Abstract
An all-atom molecular dynamics simulation of the archetype barrel-stave alamethicin (alm) pore in a 1,2-dioleoyl-sn-glycero-3-phosphocholine bilayer at 313 K indicates that ∼7 μs is required for equilibration of a preformed 6-peptide pore; the pore remains stable for the duration of the remaining 7 μs of the trajectory, and the structure factors agree well with experiment. A 5 μs simulation of 10 surface-bound alm peptides shows significant peptide unfolding and some unbinding, but no insertion. Simulations at 363 and 413 K with a −0.2 V electric field yield peptide insertion in 1 μs. Insertion is initiated by the folding of residues 3–11 into an α-helix, and mediated by membrane water or by previously inserted peptides. The stability of five alm pore peptides at 413 K with a −0.2 V electric field demonstrates a significant preference for a transmembrane orientation. Hence, and in contrast to the cationic antimicrobial peptide described in the following article, alm shows a strong preference for the inserted over the surface-bound state.
Introduction
Numerous peptides have antibacterial activity and are a focus of therapeutic research targeting bacteria resistant to traditional antibiotics (1, 2). These include a group of membrane-disrupting peptides that are <50 residues in length, bind bacterial membranes with α-helical secondary structures, and have antibacterial activities that correlate with their abilities to disrupt model membranes. While these peptides have a broad range of sequences and disruption mechanisms (see reviews by Nguyen et al. (3), Wimley and Hristova (4), and Wimley (5)), the disruptive pathways have similar steps (i.e., binding the membrane, folding, inserting to a transmembrane orientation, peptide aggregation). Understanding the stability of each step for specific peptides will help relate the primary sequence to its mode of membrane disruption. Recent advances in computational force fields and hardware have made multimicrosecond all-atom simulations possible, which can help identify key properties that stabilize a specific state. While the rates between states in the disruptive pathway may not be confidently assessed with these timescales, the relative stabilities between states can. This is demonstrated in two articles that involve simulations of two amphipathic peptides. This article (Article I) focuses on the known pore-forming peptide alamethicin (alm), which is more hydrophobic and has less favorable surface binding. The following article (Article II) focuses on the cationic peptide piscidin 1, which has a well-characterized surface structure, but an unknown disruption mechanism and structure. The peptide’s sequences and amphipathicities are shown in Fig. 1.
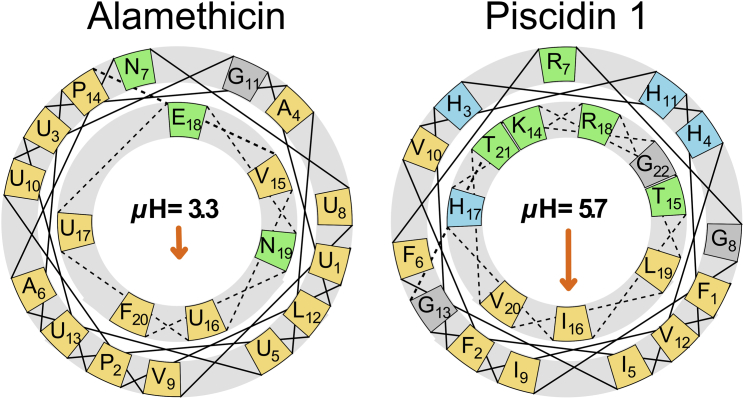
Figure 1.
The helical wheel diagrams of alm (left) and piscidin 1 (right) show positions of the polar (green) and nonpolar (orange) residues when the peptides are assumed to be α-helical, with the magnitude and direction of the hydrophobic moment (μH in kcal/mol) denoted by the orange arrow. U = α-aminoisobutyric acid.
Alamethicin (UPUAUANUVUGLUPVUUENF, where U = α-aminoisobutyric acid) is a 20-residue antibiotic peptide from the fungus Trichoderma viride. The membrane-bound alm fold consists of an α-helical segment for residues 1–12 and a 310 helical segment for residues 15–20. Early reports indicate that the α-helical content is 46%, and the C-terminal segment is primarily unstructured (6). Subsequent studies refer to the alm crystal structure (7) as a guide to the C-terminal segment structure (8). Alm forms membrane-spanning pores with barrel-stave structures (9, 10, 11, 12, 13, 14, 15) (where transmembrane peptides pack like the staves of a barrel with their hydrophilic surfaces aligned toward the center of a water-filled pore). This packing of the hydrophobic residues and aligning of the peptides’ two polar residues toward the transmembrane water stabilizes the pore. The pore serves as an ion channel that conducts an inwardly rectifying current with an external electric field (16). The reported alm tilt angles are dependent on membrane composition and the experimental method. Measurements of labeled alm by NMR or electron paramagnetic resonance are generally associated with 10–25° tilts (17, 18, 19, 20), while those calculated by sum frequency generation vibrational spectroscopy are 63° (21). The pore state was characterized by various diffraction methods, and the numbers of peptides per pore is dependent on the membrane lipid composition (22). Specifically, alm forms hexameric pores in 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) bilayers (9); these pores were simulated with all-atom molecular dynamics (MD) (23, 24, 25), but their stability could not be adequately assessed because the trajectories (all <100 ns) were too short to sample large changes in peptide orientation. More recently, the CHARMM 36 lipid force field was developed and shown to reproduce experimental properties of DOPC and numerous other lipids (26, 27, 28).
These highly studied and well-characterized alm pores serve as a reference for describing other peptide-stabilized pores. While all-atom MD simulations are ideal for determining the forces involved in the disruption of membranes by amphipathic peptides, the timescales of peptide insertion and reorientation can take microseconds to minutes (29), and it is difficult to achieve timescales longer than several microseconds on conventional computers. Fortunately, special-purpose computers, such as Anton (30), greatly enhance the speed of all-atom MD simulations, and make multimicrosecond timescales accessible. For example, a four-peptide melittin pore simulated on Anton started in a barrel-stave arrangement quickly became toroidal and remained stable for the duration of the 9-μs trajectory, in agreement with experiment (31). The same protocol was used to study Magainin 2, and PGLa-disordered toroidal pores (31, 32). However, simulations of melittin started from surface-bound or tilted conformations did not form a transmembrane orientation within 4 μs (33). These studies support the notion that long simulations of antimicrobial peptides initialized in barrel-stave pores can assess their ability to form toroidal pores. Therefore, the stability of the alm pore in DOPC with the CHARMM 36 force field over a multimicrosecond trajectory provides an essential control for simulations of less structured pores.
