Abstract
Interruption of combination antiretroviral therapy in HIV-1-infected individuals leads to rapid viral rebound. Here we report the results of a phase IIa open label clinical trial evaluating 3BNC117, a broad and potent neutralizing antibody (bNAb) against the CD4 binding site of HIV-1 Env1, in the setting of analytical treatment interruption in 13 HIV-1-infected individuals. Participants with 3BNC117-sensitive virus outgrowth cultures were enrolled. Two or four 30 mg kg−1 infusions of 3BNC117, separated by 3 or 2 weeks, respectively, are generally well tolerated. Infusions are associated with a delay in viral rebound for 5–9 weeks after two infusions, and up to 19 weeks after four infusions, or an average of 6.7 and 9.9 weeks respectively, compared with 2.6 weeks for historical controls (P < 0.00001). Rebound viruses arise predominantly from a single provirus. In most individuals, emerging viruses show increased resistance, indicating escape. However, 30% of participants remained suppressed until antibody concentrations waned below 20 μg ml−1, and the viruses emerging in all but one of these individuals showed no apparent resistance to 3BCN117, suggesting failure to escape over a period of 9–19 weeks. We conclude that administration of 3BNC117 exerts strong selective pressure on HIV-1 emerging from latent reservoirs during analytical treatment interruption in humans.
A fraction of HIV-1 infected individuals develops broad and potent serologic activity against the virus. Single-cell antibody cloning methods2 have uncovered the source of this activity as broadly neutralizing antibodies (bNAbs), which target different sites on the HIV-1 envelope spike protein, gp1601–3.
In animal models, bNAbs show potent prophylactic activity, suppress established viraemia, and delay viral rebound during analytical treatment interruption (ATI)4–8. In humans, a phase I clinical trial showed that 3BNC117 is generally safe and effective in transiently reducing viraemia in chronically HIV-1-infected individuals9. A single infusion of 3BNC117 was well tolerated, rapidly decreased viral loads in viraemic individuals by an average of 1.48 log10 copies per ml, with durable activity for 4 weeks9. In addition, 3BNC117 increased autologous antibody responses in HIV-1-infected individuals, and enhanced clearance of infected cells in humans and in humanized mice10,11. VRC01, a less potent bNAb that also targets the CD4-binding site, suppressed viraemia by 1.14 log10 (refs 12,13 and Fig. 1a, b).
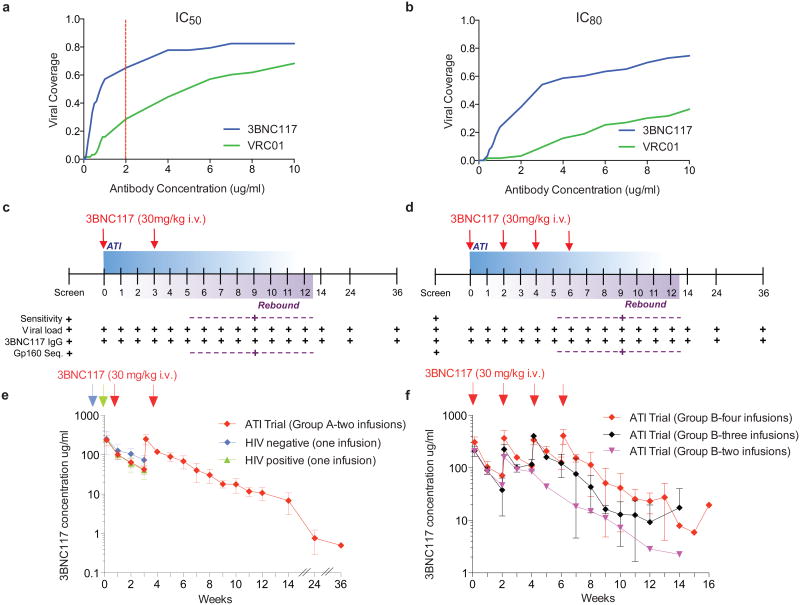
Figure 1. 3BNC117 neutralization coverage, trial design and pharmacokinetics of 3BNC117 in HIV-1-infected individuals during ATI.
a, b, Sensitivity of virus outgrowth cultures from 63 ART suppressed individuals to 3BNC117 and VRC01 (Supplementary Table 1). The y-axis shows the fraction of viral outgrowth culture supernatants neutralized by a given antibody concentration (x-axis) in Tzm-bl assays. Red line indicates cut-off IC50 (2 μg ml−1) for participation in the trial. c, d, Diagrammatic representation of study groups A and B respectively. 3BNC117 infusions indicated by the red arrows, and sampling for PK and virologic studies indicated below. Numbers indicate study weeks. e, f, 3BNC117 levels as determined by ELISA for group A (n = 6, left panel, red), group B (n = 7, right panel, red (n = 4), black (n = 2) and purple (n = 1)), HIV-1 negative (n = 3, blue) and viraemic individuals (n = 6, green)9. Curves indicate mean 3BNC117 levels, error bars the standard deviation. Arrows indicate 3BNC117 infusions.
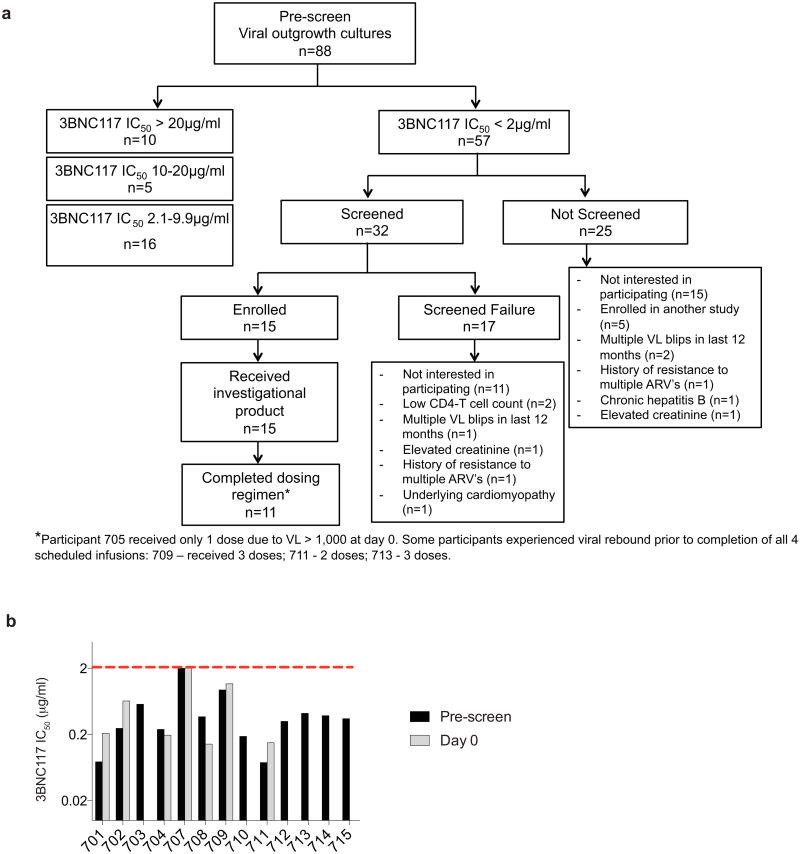
To investigate whether 3BNC117 can suppress viral rebound from the latent reservoir during ATI in chronically suppressed HIV-1 infected humans, we conducted a phase IIa open label clinical trial. To select participants with 3BNC117-sensitive viruses in their latent reservoirs, we performed bulk viral outgrowth cultures of peripheral blood mononuclear cells (PBMCs) from individuals whose viraemia was suppressed by combination antiretroviral therapy (ART). The resulting isolates were screened for sensitivity to 3BNC117 using the TZM-bl assay (Supplementary Table 1). Of 63 individuals screened, only 11% yielded viruses that were fully resistant to 3BNC117 (IC50 > 20 μg/ml), and 65% were sensitive to 3BNC117 IC50 at concentrations below 2.0 μg/ml. In contrast only 29% were similarly sensitive to VRC01 (Fig. 1a and b, Extended Data Fig. 1 and Supplementary Table 1).
We enrolled HIV-1 infected individuals who were on suppressive antiretroviral therapy (ART) with plasma viral loads <50 HIV-1 RNA copies per ml for at least 12 months, had CD4 counts >500 cells per mm3, yielded 3BNC117-sensitive outgrowth viruses (IC50 ≤ 2.0 μg ml−1), and whose viral load at screen was <20 copies per ml (Extended Data Fig. 1, Supplementary Tables 2 and 4, and Methods). Participants were enrolled in two groups: eightin group A to receive two 30 mg kg−1 infusions three weeks apart, while seven in group B received up to four 30 mg kg−1 infusions at two-week intervals (Fig. 1c, d, Supplementary Table 2). Two group A participants had viral loads >20 copies per ml at the time of infusion and were excluded from further analysis (Supplementary Tables 2 and 4).Participants are numbered 701–715 (Supplementary Table 2).
ATI was started 2 days after the first 3BNC117 infusion. ART was reinitiated and infusions were stopped after two consecutive plasma viral load measurements exceeded 200 copies per ml. All individuals on non-nucleoside reverse transcriptase inhibitors (NNRTIs) were switched to an integrase-inhibitor-based regimen (dolutegravir plus tenoforvir disoproxil fumarate/emtricitabine) four weeks before ATI owing to the long half-life of NNRTIs (Supplementary Table 2).
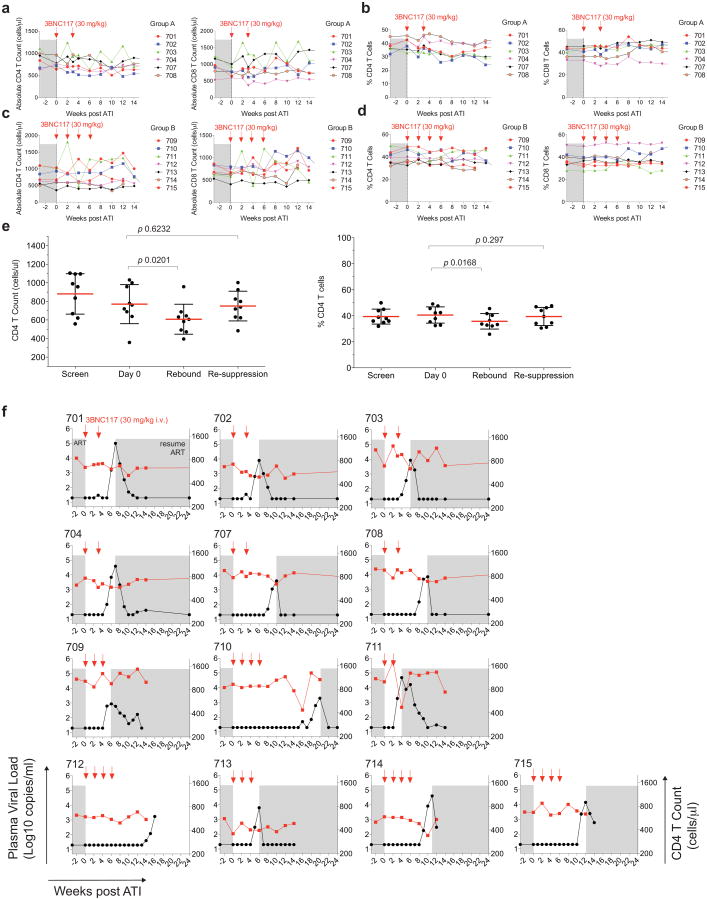
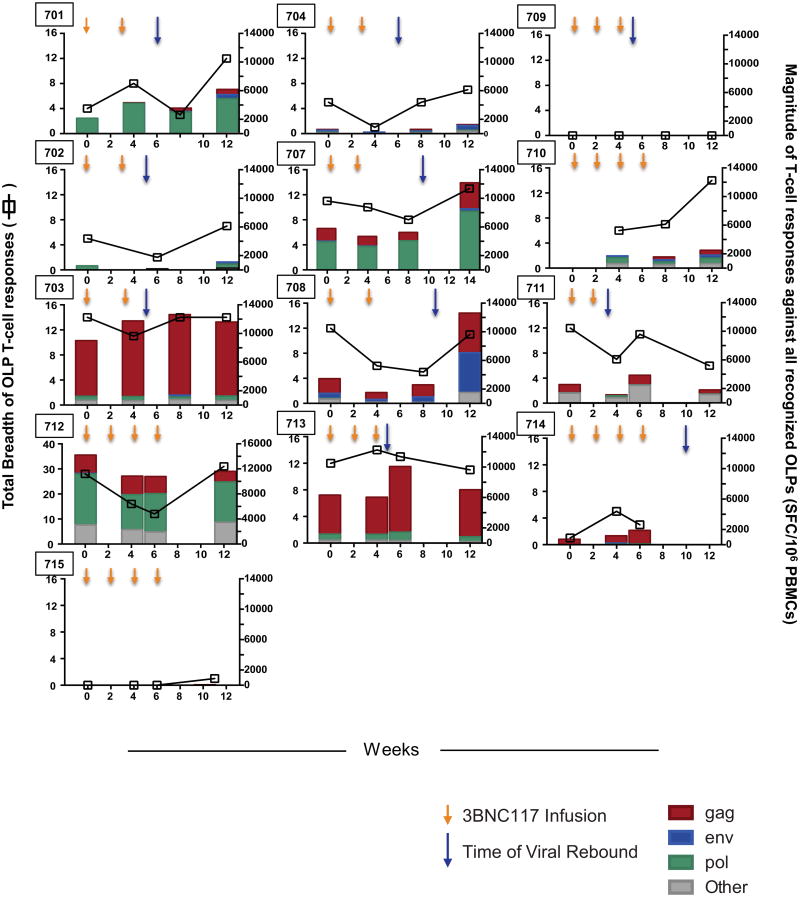
Both dosing regimens were generally well tolerated. The majority of reported adverse events were transient and grade 1 in severity (Supplementary Table 5). The mean CD4 T-cell count at baseline (day 0) was 747 cells per mm3, and the average change in CD4 T-cell counts between start of ATI and rebound was −127 cells per mm3. Although CD4T cells declined modestly during viral rebound in some participants, CD4 T-cells returned to baseline by week 12 in most participants (mean 828 cells per mm3) (Extended Data Fig. 2 and Supplementary Table 4). Of 12 individuals tested, 5 showed measurable increases in the magnitude and/or breadth of T cell responses to HIV-1 12 weeks after ATI, relative to baseline (Extended Data Fig. 3). None of the participants experienced acute retroviral syndrome during rebound, and viraemia was re-suppressed below 20 copies per ml in all participants within 2–7 weeks after restarting ART (Supplementary Table 4). We conclude that up to four 30 mg kg−1 infusions of 3BNC117 during ATI are generally safe and well tolerated.
