Abstract
Objective
Arising from common progenitors in the bone marrow, adipogenesis and osteogenesis are closely associated yet mutually exclusive during bone marrow mesenchymal stem cell (BMSC) development. Previous studies have shown that morphological changes can affect the early commitment of pluripotent BMSCs to the adipose versus osteoblastic lineage via modulation of RhoA activity. The RhoA pathway regulates actin polymerization to promote the incorporation of globular actin (G-actin) into filamentous actin (F-actin). In doing so, myocardin-related transcription factors (MRTFs) dissociate from bound G-actin and enter the nucleus to co-activate serum response factor (SRF) target gene expression. In this study, we investigated whether MRTFA/SRF is acting downstream of the RhoA pathway to regulate BMSC commitment in mice.
Methods
The effects of knocking out MRTFA on skeletal homeostasis was studied in MRTFA KO mice using micro-CT, QPCR and western blot assays. To determine how MRTFA affects the mechanisms regulating BMSC fate decisions, primary bone marrow stromal cells from WT and MRTFA KO mice as well as C3H10T1/2 cell lines were analyzed in vitro.
Results
Global MRTFA KO mice have lower whole body weight, shorter femoral and tibial lengths as well as significantly decreased bone mass in their femurs. BMSCs isolated from the KO mice show increased adipogenesis and reduced osteogenesis when compared to WT littermates. KO mice, particularly females, develop osteopenia with age, and this was enhanced by a high fat diet. Over-expression of MRTFA or SRF enhances osteogenesis in CH310T1/2 cell lines. Sca1+, CD45− cells from KO marrow express lower amounts of smooth muscle actin (SMA) and TAZ/YAP target genes compared to WT counterparts.
Conclusion
This study identified MRTFA as a novel regulator of skeletal homeostasis by regulating the balance between adipogenic and osteogenic differentiation of BMSCs. We propose that MRTFA promotes the osteogenic activity of TAZ/YAP by maintaining SMA production in BMSCs.
Keywords: Mesenchymal stem cells, Myocardin-related transcription factor A (MRTFA), Serum response factor (SRF), Osteoporosis, Bone formation, Lineage commitment
1. Introduction
Adipogenesis and osteogenesis are mutually exclusive processes during mesenchymal stem cell (MSC) differentiation and appropriate balance between them is essential for maintaining homeostasis in bone marrow [1], [2], [3], [4]. Once the precisely regulated balance is disturbed, various metabolic-related diseases develop. For example, in patients with post-menopausal osteoporosis, a significant increase of adipose tissue was observed in the bone marrow and this was accompanied by loss of bone mass [5]. Secondary osteoporosis is frequently observed in type II diabetic patients treated with Avandia, a member of the thiazolidinedione (TZD) family of insulin sensitizers and PPAR gamma agonists, resulting in an increase of adipocytes in the bone marrow in both males and females [6], [7]. These phenotypes might be caused by deregulation of early MSCs commitment by switching their fate from a bone to an adipose lineage [3], [8]. The clinical consequences of disruptions in skeletal homeostasis include susceptibility to life threatening fractures in people with osteoporosis. Changes in the fate of MSCs also impacts metabolic homeostasis since bone is an endocrine organ, which secretes osteocalcin into the circulation to control insulin secretion and glucose tolerance [9], [10]. Elucidation of the mechanisms controlling MSC fate switch will lead to a better understanding of the pathogenesis of osteoporosis and provide insights into potential therapeutic approaches [3].
MSCs are pluripotent stromal progenitor cells, which possess the ability to differentiate into chondrogenic, adipogenic, myogenic or osteogenic cells given the appropriate milieu of growth factors [11]. Multiple signaling pathways and regulatory factors are reported to strictly regulate the commitment of MSCs to each of these lineages. During the differentiation of MSCs, cell morphology changes significantly in order to serve the specialized functions of the terminally differentiated cells in different tissues. Adipocytes are round, which maximizes the cell volume for energy storage, while osteoblasts are well spread and flat for bone remodeling and mineral deposition. While differentiation of MSCs to a specific cell type dictates its morphology, it is also the situation that morphology regulates the fate of MSCs [12]. Human bone marrow stem cells (hBMSCs) have been shown to preferentially differentiate into adipocytes versus osteoblasts when attached to smaller fibronectin islands segregated by non-adhesive materials when given both adipogenic and osteogenic differentiation factors. In contrast, the hBMSCs on larger fibronectin islands allowing for extensive cell spreading differentiate into osteoblasts [13]. This switch of lineage commitment is regulated by RhoA activity through the RhoA kinase (ROCK) pathway. In fact, over-expression of RhoA in hBMSCs enhanced osteogenesis while over-expression of dominant-negative RhoA reversed this phenotype [13].
Rho GTPases regulate cell migration, adhesion, spreading and polarization [14]. When activated, Rho GTPase promotes incorporation of G-actin (globular actin) into F-actin (filamentous actin) filaments mainly through ROCK and formins. When cytoplasmic G-actin levels decrease, myocardin-related transcription factors (MRTFs) are released from their association with G-actin allowing for their translocation into the nucleus where they co-activate serum response factor (SRF) thereby strongly inducing expression of SRF target genes [15], [16]. These genes include actins such as smooth muscle actin (SMA) as well as several actin-binding proteins, which can affect actin dynamics, cell morphology, growth and differentiation [16].
