Abstract
This study aimed to derive quantitative abundance values for key hepatic transporters suitable for in vitro–in vivo extrapolation within a physiologically based pharmacokinetic modeling framework. A meta-analysis was performed whereby data on abundance measurements, sample preparation methods, and donor demography were collated from the literature. To define values for a healthy Caucasian population, a subdatabase was created whereby exclusion criteria were applied to remove samples from non-Caucasian individuals, those with underlying disease, or those with subcellular fractions other than crude membrane. Where a clinically relevant active genotype was known, only samples from individuals with an extensive transporter phenotype were included. Authors were contacted directly when additional information was required. After removing duplicated samples, the weighted mean, geometric mean, standard deviation, coefficient of variation, and between-study homogeneity of transporter abundances were determined. From the complete database containing 24 transporters, suitable abundance data were available for 11 hepatic transporters from nine studies after exclusion criteria were applied. Organic anion transporting polypeptides OATP1B1 and OATP1B3 showed the highest population abundance in healthy adult Caucasians. For several transporters, the variability in abundance was reduced significantly once the exclusion criteria were applied. The highest variability was observed for OATP1B3 > OATP1B1 > multidrug resistance protein 2 > multidrug resistance gene 1. No relationship was found between transporter expression and donor age. To our knowledge, this study provides the first in-depth analysis of current quantitative abundance data for a wide range of hepatic transporters, with the aim of using these data for in vitro–in vivo extrapolation, and highlights the significance of investigating the background of tissue(s) used in quantitative transporter proteomic studies. Similar studies are now warranted for other ethnicities.
Introduction
Physiologically based pharmacokinetic (PBPK) models are able to use in vitro data from recombinant expression systems to predict drug disposition via in vitro–in vivo extrapolation (IVIVE). In such cases, scalars are used to account for differences between in vitro and in vivo systems based on absolute or relative protein and/or activity measurements (Proctor et al., 2004; Harwood et al., 2013) and organ physiology (Johnson et al., 2005; Barter et al., 2007). Previously, IVIVE has been successfully applied to scale in vitro–derived kinetic data for hepatic cytochrome P450 (P450) (Howgate et al., 2006). The availability of recombinant P450 standards has facilitated the quantification of a wide range of P450 isoenzyme abundances in human liver samples, typically by immunoblotting techniques (Shimada et al., 1994; Rowland-Yeo et al., 2004). Until recently, such data have been lacking for non–P450-metabolizing enzymes, including UDP glucuronosyltransferases, esterases, and flavin-containing monooxygenases, as well as drug transporters. As a result, the scalars currently used for transporters have been based on relative protein measurements from semiquantitative immunoblot data (Neuhoff et al., 2013) or are empirical (Jones et al., 2012; Varma et al., 2012; Jamei et al., 2014). Transporter expression has thereby been measured via Western blotting; because of the limited availability of recombinant standards of membrane proteins, Western blotting yields relative or, at best, semiquantitative data, (Troutman and Thakker, 2003). Furthermore, robust results from Western blots rely on the use of specific antibodies, which are not always available, especially in the case of the solute carrier (SLC) family of transporters (Nies et al., 2009). More recently, proteomics techniques based on liquid chromatography (LC) with tandem mass spectrometry (MS/MS) have been developed in an effort to overcome the deficiencies in Western blotting. LC-MS/MS–based proteomic methodologies use peptides that are either metabolically (Achour et al., 2014) or chemically (Ohtsuki et al., 2011) isotope labeled and unique to the target protein as standards that act as surrogates to the whole protein. To date, the majority of quantitative transporter expression data available in the literature are based on measurements of hepatic uptake transporters due to their relevance in drug–drug interaction studies (Maeda and Sugiyama, 2013). Consequently, a relatively large abundance data set is available in the literature for transporters expressed in the human liver; hence, this organ provides a reasonable basis in which to begin generating quantitative IVIVE scalars for PBPK modeling. To generate robust IVIVE scalars that are relevant to a particular target population, the design of the currently reported quantitative proteomic studies assessing liver transporter abundance must be evaluated. This includes the choice of the subcellular fraction in which the protein was quantified and the corresponding extraction method as well as the source of liver samples, which may be derived from a multitude of disease, phenotype, age, and ethnic backgrounds. In light of the potential differences in transporter expression among individuals of different backgrounds, it is important to define selection criteria based on these differences to ensure that the quantitative scalars are based on a homogeneous baseline population. Similarly, the between-study heterogeneity should be analyzed and duplicate use of source data should be avoided. Here we aim to provide an up-to-date meta-analysis of quantitative hepatic transporter abundance data, which are readily available for quantitative abundance scaling within a PBPK framework such as the Simcyp Simulator (version 15, release 1; Simcyp Limited, Sheffield, UK), and to highlight the potential limitations that might be associated with these values.
Materials and Methods
Abundance Data.
An in-house database of published quantitative abundance data was first established in 2009. This initial database was expanded to contain published data for all known hepatic drug transporters that have been quantified in human liver tissue (n = 24 transporters). This was performed via searching the PubMed electronic database using the following keyword combination: hepatic/liver transporter abundance, absolute quantification, proteomics, and quantitative immunoblotting. A complete database including all available measurements was established (final literature search, November 2015), with background information on the methods as well as donor demographics collated where provided. In cases in which individual data were not directly reported, data were extracted via GetData Graph Digitizer (version 2.22, http://getdata-graph-digitizer.com) or authors were contacted directly to request individual donor data. A refined subdatabase was created through the use of various exclusion criteria. First, study methodologies were reviewed to ensure that abundances were quantified using LC-MS/MS or quantitative Western blotting in crude membrane (CM) fractions. Next, data for which it was stated that the corresponding human liver tissue was not from adult (aged >18 years), healthy, extensive transporter (ET) phenotype (where a clinically relevant active phenotype has been reported) or Caucasian individuals were excluded. Individuals with the ET phenotype were considered to be those that are either wild type or have similar levels of activity. Where only mean abundance data were available, measurements were included in the final database if fewer than 10% of the individuals met the exclusion criteria. Furthermore, the source of data was identified to ensure that duplicate measurements from the same tissue sample were not included in the refined database. Meta-analysis was then used to characterize the abundance of hepatic transporters in both the complete and refined databases.
IVIVE and CM Yield.