Prevailing alm research involves elucidating the pore formation mechanism. The driving force for the transition from the surface-bound to the transmembrane orientation is unclear, but is attributed to peptide concentration (34) or an external electric field (35) (such as the membrane potential). Surface-bound alm is unstable in many fluid bilayers, but is observed in the gel-phase (21). One theory is that surface-bound alm peptides aggregate as dimers and trimers, and insert with the addition of an electric field (35, 36). These inserted peptides then aggregate to form the pore. Another theory is that the surface-bound state has a low stability, and the peptides favor a transmembrane orientation that may include dimers or trimers (34). Application of an electric field aligns the transmembrane peptides into a parallel orientation and they aggregate into pores. Simulations of spontaneous peptide insertion at different external electric field strengths will contribute to refining the alm insertion mechanism. These transitions between surface-bound and transmembrane states occur on timescales generally inaccessible by atomistic simulations (even with Anton). Increasing the temperature to 413 K accelerated the insertion of polyleucine (37, 38). The simulations include a barostat that prevents water from boiling, and the simulation stability is generally dependent on the ability for the membrane to remain a bilayer. Additionally, many lipid properties were maintained at these higher temperatures (38).
This article presents a series of simulations that explore both the pore and surface-bound states of alm in a DOPC bilayer: a 14-μs trajectory of a hexameric barrel-stave pore, a 5-μs trajectory of 10 surface-bound alm, and five 1-μs trajectories at various temperatures and applied electric fields detail the mechanisms of insertion and preference for a transmembrane orientation (see Table 1). These simulations provide a counterpoint for Article II (39), where completely different behavior is observed for the cationic antimicrobial peptide piscidin.
Table 1.
Summary of Simulations Reported Here
| Simulation | Temperature |
E-field |
Time |
Primary Result |
|---|---|---|---|---|
| (K) | (V) | (μs) | ||
| Six peptides starting in pore | ||||
| Initial pore | 313 | 0.0 | 14 | Stable water-filled pore |
| Continued from IP14 | 413 | −0.2 | 1.2 | Five-peptide pore, one surface peptide |
| 10 peptides starting on surface | ||||
| Initial surface | 313 | 0.0 | 5 | Peptides remain on surface |
| Continued from initial surface | 313 | 0.2 | 1 | Peptides remain on surface |
| Continued from initial surface | 313 | −0.2 | 1 | Peptides remain on surface |
| Continued from initial surface | 363 | −0.2 | 1 | Three peptides insert and fold to helices |
| Continued from initial surface | 413 | −0.2 | 1 | All peptides insert and fold to helices |
| Continued from initial surface | 413 | 0.0 | 1 | Three peptides insert and fold to helices |
| Six peptides starting transmembrane, no pore | ||||
| Parallel | 313 | 0.0 | 1.0 | Three dimers |
| Antiparallel | 313 | 0.0 | 1.0 | Trimer, dimer, and monomer |
“IP14” refers to the 14-μs coordinate set of the initial pore system.
Materials and Methods
All MD simulations described here were performed with a constant number of atoms, pressure, and temperature (i.e., NPT). The equations of motion were determined from the Verlet algorithm with a time step of 2.0 fs (see Alamethicin Surface-Bound Simulations, below, for the one exception). The Lennard-Jones potentials were terminated at 12 Å, with a smoothing function operating between 8 and 12 Å. Simulations were performed in a tetragonal unit cell with x and y dimensions set to the same length and independent of the z dimension. The temperature was maintained at 313, 363, or 413 K by the Nosé-Hoover thermostat (40, 41). A total pressure of 1 atm was maintained by a Nosé-Hoover piston (42, 43) (using CHARMM; 44) or the Martyna-Tobias-Klein barostat (45) with semiisotropic scaling applied every 100 time steps (using the Anton supercomputer; D. E. Shaw Research, New York, NY). Long-range interactions were evaluated every time step for simulations run with CHARMM (44) and every other time step for those run on Anton.
CHARMM 38b2 (44) and the National Heart, Lung and Blood Institute LoBoS cluster were used for the preparation of simulations run on Anton (Anton requires starting trajectories with well-equilibrated velocities), the full surface-bound simulations, and analysis of all simulations. CHARMM 36 protein (46) and lipid (26) parameters with modified Lennard-Jones pairwise distances for sodium ions interacting with select lipid oxygens (47) were used in all simulations. Parameters for 2-aminoisobutyric acid were developed by modifying the CHARMM 36 alm parameters, with no CMAP correction. A stream file for 2-aminoisobutyric acid is included in the Supporting Material. The systems were solvated with TIP3P water. Initial conditions for all simulations were generated with CHARMM-GUI (48), and the details are given below. The application of an electric field on Anton is described in Jensen et al. (49).
Alamethicin pore simulations
Six alm peptides were built as ideal α-helices with extended side chains, and the helical axes aligned along z. The peptides were initialized as α-helices to be consistent with the piscidin simulations in Article II; as expected, the C-terminal residues rapidly converted to 310-helices. The peptides were centered at z = 0 with their helical axis along the perimeter of a 20 Å pore. Peptides were rotated around the helical axis so that hydrophilic side chains faced the center of the pore. One-hundred-twenty DOPCs were packed into a bilayer around the pores (60 DOPCs per leaflet). The simulation had a tetragonal unit cell with dimensions of 126.3 × 126.3 Å along the x and y directions. A quantity of 25,542 water molecules was added to reach a cell height (z) of 85.0 Å to meet the requirement of a minimum ratio of 1.5 between the longest and smallest dimensions for Anton. The system was replicated in all three dimensions by P1 boundary conditions. The system was solvated with 4,464 waters, 14 sodiums, and 8 chlorides. The system was equilibrated for 100 ns on LoBoS, and then simulated for 14 μs on Anton.
A simulation of the pore was performed at 413 K with a −0.2 V electric field. This corresponds to an inwardly aligned electric field based on the orientation of the peptides (N-termini in the –z direction and C-termini in the +z direction). The simulation began from the last snapshot of the 14-μs pore simulation.