The MRTF family of transcription factors including myocardin, MRTFA, and MRTFB plays essential roles in development of different tissues but particularly the cardiovascular system [17], [18]. Recent studies have identified MRTFA as a negative regulator of adipogenesis through its ability to affect actin dynamics [19], [20]. Additionally, we showed that MRTFA is also a regulator of beige fat formation and that MRTFA−/− mice are protected from diet induced obesity and insulin resistance [20]. In the present study, we used MRTFA deficient (MRTFA−/−) mice to demonstrate that the actin-MRTFA/SRF circuit plays an essential role in controlling BMSC fate decisions and skeletal homeostasis.
2. Results
2.1. MRTFA KO mice exhibit lower body weight, shorter femurs and tibiae, and have significantly lower bone mass
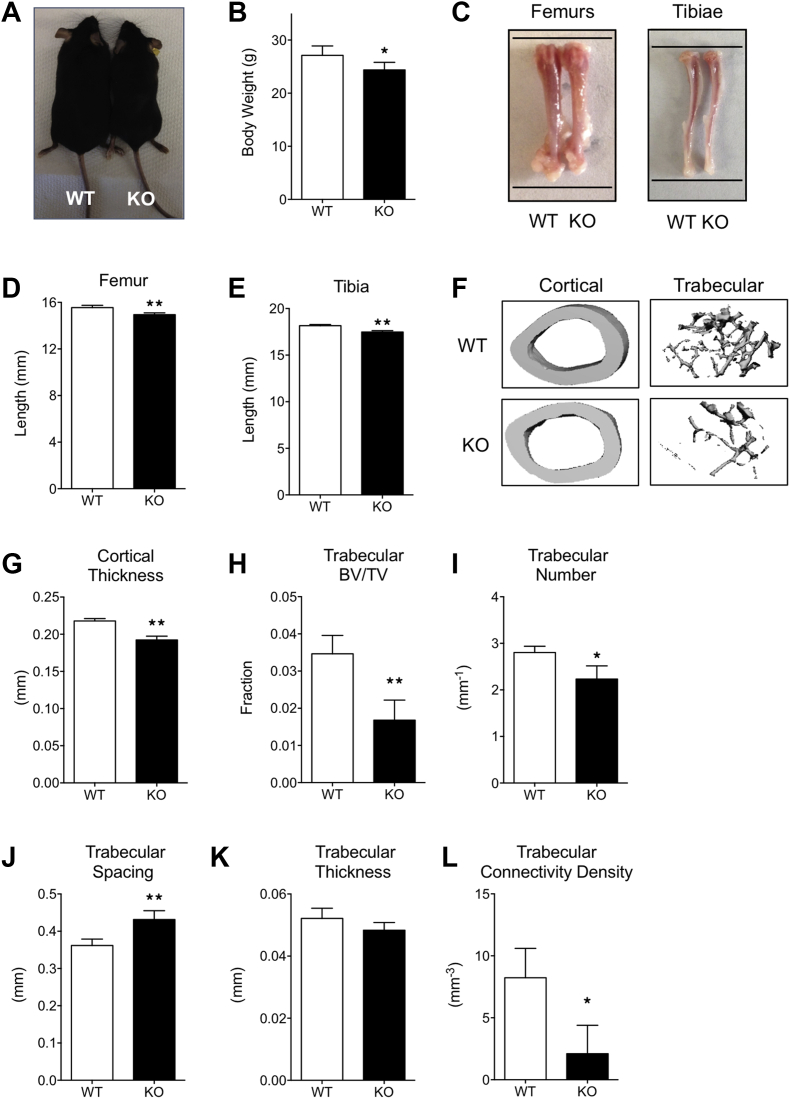
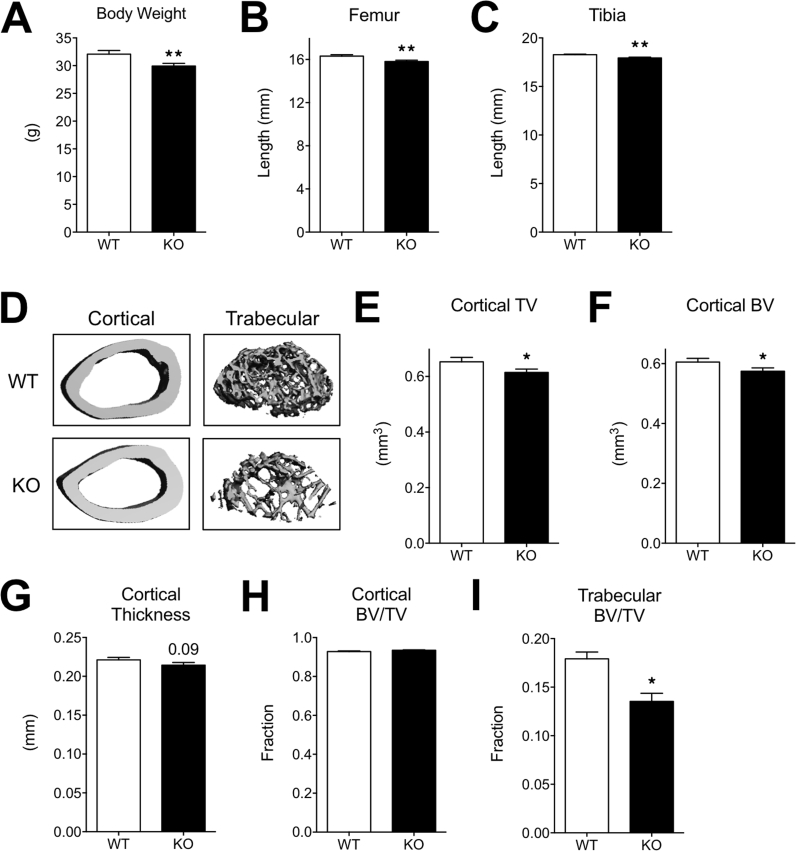
Six-week-old MRTFA KO mice are shorter and weigh less than WT littermates (Figure 1A and B and Supplementary Figure 1). This observation suggested that there might be a defect in skeletal development. To investigate a possible role of MRTFA in bone formation, lengths of the femurs and tibiae were measured with calipers. The KO mice have significantly shorter femurs and tibiae (Figure 1C,D, E and Supplementary Figure 1) when compared to WT controls. Micro-computed tomography (μCT) was performed on femurs of 24-week-old female mice (n = 7 in each group) to assess bone growth and development. μCT of the mid-diaphyseal region of femurs showed that cortical thickness of femurs from MRTFA KO mice is significantly thinner than WT controls (Figure 1F,G); however, cortical bone volume fraction (BV/TV) did not differ significantly (Table 1). μCT of the trabecular regions of proximal femurs showed a marked reduction in trabecular bone mass in the MRTFA KO mice as compared to controls (Figure 1F). Trabecular bone volume fraction is about 50% lower in KO mice (Figure 1H). Bone volume of the trabecular region is also significantly reduced, although the total tissue volume did not appear to differ (Table 1). Furthermore, average trabecular number (Figure 1I) and trabecular bone connectivity density (Figure 1L) are both lower in MRTFA deficient mice, and trabecular spacing (Figure 1J) is larger. There was no significant difference in trabecular thickness (Figure 1K), which is likely due to high bone turnover rate in rodents. Taken together, these changes in bone microstructural parameters suggest an osteopenic phenotype in female MRTFA KO mice (Table 1). Male MRTFA KO mice have a milder but similar phenotype (Supplementary Table 1).
Figure 1.
MRTFA KO mice have smaller body weight, reduced bone mass with shorter femurs and tibiae. WT (n = 7) and MRTFA KO (n = 7) female mice (24 weeks old) were euthanized and then weighed. The image of a matched pair of WT and MRTFA KO mice is shown in (A). The femurs and tibiae of the mice were then dissected (images shown in C) and the bone lengths were measured with a caliper. The upper and lower black lines in C mark the length of the WT mouse femurs to show the differences. Whole body weights (B), femur lengths (D), and tibia lengths (E) of the mice are shown in bar graphs. Representative 3D images of cortical and trabecular region of WT and MRTFA KO mice femurs generated by μCT analysis are shown in F. Cortical thickness of WT and MRTFA KO femurs are shown in bar graphs (G). Trabecular bone volume fraction (H), trabecular number (I), trabecular thickness (J), trabecular separation (K), and connectivity density (L) are shown in bar graphs. Data are expressed as mean ± SEM. *p < 0.05; **p < 0.01.
Table 1.
MRTFA KO female mice showed an osteopenic phenotype as compared to the WT mice. Micro-CT analysis of bone parameters in cortical diaphysis and trabecular region of WT and MRTFA KO mice femurs are shown. 14 female mice (WT n = 7, KO n = 7) at the age of 24 weeks were analyzed for this study. Whole body weight, length of femurs and tibiae of WT and MRTFA KO mice are also shown (*p < 0.05, **p < 0.01).
| WT (n = 7) | KO (n = 7) | p-Value | |
|---|---|---|---|
| Body weight (g)* | 28.2 ± 3.2 | 24.7 ± 3.4 | 0.04 |
| Femur length (mm)** | 15.6 ± 0.5 | 15.0 ± 0.5 | 0.006 |
| Tibia length (mm)** | 18.2 ± 0.4 | 17.5 ± 0.4 | 0.0003 |
| Cortical total volume (TV) (mm³)** | 0.50 ± 0.04 | 0.44 ± 0.05 | 0.01 |
| Cortical bone volume (BV) (mm³)** | 0.48 ± 0.03 | 0.43 ± 0.05 | 0.01 |
| Cortical BV/TV (1) | 0.98 ± 0.009 | 0.98 ± 0.001 | 0.41 |
| Cortical structure model index (1)** | 0.70 ± 0.03 | 0.61 ± 0.05 | 0.0002 |
| Cortical thickness (mm)** | 0.22 ± 0.009 | 0.19 ± 0.013 | 0.00001 |
| Cortical mean/density of TV (apparent) (mgHA/ccm)** | 1412 ± 9 | 1384 ± 20 | 0.0003 |
| Cortical mean/density of BV (material) (mg HA/ccm)** | 1531 ± 9 | 1513 ± 17 | 0.004 |
| Trabecular total volume (TV) (mm³) | 1.42 ± 0.1 | 1.37 ± 0.1 | 0.18 |
| Trabecular bone volume (BV) (mm³)** | 0.05 ± 0.1 | 0.02 ± 0.01 | 0.004 |
| Trabecular BV/TV (1)** | 0.04 ± 0.01 | 0.02 ± 0.007 | 0.004 |
| Connectivity density (1/mm³)* | 8.2 ± 6.3 | 2.1 ± 1.9 | 0.01 |
| Structure model index (1) | 3.72 ± 0.2 | 3.84 ± 0.2 | 0.14 |
| Trabecular number (1/mm)* | 2.80 ± 0.4 | 2.23 ± 0.6 | 0.02 |
