Abstract
Objective
Characterizing specific metabolites in sub-clinical phases preceding the onset of type 2 diabetes to enable efficient preventive and personalized interventions.
Research design and methods
We developed predictive models of type 2 diabetes using two strategies. One strategy focused on the probability of incidence only and was based on logistic regression (MRS1); the other strategy accounted for the age at diagnosis of diabetes and was based on Cox regression (MRS2). We assessed 293 metabolites using non-targeted metabolomics in fasting plasma samples of 1,044 participants (including 231 incident cases over 9 years) used as training population; and fasting serum samples of 128 participants (64 incident cases versus 64 controls) used as validation population. We applied a LASSO-based variable selection aiming at maximizing the out-of-sample area under the receiver operating characteristic curve (AROC) and integrated AROC.
Results
Sixteen and 17 metabolites were selected for MRS1 and MRS2, respectively, with AROC = 90% and 73% in the training and validation populations, respectively for MRS1. MRS2 had a similar performance and was significantly associated with a younger age of onset of type 2 diabetes (β = −3.44 years per MRS2 SD in the training population, p = 1.56 × 10−7; β = −4.73 years per MRS2 SD in the validation population, p = 4.04 × 10−3).
Conclusions
Overall, this study illustrates that metabolomics improves prediction of type 2 diabetes incidence of 4.5% on top of known clinical and biological markers, reaching 90% in total AROC, which is considered the threshold for clinical validity, suggesting it may be used in targeting interventions to prevent type 2 diabetes.
Keywords: Type 2 diabetes, Metabolomics, Risk prediction, High dimensional regression, LASSO
Highlights
-
•
Metabolites Risk Scores improve the prediction of type 2 diabetes on top of clinical and biological risk factors in both high and low-risk sub-populations.
-
•
Two predictive metabolites (1,5-anhydroglucitol and Dehydroisoandrosterone sulfate) were well conserved over 9 years.
-
•
Comparing two statistical approaches revealed that lipid metabolism distinguishes baseline risk from that of fast converters.
1. Introduction
Characterizing metabolic disruptions preceding the onset of type 2 diabetes is critical to identify individuals at risk, especially at the early asymptomatic stages of the disease when intervention can be most effective. Given the high rate of complications associated with long duration hyperglycemia [1], it is particularly important to prevent or at least delay type 2 diabetes in individuals in their early forties or younger. Although epidemiological studies have reported numerous risk factors for type 2 diabetes [2], [3], the predictive performances of statistical models based on these predictors still need to be improved. Different approaches such as genome-wide association studies (GWAS) have been proposed to identify new risk factors. GWAS have generated a catalog of replicated genetic loci that includes up to 100 variants [4]. However, these genetic variants only explain an unexpectedly small fraction (<15%) of type 2 diabetes estimated heritability and their inclusion only marginally improves the performances of previously existing predictive models [5], [6].
Metabolomics, defined as the comprehensive analysis of low-molecular weight metabolites produced by a system, has recently emerged for disease diagnosis and biomarker identification [7]. Several studies showed that high levels of the branched-chain amino acids (BCAA) such as leucine, isoleucine, and valine as well as high levels of the aromatic amino acids phenylalanine and tyrosine are strong predictors of insulin resistance and type 2 diabetes [8], [9], [10], [11]. Furthermore, increased plasma levels of alpha-hydroxybutyrate (AHB) and decreased levels of 1-linoleoyl-glycerophosphocholine (L-GPC) were associated with glucose intolerance [12]. Other studies also have reported carbohydrates (glucose, mannose, 1,5-anhydroglucitol) [13], [14], gamma-glutamyl derivatives (γ-glutamylphenylalanine, γ-glutamyltyrosine, γ-glutamylvaline) [15], glycine [14], and serine [15] as good predictors of type 2 diabetes.
Despite this increasing catalog of potential predictors, statistical approaches implemented to train predictive models suffer from two main limitations. The first limitation relates to the commonly admitted assumption that significantly associated (often under a regression framework) metabolites would automatically be good predictors [16]. Although partially true, this assumption ignores that predictive performances are driven not only by a significant shift in the metabolite mean level (as classically captured by a test of association) but also generally by any change in the entire distribution of the metabolites. As a consequence, if the variance of a metabolite is significantly different between incident cases and controls, despite no significant difference in means, the latter metabolite can be a rather good predictor.
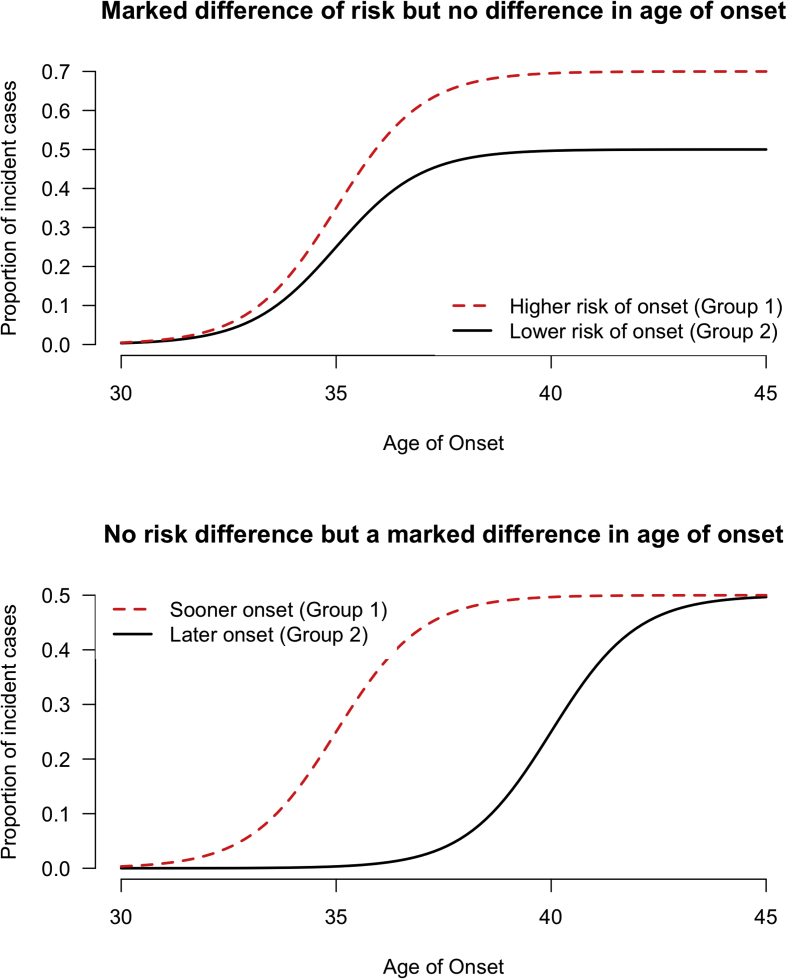
The second limitation of most implemented approaches is that the bivalent notion of incidence is often overlooked. Indeed, incidence covers two distinct, yet complementary, aspects that are: first, the probability of developing the disease in the future, and secondly the speed at which this occurs (Supplementary Figure 1). Most studies using metabolomics to study the incidence of type 2 diabetes have been focused only on characterizing the probability to develop the disease, but have mostly ignored the second aspect of incidence. This is illustrated by the recurrent use of logistic regression models in the related literature [6], [8], [10], [12], [14], [17]. Even when more suitable models such as Cox regression are used, the model performances are often assessed using static metrics such as the area under the receiver operating characteristic curve (AROC) or the net reclassification index (NRI). Instead, the use of dynamic metrics such as the integrated time-dependent AROC (iAROC) should be used to take full advantage of the time-dependent nature of the predicted outcome [18]. One expected consequence of these classical modeling choices is a sub-optimal performance in both evaluating the probability of the incidence and predicting who will develop the disease earlier or later.
