Abstract
In plants from temperate climates such as Arabidopsis thaliana low, non-freezing temperatures lead to increased freezing tolerance in a process termed cold acclimation. This process is accompanied by massive changes in gene expression and in the content of primary metabolites and lipids. In addition, most flavonols and anthocyanins accumulate upon cold exposure, along with most transcripts encoding transcription factors and enzymes of the flavonoid biosynthetic pathway. However, no evidence for a functional role of flavonoids in plant freezing tolerance has been shown. Here, we present a comprehensive analysis using qRT-PCR for transcript, LC-MS for flavonoid and GC-MS for primary metabolite measurements, and an electrolyte leakage assay to determine freezing tolerance of 20 mutant lines in two Arabidopsis accessions that are affected in different steps of the flavonoid biosynthetic pathway. This analysis provides evidence for a functional role of flavonoids in plant cold acclimation. The accumulation of flavonoids in the activation tagging mutant line pap1-D improved, while reduced flavonoid content in different knock-out mutants impaired leaf freezing tolerance. Analysis of the different knock-out mutants suggests redundancy of flavonoid structures, as the lack of flavonols or anthocyanins could be compensated by other compound classes.
Flavonoids are phenylpropanoids and a major class of secondary plant metabolites which are, unlike primary metabolites, not associated with plant survival and growth under standard controlled growth conditions1. Experimentally this has the advantage that mutants in this pathway show unaltered growth and development compared to wild type plants2. In Arabidopsis, at least 35 flavonol and 11 anthocyanin derivatives were detected in various tissues including leaves3,4,5,6,7, flowers6,8, roots4,6,9,10 and seeds11,12. The structural variety of the compounds is due to different modifications of the basic skeleton. In Arabidopsis, the flavonol aglycones (kaempferol, quercetin and isorhamnetin) can be mono-, di-, or tri-glycosylated by Glc, Rha or Ara, while the anthocyanin aglycone cyanidin can be additionally modified by xylosyl, sinapoyl, p-coumaroyl or malonyl residues.
The genes underlying the flavonoid pathway in Arabidopsis are mostly single-copy genes, making this species a convenient organism to investigate flavonoid biosynthesis2. Flavonoid biosynthesis genes include genes encoding transcription factors regulating the pathway and genes encoding different enzymes involved in the biosynthesis of the aglycones and their modifications4,7,9,13,14.
A variety of functions were reported for flavonoids ranging from flower and fruit pigmentation to regulation of polar transport of the phytohormone auxin and plant defense against pathogens and UV light. In addition, flavonoids accumulate in response to abiotic stresses such as drought, cold and high light intensity and are thought to be important for tolerance against these stresses15,16. In particular, low temperature strongly increases flavonoid content17,18,19,20, the abundance of enzymes involved in flavonoid biosynthesis21 and the expression of flavonoid biosynthesis genes20,22,23,24 in various plant species. Often a purple leaf phenotype, due to the accumulation of anthocyanins, is taken as a general indicator of plant stress exposure.
Plant species from temperate regions, among them Arabidopsis thaliana, increase in freezing tolerance during exposure to low, but non-freezing temperatures in a process termed cold acclimation25,26. This involves a massive reprogramming of the transcriptome, proteome and metabolome (see refs 27 and 28 for reviews) including flavonols and anthocyanins20. The content of different flavonols is correlated with leaf freezing tolerance in various Arabidopsis accessions and their crosses19,20 and in addition the expression of some flavonoid biosynthesis genes and freezing tolerance are correlated in diverse Arabidopsis accessions20,23. However, a direct causal relationship between the accumulation of flavonoids and freezing tolerance or the ability to cold acclimate has not been shown.
Results
We used a collection of 20 mutants in two different Arabidopsis thaliana accessions (Col-0 and Ler) that cover all major regulatory and biosynthetic steps in flavonoid biosynthesis (Fig. 1) to investigate the role of flavonols and anthocyanins in plant freezing tolerance and cold acclimation. Nineteen of these mutants are single (15 lines), double (three lines) or triple (one line) gene knock-outs. In addition, we included the activation tagging line pap1-D, which exhibits constitutive overexpression of the gene PAP1/MYB75 encoding the PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1) MYB transcription factor. The ko mutants showed no obvious phenotypic differences compared to the corresponding wild type plants under either non-acclimating or cold acclimating (14 days at 4 °C) conditions. This is in agreement with the finding that our cold acclimation conditions did not induce significant accumulation of reactive oxygen species (ROS)29 or ROS-related chilling stress that might require flavonoids for protection15,30. The pap1-D mutant, on the other hand, exhibited a purple leaf phenotype due to the activation of the whole flavonoid biosynthetic pathway31.
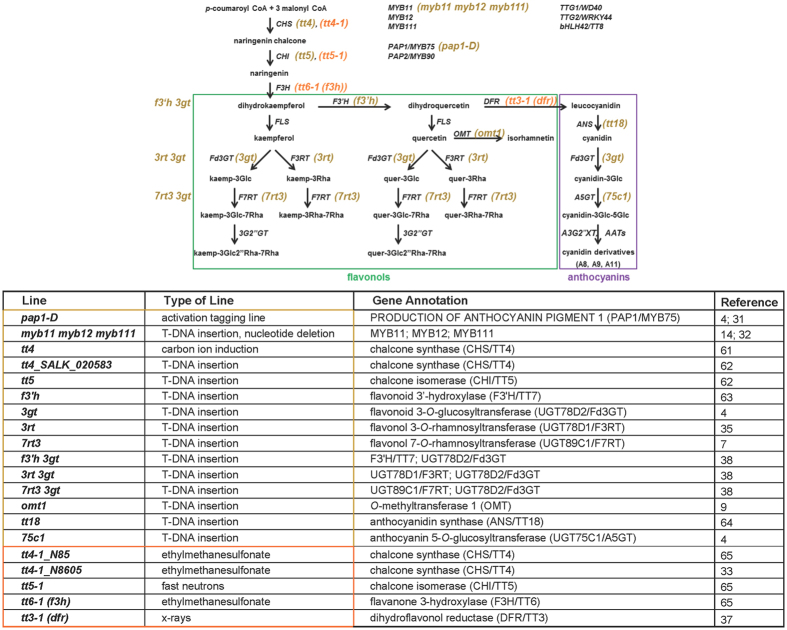
Figure 1. Flavonol and anthocyanin biosynthetic pathway indicating mutant lines used in this study.
