Abstract
Nociceptin (NCC, also known as FQ (N/OFQ)) is the 17-amino acid neuropeptide, endogenous ligand for the G-protein-coupled receptor (NOP, also known as ORL-1). In this study, starting from the recently reported x-ray structure at pH 7 of NOP in complex with an antagonist, new insights, to our knowledge, on the binding geometry of NCC to NOP have been provided in silico. After a rigid docking of NCC in an α-helix conformation, molecular dynamics (MD) and metadynamics (METAD), a method for the analysis of free-energy surfaces (FES), were performed on the protein-peptide complex. Free-energy profiles were obtained as a function of the α-helix content of different segments of the 17-mer ligand, and a structural ensemble of conformations of NCC, corresponding to the minimum of the FES, was extracted, thus representing the NCC bound to the inactive form of NOP. The structural features were compared with many known experimental data. The pose of the “message” domain (residues 1–4) of NCC differs from that of the known NOP antagonists, as being slightly slipped deeper inside the protein core. A residual α-helix content in the central part of the peptide (residues 4–9) is maintained, whereas the C-terminal segment (residues 13–17) is unstructured and highly flexible. An important stabilization due to interactions with residues D130 and D110 of the receptor has been found, in agreement with the large decrease in agonist potency reported for the D130A and D110A mutants. The importance of the extracellular domain 2 (ECL2) in the selectivity toward the endogenous ligand has been confirmed. A pivotal role for the conserved residue N133 is suggested and further supported by a study of the N133A in silico mutant. Accordingly, N133 can work as a molecular microswitch driving the change between the inactive and active NOP conformations, in the framework of an extended H-bond and water network rearrangement in the deep binding site.
Introduction
Nociceptin/orphanin FQ (N/OFQ; hereafter NCC) (1, 2) is an endogenous 17-mer peptide (with sequence FGGFTGARKSARKLANQA) involved in a wide range of physiological responses, with effects noted in the nervous system (central and peripheral), the cardiovascular system, the airways, the gastrointestinal tract, the urogenital tract, and the immune system (3, 4). Its receptor, the G-protein-coupled nociceptin/orphanin FQ receptor (NOP), is considered a drug target with broad therapeutic potential (5). Activation of the NOP results in the inhibition of cAMP synthesis and the increase of membrane permeability to K+, as in the canonical way of the overall class of opiate receptors.
Deciphering the structure of the NOP_NCC complex would be very important to unravel the details of its in vivo function, however this aim remains relatively elusive. X-ray diffraction data of the NOP in complex to some potent stabilizing antagonists have recently been published (6, 7), but neither crystallographic nor NMR data have been reported with respect to NOP-agonist complexes or the free receptor. Concerning the transition from the inactive to the active form of the receptor, actually the “ionic lock” identified in the inactive form of rhodopsin (8) by the residues forming the Asp-Arg-Tyr (DRY) motif of TM3 (conserved in the opioid receptor family) and an acidic residue of TM6, is not observed in the crystallized NOP_C24 complex (6). Concerning the NCC structure alone, NMR solution measurements are consistent with a random coil conformation in water, whereas a relatively more stable helix conformation from residues 4–17 in membrane-like environment (trifluoroethanol and SDS micelles) was observed (9). According to the received view, the N-terminal residues of NCC would constitute the “message” domain of the agonist, entering deeply in the binding pocket, and responsible for its efficacy in dose-response experiments, whereas the central and C-terminal segment of NCC constitute the “address” domain, interacting with the residues on the receptor surface and responsible for selectivity between different ligand molecules (10). A recent molecular dynamics (MD) study (11), based on the assumption that NCC interacts with the membrane environment before binding with its receptor, used an α-helix structure of the peptide as ligand in rigid-docking calculations toward the putative binding site of NOP, followed by equilibration with MD of the ligand-protein-membrane system.
However, there is no evidence that NCC interacts with the membrane environment before binding to the receptor (i.e., the access to the binding pocket is from the solvent). Thus, either the α-helix content of NCC arises just within the binding pocket, starting from a random-coil structure in extracellular environment, or NCC reaches its receptor with at least a partial α-helix content. There is no report on NCC storage and processing in axon terminals and synapses. However, by analogy with what is known for dynorphin (12), it is likely that NCC and/or its precursor pronociceptin are stored in the synaptic vesicles as oligomers, where an α-helix structure could be maintained, protected from the destabilizing polar interactions of the cytosol. Thus α-helix structure destabilization would occur in a time interval corresponding to the particle diffusion within the synaptic cleft (d ∼ 30–100 nm). This time can be estimated by the Einstein equation t = d2/D, where D is the diffusion constant (for dynorphin the measured constant D is ∼1.7 10−6 cm2/s), being ∼200 ns. We performed two preliminary 100 ns MD calculations to test the α-helix stability of NCC in water (Fig. S1 in the Supporting Material), where the fraction of α-helix content is featured using the function described in Materials and Methods. According to these calculations, the α-helix content drops to zero in less than 20 ns, hence a diffusion time of the order of hundreds of nanoseconds should be long enough to completely destabilize the NCC α-helix structure. However, in a crowded environment (i.e., 60 mM clustered NCC molecules), the secondary structure could be maintained much longer, allowing entering the binding pocket with an at least partially structured conformation.
Starting from these considerations, we investigated the docking of NCC to NOP and evaluated the energy landscape as a function of the α-helix content, i.e., using an alternative approach with respect to the one by Kothandan et al. (11), to determine the structure and dynamics of the NOP_NCC complex. We started by using a rigid docking of NCC (α-helix) into the NOP receptor, followed by a short equilibration (20 ns) by classical MD. Then, we applied the metadynamics (METAD) method to extract the free-energy minimum of the NCC conformations inside the binding pocket as a function of its α-helix degree, using biased collective variables built for this purpose (13). Finally, we tested the stability and the unbiased dynamics of a representative configuration extracted from the minimum of the free-energy landscape. Our METAD results have been compared with the known structure of NOP in complex with the antagonist compound C24 (6), and with the MD simulation of the NOP_free. A detailed analysis of the α-helix content, the hydrophobic environment, and the hydrogen bond network including the rearrangement of structural waters around the message and the address domain of NCC have been provided, suggesting the role of the asparagine 133 (N1333,35 in the Ballesteros-Weinstein notation (14)) as a molecular microswitch related to the stability between the inactive and active conformations of the receptor. This hypothesis has been further investigated by the analysis of the free-energy surfaces (FES) of the in silico mutant N133A that has been built for the purpose.
Materials and Methods
Model setup of the NOP receptor
The initial structure of the NOPR was built starting from the recently reported x-ray crystal structure of the receptor in complex with a peptide mimetic (PDB: 4EA3) (6). Residues 155–163 belonging to a coil in the intracellular side, and side-chain atoms lacking in the reported structure due to structural disorder, were added and minimized “in vacuum” by using the Modeler and Rotamer modules in the Chimera UCSF package (15).
Docking of NCC to NOP and setup of the protein-peptide complex
We set up an α-helix-structured NCC peptide by the Chimera UCSF package (15), and then we attempted a protein-protein rigid docking using the Zdock server (16). From the output of the server, according to the received view that F1(NCC) is involved in H-bond with D1303,32, a model having the minimal distance between F1(NCC) and D1303,32 (NOPR) was chosen as a starting model for MD setup. The protonation state of the ND1 and NE2 atoms of the histidines was chosen as suggested by the crystallographic structure, i.e., ND1(H79ICL1) and NE2(H154ICL2) were protonated, resulting in a charge of +6 (NOPR) and +4 (NCC). The highly conserved disulfide bond between C1233,25 and C200ECL2 of the protein was properly kept fixed during the MD runs. This disulfide bridge anchors the extracellular side of the TM3 helix near the binding site, therefore limiting the extent of conformational changes around this region during receptor activation (17).
