transients and periodic oscillation (67).
Primary cilia
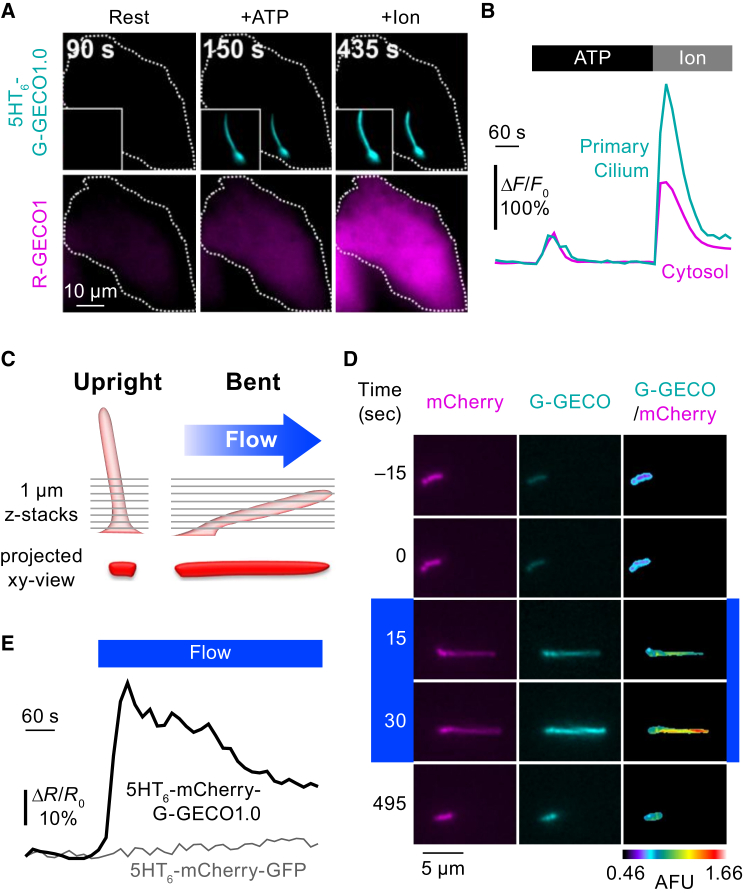
A primary cilium is a solitary hairlike organelle on the cell surface that functions as a sensor for diverse chemical and mechanical stimuli (68, 69). Emerging evidence suggests that Ca2+ acts as a principal second messenger in the signaling pathway in cilia. However, it has been difficult to selectively visualize ciliary Ca2+ signal because of its small volume, which is ∼1:10,000 of the total cell volume. Recently, four types of ciliary GECIs were created by fusing single-FP-type cytosolic GECIs with ciliary membrane proteins (28, 70, 71, 72). Ciliary Ca2+ imaging with these indicators shows that both stimulation with ATP and laminar fluid flow elicit Ca2+ elevation in primary cilia (Fig. 4).
Figure 4.
Ca2+ imaging in the primary cilia upon chemical or mechanical stimulation. (A) Ca2+ signals in a NIH-3T3 cell in the resting state (left), after 10 μM ATP application (middle) and after 2 μM ionomycin application (right), visualized with 5-HT6-G-GECO1.0 and R-GECO1 in the primary cilium (upper) and the cytosol (lower), respectively. (B) Time-courses of Ca2+ signals in the primary cilium (cyan) and cytosol (magenta) within the cell indicated in (A). (C) Schematic of flow-induced movement of a primary cilium. A primary cilium in an upright position bends in response to laminar flow. Images of the primary cilium were generated by z-projections of consecutive xy-plane images. (D) Time-lapse images of a primary cilium subjected to laminar flow (1 dyne/cm2 shear, blue bar) in a mouse inner medullary collecting duct cell expressing 5HT6-mCherry-G-GECO1.0 (left and middle). mCherry is expressed as a marker to track the spatial movement of the cilium. Time-dependent changes in G-GECO1.0/mCherry fluorescence intensities are shown in pseudo-color (right). (E) Time-courses of 5HT6-mCherry-G-GECO1.0 (black) and 5HT6-mCherry-GFP (gray, as a control). This figure is adapted from Su et al. (28). To see this figure in color, go online.