Transmembrane alamethicin simulations (without a pore)
Two sets of six peptides were built as described above, except one had peptides aligned head-to-tail. 240 DOPC lipids were randomly packed around the peptides with acyl chains between the planes of z = 14 Å and z = −14 Å, including between peptides (replacing pore water). The system was solvated with 7342 waters, 27 sodiums, and 21 chlorines. Each simulation was performed for 1 μs at 313 K.
Alamethicin surface-bound simulations
Ten alm peptides were built as ideal α-helices with extended side chains, and the helical axes aligned along the xy plane. The peptides’ centers of mass were positioned at z = 14 Å, and the peptides were rotated about the helical axes so that the three hydrophilic residues faced away from the plane at z = 0 Å. Four-hundred DOPCs were randomly packed around the peptides with acyl chains between the planes of z = 14 Å and z = −14 Å, with 180 lipids on the same leaflet as the peptides and 220 lipids on the opposing leaflet. Fig. S1 in the Supporting Material shows the starting configuration. The system was solvated with 22,363 waters, 74 sodiums, and 64 chlorides. The system was equilibrated for 100 ns on LoBoS, and then simulated for 5 μs on Anton. Four replicas were started from the last frame of the 5-μs surface-bound alm simulation. For the T = 413 K, E = −0.2 V simulation, the simulation crashed at 0.34 μs due to particles with large momenta. The time step was decreased to 1-fs time step run for the remaining 0.66 μs.
Analysis
The peptide folds were determined using the DSSP-2.2.0 (50) algorithm in MDTraj-1.5.1 (51). The structure factors were determined by calculating the atomistic electron densities for each simulation in CHARMM, and converting them to structure factors with SIMtoEXP-2.0.5 (52).
Results and Discussion
Table 1 includes a list of simulations, the conditions, and the primary result from each.
Alamethicin barrel-stave pore
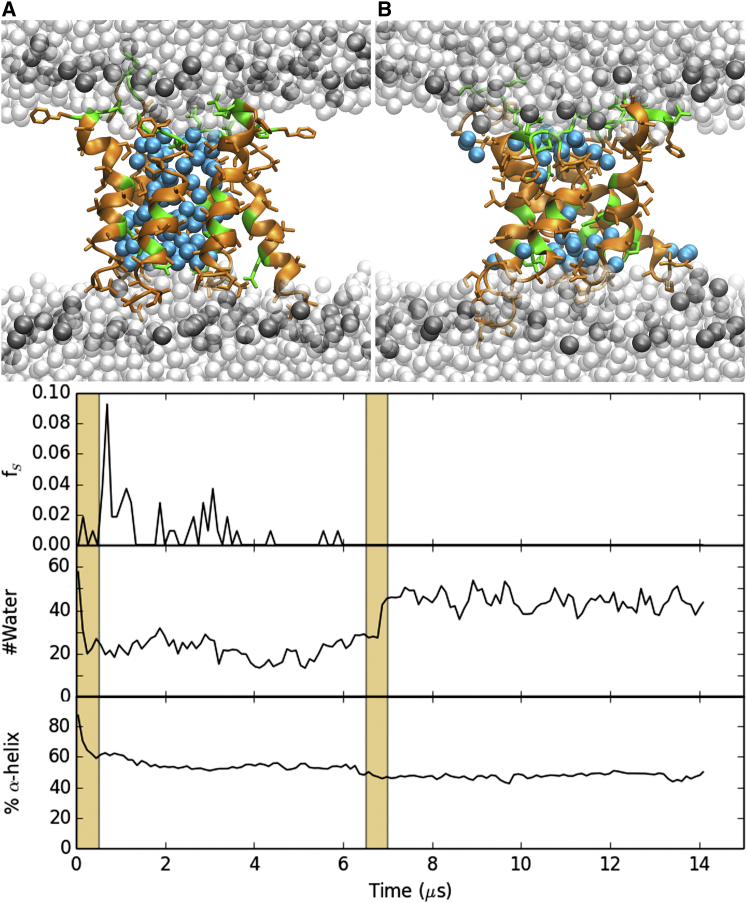
A 14-μs trajectory of a 6-peptide alm pore in DOPC demonstrates the ability of an amphipathic peptide to stabilize a transmembrane water channel. Within 200 ns, the alm peptides formed a tight pore that sealed off the lipid acyl chains from the pore water and left the lipids relatively unperturbed. All but one peptide remained transmembrane during the entire 14 μs simulation. This peptide partially slid out of the bilayer interior and residues 14–20 sampled surface-bound orientations during the first 6.5 μs (top time series in Fig. 2). The pore completely reannealed between 6.5 and 7.5 μs to include all six peptides (consistent with the x-ray diffraction results in DOPC (9, 23)), and remained stable for the remainder of the trajectory; Fig. 2 B shows the 14 μs configuration. The reformation of the pore doubled the number of water molecules (22 ± 1 to 44 ± 1; middle time series in Fig. 2), and was accompanied by an increased frequency at which one or two sodium ions enter the pore. Waters and ions are defined as being within the pore if they are within 10 Å of the bilayer midplane.
Figure 2.
(Top) Snapshot of the alm pore at 0 (A) and 14 μs (B). Peptides are colored by residue type (orange for hydrophobic residues; green for polar and charged), waters within 10 Å of the midplane are in cyan, waters further than 10 Å from the midplane are white, and the DOPC phosphorus atoms are dark gray. The rest of the lipid atoms are omitted for clarity. (Bottom) Fraction of surface-bound (S) peptides (top plot), the number of pore water molecules (middle), and the fraction of α-helical content (bottom) as a function of time. Note that the scale for the fraction S is from 0 to 0.1. Areas with an orange background highlight times of significant changes in the fraction of S and the number of pore waters.
