Abstract
Background
Culex Flavivirus (CxFV) is an insect-specific virus that is widely distributed and primarily infects mosquito species from the genus Culex. Its hosts include Culex tritaeniorhynchus, Culex quinquefasciatus, and Anopheles sinensis mosquitoes. Since its original identification, CxFV has been reported in several countries. Despite the increasing number of reports on CxFV, little is known about its genomic characteristics. It is unclear whether the phylogenetic relationships between the strains are influenced by host species and geographic location.
Results
We characterized the Brazilian CxFV strain and performed a comprehensive genetic and phylogenetic characterization of CxFV based on all ORF sequences described so far. Our results revealed that the Brazilian strain is in a monophyletic clade with the Mexican strain. Overall, selective pressure indicates that the ORF is undergoing purifying selection.
Conclusions
The phylogenetic analysis revealed a strong association between climate and CxFV ancestry. Also, based on phylogeny and the genetic distance between the main branches of the tree, we propose the classification of the available sequences into two different genotypes. We also suggest the existence of two different subtypes within Genotype 1.
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-016-0614-3) contains supplementary material, which is available to authorized users.
Keywords: Culex flavivirus, Phylogeny, Genotypes
Background
Flaviviruses are well known for causing important mosquito-borne human diseases. They are transmitted between arthropods and vertebrates, and are capable of replicating in both hosts. In 1975, a new member of this genus, incapable of replicating in vertebrate cells, was isolated from mosquito cell cultures and named Cell Fusing Agent Virus (CFAV) [1]. More recently, the isolation of other insect-specific viruses has been reported [2–5]. One of these insect-specific viruses is Culex Flavivirus (CxFV), an insect-specific virus that was first identified in 2007 in Culex pipiens mosquitoes [6]. Other hosts include Culex tritaeniorhynchus, Culex quinquefasciatus, and Anopheles sinensis mosquitoes [5, 7, 8].
Similar to other flaviviruses, CxFV encodes a polyprotein from a single-strand positive RNA open reading frame (ORF), flanked by 3′ and 5′-untranslated regions (UTR). The polyprotein of 3,364 aa is cleaved during and after translation into structural and non-structural proteins in the following order: C-prM(M)-E-NS1-NS2A-NS2B-NS3-NS4A-2K-NS4B-NS5 [6].
Since its identification, Culex Flavivirus has been reported in the United States, Mexico, Guatemala, Trinidad, Uganda, Indonesia, Japan, China, and Brazil, indicating that it is widely distributed [5, 6, 9–14]. Despite the increasing number of CxFV sequences deposited in GenBank, it is not clear if the phylogenetic relationships between the strains are influenced by the host species or geographic location. Moreover, little is known about the genetic profile of the CxFV genome.
Previously, we reported the first identification of CxFV in Brazil [14]. Here, we perform a genetic and phylogenetic analysis of the complete ORF of the Brazilian CxFV strain. In addition, we perform the genetic characterization of CxFV based on the ORF sequences described so far, and we propose a novel genotype classification.
Methods
Culex Flavivirus (CxFV) strain
According to report published previously by this research group [14], The CxFV strain obtained in this study was derived from Culex quinquefasciatus mosquitoes, collected between April 2007 and January 2008, in the city of São José do Rio Preto (SJRP). The mosquitoes were pooled according to genus, gender and geographic location, for RNA extraction using Trizol protocol (Invitrogen).
The presence of Flavivirus was detected for each pool by Multiplex-Nested_RT-PCR [15]. Ten positives pools were inoculated three times passages on C6/36. After the third passage pools were tested with primers specific for Culex Flavivirus (CxFV), Saint Louis Encephalitis Virus (SLEV), and West Nile Virus (WNV) [14]. One CxFV isolated was used for sequence analysis in this manuscript.