The present study aimed to overcome these two limitations by calibrating two predictive models; one focused on the probability to develop type 2 diabetes in the future regardless of the time scale (Strategy 1: Metabolomic Risk Score 1; MRS1) and the other trying to simultaneously predict the risk and the age of onset (Strategy 2: Metabolomic Risk Score 2; MRS2). To complete this aim, we used a comprehensive profiling of metabolites in plasma and serum samples from middle-aged participants of prospective cohorts. The comparison of the two prediction strategies is an underlying aim of this study that would bring to light metabolites simultaneously and/or specifically contributing to type 2 risk and early onset of diabetes. We also aimed to evaluate the stability over time of the metabolites found through both strategies, which is a key element in their clinical use. Indeed, targeting metabolites conserved in time is mandatory to implement any measurable preventive intervention. Finally, we aimed to investigate the capacity of the calibrated predictive models to improve risk prediction on top of known clinical and biological risk factors.
2. Research design and methods
2.1. Training population
We studied men and women who participated in the nine-year follow-up study D.E.S.I.R., a middle-aged, European cohort [5], [19], [20]. A case-cohort design was used to include 231 cases of incident type 2 diabetes and 836 participants randomly sampled from the entire cohort. Baseline and follow-up clinical characteristics of participants included in the training population are shown in Supplementary Table 1. Type 2 diabetes was defined using one of the following criteria: use of glucose lowering medication, fasting plasma glucose [FG] ≥7 mmol/L, or glycated hemoglobin A1c [HbA1c] ≥6.5% (48 mmol/mol) [21]. Clinical and biological evaluations were performed at inclusion and after three, six, and nine years, as previously described [22], [23]. All participants provided written informed consent and the study protocol was approved by the Ethics Committee for the Protection of Subjects for Biomedical Research of Bicêtre Hospital, France.
2.2. Validation population
To provide an external assessment of the predictive models from the training population, we selected 64 incident type 2 diabetes cases and 64 controls (matched on age at inclusion, sex and body mass index [BMI]) from French families with type 2 diabetes or obesity recruited by the CNRS UMR8199 unit (Lille, France) [24], [25], [26]. Among the recruited participants we selected those with baseline characteristics (age, sex, BMI, fasting glucose, 2-hour glucose and glucose lowering treatment) available, with a follow-up including at least two measurements and with at least 100 μL of fasting serum available. Baseline clinical characteristics of participants included in the validation population are shown in Supplementary Table 1. Type 2 diabetes was defined using the following criteria: use of glucose lowering medication, fasting plasma glucose [FG] ≥7 mmol/L, or 2-hour glucose ≥ 11 mmol/L. The average follow-up length was 8.6 years (standard deviation: 4.6 years) in the validation population. Informed consent was obtained from all subjects, and the study was approved by the ethics committees from Lille, France.
2.3. Metabolite measurements
Metabolomic measurements were performed in fasting plasma samples from D.E.S.I.R. participants and in fasting serum samples from those included in the validation population. All fasting plasma and serum samples were processed by the Metabolon (Durham, NC) platform using GC/MS and LC/MS/MS as previously described [27], [28]. Since the analysis spanned a number of days, a data normalization step was applied to correct inter-day variations. Each compound was therefore corrected in run-day blocks, medians were equated to one (1.00), and each data point was normalized. We analyzed 293 metabolites (intersection between 491 detected in plasma and 625 detected in serum) that were detected (missing value rate <20%) in both plasma and serum samples. Metabolites were divided into two categories according to their missing value rate. The first category involved 255 metabolites with missing value rate <5% in either plasma or serum samples. For these metabolites, missing values were imputed with the smallest detected value. The second category involved 38 metabolites, for which the missing value rate ranged from 5% to 80%. These metabolites were analyzed as binary exposures (presence vs absence) and observed values were coded “1” and missing values “0”.
2.4. Clinical and biological risk factors
We used several clinical and biological type 2 diabetes risk factors to compare the discriminative performances of metabolomic markers with established predictors. We restricted the set of clinical and biological risk factors assessed in this study to risk factors available in both training and validation populations. Listed below, the latter risk factors were dichotomized so as to define a stratum at higher risk vs a stratum at lower risk: sex (men vs women), age (≥45 vs <45 years), body mass index (BMI: ≥25 vs <25 kg/m2), fasting glucose (FG ≥5.6 vs <5.6 mmol/L), blood pressure (BP: diagnosed hypertension or systolic BP ≥ 130 or diastolic BP ≥ 85 mm Hg vs no hypertension and systolic BP < 130 and diastolic BP < 85 mm Hg), triglycerides (TG: TG ≥ 1.7 vs < 1.7 mmol/L), high density lipoprotein (HDL) cholesterol (≤1.03 in men or ≤1.29 mmol/L in women vs >1.03 in men and >1.29 mmol/L in women), smoking status (current smoker vs current non-smoker), waist circumference (WC: WC ≥ 94 in men and ≥80 cm in women vs <94 in men and <80 cm in women). The thresholds used to dichotomize continuous risk factors were chosen from the harmonized definition of the metabolic syndrome [29].
