Abstract
The cardiac sarco/endoplasmic reticulum calcium ATPase (SERCA) establishes the intracellular calcium gradient across the sarcoplasmic reticulum membrane. It has been proposed that SERCA forms homooligomers that increase the catalytic rate of calcium transport. We investigated SERCA dimerization in rabbit left ventricular myocytes using a photoactivatable cross-linker. Western blotting of cross-linked SERCA revealed higher-molecular-weight species consistent with SERCA oligomerization. Fluorescence resonance energy transfer measurements in cells transiently transfected with fluorescently labeled SERCA2a revealed that SERCA readily forms homodimers. These dimers formed in the absence or presence of the SERCA regulatory partner, phospholamban (PLB) and were unaltered by PLB phosphorylation or changes in calcium or ATP. Fluorescence lifetime data are compatible with a model in which PLB interacts with a SERCA homodimer in a stoichiometry of 1:2. Together, these results suggest that SERCA forms constitutive homodimers in live cells and that dimer formation is not modulated by SERCA conformational poise, PLB binding, or PLB phosphorylation.
Introduction
Sarco/endoplasmic reticulum calcium ATPase (SERCA) is an ion-motive ATPase that couples the hydrolysis of ATP with calcium transport from the cytosol into the endoplasmic reticulum. In the heart, the cardiac isoform SERCA2a is critical for cardiomyocyte relaxation and reloading sarcoplasmic reticulum (SR) calcium stores for subsequent contractions. Cardiac calcium transport activity is regulated by a 52-amino-acid membrane protein, phospholamban (PLB), which inhibits SERCA by reducing the apparent affinity of the pump for calcium. SERCA inhibition is relieved when PLB is phosphorylated by protein kinase A or calcium-calmodulin kinase II (1, 2, 3). As the main calcium-handling pathway during relaxation of myocytes, SERCA plays a key role in cardiomyocyte function and presents a valuable target for therapies aimed at treating heart failure (HF). Alterations to SERCA expression and regulation by PLB are associated with HF, underscoring its importance in cardiac function (4, 5). Moreover, overexpression of SERCA in various animal models has been shown to improve cardiac function and survival (6, 7, 8), creating interest in targeting calcium handling for treatment of heart disease. However, a recent clinical trial utilizing SERCA overexpression in patients with HF did not improve clinical outcome, so alternative approaches to improving cardiac calcium handling are needed. Development of such future therapies will require a better understanding of SERCA function and regulation.
Multiple studies since the 1970s have provided evidence for the assembly of skeletal isoform SERCA1a in homooligomeric complexes (9, 10, 11, 12). The functional significance of this was unclear, but early studies suggested SERCA-SERCA interactions altered SERCA responsiveness to Mg (12) or ATP (9). Some recent functional studies correlated oligomerization/aggregation with decreased SERCA catalytic activity (13, 14, 15). However, other data suggest that SERCA oligomerization may be a mechanism for increased transport activity after relief of inhibition by adrenergic stimulation. In particular, saturation transfer electron paramagnetic resonance experiments indicated that phosphorylation of PLB reduced the rotational mobility of SERCA2a, consistent with SERCA homooligomerization (16). This interpretation is also supported by kinetic measurements of calcium transport that indicated that SERCA2a dimerizes in the presence of phosphorylated PLB (17). Significantly, the kinetic data suggested conformational coupling of SERCA protomers into a single functional unit. In this model, energetically unfavorable structural transitions of one SERCA protomer are assisted by coupling them to energetically favorable conformational changes of the other protomer. Thus, by reducing the barrier of the slow step, the overall rate of calcium transport is increased. Other studies also support the hypothesis that oligomerization enhances SERCA catalytic activity. Chen et al. showed that dilution of active wild-type SERCA2a with inactivated SERCA2a reduced ATPase activity by more than would be expected from the molar ratio of wild-type to inactive species (18). An analogous experiment was performed by Chamberlain et al. (19), who demonstrated that irradiation of SERCA2a produced a nonlinear reduction of activity. The shape of this irradiation curve was suggestive of a functional dimer. Together, these studies suggest that the activity of the SERCA oligomer is greater than the sum of its parts.
In this study, we directly tested this theory of SERCA functional oligomerization that was developed from in vitro observations. We used photoactivatable cross-linking to determine whether the cardiac isoform SERCA2a is oligomeric in cardiac myocytes. Then, using spectroscopic approaches, we quantified the stoichiometry of the putative oligomer and investigated whether oligomerization was regulated by mechanisms known to be important determinants of SERCA activity. Specifically, we sought to determine whether the presence of PLB or PLB phosphorylation alter SERCA-SERCA physical coupling. That is, does PLB control the functional coupling of the putative dimer complex by altering SERCA-SERCA binding affinity or are physical and functional coupling distinct facets? We report detection of dimeric SERCA in the membranes of living cells, including native SERCA2a in cardiac myocytes. The data suggest that SERCA-SERCA dimers are a constitutive feature that is not altered by SERCA conformation or PLB phosphorylation.
Materials and Methods
Molecular biology and cell culture
Enhanced green fluorescent protein (GFP) or mCerulean (Cer) was fused to the N-terminus of canine SERCA2a as described previously (20, 21). Enhanced yellow fluorescent protein (YFP) was fused to the N-terminus, between amino acids 508 and 509, or to the C-terminus of canine SERCA2a (22). We have previously demonstrated that attachment of the fluorescent proteins at these sites does not interfere with the activity of SERCA (22, 23). Canine PLB mutants were designed by mutating the serine at position 16 to alanine (S16A; nonphosphorylatable) or glutamate (S16E; phosphomimetic). We used as positive controls two fluorescence resonance energy transfer (FRET) standard constructs in which Cer is fused to Venus fluorescent protein with a 32-amino-acid linker (C32V) or a Cer fused between two Venus fluorescent proteins with linker sizes of five amino acids (VCV) (24, 25). All plasmids were validated by nucleotide sequencing. AAV-293 cells (Agilent Technologies, Santa Clara, CA) were transiently transfected with the MBS mammalian transfection kit (Stratagene, San Diego, CA) as previously described (21). After 24 h, transfected cells were trypsinized for 1 min and applied to glass-bottom multichambered slides (Matek, FPVHQ, Watsonville, CA) coated with poly-D-lysine. Cells were allowed to adhere for 2 h before microscopy experiments were carried out.
Photoactivatable cross-linking
Left ventricular cardiomyocytes were isolated from adult New Zealand white rabbits as previously described (26). All animal protocols were approved by the Loyola University Institutional Animal Care and Use Committee. Myocytes were permeabilized for 2 min with 100 μg/mL saponin (Sigma-Aldrich, St. Louis, MO) in a potassium-free relaxing solution containing 100 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 10 mM imidazole, 4 mM ATP, complete protease inhibitor cocktail (Santa Cruz Biotechnology, Dallas, TX), and 50 mM tris(2-carboxyethyl)phosphine at pH 7.0. The permeabilized myocytes were centrifuged for 5 min at 500 G at 4°C. The supernatant was removed and replaced with potassium-free relaxing solution containing the photoactivatable cross-linker benzophenone-4-maleimide (BPM) (Sigma-Aldrich), which contains a sulfhydryl-specific group and a photoactivatable group. The pellet was resuspended and incubated for 10 min at room temperature. The suspension was then placed on ice and irradiated for 20 min by a long-wave ultraviolet lamp set at 365 nm (UVP, Upland, CA). A 2× cell lysis solution (300 mM NaCL, 100 mM Tris, and 2% SDS, pH 8.0) was added in equal volume to the suspension in preparation for Western blotting.