Next generation sequencing
The CxFV RNA extracted from the C6/36 supernatant was quantified using a UV/Vis spectrophotometer (PicoDrop P200, Picodrop, Hinxton, Cambridgeshire, United Kingdom) and treated with Deoxyribonuclease (DNase I, Invitrogen, Carlsbad, CA, USA) to digest single and double-stranded DNA. cDNA was obtained using commercial SuperScript® III Reverse Transcriptase (Thermo Scientific, San Jose, CA, USA) according to the manufacturer’s protocol. A multiplex nested PCR was used once again to confirm CxFV positive sample [14, 15].
Double-stranded DNA was obtained using the SuperScript® Double-Stranded cDNA Synthesis Kit (Thermo Scientific, San Jose, CA, USA), purified with commercial magnetic beads (Agencourt AMPure XP, Beckman Coulter Inc., Indianapolis, IN, USA), and quantified. Sample were processed with the Nextera XT DNA Library Preparation Kit and sequenced using a commercial kit (MiSeq System, Illumina Inc., San Diego, CA, USA).
The assembly was performed by Geneious R6 (Biomatters, Auckland, New Zealand), using sequence of Toyama740/2005 CxFV strain (gb AB701772) as reference to assemble it.
Culex flavivirus sequences
All the analyses in this study were based on a 26-sequence dataset composed of all complete Culex Flavivirus ORF sequences (10,092 nucleotides) deposited in GenBank at the time of the study, plus the Brazilian sequence described here (KT726939). Their accession numbers are: AB701776.1, AB701775.1, AB701774.1, AB701773.1, AB701772.1, AB701771.1, AB701770.1, AB701769.1, AB701768.1, AB701767.1, AB701766.1, JQ308190.1, JQ308189.1, JQ308188.1, JQ308187.1, JQ308186.1, HQ678513.1, FJ663034.1, GQ165808.1, FJ502995.1, NC_008604.2, JQ518484.1, EU879060.1, AB377213.1, and AB262759.2. All Sequences were aligned using Muscle in the SeaView 4.4.2 package [16, 17].
Similarity analysis
Similarity analysis was carried out using the Sequence Identity Matrix tool from the BioEdit 7.0.5.3 package [18].
Genetic distance
Mean genetic distances for the CxFV ORF and each specific region were calculated by the Kimura 2-parameters substitution model using Mega 6 [19].
The site-by-site genetic distances between CxFV_BR-RP01/2007 (KT726939) and the sequences identified in other countries were calculated in SimPlot 3.5.1 [20]. The analysis was carried out using the Kimura 2-parameter distance model and a 200-bp sliding window. Sequences were grouped by geographic location.
Selective pressure analysis
Global and site-by-site ω were estimated in HyPhy [21]. Global ω was calculated for the full ORF and for each genomic region using the HKY85_3x4 substitution model and global parameters. Site-specific selection was calculated Single Likelihood Ancestor Counting (SLAC) method, the MG94xHKY85 model, and a p-value of 0.05.
As HyPhy requires a starting tree for all analysis, a maximum-likelihood tree was constructed for the entire ORF and for specific regions by PhyML in the SeaView package [22]. The GTR model was used, with optimized nucleotide equilibrium frequencies and site rate variation.
Phylogenetic analysis
A Bayesian Markov Chain Monte Carlo (MCMC) analysis was used to estimate the phylogenetic tree using BEAST v1.8 [23]. The analysis was performed under strict molecular clock using as tree prior a constant population size coalescent and MCMC was run for 10,000,000 steps and sampled every 1000 steps. Trees were summarized using Tree Annotator 1.8 available on Beast package and edited using FigTree 1.4 [24]. BaTS 0.9 was used to statistically test for association between tree topology and geography, host species or climate [25].
The mean genetic distance between the sequences in specific branches of the phylogenetic tree was calculated in Mega 6 [19]. Sequences were divided into groups according to their distribution in the phylogenetic tree and the mean base substitution per site between groups was conducted using the Kimura 2-parameters substitution model [26].
Results
Culex Flavivirus (CxFV) identification
After collection, the mosquitoes were identified by gender and grouped into pools. A total of 83 pools of female Culex quinquefasciatus, ten mosquitoes per pool, were analyzed. Among the samples, 41 (49.4 %) were positive for CxFV in multiplex RT-PCR and ten were isolated in C6/36. After three passages in C6/36 cells isolation was confirmed from pool 73C by Multiplex-RT-PCR. The complete genome of the isolate from pool 73C was perform by Illumina Plataform.