Classical molecular dynamics
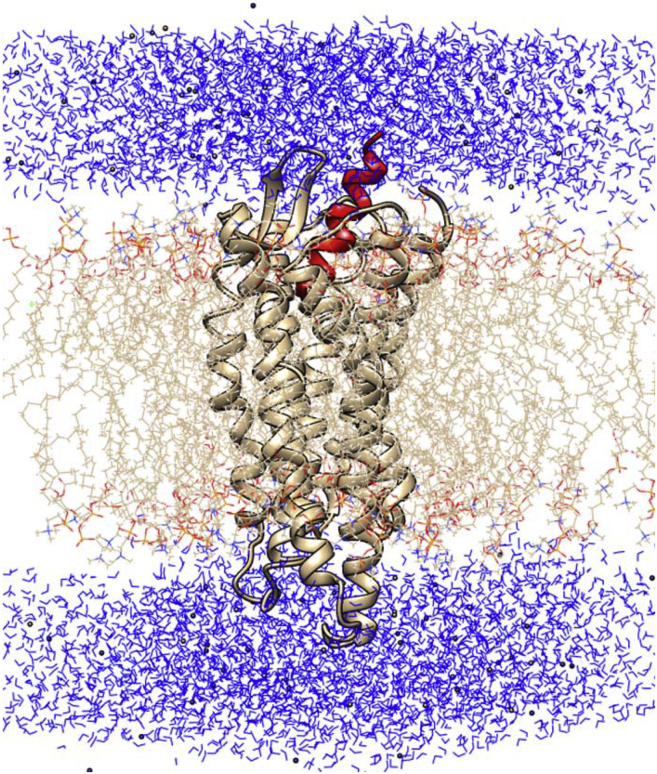
Classical MD simulations were performed with the GROMACS 4.6 package (18, 19) under the AMBER parm99sb force field (20) at the full atomistic level using a TIP3P water solvent and an explicit preequilibrated phospholipid bilayer of 128 POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) molecules obtained from P. Tieleman’s University of Calgary website (http://moose.bio.ucalgary.ca). Lipids were packed and energy-minimized around an embedded, strongly constrained protein structure, at the proper density (area per lipid) of ∼0.63 nm2 using the INFLATEGRO procedure, following the same steps as described in (11). The protein-peptide-membrane system was then solvated in a triclinic water box (having basis vectors lengths of 7, 7.4, and 9.3 nm) under periodic boundary conditions, for a total number of 40,000 atoms (6400 solvent molecules) and a total charge +10. This positive charge was neutralized by randomly substituting water molecules with 44 Na+ ions and 54 Cl- ions, to obtain neutrality with 0.15M salt concentration. A sketch of the full minimized system is displayed in Fig. 1.
Figure 1.
Setup of the starting system (State_0) obtained after rigid docking of NCC for the simulation: NOP receptor (beige ribbons), NCC (red ribbon), the lipid (beige wires), and water (blue). To see this figure in color, go online.
Following a steepest descent minimization, the system was equilibrated in canonical ensemble (NVT) conditions for 300 ps, using a V-rescale, modified Berendsen thermostat (21) with position restrains for both the protein-peptide complex and the lipids, and thereafter in a isothermal-isobaric ensemble (NPT) for 500 ps, applying position restraints to the heavy atoms of the protein-peptide complex, and using a Nose-Hoover thermostat (22, 23) and a Parrinello-Rahman barostat (24) at 1 Atm with a relaxation time of 2.0 ps. Finally, all restraints were removed, and MD runs were performed under NPT conditions at 300 K with a T-coupling constant of 1 ps. Van der Waals interactions were modeled using 6–12 Lennard-Jones potential with a 1.2 nm cutoff. Long-range electrostatic interactions were calculated, with a cutoff for the real space term of 1.2 nm. All covalent bonds were constrained using the LINCS algorithm. The time step employed was 2 fs, and the coordinates were saved every 5 ps for analysis, which was performed using the standard GROMACS tools.
Metadynamics
The conformational space of the docked NCC was explored by METAD (25) in the well-tempered ensemble (WTE) (26). The METAD algorithm uses a set of collective variables (CVs) si (i = 1,…,NCVs), which are functions of the coordinates of the system. Such coordinates are evolved along a standard MD trajectory, supplemented by a history-dependent potential that add penalties to the system discouraging it from visiting previously sampled conformations. The WTE is an ensemble designed to have the same energy of the canonical ensemble, but with much larger fluctuations. It has been demonstrated that the use of the WTE avoids bias overloading, and accelerates convergence of the METAD runs by even orders of magnitude (26).
The PLUMED 2.1 package (21) was patched to the GROMACS engine to carry out our METAD runs. As in the standard implementation, the history-dependent potential is built by using Gaussians of NCVs-th dimension, height w, and widths δsi, and deposed at time intervals τG along the CVs trajectory. The choice of the w, δsi, and τG parameters is crucial to obtain an accurate reproduction of the FES in reasonable time, and their proper values are given just after the definition of the chosen collective variables. As long as the METAD simulation runs, the sum of these penalty terms tends to compensate exactly the underlying FES in the reduced space, thus allowing a reconstruction of the FES explored up to time t (27).
We selected as collective variables (CVs) the α-helix content of NCC between residues 2–9 (CV1) and 10–16 (CV2). This kind of collective variables is based on the work by Pietrucci and Laio (13), using the set of root mean square (RMS) distances of every six contiguous residues in a chosen segment of a protein chain from an ideal α-helix configuration. This is done by calculating the following function:
with the i index running over all possible segments of α-helix defined in the chosen sequences (3 for the 2–9 segment, and 2 for the 10–16 segment), and r0 = 0.08 nm. Thus the CVs go to 0 when the root mean square deviation (RMSD) values approach r0, go to 3(CV1) or 2(CV2) when these segments form a full α-helix structure, and range from 0 to 1 for every segment of six residues. The height w of the Gaussian parameter was chosen to be 0.32 kJ/mol according to values found in literature (28) as a compromise between accuracy and speed of the FES scanning; the widths δs1 and δs2, as an empirical rule were taken to be one-third of the fluctuations of each CV in a free MD run; hence they were chosen to be 0.1 for both CV1 and CV2. The pace time τG for deposition of the bias was 0.5 ps.
Results
MD simulation of State_0
After the rigid docking of NCC (α-helix) into the NOP receptor, as described in the Materials and Methods, the resulting configuration of the NOP_NCC complex (hereafter “State_0”) was subjected to a MD protocol.