Conclusions and perspectives
Fluorescent GECIs are valuable tools for the measurements of the dynamics and functions of Ca2+ signaling in intracellular organelles. Extensive and continuous improvements have made it possible to localize indicators specifically to the desired organelles, to adjust their properties to match the organellar environment, and to resolve the spatiotemporal dynamics of organellar Ca2+ signals. However, the currently available organellar GECIs still have several limitations, one of which is in the selection of a signal sequence. As discussed above, the selective and efficient localization of GECIs with organelle-specific targeting sequences is one of the major advantages in their use to image intraorganellar Ca2+, but the organelle-targeting efficiency is affected by several factors. Filippin et al. (13) evaluated the targeting efficiency of mitochondrial targeting sequences in several types of GECIs, and they pointed out that the expression level and maturation speed of GECIs are critical for their targeting efficiency. They also showed that the localization efficiency depends on which GECIs are fused, even when the same signal sequence is used. Additionally, the properties of GECIs, such as Ca2+ affinity, can be altered by the signal sequence. Therefore, when imaging intraorganellar Ca2+ with GECIs, it is indispensable to carefully evaluate whether the attached signal sequence effectively works for their specific localization in the desired organelle. In situ Ca2+ titration of organellar GECIs would be also useful to confirm whether GECIs targeted in the organelle retain the properties, that were measured in vitro, such as Kd and dynamic range.
The kinetics of indicators is also a critical parameter for the further improvement of organellar GECIs. Some types of Ca2+ signals—for example, the local Ca2+ concentration in the subplasmamembrane domain—can rise and fall within a millisecond. However, GECIs generally exhibit slow kinetics, which limit the detection of such fast Ca2+ responses. Much effort has been dedicated to creating cytosolic GECIs that can resolve fast Ca2+ signals, especially Ca2+ responses generated by action potentials in neurons. Similarly, intraorganellar GECIs with relatively fast kinetics have been developed by several groups. One is CatchER, which successfully detects repetitive Ca2+ release from the SR in muscle fibers. G-CEPIA1er and R-CEPIA1er also exhibit fast kinetics, which can resolve wavelike propagations of ER Ca2+ release (Fig. 2). To clarify whether there are any fast Ca2+ signals in other organelles and if fast Ca2+ signals play any physiological role in organellar functions, further improvement of the kinetics of organellar GECIs will be required.
Another limitation of organellar GECIs is associated with in vivo imaging. Needless to say, in vivo imaging is necessary to fully understand the biological functions of organellar Ca2+ signals. However, as compared with imaging in culture dishes, in vivo Ca2+ imaging has several technical difficulties, such as severe light scattering by tissues and motion artifacts. Whereas GECIs are now widely used for in vivo cytosolic Ca2+ imaging, especially in neuronal cells (73), in vivo organellar Ca2+ imaging with GECIs has so far been limited to the ER and mitochondria in zebrafish or mouse skeletal muscles (74, 75, 76, 77), possibly due to problems in brightness, signal/noise, kinetics, or affinity for Ca2+. Undoubtedly, improvements in the performance of GECIs, including brightness, dynamic range, kinetics, and affinity, are indispensable to advance the frontiers of in vivo organellar Ca2+ imaging. Ratiometric imaging with FRET-typed GECIs may have an advantage in in vivo imaging because it is able to cancel out movement artifacts. At the same time, there are additional promising strategies. One such strategy is the use of near-infrared FPs such as IFP1.4 and iRFP that emit light over 650 nm, which penetrates biological tissues, although these indicators currently require the addition of exogenous biliverdin to obtain bright fluorescent signals (78, 79, 80). An alternative strategy is the use of bright LP-based GECIs such as BRAC and nano-lantern (Ca2+) (81, 82, 83). These comprise three components: a Ca2+-responsive element, a FP and a LP. Using the process of bioluminescence resonance energy transfer between the LP and the FP, these indicators have successfully detected cytosolic Ca2+ dynamics at single-cell resolution, although continuous supply of the cofactor is required to obtain bright images.