By anti-idiotype ELISA9 the half-life of 3BNC117 during ATI was 19.6 days among group A participants, and 14.1 days among those in group B (Fig. 1e, f and Supplementary Table 4). These measurements are similar to previously reported values for 3BNC117 in HIV-1-infected individuals on ART9 (Fig. 1e).
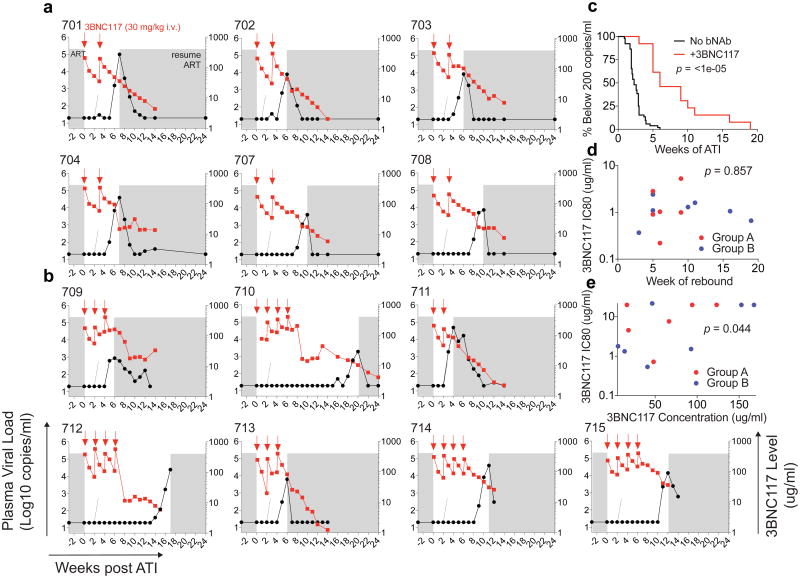
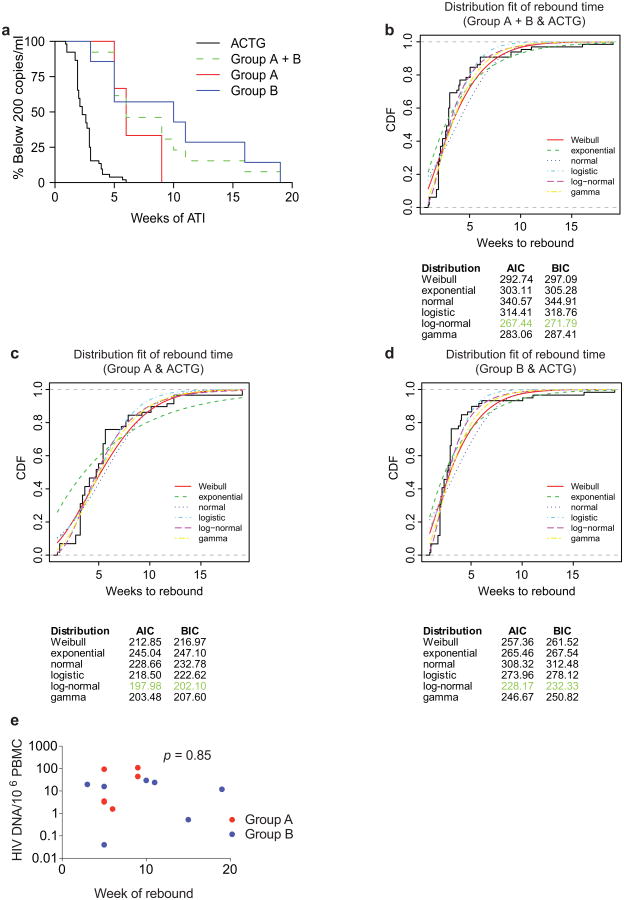
All six group A participants maintained viral loads below 200 copies per ml during the first 4 weeks, with rebound 5–9 weeks after ART interruption (Fig. 2a and Supplementary Table 4a). In group B, rebound occurred 3–19 weeks after ATI, with four out of seven (57%) participants remaining suppressed for at least 10 weeks (Fig. 2b and Supplementary Table 4b). The average time to rebound was 6.7 weeks in group A, 9.9 weeks in group B, and 8.4 weeks for all participants together, compared with 2.6 weeks for matched historical non-infused control individuals (Fig. 2c, Extended Data Fig. 4a, Supplementary Tables 4, 6 and 7). Altogether, 6 of the 13 infused individuals (46%) remained suppressed until at least 9 weeks after ATI. Relative to matched historical control individuals, the delay to rebound among all 3BNC117-infused participants was highly significant (P < 0.00001, weighted log-rank test, Fig. 2c, Extended Data Fig. 4, Supplementary Tables 4, 6 and 7 and Methods). We conclude that repeated infusions of 3BNC117 are generally safe and significantly delay HIV-1 rebound from the latent reservoir during ATI.
Figure 2. Delay in viral rebound in the presence of 3BNC117.
a, b, Plasma viral loads and 3BNC117 levels in group A and group B participants respectively. 3BNC117 infusions are indicated with red arrows. The left y-axis shows plasma viral loads in RNA copies per ml (black curves), and right y-axis shows antibody levels measured by ELISA (red curves). Average rebound time point (2.6 weeks, Supplementary Table 6) in 52 ACTG trial participants who underwent ATI without antibody treatment29 is shown with dotted lines. Grey areas indicate ART therapy. c, Kaplan–Meier plot summarizing viral rebound in 52 ACTG trial participants who underwent ATI without antibody treatment (black, Supplementary Table 6), and the combination of all 13 participants (red) who underwent ATI with 3BNC117 infusions. The y-axis indicates the percentage of participants with viral levels below 200 RNA copies per ml, x-axis indicates weeks after ATI initiation. The P value is based on a bootstrap version of the weighted log-rank test adjusting for the potential confounders ‘years on ART’, and ‘age’ (Supplementary Table 7, Methods Statistical Analyses). d, Dot plot indicating the relationship between 3BNC117 sensitivity of pre-infusion outgrowth cultures at screening (y-axis, IC80 in μg ml−1) and the week of rebound (x-axis). Group A (n = 6) and group B (n = 7) participants are coloured red and blue respectively. The P value was derived from calculating the Pearson correlation coefficients. e, Dot plot indicating the relationship between 3BNC117 sensitivity of rebound outgrowth cultures (y-axis, IC80 in μg ml−1) and the 3BNC117 serum concentration at rebound (x-axis, in μg ml−1). 704, 708, 709 and 713 did not reach IC80 at the concentrations tested and were assigned a value of 22 μg ml−1. Group A (n = 6) and group B (n = 7) participants are coloured red and blue, respectively. The P value was derived from calculating the Pearson correlation coefficients.
Time to viral rebound did not correlate with pre-ATI viral culture sensitivity to 3BNC117, nor to baseline levels of cell-associated HIV-1 DNA (Fig. 2d and Extended Data Fig. 4e). Therefore the significance of viral outgrowth sensitivity as an inclusion criterion is not clear. 3BNC117 levels at rebound were also variable, ranging from 6–168 μg ml−1, but were directly correlated to the IC80 of the emerging virus (Fig. 2a, b, e)
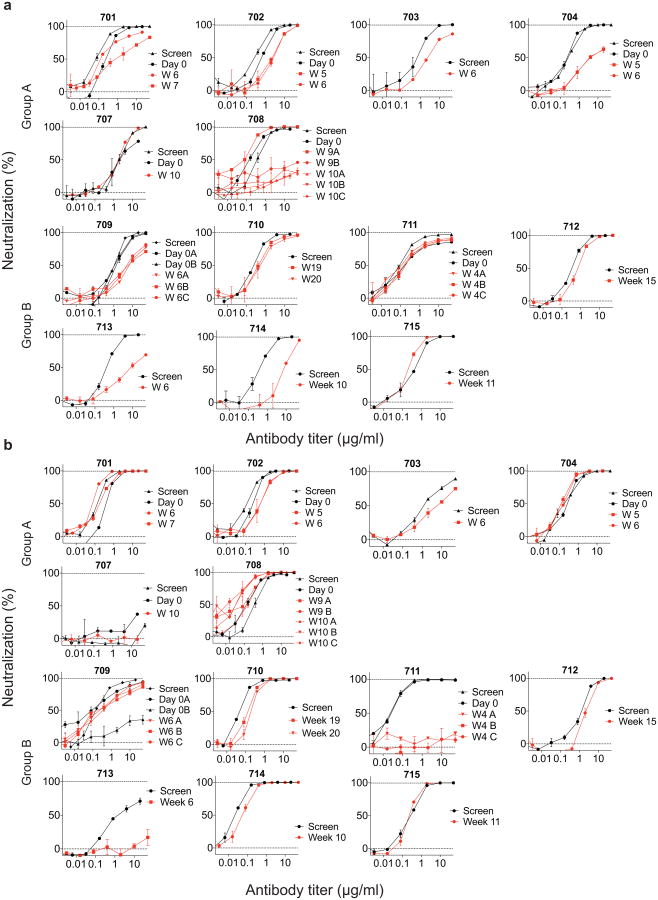
To determine whether rebound was associated with resistance to 3BNC117, we compared pre-infusion and rebound viral outgrowth cultures. A majority (8/13) of participants had rebound viruses that were more resistant to 3BNC117 (IC80 > threefold higher, Fig. 3a, c, Extended Data Fig. 5a, Supplementary Table 3). Among group A participants, all but one (707) had more resistant rebound viruses; however, among group B participants, four of seven (710, 711, 712 and 715) showed similar pre-infusion and rebound sensitivity to 3BNC117 (Fig. 3a, c, Extended Data Fig. 5a, Supplementary Table 3). Among these five individuals, 711 was the earliest to rebound at 3 weeks, despite having viruses that were surprisingly sensitive to 3BNC117 as measured by IC50 (Fig. 2b, Extended Data Fig. 5a, Supplementary Tables 3 and 4). However, 100% neutralization was not achieved against 711 rebound or pre-infusion viruses, even at high (50 μg ml−1) antibody concentrations, suggesting that 3BNC117 was not fully therapeutic (Extended Data Fig. 5, Supplementary Table 3). Thus, the only participant in the study to rebound within 3 weeks of ATI may have done so because of pre-existing resistance to 3BNC117 by the dominant virus in the reservoir.
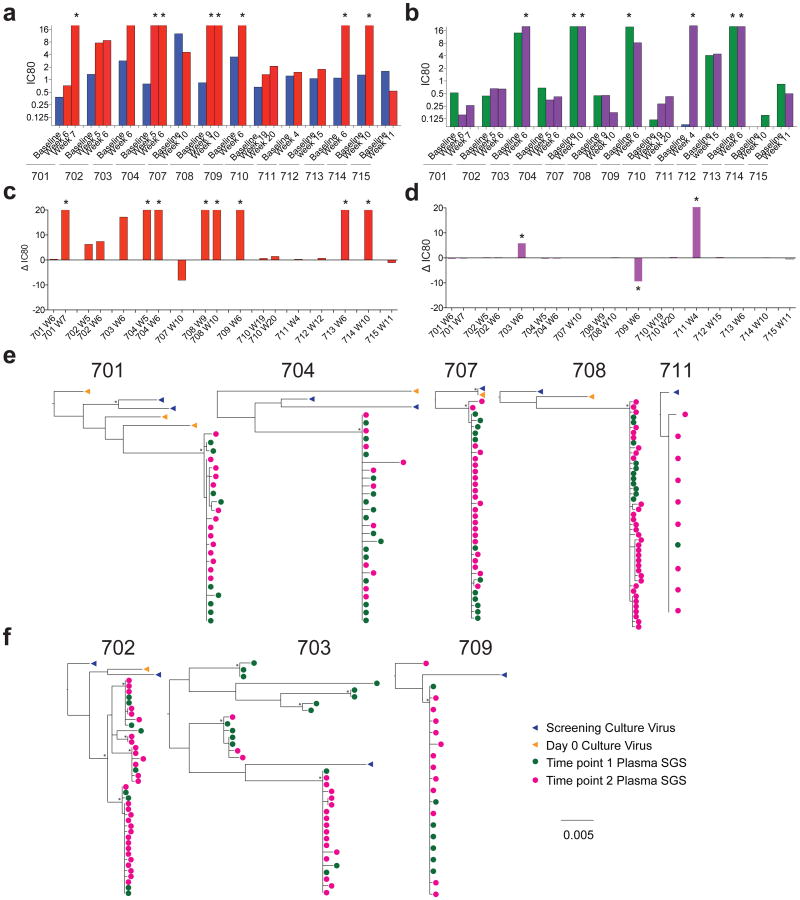
Figure 3. Viral rebound during ATI and 3BNC117 treatment.
