Abstract
The voltage-dependent anion channel 1 (VDAC-1) is an important protein of the outer mitochondrial membrane that transports energy metabolites and is involved in apoptosis. The available structures of VDAC proteins show a wide β-stranded barrel pore, with its N-terminal α-helix (N-α) bound to its interior. Electrophysiology experiments revealed that voltage, its polarity, and membrane composition modulate VDAC currents. Experiments with VDAC-1 mutants identified amino acids that regulate the gating process. However, the mechanisms for how these factors regulate VDAC-1, and which changes they trigger in the channel, are still unknown. In this study, molecular dynamics simulations and single-channel experiments of VDAC-1 show agreement for the current-voltage relationships of an “open” channel and they also show several subconducting transient states that are more cation selective in the simulations. We observed voltage-dependent asymmetric distortions of the VDAC-1 barrel and the displacement of particular charged amino acids. We constructed conformational models of the protein voltage response and the pore changes that consistently explain the protein conformations observed at opposite voltage polarities, either in phosphatidylethanolamine or phosphatidylcholine membranes. The submicrosecond VDAC-1 voltage response shows intrinsic structural changes that explain the role of key gating amino acids and support some of the current gating hypotheses. These voltage-dependent protein changes include asymmetric barrel distortion, its interaction with the membrane, and significant displacement of N-α amino acids.
Introduction
Ion channels are membrane proteins important for controlling the passage of solutes between cellular compartments (1). The mechanism(s) of opening and closing of channels—gating—can be modulated by several factors, including voltage changes, membrane composition, mechanical stress, ions, other molecules, and proteins. One of the challenges in understanding ion channels is to connect electrophysiological with structural information. In this respect, recent developments in computational simulations of ion channels have contributed to the understanding of the permeation and selectivity of several channels (2, 3, 4) and porins from the VDAC family, like VDAC-1 (5, 6, 7, 8).
VDAC-1 is mainly present in the outer mitochondrial membrane (OMM) of eukaryotic cells, and is the most studied isoform from the VDAC family. It is important for the traffic regulation of ATP, ions, and other metabolites between the mitochondria and cytosol. During apoptosis, VDAC-1 constitutes the mitochondrial permeability transition pore, which leads to mitochondrial swelling (9, 10), and in several cancer types, VDAC-1 is overexpressed (11, 12). From the available electrophysiological data, it is concluded that the main characteristics of VDAC channels are shared across species and isoforms (13, 14). For the last 40 years, VDAC proteins of several organisms have been mainly studied by reconstitution in black lipid membranes (BLMs). These experiments have shown that VDAC has an “open” anion-selective state ( of 2–5) of 4 nS at 1 M NaCl or KCl (15, 16), whereas the “closed” states are cation selective, with approximately half the conductance of the “open” state (17). It was shown in early electrophysiological experiments (18) that VDAC voltage dependence of normalized conductances follows a bell-shaped curve with the maximum around zero volts.
In addition, the lipid composition influences the voltage dependence of the open probability of Neurospora crassa VDAC (ncVDAC) (19). ncVDAC channels inserted in dioleoylphosphatidylethanolamine (DOPE) membranes show lower conductances at negative voltages compared with those in dioleoylphosphatidylcholine (DOPC). Asymmetric versus random ncVDAC channel insertions in different membranes may explain these differences at negative voltages. However, phosphorylation (20), caspase cleavage (21), and acidic susceptibility experiments combined with molecular dynamics (MD) simulations (22) suggest that the cis side in BLM experiments corresponds to the cytosolic side of the VDAC. This evidence means that asymmetric channel insertion in vivo and in vitro might explain why some VDAC proteins respond differently to the voltage polarity (23).
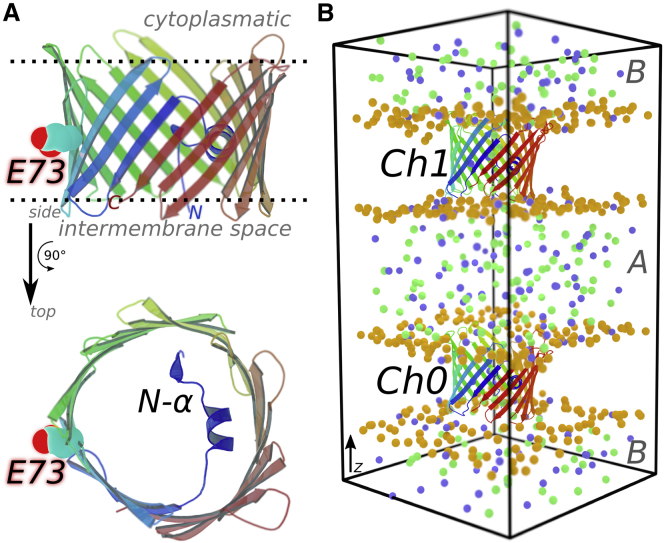
The available structures of human and mouse VDAC-1 (hVDAC-1 and mVDAC-1, respectively) (24, 25, 26) and VDAC-2 from zebrafish (27) show a 19-stranded β-barrel forming a large pore of 2.5–3 nm in diameter (Fig. 1 A). The folded VDAC structure is stable enough to be characterized in detergent micelles (24, 26) or in bicelles (25). hVDAC-1 and mVDAC-1 share 98% sequence identity and both are 29% identical to ncVDAC. The N-terminal part of VDAC shows an amphipathic α-helix structure (termed N-α) attached to the inside of the barrel (Fig. 1 A). N-α narrows the pore diameter to 1.4 nm (25). The position and orientation of N-α with respect to the barrel in the different VDAC structures is slightly different. This geometrical information supports a closing mechanism where the N-α displacement may obstruct the pore cavity (25). In addition, the barrel geometry of the VDAC NMR structures shows moderate cylindrical distortions (24), which might be another mechanism for how VDAC channels access more occluded states (14). It has been shown by Brownian and MD simulations (5, 6, 28, 29, 30) that the available VDAC structures show conductance levels (≈4 nS at 1 M [Cl−] and 2 nS at 0.5 M [Cl−]) similar to those in the experimental “open” VDAC states (6, 31). How such a large open pore inside the OMM can be gated is still unclear, however.
Figure 1.
Structure and computational setup of mVDAC-1. The mVDAC-1 structure was taken from PDB: 3EMN (25). The cartoon representations show mVDAC-1 β-strands colored from the N- to the C-terminus in a blue-green-red scale. (A) Side and top perspectives of the mVDAC-1 structure, where glutamate 73 (E73) and the N-terminal α-helix (N-α) are highlighted, with E73 pointing toward the membrane, opposite to the N-α. (B) Computational electrophysiology setup (53) for simulating mVDAC-1 SC currents. One mVDAC-1 channel is inserted in each membrane bilayer (labeled Ch0 and Ch1). The bilayers separate solution compartments A and B due to periodic boundary conditions. The phosphorus atoms from phospholipids and chloride and sodium ions are shown as orange, green, and blue spheres, respectively. Water molecules and lipid acyl chains are not shown. The currents are triggered by setting up a constant ion difference between the compartments. To see this figure in color, go online.
Concerning VDAC regions that participate in its regulation, Zachariae et al. (29) have found (by combining NMR and MD simulations) that in the absence of the N-α, the deletion mutant, Δ(1–20)-hVDAC-1, displayed distorted barrel states and the currents changed from anion- to cation-selective. The L10N mutation, which destabilizes the helix contact with the barrel, showed intermediate conductance behavior between wild-type (wt) and Δ(1–20)-hVDAC-1. This evidence indicates that collapsed barrel states might be part of the “closed” VDAC-1 states (14).
The crystallographic structure and the microsecond simulations of Choudhary et al. (6) showed that N-α attached to the pore interior of mVDAC-1 formed a stable fold that remained anion selective. They also showed that the positively charged amino acids K12, R15, and K20 (termed the KRK motif) of N-α are important for chloride permeation and define the ATP binding site. Further, the interaction of K20 with ATP has been confirmed by NMR (32) and chromatography assays (33). In yeast VDAC, charge-inverting mutations of the amino acids equivalent to D16 and K20 in VDAC-1 are able to change the voltage-dependent gating (34). The KRK motif, including K119 and R218, forms part of the “charged brush” (7), which facilitates the passage of chloride and inorganic phosphate ions through mVDAC-1. From other simulation studies (5, 35), it was concluded that several negative amino acids in the channel wall facing N-α are involved in the permeation of potassium ions. This area of the protein is of particular interest because it includes E73, whose side chain points to the middle plane of the membrane in the VDAC structures (Fig. 1 A).
