Abstract
Objective
Plasma levels of branched-chain amino acids (BCAA) are consistently elevated in obesity and type 2 diabetes (T2D) and can also prospectively predict T2D. However, the role of BCAA in the pathogenesis of insulin resistance and T2D remains unclear.
Methods
To identify pathways related to insulin resistance, we performed comprehensive gene expression and metabolomics analyses in skeletal muscle from 41 humans with normal glucose tolerance and 11 with T2D across a range of insulin sensitivity (SI, 0.49 to 14.28). We studied both cultured cells and mice heterozygous for the BCAA enzyme methylmalonyl-CoA mutase (Mut) and assessed the effects of altered BCAA flux on lipid and glucose homeostasis.
Results
Our data demonstrate perturbed BCAA metabolism and fatty acid oxidation in muscle from insulin resistant humans. Experimental alterations in BCAA flux in cultured cells similarly modulate fatty acid oxidation. Mut heterozygosity in mice alters muscle lipid metabolism in vivo, resulting in increased muscle triglyceride accumulation, increased plasma glucose, hyperinsulinemia, and increased body weight after high-fat feeding.
Conclusions
Our data indicate that impaired muscle BCAA catabolism may contribute to the development of insulin resistance by perturbing both amino acid and fatty acid metabolism and suggest that targeting BCAA metabolism may hold promise for prevention or treatment of T2D.
Keywords: Insulin sensitivity, BCAA, Fatty acid oxidation, TCA cycle
Highlights
-
•
Human insulin resistance is associated with perturbed muscle BCAA metabolism.
-
•
Experimental modulation of BCAA metabolic flux alters fatty acid oxidation in vitro.
-
•
Mut heterozygosis leads to increased body weigh and muscle TAG accumulation in mice.
1. Introduction
The global epidemic of type 2 diabetes (T2D) impacts both individual and public health and highlights the urgent need for improved understanding of T2D pathophysiology to facilitate new approaches to prevention and therapy. A key feature of T2D risk is insulin resistance, a metabolic phenotype which can both precede and predict the development of diabetes decades later in high-risk individuals [1], [2]. Emerging data underscore important links between energy metabolism and insulin resistance [3]. Potential contributors to insulin resistance include alterations in lipid oxidative metabolism leading to accumulation of acylcarnitines and other lipid intermediates [4], [5], ceramides [6], reactive oxygen species [7], pro-inflammatory signals [8], and endoplasmic reticulum stress [9].
Branched-chain amino acids (BCAAs) have been identified as potential contributors to insulin resistance [10]. Plasma concentrations of amino acids, including BCAA, are increased in obesity [11] and in parallel with glycemia in patients with T2D [12]. Unbiased metabolomics approaches have confirmed the association between plasma amino acids and metabolic disorders [10], [13], [14], [15]. Newgard and colleagues reported elevated plasma BCAAs in obese insulin resistant humans and demonstrated that increased BCAA intake contributes to insulin resistance in high fat diet-fed rodents [10]. Conversely, BCAA levels are reduced after bariatric surgery or weight loss, paralleling improvements in insulin sensitivity [16], [17], and changes in BCAA are correlated with improved insulin sensitivity after exercise training [18]. Elevations in branched-chain and aromatic amino acids can predict T2D [19] and are also associated with progression of insulin resistance in children in longitudinal cohort studies. Despite these intriguing associations, it remains incompletely understood whether elevations in BCAA in insulin resistant humans reflect chronically increased intake, altered gut microbiome [20], resistance to insulin's ability to suppress proteolysis, or alterations in their metabolism [21], possibly mediated in part by hypothalamic insulin resistance [22]. Furthermore, increases in BCAA may directly modulate insulin action or Tor-dependent nutrient signaling, as seen in cultured cells and animals [23], [24].
Emerging data suggest that perturbations in metabolism of BCAA may not only promote increases in plasma BCAA levels but also contribute to insulin resistance [10], [21], [25]. BCAA are converted into their corresponding branched-chain α-ketoacids (BCKA) by the BCAA transaminase. BCKA are further catabolized within the mitochondrial matrix by the branched-chain α-ketoacid dehydrogenase (BCKDH) complex; a subsequent series of metabolic reactions yield acetyl-CoA (from isoleucine and leucine) and methylmalonyl-CoA (from valine and isoleucine). Methylmalonyl-CoA is then converted by methylmalonyl-CoA mutase (MUT) to succinyl-CoA, which can be incorporated into the tricarboxylic acid (TCA) cycle or enter complex II of the electron transport chain. Thus, altered BCAA metabolism may impact anaplerotic flux into the TCA. Indeed, Adams and colleagues demonstrated elevations in BCAA, but reductions in amino acid-derived carnitine species, in plasma from insulin resistant women and suggested that “anaplerotic stress” could contribute to incomplete fatty acid oxidation [12], [26]. Interestingly, recent metabolomics studies revealed correlations between plasma levels of the three BCKA derived from BCAA and both insulin resistance and elevated fasting plasma glucose [27]. Thus, alterations in BCAA oxidative metabolism rather than BCAA concentrations per se may be more closely associated with insulin resistance.
Using an integrative approach to analyze gene expression in parallel with metabolite profiling in skeletal muscle of insulin resistant humans, we now demonstrate that insulin resistance is associated with alterations in both BCAA and lipid metabolism. Moreover, experimental modulation of BCAA metabolism in cultured cells and in mice also perturbs lipid metabolism. Together these studies provide novel insights into the mechanisms underlying the association of BCAA with dysregulated metabolism in skeletal muscle.
2. Methods
2.1. Study design
The aims of the observational human study were to test the hypothesis that muscle gene expression patterns would differ in individuals with established T2D in comparison to healthy controls. Participants with or without T2D were recruited based on family history of T2D (either or both parents with T2D). Results from these analyses were previously reported [28]. In secondary analyses, we aimed to identify genes and pathways correlating with insulin sensitivity and thus potentially pathogenic for T2D risk. We now report data from these analyses, which formed the basis of the hypotheses further tested in animal and cell culture models in the current report. The demographic and clinical characteristics of the participants are shown in Table 1. The Joslin Diabetes Center Institutional Review Board approved the human study; written informed consent was received from participants prior to inclusion in the study.
Table 1.
Clinical characteristics of study subjects.
| IS | IR | T2D | p value | |
|---|---|---|---|---|
| Subjects | n = 18 | n = 22 | n = 11 | – |
| Age (years) | 35.7 ± 2.3 | 39.4 ± 2.6 | 50.7 ± 3.9c | 0.005 |
| Females | 10 (56%) | 12 (55%) | 7 (64%) | – |
| BMI (kg/m2) | 25.8 ± 1.0 | 27.9 ± 1.2 | 31.5 ± 2.7 | 0.067 |
| Fasting Glucose (mmol/l) | 4.89 ± 0.07 | 5.27 ± 0.09 | 7.25 ± 0.85d | <0.0001 |
| Fasting Insulin (pmol/l) | 28 ± 3 | 58 ± 7 | 137 ± 46c | 0.001 |
| HbA1c (%) | 5.0 ± 0.1 | 5.1 ± 0.1 | 6.9 ± 0.4d | <0.0001 |
| Triglycerides (mmol/l) | 0.69 ± 0.09 | 1.07 ± 0.10a | 1.16 ± 0.17a | 0.013 |
| Cholesterol (mmol/l) | 4.42 ± 0.19 | 4.43 ± 0.14 | 4.05 ± 0.24 | 0.333 |
| HDL-Cholesterol (mmol/l) | 1.48 ± 0.08 | 1.23 ± 0.09 | 1.00 ± 0.07c | 0.004 |
| LDL-Cholesterol (mmol/l) | 2.60 ± 0.24 | 2.94 ± 0.14 | 2.58 ± 0.27 | 0.356 |
| SI (IVGTT) | 8.12 ± 0.56 | 2.81 ± 0.27d | 1.69 ± 0.45d | <0.0001 |
| Leucine | 0.97 ± 0.05 | 1.02 ± 0.03 | 0.97 ± 0.04 | 0.560 |
| Isoleucine | 1.03 ± 0.06 | 1.14 ± 0.04 | 1.00 ± 0.05 | 0.135 |
| Valine | 0.98 ± 0.04 | 0.98 ± 0.05 | 1.00 ± 0.04 | 0.793 |
Subjects were metabolically characterized and insulin sensitivity (SI) was determined by intravenous glucose tolerance test (IVGTT). Subjects without T2D were categorized as insulin sensitive (IS) or resistant (IR) based on SI values above or below the median value of 4.78 for a larger population of normoglycemic individuals at the Joslin Diabetes Center. Relative quantitation of fasting plasma BCAA was performed; levels are relative to the median of the IS group. Data are mean ± SEM. One-Way ANOVA was applied to detect statistical significance between groups; statistical significant features were further analyzed by post-hoc Dunnet's test: ap < 0.05, bp < 0.01, cp < 0.005, dp < 0.0001 vs IS.