Coimmunoprecipitation
AAV-293 cells were transiently transfected for 24 h with GFP-SERCA2a and cMyc-SERCA2a at a molar plasmid ratio of 1:1. The cells were resuspended in cell lysis buffer containing: 150 mM NaCl, 50 mM Tris, 1% Nonidet-P40, and EDTA-free protease inhibitor cocktail (Santa Cruz Biotechnology) at pH 8.0. The lysed product was placed on a shaker for 20 min at 4°C followed by centrifugation at 13,000 × g for 10 min. The supernatant was incubated overnight with mouse anti-cMyc antibody (cat. no. 631206, Clontech Laboratories, Mountain View, CA) or nonspecific mouse IgG antibody cat. no. I5381, Sigma-Aldrich). The resulting solution was incubated with protein G Dynabeads (Life Technologies, Carlsbad, CA) for 1 h at room temperature and then washed four times with cell lysis buffer. The bound protein was eluted for 10 min at 55°C in 1% SDS with β-mercaptoethanol. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as described below.
Western blotting
Samples were separated on a 4–15% polyacrylamide gradient gel (Bio-Rad, Hercules, CA) and transferred to polyvinylidene difluoride membrane. The membrane was blocked in milk followed by overnight incubation of the primary antibody rabbit anti-GFP (1:2000; Life Technologies), mouse anti-SERCA2 (1:2000, cat. no. ab2817, AbCam, Cambridge, United Kingdom), or mouse anti-cMyc (1:2000; cat. no. 631206, Clontech Laboratories). Blots were incubated with anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase for 2 h at room temperature (1:20,000 dilution). The blots were developed using an electrochemiluminescence substrate (Perkin Elmer, Waltham, MA) and imaged using a ChemiDoc XRS+ (Bio-Rad).
Photon counting histogram analysis-
AAV-293 cells transiently transfected with GFP-SERCA2a were subjected to cross-linking with 300 μM BPM followed by SDS-PAGE, as described above. The SERCA2 immunoreactive bands were excised from the polyacrylamide gel and soaked in phosphate-buffered saline (PBS) for 3 h at room temperature. Photon counting was performed on a Nikon Eclipse Ti inverted microscope (Nikon Instruments, Melville, NY) equipped with a 60× water immersion objective (NA 1.2). GFP was excited using a 20 MHz supercontinuum laser (Fianium Ltd) and a 482/18 nm bandpass filter (Semrock Inc). The fluorescence emission was passed through a dual band bandpass filter (523/610) and dichroic filter in the microscope (488/561 nm) and a 50 μm confocal pinhole. The emission was then directed through a 525/50 nm bandpass filter to an avalanche photodiode (Micro Photon Devices) using a dichroic beamsplitter (565 nm, Chroma). Photon counting was performed with a PicoHarp 300 time-correlated single-photon counting module (PicoQuant Photonics, West Springfield, MA). Fluorescence correlation spectroscopy was used to determine the concentration of GFP-SERCA of the gel eluants and the samples were diluted to compare equal concentrations of the eluted species in the subsequent photon-counting histogram (PCH) analysis. Histograms were generated using 2 ms bin width in SymPhoTime 64 software (PicoQuant Photonics) and exported to Globals Software for Spectroscopy and Images (Laboratory for Fluorescence Dynamics, University of California, Irvine, CA) for PCH analysis. PCH molecular brightness analysis was performed using a three-dimensional Gaussian-Lorentzian model with one- or two-species minimization.
Time-correlated single-photon counting
Fluorescence lifetime measurements were obtained from AAV-293 cells expressing GFP-SERCA alone or GFP-SERCA with mCherry-PLB using the experimental setup described above. The level of expression of fluorescently labeled proteins in each cell was determined by using a 500-mm-focal-length planoconvex lens in a flip mount to defocus the excitation beam (560/14 nm) of the supercontinuum laser to excite the entire cell. The emitted fluorescence of mCherry-PLB was detected using a 640/50 nm bandpass filter and a CCD camera (CoolSNAP K4, Photometrics, Tucson, AZ). After each image was obtained, the lens was removed from the path and time-correlated single-photon counting was performed using single-point excitation and confocal detection using the avalanche photodiode. The magic angle condition was not used in these experiments because the supercontinuum excitation laser is not polarized. Moreover, it is not recommended that a high-numerical-aperture lens be combined with a polarizer under magic angle conditions, because this can create or exacerbate polarization effects (27, 28). We noted that the decay of the donor-only sample was monoexponential, suggesting that additional lifetimes in the presence of the acceptor were not due to polarization artifacts and could be attributed to FRET. Fluorescence decays obtained from each cell were analyzed independently by tail fitting with a one- or two-component exponential decay from 3.5 ns using the SymPhoTime 64 software. The amplitude parameter was constrained by a lower limit of zero. The tau parameter was compared to the average fluorescence intensity of mCherry-PLB for each cell measured with the charge-coupleld device camera to determine the dependence of fluorescence lifetime on protein expression. Two-component analysis of GFP-SERCA fluorescence decays was performed by fixing one lifetime to the donor-only tau to limit the number of floating parameters. Distance measurements were calculated from the relationship R = R0(E−1 – 1)1/6, where R0 is the Förster distance for the GFP/mCherry pair (5.31 nm) and E is the average FRET value from the lifetime measurements, assuming a random orientation factor (κ2 = 2/3).
Acceptor sensitization FRET and cell scoring
Sensitized donor/acceptor emission FRET was performed as previously described (29). Each cell that expressed both Cer and YFP was plotted as the percent FRET over the range of expressed protein concentration, as determined by the fluorescence intensity of the yellow fluorescent protein (YFP) channel (expressed in arbitrary units). To demonstrate specificity of FRET, we performed competition of the SERCA-SERCA complex using unlabeled SERCA protein. Just as “cold” unlabeled protein can be used to compete for “hot” labeled protein in a conventional radioisotope binding assay, specific FRET is decreased by competition, allowing quantification of the remainder as nonspecific FRET.
Progressive acceptor photobleaching
Acceptor photobleaching was performed as previously described (24). Briefly, AAV-293 cells transfected with Cer-SERCA and YFP-SERCA in a 1:5 molar plasmid ratio. Cells were placed on an inverted microscope (Nikon Instruments) equipped with a 60× 1.49 NA oil objective CCD camera (iXon 887, Andor Technology, Belfast, United Kingdom), Lumen 200 metal halide lamp (Prior, Rockland, MA), and cyan fluorescent protein (CFP)/YFP emission and excitation filters (Semrock, Rochester, NY). A field of cells was selected and imaged every 10 s for 500 s. YFP was selectively photobleached by exposure to YFP excitation between each image. The photobleaching power at the sample was ∼1.59 nW/μm2.The change in average fluorescence intensity of each cell was calculated after subtraction of background fluorescence.