Previous studies have indicated the fundamental role of E73. The E73Q mutation in mVDAC-1 prevents hexokinase-mediated protection against apoptosis (9, 36). E73 in hVDAC-1 is involved in VDAC-1’s calcium binding ability (37, 38), hexokinase regulation (39), and VDAC oligomerization (40). Dicyclohexylcarbodiimide modification experiments (41) suggest that this amino acid might be unprotonated in the membrane. The local mobility of mVDAC-1 (Protein Data Bank (PDB): 3EMN) around E73, in terms of the temperature factors (25), is comparable to the fluctuations of unprotonated E73 in MD simulations (42). mVDAC-1 simulations of charged E73 showed larger membrane distortion, in terms of local thickness, and the mVDAC-1 barrel accessed more elliptically distorted states compared with neutral residues at this position, E73Q and E73V (42).
From the available experimental and computational studies of VDAC channels, several gating models have been proposed. Some models propose large and complex conformational changes, and other models propose more moderate ones (13). The more moderate conformational changes include N-α displacement from the barrel interior, barrel deformation, or a combination of the two, which may cause pore collapse or partial pore blockage (13, 14). However, there is no consensus as to which conformational changes must occur during VDAC transitions to closed states. It is still a matter of debate how to reconcile some of the available electrophysiological evidence with the VDAC structures (43). Making the VDAC gating story more complex, single-channel (SC) VDAC shows several subconducting “closed” states (22, 44, 45, 46), which are more challenging to characterize in experiments with multiple channels per patch (47). The multiplicity of “closed” states might account for the experimental accessibility of N-α from both the cis and trans sides in BLM setups (44, 48). However, tethering the N-α to the mVDAC-1 channel wall by cysteine cross-linking does not prevent its voltage gating (49). This experimental evidence suggests that there is a mVDAC-1 closing transition that does not necessarily require complete N-α displacement from the pore lumen. Moderate voltage-dependent conformational changes that distort the barrel structure or involve amino acid reorganization in the pore are suitable for study by MD simulations (4).
Considering the current gating hypotheses, large and complex conformational changes (13) are beyond the actual capabilities of MD simulations. However, barrel changes and amino acid reorganization—including N-α amino acids—have been proposed as VDAC closing mechanisms (13, 14). In this work, by simulating an mVDAC-1 channel in the presence of voltage, we calculated its current-voltage (I/V) relationships and selectivity. We also had access to submicrosecond voltage-dependent structural changes of mVDAC-1 that revealed transient subconducting states. By performing SC hVDAC-1 experiments, we showed that hVDAC-1 displays defined closed states (45). Voltage and its polarity were correlated with global or local mVDAC-1 structural changes. Local geometrical parameters that describe the barrel distortion, the position of N-α, and the changes in the pore lumen were used to quantify their contribution to the voltage dependence of VDAC-1.
Materials and Methods
MD simulations
MD simulations in the isothermal-isobaric ensemble (NPT) (310 K, 1 bar) were performed with the MD software package Gromacs 4.5 (50). The mVDAC-1 protein (PDB: 3EMN) was inserted in preequilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phospatidylethanolamine (POPE; 478 lipids) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC; 472 lipids) bilayer patches, respectively. The mVDAC-1 protein was modeled using the CHARMM27 force field (51) with E73 in its unprotonated form. The CHARMM36 force field was used for the lipids (52) together with the charmm tip3p water model. Double mVDAC-1-bilayer systems (under tinfoil boundary conditions) were simulated, keeping a fixed ion difference (Δq of 2–14q) between compartments, in the computational electrophysiology setup (53) (Fig. 1 B). Ten replicas of each Δq were simulated for 40 ns, for a total time of ∼3 μs. The first 10 ns of each simulation were discarded as equilibration. To monitor the equilibration of the membrane-protein system, we used g_lomepro (54) to calculate the average area per lipid (APL) together with the cytoplasmic and intermembrane-space (IMS) protein areas. From the computational electrophysiology simulations, we counted the permeation events and transformed them to currents. The average voltage across the simulation box (along the z axis in Fig. 1 B) was calculated by first summing the charges per slice (200 used) and then integrating the charge distribution twice along the x and y axes using the g_potential software tool from Gromacs. The trajectories were binned in 1- or 10-ns time windows (Fig. S1 in the Supporting Material) The pore-radius profile of the protein, excluding the hydrogens, was calculated by running the trj_cavity software tool (55). It covered a z-section of 3.2 nm that includes the cytosolic and the IMS entrance of mVDAC-1 (Fig. S4). The most constricted region of the pore profiles defined the minimum-pore radius (termed ). Areas and ellipticity changes of the channel barrel were computed by approximating the C-α coordinates of barrel slices as ellipses (56). The used C-α excluded the N-α helix (amino acids 1–26) and the longest cytoplasmic loop (amino acids 264–271). Linear conformational models between mVDAC-1 atomic coordinates and the voltage or the were built with the partial-least-squares (PLS) implementation of PLS functional-mode analysis (57). We tested the robustness of the models using half of the data for training the models and the other half for cross-validation (Fig. S5). Data analysis and plots were generated using IPython (58). Rendering of molecular images was done with the software Pymol (59) and Blender (60).
NMR spectroscopy
wt hVDAC-1 was overexpressed in Escherichia coli, purified, and refolded into micelles consisting of the detergent N-lauryl-N,N-dimethylamine-N-oxide (LDAO) as previously described (26). Solution-state NMR spectra were recorded on a 900 MHz spectrometer (Bruker, Billerica, MA) equipped with a cryogenic probe head using 0.5–0.76 mM 2H(75%)–15N or 2H(75%)-13C-15N labeled hVDAC-1 in 25 mM BisTris, pH 6.8, ∼120 mM LDAO, and 5% D2O at 37°C. Spectra were processed with NMRPipe (61). 13C-α chemical shifts were extracted from transverse-relaxation-optimized-spectroscopy-based HNCA experiments. For solid-state NMR experiments, a 13C-15N-labeled hVDAC-1 sample was reconstituted into 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine liposomes at a protein/lipid ratio of 1:50 (mol/mol). Spectra were recorded using a 3.2 mm triple-resonance (1H, 13C, 15N) magic-angle spinning probe head at a static magnetic field of 20.0 T (Bruker Biospin, Karlsruhe, Germany). Sample temperature and magic-angle spinning speed were set to +5°C and 10.6 kHz, respectively. 13C-13C mixing was accomplished by proton-driven spin diffusion for 15 ms.
hVDAC-1 reconstitution
Giant unilamellar vesicles (GUVs) with diameters of up to 100 μm were prepared by electroformation. A 10 mM solution of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC, 15 μL; Avanti Polar Lipids, Alabaster, AL) and cholesterol (Sigma-Aldrich, Taufkirchen, Germany) dissolved in chloroform at a molar ratio of 9:1 were applied to indium tin-oxide-coated glass slides and allowed to dry. The lipid was rehydrated in 1 M sorbitol and treated with a 3 V AC potential (peak to peak) at a frequency of 5 Hz for 2 h. The resulting GUVs were incubated overnight at 4°C with hVDAC-1 in 0.3% (v/v) LDAO micelles that were prepared as described previously (62) to give a final protein concentration of 25–250 nM.
Electrophysiology experiments
Electrophysiological experiments on hVDAC-1 were carried out using the port-a-patch technique (Nanion, Munich). GUVs with reconstituted hVDAC-1 were added to a droplet of buffer on a planar borosilicate chip containing a single pore with a diameter of 1–3 μm. The setup was attached to a pump to apply negative pressure from the trans side to spread a GUV resulting in the formation of a protein-containing membrane with resistances in the GΩ regime. Electrical recordings were performed in 1 M KCl, 1 mM CaCl2, and 5 mM HEPES, pH 7.4, using the Axopatch 200B integrating patch-clamp amplifier equipped with a CV 203BU head stage (Molecular Devices, Sunnyvale, CA). The resulting signals were recorded with sampling rates of 10 or 50 kHz, prefiltered with a 1 kHz 4-pole Bessel filter (–80 dB/decade), digitized via a 1322A Digidata 16-bit digitizer and evaluated using the electrophysiology data acquisition and analysis software package pCLAMP 9 (Axon Instruments, Union City, CA). The open probability of the experiments was calculated as described in Carl and Sanders (63) by
| (1) |
where is the number of data points at membrane potential , is the channel in the “open” state, and is the number of all data points at , which is equivalent to the number of voltage ramps that were recorded with one reconstituted hVDAC-1. The data of each recorded current trace were converted to conductance according to and a threshold was set to distinguish between “open” and “closed” states.