| Trabecular thickness (mm) | 0.052 ± 0.01 | 0.048 ± 0.01 | 0.25 |
| Trabecular separation (mm)** | 0.36 ± 0.05 | 0.43 ± 0.04 | 0.006 |
| Trabecular mean/density of TV (apparent) (mg HA/ccm)** | 123 ± 16 | 98 ± 18 | 0.009 |
| Trabecular mean/density of BV (material) (mg HA/ccm) | 1032 ± 31 | 1017 ± 76 | 0.32 |
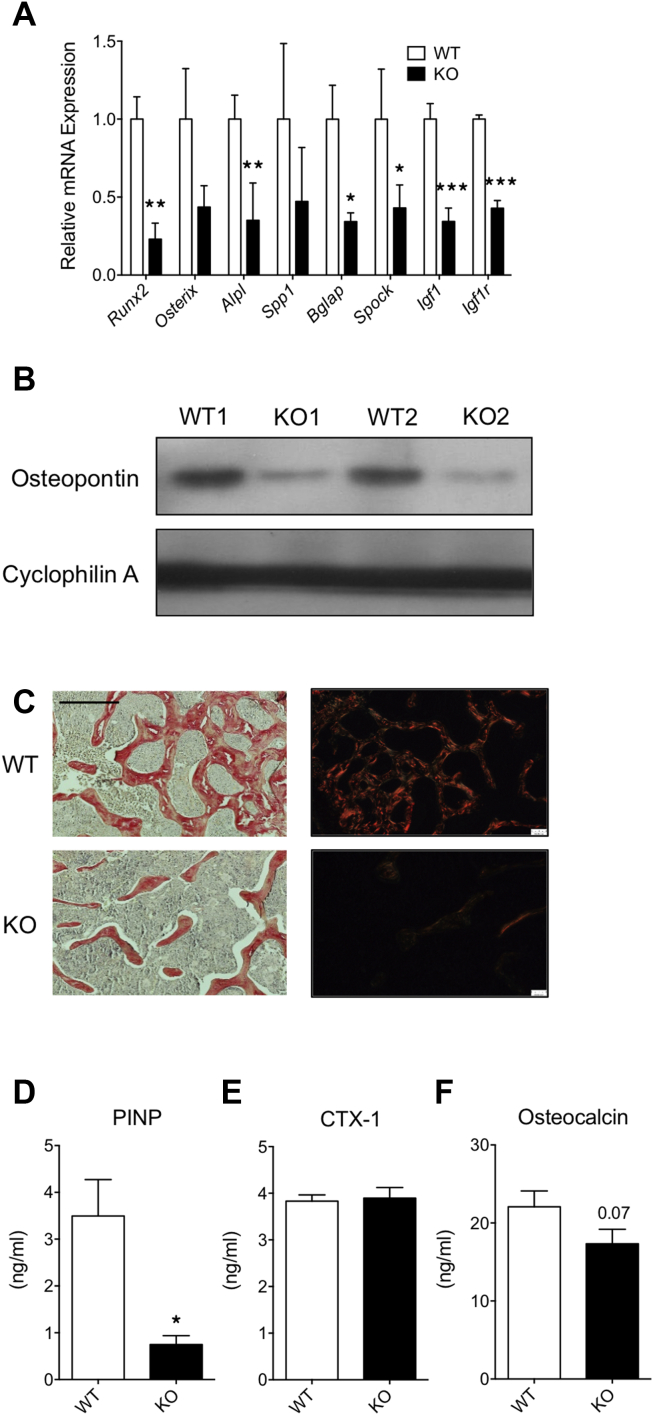
2.2. Osteogenic gene expression is compromised in MRTFA KO femurs
To study whether loss of bone mass in MRTFA KO mice is due to inadequate osteogenesis, we compared expression of osteogenic genes in WT and KO femurs. Messenger RNAs coding for several osteogenic proteins including Runx2, Osterix, Alpl, osteopontin (Spp1), osteocalcin (Bgal), osteonectin (Spock), Igf1, and Igfr were expressed at consistently lower levels in MRTFA KO than in WT femurs (Figure 2A). The western blot displayed in Figure 2B demonstrates a dramatic reduction in osteopontin protein expression in KO femurs. To analyze deposition of collagens, the trabecular region of proximal femurs were sectioned and stained with picrosirius red and visualized under normal and polarized light (red color represents thick fibers whereas green/yellow color represents thinner fibers). MRTFA-KO femurs have lower amounts of type I and III collagens within thinner fibers compared to WT femurs (Figure 2C). Procollagen intact N-terminal peptide (PINP) is a cleaved by-product produced during type I collagen synthesis, and it is positively correlated with osteoblast activity. Circulating levels of PINP are significantly lower in MRTFA KO mice (Figure 2D). As a systemic osteogenic marker, it indicates that osteoblast activity in other types of bone such as vertebrae and tibiae might also be compromised in KO mice. C-terminal telopeptide (CTX-1) is a by-product of enzymatic cleavage of type 1 collagen and serves as a marker for bone breakdown. There is no significant difference in CTX-1 levels between WT and MRTFA KO mice (Figure 2E), which suggests the osteopenic phenotype in MRTFA KO mice is not due to excessive bone resorption by osteoclasts. Osteocalcin is an osteoblast-secreted protein that can act on adipose tissue and pancreas to enhance insulin action [21]. The serum levels of osteocalcin are trending lower in MRTFA KO mice (Figure 2F). Taken together these three parameters show that the observed osteopenia is likely due to a decrease in osteoblasts activity.
Figure 2.
MRTFA KO mice have lower osteoblastic gene expression and lower serum osteoblast activity markers. Femurs of WT (n = 3) and MRTFA KO (n = 3) female mice were pulverized in liquid nitrogen for RNA extraction and then RNA was reversed transcribed for RT-PCR analysis for measuring osteogenic gene expression (A). p-values were determined by paired t-test analysis of KO and control littermates. Western blot of Osteopontin in WT and MRTFA KO mice femurs is shown in B. Decalcified histological sections of proximal femur in 12 weeks old littermates were stained with picrosirius red to visualize bone structures and Type I and II collagen in WT (n = 3) and MRTFA KO (n = 3) mice femurs in C. The left panels are images captured under light microscope with 10× magnification and the right panels are captured under polarized light with 10× magnification (Scale bar = 0.3 mm). Blood samples were collected from hearts of matched WT (n = 6) and MRTFA KO (n = 7) mice. ELISA assays were performed in diluted serum to measure the levels of procollagen I N-terminal propeptide (PINP) (D), cross linked C-telopeptide of Type I collagen (CTX-1) (E), and osteocalcin (F). Data are expressed as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
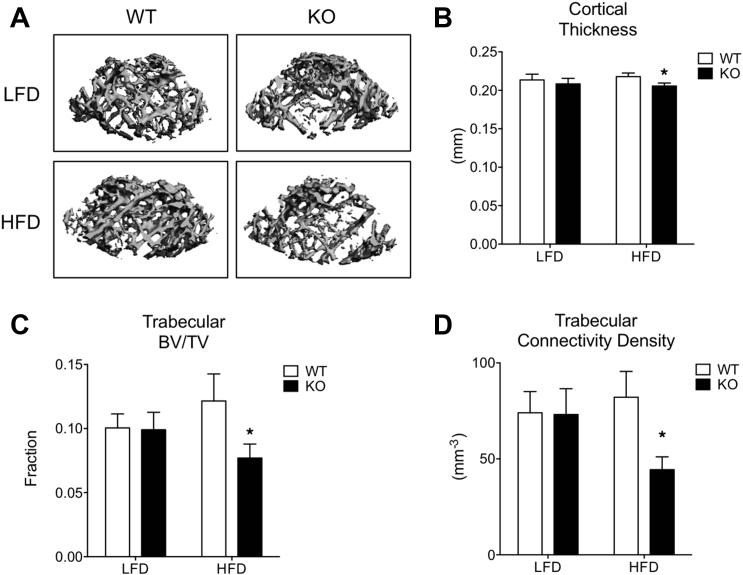
2.3. MRTFA deficiency potentiated bone loss in younger mice on a high fat diet
In contrast to the osteopenic phenotype observed in 24-week-old female MRTFA KO mice (Figure 1), the bone phenotypes of younger, 12-week-old KO and WT male mice fed a low fat diet for 6 weeks was indistinguishable (Figure 3A,B). These data suggest that osteopenia due to MRTFA deficiency manifests itself with age. Recent studies have demonstrated that a high fat diet (HFD) can adversely affect bone health [22], [23]. We questioned, therefore, whether a HFD could affect the rate at which osteopenia develops in MRTFA KO mice. The data in Figure 3B–D shows that cortical thickness, bone volume fraction, and connectivity density are significantly lower in femurs of MRTFA KO male mice on a HFD but not if they were fed a low fat diet (LFD). There is no change in bone health parameters in WT mice whether they were fed a LFD or HFD (Figure 3). This observation is consistent with recent studies by Rosen and collaborators showing that a HFD has no effect on bone mass in normal male C57BL/6 mice [24]. Together, these results demonstrate that MRTFA KO mice develop an osteopenic phenotype at a younger age when challenged with an obesogenic diet (Supplementary Table 1).
Figure 3.
MRTFA KO mice develop an osteopenic phenotype at an earlier age when challenged with a high fat diet (HFD). Representative 3D images of cortical region (mid-shaft of femurs) and trabecular region of femurs of MRTFA KO or WT male mice on LFD (10% fat) or HFD (60% fat) generated by μCT analysis are shown in A. Twenty five mice (WT LFD n = 6, KO LFD n = 8, WT HFD n = 5, KO HFD n = 6) at the age of 12 weeks were analyzed for this study. The cortical thickness (B), connectivity density (C), and trabecular volume/total volume ratio (D). Data are expressed as mean ± SEM. *p < 0.05.