Bold indicates p < 0.05.
2.2. Human metabolic characterization and muscle biopsies
Written informed consent was obtained from participants. All subjects had normal coagulation, liver function, and no other major systemic illness. Individuals with and without established T2D were recruited for the study. Subjects with diabetes were treated with dietary measures and exercise, and were drug-naïve for diabetes medication. All participants without a history of diabetes underwent a 75 g glucose tolerance test to exclude impaired glucose tolerance or diabetes, analyzed according to World Health Organization criteria [29]. An intravenous glucose tolerance test was performed on a non-sequential day on all subjects without diabetes; data were analyzed using Minimal Model software (MINMOD) [30] for calculation of SI. Subjects without diabetes were classified as insulin sensitive or insulin resistant on the basis of SI values above or below median (4.79) for a larger population of normal glucose tolerant individuals studied at Joslin [31]. Since SI values were not normally distributed over the subject population, values were log-transformed, and log SI was chosen as the primary metabolic variable for correlation analyses. Fasting blood samples were obtained for insulin, glucose, cholesterol and liver enzymes. Following local anesthesia with 1% lidocaine, percutaneous biopsies of vastus lateralis muscle were performed, using a triport cannula and Bergstrom needle. Samples were blotted free of blood, connective tissue and visible fat, frozen in liquid nitrogen, and stored at −80 °C until further analysis.
2.3. RNA isolation and microarray analysis
Total RNA was isolated from frozen muscle after homogenization with a Polytron (Brinkmann Instruments) in TRIzol® (Invitrogen, Carlsbad, CA), using high-salt precipitation modification. RNA was purified using RNeasy® columns (Qiagen, Chatsworth, CA). An equal number of samples from all three groups were included at all steps to minimize risk of technical bias. Five μg of DNAse I-treated total RNA from each of these samples was used to generate double-stranded cDNA (SuperScript Choice, Invitrogen), followed by in vitro transcription (ENZO BioArray RNA labeling kit, Affymetrix, Santa Clara, CA). Five μg of adjusted, fragmented cRNA were hybridized to Affymetrix HG-U133 2.0 Plus arrays. Arrays were washed, stained, and scanned (GeneChip® Scanner 3000). Remaining total RNA was utilized for cDNA synthesis and subsequent qRT-PCR. Microarrays were preprocessed and scaled using Affymetrix MAS 5.0, and log2-transformed. Primary data from microarrays have been deposited into GEO (GSE25462). Pearson correlation of probe sets to log SI was performed in the R software (www.r-project.org). Gene Set Enrichment Analysis (http://www.broad.mit.edu/gsea/) was used to identify gene sets overrepresented in correlation with logSI.
2.4. Animal care and treatment
Mice were housed 4 per cage in an OLAW-certified animal facility, with 12 h light cycle. For dietary analyses, six week old C57BL/6 male mice were obtained from Jackson Laboratory and placed on a chow diet (10% calories from fat, D12450, Research Diets, Inc.) or HF diet (45% calories from fat, D12451, Research Diets, Inc.) for 4 months. Mice were fasted overnight for 16 h before anesthesia (intraperitoneal pentobarbital, 50 mg/kg) and harvest of skeletal muscle. For exercise training experiments, Sprague–Dawley rats were maintained with or without wheel cage for 4 weeks on a chow diet as previously described [32]; triceps muscle was harvested after anesthesia with pentobarbital (50 mg/kg, intraperitoneal). For denervation studies, C57BL/6J mice (age 16 weeks) were denervated by surgical removal of 5–6 mm of sciatic nerve from one hind limb; the intact contralateral limb was used as a control. After nine days of daily monitoring, mice were anesthetized with pentobarbital (50 mg/kg, intraperitoneal) prior to dissection of muscle from both limbs. For analysis of pharmacologically-induced insulin resistance, mice were treated with vehicle (saline) or the insulin receptor antagonist S961 (10 nmol), infused via osmotic pump (Alzet 2001) implanted subcutaneously in the dorsal interscapular region. S961-treated mice were hyperinsulinemic as compared with saline-infused mice (108 ng/ml vs. 5 ng/ml, p = 0.004), consistent with prior studies demonstrating insulin resistance with this protocol [33]. After 1 week, mice were fasted overnight for 16 h and refed for 5 h prior to anesthesia with pentobarbital (50 mg/kg, intr) and muscle isolation. All animal protocols were approved by the Joslin Institutional Animal Care and Use Committee and the National Human Genome Research Institute Animal Care and Use Committee.
2.5. Mut mouse generation and analysis
Generation of Mut+/− and Mut−/− mice has been previously described [34]. Genotyping was performed by PCR using tail DNA and two primer pairs. Littermate wild-type and Mut+/− 5 week-old male mice were fed a HF diet (45% kcal from fat, D12451, Research Diets, Inc.) for 4 months. Blood glucose was measured and insulin levels determined by ELISA (Crystal Chem, Inc.). Glucose tolerance was analyzed after an overnight fast by intraperitoneal injection of glucose (1.5 g/kg). Insulin tolerance testing was performed after a 4 h fast (insulin 0.65 U/kg, intraperitoneally). For tissue collection, mice were fasted overnight and anesthetized prior to dissection of quadriceps muscle. RNA was isolated and cRNA was prepared for hybridization to Affymetrix M430 2.0 microarrays (GEO, GSE66766).
2.6. Cell culture
Mouse C2C12 myoblasts were maintained in DMEM supplemented with 10% FBS. To initiate differentiation, confluent C2C12 cells were incubated in DMEM containing 2% horse serum (HS). Full differentiation was observed by day 5. Mouse embryonic fibroblasts (MEFs) were isolated from wild-type and Mut−/− mice as described [34]. MEFs were maintained in DMEM with 10% FBS. For fatty acid exposure experiments, palmitate and oleate (Alltech) were complexed with BSA (3:1 ratio) and dissolved in DMEM at the indicated final concentration.
2.7. RNA isolation and expression analysis
Total RNA was isolated from cells and mouse tissues with Trizol (Invitrogen). cDNA was synthesized and analyzed by RT-PCR (ABI Prism 7000 or 7700 Sequence Detection System, Applied Biosystems), using SYBR Green. Primer sequences are available upon request.
2.8. Comprehensive metabolic profile
Untargeted metabolomic profiling was performed using multi-platform mass spectroscopy (Metabolon, Durham, NC). Detailed descriptions of sample preparation, mass spectroscopy, and automated metabolite identification procedures have been previously published [27]. We obtained semi-quantitative concentrations (expressed in arbitrary units) for 348 biochemicals in human muscle biopsies. We excluded from analysis 33 metabolites with undetectable values in >75% of samples in any of the groups, metabolites with undetectable values, or outliers (defined as values deviating more than two standard deviations from the mean).
2.9. Valine and fatty acid oxidation and TAG accumulation
For valine oxidation, cells were incubated with Dulbecco's Phosphate Buffered Saline (dPBS) with 0.2% BSA in the presence of 0.1 mM 0.5 mCi/ml L-[U–14C]Valine for 2 h. For fatty acid oxidation (FAO) experiments, cells were grown in 12-well plates and incubated overnight with DMEM with 5 mM glucose, 1 mM carnitine and 0.1 mM oleic acid; [1–14C]-oleic acid was then added at a concentration of 1 μCi/ml (5 mCi/mmol) for 2 h. FAO assay in MEFs was performed by incubating cells with dPBS supplemented with 2.5 mM glucose, 0.5 mM carnitine, 0.1 mM oleic acid, and 1 μCi/ml (5 mCi/mmol) [1–14C]-oleic acid for 4 h. For all oxidation experiments, the reaction was terminated by addition of 100 μl of perchloric acid to each well, and a filter paper soaked with 100 μl of phenylethylamine was placed over each well to capture 14CO2. For analysis of cellular TAG content, lipids were extracted using chloroform/methanol and TAG content was determined using the Triglyceride Assay Kit (Sigma).