Results
Photoactivatable cross-linking of SERCA yields high-molecular-weight species consistent with a dimer complex
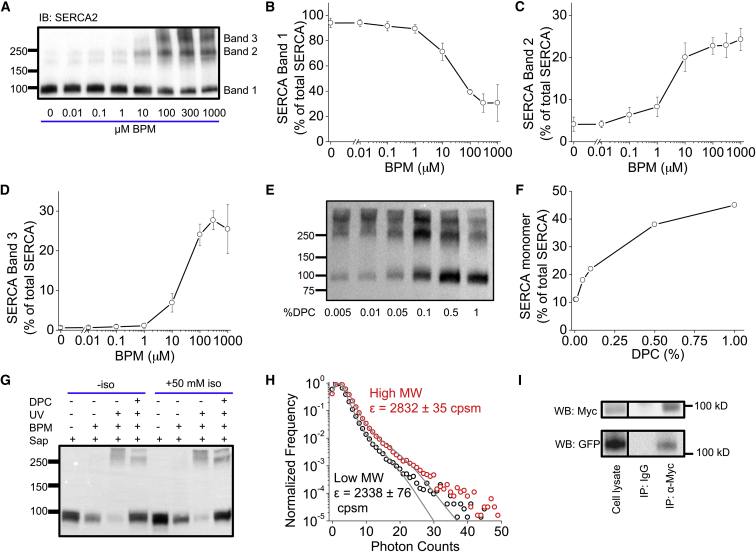
Other studies have shown that SERCA is capable of forming homooligomers in reconstituted lipid bilayers. To determine whether SERCA is oligomeric in the biosynthetic membranes of ventricular cells, we treated isolated cardiac left ventricular myocytes with saponin and performed photoactivatable cross-linking with BPM. Western blots of the cross-linked material were probed with a SERCA2 primary antibody, revealing a single band at ∼99 kDa (Fig. 1 A, Band 1), in keeping with the molecular weight of a SERCA monomer. Increasing concentrations of BPM caused a decrease in the relative amount of the SERCA monomer (Fig. 1 B) and yielded a new prominent band at 230 kDa (Fig. 1 A, Band 2). At the highest BPM concentrations (>100 μM) (Fig. 1 D), we observed a third band at 315 kDa (Fig. 1 A, Band 3). The relative amount of the SERCA monomer increased with preincubation of increasing amounts of the detergent n-dodecylphosphocholine (Fig. 1, E and F). It was noteworthy that high-molecular-weight species were still produced even after solubilization with up to 1% n-dodecylphosphocholine, suggesting that some oligomeric interactions were preserved in detergent before the addition of cross-linker. Previous studies have suggested that phosphorylation of PLB at serine 16 may play an essential role in SERCA homodimer formation (18, 30). However, we did not observe a significant change in cross-linking after treatment of myocytes with 50 nM of the β-adrenergic agonist isoproterenol (iso) (Fig. 1 G). The data suggest that PLB phosphorylation did not alter BPM cross-linking of SERCA.
Figure 1.
Oligomerization of SERCA probed with cross-linking and co-immunoprecipitation (co-IP). (A) Western blot analysis of rabbit LV myocytes. Increasing concentrations of BPM decreased the amount of monomeric SERCA (Band 1) and gave rise to two additional bands (Bands 2 and 3). (B–D) Quantification of Bands 1–3 relative to total SERCA. Values are the mean ± SD from three independent blots. (E) Preincubation with the detergent n-dodecylphosphocholine in 300 μM BPM increased the relative amount of monomeric SERCA (Band 1). (F) Quantification of (E). (G) Pretreatment of myocytes with 50 nM isoproterenol (iso) did not appreciably alter the SERCA electrophoretic pattern. (H) PCH analysis of GFP-SERCA isolated from low-molecular-weight (black) and high-molecular-weight (red) fractions revealed an increased molecular brightness (ε) for the high-molecular-weight species (fits shown in gray). Molecular brightness values are the mean ± SD from N = 3 individual gel slices (unpaired t-test, p < 0.001). (I) Co-IP of GFP-SERCA with Myc-SERCA. To see this figure in color, go online.
To rule out the possibility that the high-molecular-weight species is solely due to a single SERCA protomer becoming cross-linked to one of its other (non-SERCA) binding partners (31, 32), we performed cross-linking of GFP-SERCA expressed in AAV-293 cells. The non-cross-linked control sample produced a band consistent with GFP-SERCA monomer, with a molecular weight of 135 kDa (note the decreased mobility of the monomer due to the GFP tag). The cross-linked sample produced this band plus a second band at 301 kDa, a mobility that is compatible with the expected molecular weight of two GFP-SERCA molecules. The two SDS-PAGE bands were excised and eluted in PBS, and diluted to equal concentrations (as quantified by fluorescence correlation spectroscopy). PCH brightness analysis revealed that the 135 kDa section of the gel-containing monomeric GFP-SERCA had an average molecular brightness of 2338 ± 76 counts/s/molecule (cpsm), whereas the higher-molecular-weight species (301 kDa) had an increased molecular brightness of 2832 ± 35 cpsm (mean ± SD) (Fig. 1 H). (The deviation of the fit from the data on the right side of the distribution is expected; it is a consequence of a low concentration of fluorophores (33)). We noted that the brightness was not increased by a factor of twofold as would be expected for a dimer. Many factors may contribute to the apparently decreased brightness of the cross-linked species. 1) BPM cross-linking may covalently modify some GFP and decrease or eliminate its fluorescence. 2) Ultraviolet exposure (for BPM activation) photobleaches some GFP molecules. 3) Competition from endogenous SERCA creates some complexes where only a single SERCA has a GFP tag. 4) Background fluorescence adds to the apparent molecular brightness of both the high- and low-molecular-weight samples. 5) SDS detergent presumably denatures a large number of the GFP molecules. All of these factors are expected to reduce the apparent difference between the low- and the high-molecular-weight bands. As a consequence of these uncontrolled influences, it is not possible to perform the customary quantitative PCH analysis to determine stoichiometry of the bound complex (34). Instead, we interpret the results as a qualitative indication of increased molecular brightness of the high-molecular-weight species. We consider this supporting experiment to be compatible with our principal hypothesis that SERCA protomers can form homooligomeric complexes.
As an alternative approach, we investigated the interaction of SERCA protomers by coimmunoprecipitation of GFP-SERCA with cMyc-SERCA. Immunoprecipitation with anti-cMyc pulled down GFP-SERCA (Fig. 1 I). A control IgG antibody did not pull down either cMyc- or GFP-labeled SERCA.
SERCA forms oligomers in the absence or presence of PLB, regardless of PLB serine 16 phosphorylation
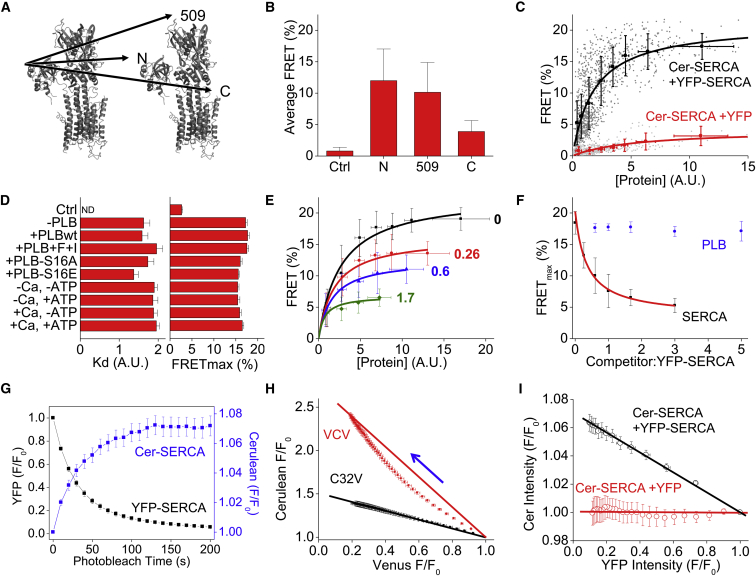
To investigate the quaternary structure and regulation of SERCA homooligomerization, we transiently transfected AAV-293 cells with Cer-SERCA and YFP-SERCA. As diagrammed in Fig. 2 A, FRET was measured from Cer-SERCA to SERCA labeled with YFP at the N-terminus (N), inserted before residue 509 (509), or fused to the C-terminus (C) of SERCA (PDB: 4KYT) (35). We observed FRET for all labeled positions (Fig. 2 B) and selected the N-terminal labeling strategy for a more detailed analysis of the SERCA-SERCA interaction. Fig. 2 C shows the FRET values obtained from individual cells (dark gray) plotted as a function of whole-cell average fluorescence intensity, taken as an index of protein concentration. We used cotransfection of Cer-SERCA and untethered YFP as a negative control (light gray). (This control yielded a nonzero FRET value, but we regard this as the trivial consequence of the difficulty in fully correcting for background, bleed-through, and direct acceptor excitation over such a wide range of expression levels. For experiments where the precise value of FRET is important, we performed acceptor photobleaching or fluorescence-lifetime FRET quantification. Nonspecific FRET was quantified below.) For clarity, the cell-by-cell FRET measurements were pooled and the values are superimposed in Fig. 2 C with error bars representing the mean ± SE. FRET efficiency increased with increasing protein concentration, and this relationship was well described by a hyperbolic fit (black line), yielding two important parameters, the apparent dissociation constant (Kd) and maximum FRET (FRETmax). These two parameters are quantified as indices of the Cer-YFP separation distance and the complex binding affinity, respectively. Neither parameter was significantly different in the absence or presence of PLB (Fig. 2 D). Activation of the protein kinase A pathway by application of 100 μM forskolin (activator of adenylate cyclase) did not alter FRETmax or Kd, even with blockade of phosphatase activity with 100 μM 3-isobutyl-1-methylxanthine (Fig. 2 D, +PLB+F+I). Mutations to PLB at serine 16 to mimic (S16E) or prevent (S16A) phosphorylation also had no effect on these two parameters. Thus, the data presented here are not consistent with previous studies indicating that PLB serine 16 phosphorylation alters SERCA homooligomerization. To investigate whether oligomerization is affected by SERCA conformational poise (36), we measured FRET in cells permeabilized with saponin and bathed in solutions of defined composition. We did not observe any significant changes to FRETmax or Kd when calcium and ATP were removed or when calcium (2 mM) and/or ATP (4 mM) were present (Fig. 2 D). These results suggest that SERCA conformational poise does not alter the oligomeric structure or affinity.