Results
In MD simulations (see Materials and Methods), the mVDAC-1 structure (PDB: 3EMN, 2.3 Å resolution) was conserved, although thermal fluctuations occurred during the simulations in all conditions. The computational electrophysiology method (53), which utilizes a double-membrane system (Fig. 1 B) with an mVDAC-1 in each membrane, was used to study mVDAC-1 I/V properties. Briefly, after an ion permeates between compartments A and B, a predefined ion difference is kept constant (Δq) by swapping back an ion for a water molecule. Ion differences from 2 to 14 charges created increasing voltage ranges varying from – 400 mV (Fig. S1, A and B). The reported currents were mostly chloride selective and proportional to the applied Δq.
In the simulation setup, mVDAC-1 was inserted in preequilibrated POPE and POPC membrane patches at opposite polarities (Fig. 1 B, Ch0 and Ch1). This arrangement allowed us to access channel currents at negative and positive voltage polarities within the same simulation. Details of the simulated currents in each membrane are shown in Fig. S1. The mVDAC-1-POPE and mVDAC-1-POPC systems were simulated at constant semi-isotropic pressure (1 bar). To monitor the stability of the membrane-protein setups, the average APL and the protein area were calculated with the grid approach implemented in the local membrane property analysis software g_lomepro (54). The average APLs for the mVDAC-1-POPE and mVDAC-1-POPC systems were insensitive to the Δq and gave stable averages of 0.60 and 0.65 nm2, respectively. Similarly, the cytosolic and IMS protein average areas remained insensitive to voltage. In POPE bilayers, the cytoplasmic and IMS protein areas were 19 and 15 nm2, and in POPC bilayers, these areas were 20 and 17 nm2 (data not shown).
SC computational and experimental current-voltage (I/V) relationships
Computational electrophysiology
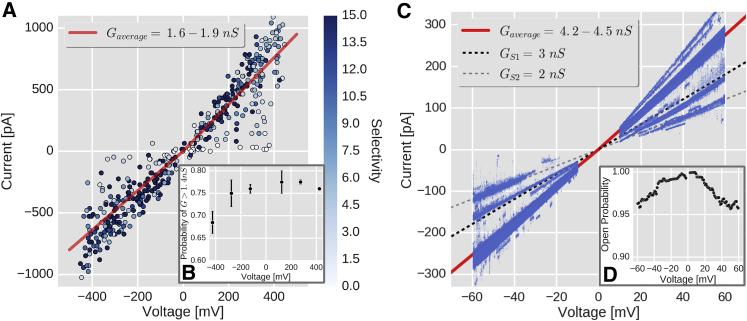
Using MD simulations, we had access to the atomic details of the permeation events with picosecond resolution. A 2 nS conductance of an “open” VDAC channel (at 0.5 M [Cl−]) should translate into an average total current of 13 elementary charges/ns·V. MD simulations of hundreds of nanoseconds at this current rate allowed us to collect a total of ∼75,000 permeation events. Although MD timescales are smaller compared with the ones in electrophysiology experiments, the number of permeation events allowed us to study the precise anionic and cationic components of the currents under the influence of a time-averaged voltage. Because the computational electrophysiology method keeps a constant ion difference between compartments, we investigated the influence of the arbitrary binning size, which mainly affects the fluctuations in the voltage estimates (64). By plotting the total currents through the channel and the voltage from the simulations binned in 10 ns time windows, we obtained the I/V curve of the mVDAC-1-POPE system shown in Fig. 2 A. The effect of the binning was explored by reducing the time windows to 1 ns of each simulation replica. The 1 ns or the 10 ns binning sizes made the I/V distributions broader and narrower, respectively. The mean conductances at negative and positive voltages were estimated by the slope of a linear regression fit. In this case, the binning choice did not significantly affect the calculated mean conductances (legend of Fig. S1).
Figure 2.
SC current-voltage (I/V) relationships of wt VDAC-1. (A) Computational I/V relationship of mVDAC-1 simulated in 0.5 M NaCl, and at ion imbalances (Δq) from 2 to 14 (only POPE is shown; for POPC results, see Fig. S1 C). Each I/V data point (circles) is colored according to selectivity, defined as the permeation ratio between chloride and sodium. The mean conductance values (± SE) of 1.63 (± 0.08) and 1.96 (± 0.11) nS are shown as red lines for the negative and positive voltages, respectively. (B) Voltage-dependent pseudo-open probability of a single mVDAC-1 channel. Each data point represents the average probability (± SD) of finding the channel at a conductance value >1.4 and 1.5 nS. The I/V data were binned every 150 mV from −500 to 500 mV and excluded the voltage range from −50 to 50 mV. (C) I/V relationship of hVDAC-1 inserted in a solvent-free DPhPC/cholesterol (9:1) membrane bathed in 1 M KCl, 1 mM CaCl2, and 5 mM HEPES, pH 7.4. The voltage-ramp data (blue dots) were taken after the insertion of a single hVDAC-1 channel was proven by a mean conductance value of 4–4.5 nS. The dashed lines mark the 3 and 2 nS conductance values of the most frequent “closed” subconducting states visited by hVDAC-1 ( and ). The red line shows the mean conductance value averaged over 185 voltage ramps, which was 4.2 and 4.5 nS (mean ± SE = 0.006) in the negative and positive voltage sides, respectively. (D) Voltage-dependent open probability of an SC hVDAC-1 for the voltage ramps shown in (C). To see this figure in color, go online.
I/V linear regressions gave consistent mean conductances of 1.6–1.9 nS, close to the experimentally determined “open” VDAC conductance of 1.85–2 nS extrapolated to 0.5 M NaCl (6, 65). The agreement between experiment and simulation for I/V confirms that the mVDAC-1 structure (PDB: 3EMN) used in the simulations corresponds to its “open” state (6), as similarly reported by Rui et al. (5) for hVDAC-1, although the NMR structures (PDB: 2K4T) have different N-α conformations (24).
The mean conductances were similar in both membranes (shown in Fig. 2 A for PE and in Fig. S1 C for PC). The mVDAC-1 channel showed transient subconducting time windows at larger voltages. The channels under negative voltage (arbitrarily set to Ch0) gave lower mean conductances in both lipids. The lower slopes at one voltage polarity is an indication of mVDAC-1 asymmetry and suggest that this mVDAC-1 orientation was more sensitive to voltage changes.
The outliers in the subconductance regions also showed lower selectivity for chloride. By binning the I/V data from Fig. 2 A and counting the proportion of values >1.4 and >1.5 nS, we estimate a pseudo-open probability, as shown in Fig. 2 B. Although not converged, probably due to sampling and timescales, this figure shows that mVDAC-1 (in a submicrosecond timescale) resembles the characteristic bell-shaped VDAC-1 profile as a function of the voltage.
Experimental electrophysiology
SC electrophysiology experiments on hVDAC-1 that complemented the MD simulations were also performed. hVDAC-1 and mVDAC-1 show 98% identity and share a similar electrophysiological behavior (14). SC experiments on mVDAC-1 have shown that the wt channel visited several well-defined “closed” subconducting states (45). However, mVDAC-1 variants, capable of cross-linking the N-α to the barrel interior, showed clear preference for only one “closed” state (45). For wt hVDAC-1, the SC experiments of Zachariae et al. (29) showed a broad distribution of conductances, where the conductance at 4 nS was predominantly observed. Because different VDAC-1 isoforms do not necessarily have to behave identically, it was interesting to know whether hVDAC-1 shows an electrophysiological behavior similar to that of mVDAC-1 when inserted in DPhPC (45).
