Abstract
Objective
Genetic background largely contributes to the complexity of metabolic responses and dysfunctions. Induction of brown adipose features in white fat, known as brown remodeling, has been appreciated as a promising strategy to offset the positive energy balance in obesity and further to improve metabolism. Here we address the effects of genetic background on this process.
Methods
We investigated browning remodeling in a depot-specific manner by comparing the response of C57BL/6J, 129/Sv and FVB/NJ mouse strains to cold.
Results
Surprisingly, 129/Sv and FVB/NJ mice showed distinct brown remodeling features despite their similar resistance to metabolic disorders in comparison to the obesity-prone C57BL/6J mice. FVB/NJ mice demonstrated a preference of brown remodeling in inguinal subcutaneous white adipose tissue (iWAT), whereas 129/Sv mice displayed robust brown remodeling in visceral epididymal fat (eWAT). We further compared gene expression in different depots by RNA-sequencing and identified Hoxc10 as a novel “brake” of brown remodeling in iWAT.
Conclusion
Rodent genetic background determines the brown remodeling of different white fat depots. This study provides new insights into the role of genetic variation in fat remodeling in susceptibility to metabolic diseases.
Keywords: Genetic background, White adipose tissue, Brown remodeling, Hoxc10, Cold exposure, Browning brake
Highlights
-
•
Rodent genetic background determines the physiological responses to cold.
-
•
Genetic background determines browning capacity of white adipose tissue.
-
•
The brown remodeling is affected by genetic background in a depot-specific manner.
-
•
Hoxc10 is a novel repressor of browning in subcutaneous white adipose tissue.
1. Introduction
Obesity is a worldwide public health problem [1], [2], [3] and a risk factor for many chronic diseases including type 2 diabetes, cardiovascular disorders, atherosclerosis, non-alcoholic fatty liver disease, and several forms of cancer [4], [5]. Obesity arises as a consequence of excess fat storage and expansion of white adipose tissue (WAT) over energy expenditure. WAT functions primarily to store energy while brown adipose tissue (BAT) catabolizes free fatty acids and dissipates energy in the form of heat. Therefore, it is appealing to introduce BAT features into WAT, namely brown remodeling, to combat obesity and its co-morbidities.
With the discovery of brown remodeling, there has been growing interest in exploring this phenomenon to manage the consequences of obesity. Brown remodeling is characterized by the emergence of clusters of brown-like UCP1-positive multilocular cells (also referred to as beige or brite cells) interspersed within WAT. In rodents, brown remodeling can be induced by cold exposure or pharmacological treatments such as β-adrenergic receptor agonists and thiazolidinediones (PPARγ agonists) [6], [7], [8], [9], [10]. Recently activated human brown fat has been shown to have a similar expression profile to beige fat, and beige adipocytes have been identified in human WAT [11], [12], [13]. The origin of beige cells has been explained by either transdifferentiation, the direct conversion of white adipocytes into brown-like cells [7], [14] or the differentiation of unique beige precursors [15], [16]. Several studies in mouse models illustrate that brown remodeling contributes to an improved metabolic profile marked by protection against diet-induced obesity, improved glucose homeostasis and increased energy expenditure [6], [17], [18], [19], [20], [21]. While brown remodeling of WAT offers a promising strategy to alleviate obesity, more knowledge is still needed to understand the genetic susceptibility of adipose plasticity to environmental cues.
The interaction of genetic and environmental factors plays a significant role in propagating obesity. It has been established that genetic background is a major contributing factor in the susceptibility to diet-induced obesity and type 2 diabetes. Among the commonly used inbred mouse strains, the C57BL/6J strain is more susceptible to obesity and type 2 diabetes while the 129/Sv strain is protected against these traits [22], [23] as is the FVB/NJ strain, which is hyperactive and has higher body temperature [24], [25], [26].
Previous studies have integrated histological and quantitative trait loci analysis to identify potential browning genes and found that multiple loci on 8 different chromosomes function synergistically to increase Ucp1 and Pgc-1α expression in retroperitoneal WAT in the A/J mice but not in the C57BL/6J strain [27], [28], [29]. However, there was no significant difference in Ucp1 expression in BAT and epididymal WAT between the two strains [17]. These results highlight the contribution of genetic variation to strain-specific differences together with depot-dependent browning potential of WAT. Given the pressing need to identify genetic mediators of adipose tissue remodeling, in this study, we examined the browning response in the 3 commonly used mouse strains, C57BL/6J, 129/Sv and FVB/NJ, coupled with RNA sequencing to identify genes that account for the adipose plasticity upon chronic cold exposure.
2. Methods
2.1. Animal studies
Five-week old male C57BL/6J (000664), 129/Sv (002448), FVB/NJ (001800) mice were purchased from Jackson Laboratory (Bar Harbor, ME) and exposed to 4 °C after two-week adaptation in the Columbia University animal barrier. Mice had ad libitum access to regular chow diet (PicoLab rodent diet 20 (LabDiet 5053)). Body temperature was measured by a subcutaneously implanted IPTT-300 transponder (DMDS). During chronic cold exposure, body temperature was measured in light phase at the same time on each day. Plasma glucose was measured with the One Touch Ultra II glucometer. The mice were gently handled by the same experienced animal technician to minimize stress-induced measuring variations. The Columbia University Animal Care and Utilization Committee approved all procedures.
2.2. RNA analysis
One entire side of each respective fat depot was used to isolate total RNA by using RNeasy Lipid Tissue kit (QIAGEN) following the manufacturer's instructions. 1 μg RNA was subjected to cDNA synthesis by using High-capacity cDNA Reverse Transcription kit (Applied Biosystems). qPCR was performed on the CFX96 Real-Time PCR system (Bio-Rad) by using GoTaq qPCR Master Mix (Promega). The relative gene expression levels were calculated by ddCt method with TBP as the reference gene. qPCR primer sequences are available upon request.
2.3. RNA-seq analysis
mRNA was enriched by poly-A pull-down from 1 μg total RNA and used to generate proceed library with using Illumina TruSeq RNA prep kit. Libraries were then sequenced using Illumina HiSeq2000 at Columbia Genome Center. Sequencing reads were aligned to the mouse genome (UCSC/mm9) using Tophat [30] with 4 mismatches and 10 maximum multiple hits. Differential gene expression across strains and tissues was found using cuffdiff [31] with cutoff FPKM > 5, FDR < 0.05 and fold-change > 2. K-means clustering, heatmaps, and scatter plots were generated in R [32]. Genes coding for transcription factors were identified among other regulated genes using “Animal TFDB” database [33].