Genes and metabolites are shown in black, mutant lines in Col-0 background in yellow and in Ler background in orange. Details of the mutant lines are listed in the bottom panel4,7,9,14,31,32,33,35,37,38,61,62,63,64,65. MYB11, MYB12, MYB111, R2R3 MYB transcription factors; TTG1/WD40, TTG2/WRKY44, transparent testa glabra; bHLH42/TT8, basic helix-loop-helix protein 42; PAP1/MYB75, PAP2/MYB90, production of anthocyanin pigment proteins 1 and 2; CHS/TT4, chalcone synthase; CHI/TT5, chalcone isomerase; F3H/TT6, flavanone 3-hydroxylase; F3′H/TT7, flavonoid 3′-hydroxylase; FLS, flavonol synthase; OMT1, O-methyltransferase 1; DFR/TT3, dihydroflavonol 4-reductase; ANS/TT18, anthocyanidin synthase; UGT78D2/Fd3GT, UGT78D1/F3RT, UGT89C1/F7RT, UGT75C1/A5GT, A3G2″XT, UDP-glucoronosyl/UDP-glucosyl transferase family proteins; AAT, anthocyanin acyl-transferase; TT, transparent testa.
Effects of mutations on the expression of flavonoid biosynthesis genes
We used quantitative RT-PCR to characterize the effects of the mutations on transcript levels of 26 genes encoding transcription factors regulating flavonoid biosynthesis and enzymes in the flavonoid biosynthetic pathway (Fig. 2; Suppl. Table 1). As described in detail recently, this analysis revealed a strong cold-induction of the pathway at the transcript level, with the exception of TTG1/WD40 and OMT1, which showed high constitutive expression20. The ko mutant lines generally showed a significant decrease in the abundance of the transcripts encoded by the affected genes compared to the wild type under both cold acclimated and non-acclimated conditions (Figs 3 and 4; Suppl. Table 4). Notable exceptions were the lines f3′h, f3′h 3gt and myb11 myb12 myb111 in Col-0, and both tt4-1 lines in Ler. The T-DNA insertion in the F3′H/TT7 gene is localized in the fourth exon immediately upstream of the 3′UTR region, rendering the design of qRT-PCR primers that bind beyond the T-DNA impossible. This led to the detection of partial transcripts with our primers binding in the second exon. The MYB12 transcript in the triple mutant contains a single base pair deletion (myb12-1f) and is measurable by qRT-PCR, but results in premature termination of translation32. Similarly, the detected CHS/TT4 trancripts in the tt4-1 mutants in Ler are nonfunctional due to point mutations33. It remains unclear why the expression of UGT89C1/F7RT was significantly reduced in the 7rt3 3gt double mutant, but was almost unchanged in the 7rt3 single mutant line.
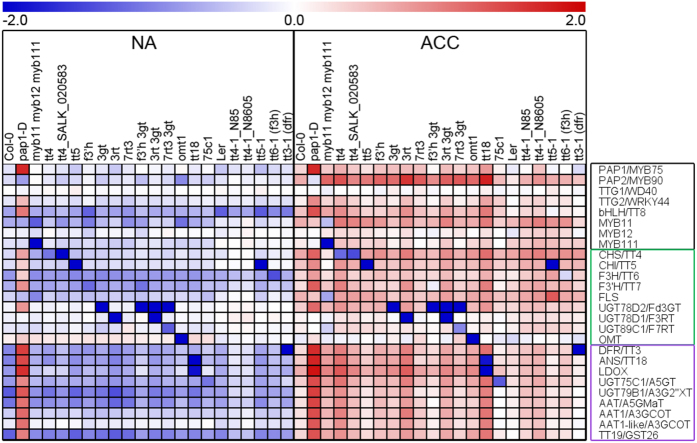
Figure 2. Relative transcript abundance (2−ΔCt) of flavonoid biosynthesis genes in mutant lines and the corresponding wild types under non-acclimated (NA) and cold acclimated (ACC) conditions.
Mutant lines are sorted according to their positions in the flavonoid biosynthetic pathway. Data represent averages of three biological replicates per line and condition, log10 median transformed over all lines and both conditions for every gene with transcript abundance above median in red and below median in blue, as indicated by the scale bar. Genes encoding transcription factors are framed in black, flavonol biosynthesis genes in green and anthocyanin biosynthesis genes in purple (see also Suppl. Table 1).