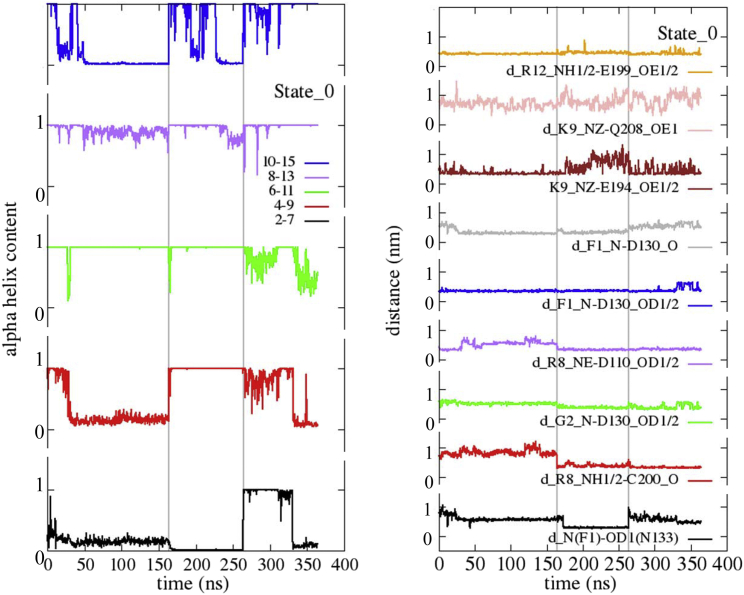
The C-alpha RMSD plot of two 100 ns MD simulations relative to the free (NOP_free) and NCC docked (NOP_NCC) receptor show stability of the backbone after 30–40 ns with a RMSD of 0.2–0.25 nm maximum for both the trajectories (Fig. S2, upper panel). The presence of NCC induces a decrease of the flexibility of ECL1, ICL3, and TM7, and an increase for ICL2 as shown by the calculated root mean square fluctuation (RMSF) per residue (C-alpha) (Fig. S2, lower panel). However, the docked NCC also shows a high degree of flexibility, undergoing multiple changes in the α-helix content along the trajectory, and exchanging its interactions with the external residues placed on the surface of the receptor. In Fig. 2 (left panel), the α-helix content of five segments of NCC, relative to residues 2–7, 4–9, 6–11, 8–13, and 10–15 are monitored along three independent 100–160 ns MD trajectories, shown as concatenated ones in the figure. It is evident, by inspecting the figure, that all segments show a certain degree of instability of the secondary structure, the central one being more stable with respect to the C-terminal and N-terminal segments. The evolution of several important hydrogen bonds is displayed on the right panel of Fig. 2, showing for example the interaction of N(F1) of NCC with OD1/2 (D1303,32) and OD1(N1333,35) of NOP. As a general procedure, we have considered the flipping of equivalent donor or acceptor atoms forming the hydrogen bond, by monitoring the carbon atom of the same functional group. In the former case the contribution of OD1 and OD2 atoms of aspartate to the hydrogen bonds with N(F1) is averaged by considering the distance with the CG atom of the aspartate (Fig. 2, right panel). It emerges from the figure that several hydrogen bonds are unstable all along the trajectories; the formation/breaking of several of them being related to thermal fluctuations of the local α-helix content of NCC; moreover, the tendency to interact more deeply inside the receptor, reaching sometimes the very central N1333,35 residue of NOP (black curve), was intriguing, as this residue is the closest one to the intracellular side that directly interacts with the agonist.
Figure 2.
(Left panel) α-helix content of different segments of NCC after rigid docking to NOP (State_0) and along classical MD trajectories. Three independent MD trajectories (100–160 ns long, concatenated in the figures) were followed, and the α-helix content was calculated for residues 2–7, 4–9, 6–11, 8–13, and 10–15. The α-helix content of each segment varies from 0 to 1. The vertical lines indicate the restart of the trajectories. (Right panel) Formation of important hydrogen bonds along the same trajectories are shown. To see this figure in color, go online.
These results are different from those by Kothandan et al. (11), which showed a more external position of the message domain, probably due to a different choice between the resulting poses of the rigid docking procedure. Interestingly, in both calculations the message domains move during the MD trajectory from the initial position going inner inside the binding site. As explained in Materials and Methods, the pose we chose, with the message domain of the peptide being as close as possible to the inner core of the binding pocket, and thereafter subjected to a short MD equilibration, constituted only the starting conformation of the NOP_NCC complex that was further investigated by the METAD protocol. By the way, these preliminary MD results support a dynamic picture of the NOP_NCC interaction and resemble the recently reported liquid-state NMR spectroscopy data on the structure of the dynorphin (1–13) peptide bound to the human k-opioid receptor (KOR) (29).
Metadynamics
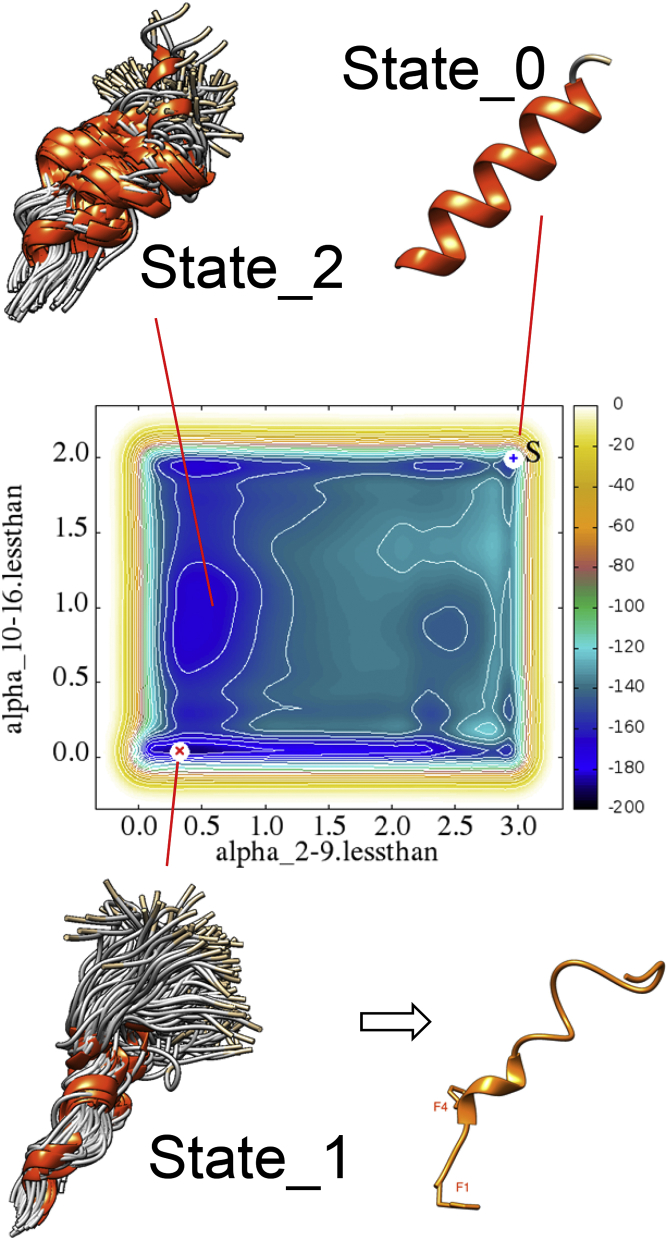
A deeper investigation of the conformational landscape of the docked NCC in complex with NOP was undertaken using METAD. A well-tempered protocol (described in Materials and Methods) was attempted in a bidimensional space. We used the α-helix content of the NCC segments 2–9 (CV1) and 10–16 (CV2) as two independent biased collective variables. In the Fig. 3 diagram, the α-helix content of segment 2–9 can vary from 0 to 3, and the α-helix content of segment 10–16 can vary from 0 to 2. We started from the structure obtained after an early equilibration (20 ns) of the NOP_NCC complex (State_0), where NCC maintains a full α-helix content (position [2, 3] of the bidimensional FES diagram). The protocol lasted 300 ns; the Gaussian heights deposited along the trajectory are reported in Fig. S3 A; the added biases tend to zero indicating that the calculation in the WTE has converged and the FES has been fully reproduced. In Fig. 3, the final bidimensional FES diagram is reported as a function of the two collective variables. It shows the absolute minimum at position [0.3, 0.03] that is identified as State_1. The structural ensemble of bound NCC structures, belonging to State_1, are depicted in the figure having energies corresponding to the calculated free-energy minimum, (−190.4 ± 2RT) kJ/mol where RT = 2.4794 kJ/mol. The projections of the averaged FES along CV1 (Fig. S3 B) and CV2 (Fig. S3 C), obtained as a function of the simulation time, show again the convergence of the METAD protocol: in fact, the two free-energy profiles at 270 ns (blue curves) and 300 ns (red curves) are almost indistinguishable for both CVs. From the visual inspection of the FES and its projections, it is also evident that the conformational landscape of the two CVs around the minimum is quite different. A deep minimum exists for the external segment (Fig. S3 C), residues 10–16, with α-helix content 0.03 ± 0.03, the error being evaluated taking all the structures with energy values around the minimum + 2RT). This result clearly points toward an absence of α-helix content in secondary structure for this segment. On the contrary a certain degree of secondary structure of the internal segment (residues 2–9) is present, as demonstrated (Fig S3 B) by a flat minimum around an α-helix content of 0.3 ± 0.2, ten times higher than the former one. A third minimum (State_2) is also depicted in Fig. 3 at position [0.5, 0.96]; however, this state seems not reachable from the absolute minimum (State_1) through thermal fluctuations, as it is ∼10 RT higher in energy (about −166 kJ/mol). Finally, State_0 is also a local minimum of the FES, with an energy of about −145 kJ/mol, a fact that explains the partial stability of the NCC α-helix observed in the aforementioned MD simulations (Fig. 2, left panel; (11)). Moreover, the energy difference between State_0 and State_1, +45 kJ/mol, is reasonable as compared with the estimated affinity of NCC to NOP, i.e., Ki = 0.93 nmol, ΔG = −51.8 kJ/mol (30).