Next generation sequencing
NGS by Illumina generated a total of 69,242 reads of coverage, with 60,405 of CxFV reads, that generated 10,706 nucleotides. The sequence obtained was analyzed on BLAST, for confirmation of its contents and was deposited on GenBank under the accession number KT726939 and is here referred as CxFV_BR-RP01/2007.
Similarity analysis
The sequence of the open reading frame (ORF) of the Brazilian strain was aligned with all available CxFV full genome sequences deposited on GenBank. The percentage of identity between the sequences was determined with the BioEdit package using CxFV_BR-RP01/2007 as the query sequence [18]. The highest similarity was observed between the Brazilian sequence and the sequence from Mexico (98.5 %) followed by that of Uganda (98.2 %) (Additional file 1: Table S1). All other sequences were approximately 90 % identical to CxFV_BR-RP01/2007.
Genetic distance
Overall mean genetic distance was calculated for the complete ORF and for each specific genomic region. The ORF mean distance among all deposited sequences was 0.046. The mean genetic distance for each specific region is shown in Fig. 1. The NS2A region presented the lowest genetic distance and NS4B the highest.
Fig. 1.
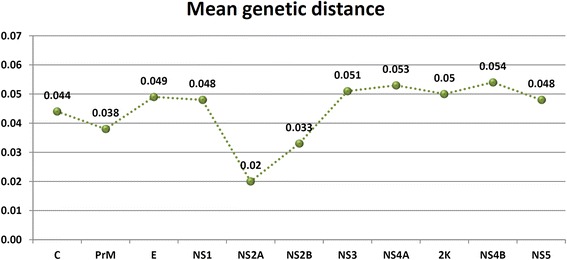
Overall mean genetic distance for CxFV specific genomic regions
A site-by-site genetic distance analysis was performed on SimPlot in order to compare the Brazilian sequence with those from other countries. Sequences were grouped by geographic location. The resulting plot is shown in Fig. 2, which also depicts each genomic region. The plot reveals a lower genetic distance between the Brazilian and the Mexican and Ugandan isolates. All other sequences show similar genetic distances along the genome. The genomic region with the lowest genetic distance for all geographic locations is the NS2A region.
Fig. 2.
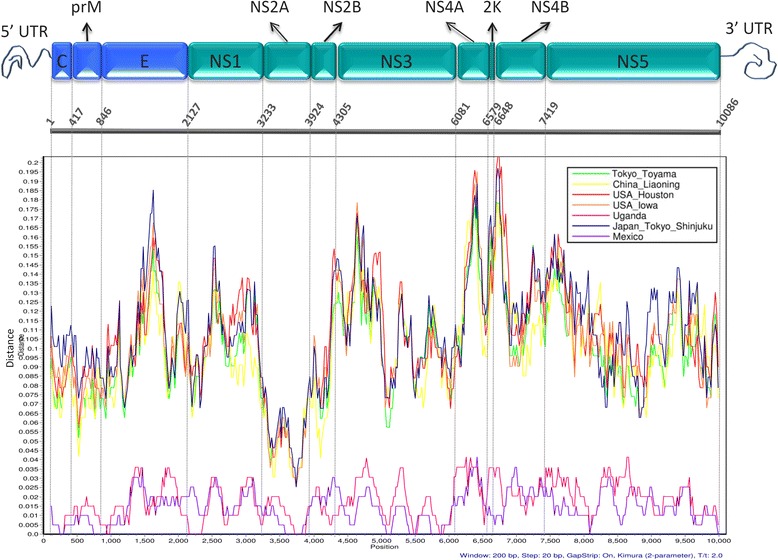
Genetic distance between the Brazilian sequence and isolates from other countries along the CxFV ORF. Analysis carried out on Simplot 3.5.1 with a 200 bp sliding window, Kimura 2-parameters distance model with CxFV_BR-RP01/2007 as query. Sequences were grouped by geographic location as specified in the legend. Schematic representation of CxFV genome based on the information provided by GenBank NC_008604.2. Structural proteins represented in blue and non-structural proteins represented in green
Selective pressure analysis
Global ω was estimated for the entire CxFV ORF and for each genomic region separately using HyPhy [21]. Our results indicate that the whole ORF is under strong purifying selection (ω = 0.107). The same applies for the individual genes encoding proteins as shown in Fig. 3. Interestingly, the NS2A region presented the highest ω (0.14), which is in contrast with the site-by-site genetic distance results, where this region presented the lowest values.