2.4. In vitro cell culture analysis
The C3H/10T1/2 cell line was purchased from ATCC and cultured and differentiated as described [34]. Myc-tagged Human HOXC10 cDNA was cloned into pTRIPZ inducible expression vector (Thermo Open Biosystems) [35] and used to generate stable cell line into C3H/10T1/2 cell under selection by 1 μg/mL puromycin. On Day 7 of differentiation, HOXC10 overexpression was induced by adding 0.1 μg/mL doxycycline. Cells were treated with isoproterenol (10 μM) and CL-316,243 (1 μg/mL) for 4 h before harvesting for RNA analysis. HOXC10 expression was validated by probing with anti-Hoxc10 antibody HoxC10 (L-17) (Santa Cruz).
2.5. Morphological analysis
Fat tissues were fixed in 10% formalin, embedded in paraffin, and stained with Hematoxylin and Eosin (H&E). Lipid content in differentiated C3H/10T1/2 adipocytes was assessed by Oil Red O staining.
2.6. Statistical analysis
All values were presented as means ± standard error of means (SEM). Unpaired 2-tailed Student's t tests were used to evaluate statistical significance and p < 0.05 was declared as a statistically significant change.
3. Results
3.1. Genetic background-dependent metabolic responses to cold challenge
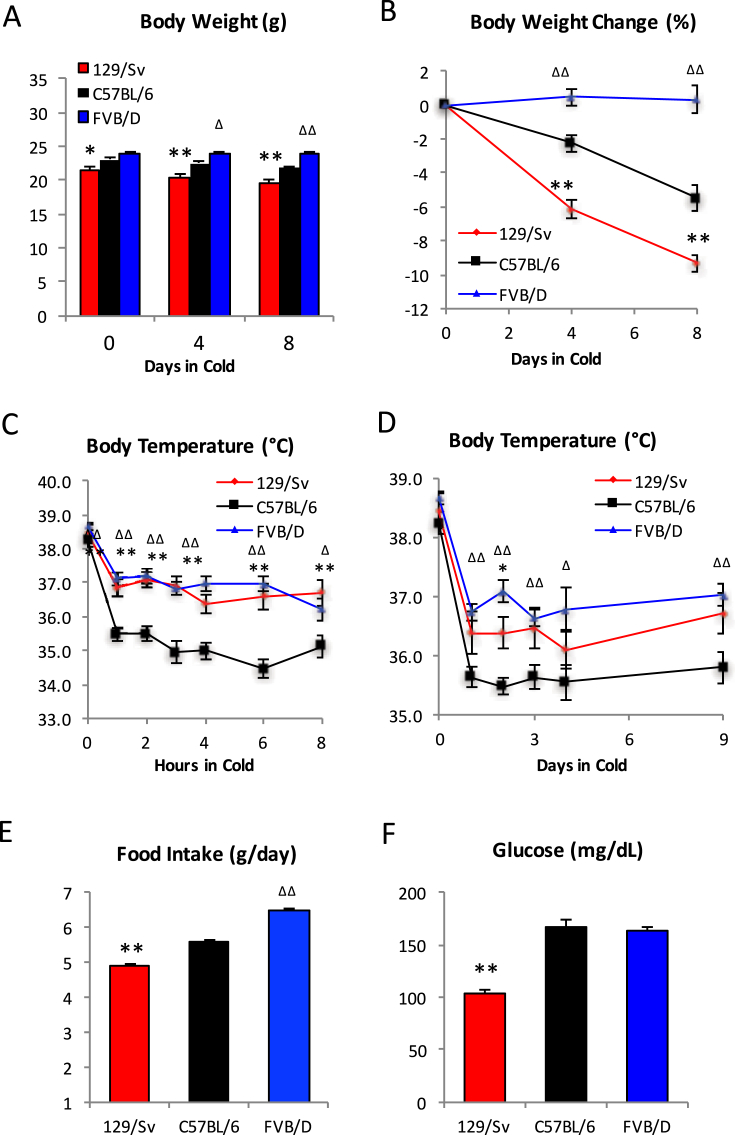
To test whether genetic background impacts thermogenic response as well as physiological adaptation to cold, we compared the cold response among 129/Sv, C57BL/6J and FVB/NJ strains. During an 8-day cold challenge at 4 °C, both C57BL/6J and 129/Sv mice lost body weight (5.5% and 9.3%, respectively) while, surprisingly, FVB/NJ mice were able to maintain their body weight (Figure 1A–B). We also monitored their cold responses by measuring body temperature. 129/Sv and FVB/NJ mice were better to maintain their body temperature than C57BL/6J mice during both acute (Figure 1C) and prolonged (Figure 1D) cold challenge, indicating higher thermogenesis in these two strains. During cold challenge, body weight change is determined by the compromise between thermogenesis-induced fat consumption and food intake. The food intake of FVB/NJ mice was significantly higher than that of C57BL/6J and 129/Sv mice (Figure 1E), in support of their maintenance of body mass and preservation of body temperature. 129/Sv mice had less food intake compared to C57BL/6J mice, but also experienced elevated thermogenesis, accounting for their greater body weight loss during cold challenge. Lastly, we measured the blood glucose levels at the end of the cold challenge. The 129/Sv strain had much lower glucose levels than the other stains (Figure 1F). Taken together, these data indicate that genetic backgrounds determine physiological responses to cold exposure, consistent with previous work [17], [27], [36]
Figure 1.
Metabolic response of three mouse strains in response to cold challenge. (A–B) Body weight (A) and body weight change (B) during cold challenge. (C–D) Core body temperature during acute (C) and chronic cold challenge (D). (E) Food intake during cold exposure. (F). Ad libitum plasma glucose levels in the cold. *: p < 0.05, **: p < 0.01 for 129/Sv vs. C57BL/6; Δ: p < 0.05, ΔΔ: p < 0.01 for FVB/D vs. C57BL/6, n = 8, 8, 8.