Figure 2.
Oligomerization of SERCA quantified by FRET. (A) Fluorescent protein fusion sites for FRET experiments. (B) Average sensitized emission FRET ± SD measured from the N-terminally labeled Cer-SERCA2a to YFP (Ctrl) or YFP-SERCA2a labeled at the N-terminus (N), between amino acids 508 and 509 (509), or at the C-terminus (C). (C) Protein concentration dependence of sensitized emission FRET from Cer-SERCA2a to YFP-SERCA2a (N) (black) or negative control Cer-SERCA2a to nonfusion YFP (red). Individual cells are shown in gray, with pooled data (mean ± SE) in red or black and hyperbolic fits shown as lines. (D) Apparent dissociation constants (Kd) and maximum FRET (FRETmax) values calculated from the hyperbolic fits of FRET data in the absence (−PLB) or presence (+PLBwt) of PLB. Application of 100 μM forskolin and 100 μM 3-isobutyl-1-methylxanthine (+PLB+F+I) did not alter the apparent affinity of the dimer, nor was it altered by coexpression with nonphosphorylatable (+PLB-S16A) or phosphomimetic (+PLB-S16E) PLB mutants. Alterations to calcium (2 mM) or ATP (4 mM) did not elicit any changes. Values are the mean ± SE of N = 3 fits (there were no statistically significant differences between groups by one-way ANOVA). (E) Sensitized emission FRET from Cer-SERCA and YFP-SERCA was reduced by coexpression of nonfluorescent SERCA (data pooled and fit as in (C)). Values reflect the molar ratio of competitor to YFP-SERCA. (F) FRETmax was decreased by competition with nonfluorescent SERCA (black), but not by PLB (blue). The hyperbolic fit is shown in red. (G) Progressive acceptor photobleaching of YFP-SERCA (black) resulted in increased Cer-SERCA fluorescence (blue), indicating FRET. Values are the mean ± SE for N = 28 cells total from three independent experiments. (H) The relationship of donor/acceptor fluorescence intensities during the process of photobleaching for a construct comprising one acceptor and one donor (C32V, black) and a construct comprising two acceptors and one donor (VCV, red). Values are the mean ± SD for N = 12 cells. Lines connect the first and last data points. (I) Photobleaching of the SERCA homooligomer resulted in a linear increase in Cer intensity with decreasing YFP intensity (black), suggesting a homodimer. No FRET was observed for nonfusion control. To see this figure in color, go online.
To determine whether the measured FRET was due to specific protein-protein interactions or nonspecific protein crowding in the membrane we measured the decrease in SERCA-SERCA FRET with competition by unlabeled SERCA (Fig. 2 E). Increasing unlabeled SERCA progressively decreased FRETmax (Fig. 2 F, black data points). This reduction in FRETmax was well described by a hyperbolic fit (red), consistent with simple competition of unlabeled SERCA for the fluorescently labeled SERCA protomers in the oligomeric complex. The data suggest that the FRET observed here is due to a specific interaction between SERCA molecules. The residual FRET taken from the asymptote of the hyperbolic fit was 3.5%. As previously described (24), we take this value as an estimate of nonspecific FRET, that is, the fraction of FRET that arises from donors and acceptors that are in close proximity in the bilayer but not physically bound to one another. Notably, using PLB as a competitor did not alter the measured SERCA-SERCA FRET at any ratio tested (Fig. 2 F, blue data points), consistent with the experiment shown in Fig. 2 D. We conclude that the presence or absence of PLB did not alter the constitutive oligomerization of SERCA under the experimental conditions in this study.
This competition experiment was also useful for approximating the density of FRET acceptors using the relationship described by Fung and Stryer (37). For a probe with an ∼50 Å Förster distance, 3.5% nonspecific FRET between unbound donor and acceptors would arise from a lipid/acceptor ratio in the range of 2000:1 to 4000:1. Significantly, native SR protein expression levels are on the order of 750:1 lipid/protein ratio (38, 39), which is three- to fivefold more concentrated than the levels achieved in this study. Thus, we conclude that native SERCA in muscle is mostly oligomeric.
SERCA forms homodimers
The rationale of the experiments depicted in Fig. 1, A–G, was to demonstrate that SERCA oligomers exist natively in intact membranes of living cardiac myocytes. To achieve this, we used a panspecific cross-linking methodology that does not require engineering specific cross-linkable residues into the proteins. However, because the photoaffinity label also cross-links other bound proteins (e.g,. PLB, or Hax1, or PP1, etc.), it is difficult to determine the stoichiometry from electrophoretic mobility. Thus, the unresolved question of regulatory complex stoichiometry was pursued using complementary spectroscopic approaches. We performed progressive acceptor photobleaching with AAV-293 cells transiently expressing Cer-SERCA and YFP-SERCA. Fig. 2 G shows the progressive decrease in YFP-SERCA emission (black) with selective photobleaching with a concomitant increase in Cer-SERCA emission (blue). The stoichiometry of the donor-acceptor complex is reflected in a comparison of the donor and fluorescence intensities (40). As an example, Fig. 2 H shows that Cer fluorescence (plotted on the ordinate) increases linearly with the decrease in Venus acceptor fluorescence (plotted on the abscissa) for a control construct (C32V) with a 1:1 donor/acceptor stoichiometry. In contrast, a construct comprising a donor tethered to two acceptors (VCV) displayed significant curvature, since FRET persists after ablation of a single acceptor, and donor brightness only fully increases late in the photobleaching process when the last acceptors are photobleached. We have observed similar curvature with FRET between PLB protomers in a pentameric complex (24). Fig. 2 I shows a linear Cer-versus-YFP relationship for the SERCA homooligomer. The data are consistent with a single acceptor in the oligomeric complex, suggesting that SERCA forms homodimers. This relationship was linear over a wide range of protein concentrations. Untethered YFP control showed no increase in Cer intensity (Fig. 2 I, red).