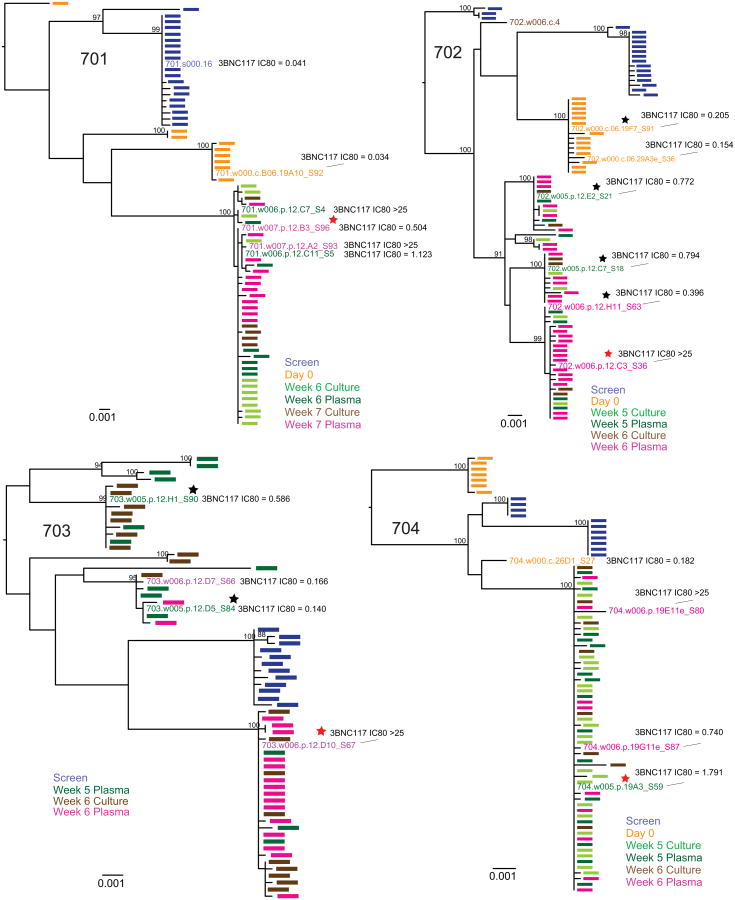
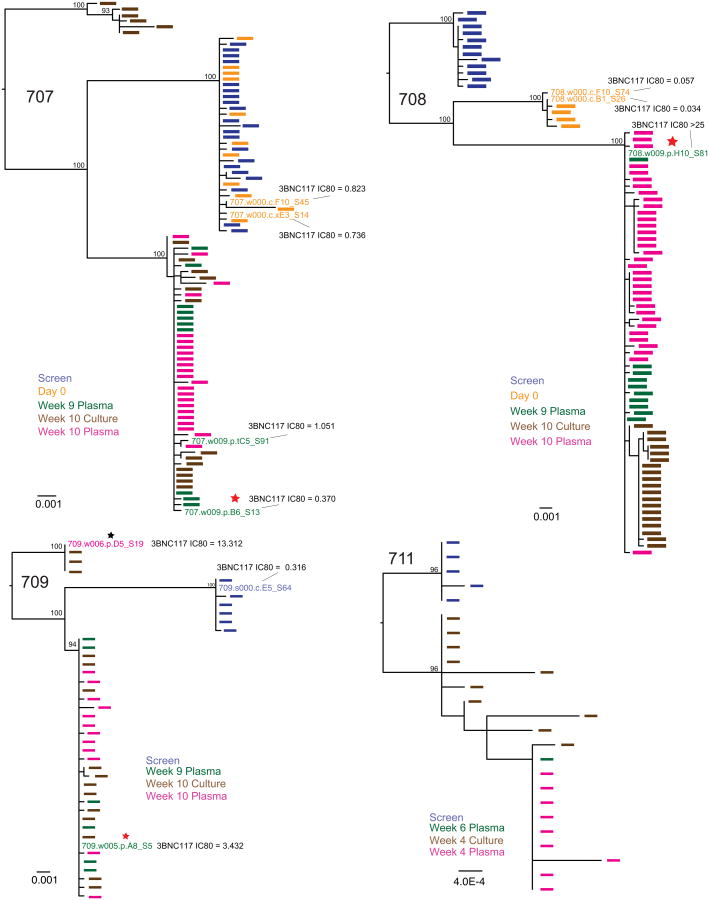
a, b, Graph of 3BNC117 (a) or 10-1074 (b) IC80 titres against baseline and rebound outgrowth cultures. Blue and green bars represent average IC80 titres against screen and day 0 outgrowth cultures; red and purple bars represent IC80 titres against rebound outgrowth cultures from the indicated time points. Asterisks indicate cultures failing to reach an IC80 up to 20 μg ml−1. c, d, Difference between rebound and pre-infusion culture IC80 titres (from a, b) for 3BNC117 (c) or 10-1074 (d). For cultures failing to reach an IC80 up to 20 μg ml−1, a value of 20 μg ml−1 was assigned, and such cultures are marked with an asterisk. e, f, Clonality of the rebound virus. Maximum likelihood phylogenetic trees comparing pre-ATI single genome derived env sequences (blue and orange) to rebound plasma env sequences (green and pink) are shown for participants whose rebound comprised single (e) versus multiple (f) viruses (Supplementary Table 8). Pre-ATI culture sequences were inferred as described in the Methods section ‘Statistical analyses’.
The other 4 participants that showed no change between pre- and post-infusion culture sensitivity to 3BNC117, 707, 710, 712, and 715, rebounded relatively late at 9, 19, 16 and 11 weeks after ATI, respectively (Figs 2a, b, 3a, c, Extended Data Fig. 5, Supplementary Tables 3 and 4). In all of these individuals rebound was associated with relatively low antibody concentrations ranging from 6–41 μg ml−1 (mean 19.7 μg ml−1). This antibody concentration represents 9.6-fold the mean IC80 for the rebounding viruses, which is consistent with previous reports on the relationship between suppressive 3BNC117 concentration and neutralization titre in macaques14 (Fig. 2, Extended Data Fig. 5, Supplementary Tables 3 and 4).
To determine whether viral rebound during ATI was associated with resistance to other bNAbs undergoing clinical testing, we examined sensitivity to 10-1074 (ref. 15), which targets a different and non-overlapping epitope on the HIV-1 trimer (Fig. 3b, d, Extended Data Fig. 5, and Supplementary Table 3). With the exception of 703 and 711, the participants' rebound cultures did not show increased resistance to 10-1074. We conclude that rebound during ATI in the presence of 3BNC117 is infrequently associated with increased resistance to 10-1074.
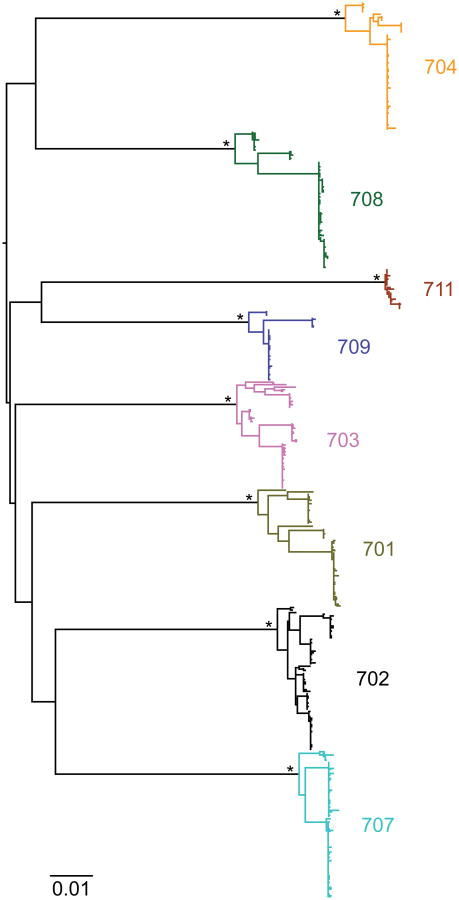
To characterize viruses emerging from the latent reservoir further, we performed single genome sequencing (SGS) of viral RNA from the plasma and viral outgrowth cultures from eight individuals. Phylogenetic analysis of these sequences indicated that all of these eight trial participants were infected with epidemiologically unrelated viruses (Extended Data Fig. 6). Given the limited sampling of the pre-infusion reservoir, rebound viruses did not always fall within the radiation of pre-infusion viral isolates (Fig. 3e, f, Extended Data Figs 7 and 8, Supplementary Figs 1 and 2).
Remarkably, in five of eight participants, all rebounding virus sequences clustered within a low diversity lineage, consistent with the clonal expansion of a single recrudescent virus (Fig. 3e, f, Extended Data Figs 7 and 8, Supplementary Table 8). These data contrast with individuals undergoing ATI in the absence of antibody infusion, where virus rebound is consistently polyclonal, indicating the activation of multiple latently infected cells16–19. Thus, in addition to delaying rebound, 3BNC117 appears to restrict the outgrowth of viral genotypes from the latent reservoir.
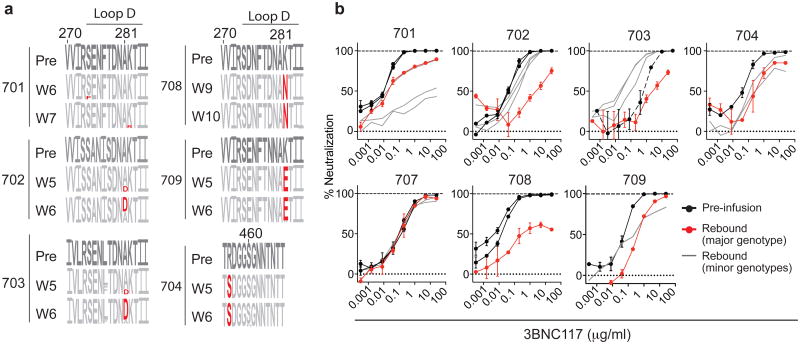
Six of eight participants sequenced had rebound viral outgrowth culture and/or plasma sequences that indicated 3BNC117 resistance. For example, in 704, all rebound viruses carried a serine at position 456 (Fig. 4a and Supplementary Figs 1 and 2), which may disrupt a highly conserved salt bridge that maintains the V5 loop's position and conformation20–22. Similarly, in 708 and 709, nearly all rebound viruses carried atypical residues at position 282, where a lysine residue typically forms a salt bridge with 3BNC117 (Fig. 4a and Supplementary Figs 1 and 2)23. However, documented 3BNC117 resistance mutations24 were not universally identified among rebound viral strains (Fig. 4a and Supplementary Figs 1 and 2). Only a minor fraction (3 of 23) of sequences in the rebound population of participant 701 had potential resistance-conferring residues in Loop D (274F, 282R), while the remaining sequences did not (Fig. 4a and Supplementary Figs 1 and 2). Similarly, in 702 and 703, only a subset of rebound viruses carried a putative resistance-conferring A281D change1,23. Nevertheless, the frequency of this change increased markedly with time in both participants, indicating continued selection for 3BNC117 resistance (Fig. 4a and Supplementary Figs 1 and 2). For participants 707 and 711, no sequence features were identified that would indicate 3BNC117 resistance.
Figure 4. 3BNC117 resistance in rebound viruses.
a, Logogram shows env gp120 regions (amino acid positions; 270–285 and 455–467, according to HXBc2 numbering) indicating sequence changes from pre-infusion culture(s) (first row) to rebound sequences derived from plasma SGS at the indicated time points. The frequency of each amino acid is indicated by its height. Red residues represent mutations predicted to affect neutralization30. b, 3BNC117 neutralization sensitivity of pseudoviruses derived from pre-infusion or rebound SGS. Black lines represent pre-infusion virus envs; red lines represent the major env at rebound for each participant (Extended Data Figs 7 and 8, Supplementary Tables 8 and 9 and Methods); grey lines represent minor rebound envs in participants with multiple rebound viruses or variants that evolved after rebound (Extended Data Figs 7 and 8, Supplementary Table 9). Symbols reflect the means of two technical replicates; error bars denote standard deviation.
To determine the sensitivity of rebound viruses to 3BNC117, we performed TZM-bl neutralization assays using pseudoviruses typed with SGS Env genotypes (Fig. 4b, Extended Data Figs 7 and 8, Supplementary Figs 1 and 2 and Supplementary Table 9). With the exception of participant 707, who rebounded 9 weeks after ATI at very low 3BNC117 titres, Env genotypes at rebound were more resistant to 3BNC117 than pre-ATI (Fig. 4b, Extended Data Figs 7 and 8 and Supplementary Table 9). We conclude that viral rebound during ATI in the presence of 3BNC117 selects for the emergence of resistant variants, indicating strong selection pressure by this antibody on viral populations arising from the reservoir.
Antibody potency and half-life are directly correlated with HIV-1 prophylaxis in pre-clinical models. For example, VRC01, a CD4bs antibody that is less potent than 3BNC1171, is less effective than 3BNC117 in preventing SHIVAD8 infection in macaques8,25. Consistent with these observations, clinical trials with combinations of three less-potent first-generation bNAbs showed limited effects on viral rebound in the setting of ATI in chronically infected individuals26,27. In addition, selective pressure as evidenced by escape mutations was only observed for one of the three antibodies used in the combination, 2G1226,27. In contrast, 3BNC117 alone significantly delayed viral rebound with nearly half of all individuals remaining below 200 copies per ml until at least 9 weeks, including four individuals who failed to develop resistance and only rebounded at low antibody concentrations. We speculate that the difference in efficacy between 3BNC117 and less potent bNAbs in the setting of ATI is due to increased potency and/or a longer half-life1,9,13.
Nevertheless, the majority of the individuals we studied rebounded at high 3BNC117 serum concentrations. A single viral genotype displaying increased resistance to 3BNC117 established rebound in most cases. These viruses represent pre-existing dormant variants that emerged from the latent reservoir. The time to rebound did not correlate with the amount of viral DNA in circulating PBMCs; however, this is a poor measure of the HIV-1 reservoir, since most integrated proviruses in patients on ART are defective28. Instead, the delay in viral rebound may represent a measure of the frequency of 3BNC117-resistant variants in the latent reservoir.
Combinations of drugs are needed to maintain HIV-1 suppression in effective ART regimens. Similarly, combinations of antibodies were required to suppress viraemia in humanized mice6,7. We speculate that combinations of bNAbs will also be needed to increase the frequency of individuals that remain suppressed by antibody during ATI.
Whether 3BNC117 can also impact the size and composition of the latent reservoir during ATI will require additional studies.