3.2. Distinct brown remodeling in subcutaneous WAT
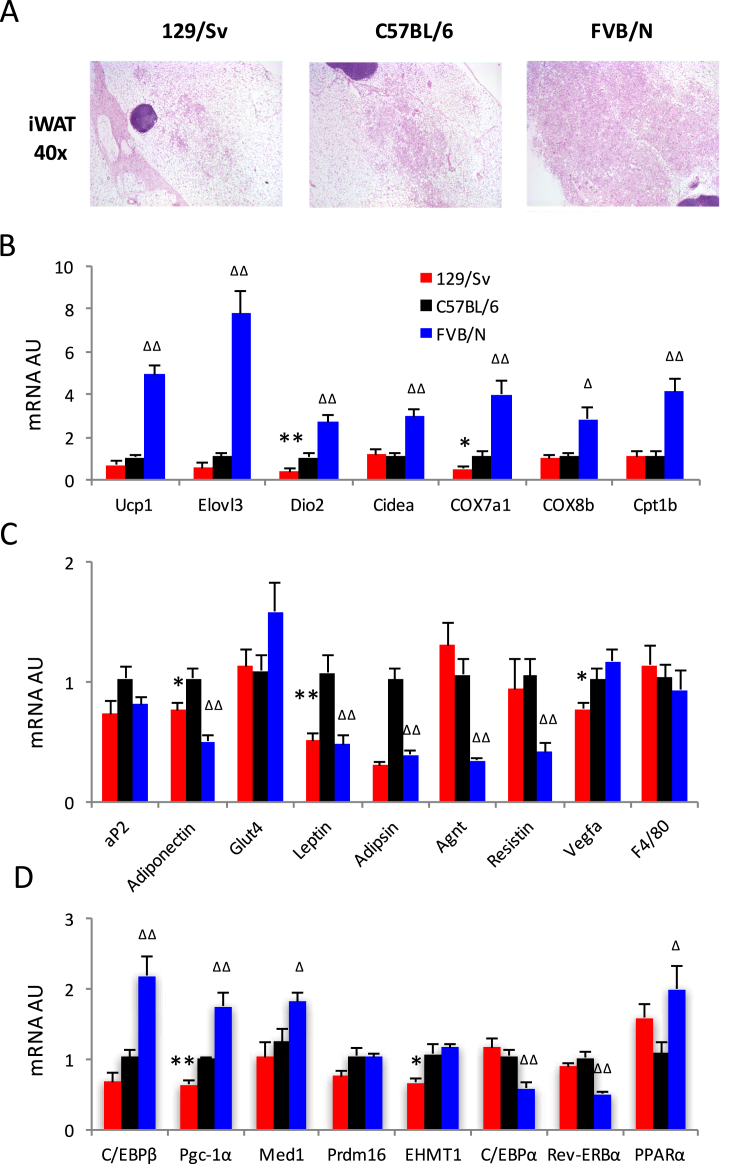
Prolonged cold exposure is the classic method of inducing brown remodeling of subcutaneous WAT. Given their distinct cold responses, we hypothesized that the subcutaneous WAT from the three strains would have different extents of brown remodeling. Indeed, histological examination of their inguinal fat depots (iWAT) showed a strikingly higher prevalence of multilocular adipocytes, that is an increased browning morphology, in FVB/NJ mice than in C57BL/6J mice (Figure 2A). To our surprise, we observed fewer multilocular adipocytes in iWAT of 129/Sv mice than in C57BL/6J mice (Figure 2A) regardless of their superior defense of temperature loss in the cold (Figure 1D).
Figure 2.
Comparisons of browning in subcutaneous white adipose tissue. (A) Histological analyses of inguinal WAT by H&E staining. (B) qPCR analyses of representative brown genes expression in iWAT (AU: relative gene expression arbitrary unit). (C) qPCR analyses of adipocyte markers and representative white genes expression in iWAT. (D) qPCR analyses of expression of browning regulators in iWAT. *: p < 0.05, **: p < 0.01 for 129/Sv vs. C57BL/6; Δ: p < 0.05, ΔΔ: p < 0.01 for FVB/D vs. C57BL/6, n = 8, 8, 8.
Consistent with the morphological analysis, the expression of representative brown genes and brown gene regulators was significantly higher in FVB/NJ mice as compared to C57BL/6J. Indeed, there was a significantly increased expression of brown markers (e.g., Ucp1, Elovl3, Dio2, Cidea), mitochondrial respiratory chain enzymes (e.g., Cox7a1, Cox8b), and genes involved in fatty acid oxidation pathway (e.g., Cpt1b) as well as regulatory genes involved in brown adipocyte differentiation and activation (e.g., C/ebpβ, Pgc-1α, Med1 and Pparα) (Figure 2B,D). 129/Sv mice had relatively lower brown gene expression compared to C57BL/6J mice, in line with less angiogenesis as indicated by lower Vegfa expression (Figure 2C). On the other hand, the expression of representative white adipocyte genes (e.g., Leptin, Adipsin, Angiotensinogen (Agnt) and Resistin) was significantly lower in FVB/NJ mice compared to C57BL/6J mice (Figure 2C). Thus, FVB/NJ genetic background was predisposed to more brown remodeling in subcutaneous fat relative to the two other strains.