Transporter protein absolute abundance values are reported in moles of transporter protein per mass of total protein, where total protein relates to the subcellular fraction under study, typically CM (or total membrane). In vitro transporter activity expressed as an intrinsic clearance (CLint) can be scaled to a whole liver clearance (in liters per hour) either via relative transporter expression (eq. 1) or via absolute transporter expression (eq. 2). In either equation, the value of CLint,j may be dependent on the concentration at the transporter binding site if nonlinear kinetics (Jmax and Km) are defined.
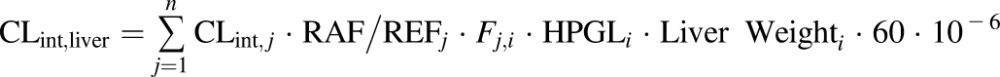 |
(1) |
where CLint,j is the in vitro intrinsic transport clearance for transporter j (in microliters per minute per million hepatocytes), REF or RAF is relative expression factor/relative activity factor in vivo compared with in vitro, Fi is the relative abundance for the phenotype of individual i (i.e., Fi = 1 for an ET), HPGL is the number of hepatocytes per gram of liver, liver weight is the subject’s liver weight (in grams), and CLint, liver is the whole liver intrinsic transport clearance (in liters per hour).
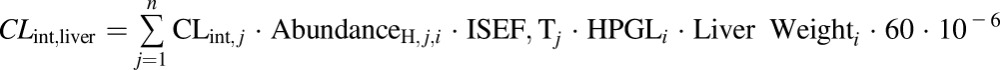 |
(2) |
where CLint,j is the in vitro intrinsic transport clearance for transporter j (in microliters per minute per picomoles of transporter), ISEF,T is the intersystem activity/abundance ratio between the in vitro system and in vivo (Harwood et al., 2013) and AbundanceH,j,i is the absolute hepatic transporter abundance in the target individual (in picomoles of transporter per million hepatocytes).
To be compatible with the IVIVE approach defined by eq. 2, the reported abundance values were converted from units of picomoles of transporter per milligram of CM protein to picomoles of transporter per million hepatocytes. This conversion requires an estimate of the yield of CM protein (in milligrams) per gram of liver tissue that was obtained when using the relevant membrane extraction method. Because such a yield is not routinely reported in transporter abundance studies, authors of potential sources of these data were contacted individually. The exclusion criteria that were applied to abundance values were also applied to obtained values of CM yield.
Transporter Genotype Analysis.
Relative hepatic abundances for individuals with several different organic anion transporting polypeptide OATP1B1 diplotypes were available from Nies et al. (2013). These diplotypes were classified into phenotypes based on Donnelly et al. (2011) for those involving *1a and *1b alleles, Katz et al. (2006) for those involving *5 and *15 alleles, and, finally, Ramsey et al. (2012) and Couvert et al. (2008) for those involving *14 and *35 alleles. The diplotypes are outlined in Table 1.
TABLE 1.
Phenotype data for OATP1B1 (SLCO1B1)
| Phenotype | Diplotype | Relative Abundance | CV |
|---|---|---|---|
| % | |||
| ET | *1a/*1a | 1 | 74 |
| *1a/*1b | |||
| *1a/*14 | |||
| *1a/*35 | |||
| IT | *1a/*5 | 0.68 | 54 |
| *1a/*15 | |||
| *1b/*15 | |||
| *5/*14 | |||
| *14/*15 | |||
| PT | *5/*5 | 0.37 | 35 |
| *15*15 | |||
| *5/*15 | |||
| UT | *1b/*35 | 1.47 | 46 |
| *1b/*14 | |||
| *14/*35 |
Relative abundance and the related population variability of each phenotype was obtained from in-house meta-analysis of published studies (Tirona et al., 2001; Iwai et al., 2004; Kameyama et al., 2005; Nozawa et al., 2005; Ho et al., 2006, 2007; Couvert et al., 2008; Deng et al., 2008; Choi et al., 2011; de Graan et al., 2012; Lancaster et al., 2012; Nies et al., 2013). Diplotypes associated with each phenotype definition are also summarized. All combinations of a given diplotype have been included in the model (e.g., *1a/*1b and *1b/*1a) but for simplicity are not included in this table.
Mean values and coefficients of variation (CVs) for OATP1B1 relative abundance in Caucasian ET and ultrarapid transporter (UT) phenotypes were taken directly from the relative abundances outlined by Nies at al. (2013). In the case of a poor transporter (PT) phenotype, a mean and CV for relative abundance was assigned on the basis of the difference in activity compared with an ET phenotype, because low transporter activity in transfected cells was not linked to decreased total cellular protein (Tirona et al., 2001; Seithel et al., 2008b) and no significant difference in protein expression between individuals with the ET phenotype and those with the PT phenotype was observed in obese subjects (Ulvestad et al., 2013). An in-house literature meta-analysis was derived using the in vitro relative activity of the *5 and *15 allelic variants compared with wild type with the OATP1B1 substrates estrone-3-sulfate and estradiol-17β-glucuronide. Relative abundance for the intermediate transporter (IT) phenotype was estimated from PT in vitro activity data and ET protein abundance data. Currently, it is not clear whether the in vivo activity of OATP1B1 in individuals with the IT phenotype is linked to protein abundance (as has been shown for ET and UT) or only in vitro activity (as has been shown for PT). Therefore, a midpoint value of relative abundance between PT and ET was assumed with the CV calculated from the abundance values reported previously (Nies et al., 2013). An estimate of the population frequencies of ET, PT, IT, and UT phenotypes in Caucasians was previously defined from a literature meta-analysis (Emami Riedmaier et al., 2016).
Data Analysis.
A relationship between age and CM yield obtained using the ProteoExtract native membrane extraction kit (MPEK) (Calbiochem, Billerica, MA) was investigated similarly to the manner in which the relationship between age and microsomal protein per gram of liver (MPPGL) was previously defined (Barter et al., 2008). Polynomial functions of two to five orders were fitted to the log-transformed CM yield (assuming a log-normal distribution of data) and goodness of fit was evaluated via visual inspection of the data and calculation of the Akaike information criterion.