Fig. 3.
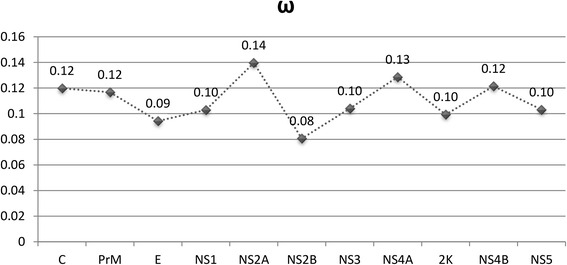
Global ω estimated for each genomic region separately using HyPhy
Site-specific selection analysis revealed 106 negatively selected and no positively selected sites. Results are graphically represented in Fig. 4. NS3 presented more negatively selected sites (6.3 %). The NS2A (1.3 %) protein is the one with less negatively selected sites followed by Core (2.2 %).
Fig. 4.
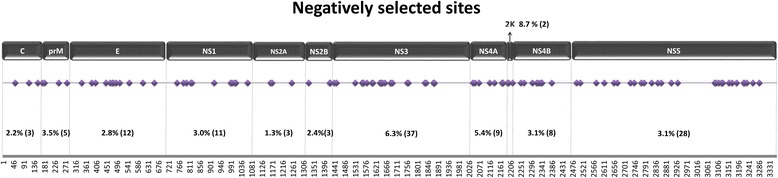
CxFV amino acid sites under negative selection. Percentage of negatively selected sites and (n) are indicated for each region. Schematic representation of CxFV genome based on the reference sequence NC_008604.2
Phylogenetic analysis
The phylogenetic tree presented in Fig. 5 was reconstructed based on the 26 full ORF sequence. It is composed of two main monophyletic Clades with high confidence (Posterior probablity: 1). Cade 1 includes most sequences, which are distributed in two groups, also supported high posterior probability (1). Clade 2 comprises viruses from Brazil, Mexico, and Uganda. All members of this clade share the same host, Culex quinquefasciatus. However, this is the same host species as that of the strain collected in Houston, USA, which groups in Clade 1.
Fig. 5.
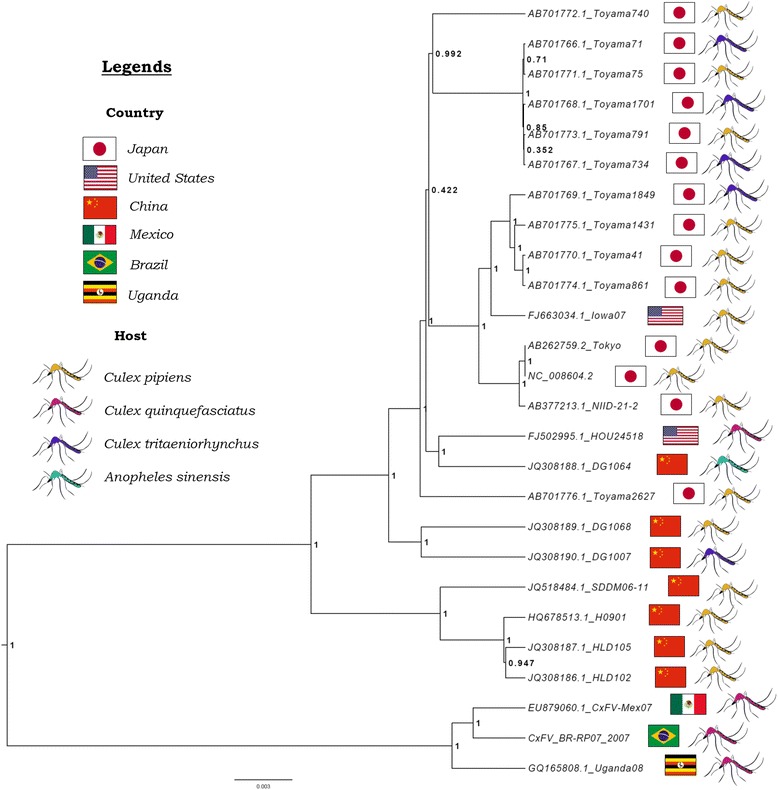
Bayesian phylogenetic tree of 26 sequences from CxFV ORF. Node labels represent the posterior probability. Country and host information is presented beside sequence name according to the legend above
In order to test for association of phylogeny with either host, geographical location or climate the posterior set of trees estimated by BEAST were analyzed by BaTS, which quantifies and statistically tests the phylogeny-trait correlation. Evidence of strong association was detected between phylogeny and climate (p < 0.05). Also, Association index (AI) and parsimony score (PS) indicate association for both host and geographical location (p < 0.05). However monophyletic clade (MC) significance reveals that it only applies for Japan and China (location) and Culex quinquefasciatus (host) (Additional files 2–4: Tables S2–S4).
In order to estimate the evolutionary divergence between the main clades of the phylogenetic tree, the mean genetic distances between these groups were calculated. Our results show that Clade 1 and Clade 2 diverge 10.4 %, while the two main branches that compose Clade 1 diverge 5.1 %.
