Abstract
Optogenetics describes the use of genetically encoded photosensitive proteins to direct intended biological processes with light in recombinant and native systems. While most of these light-responsive proteins were originally discovered in photosynthetic organisms, the past few decades have been punctuated by experiments that not only commandeer but also engineer and enhance these natural tools to explore a wide variety of physiological questions. In addition, the ability to tune dynamic range and kinetic rates of optogenetic actuators is a challenging question that is heavily explored with computational methods devised to facilitate optimization of these systems. Here, we explain the basic mechanisms of a few popular photodimerizing optogenetic systems, discuss applications, compare optogenetic tools against more traditional chemical methods, and propose a simple quantitative understanding of how actuators exert their influence on targeted processes.
Main Text
Optogenetic systems in their broader context
By harnessing the power of genetically encoded light-sensitive proteins, optogenetics seeks to interrogate biological processes via two complementary methods: sensing and actuating (1). Sensors, derived mainly from fluorescent proteins, measure a plethora of outputs. Actuators, composed chiefly of photoreceptor proteins, exert distinctive and diverse cellular functions.
Two major approaches for designing sensors are manipulating fluorescent protein spectral properties and inducing Förster resonance energy transfer efficiency changes under specified conditions (2). While both engineering blueprints translate protein conformational changes into functional measurements of fluorescence, the general principle of design for the first type of sensor relies upon the signal changes of a single fluorescent protein typically arising from β-barrel structural rearrangements when it senses the intended target (3). In contrast, the second type of sensor correlates its conformational changes in the absence or presence of the intended target to an effective measurement of Förster resonance energy transfer efficiency between two fluorescent proteins (4, 5). Well-known sensors report changes in intracellular calcium (Ca2+) (6), voltage (7), and secondary messengers (8).
Actuators may also be subdivided into two major groups: microbial opsins (9) and photomultimerizing proteins. From microbial opsins, rhodopsins have come to the forefront of optogenetics with channelrhodopsins (10) and halorhodopsins, which depolarize to activate (11) and hyperpolarize to inhibit action potential firing in the cell, respectively (12). Pioneering work with these Chlamydomonas reinhardtii proteins show that they can fine-tune neuronal responses through single-cell excitation (13). Other design tactics with photoswitchable receptors (14) also corroborate the usefulness of optogenetics for many purposes including brain-mapping and studying neurological diseases (15).
While the combinatorial use of sensors and microbial opsins revolutionized many aspects of neuroscience and basic science research, this article will not proceed beyond citing comprehensive reviews for these topics. We will also offer little practical guidance for using optogenetic switches past citing relevant reviews as there are many interesting discussions on how to choose an optogenetic system based upon dynamic range (16), kinetics (17), and experimental requirements (18). Instead, we focus on understanding broader concepts for the various categories of uses for photomultimerizing proteins in probing and engineering biology and medicine today. More specifically, we will discuss the origin and development of optogenetic tools and then present a simple quantitative understanding of optogenetic systems derived from these proteins.
Development of optogenetic designs
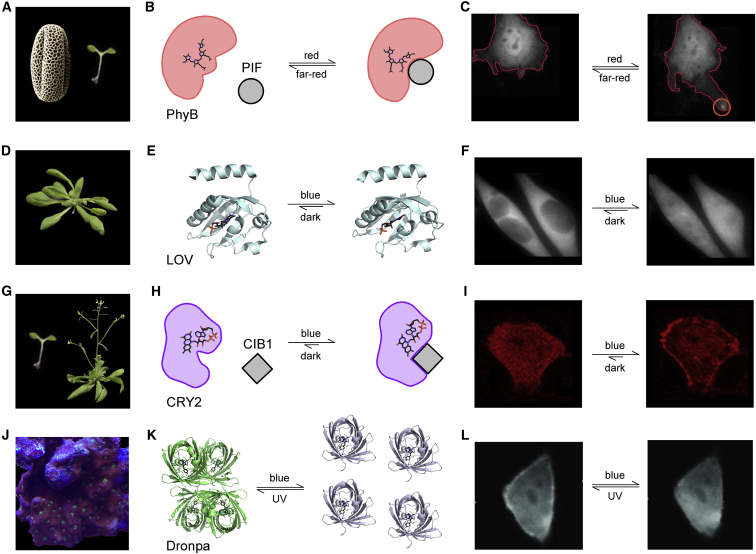
Since the dawn of evolution, primary producers have been translating light into cellular signals. Accordingly, the more primitive forms of these photomultimerizing actuators originated from scientists studying the regulation of transcription pathways in plants as well as similar mechanisms in other photosynthetic organisms such as algae and cyanobacteria. The mustard plant Arabidopsis thaliana is recognized for phytochrome B (PhyB)/phytochrome-interacting factor (PIF) (Fig. 1 A), light-oxygen-voltage (LOV) domain (Fig. 1 D), and cryptochrome2 (CRY2)/cryptochrome-interacting basic-helix-loop-helix (CIB1)-based (Fig. 1 G) optogenetic systems while isolation of mutant GFP-like proteins from the Pectiniidae coral family, also known as chalice coral, brought about Dronpa (Fig. 1 J)-based systems.