Abundances were converted from picomoles of transporter protein per milligram of CM protein to picomoles per million hepatocytes using eq. 3.
| (3) |
where Abundancey is the abundance in picomoles of transporter per million hepatocytes, Abundancex is the abundance reported in the literature (picomoles per milligram of protein), Yieldage is the CM yield for donor age (milligrams of protein per gram of liver), and HPGLage is the hepatocellularity (106 hepatocytes per gram of liver) for donor age (Barter et al., 2007). Where individual abundance measurements were available, Yieldage and HPGLage were calculated for each donor, whereas the mean age of the donors was used for mean abundance measurements. The relationship between yield and age that was applied in eq. 3 was specific to the extraction method used. In cases in which the CM was generated using differential centrifugation, yield was determined from the MPPGL relationship with age (Barter et al., 2008), whereas the relationship defined in our study was applied if the MPEK was used. In cases in which the filter-aided sample preparation method was used (Vildhede et al., 2014), a yield similar to that obtained by an MPEK was assumed.
After applying the exclusion criteria to the complete database, the collated individual abundance values for the healthy, Caucasian adult subdatabase were combined for a given transporter to generate weighted mean, geometric mean, standard deviation, and weighted coefficient of variation based on the equations described previously (Perrett et al., 2007). The abundance values were further tested for between-study heterogeneity using the Cochran X2-based Q test (Perrett et al., 2007), whereby heterogeneity was assigned as low (P > 0.05), moderate (P < 0.05 and P > 0.001), or high (P < 0.001).
Using cases where more than one transporter was quantified in the same liver sample, correlations between transporters were tested in both the complete and refined databases. A Shapiro–Wilk normality test was used to test for Gaussian distributions of the abundance data for OATP1B1, OATP1B3, OATP2B1, and P-glycoprotein (P-gp).
The relationship between the expression of OATP transporters and age was examined in the subdatabase in which case samples from individuals aged younger than 18 years were also included (all other exclusion factors applied). In addition, the effect of fatty liver disease on the expression of OATPs, P-gp, multidrug resistance protein MRP2, and breast cancer resistance protein (BCRP) and sex-associated differences in expression for OATPs and P-gp were investigated by including such samples in the subdatabase with the other exclusion factors retained.
Simulations.
To assess the number of livers required to replicate the reported subset of transporter abundance and variability using the Simcyp Simulator (version 15, release 1), simulations were run in 100, 200, 500, 1000, and 2000 healthy virtual individuals of North European Caucasian background and the population abundance values were compared with values used to create the population library. The Mersenne Twister MT19937 random number generator was used to generate individuals.
Results
Genotype Data and Frequencies.
Table 1 shows the classification of OATP1B1 genotypes and phenotype-linked relative abundances.
Abundance Database.
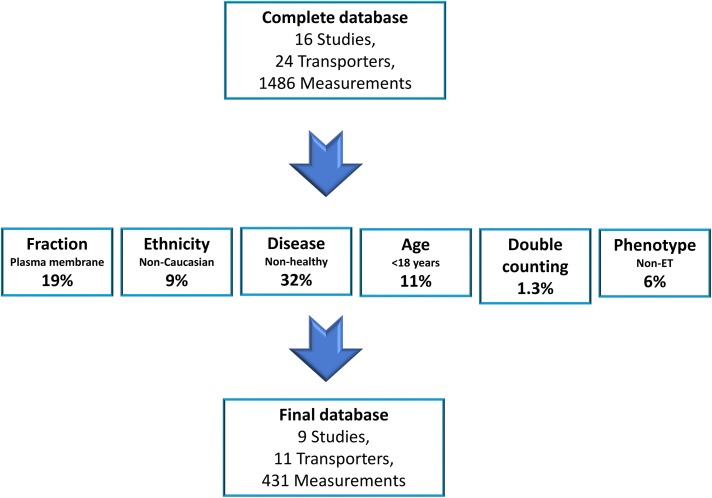
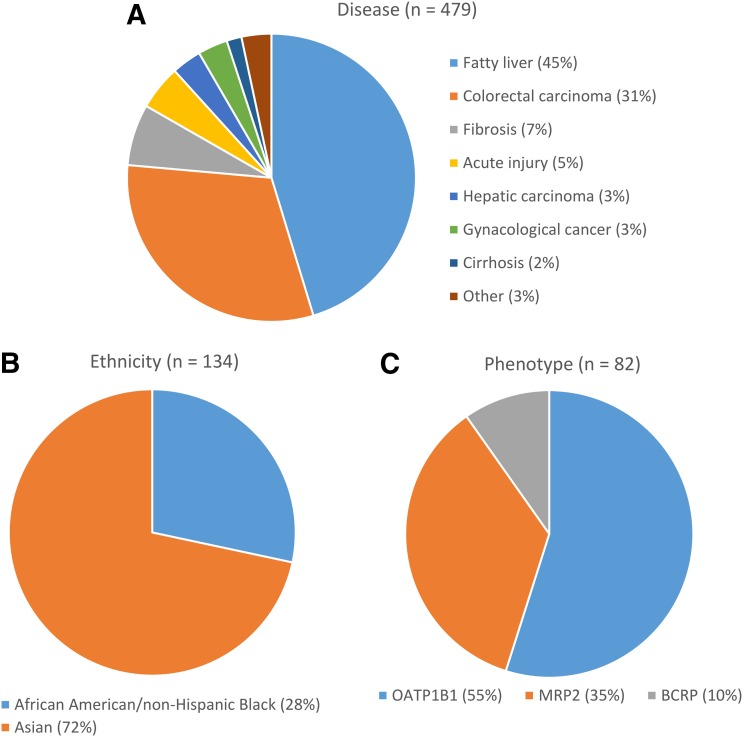
In this study, a total of 1486 measurements for 24 transporters were collated from 16 independent studies (Li et al., 2009a,b; Balogh et al., 2012; Bi et al., 2012; Deo et al., 2012; Kimoto et al., 2012; Ohtsuki et al., 2012; Tucker et al., 2012; Prasad et al., 2013, 2014; Qiu et al., 2013; Bosgra et al., 2014; Kunze et al., 2014; Vildhede et al., 2014; Peng et al., 2015; Wang et al., 2015). Of this complete database, only 431 human liver measurements for 11 transporters matched our inclusion criteria and were thus included in the sub–data set for adult healthy Caucasians, which were obtained from nine independent studies (Fig. 1) (Li et al., 2009a,b; Balogh et al., 2012; Deo et al., 2012; Kimoto et al., 2012; Tucker et al., 2012; Prasad et al., 2013, 2014; Peng et al., 2015). The most common reason (32% of the complete database) for the exclusion of abundance data were the use of samples from individuals with underlying disease conditions. The second most common reason for exclusion of abundance data were quantification in plasma membrane fractions (PMs) instead of CMs (19% of complete database), followed by the exclusion of samples from individuals aged <18 years, which resulted in the omission of 11% of samples from the subdatabase. Non-Caucasian ethnicities resulted in 9% of the complete database samples being excluded, and a further 6% of samples in the complete database were excluded on the basis of individuals having a non-ET phenotype. Finally, 1.3% of samples were excluded from the subdatabase due to double counting. A number of samples were excluded on the basis of two or more reasons; therefore, the sum of the percentages quoted above (78%) does not match the total percentage of the complete database that was excluded (71%). Of the sample excluded because of underlying disease, fatty liver disease (45% of samples excluded due to disease) and colorectal carcinoma (31%) were the most frequent diseases (Fig. 2A). Of the samples excluded because of non-Caucasian ethnicity, 72% were Asian and 28% were African American or non-Hispanic black (Fig. 2B). Finally, of the samples excluded due to a non-ET phenotype, the transporter was OATP1B1, MRP2, or BCRP in 55%, 35%, and 10% of cases, respectively (Fig. 2C).