3.3. Brown remodeling of visceral WAT in 129/Sv mice
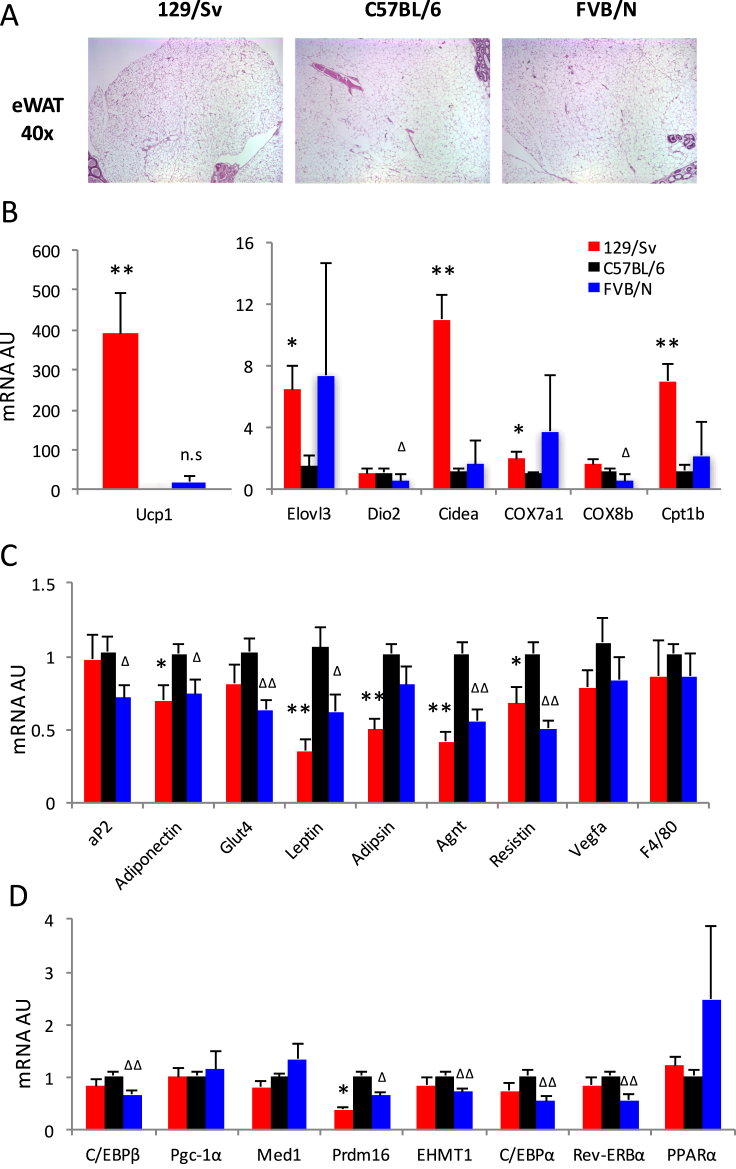
Visceral fat accumulation causes abdominal obesity, aggravating the risk for the metabolic syndrome, and it is much more resistant to browning compared to subcutaneous fat. Indeed, there was a notable absence of multilocular adipocytes in visceral epididymal fat (eWAT) across all three strains (Figure 3A). Surprisingly, in spite of their nearly identical eWAT morphology (Figure 3A), 129/Sv mice showed a much higher level of brown remodeling than C57BL/6J and FVB/NJ mice (Figure 3B). The representative brown adipocyte marker Ucp1 was expressed ∼400-fold higher in 129/Sv mice than in C57BL/6J mice, as were other brown adipocyte markers, including Elovl3, Cidea, COX7a1 and Cpt-1b (Figure 3B). The pan-adipocyte genes aP2 and Glut4 were comparable between 129/Sv and C57BL/6J mice, but the white adipocyte genes Leptin, Adipsin, Agnt, and Resistin were all down-regulated in 129/Sv mice, in line with their more pronounced brown remodeling (Figure 3C). The enhanced brown remodeling in 129/Sv mice was unlikely to have been caused by the classic browning regulators since we failed to observe higher expressions of C/EBPβ, Pgc-1α, Med1, Prdm16 and PPARα compared to C57BL/6J mice (Figure 3D). These data indicate that genetic backgrounds have differential impacts on brown remodeling of eWAT, and 129/Sv strain has a stronger brown remodeling in metabolically deleterious visceral fat.
Figure 3.
Comparisons of browning in visceral white adipose tissue. (A) Histological analyses of epididymal WAT (eWAT) by H&E staining. (B) qPCR analyses of representative brown genes expression in eWAT. (C) qPCR analyses of adipocyte markers and representative white genes expression in eWAT. (D) qPCR analyses of expression of browning regulators in eWAT. *: p < 0.05, **: p < 0.01 for 129/Sv vs. C57BL/6; Δ: p < 0.05, ΔΔ: p < 0.01 for FVB/D vs. C57BL/6, n = 8, 8, 8.
3.4. Marginal impact of genetic backgrounds on BAT
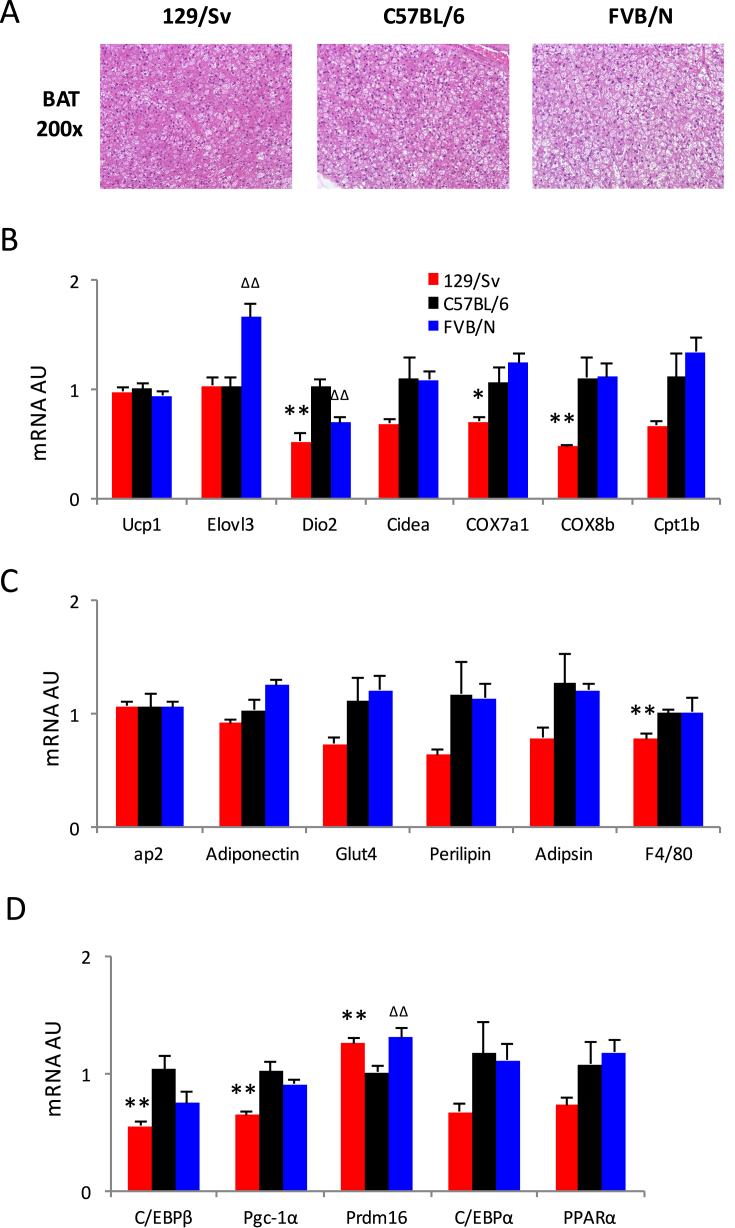
Histological analyses of BAT in the three strains endorsed the preponderance of lipid contents in FVB/NJ mice, followed by C57BL/6J and lastly 129/Sv mice (Figure 4A). qPCR analyses of the expression of representative and regulatory brown genes demonstrated a weaker BAT activation in 129/Sv than in FVB/NJ and C57BL/6J mice (Figure 4B,D). No statistically relevant difference was found regarding adipocyte markers and representative white genes expression in BAT (Figure 4C). These data together with analyses in iWAT and eWAT suggest that the physiological responses to cold exposure are unlikely caused by the variations of BAT activation but rather by brown remodeling in white fat.