The imaging tools discussed above have provided us with useful information on organellar Ca2+ dynamics. Additionally, a method allowing organellar-specific manipulation of Ca2+ signals will promote research on intraorganellar Ca2+ signals. Caged Ca2+ compounds, which release Ca2+ upon irradiation of UV light, were first used to manipulate intracellular Ca2+ signals (84). However, it is difficult to localize caged Ca2+ within specific organelles. Recently, several groups successfully generated optogenetic manipulators of cytosolic Ca2+ concentrations. One of them is based on a photoactivatable Ca2+ releasing protein, named PACR, which is a fusion protein of CaM, M13, and a photosensitive protein, LOV2 (85). Light irradiation changes its conformation and affinity for Ca2+, resulting in release of Ca2+. The others are LOVS1K, OptoSTIM1, and Opto-CRAC, all of which utilize light-dependent activation of the Ca2+ channel Orai1 on the plasma membrane, although there are differences among their activation mechanisms (86, 87, 88). At present, these methods are used only in the regulation of cytosolic Ca2+ concentrations, and none of them have been successfully used in manipulating intraorganellar Ca2+ concentrations. Development of methods to control intraorganellar Ca2+ levels in a specific and reversible manner would be useful.
Thus, further development of GECIs and genetically encoded Ca2+ manipulators are expected to advance our understanding of the functions and roles of organellar Ca2+ signals in health and disease.
Author Contributions
J.S., K.K., and M.I. wrote the article.
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Nos. 21229004, 25221304, and 25117002 to M.I.; No. 15H05648 to K.K.), and the Takeda Science Foundation to J.S.
Editor: Mark Cannell.
References
- 1.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Berridge M.J., Bootman M.D., Roderick H.L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Mekahli D., Bultynck G., Missiaen L. Endoplasmic-reticulum calcium depletion and disease. Cold Spring Harb. Perspect. Biol. 2011;3:3. doi: 10.1101/cshperspect.a004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzuto R., De Stefani D., Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi A., Camacho P., Herman B. Measurement of intracellular calcium. Physiol. Rev. 1999;79:1089–1125. doi: 10.1152/physrev.1999.79.4.1089. [DOI] [PubMed] [Google Scholar]
- 6.Kendall J.M., Dormer R.L., Campbell A.K. Targeting aequorin to the endoplasmic reticulum of living cells. Biochem. Biophys. Res. Commun. 1992;189:1008–1016. doi: 10.1016/0006-291x(92)92304-g. [DOI] [PubMed] [Google Scholar]
- 7.Rizzuto R., Simpson A.W., Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 8.Bonora M., Giorgi C., Pinton P. Subcellular calcium measurements in mammalian cells using jellyfish photoprotein aequorin-based probes. Nat. Protoc. 2013;8:2105–2118. doi: 10.1038/nprot.2013.127. [DOI] [PubMed] [Google Scholar]
- 9.Miyawaki A., Llopis J., Tsien R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 10.Arnaudeau S., Kelley W.L., Demaurex N. Mitochondria recycle Ca2+ to the endoplasmic reticulum and prevent the depletion of neighboring endoplasmic reticulum regions. J. Biol. Chem. 2001;276:29430–29439. doi: 10.1074/jbc.M103274200. [DOI] [PubMed] [Google Scholar]
- 11.Griesbeck O., Baird G.S., Tsien R.Y. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 12.Palmer A.E., Jin C., Tsien R.Y. Bcl-2-mediated alterations in endoplasmic reticulum Ca2+ analyzed with an improved genetically encoded fluorescent sensor. Proc. Natl. Acad. Sci. USA. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filippin L., Abad M.C., Pozzan T. Improved strategies for the delivery of GFP-based Ca2+ sensors into the mitochondrial matrix. Cell Calcium. 2005;37:129–136. doi: 10.1016/j.ceca.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Palmer A.E., Giacomello M., Tsien R.Y. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Drago I., Giacomello M., Pozzan T. Calcium dynamics in the peroxisomal lumen of living cells. J. Biol. Chem. 2008;283:14384–14390. doi: 10.1074/jbc.M800600200. [DOI] [PubMed] [Google Scholar]