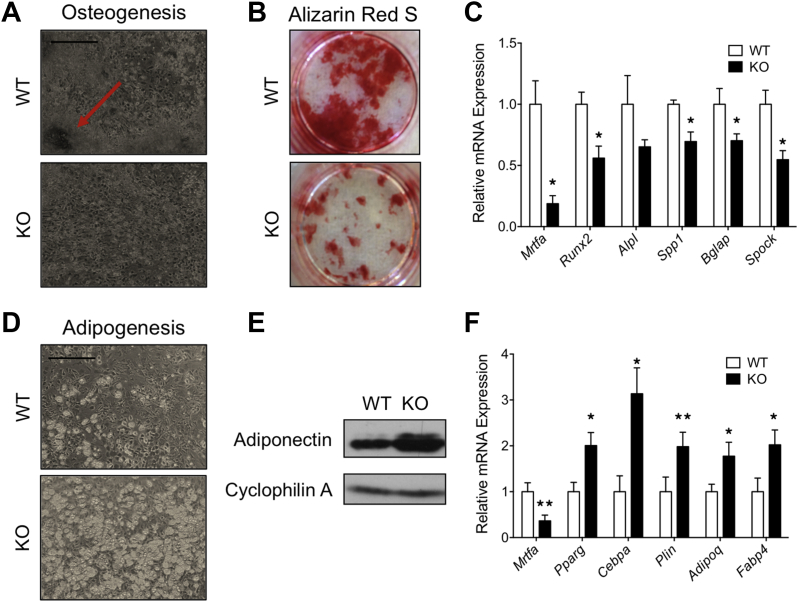
2.4. MRTFA deficiency reduces osteogenesis but enhances adipogenesis in bone marrow stromal cells
To determine whether the defect in bone development in MRTFA KO mice occurs in a cell autonomous manner, primary stromal cells isolated from the bone marrow of WT and KO mice were compared for their osteogenic potential in culture. MRTFA KO stromal cells showed compromised osteogenic differentiation with decreased bone nodule formation, as marked by the arrow in WT cells (Figure 4A), and less mineral deposition (stained with Alizarin Red S) (Figure 4B). The decreased expression of osteogenic genes in MRTFA KO cells further validated these observations (Figure 4C). On the other hand, the MRTFA-KO stromal cells produced more lipid-laden adipocytes when treated with adipogenic cocktail for 10 days (Figure 4D). The adiponectin protein expression and relative mRNA expression of adipogenic genes, including Pparg, Cebpa, AdipoQ (adiponectin), and Fabp4 was also higher in MRTFA KO differentiated adipocytes (Figure 4E,F). MRTFA transcripts are detected in the KO mice, but this is due to aberrant splicing of the mutant MRTFA gene generating an in frame fusion transcript. The fusion protein resulting from the transcript is non-functional since it lacks the SRF binding region [25]. In summary, these results suggest that the differentiation of stromal cells is shifted to adipogenesis at the expense of osteogenesis when MRTFA is deleted.
Figure 4.
Bone marrow stromal cells from MRTFA KO mice showed enhanced adipogenic but reduced osteoblastic differentiation. 12 weeks old WT and MRTFA KO mice were euthanized, and their femur and tibia bone marrow cavity contents were flushed out with αMEM growth media containing 10% fetal bovine serum (FBS). Cells were cultured with osteogenic inducers for 21 days until fully differentiated (A–C). Light microscopy images of osteogeneic cultures at day 21 (scale bar = 0.3 mm) (A). Osteogenic gene mRNA expression of WT and KO osteogeneic cultures (B). Alizarin Red S staining of WT and KO cells visualizing mineral deposition during osteogenesis. Bone marrow derived cells were cultured with adipogenic inducers for 10 days (D–F). Light microscopy images of adipogenic cultures at day 10 (scale bar = 0.3 mm) (E). Cells were then harvested for western blot and RT-PCR analysis. Adiponectin protein expression and mRNA expression of adipogenic makers (E–F) of WT and KO cells. Data are expressed as mean ± SEM (n = 3). *p < 0.05; **p < 0.01.
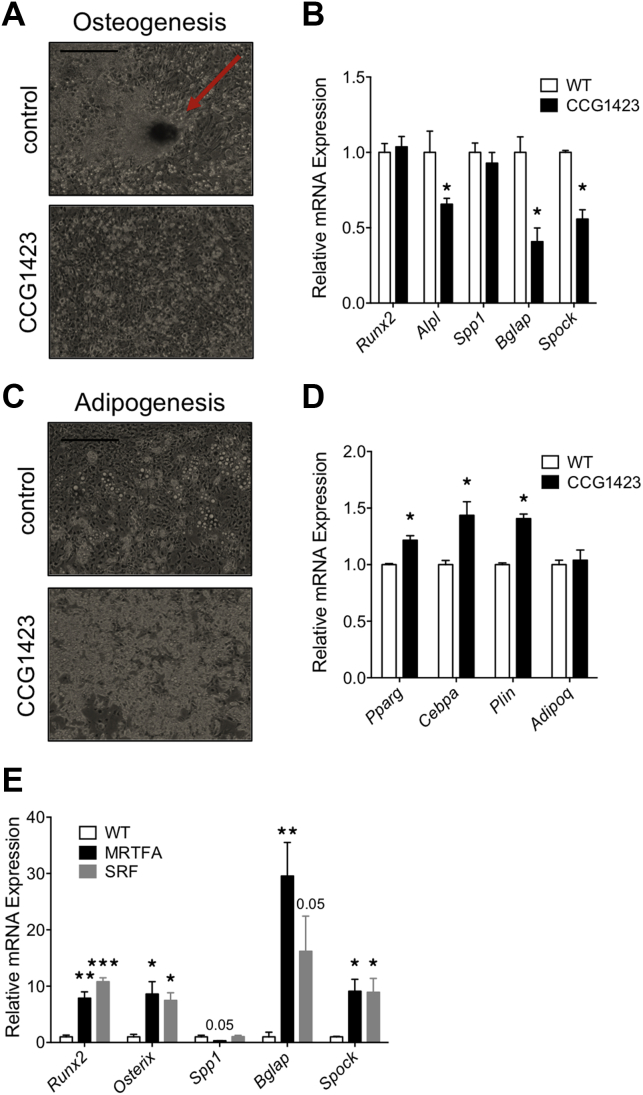
2.5. Treatment of WT bone marrow stromal cells with the MRTFA/SRF inhibitor CCG1423 mimics the effects of knocking out MRTFA
To understand the importance of MRTFA/SRF in regulating the fate of bone marrow stromal cells, we treated WT cells with CCG1423, a Rho/SRF pathway inhibitor [26], [27]. Exposure of cells to CCG1423 during the 21-day period of osteogenesis reduced bone nodule formation compared to formation of multiple nodules (marked by arrow) in control cells (Figure 5A) and reduced the expression of Alpl, osteocalcin (Bgal), and osteonectin (Spock), mRNAs (Figure 5B). These data suggest that MRTFA/SRF activity facilitates osteogenesis. In the case of adipogenesis, stromal cells treated with CCG1423 for 2 days prior to adipogenic induction accumulated more lipid (Figure 5C) and expressed higher levels of the adipogenic genes Pparg, Cepba, Plin, and Fabp4 than untreated cells (Figure 5D). These data are consistent with earlier studies [20] showing that suppression of MRTFA/SRF activity commits mesenchymal progenitors to the adipogenic lineage, and the resulting preadipocytes are more responsive to the adipogenic inducers than uncommitted cells.
Figure 5.
Treatment of WT bone marrow derived MSCs with the SRF inhibitor CCG1423 mimicked the effects of knocking out MRTFA. DMSO (vehicle) or 1 mM SRF inhibitor CCG1423 was added to the WT BMSCs when cells reached confluence, together with adipogenic or osteoblastic inducers as described in Experimental Procedures. Representative light microscopy images of osteogenesis at day 21 (scale bar = 0.3 mm) (A). mRNA expression of osteogenic markers measured by RT-PCR (B). Similarly, representative light microscopy images of adipogenesis were captured at day 10 (scale bar = 0.3 mm) (C). Markers of adipogenesis were measured by RT-PCR (D). Post-confluent C3H10T1/2 cell lines over-expressing MRTFA, SRF, or control vector were treated with osteogenic inducing media for 21 days. The RNA was then extracted from these cells for RT-PCR experiments in (E). Data are expressed as mean ± SEM (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001.