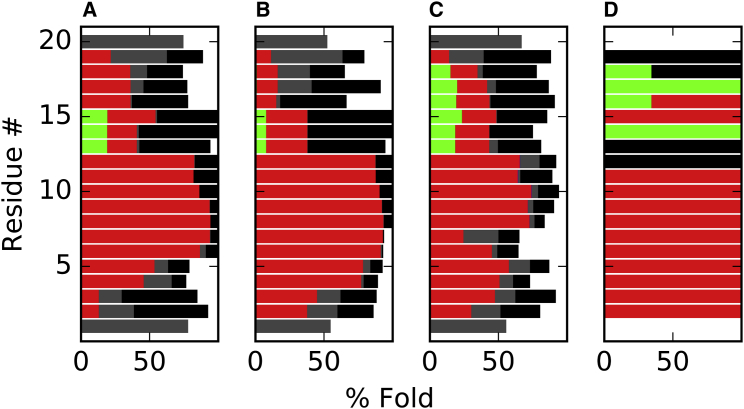
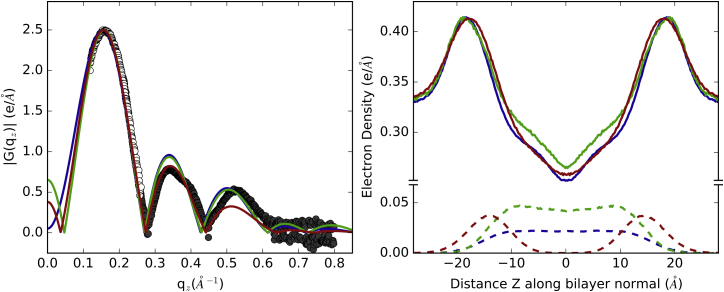
In addition to the increase of water content, the α-helical content of alm dropped from 100% to 47 ± 3% for the last 6.5 μs (bottom time series in Fig. 2). This agrees with the experimental helicity of ∼50% as measured by circular dichroism in fluid-phase DOPC bilayers (53). Residues 4–13 remained α-helical, consistent with the purely α-helical segment from the x-ray crystal structure of alm in a nonaqueous solvent (7). The C-terminal residues (14–20) were significantly more disordered in the simulation than in the crystal structure (Fig. 3); however, these residues in the three monomers of the crystal structure asymmetric unit have different conformations. It is plausible that such structural inhomogeneity in the crystal could be reflected by disorder in the fluid lipid bilayer. Hence, the second state of the alm pore (after 7.5 μs) is in excellent agreement with the experimental observations of a 6-peptide barrel-stave pore, and appears to be an equilibrated structure. The superhelical pore structure does not show a significant chirality unlike previously simulations and measurements. The tilts for the α-helical peptide segments are 27 ± 13° with large deviations during the simulation. The tilts for the C-terminal helices were not computed because they were transitioning between α- and 310-helices (see Figs. S2–S4). The computed x-ray scattering profile agrees well with published values (Fig. 4).
Figure 3.
Average percent fold of all alm peptides at 313 K for the last microsecond of the pore (A), transmembrane (B), and surface-bound (C) simulations. (D) Fold for the alm crystal structure. The peptide fold consists of α-helix (red), 3-10-helix (green), hydrogen-bonded turn (black), bend (gray), and unstructured loop (white).
Figure 4.
(Left) Experimental x-ray form factors for 1:20 alm per DOPC (black), and those calculated from simulations of surface-bound (red), pore (green), and transmembrane (blue). (Right) Electron density profiles for preceding systems (with the peptide-only densities in dashes).
Two 1-μs trajectories of nonpore transmembrane alm peptides provide a reference for the pore peptides. Each simulation contained six peptides in parallel or antiparallel orientations. The peptides remained transmembrane throughout the simulation with a tilt angle of 25 ± 12°, similar to the pore peptides. Residues 2–12 of the nonpore peptides were more α-helical than the pore peptides, while residues 16–20 were less structured at 313 K (Fig. 3). The pore and transmembrane structure factors are nearly indistinguishable between their first three peaks (Fig. 4). The electron density profiles are similar for the pore and nonpore peptides; however, the central trough for the pore profile is shallower due to the water density. A majority of the peptides was dimers or tetramers, and there was no preference for parallel or antiparallel alignment. Fig. S5 shows each simulation at 1 μs.
Structure of surface-bound alamethicin and insertion to transmembrane
Ten α-helical alm peptides were placed on the surface of a DOPC lipid bilayer. Within 200 ns, all peptides migrated toward the water layer with most undergoing significant unfolding (Fig. S4 shows the fold of each peptide is plotted against time). On three occasions, one peptide left the membrane and became completely solvated for >5 ns. After 5 μs, all peptides were loosely bound to the membrane (Fig. 5 A), but not significantly inserted. The alm peptides are less structured on the membrane surface than in the transmembrane or pore orientation (Fig. 3). The total helical content for all peptides in the last microsecond of the surface-bound simulations is 39% compared to 51 and 54% for the last microsecond of the pore and nonpore transmembrane (TM) simulations. This drop in α-helicity primarily occurs at residues 5 and 6, while the three N-terminal residues are more α-helical than the pore state (the membrane surface provides a less solvated environment and terminal residues in the pore must interact with pore water and water on the membrane surface). The α-helical content was measured by circular dichroism for both the surface-bound and TM states (54), and is 40% without a length effect and up to 60–70% with a length effect for either state. The simulation results presented here fall within the measured range, with the α-helicity of the inserted states 12–15% greater than that of the surface states. The C-terminal residues (12–20) have an increase in the 310-helix population than in either transmembrane state; however, this section is predominantly unstructured. The electron density profile for surface-bound peptides in DOPC has a smaller peak-to-peak distance than that of the pore or transmembrane simulations (36.2 vs. 37.8 for the pore and 36.6 for the transmembrane states). This is reflective of significant membrane thinning by the surface-bound peptides. The trough at the midplane is shallower than for the transmembrane peptides due to the lower peptide concentration.
Figure 5.