- 16.Giacomello M., Drago I., Pozzan T. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell. 2010;38:280–290. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Dickson E.J., Duman J.G., Hille B. Orai-STIM-mediated Ca2+ release from secretory granules revealed by a targeted Ca2+ and pH probe. Proc. Natl. Acad. Sci. USA. 2012;109:E3539–E3548. doi: 10.1073/pnas.1218247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heim N., Griesbeck O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J. Biol. Chem. 2004;279:14280–14286. doi: 10.1074/jbc.M312751200. [DOI] [PubMed] [Google Scholar]
- 19.Nagai T., Sawano A., Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc. Natl. Acad. Sci. USA. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y., Araki S., Campbell R.E. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Garcia A., Rojo-Ruiz J., Alonso M.T. GAP, an aequorin-based fluorescent indicator for imaging Ca2+ in organelles. Proc. Natl. Acad. Sci. USA. 2014;111:2584–2589. doi: 10.1073/pnas.1316539111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J., Abdelfattah A.S., Campbell R.E. A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging. Nat. Commun. 2014;5:5262. doi: 10.1038/ncomms6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang S., Wong H.C., Yang J.J. Design and application of a class of sensors to monitor Ca2+ dynamics in high Ca2+ concentration cellular compartments. Proc. Natl. Acad. Sci. USA. 2011;108:16265–16270. doi: 10.1073/pnas.1103015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki J., Kanemaru K., Iino M. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 2014;5:4153. doi: 10.1038/ncomms5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J., Prole D.L., Campbell R.E. Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. Biochem. J. 2014;464:13–22. doi: 10.1042/BJ20140931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csordás G., Várnai P., Hajnóczky G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 2010;39:121–132. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albrecht T., Zhao Y., Johnson J.D. Fluorescent biosensors illuminate calcium levels within defined β-cell endosome subpopulations. Cell Calcium. 2015;57:263–274. doi: 10.1016/j.ceca.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Su S., Phua S.C., Inoue T. Genetically encoded calcium indicator illuminates calcium dynamics in primary cilia. Nat. Methods. 2013;10:1105–1107. doi: 10.1038/nmeth.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang W., Lu F., Cheng H. Imaging Ca2+ nanosparks in heart with a new targeted biosensor. Circ. Res. 2014;114:412–420. doi: 10.1161/CIRCRESAHA.114.302938. [DOI] [PubMed] [Google Scholar]
- 30.Shigetomi E., Kracun S., Khakh B.S. Monitoring astrocyte calcium microdomains with improved membrane targeted GCaMP reporters. Neuron Glia Biol. 2010;6:183–191. doi: 10.1017/S1740925X10000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prins D., Michalak M. Organellar calcium buffers. Cold Spring Harb. Perspect. Biol. 2011;3:3. doi: 10.1101/cshperspect.a004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson M.J., Baldwin H.A., Harvey B.K. A low affinity GCaMP3 variant (GCaMPer) for imaging the endoplasmic reticulum calcium store. PLoS One. 2015;10:e0139273. doi: 10.1371/journal.pone.0139273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou J., Hofer A.M., Yang J.J. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry. 2007;46:12275–12288. doi: 10.1021/bi7007307. [DOI] [PubMed] [Google Scholar]
- 34.Osibow K., Malli R., Graier W.F. A new type of non-Ca2+-buffering Apoa-based fluorescent indicator for intraluminal Ca2+ in the endoplasmic reticulum. J. Biol. Chem. 2006;281:5017–5025. doi: 10.1074/jbc.M508583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishii K., Hirose K., Iino M. Ca2+ shuttling between endoplasmic reticulum and mitochondria underlying Ca2+ oscillations. EMBO Rep. 2006;7:390–396. doi: 10.1038/sj.embor.7400620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravier M.A., Daro D., Gilon P. Mechanisms of control of the free Ca2+ concentration in the endoplasmic reticulum of mouse pancreatic β-cells: interplay with cell metabolism and [Ca2+]c and role of SERCA2b and SERCA3. Diabetes. 