Highly pure samples of wt hVDAC-1 were obtained from recombinant protein expressed in Escherichia coli (26). The structural integrity of the protein was assessed by reconstitution in both LDAO micelles and 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine liposomes and characterized by solution and solid-state NMR spectroscopy, respectively. The two-dimensional 1H-15N transverse relaxation-optimized spectrum of hVDAC-1 in LDAO showed a well-dispersed set of sharp NMR signals, demonstrating that most of the protein is rigidly folded in micelles or liposomes (Fig. S2). A large number of NMR resonances downfield of 8.5 ppm further supported the presence of β-strands, in agreement with the 3D structure of hVDAC-1 (25, 26, 66). In addition, supporting a natively folded state after reconstitution, 13C-13C correlation spectra of hVDAC-1 in liposomes showed a large number of well-defined NMR resonances originating from both the N-terminal α-helix and the barrel (Fig. S2 B).
Functionality of the channels was accessed by electrophysiology. To ensure that membrane preparations contained only one active hVDAC-1 channel, the evaluated current traces were only those that showed an “open” conductance state of 4–4.5 nS and a “closure” with a conductance of <3 nS. Voltage-clamp and voltage-ramp electrophysiology protocols were applied. Electrophysiological experiments with a single hVDAC-1 inserted into solvent-free DPhPC/cholesterol (9:1) membranes were performed to ensure full functionality of the protein and to monitor its conductance values in the “open” and the “closed” states. The insertion of a single hVDAC-1 into the freestanding membrane in the “open” state resulted in an average SC conductance of 4–4.5 nS in 1 M KCl (Fig. S3 A), similar to that described by Shanmugavadivu et al. (67) and Hiller et al. (24). If the incorporated hVDAC-1 gated from the “open” to a “closed” state as a result of a transmembrane potential exceeding mV, the conductance decreased in a stepwise fashion to values of ∼40–70% of the “open” conductance (Fig. S3 B). These values are in accordance with reported values from the literature for mVDAC-1 (45) and rat VDAC-1 (rVDAC-1) (22) in experiments (Fig. S3 C), and they demonstrate the full functionality of the protein in solvent-free DPhPC/cholesterol membranes. As SC activity was unambiguously proven, multiple experiments were performed. Fig. 2 C shows all current-voltage traces and corresponding mean values at each voltage polarity, averaged over 185 curves. The interval between −10 and +10 mV was excluded due to the low signal/noise ratio. The red line shows the calculated mean conductance for the negative (4.2 nS) and positive voltages (4.5 nS). The dashed lines mark conductance levels of 3 and 2 nS where the “closed” substates ( and ) were observed, as similarly described by Mertins et al. (45) for mVDAC-1. At larger voltage, the closing probability increases, resulting in a bell-shaped voltage dependency. The can be calculated from the voltage-ramp data with one inserted hVDAC-1, as described by Carl and Sanders (63) (see Materials and Methods). Fig. 2 D shows the result of the procedure, which indicates that the channel resides in “closed” states rather infrequently (a maximum of ∼5% of the time).
Conformational models of the voltage and the minimum-pore radius (minr)
To understand the voltage-dependent mVDAC-1 structural changes, we investigated whether the voltage (an external property, fe) was correlated with the ensemble of simulated mVDAC-1 structures. We also correlated the ensemble of structures with an mVDAC-1 geometrical parameter (a single valued internal property, fi), which indicated the degree of pore collapse. To quantify the degree of pore collapse, we calculated the pore radii (r) along the z-coordinate for each MD frame (rz) by using the protein cavity and tunnel analysis tool trj_cavity (55). The trj_cavity method performs a grid-based tunnel search to analyze the mean pore profiles of channels and other proteins. Pore profiles of a chloride channel (57) and aquaporins (68) have been successfully used to understand global or local conformational changes. Within the pore-radius profiles (Fig. S4), the most constricted region (zmin) was consistently located at the z-coordinate range of N-α. In this way, the minimum-pore radius (termed ) was defined and its value reflected the degree of constriction around the N-α.
The instantaneous voltage (fe) and the channel (fi) values were correlated with their corresponding protein coordinates (X), using PLS functional-mode analysis (57). The goal of the PLS algorithm is to obtain a relation (a conformational model, β), i.e., a linear combination of the protein coordinates that correlate best with f,
| (2) |
but at the same time allows us to identify the dynamics information in the input coordinates, which contribute the most to the fluctuation in f. The PLS components (included in β) are defined iteratively such that each component is a linear combination of the original coordinates with maximal covariance with f (i.e., minimizing the residuals, ε) while being uncorrelated to each previous component. This method has proven to be useful to detect complex protein rearrangements related to external and internal properties (57, 68). The quality of the models was assessed using the Pearson correlation coefficient (R) as a function of the number of PLS components. To avoid model overfitting, we cross-validated our models using half of the data and chose those models that maximized R in the cross-validation part. Validation details of the conformational models that describe the V and are shown in Fig. S5. The details include the convergence of the correlations (Fig. S5, A and D) and the prediction versus actual values (Fig. S5, B and E). Fig. S5, C and F, show a representation of the root mean-square fluctuation of the amino acids that were displaced the most along the conformational models.
Voltage and polarity
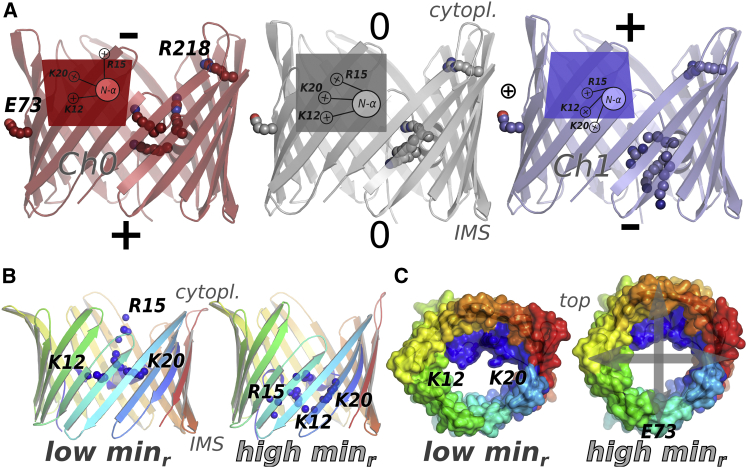
By using the calculated voltages and the protein coordinates, we were able to construct a linear conformational model with reasonable predictive power (Fig. S5, A and B). The model training was performed using the mVDAC-1–POPE simulations and the cross-validation, with the mVDAC-1–POPC simulations. The model described the main protein conformations associated with the instantaneous calculated voltage and its polarity. It also indicates that the voltage-dependent conformations of mVDAC-1 were similar, either in POPE or POPC membranes. Fig. 3 A shows the protein conformations predicted by the model, at inverse polarities of ±400 mV and at ∼0 mV. This model predicts that the mVDAC-1 structure slightly expands the cytoplasmic side of the barrel at negative polarity (Ch0). An expansion in the IMS side of the barrel occurs at positive polarity (Ch1).
Figure 3.
mVDAC-1 global conformational models of the voltage (V) and the channel minimum-pore radius . All models were generated using the protein atoms excluding the hydrogens. Model validation details are shown in Fig. S5. (A) Cartoon and schematic representations of mVDAC-1 conformational model changes in response to V. The predicted structures under negative voltage (Ch0) are in red, those under neutral voltage in gray, and those under positive voltage in blue (Ch1). Labels indicate the cytoplasmic (cytopl.) and the IMS sides. In the positive polarity, sodium ions (circled plus signs) were observed closer to E73. The cartoon shows the β-barrel mVDAC-1 structure. K12, R15, and K20 (together termed KRK), E73, and R218 are shown as spheres using the same color code. The schematic insets show the deformations of the barrel, the position of the N-terminal α-helix, and the relative conformation of the KRK, E73, and R218 side chains. (B and C) Extreme structures of the conformational model that correlate with the changes of the channel under negative voltage (Ch0) in the mVDAC-1 MD simulations. The structures are shown as cartoons (β-sheets in a blue-green-red scale) (B) and surface representations (C) of the conformations that show the lowest and highest values (from 4 to 11 Å) in the model. Residues K12, R15, and K20 are shown as blue spheres. These positively charged amino acids show larger contributions to the narrowing of the pore in this conformational model. The conformational changes depicted describe motions that distorted the barrel either parallel or perpendicular to the position of N-α. The distortion axes of the barrel are illustrated as gray arrows in (C). To see this figure in color, go online.