Methods
Study design
An open-label, dose-escalation phase 2a study was conducted in HIV-1-infected participants (http://www.clinicaltrials.gov; NCT02446847). Study participants were enrolled sequentially according to eligibility criteria. Group A received 3BNC117 on days 0 and 21 at a dose of 30 mg/kg body weight at a rate of 250 ml/hour. Group B received 3BNC117 on days 0, 14, 28 and 42 at a dose of 30 mg/kg, as long as viral rebound did not occur. Antiretroviral therapy (ART) was discontinued 2 days after the first 3BNC117 infusion (day 2). Plasma HIV-1 RNA levels were monitored weekly, and ART was resumed when viral load increased to ≥200 c.p.m. in two consecutive weekly measurements.
Study participants were followed for 36 weeks after the first infusion. Safety data are reported until week 36 for participants enrolled in group A and until week 14 for participants enrolled in group B. All participants provided written informed consent before participation in the study and the study was conducted in accordance with Good Clinical Practice. The protocol was approved by the Federal Drug Administration in the USA and the Institutional Review Board at the Rockefeller University.
Study participants
All study participants were recruited at the Rockefeller University Hospital, New York, USA. Eligible participants were adults aged 18–65 years, HIV-1-infected and before enrolment had plasma HIV-1 RNA levels <50 c.p.m. for at least 12 months while on combination ART and <20 c.p.m. at the screening visit, and current CD4 count >500/μl. In addition, participant-derived HIV-1 isolates produced by co-culture of participant PBMCs with HIV-uninfected donor PBMCs were required to be neutralized by 3BNC117 with an IC50 <2 μg/ml in TZM-bl neutralization assays, as previously described31. An IC50 of <2 ug/ml was chosen as a cut-off based on previous PK data of 3BNC117 in humans9 and data in macaques showing that antibody levels 10–100 times the IC50 value against infecting viral strains are necessary to control viral rebound14. However, given the limited diversity and representation of the latent reservoir in outgrowth cultures (Supplementary Fig. 2) and the fact that no correlation between pre-infusion IC50 and delay of viral rebound was found in this study, the significance of this criterion is unclear. Participants on an NNRTI-based ART regimen were switched to a study-provided integrase-inhibitor-based regimen (dolutegravir (Tivicay, ViiV Pharmaceuticals) + tenofovir disoproxil fumarate/emtricitabine (Truvada, Gilead Sciences) 4 weeks before treatment interruption due to the prolonged half-life of NNRTIs. Exclusion criteria included history of CD4 nadir <200 cells/μl, concomitant hepatitis B or C infections, previous receipt of a monoclonal antibody of any kind, or clinically relevant physical findings, medical conditions or laboratory abnormalities. Pregnant and breastfeeding women were not eligible.
Historical controls (ACTG trial participants)
Viral rebound data from 52 participants who participated in four ACTG ATI studies without additional interventions (ACTG 37132, A502433, A506834, and A519732) were compared with viral rebound data in this study. Historical controls were selected based on similar inclusion criteria: age 18–65, Plasma HIV-1 RNA <50 c.p.m. for at least 12 months before ATI while on combination ART, CD4 count at time of ATI >500 cells/μl, CD4 nadir >200 cells/μl, weekly viral load measurements at least until viral rebound occurred.
Study procedures
The appropriate volume of 3BNC117 was calculated according to body weight, diluted in sterile normal saline to a total volume of 250 ml, and administered intravenously over 60 min. Study participants received 3BNC117 on days 0 and 21, or 0, 14, 28, and 42 and remained under monitoring at the Rockefeller University Hospital for 4 h after each infusion. Participants returned for frequent follow up visits for safety assessments, which included physical examination, measurement of clinical laboratory parameters such as haematology, chemistries, urinalysis, and pregnancy tests (for women). Plasma HIV-1 RNA levels were monitored weekly during the ATI period and CD4 counts were measured every other week (Supplementary Table 4). Study investigators evaluated and graded adverse events according to the DAIDS AE Grading Table and determined causality. Blood samples were collected before and at multiple times after 3BNC117 infusions. Samples were processed within 4 h of collection, and serum and plasma samples were stored at −80 °C. PBMCs were isolated by density gradient centrifugation. The absolute number of peripheral blood mononuclear cells was determined by an automated cell counter (Vi-Cell XR; Beckman Coulter), and cells were cryopreserved in fetal bovine serum plus 10% DMSO.
ART re-initiation criteria
Antiretroviral therapy was discontinued 2 days after the first 3BNC117 infusion (day 2). ART was re-initiated when HIV-1 RNA levels were found to be ≥200 c.p.m. and/or CD4 T cell counts decreased to <350 cells/μl and the result was confirmed with a repeat measurement.
Plasma HIV-1 RNA Levels
HIV-1 RNA levels in plasma were measured at the time of screening (within 49 days before the first infusion), day 0 (before infusion), and weekly until week 12, then at weeks 14, 24 and 36. Participants that remained virologically suppressed to <20 c.p.m. off ART beyond week 12, returned for weekly measurements of plasma HIV-1 RNA levels. HIV-1 RNA levels were determined using the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 Assay, Version 2.0, which detects between 20 and 1 × 107 c.p.m. This assay was performed at LabCorp.
CD4+ and CD8+ T cells
CD4+ and CD8+ T-cell counts were determined at screening, on day 0 (before infusion), and weeks 2, 3, 4, 6, 8, 10, 12, 14, and 36 by a clinical flow cytometry assay, performed at LabCorp. Cells were analysed by flow cytometry. Leukocytes were determined as CD45+ cells. Percentage of cells positively stained for CD3, CD4, CD8 as well as the CD4/CD8 ratio were analysed with the BD Multiset software (BD Biosciences).
3BNC117 study drug
3BNC117 is a recombinant, fully human IgG1κ mAb recognizing the CD4 binding site on the HIV-1 envelope1. The antibody was cloned from an HIV-1-infected viraemic controller in the International HIV Controller Study1,35, expressed in Chinese hamster ovary cells (clone 5D5-5C10), and purified using standard methods. The 3BNC117 drug substance was produced at Celldex Therapeutics Fall River (MA) GMP facility, and the drug product was fill-finished at Gallus BioPharmaceuticals (NJ). The resulting purified 3BNC117 was supplied as a single use sterile 20 mg/ml solution for intravenous injection in 8.06 mM sodium phosphate, 1.47 mM potassium phosphate, 136.9 mM sodium chloride, 2.68 mM potassium chloride, and 0.01% polysorbate 80. 3BNC117 vials were shipped and stored at 4°C.
Measurement of 3BNC117 serum levels
Serum levels of 3BNC117 were determined by a validated sandwich ELISA at Celldex Theapeutics as described previously9. Plates (Sigma-Aldrich PN: CLS3590 96-well, High Bind, polystyrene) were coated with 4 μg/ml of an anti-idiotypic antibody specifically recognizing 3BNC117 (anti-ID 1F1 mAb), and incubated overnight at 2–8 °C. After washing, plates were blocked for 1 h with 5% BSA. Serum samples, QCs and standards were added (1:50 minimum dilution in 5% BSA) and incubated for 1 h at room temperature. 3BNC117 was detected using an HRP-conjugated mouse anti-human IgG kappa chain specific antibody (Abcam PN: ab79115) and the HRP substrate tetra-methylbenzidine. 3BNC117 concentrations were then interpolated from a standard curve of 3BNC117 using a four-parameter logistic curve-fitting algorithm. The reference standard and positive controls were created from the drug product lot of 3BNC117 used in the clinical study.
Pharmacokinetic analysis
Blood samples were collected immediately before and at the end of infusions as well as on the day after infusion, weekly during the ATI period and at weeks 14, 24 and 36. 3BNC117 serum levels were measured by ELISA (Celldex Therapeutics).
Neutralization assay
Serum samples, viral supernatants, and control antibodies were tested against HIV-1 envelope pseudoviruses as previously described36,37.
Cell-associated HIV-1 DNA
Participant's CD4+ T-cells were isolated from 10 million cryopreserved PBMCs by negative magnetic selection (Miltenyi). Total DNA was extracted and quantitative PCR performed using pol- and CCR5-directed primers as previously described7.
Virus cultures
Autologous virus was retrieved from HIV-1 infected individuals as previously described31. Briefly, healthy donor PBMCs were obtained by leukapheresis from a single donor. Cells were cultured at a concentration of 5 × 106/ml in Iscove's Modified Dulbecco's Medium (IMDM; Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Scientific), 1% penicillin/streptomycin (Gibco), and 1 μg/ml phytohemagglutinin (PHA; Life Technologies) at 37 °C and 5% CO2. After 2–3 days, 5 × 106 CD8+ depleted cells were transferred into IMDM supplemented with 10% FBS, 1% penicillin/streptomycin, 5 μg/ml polybrene (Sigma), and 100 U/ml of IL-2. Cells were then co-incubated with 4–8 × 106 CD4+ T cells from the study participants and 10 million irradiated healthy donor PBMCs that had been cultured together for 24 h prior in IMDM supplemented with 10% FBS, 1% penicillin/streptomycin, 100 U/ml IL-2 and 1 μg/ml PHA at 37°C and 5% CO2. Lymphoblasts were replenished weekly by adding 3 million healthy donor PHA stimulated CD8+ depleted lymphoblasts. Culture supernatants were quantified using the Alliance HIV-1 p24 Antigen ELISA kit (PerkinElmer) according to the manufacturer's instructions. TCID50s were determined for all HIV-1 containing supernatants36,37 and then tested for sensitivity against 3BNC117 and other bNAbs in a TZM.bl neutralization assay. Blood samples and leukapheresis were collected under separate IRB-approved protocols and after volunteers provided informed consent.
Sequence analysis
HIV-1 RNA extraction and single genome amplification was performed as described previously38. In detail, HIV-1 RNA was extracted from plasma samples using the Qiagen MinElute Virus Spin kit (Qiagen) followed by first strand cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen Life Technologies) and the antisense primer env3out 5′-TTGCTACTTGTGATTGCTCCATGT-3′. gp160 env was amplified using envB5out 5′-TAGAGCCCTGGAAGCATCCAGGAAG-3′ and envB3out 5′-TTGCTACTTGTGATTGCTCCATGT-3′ in the first round and second round nested primers envB5in 5′-CACCTTAGGCATCTCCTATGGCAGGAAGAAG-3′ and envB3in 5′-GTCTCGAGATACTGCTCCCACCC-3′. PCRs were performed using a High Fidelity Platinum Taq (Invitrogen) at 94 °C, 2 min; (94 °C, 15 s; 55 °C 30 s; 68 °C, 4 min) × 35; 68°C, 15 min. Second round PCR was performed with 1 μl of first PCR product as template and High Fidelity Platinum Taq at 94 °C, 2 min; (94 °C, 15 s; 55 °C 30 s; 68 °C, 4 min) × 35; 68 °C, 15 min. Sequence alignments, phylogenetic trees and mutation analysis of gp160 was performed by using Geneious Pro software, version 8.1.6 (Biomatters Ltd.)39. Sequence analysis was performed using Antibody database by A. West30. Logograms were generated using the Weblogo 3.0 tool40.
Pseudovirus generation
Selected SGS from virus culture supernatants and plasma were used to generate pseudoviruses and tested for sensitivity to bNAbs in a TZM.bl assay41. To produce the pseudoviruses, plasmid DNA containing the cytomegalovirus (CMV) promoter was amplified by PCR using forward primer 5′-GTTGACATTGATTATTGACTAG and reverse primer 5′-CTTCCTGCCATAGGAGATGCCTAAAGCTCTGCTTATATAGAC-CTC. The CMV promoter amplicon was fused to individual env SGS amplicons by PCR using forward primer 5′-AGTAATCAATTACGGGGTCATTAGTTCAT and reverse primer 5′-ACTTTTTGACCACTTGCCACCCAT. Fusion PCR was carried out using the Expand Long Template PCR System (Roche) in a 60μl reaction consisting of 1 ng purified CMV promoter amplicon, 0.125 μl unpurified env SGA amplicon, 200 nM forward and reverse primers, 200 μM dNTP mix, 1 × Buffer 1, and 1 μl DNA polymerase mix. PCR was run at 94 °C for 2 min; 25 cycles (94 °C for 12 s, 55 °C for 30 s, 68 °C for four minutes); and 72 °C for 10 min. Resulting amplicons were analysed by gel electrophoresis, purified without gel extraction, and co-transfected with pSG3Δenv into HEK293T cells to produce pseudoviruses as described previously41.
Statistical analyses
Adverse events were summarized by the number of participants who experienced the event, by severity grade according to the DAIDS AE Grading Table and by relationship to 3BNC117 as determined by the investigator. PK-parameters were estimated by performing a non-compartmental analysis (NCA) using WinNonlin 6.3. Kaplan–Meier survival curves were used to compare time to rebound in trial participants to participants in previous ATI studies conducted by ACTG29. To exclude the possibility that the observed delay in rebound is confounded by clinical factors, we compared the clinical variables between the control (ACTG trial participants) and treated group using a two-sided Fisher's Exact test for categorical variables (gender and CD4 Nadir) and an unpaired Wilcoxon test (two-sided) for continuous variables (age, years on ART and CD4 count before ATI initiation) (Supplementary Table 7). Additionally, we tested for each potential confounder whether the variable is predictive for the rebound time. Therefore, we built a univariate survival regression model for each potential confounder and compared those models to a null model using a likelihood ratio test (LRT), which determines how much better the more complex model explains the data than the less complex model. Confounders were considered significant if the model with the potential confounder had an LRT P value of 0.05 or less, which was the case for ‘years on ART’ as well as ‘age’ for the comparison between the controls and the combined treatment group (Supplementary Table 7). We did not perform a standard Cox regression, since the proportional hazards assumption was not fulfilled for some of the variables. Rebound time was modelled using a log-normal distribution, which resulted in the best model fit as measured by Akaike information criterion (AIC) among several different distributions (Extended Data Fig. 4b–d). To determine the effect of the treatment after adjusting for the discovered confounders, we performed a weighted log-rank test42. Therefore, for each sample inverse probability weights based on the discovered confounders were estimated, which were used to re-weigh the variables of the log-rank statistic. We performed a bootstrapped version of the weighted log-rank test, as recommended in ref. 42 owing to the small sample size. We estimated the class probabilities using a lasso logistic regression model trained with the Matlab function lassoglm with five lambda values in a threefold cross-validation. To improve stability, the optimal lambda for the lasso logistic regression was determined only once using the original labels and used in all bootstrap runs to train the models that estimate the class probabilities.