Figure 3.
Well-tempered METAD of the NOP_NCC complex as a function of NCC α-helix content. Clusters corresponding to the absolute minimum (State_1) and a local minimum (State_2) are shown, together with the starting structure (State_0), and the representative structure of State_1 extracted by clustering analysis (bottom right). To see this figure in color, go online.
The representative structure in State_1 (Fig. 3, bottom right), chosen as central structure after single linkage clustering method, has been further investigated as well as State_0, by classical MD.
Molecular dynamics of State_1
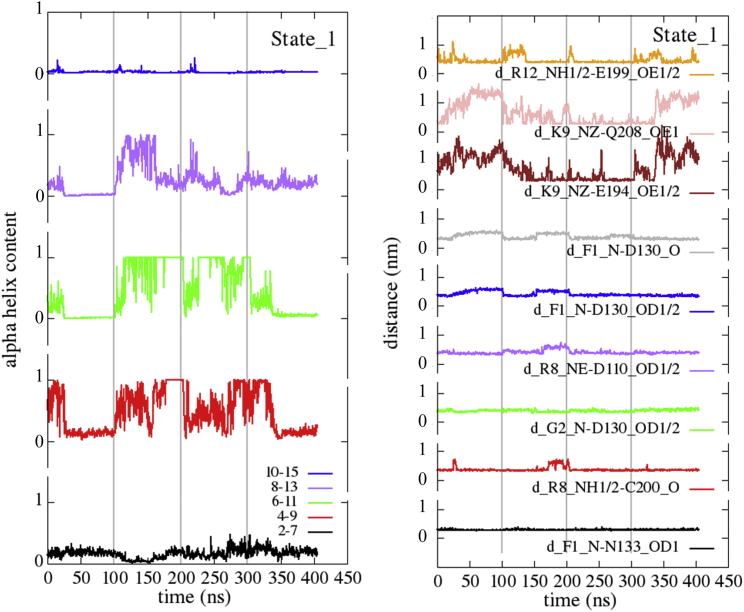
In Fig. 4 (left panel), the evolution of the α-helix content of the same NCC segments 2–7, 4–9, 6–11, 8–13, and 10–15 as in Fig. 2 is calculated starting from State_1, along each one of the repeated MD trajectories (concatenated in the figure). Rapid exchanges between multiple α-helix configurations of the bound peptide are still observed in agreement with the flat minimum calculated in the FES; as expected, the behavior of the curves is similar to those relative to State_0, with the main fraction of the α-helix content placed in the central segment between F4 and A11.
Figure 4.
(Left panel) α-helix content of different segments of NCC along classical MD trajectories starting from State_1. Four independent MD trajectories (100 ns long, concatenated in the figures) were calculated starting from State_1 and displayed as in Fig. 2. (Right panel) Evolution of the most stable hydrogen bonds along the same trajectories are shown. To see this figure in color, go online.
In Fig. 4 (right panel), the evolution of the more stable hydrogen bonds formed between NCC and NOP along the four repeated trajectories starting from State_1 are shown, and a more extensive list of them is given in Table 1. Not surprisingly, residues 14–17 of NCC interact weakly with NOP; in fact, they do not form H-bonds present more than 15% of the simulation time. This result is in agreement with the experimental finding that the synthetic fragment NCC(1–13)-NH2 is active as well as NCC (31).
Table 1.
List of the Most Stable Hydrogen Bonds between NOP and NCC Formed along Four Independent MD Trajectories Calculated Starting from State_1
| NCC | NOP | Protein Location | Stability (Fraction) | ||
|---|---|---|---|---|---|
| PHE-1 | N | ASN-133 | OD1 | TM3_35 | 0.999 |
| ASP-130 | OD1/2 | TM3_32 | 0.562 | ||
| ASP-130 | O | TM3_32 | 0.504 | ||
| GLY-308 | O | TM7_42 | 0.172 | ||
| TYR-309 | OH | TM7_43 | 0.166 | ||
| GLY-2 | N | ASP-130 | OD1/2 | TM3_32 | 0.736 |
| GLY-3 | N | ASP-130 | OD1/2 | TM3_32 | 0.289 |
| TYR-309 | OH | TM7_43 | 0.224 | ||
| PHE-4 | N | TYR-309 | OH | TM7_43 | 0.184 |
| GLY-6 | O | GLN-286 | NE2 | TM6_58 | 0.252 |
| ALA-7 | O | ARG-302 | NH1/2 | TM7_36 | 0.160 |
| ARG-8 | NH1/2 | CYS-200 | O | ECL2 | 0.889 |
| NE | ASP-110 | OD1/2 | TM2_63 | 0.642 | |
| LYS-9 | NZ | GLU-194 | OE1/2 | ECL2 | 0.448 |
| GLN-208 | OE1 | ECL2 | 0.430 | ||
| GLN-208 | NE2 | ECL2 | 0.180 | ||
| SER-10 | OG | PRO-292 | O | ECL3 | 0.199 |
| ARG-12 | NH1/2 | GLU-199 | OE1/2 | ECL2 | 0.389 |
| NE | GLU-199 | OE1/2 | ECL2 | 0.199 | |
| LYS-13 | NZ | GLU-194 | OE1/2 | ECL2 | 0.240 |
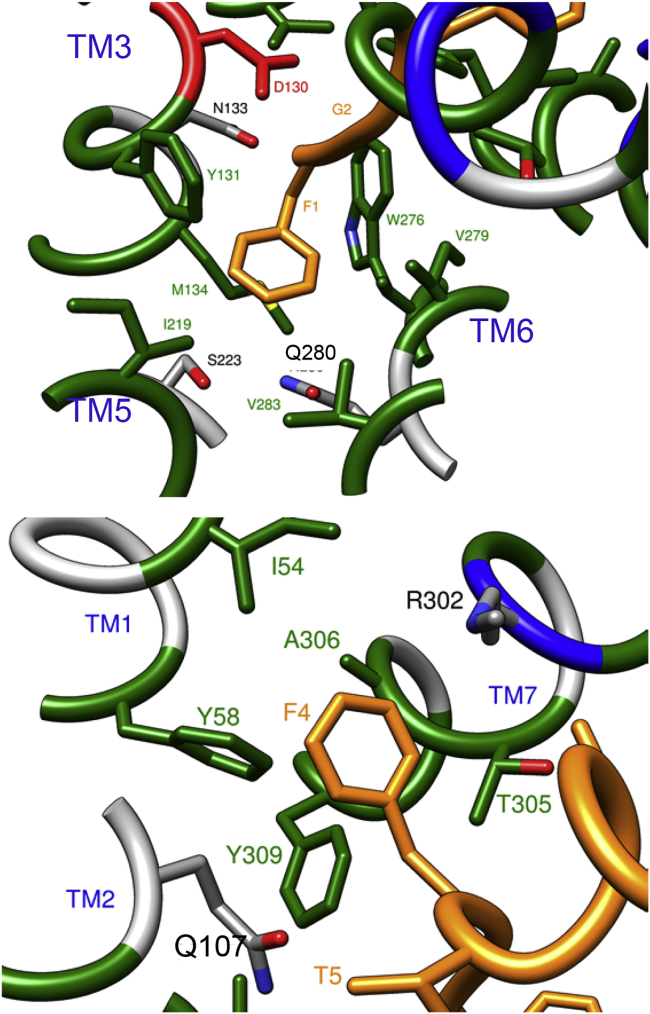
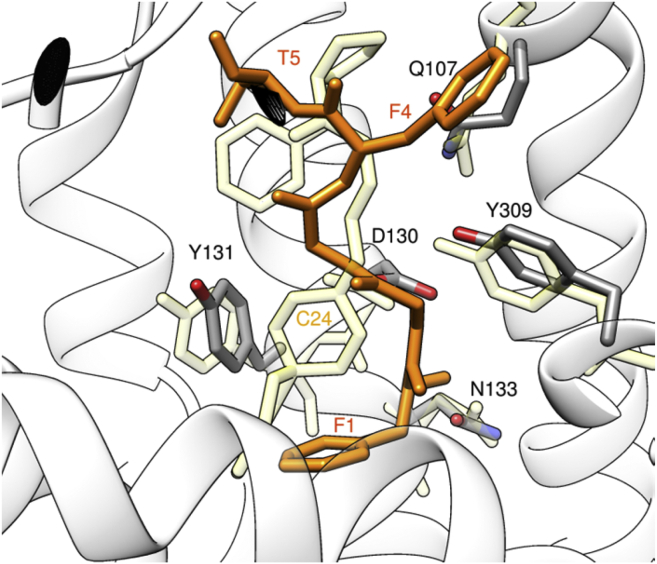
In the following we will discuss the representative structure of State_1, depicted in the bottom right of Fig. 3. The environment around the message domain, i.e., the N-terminal residues 1–4 (FGGF) of NCC, is depicted in Fig. 5 (where NCC is orange) in two different orientations, focused on residues F1 (upper panel) and F4 (lower panel). The message domain of the endogenous agonist, thought to be mainly responsible for efficacy in dose-effect experiments, is buried in the same hydrophobic pocket created by residues of helices TM3, TM5, and TM6 that surround the C24 and C35 antagonists as observed by crystallography (6, 7), the latter two molecules being