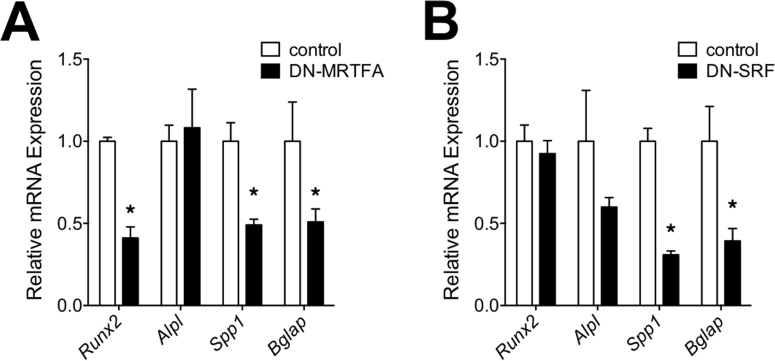
The possibility that some of the effects of CCG1423 might be independent of MRTFA or SRF caused us to further investigate these effects in C3H10T1/2 cell lines expressing MRTFA or SRF or their dominant negative forms (DN-MRTFA or DN-SRF). The qPCR results showed that over-expression of MRTFA or SRF enhanced expression of osteogenic genes (Figure 5 E) and DN-MRTFA and DN-SRF had the opposite effects (Supplementary Figure 2A,B). On the other hand, over-expression of MRTFA or SRF inhibited adipogenesis and adipogenic gene expression as previously shown by us [20].
2.6. MRTFA regulates smooth muscle actin and TAZ/YAP target gene expression in BMSCs
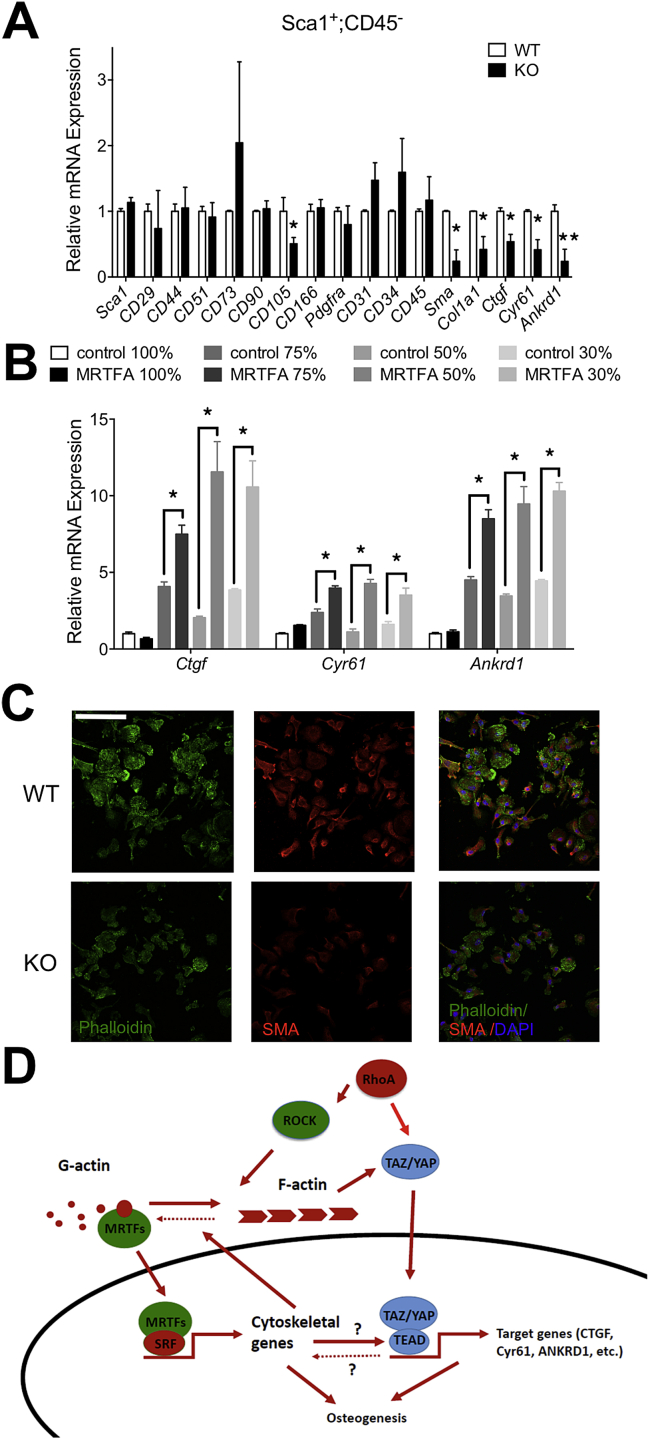
Until now, we have investigated MRTFA activity in total bone marrow stromal cells following plating on tissue culture surfaces that selects for non-hematopoetic cells. To determine whether MRTFA regulates the fate of skeletal progenitors, we used magnetic-activated cell sorting (MACS) to enrich for Sca1+:CD45− cells from the stromal vascular compartment of WT and MRTFA KO femoral marrow. Scal1+:CD45− cells from MRTFA KO mice exhibited lower collagen type 1 alpha 1 (Col1a1) mRNA expression than WT cells (Figure 6A). They also had reduced expression of SRF target genes including Sma and Ctgf (Figure 6A). Studies by others have identified TAZ as a principal regulator of cell fate driving osteogenesis versus adipogenesis in MSCs [28]. TAZ transcriptional activity, like MRTFA, is regulated by the morphology of cells and the SMA cytoskeleton [29], [30]. Therefore, we questioned whether MRTFA is a determinant of TAZ/YAP target gene expression. Figure 6 shows significantly lower expression of TAZ target genes in MRTFA KO cells than WT cells (Figure 6A). Similarly, in C3H10T1/2 cell lines over-expressing MRTFA, TAZ/YAP target genes Ctgf, Cyr61, and Ankrd1 are expressed at higher levels compared to the controls when cells are sub-confluent (Figure 6B). Interestingly, MRTFA is incapable of inducing TAZ/YAP targets when cells reach confluence (Figure 6B). Additionally, MRTFA facilitates extensive spreading of 10T1/2 cells on soft surfaces consistent with it driving a morphology that favors osteogenesis (Supplementary Figure 3). Immunofluorescence staining for F-actin with phalloidin (green) or an antibody against SMA (red) in Sca1+:CD45− cells showed a significant decrease in SMA levels in the MRTFA KO cells, with only a minor decrease in F-actin staining (Figure 6C). These data show that MRTFA regulates SMA expression and TAZ transcriptional activity in Sca1+:CD45− cells.
Figure 6.
MRTFA regulates bone marrow MSCs fate commitment via regulating TAZ/YAP transcriptional activity. WT and MRTFA KO bone marrow stromal cells were sorted with MACS magnetic beads to amplify the mesenchymal stem cells population. Sca1 positive and CD45 negative population of bone marrow stromal cells were isolated and harvested for RT-PCR to measure stem cell markers, osteogenic genes, and TAZ/YAP target genes (A). p-values were determined by paired t-test analysis of KO and control littermates. C3H10T1/2 cell lines over-expressing MRTFA and vector controls were seeded at different densities (100%, 75%, 50% and 30% confluence) and harvested after attaching for RT-PCR to measure the mRNA levels of TAZ/YAP target genes (B). WT and MRTFA KO Scal1+ CD45-bone marrow stem cells were plated and stained with phallodin, SMA antibody and DAPI (C) (scale Bar = 0.04 mm). A schematic model of how MRTFA inhibits adipogenesis and enhances osteogenesis is shown in (D). MRTFA is regulated by actin cytoskeleton dynamics through RhoA-Rock signaling pathway. Data are expressed as mean ± SEM (n = 3). *p < 0.05; **p < 0.01.
3. Discussion
Osteoporosis is manifested by excessive loss of bone mass and increased tendency for pathological fractures. Previous studies showed osteoporotic bones often have increased bone marrow adiposity in the trabecular region [5], [31], [32]. Since adipocytes and osteoblasts originate from common progenitors, disruption of fate commitment between these two lineages can lead to decreased osteogenesis, enhanced adipogenesis, and, subsequently, osteoporosis [32]. It is very important to understand the underlying mechanisms regulating MSC commitment for development of therapeutic strategies. In this study, we demonstrate that MRTFA KO mice exhibit a significant osteopenic phenotype with decreased bone mass in trabecular femurs and thinning of cortical femurs. This reduction in bone size and mass likely contributes to lower body weight of MRTFA KO mice, but we have additionally shown that these mice also have a reduced amount of adipose tissue [20]. The data in Figure 2 suggest that the bone defects observed in KO mice result from impairment of osteoblast development and function, with limited contribution from osteoclasts and bone resorption. It is possible that a decrease in chondrogenesis, as suggested by the lower levels of Runx2 expression, could also be contributing to the defects in longitudinal growth of the limbs in KO mice [33].