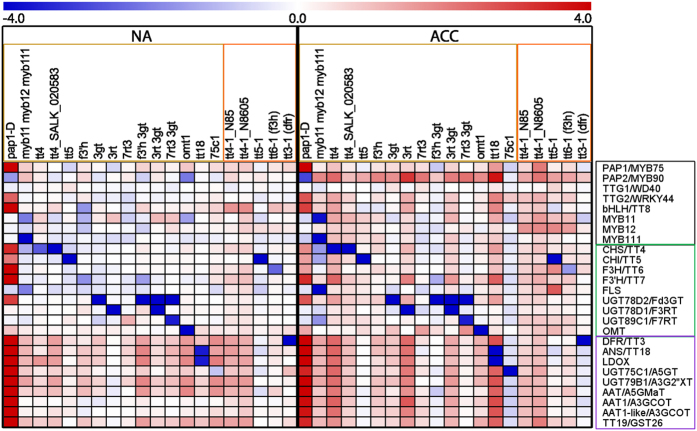
Figure 3. Log2 fold change of relative transcript abundance (2−ΔCt) of flavonoid biosynthesis genes in mutant lines relative to Col-0 or Ler wild type under non-acclimated (NA) and cold acclimated (ACC) conditions.
Mutant lines in Col-0 (yellow framed) and Ler (orange framed) are sorted according to their positions in the flavonoid biosynthetic pathway. Data represent averages of three biological replicates per line and condition with higher relative expression in mutant lines compared to wild type in red and lower expression in blue, as indicated by the scale bar. Genes encoding TFs are framed in black, flavonol biosynthesis genes in green and anthocyanin biosynthesis genes in purple (see also Suppl. Table 1).
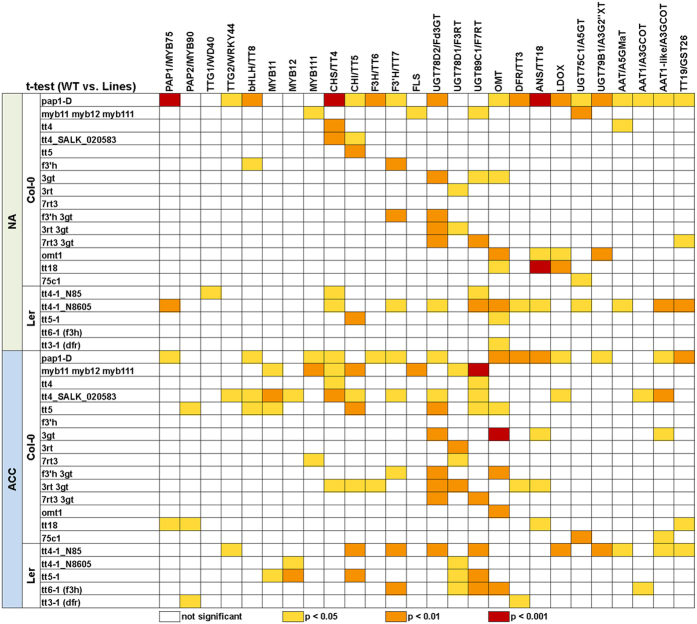
Figure 4. Statistical analysis of gene expression data (log2-fold change) shown inFig. 3.
t-tests (two-sided with unequal variance) were used to determine the significance of expression changes between mutant lines and their wild types Col-0 or Ler under non-acclimated (NA) and acclimated condition (ACC). p-values were corrected using the Benjamini-Hochberg procedure and color coded as indicated in the figure (see also Suppl. Table 4).
In general, mutations in the genes encoding enzymes had only minor effects on the abundance of all other investigated transcripts (Fig. 4; Suppl. Table 4). On the other hand, knock-out of the MYB11, MYB12 and MYB111 transcription factors resulted, as expected14, in strongly reduced expression of key flavonol biosynthesis genes such as CHS/TT4, CHI/TT5 and FLS, especially after cold acclimation. The activation tagging line pap1-D showed high PAP1/MYB75 transcript abundance under both non-acclimated and acclimated conditions and constitutive activation of down-stream genes encoding the transcription factors TTG2/WRKY44 and bHLH42/TT8, some flavonol and most anthocyanin biosynthetic enzymes. In addition, two of the four tt4 lines showed a significant upregulation of several of the genes encoding down-stream enzymes.
Effects of mutations on flavonoid content
We further evaluated by LC-MS analysis, whether the observed changes in gene expression in the mutant lines also resulted in corresponding changes in flavonoid content (Fig. 5; Suppl. Table 2). Three kaempferol and two quercetin derivatives, three major anthocyanins (A8, A9 and A11), four minor flavonols and one minor anthocyanin (A7) as well as seven unidentified flavonols and two unidentified anthocyanins could be quantified. A preliminary identification of these compounds based on MS/MS fragmentation analysis is provided in Suppl. Table 2. The significance of differences in flavonoid content between mutants and wild types (Fig. 6; Suppl. Table 5) was evaluated by t-tests corrected for multiple testing errors34. Changes in Arabidopsis flavonoid content during cold acclimation have been reported recently20 and will not be considered here.
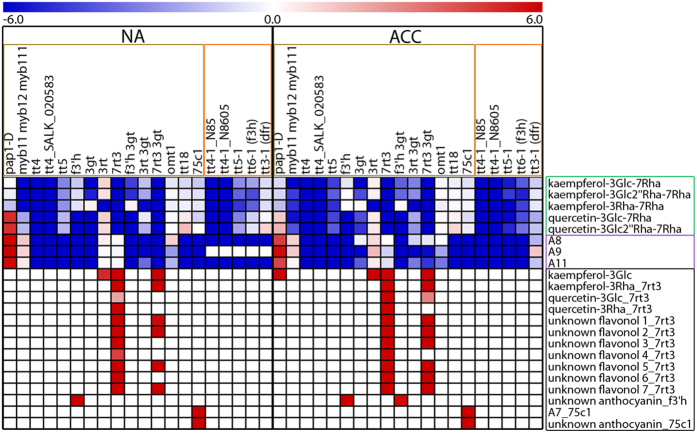
Figure 5. Flavonoid content (relative peak area) in mutant lines changed in flavonoid metabolism under non-acclimated (NA) and acclimated (ACC) conditions.