2.5. Statistical analyses
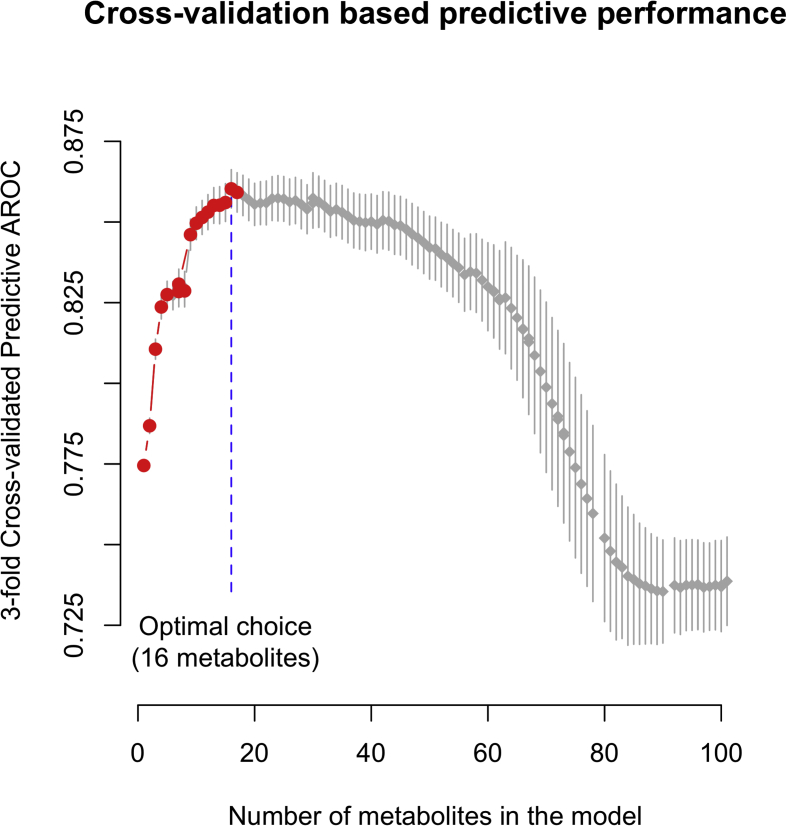
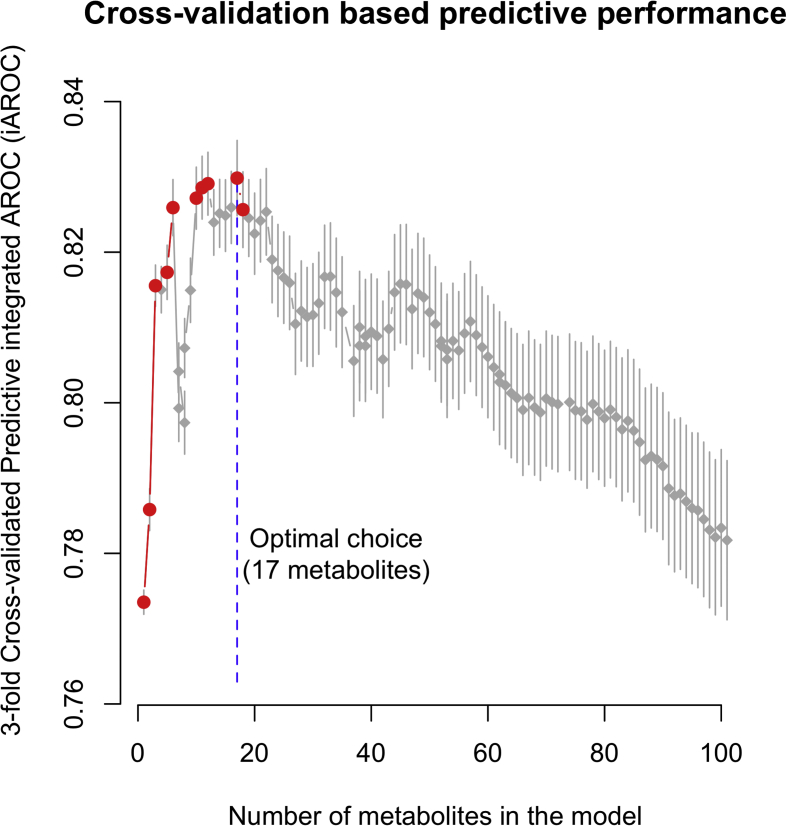
The characteristics of participants are described by mean (SD) and n (%) in Supplementary Table 1. Two strategies for predicting incident type 2 diabetes were implemented. The first one relies on multivariable logistic regression only modeling the probability of developing type 2 diabetes, while the second, based on multivariable Cox regression with age as the time scale, tries to simultaneously identify those with an early age at diagnosis. These two models used 293 metabolites as explanatory variables and the Least Absolute Shrinkage and Selection Operator (LASSO) regularization was applied [30] to select the most relevant metabolites. We used 3-fold cross-validation to select the number of metabolites to include in the logistic regression (strategy 1) and Cox regression (strategy 2) models. The number of metabolites in each model was selected to maximize the averaged AROC for logistic regression, and the averaged integrated AROC [18] (iAROC) for Cox regression, over 10,000 replications (Supplementary Figures 2 and 3). For any given number of metabolites, 95% confidence intervals for averaged AROC and iAROC were calculated as the intervals centered on the averaged values and containing 95% of the generated AROC values over the 10,000 replications. All models were fitted using two thirds of the training population only.
To assess the stability of metabolites between baseline and year nine, we compared the average values between these two time points using paired t-tests as well as the correlation between these two measurements. This comparison was performed for each of the identified metabolites in the 778 D.E.S.I.R participants included in the random sample cohort and who remained non-diabetic during the follow-up. The statistical significance for this analysis was set at p < 0.05.
Statistical analyses used R version 3.1.0 (http://www.r-project.org/) with the R packages survival, pROC and glmnet.
3. Results
3.1. Strategy 1: Predicting the incidence of type 2 diabetes regardless of the age at diagnosis
Using 3-fold cross-validation in D.E.S.I.R. samples, we identified 16 metabolites which produced a combined score that best discriminated type 2 diabetes cases from controls (Supplementary Figure 2; Table 1), regardless of age at diagnosis. This score subsequently referred to as Metabolomic Risk Score 1 (MRS1) includes six amino acids or derivatives (isoleucine, isovalerylcarnitine, phenylalanine, pro-hydroxy-pro, serine, tyrosine), four carbohydrates (fructose, mannose, glucose and 1,5-anhydroglucitol), two lipids (L-GPC and 1-palmitoylglycerol), two peptides (γ-glutamylphenylalanine and γ-glutamyltyrosine), and two xenobiotics (cotinine and piperine) (Table 1). Cotinine was analyzed as a binary exposure (presence versus absence) since it was undetected in >50% of the study participants. Importantly, we found a concordance of 96% (Fisher's exact test p < 10−10) between dichotomized cotinine that is a biomarker of exposure to tobacco smoke and self-reported smoking habits.
Table 1.
Metabolites contributing to MRS1 and MRS2. The first nine metabolites contribute to both scores and the 15 others are specific to each score. Relative contributions ≥1/16 = 6.25% for MRS1 and ≥1/17 ≈ 5.88% for MRS2 are highlighted in bold font. Relative contribution ratios for the two scores that are above 2 are also highlighted in bold font. References are given for metabolites reported in the literature for associations with insulin resistance or prevalent and incident type 2 diabetes.