Figure 1.
Organism of origin and relevant lifecycle is shown for various optogenetic actuators. (A) PhyB is known to be involved in germination and flowering of Arabidopsis thaliana (84). (B) Cartoon depiction of PhyB/PIF photoswitching mechanism with chromophore structure (85). (de-etioliation C) PhyB/PIF-induced lamellipodia formation (red circle) (86). (D) LOV domain of phototropin is a key component of phototropism and stomatal opening in Arabidopsis (84). (E) Atomic structures show two light-dependent configurations of LOV domain isolated from Avena sativa (left, Protein Data Bank (PDB): 2V0U; right, PDB: 2V0W) without the Jα helix (87). An alternate mechanism is LOV photodimerization with an EL222 mutation and VVD. (F) Lov2-Jα mediated nuclear localization (88). (G) CRY2 is an important player in flowering and deetiolation of Arabidopsis (84). (H) Cartoon rendition of CRY2/CIB1 dimerization with blue light (89). Light-dependent oligomerization of CRY2 is another powerful optogenetic mode of operation. (I) Membrane localization of CRY2-Akt in response to blue light (44). (J) Dronpa is a homolog of GFP found in coral Pectiniidae. (K) Crystal structures show reverse dimerization of Dronpa (left, tetramer PDB: 2POX (32); right, monomers PDB: 2IOV (90)). (Green) Fluorescent state; (gray) dark state of Dronpa. (L) Functional exemplar of light-regulated photoswitching of Dronpa (30). For visualization of relocalization, the fluorescence of mNeptune tagged to Dronpa is monitored. Images in (A), (D), and (G) are reproduced from Krämer (91). To see this figure in color, go online.
The oldest characterized system from the Arabidopsis group is the PhyB/PIF system, which may be traced as far back as the 1950s. Lettuce seeds were observed to germinate at differing speeds; red light promoted germination while infrared light repressed germination (19). From these findings, it was assumed that a pigment was chemically changing depending on the wavelength of light. Approximately four decades later, this pigment was identified as PhyB (20). Upon light stimulation, PhyB binds to its partner PIF3, which is a transcription factor with a helix-loop-helix structure, and begins DNA transcription to signal seed germination (Fig. 1 B) (21).
Similarly, the LOV domain was found to be an important photosensing protein when it was discovered that exposure to blue light resulted in heavy phosphorylation of the protein encoded in the NPH1 gene of Arabidopsis (22). When LOV is irradiated with blue light, the flavin cofactor forms a covalent bond with a key cysteine residue and changes the conformation of the LOV domain (Fig. 1 E). Many studies focus on optimizing this system as the different types of LOV domains offer a large selection of kinetic rates from highly stable conformations in both the dark state and light state as seen with FKF1 (23) to fast on- and off-rates as with AsLOV2 (24).
CRY2 is a member of the cryptochrome family, and some argue that it takes part in regulating the circadian rhythm like its homolog CRY1 (25). More important for the points discussed here, however, is CRY2’s integral role with its partner CIB1 (Fig. 1 H) in modulating transcription leading to Arabidopsis flowering (26). Recently, the pair garnered a lot of attention due to their intermediate kinetic rates with an on-rate of milliseconds and an off-rate of minutes (27). The CRY2 mutation E490G also dramatically increases the oligomerization affinity of CRY2 (CRY2olig) and induces clustering at the expense of slower kinetics (28). Nonetheless, CRY2olig provides another powerful flavor of optical signaling through clustering compared to colocalization of CRY2 with its binding partner CIB1.
Lastly, Dronpa was initially found as a homolog of GFP in the coral family Pectiniidae (Fig. 1 J) (29). Monomers of Dronpa bind to one another to form dimers, and then these dimers come together to form tetramers. A point mutation, K145N, was introduced to weaken tetramer formation and thereby enhance the probability of finding Dronpa dimers (30). With blue light, Dronpa switches from a fluorescent multimer to dark monomers and reverses with ultraviolet light (Fig. 1 K). Unlike the PhyB/PIF system, Dronpa reverses dimerization, allowing it to photo-uncage enzymes (Fig. 1 L). In addition, the fascinating property of fluorophore switching (31, 32) makes Dronpa unique from its fellow optogenetic proteins in it can also serve as a sensor of its own activity (30).