SERCA homodimers interact with a single PLB protomer
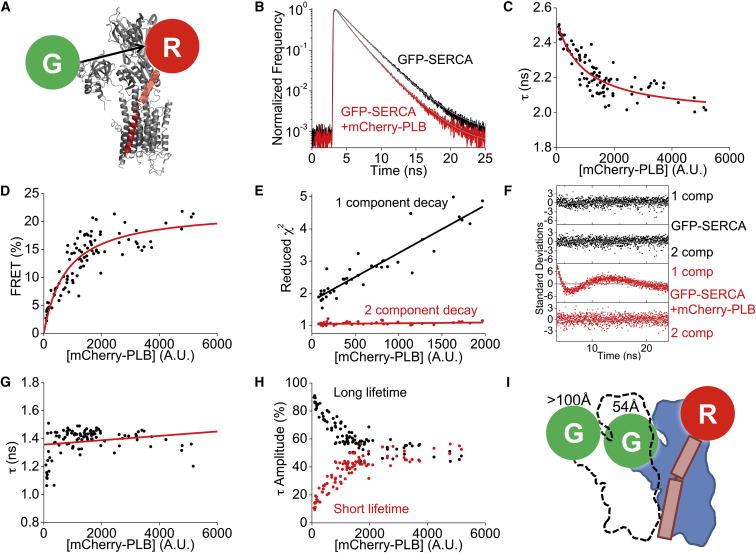
We have previously used acceptor photobleaching to quantify Cer-SERCA to YFP-PLB regulatory complex. We observed a linear donor/acceptor relationship consistent with a single YFP-PLB acceptor in the complex, but this assay cannot rule out the possibility of multiple Cer-SERCA donors in the complex. To test this directly, one could swap the fluorophores and see whether the reciprocal experiment also shows a linear relationship. However, this was impractical for this system, as PLB forms avid homopentamers. Consequently, most donor-labeled PLB donors would not participate in FRET, complicating the analysis. As an alternative we probed the stoichiometry of the SERCA-PLB complex with fluorescence-lifetime analysis, which can resolve subpopulations of donors with differential FRET efficiencies. Fig. 3 A depicts the FRET strategy, with a GFP donor fused to the N-terminus of SERCA and an mCherry acceptor fused to the N-terminus of PLB. The crystal structure of the regulatory complex did not reveal the PLB cytoplasmic domain (35), so the actual position of the PLB N-terminus and fluorescent protein tag are unknown. Fig. 3 B shows the fluorescence decay of GFP-SERCA in the absence (black) or presence (red) of mCherry-PLB. The decrease in the GFP fluorescence lifetime in the presence of mCherry-PLB is consistent with FRET from GFP to mCherry. Decays were obtained from 99 cells and analyzed by fitting to a single-component exponential decay. The fluorescence lifetime (τD) of GFP-SERCA alone was 2.56 ± 0.02 ns. The fluorescence lifetime (τDA) of GFP-SERCA decreased with increasing mCherry-PLB protein expression to a minimum value (Fig. 3 C). FRET was quantified according to the relationship FRET = 100 × (1 − τDA/τD). Fig. 3 D shows that FRET increased with acceptor expression in a hyperbolic relationship to a maximum of 22.2% FRET (Fig. 3 D), a value that is in harmony with previous steady-state FRET measurements (20, 29). However, we observed that the single-component exponential-decay goodness of fit (reduced χ2) became progressively worse as the level of protein expression increased (Fig. 3 E, black), suggesting that a single-species model was a poor description of the complex at higher protein-expression levels. Reduced χ2 was significantly improved by a two-component exponential model, which was a good description of the data over a wide range of concentrations (Fig. 3 E, red). Fit residuals for one- and two-component models show that the GFP-SERCA/mCherry-PLB samples are well described by the two-component model (Fig. 3 F). Interestingly, the two-component exponential fit yielded a long lifetime (τL) of 2.48 ± 0.09 ns that was very similar to the “donor-alone” lifetime and a short lifetime (τS) that averaged 1.37 ± 0.09 ns. The data are consistent with the existence of a subpopulation of high FRET donors (τS) and another subpopulation of non-FRET donors (τL). Fig. 3 G is a cell-by-cell measurement of the short-lifetime component, revealing an apparent FRET efficiency of 46.3 ± 3.9% that did not change with protein expression level. This average FRET value corresponds to an apparent probe separation distance of 54 Å. Interestingly, the estimate of SERCA-PLB FRET efficiency presented here is approximately twice the value we previously determined using sensitized emission FRET measurements (20, 29). Those previous steady-state measurements (which measure average FRET without distinguishing subpopulations of donors) yielded distance estimates ranging from 4 to 10 Å longer than the time-resolved quantification presented here. The significance of this discrepancy between the two methods is discussed below. We also performed fluorescence-lifetime measurements of SERCA bound to a nonphosphorylatable PLB mutant, S16A, and obtained 44.6% FRET, which was similar to the wild-type value. Measurements with PLB-S16E gave a FRET value of 32.6% FRET. This decrease compared to nonphosphorylatable PLB is consistent with previous acceptor-sensitization FRET measurements (41).
Figure 3.
Fluorescence-lifetime FRET analysis of the SERCA-PLB regulatory complex. (A) Labeling strategy for FRET from GFP-SERCA (gray) to mCherry-PLB (red). (B) Representative fluorescence decay of GFP-SERCA shortened by coexpression of mCherry-PLB, indicating FRET. Exponential tail fitting is shown in gray. (C) A single-component exponential fit of the fluorescence lifetime shows a decrease in τ with increasing protein expression. The hyperbolic model is in red. (D) FRET (calculated from the data in (C)) increased with protein expression. The hyperbolic model is in red. (E) Reduced-χ2 values for one-component fits worsened as protein expression increased. The reduced χ2 value was improved by a two-component fit. Linear fits appear as lines. (F) Residual plots of (B) from one- or two-component exponential decay fitting of the GFP-SERCA alone (black) or in the presence of mCherry-PLB (red). (G) The short-lifetime values from two-component exponential decays were consistent across a large range of protein expression levels. The linear fit is shown in red. (H) The relative contributions (amplitude) of the short and long lifetimes. (I) A model of the SERCA-SERCA-PLB regulatory complex in which one of the donor-labeled SERCA protomers (dotted outline) is too distant to participate in FRET with PLB. To see this figure in color, go online.
Two-component fitting of GFP-SERCA decays also revealed the relative proportion of the high- and low-FRET populations. As expected, the percentage of donors with short lifetimes (high FRET) increased with protein expression at the expense of the long-lifetime (non-FRET) species. Significantly, the percentage of high-FRET donors approached but did not exceed 50% of the total (Fig. 3 H). Thus, the numbers of high-FRET and non-FRET donors converged to approximately equal-size populations. This phenomenon may be explained by a model in which a single SERCA protomer undergoes FRET with PLB, whereas a second bound SERCA protomer does not (Fig. 3 I).
Discussion
Diverse experimental approaches have provided evidence for SERCA homooligomerization (10, 19). Previous studies have provided various estimates of the stoichiometry of the SERCA oligomeric complex, with evidence supporting dimers (17, 18, 42, 43), trimers (44), or higher-order oligomers (13, 14, 45). The results presented here are consistent with a SERCA homodimer, as we observe a linear increase in donor fluorescence with the progressive photobleaching of the acceptor (Fig. 2 I). This linear relationship was preserved even at the highest levels of protein expression, suggesting that the dimer does not proceed to higher-order oligomerization. The functional significance of the SERCA-SERCA interaction is not clear. Detergent-solubilized SERCA1a is still catalytically active (11), suggesting that dimerization is not obligatory for function. However, several key functional studies suggested that SERCA dimer activity is greater than the sum of two monomers (18, 19). Although in vitro studies have suggested that SERCA coupling is regulated by PLB phosphorylation (16, 17), we did not observe a change in either the structure or the binding affinity of the homodimer in response to PLB binding or PLB phosphorylation or Ca binding (Fig. 2 D). It is possible that SERCA is always physically coupled, but PLB phosphorylation engages functional coupling of the protomers. This hypothesis is compatible with reports that indicate that phosphorylated PLB increases SERCA Vmax (46, 47, 48) (though other groups did not observe a Vmax change (49, 50, 51)). Importantly, PLB-dependent functional coupling implies that phosphorylated PLB stays bound to SERCA throughout the calcium transport cycle, a concept that is supported by many studies (29, 38, 52) but not all (53, 54). Our lab has previously reported spectroscopic evidence for a particularly compact and ordered conformation of SERCA when bound to phosphorylated PLB and Ca (23). This conformation may be the structural signature of functionally coupled SERCA dimers.