Figure 4.
Comparisons of brown adipose tissue (BAT) of three strains during cold challenge. (A) Histological analyses of BAT by H&E staining. (B) qPCR analyses of representative brown genes expression in BAT. (C) qPCR analyses of adipocyte markers and representative white genes expression in BAT. (D) qPCR analyses of expression of browning regulators in BAT. *: p < 0.05, **: p < 0.01 for 129/Sv vs. C57BL/6; Δ: p < 0.05, ΔΔ: p < 0.01 for FVB/D vs. C57BL/6, n = 8, 8, 8.
3.5. Gene expression profiling of WAT by RNA-seq analysis
To understand the distinct effects of genetic background on brown remodeling patterns, we compared WAT gene expression profiles across the three strains by performing RNA sequencing. The eWAT of 129/Sv mice showed a distinct expression from that of C57BL/6J and FVB/NJ mice with a large number of brown markers up-regulated (Figure 5A), consistent with the qPCR analysis (Figure 3). In iWAT, FVB/NJ mice demonstrated a consistently higher thermogenic gene expression than the other two stains (Figure 5C). In an effort to identify potential browning regulators, we focused on the transcriptional factors that showed significant changes of their expression among the three stains (Figure 5B,D). Among them, C/ebpβ expression was the highest while Rev-erbα (Nr1d1) was the lowest in iWAT of FVB/NJ mice (Figure 5D). It's known that C/ebpβ promotes brown adipogenesis (7) and Rev-erbα has been demonstrated as a repressor of thermogenic response (8). Thus the comparison of differential brown remodeling on different genetic backgrounds could provide a useful tool to discover novel factors regulating this process.
Figure 5.
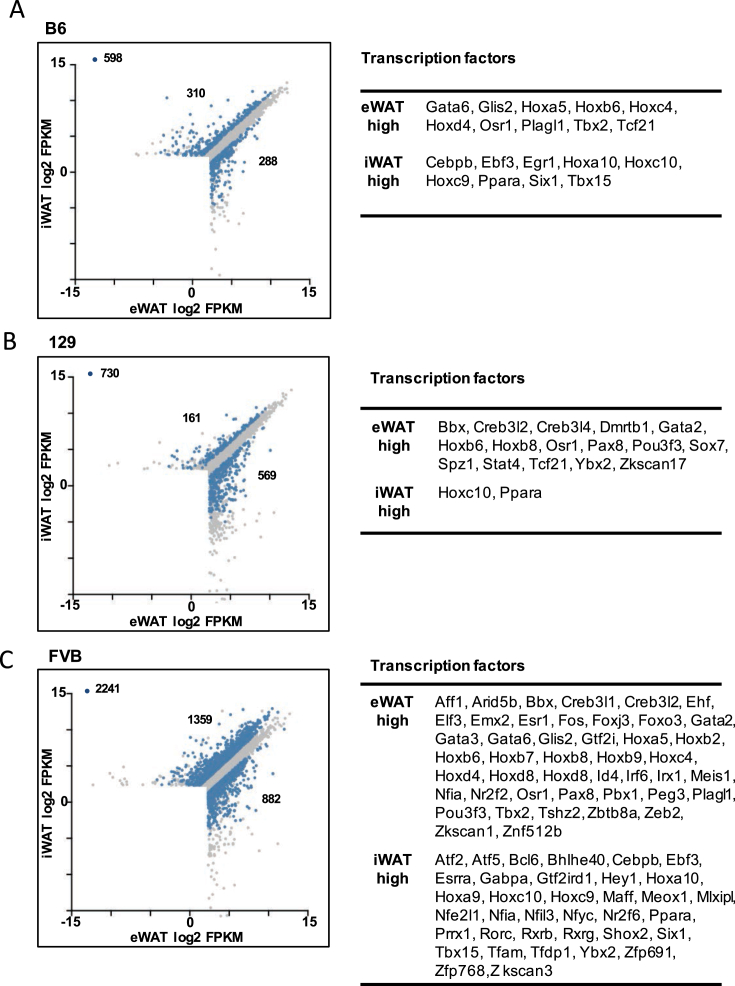
WAT gene expression profiling by RNA-seq analysis. Heatmaps showing genes expressed differentially across three strains (B6, 129 and FVB) in eWAT (A–B) and iWAT (C–D). Genes with FPKM > 5, FDR < 0.05 and fold-change > 2 are represented in the heatmaps using relative expression across three tissues. The genes were k-means clustered with name and size of each cluster labeled on the left and right, respectively. Each heatmap is accompanied by a list of transcription factors found in genes with high or low expression in B6, 129 or FVB.
3.6. Identification of Hoxc10 as a novel “brake” for brown remodeling
A search for transcription factors sharing the same expression patterns in iWAT and eWAT across all three strains identified Pparα and Hoxc10 as the only two transcription factors expressed at higher level in subcutaneous fat in all three strains (Figure 6). Pparα is well known to regulate lipid oxidation and brown adipocyte activity, and Hoxc10 has been found to be enriched in subcutaneous fat compared with visceral fat in humans [37], raising the possibility of Hoxc10 in regulating adipose tissue remodeling.
Figure 6.
Subcutaneous and visceral fat gene expression profiling in three strains. Scatter plots showing genes expressed differentially across two tissues (eWAT and iWAT) in three strains, B6 (A), 129 (B) and FVB (C). Genes with FPKM > 5 and q-value < 0.05 are represented in the plots. The blue dots represent genes changing more than two-fold. Each plot is accompanied by a list of transcription factors found in the genes with higher expression in eWAT or iWAT.