Topics of achievement in discovering and constructing cellular processes
With the elucidation of these systems, many have selected their favorite optogenetic tool to examine their preferred biological problems. Some common themes that emerge through nature and converge with other multimerization systems, such as chemically inducible dimerization (CID) systems (33, 34), include manipulating gene expression and toggling molecular concentrations with localization and clustering-based logic. Classic problems that have been explored include activation of DNA transcription and other well-known molecular pathways, while newer ventures focus on other modes of intracellular signaling such as Ca2+ regulation.
There is much success associated with using these tools to control gene expression as many of these systems were derived from light-sensitive transcription pathways. In one case, the EL222 transcription factor, which contains a LOV domain and helix-turn-helix (HTH) motif, was used to upregulate the production of luciferase in HEK293T cells. While LOV binds HTH in the dark, blue light stimulation causes LOV to undock HTH, which leads to HTH dimerization, then HTH dimer-DNA binding, and finally, DNA transcription (35). Another example is using CRY2/CIB1 with epigenetic modifiers such as LITE (36) or permanent genetic modifiers such as TALEN and CRISPR-Cas9 to pattern perpetual transcription profiles in cells spatially (37, 38). Similar experiments have also been performed with split-Cas9 and Magnets, a variant of LOV domains, so that upon blue light stimulation, magnets dimerize and produce a functional Cas9 (39). Outside of single-cell applications, these tools have also been implemented in model organisms. The CRY2/CIB1 system has been shown to drive luciferase expression in zebrafish (40) as well as increase GFP levels in Drosophila (41) under their respective transcription systems.
More phenotypical assessments of cellular output with optogenetic systems target well-understood molecular pathways such as Rac activation in membrane ruffling and cell motility. A photoactivatable version of Rac1 with LovJα-directed cell movement with blue light stimulation (42). Comparably, the PhyB/PIF system was also used to show that it is not only possible to direct lamellipodia and filopodia formation (Fig. 1 C) (43) but also possible to generate a train of signals and subsequently evaluate how the functional output of cell survival and cell growth changes with stimulation frequency of the Ras/ERK pathway (18). Moreover, the achievements of optogenetic techniques have not been limited to cell motility. Akt localization to the membrane with CRY2/CIB1 (Fig. 1 I) uncovered the importance of Akt in mechanistic target of the rapamycin growth pathway and the forkhead box O metabolism pathway for the generation of Atrogin1 implicated in muscle atrophy (44).
One powerful notion is using optogenetics to control Ca2+ signaling. Ca2+ regulation is important for numerous yet dissimilar physiological processes including transcription activation, cell death, and neurological development (45, 46). Ca2+ release-activated channels (later mapped to the ORAI gene) and stromal interaction molecules (STIM) were first discovered because of their importance in T cell activation. When Ca2+ stores are depleted, it is believed that STIM EF-hands undergo a conformational change that allows aggregation of the C-terminal domain of STIM (CCD) to prompt the opening of ORAI (47). Recently, multiple groups confirmed that it is possible to increase cytosolic Ca2+ levels by opening ORAI channels through either clustering CCD with CRY2olig (48) or releasing CCD with the LOV domain (49). Thus, these studies provide additional steps toward examining more complex processes involving Ca2+ signaling.
Explicit differences between optogenetics and CID techniques
One established rival technology we would like to consider is CID (34). A few points of contemplation important for understanding the differences between optogenetics and CID include inherent properties of the systems themselves and experimental equipment. Explicitly, these two factors influence the reversibility, spatial specificity, temporal precision, and dynamic range of biological signals generated by these systems during experiments.
Generally, after light-sensitive proteins absorb photons, they enter a high-energy state with a different conformation due to energy transfer. Then, when the light source is removed, they relax and revert to their original conformation. As most CID processes appear irreversible under experimental conditions, optogenetic systems are attractive for their transient responses that enable reversible cycling of signals. One possible resolution for the irreversibility of CID systems, however, is the idea of using a mixture of different CID systems to create antagonistic reversal of the original signal (50). A process that has been highlighted with both optogenetics and CID is KCNQ2/3 channel activity. In 2006, it was proved with CID that PIP2 levels in the plasma membrane directly modulate KCNQ2/3 so that current decreases upon PIP2 depletion by an engineered PIP2 phosphatase (51). Shortly after the popularization of optogenetic techniques, the CRY2/CIB1 system was also applied to the regulation of KCNQ2/3 channels. In addition to replicating the attenuation of current from the original CID experiment, a measurement of the recovery of current and PIP2 concentrations at the membrane could also be made within 10 min (52).