In this study, we investigated the regulatory complex in detail using fluorescence lifetime analysis to quantify FRET from SERCA to PLB. We observed that there are always donors that do not participate in FRET, even at the highest protein concentrations where binding interactions appear to approach saturation. One may consider several possible interpretations of this result. There may simply be a fraction of SERCA that is inaccessible to PLB. We did not observe any mislocalization of SERCA fluorescence, which was 100% colocalized with PLB, so any such sequestration of SERCA would have to be in membrane microdomains that are below the resolution limit of fluorescence microscopy. However, it seems implausible that the proportion of inaccessible SERCA would decrease with increasing protein expression down to a minimum of almost exactly 50% of total SERCA (Fig. 3 H). A second alternative hypothesis is that only SERCA molecules with the appropriate conformational poise can interact with PLB (55). Again, it seems surprising that this proportion of pumps in the accessible state would titrate to 50%. Moreover, we have previously shown that Ca only modestly alters the PLB-SERCA binding affinity (29), suggesting that PLB can bind to SERCA without regard to conformational poise. Therefore, we interpret the data presented here as indicative of a model in which the SERCA dimer interacts with PLB such that only one of the two donors in the complex may perform FRET with the acceptor on PLB. Several structural schemes could satisfy this; one simple arrangement is diagrammed in Fig. 3 I, where a dimer of SERCA protomers interacts with a single PLB. The leftmost donor (Fig. 3 I, G), which is bound to the left/front SERCA protomer (dotted outline), is >100 Å away from the mCherry (R), too far to participate in FRET. The donor bound to the right SERCA protomer is 54 Å from the mCherry, supporting robust FRET. Thus, no matter how much increasing protein expression drives the equilibrium toward the bound complex, the population of donors participating in FRET can never exceed 50% of the total. Thus, the maximal FRET measured by steady-state methods (Fig. 2, C and D) (20, 24) is half of that quantified from the short-lifetime component (τS) of a FRET donor fluorescence decay (Fig. 3 G). In addition to the tail-fit fluorescence lifetime analysis presented here, we performed reconvolution fitting with an inferred instrument-response function. In our implementation, this analysis gave a reduced-χ2 value of 2.4 ± 0.9, significantly worse than tail fits that gave reduced-χ2 values of ∼1 (Fig. 3 E). The χ2 value for reconvolution could be improved by adding a third species to the model, but in our hands, this produced spurious results, such as negative amplitudes and implausible lifetimes (e.g., 170 ps) for the third species. Nevertheless, we did a side-by-side comparison of two-species reconvolution and two-species tail-fit analyses and obtained qualitatively similar results: cells with low protein expression had a dominant low-FRET (long-lifetime) population, whereas the highest-expressing cells had a balanced mix of high-FRET and low-FRET components. The data are consistent with our hypotheses that there are two GFP-SERCA donors in the regulatory complex, one that participates in FRET with mCherry-PLB, and one that is too far away.
The functional implication of a single PLB regulating a pair of SERCAs is unclear, however one may speculate that this stoichiometry would make PLB relatively more potent in modulating transport activity. In particular, estimates of the in vivo expression ratio of PLB to SERCA range from 1:5 to 4:1 (56, 57), and a substantial portion (∼80% (58)) of the PLB is pentameric and therefore inaccessible to SERCA. Regulation of two SERCA pumps by a single PLB would explain this apparent regulatory-protein expression mismatch. The stoichiometry may also account for the greater cooperativity of Ca-ATPase activity in cardiac muscle compared to skeletal muscle lacking PLB. If PLB engages functional coupling of the two SERCA molecules, the number of Ca-binding sites per transport unit is effectively increased. Indeed, some superinhibitory PLB mutants can deliver Hill coefficients that exceed 2 (59). Finally, these results have implications for human disease, particularly heart failure, in which SERCA activity is decreased as a result of changes in SERCA/PLB ratios, SERCA expression levels (60), or changes in SERCA specific activity (61, 62). These results suggest that deviation from optimal protein expression level or damage to a fraction of the SERCA population may yield a larger-than-expected functional deficit as a consequence of altered conformational coupling.
Conclusions
Our data support a model of a quaternary regulatory complex consisting of a single PLB bound to a pair of SERCA protomers. We observe SERCA2a dimers in the absence/presence of PLB and the dimers do not require PLB phosphorylation to assemble. Moreover, cross-linking experiments suggest that SERCA2a readily forms dimers in the native environment of the sarcoplasmic reticulum membrane of the cardiac muscle cell. Overall, we conclude that SERCA dimers are always physically coupled, but we speculate that they become functionally coupled only after PLB phosphorylation.
Author Contributions
D.J.B. performed all the experiments in the study and wrote the manuscript. T.J.Z. performed the P.C.H. experiments and analysis and helped revise the article. S.L.R. conceived the project, supervised the work, and helped write the article. All authors reviewed the results and approved the final version of this manuscript.
Acknowledgments
The authors are grateful for helpful conversations with Prof. David D. Thomas, Prof. Pieter de Tombe, and Prof. Howard S. Young. We also appreciate technical assistance from Zhanjia Hou and Jollyn Tyryfter. Cardiac myocytes were kindly provided by Elisa Bovo and Prof. Aleksey Zima and the Department of Cell and Molecular Physiology cell isolation facility.
This research was supported by the American Heart Association (13PRE16840014, to D.J.B.) and the National Institutes of Health (HL092321 and HL106189, to S.L.R.).
Editor: Paul Wiseman.