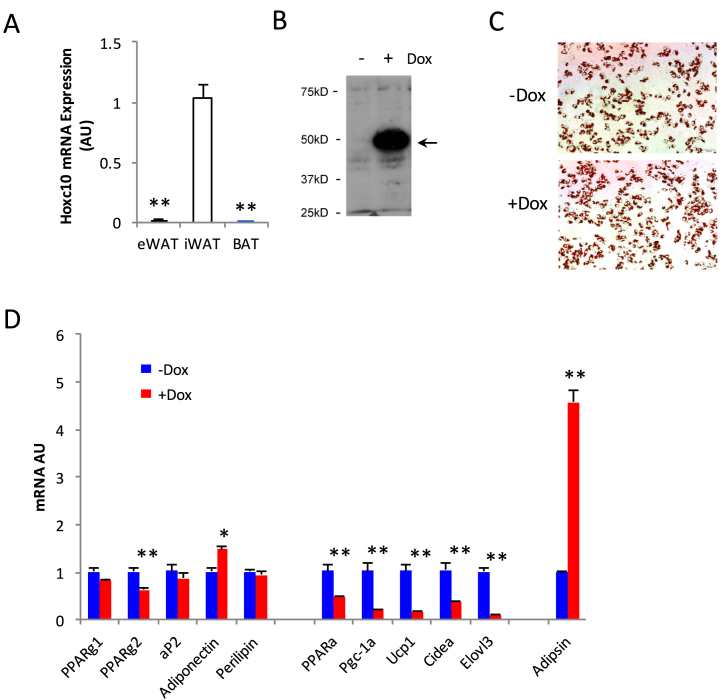
We then set out to investigate Hoxc10's function in adipocytes. We first confirmed the expression of Hoxc10 in iWAT but at background levels in BAT and eWAT (Figure 7A). HOXC10 is expressed at higher level in stromal vascular fraction (SVF) cells than in adipocytes [37], and we did observe a down-regulation of Hoxc10 during adipogenesis (data not shown). To avoid any possible involvement of Hoxc10 in adipogenesis, we then employed doxycycline-inducible HOXC10 overexpression in C3H/10T1/2 cells after differentiation (Figure 7B). After the induction of HOXC10 for two days, we observed little to no effects on lipid contents in C3H/10T1/2 cells (Figure 7C), neither on pan-adipocyte gene expression (Figure 7D). Strikingly, brown adipocyte regulators and markers represented by Pparα, Pgc-1α, Ucp1, Cidea, and Elovl3 were significantly repressed. On the other hand, the white adipocyte gene Adipsin was up-regulated to above four-fold. Taken together, we have identified Hoxc10 as a specific repressor for brown adipocyte gene expression that does not affect basic adipocyte activity.
Figure 7.
Identification of Hoxc10 as a novel repressor for browning. (A) Hoxc10 expression in 3 adipose tissues from C57BL/6 mice, **: p < 0.01 vs. iWAT (n = 8, 8, 8). (B) Overexpression of HOXC10 in C3H/10T1/2 cells. Doxycycline was added on D7 of differentiation for two days. Arrow indicates the Myc-tagged HOXC10. (C) Oil Red O staining of differentiated C3H/10T1/2 adipocyte after two days' doxycycline treatment (D7-D9). (D) qPCR analysis of gene expression in fully differentiated C3H/10T1/2 cells after induction of HOXC10 expression for two days. *: p < 0.05, **: p < 0.01 for -Dox vs. +Dox (n = 3–4).
4. Discussion
A number of studies in rodents have shown that brown remodeling of WAT confers metabolic benefits by protecting against diet-induced obesity and type 2 diabetes [18], [38], [39], [40]. In order to fully harness the therapeutic potential of brown remodeling, we need to better understand the genetic factors and mechanisms that regulate adipose plasticity, particularly, the recruitment of brown-like cells in WAT. To this end, we compared the physiological response and browning capacity of different WAT depots in C57BL6/J, FVB/NJ, and 129/Sv mice upon cold exposure. We found a surprising depot-specific browning preference in these commonly used mouse strains. Brown remodeling has been shown to predominantly occur in the subcutaneous depot [7], [38], [41], [42]. Our results were in overall agreement with this notion but specified that brown remodeling occurs in a strain-dependent manner.
Relative to the C57BL6/J mice, the 129/Sv strain showed preferential brown remodeling in the visceral depot whereas the FVB/NJ strain showed brown remodeling predominantly in the subcutaneous depot. Other studies have reported the C57BL6/J mice to have lower browning potential of the retroperitoneal WAT and subcutaneous depot when compared to the A/J and the 129 strains, respectively [28], [29], [36], [43], and this distinction may contribute to the latter's obesity-resistance phenotype [10], [42], [44]. Despite the similar obesity-resistance and better defense of body temperature loss in 129/Sv and FVB/NJ strains (Figure 1), our study demonstrates that genetic background determines the preference of brown remodeling in specific WAT depots during cold stimulation, which could be achieved through SNPs in the regulatory regions of downstream genes [45] and/or differential expression of regulatory factors.
A key addition from our study is that we used an unbiased approach to identify novel regulators of brown remodeling (Figure 5). Beyond elaborating the specific expression of representative brown and white genes among the three strains, we also investigated potential mechanisms that differentially drive WAT remodeling in visceral and subcutaneous fat. For example, we found that C/ebpβ, Pgc1-α, and Med1 positively correlated with brown gene expression in inguinal WAT, and they have been identified as positive regulators of brown remodeling [46], [47]. In contrast, Rev-ERBα negatively correlates with the brown remodeling in this depot by suppressing the expression of brown genes [48]. Herein, analyses of brown remodeling on different genetic backgrounds, as we employed in the current study, can be informative to understand the mechanisms involved in regulating adipose plasticity and, to take a step further, targeting a specific fat depot. Moreover, besides the intrinsic regulations within adipocytes, it will be of interest to investigate the behavioral differences associated with different genetic backgrounds exposed to cold. For example, we observed that 129/Sv mice were less active and FVB/NJ showed higher anxiety during cold challenge. These behavioral differences together with intrinsic regulations within adipocytes largely determine their corresponding responses to the cold.