bound in a very similar fashion. More specifically, the F1 residue in our simulation is surrounded by Y1313,33 and M1343,36 from TM3, I2195,42 from TM5, W2766,48, V2796,51, and V2836,55 from TM6 (Fig. 5, upper panel). The poses of C24 (found by crystallography) and NCC (1–5) are compared in Fig. 6. The F1 of NCC lies approximately in the same location of the benzofuran-piperidine rings of C24, as expected, but the backbone nitrogen N(F1) is 5 Å closer to the center of the overall TM bundle than the N1 piperidine nitrogen of C24, making it capable to make a stable hydrogen bond with the deeply buried OD1 of N1333,35, which is a residue included in the conserved 129–141 TM3 sequence (IDYYNMFTSTFTL) of the human opioid receptors. This arrangement of the message region of NCC also differs from the docking results shown by Thompson et al. (6) for the N-terminal tetrapeptide of the [Nphe1, Arg14, Lys15]-NCC peptide (named UFP-101), which is a known NOP antagonist. According to their docking calculations, the tetrapeptide (NPhe1-Gly2-Gly3-Phe4) portion of UFP-101, binds with a conformation very similar to C24, having the two aromatic rings of NPhe1 and Phe4 overlapped to the external aromatic groups of C24. The NPhe1 chemical modification is known to eliminate efficacy, whereas the L14R and N15K mutation are reported to increase potency and duration of action (32). Thus it is reasonable that the ability of NPhe1 to abolish efficacy would be in reinforcing its anchoring to D1303,32 and avoiding to reach N1333,35, thus forming a complex so similar in structure, but also in lack of activity, to the known antagonists C24 and C35.
Figure 5.
The hydrophobic environment where the message domain of NCC is docked (State_1): NCC (orange), residues involved in hydrophobic interaction (green), positive residues (blue), and negative residues (red). To see this figure in color, go online.
Figure 6.
Comparison between the binding poses of the C24 mimetic compound seen by crystallography (light yellow; (6)) and NCC (orange; only residues 1–5, this study). To see this figure in color, go online.
In this framework, the NCC interaction with NOP is different and with a longer segment of the TM3 helix than the C24 compound leading to interact with N1333,35 beside D1303,32.
Comparing the poses of C24 and NCC (Fig. 6), the position of pirrolidine ring corresponding to the tail end of the C24 compound is closer to T5 of NCC rather than F4, which is oriented in an opposite direction, as a consequence of the sliding of the message domain deep inside the receptor during the METAD protocol. The aromatic group of F4 of NCC is maintained in this position pointing toward helices TM1, TM2, and TM7, within an hydrophobic environment including Y581,39, Y3097,43, I541,35, and A3067,40 (Fig. 5, lower panel).
According to our calculations reported in Table 1, the donor backbone nitrogens of residues 1–4 of NCC are surrounded by the acceptor oxygens of N1333,35, D1303,32, G3087,42, and Y3097,43 and form hydrogen bonds with high stability. Moreover, the more stable bonds are formed between N(F1) and OD1 (N1333,35), 100% stable; NH1/2(R8) and O(C200ECL2), 89% stable; N(G2) and OD1/2(D1303,32), 74% stable; NE(R8) and OD1/2(D1102,63), 64% stable. The key aspartate D1303,32 is further involved in hydrogen bonds with the N-terminal phenylalanine, N(F1)-OD1/OD2(D1303,32), 56% stable; and N(F1)-O(D130 (3,32)), 50% stable. Finally, the positively charged lysine K9 is involved in alternative H-bonds with two negative glutamate residues of ECL2: NZ(K9)-OE1/2(E194ECL2), 44% stable; and NZ(K9)-OE1(Q208ECL2), 44% stable. Fig. 7 shows the more stable hydrogen bonds in two different snapshots: the representative structure of State_1 (t = 0, upper panel), and the structure corresponding to t = 260 ns of the concatenated trajectory (lower panel) giving rise to the distance plot of Fig. 4 (when all of the more stable hydrogen bonds are formed).
Figure 7.