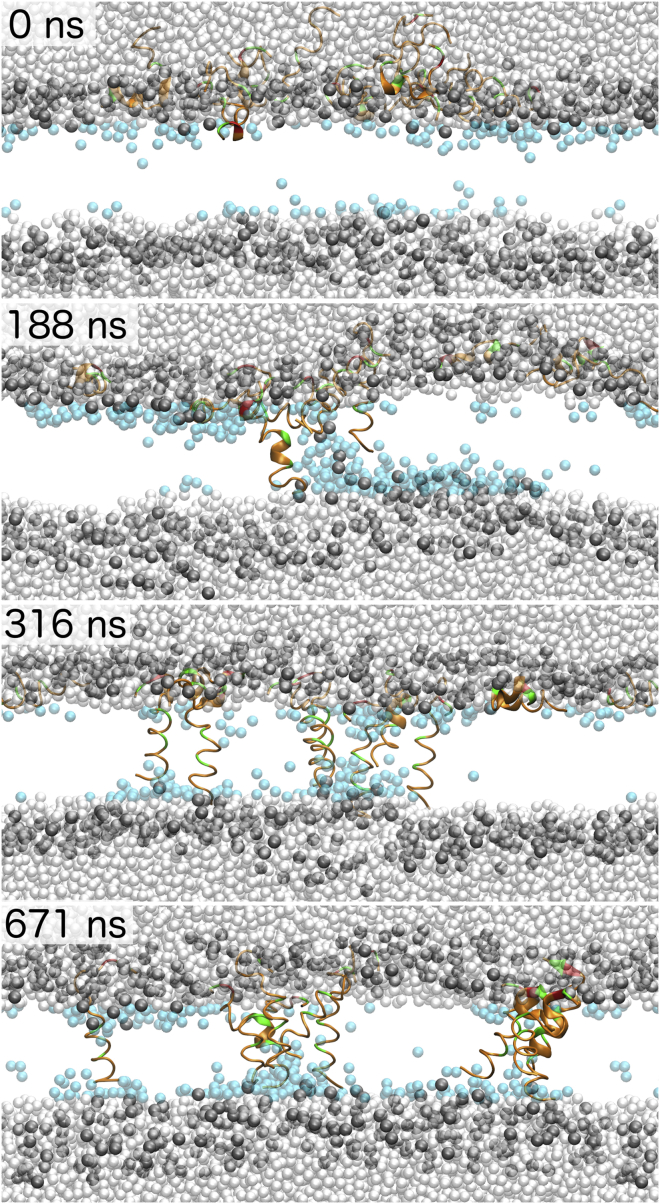
Snapshots of alm insertion at 0, 188, 316, and 671 ns. Peptides are colored by residue type (orange for hydrophobic residues; green for polar and charged), waters within 10 Å of the midplane are in cyan, waters further than 10 Å from the midplane are white, and the DOPC phosphates are gray. The rest of the lipid atoms are omitted for clarity.
To accelerate insertion of the peptides, the temperature was raised to 413 K and a −200 mV electric field was applied after the 5-μs simulation. The first peptide inserted at 84 ns; all peptides were transmembrane at 514 ns, and remained transmembrane over the remaining 500 ns. To test the role of the temperature and voltage on alm insertion, simulations were performed at 313 K with a −200 mV potential, 313 K with a +200 mV potential, 363 K with a −200 mV potential, and 413 K with no potential. The first two simulations had no insertion within 1 μs, while the last two had three peptides insert within 1 μs (at 213, 626, and 680 ns for 363 K; and 650, 910, and 980 ns for 413 K). The applied electric field is more significant to accelerating insertion; however, higher temperatures are necessary for insertion within a microsecond.
The preceding three simulations provide 16 examples of insertion. All of peptides insert with their N-terminus. Residues 3–12 must fold into α-helices before insertion. The alamethicin peptides inserted with a preference for a left-handed conformation. This bias was recently demonstrated for the CHARMM 36 protein force field (55). Ten of the peptides insert with the assistance of a previously inserted peptide. For the six peptides that self-insert, insertion is mediated by water in the hydrophobic core of the membrane. This mediation involves hydrogen bonding between N-terminal amine and water (Fig. 6). This is a contributing factor to accelerated insertion at high temperatures, because the membrane becomes more porous and increases the probability of water chains to guide insertion. Membrane water is further increased by inward bulging of the opposing leaflet (Fig. 6). Self-insertion took ∼5 ns between reaching the midplane and a TM orientation. The 10 peptide-mediated insertions involve the N-terminus of one surface-bound peptide passing down a monomer or dimer of previously inserted peptides. Bulging is not observed for the peptides that insert with the assistance of a transmembrane alm, as transmembrane peptides stiffen the membrane. However, significant water is brought into the membrane with insertion (Fig. 6). After insertion, the peptides associate as dimers or trimers for the remainder of the simulation. These insertion mechanisms are consistent among the three simulations, and are independent of temperature or the applied electric field strength.
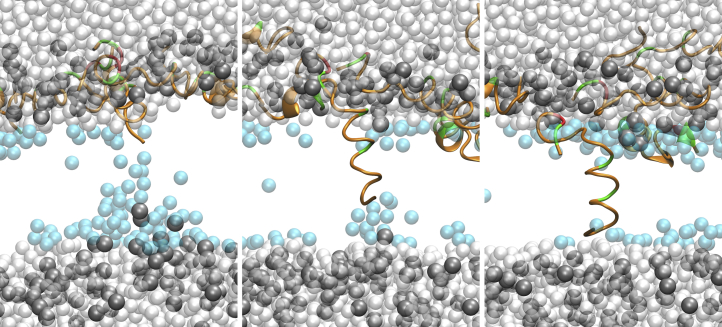
Figure 6.
Snapshots of the first peptide to insert at 413 K and −0.2 V electric field strength. Snapshots are at 81.1, 83.5, and 85.9 ns. Peptides are colored by residue type (orange for hydrophobic residues; green for polar and charged), waters within 10 Å of the midplane are in cyan, waters further than 10 Å from the midplane are white, and the DOPC phosphates are gray. The rest of the lipid atoms are omitted for clarity.
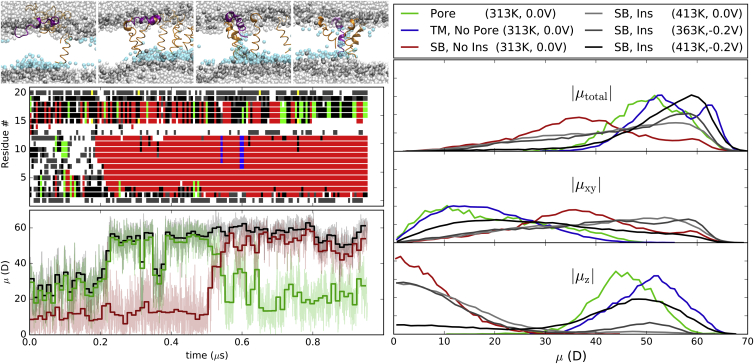
The electric field accelerates insertion by increasing and orienting the dipole moment of the peptide. This is demonstrated by comparing the peptide fold and dipole moments for a single peptide as shown in the left panel of Fig. 7. This figure is representative of all 16 peptides that inserted in the membrane at higher temperatures. Before each insertion, residues 4–11 folded into an α-helix (∼0.2 μs in Fig. 7, left). Upon insertion, residues 2 and 3 join the α-helix, and the fold of residues 14–17 still fluctuates, but more time is spent in a helical state. The distribution of dipole moments for all peptides is shown in the right panel of Fig. 7.