Another set of experiments for comparison is probing processes that require spatial signaling differences such as cell polarity. Creating molecular concentration gradients is a challenging problem that requires proper equipment for both CID and optogenetic systems. For CID, careful design of microfluidic devices or biomimetic scaffolding is imperative to correctly shape a cell’s chemical environment such as with graded rapamycin activation of Rac1 (43). On the other hand, technological advances of microscope setups with optical filters, pinholes (53), digital micromirror devices (54), and even endoscopy tools (55) provide a variety of effective solutions for defining the field of view stimulated with light. Examples include controlling the polarity of yeast for budding with Bem1 (56), using CRY2olig to cluster receptor tyrosine kinases (57), and using CRY2/CIB1 to manipulate G-protein-coupled receptors that promote lamellipodia and cell migration (58). LOV domains have also been used to optogenetically control protein degradation (59).
Speed of stimulus delivery is an area in which optogenetics exceeds its chemical competitors. Most optogenetic systems produce the desired effect within seconds while CID systems require tens of seconds due to limited delivery of rapamycin. One example of this phenomenon is seen with organelle transport by kinesins and dynein where directed migration of peroxisomes using the LOV domain linked to the respective cytoskeleton motor (60) resulted in an approximately twofold faster recruitment compared to its CID analog (61). Regardless, both systems offer great promise for answering a myriad of biological questions and contributing to the synthetic cell biology toolbox (62), but still require optimization and further specific design to reduce unwanted chemical and phototoxicity effects as well as undesired basal cellular perturbations.
Dynamic range in relation to kinetics
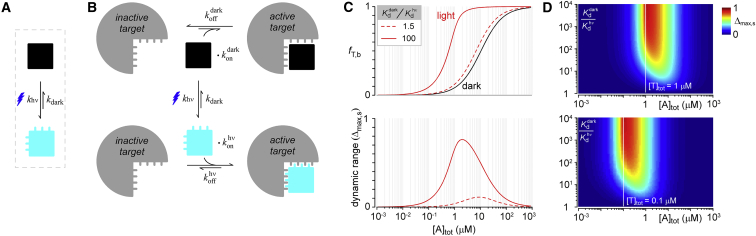
On the topic of dynamic range and speed, it may be useful to contemplate a general scheme for how photomultimerizing proteins behave as optogenetic actuators. Upon photoexcitation, light-sensitive proteins undergo conformational changes (Fig. 2 A) that result in functional outputs. For the photomultimerizing optogenetic tools described above, this conformational change often exposes a hidden surface on the light-sensitive protein that allows it to interact with its target. This bimolecular reaction converts the target into its active form (Fig. 2 B). These actuators may also interact with their target in the dark, albeit with a substantially weaker affinity, leading to basal activity. Thus, the output of target’s activity in a cell is directly proportional to the fraction of targets bound to an actuator (fT,b). If we assume rapid and complete photoconversion of the optogenetic actuator (free concentration, ), then after light activation is governed by
| (1) |
Solving Eq. 1 for steady state yields
| (2) |
where is off-rate, is on-rate, and the ratio represents the affinity of the actuator for the target molecule in the presence of light. Because the free concentration of optogenetic actuator in a cell is unknown, a more convenient formulation of Eq. 2 assesses in terms of total concentrations of actuator and target molecules:
| (3) |
Given that the actuator can also bind to the target in darkness, its dynamics is determined by
| (4) |
assuming that the on/off-rate constants change instantaneously. Similar to Eq. 2, the steady-state solution of Eq. 4 is
| (5) |
where the ratio corresponds to affinity of the actuator for the target in the absence of light. Once again, for convenience, can be determined in terms of total concentrations of actuator and target molecules,
| (6) |
With this framework, the dynamics of the system can be viewed as a simple transition from one steady state to the other in the absence and presence of light with the time-course of relaxations determined by the appropriate on-off rate constants. The system’s maximal dynamic range (Δmax,s) considering only the signaling aspects of the system is then the difference between the dark and light steady-state solutions,
| (7) |
Note that the dynamic range defined here assumes a complete photoconversion of all actuator molecules. In practice, depending on the wavelength of the incident light and the optical fluence of the sample (63, 64), expressing the optogenetic dimerizing tool, the actual dynamic range may be substantially lower.
Figure 2.