Additionally, we performed an LRT at significance level α = 0.05 based on a parametric survival regression model adjusted for the discovered confounders. In this analysis the treatment group still significantly predicted the delay in rebound (Supplementary Table 7). For the analyses, the R (version 3.2.1) packages survival (version 2.38-3) and fitdistrplus (version 1.0-6) were used and Matlab (version R2015b) for implementation of the weighted log-rank test.
Sequence and phylogenetic analysis
Nucleotide alignments were generated using ClustalW (v.2.11)43 and manually adjusted using Geneious R8 (v.8.1.6)39 and MacClade (v.4.08a)44. Sites that could not be unambiguously aligned were removed for all phylogenetic analyses. Optimal evolutionary model classes were determined using jModelTest (v.2.1.4)45. Maximum likelihood phylogenetic trees were generated using PhyML (v.3)46 with joint estimation of evolutionary model parameter values and phylogenies. The tree comparing all participants was midpoint rooted and each within-subject tree was rooted on the basal branch as determined by the between-subject tree. Sequences with premature stop codons and frameshift mutations that fell in the gp120 surface glycoprotein region were excluded from all deduced protein analyses.
Sequences generated from the supernatants of viral outgrowth assays represented viruses that were present in the latent reservoir. Per assay, 4–8 million CD4+ T cells were activated. In an HIV-infected person who is completely suppressed on antiretroviral therapy, it has been determined that 1 × 10−6 resting CD4 cells are latently infected with replication competent virus47. Thus, one would expect to identify up to eight distinct viral isolates per individual culture. Single genome sequencing of the culture supernatants revealed sets of clonally related sequences, which appear as ‘rakes’ in a phylogenetic tree (Extended Data Figs 7 and 8). The most recent common ancestor of these rakes represents the reactivated virus that was present in the host (similar to the inference of infectious molecular clones as described in ref. 48). As shown in Extended Data Figs 7 and 8, sequences from culture reactions fall in 1–3 rakes within a given individual. We inferred each rake's most recent common ancestor (MRCA) by building a majority-rule consensus and treated it as a single virus from the participant's latent pool. These MRCAs were used to build the phylogenetic trees shown in Fig. 3.
Because mixed culture isolates replicated for 14 or more days, in vitro recombinants were observed. In vitro recombinants from culture reactions were identified and removed from the data set if they: (i) had two identifiable parental sequences within the same culture reaction; and (ii) exhibited three consecutive informative sites relative to one parent followed by three consecutive informative sites relative to another. We independently verified that a subset of these sequences showed evidence of recombination using the Recco tool (v.0.93)49.
Assessment of rebound virus clonality
The Poisson Fitter v2 tool50 is designed to test if a set of homogeneous sequences exhibits random diversification. If such a set of sequences exhibits a star-like phylogeny and a Poisson distribution of pairwise differences (Hamming distances), it can be inferred that a single virus gave rise to those sequences. Poisson Fitter v2 tests these and other parameters using maximum likelihood methods and performs a χ2 goodness of fit test to obtain a P value. A non-significant P value signifies that the observed Hamming distances adhere to a Poisson distribution and it can be inferred that a single virus gave rise to rebound. Single genome derived env sequences from the plasma at the earliest time point post-rebound from each participant were tested using Poisson Fitter (Supplementary Table 8).
ELISPOT T-cell response analysis
Interferon gamma Elispot assays were performed as described51. Briefly, 96-well polyvinylidene plates (Millipore, Bedford, Mass.) were precoated with 0.5 g/ml of anti-IFNγ monoclonal antibody, 1-DIK (Mabtech, Stockholm, Sweden) and previously frozen PBMCs were plated at a concentration of 50,000 to 100,000 cells per well in a volume of 100 μl of R10 medium (RPMI 1640 (Sigma), 10% fetal calf serum (Sigma), 10 mM HEPES buffer (Sigma)) with antibiotics (2 mM l-glutamine, 50 U of penicillin-streptomycin/ml). Plates were incubated overnight at 37°C, 5% CO2, and developed as described51. Cells were tested against a panel of 410 B-clade overlapping 18-mer peptides (OLPs) spanning the entire HIV-1 genome (consensus sequence from 2001). These peptides were used in a matrix system of 11–12 peptides per pool to screen study participants for HIV-specific T cell responses. Confirmation of recognized individual peptides within a peptide pool was undertaken in an additional Elispot assay, as described52. Wells containing PBMCs and R10 medium alone were used as negative controls and were run in duplicate on each plate. Wells containing PBMCs and phytohemagglutinin (PHA) served as positive controls. The numbers of spots per well were counted using an automated Elispot plate reader (ImmunoSpot Reader System; Cellular Technology Limited, Shaker Heights, OH). Responses were regarded as positive if they had at least three times the mean number of spot forming cells (SFC) in the two negative control wells and had to be >50 SFC/106 PBMCs (ref. 51,52).
Extended Data
Extended Data Figure 1. Study participant selection and neutralization of pre-infusion cultures by 3BNC117.
a, Flow diagram showing the selection of study participants. b, Bar diagrams showing IC50 values (μg ml−1) in TZM-bl assays for 3BNC117 against bulk virus outgrowth culture supernatants from the indicated time point pre-infusion for each participant (Supplementary Table 3). For some participants both screen and day 0 cultures were obtained and showed less than threefold variation in IC50 values. The red dotted line indicates an IC50 of 2 μg ml−1 which was used as a threshold for inclusion in the study.
Extended Data Figure 2. CD4+ and CD8+ T cells during study period in participants.
a–d, Absolute T cell counts (a, c) and percentage of CD4+ and CD8+ T cells among CD3+T cells (b, d) for group A and B, respectively (Supplementary Table 4). 3BNC117 infusions are indicated with red arrows. e, Comparison of absolute CD4+ T cell counts and percentage of CD4+ T cells among CD3+T cells at screen, day 0, rebound and after re-suppression. Shown is the data from participants 701, 702, 703, 704, 707, 708, 709, 711 and 713 for whom re-suppression CD4 counts were available (Supplementary Table 4). The last available time point was used as re-suppression time point. Red lines indicate the mean value and error bars indicate standard deviation. P values were obtained using a paired t-test comparing the indicated time points. f, Plasma viral loads and CD4 counts in all study participants. 3BNC117 infusions are indicated with red arrows. The left y-axis shows plasma viral loads in RNA copies per ml (black curves), and right y-axis shows absolute CD4 counts in cells per μl (red curves). Grey areas indicate ART therapy.
Extended Data Figure 3. HIV-specific T-cell responses.
Total breadth (open squares) and magnitude (bars) of T-cell responses against HIV-1 overlapping peptides (OLPs) at the designated time points following administration of 3BNC117 (yellow arrows indicate infusions of 3BNC117 at 30 mg kg−1). For all study participants, antiretroviral therapy was discontinued on day 2 after the first 3BNC117 administration. Blue arrows indicate the time of viral rebound. For study participants 710, 712 and 715 rebound occurred at week 19, 16 and 11, respectively. Baseline samples for study participant 710 and week 12 samples for study participant 714 were not available for ELISpot analysis. Overall, breadth, magnitude and protein specificity were heterogeneous among the study participants.
Extended Data Figure 4. Viral rebound in ACTG control subjects and trial participants.
a, Kaplan–Meier plot summarizing viral rebound in 52 ACTG trial participants who underwent ATI without antibody treatment (black, Supplementary Table 6) and trial participants (Fig. 2a, b, Supplementary Table 4). Six group A participants are shown in red, seven Group B participants in blue and the combination in green as indicated. The y-axis indicates the percentage of participants with viral levels below 200 RNA copies per ml, x-axis indicates weeks after ATI initiation. The survival curves of all considered partitions of the trial participants (group A, group B and group A + B) differed significantly at significance level α = 0.05 from the survival curve of the ACTG trial participants. For the comparison of group A (group A + B) with the ACTG trial participants, we performed a weighted log-rank test adjusting for the clinical variables ‘years on ART’ and ‘age’ to correct for possible confounding factors (Supplementary Table 7 P <0.00001). We identified those potential confounders by univariate parametric survival regression using a likelihood ratio test (Statistical Methods). Since we did not discover any confounders with the same analysis among all available clinical variables for the comparison between group B participants and the ACTG trial participants, we performed a standard log-rank test in that setting (P < 0.0001). b–d, In order to perform a survival regression, the distribution of the rebound times has to be determined. Therefore, we compared the empirical cumulative distribution function (CDF) of the rebound times (black, solid line) with the CDF of the rebound times to a fitted distribution (Weibull, exponential, normal, logistic, log-normal, and gamma) for each comparison group (combined trial participants, group A or group B with ACTG control patients). Since the Akaike information criterion (AIC) and the Bayesian information criterion (BIC) were smallest for the log-normal distribution (green), we have chosen to model the rebound times with the log-normal distribution. e, Dot plot indicating the relationship between cell associated HIV DNA in pre-infusion PBMCs (y-axis) and the week of rebound (x-axis). Group A and group B participants are coloured red and blue respectively. The P value was derived from calculating the Pearson correlation coefficient.
Extended Data Figure 5. In vitro neutralization of pre-infusion and rebound virus outgrowth cultures by 3BNC117 or 10-1074.
a, b, TZM-bl assay neutralization by 3BNC117 (a) and 10-1074 (b) are shown for individual virus outgrowth cultures derived from pre-infusion (black lines/symbols) or rebound (red lines/symbols) time points for each participant. In some cases, multiple independent cultures were grown from a single time point and assayed for neutralization (Supplementary Table 3). ‘Screen’ refers to cultures of PBMC samples taken weeks before infusion during screening, while ‘Day 0’ refers to cultures of PBMCs collected immediately before the first 3BNC117 infusion. Rebound culture time points are denoted by the week (W) at which the samples were collected. Symbols reflect the means of two technical replicates; error bars denote standard deviation.
Extended Data Figure 6. Phylogenetic tree of env nucleotide sequences from trial participants.
A maximum likelihood phylogenetic tree was constructed from single-genome-derived viral env sequences from outgrowth culture supernatants as well as plasma from participants 701 (olive), 702 (black), 703 (pink), 704 (yellow), 707 (light blue), 708 (green), 709 (dark blue) and 711 (brown). Hypervariable (as defined in http://www.hiv.lanl.gov/content/sequence/VAR_REG_CHAR/) and other poorly aligned regions were excluded from the analysis. The tree was constructed using PhyML with a GTR+I+G substitution model and midpoint rooted. Asterisks indicate 100% bootstrap support (only values for major nodes are shown). The scale bar indicates 0.01 substitutions per site.
Extended Data Figure 7. Rebound virus clonality and neutralization sensitivity to 3BNC117.
Maximum likelihood phylogenetic trees of plasma and culture-derived env sequences are shown for participants 701, 702, 703, 704. Sequences obtained at screening, on Day 0, and consecutive rebound time points (plasma and cultures) are colour coded as indicated. The trees were rooted based on the branch insertion identified in the between-subject tree (Extended Data Fig. 6). Bootstrap values ≥90% are shown. Names of env sequences used to generate pseudoviruses for 3BNC117 neutralization analysis are indicated along with the respective IC80 titres in μg ml−1. Representative rebound viruses selected in Fig. 4b are marked with red stars (Fig. 4b, Supplementary Table 9). Zero branch length viruses in multi-rebounders 702 and 703 are marked with black stars.
Extended Data Figure 8. Rebound virus clonality and neutralization sensitivity to 3BNC117.
Maximum likelihood phylogenetic trees of plasma- and culture-derived env sequences are shown for participants 707, 708, 709 and 711. Sequences obtained at screening, on day 0, and consecutive rebound time points (plasma and cultures) are colour coded as indicated. The trees were rooted based on the branch insertion identified in the between-subject tree (Extended Data Fig. 6). Bootstrap values ≥90% are shown. Names of env sequences used to generate pseudoviruses for 3BNC117 neutralization analysis are indicated along with the respective IC80titres in μg ml−1. Representative rebound viruses selected in Fig. 4b are marked with red stars (Fig. 4b, Supplementary Table 9). Zero branch length viruses in multi-rebounder 709 are marked with black stars.