Mutant lines in Col-0 (yellow framed) and Ler (orange framed) are sorted according to their positions in the flavonoid biosynthetic pathway. Data represent averages of three or six (only Col-0 wild type and single mutants) biological replicates with higher flavonoid content in mutant lines in comparison to corresponding wild type in red and lower content in blue, as indicated by the scale bar. Central flavonols and anthocyanins are framed in green and purple, minor flavonoids and flavonols with unknown structure in black (see also Suppl. Table 2).
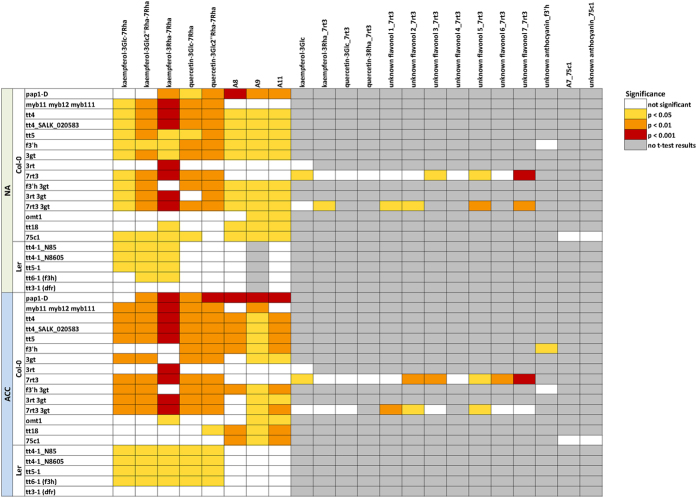
Figure 6. Statistical analysis of differences in flavonoid content between mutant lines and their wild types Col-0 or Ler under non-acclimated (NA) and acclimated condition (ACC).
t-tests (two-sided with unequal variance) were used to determine the significance of expression changes. p-values were corrected using the Benjamini-Hochberg procedure and color coded as indicated in the figure. Gray fields indicate that no t-test could be performed because the metabolite was undetectable in these samples (see also Suppl. Table 5).
Figure 5 shows that most mutations resulted in the absence or a strong reduction of major flavonoids, while only pap1-D showed a strong accumulation, particularly of anthocyanins, but also of the two major quercetin derivatives. Additionally, kaempferol-3Glc increased massively in the acclimated state in this mutant. On the other hand, myb11 myb12 myb111 showed a strong reduction in the content of all flavonols, but not of anthocyanins. All four investigated tt4 lines in both accessions completely lacked flavonoids, while the tt5 and tt6-1 lines, also affected in the initial steps of the pathway, still contained low, significantly reduced levels of flavonols and anthocyanins, particularly under cold acclimated conditions. Similarly, the lines 75c1 and tt18 in Col-0 and tt3-1 in Ler were specifically reduced in anthocyanin content.
All other mutant lines showed more specific effects on the content of particular flavonoids. For example, the f3′h lines lacked all major quercetin and anthocyanin derivatives, whereas the f3′h 3gt line additionally showed reduced levels of glucosylated kaempferols that were also affected in line 3gt, while 3rt was specifically reduced in its kaempferol-3Rha-7Rha content. The line 7rt3 showed a strong reduction of all central 7-O-rhamnosylated flavonols. Noteworthy, 7rt3 and f3′h single and double mutant lines as well as 75c1 accumulated mutant-specific flavonol or anthocyanin derivatives (see Suppl. Table 2 for preliminary structure assignments) which might be evidence of compensatory effects. These findings are consistent with previous studies under normal (non-acclimated) growth conditions4,7,9,31,35,36,37,38.
Effects of mutations on leaf freezing tolerance
To assess whether flavonoids play a functional role in leaf freezing tolerance, we compared wild type and mutant lines both before and after cold acclimation (Fig. 7), using an electrolyte leakage assay39. Only the pap1-D line showed significantly improved freezing tolerance under acclimated conditions, which can be attributed to the accumulation of quercetin and anthocyanin derivatives, while all ko lines showed either reduced freezing tolerance or were not significantly affected.
Figure 7. Freezing tolerance of all mutant lines (see Fig. 1) compared to the corresponding wild types, of non-acclimated and cold acclimated plants.
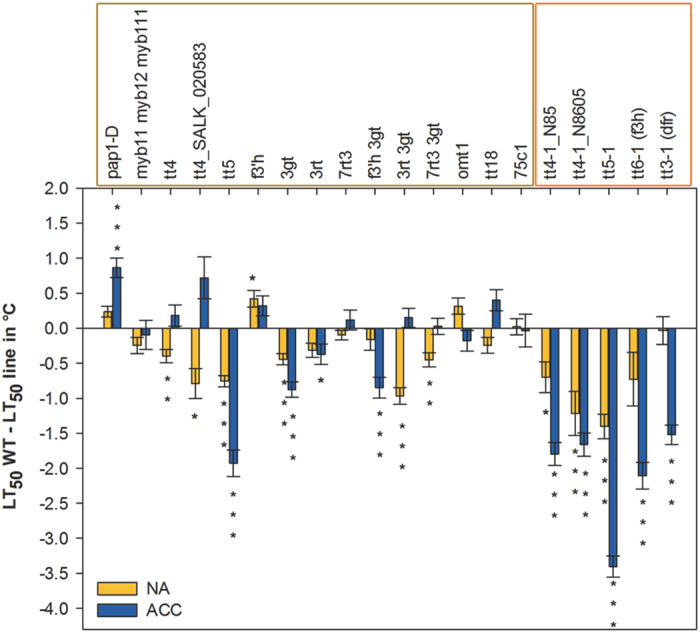
Mutant lines in Col-0 (framed in yellow) and Ler (framed in orange) are sorted according to their position in the flavonoid biosynthetic pathway. The bars indicate the differences in leaf freezing tolerance between wild type and mutant expressed as LT50 (temperature at which 50% electrolyte leakage occurred). Positive values indicate improved freezing tolerance, negative values impaired freezing tolerance compared to the respective wild type. Data are means from two to four independent plant cultures, each with four biological replicates per line and condition. Significant differences, evaluated by Welch’s unpaired t-test, are marked by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001.