In order to overcome the limitation of studying brown remodeling in one strain, we compared iWAT with eWAT across the three strains (Figure 6). The rationale was that if we identified regulators, here with a focus on transcription factors, that share the same expression pattern (iWAT vs. eWAT) among the three strains, they are more likely to be involved in brown remodeling. Indeed, only four such factors were identified: Hoxb6 and Osr1 were abundantly expressed in eWAT, and Pparα and Hoxc10 were highly expressed in iWAT. We chose to test whether Hoxc10 is a novel browning factor. To our surprise, we discovered that it had minimal effects on adipocyte housekeeping gene expression but markedly repressed brown markers Ucp1, Cidea, and Elovl3, possibly through repressing brown regulators Pparα and Pgc-1α (Figure 7). To date, many transcriptional regulators of fat remodeling have been identified (comprehensively reviewed in [49], [50]) and among them, only a few are repressors of brown remodeling in subcutaneous fat depot, including Foxa3 [51], p107 [52], RIP140 [53], [54], and Rev-erbα [48].
Our in vitro studies suggest that Hoxc10 likely represses brown remodeling in iWAT in vivo. Since thermogenic activity is catabolic, which acts against the native energy-storing function of white adipocyte, brown remodeling in white fat should be tightly controlled. The energy wasting character of beige adipocytes is evolutionarily disadvantageous unless it is necessary to defend body temperature or to adapt to environmental stimulations. From this stand point, Hoxc10 is important to be quickly turned on by activators sensitive to the corresponding stimuli, but it should also be equally important to be turned off when not necessary. Therefore, a ready-to-use brake system is desired. Given its abundance in brown-prone iWAT, but not in brown-resistant eWAT, neither in thermogenic BAT, Hoxc10 is an ideal candidate to function as a “browning brake” in iWAT. Further investigation is required on the detailed repression mechanism of Hoxc10 on brown remodeling as well as the regulation of Hoxc10 in cold response and in the pathogenesis of obesity.
In summary, our study has revealed that genetic background is a significant factor in regulating brown remodeling of WAT in a strain-specific and depot-specific manner. We have identified novel candidates in regulating adipose tissue plasticity, particularly Hoxc10 as a bona fide “browning brake” in iWAT. Our expectation is that the coordination between activators and repressors in brown remodeling determines the physiological consequences of adipose tissue and thus may provide novel insights for obesity treatment and in a depot-specific manner.
Acknowledgments
We would like to thank Thomas Kolar for technical assistance. This work was supported by National Institutes of Health grants R00DK97455 to L. Q., Pilot and Feasibility funding from the Diabetes Research Center to L.Q. (P30 DK063608), R01DK49780 to M. L. and by the JPB Foundation.
Conflict of interest
None declared.
References
- 1.Cawley J.J., Meyerhoefer C.C. The medical care costs of obesity: an instrumental variables approach. Journal of Health Economics. 2012;31(1):219–230. doi: 10.1016/j.jhealeco.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Yang L., Colditz G.A. Prevalence of overweight and obesity in the United States, 2007–2012. JAMA Internal Medicine. 2015;175(8):1412–1413. doi: 10.1001/jamainternmed.2015.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaboration, N.R.F Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19.2 million participants. The Lancet. 2016 doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti K.G.M.M., Zimmet P., Shaw J., IDF Epidemiology Task Force Consensus Group The metabolic syndrome–a new worldwide definition. Lancet. 2005;366(9491):1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 5.Guh D.P., Zhang W., Bansback N., Amarsi Z., Birmingham C.L., Anis A.H. The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC Public Health. 2009;9:88. doi: 10.1186/1471-2458-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himms-Hagen J., Cui J., Danforth E.J., Taatjes D.J., Lang S.S., Waters B.L. Effect of CL-316,243, a thermogenic beta 3-agonist, on energy balance and brown and white adipose tissues in rats. American Journal of Physiology. 1994;266(4 Pt 2):R1371–R1382. doi: 10.1152/ajpregu.1994.266.4.R1371. [DOI] [PubMed] [Google Scholar]
- 7.Barbatelli G., Murano I., Madsen L., Hao Q., Jimenez M., Kristiansen K. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. American Journal of Physiology, Endocrinology and Metabolism. 2010;298(6):E1244–E1253. doi: 10.1152/ajpendo.00600.2009. [DOI] [PubMed] [Google Scholar]
- 8.Petrovic N., Walden T.B., Shabalina I.G., Timmons J.A., Cannon B., Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. Journal of Biological Chemistry. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohno H., Shinoda K., Spiegelman B.M., Kajimura S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metabolism. 2012;15(3):395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitali A., Murano I., Zingaretti M.C., Frontini A., Ricquier D., Cinti S. The adipose organ of obesity-prone C57BL/6J mice is composed of mixed white and brown adipocytes. Journal of Lipids & Research. 2012;53(4):619–629. doi: 10.1194/jlr.M018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp L.Z., Shinoda K., Ohno H., Scheel D.W., Tomoda E. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS One. 2012 doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frontini A., Vitali A., Perugini J., Murano I., Romiti C., Ricquier D. White-to-brown transdifferentiation of omental adipocytes in patients affected by pheochromocytoma. Biochimica et Biophysica Acta. 2013;1831(5):950–959. doi: 10.1016/j.bbalip.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Sidossis L.S., Porter C., Saraf M.K., Børsheim E., Radhakrishnan R.S., Chao T. Browning of subcutaneous white adipose tissue in humans after severe adrenergic stress. Cell Metabolism. 2015;22(2):219–227. doi: 10.1016/j.cmet.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenwald M., Perdikari A., Rülicke T., Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nature Cell Biology. 2013;15(6):659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 15.Wu J., Boström P., Sparks L.M., Ye L., Choi J.H., Giang A.-H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerra C., Koza R.A., Yamashita H., Walsh K., Kozak L.P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. The Journal of Clinical Investigation. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vegiopoulos A., Müller-Decker K., Strzoda D., Schmitt I., Chichelnitskiy E., Ostertag A. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science. 2010;328(5982):1158–1161. doi: 10.1126/science.1186034. [DOI] [PubMed] [Google Scholar]
- 19.Cao L., Choi E.Y., Liu X., Martin A., Wang C., Xu X. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell Metabolism. 2011;14(3):324–338. doi: 10.1016/j.cmet.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emanuelli B., Vienberg S.G., Smyth G., Cheng C., Stanford K.I., Arumugam M. Interplay between FGF21 and insulin action in the liver regulates metabolism. The Journal of Clinical Investigation. 2014;124(2):515–527. doi: 10.1172/JCI67353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Li R., Meng Y., Li S., Donelan W., Zhao Y. Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes. 2014;63(2):514–525. doi: 10.2337/db13-1106. [DOI] [PubMed] [Google Scholar]