Discussion
Insect-specific flaviviruses, including CxFV, are a highly divergent group within the Flavivirus genus that shares a common ancestor with all other members, including the disease-causing ones. Understanding the genetic characteristics of CxFV is important not only to understand its biology and evolutionary history, but also to clarify some aspects of the whole genus.
Culex Flavivirus primarily infects globally distributed mosquito species of the genus Culex, which are vectors for pathogenic flaviviruses like WNV, SLEV, and JEV [27–29]. The most represented hosts in this work, Culex pipiens and Culex quinquefasciatus, are members of the Culex pipiens complex that consists of closely related species that are difficult to distinguish morphologically [30]. Studying the geographical distribution of the host is important to understand the evolutionary relationships between the identified CxFV strains. Two main monophyletic branches, Clade 1 and Clade 2, can be observed in the phylogenetic tree with high branch support. Regarding countries, all Asian strains are grouped in Clade 1 along with some sequences from USA. Clade 2 is composed by sequences from different countries with only one sequence each. When considering host species, Clade 2 is composed only of sequences derived from Culex quinquefasciatus while Clade 1 as sequences from different hosts. This scenario prevents a clear association by simply analyzing the ancestry. Statistical analysis however revealed significant association between phylogeny and climate (temperate x tropical). Clade 1 consists of virus from temperate climate regions while Clade 2 is composed from sequences from region of tropical climate. Climate association is related with host species distribution. Although Culex pipiens occurs in temperate regions and Culex quinquefasciatus in tropical and subtropical regions their range overlaps and it has been proven they can hybridize [30–32]. When testing for host-phylogeny association only Culex quinquefasciatus was significantly associated. Moreover, country association was only found for Japan and China. Considering that both country and host are related to climate these factors may also be associated with CxFV ancestry, however the limited number of sequences from some hosts and geographic regions is probably preventing a more consistent conclusion.