References
- 1.Plank B., Pifl C., Suko J. Correlation between calmodulin-dependent increase in the rate of calcium transport and calmodulin-dependent phosphorylation of cardiac sarcoplasmic reticulum. Characterization of calmodulin-dependent phosphorylation. Eur. J. Biochem. 1983;136:215–221. doi: 10.1111/j.1432-1033.1983.tb07729.x. [DOI] [PubMed] [Google Scholar]
- 2.Simmerman H.K., Collins J.H., Jones L.R. Sequence analysis of phospholamban. Identification of phosphorylation sites and two major structural domains. J. Biol. Chem. 1986;261:13333–13341. [PubMed] [Google Scholar]
- 3.Tada M., Kirchberger M.A., Katz A.M. Phosphorylation of a 22,000-dalton component of the cardiac sarcoplasmic reticulum by adenosine 3′:5′-monophosphate-dependent protein kinase. J. Biol. Chem. 1975;250:2640–2647. [PubMed] [Google Scholar]
- 4.Kiss E., Ball N.A., Walsh R.A. Differential changes in cardiac phospholamban and sarcoplasmic reticular Ca2+-ATPase protein levels. Effects on Ca2+ transport and mechanics in compensated pressure-overload hypertrophy and congestive heart failure. Circ. Res. 1995;77:759–764. doi: 10.1161/01.res.77.4.759. [DOI] [PubMed] [Google Scholar]
- 5.Zarain-Herzberg A., Afzal N., Dhalla N.S. Decreased expression of cardiac sarcoplasmic reticulum Ca2+-pump ATPase in congestive heart failure due to myocardial infarction. Mol. Cell. Biochem. 1996;163-164:285–290. doi: 10.1007/BF00408669. [DOI] [PubMed] [Google Scholar]
- 6.Baker D.L., Hashimoto K., Periasamy M. Targeted overexpression of the sarcoplasmic reticulum Ca2+-ATPase increases cardiac contractility in transgenic mouse hearts. Circ. Res. 1998;83:1205–1214. doi: 10.1161/01.res.83.12.1205. [DOI] [PubMed] [Google Scholar]
- 7.Davia K., Bernobich E., Harding S.E. SERCA2A overexpression decreases the incidence of aftercontractions in adult rabbit ventricular myocytes. J. Mol. Cell. Cardiol. 2001;33:1005–1015. doi: 10.1006/jmcc.2001.1368. [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto M.I., del Monte F., Hajjar R.J. Adenoviral gene transfer of SERCA2a improves left-ventricular function in aortic-banded rats in transition to heart failure. Proc. Natl. Acad. Sci. USA. 2000;97:793–798. doi: 10.1073/pnas.97.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Møller J.V., Lind K.E., Andersen J.P. Enzyme kinetics and substrate stabilization of detergent-solubilized and membraneous (Ca2+ + Mg2+)-activated ATPase from sarcoplasmic reticulum. Effect of protein-protein interactions. J. Biol. Chem. 1980;255:1912–1920. [PubMed] [Google Scholar]
- 10.Vanderkooi J.M., Ierokomas A., Martonosi A. Fluorescence energy transfer between Ca2+ transport ATPase molecules in artificial membranes. Biochemistry. 1977;16:1262–1267. doi: 10.1021/bi00626a003. [DOI] [PubMed] [Google Scholar]
- 11.Vilsen B., Andersen J.P. Effect of phospholipid, detergent and protein-protein interaction on stability and phosphoenzyme isomerization of soluble sarcoplasmic reticulum Ca-ATPase. Eur. J. Biochem. 1987;170:421–429. doi: 10.1111/j.1432-1033.1987.tb13716.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto T., Yantorno R.E., Tonomura Y. Comparative study of the kinetic and structural properties of monomeric and oligomeric forms of sarcoplasmic reticulum ATPase. J. Biochem. 1984;95:1783–1791. doi: 10.1093/oxfordjournals.jbchem.a134791. [DOI] [PubMed] [Google Scholar]
- 13.Mersol J.V., Kutchai H., Thomas D.D. Self-association accompanies inhibition of Ca-ATPase by thapsigargin. Biophys. J. 1995;68:208–215. doi: 10.1016/S0006-3495(95)80176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voss J., Birmachu W., Thomas D.D. Effects of melittin on molecular dynamics and Ca-ATPase activity in sarcoplasmic reticulum membranes: time-resolved optical anisotropy. Biochemistry. 1991;30:7498–7506. doi: 10.1021/bi00244a019. [DOI] [PubMed] [Google Scholar]
- 15.Voss J., Jones L.R., Thomas D.D. The physical mechanism of calcium pump regulation in the heart. Biophys. J. 1994;67:190–196. doi: 10.1016/S0006-3495(94)80469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negash S., Chen L.T., Squier T.C. Phosphorylation of phospholamban by cAMP-dependent protein kinase enhances interactions between Ca-ATPase polypeptide chains in cardiac sarcoplasmic reticulum membranes. Biochemistry. 1996;35:11247–11259. doi: 10.1021/bi960864q. [DOI] [PubMed] [Google Scholar]
- 17.Mahaney J.E., Albers R.W., Froehlich J.P. Intermolecular conformational coupling and free energy exchange enhance the catalytic efficiency of cardiac muscle SERCA2a following the relief of phospholamban inhibition. Biochemistry. 2005;44:7713–7724. doi: 10.1021/bi048011i. [DOI] [PubMed] [Google Scholar]
- 18.Chen L.T., Yao Q., Bigelow D.J. Phospholamban modulates the functional coupling between nucleotide domains in Ca-ATPase oligomeric complexes in cardiac sarcoplasmic reticulum. Biochemistry. 2009;48:2411–2421. doi: 10.1021/bi8021526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamberlain B.K., Berenski C.J., Fleischer S. Determination of the oligomeric structure of the Ca2+ pump protein in canine cardiac sarcoplasmic reticulum membranes using radiation inactivation analysis. J. Biol. Chem. 1983;258:11997–12001. [PubMed] [Google Scholar]
- 20.Hou Z., Robia S.L. Relative affinity of calcium pump isoforms for phospholamban quantified by fluorescence resonance energy transfer. J. Mol. Biol. 2010;402:210–216. doi: 10.1016/j.jmb.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robia S.L., Campbell K.S., Thomas D.D. Förster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ. Res. 2007;101:1123–1129. doi: 10.1161/CIRCRESAHA.107.159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou Z., Hu Z., Robia S.L. 2-Color calcium pump reveals closure of the cytoplasmic headpiece with calcium binding. PLoS One. 2012;7:e40369. doi: 10.1371/journal.pone.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallikkuth S., Blackwell D.J., Robia S.L. Phosphorylated phospholamban stabilizes a compact conformation of the cardiac calcium-ATPase. Biophys. J. 2013;105:1812–1821. doi: 10.1016/j.bpj.2013.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly E.M., Hou Z., Robia S.L. Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J. Biol. Chem. 2008;283:12202–12211. doi: 10.1074/jbc.M707590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koushik S.V., Chen H., Vogel S.S. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys. J. 2006;91:L99–L101. doi: 10.1529/biophysj.106.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domeier T.L., Blatter L.A., Zima A.V. Alteration of sarcoplasmic reticulum Ca2+ release termination by ryanodine receptor sensitization and in heart failure. J. Physiol. 2009;587:5197–5209. doi: 10.1113/jphysiol.2009.177576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisz J.J. Another look at magic-angle-detected fluorescence and emission anisotropy decays in fluorescence microscopy. J. Phys. Chem. A. 2007;111:12867–12870. doi: 10.1021/jp0775855. [DOI] [PubMed] [Google Scholar]
- 28.Fisz J.J. Fluorescence polarization spectroscopy at combined high-aperture excitation and detection: application to one-photon-excitation fluorescence microscopy. J. Phys. Chem. A. 2007;111:8606–8621. doi: 10.1021/jp072113b. [DOI] [PubMed] [Google Scholar]
- 29.Bidwell P., Blackwell D.J., Robia S.L. Phospholamban binds with differential affinity to calcium pump conformers. J. Biol. Chem. 2011;286:35044–35050. doi: 10.1074/jbc.M111.266759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahaney J.E., Albers R.W., Froehlich J.P. Phospholamban inhibits Ca2+ pump oligomerization and intersubunit free energy exchange leading to activation of cardiac muscle SERCA2a. Ann. N. Y. Acad. Sci. 2003;986:338–340. doi: 10.1111/j.1749-6632.2003.tb07208.x. [DOI] [PubMed] [Google Scholar]