| Metabolites | Associated pathways | MRS1 |
MRS2 |
References | ||
|---|---|---|---|---|---|---|
| Regression coefficient | Relative contribution to the score | Regression coefficient | Relative contribution to the score | |||
| 1,5-Anhydroglucitol | Glycolysis, gluconeogenesis, Pyruvate Metabolism | −0.50 | 9.77% | −0.26 | 7.13% | [13], [14] |
| 1-Linoleoyl-GPC | Lysolipid | −0.31 | 5.97% | −0.07 | 1.92% | [12] |
| 1-Palmitoylglycerol | Monoacylglycerol | 0.16 | 3.10% | 0.25 | 6.96% | [29] |
| Cotinine | Tobacco Metabolite | 0.33 | 6.34% | 0.32 | 8.68% | [3] |
| γ-Glutamylphenylalanine | Gamma-glutamyl Amino Acid | 0.17 | 3.34% | 0.09 | 2.61% | [15] |
| Glucose | Glycolysis, Gluconeogenesis, Pyruvate Metabolism | 1.03 | 20.0% | 0.51 | 13.8% | [13], [14] |
| Isoleucine | Leucine, Isoleucine, Valine Metabolism | 0.28 | 5.39% | 0.27 | 7.33% | [13], [14] |
| Mannose | Fructose, Mannose, Galactose Metabolism | 0.37 | 7.26% | 0.13 | 3.48% | [13], [14] |
| Pro-hydroxy-pro | Urea cycle; Arginine, Proline Metabolism | −0.30 | 5.85% | −0.16 | 4.40% | |
| Fructose | Fructose, Mannose, Galactose Metabolism | 0.27 | 5.21% | [14] | ||
| γ-Glutamyltyrosine | Gamma-glutamyl Amino Acid | 0.29 | 5.59% | [15] | ||
| Isovalerylcarnitine | Leucine, Isoleucine, Valine Metabolism | 0.19 | 3.73% | |||
| Phenylalanine | Phenylalanine, Tyrosine Metabolism | 0.28 | 5.48% | [10], [13] | ||
| Piperine | Food Component/Plant | 0.30 | 5.91% | |||
| Serine | Glycine, Serine, Threonine Metabolism | −0.31 | 6.08% | [15] | ||
| Tyrosine | Phenylalanine, Tyrosine Metabolism | −0.05 | 0.97% | [10] | ||
| 1-Stearoyl-GPI | Lysolipid | −0.26 | 7.11% | |||
| 3-Hydroxyisobutyrate | Leucine, Isoleucine, Valine Metabolism | 0.15 | 4.03% | [13] | ||
| Dehydroisoandrosterone sulfate | Steroid | 0.30 | 8.27% | |||
| γ-Glutamylvaline | Gamma-glutamyl Amino Acid | 0.12 | 3.35% | [15] | ||
| Glycine | Glycine, Serine, Threonine Metabolism | −0.13 | 3.45% | [8], [17] | ||
| Palmitoyl sphingomyelin | Sphingolipid Metabolism | −0.14 | 3.93% | [14] | ||
| Stearoylcarnitine | Fatty Acid Metabolism (Acyl Carnitine) | −0.19 | 5.12% | |||
| Urea | Urea cycle; Arginine, Proline Metabolism | −0.31 | 8.40% | [14], [15] | ||
MRS1 was successful at discriminating incident cases from controls with high accuracy (mean cross-validated AROC among D.E.S.I.R participants: 86.0% [84.8%−87.2%]95%CI; mean cross-validated AROC in the validation population: 71.2% [70.2%−72.2%]95%CI). Moreover, we found that in D.E.S.I.R. participants, the proportion of incident cases above the second tertile of MRS1 was 21.5-fold larger than below the first tertile. This finding was confirmed in the validation population although with a smaller proportion (HR = 3.39, p = 4 × 10−3; Table 2). Finally, we did not find any significant association between MRS1 and the age at diagnosis of type 2 diabetes in D.E.S.I.R. participants or in the validation population (D.E.S.I.R participants: β = 0.08 year per MRS1 SD; p = 0.91; Validation population: β = 0.99 years per MRS1 SD of MRS1; p = 0.58 Table 2).
Table 2.
Association between MRS1/MRS2 (continuous score or categorized score) with incidence of type 2 diabetes measured with hazard and odds ratios; and with age at diagnosis.
| Training population (D.E.S.I.R. participants) |
Validation population |
|||||
|---|---|---|---|---|---|---|
| Hazard Ratio (p-value) |
Odds Ratio (p-value) |
Regression coefficient for association with at diagnosis (p-value) |
Hazard Ratio (p-value) |
Odds Ratio (p-value) |
Regression coefficient for association with at diagnosis (p-value) |
|
| Continuous MRS1 (unit: per standard deviation of MRS1) |
2.88 (2 × 10−16) | 8.44 (6 × 10−47) |
0.08 year (0.91) |
1.49 (8 × 10−4) |
3.3 (5 × 10−5) |
1 year (0.57) |
| Categorized MRS1 | ||||||
| 1st tertile groups vs 2nd tertile group 1st tertile groups vs 3rd tertile group |
4.13 (6 × 10−4) 21.5 (2 × 10−15) |
1.78 (2 × 10−5) 4.02 (3 × 10−24) |
7.16 years (0.06) 5.43 years (0.14) |
1.52 (0.30) 3.39 (3 × 10−3) |
1.95 (0.17) 8.46 (2 × 10−4) |
4.08 years (0.38) 0.80 year (0.86) |
| Continuous MRS2 (unit: per standard deviation of MRS2) |
2.72 (2 × 10−16) |
3.63 (6 × 10−43) |
−2.7 years (2 × 10−7) |
1.63 (1 × 10−7) |
1.78 (9 × 10−4) |
−3.75 years (4 × 10−3) |
| Categorized MRS2 | ||||||
| 1st tertile groups vs 2nd tertile group 1st tertile groups vs 3rd tertile group |
3.35 (6 × 10−4) 15.2 (2 × 10−15) |
3.04 (2 × 10−4) 18.0 (1 × 10−26) |
−1.12 years (0.67) −6.06 years (0.01) |
1.97 (0.06) 5.85 (2 × 10−6) |
2.01 (0.13) 4.71 (1 × 10−3) |
−0.81 year (0.83) −10.9 years (5 × 10−3) |
3.2. Strategy 2: Predicting the incidence of type 2 diabetes accounting for the age at diagnosis
Using 3-fold cross-validation in D.E.S.I.R samples, we found that 17 metabolites could discriminate incident cases from controls while simultaneously accounting for the age of onset (Table 1). Including more metabolites led to over-fitting and, consequently, to reduced out-of-sample discriminative performances (Supplementary Figure 3). Among these 17 metabolites we found six lipids (L-GPC, 1-palmitoylglycerol, 1-stearol-GPI, dehydroisoandrosterone sulfate (DHEA-S), palmitoyl sphingomyelin, and stearoylcarnitine), five BCAA derivatives (3-hydroxyisobutyrate, glycine, isoleucine, pro-hydroxy-pro, and urea), three carbohydrates (mannose, glucose, and 1,5-anhydroglucitol), one peptide (γ-glutamylphenylalanine), and one xenobiotic (cotinine) (Table 1).
These 17 metabolites (Table 1) were combined into a Metabolomic Risk Score 2 (MRS2) that was highly discriminant between incident cases and controls (mean cross-validated iAROC among D.E.S.I.R participants: 83% [82%−84%]95%CI; mean cross-validated iAROC in the validation population: 67.2% [66.5%−67.8%]95%CI) and was also significantly associated with a younger onset of type 2 diabetes (among D.E.S.I.R participants: β = −3.44 years per MRS2 SD, p = 2 × 10−7; in the validation population: β = −4.73 years per MRS2 SD, p = 4 × 10−3; Table 2). On average, D.E.S.I.R. participants above the second MRS2 tertile developed type 2 diabetes at 56 years while diabetes occurred at 62 years in the first tertile group (β = −6.06, p = 0.01; Table 2). We confirmed this significant difference in the validation population, in which type 2 diabetes occurred 11 years earlier (p = 5 × 10−3; Table 2) in the third compared to the first tertile group.
3.3. Comparison of MRS1 and MRS2
Strategies 1 and 2 led to different sets of metabolites to be included in MRS1 and MRS2. Nonetheless, nine metabolites were common to both strategies: 1,5-anhydroglucitol, L-GPC, 1-palmitoylglycerol, cotinine, γ-glutamylphenylalanine, glucose, isoleucine, mannose and pro-hydroxy-pro (Table 1). The regression coefficients associated with these metabolites were sign consistent in each risk score (Table 1). However, the relative contributions of lipids were different in MRS2 and MRS1. Indeed, the contribution of 1-palmitoylglycerol was 2.2-fold (2.2 ≈ 6.96/3.1; Table 1) larger than other metabolites in MRS2, while the contribution of L-GPC was 3.1-fold larger in MRS1 (3.1 ≈ 5.97/1.92; Table 1).