We analyzed the genetic distance between the main branches of the phylogenetic tree that was reconstructed based on all CxFV ORF sequences described so far, including the Brazilian strain described in this work. Clades 1 and 2 consist of strong groups with robust branch support, which are 10.4 % genetically distant. These observations sustain the idea that they could be two different genotypes of CxFV. For Japanese Encephalitis Virus, another Culex borne virus, genotypes are defined by 12 % difference in nucleotide composition of the highly divergent PrM gene [33, 34]. Genotypes of DENV, also a flavivirus, are defined by 6 % difference within a serotype based on the E region [35, 36]. In both cases, the limits were determined arbitrarily but supported by strong phylogenetic evidence such as ours. Other researchers have previously suggested that CxFV should be classified into different genotypes, based on the phylogenetic analysis of the E genomic region [37]. According to those authors, Clade 1 described here would consist of Genotype 1 and Clade 2 of Genotype 2. Our analysis also evidenced that the two main branches, with strong branch support (1000), in Clade 1 diverge 5.1 %. These two branches could be considered different subtypes of CxFV Genotype 1 (1a and 1b). Similar results were obtained using the E region (Additional file 5: Table S5), suggesting it is a suitable region for CxFV genotyping and subtyping. We propose that groups of CxFV that are more than 9 % genetically distant could be classified as different genotypes and that differences higher than 4 % within these groups could be considered different subtypes. Although the cut-off values are arbitrarily defined, based on the genetic data available so far, they seem suitable.
Overall, selective pressure analysis indicates that the entire ORF, as well as specific regions, are undergoing purifying selection. Accordingly, site-by-site analysis found no positively selected sites and 106 sites under negative selection. This suggests that the virus is well adapted to its host and evolutionary forces are working against amino acid change. The infection of Culex mosquitoes by CxFV apparently does not result in disease. This observation is supported by the fact that the infection of C6/36 and AeAl-2, two Aedes albopictus cell lines, results only occasionally in moderate cytopathic effects [5, 6, 12]. If this is the case, any change in the genomic composition could disrupt the fine balance between what is advantageous for the virus and at the same time does not negatively affect the host, supporting the purifying selection.
Although the differences are not substantial, the NS2A region presents higher ω values and lower percentages of negatively selected sites. It is interesting, however, to notice that the same region also showed the lowest genetic distance. At first, these data might seem in disagreement. However, it suggests that NS2A displays low genetic variability in general, but a more relaxed pressure against amino acid change when compared to other regions. CxFV and other Flaviviruses from the insect-specific group present a frameshift signal that produces an alternate reading frame called fifo [38]. The signal consists of a stem-loop structure in the RNA genome that is harbored inside the NS2A region [39]. In this case, the selection is acting not only on the protein, but also on the RNA structure to avoid disturbing the frameshift signal. It is important to highlight that different amino acids might have similar chemical properties and a change would not necessarily disrupt the tertiary structure. This could explain why, despite the low genetic distance, this region is more permissive to amino acid change.
Conclusion
There are still many aspects of Culex Flavivirus and other insect-specific flaviviruses that needs to be clarified. Culex ssp. mosquitoes are distributed worldwide suggesting that CxFV should be present in more places than currently reported. Here we report an association between the ancestry of CxFV and climate. The identification of new strains from different countries and hosts is important for elucidating the ecology and evolutionary history of this virus. Based on the available sequences, we propose the classification of Culex Flavivirus into two genotypes (1 and 2) and the existence of two subtypes within genotype 1.
Acknowledgements
The authors acknowledge CNPq, CAPES and FAPESP for the financial support of MLN (2013/21719-3), CB (165802/2015-4; 2012/028560) and DV (PNPD/Microbiologia 1278383/2012).
Authors’ contributions
CB carried out the genome analysis, analyzed the data and wrote the first draft of the manuscript. DCM isolated the virus and participate in the genome sequencing and analysis of the data. DV and LSU carried out the genome sequence and genome annotation, and analysis of the data. PR, JPAJ, MLN designed the study, analyzed the data and reviewed the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional file
Percentage of identity between CxFV_BR-RP01/2007 and sequences from GenBank. Data determined by Sequence Identity Matrix tool on BioEdit package. Table S2. Results from BaTS when testing for country association. State 0 – Japan; State 1 – Brazil; State 2 – Mexico; State 3 – USA; State 4 – Uganda; State 5 – China; AI – Association Index; PS – Parsimony Score; MC – Monophyletic clade. Table S3. Results from BaTS when testing forhostassociation. State 0 – Culexpipiens; State 1 – Culextritaeniorhynchus; State 2 – Culexquinquefasciatus; State 3 – Anopheles sinensis; AI – Association Index; PS – Parsimony Score; MC – Monophyletic clade. Table S4. Results from BaTS when testing for climate association. State 0 – Temperate; State 1 – Tropical; AI – Association Index; PS – Parsimony Score; MC – Monophyletic clade. Table S5. Mean genetic distance between the sequences from envelope E region. Sequences were divided into groups according to their distribution in the phylogenetic tree and the mean base substitution per site between groups was conducted using the Kimura 2-parameters substitution model. Cluster 1 - KT726939, GQ165808.1, EU879060.1; Cluster 2 - JQ308187.1, JQ308186.1, JQ518484.1, HQ678513.1; Cluster 3 – All others. (DOCX 83.3 kb)
Contributor Information
Cíntia Bittar, Email: cibittar@gmail.com.