Most stable H-bonds between NCC (orange) and NOP. (Upper panel) State_1 (representative structure) is shown. (Lower panel) t = 260 ns of the concatenated trajectory giving rise to the distance plot of Fig. 4 is shown. At this time all the most stable H-bonds of Table 1 are formed. To see this figure in color, go online.
Interestingly, in both the conformations represented in Fig. 7, the NCC fragment 8–13 containing the positively charged R8, K9, R12, and K13 strongly interacts with D1102,63, and with an electrostatic trap constituted by several negatively charged NOP residues within ECL2 (sequence 194–199, EDEEIE, specific for NOP), in agreement with fluorescence and NMR experiments on a model system of NOP_NCC, based on synthetic peptides, reporting that ECL2 contributes to selective ligand binding with low affinity, essentially due to ionic interactions (33). The electrostatic field is stabilized by direct hydrogen bonds with C200ECL2, N208ECL2, and salt bridges with E194ECL2 and E199ECL2. In particular, the H-bond between NH1/2(R8) of NCC and O(C200ECL2) is 90% stable along the repeated MD trajectories. The stacked positioning of the side chains of the acidic residue D1102,63 and of the electrostatic trap of ECL, interacting with the positive residues of NCC (Fig. 7) favor an α-helix conformation of the 6–12 segment of the agonist. Actually the α-helix content of this segment interconverts several times during our calculated trajectories between a fully formed and a destructured one (compare upper panel and lower panel of Fig. 7; see also Fig. 4, left panel)
Thus, interactions of the address region of NCC with TM2 and ECL2 rather than with TM3,6,7 is in agreement with those of “nonclassical” opioids (10). According to this result, specificity of the ECL domains should contribute to NOP selectivity for NCC, even if further contributions from the so-called negative determinacy, i.e., selectivity due to negative interaction with other extracellular domains of the receptor (34, 35) cannot be ruled out. Indeed, the comparison between the sequences of NCC and of the classical k-opioid dynorphin A (YGGFLRRIRPKLKWDNQ) shows a proline residue at position 10 in the latter one, which has significant effects on the backbone conformation; thus it is likely to be crucial in lowering affinity for NOP with respect to the endogenous agonist. However, even the chimeric peptide obtained by substituting the six C-terminal residues of NCC (RKLANQ) with those of dynorphin (LKWDNQ) markedly impairs the affinity and activity profile toward NOP (36). As previously mentioned, the absence of the last four N-terminal residues in the synthetic fragment NCC(1–13)-NH2 does not change its activity (31), thus the affinity impairment due to C-terminal substitution could be a negative-determinacy effect due to the presence of the bulky aromatic side chain of W14 and of the acidic side chain of D15, both of them disfavoring the interactions with the electrostatic trap of ECL2.
Our trajectories of State_1 show a strong stability of the H-bonds between K9 and both C200ECL2 and D1102,63 (Table 1). The latter one could explain why the D110A point mutation affects the NCC potency the most with respect to other measured ones, e.g., D130A (6).
The role of structural water
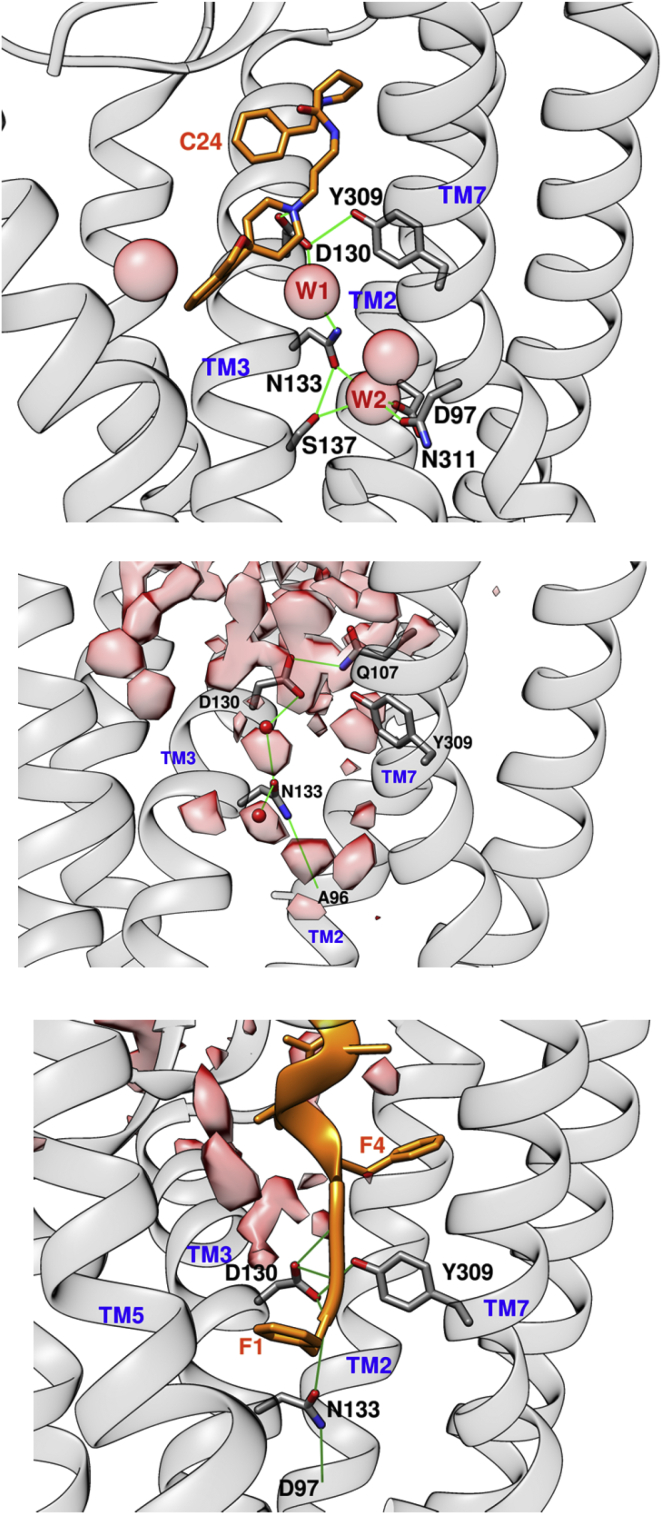
We have searched for the presence of structural waters in the binding pocket in the NOP_NCC complex and in the NOP_free, and compared our results in light of the reported x-ray structure of NOP_C24 (PDB: 4EA3), in which four waters were found in proximity of the small molecule ligand (6) (Fig. 8, upper panel). We have calculated the water density around NCC along the four concatenated trajectories of Fig. 4 for State_1 (Fig. 8, lower panel) and along a 100 ns trajectory of the NOP_free structure (Fig. 8, central panel). Several density spots are visible as pink blobs in both simulation data sets, even though, as expected, in the absence of the endogenous ligand the wider space available in the pocket is more homogenously filled with regions of high density of water. The water density in the NOP_free simulation indicates the presence of some of the water molecules in the same positions as in the NOP_C24 complex, including the one (W1) bridging N1333,35 to D1303,32, with the important difference that the orientation of the amide group of N1333,35 is flipped with respect to the crystallographic data. On the contrary the results of the simulation of the NOP_NCC complex show a “dried” deep site where no water density is found below the F1 N-terminal head of NCC, and the orientation of the amide group of asparagine N1333,35 is the same as in the NOP_free receptor.
Figure 8.
(Upper panel) H-bond network connecting C24 (PDB: 4EA3) to TM2,3,7. Water is depicted as pink spheres. (Central panel) Water-density (pink) and H-bond network calculated for NOP_free are shown. (Lower panel) Water density and H-bond network for the NOP_NCC complex are shown. A flip between the oxygen and nitrogen amide atoms of N1333,35 could act as a microswitch for the equilibrium between the active and inactive form of the receptor. To see this figure in color, go online.