Similarly, mannose had 2.1-times more weight in MRS1 than in MRS2 (2.1 ≈ 7.26/3.48; Table 1). In addition, we found fructose, γ-glutamyltyrosine, isovalerylcarnitine, phenylalanine, piperine, serine, and tyrosine to be specific for MRS1; while 1-stearoyl-GPI, 3-hydroxyisobutyrate, DHEA-S, γ-glutamylvaline, glycine, palmitoyl sphingomyelin, stearoylcarnitine, and urea were only contributing to MRS2.
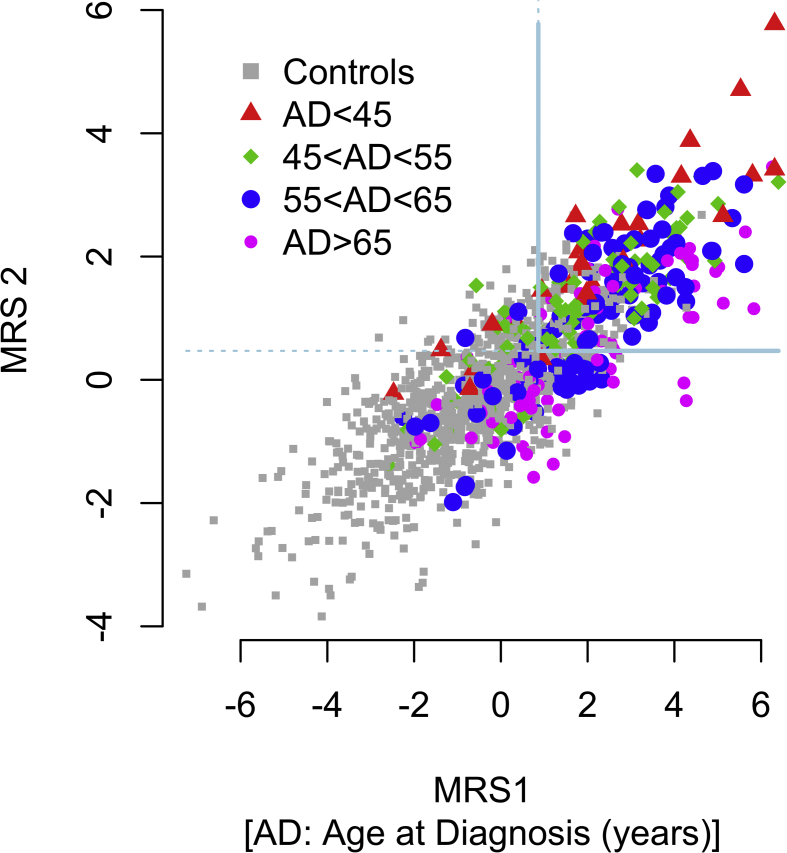
Moreover, we assessed the value of combining both MRS1 and MRS2 to stratify individuals at higher risk to develop T2D at an earlier age (Supplementary Figure 4). We observed that participants with both MRS1 and MRS2 scores above the 2nd tertile of each score not only had a higher risk to develop type 2 diabetes (61.5% of all incident cases) but also developed type 2 diabetes at 56 years, on average, 4 years before the average age at diagnosis in the training and the validation populations (p = 3.4 × 10−4; data not shown).
3.4. MRS1/MRS2 versus clinical and biological risk factors of glucose intolerance
For each clinical and biological risk factor, we defined a stratum at higher risk versus a stratum at lower risk according to the dichotomization proposed in the Clinical and biological risk factors section. When assessing the predictive power of MRS1 and MRS2 in each stratum, we found that the discrimination accuracy of MRS2 was larger in younger individuals (iAROC in individuals <45 years: 86.5% vs iAROC in individuals ≥45 years: 72.5%; p = 1.26 × 10−6; Supplementary Table 2), and in individuals with mild impaired fasting glucose (iAROC in individuals with FG < 5.6 mmol/L: 74.4% vs iAROC in individuals with FG ≥ 5.6 mmol/L: 82.2%, p = 0.03; Supplementary Table 2). This finding was only statistically significant in the training population. In addition, the performances of MRS1 were not different in strata at lower risk compared to strata at higher risk (Supplementary Table 2).
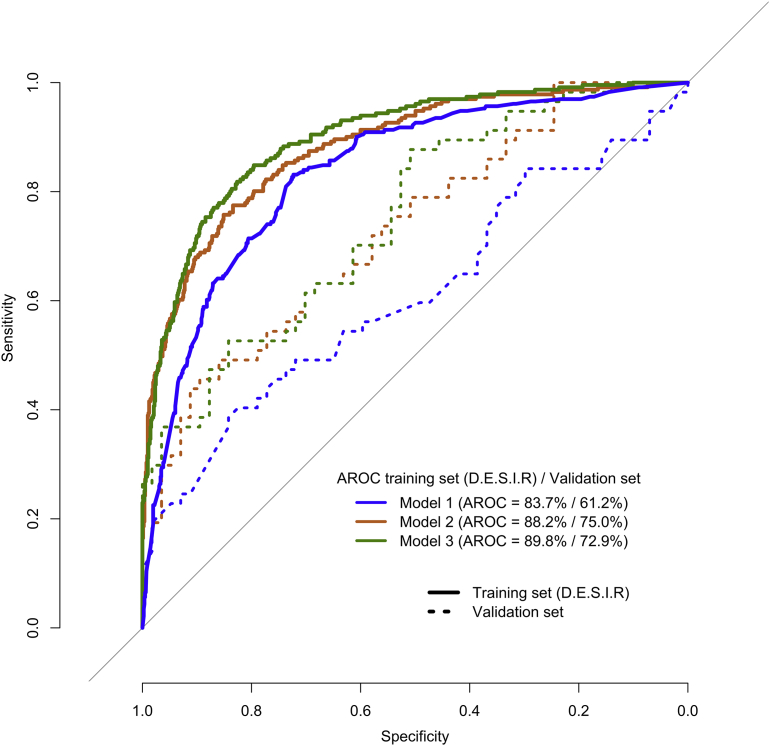
To assess the relative predictive performances of MRS1 and MRS2 in comparison with classic clinical and biological risk factors, we considered three predictive models: Model 1 included all clinical and biological risk factors listed in the Clinical and biological risk factors section; Model 2 included only MRS1 when the metrics used for comparison is AROC; or only MRS2 when the metrics used for comparison is iAROC; and Model 3 included all predictors in Model 1 plus MRS1 or MRS 2. The ROC curves for all models are shown in Figure 1. In D.E.S.I.R. participants, Model 1 yielded an AROC and an iAROC of 83.7% and 60.5%, respectively. In the validation population, however, the performances of these models were lower: AROC = 61.2% and iAROC = 52.5% (Table 3). MRS1 and MRS2 alone (Model 2) had better performances than Model 1 in D.E.S.I.R. participants, both in terms of AROC and iAROC (p < 5 × 10−8; Table 3). In the validation population however, only MRS2 yielded a statistically better iAROC than Model 1 (+15.4%; p = 9 × 10−3; Table 3). Finally, the most comprehensive model, namely Model 3, had significantly better predictive performances than Model 1 and Model 2 in D.E.S.I.R. participants (largest AROC = 89.8%; p < 5 × 10−3; Table 3) but was less predictive than Model 2 in the validation population.
Figure 1.
ROC Receiver operating characteristic (ROC) curves and area under these curves (AROC) statistics for three predictive models: Model 1 with clinical and biological risk factors only, Model 2 with MRS1 only, and Model 3 including clinical and biological risk factors + MRS1.