- 22.Surwit R.S., Feinglos M.N., Rodin J., Sutherland A., Petro A.E., Opara E.C. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44(5):645–651. doi: 10.1016/0026-0495(95)90123-x. [DOI] [PubMed] [Google Scholar]
- 23.Almind K., Kahn C.R. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53(12):3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 24.Taketo M., Schroeder A.C., Mobraaten L.E., Gunning K.B., Hanten G., Fox R.R. FVB/N: an inbred mouse strain preferable for transgenic analyses. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(6):2065–2069. doi: 10.1073/pnas.88.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petkov P.M., Cassell M.A., Sargent E.E., Donnelly C.J., Robinson P., Crew V. Development of a SNP genotyping panel for genetic monitoring of the laboratory mouse. Genomics. 2004;83(5):902–911. doi: 10.1016/j.ygeno.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Abreu-Vieira G., Xiao C., Gavrilova O., Reitman M.L. Integration of body temperature into the analysis of energy expenditure in the mouse. Molecular Metabolism. 2015;4(6):461–470. doi: 10.1016/j.molmet.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koza R.A., Hohmann S.M., Guerra C., Rossmeisl M., Kozak L.P. Synergistic gene interactions control the induction of the mitochondrial uncoupling protein (Ucp1) gene in white fat tissue. Journal of Biological Chemistry. 2000;275(44):34486–34492. doi: 10.1074/jbc.M002136200. [DOI] [PubMed] [Google Scholar]
- 28.Coulter A.A., Bearden C.M., Liu X., Koza R.A., Kozak L.P. Dietary fat interacts with QTLs controlling induction of Pgc-1 alpha and Ucp1 during conversion of white to brown fat. Physiological Genomics. 2003;14(2):139–147. doi: 10.1152/physiolgenomics.00057.2003. [DOI] [PubMed] [Google Scholar]
- 29.Xue B., Coulter A., Rim J.S., Koza R.A., Kozak L.P. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Molecular and Cellular Biology. 2005;25(18):8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Team R. R Foundation for Statistical Computing; Vienna, Austria: 2008. Book R: a language and environment for statistical computing. [Google Scholar]
- 33.Zhang H.-M., Chen H., Liu W., Liu H., Gong J., Wang H. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Research. 2012;40(Database issue):D144–D149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald M.E., Li C., Bian H., Smith B.D., Layne M.D., Farmer S.R. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell. 2015;160(1–2):105–118. doi: 10.1016/j.cell.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L., Kon N., Li T., Wang S.-J., Su T., Hibshoosh H. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue B., Rim J.S., Hogan J.C., Coulter A.A., Koza R.A., Kozak L.P. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. Journal of Lipids & Research. 2007;48(1):41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Brune J.E., Kern M., Kunath A., Flehmig G., Schön M.R., Lohmann T. Fat depot-specific expression of HOXC9 and HOXC10 may contribute to adverse fat distribution and related metabolic traits. Obesity (Silver Spring) 2016;24(1):51–59. doi: 10.1002/oby.21317. [DOI] [PubMed] [Google Scholar]
- 38.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. The Journal of Clinical Investigation. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher F.M., Kleiner S., Douris N., Fox E.C., Mepani R.J., Verdeguer F. FGF21 regulates PGC-1{alpha} and browning of white adipose tissues in adaptive thermogenesis. Genes and Development. 2012;26(3):271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiefer F.W., Vernochet C., O'Brien P., Spoerl S., Brown J.D., Nallamshetty S. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nature Medicine. 2012;18(6):918–925. doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walden T.B., Hansen I.R., Timmons J.A., Cannon B., Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. American Journal of Physiology, Endocrinology and Metabolism. 2012;302(1):E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Bolze F., Fromme T., Klingenspor M. Intrinsic differences in BRITE adipogenesis of primary adipocytes from two different mouse strains. Biochimica et Biophysica Acta. 2014;1841(9):1345–1352. doi: 10.1016/j.bbalip.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Collins S., Daniel K.W., Petro A.E., Surwit R.S. Strain-specific response to beta 3-adrenergic receptor agonist treatment of diet-induced obesity in mice. Endocrinology. 1997;138(1):405–413. doi: 10.1210/endo.138.1.4829. [DOI] [PubMed] [Google Scholar]
- 44.Lasar D., Julius A., Fromme T., Klingenspor M. Browning attenuates murine white adipose tissue expansion during postnatal development. Biochimica et Biophysica Acta. 2013;1831(5):960–968. doi: 10.1016/j.bbalip.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Soccio R.E., Chen E.R., Rajapurkar S.R., Safabakhsh P., Marinis J.M., Dispirito J.R. Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell. 2015;162(1):33–44. doi: 10.1016/j.cell.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kajimura S., Seale P., Kubota K., Lunsford E., Frangioni J.V., Gygi S.P. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W., Yang Q., Roeder R.G. Dynamic interactions and cooperative functions of PGC-1alpha and MED1 in TRalpha-mediated activation of the brown-fat-specific UCP-1 gene. Molecular Cell. 2009;35(6):755–768. doi: 10.1016/j.molcel.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerhart-Hines Z., Feng D., Emmett M.J., Everett L.J., Loro E., Briggs E.R. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature. 2013;503(7476):410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu J., Cohen P., Spiegelman B.M. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes & Development. 2013;27(3):234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller E. Browning and graying: novel transcriptional regulators of brown and beige fat tissues and aging. Frontiers in Endocrinology. 2016;7:19. doi: 10.3389/fendo.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma X., Xu L., Gavrilova O., Mueller E. Role of forkhead box protein A3 in age-associated metabolic decline. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(39):14289–14294. doi: 10.1073/pnas.1407640111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scimè A., Grenier G., Huh M.S., Gillespie M.A., Bevilacqua L., Harper M.-E. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metabolism. 2005;2(5):283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Leonardsson G., Steel J.H., Christian M., Pocock V., Milligan S., Bell J. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hallberg M., Morganstein D.L., Kiskinis E., Shah K., Kralli A., Dilworth S.M. A functional interaction between RIP140 and PGC-1alpha regulates the expression of the lipid droplet protein CIDEA. Molecular and Cellular Biology. 2008;28(22):6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]