2.10. Oxygen consumption
Oxygen consumption rates (OCR) were measured using a Seahorse XF24 Flux Analyzer. MEF cells were grown for 24 h in DMEM with 10% FBS. The following day, cells were incubated with DMEM with 1% FBS and basal OCR was measured. C2C12 cells were incubated in KRB buffer supplemented with 2.5 mM glucose and 0.5 mM carnitine for 1 h. Half of the samples were then incubated with 0.2 mM Cl-IC, and OCR was measured. Finally, 0.2 mM oleate was injected into each well, and OCR was monitored.
2.11. Statistics
Unless otherwise stated, data are presented as mean ± SEM. Two-tailed Student t-test was used to assess significance. Where three or more groups were present, a one-way ANOVA test was applied. p values < 0.05 were considered statistically significant. Partial least squares discriminant analysis was implemented in XLSTAT (version 2013.6.03) using autoscaled data and leave-one-out cross-validation. Variable Importance to Projection (VIP) scores higher than 1.5 and p < 0.05 in the IR vs IS comparison were used to identify metabolic classifiers between groups.
3. Results
3.1. Expression of genes related to BCAA oxidation correlates positively with insulin sensitivity in human subjects
To identify gene expression and metabolomic patterns associated with insulin resistance, we recruited 40 individuals with normal glucose tolerance verified by oral glucose tolerance testing, and 11 with T2D (Table 1). All subjects underwent insulin-modified intravenous glucose tolerance tests analyzed using the Bergman minimal model (MinMod) [30] to quantify insulin sensitivity (SI) (range 0.49–14.28). Participants with normal glucose tolerance were categorized as insulin sensitive (IS) or resistant (IR) based on SI values above or below the median value of 4.78 for a larger population of normoglycemic individuals studied at the Joslin Diabetes Center [31]. Consistent with their insulin resistant phenotype, IR subjects had higher fasting plasma glucose, insulin, and triglyceride concentrations; age, BMI, and HbA1c levels did not differ from insulin sensitive individuals (Table 1). Individuals with T2D were older, had higher BMI and, as expected, had increased HbA1c levels, higher fasting plasma glucose and insulin concentrations, and lower insulin sensitivity (SI) as compared with IS and IR subjects. Plasma levels of BCAA did not differ in this population with only modestly increased BMI. Quadriceps muscle biopsies were performed after an overnight fast in all subjects and processed for microarray-based gene expression and mass spectrometry-based metabolomic analyses.
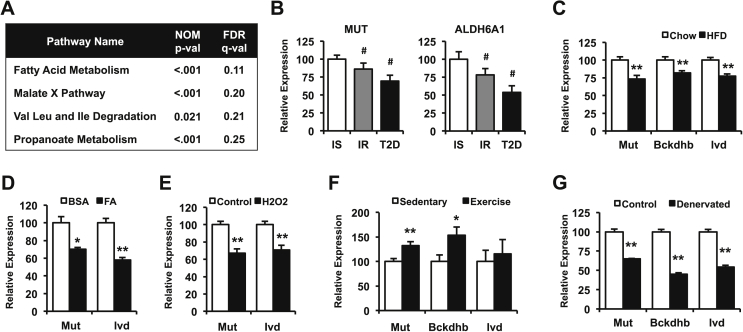
To identify potential pathways affecting insulin sensitivity, gene expression was correlated with insulin sensitivity (log SI) for individual patients. Correlation coefficients for all probe sets were used as a ranking metric, and gene set enrichment analysis (GSEA) was performed to identify pathways correlating with insulin sensitivity. Pathways related to inflammation and immune activation were inversely correlated with insulin sensitivity (FDR ≤ 0.25, Table S1), consistent with the established links between inflammation and insulin resistance [8]. Conversely, pathways positively correlating with insulin sensitivity (FDR ≤ 0.25) included fatty acid metabolism, malate exchange, BCAA degradation, and propanoate/propionate metabolism (Figure 1A). BCAA degradation genes contributing to pathway enrichment included proximal (BCKDHB, DBT, DLD), middle (IVD, ACADSB, MCCC1/2, ECHS1, EHHADH, ACAA2), and distal genes (HMGCS2, OXCT1, ALDH6A1/ALDH3A2, PCCA/B, MUT), indicating downregulation in multiple steps of the pathway in insulin resistant individuals (Figure S1). Furthermore, both BCAA and propanoate pathways showed a significant overlap; most of the distal BCAA metabolic genes contributed to propanoate pathway enrichment (Figure S1), including MUT and ALDH6A1. These genes also showed the highest correlation with insulin sensitivity among BCAA degradation genes (Table S2). Given the strong associations between plasma BCAA and insulin resistance, we analyzed expression of genes within the BCAA oxidative pathway by RT-PCR. MUT and ALDH6A1 were downregulated in both insulin resistance and established T2D (Figure 1B), confirming the downregulation of these genes observed in the microarray analysis.
Figure 1.
Skeletal muscle BCAA gene expression correlates with insulin sensitivity. (A) Pathways positively correlating with logSI (false discovery rate (FDR) q < 0.25), identified through gene set enrichment analysis of microarray data from human skeletal muscle. (B) mRNA levels from human muscle biopsies of insulin-sensitive (IS, white bars), insulin-resistant (IR, gray bars) and subjects with diabetes (T2D, black bars) normalized to cyclophilin levels. (C) mRNA levels from mouse gastrocnemius muscle after 4 months on a HFD (black bars, n = 8) compared to chow diet (white bars, n = 8). (D) C2C12 myotubes were incubated in the presence (black bars) or absence (white bars) of a mixture of oleic and palmitic acid 0.25 mM each for 18 h, before mRNA levels were determined (data from one representative experiment of three independent experiments, each with n = 4 replicates). (E) mRNA levels from C2C12 myotubes treated with (black bars) or without (white bars) 0.5 mM hydrogen peroxide (H2O2) for 2 h, followed by 2 h recovery in differentiation media (one representative of two independent experiments, each with n = 4 replicates). (F) mRNA levels from rat triceps muscle after 1 month of voluntary wheel training (black bars, n = 10) compared to sedentary controls (white bars, n = 9). (G) mRNA levels from mice subjected to unilateral denervation, comparing control (white bars, n = 3) and denervated limbs (black bars, n = 3). Data are presented as mean ± SEM. # indicates one-way ANOVA p < 0.05. *, p < 0.05 and **, p < 0.01 after a two-tail t-test.
We next examined whether BCAA gene expression patterns are also altered in skeletal muscle of mice with diet-induced insulin resistance. Mut mRNA levels were downregulated by 27% in high fat (HFD)-fed mice (p < 0.005), and expression of other BCAA metabolism genes Bckdhb and Ivd was also decreased (Figure 1C). Furthermore, exposure of C2C12 myotubes to a mixture of palmitate and oleate (0.25 mM each) downregulated expression of Mut and Ivd by 30% and 40% (p < 0.05 and p < 0.01, respectively, Figure 1D). Interestingly, incubation of myotubes with H2O2, mimicking an increase in reactive oxygen species, also decreased expression of these BCAA degradation pathway genes (Figure 1E), suggesting a role for oxidative stress in the downregulation of these genes. However, insulin resistance induced by the insulin receptor blocker S961 did not decrease expression of BCAA pathway genes in skeletal muscle (Figure S2), suggesting systemic insulin resistance and hyperinsulinemia per se does not drive regulation of BCAA pathway expression. Expression of genes within this pathway was also modulated in response to activity. One month of voluntary exercise training, an intervention known to improve metabolic health [32], increased skeletal muscle mRNA levels of Bckdhb and Mut by 50% and 30% (p < 0.05 and p < 0.005, respectively) in rats compared to sedentary controls (Figure 1F). Conversely, denervation of mouse hind limb markedly decreased expression of Mut, Bckdhb, and Ivd in soleus muscle as compared with the control limb (Figure 1G). Together, these results indicate that muscle expression of BCAA degradation genes can be downregulated by lipid excess (both in vivo and in vitro) and inactivity, and upregulated in response to exercise.