Table 3.
Discriminative performances of different models; model 1 including only classic clinical and biological risk factors; model 2 including MRS1 (when comparison is made using AUC) or MRS2 (when comparison is made using integrated AROC or iAROC) and model 3 including all risk factors + MRS1 or MRS2. MRS1 and MRS 2 were never added simulatneously in any models. When MRS1 or MRS2 were added in a model, impaired fasting glucose and current smoking status were not included as clinical and biological risk factors to avoid redundancy.
| Predictive models | Training population (D.E.S.I.R. participants) |
Validation population |
||||
|---|---|---|---|---|---|---|
| AROC | iAROC | p-Value for AROC comparison/p-value for iAROC comparison | AROC | iAROC | p-Value for AROC comparison/p-value for iAROC comparison | |
| Model 1: clinical and biological risk factors only | 83.7% | 60.5% | Model 1 vs Model 2 2 × 10−9/2 × 10−8 |
61.2% | 52.5% | Model 1 vs Model 2 0.08/9 × 10−3 |
| Model 2: MRS1/MRS2 only | 88.2% | 84.4% | Model 2 vs Model 3 5 × 10−4/2 × 10−14 |
75.0% | 67.9% | Model 2 vs Model 3 0.41/5 × 10−3 |
| Model 3: clinical, biological risk factors and MRS1/MRS 2 | 89.8% | 70.0% | Model 3 vs Model 1 3 × 10−3/3 × 10−3 |
72.9% | 52.9% | Model 3 vs Model 1 0.01/0.92 |
3.5. Time conservation of identified metabolites
We assessed the time conservation of the 24 metabolites (16 in MRS1, 17 in MRS2 but 9 in common) involved in MRS1 and/or MRS2 by comparing baseline to follow-up (nine years after) levels as well as by estimating correlation coefficients between these two measurements in D.E.S.I.R. participants. We found that correlations between baseline and follow-up were strongly significant (p < 5 × 10−7; Supplementary Table 3) for all metabolites, except for 1-stearol-GPI (r = 0.08, p = 0.02; Supplementary Table 3) and fructose (r = 0.05, p = 0.15; Supplementary Table 3). However, we observed that 12 metabolites significantly increased and seven decreased with age during the nine years follow up (p < 0.05; Supplementary Table 3). Two metabolites, 1,5-AG and DHEA-S, were particularly well conserved, as their between-measurements correlation was above 0.73 (p < 10−10) which is larger than HbA1c (r = 0.63 [0.58–0.67]95%CI; data not shown). Among the metabolites analyzed as binary predictors (detected vs not-detected), we found that cotinine, piperine and stearoylcarnitine were the most stable with a concordance of >79% (data not shown) between baseline and follow-up measurements.
4. Conclusions
This study proposes two strategies for predicting incident type 2 diabetes. The first one relies on multivariable logistic regression, modeling only the probability of developing type 2 diabetes, while the second, based on multivariable Cox regression, tries also to identify those with an early age at diagnosis. The performances of these two strategies were assessed using both out-of-sample cross validation and an actual validation sample, which emphasizes their applicability to external populations.
This study also illustrates the complementarity of these two approaches especially since identifying early type 2 diabetes converters has a major impact on their overall mortality risk as previously reported [31]. We found that some metabolites only contributed to one model, and for those shared by the two models, their relative contributions could vary. Indeed, metabolites involved in steroid, lysolipid, and fatty acid metabolism were specifically identified when the age at diagnosis was accounted for in the Cox model. Moreover, when focusing on metabolites selected in both models, we observed that relative weights of lipids (1-palmitoylglycerol and L-GPC) differed between the two scores. This underlines the important role of lipid metabolism in accelerating the onset of type 2 diabetes.
The complementarity between those two modeling strategies was emphasized by the comparison of MRS1 and MRS2 with clinical and biological predicting risk factors. Our study strongly confirms that metabolomic markers have a significant added-value on top of classic type 2 diabetes predictors (including glucose) as previously reported [6]. Importantly, in our study, the improvement in the AROC brought by metabolomics is larger (+4.5%) than previously reported [6] with an AROC close to 90% when metabolomic, biological, and clinical factors are used together. This illustrates that such a combined score could be clinically valid to discriminate those who will and will not become diabetic. In contrast, for the second modeling strategy taking into analysis the age at diagnosis, the discriminative power of MRS2 alone was better than when combined with classic predictors.
Our data may be useful to better design preventive intervention by stratifying and further targeting individuals with both large MRS1 and MRS2 scores as illustrated in Supplementary Figure 4. We observed that the discriminative performances of MRS1 and MRS2 were lower in the validation sample than in the training sample. Given that a reduction in discriminative performances was similarly observed when using clinical and biological risk factors only, we assume that the reduced performances of MRS1 and MRS2 are not due to over-fitting. Instead, the reduced performances of MRS1 and MRS2 in the validation population can be explained by marked differences regarding clinical parameters between the two populations. Indeed, participants in the validation population all had a family history of type 2 diabetes and/or obesity and were themselves mostly obese (Supplementary Table 1). Despite their reduced performances, MRS1 and MRS2 remained more predictive than known risk factors (Table 3) in this population already at high risk for type 2 diabetes. Although MRS1 and MRS2 also improve the specificity of type 2 diabetes prediction here, other risk factors, possibly rare family shared mutations or other metabolites not detected, remain to be identified.
We showed that MRS2 was simultaneously more predictive in younger individuals and in those with very mild impaired fasting glucose (defined by fasting glucose at baseline higher than 5.6 mmol/L which is far lower than the alternative definition of prediabetes – 6.1 mmol/L). This important finding reinforces the relevance of aiming for early preventive intervention. Indeed, as previously pointed out in the Whitehall II study [32], future incident diabetes cases often present fasting glucose above 5.6 mmol/L up to 10 years before the onset of type 2 diabetes. At that time the identification of people at risk of diabetes and preventive intervention are the most useful to prevent diabetes onset.
In contrast to genetic studies, the number and nature of the metabolites accurately measured by the different available technical platforms and the reproducibility of the metabolomic data from these platforms is still an unresolved issue. Despite that limitation, the vast majority of the 24 metabolites highlighted in this paper had previously been shown to be associated with type 2 diabetes or with insulin resistance [10], [12], [13], [14], [15], [17], [33] (Table 1). We are therefore confident that they are truly predictive of diabetes.
One strength of our study was the analysis of the conservation over time of the 24 identified type 2 diabetes metabolites. The most stable metabolites were 1,5-AG and DHEA-S. Stability over shorter spans of time (1 and 7 years) of 1,5-AG and DHEA-S was previously reported in the study by Yousri et al. (2014) [34]. The latter study also reported a relatively good time conservation (0.4 < r < 0.5) of glycine, isoleucine, isovalerylcarnitine and γ-glutamylvaline.1,5-AG, DHEA-S glycine, isoleucine, isovalerylcarnitine and γ-glutamylvaline. Furthermore 1,5-AG levels in saliva were associated with type 2 diabetes risk [35]. In addition, we also reported the stability of two xenobiotics, cotinine (tobacco consumption) and piperine (pepper consumption), which suggests the stability of the environment contributing to diabetes onset. Finally, we confirmed that urea and serine, previously reported for their association with chronological age [36], significantly varied with age during the 9 year follow-up. Altogether, our data suggest that these stable biomarkers can be safely used for large scale type 2 diabetes risk prediction.