Most of the barrel changes observed in the model (in terms of atomic fluctuations) occurred in the cytoplasmic loops (amino acids 104–107 and 266–268) and in β-sheets 1, 3, and 7, close to E73. This conformational model described a significant displacement of the positively charged amino acids K12, R15, and K20 (the KRK motif). It also predicts the contributions of K33 and R62 in the IMS part of the barrel and of K107 and K111 in the cytoplasmic part (Fig. S5 C). The model showed the favorable reorientation of charged amino acid side chains with respect to the voltage. Not all charged amino acids displayed increased mobility, R218 (amino acid that belongs to the “charged brush” (7)), remained in a similar orientation at all voltages. As the conformational model showed, R15 had access to the cytoplasmic side of the pore lumen at negative voltages, whereas K20 was directed to the IMS at positive ones. A small change in the relative position of N-α with respect to the barrel was observed in this model: Amino acids 12–20 of N-α were displaced 3–4 Å along the pore axis.
In the simulations, the average local electric field acting on the KRK and R218 side chains (Table S1) showed large and opposite values for these amino acids at ±0.4 V (especially for R15 and K20), compared with the ones at 0 V. Locally, however, Lennard-Jones and bonded interactions compensated these forces.
In the study of Teijido et al. (22), the distribution of negatively charged amino acids and their salt-bridging probability explained the pH sensitivity of mVDAC-1. A salt-bridge analysis of the KRK, R218, R120, K109, K53, and K113 amino acids (Table S2) showed that when the voltage was increased, the salt-bridge probability and the negative amino acid partners remained constant or changed considerably. For instance, K12 and R218 conserved their salt-bridging probability and partners. R15 had the lowest salt-bridging probability and conserved D9 and D16 partners. At negative voltages, R15 reduced the salt-bridging probability and also added E189 as a bridging partner. K20 showed moderate salt-bridging probabilities at 0 V with D16 and E280 as partners. At negative voltages, K20 reduced its bridging probability and lost E280 as a partner. At positive voltages, the bridging probabilities of K20 are reduced compared with the probabilities at ∼0 V, and it incorporates E203 as a salt-bridging partner. The mean position of the side-chain nitrogens of R15 and K20 (guanidinium and amino groups) was significantly shifted in the simulations at higher voltages, and confirms the observed displacement of these amino acids in the voltage-dependent conformational model (Fig. 3 A). Interestingly, a sodium ion was frequently observed close to E73 at positive voltages during the simulations.
Pore radius profiles and minr
Pore-radius profiles along the z-coordinate were calculated with trj_cavity (55). From each of the profiles, we extracted the minimum-pore radius and used these values as a proxy of the degree of mVDAC-1 pore collapse (details above and in Fig. S4). Using mVDAC-1 atoms (excluding the hydrogens) and the of mVDAC-1 computed at each frame, we created conformational models for both channel polarities, Ch0 and Ch1, in PE and PC lipids. The model predictions showed higher correlations for Ch0 (negative voltages) than for Ch1 (positive voltage). This result indicated that the protein changes that lead to pore constriction were more similar at negative voltages than at positive ones. Then, by using the values and the protein coordinates from 42,000 MD frames, we calculated a global conformational model that explained the local pore changes at negative voltages. The -Ch0 models converged to for the training set and to 0.6 for the cross-validation part. Cross-validation details are shown in Fig. S5, D and E. The motions described by the model (Fig. 3 B) indicated that the pore constriction-expansion dynamics was dominated by the displacement of charged mVDAC-1 amino acids and a moderate barrel deformation. The global conformational model predicts the changes in the values from 11 to 4 Å (Fig. 3 C). The larger fluctuations that contributed to pore narrowing or expansion included K12 and R15 from the KRK motif (Fig. S5 F). Transient displacement of the KRK side chains and barrel distortion were able to reduce the pore lumen significantly.
Interestingly, in the simulations, an overlay of pore profiles showed that the variability of increases at higher voltages (Fig. S4).
mVDAC-1 barrel deformation
The previous conformational models included all nonhydrogen atoms of the protein, they predicted structural changes that slightly distorted the barrel geometry and showed significant amino acid side-chain orientations. However, to separate the pure barrel contribution from the voltage response, we calculated the time-averaged structural barrel deformations at different simulated voltages. We fitted ellipses to the C-α atoms solely from the barrel (See Materials and Methods).
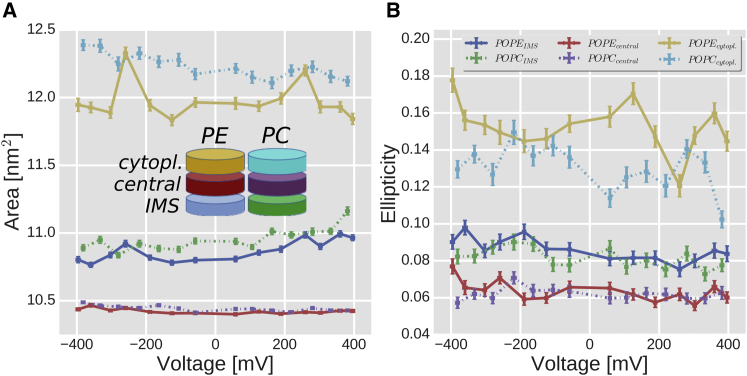
The fitted ellipse axes (a, major axis; b, minor axis) were oriented either perpendicular or parallel to the N-α axis (data not shown). These orientations indicate that the barrel showed two preferential deformation axes, which is consistent with other simulations performed for several VDAC channels (35). We calculated the average areas (±SE) of these barrel ellipses and their degree of ellipticity . Higher ellipticities mean more distorted barrels and values closer to zero more cylindrical ones. The mVDAC-1 barrel was divided in three slices to monitor its IMS, middle, and cytoplasmic parts.
The areas of the fitted ellipses for each slice (Fig. 4 A) and for PE and PC lipids showed larger values for the cytoplasmic part than for the IMS, and the IMS exhibited larger values than the central part. The barrel central area remained constant at all voltages. When moving from negative to positive voltage in PE and PC membranes, the barrel area of the cytosolic part decreased, whereas the area of the IMS side increased. This trend was more pronounced in PC membranes.
Figure 4.
mVDAC-1 β-barrel deformations. (A) Changes of the barrel area (mean ± SE) as a function of the voltage and lipid composition. Each colored curve represents the area change for one of three barrel slices, depicted by the cartoon cylinder inset. (B) Changes of ellipticity (, where are the ellipse axes) ± SE as a function of the voltage for the same barrel slices as in (A). In both (A) and (B), the dashed lines indicate the plots for PC lipids. To see this figure in color, go online.
In terms of barrel ellipticities (Fig. 4 B), the values followed a similar trend compared to the areas. Although the changes were small, they were significant beyond the error estimates. mVDAC-1 in PE membranes and at negative V showed higher ellipticity values. The distortions were either aligned or perpendicular to the barrel area where E73 is located, in line with the changes observed in the global voltage-dependent conformational model (Fig. 3 A). These results show that the mVDAC-1 barrel inserted in membranes gets distorted in an asymmetric way in response to V. The central part is more stable and cylindrical, whereas the cytoplasmic part shows more structural flexibility. The longest loops in the mVDAC-1 structures are located in the cytoplasmic side.
Discussion
One of the challenges of understanding the gating mechanisms of ion channels by MD simulations is connected to the timescales involved in these processes (milliseconds to seconds), which are larger than the timescales accessible in state-of-the-art MD simulations (nanoseconds to microseconds). This problem is particularly challenging in the VDAC case, where slow conformational changes are expected. However, combined insights obtained from experiments and simulations have helped to unravel some of its mechanisms (5, 8, 29). In our computational approach, we extended the voltage range at which VDAC-1 channels have been experimentally measured to access slow possible voltage-dependent conformational changes.
As previously reported by Choudhary et al. (6), no significant displacement of N-α was observed in our mVDAC-1 MD simulations. However, the first 10 amino acids of N-α in the hVDAC-1 MD simulations of Rui et al. (5) showed higher flexibility, which might be due to the different N-α starting conformations of both structures (24, 25).
In terms of the mVDAC-bilayer stability, the simulations showed that the voltage-range used had no significant influence on the average APL and the protein areas. The average APL values were close to the bulk experimental and simulated values for both phospholipids (52). The protein areas on the cytoplasmatic and IMS sides were 19 and 15 nm2 in POPE and 20 and 17 nm2 in POPC. Larger cytoplasmatic protein areas are a clear indication of the mVDAC-1 asymmetry in membranes. The slightly larger protein areas in POPC membranes probably reflect a higher flexibility of mVDAC-1 in the less dense POPC bilayer compared with POPE.