Fig. 1.
Exclusion criteria applied to the complete database. Percentages for each exclusion criterion refer to the fraction of the samples in the complete database that were excluded on its basis. Note, some samples were excluded on the basis of more than one factor; therefore, the sum of percentages is less than the total percentage of the complete databases that was excluded.
Fig. 2.
(A) Relative proportion of the different diseases associated with samples that were excluded from the complete database on the basis of disease. (B) Relative proportion of the different ethnicities associated with samples that were excluded from the complete database on the basis of non-Caucasian ethnicity. (C) Relative proportion of the transporters associated with samples excluded from the complete database on the basis of a non-ET phenotype. n represents the number of samples excluded.
Of the 24 transporters for which abundance values were collated, suitable data could not be obtained for 13 at the time of this meta-analysis due to one or more of the criteria outlined in Fig. 1. These transporters were as follows: equilibrative nucleoside transporter 1 (SLC29A1); monocarboxylate transporter 1 (SLC16A1); organic anion transporters 2 and 7 (SLC22A7 and SLC22A9); multidrug resistance-associated proteins 1, 4, 5 and 6 (ABCC1, ABCC4, ABCC5 and ABCC6); organic cation transporter 3 (SLC22A3); multidrug resistance protein 3 (ABCB4); concentrative nucleoside transporter 1 (SLC28A1); ABCA6 and ABCA8. Thus, only 24% of the total data set was deemed suitable for characterizing transporter abundance in healthy Caucasian adults.
CM Yield.
A mean CM yield of 35.8 ± 14.3 mg membrane protein/g liver using a differential centrifugation method was confirmed via personal communication with Tucker et al. (2012) (n = 13 livers). This value is consistent with the MPPGL value that is obtained via a similar method (Barter et al., 2007). Corresponding mean CM yields of 38.6 ± 8.3 and 33.4 ± 4.6 mg membrane protein/g liver using MPEK were obtained via personal communication with investigators at the University of Washington (Seattle, WA) (n = 65 livers) and Eli Lilly (Indianapolis, IN)/University of Kansas (Lawrence, KS) (n = 141 livers), respectively.
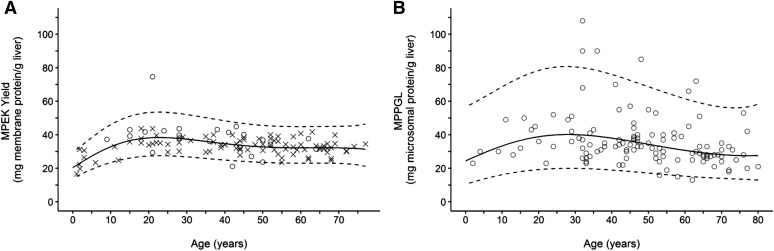
Because demographic data were available, the University of Washington and Eli Lilly/University of Kansas data sets were subsequently refined with the same exclusion criteria applied to abundance data with the exception of donor ages to create a specific healthy, Caucasian data set. In this refined data set, the relationship established between age and MPEK yield (Fig. 3A) was similar to that obtained for MPPGL (Barter et al., 2008) (eq. 4; Fig. 3B) and was best described by the fourth-order polynomial (eq. 5). However, the wider 95% prediction intervals (Fig. 3) and 2.4-fold higher root mean squared error value for the MPPGL data set indicated considerably higher variability around the central tendency compared with the MPEK yield.
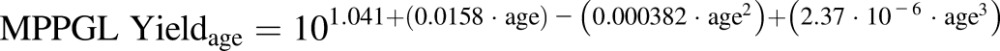 |
(4) |
| (5) |
As described in the Materials and Methods, eqs. 4 and 5 were used for the conversion of abundance units obtained using differential centrifugation and MPEK CM extraction methods, respectively (eq. 3). In the final subdatabase, three values were obtained using the differential centrifugation method, whereas the remaining 428 values used MPEK for CM extraction.
Fig. 3.
The relationship between MPEK yield and age defined in this study after the application of exclusion criteria (A) and the corresponding relationship between MPPGL and age defined by Barter et al. (2008) (B). MPEK yield data from the University of Washington database (n = 23) are displayed as circles and data from the University of Kansas/Eli Lilly database (n = 99) are displayed as crosses. Dashed lines represent the 95% prediction interval.
Abundance Data Analysis.