Supplementary Material
Table 1.
| a | |||||||
|---|---|---|---|---|---|---|---|
| Study ID | 3BNC117 dose | Age | Years since HIV Diagnosis |
Current ART regimen |
Clade | HIV-RNA level (copies/ml) |
abs. CD4+ T cell count (day 0; cells/mm3) |
| 2A1 | 1 mg/kg | 35 | 11 | ART naïve | B | 3,210 | 674 |
| 2A3 | 1 mg/kg | 39 | 14 | Off ART | B | 43,650 | 520 |
| 2A4 | 1 mg/kg | 42 | 8 | ART naïve | B | 5,340 | 607 |
| 2B1 | 3 mg/kg | 20 | 1 | Off ART | ND | 4,090 | 264* |
| 2B2 | 3 mg/kg | 48 | 20 | DRV/r/TDF/FTC** | ND | 100 | 706 |
| 2B3 | 3 mg/kg | 20 | 1 | ART naïve | B | 38,190 | 777 |
| 2C2 | 10 mg/kg | 51 | 12 | ATV/r/3TC/ZDV** | ND | 30 | 728 |
| 2C4 | 10 mg/kg | 54 | 23 | Off ART | ND | 820 | 805 |
| 2C5 | 10 mg/kg | 50 | 4 | ART naïve | B | 9,260 | 245* |
| 2D1 | 30 mg/kg | 33 | 3 | ART naïve | B | 53,470 | 980 |
| 2C1 | 30 mg/kg | 51 | 17 | Off ART | B | 47,650 | 1129 |
| 2D3 | 30 mg/kg | 33 | 0.5 | ART naïve | ND | 640 | 618 |
| 2E1 | 30 mg/kg | 21 | 2 | ART naïve | B | 15,780 | 847 |
| 2E2 | 30 mg/kg | 46 | 1.5 | ART naïve | B | 6,990 | 513 |
| 2E3 | 30 mg/kg | 23 | 1.5 | ART naïve | BF | 22,030 | 590 |
| 2E4 | 30 mg/kg | 38 | 1 | ART naïve | ND | 32,220 | 603 |
| 2E5 | 30 mg/kg | 30 | 1 | ART naïve | ND | 3,610 | 532 |
| b | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uninfected (No. of AEs) | HIV-1-infected (No. of AEs) | |||||||||||||
| Adverse Events | No. AEs |
% of reported AEs |
No. possibly related |
No. Mild |
No. Moderate |
No. Severe |
1 mg/kg |
3 mg/kg |
10 mg/kg |
30 mg/kg |
1 mg/kg |
3 mg/kg |
10 mg/kg |
30 mg/kg |
| Rhinorrhea and/or cough | 10 | 16.9 | 3 | 8 | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 6 |
| Malaise | 7 | 11.9 | 5 | 5 | 2 | 0 | 1 | 1 | 2 | 0 | 1 | 2 | 0 | 0 |
| Headache | 6 | 10.2 | 3 | 5 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 3 |
| Diarrhea | 5 | 8.5 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 |
| Myalgia/arthralgia (localized) | 4 | 6.8 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 |
| Sore throat | 4 | 6.8 | 2 | 4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Tenderness | 3 | 5.1 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 |
| Increased Lacrimation | 2 | 3.4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Myalgia | 2 | 3.4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Chills | 2 | 3.4 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Conjunctival erythema | 2 | 3.4 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Fevershiness | 2 | 3.4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Nausea | 2 | 3.4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Pruritus | 2 | 3.4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Blurry vision | 1 | 1.7 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Decreased appetite | 1 | 1.7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Erythema | 1 | 1.7 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Paresthesia upper extremity | 1 | 1.7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Shingles | 1 | 1.7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Vomiting | 1 | 1.7 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
Table 2.
| a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2A1 | HIV-1 RNA Level | 2A3 | HIV-1 RNA Level | 2A4 | HIV-1 RNA Level | |||||||
| Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | |
| 1 mg/kg | Screen | 1,340 | 3.13 | - | Screen | 41,670 | 4.62 | - | Screen | 5,350 | 3.73 | - |
| Pre | 1,100 | 3.04 | - | Pre | 48,100 | 4.68 | - | Pre | 6,490 | 3.81 | - | |
| Day 0 | 3,210 | 3.51 | 0.00 | Day 0 | 43,560 | 4.64 | 0.00 | Day 0 | 5,340 | 3.73 | 0.00 | |
| Day 1 | 10,810 | 4.03 | 0.52 | Day 1 | 50,180 | 4.70 | 0.06 | Day 1 | 10,880 | 4.04 | 0.31 | |
| Day 4 | 4,970 | 3.70 | 0.19 | Day 4 | 24,050 | 4.38 | -0.26 | Day 4 | 8,430 | 3.93 | 0.20 | |
| Day 7 | 3,190 | 3.50 | -0.01 | Day 7 | 34,850 | 4.54 | -0.10 | Day 7 | 4,870 | 3.69 | -0.04 | |
| Day 14 | 2,600 | 3.41 | -0.10 | Day 14 | 31,770 | 4.50 | -0.14 | Day 14 | 11,000 | 4.04 | 0.31 | |
| Day 21 | 3,820 | 3.58 | 0.07 | Day 21 | 35,840 | 4.55 | -0.09 | Day 21 | 7,470 | 3.87 | 0.14 | |
| Day 28 | 5,070 | 3.71 | 0.20 | Day 28 | 38,500 | 4.59 | -0.05 | Day 28 | 7,880 | 3.90 | 0.17 | |
| Day 42 | 3,570 | 3.55 | 0.04 | Day 42 | 59,780 | 4.78 | 0.14 | Day 42 | 12,150 | 4.08 | 0.35 | |
| Day 56 | 6,370 | 3.80 | 0.29 | Day 56 | 63,600 | 4.80 | 0.16 | Day 56 | 14,020 | 4.15 | 0.42 | |
| 2B1 | HIV-1 RNA Level | 2B2 | HIV-1 RNA Level | 2B3 | HIV-1 RNA Level | |||||||
| Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | |
| 3 mg/kg | Screen | 6,420 | 3.81 | - | Screen | 80* | 1.90 | - | Screen | 53,660 | 4.73 | - |
| Pre | 4,950 | 3.69 | - | Pre | 30 | 1.48 | - | Pre | 28,810 | 4.46 | - | |
| Day 0 | 4,090 | 3.61 | 0.00 | Day 0 | 100 | 2.00 | 0.00 | Day 0 | 38,190 | 4.58 | 0.00 | |
| Day 1 | 6,470 | 3.81 | 0.20 | Day 1 | 140 | 2.15 | 0.15 | Day 1 | 31,950 | 4.50 | -0.08 | |
| Day 4 | 2,690 | 3.43 | -0.18 | Day 4 | 190 | 2.28 | 0.28 | Day 4 | 11,540 | 4.06 | -0.52 | |
| Day 7 | 1,610 | 3.21 | -0.40 | Day 7 | 20 | 1.30 | -0.70 | Day 7 | 20,500 | 4.31 | -0.27 | |
| Day 14 | 4,880 | 3.69 | 0.08 | Day 14 | 130 | 2.11 | 0.11 | Day 14 | 35,830 | 4.55 | -0.03 | |
| Day 21 | 4,160 | 3.62 | 0.01 | Day 21 | 90 | 1.95 | -0.05 | Day 21 | 41,570 | 4.62 | 0.04 | |
| Day 28 | 3,800 | 3.58 | -0.03 | Day 28 | 70 | 1.85 | -0.15 | Day 28 | 30,810 | 4.49 | -0.09 | |
| Day 42 | 4,970 | 3.70 | 0.09 | Day 42 | 90 | 1.95 | -0.05 | Day 42 | 29,020 | 4.46 | -0.12 | |
| Day 56 | 3,550 | 3.55 | -0.06 | Day 56 | 30 | 1.48 | -0.52 | Day 56 | 27,700 | 4.44 | -0.14 | |
| 2C2 | HIV-1 RNA Level | 2C4 | HIV-1 RNA Level | 2C5 | HIV-1 RNA Level | |||||||
| Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | |
| 10 mg/kg | Screen | 20 | 1.30 | - | Screen | 2,510 | 3.41 | - | Screen | 19,970 | 4.30 | - |
| Pre | 30 | 1.48 | - | Pre | 1,100 | 3.04 | - | Pre | 10,860 | 4.04 | - | |
| Day 0 | 30 | 1.48 | 0.00 | Day 0 | 820 | 2.91 | 0.00 | Day 0 | 9,260 | 3.97 | 0.00 | |
| Day 1 | 60 | 1.78 | 0.30 | Day 1 | 690 | 2.84 | -0.07 | Day 1 | 5,300 | 3.72 | -0.25 | |
| Day 4 | 50 | 1.70 | 0.22 | Day 4 | 980 | 2.99 | 0.08 | Day 4 | 2,340 | 3.37 | -0.60 | |
| Day 7 | 9 | 0.96 | -0.52 | Day 7 | 990 | 3.00 | 0.09 | Day 7 | 410 | 2.61 | -1.36 | |
| Day 14 | 6 | 0.79 | -0.69 | Day 14 | 700 | 2.85 | -0.06 | Day 14 | 1,070 | 3.03 | -0.94 | |
| Day 21 | 20 | 1.30 | -0.18 | Day 21 | 1,420 | 3.15 | 0.24 | Day 21 | 8,900 | 3.95 | -0.02 | |
| Day 28 | 2 | 0.23 | -1.25 | Day 28 | 630 | 2.80 | -0.11 | Day 28 | 8,550 | 3.93 | -0.04 | |
| Day 42 | 3 | 0.45 | -1.03 | Day 42 | 980 | 2.99 | 0.08 | Day 42 | 12,410 | 4.09 | 0.12 | |
| Day 56 | 21 | 1.32 | -0.16 | Day 56 | 940 | 2.97 | 0.06 | Day 56 | 12,840 | 4.11 | 0.14 | |
| 2D1 | HIV-1 RNA Level | 2C1 | HIV-1 RNA Level | 2D3 | HIV-1 RNA Level | |||||||
| Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | |
| 30 mg/kg | Screen | 5,730 | 3.76 | - | Screen | 31,870 | 4.50 | - | Screen | 3,620 | 3.56 | - |
| Pre | 34,840 | 4.54 | - | Pre | 31,870 | 4.50 | - | Pre | 3,280 | 3.52 | - | |
| Day 0 | 53,470 | 4.73 | 0.00 | Day 0 | 47,650 | 4.68 | 0.00 | Day 0 | 640 | 2.81 | 0.00 | |
| Day 1 | 84,450 | 4.93 | 0.20 | Day 1 | 46,040 | 4.66 | -0.02 | Day 1 | 450 | 2.65 | -0.16 | |
| Day 4 | 36,140 | 4.56 | -0.17 | Day 4 | 7,790 | 3.89 | -0.79 | Day 4 | 210 | 2.32 | -0.49 | |
| Day 7 | 5,980 | 3.78 | -0.95 | Day 7 | 7,470 | 3.87 | -0.81 | Day 7 | 80 | 1.90 | -0.91 | |
| Day 14 | 4,830 | 3.68 | -1.05 | Day 14 | 19,360 | 4.29 | -0.39 | Day 14 | 50 | 1.70 | -1.11 | |
| Day 21 | 15,180 | 4.18 | -0.55 | Day 21 | 30,380 | 4.48 | -0.20 | Day 21 | 3 | 0.52 | -1.77 | |
| Day 28 | 10,960 | 4.04 | -0.69 | Day 28 | 25,610 | 4.41 | -0.27 | Day 28 | 40 | 1.60 | -1.21 | |
| Day 42 | 7,650 | 3.88 | -0.85 | Day 42 | 22,830 | 4.36 | -0.32 | Day 42 | 50 | 1.70 | -1.11 | |
| Day 56 | 9,820 | 3.99 | -0.74 | Day 56 | 32,310 | 4.51 | -0.17 | Day 56 | 11 | 1.04 | -1.77 | |