Schematic depiction of the behavior of an optogenetic actuator. (A) Light-activation leads to conformational changes in the actuator. (B) Four molecular conformations are possible: unbound dark actuator, bound dark actuator, unbound lit actuator, and bound lit actuator. Physical changes resulting from light allow the protein to interact better with its target under illumination and bias it to reside as a bound lit actuator. (C) The top graph shows a simulation for the changes in fraction bound of actuator-target complexes as a function of actuator concentration due to different fold changes in actuator binding affinity before and after light. Similarly, the bottom graph illustrates the dynamic range of actuator-target complexes for these various conditions. (D) Top graph color maps dynamic range at a total target concentration of 1 μM as a function of total actuator concentration and binding affinity fold enhancement for lit versus dark states. An analogous graph is drawn on the bottom for a total target concentration 0.1 μM. (Open line) Total concentration of actuator and target are equal, which is also the optimal total actuator concentration for maximizing dynamic range. To see this figure in color, go online.
Two key outcomes follow from Eq. 7: (1) The dynamic range of the system relies upon the fold enhancement in the actuator’s binding affinity after light activation (Fig. 2 C). For example, if photoexcitation enhances the actuator’s binding affinity minimally (∼50%), then the fraction of target molecules (fT,b) bound to the actuator shifts subtly to the left (compare black curve to red dashed curve, Fig. 2 C, top) resulting in a limited dynamic range of the system . By contrast, a large increase in affinity results in a marked shift in fT,b (compare black curve to red solid curve, Fig. 2 C, top) and a substantial enhancement in the dynamic range . (2) The dynamic range of the system is also sensitive to the total concentration of the actuator in the cell—the “Goldilocks problem”. If the concentration of the actuator is very low, then only a small fraction of target molecules will become bound to an actuator after light stimulation (Fig. 2 C, bottom). Similarly, if the actuator is abundant in the cell, then a significant fraction of targets will be prebound to the actuator, resulting in high basal activity and minimal subsequent light-dependent recruitment (Fig. 2 C, bottom). By contrast, at intermediate actuator concentrations, there is minimal prebinding in the dark, but light-activation results in substantial translocation of the actuator to its target (Fig. 2 C, bottom). Further analysis of the conditions that improve the dynamic range of the optogenetic actuator system can be visualized in Fig. 2 D for a given concentration of target molecules in the cell (top, = 1 μM; bottom, = 0.1 μM). In both cases, the dynamic range improves if the fold enhancement in binding affinity upon photoexcitation is increased. In addition, the dynamic range is also improved when the total concentration of actuators in the cell matches the concentration of targets (Fig. 2 D; top, white line ∼ ∼1 μM; bottom, white line, ∼ ∼0.1 μM), akin to impedance matching in electronic circuits.
To optimize the dynamic range of the optogenetic system, one strategy is to maximize the actuator’s affinity for its target in the presence of light, which also broadens the system’s operating range. Strengthening the interactions that form at the interface between the two partners stabilizes the actuator-target bound complex. This manipulation decreases to reduce and reflects the enhanced affinity for the actuator-target interaction in the presence of light. An important shortcoming of this approach becomes apparent, however, if we consider the time constant of the optogenetic system in the presence of light,
| (8) |
where reducing inevitably increases , the time required to attain 63% recruitment. Thus, the net improvement in the affinity and the dynamic range comes at the cost of a kinetically slower system. Another complexity for this approach arises from the fact that the actuator binds the target in darkness. To fulfill the conditions of detailed balance in kinetic theory, the strengthened actuator-target interface also enhances the binding of the two proteins in the dark (65), resulting in increased basal activity of the actuator. An alternative strategy to optimize the system is to increase by destabilizing the unbound configuration in the presence of light. This process enhances and decreases , resulting in a system with a potentially large dynamic range and fast kinetics.
Although promising, such rational approaches to optimize optogenetic systems remain challenging without extensive structural information. Moreover, the schematic presented here renders the optogenetic process as a single-step reaction with a single time-constant limited with the actuator-target interaction being the limiting step. In most cases, however, the system is more complex with multiple steps that reflect the photophysics of the actuator chromophore and other intermediate relaxation steps. In addition, the dynamics of some optogenetic systems may be limited by the kinetics of downstream signaling by the target. For CRY2, studies argue an intermediate step may be due to stabilization through ATP binding (66). Nevertheless, ongoing studies are actively continuing to address these issues. Examples of specific models that incorporate further details include engineering of light-regulated histidine kinases (67), photocontrolled delivery of toxins to ion channels (68), light-gated transcription factors (35), and analysis of the channelrhodopsin photocycle (10, 69). These models have proven insightful for refinement and optimization of various optogenetic tools and may pave the path toward rationally designed synthetic actuators to manipulate biological function.