- 31.Arvanitis D.A., Vafiadaki E., Kranias E.G. Histidine-rich Ca-binding protein interacts with sarcoplasmic reticulum Ca-ATPase. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H1581–H1589. doi: 10.1152/ajpheart.00278.2007. [DOI] [PubMed] [Google Scholar]
- 32.Vafiadaki E., Arvanitis D.A., Kranias E.G. The anti-apoptotic protein HAX-1 interacts with SERCA2 and regulates its protein levels to promote cell survival. Mol. Biol. Cell. 2009;20:306–318. doi: 10.1091/mbc.E08-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., Müller J.D., Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Q., Pallikkuth S., Robia S.L. Phosphomimetic mutations enhance oligomerization of phospholemman and modulate its interaction with the Na/K-ATPase. J. Biol. Chem. 2011;286:9120–9126. doi: 10.1074/jbc.M110.198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akin B.L., Hurley T.D., Jones L.R. The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum. J. Biol. Chem. 2013;288:30181–30191. doi: 10.1074/jbc.M113.501585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen J.P., Vilsen B. Equilibrium between monomers and oligomers of soluble Ca2+-ATPase during the functional cycle. FEBS Lett. 1985;189:13–17. doi: 10.1016/0014-5793(85)80832-2. [DOI] [PubMed] [Google Scholar]
- 37.Fung B.K., Stryer L. Surface density determination in membranes by fluorescence energy transfer. Biochemistry. 1978;17:5241–5248. doi: 10.1021/bi00617a025. [DOI] [PubMed] [Google Scholar]
- 38.Ferrington D.A., Yao Q., Bigelow D.J. Comparable levels of Ca-ATPase inhibition by phospholamban in slow-twitch skeletal and cardiac sarcoplasmic reticulum. Biochemistry. 2002;41:13289–13296. doi: 10.1021/bi026407t. [DOI] [PubMed] [Google Scholar]
- 39.Mueller B., Karim C.B., Thomas D.D. Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfer. Biochemistry. 2004;43:8754–8765. doi: 10.1021/bi049732k. [DOI] [PubMed] [Google Scholar]
- 40.Li M., Reddy L.G., Thomas D.D. A fluorescence energy transfer method for analyzing protein oligomeric structure: application to phospholamban. Biophys. J. 1999;76:2587–2599. doi: 10.1016/S0006-3495(99)77411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou Z., Kelly E.M., Robia S.L. Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J. Biol. Chem. 2008;283:28996–29003. doi: 10.1074/jbc.M804782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Q., Chen L.T., Bigelow D.J. Oligomeric interactions between phospholamban molecules regulate Ca-ATPase activity in functionally reconstituted membranes. Biochemistry. 2001;40:6406–6413. doi: 10.1021/bi002891t. [DOI] [PubMed] [Google Scholar]
- 43.Young H.S., Jones L.R., Stokes D.L. Locating phospholamban in co-crystals with Ca2+-ATPase by cryoelectron microscopy. Biophys. J. 2001;81:884–894. doi: 10.1016/S0006-3495(01)75748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahaney J.E., Thomas D.D., Froehlich J.P. Intermolecular interactions in the mechanism of skeletal muscle sarcoplasmic reticulum Ca2+-ATPase (SERCA1): evidence for a triprotomer. Biochemistry. 2008;47:13711–13725. doi: 10.1021/bi801024a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voss J.C., Mahaney J.E., Thomas D.D. Mechanism of Ca-ATPase inhibition by melittin in skeletal sarcoplasmic reticulum. Biochemistry. 1995;34:930–939. doi: 10.1021/bi00003a027. [DOI] [PubMed] [Google Scholar]
- 46.Antipenko A.Y., Spielman A.I., Kirchberger M.A. Comparison of the kinetic effects of phospholamban phosphorylation and anti-phospholamban monoclonal antibody on the calcium pump in purified cardiac sarcoplasmic reticulum membranes. Biochemistry. 1997;36:12903–12910. doi: 10.1021/bi971109v. [DOI] [PubMed] [Google Scholar]
- 47.Lu Y.Z., Kirchberger M.A. Effects of a nonionic detergent on calcium uptake by cardiac microsomes. Biochemistry. 1994;33:5056–5062. doi: 10.1021/bi00183a008. [DOI] [PubMed] [Google Scholar]
- 48.Reddy L.G., Cornea R.L., Thomas D.D. Defining the molecular components of calcium transport regulation in a reconstituted membrane system. Biochemistry. 2003;42:4585–4592. doi: 10.1021/bi026995a. [DOI] [PubMed] [Google Scholar]
- 49.Morris G.L., Cheng H.C., Wang J.H. Phospholamban regulation of cardiac sarcoplasmic reticulum (Ca2+-Mg2+)-ATPase. Mechanism of regulation and site of monoclonal antibody interaction. J. Biol. Chem. 1991;266:11270–11275. [PubMed] [Google Scholar]
- 50.Reddy L.G., Jones L.R., Stokes D.L. Functional reconstitution of recombinant phospholamban with rabbit skeletal Ca2+-ATPase. J. Biol. Chem. 1995;270:9390–9397. doi: 10.1074/jbc.270.16.9390. [DOI] [PubMed] [Google Scholar]
- 51.Trieber C.A., Douglas J.L., Young H.S. The effects of mutation on the regulatory properties of phospholamban in co-reconstituted membranes. Biochemistry. 2005;44:3289–3297. doi: 10.1021/bi047878d. [DOI] [PubMed] [Google Scholar]
- 52.Li J., Bigelow D.J., Squier T.C. Conformational changes within the cytosolic portion of phospholamban upon release of Ca-ATPase inhibition. Biochemistry. 2004;43:3870–3879. doi: 10.1021/bi036183u. [DOI] [PubMed] [Google Scholar]
- 53.Jones L.R., Cornea R.L., Chen Z. Close proximity between residue 30 of phospholamban and cysteine 318 of the cardiac Ca2+ pump revealed by intermolecular thiol cross-linking. J. Biol. Chem. 2002;277:28319–28329. doi: 10.1074/jbc.M204085200. [DOI] [PubMed] [Google Scholar]
- 54.Kimura Y., Kurzydlowski K., MacLennan D.H. Phospholamban inhibitory function is activated by depolymerization. J. Biol. Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 55.Chen Z., Stokes D.L., Jones L.R. Spatial and dynamic interactions between phospholamban and the canine cardiac Ca2+ pump revealed with use of heterobifunctional cross-linking agents. J. Biol. Chem. 2003;278:48348–48356. doi: 10.1074/jbc.M309545200. [DOI] [PubMed] [Google Scholar]
- 56.Ceholski D.K., Trieber C.A., Young H.S. Hydrophobic imbalance in the cytoplasmic domain of phospholamban is a determinant for lethal dilated cardiomyopathy. J. Biol. Chem. 2012;287:16521–16529. doi: 10.1074/jbc.M112.360859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Louis C.F., Turnquist J., Jarvis B. Phospholamban stoichiometry in canine cardiac muscle sarcoplasmic reticulum. Neurochem. Res. 1987;12:937–941. doi: 10.1007/BF00966316. [DOI] [PubMed] [Google Scholar]
- 58.Cornea R.L., Jones L.R., Thomas D.D. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry. 1997;36:2960–2967. doi: 10.1021/bi961955q. [DOI] [PubMed] [Google Scholar]
- 59.Trieber C.A., Afara M., Young H.S. Effects of phospholamban transmembrane mutants on the calcium affinity, maximal activity, and cooperativity of the sarcoplasmic reticulum calcium pump. Biochemistry. 2009;48:9287–9296. doi: 10.1021/bi900852m. [DOI] [PubMed] [Google Scholar]
- 60.Hasenfuss G., Reinecke H., Drexler H. Relation between myocardial function and expression of sarcoplasmic reticulum Ca2+-ATPase in failing and nonfailing human myocardium. Circ. Res. 1994;75:434–442. doi: 10.1161/01.res.75.3.434. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt U., Hajjar R.J., Gwathmey J.K. Human heart failure: cAMP stimulation of SR Ca2+-ATPase activity and phosphorylation level of phospholamban. Am. J. Physiol. 1999;277:H474–H480. doi: 10.1152/ajpheart.1999.277.2.H474. [DOI] [PubMed] [Google Scholar]
- 62.Schwinger R.H., Böhm M., Erdmann E. Unchanged protein levels of SERCA II and phospholamban but reduced Ca2+ uptake and Ca2+-ATPase activity of cardiac sarcoplasmic reticulum from dilated cardiomyopathy patients compared with patients with nonfailing hearts. Circulation. 1995;92:3220–3228. doi: 10.1161/01.cir.92.11.3220. [DOI] [PubMed] [Google Scholar]