| 2E1 | HIV-1 RNA Level | 2E2 | HIV-1 RNA Level | 2E3 | HIV-1 RNA Level | |||||||
| Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | |
| 30 mg/kg | Screen | 8,742 | 3.94 | - | Screen | 3,394 | 3.53 | - | Screen | 23,168 | 4.36 | - |
| Pre | 12,031 | 4.08 | - | Pre | 1,802 | 3.26 | - | Pre | 30,570 | 4.49 | - | |
| Day 0 | 15,780 | 4.20 | 0.00 | Day 0 | 6,990 | 3.84 | 0.00 | Day 0 | 22,030 | 4.34 | 0.00 | |
| Day 1 | 14,790 | 4.17 | -0.03 | Day 1 | 6,450 | 3.81 | -0.03 | Day 1 | 38,620 | 4.59 | 0.25 | |
| Day 4 | 3,560 | 3.55 | -0.65 | Day 4 | 2,340 | 3.37 | -0.47 | Day 4 | 10,540 | 4.02 | -0.32 | |
| Day 7 | 404 | 2.61 | -1.59 | Day 7 | 1,663 | 3.22 | -0.62 | Day 7 | 1,308 | 3.12 | -1.22 | |
| Day 14 | 468 | 2.67 | -1.53 | Day 14 | 253 | 2.40 | -1.44 | Day 14 | 3,901 | 3.59 | -0.75 | |
| Day 21 | 8,557 | 3.93 | -0.27 | Day 21 | 917 | 2.96 | -0.88 | Day 21 | 23,717 | 4.38 | 0.04 | |
| Day 28 | 8,159 | 3.91 | -0.29 | Day 28 | 4,273 | 3.63 | -0.21 | Day 28 | 33,370 | 4.52 | 0.18 | |
| Day 42 | 6,671 | 3.82 | -0.38 | Day 42 | 15,721 | 4.20 | 0.36 | Day 42 | 22,699 | 4.36 | 0.01 | |
| Day 56 | 13,486 | 4.13 | -0.07 | Day 56 | 3,465 | 3.54 | -0.30 | Day 56 | 27,998 | 4.45 | 0.10 | |
| 2E4 | HIV-1 RNA Level | 2E5 | HIV-1 RNA Level | |||||||||
| Timepoint | c/ml | log10 | Δ log10 | Timepoint | c/ml | log10 | Δ log10 | |||||
| 30 mg/kg | Screen | 45,311 | 4.66 | - | Screen | 4,720 | 3.67 | - | ||||
| Pre | 66,889 | 4.83 | - | Pre | 4,244 | 3.63 | - | |||||
| Day 0 | 32220 | 4.51 | 0.00 | Day 0 | 3610 | 3.56 | 0.00 | |||||
| Day 1 | 44980 | 4.65 | 0.14 | Day 1 | 5300 | 3.72 | 0.16 | |||||
| Day 4 | 19,290 | 4.29 | -0.22 | Day 4 | 1,000 | 3.00 | -0.56 | |||||
| Day 7 | 3,642 | 3.56 | -0.95 | Day 7 | 181 | 2.26 | -1.30 | |||||
| Day 14 | 983 | 2.99 | -1.52 | Day 14 | 77 | 1.89 | -1.67 | |||||
| Day 21 | 5,726 | 3.76 | -0.75 | Day 21 | 11 | 1.04 | -2.52 | |||||
| Day 28 | 18,005 | 4.26 | -0.25 | Day 28 | 148 | 2.17 | -1.39 | |||||
| Day 42 | 40,079 | 4.60 | 0.09 | Day 42 | 1,064 | 3.03 | -0.53 | |||||
| Day 56 | 22,100 | 4.34 | -0.16 | Day 56 | 589 | 2.77 | -0.79 | |||||
| b | |||||
|---|---|---|---|---|---|
| Contrast | Dose | Average | SE | p.value | |
| 1 mg/kg | Screen - Day 0 | 1mg/kg | -0.133 | 0.258 | 0.607 |
| Day -7 - Day 0 | 1mg/kg | -0.112 | 0.200 | 0.573 | |
| Day 1 - Day 0 | 1mg/kg | 0.299 | 0.200 | 0.134 | |
| Day 4 - Day 0 | 1mg/kg | 0.043 | 0.258 | 0.866 | |
| Day 7 - Day 0 | 1mg/kg | -0.047 | 0.290 | 0.873 | |
| Day 14 - Day 0 | 1mg/kg | 0.028 | 0.310 | 0.927 | |
| Day 21 - Day 0 | 1mg/kg | 0.046 | 0.323 | 0.888 | |
| Day 28 - Day 0 | 1mg/kg | 0.105 | 0.331 | 0.752 | |
| Day 42 - Day 0 | 1mg/kg | 0.180 | 0.337 | 0.592 | |
| Day 56 - Day 0 | 1mg/kg | 0.294 | 0.340 | 0.388 | |
| Contrast | Dose | Average | SE | p.value | |
| 3 mg/kg | Screen - Day 0 | 3mg/kg | 0.172 | 0.316 | 0.587 |
| Day -7 - Day 0 | 3mg/kg | -0.020 | 0.244 | 0.936 | |
| Day 1 - Day 0 | 3mg/kg | 0.061 | 0.244 | 0.803 | |
| Day 4 - Day 0 | 3mg/kg | -0.351 | 0.316 | 0.267 | |
| Day 7 - Day 0 | 3mg/kg | -0.338 | 0.356 | 0.343 | |
| Day 14 - Day 0 | 3mg/kg | 0.024 | 0.380 | 0.949 | |
| Day 21 - Day 0 | 3mg/kg | 0.022 | 0.396 | 0.955 | |
| Day 28 - Day 0 | 3mg/kg | -0.063 | 0.406 | 0.877 | |
| Day 42 - Day 0 | 3mg/kg | -0.017 | 0.412 | 0.966 | |
| Day 56 - Day 0 | 3mg/kg | -0.100 | 0.416 | 0.809 | |
| Contrast | Dose | Average | SE | p.value | |
| 10 mg/kg | Screen - Day 0 | 10mg/kg | 0.415 | 0.316 | 0.189 |
| Day -7 - Day 0 | 10mg/kg | 0.098 | 0.244 | 0.687 | |
| Day 1 - Day 0 | 10mg/kg | -0.159 | 0.244 | 0.516 | |
| Day 4 - Day 0 | 10mg/kg | -0.260 | 0.316 | 0.410 | |
| Day 7 - Day 0 | 10mg/kg | -0.636 | 0.356 | 0.074 | |
| Day 14 - Day 0 | 10mg/kg | -0.503 | 0.380 | 0.186 | |
| Day 21 - Day 0 | 10mg/kg | 0.111 | 0.396 | 0.780 | |
| Day 28 - Day 0 | 10mg/kg | -0.075 | 0.406 | 0.854 | |
| Day 42 - Day 0 | 10mg/kg | 0.102 | 0.412 | 0.804 | |
| Day 56 - Day 0 | 10mg/kg | 0.101 | 0.416 | 0.809 | |
| Contrast | Dose | Average | SE | p.value | |
| 30 mg/kg | Screen - Day 0 | 30mg/kg | -0.084 | 0.158 | 0.593 |
| Day -7 - Day 0 | 30mg/kg | 0.022 | 0.122 | 0.860 | |
| Day 1 - Day 0 | 30mg/kg | 0.065 | 0.122 | 0.593 | |
| Day 4 - Day 0 | 30mg/kg | -0.458 | 0.158 | 0.004 | |
| Day 7 - Day 0 | 30mg/kg | -1.043 | 0.178 | < 0.001 | |
| Day 14 - Day 0 | 30mg/kg | -1.181 | 0.190 | < 0.001 | |
| Day 21 - Day 0 | 30mg/kg | -0.866 | 0.198 | < 0.001 | |
| Day 28 - Day 0 | 30mg/kg | -0.515 | 0.203 | 0.011 | |
| Day 42 - Day 0 | 30mg/kg | -0.339 | 0.206 | 0.099 | |
| Day 56 - Day 0 | 30mg/kg | -0.371 | 0.213 | 0.081 | |
Table 3.
| Autologous virus isolates | HIV-1 envelopes cloned from plasma | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Dose | ID | Day post infusion | 3BNC117 IC50 (μg/ml) | 3BNC117 IC50 | Clone | Cloning procedure | Vector backbone | 3BNC117 (IC50; μg/ml) | Average (geo. mean) |
|
| |||||||||
| 1 mg/kg | 2A1 | Day 0 | ND | ND | ND | ||||
|
| |||||||||
| Day 28 | 0.90 | ||||||||
|
| |||||||||
| 2A3 | Day 0 | 0.11 | 35.3 | ND | |||||
|
| |||||||||
| Day 28 | 3.78 | ||||||||
|
| |||||||||
| 2A4 | Day 0 | 0.07 | 13.5 | ND | |||||
|
| |||||||||
| Day 28 | 0.94 | ||||||||
|
| |||||||||
| 3 mg/kg | 2B1 | Day 0 | 0.77 | 25.9 | ND | ||||
|
| |||||||||
| Day 28 | >20 | ||||||||
|
| |||||||||
| 2B2 | Day 0 | >20 | ND | ND | |||||
|
| |||||||||
| Day 28 | ND | ||||||||
|
| |||||||||
| 2B3 | Day 0 | 0.20 | 1.5 | ND | |||||
|
| |||||||||
| Day 28 | 0.30 | ||||||||
|
| |||||||||
| 10 mg/kg | 2C2 | Day 0 | 0.49 | 0.1 | ND | ||||
|
| |||||||||
| Day 56 | 0.03 | ||||||||
|
| |||||||||
| 2C4 | Day 0 | >20 | No change | ND | |||||
|
| |||||||||
| Day 28 | >20 | ||||||||
|
| |||||||||
| 2C5 | Day 0 | 0.09 | 167.0 | 2C5_D0_12 | gp120 | pSVIII | 0.015 | 0.02 | |
| 2C5_D0_21 | gp120 | pSVIII | 0.017 | ||||||
| 2C5_D0_27 | gp120 | pSVIII | 0.017 | ||||||
|
|
|
||||||||
| Day 28 | 15.36 | 2C5_W4_59 | gp120 | pSVIII | 11.543 | 7.09 | |||
| 2C5_W4_22 | gp120 | pSVIII | 6.737 | ||||||
| 2C5_W4_27 | gp120 | pSVIII | 7.514 | ||||||
| 2C5_W4_28 | gp120 | pSVIII | 3.495 | ||||||
| 2C5_W4_34 | gp120 | pSVIII | 8.758 | ||||||
|
| |||||||||
| 30 mg/kg | 2C1 | Day -7 | 0.54 | 1.8 | 2C1_D0_12 | gp120 | pSVIII | 0.209 | 0.06 |
| 2C1_D0_22 | gp120 | pSVIII | 0.158 | ||||||
| 2C1_D0_32 | gp120 | pSVIII | 0.006 | ||||||
|
|
|
||||||||
| Day 28 | 1.78 | 2C1_W4_12 | gp120 | pSVIII | 0.043 | 0.14 | |||
| 2C1_W4_18 | gp120 | pSVIII | 0.225 | ||||||
| 2C1_W4_31 | gp120 | pSVIII | 0.262 | ||||||
|
| |||||||||
| 2D1 | Day 0 | 0.68 | 1.3 | 2D1_D0_D5 | gp160 | pcDNA3.1 | 0.165 | 0.15 | |
| 2D1_D0_B3.1 | gp160 | pcDNA3.1 | 0.128 | ||||||
| 2D1_D0_B10 | gp160 | pcDNA3.1 | 0.172 | ||||||
|
|
|
||||||||
| Day 28 | 0.90 | 2D1_W4_37 | gp120 | pSVIII | 0.578 | 0.52 | |||
| 2D1_W4_40 | gp120 | pSVIII | 0.501 | ||||||
| 2D1_W4_69 | gp120 | pSVIII | 0.465 | ||||||
| 2D1_W4_71 | gp120 | pSVIII | 0.523 | ||||||
|
| |||||||||
| 2D3 | Day 0 | 0.13 | 2.7 | ND | |||||
|
| |||||||||
| Day 28 | 0.35 | ||||||||
|
| |||||||||
| 2E1 | Screen | 0.40 | ND | 2E1_D0_12 | gp160 | pcDNA3.1 | 0.103 | 0.09 | |
| 2E1_D0_20 | gp160 | pcDNA3.1 | 0.115 | ||||||
| 2E1_D0_34 | gp160 | pcDNA3.1 | 0.068 | ||||||
|
|
|
||||||||
| Day 28 | ND | 2E1_W4_23 | gp160 | pcDNA3.1 | 0.041 | 0.23 | |||
| 2E1_W4_E1 | gp160 | pcDNA3.1 | 0.590 | ||||||
| 2E1_W4_F6 | gp160 | pcDNA3.1 | 0.496 | ||||||
|
| |||||||||
| 2E2 | Screen | 0.18 | 12.3 | 2E2_D0_A10 | gp160 | pcDNA3.1 | 0.017 | 0.01 | |
| 2E2_D0_C3 | gp160 | pcDNA3.1 | 0.010 | ||||||
| 2E2_D0_E9 | gp160 | pcDNA3.1 | 0.018 | ||||||
|
|
|
||||||||
| Day 28 | 2.23 | 2E2_W4_B9 | gp160 | pcDNA3.1 | 0.017 | 0.03 | |||
| 2E2_W4_C11 | gp160 | pcDNA3.1 | 0.020 | ||||||
| 2E2_W4_D5 | gp160 | pcDNA3.1 | 0.057 | ||||||
|
| |||||||||
| 2E3 | Screen | 0.18 | 6.1 | ND | |||||
|
| |||||||||
| Day 28 | 1.10 | ||||||||
|
| |||||||||
| 2E4 | Screen | 0.24 | ND | ND | |||||
|
| |||||||||
| Day 28 | ND | ||||||||
|
| |||||||||
| 2E5 | Screen | 0.30 | ND | ND | |||||
|
|
|||||||||
| Day 28 | ND | ||||||||
Table 4.