Mutants in the three earliest steps of the pathway (tt4, tt5, tt6) which showed a massive reduction in both flavonols and anthocyanins, exhibited a strong reduction in freezing tolerance in the Ler background, in particular after cold acclimation. Interestingly, while the tt5 mutant behaved similarly in Col-0, this was not found for the tt4 mutants in this accession, which only showed reduced freezing tolerance in the non-acclimated state. On the other hand, the tt3-1 mutant in Ler showed a significant reduction in cold acclimated freezing tolerance, although only small, non-significant differences in flavonoid content could be detected compared to the wild type (Figs 5 and 6).
No changes in freezing tolerance were observed in mutants where only anthocyanin (tt18, 75c1) or only flavonol contents (myb111 myb12 myb111) were reduced. However, when all flavonoids were strongly reduced (tt5, tt6-1 and 3rt 3gt) freezing tolerance was impaired at least under one condition. The reduction of all 3-O-glucosylated flavonoids in 3gt reduced freezing tolerance under both conditions, while a complete lack of only kaempferol-3Rha-7Rha in 3rt resulted in slightly impaired freezing tolerance only under non-acclimated conditions. The absence of all major flavonols in 7rt3 accompanied by the occurrence of flavonols with unknown structure at unchanged anthocyanin levels had no impact on freezing tolerance. The additional reduction of anthocyanins in the 7rt3 3gt double mutant led to impaired non-acclimated freezing tolerance. In addition, a complete lack of quercetin and anthocyanin derivatives with unchanged kaempferol levels in f3′h had no significant effect on freezing tolerance. The f3′h 3gt double mutant line was additionally reduced in kaempferol-3Glc-7Rha and kaempferol-3Glc2″7Rha-7Rha and therefore only unaffected in the content of kaempferol-3Rha-7Rha resulting in impaired freezing tolerance after cold acclimation.
Effects of mutations on primary metabolism
Cold acclimation is accompanied by a massive accumulation of many primary metabolites such as sugars and amino acids that may act as compatible solutes to improve cellular freezing tolerance27,28. We therefore checked whether the mutations in secondary metabolism had additional effects on primary metabolism. We compared the relative pool sizes of 114 primary metabolites (34 organic, amino and hydroxyl acids, four N-compounds, six P-compounds, 15 sugars and 55 unknown putative primary metabolites) measured by GC-MS (Suppl. Table 6) between wild type and mutant plants under both non-acclimated and acclimated conditions. This analysis showed only very little effect of mutations in the flavonoid biosynthetic pathway on primary metabolism under either condition, with only eight significantly different pool sizes among 114 metabolites in 40 comparisons (Suppl. Table 6) in agreement with an earlier study that was conducted only under non-acclimating conditions4.
Discussion
Comprehensive characterization of the flavonoid mutants with respect to their freezing tolerance, gene expression and metabolite profiles indicates a role of flavonoids in freezing tolerance and cold acclimation. Previous work has implicated flavonoids in the defence against ROS (e.g. ref. 15), although the importance of flavonoids for ROS scavenging in vivo has been questioned40. This ROS-scavenging activity has been invoked mainly for plant chilling tolerance, i.e. tolerance against low temperatures above 0 °C, while here we investigated the tolerance of plants against freezing, i.e. the stresses associated with apoplastic ice formation at sub-zero temperatures. A clear physiological role for flavonoids, on the other hand, has been established for their UV-B screening function18,30,41. A function of flavonoids in plant freezing tolerance has been hypothesized in Arabidopsis due to the correlation of both metabolite and transcript abundances with freezing tolerance19,20,23, but a clear causal relationship had not been established previously.
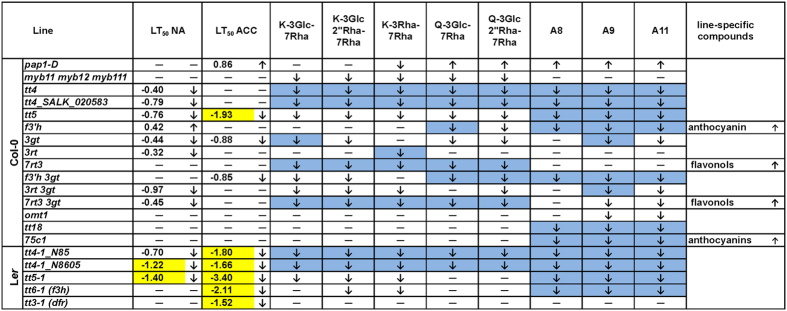
In contrast to the overexpression line pap1-D with improved freezing tolerance, most of the ko mutant lines showed reduced freezing tolerance. In general, the complete loss or strong reduction of flavonoid compounds resulted in impairment of freezing tolerance or in the accumulation of other known or unknown flavonoids with potential compensatory effects, while the accumulation of predominantly quercetin and anthocyanin compounds in the pap1-D mutant enhanced freezing tolerance (see Fig. 8 for a summary of these results). This is in good agreement with our previous finding that the content of all three quercetin derivatives and of the anthocyanins A5 and A9 were correlated with acclimated freezing tolerance in a collection of Arabidopsis accessions20. Interestingly, the reduction of either flavonols or anthocyanins did not result in a decline in freezing tolerance, indicating that the two groups of compounds may have at least partially redundant functions. As the presence of kaempferols, flavonols in general or anthocyanin derivatives preserved freezing tolerance, the combined contribution of a set of different compounds is more likely than the specific action of a particular molecule.