3.2. Human insulin resistance is associated with alterations in skeletal muscle BCAA metabolism, TCA cycle intermediates, and acylcarnitine levels
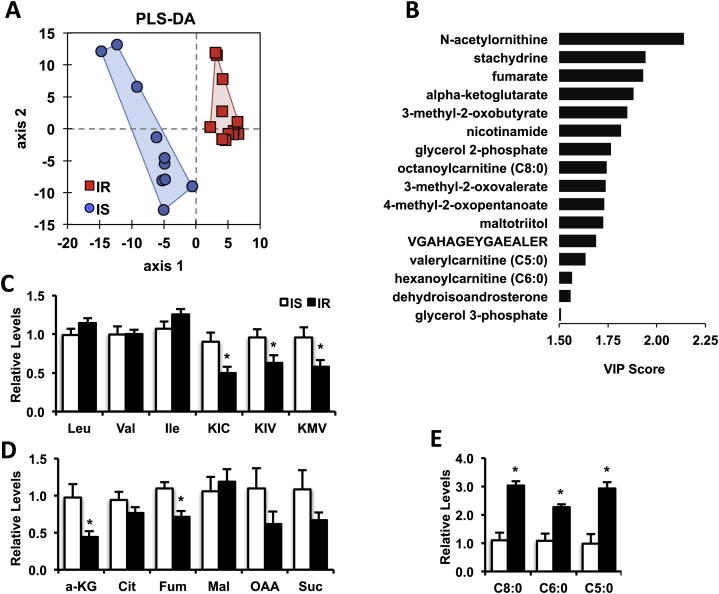
To identify new molecular signatures of early insulin resistance, we performed comprehensive non-targeted mass spectrometry to analyze the muscle metabolome of IS and IR humans. Limited amount of muscle sample was available; thus, this analysis was done on a subset of the larger cohort for which adequate biopsy material was available (10 IS and 13 IR subjects; demographic and metabolic characteristics provided in Table S3). Using a combination of UHPLC-MS/MS and GC–MS, 315 metabolites were detected, with 145 unidentified and 170 named metabolites (complete data provided in Data File S1). Unsupervised principal component analysis revealed no discrimination between IS and IR groups. Supervised partial least squares discriminant analysis (PLS-DA) was next employed to identify metabolic variables discriminating between IS and IR groups (Figure 2A), and variable importance to projection (VIP) scores to identify metabolites contributing to between-group discrimination. Known compounds with a VIP score >1.5 and p < 0.05 for comparison of IS vs. IR groups are shown in Figure 2B. Consistent with the gene expression analysis suggesting alterations in skeletal muscle BCAA metabolism, all three branched chain ketoacids (BCKA), ketoisovaleric acid (KIV), ketomethylvalerate (KMV), and ketoisocaproate (KIC) contributed to group discrimination (VIP scores of 1.85, 1.74, and 1.73, respectively). While tissue BCAA (Val, Ile, and Leu) were similar between groups, all three BCKA were significantly decreased in IR compared with IS muscle (Figure 2C).
Figure 2.
Skeletal muscle metabolomic analysis reveals reduced BCKA and TCA cycle intermediates and altered acylcarnitine levels. (A) PLS-DA score plot (R2Y = 0.897, Q2 = 0.574) of the metabolomics dataset with IS (blue circles) and IR subjects (red squares). (B) Known metabolites arising from the PLS-DA model with a VIP score > 1.5 and p < 0.05 for the IR vs. IS group comparison. Levels of BCAAs and BCKAs (C), TCA cycle intermediates (D) and acylcarnitines (E) from IS (white bars) and IR (black bars) subjects are shown. Data are mean ± SEM, normalized to the median of the IS group. *p < 0.05 (t-test). KIC, ketoisocaproate; KIV, ketoisovalerate; KMV, ketomethylvalerate; a-KG, alpha ketoglutarate; Cit, citrate; Fum, fumarate; Mal, Malate; OAA, oxaloacetate; Suc, succinate.
Additional oxidative pathways were also altered in insulin resistant vs. sensitive muscle. The TCA cycle intermediates fumarate and α-ketoglutarate also contributed to group separation (VIP scores 1.93 and 1.88, respectively), and were significantly reduced in insulin resistance, with similar trends for oxaloacetate and succinate (Figure 2D). Interestingly, skeletal muscle levels of fumarate, α-ketoglutarate, KIC, and KMV also correlated with insulin sensitivity (Table S4). Moreover, and consistent with data from high fat-fed rodents [5], the medium/short-chain acylcarnitines octanoylcarnitine, valerylcarnitine, and hexanoylcarnitine, were also selected in the PLS-DA model (VIP scores of 1.74, 1.63, and 1.57, respectively). These medium-chain acylcarnitines were increased by 2–3 fold in IR muscle (Figure 2E), suggesting defects in complete fatty acid oxidation. Together, these data indicate that both BCAA and lipid metabolism are altered in skeletal muscle from insulin resistant humans.
3.3. Alterations in BCAA oxidative metabolism contribute to impaired fatty acid oxidation in cultured cells
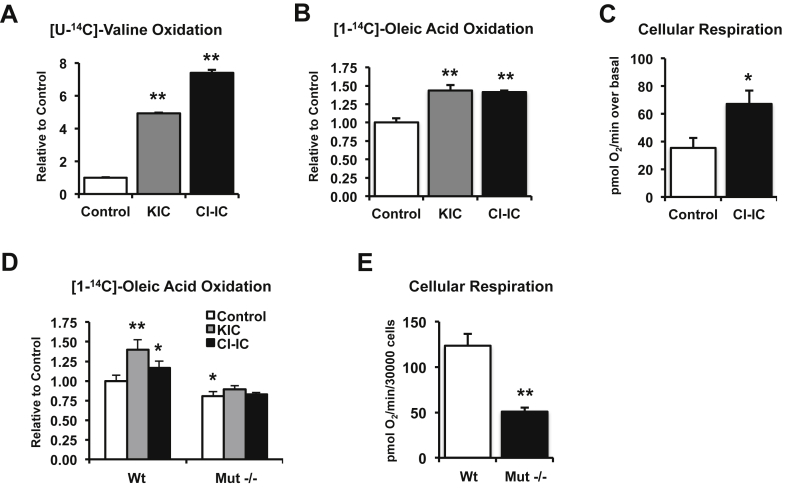
Given the emerging and robust links between BCAA levels and insulin resistance, the major role for skeletal muscle in mediating BCAA catabolism, and the association of muscle BCAA and lipid oxidative gene expression with insulin sensitivity in our cohort, we asked whether alterations in BCAA flux might be a primary contributor to disordered lipid metabolism in muscle. We first tested whether modulating the Bckdh complex would alter fatty acid oxidation in cultured skeletal muscle cells. C2C12 myotubes were treated with either KIC or chloroisocaproic acid (Cl-IC), a non-metabolized KIC mimetic. Both KIC and Cl-IC are inhibitors of the branched-chain ketoacid dehydrogenase kinase (Bckdk), resulting in strong activation of the Bckdh complex and increased flux through the BCAA oxidative pathway [35]. Treatment with either KIC or Cl-IC increased oxidation of valine in C2C12 myotubes by 5- and 7-fold respectively, as predicted (Figure 3A). In parallel, KIC and Cl-IC both increased complete fatty acid oxidation by 1.5 fold (p < 0.05), as measured by CO2 production after incubation with [1–14C]-oleic acid (Figure 3B). Cl-IC also resulted in an 89% higher increase in oxygen consumption in response to oleic acid (p < 0.05, Figure 3C), indicating that increasing BCAA oxidative metabolism enhances both fatty acid oxidation and cellular respiration.
Figure 3.
Alteration of BCAA pathway flux modulates complete fatty acid oxidation. (A) C2C12 myotubes were treated with 2 mM ketoisocaproate (KIC, gray bar) or 0.5 mM chloroisocaproate (Cl-IC, black bar) for 30 min and valine oxidation was measured as 14CO2 produced from [U–14C]-valine (n = 4 replicates). (B) C2C12 myotubes were treated with 2 mM KIC or 0.2 mM Cl-IC for 30 min before complete fatty acid oxidation was measured as 14CO2 produced from [1–14C]-oleic acid. Shown are data from one representative experiment, from two independent experiments, each with n = 4 replicates. (C) C2C12 cells were incubated in the presence (black bar) or absence (white bar) of 0.2 mM Cl-IC. Oxygen consumption rate was assessed before and after cells were exposed to 0.1 mM oleic acid (n = 5 replicates). (D) Mouse embryonic fibroblasts (MEFs) derived from wild-type or Mut−/− mice were incubated with 2 mM KIC or 0.2 mM Cl-IC and fatty acid oxidation was measured. Shown are data from one representative experiment, from two independent experiments, each with n = 4 replicates. (E) Basal oxygen consumption rate from wild-type (white bars) and Mut−/− (black bars) MEFs was measured (n = 9 replicates). Data are mean ± SEM. One-Way ANOVA was applied when three or more groups are compared. *p < 0.05 **, p < 0.005 vs control.