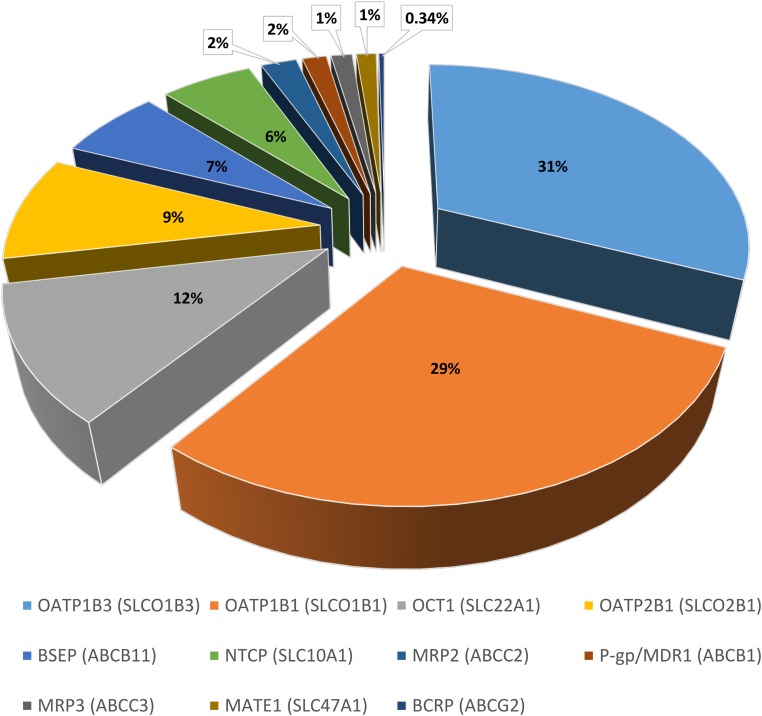
Table 2 provides a summary of the abundances for the 11 transporters in the healthy Caucasian adult subdatabase. The SLC uptake transporters OATP1B1 and OATP1B3 showed the highest abundance in the human liver samples, with a weighted mean abundance of 3.83 ± 2.5 and 4.13 ± 2.9 pmol transporter/million hepatocytes, respectively. The highest variability was also associated with OATP1B1 and OATP1B3, with CV values of 66% and 70%, respectively (Table 2). Of the ATP binding cassette (ABC) transporters, the bile salt efflux pump canalicular efflux transporter was the most abundant, with a weighted mean abundance of 0.839 ± 0.35 pmol transporter/million hepatocytes. The relative proportion of abundance for each hepatic transporter in the subdatabase is shown in Fig. 4.
TABLE 2.
Weighted mean, CV, and geometric mean of total membrane protein abundance of hepatic drug transporters obtained from meta-analysis of measurements in liver tissue of healthy Caucasian adults
| Transporter | Meana | Mean Percent Differenceb | CV Percent | CV Percent Differenceb | Geometric Meana | No. of Samples | No. of Studies | Heterogeneity |
References | |
|---|---|---|---|---|---|---|---|---|---|---|
| P | Class | |||||||||
| OATP1B1 (SLCO1B1) | 3.83 | 20 | 66 | −29 | 3.2 | 64 | 3 | 0.000442 | High | Kimoto et al. (2012), Prasad et al. (2014), Peng et al. (2015) |
| OATP1B3 (SLCO1B3) | 4.13 | 51 | 70 | −41 | 3.39 | 100 | 3 | 0.0225 | Moderate | Kimoto et al. (2012), Prasad et al. (2014), Peng et al. (2015) |
| OATP2B1 (SLCO2B1) | 1.2 | 26 | 40 | −32 | 1.12 | 100 | 3 | 0.271 | Low | Kimoto et al. (2012), Prasad et al. (2014), Peng et al. (2015) |
| OCT1 (SLC22A1) | 1.53 | 10.9 | 43 | −16 | 1.41 | 55 | 1 | N/A | N/A | Wang et al. (2015) |
| NTCP (SLC10A1) | 0.757 | −19 | 41 | −39 | 0.7 | 55 | 1 | N/A | N/A | Wang et al. (2015) |
| MATE1 (SLC47A1) | 0.165 | −16 | 31 | −50 | 0.158 | 55 | 1 | N/A | N/A | Wang et al. (2015) |
| P-gp/MDR1 (ABCB1) | 0.201 | 0.50 | 46 | −29 | 0.183 | 109 | 3 | 0.252 | Low | Tucker et al. (2012), Prasad et al. (2014), Peng et al. (2015) |
| BSEP (ABCB11) | 0.839 | 2.9 | 42 | −8 | 0.774 | 69 | 2 | 0.345 | Low | Li et al. (2009a) |
| MRP2 (ABCC2) | 0.296 | −35 | 65 | 28 | 0.249 | 37 | 3 | 0.509 | Low | Wang et al. (2015) |
| MRP3 (ABCC3) | 0.176 | 2.9 | 29 | −8 | 0.168 | 55 | 1 | N/A | N/A | Li et al. (2009b), Deo et al. (2012), Tucker et al. (2012) |
| BCRP (ABCG2) | 0.0442 | −24 | 40 | −48 | 0.041 | 38 | 3 | 0.116 | Low | Wang et al. (2015) |
BSEP, bile salt efflux pump; MATE, multidrug and toxin extrusion protein; MDR, multidrug resistance gene; N/A, not applicable; NTCP, sodium taurocholate cotransporting polypeptide; OCT, organic cation transporter.
Values are given as picomoles per 106 hepatocytes.
Percent difference compared with the corresponding value in the complete database without exclusion criteria applied.
Fig. 4.
Hepatic drug transporter pie chart. Relative protein abundance for 11 hepatic transporters as a percentage of the total abundance in the final subdatabase. BSEP, bile salt efflux pump; MATE, multidrug and toxin extrusion protein; MDR, multidrug resistance gene; NTCP, sodium taurocholate cotransporting polypeptide; OCT, organic cation transporter.
A summary of the abundances for the 24 transporters in the complete database without exclusion criteria is provided in Table 3. The greatest difference in mean abundance between the complete and subdatabases was observed for OATP1B3, which had a 51% higher abundance in the subdatabase. The largest decrease was observed for MRP2, which had a 35% lower abundance in the subdatabase. Although there was no clear trend for the difference in mean abundances between the subdatabase compared with the complete database (an increase was observed for six transporters, whereas a decrease was observed for five), there was a clear trend for a decrease in variability in the subdatabase. For example, the CV of OATP1B3 abundance was reduced from 117% (n = 289) to 70% (n = 100). Across transporters, a median 29% decrease in the abundance CV was observed when the exclusion criteria were applied to the complete database. The only transporter for which the variability increased was MRP2, in which case the CV increased from 51% to 65% in the subdatabase.
TABLE 3.