Figure 7.
(Left, top) Snapshots of an alm peptide (purple) at 86, 512, 537, and 649 ns; left, middle) time series of the fold for that same peptide (α-helix (red), 3-10-helix (green), hydrogen-bonded turn (black), bend (gray), and unstructured loop (white)); and (left, bottom) the peptide’s total dipole moment magnitude (|μtotal|, black), and the dipole moment parallel (|μz|, red) or perpendicular (|μxy|, green) to the membrane normal. The average μ is plotted for blocks of 12 ns. (Right) Distribution of the total alm dipole moments (|μtotal|) for peptides in the pore (green), transmembrane (blue), and surface-bound orientations (red for 313 K, 0.0 V; light gray for 413 K, 0.0 V; dark gray for 363 K, −0.2 V; and black for 413 K, −0.2 V). Dipole moments are decomposed into the magnitudes perpendicular (|μxy|) and parallel (|μz|) to the membrane normal.
The average dipole moment for surface-bound alm (|μtotal|) is 35 Da with a range of nearly 70 Da and oriented parallel to the membrane surface. The dipole moment along the membrane normal is near zero (|μz|). This polarity comes from the few helical segments of the peptide. Alms in a pore or nonpore transmembrane state have a larger |μtotal| of 50–65 Da, with a majority aligned along the membrane normal. The component of the μtotal parallel to the membrane (|μxy|) is nonzero (∼10 Da), signifying a small tilt. The peptide tilt is greater in the pore state than in the nonpore state. This increase in |μtotal| and |μz| with insertion of peptide is demonstrated by the change in the curves for surface-bound alms at 413 K, 0.0 V; to 363 K, −0.2 V; to 413 K, −0.2 V. This trend follows the rate of peptide insertion, and therefore the averages over the trajectory show the change in |μtotal| from predominantly parallel to the surface to along the membrane normal. The larger |μtotal| for a transmembrane peptide is consistent with the greater percent helical peptide in Fig. 3.
Pore at 413 K and an electric field
In the final simulation, the 14-μs alm pore was simulated at 413 K and a −0.2 V potential to compare pore stability to the insertion simulations. Over the 1 μs trajectory, one peptide was expelled from the pore within 30 ns and returned to the surface for the remainder of the simulation. The remaining five peptides sampled a variety of unfolded states and orientations but remained as an aggregate with a persistent water channel. Fig. S6 shows the pore at 1.2 μs.
Conclusions
The alm pore simulation convincingly illustrates the timescales necessary to sample the rearrangement of peptides in the viscous environment of a lipid membrane; i.e., 7.5 μs were required to reach equilibration from reasonable starting conditions (Fig. 2). It is reassuring that after 7.5 μs (and up to 14 μs), the alm pore packed into a tight barrel-stave pore structure in agreement with experiment, indicating that the CHARMM 36 protein (46) and lipid (26) force fields are capable of overcoming nonideal starting conditions. This is a necessary validation for a critical evaluation of antimicrobial peptide pore stability. The alm pore and the aforementioned 9-μs melittin pore simulations provided direct evidence of stability: alm reincorporated a peptide into the pore (Fig. 2), and the trajectory of melittin included at least one instance of a peptide transitioning from TM to S and back to TM (31). However, structural information pertaining to the toroidal pore of melittin is admittedly sparse. The extensive experimental evidence for the alm pore structure is why it was chosen for this study. The preference for helical transmembrane alm is reinforced by the spontaneous insertion of the surface-bound peptide at higher temperatures, with or without an applied electric field. Under the most extreme condition discussed here (413 K and −0.2 V), the alm pore was stable for 1 μs, with only one peptide transitioning from pore to surface-bound. This was the only instance of a peptide leaving the inserted state among the simulations in Table 1.
The surface-bound and inserted alm structures demonstrated a broad range of disorder. First, insertion of the peptide stabilizes the α-helical region; however, residues 13–19 fluctuate between unstructured, α-helix, and 310-helix for all orientations. This is consistent with the varied hydrogen-bonding patterns in the crystal structure of Fox and Richards (7), and informs assumptions on the peptide orientation derived from sparsely labeled peptides. The helical N-terminal segment of alm is consistently ∼25–30° despite the disordered C-terminal segment or if the peptide is part of a pore. The 12° fluctuation in the tilt is also independent of pore state. These results are not unexpected, as alm functions as an ion channel with an external voltage; an applied voltage will align and pack the pore peptides and may reduce the tilt and its fluctuations compared to the nonpore transmembrane peptides.
An important result of this article regards the instability of the surface-bound state of alm, especially compared to piscidin 1 (see Article II). Simulations of the surface-bound state of both peptides were initiated from α-helical conformations with the center of mass 14 Å above the lipid midplane, just below the headgroups. Piscidin 1 remained helical at this height, and never entered the water above the bilayer; the distance between the center of the peptide and phosphate peaks in the electron density ranges from 5.4 to 8.2 Å, depending on the lipid composition (see Fig. 1 of Perrin et al. (56)). In contrast, alm quickly lost helical content, and associated with the headgroups; from Fig. 4, the distance between the peptide and phosphate peaks in the electron density is only 4.4 Å. Furthermore, one of the peptides entered the water phase for several periods during the trajectory. Hence, the large fraction of hydrophobic residues evident in Fig. 1 does not stabilize a surface-bound α-helical conformation for alm. The recently demonstrated preference for the CHARMM 36 protein force field to overpopulate the left-handed helical conformation (55) is an artifact that contributed to alamethicin unfolding. It was not previously observed in simulations of piscidin (39, 56) or melittin (31, 33) because these peptides did not unfold. Nevertheless, this flaw should not substantially impact the inherent preference for alamethicin insertion given the fluidity and disorder of the membrane surface.