To confirm that these effects were indeed mediated by increased flux through BCAA oxidative pathways, we analyzed metabolism in mouse embryonic fibroblasts (MEFs) derived from wild type mice or mice null for the mutase (Mut) gene, the distal step in oxidative metabolism of Val and Ile. Both KIC and Cl-IC increased complete fatty acid oxidation in wild type MEFs, as in myotubes (Figure 3D). By contrast, MEFs derived from Mut−/− mice had decreased complete fatty acid oxidation in the basal state, and no increase was observed after incubation with KIC or Cl-IC (Figure 3D). Furthermore, basal oxygen consumption was reduced by 60% (p < 0.01) in Mut−/− MEFs, as compared with wild type (Figure 3E). Together, these data indicate altered flux through BCAA oxidative pathways and reduced expression of Mut may contribute to impaired fatty acid oxidation and reduced total cellular respiration.
3.4. Mut heterozygous mice have increased body weight and impaired muscle lipid metabolism
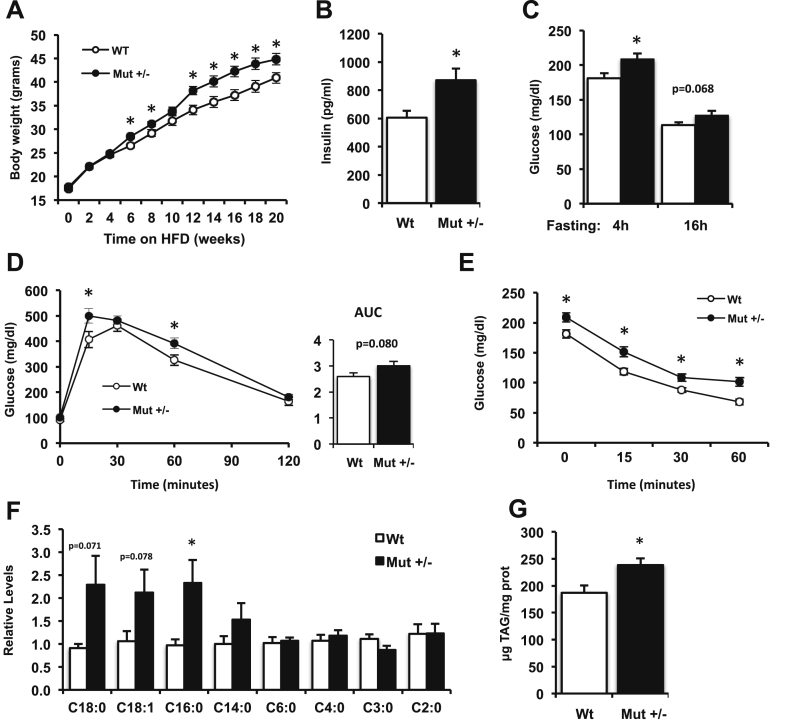
To test whether alterations in BCAA metabolism could affect systemic metabolism in vivo, we analyzed mice with heterozygous disruption of Mut. Homozygous mutations in this gene cause methylmalonic aciduria (MMA), a severe metabolic disease characterized by increased plasma concentrations of methylmalonic acid. Deletion of the Mut gene in mice results in early neonatal lethality and induces significant mitochondrial dysfunction [36], [37]. In contrast, heterozygous Mut+/− mice are viable and show normal growth. We evaluated whether Mut+/− mice would develop metabolic defects when challenged with a high-fat diet (45% calories from fat). Mut+/− mice gained more weight in response to high-fat feeding than their wild-type littermates (Figure 4A). In addition to higher body weight, these mice had a 43% increase in fasting insulin concentrations as compared with wild type controls (p < 0.05, Figure 4B) and increased fasting glucose (Figure 4C). Glucose tolerance tended to be impaired in Mut+/− mice compared to wild-type littermates (Figure 4D). Interestingly, insulin tolerance testing revealed significantly higher blood glucose at each time point in Mut+/− mice, but no difference in glycemic response (% decrease) following insulin (Figure 4E).
Figure 4.
Mut+/−mice show increased weight, impaired glucose homeostasis, and increased triglyceride accumulation after high fat diet. (A) Wild-type (Wt, white circles, n = 16) and Mut+/− (black circles, n = 16) male mice were fed a 45% high fat diet for 20 weeks and body weight was monitored. (B) Overnight (16 h) fasting insulin and (C) 4 h and overnight (16 h) fasting glucose levels were determined. (D) Intraperitoneal glucose (1.5 g/kg) and (E) insulin tolerance tests (0.65 U/kg) were performed in Wt and Mut+/− mice. (F) Acylcarnitine levels in skeletal muscle, normalized to the median of the Wt group (n = 8 per group). (G) Triglyceride content was measured in quadriceps of Wt and Mut+/− mice (n = 16 per group). Data are mean ± SEM. *p < 0.05 (t-test). AUC, area under the curve.
To further examine the effects of decreased Mut expression, we analyzed skeletal muscle gene expression in wild-type and Mut+/− mice. As anticipated from the deletion of one copy of Mut, expression of Mut was reduced by 50% in heterozygous muscle (Figure S3A). Reduced Mut also decreased expression of multiple mitochondrial genes regulating oxidative metabolism, including Sdha, Cs, Acadl, and the ATP synthase subunit Atp5d (Figure S3A), with a trend for reduced Pgc1a (Ppargc1a). In contrast, the lipid synthesis gene Scd2 was increased, with similar trends for upregulation of Fasn, Fads3, and Fabp5 (Figure S3B).
To evaluate additional metabolic effects of decreased Mut expression, we analyzed metabolites in muscle in the fasting state. Although we observed no significant changes in muscle BCAA levels (Figure S4), acylcarnitine patterns were altered in response to Mut heterozygosity. Palmitoylcarnitine (C16:0) levels were more than 2-fold higher in muscle from Mut+/− versus wild-type mice (p < 0.05), and levels of other acylcarnitines tended to be increased in Mut+/− as well (Figure 4F), indicating disordered fatty acid oxidation. Finally, muscle triglyceride content was 30% higher in Mut+/− mice (Figure 4G), further confirming that downregulation of Mut alters lipid metabolism in muscle.
4. Discussion
Recent unbiased metabolomics analyses have demonstrated robust links between insulin resistance, obesity, and T2D and alterations in circulating metabolites, indicating major dysregulation of systemic metabolism. These metabolic disorders are associated with changes in plasma amino acids [11], including BCAA, phenylalanine, tyrosine, and glycine, branched-chain ketoacids, and other metabolites such as 2-hydroxybutyrate, 2-aminoadipidic acid, lysophosphatidylcholine (18:2), and medium- and short-chain acylcarnitines [10], [12], [27], [38], [39], [40], [41], [42], [43]. Among these, increased plasma BCAA concentrations also predict progressive insulin resistance and T2D [19]. In this manuscript, we identify a muscle BCAA-related dual transcriptomic and metabolomics signature of human insulin resistance using unsupervised analysis of muscle biopsies from a normoglycemic population across a broad range of insulin sensitivity. mRNA for multiple genes regulating BCAA metabolism are inversely correlated with insulin sensitivity. These gene expression patterns are most striking for distal steps of valine and isoleucine metabolism, including propanoate/propionic acid and methylmalonyl-CoA metabolism. Similar reductions in genes related to BCAA metabolism were observed in mice made insulin resistant by high-fat feeding, indicating these changes can be acquired.