Weighted mean, CV, and geometric mean of total membrane protein abundance of hepatic drug transporters obtained from meta-analysis of measurements in liver tissue without exclusion criteria applied
| Transporter | Meana | CV Percent | Geometric Meana | No. of Samples | No. of Studies | Heterogeneity |
|
|---|---|---|---|---|---|---|---|
| P | Class | ||||||
| OATP1B1 (SLCO1B1) | 3.18 | 92.1 | 2.34 | 280 | 7 | 0.0195 | Moderate |
| OATP1B3 (SLCO1B3) | 2.73 | 117 | 1.78 | 289 | 7 | 0.177 | Low |
| OATP2B1 (SLCO2B1) | 0.956 | 58.6 | 0.825 | 277 | 7 | 0.658 | Low |
| ENT1 (SLC9A1) | 0.489 | 24.3 | 0.475 | 17 | 1 | N/A | N/A |
| NTCP (SLC10A1) | 0.935 | 66.7 | 0.778 | 94 | 4 | 0.121 | Low |
| MCT1 (SLC16A1) | 0.58 | 39 | 0.541 | 17 | 1 | N/A | N/A |
| OCT1 (SLC22A1) | 1.38 | 50.7 | 1.23 | 67 | 2 | 0.287 | Low |
| OAT2 (SLC22A7) | 0.471 | 75.5 | 0.376 | 25 | 2 | 0.00268 | Moderate |
| OAT7 (SLC22A9) | 0.41 | 50.9 | 0.365 | 12 | 1 | N/A | N/A |
| MATE1 (SLC47A1) | 0.196 | 63 | 0.166 | 83 | 3 | 0.000302 | High |
| OCT3 (SLC22A3) | 0.055 | 60.6 | 0.047 | 11 | 1 | N/A | N/A |
| CNT1 (SLC28A1) | 0.299 | 33 | 0.284 | 17 | 1 | N/A | N/A |
| ABCA6 | 2.58 | 41.8 | 2.38 | 17 | 1 | N/A | N/A |
| ABCA8 | 0.486 | 33.3 | 0.461 | 16 | 1 | N/A | N/A |
| P-gp/MDR1 (ABCB1) | 0.2 | 64.2 | 0.168 | 290 | 6 | 0.216 | Low |
| MDR3 (ABCB4) | 0.668 | 0.422 | 63.1 | 17 | 1 | N/A | N/A |
| BSEP (ABCB11) | 0.815 | 45.6 | 0.741 | 99 | 4 | 0.326 | Low |
| MRP1 (ABCC1) | 0.568 | 111 | 0.38 | 27 | 2 | 0.0492 | Moderate |
| MRP2 (ABCC2) | 0.452 | 50.6 | 0.403 | 162 | 6 | 0.419 | Low |
| MRP3 (ABCC3) | 0.171 | 31.8 | 0.163 | 67 | 2 | 0.76 | Low |
| MRP4 (ABCC4) | 0.0546 | 148 | 0.0305 | 8 | 2 | 0.0677 | Low |
| MRP5 (ABCC5) | 0.719 | 8.63 | 0.716 | 3 | 1 | N/A | N/A |
| MRP6 (ABCC6) | 0.679 | 46.6 | 0.615 | 29 | 2 | 0.466 | Low |
| BCRP (ABCG2) | 0.0582 | 76.1 | 0.0463 | 146 | 5 | 0.00103 | Moderate |
References for each transporter are provided in the Supplemental Material. BSEP, bile salt efflux pump; MATE, multidrug and toxin extrusion protein; MDR, multidrug resistance gene; N/A, XXX Not applicable (only 1 study); NTCP, sodium taurocholate cotransporting polypeptide; OCT, organic cation transporter.
Values are given as picomoles per 106 hepatocytes.
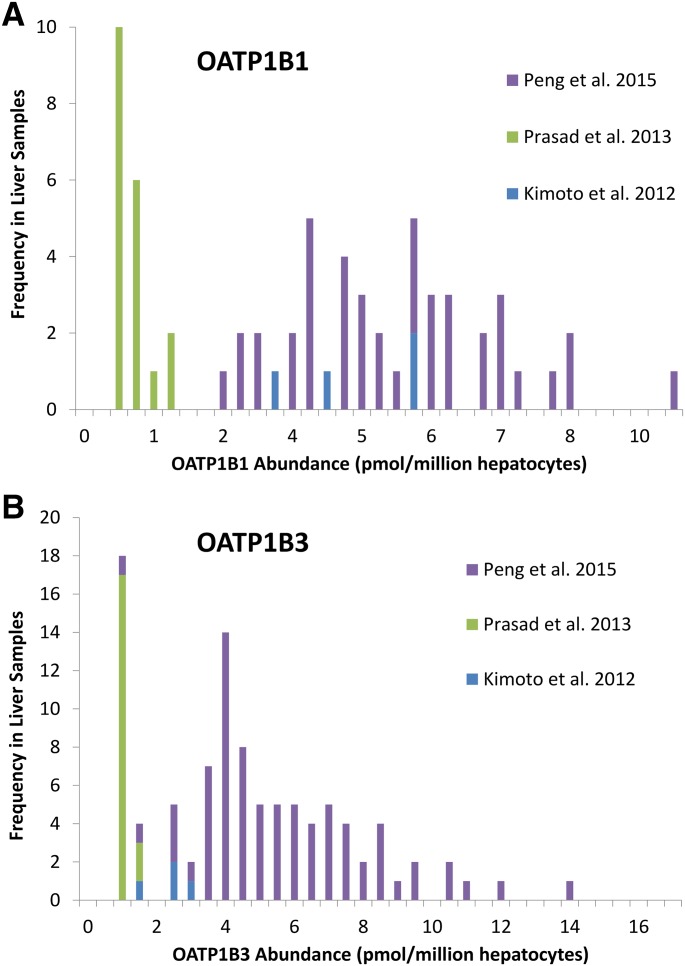
In the subdatabase, the abundance data for OATP1B1 and OATP1B3 were heterogeneous (Fig. 5; Table 2), which appears to be due to lower abundances determined in the study of Prasad et al. (2013) compared with Peng et al. (2015) and Kimoto et al. (2012). In the complete database, heterogeneity for OATP1B1 was of a lower severity and was not observed for OATP1B3 (Table 3).
Fig. 5.
Distribution of the individual absolute protein abundance values obtained for OATP1B1 (A) and OATP1B3 (B) by three independent studies (Kimoto et al., 2012; Prasad et al., 2014; Peng et al., 2015).