Figure 8. Overview of the effects of all investigated mutations on non-acclimated (LT50 NA) and acclimated freezing tolerance (LT50 ACC) and on flavonoid content.
Significantly changed LT50 values are shown (LT50 wild type – LT50 mutant) and decrease (↓), increase (↑), and no changes (—) are indicated. Changes in LT50 of more than 1 °C are highlighted in yellow. Significantly decreased (↓) or increased (↑) contents of kaempferols (K), quercetins (Q), anthocyanins (A) and mutant-specific flavonoids are indicated for both NA and ACC conditions combined. Complete absence of a compound is indicated in blue.
Differences in flavonoid composition between Col-0 and Ler have been reported previously for both plants exposed to non-acclimating and cold acclimating conditions20,42. The effects of mutations in the flavonoid biosynthesis pathway on freezing tolerance were generally more pronounced in the Ler than in the Col-0 background, suggesting that the contribution of flavonoids to freezing tolerance may be genotype-dependent. This is supported by the fact that tt3-1 in Ler, with only a moderate reduction in anthocyanin levels, was affected in its acclimated freezing tolerance, while tt18 and 75c1 in Col-0, with much stronger reductions in anthocyanin contents, were not affected. In addition, the Ler tt5-1 and both tt4-1 lines were more strongly affected in their freezing tolerance than the corresponding lines in Col-0. This accession effect is particularly striking for the tt4 mutants, which showed a significant reduction in anthocyanin content after cold acclimation in Col-0, but not in Ler, while freezing tolerance was only reduced in Ler (compare Fig. 8). The physiological basis of this difference between the accessions is currently unclear but it strongly underscores the fact that plant freezing tolerance is a quantitative trait that is based on the concerted action of many proteins and metabolites. This makes it difficult to estimate the contribution of single components to the total phenotype. For example, while raffinose content correlates strongly with acclimated freezing tolerance in Arabidopsis accessions, a ko mutant in the raffinose synthase gene that is completely devoid of raffinose shows no measurable impairment in freezing tolerance43.
Due to their antioxidant activity, a possible protective mechanism of flavonoids during freezing may be the scavenging of ROS44. In our experiments, however, this is rather unlikely, as freezing of leaves was performed in the dark to exclude ROS formation in the chloroplast. In addition, the cold acclimation treatment was performed at a significantly lower light intensity than growth under control conditions, also reducing ROS accumulation29. Flavonoids could also modulate stress response pathways through phytohormones as they negatively regulate auxin transport45 and jasmonate levels46. However, we find such a pleiotropic effect unlikely, as plant growth, development and primary metabolism were unaffected in all mutants used in this study.
The direct protection of membranes and/or proteins from freezing damage are other possible mechanisms through which flavonoids could increase leaf freezing tolerance, in particular since flavonoids have been localized in the vacuole, nucleus and chloroplast, which may all be protected by flavonoids during freezing47. Membranes have long been recognized as major sites of freezing injury48 and soluble cold-induced proteins that specifically stabilize membranes during freezing in Arabidopsis have been characterized49,50. A similar function of flavonoids seems possible as they can partition into both model and plant membranes in vitro and influence their stability51. In addition, enzymes can be inactivated when Arabidopsis leaves are frozen to temperatures that also induce electrolyte leakage and the inactivation of photosynthesis50. Flavonoids are able to stabilize proteins in vitro by preventing their aggregation52,53,54. Clearly, further research is needed to understand the functional mechanisms that link flavonoid content to plant freezing tolerance.
Methods
Plant material
The mutant lines used in this study included one activation tagging line and 13 T-DNA insertion lines of which nine were single, three double and one triple gene knock-outs in Columbia-0 (Col-0). Five mutants in Landsberg erecta (Ler) were generated by ethylmethansulfonate (EMS), fast neutron or x-ray mutagenesis and one line in Col-0 by carbon ion induction. In addition we used one activation tagging line. Details to all lines are given in Fig. 1. Plants were grown in a greenhouse at 16 h day length with light supplementation to reach at least 200 μE m−2 s−1 and a temperature of 20 °C during the day, 18 °C during the night until 42 days after sowing (non-acclimated plants). For cold acclimation, plants were transferred to a 4 °C growth cabinet at 16 h day length with 90 μE m−2 s−1 for an additional 14 days23,55.
Determination of leaf freezing tolerance
Freezing damage was determined as LT50 (temperature of 50% electrolyte leakage) from electrolyte leakage measurements after freezing of detached leaves to temperatures ranging from −1 to −21 °C at 4 °C h−1 as described in detail in previous publications39,56. Per line and condition four temperature curves were analyzed in each experiment, using leaves from three to six individual plants each. Due to the number of mutant lines, several experiments had to be performed, which each contained samples from the respective wild type. Data were collected for every mutant line from two to four independent sowings.
qRT-PCR analysis of gene expression
Quantitative RT-PCR was performed according to ref. 20, where also the sequences of all primers can be found. Ct values for the genes of interest were normalized by subtracting the mean Ct of four reference genes and averages of three biological replicates were determined (Suppl. Table 1).