To identify metabolic pathways associated with insulin sensitivity, the metabolome was analyzed in IS and IR human subjects without T2D. Notably, this cohort of insulin resistant subjects had normal glucose tolerance, fasting glucose and HbA1c levels, and only slightly higher BMI compared to insulin sensitive controls. Despite this mild degree of metabolic dysfunction, key differences in muscle metabolic profiles concordant with gene expression pathway differences in insulin resistance were observed. All three BCKA were reduced in insulin resistant muscle, despite similar tissue and plasma BCAA levels. This pattern may be potentially due to altered enzymatic activity of the branched-chain aminotransferase or the reduced levels of its substrate α-ketoglutarate. Indeed, both α-ketoglutarate and fumarate were reduced in insulin resistant muscle, with similar trends for several other TCA cycle metabolites. Conversely, medium-chain acylcarnitines (including C5, C6, and C8) were more than 2-fold higher in insulin resistant muscle; increases in C5 species may reflect dysregulation of BCAA and propanoate metabolism. These patterns are also concordant with those observed with BCAA supplementation in high-fat diet-fed mice [10], where BCAA-driven elevations in muscle short-chain acylcarnitines of both odd and even chain length were especially prominent. Increased C5 acylcarnitine levels also suggest alterations in amino acid metabolism, as these metabolites are markedly elevated in humans with reduced activity of IVD or short/branched chain acyl CoA dehydrogenase [44], [45]. While cross-sectional tissue metabolite data cannot allow us to determine which of these defects are primary to the insulin resistant phenotype in humans, it is possible that reduced valine and isoleucine catabolism could reduce TCA anaplerosis and flux, further decreasing oxidative capacity and fatty acid oxidation, and promoting incomplete lipid oxidation, as proposed by Adams et al. [46], [47]. Furthermore, acylcarnitines resulting from incomplete oxidation or other perturbations in both amino acid and fatty acid metabolism could promote oxidative stress and insulin resistance [4]. Taken together, our data support the hypothesis that multiple steps of muscle BCAA metabolism are impaired in insulin resistant humans, and that these defects may not only contribute to concomitant alterations in TCA cycle activity but also interact to perturb lipid metabolism.
To determine whether changes in BCAA metabolism observed in human muscle could contribute to the pathogenesis of dysregulated metabolism associated with insulin resistance, we experimentally modulated BCAA oxidative metabolism and analyzed the metabolic impact. While our studies in cultured myotubes and MEFs cannot fully recapitulate the complexity or tissue-specific regulatory patterns of intact skeletal muscle, these studies demonstrate that experimental modulation of BCAA oxidation exerts robust effects on cellular metabolism. Firstly, activation of the BCKDH complex by either KIC or chloroisocaproic acid not only increases BCAA catabolic flux, as predicted, but also increases fatty acid oxidation, potentially via increases in TCA activation. Conversely, reduced flux (e.g. deletion of Mut) impairs fatty acid oxidation and reduces cellular respiration. These data are also consistent with recognized effects of exercise to promote increases in both BCAA and fatty acid oxidation [48], [49]. While the mechanisms responsible for these effects are still not fully understood, our data support the hypothesis that reduced flux through the BCAA pathway leads to incomplete oxidation and reduced availability of BCAA-derived anaplerotic TCA intermediates; in turn, this may reduce complete fatty acid oxidation and increase accumulation of acylcarnitines and mitochondrial stress [26], [47].
We now demonstrate that chronic alterations in tissue BCAA metabolism also impact systemic metabolism in vivo. Indeed, mice with heterozygous deletion of the Mut gene, which have decreased expression of Mut, have increased weight gain, hyperinsulinemia, elevated blood glucose levels, and impaired lipid metabolism in skeletal muscle in response to high-fat feeding. These phenotypes are especially striking given that Mut heterozygous mice have only a 50% reduction in a single enzyme within the BCAA metabolic pathway. Furthermore, MUT deficiency affects degradation of only two out of the three BCAA (Ile and Val, but not Leu). Thus, it is likely that alterations in expression and metabolic flux at multiple steps of the BCAA and other key pathways, as observed in our human cohort, will be necessary to fully recapitulate human disease phenotypes. Furthermore, lifestyle factors such as increased dietary BCAA [10] or inactivity, could further magnify the metabolic impact of isolated MUT deficiency or more global alterations in BCAA oxidative flux. In addition to Val and Ile, catabolism of Thr, Met, and odd-chain fatty acids can also provide methylmalonyl-CoA, the substrate for Mut; therefore, we cannot exclude the possibility that Mut heterozygosity could also affect metabolism of these compounds. Finally, we recognize that the Mut+/− mice harbor a whole-body knockout, and effects in tissues other than skeletal muscle (e.g. adipose, liver) may also contribute to impaired glucose tolerance and lipid metabolism. Collectively, however, these data demonstrate that alterations in distal BCAA oxidative metabolism, when combined with lipid excess, can not only perturb cellular oxidative mitochondrial metabolism, but also contribute to dysregulation of systemic metabolism in vivo.
Perturbations in cellular metabolism can result from dysregulation at multiple levels. These include not only transcriptional dysregulation but also allosteric regulation, substrate availability, and posttranslational modifications (e.g. phosphorylation and acetylation), all of which have been shown to be important contributors to regulation of BCAA metabolism [50], [51]. At a transcriptional level, perturbed expression of genes related to BCAA metabolism has now been identified in both adipose and muscle tissue from several distinct cohorts. For example, genes regulating BCAA catabolism are downregulated in omental adipose from obese insulin resistant individuals, as compared with obese, yet insulin sensitive, individuals [52] and in mice with transgenic expression of GLUT4 in adipose [21]. Downregulation of BCAA catabolic genes in the heart was recently reported in a mouse model of experimental pressure overload-linked heart failure [53].
The importance of environmental factors independent of DNA sequence has been underscored by studies of human monozygotic twins; obese [54] and less active [55] members of twin pairs have reduced BCAA gene expression, as compared with their non-obese co-twin. Similar patterns are observed at the protein level, with reduced ALDH6A and PCCB in mitochondria isolated from muscle of lean insulin resistant individuals. Moreover, interventions which modify insulin sensitivity or metabolism also impact BCAA expression in humans. For example, BCAA-related gene expression is increased (including ALDH6A1) in adipose tissue after weight loss induced by gastric bypass [25], [56] and in response to the thiazolidinedione class of insulin sensitizers [52], [57].
Our data demonstrate that diet-induced obesity or denervation-induced inactivity in mice, and exposure to fatty acids or H2O2 in cultured myocytes, can potently downregulate expression of BCAA metabolic genes. Conversely, expression of BCAA metabolic genes can be increased by exercise training in rodents. BCAA-related expression patterns also can be modulated in response to acute CNS infusion of insulin [22]. Given that each of these stimuli modulate insulin sensitivity (e.g. reduced with obesity, inactivity, lipid excess, and ROS; improvement with exercise, weight loss, and centrally-delivered insulin), it is possible that transcriptional regulation of muscle BCAA gene expression is mediated by insulin resistance per se. Interestingly, incubation with 2-deoxyglucose, but not insulin, decreased expression of BCAA metabolism genes in 3T3 adipocytes [52]. Furthermore, these patterns are not observed in mice made insulin resistant by genetic deletion of the insulin receptor in muscle (MIRKO) [58] or by infusion with the insulin receptor antagonist peptide S961, suggesting receptor-mediated impairments in insulin action are not the sole determinant of these patterns. Rather, we hypothesize that downregulation of this pathway in muscle could represent a regulated transcriptional response to excessive oxidative stress under conditions of sustained nutrient load or inactivity, thus serving to redirect BCAA flux away from oxidative metabolism. Unfortunately, reduced flux through this pathway may ultimately be detrimental, reducing maximal anaplerotic flux and TCA cycle activity and overall cellular oxidative capacity. Consistent with this hypothesis, mice with Mut mutations have reduced tissue succinate levels [59] while mice with knockout of Ppm1k, resulting in reduced Bckdh activity and BCAA oxidative flux, have reduced oxidative capacity and increased reliance on glycolytic metabolism, ultimately contributing to contractile failure [53]. Reductions in alpha-ketoglutarate may further limit capacity for transamination, thus perpetuating a vicious cycle of impaired amino acid oxidative metabolism and reduced capacity for ATP production.