The lack of alm insertion in the surface-bound simulation at 313 K indicates that spontaneous insertion is unlikely on the 5-μs timescales. It was not until the system temperature was increased that the peptides inserted. The inserted peptides did not aggregate to form a water channel, and the nonpore TM peptides formed dimers a trimer, but no pore within 1 μs. The mechanism of alamethicin pore formation and the necessity of an electric field to form a pore are open questions in Guidelli and Becucci (57). This study illustrates the critical need for enhanced sampling methods to overcome large barriers to membrane insertion, and to study pore formation by predominantly hydrophobic peptides.
In conclusion, the alm simulations presented here demonstrate the timescales and computer resources necessary to simulate peptide rearrangement in the viscous environment of a biological membrane. Multiple microseconds were required to overcome the starting configurations and recapture the pore structure (100 ns would have missed the peptide leaving, and the pore would have appeared to be unstable at 1 μs). The stability of the alm pores for 14 μs in DOPC provides essential support for the results in Article II on the membrane-bound piscidin structure. Alm has a strong preference for an inserted structure with a predominantly α-helical segment N-terminal to Pro14 with tilt 20–30° from the membrane normal. The C-terminus of the peptide is intrinsically disordered from the simulations, with a significant population of α- and 310-helical bonding patterns; however, this segment is rarely a straight helix.
Author Contributions
B.S.P. and R.W.P. designed the research and wrote the article, and B.S.P. carried out the simulations.
Acknowledgments
The Anton machine at the Pittsburgh Supercomputing Center was generously made available by D. E. Shaw Research. We thank Stephanie Tristam-Nagle for providing the Alamethicin form factors.
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Heart, Lung and Blood Institute, and utilized the high performance computational capabilities at the National Institutes of Health, Bethesda, MD (National Heart, Lung and Blood Institute LoBoS cluster). Anton computer time was provided by the National Center for Multiscale Modeling of Biological Systems through grant No. P41GM103712-S1 from the National Institutes of Health and the Pittsburgh Supercomputing Center.
Editor: Scott Feller.
Footnotes
Supporting Materials and Methods and six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30702-0.
Supporting Material
References
- 1.Boman H.G. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 2.Hancock R.E.W., Chapple D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen L.T., Haney E.F., Vogel H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Wimley W.C., Hristova K. Antimicrobial peptides: successes, challenges and unanswered questions. J. Membr. Biol. 2011;239:27–34. doi: 10.1007/s00232-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimley W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010;5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel H. Comparison of the conformation and orientation of alamethicin and melittin in lipid membranes. Biochemistry. 1987;26:4562–4572. doi: 10.1021/bi00388a060. [DOI] [PubMed] [Google Scholar]
- 7.Fox R.O., Jr., Richards F.M. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution. Nature. 1982;300:325–330. doi: 10.1038/300325a0. [DOI] [PubMed] [Google Scholar]
- 8.Ye S., Li H., Chen Z. Observing a model ion channel gating action in model cell membranes in real time in situ: membrane potential change induced alamethicin orientation change. J. Am. Chem. Soc. 2012;134:6237–6243. doi: 10.1021/ja2110784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan J., Tristram-Nagle S., Nagle J.F. Alamethicin aggregation in lipid membranes. J. Membr. Biol. 2009;231:11–27. doi: 10.1007/s00232-009-9199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He K., Ludtke S.J., Huang H.W. Neutron scattering in the plane of membranes: structure of alamethicin pores. Biophys. J. 1996;70:2659–2666. doi: 10.1016/S0006-3495(96)79835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He K., Ludtke S.J., Worcester D.L. Antimicrobial peptide pores in membranes detected by neutron in-plane scattering. Biochemistry. 1995;34:15614–15618. doi: 10.1021/bi00048a002. [DOI] [PubMed] [Google Scholar]
- 12.Qian S., Wang W., Huang H.W. Structure of the alamethicin pore reconstructed by x-ray diffraction analysis. Biophys. J. 2008;94:3512–3522. doi: 10.1529/biophysj.107.126474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee M.-T., Chen F.-Y., Huang H.W. Energetics of pore formation induced by membrane active peptides. Biochemistry. 2004;43:3590–3599. doi: 10.1021/bi036153r. [DOI] [PubMed] [Google Scholar]
- 14.Nagao T., Mishima D., Naito A. Structure and orientation of antibiotic peptide alamethicin in phospholipid bilayers as revealed by chemical shift oscillation analysis of solid state nuclear magnetic resonance and molecular dynamics simulation. Biochim. Biophys. Acta. 2015;1848(11 Pt A):2789–2798. doi: 10.1016/j.bbamem.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Bechinger B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta. 1999;1462:157–183. doi: 10.1016/s0005-2736(99)00205-9. [DOI] [PubMed] [Google Scholar]
- 16.Woolley G.A., Biggin P.C., Sansom M.S. Intrinsic rectification of ion flux in alamethicin channels: studies with an alamethicin dimer. Biophys. J. 1997;73:770–778. doi: 10.1016/S0006-3495(97)78109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salnikov E.S., Friedrich H., Bechinger B. Structure and alignment of the membrane-associated peptaibols ampullosporin A and alamethicin by oriented 15N and 31P solid-state NMR spectroscopy. Biophys. J. 2009;96:86–100. doi: 10.1529/biophysj.108.136242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bak M., Bywater R.P., Nielsen N.C. Conformation of alamethicin in oriented phospholipid bilayers determined by 15N solid-state nuclear magnetic resonance. Biophys. J. 2001;81:1684–1698. doi: 10.1016/S0006-3495(01)75822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.North C.L., Barranger-Mathys M., Cafiso D.S. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys. J. 1995;69:2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marsh D., Jost M., Toniolo C. Lipid chain-length dependence for incorporation of alamethicin in membranes: electron paramagnetic resonance studies on TOAC-spin labeled analogs. Biophys. J. 2007;92:4002–4011. doi: 10.1529/biophysj.107.104026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye S., Nguyen K.T., Chen Z. Interactions of alamethicin with model cell membranes investigated using sum frequency generation vibrational spectroscopy in real time in situ. J. Phys. Chem. B. 2010;114:3334–3340. doi: 10.1021/jp911174d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K.F., Nagarajan R., Camesano T.A. Antimicrobial peptide alamethicin insertion into lipid bilayer: a QCM-D exploration. Colloids Surf. B Biointerfaces. 2014;116:472–481. doi: 10.1016/j.colsurfb.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 23.Pan J., Tieleman D.P., Tristram-Nagle S. Alamethicin in lipid bilayers: combined use of x-ray scattering and MD simulations. Biochim. Biophys. Acta. 2009;1788:1387–1397. doi: 10.1016/j.bbamem.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breed J., Biggin P.C., Sansom M.S.P. Alamethicin channels—modelling via restrained molecular dynamics simulations. Biochim. Biophys. Acta. 1997;1325:235–249. doi: 10.1016/s0005-2736(96)00262-3. [DOI] [PubMed] [Google Scholar]
- 25.Tieleman D.P., Berendsen H.J.C., Sansom M.S.P. An alamethicin channel in a lipid bilayer: molecular dynamics simulations. Biophys. J. 1999;76:1757–1769. doi: 10.1016/s0006-3495(99)77337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastor R.W., MacKerell A.D., Jr. Development of the CHARMM force field for lipids. J. Phys. Chem. Lett. 2011;2:1526–1532. doi: 10.1021/jz200167q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venable R.M., Brown F.L.H., Pastor R.W. Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem. Phys. Lipids. 2015;192:60–74. doi: 10.1016/j.chemphyslip.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fantner G.E., Barbero R.J., Belcher A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 2010;5:280–285. doi: 10.1038/nnano.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw, D. E., R. O. Dror, …, B. Towles. 2009. Millisecond-scale molecular dynamics simulations on Anton. In Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis. ACM, Portland, OR. 1–11.