We found an excellent agreement of the MD conductances with the experimental and simulated ones (scaled by one-half to account for the difference in chloride concentration used: 0.5 M in the simulations and 1 M in the experiments (6)). In the simulations at higher potentials, we found also some subconducting time windows. The transiently less conductive channel in these time windows, interestingly, also showed lower selectivity for chloride, similar to what has been observed experimentally (17). Whether these less conductive states may lead to more permanent “closed” conformations remains unknown at this point.
The SC MD simulations presented in this work showed that the submicrosecond I/V relationships were invariant to the binning size and demonstrated a degree of asymmetry. The different time windows had a negligible effect on the mean conductances (Fig. S1). This invariance showed us that the voltage estimates had no or little bias, and it allowed us to extract detailed permeation information with better statistics. The I/V curves showed lower mean conductances at negative voltages. Also, we observed more structural changes occurring at the cytoplasmic side of mVDAC-1. The voltage affected the cytoplasmic side of mVDAC-1, especially the Ch0 channel, in agreement with the orientation evidence regarding VDAC in membranes (20, 21, 22, 23). Because the VDAC orientation with the cytosolic loops pointing to the negative potential also shows more sensitivity for the potential, as shown by experiments (20, 21, 22) and simulations (22), our observations support the hypothesis that the cytosolic side of the channel is the one exposed (at least preferentially) to the cis side in BLM experiments.
Rostovtseva et al. (19) investigated the influence of the lipid composition on the open probability of the fungal ncVDAC. They found that in PC membranes, the current response to triangular voltage waves was symmetric, resulting in a symmetric open-probability curve. However, in PE membranes, the current is significantly lower at applied negative voltages than at positive ones, suggesting that “closed” states were reached more frequently. To analyze the impact of the lipid composition and the probability of accessing a “closed” state, we performed MD simulations of mVDAC-1 in PC and in PE membranes. In this work, the lipid composition showed no considerable effect on the I/V relationships, although it moderately influenced the magnitude of the protein areas (as discussed above) and barrel distortions (Fig. 4). The lipid-dependent barrel changes probably reflect the lipid packing influence on the overall protein structure. Less compact lipids such as POPC (in terms of APL) presumably allow higher barrel mVDAC-1 areas and lower ellipticity distortions compared with the more compact POPE bilayer. In addition, the conformational model for mVDAC-1 voltage dependence was trained with the mVDAC-1 structures in PE lipids and cross-validated with those in PC . These observations indicate that the conformational space sampled by mVDAC-1 in our MD simulations was similar, irrespective of the phospholipid headgroup. Nevertheless, we cannot exclude the possibility that mammalian VDAC-1 proteins behave differently than ncVDAC (19) or that the phospholipid effects take longer than the simulation timescales explored in this work. Another aspect to consider is the number of VDAC molecules in the membrane. In this study, SC VDAC-1 simulations and experiments were analyzed, whereas Rostovtseva et al. (19) investigated membranes with multiple channels, i.e., membranes containing 10–125 channels. It is known that VDAC channels can oligomerize. Some lipids, such as phosphatidylglycerol, promote VDAC oligomerization, whereas cardiolipin reduces oligomer formation (69). Moreover, VDAC oligomerization increases during apoptosis (40, 70). An interaction among VDAC proteins might influence the gating behavior, and hence, the SC behavior (45, 47) might be different compared with behavior with multiple channels. Increased voltage dependence for multimeric channels also has been observed for the potassium channel KcsA (71).
The SC experiments ( and ) of hVDAC-1 clearly confirm the known conductance of 4–4.5 nS in the open state (at 1 M KCl) (67). These experiments also showed closing of the channel at higher voltages, with apparently two subconducting values of ∼40–70% of the “open” state. These conductance states are also in agreement with the ones observed for mVDAC-1 in DPhPC (45). The voltage dependence of VDAC-1 has been described as a bell-shaped curve with lower conductance values at larger potential differences (Fig. 2 D). It has been further shown that the opening and closing rates for hVDAC-1 are very different. Whereas the opening rate is in the millisecond timescale, the closing rate is 2 to 5 orders of magnitude slower (72). In our experiments, we have chosen voltage ramps with a frequency of 100 mHz, i.e., voltage changes with 24 mV s−1, which are fast compared to the closing-rate constants for hVDAC-1. Hence, it is expected that the channel resided most of the time in the “open” state, in accordance with our mVDAC-1 MD simulations. In SC experiments and MD simulations, VDAC-1 accessed infrequently subconducting states, i.e., VDAC-1 “closed” states. By applying voltage for longer time periods ( for 10 s; see Fig. S3 C), we accessed “closed” states and the conductance values obtained from the curves make us confident that the lower conductance values measured in the experiments are indeed a result of the channel being in a “closed” state.
The voltage-dependent conformational dynamics of mVDAC-1 by MD simulations showed that the voltage distorts the barrel asymmetrically by reorganization of its charged amino acids distributed throughout the channel. It is interesting that changes showed a clear involvement of the charged amino acids (K12, R15, and K20), which are part of the N-terminal α-helix. These changes also involved β-sheet zones close to E73. The barrel distortion around E73 is related to membrane thinning observed in this area (42). At positive voltages, sodium ions were frequently observed near E73 on the membrane side outside of the pore. Because of the calcium (37) and hexokinase (39) binding ability of E73, it might well be that these factors decrease the barrel perturbation, as shown by Ge et al. (38). We speculate that reducing the bilayer thickness around this area might help to stabilize an “open” VDAC state. In simulations, the elliptical distortions were smaller at positive voltages (Fig. 4 B), polarity at which sodium ions were observed close to E73.
The cross-validation of the conformational models also showed that the channel perturbations were similar in PE and PC membranes. The KRK amino acids showed a large contribution to the predicted conformational changes, which explained the changes in the minimum-pore-radius of mVDAC-1 at negative polarities. By comparing the pore profiles at zero and ∼–0.4 V, we observed that the N-α region shows more variability, which is increased at higher voltages. The variability of suggest that the pore collapse in negative voltages might also be voltage-dependent (Fig. S4).
The N-α amino acids D16 and K20 are involved in VDAC gating and selectivity (14, 34) and K20 also forms part of the ATP binding site (6, 32). The displacement of these amino acids was also found by Krammer et al. (7) in mVDAC-1 simulations, where sweeping of these side-chains was reported to help the permeation of anions across the channel lumen. It is interesting to observe that KRK side chains were exposed to the IMS or the cytoplasmic side of the pore. Based on the salt-bridge analysis (Table S2) and the pH sensitivity of mVDAC-1 (22), we think that a reorganization of the salt-bridging patterns within the VDAC structure help to stabilize the conformations at opposite voltages. This charge reorganization seems to be voltage regulated and it might be related to gating, the stabilization of “closed” states, or, alternatively, a twisting type of motion suggested for N-α (14, 45).
The nanoscopic picture from our work shows that mVDAC-1 is—at least—able to distort asymmetrically upon opposite voltage polarities by perturbing the cytoplasmic and IMS barrel parts around the E73-N-α axis and significantly reorienting the KRK amino acids of the N-α. We think these motions sample part of the conformational space of mVDAC-1 in the presence of voltage, which, in an extreme version, or at longer timescales, might lead to the necessary changes to access “closed” subconducting state(s). The structural nature of a permanent VDAC “closed” state is still elusive.
Conclusions
In this work, by simulating the SC mVDAC-1 responses at several potential differences (V) inserted in PE and PC membranes, we connected SC VDAC experiments with VDAC structural dynamic information to get a better understanding of the channel voltage dependence. We found that simulations are mostly compatible with the “open” channel currents and a quasi-ohmic conductance holds for an extended V range. The simulated conductances of mVDAC-1 were insensitive to the phospholipid composition, although mVDAC-1 barrel geometrical distortions were different. In a set of complementary and extensive SC experiments on the highly homologous hVDAC-1, defined subconducting states were observed, as previously reported for mVDAC-1 (45). In the simulations, the time windows of reduced conductance (which we called transient subconducting states) were more often cation selective. We found significant changes of the barrel conformation and displacement of N-α amino acids, including K20. The protein conformations at the investigated voltages allowed us to construct highly correlated conformational models that show how mVDAC-1 accesses asymmetric, semicollapsed, and subconducting states in response to opposite voltage polarities. The submicrosecond voltage-dependent conformational dynamics of VDAC-1 support the hypotheses that barrel distortion, displacement of N-α amino acids, and interaction of E73 with the membrane and ions are involved in VDAC regulation and closing mechanisms.