Daiane Cristina Machado, Email: daiane_cris1@hotmail.com.
Danila Vedovello, Email: danvedo@hotmail.com.
Leila Sabrina Ullmann, Email: leila_ullmann@yahoo.com.br.
Paula Rahal, Email: rahalp@yahoo.com.br.
João Pessoa Araújo Junior, Email: jpessoa@ibb.unesp.br.
Maurício Lacerda Nogueira, Phone: +55 17 32015731, Email: mnogueira@famerp.br.
References
- 1.Stollar V, Thomas VL. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology. 1975;64:367–77. doi: 10.1016/0042-6822(75)90113-0. [DOI] [PubMed] [Google Scholar]
- 2.Huhtamo E, Moureau G, Cook S, Julkunen O, Putkuri N, Kurkela S, Uzcategui NY, Harbach RE, Gould EA, Vapalahti O, de Lamballerie X. Novel insect-specific flavivirus isolated from northern Europe. Virology. 2012;433:471–8. doi: 10.1016/j.virol.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobson-Peters J, Yam AW, Lu JW, Setoh YX, May FJ, Kurucz N, Walsh S, Prow NA, Davis SS, Weir R, et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One. 2013;8:e56534. doi: 10.1371/journal.pone.0056534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crabtree MB, Sang RC, Stollar V, Dunster LM, Miller BR. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch Virol. 2003;148:1095–118. doi: 10.1007/s00705-003-0019-7. [DOI] [PubMed] [Google Scholar]
- 5.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, Dorman KS, Platt KB, Bartholomay LC, Soto V, et al. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, Yuda M, Takasaki T, Kobayashi M, Sawabe K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–14. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Guo J, An S: JQ308188.1 - Culex flavivirus was isolated from Culex pipiens and Anopheles in liaoning province. In Nucleotide. GenBank: NCBI; 2011.
- 8.Obara-Nagoya M, Yamauchi T, Watanabe M, Hasegawa S, Iwai-Itamochi M, Horimoto E, Takizawa T, Takashima I, Kariwa H. Ecological and genetic analyses of the complete genomes of Culex flavivirus strains isolated from Culex tritaeniorhynchus and Culex pipiens (Diptera: Culicidae) group mosquitoes. J Med Entomol. 2013;50:300–9. doi: 10.1603/ME12159. [DOI] [PubMed] [Google Scholar]
- 9.Blitvich BJ, Lin M, Dorman KS, Soto V, Hovav E, Tucker BJ, Staley M, Platt KB, Bartholomay LC. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa. J Med Entomol. 2009;46:934–41. doi: 10.1603/033.046.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook S, Moureau G, Harbach RE, Mukwaya L, Goodger K, Ssenfuka F, Gould E, Holmes EC, de Lamballerie X. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J Gen Virol. 2009;90:2669–78. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DY, Guzman H, Bueno R, Jr, Dennett JA, Auguste AJ, Carrington CV, Popov VL, Weaver SC, Beasley DW, Tesh RB. Characterization of Culex Flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology. 2009;386:154–9. doi: 10.1016/j.virol.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 12.Morales-Betoulle ME, Monzon Pineda ML, Sosa SM, Panella N, Lopez MR, Cordon-Rosales C, Komar N, Powers A, Johnson BW. Culex flavivirus isolates from mosquitoes in Guatemala. J Med Entomol. 2008;45:1187–90. doi: 10.1603/0022-2585(2008)45[1187:CFIFMI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Saiyasombat R, Dorman KS, Garcia-Rejon JE, Lorono-Pino MA, Farfan-Ale JA, Blitvich BJ. Isolation and sequence analysis of Culex flavivirus from Culex interrogator and Culex quinquefasciatus in the Yucatan Peninsula of Mexico. Arch Virol. 2010;155:983–6. doi: 10.1007/s00705-010-0665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machado DC, Mondini A, dos Santos Santana V, Yonamine PT, Chiaravalloti Neto F, Zanotto PM, Nogueira ML. First identification of Culex flavivirus (Flaviviridae) in Brazil. Intervirology. 2012;55:475–83. doi: 10.1159/000337166. [DOI] [PubMed] [Google Scholar]