Particular successes for intentional design of photomultimerizing optogenetic tools include manipulating LOV domains, which have available structures (70). Furthermore, natural processes provide a library of diverse LOV proteins (71, 72). A few specific methods that have appeared to tackle this problem are computational modeling and crystal structure analysis with programs such as Rosetta (73, 74). By first predicting more stable dark forms of LOV, screening of these mutated proteins revealed a faster system with a larger dynamic range (17, 75). These concepts are also generalizable to describe coarse behavior of other systems including CID actuators. Overall, the quest for designing and optimizing optogenetic tools to manipulate various facets of biological function will be greatly facilitated by the increasing number of atomic structures that are available and through in-depth biophysical and kinetic analysis.
Future directions extrapolated from the past and present
Light-sensitive proteins are intriguing and useful for many reasons. Besides their application to basic sciences, medical fields have also employed optogenetics in retinal therapies (76), maintaining blood glucose homeostasis (77), and even adjusting transgene expression with brain activity (78). An idea that has resurfaced on the horizon is directing axonal growth with light. While a study performed more than a decade ago used caged Ca2+ to show light directing the growth cone of neurons (79), more recent studies used LOV (60) and CRY2/CIB1 (80) to guide neuronal growth. Hence, these methods may be very powerful for studying and understanding the formation of synapses and possibly memories.
Nonetheless, one should consider the contrasting advantages between optogenetics and other techniques, as this decision is dependent on a few criteria including equipment, time, space, signal effect, and available wavelengths of light for stimuli and sensor measurements. Merging optogenetics and other orthogonal systems such as CID may also provide extended flexibility for honing cellular control with more complex logics. All in all, these systems provide many tools important for advancing scientific knowledge. As crystal structures emerge in tandem with further understanding of important signaling cascades, the engineer’s dream of meticulously designing light-responsive cellular components may be closer than imagined (81, 82, 83). Ultimately, this new modality of interacting and manipulating biological systems echoes the brave new world we face.
Acknowledgments
This review is dedicated to David T. Yue, MD, Ph.D., who was not only an incredible scientist but also an amazing mentor—a man whose passion for science shed much light on his field of study as well as inspired many more around him.
Editor: Brian Salzberg.
Contributor Information
Jacqueline Niu, Email: jniu4@jhu.edu.
Takanari Inoue, Email: jctinoue@jhmi.edu.
References
- 1.Miesenböck G. The optogenetic catechism. Science. 2009;326:395–399. doi: 10.1126/science.1174520. [DOI] [PubMed] [Google Scholar]
- 2.Broussard G.J., Liang R., Tian L. Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 2014;7:97. doi: 10.3389/fnmol.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird G.S., Zacharias D.A., Tsien R.Y. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyawaki A., Llopis J., Tsien R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 5.Paramonov V.M., Mamaeva V., Rivero-Müller A. Genetically-encoded tools for cAMP probing and modulation in living systems. Front. Pharmacol. 2015;6:196. doi: 10.3389/fphar.2015.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakai J., Ohkura M., Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 7.Siegel M.S., Isacoff E.Y. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 8.Adams S.R., Harootunian A.T., Tsien R.Y. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 9.Grote M., Engelhard M., Hegemann P. Of ion pumps, sensors and channels—perspectives on microbial rhodopsins between science and history. Biochim. Biophys. Acta. 2014;1837:533–545. doi: 10.1016/j.bbabio.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Nagel G., Szellas T., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyden E.S., Zhang F., Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F., Wang L.-P., Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 13.Warden M.R., Cardin J.A., Deisseroth K. Optical neural interfaces. Annu. Rev. Biomed. Eng. 2014;16:103–129. doi: 10.1146/annurev-bioeng-071813-104733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiner A., Levitz J., Isacoff E.Y. Controlling ionotropic and metabotropic glutamate receptors with light: principles and potential. Curr. Opin. Pharmacol. 2015;20:135–143. doi: 10.1016/j.coph.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lüscher C., Pascoli V., Creed M. Optogenetic dissection of neural circuitry: from synaptic causalities to blue prints for novel treatments of behavioral diseases. Curr. Opin. Neurobiol. 2015;35:95–100. doi: 10.1016/j.conb.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Pathak G.P., Strickland D., Tucker C.L. Benchmarking of optical dimerizer systems. ACS Synth. Biol. 2014;3:832–838. doi: 10.1021/sb500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallett R., Zimmerman S.P., Kuhlman B. Correlating in vitro and in vivo activities of light inducible dimers: a cellular optogenetics guide. ACS Synth. Biol. 2015;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toettcher J.E., Weiner O.D., Lim W.A. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borthwick H.A., Hendricks S.B., Toole V.K. A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA. 1952;38:662–666. doi: 10.1073/pnas.38.8.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinomura T., Nagatani A., Furuya M. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome-b and secondarily by phytochrome-a. Plant Physiol. 1994;104:363–371. doi: 10.1104/pp.104.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni M., Tepperman J.M., Quail P.H. Binding of phytochrome b to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 22.Huala E., Oeller P.W., Briggs W.R. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 23.Imaizumi T., Tran H.G., Kay S.A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 24.Harper S.M., Neil L.C., Gardner K.H. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 25.Griffin E.A., Jr., Staknis D., Weitz C.J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 26.Liu H., Yu X., Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 27.Kennedy M.J., Hughes R.M., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taslimi A., Vrana J.D., Tucker C.L. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ando R., Mizuno H., Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X.X., Chung H.K., Lin M.Z. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habuchi S., Ando R., Hofkens J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc. Natl. Acad. Sci. USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andresen M., Stiel A.C., Jakobs S. Structural basis for reversible photoswitching in Dronpa. Proc. Natl. Acad. Sci. USA. 2007;104:13005–13009. doi: 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fegan A., White B., Wagner C.R. Chemically controlled protein assembly: techniques and applications. Chem. Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 34.DeRose R., Miyamoto T., Inoue T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013;465:409–417. doi: 10.1007/s00424-012-1208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motta-Mena L.B., Reade A., Gardner K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann S., Brigham M.D., Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polstein L.R., Gersbach C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nihongaki Y., Yamamoto S., Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Nihongaki Y., Kawano F., Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 40.Liu H., Gomez G., Lin C. Optogenetic control of transcription in zebrafish. PLoS One. 2012;7:e50738. doi: 10.1371/journal.pone.0050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan Y.-B., Alekseyenko O.V., Kravitz E.A. Optogenetic control of gene expression in Drosophila. PLoS One. 2015;10:e0138181. doi: 10.1371/journal.pone.0138181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y.I., Frey D., Hahn K.M. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin B., Holmes W.R., Levchenko A. Synthetic spatially graded Rac activation drives cell polarization and movement. Proc. Natl. Acad. Sci. USA. 2012;109:E3668–E3677. doi: 10.1073/pnas.1210295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katsura Y., Kubota H., Ozawa T. An optogenetic system for interrogating the temporal dynamics of Akt. Sci. Rep. 2015;5:14589. doi: 10.1038/srep14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clapham D.E. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 46.Greer P.L., Greenberg M.E. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Prakriya M., Lewis R.S. Store-operated calcium channels. Physiol. Rev. 2015;95:1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kyung T., Lee S., Heo W.D. Optogenetic control of endogenous Ca2+ channels in vivo. Nat. Biotechnol. 2015;33:1092–1096. doi: 10.1038/nbt.3350. [DOI] [PubMed] [Google Scholar]
- 49.Ishii T., Sato K., Nakata T. Light generation of intracellular Ca2+ signals by a genetically encoded protein BACCS. Nat. Commun. 2015;6:8021. doi: 10.1038/ncomms9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto T., DeRose R., Inoue T. Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nat. Chem. Biol. 2012;8:465–470. doi: 10.1038/nchembio.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suh B.-C., Inoue T., Hille B. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 2006;314:1454–1457. doi: 10.1126/science.1131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Idevall-Hagren O., Dickson E.J., De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swedlow J.R. Innovation in biological microscopy: current status and future directions. BioEssays. 2012;34:333–340. doi: 10.1002/bies.201100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin S., Kim K., Park Y. Active illumination using a digital micromirror device for quantitative phase imaging. Opt. Lett. 2015;40:5407–5410. doi: 10.1364/OL.40.005407. [DOI] [PubMed] [Google Scholar]
- 55.Klimas A., Entcheva E. Toward microendoscopy-inspired cardiac optogenetics in vivo: technical overview and perspective. J. Biomed. Opt. 2014;19 doi: 10.1117/1.JBO.19.8.080701. 08070-1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jost A.P.-T., Weiner O.D. Probing yeast polarity with acute, reversible, optogenetic inhibition of protein function. ACS Synth. Biol. 2015;4:1077–1085. doi: 10.1021/acssynbio.5b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bugaj L.J., Spelke D.P., Schaffer D.V. Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat. Commun. 2015;6:6898. doi: 10.1038/ncomms7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Neill P.R., Gautam N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol. Biol. Cell. 2014;25:2305–2314. doi: 10.1091/mbc.E14-04-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Renicke C., Schuster D., Taxis C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 2013;20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 60.van Bergeijk P., Adrian M., Kapitein L.C. Optogenetic control of organelle transport and positioning. Nature. 2015;518:111–114. doi: 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapitein L.C., Schlager M.A., Hoogenraad C.C. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J. 2010;99:2143–2152. doi: 10.1016/j.bpj.2010.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim A.K., DeRose R., Inoue T. Toward total synthesis of cell function: reconstituting cell dynamics with synthetic biology. Sci. Signal. 2016;9:re1. doi: 10.1126/scisignal.aac4779. [DOI] [PubMed] [Google Scholar]