2011;60:2533–2545. doi: 10.2337/db10-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldeck-Weiermair M., Bischof H., Malli R. Generation of red-shifted cameleons for imaging Ca2+ dynamics of the endoplasmic reticulum. Sensors (Basel) 2015;15:13052–13068. doi: 10.3390/s150613052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okubo Y., Suzuki J., Iino M. Visualization of mechanisms for depletion and filling of Ca2+ within the endoplasmic reticulum upon synaptic inputs in cerebellar Purkinje cells. J. Neurosci. 2015;35:15837–15846. doi: 10.1523/JNEUROSCI.3487-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murayama T., Kurebayashi N., Sakurai T. Divergent activity profiles of Type 1 ryanodine receptor channels carrying malignant hyperthermia and central core disease mutations in the amino-terminal region. PLoS One. 2015;10:e0130606. doi: 10.1371/journal.pone.0130606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casey J.R., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 41.Pinton P., Pozzan T., Rizzuto R. The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 1998;17:5298–5308. doi: 10.1093/emboj/17.18.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lissandron V., Podini P., Pozzan T. Unique characteristics of Ca2+ homeostasis of the trans-Golgi compartment. Proc. Natl. Acad. Sci. USA. 2010;107:9198–9203. doi: 10.1073/pnas.1004702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong A.K., Capitanio P., Pizzo P. Heterogeneity of Ca2+ handling among and within Golgi compartments. J. Mol. Cell Biol. 2013;5:266–276. doi: 10.1093/jmcb/mjt024. [DOI] [PubMed] [Google Scholar]
- 44.McCormack J.G., Halestrap A.P., Denton R.M. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol. Rev. 1990;70:391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- 45.Sheng Z.H., Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat. Rev. Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Logan C.V., Szabadkai G., Sheridan E. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 2014;46:188–193. doi: 10.1038/ng.2851. [DOI] [PubMed] [Google Scholar]
- 47.Pan X., Liu J., Finkel T. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 2013;15:1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haviland S., Cleemann L., Morad M. Diversity of mitochondrial Ca2+ signaling in rat neonatal cardiomyocytes: evidence from a genetically directed Ca2+ probe, mitycam-E31Q. Cell Calcium. 2014;56:133–146. doi: 10.1016/j.ceca.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santo-Domingo J., Giacomello M., Demaurex N. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J. 2013;32:1927–1940. doi: 10.1038/emboj.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poburko D., Santo-Domingo J., Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J. Biol. Chem. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins T.J., Lipp P., Bootman M.D. Mitochondrial Ca2+ uptake depends on the spatial and temporal profile of cytosolic Ca2+ signals. J. Biol. Chem. 2001;276:26411–26420. doi: 10.1074/jbc.M101101200. [DOI] [PubMed] [Google Scholar]
- 52.Mellström B., Savignac M., Naranjo J.R. Ca2+-operated transcriptional networks: molecular mechanisms and in vivo models. Physiol. Rev. 2008;88:421–449. doi: 10.1152/physrev.00041.2005. [DOI] [PubMed] [Google Scholar]
- 53.Carrión A.M., Link W.A., Naranjo J.R. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 54.Ikeda M., Sugiyama T., Allen C.N. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 55.Robert V., Gurlini P., Pozzan T. Beat-to-beat oscillations of mitochondrial [Ca2+] in cardiac cells. EMBO J. 2001;20:4998–5007. doi: 10.1093/emboj/20.17.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bengtson C.P., Freitag H.E., Bading H. Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys. J. 2010;99:4066–4077. doi: 10.1016/j.bpj.2010.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding Y., Ai H.W., Campbell R.E. Förster resonance energy transfer-based biosensors for multiparameter ratiometric imaging of Ca2+ dynamics and caspase-3 activity in single cells. Anal. Chem. 2011;83:9687–9693. doi: 10.1021/ac202595g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lodhi I.J., Semenkovich C.F. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tovey S.C., de Smet P., Bootman M.D. Calcium puffs are generic InsP3-activated elementary calcium signals and are downregulated by prolonged hormonal stimulation to inhibit cellular calcium responses. J. Cell Sci. 2001;114:3979–3989. doi: 10.1242/jcs.114.22.3979. [DOI] [PubMed] [Google Scholar]