Our data and recent mouse studies demonstrate that alterations in both muscle and adipose tissue BCAA utilization can perturb systemic metabolism. For example, reduced adipose tissue BCAA gene expression not only reduces BCAA oxidation, but also results in increased circulating concentrations of BCAA in vivo [21]. BCAA likewise play important roles in skeletal muscle, serving as substrates for the TCA cycle and ultimately as fuels for the electron transport chain (e.g. via ETFDH). Interestingly, we recently identified ETFDH as a key contributor to altered energetics in a computational model of muscle insulin resistance [60], while BCKDHA was also predicted as an obesity and diabetes candidate gene using a suite of bioinformatics predictive tools [61]. Furthermore, genetic disruption of the first step of BCAA catabolism (Bcat2) alters metabolism during exercise [62]. Conversely, dietary supplementation with BCAA mixtures improves exercise performance [63], and activation of BCKDH activity improves cardiac performance in the failing heart, even without changing systemic levels of BCAA [53]. Thus, when energy demands are high (e.g. exercise training, sustained contraction), this pathway may play a pivotal role in metabolic adaptation. Thus, increases in either expression of BCAA oxidative enzymes and/or flux may be an important mechanism regulating oxidative (aerobic) capacity.
We acknowledge several limitations of our study. Firstly, gene expression is one of many determinants of pathway flux [50], [51]. Secondly, we utilized a nontargeted metabolomics platform to discover perturbations in metabolic pathways; while coverage of the metabolome is extensive, identification of all potential cellular metabolites is not available. Thirdly, our primary analysis was conducted in biopsies of the vastus lateralis (quadriceps muscle), a mixed fiber muscle, obtained in the fasting state; as a result, we cannot fully assess the role of fiber type or postprandial regulation to observed transcriptomic and metabolomic patterns [64], [65]. Moreover, muscle patterns may differ from those in other tissues also contributing to whole-body BCAA metabolism (e.g. liver). Finally, while our metabolomics data do provide support for alterations in flux within this pathway in insulin resistant states, future targeted metabolomics analyses will be required to quantify metabolites of interest and to assess pathway flux in muscle.
In conclusion, this study demonstrates defects in skeletal muscle BCAA-related gene expression and metabolism in insulin resistant humans. Furthermore, experimentally-induced defects in BCAA metabolic flux can reduce cellular respiration and lipid oxidation in cultured cells, particularly in the setting of lipid excess, and promote weight gain, muscle lipid accumulation, and insulin resistance in vivo. Taken together, these data suggest that perturbations in BCAA metabolism may contribute to metabolic phenotypes associated with insulin resistance and T2D risk. This study also suggests that searching for metabolic phenotypes in carriers of MUT mutations would be relevant for understanding disease pathogenesis and guiding clinical management strategies. Furthermore, targeting BCAA metabolism may hold promise for prevention or treatment of T2D.
Acknowledgments
We gratefully acknowledge research grant support from the Graetz Foundation and the American Diabetes Association (to MEP), NIH K23 DK02795 (to AG), NIH RO1DK101043 (to LG), K99 HD064793 (to EI), NIH U54-LM008748 (to SK), and NIH P30DK 036836 (Diabetes Research Center, Joslin). CL was also supported by grant RYC-2010-06789 from the Ministerio de Ciencia e Innovación (MICINN, Spain) and a Marie Curie FP7-PEOPLE-2011-CIG. IM, JRS and CPV were supported by the intramural research program of the National Human Genome Research Institute at the National Institutes of Health. We also thank Dr. Peng Yi for assistance with pump insertion and Dr. Robert Harris (Indiana University) for providing the BCKDK inhibitor chloroisocaproate.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.08.001.
Contributor Information
Carles Lerin, Email: clerin@fsjd.org.
Mary-Elizabeth Patti, Email: mary.elizabeth.patti@joslin.harvard.edu.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Martin B.C., Warram J.H., Krolewski A.S., Bergman R.N., Soeldner J.S., Kahn C.R. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340:925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- 2.Tabak A.G., Jokela M., Akbaraly T.N., Brunner E.J., Kivimaki M., Witte D.R. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patti M.E., Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocrine Reviews. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguer C., McCoin C.S., Knotts T.A., Thrush A.B., Ono-Moore K., McPherson R. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB Journal. 2015;29:336–345. doi: 10.1096/fj.14-255901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koves T.R., Ussher J.R., Noland R.C., Slentz D., Mosedale M., Ilkayeva O. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metabolism. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Bikman B.T., Summers S.A. Ceramides as modulators of cellular and whole-body metabolism. The Journal of Clinical Investigation. 2011;121:4222–4230. doi: 10.1172/JCI57144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson E.J., Lustig M.E., Boyle K.E., Woodlief T.L., Kane D.A., Lin C.T. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. The Journal of Clinical Investigation. 2009;119:573–581. doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoelson S.E., Herrero L., Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132:2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- 9.Hotamisligil G.S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newgard C.B., An J., Bain J.R., Muehlbauer M.J., Stevens R.D., Lien L.F. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felig P., Marliss E., Cahill G.F., Jr. Plasma amino acid levels and insulin secretion in obesity. The New England Journal of Medicine. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- 12.Fiehn O., Garvey W.T., Newman J.W., Lok K.H., Hoppel C.L., Adams S.H. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African–American women. PLoS One. 2010;5:e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu F., Tavintharan S., Sum C.F., Woon K., Lim S.C., Ong C.N. Metabolic signature shift in type 2 diabetes mellitus revealed by mass spectrometry-based metabolomics. Journal of Clinical Endocrinology & Metabolism. 2013;98:E1060–E1065. doi: 10.1210/jc.2012-4132. [DOI] [PubMed] [Google Scholar]
- 14.Wurtz P., Soininen P., Kangas A.J., Ronnemaa T., Lehtimaki T., Kahonen M. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36:648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huffman K.M., Shah S.H., Stevens R.D., Bain J.R., Muehlbauer M., Slentz C.A. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32:1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laferrere B., Reilly D., Arias S., Swerdlow N., Gorroochurn P., Bawa B. Differential metabolic impact of gastric bypass surgery versus dietary intervention in obese diabetic subjects despite identical weight loss. Science Translational Medicine. 2011;3:80re82. doi: 10.1126/scitranslmed.3002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah S.H., Crosslin D.R., Haynes C.S., Nelson S., Turer C.B., Stevens R.D. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia. 2012;55:321–330. doi: 10.1007/s00125-011-2356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glynn E.L., Piner L.W., Huffman K.M., Slentz C.A., Elliot-Penry L., AbouAssi H. Impact of combined resistance and aerobic exercise training on branched-chain amino acid turnover, glycine metabolism and insulin sensitivity in overweight humans. Diabetologia. 2015;58:2324–2335. doi: 10.1007/s00125-015-3705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T.J., Larson M.G., Vasan R.S., Cheng S., Rhee E.P., McCabe E. Metabolite profiles and the risk of developing diabetes. Nature Medicine. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman M.A., She P., Peroni O.D., Lynch C.J., Kahn B.B. Adipose tissue branched chain amino acid (BCAA) metabolism modulates circulating BCAA levels. Journal of Biological Chemistry. 2010;285:11348–11356. doi: 10.1074/jbc.M109.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin A.C., Fasshauer M., Filatova N., Grundell L.A., Zielinski E., Zhou J.Y. Brain insulin lowers circulating BCAA levels by inducing hepatic BCAA catabolism. Cell Metabolism. 2014;20:898–909. doi: 10.1016/j.cmet.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patti M.E., Brambilla E., Luzi L., Landaker E.J., Kahn C.R. Bidirectional modulation of insulin action by amino acids. Journal of Clinical Investigation. 1998;101:1519–1529. doi: 10.1172/JCI1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macotela Y., Emanuelli B., Bang A.M., Espinoza D.O., Boucher J., Beebe K. Dietary leucine–an environmental modifier of insulin resistance acting on multiple levels of metabolism. PLoS One. 2011;6:e21187. doi: 10.1371/journal.pone.0021187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She P., Van Horn C., Reid T., Hutson S.M., Cooney R.N., Lynch C.J. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. American Journal of Physiology, Endocrinology and Metabolism. 2007;293:E1552–E1563. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams S.H., Hoppel C.L., Lok K.H., Zhao L., Wong S.W., Minkler P.E. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African–American women. Journal of Nutrition. 2009;139:1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gall W.E., Beebe K., Lawton K.A., Adam K.P., Mitchell M.W., Nakhle P.J. A-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin W., Goldfine A.B., Boes T., Henry R.R., Ciaraldi T.P., Kim E.-Y. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. Journal of Clinical Investigation. 2011;121:918–929. doi: 10.1172/JCI41940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization . vol. 50. 2006. (Definition and diagnosis of diabetes mellitus and intermediate hyperglycaemia). [Google Scholar]