- 63.Cheong W.F., Prahl S.A., Welch A.J. A review of the optical properties of biological tissues. IEEE J. Quantum Electron. 1990;26:2166–2185. [Google Scholar]
- 64.Stujenske J.M., Spellman T., Gordon J.A. Modeling the spatiotemporal dynamics of light and heat propagation for in vivo optogenetics. Cell Reports. 2015;12:525–534. doi: 10.1016/j.celrep.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson M.B. Cambridge University Press; Cambridge, UK: 2006. Molecular and Cellular Biophysics. [Google Scholar]
- 66.Cailliez F., Müller P., de la Lande A. ATP binding and aspartate protonation enhance photoinduced electron transfer in plant cryptochrome. J. Am. Chem. Soc. 2014;136:12974–12986. doi: 10.1021/ja506084f. [DOI] [PubMed] [Google Scholar]
- 67.Möglich A., Ayers R.A., Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J. Mol. Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt D., Tillberg P.W., Boyden E.S. A fully genetically encoded protein architecture for optical control of peptide ligand concentration. Nat. Commun. 2014;5:3019. doi: 10.1038/ncomms4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider F., Grimm C., Hegemann P. Biophysics of channelrhodopsin. Annu. Rev. Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 70.Halavaty A.S., Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 71.Pudasaini A., El-Arab K.K., Zoltowski B.D. LOV-based optogenetic devices: light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci. 2015;2:18. doi: 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strickland D., Lin Y., Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Das R., Baker D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 74.Kaufmann K.W., Lemmon G.H., Meiler J. Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry. 2010;49:2987–2998. doi: 10.1021/bi902153g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guntas G., Hallett R.A., Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. USA. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomita H., Sugano E., Tamai M. Channelrhodopsin-2 gene transduced into retinal ganglion cells restores functional vision in genetically blind rats. Exp. Eye Res. 2010;90:429–436. doi: 10.1016/j.exer.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 77.Ye H., Daoud-El Baba M., Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 78.Folcher M., Oesterle S., Fussenegger M. Mind-controlled transgene expression by a wireless-powered optogenetic designer cell implant. Nat. Commun. 2014;5:5392. doi: 10.1038/ncomms6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng J.Q. Turning of nerve growth cones induced by localized increases in intracellular calcium ions. Nature. 2000;403:89–93. doi: 10.1038/47501. [DOI] [PubMed] [Google Scholar]
- 80.Kakumoto T., Nakata T. Optogenetic control of PIP3: PIP3 is sufficient to induce the actin-based active part of growth cones and is regulated via endocytosis. PLoS One. 2013;8:e70861. doi: 10.1371/journal.pone.0070861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Volgraf M., Gorostiza P., Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat. Chem. Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Airan R.D., Thompson K.R., Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 83.Xu Y., Hyun Y.-M., Kim M. Optogenetic control of chemokine receptor signal and T-cell migration. Proc. Natl. Acad. Sci. USA. 2014;111:6371–6376. doi: 10.1073/pnas.1319296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sullivan J.A., Deng X.W. From seed to seed: the role of photoreceptors in Arabidopsis development. Dev. Biol. 2003;260:289–297. doi: 10.1016/s0012-1606(03)00212-4. [DOI] [PubMed] [Google Scholar]
- 85.Burgie E.S., Bussell A.N., Vierstra R.D. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. USA. 2014;111:10179–10184. doi: 10.1073/pnas.1403096111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levskaya A., Weiner O.D., Voigt C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mitra D., Yang X., Moffat K. Crystal structures of Aureochrome1 LOV suggest new design strategies for optogenetics. Structure. 2012;20:698–706. doi: 10.1016/j.str.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Niopek D., Benzinger D., Di Ventura B. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat. Commun. 2014;5:4404. doi: 10.1038/ncomms5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xing W., Busino L., Zheng N. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stiel A.C., Trowitzsch S., Wahl M.C. 1.8 Å bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem. J. 2007;402:35–42. doi: 10.1042/BJ20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krämer U. Planting molecular functions in an ecological context with Arabidopsis thaliana. eLife. 2015;4:1–13. doi: 10.7554/eLife.06100. [DOI] [PMC free article] [PubMed] [Google Scholar]