NCC binding to the in silico mutant N133A of NOP
We have tested the effect of the N133A point mutation on the FES. In Fig. S4 the convergence of the METAD trajectory (upper panel), and the final 2D FES diagram (lower panel) are displayed as a function of the same variables, i.e., the α-helix content of fragments 2–9 and 10–16 of NCC. The absolute minimum of the FES (State_1 of the N133A mutant) is −202 kJ/mol at position [0.12, 0.04] with an error of 0.04, i.e., the α-helix content is negligible for the external segment, and very low for the internal one. The representative structures of State_1 for the wild-type and the N133A mutant are compared in Fig. S5. Notably, the direct interaction of F1(NCC) with TM3 is reduced to a single H-bond with D1303,32, while the interaction between R8(NCC) and the D1102,63, E199ECL2, and C200ECL2 of the electrostatic trap is conserved. The distributions of the distances d_F1(NCC)-CG(D130) and d_F1(NCC)-CA(N/A1333,35) along the two overall METAD runs leading to the FES diagrams are compared for the WT and the mutant (Fig. S5, lower panel), providing evidence that in the mutant, although NCC still forms one H-bond with D1303,32, the NCC backbone penetrates ∼5 Å less inside the binding pocket, forming alternative H-bonds with V2796,51, R3027,36, and T3057,39. In Fig. S6, the water density calculated for a cluster of ∼100 structures around the absolute minimum of the FES diagram (State_0 of the N133A mutant) is shown to be comparable with analogous pictures of Fig. 8: without the formation of the N(F1)-OD1(N1333,35) bond, water penetrates deeper than in the WT NOP-NCC complex (Fig. 8, lower panel), restoring part of the water-bridge interactions observed for the NOP_free receptor (Fig. 8, central panel), and the NOP-C24 complex (Fig. 8, upper panel).
Discussion
In this study, a dynamic representation of NCC binding to the inactive form of NOP has been obtained. From this picture, clues to the very first events of the inactive to active transition can be searched for, the action mechanism being still unknown. The results of our simulations suggest that residue N1333,35 could have a role in modulating the equilibrium between inactive and active conformations of NOP. In the crystal structure of the NOP_C24 complex, N1333,35 lies in an extended hydrogen bonding network, including direct interactions with S1373,39, T1363,38, and I1293,31, and water-bridged to D1303,32, S1373,39, N3117,45, S3127,46, D972,50, and G3087,42. As shown in Fig. 8 (upper panel), a water bridge (molecule W1) between D1303,32 and N1333,35 maintains the amidic nitrogen of N1333,35 pointing toward the extracellular side, while the amidic oxygen, hydrogen-bonded to the donor oxygen of the side chain of S1373,39 and to another water molecule (W2), points toward the center of the TM bundle. In particular this second water molecule is hydrogen-bonded to other well-conserved residues, S1373,39, D972,50, and N3117,45, in a position that in the structure of the delta opioid receptor reported by crystallography, is occupied by a sodium ion (37) (PDB: 4N6H). The role of sodium as negative modulator of agonist binding in opioid receptors (38), and in particular in NOP (39, 40) is well known and its positioning within NOP, in place of water W2, is therefore very likely.
The F1 of NCC occupies a position closer to N1333,35, approximately the same as water W1 as shown in the crystal structure of NOP_C24, and compatible with the water density calculated along the simulation of the NOP_free; the amidic oxygen of N1333,35 is directly hydrogen-bonded to N(F1) and the asparagine is no longer water-bridged to D1303,32. Therefore, we suggest that the orientation of the amide group of N1333,35 is a molecular microswitch that triggers the readjustment of the overall H-bond network between active and inactive conformations of the NOP receptor. This perturbation includes a destabilization of the segment 134–137 of TM3, conserved in the opioid receptor family, as shown by the analysis of the fraction of formation of the hydrogen bonds in NOP_C24, NOP_free, and NOP_NCC (Fig. S7). Thus, although there is no direct link between N1333,35 and the helices 5,6,7 (thought to be co-involved in the transition toward the active conformation), the perturbation of an extended segment of TM3 helix in the inner core, and a rearrangement of water-mediated hydrogen bond network close to the extracellular side of the receptor, could act as a trigger as well.
These results, together with the N133A in silico mutant data that show a higher distance between F1(NCC) and the mutated residue, as compared with the WT receptor, further support the proposed central role of N1333,35 as a microswitch involved in balancing the equilibrium between inactive and active states of the receptor. However, our investigations are limited to the nanosecond timescale, and further evidence can be given only by either experiments providing functional data on this mutant, or longer MD trajectories in the microsecond time window or more.
According to the microswitch hypothesis, we can provide a mechanistic explanation of the negative allosteric effect of sodium for agonist binding to NOP. In the simulation of NOP_free, the amide group of N1333,35 is oriented similar to the agonist bound form, with the amidic nitrogen ND2 hydrogen-bonded to A962,49. The replacement of the water W2 with the positively charged Na+ ion positioned in the site surrounded by S1373,39, D972,50, and N1333,35 (conserved among human opioid receptors) should stabilize the inactive configuration by attracting the partially negative charge of the amidic OD1, and repelling the partially positive charge of ND2, i.e., by inverting the orientation of the amide group of N1333,35. The rotation of the amide group and the readjustment of the overall surrounding water network would impair the formation of the N(F1)-OD1(N1333,35) hydrogen bond leading to the NOP-agonist complex. Our proposed explanation of the allosteric mechanism is fully supported by a 100 ns MD simulation of the NOP_C24 complex where water W2 has been replaced by a sodium ion (Fig. S8). The sodium ion is quite stable in this position, forming several salt bridges as illustrated in the figure, and especially, as supposed, a 91% stable salt bridge with OD1(N1333,35). On the contrary in the presence of the agonist and in the absence of the Na+ ion, the binding of the message domain of the agonist deeply inside the hydrophobic pocket would be favored, leading to collapse of the binding pocket with the extrusion of the inner structural waters, and with a possible shift of the equilibrium between the inactive and active conformations of the receptor toward the active one.
Conclusions
In conclusion, we have obtained a dynamical picture of the binding of NCC to the inactive state of NOP by the free-energy profiles as a function of the α-helix content of different segments of NCC, provided by METAD. The main structural features we have found are in agreement with the overall set of functional data derived after point mutations. The pose of the message domain (residues 1–4) of NCC has been found as slightly slipped deeper inside the TM bundle with respect to the known antagonists, and the new position allows a mechanistic explanation of the negative allosteric effect by Na+ ion, for which a very likely location inside the TM bundle has been identified. A residual α-helix content in the central part of the NCC peptide (residues 4–9) is maintained, whereas the C-terminal segment (residues 13–17) is unstructured, highly flexible but dynamically connected to the ECL2 domain in agreement to the importance of this external loop in the selection of the endogenous ligand. A pivotal role for residue N1333,35 is suggested and corroborated by the results of the in silico mutant N133A, as a molecular microswitch that could perturb the equilibrium between the active and inactive NOP conformations, in the framework of an extended H-bond network rearrangement, including several solvent waters present in the deep binding site. Clearly, single-point mutation experiments on the key residue identified by this study could prove useful.
Author Contributions
S.D.L. conceived the idea for the project, conducted all the bioinformatic tools, analyzed the results, and wrote the article; A.A. analyzed the results and wrote the article.