LC-MS analysis of flavonoids
Secondary metabolite profiling by LC-MS was carried out as described recently20,57. Peak identification and annotation was performed with Xcalibur (Thermo Finnigan) based on previously published reference data4,58,59. Relative peak areas representing mass spectral ion currents were normalized to fresh weight and peak area of the internal standard. The data represent the averages of three or six (Col-0 wild type and single mutants) biological replicates (Suppl. Table 2). The relative peak area obtained for kaempferol-3Glc-7Rha also represents a minor contribution from quercetin-3Rha-7Rha, as these compounds were not completely separated.
Primary metabolite profiling by GC-MS
Polar metabolites were extracted from100 mg frozen plant material and analyzed by gas chromatography/electron impact ionization-time of flight mass spectrometry (GC/EI-TOF-MS) as described in detail recently60. For every metabolite, relative mean intensities were calculated for each genotype as the average of five biological replicates (Suppl. Table 3).
Additional Information
How to cite this article: Schulz, E. et al. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 6, 34027; doi: 10.1038/srep34027 (2016).
Supplementary Material
Acknowledgments
Some of the mutant lines were kindly provided by Dr. A. Schäffner (Helmholtz Zentrum München, Germany) and Dr. R. Stracke (Bielefeld University, Germany). We would like to thank Dr. Joachim Kopka and his team at the Max-Planck-Institute of Molecular Plant Physiology for their excellent GC-MS service. ES was supported through a PhD fellowship from the Deutsche Bundesstiftung Umwelt.
Footnotes
Author Contributions E.S., E.Z. and D.K.H. designed the experiments; E.S. performed the experiments including freezing tolerance measurements and qRT-PCR; T.T. performed the LC-MS analyses; E.Z., A.R.F. and D.K.H. provided advice about the experiments; E.S. and D.K.H. wrote the manuscript; all authors read the manuscript, edited and commented on it before submission.
References
- Buer C. S., Imin N. & Djordjevic M. A. Flavonoids: new roles for old molecules. J. Integr. Plant Biol. 52, 98–111 (2010). [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobiecki M. A. et al. Profiling of phenolic glycosidic conjugates in leaves of Arabidopsis thaliana using LC/MS. Metabolomics, 197–219 (2006). [Google Scholar]
- Tohge T. et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235 (2005). [DOI] [PubMed] [Google Scholar]
- Veit M. & Pauli G. F. Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62, 1301–1303 (1999). [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K. et al. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20, 2160–2176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Niida R. & Saito K. Identification of a flavonol-7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282, 14932–14941 (2007). [DOI] [PubMed] [Google Scholar]
- Matsuda F. et al. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 57, 555–577 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Yonekura-Sakakibara K., Niida R., Watanabe-Takahashi A. & Saito K. Phytochemical genomics in Arabidopsis thaliana: a case study for functional identification of flavonoid biosynthesis genes. Pure Appl. Chem. 79, 811–823 (2007). [Google Scholar]
- Vallabhaneni P., Ray W. K., Winkel B. S. J. & Helm R. F. Characterization of flavonol glycosides in individual Arabidopsis root tips by flow injection electrospray mass spectrometry. Phytochemistry 73, 114–118 (2012). [DOI] [PubMed] [Google Scholar]
- Kerhoas L. et al. Structural characterization of the major flavonoid glycosides from Arabidopsis thaliana seeds. J. Agric. Food Chem. 54, 6603–6612 (2006). [DOI] [PubMed] [Google Scholar]
- Routaboul J. M. et al. Flavonoid diversity and biosynthesis in seed of Arabodpsis thaliana. Planta 224, 96–107 (2006). [DOI] [PubMed] [Google Scholar]
- Liu J., Osbourn A. & Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708 (2015). [DOI] [PubMed] [Google Scholar]
- Stracke R. et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R. et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223 (2002). [DOI] [PubMed] [Google Scholar]
- Becker C., Klaering H. P., Kroh L. W. & Krumbein A. Cool-cultivated red leaf lettuce accumulates cyanidin-3-O-(6”-O-malonyl)-glucoside and caffeoylmalic acid. Food Chem. 146, 404–411 (2014). [DOI] [PubMed] [Google Scholar]
- Bilger W., Rolland M. & Nybakken L. UV screening in higher plants induced by low temperature in the absence of UV-B radiation. Photochem. Photobiol. Sci. 6, 190–195 (2007). [DOI] [PubMed] [Google Scholar]
- Korn M., Peterek S., Mock H.-P., Heyer A. G. & Hincha D. K. Heterosis in the freezing tolerance, and sugar and flavonoid contents of crosses between Arabidopsis thaliana accessions of widely varying freezing tolerance. Plant Cell Environ. 31, 813–827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E., Tohge T., Zuther E., Fernie A. R. & Hincha D. K. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 38, 1658–1672 (2015). [DOI] [PubMed] [Google Scholar]
- Koehler G. et al. Proteomic study of low-temperature responses in strawberry cultivars (Fragaria x ananassa) that differ in cold tolerance. Plant Physiol. 159, 1787–1809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crifo T., Puglisis I., Petrone G., Recupero G. R. & Lo Piero A. R. Expression analysis in response to low temperature stress in blood oranges: implication of the flavonoid biosynthetic pathway. Gene 476, 1–9 (2011). [DOI] [PubMed] [Google Scholar]
- Hannah M. A. et al. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 142, 98–112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F. et al. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 50, 967–981 (2007). [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599 (1999). [DOI] [PubMed] [Google Scholar]
- Xin Z. & Browse J. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ. 23, 893–902 (2000). [Google Scholar]