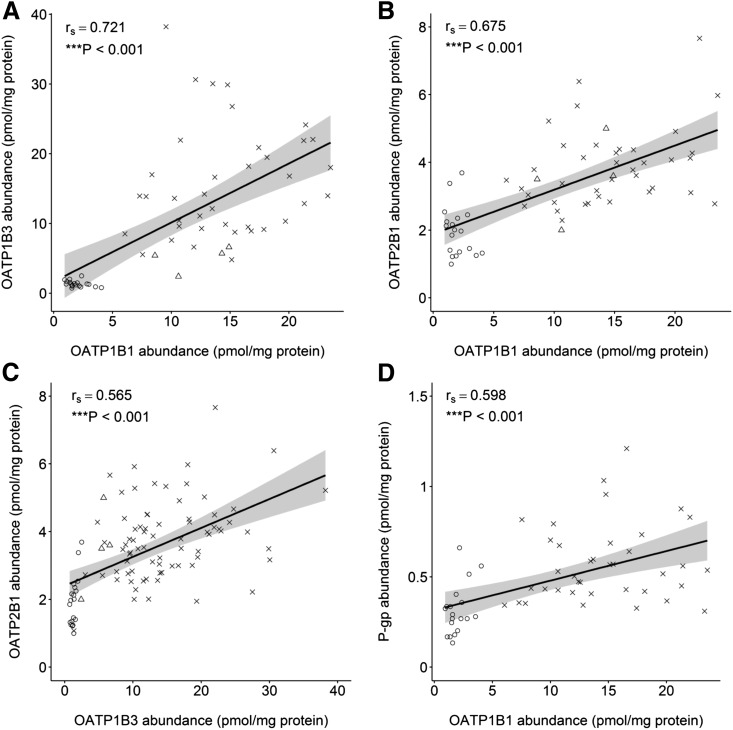
For all transporters, abundance estimates were not normally distributed; therefore, the nonparametric Spearman’s rank-order analysis was applied to define correlation coefficients (rs). Correlation in abundance between transporters was tested in both the complete and refined databases using cases where more than one transporter was quantified in the same liver tissue sample. This was possible for OATP1B1, OATP1B3, OATP2B1, P-gp, MRP2, and BCRP, whereby individual abundance data from the University of Washington (Deo et al., 2012; Prasad et al., 2013, 2014), University of Kansas and Eli Lilly (Peng et al., 2015), and Pfizer (Groton, CT) (Kimoto et al., 2012) could be combined. This allowed comparison of between 50 to 213 and 8 to 97 livers (depending on the transporter) for the complete and subdatabases, respectively. In the subdatabase, significant (P ≤ 0.05, rs > 0.5) correlations were observed between all OATPs and between OATP1B1 and P-gp. A significant correlation was not observed between the abundance of MRP2 or BCRP and any other transporter. However, on further inspection of the data, the heterogeneity between studies was an underlying factor in the observed correlations (Fig. 6). When data from each source were analyzed for correlation independently, the only significant (P ≤ 0.05, rs > 0.5) correlation was observed between OATP1B3 and OATP2B1 in the University of Washington data set (Fig. 6C, circles).
Fig. 6.
A comparison of hepatic abundance data for individuals with matched measurements for OATP1B1 and OATP1B3 (A), OATP1B1 and OATP2B1 (B), OATP1B3 and OATP2B1 (C), and OATP1B1 and P-gp (D). The solid line represents a linear regression line and the shaped area represents the 95% confidence interval around the linear regression. Circles represent data from the University of Washington database, crosses represent data from the University of Kansas/Eli Lilly database, and triangles represent data from the Pfizer database.
In the subdatabase, there were more than 50 individual samples with matching age demographics for OATP1B1, OATP1B3, OATP2B1, and P-gp, which permitted an evaluation of the relationship between age and abundance. This was performed using abundance values in units of picomoles per milligram of protein, prior to unit conversion with age-dependent yield and HPGL factors. For this analysis, samples from donors aged <18 years were included in the subdatabase with all other exclusion factors maintained. Of the transporters investigated, a significant (P ≤ 0.05, rs > 0.5) correlation was not found for any transporter in either the subdatabase or complete database.
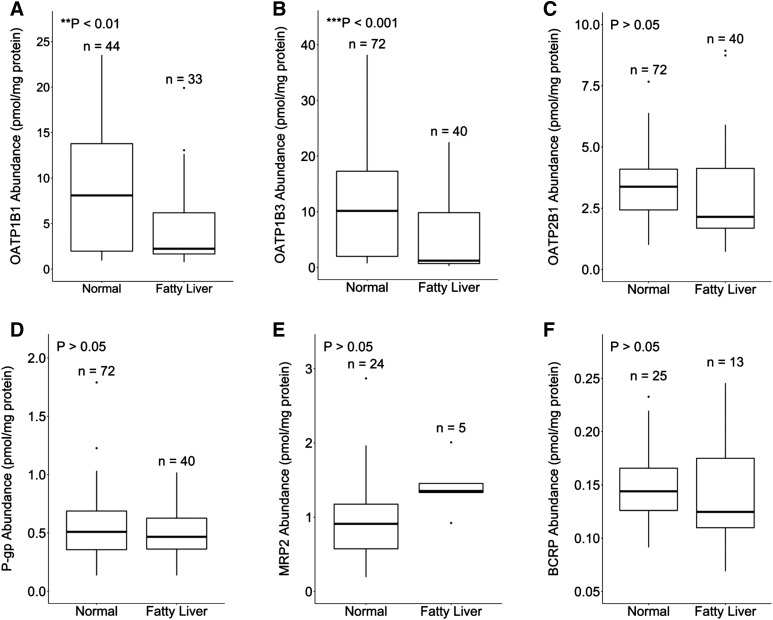
The difference in abundance between individuals with livers of normal appearance and those with fatty livers was evaluated by including individuals with fatty liver disease into the subdatabase, with all other exclusion factors maintained. A significantly lower abundance of OATP1B1 (P ≤ 0.01) and OATP1B3 (P ≤ 0.001) was observed in fatty livers compared with normal livers (Fig. 7, A and B). The median abundances were 3.6- and 8.4-fold lower for OATP1B1 and OATP1B3 in individuals with fatty livers compared with those with normal livers, respectively. No significant difference (P ≥ 0.05) was observed for OATP2B1, P-gp, MRP2, and BCRP (Fig. 7, C–F).
Fig. 7.
The relationship between hepatic abundance of OATP1B1 (A), OATP1B3 (B), OATP2B1 (C), P-gp (D), MRP2 (E), and BCRP (F) in individuals with normal and fatty livers. Data are from the final subdatabase with all exclusion criteria applied with the exception of fatty livers. The central band represents the median. The bottom and top of the box represents the first and third quartiles, respectively. Whiskers extend to the highest and lowest data points within 1.5-fold of the distance between the first and third quartiles and data outside of this range are considered outliers (points).