To identify possible mechanisms responsible for the osteopenic phenotype, we studied the differentiation of primary bone marrow stromal cells in culture. Genetic deletion of MRTFA enhanced the adipogenic potential of these cells at the expense of osteogenesis. These data help to rule out the possibility that the osteopenia is caused by systemic changes in hormones that regulate bone growth and regeneration such as IGF1. This is particularly relevant since Chen et al., 2012, recently demonstrated that SRF regulates bone formation via IGF-1 and Runx2 signaling [34]. It is important to mention, however, that the changes in circulating hormones, which occur in the absence of MRTFA, are in a direction favoring bone growth. Specifically, leptin has recently been shown to inhibit osteogenesis and promote adipogenesis [35], and we showed leptin is lower in MRTFA KO mice [20]. In the case of adiponectin and insulin, they are reported to be pro-osteogenic [36], [37], and their levels are higher in KO mice compared to WT littermates [20].
To provide even greater support for an autonomous process regulating the bone phenotype, we isolated by MACS Sca1+:CD45− progenitors from the bone marrow of WT and KO mice and analyzed their expression of select genes. The data show that MRTFA is required for optimum expression of SMA and select TAZ/YAP target genes in the progenitors. TAZ has previously been shown to drive MSC commitment toward the osteogenic lineage [28]. Hinz and colleagues have recently suggested that SMA-mediated contraction plays a critical role in mechanically regulating MSC fate by controlling TAZ/YAP activation [30]. Taken together, the data above support an autonomous role for MRTFA in regulating the fate of BMSCs by ensuring the formation of a SMA-cytoskeleton to support TAZ/YAP activity (Figure 6D). We acknowledge however that some identified mechanism could also be contributing to the phenotype of the MRTFA−/− mice, particularly since the deficiency has such a major effect on overall metabolic homeostasis.
HFD appears to exacerbate the osteopenia caused by the absence of MRTFA. Possible reasons for this include activation of PPARγ activity in the pro-adipogenic BMSCs due to the elevated circulating levels of lipids that may provide ligands for this nuclear receptor. It is well documented that individuals taking Avandia, a synthetic PPARγ ligand for their insulin resistance or diabetes often develop osteoporosis [6], [7]. Other possibilities are that HFD alters the composition of the stromal vascular compartment (SVC) in which BMSCs reside due to deposition of marrow fat that accompanies whole body obesity [24], [38]. It is important to mention, however, that our earlier studies have shown that the KO mice are protected, to some extent, from obesity and insulin resistance [20]. Consequently, the HFD might have more drastic effects on bone health if MRTFA deficiency is localized to bone progenitors. A remodeling of the marrow SVC could alter the fate of the BMSCs by affecting how they respond to various osteogenic factors. Bone morphogenetic proteins (BMPs), for instance, are members of the TGFβ superfamily and play a major role in skeletal development and bone formation [39], [40]. BMP signaling also regulates adipocyte formation in vivo as well as in vitro [41], [42], [43]. The fact that adipogenesis and osteogenesis are mutually exclusive raises an interesting question of how BMPs can induce both processes. We have shown that under adipogenic conditions BMPs suppress the ROCK-MRTFA pathway [20]. In other situations it activates the pathway [44], this will likely be the case for osteogenesis. We propose, therefore, that the bone marrow SVC can influence the fate of MSCs by specifying whether BMPs suppress or activate MRTFA/SRF transcriptional activity.
4. Conclusion
This study identifies MRTFA as a novel regulator of skeletal homeostasis by controlling the balance between adipogenic and osteogenic differentiation in bone marrow stem cells, and thus holds promise as a potential target for therapeutic intervention for osteoporosis.
5. Experimental procedures
5.1. Animals
All animal experiments were conducted following Boston University Laboratory Animal Science Center experimental protocols approved by Institutional Animal Care and Use Committee. MRTFA+/− and wild type mice (mixed C57BL/6J 129 genetic background) were originally created by Dr. Eric Olson, UT Southwestern Medical Center [25] and were given to us as a kind gift. MRTFA global KO mice are viable and fertile except for mammary gland developmental defects in female mice [25]. Age-, strain- and sex- matched WT and MRTFA KO mice were used in all experiments. Male mice were between 15 and 17 weeks old, and female mice were between 23 and 24 weeks for the Micro-CT studies. For diet studies, 4–6 weeks-old male WT and MRTFA KO mice were fed a diet with 10% kcal% fat (low fat diet, Research Diets Inc, D12450B) or diet with 60% kcal% fat (high fat diet, Research Diets Inc, D12492) for 6 weeks.
5.2. Cell culture and differentiation induction conditions
C3H/10T1/2 cells were obtained from American Type Culture Collection. Bone marrow stromal cells were isolated from WT and MRTFA KO mice. For adipogenic induction, at confluence, cells were treated with DMEM supplemented with 10% FBS, 5 μM dexamethasone, 0.5 mM isobutylmethylxanthine, 860 nM insulin, 1 nM 3, 3, 5-triiodo-l-thyronine (T3), and 125 μM indomethacin. 48 h post induction, the cells were maintained in medium containing 10% FBS, 860 nM insulin, and 1 nM T3 for another 8 days. For osteogenic induction, cells after reaching reach confluence were treated with DMEM supplemented with 10% FBS, 5 nM dexamethasone, 70 ng/ml l-ascorbic acid, and 8 mM β-glycerophosphate, disodium salt for 21 days. For studies of SRF inhibition, sub-confluent cells were exposed to 1 mM CCG1423 (Cayman Chemicals) for 3 days prior to induction of adipogenesis or for the entire 21-days of osteogenesis.
5.3. Plasmids and viruses
MRTFA, DN-MRTFA, SRF, and DN-SRF cDNAs [45], [46] were sub-cloned into pMSCV retroviral vector (Clontech). For retrovirus production, EcoPack (Clontech) packaging cells were transfected at 70% confluence using Lipofectamine LTX and Plus Reagent (Invitrogen) and 8 μg of pMSCV vectors. After 48 h, viral supernatant was harvested and filtered. C3H/10T1/2 cells were incubated overnight with viral supernatant and supplemented with 10 μg/mL polybrene. Cell lines were established by selection with 350 μg/mL hygromycin.
5.4. Western blot analysis
Total cellular protein was extracted from primary bone marrow stromal cells or C3H10T1/2 cells and subjected to western blot analysis as previously described [47]. Total protein was also extracted from mice femur as previously described [48]. Antibodies against the following proteins were obtained from indicated vendors: MRTFA, Osteopontin, C/EBPα (Santa Cruz, CA), adiponectin (Pierce, Rockford, IL) and cyclophilin A (Cyc A) (Millipore, Billerica, MA).
5.5. Primary bone marrow stem cells isolation, sorting and immunofluorescence
Primary bone marrow stromal cells were used as an ex-vivo model to further study the role of MRTFA in the differentiation potential of these cells to adipose versus osteoblastic lineage. WT and MRTFA KO mice were euthanized and their femur and tibia bone marrow cavity contents were flushed out with α-MEM growth media containing 10% FBS. Cells were re-suspended, counted, and then plated without being disturbed until day 4 of culture when half of the media was replaced. On day 6 of culture, the cells reached confluence and were treated with adipogenic or osteogenic induction media. The sorting of the bone marrow stromal cells was conducted with MACS magnetic microbeads sorting kit (Scal1 and CD45) (purchased from Miltenyi Biotec). All the sorting experiments were conducted according to manufacturer's instructions. The sorted cells were plated on plastic surface for 4–6 days and then stained with SMA antibody overnight and counter stained with DAPI and phalloidin during secondary antibody incubation.
5.6. Specimen harvesting
MRTFA-KO and WT femurs were snap-frozen in liquid nitrogen at the time of collection and then pulverized in liquid nitrogen using a mortar and pestle in either 600 ul RNA extraction RLT buffer (RNeasy Mini Kit) or 600 ul RIPA buffer [48]. The samples were then homogenized and lysates were centrifuged. Supernatant was collected from these centrifuged samples for RNA extraction or protein extraction for RT-PCR or Western Blot analysis, respectively.
5.7. Histology
Mice femurs and tibiae were fixed in 4% formaldehyde for 1–5 days depending on the age of the mice and then decalcified in 14% w/v EDTA dissolved in water for 5 days. These samples were then sent to Boston University's Experimental Pathology Laboratory Services Core for embedding in paraffin and sectioning. Sections of the femurs and tibiae were then stained with Picrosirius Red (Polyscientifics Inc.) according to manufacturer's instructions to visualize Type I and III collagen.