- 15.de Morais Bronzoni RV, Baleotti FG, Ribeiro Nogueira RM, Nunes M, Moraes Figueiredo LT. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol. 2005;43:696–702. doi: 10.1128/JCM.43.2.696-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 17.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–60. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–9. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 22.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 23.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.FigTree 1.4 [http://tree.bio.ed.ac.uk/software/figtree/].
- 25.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8:239–46. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.WHO . Fact sheet N°354. Geneva: Word Health Organization; 2011. West Nile virus. [Google Scholar]
- 28.WHO . Fact sheet No 386. Geneva: World Health Organization; 2014. Japanese encephalitis. [Google Scholar]
- 29.CDC . Technical Fact Sheet. Atlanta: Centers for Disease Control and Prevention; 2009. Saint Louis Encephalitis. [Google Scholar]
- 30.Farajollahi A, Fonseca DM, Kramer LD, Marm Kilpatrick A. “Bird biting” mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect Genet Evol. 2011;11:1577–85. doi: 10.1016/j.meegid.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonseca DM, Smith JL, Kim HC, Mogi M. Population genetics of the mosquito Culex pipiens pallens reveals sex-linked asymmetric introgression by Culex quinquefasciatus. Infect Genet Evol. 2009;9:1197–203. doi: 10.1016/j.meegid.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fonseca DM, Smith JL, Wilkerson RC, Fleischer RC. Pathways of expansion and multiple introductions illustrated by large genetic differentiation among worldwide populations of the southern house mosquito. Am J Trop Med Hyg. 2006;74:284–9. [PubMed] [Google Scholar]
- 33.Solomon T, Ni H, Beasley DW, Ekkelenkamp M, Cardosa MJ, Barrett AD. Origin and evolution of Japanese encephalitis virus in southeast Asia. J Virol. 2003;77:3091–8. doi: 10.1128/JVI.77.5.3091-3098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WR, Tesh RB, Rico-Hesse R. Genetic variation of Japanese encephalitis virus in nature. J Gen Virol. 1990;71(Pt 12):2915–22. doi: 10.1099/0022-1317-71-12-2915. [DOI] [PubMed] [Google Scholar]
- 35.Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–93. doi: 10.1016/0042-6822(90)90102-W. [DOI] [PubMed] [Google Scholar]
- 36.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–41. doi: 10.1016/S0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huanyu W, Haiyan W, Shihong F, Guifang L, Hong L, Xiaoyan G, Lizhi S, Rayner S, Aiqiang X, Guodong L. Isolation and identification of a distinct strain of Culex Flavivirus from mosquitoes collected in Mainland China. Virol J. 2012;9:73. doi: 10.1186/1743-422X-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Firth AE, Blitvich BJ, Wills NM, Miller CL, Atkins JF. Evidence for ribosomal frameshifting and a novel overlapping gene in the genomes of insect-specific flaviviruses. Virology. 2010;399:153–66. doi: 10.1016/j.virol.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blitvich BJ, Firth AE. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses. 2015;7:1927–59. doi: 10.3390/v7041927. [DOI] [PMC free article] [PubMed] [Google Scholar]