Acknowledgments
We thank Dr. Isabella Baccarelli and Dr. Nicola Spallanzani of the CINECA supercomputing centers for their help in the initial setup of calculations.
This work was supported in whole or part by Italian Ministry of University and Research (LINEA D1 Università Cattolica del Sacro Cuore) and by the CINECA supercomputing centers through the grant IsC34 (n.HP10CWX60F).
Editor: Amedeo Caflisch
Footnotes
Eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30521-5.
Supporting Material
References
- 1.Meunier J.C., Mollereau C., Costentin J. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 1995;377:532–535. doi: 10.1038/377532a0. [DOI] [PubMed] [Google Scholar]
- 2.Reinscheid R.K., Nothacker H.P., Civelli O. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- 3.Chiou L.C., Liao Y.Y., Prinssen E.P. Nociceptin/orphanin FQ peptide receptors: pharmacology and clinical implications. Curr. Drug Targets. 2007;8:117–135. doi: 10.2174/138945007779315605. [DOI] [PubMed] [Google Scholar]
- 4.Mogil J.S., Pasternak G.W. The molecular and behavioral pharmacology of the orphanin FQ/nociceptin peptide and receptor family. Pharmacol. Rev. 2001;53:381–415. [PubMed] [Google Scholar]
- 5.Lambert D.G. The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat. Rev. Drug Discov. 2008;7:694–710. doi: 10.1038/nrd2572. [DOI] [PubMed] [Google Scholar]
- 6.Thompson A.A., Liu W., Stevens R.C. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller R.L., Thompson A.A., Stevens R.C. The importance of ligand-receptor conformational pairs in stabilization: spotlight on the N/OFQ G protein-coupled receptor. Structure. 2015;23:2291–2299. doi: 10.1016/j.str.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobilka B.K., Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Orsini M.J., Nesmelova I., Mayo K.H. The nociceptin pharmacophore site for opioid receptor binding derived from the NMR structure and bioactivity relationships. J. Biol. Chem. 2005;280:8134–8142. doi: 10.1074/jbc.M406405200. [DOI] [PubMed] [Google Scholar]
- 10.Filizola M., Devi L.A. Structural biology: how opioid drugs bind to receptors. Nature. 2012;485:314–317. doi: 10.1038/485314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kothandan G., Gadhe C.G., Cho S.J. The nociceptin receptor (NOPR) and its interaction with clinically important agonist molecules: a membrane molecular dynamics simulation study. Mol. Biosyst. 2014;10:3188–3198. doi: 10.1039/c4mb00323c. [DOI] [PubMed] [Google Scholar]
- 12.Yakovleva T., Bazov I., Bakalkin G. Prodynorphin storage and processing in axon terminals and dendrites. FASEB J. 2006;20:2124–2126. doi: 10.1096/fj.06-6174fje. [DOI] [PubMed] [Google Scholar]
- 13.Pietrucci F., Laio A. A collective variable for the efficient exploration of protein beta-structures with metadynamics: application to SH3 and GB1. J. Chem. Theory Comput. 2009;5:2197–2201. doi: 10.1021/ct900202f. [DOI] [PubMed] [Google Scholar]
- 14.Ballesteros J., Weinstein H. Academic Press; San Diego, CA: 1995. Integrated Methods for the Construction of Three Dimensional Models and Computational Probing of Structure Function Relations in G Protein-Coupled Receptors. [Google Scholar]
- 15.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 16.Pierce B.G., Wiehe K., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkatakrishnan A.J., Deupi X., Babu M.M. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 18.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 19.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 20.Chen C., Ke J., Melcher K. Structural basis for molecular recognition of folic acid by folate receptors. Nature. 2013;500:486–489. doi: 10.1038/nature12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tribello G.A., Bonomi M., Bussi G. Plumed 2.0: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 22.Nosé S. A unified formulation of the constant temperature molecular-dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 23.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 24.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 25.Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonomi M., Parrinello M. Enhanced sampling in the well-tempered ensemble. Phys. Rev. Lett. 2010;104:190601. doi: 10.1103/PhysRevLett.104.190601. [DOI] [PubMed] [Google Scholar]
- 27.Laio A., Rodriguez-Fortea A., Parrinello M. Assessing the accuracy of metadynamics. J. Phys. Chem. B. 2005;109:6714–6721. doi: 10.1021/jp045424k. [DOI] [PubMed] [Google Scholar]
- 28.Daura X., Gademann K., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. 1999;38:236–240. [Google Scholar]
- 29.O’Connor C., White K.L., Milon A. NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc. Natl. Acad. Sci. USA. 2015;112:11852–11857. doi: 10.1073/pnas.1510117112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butour J.L., Moisand C., Meunier J.C. Recognition and activation of the opioid receptor-like ORL 1 receptor by nociceptin, nociceptin analogs and opioids. Eur. J. Pharmacol. 1997;321:97–103. doi: 10.1016/s0014-2999(96)00919-3. [DOI] [PubMed] [Google Scholar]
- 31.Guerrini R., Calo G., Regoli D. Address and message sequences for the nociceptin receptor: a structure-activity study of nociceptin-(1-13)-peptide amide. J. Med. Chem. 1997;40:1789–1793. doi: 10.1021/jm970011b. [DOI] [PubMed] [Google Scholar]
- 32.Calo G., Guerrini R., Regoli D. UFP-101, a peptide antagonist selective for the nociceptin/orphanin FQ receptor. CNS Drug Rev. 2005;11:97–112. doi: 10.1111/j.1527-3458.2005.tb00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent B., Mouledous L., Demange P. Description of the low-affinity interaction between nociceptin and the second extracellular loop of its receptor by fluorescence and NMR spectroscopies. J. Pept. Sci. 2008;14:1183–1194. doi: 10.1002/psc.1057. [DOI] [PubMed] [Google Scholar]
- 34.Moyle W.R., Campbell R.K., Wang X. Co-evolution of ligand-receptor pairs. Nature. 1994;368:251–255. doi: 10.1038/368251a0. [DOI] [PubMed] [Google Scholar]
- 35.Metzger T.G., Ferguson D.M. On the role of extracellular loops of opioid receptors in conferring ligand selectivity. FEBS Lett. 1995;375:1–4. doi: 10.1016/0014-5793(95)01185-h. [DOI] [PubMed] [Google Scholar]
- 36.Lapalu S., Moisand C., Meunier J.C. Comparison of the structure-activity relationships of nociceptin and dynorphin A using chimeric peptides. FEBS Lett. 1997;417:333–336. doi: 10.1016/s0014-5793(97)01318-5. [DOI] [PubMed] [Google Scholar]
- 37.Granier S., Manglik A., Kobilka B.K. Structure of the δ-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pert C.B., Pasternak G., Snyder S.H. Opiate agonists and antagonists discriminated by receptor binding in brain. Science. 1973;182:1359–1361. doi: 10.1126/science.182.4119.1359. [DOI] [PubMed] [Google Scholar]
- 39.Ardati A., Henningsen R.A., Monsma F.J., Jr. Interaction of [3H]orphanin FQ and 125I-Tyr14-orphanin FQ with the orphanin FQ receptor: kinetics and modulation by cations and guanine nucleotides. Mol. Pharmacol. 1997;51:816–824. doi: 10.1124/mol.51.5.816. [DOI] [PubMed] [Google Scholar]
- 40.Mahmoud S., Margas W., Ruiz-Velasco V. Modulation of silent and constitutively active nociceptin/orphanin FQ receptors by potent receptor antagonists and Na+ ions in rat sympathetic neurons. Mol. Pharmacol. 2010;77:804–817. doi: 10.1124/mol.109.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.