5.8. Quantitative micro computated tomography (Micro-CT)
Mice femurs were fixed in 4% formaldehyde for 1–5 days and then stored in PBS at 4 °C. Scans were performed using a Scanco micro-CT 40 system (Scanco Medical, Basserdorf, Switzerland) located in Orthopedic and Developmental Biomechanics Laboratory at Boston University. These scans were performed using 12 micron voxel size resolution with 200 ms integration time, under conditions of 55 E (KVp) and 145 I (μA). Transverse images scanned by the micro-CT were then traced manually with a computer program and stacked to render a 3-D image of the cortical and trabecular regions of the femurs extracted from WT and MRTFA-KO mice. The mid-diaphyseal region scanned was in the femur mid-point, whereas the trabecular region scanned was right above the growth plate for each femur. The lengths (number of μCT slices) of cortical and trabecular bone scanned are proportional to the lengths of the bone to ensure the same regions are compared in WT and MRTFA KO mice. Image analysis was conducted as previously described [49].
5.9. Quantitative RT-PCR
Total RNA was isolated from cells or tissues using TRIzol reagent (Life Technologies), following manufacturers' protocol. Total RNA was isolated from mice femurs with RNA extraction buffer RLT buffer (RNeasy Mini Kit) according to manufacturer's instruction. Reverse Transcriptase (RT) reactions were performed using 1 μg RNA and a high-capacity cDNA RT Kit (Applied Biosystems) according to the manufacturer's instructions. Analysis of gene expression was performed using Maxima SYBR Green qPCR Master Mix (Fermentas Life Sciences) in the ABI Prism 7300 sequence detector as previously described [50]. Primer sequences used will be provided on request.
5.10. Statistical analysis
Unpaired 2-tail Student t test was used to evaluate statistical significance and p ≤ 0.05 being considered significant. All values are presented as means ± standard deviation (SD). All experiments were repeated at least three times.
Author contribution
S.R.F. and H.B. conceived of the project, designed all experiments and wrote manuscript. H.B. performed all experiments from Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6. J.Z.L. revised the manuscript and provided tissues and serum for Figure 2 and Figure 6. C.L. assisted with the transgenic mice genotyping and colony management.
Acknowledgements
This work was supported by National Institutes of Health grants DK51586, DK098830 and DK102199. JZL was supported from a National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health training grant T32DK007201. We are grateful to Dr. Elise Morgan and Zach Webster for providing technical support in designing studies and interpreting results for Micro-CT studies. We thank Dr. Shannon H. Carroll for assisting with MRTFA KO mice bone phenotyping. We appreciate the advice of Drs. Matthew D. Layne and Barbara D. Smith in generating cell lines and management of the mice colony. We are grateful to Eric Olson, UT Southwestern Medical Center, TX, for providing the MRTFA+/− mice. We thank Dr. Taylor Evan English for editing the manuscript.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.08.012.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Supplemental Table 1. MRTFA KO male mice have significant trabecular bone loss as compared to the WT mice. Micro-CT analysis of bone parameters in cortical diaphysis and trabecular region of WT and MRTFA KO mice femurs are shown in this table. 14 male mice (WT n = 7, KO n = 7) at the age of 17 weeks were analyzed for this study. The whole body weight, length of femurs and tibiae of the mice are also shown in this table (*p < 0.05, **p < 0.01).
Supplemental Table 2. MRTFA KO developed an osteoporotic phenotype at an earlier age when challenged with HFD. Micro-CT analysis of bone parameters in cortical diaphysis and trabecular region of male WT and MRTFA KO mice (on HFD or LFD) femurs are shown in this table. 25 male mice (WT LFD n = 6, KO LFD n = 8, WT HFD n = 5, KO HFD n = 6) at the age of 12 weeks were analyzed for this study. The whole body weight, length of femurs and tibiae of the mice are also shown in this table.
Supplemental Figure 1Male MRTFA KO mice exhibited mild osteopenic phenotype. WT (n = 7) and MRTFA KO (n = 7) male mice (17 weeks old) were euthanized and then weighed. The femurs and tibiae of the mice were then dissected, and the bone lengths were measured with a caliper. Whole body weights (A), femur lengths (B), and tibia lengths (C) of WT and KO mice Transverse images scanned by the micro-CT were traced manually with a computer program and stacked to render 3D images. Representative three-dimensional images of cortical region (mid-shaft of femurs) and trabecular region of femurs generated by μCT analysis (D). The cortical total volume (E), cortical bone volume (F), cortical thickness (G), cortical bone volume/total volume ratio (BV/TV) (H), and trabecular bone volume fraction (BV/TV) (I). Data are expressed as mean ± SEM (n = 3). *p < 0.05; **p < 0.01.
Supplemental Figure 2.
Over-expressing dominant negative variants of MRTFA or SRF in C3H10T1/2 cells inhibited osteogenesis but enhanced adipogenesis. Post-confluent C3H10T1/2 cell lines over-expressing DN-MRTFA, DN-SRF, and controls were treated with osteogenic inducing media for 21 days. The RNA was then extracted from these cells for RT-PCR experiments to measure the fold change of osteoblastic genes in A and B. Data are expressed as mean ± SEM (n = 3). *p < 0.05.
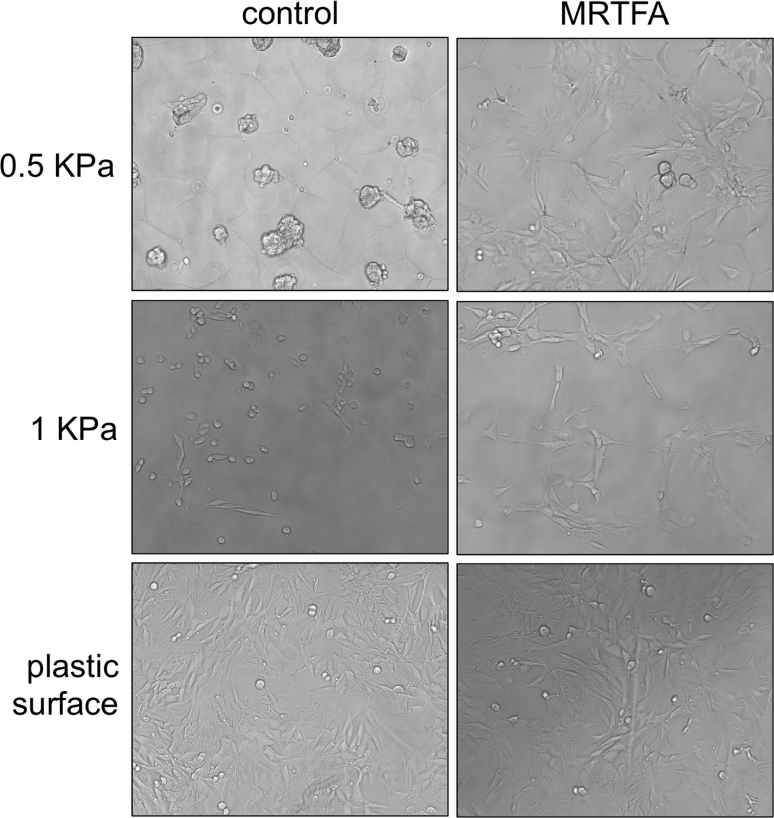
Supplemental Figure 3MRTFA enhances cell spreading on soft surfaces. MRTFA-expressing or control C3HT101/2 cells were cultured on soft hydrogel matrix (0.5 and 1 KPa) and standard tissue culture plastic in 10% FBS supplemented DMEM for 48 h.