- Guy C. L., Kaplan F., Kopka J., Selbig J. & Hincha D. K. Metabolomics of temperature stress. Physiol. Plant. 132, 220–235 (2008). [DOI] [PubMed] [Google Scholar]
- Hincha D. K., Espinoza C. & Zuther E. In Improving Crop Resistance to Abiotic Stress Vol. 1 (eds Tuteja N., Gill S. S., Tiburcio A. F. & Tuteja R.) 255–287 (Wiley-Blackwell, 2012). [Google Scholar]
- Juszak I., Cvetkovic J., Zuther E., Hincha D. K. & Baier M. Natural variation of cold deacclimation correlates with variation of cold-acclimation of the plastid antioxidant system in Arabidopsis thaliana accessions. Front. Plant Sci. 7, 305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra M. L. F., Rius S. P. & Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Frontiers Plant Sci. 3, 222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J. O., Xia Y., Blount J., Dixon R. A. & Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P. & Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan J. J. & Rechnitz G. A. Differential visualization of transperant testa mutants in Arabidopsis thaliana. Anal. Chem. 65, 961–963 (1993). [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- Jones P., Messner B., Nakajima J., Schäffner A. R. & Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278, 43910–43918 (2003). [DOI] [PubMed] [Google Scholar]
- Owens D. K., Crosby K. C., Runac J., Howard B. A. & Winkel B. S. Biochemical and genetic characterization of Arabidopsis flavanone 3ß-hydroxylase. Plant Physiol. Biochem. 46, 833–843 (2008). [DOI] [PubMed] [Google Scholar]
- Shirley B. W. et al. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8, 659–671 (1995). [DOI] [PubMed] [Google Scholar]
- Yin R. H. et al. Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 201, 466–475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A., Hincha D. K. & Zuther E. In Methods in Molecular Biology Vol. 1166 (eds Hincha D. K. & Zuther E.) 15–24 (Springer, 2014). [DOI] [PubMed] [Google Scholar]
- Hernandez I., Alegre L., van Breusegem F. & Munne-Bosch S. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 14, 125–132 (2009). [DOI] [PubMed] [Google Scholar]
- Emiliani J., Grotewold E., Falcone Ferreyra M. L. & Casati P. Flavonols protect Arabidopsis plants against UV-B deleterious effects. Mol. Plant 6, 1376–1379 (2013). [DOI] [PubMed] [Google Scholar]
- Ishihara H. et al. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. J. Exp. Bot. doi: 10.1093/jxb/erv546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther E. et al. The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett. 576, 169–173 (2004). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Distinctive antioxidant and antiinflammatory effects of flavonols. J. Agric. Food Chem. 54, 9798–9804 (2006). [DOI] [PubMed] [Google Scholar]
- Buer C. S. & Munday G. K. The transperant testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16, 1191–1205 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L. et al. A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 74, 383–397 (2013). [DOI] [PubMed] [Google Scholar]
- Agati G., Azzarello E., Pollastri S. & Tattini M. Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 196, 67–76 (2012). [DOI] [PubMed] [Google Scholar]
- Steponkus P. L. Role of the plasma membrane in freezing injury and cold acclimation. Annu. Rev. Plant Physiol. 35, 543–584 (1984). [Google Scholar]
- Artus N. N. et al. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 93, 13404–13409 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A., Bryant G., Sulpice R. & Hincha D. K. Disordered Cold Regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 166, 190–201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska-Pawlega B. et al. Characteristics of quercetin interaction with liposomal and vacuolar membranes. Biochim. Biophys. Acta 1838, 254–265 (2014). [DOI] [PubMed] [Google Scholar]
- Hamaguchi T., Ono K., Murase A. & Yamada M. Phenolic compounds prevent Alzheimer′s pathology through different effects on the amyloid-beta aggregation pathway. Am. J. Pathol. 175, 2557–2565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K. et al. Phenolic compounds prevent amyloid β-protein oligomerization and synaptic dysfunction by site-specific binding. J. Biol. Chem. 287, 14631–14643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Grape-derived polyphenolics prevent a β oligomerization and attenuate cognitive deterioration in a mouse model of Alzheimer’s disease. J. Neurosci. 28, 6388–6392 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde P., Hincha D. K. & Heyer A. G. Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 38, 790–799 (2004). [DOI] [PubMed] [Google Scholar]
- Zuther E., Schulz E., Childs L. H. & Hincha D. K. Natural variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ. 35, 1860–1878 (2012). [DOI] [PubMed] [Google Scholar]
- Tohge T. & Fernie A. R. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protocols 5, 1210–1227 (2010). [DOI] [PubMed] [Google Scholar]
- Nakabayashi R. et al. Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana. Phytochemistry 70, 1017–1029 (2009). [DOI] [PubMed] [Google Scholar]
- Saito K. et al. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 72, 21–34 (2013). [DOI] [PubMed] [Google Scholar]
- Dethloff F. et al. In Plant Cold Acclimation Vol. 1166 Methods in Molecular Biology (eds Hincha D. K. & Zuther E.) 171–198 (Springer, 2014). [DOI] [PubMed] [Google Scholar]
- Shikazono N., Tanaka A., Watanabe H. & Tano S. Rearrangements of the DNA in carbon ion-induced mutants of Arabidopsis thaliana. Genetics 157, 379–387 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). [DOI] [PubMed] [Google Scholar]
- Rosso M. G. et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53, 247–259 (2003). [DOI] [PubMed] [Google Scholar]
- Abrahams S. et al. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 35, 624–636 (2003). [DOI] [PubMed] [Google Scholar]
- Koornneef M. et al. Linkage map of Arabidopsis thaliana. J. Hered. 74, 265–272 (1983). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.