| ID | HIV-1/ART | 3BNC117 (mg/kg) |
Method | Cmax (μg/ml) |
adjusted R-squared |
Estimated T1/2 (days) |
Lambda (lower)* |
Lambda (upper)* |
AUC (INF_pred) |
Tlast* | Clast (pg/ml) |
AUC_% Extrap_pred |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A1 | Negative | 1 | ELISA | 27.4 | n.d. | n.d. | - | - | n.d. | - | - | n.d. |
| TZM.bl | 27.4 | 0.936 | 21.32 | 14 | 42 | 184.0 | 42 | 1.2 | 19.2 | |||
| 1A2 | Negative | 1 | ELISA | 18.8 | n.d. | n.d. | - | - | n.d. | - | - | n.d. |
| TZM.bl | 27.8 | 0.893 | 12.89 | 7 | 42 | 181.7 | 42 | 1.0 | 9.0 | |||
| 1A3 | Negative | 1 | ELISA | 11.2 | n.d. | n.d. | - | - | n.d. | - | - | n.d. |
| TZM.bl | 23.5 | 0.976 | 10.38 | 4 | 28 | 138.0 | 28 | 1.3 | 14.0 | |||
| 2A1 | Positive/Off | 1 | ELISA | 15.7 | n.d. | n.d. | - | - | n.d. | - | - | n.d. |
| 2A3 | Positive/Off | 1 | ELISA | 22.7 | n.d. | n.d. | - | - | n.d. | - | - | n.d. |
| 2A4 | Positive/Off | 1 | ELISA | 33.8 | n.d. | n.d. | - | - | n.d. | - | - | n.d. |
| TZM.bl | 15.6 | n.d. | n.d. | - | - | n.d. | - | - | n.d. | |||
|
| ||||||||||||
| 1B1 | Negative | 3 | ELISA | 89.9 | 0.985 | 19.64 | 7 | 56 | 685.2 | 56 | 3.1 | 12.1 |
| TZM.bl | 70.9 | 0.881 | 24.30 | 7 | 56 | 483.7 | 56 | 2.7 | 17.9 | |||
| 1B2 | Negative | 3 | ELISA | 90.1 | 0.878 | 12.91 | 7 | 42 | 523.5 | 42 | 2.2 | 8.8 |
| TZM.bl | 116.0 | 0.936 | 10.04 | 7 | 42 | 481.2 | 42 | 1.4 | 4.6 | |||
| 1B3 | Negative | 3 | ELISA | 243.4 | 0.954 | 21.54 | 14 | 56 | 1017.3 | 56 | 4.2 | 12.1 |
| TZM.bl | 74.5 | 0.954 | 21.70 | 7 | 56 | 706.6 | 56 | 3.9 | 15.6 | |||
| 2B1 | Positive/Off | 3 | ELISA | 90.8 | 0.942 | 9.60 | 4 | 21 | 219.4 | 21 | 2.8 | 19.2 |
| TZM.bl | 57.4 | 0.994 | 10.28 | 7 | 28 | 245.9 | 28 | 1.9 | 12.1 | |||
| 2B2 | Positive/On | 3 | ELISA | 97.6 | 0.928 | 8.74 | 7 | 21 | 200.7 | 21 | 2.5 | 16.5 |
| 2B3 | Positive/Off | 3 | ELISA | 60.0 | 0.981 | 9.08 | 4 | 21 | 251.2 | 21 | 3.5 | 17.0 |
| TZM.bl | 32.6 | 0.994 | 9.19 | 4 | 28 | 129.8 | 28 | 1.0 | 10.1 | |||
|
| ||||||||||||
| 1C1 | Negative | 10 | ELISA | 347.8 | 0.942 | 16.33 | 7 | 56 | 1667.9 | 56 | 5.9 | 7.5 |
| TZM.bl | 281.8 | 0.964 | 15.79 | 7 | 56 | 1598.9 | 56 | 5.6 | 7.3 | |||
| 1C3 | Negative | 10 | ELISA | 308.9 | 0.860 | 29.01 | 7 | 56 | 2529.4 | 56 | 15.8 | 23.5 |
| TZM.bl | 244.7 | 0.700 | 26.59 | 14 | 56 | 2228.5 | 56 | 13.1 | 19.3 | |||
| 1C4 | Negative | 10 | ELISA | 180.6 | 0.835 | 10.73 | 7 | 42 | 859.1 | 42 | 4.0 | 5.5 |
| TZM.bl | 230.7 | 0.990 | 10.56 | 14 | 56 | 1022.0 | 56 | 1.4 | 2.2 | |||
| 2C2 | Positive/On | 10 | ELISA | 326.8 | 0.866 | 10.87 | 7 | 42 | 904.5 | 42 | 2.7 | 5.6 |
| 2C4 | Positive/Off | 10 | ELISA | 178.2 | 0.944 | 6.92 | 4 | 28 | 550.2 | 28 | 2.8 | 5.1 |
| TZM.bl | 193.0 | 0.979 | 6.14 | 4 | 28 | 417.9 | 28 | 1.8 | 3.4 | |||
| 2C5 | Positive/Off | 10 | ELISA | 201.1 | 0.997 | 6.60 | 7 | 28 | 592.6 | 28 | 2.6 | 4.1 |
| TZM.bl | 174.4 | 0.994 | 6.50 | 7 | 28 | 431.2 | 28 | 1.8 | 3.7 | |||
|
| ||||||||||||
| 1E2 | Negative | 30 | ELISA | 360.8 | 0.930 | 14.48 | 7 | 56 | 4259.4 | 56 | 13.6 | 5.9 |
| TZM.bl | 1166.3 | 0.978 | 16.19 | 14 | 56 | 5901.2 | 56 | 16.1 | 6.2 | |||
| 1E3 | Negative | 30 | ELISA | 361.8 | 0.990 | 16.26 | 7 | 56 | 3177.5 | 56 | 11.7 | 8.2 |
| TZM.bl | 606.2 | 0.989 | 17.85 | 7 | 56 | 3422.2 | 56 | 12.3 | 9.3 | |||
| 1E5 | Negative | 30 | ELISA | 765.0 | 0.992 | 14.15 | 4 | 56 | 5874.2 | 56 | 16.3 | 5.7 |
| TZM.bl | 939.6 | 0.959 | 15.22 | 14 | 56 | 6424.5 | 56 | 19.1 | 6.1 | |||
| 2C1 | Positive/Off | 30 | ELISA | 410.2 | 0.991 | 5.80 | 7 | 28 | 1707.8 | 28 | 5.1 | 2.6 |
| TZM.bl | 717.4 | 0.993 | 5.99 | 7 | 42 | 2186.4 | 42 | 1.6 | 0.6 | |||
| 2D1 | Positive/Off | 30 | ELISA | 976.4 | 0.996 | 6.86 | 7 | 42 | 2494.9 | 42 | 3.4 | 1.3 |
| TZM.bl | 849.3 | 0.990 | 8.83 | 7 | 56 | 1825.5 | 56 | 1.1 | 0.8 | |||
| 2D3 | Positive/Off | 30 | ELISA | 571.0 | 0.962 | 13.39 | 7 | 56 | 3616.5 | 56 | 9.9 | 4.8 |
| TZM.bl | 953.7 | 0.993 | 11.23 | 7 | 42 | 4346.2 | 42 | 16.0 | 6.0 | |||
| 2E1 | Positive/Off | 30 | ELISA | 712.4 | 0.970 | 11.14 | 14 | 56 | 3028.5 | 56 | 4.6 | 2.3 |
| 2E2 | Positive/Off | 30 | ELISA | 789.4 | 0.920 | 11.14 | 7 | 56 | 3596.1 | 56 | 6.7 | 2.4 |
| 2E3 | Positive/Off | 30 | ELISA | 559.6 | 0.821 | 8.54 | 7 | 28 | 2495.6 | 28 | 13.7 | 8.4 |
Table 5.
| Dose | HIV-1-status | Subjects | Method (subjects analyzed) | Cmax (μg/ml) | t1/2 (days) (1) | ||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Mean | SD | Range | Mean | SD | Range | ||||
|
|
|
||||||||
| 1 mg | Neg. | 3 | ELISA (3) | 19.1 | 8.1 | 11.2 - 27.4 | n.d. | n.d. | n.d. |
| TZM.bl (3) | 26.2 | 2.4 | 23.5 - 27.8 | n.d. | n.d. | n.d. | |||
| 1 mg | Pos. | 3 | ELISA (3) | 24.1 | 9.1 | 15.7 - 33.8 | n.d. | n.d. | n.d. |
| TZM.bl (1) | 23.5 | n.d. | n.d. | n.d. | n.d. | n.d. | |||
| 3 mg | Neg. | 3 | ELISA (3) | 141.1 | 88.6 | 89.9 - 243.4 | 18.0 | 4.5 | 12.9 - 21.5 |
| TZM.bl (3) | 87.1 | 25.1 | 70.9 - 116.0 | 18.7 | 7.6 | 10.0 - 24.3 | |||
| 3 mg | Pos. | 3 | ELISA (3) | 73.7 | 35.7 | 32.6 - 97.6 | 9.2 | 0.4 | 8.7 - 9.6 |
| TZM.bl (3) | 50.6 | 15.7 | 32.6 - 61.7 | 11.2 | 2.2 | 9.5 - 13.73 | |||
| 10 mg | Neg. | 3 | ELISA (3) | 279.1 | 87.5 | 180.6 - 347.8 | 18.7 | 9.4 | 10.7 - 29.0 |
| TZM.bl (3) | 252.4 | 26.4 | 230.7 - 281.8 | 17.7 | 8.1 | 10.7 - 26.6 | |||
| 10 mg | Pos. | 3 | ELISA (3) | 235.4 | 80.0 | 178.2 - 326.8 | 8.1 | 2.4 | 6.6 - 10.9 |
| TZM.bl (3) | 213.6 | 52.6 | 174.4 - 273.4 | 8.8 | 4.3 | 6.5 - 13.7 | |||
| 30 mg | Neg. | 3 | ELISA (3) | 495.9 | 233.1 | 360.8 - 765.0 | 15.0 | 1.1 | 14.2 - 16.3 |
| TZM.bl (3) | 904.0 | 281.7 | 1166.3 - 606.2 | 16.4 | 1.3 | 15.2 -17.9 | |||
| 30 mg | Pos. | 6 | ELISA (6) | 669.8 | 199.7 | 410.2 - 976.4 | 9.9 | 3.3 | 5.8 - 13.7 |
| TZM.bl (3) | 840.1 | 118.4 | 717.4 - 953.7 | 8.9 | 2.3 | 6.7 - 11.2 | |||
| All | Neg. | 12 | ELISA (9) | - | - | - | 17.2 | 5.5 | 10.7 - 29.0 |
| TZM.bl (9) | - | - | - | 17.6 | 5.7 | 10.0 - 26.6 | |||
| All | Pos. | 15 | ELISA (12) | - | - | - | 9.3 | 2.6 | 5.7 - 13.7 |
| TZM.bl (9) | - | - | - | 9.6 | 2.9 | 6.1 - 13.7 | |||
Acknowledgments
We would like to thank the trial participants for their invaluable support. We thank the Rockefeller University Hospital Clinical Research Support Office and nursing staff for help with recruitment and study implementation, especially N. Buckley, A. Hurley, S. B. A. Shulman and L. Corregano; all members of the Nussenzweig laboratory, especially T. Schoofs, A. Halper-Stromberg, M. and Z. Jankovic; C. Unson-O'Brien, J. Dizon, R. Baptiste and R. Levin for sample processing and study coordination; A. Louie for regulatory support; P. Fast and H. Park for clinical monitoring; E. Giorgi and W. Fischer from Los Alamos National Laboratory; R. T. Gandhi, J. Li and The AIDS Clinical Trials Group (grant UM1 AI068636) and its Statistical and Data Management Center (grant UM1 AI068634). This study was supported by the following grants: Collaboration for AIDS Vaccine Discovery grant OPP1033115 (M.C.N.) and OPP1032144 (M.S.S.). Grant 8 UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS); NIH Clinical and Translational Science Award (CTSA) program; NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-01 (M.C.N.) and 5UM1 AI100645-03 (B.H.H.); Bill and Melinda Gates Foundation grants OPP1092074 and OPP1124068 (M.C.N.); NIH HIVRAD P01 AI100148 (P.J.B. and M.C.N.); the Robertson Foundation to M.C.N. M.C.N. is a Howard Hughes Medical Institute Investigator. Ruth L. Kirschstein National Research Service Award F30 AI112426 (E.F.K.); F31 AI118555 (J.A.H.); The NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI00645 (B.H.H.); The University of Pennsylvania Center for AIDS Research (CFAR) Single Genome Amplification Service Center P30 AI045008 (B.H.H.); The NIH Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID and 1UM1-AI100663) (B.D.W.).
Reviewer Information Nature thanks S. Deeks, D. Richman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions M.C.N., J.F.S., J.A.H. and M.C. wrote the manuscript; J.F.S., M.C. and M.C.N. designed the trial; J.F.S., J.A.H., Y.B., J.C.C.L., L.N., Y.Z.C., C.-L.L. and M.B. performed tissue culture experiments and SGS amplifications; M.S.S. performed TZM-bl assays; J.F.S., J.A.H., Y.B., E.F.K., T.O., A.P.W., G.H.L., P.J.B., F.K., S.J.S., B.H.H., M.C.N. and M.C. analysed the data; E.F.K., G.H.L. and B.H.H. performed SGA analysis; I.S., R.P. and J.F.S. processed patient samples; L.B., S.H., A.S., M.W.-P., B.Z., R.M.G., S.J.S. and M.C. performed patient recruitment; A.F. and N.P. performed statistical analyses; B.J. and B.D.W. performed antigen-specific T cell experiments; T.K. and T.H. produced 3BNC117 and provided PK data.
Author Information The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
References
- 1.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 3.Klein F, et al. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halper-Stromberg A, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein F, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz JA, et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci USA. 2013;110:16538–16543. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shingai M, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caskey M, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoofs T, et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu CL, et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science. 2016;352:1001–1004. doi: 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong R, et al. Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol. 2015;89:2659–2671. doi: 10.1128/JVI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch RM, et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shingai M, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothenberger MK, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci USA. 2015;112:E1126–E1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearney MF, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearney MF, et al. Origin of rebound plasma HIV includes cells with identical proviruses that are transcriptionally active before stopping of antiretroviral therapy. J Virol. 2015;90:1369–1376. doi: 10.1128/JVI.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salantes BSB. Bar, Katharine. Potency and Kinetics of Autologous HIV-1 Neutralizing Antibody Responses During ATI. CROI Conference Abstracts. 2016;Abstract #92 [Google Scholar]
- 20.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci USA. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diskin R, et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J Exp Med. 2013;210:1235–1249. doi: 10.1084/jem.20130221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch RM, et al. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol. 2015;89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou T, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautam R, et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trkola A, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 27.Mehandru S, et al. Adjunctive passive immunotherapy in human immunodeficiency virus type 1-infected individuals treated with antiviral therapy during acute and early infection. J Virol. 2007;81:11016–11031. doi: 10.1128/JVI.01340-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JZ, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS. 2016;30:343–353. doi: 10.1097/QAD.0000000000000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West AP, Jr, et al. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Natl Acad Sci USA. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volberding P, et al. Antiretroviral therapy in acute and recent HIV infection: a prospective multicenter stratified trial of intentionally interrupted treatment. AIDS. 2009;23:1987–1995. doi: 10.1097/QAD.0b013e32832eb285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilby JM, et al. A randomized, partially blinded phase 2 trial of antiretroviral therapy, HIV-specific immunizations, and interleukin-2 cycles to promote efficient control of viral replication (ACTG A5024) J Infect Dis. 2006;194:1672–1676. doi: 10.1086/509508. [DOI] [PubMed] [Google Scholar]
- 34.Jacobson JM, et al. Evidence that intermittent structured treatment interruption, but not immunization with ALVAC-HIV vCP1452, promotes host control of HIV replication: the results of AIDS Clinical Trials Group 5068. J Infect Dis. 2006;194:623–632. doi: 10.1086/506364. [DOI] [PubMed] [Google Scholar]
- 35.Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12 doi: 10.1002/0471142735.im1211s64. Unit 12 11. [DOI] [PubMed] [Google Scholar]
- 37.Li M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salazar-Gonzalez JF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirchherr JL, et al. High throughput functional analysis of HIV-1 env genes without cloning. J Virol Methods. 2007;143:104–111. doi: 10.1016/j.jviromet.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie J, Liu C. Adjusted Kaplan-Meier estimator and log-rank test with inverse probability of treatment weighting for survival data. Stat Med. 2005;24:3089–3110. doi: 10.1002/sim.2174. [DOI] [PubMed] [Google Scholar]
- 43.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Maddison WP, Maddison DR. MacClade – Analysis of Phylogeny and Character Evolution – Version 4. Sinauer Associates, Inc.; 2001. [Google Scholar]
- 45.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 47.Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 48.Parrish NF, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci USA. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maydt J, Lengauer T. Recco: recombination analysis using cost optimization. Bioinformatics. 2006;22:1064–1071. doi: 10.1093/bioinformatics/btl057. [DOI] [PubMed] [Google Scholar]
- 50.Giorgi EE, Bhattacharya T. A note on two-sample tests for comparing intra-individual genetic sequence diversity between populations. Biometrics. 2012;68:1323–1326. doi: 10.1111/j.1541-0420.2012.01775.x. author reply 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altfeld MA, et al. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/jvi.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Addo MM, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.