- 60.Rizzuto R., Pozzan T. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol. Rev. 2006;86:369–408. doi: 10.1152/physrev.00004.2005. [DOI] [PubMed] [Google Scholar]
- 61.Shigetomi E., Kracun S., Khakh B.S. A genetically targeted optical sensor to monitor calcium signals in astrocyte processes. Nat. Neurosci. 2010;13:759–766. doi: 10.1038/nn.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee M.Y., Song H., Blaustein M.P. Local subplasma membrane Ca2+ signals detected by a tethered Ca2+ sensor. Proc. Natl. Acad. Sci. USA. 2006;103:13232–13237. doi: 10.1073/pnas.0605757103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohamed T.M., Abou-Leisa R., Oceandy D. Development and characterization of a novel fluorescent indicator protein PMCA4-GCaMP2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2013;63:57–68. doi: 10.1016/j.yjmcc.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 64.Pinton P., Tsuboi T., Rutter G.A. Dynamics of glucose-induced membrane recruitment of protein kinase C β II in living pancreatic islet β-cells. J. Biol. Chem. 2002;277:37702–37710. doi: 10.1074/jbc.M204478200. [DOI] [PubMed] [Google Scholar]
- 65.Richler E., Chaumont S., Khakh B.S. Tracking transmitter-gated P2X cation channel activation in vitro and in vivo. Nat. Methods. 2008;5:87–93. doi: 10.1038/nmeth1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tay L.H., Dick I.E., Yue D.T. Nanodomain Ca2+ of Ca2+ channels detected by a tethered genetically encoded Ca2+ sensor. Nat. Commun. 2012;3:778. doi: 10.1038/ncomms1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dynes J.L., Amcheslavsky A., Cahalan M.D. Genetically targeted single-channel optical recording reveals multiple Orai1 gating states and oscillations in calcium influx. Proc. Natl. Acad. Sci. USA. 2016;113:440–445. doi: 10.1073/pnas.1523410113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singla V., Reiter J.F. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 69.Whitfield J.F. The solitary (primary) cilium—a mechanosensory toggle switch in bone and cartilage cells. Cell. Signal. 2008;20:1019–1024. doi: 10.1016/j.cellsig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Delling M., DeCaen P.G., Clapham D.E. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan S., Zhao L., Sun Z. Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr. Biol. 2015;25:556–567. doi: 10.1016/j.cub.2014.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin X., Mohieldin A.M., Nauli S.M. Cilioplasm is a cellular compartment for calcium signaling in response to mechanical and chemical stimuli. Cell. Mol. Life Sci. 2014;71:2165–2178. doi: 10.1007/s00018-013-1483-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grienberger C., Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 74.Mizuno H., Sassa T., Miyawaki A. Transgenic zebrafish for ratiometric imaging of cytosolic and mitochondrial Ca2+ response in teleost embryo. Cell Calcium. 2013;54:236–245. doi: 10.1016/j.ceca.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 75.Jiménez-Moreno R., Wang Z.M., Delbono O. Sarcoplasmic reticulum Ca2+ depletion in adult skeletal muscle fibres measured with the biosensor D1ER. Pflugers Arch. 2010;459:725–735. doi: 10.1007/s00424-009-0778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudolf R., Magalhães P.J., Pozzan T. Direct in vivo monitoring of sarcoplasmic reticulum Ca2+ and cytosolic cAMP dynamics in mouse skeletal muscle. J. Cell Biol. 2006;173:187–193. doi: 10.1083/jcb.200601160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rudolf R., Mongillo M., Pozzan T. In vivo monitoring of Ca2+ uptake into mitochondria of mouse skeletal muscle during contraction. J. Cell Biol. 2004;166:527–536. doi: 10.1083/jcb.200403102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filonov G.S., Piatkevich K.D., Verkhusha V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu D., Baird M.A., Shu X. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods. 2015;12:763–765. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin L.T., Wang B.S., Lee Y.J. mPlum-IFP 1.4 fluorescent fusion protein may display Förster resonance energy transfer associated properties that can be used for near-infrared based reporter gene imaging. J. Biomed. Opt. 2013;18:126013. doi: 10.1117/1.JBO.18.12.126013. [DOI] [PubMed] [Google Scholar]