- 31.Leveritt J.M., 3rd, Pino-Angeles A., Lazaridis T. The structure of a melittin-stabilized pore. Biophys. J. 2015;108:2424–2426. doi: 10.1016/j.bpj.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pino Angeles A., Leveritt J.M., III, Lazaridis T. Structure of transmembrane pores stabilized by antimicrobial peptides magainin and PGLa. Biophys. J. 2015;108:549a–550a. [Google Scholar]
- 33.Upadhyay S.K., Wang Y., Ulmschneider J.P. Insights from micro-second atomistic simulations of melittin in thin lipid bilayers. J. Membr. Biol. 2015;248:497–503. doi: 10.1007/s00232-015-9807-8. [DOI] [PubMed] [Google Scholar]
- 34.Boheim G., Hanke W., Jung G. Alamethicin pore formation: voltage-dependent flip-flop of α-helix dipoles. Biophys. Struct. Mech. 1983;9:181–191. [Google Scholar]
- 35.Sakmann B., Boheim G. Alamethicin-induced single channel conductance fluctuations in biological membranes. Nature. 1979;282:336–339. doi: 10.1038/282336a0. [DOI] [PubMed] [Google Scholar]
- 36.Kolb H.-A., Boheim G. Analysis of the multi-pore system of alamethicin in a lipid membrane. J. Membr. Biol. 1978;38:151–191. [Google Scholar]
- 37.Ulmschneider M.B., Ulmschneider J.P., White S.H. Spontaneous transmembrane helix insertion thermodynamically mimics translocon-guided insertion. Nat. Commun. 2014;5:4863. doi: 10.1038/ncomms5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulmschneider J.P., Smith J.C., Ulmschneider M.B. In silico partitioning and transmembrane insertion of hydrophobic peptides under equilibrium conditions. J. Am. Chem. Soc. 2011;133:15487–15495. doi: 10.1021/ja204042f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perrin B.S., Jr., Fu R., Pastor R.W. Simulations of membrane-disrupting peptides II: AMP piscidin 1 favors surface defects over pores. Biophys. J. 2016;111:1258–1266. doi: 10.1016/j.bpj.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 41.Nosé S. A unified formulation of the constant temperature molecular-dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 42.Nosé S., Klein M.L. A study of solid and liquid carbon tetrafluoride using the constant pressure molecular dynamics technique. J. Chem. Phys. 1983;78:6928. [Google Scholar]
- 43.Andersen H.C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 1980;72:2384. [Google Scholar]
- 44.Brooks B.R., Brooks C.L., III, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 46.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 47.Venable R.M., Sodt A.J., Klauda J.B. CHARMM all-atom additive force field for sphingomyelin: elucidation of hydrogen bonding and of positive curvature. Biophys. J. 2014;107:134–145. doi: 10.1016/j.bpj.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 49.Jensen M.Ø., Jogini V., Shaw D.E. Atomic-level simulation of current-voltage relationships in single-file ion channels. J. Gen. Physiol. 2013;141:619–632. doi: 10.1085/jgp.201210820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 51.McGibbon R.T., Beauchamp K.A., Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kučerka N., Katsaras J., Nagle J.F. Comparing membrane simulations to scattering experiments: introducing the SIMtoEXP software. J. Membr. Biol. 2010;235:43–50. doi: 10.1007/s00232-010-9254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizzo V., Stankowski S., Schwarz G. Alamethicin incorporation in lipid bilayers: a thermodynamic study. Biochemistry. 1987;26:2751–2759. doi: 10.1021/bi00384a015. [DOI] [PubMed] [Google Scholar]
- 54.Huang H.W., Wu Y. Lipid-alamethicin interactions influence alamethicin orientation. Biophys. J. 1991;60:1079–1087. doi: 10.1016/S0006-3495(91)82144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rauscher S., Gapsys V., Grubmüller H. Structural ensembles of intrinsically disordered proteins depend strongly on force field: a comparison to experiment. J. Chem. Theory Comput. 2015;11:5513–5524. doi: 10.1021/acs.jctc.5b00736. [DOI] [PubMed] [Google Scholar]
- 56.Perrin B.S., Jr., Sodt A.J., Pastor R.W. The curvature induction of surface-bound antimicrobial peptides piscidin 1 and piscidin 3 varies with lipid chain length. J. Membr. Biol. 2014;248:455–467. doi: 10.1007/s00232-014-9733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guidelli R., Becucci L. Mechanism of voltage-gated channel formation in lipid membranes. Biochim. Biophys. Acta. 2016;1858:748–755. doi: 10.1016/j.bbamem.2015.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.