In conclusion, the present study highlights that few biomarkers with an efficient combination as risk scores can improve the identification of incident type 2 diabetes cases, especially in those poorly recognized by classical clinical risk factors. The clinical use of such biomarkers are important for the development of early interventions for the prevention of type 2 diabetes, involving changes in life style and pharmacotherapy. A comprehensive list of metabolomic biomarkers, as well as an assessment of their predictive capacity, is under construction through a number of research studies. Our study contributes to this effort. However, to complement our findings, additional research is needed to understand the potential causality relating metabolomic biomarkers and other known risk factors to the onset of type 2 diabetes. For this, the use of statistical methodologies such as mediation analyses [37], [38] and Mendelian randomization [39] could provide avenues for further improvement.
Contribution statement
MF, BB and PF designed the study. LY performed data acquisition, data analysis, drafted and wrote the manuscript. LY, AA, AB and PF interpreted the data. AB and PF contributed to writing the manuscript. AA, MM, RR, MV, AH and BB reviewed the manuscript. All authors have read and approved the final version of the manuscript.
Acknowledgments
We are grateful to all participants of this study. The D.E.S.I.R. Study Group is composed of Inserm-U1018 (Paris: B. Balkau, P. Ducimetière, E. Eschwège), Inserm-U367 (Paris: F. Alhenc-Gelas), CHU d'Angers (A. Girault), Bichat Hospital (Paris: F. Fumeron, M. Marre, R. Roussel); CHU de Rennes (F. Bonnet), CNRS UMR-8199 (Lille: S. Cauchi, P. Froguel), Medical Examination Services (Alençon, Angers, Blois, Caen, Chartres, Chateauroux, Cholet, Le Mans, Orléans and Tours), Research Institute for General Medicine (J. Cogneau), General practitioners of the region, and Cross- Regional Institute for Health (C. Born, E. Caces, M. Cailleau, N. Copin, O. Lantieri, J.G. Moreau, F. Rakotozafy, J. Tichet, S. Vol).
This study was supported by Qatar Foundation and the Centre National de la Recherche Scientifique (CNRS). The D.E.S.I.R. study has been supported by Inserm contracts with CNAMTS, Lilly, Novartis Pharma and Sanofi-aventis, and by Inserm (Réseaux en Santé Publique, Interactions entre les déterminants de la santé, Cohortes Santé TGIR 2008), the Association Diabète Risque Vasculaire, the Fédération Française de Cardiologie, La Fondation de France, ALFEDIAM, ONIVINS, Société Francophone du Diabète, Ardix Medical, Bayer Diagnostics, Becton Dickinson, Cardionics, Merck Santé, Novo Nordisk, Pierre Fabre, Roche and Topcon.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.08.011.
Conflicts of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Baseline and follow-up characteristics of the study participants. Data are mean (Standard Deviation) or n (%). Missing data are represented by “-”.
Discriminative performances of metabolomic risk scores (AROC for MRS1 and integrated AROC or iAROC for MRS2) when study participants were stratified as being at lower or higher risk to develop type 2 diabetes according to 9 classical risk factors. *Elevated blood pressure (BP) is defined as diagnosed hypertension or systolic BP ≥ 130 mm Hg or diastolic BP ≥ 85 mm Hg; and ‡reduced HDL cholesterol by: ≤1.03 mmol/L in men, ≤1.29 mmol/L in women.
T-statistics and p-values compare average levels in metabolites contributing to MRS1 and MRS2, at baseline and at 9 years. Pearson correlation coefficients (p-values) are also provided. ↘ and ↗ mean that metabolite levels have decreased or increased respectively nine years after inclusion. The D.E.S.I.R. study.
Supplementary Figure 1.
The two components in the notion of incidence. A: On average, onset will happen at the same age between group 1 (dotted line) and group 2 (plain line), but the risk is larger in group 1 versus group 2. B: There is no risk difference between group 1 and group 2 despite a markedly delayed onset in group 2 versus group 1.
Supplementary Figure 2.
AUC Areas under receiver operational characteristic curves (AROC) calculated using 3-fold cross-validation to determine the number of metabolites to include in the model. The optimal choice is 16 metabolites for inclusion in the Metabolomic score 1 (MRS1) to provide the best discrimination between incident cases and controls.
Supplementary Figure 3.
Areas under the integrated receiver operational characteristic curves (iAROC) calculated using 3-fold cross-validation to determine the number of metabolites to include in the model. The optimal choice is 16 metabolites for inclusion in the Metabolomic score 2 (MRS2) to provide the best discrimination between incident cases and controls.
Supplementary Figure 4.
Scatter plot showing the distribution of study participants according to their values of MRS1 (x-axis) and MRS2 (y-axis). The upper-right corner represents participants with a MRS1 and MRS2>second tertile and having thus a large risk of early type 2 diabetes onset.
References
- 1.Vijan S., Sussman J.B., Yudkin J.S., Hayward R.A. Effect of patients' risks and preferences on health gains with plasma glucose level lowering in type 2 diabetes mellitus. JAMA Internnal Medicine. 2014;174(8):1227–1234. doi: 10.1001/jamainternmed.2014.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herder C., Karakas M., Koenig W. Biomarkers for the prediction of type 2 diabetes and cardiovascular disease. Clinical Pharmacology & Therapeutics. 2011;90(1):52–66. doi: 10.1038/clpt.2011.93. [DOI] [PubMed] [Google Scholar]
- 3.Balkau B., Lange C., Fezeu L., Tichet J., de Lauzon-Guillain B., Czernichow S. Predicting diabetes: clinical, biological, and genetic approaches: data from the Epidemiological Study on the Insussslin Resistance Syndrome (DESIR) Diabetes Care. 2008;31(10):2056–2061. doi: 10.2337/dc08-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics. 2012;44(9):981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaxillaire M., Yengo L., Lobbens S., Rocheleau G., Eury E., Lantieri O. Type 2 diabetes-related genetic risk scores associated with variations in fasting plasma glucose and development of impaired glucose homeostasis in the prospective DESIR study. Diabetologia. 2014;57(8):1601–1610. doi: 10.1007/s00125-014-3277-x. [DOI] [PubMed] [Google Scholar]
- 6.Walford G.A., Porneala B.C., Dauriz M., Vassy J.L., Cheng S., Rhee E.P. Metabolite traits and genetic risk provide complementary information for the prediction of future type 2 diabetes. Diabetes Care. 2014;37(9):2508–2514. doi: 10.2337/dc14-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteiro M.S., Carvalho M., Bastos M.L., Guedes de Pinho P. Metabolomics analysis for biomarker discovery: advances and challenges. Current Medicinal Chemistry. 2013;20(2):257–271. doi: 10.2174/092986713804806621. [DOI] [PubMed] [Google Scholar]