Author Contributions
B.L.d.G., R.B., and C.S. developed the concept of the study. L.P. prepared the simulations and initial MD analysis. R.B. performed MD analysis, scripting, and figure preparation. R.B., C.S., and B.L.d.G., wrote the article. C.W. performed the SC experiments. I.M. performed the SC experiment analysis. M.Z. and C.G. designed solution NMR experiments. S.V. performed the solution NMR experiments. K.G. and S.B. prepared the protein samples. A.L. performed the solid-state NMR experiments.
Acknowledgments
We thank Dr. Manuel Etzkorn for solid-state NMR measurements and Claudio Briones for help with Blender.
This work was partially supported by the National Science Foundation (L.P.) and the Deutsche Forschungsgesellschaft (DFG) Sonderforschungsbereich (SFB) 803 (project A03 to R.B. and B.L.d.G., project A01 to C.S., and projects A11 and A4 to A.L, M.Z., and C.G.), and by a European Research Council consolidator grant to M.Z. (DYNAMOM, grant agreement no. 282008).
Editor: Jose Faraldo-Gomez.
Footnotes
Five figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30665-8.
Contributor Information
Rodolfo Briones, Email: rbrione@gwdg.de.
Claudia Steinem, Email: csteine@gwdg.de.
Bert L. de Groot, Email: bgroot@gwdg.de.
Supporting Material
References
- 1.Hille B. 3rd ed. Sinauer; Sunderland, MA: 2001. Ion Channels of Excitable Membranes. [Google Scholar]
- 2.Shrivastava I.H., Tieleman D.P., Sansom M.S. K+ versus Na+ ions in a K channel selectivity filter: a simulation study. Biophys. J. 2002;83:633–645. doi: 10.1016/s0006-3495(02)75197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köpfer D.A., Song C., de Groot B.L. Ion permeation in K+ channels occurs by direct Coulomb knock-on. Science. 2014;346:352–355. doi: 10.1126/science.1254840. [DOI] [PubMed] [Google Scholar]
- 4.Machtens J.-P., Kortzak D., Fahlke C. Mechanisms of anion conduction by coupled glutamate transporters. Cell. 2015;160:542–553. doi: 10.1016/j.cell.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Rui H., Lee K.I., Im W. Molecular dynamics studies of ion permeation in VDAC. Biophys. J. 2011;100:602–610. doi: 10.1016/j.bpj.2010.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhary O.P., Paz A., Grabe M. Structure-guided simulations illuminate the mechanism of ATP transport through VDAC1. Nat. Struct. Mol. Biol. 2014;21:626–632. doi: 10.1038/nsmb.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer E.-M., Vu G.T., Prévost M. Dual mechanism of ion permeation through VDAC revealed with inorganic phosphate ions and phosphate metabolites. PLoS One. 2015;10:e0121746. doi: 10.1371/journal.pone.0121746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noskov S.Y., Rostovtseva T.K., Bezrukov S.M. Current state of theoretical and experimental studies of the voltage-dependent anion channel (VDAC) Biochim. Biophys. Acta. 2016;1858:1778–1790. doi: 10.1016/j.bbamem.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaid H., Abu-Hamad S., Shoshan-Barmatz V. The voltage-dependent anion channel-1 modulates apoptotic cell death. Cell Death Differ. 2005;12:751–760. doi: 10.1038/sj.cdd.4401599. [DOI] [PubMed] [Google Scholar]
- 10.Shoshan-Barmatz V., Keinan N., Zaid H. Uncovering the role of VDAC in the regulation of cell life and death. J. Bioenerg. Biomembr. 2008;40:183–191. doi: 10.1007/s10863-008-9147-9. [DOI] [PubMed] [Google Scholar]
- 11.Shoshan-Barmatz V., Mizrachi D. VDAC1: from structure to cancer therapy. Front. Oncol. 2012;2:164. doi: 10.3389/fonc.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koren I., Raviv Z., Shoshan-Barmatz V. Downregulation of voltage-dependent anion channel-1 expression by RNA interference prevents cancer cell growth in vivo. Cancer Biol. Ther. 2010;9:1046–1052. doi: 10.4161/cbt.9.12.11879. [DOI] [PubMed] [Google Scholar]
- 13.Colombini M., Blachly-Dyson E., Forte M. VDAC, a channel in the outer mitochondrial membrane. Ion Channels. 1996;4:169–202. doi: 10.1007/978-1-4899-1775-1_5. [DOI] [PubMed] [Google Scholar]
- 14.Shuvo S.R., Ferens F.G., Court D.A. The N-terminus of VDAC: structure mutational analysis, and a potential role in regulating barrel shape. Biochim. Biophys. Acta. 2016;1858:1350–1361. doi: 10.1016/j.bbamem.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Colombini M. Voltage gating in the mitochondrial channel, VDAC. J. Membr. Biol. 1989;111:103–111. doi: 10.1007/BF01871775. [DOI] [PubMed] [Google Scholar]
- 16.Hodge T., Colombini M. Regulation of metabolite flux through voltage-gating of VDAC channels. J. Membr. Biol. 1997;157:271–279. doi: 10.1007/s002329900235. [DOI] [PubMed] [Google Scholar]
- 17.Colombini M. Structure and mode of action of a voltage dependent anion-selective channel (VDAC) located in the outer mitochondrial membrane. Ann. N. Y. Acad. Sci. 1980;341:552–563. doi: 10.1111/j.1749-6632.1980.tb47198.x. [DOI] [PubMed] [Google Scholar]
- 18.Schein S.J., Colombini M., Finkelstein A. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J. Membr. Biol. 1976;30:99–120. doi: 10.1007/BF01869662. [DOI] [PubMed] [Google Scholar]
- 19.Rostovtseva T.K., Kazemi N., Bezrukov S.M. Voltage gating of VDAC is regulated by nonlamellar lipids of mitochondrial membranes. J. Biol. Chem. 2006;281:37496–37506. doi: 10.1074/jbc.M602548200. [DOI] [PubMed] [Google Scholar]
- 20.Sheldon K.L., Maldonado E.N., Bezrukov S.M. Phosphorylation of voltage-dependent anion channel by serine/threonine kinases governs its interaction with tubulin. PLoS One. 2011;6:e25539. doi: 10.1371/journal.pone.0025539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomasello M.F., Guarino F., De Pinto V. The voltage-dependent anion selective channel 1 (VDAC1) topography in the mitochondrial outer membrane as detected in intact cell. PLoS One. 2013;8:e81522. doi: 10.1371/journal.pone.0081522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teijido O., Rappaport S.M., Bezrukov S.M. Acidification asymmetrically affects voltage-dependent anion channel implicating the involvement of salt bridges. J. Biol. Chem. 2014;289:23670–23682. doi: 10.1074/jbc.M114.576314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques E.J., Carneiro C.M., Krasilnikov O.V. Does VDAC insert into membranes in random orientation? Biochim. Biophys. Acta. 2004;1661:68–77. doi: 10.1016/j.bbamem.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 24.Hiller S., Garces R.G., Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ujwal R., Cascio D., Abramson J. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayrhuber M., Meins T., Zeth K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schredelseker J., Paz A., Abramson J. High resolution structure and double electron-electron resonance of the zebrafish voltage-dependent anion channel 2 reveal an oligomeric population. J. Biol. Chem. 2014;289:12566–12577. doi: 10.1074/jbc.M113.497438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K.I., Rui H., Im W. Brownian dynamics simulations of ion transport through the VDAC. Biophys. J. 2011;100:611–619. doi: 10.1016/j.bpj.2010.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zachariae U., Schneider R., Lange A. β-Barrel mobility underlies closure of the voltage-dependent anion channel. Structure. 2012;20:1540–1549. doi: 10.1016/j.str.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krammer E.-M., Hombl F., Prvost M. Molecular origin of VDAC selectivity towards inorganic ions: a combined molecular and Brownian dynamics study. Biochim. Biophys. Acta. 2013;1828:1284–1292. doi: 10.1016/j.bbamem.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Szabó I., De Pinto V., Zoratti M. The mitochondrial permeability transition pore may comprise VDAC molecules. II. The electrophysiological properties of VDAC are compatible with those of the mitochondrial megachannel. FEBS Lett. 1993;330:206–210. doi: 10.1016/0014-5793(93)80274-x. [DOI] [PubMed] [Google Scholar]