- 30.Bergman R.N., Prager R., Volund A., Olefsky J.M. Equivalence of the insulin sensitivity index in man derived by the minimal model method and the euglycemic glucose clamp. The Journal of Clinical Investigation. 1987;79:790–800. doi: 10.1172/JCI112886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldfine A.B., Gerwien R.W., Kolberg J.A., O'Shea S., Hamren S., Hein G.P. Biomarkers in fasting serum to estimate glucose tolerance, insulin sensitivity, and insulin secretion. Clinical Chemistry. 2011;57:326–337. doi: 10.1373/clinchem.2010.156133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gollisch K.S.C., Brandauer J., Jessen N., Toyoda T., Nayer A., Hirshman M.F. Effects of exercise training on subcutaneous and visceral adipose tissue in normal- and high-fat diet-fed rats. American Journal of Physiology – Endocrinology and Metabolism. 2009;297:E495–E504. doi: 10.1152/ajpendo.90424.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi P., Park J.S., Melton Douglas A. Betatrophin: a hormone that controls pancreatic β cell proliferation. Cell. 2013;153:747–758. doi: 10.1016/j.cell.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Chandler R.J., Sloan J., Fu H., Tsai M., Stabler S., Allen R. Metabolic phenotype of methylmalonic acidemia in mice and humans: the role of skeletal muscle. BMC Medical Genetics. 2007;8:64. doi: 10.1186/1471-2350-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris R.A., Paxton R., DePaoli-Roach A.A. Inhibition of branched chain alpha-ketoacid dehydrogenase kinase activity by alpha-chloroisocaproate. Journal of Biological Chemistry. 1982;257:13915–13918. [PubMed] [Google Scholar]
- 36.Chandler R.J., Zerfas P.M., Shanske S., Sloan J., Hoffmann V., DiMauro S. Mitochondrial dysfunction in mut methylmalonic acidemia. FASEB Journal. 2009;23:1252–1261. doi: 10.1096/fj.08-121848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters H., Nefedov M., Sarsero J., Pitt J., Fowler K.J., Gazeas S. A knock-out mouse model for methylmalonic aciduria resulting in neonatal lethality. Journal of Biological Chemistry. 2003;278:52909–52913. doi: 10.1074/jbc.M310533200. [DOI] [PubMed] [Google Scholar]
- 38.Wang T.J., Ngo D., Psychogios N., Dejam A., Larson M.G., Vasan R.S. 2-Aminoadipic acid is a biomarker for diabetes risk. Journal of Clinical Investigation. 2013;123:4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menni C., Fauman E., Erte I., Perry J.R., Kastenmuller G., Shin S.Y. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suhre K., Meisinger C., Doring A., Altmaier E., Belcredi P., Gieger C. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kien C.L., Bunn J.Y., Poynter M.E., Stevens R., Bain J., Ikayeva O. A lipidomics analysis of the relationship between dietary fatty acid composition and insulin sensitivity in young adults. Diabetes. 2012;62:1054–1063. doi: 10.2337/db12-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang-Sattler R., Yu Z., Herder C., Messias A.C., Floegel A., He Y. Novel biomarkers for pre-diabetes identified by metabolomics. Molecular Systems Biology. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perng W., Gillman M.W., Fleisch A.F., Michalek R.D., Watkins S.M., Isganaitis E. Metabolomic profiles and childhood obesity. Obesity (Silver Spring) 2014;22:2570–2578. doi: 10.1002/oby.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ensenauer R., Vockley J., Willard J.M., Huey J.C., Sass J.O., Edland S.D. A common mutation is associated with a mild, potentially asymptomatic phenotype in patients with isovaleric acidemia diagnosed by newborn screening. American Journal of Human Genetics. 2004;75:1136–1142. doi: 10.1086/426318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams S.H., Hoppel C.L., Lok K.H., Zhao L., Wong S.W., Minkler P.E. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid β-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African–American women. The Journal of Nutrition. 2009;139:1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams S.H. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Advances in Nutrition. 2011;2:445–456. doi: 10.3945/an.111.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimomura Y., Fujii H., Suzuki M., Murakami T., Fujitsuka N., Nakai N. Branched-chain alpha-keto acid dehydrogenase complex in rat skeletal muscle: regulation of the activity and gene expression by nutrition and physical exercise. Journal of Nutrition. 1995;125:1762S–1765S. doi: 10.1093/jn/125.suppl_6.1762S. [DOI] [PubMed] [Google Scholar]
- 49.Shimomura Y., Murakami T., Nakai N., Nagasaki M., Harris R.A. Exercise promotes BCAA catabolism: effects of BCAA supplementation on skeletal muscle during exercise. Journal of Nutrition. 2004;134:1583S–1587S. doi: 10.1093/jn/134.6.1583S. [DOI] [PubMed] [Google Scholar]
- 50.Brosnan J.T., Brosnan M.E. Branched-chain amino acids: enzyme and substrate regulation. Journal of Nutrition. 2006;136:207S–211S. doi: 10.1093/jn/136.1.207S. [DOI] [PubMed] [Google Scholar]
- 51.Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lackey D.E., Lynch C.J., Olson K.C., Mostaedi R., Ali M., Smith W.H. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. American Journal of Physiology, Endocrinology and Metabolism. 2013;304:E1175–E1187. doi: 10.1152/ajpendo.00630.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H., Olson K.C., Gao C., Prosdocimo D.A., Zhou M., Wang Z. Catabolic defect of branched-chain amino acids promotes heart failure. Circulation. 2016;133:2038–2049. doi: 10.1161/CIRCULATIONAHA.115.020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pietilainen K.H., Naukkarinen J., Rissanen A., Saharinen J., Ellonen P., Keranen H. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Medicine. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leskinen T., Rinnankoski-Tuikka R., Rintala M., Seppanen-Laakso T., Pollanen E., Alen M. Differences in muscle and adipose tissue gene expression and cardio-metabolic risk factors in the members of physical activity discordant twin pairs. PLoS One. 2010;5:e12609. doi: 10.1371/journal.pone.0012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mardinoglu A., Heiker J.T., Gärtner D., Björnson E., Schön M.R., Flehmig G. Extensive weight loss reveals distinct gene expression changes in human subcutaneous and visceral adipose tissue. Scientific Reports. 2015;5:14841. doi: 10.1038/srep14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsiao G., Chapman J., Ofrecio J.M., Wilkes J., Resnik J.L., Thapar D. Multi-tissue, selective PPAR modulation of insulin sensitivity and metabolic pathways in obese rats. AJP: Endocrinology and Metabolism. 2010;300:E164–E174. doi: 10.1152/ajpendo.00219.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yechoor V.K., Patti M.E., Ueki K., Laustsen P.G., Saccone R., Rauniyar R. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proceedings of the National Academy of Sciences United States of America. 2004;101:16525–16530. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caterino M., Chandler R.J., Sloan J.L., Dorko K., Cusmano-Ozog K., Ingenito L. The proteome of methylmalonic acidemia (MMA): the elucidation of altered pathways in patient livers. Molecular BioSystems. 2016;12:566–574. doi: 10.1039/c5mb00736d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nogiec C., Burkart A., Dreyfuss J.M., Lerin C., Kasif S., Patti M.E. Metabolic modeling of muscle metabolism identifies key reactions linked to insulin resistance phenotypes. Molecular Metabolism. 2015;4:151–163. doi: 10.1016/j.molmet.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tiffin N., Adie E., Turner F., Brunner H.G., van Driel M.A., Oti M. Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Research. 2006;34:3067–3081. doi: 10.1093/nar/gkl381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.She P., Zhou Y., Zhang Z., Griffin K., Gowda K., Lynch C.J. Disruption of BCAA metabolism in mice impairs exercise metabolism and endurance. Journal of Applied Physiology (1985) 2010;108:941–949. doi: 10.1152/japplphysiol.01248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Antona G., Ragni M., Cardile A., Tedesco L., Dossena M., Bruttini F. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell Metabolism. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 64.Oberbach A., Bossenz Y., Lehmann S., Niebauer J., Adams V., Paschke R. Altered fiber distribution and fiber-specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes Care. 2006;29:895–900. doi: 10.2337/diacare.29.04.06.dc05-1854. [DOI] [PubMed] [Google Scholar]
- 65.He J., Watkins S., Kelley D.E. Skeletal muscle lipid content and oxidative enzyme activity in relation to muscle fiber type in type 2 diabetes and obesity. Diabetes. 2001;50:817–823. doi: 10.2337/diabetes.50.4.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.