- 8.Floegel A., Stefan N., Yu Z., Mühlenbruch K., Drogan D., Joost H.G. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newgard C.B., An J., Bain J.R., Muehlbauer M.J., Stevens R.D., Lien L.F. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metabolism. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T.J., Larson M.G., Vasan R.S., Cheng S., Rhee E.P., McCabe E. Metabolite profiles and the risk of developing diabetes. Nature Medicine. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Würtz P., Mäkinen V.P., Soininen P., Kangas A.J., Tukiainen T., Kettunen J. Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes. 2012;61(6):1372–1380. doi: 10.2337/db11-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrannini E., Natali A., Camastra S., Nannipieri M., Mari A., Adam K.P. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62(5):1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousri N.A., Mook-Kanamori D.O., Selim M.M., Takiddin A.H., Al-Homsi H., Al-Mahmoud K.A. A systems view of type 2 diabetes-associated metabolic perturbations in saliva, blood and urine at different timescales of glycaemic control. Diabetologia. 2015;58(8):1855–1867. doi: 10.1007/s00125-015-3636-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menni C., Fauman E., Erte I., Perry J.R., Kastenmüller G., Shin S.Y. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62(12):4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suhre K., Meisinger C., Döring A., Altmaier E., Belcredi P., Gieger C. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5(11):e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo A., Chernoff H., Zheng T., Lo S.-H. Why significant variables aren't automatically good predictors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(45):13892–13897. doi: 10.1073/pnas.1518285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang-Sattler R., Yu Z., Herder C., Messias A.C., Floegel A., He Y. Novel biomarkers for pre-diabetes identified by metabolomics. Molecular Systems Biology. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid M., Kestler H.A., Potapov S. On the validity of time-dependent AUC estimators. Briefings in Bioinformatics. 2015;16(1):153–168. doi: 10.1093/bib/bbt059. [DOI] [PubMed] [Google Scholar]
- 19.Balkau B. An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome. Revue d'Épidémiologie et de Santé Publique. 1996;44(4):373–375. [PubMed] [Google Scholar]
- 20.Bonnet F., Roussel R., Natali A., Cauchi S., Petrie J., Saville M. Parental history of type 2 diabetes, TCF7L2 variant and lower insulin secretion are associated with incident hypertension. Data from the DESIR and RISC cohorts. Diabetologia. 2013;56(11):2414–2423. doi: 10.1007/s00125-013-3021-y. [DOI] [PubMed] [Google Scholar]
- 21.American Diabetes Association Standards of medical care in diabetes–2014. Diabetes Care. 2014;37(Suppl 1):S14–S80. doi: 10.2337/dc14-S014. [DOI] [PubMed] [Google Scholar]
- 22.Balkau B., Eschwege E., Tichet J., Marre M. Proposed criteria for the diagnosis of diabetes: evidence from a French epidemiological study (D.E.S.I.R.) Diabetes & Metabolism. 1997;23(5):428–434. [PubMed] [Google Scholar]
- 23.Vaxillaire M., Veslot J., Dina C., Proença C., Cauchi S., Charpentier G. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes. 2008;57(1):244–254. doi: 10.2337/db07-0615. [DOI] [PubMed] [Google Scholar]
- 24.Bell C.G., Benzinou M., Siddiq A., Lecoeur C., Dina C., Lemainque A. Genome-wide linkage analysis for severe obesity in french caucasians finds significant susceptibility locus on chromosome 19q. Diabetes. 2004;53(7):1857–1865. doi: 10.2337/diabetes.53.7.1857. [DOI] [PubMed] [Google Scholar]
- 25.Meyre D., Lecoeur C., Delplanque J., Francke S., Vatin V., Durand E. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31–q23.2. Diabetes. 2004;53(3):803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 26.Vionnet N., Hani E.H., Lesage S., Philippi A., Hager J., Varret M. Genetics of NIDDM in France: studies with 19 candidate genes in affected sib pairs. Diabetes. 1997;46(6):1062–1068. doi: 10.2337/diab.46.6.1062. [DOI] [PubMed] [Google Scholar]
- 27.Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical Chemistry. 2009;81(16):6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 28.Cheng J., Joyce A., Yates K., Aouizerat B., Sanyal A.J. Metabolomic profiling to identify predictors of response to vitamin E for non-alcoholic steatohepatitis (NASH) PLoS One. 2012;7(9):e44106. doi: 10.1371/journal.pone.0044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alberti K.G., Eckel R.H., Grundy S.M., Zimmet P.Z., Cleeman J.I., Donato K.A. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120(16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 30.Tibshirani R. The lasso method for variable selection in the Cox model. Statistics in Medicine. 1997;16(4):385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Tancredi M., Rosengren A., Svensson A.M., Kosiborod M., Pivodic A., Gudbjörnsdottir S. Excess mortality among persons with type 2 diabetes. The New England Journal of Medicine. 2015;373(18):1720–1732. doi: 10.1056/NEJMoa1504347. [DOI] [PubMed] [Google Scholar]
- 32.Tabák A.G., Jokela M., Akbaraly T.N., Brunner E.J., Kivimäki M., Witte D.R. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet (London, England) 2009;373(9682):2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng M., Che Z., Liang Y., Wang B., Chen X., Li H. GC–MS based plasma metabolic profiling of type 2 diabetes mellitus. Chromatographia. 2009;69(9–10):941–948. [Google Scholar]
- 34.Yousri N.A., Kastenmüller G., Gieger C., Shin S.Y., Erte I., Menni C. Long term conservation of human metabolic phenotypes and link to heritability. Metabolomics. 2014;10(5):1005–1017. doi: 10.1007/s11306-014-0629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mook-Kanamori D.O., Selim M.M., Takiddin A.H., Al-Homsi H., Al-Mahmoud K.A., Al-Obaidli A. 1,5-Anhydroglucitol in saliva is a noninvasive marker of short-term glycemic control. The Jourdnal of Clinical Endocrinology & Metabolism. 2014;99(3):E479–E483. doi: 10.1210/jc.2013-3596. [DOI] [PubMed] [Google Scholar]
- 36.Menni C., Kastenmüller G., Petersen A.K., Bell J.T., Psatha M., Thai P.C. Metabolomic markers reveal novel pathways of ageing and early development in human populations. International Journal of Epidemiology. 2013;42(4):1111–1119. doi: 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanderweele T.J., Vansteelandt S. Odds ratios for mediation analysis for a dichotomous outcome. American Journal of Epidemiology. 2010;172(12):1339–1348. doi: 10.1093/aje/kwq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.InterAct Consortium The link between family history and risk of type 2 diabetes is not explained by anthropometric, lifestyle or genetic risk factors: the EPIC-InterAct study. Diabetologia. 2013;56(1):60–69. doi: 10.1007/s00125-012-2715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abbasi A., Deetman P.E., Corpeleijn E., Gansevoort R.T., Gans R.O., Hinlege H.L. Bilirubin as a potential causal factor in type 2 diabetes risk: a Mendelian randomization study. Diabetes. 2014 doi: 10.2337/db14-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baseline and follow-up characteristics of the study participants. Data are mean (Standard Deviation) or n (%). Missing data are represented by “-”.
Discriminative performances of metabolomic risk scores (AROC for MRS1 and integrated AROC or iAROC for MRS2) when study participants were stratified as being at lower or higher risk to develop type 2 diabetes according to 9 classical risk factors. *Elevated blood pressure (BP) is defined as diagnosed hypertension or systolic BP ≥ 130 mm Hg or diastolic BP ≥ 85 mm Hg; and ‡reduced HDL cholesterol by: ≤1.03 mmol/L in men, ≤1.29 mmol/L in women.
T-statistics and p-values compare average levels in metabolites contributing to MRS1 and MRS2, at baseline and at 9 years. Pearson correlation coefficients (p-values) are also provided. ↘ and ↗ mean that metabolite levels have decreased or increased respectively nine years after inclusion. The D.E.S.I.R. study.