- 32.Villinger S., Giller K., Zweckstetter M. Nucleotide interactions of the human voltage-dependent anion channel. J. Biol. Chem. 2014;289:13397–13406. doi: 10.1074/jbc.M113.524173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yehezkel G., Hadad N., Shoshan-Barmatz V. Nucleotide-binding sites in the voltage-dependent anion channel: characterization and localization. J. Biol. Chem. 2006;281:5938–5946. doi: 10.1074/jbc.M510104200. [DOI] [PubMed] [Google Scholar]
- 34.Thomas L., Blachly-Dyson E., Forte M. Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc. Natl. Acad. Sci. USA. 1993;90:5446–5449. doi: 10.1073/pnas.90.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amodeo G.F., Scorciapino M.A., Ceccarelli M. Charged residues distribution modulates selectivity of the open state of human isoforms of the voltage dependent anion-selective channel. PLoS One. 2014;9:e103879. doi: 10.1371/journal.pone.0103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Israelson A., Zaid H., Shoshan-Barmatz V. Mapping the ruthenium red-binding site of the voltage-dependent anion channel-1. Cell Calcium. 2008;43:196–204. doi: 10.1016/j.ceca.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Gincel D., Zaid H., Shoshan-Barmatz V. Calcium binding and translocation by the voltage-dependent anion channel: a possible regulatory mechanism in mitochondrial function. Biochem. J. 2001;358:147–155. doi: 10.1042/0264-6021:3580147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge L., Villinger S., Zweckstetter M. Molecular plasticity of the human voltage-dependent anion channel embedded into a membrane. Structure. 2016;24:585–594. doi: 10.1016/j.str.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastorino J.G., Hoek J.B. Regulation of hexokinase binding to VDAC. J. Bioenerg. Biomembr. 2008;40:171–182. doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keinan N., Pahima H., Shoshan-Barmatz V. The role of calcium in VDAC1 oligomerization and mitochondria-mediated apoptosis. Biochim. Biophys. Acta. 2013;1833:1745–1754. doi: 10.1016/j.bbamcr.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 41.De Pinto V., al Jamal J.A., Palmieri F. Location of the dicyclohexylcarbodiimide-reactive glutamate residue in the bovine heart mitochondrial porin. J. Biol. Chem. 1993;268:12977–12982. [PubMed] [Google Scholar]
- 42.Villinger S., Briones R., Zweckstetter M. Functional dynamics in the voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA. 2010;107:22546–22551. doi: 10.1073/pnas.1012310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colombini M. The published 3D structure of the VDAC channel: native or not? Trends Biochem. Sci. 2009;34:382–389. doi: 10.1016/j.tibs.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Song J., Midson C., Colombini M. The topology of VDAC as probed by biotin modification. J. Biol. Chem. 1998;273:24406–24413. doi: 10.1074/jbc.273.38.24406. [DOI] [PubMed] [Google Scholar]
- 45.Mertins B., Psakis G., Essen L.-O. Flexibility of the N-terminal mVDAC1 segment controls the channel’s gating behavior. PLoS One. 2012;7:e47938. doi: 10.1371/journal.pone.0047938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thinnes F.P. After all, plasmalemmal expression of type-1 VDAC can be understood. Phosphorylation, nitrosylation, and channel modulators work together in vertebrate cell volume regulation and either apoptotic pathway. Front. Physiol. 2015;6:126. doi: 10.3389/fphys.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Báthori G., Szabó I., Zoratti M. Novel aspects of the electrophysiology of mitochondrial porin. Biochem. Biophys. Res. Commun. 1998;243:258–263. doi: 10.1006/bbrc.1997.7926. [DOI] [PubMed] [Google Scholar]
- 48.De Pinto V., Reina S., Messina A. Structure of the voltage dependent anion channel: state of the art. J. Bioenerg. Biomembr. 2008;40:139–147. doi: 10.1007/s10863-008-9140-3. [DOI] [PubMed] [Google Scholar]
- 49.Teijido O., Ujwal R., Abramson J. Affixing N-terminal α-helix to the wall of the voltage-dependent anion channel does not prevent its voltage gating. J. Biol. Chem. 2012;287:11437–11445. doi: 10.1074/jbc.M111.314229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bjelkmar P., Larsson P., Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 52.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kutzner C., Grubmüller H., Zachariae U. Computational electrophysiology: the molecular dynamics of ion channel permeation and selectivity in atomistic detail. Biophys. J. 2011;101:809–817. doi: 10.1016/j.bpj.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gapsys V., de Groot B.L., Briones R. Computational analysis of local membrane properties. J. Comput. Aided Mol. Des. 2013;27:845–858. doi: 10.1007/s10822-013-9684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paramo T., East A., Bond P.J. Efficient characterization of protein cavities within molecular simulation trajectories: trj_cavity. J. Chem. Theory Comput. 2014;10:2151–2164. doi: 10.1021/ct401098b. [DOI] [PubMed] [Google Scholar]
- 56.Fitzgibbon A., Pilu M., Fisher R.B. Direct least square fitting of ellipses. IEEE Trans. Pattern Anal. Mach. Intell. 1999;21:476–480. [Google Scholar]
- 57.Krivobokova T., Briones R., de Groot B.L. Partial least-squares functional mode analysis: application to the membrane proteins AQP1, Aqy1, and CLC-ec1. Biophys. J. 2012;103:786–796. doi: 10.1016/j.bpj.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez F., Granger B.E. IPython: a system for interactive scientific computing. Comput. Sci. Eng. 2007;9:21–29. http://ipython.org [Google Scholar]
- 59.Schrödinger, LLC, 2015. The PyMOL Molecular Graphics System, Version 1.8.
- 60.Community, B. O. 2015. Blender - a 3D modelling and rendering package. Blender Foundation, Blender Institute, Amsterdam, the Netherlands. http://www.blender.org.
- 61.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 62.Engelhardt H., Meins T., Zeth K. High-level expression, refolding and probing the natural fold of the human voltage-dependent anion channel isoforms I and II. J. Membr. Biol. 2007;216:93–105. doi: 10.1007/s00232-007-9038-8. [DOI] [PubMed] [Google Scholar]
- 63.Carl A., Sanders K.M. Measurement of single channel open probability with voltage ramps. J. Neurosci. Methods. 1990;33:157–163. doi: 10.1016/0165-0270(90)90019-c. [DOI] [PubMed] [Google Scholar]
- 64.Roux B. Computational electrophysiology: the molecular dynamics of ion channel permeation and selectivity in atomistic detail. Biophys. J. 2011;101:755–756. doi: 10.1016/j.bpj.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gincel D., Vardi N., Shoshan-Barmatz V. Retinal voltage-dependent anion channel: characterization and cellular localization. Invest. Ophthalmol. Vis. Sci. 2002;43:2097–2104. [PubMed] [Google Scholar]
- 66.Hiller S., Wagner G. The role of solution NMR in the structure determinations of VDAC-1 and other membrane proteins. Curr. Opin. Struct. Biol. 2009;19:396–401. doi: 10.1016/j.sbi.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shanmugavadivu B., Apell H.-J., Kleinschmidt J.H. Correct folding of the β-barrel of the human membrane protein VDAC requires a lipid bilayer. J. Mol. Biol. 2007;368:66–78. doi: 10.1016/j.jmb.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 68.Kaptan S., Assentoft M., de Groot B.L. H95 is a pH-dependent gate in aquaporin 4. Structure. 2015;23:2309–2318. doi: 10.1016/j.str.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 69.Betaneli V., Petrov E.P., Schwille P. The role of lipids in VDAC oligomerization. Biophys. J. 2012;102:523–531. doi: 10.1016/j.bpj.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zalk R., Israelson A., Shoshan-Barmatz V. Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem. J. 2005;386:73–83. doi: 10.1042/BJ20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molina M.L., Barrera F.N., González-Ros J.M. Clustering and coupled gating modulate the activity in KcsA, a potassium channel model. J. Biol. Chem. 2006;281:18837–18848. doi: 10.1074/jbc.M600342200. [DOI] [PubMed] [Google Scholar]
- 72.Colombini M. VDAC structure, selectivity, and dynamics. Biochim. Biophys. Acta. 2012;1818:1457–1465. doi: 10.1016/j.bbamem.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.