- 81.Takai A., Nakano M., Nagai T. Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging. Proc. Natl. Acad. Sci. USA. 2015;112:4352–4356. doi: 10.1073/pnas.1418468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saito K., Chang Y.F., Nagai T. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saito K., Hatsugai N., Nagai T. Auto-luminescent genetically-encoded ratiometric indicator for real-time Ca2+ imaging at the single cell level. PLoS One. 2010;5:e9935. doi: 10.1371/journal.pone.0009935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellis-Davies G.C. Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods. 2007;4:619–628. doi: 10.1038/nmeth1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fukuda N., Matsuda T., Nagai T. Optical control of the Ca2+ concentration in a live specimen with a genetically encoded Ca2+-releasing molecular tool. ACS Chem. Biol. 2014;9:1197–1203. doi: 10.1021/cb400849n. [DOI] [PubMed] [Google Scholar]
- 86.Pham E., Mills E., Truong K. A synthetic photoactivated protein to generate local or global Ca2+ signals. Chem. Biol. 2011;18:880–890. doi: 10.1016/j.chembiol.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 87.Kyung T., Lee S., Heo W.D. Optogenetic control of endogenous Ca2+ channels in vivo. Nat. Biotechnol. 2015;33:1092–1096. doi: 10.1038/nbt.3350. [DOI] [PubMed] [Google Scholar]
- 88.He L., Zhang Y., Zhou Y. Near-infrared photoactivatable control of Ca2+ signaling and optogenetic immunomodulation. eLife. 2015;4:4. doi: 10.7554/eLife.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ai H.W., Henderson J.N., Campbell R.E. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abell E., Ahrends R., Teruel M.N. Parallel adaptive feedback enhances reliability of the Ca2+ signaling system. Proc. Natl. Acad. Sci. USA. 2011;108:14485–14490. doi: 10.1073/pnas.1018266108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Palmer A.E., Tsien R.Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 92.Zampese E., Fasolato C., Pizzo P. Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc. Natl. Acad. Sci. USA. 2011;108:2777–2782. doi: 10.1073/pnas.1100735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waldeck-Weiermair M., Alam M.R., Malli R. Spatiotemporal correlations between cytosolic and mitochondrial Ca2+ signals using a novel red-shifted mitochondrial targeted cameleon. PLoS One. 2012;7:e45917. doi: 10.1371/journal.pone.0045917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iguchi M., Kato M., Akao M. Direct monitoring of mitochondrial calcium levels in cultured cardiac myocytes using a novel fluorescent indicator protein, GCaMP2-mt. Int. J. Cardiol. 2012;158:225–234. doi: 10.1016/j.ijcard.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 95.Isshiki M., Nishimoto M., Fujita T. FRET-based sensor analysis reveals caveolae are spatially distinct Ca2+ stores in endothelial cells. Cell Calcium. 2013;54:395–403. doi: 10.1016/j.ceca.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 96.Li H., Wang X., Ding S. Imaging of mitochondrial Ca2+ dynamics in astrocytes using cell-specific mitochondria-targeted GCaMP5G/6s: mitochondrial Ca2+ uptake and cytosolic Ca2+ availability via the endoplasmic reticulum store. Cell Calcium. 2014;56:457–466. doi: 10.1016/j.ceca.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lynes E.M., Raturi A., Simmen T. Palmitoylation is the switch that assigns calnexin to quality control or ER Ca2+ signaling. J. Cell Sci. 2013;126:3893–3903. doi: 10.1242/jcs.125856. [DOI] [PubMed] [Google Scholar]
- 98.Frieden M., James D., Demaurex N. Ca2+ homeostasis during mitochondrial fragmentation and perinuclear clustering induced by hFis1. J. Biol. Chem. 2004;279:22704–22714. doi: 10.1074/jbc.M312366200. [DOI] [PubMed] [Google Scholar]
- 99.Kettlewell S., Cabrero P., Smith G.L. Changes of intra-mitochondrial Ca2+ in adult ventricular cardiomyocytes examined using a novel fluorescent Ca2+ indicator targeted to mitochondria. J. Mol. Cell. Cardiol. 2009;46:891–901. doi: 10.1016/j.yjmcc.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 100.Chang K.T., Niescier R.F., Min K.T. Mitochondrial matrix Ca2+ as an intrinsic signal regulating mitochondrial motility in axons. Proc. Natl. Acad. Sci. USA. 2011;108:15456–15461. doi: 10.1073/pnas.1106862108. [DOI] [PMC free article] [PubMed] [Google Scholar]