References
- 1.Beresford J.N., Bennett J.H., Devlin C., Leboy P.S., Owen M.E. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. Journal of Cell Science. 1992;102(Pt 2):341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 2.Muruganandan S., Roman A.A., Sinal C.J. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: cross talk with the osteoblastogenic program. Cellular and Molecular Life Sciences. 2009;66(2):236–253. doi: 10.1007/s00018-008-8429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takada I., Kouzmenko A.P., Kato S. Molecular switching of osteoblastogenesis versus adipogenesis: implications for targeted therapies. Expert Opinion on Therapeutic Targets. 2009;13(5):593–603. doi: 10.1517/14728220902915310. [DOI] [PubMed] [Google Scholar]
- 4.Gimble J.M., Zvonic S., Floyd Z.E., Kassem M., Nuttall M.E. Playing with bone and fat. Journal of Cellular Biochemistry. 2006;98(2):251–266. doi: 10.1002/jcb.20777. [DOI] [PubMed] [Google Scholar]
- 5.Justesen J., Stenderup K., Ebbesen E.N., Mosekilde L., Steiniche T., Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2(3):165–171. doi: 10.1023/a:1011513223894. [DOI] [PubMed] [Google Scholar]
- 6.Ali A.A., Weinstein R.S., Stewart S.A., Parfitt A.M., Manolagas S.C., Jilka R.L. Rosiglitazone causes bone loss in mice by suppressing osteoblast differentiation and bone formation. Endocrinology. 2005;146(3):1226–1235. doi: 10.1210/en.2004-0735. [DOI] [PubMed] [Google Scholar]
- 7.McDonough A.K., Rosenthal R.S., Cao X., Saag K.G. The effect of thiazolidinediones on BMD and osteoporosis. Nature Clinical Practice, Endocrinology & Metabolism. 2008;4(9):507–513. doi: 10.1038/ncpendmet0920. [DOI] [PubMed] [Google Scholar]
- 8.Duque G. Bone and fat connection in aging bone. Current Opinion in Rheumatology. 2008;20(4):429–434. doi: 10.1097/BOR.0b013e3283025e9c. [DOI] [PubMed] [Google Scholar]
- 9.Karsenty G., Olson E.N. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell. 2016;164(6):1248–1256. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riddle R.C., Clemens T.L. Insulin, osteoblasts, and energy metabolism: why bone counts calories. Journal of Clinical Investigation. 2014;124(4):1465–1467. doi: 10.1172/JCI75554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 12.Ivanovska I.L., Shin J.W., Swift J., Discher D.E. Stem cell mechanobiology: diverse lessons from bone marrow. Trends in Cell Biology. 2015;25(9):523–532. doi: 10.1016/j.tcb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Developmental Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 14.Jaffe A.B., Hall A. Rho GTPases: biochemistry and biology. Annual Review of Cell Developmental Biology. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 15.Olson E.N., Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nature Reviews Molecular Cell Biology. 2010;11(5):353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98(2):159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang D.Z., Li S., Hockemeyer D., Sutherland L., Wang Z., Schratt G. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proceedings of the National Academy of Sciences United States of America. 2002;99(23):14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parmacek M.S. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circulation Research. 2007;100(5):633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 19.Nobusue H., Onishi N., Shimizu T., Sugihara E., Oki Y., Sumikawa Y. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nature Communication. 2014;5:3368. doi: 10.1038/ncomms4368. [DOI] [PubMed] [Google Scholar]
- 20.McDonald M.E., Li C., Bian H., Smith B.D., Layne M.D., Farmer S.R. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell. 2015;160(1–2):105–118. doi: 10.1016/j.cell.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee N.K., Sowa H., Hinoi E., Ferron M., Ahn J.D., Confavreux C. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130(3):456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wohl G.R., Loehrke L., Watkins B.A., Zernicke R.F. Effects of high-fat diet on mature bone mineral content, structure, and mechanical properties. Calcified Tissue International. 1998;63(1):74–79. doi: 10.1007/s002239900492. [DOI] [PubMed] [Google Scholar]
- 23.Corwin R.L., Hartman T.J., Maczuga S.A., Graubard B.I. Dietary saturated fat intake is inversely associated with bone density in humans: analysis of NHANES III. Journal of Nutrition. 2006;136(1):159–165. doi: 10.1093/jn/136.1.159. [DOI] [PubMed] [Google Scholar]
- 24.Doucette C.R., Horowitz M.C., Berry R., MacDougald O.A., Anunciado-Koza R., Koza R.A. A high fat diet increases bone marrow adipose tissue (MAT) but does not alter trabecular or cortical bone mass in C57BL/6J mice. Journal of Cellular Physiology. 2015;230(9):2032–2037. doi: 10.1002/jcp.24954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S., Chang S., Qi X., Richardson J.A., Olson E.N. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Molecular and Cellular Biology. 2006;26(15):5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evelyn C.R., Wade S.M., Wang Q., Wu M., Iniguez-Lluhi J.A., Merajver S.D. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Molecular Cancer Therapeutics. 2007;6(8):2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- 27.Lundquist M.R., Storaska A.J., Liu T.C., Larsen S.D., Evans T., Neubig R.R. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell. 2014;156(3):563–576. doi: 10.1016/j.cell.2013.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J.H., Hwang E.S., McManus M.T., Amsterdam A., Tian Y., Kalmukova R. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 29.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 30.Talele N.P., Fradette J., Davies J.E., Kapus A., Hinz B. Expression of alpha-smooth muscle actin determines the fate of mesenchymal stromal cells. Stem Cell Reports. 2015;4(6):1016–1030. doi: 10.1016/j.stemcr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meunier P., Aaron J., Edouard C., Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clinical Orthopaedics and Related Research. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- 32.Rosen C.J., Bouxsein M.L. Mechanisms of disease: is osteoporosis the obesity of bone? Nature Clinical Practice, Rheumatology. 2006;2(1):35–43. doi: 10.1038/ncprheum0070. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida C.A., Yamamoto H., Fujita T., Furuichi T., Ito K., Inoue K. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes & Development. 2004;18(8):952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Yuan K., Mao X., Miano J.M., Wu H., Chen Y. Serum response factor regulates bone formation via IGF-1 and Runx2 signals. Journal of Bone and Mineral Research. 2012;27(8):1659–1668. doi: 10.1002/jbmr.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue R., Zhou B.O., Shimada I.S., Zhao Z., Morrison S.J. Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell. 2016;18(6):782–796. doi: 10.1016/j.stem.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Naot D., Watson M., Callon K.E., Tuari D., Musson D.S., Choi A.J. Reduced bone density and cortical bone indices in female adiponectin-knockout mice. Endocrinology. 2016:en20161059. doi: 10.1210/en.2016-1059. [DOI] [PubMed] [Google Scholar]
- 37.Fulzele K., Riddle R.C., DiGirolamo D.J., Cao X., Wan C., Chen D. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell. 2010;142(2):309–319. doi: 10.1016/j.cell.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheller E.L., Cawthorn W.P., Burr A.A., Horowitz M.C., MacDougald O.A. Marrow adipose tissue: trimming the fat. Trends in Endocrinology & Metabolism. 2016;27(6):392–403. doi: 10.1016/j.tem.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogan B.L. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes & Development. 1996;10(13):1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 40.Zhao G.Q. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35(1):43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- 41.Schulz T.J., Huang P., Huang T.L., Xue R., McDougall L.E., Townsend K.L. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495(7441):379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tseng Y.H., Kokkotou E., Schulz T.J., Huang T.L., Winnay J.N., Taniguchi C.M. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454(7207):1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowers R.R., Lane M.D. A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle. 2007;6(4):385–389. doi: 10.4161/cc.6.4.3804. [DOI] [PubMed] [Google Scholar]
- 44.Wang D., Prakash J., Nguyen P., Davis-Dusenbery B.N., Hill N.S., Layne M.D. Bone morphogenetic protein signaling in vascular disease: anti-inflammatory action through myocardin-related transcription factor A. The Journal of Biological Chemistry. 2012;287(33):28067–28077. doi: 10.1074/jbc.M112.379487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y.F., Wei J., Liu X., Chen Y.H., Layne M.D., Yet S.F. Identification of a CArG-independent region of the cysteine-rich protein 2 promoter that directs expression in the developing vasculature. American Journal of Physiology, Heart and Circulatory Physiology. 2003;285(4):H1675–H1683. doi: 10.1152/ajpheart.00165.2003. [DOI] [PubMed] [Google Scholar]
- 46.Luchsinger L.L., Patenaude C.A., Smith B.D., Layne M.D. Myocardin-related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. Journal of Biological Chemistry. 2011;286(51):44116–44125. doi: 10.1074/jbc.M111.276931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vernochet C., Peres S.B., Davis K.E., McDonald M.E., Qiang L., Wang H. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Molecular and Cellular Biology. 2009;29(17):4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kon T., Cho T.J., Aizawa T., Yamazaki M., Nooh N., Graves D. Expression of osteoprotegerin, receptor activator of NF-kappaB ligand (osteoprotegerin ligand) and related proinflammatory cytokines during fracture healing. Journal of Bone and Mineral Research. 2001;16(6):1004–1014. doi: 10.1359/jbmr.2001.16.6.1004. [DOI] [PubMed] [Google Scholar]
- 49.Bouxsein M.L., Boyd S.K., Christiansen B.A., Guldberg R.E., Jepsen K.J., Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. Journal of Bone and Mineral Research. 2010;25(7):1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 50.Pino E., Wang H., McDonald M.E., Qiang L., Farmer S.R. Roles for peroxisome proliferator-activated receptor gamma (PPARgamma) and PPARgamma coactivators 1alpha and 1beta in regulating response of white and brown adipocytes to hypoxia. Journal of Biological Chemistry. 2012;287(22):18351–18358. doi: 10.1074/jbc.M112.350918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.