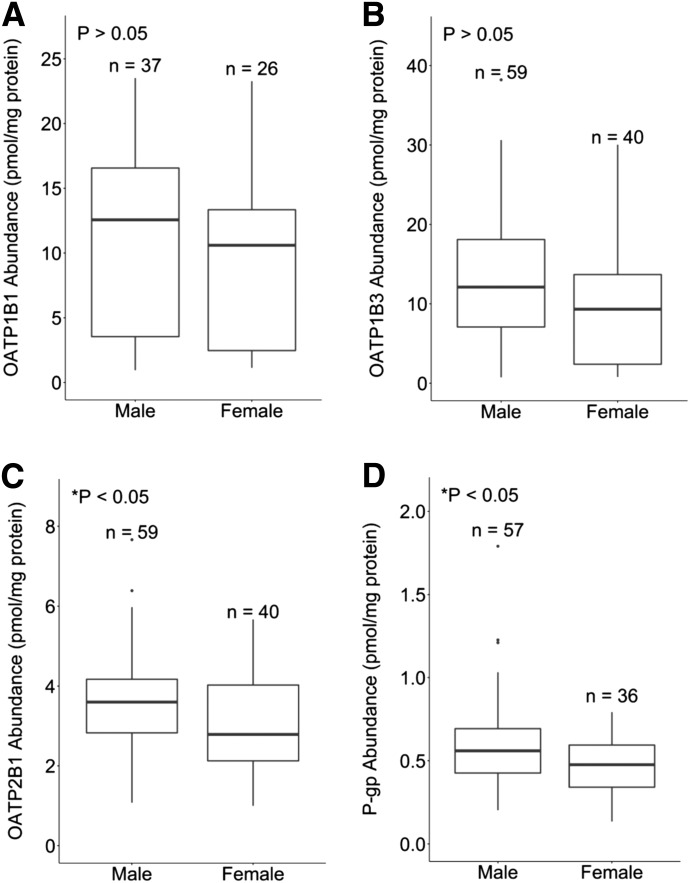
Finally, there were no significant (P < 0.05) sex-related differences in the hepatic abundance of OATP1B1 and OATP1B3. However, a significantly (P ≤ 0.05) higher abundance of OATP2B1 and P-gp was found in male individuals, with 1.3- and 1.2-fold higher median values, respectively (Fig. 8).
Fig. 8.
The relationship between hepatic abundance of OATP1B1 (A), OATP1B3 (B), OATP2B1 (C), and P-gp (D) in male and female individuals. Data are from the final subdatabase with all exclusion criteria applied.
Simulations.
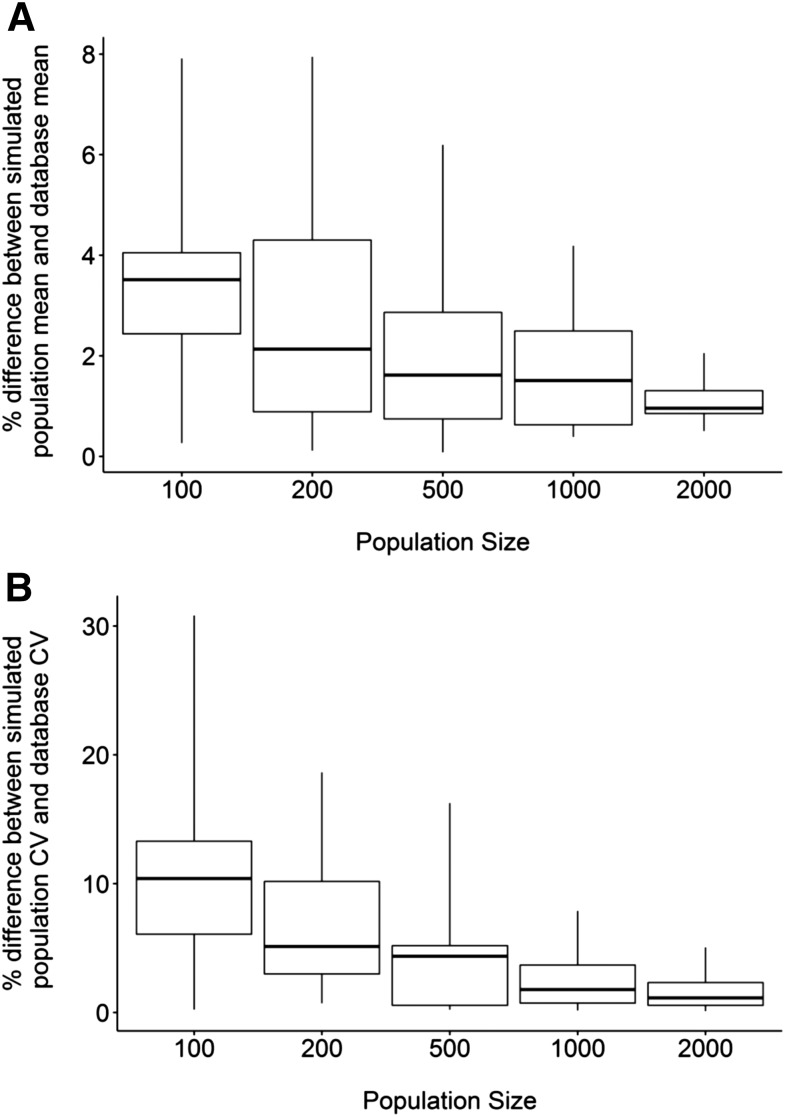
After implementation of the quantitative abundance values into the Simcyp Simulator database and population library (version 15), simulations were run in North European Caucasian virtual individuals to determine the population size required to replicate the reported mean abundance values and population variability for the 11 hepatic transporters for which suitable data were available. The mean hepatic transporter abundance values were less than 10% different from the subdatabase values for all of the investigated population sizes (100–2000 individuals), whereas the median difference was less than 2% when the population contained more than 500 individuals (Fig. 9A). The CVs for hepatic transporter abundances were up to 30% different to the subdatabase values with a population of 100 individuals, which was reduced to less than 10% with a population of 1000 individuals or more (Fig. 9B). The median difference in CV was less than 5% at populations greater than 500 individuals.
Fig. 9.
A comparison between the percent difference of the mean (A) and CV (B) for 11 different hepatic transporters when virtual populations of varying size were simulated and their corresponding values in the final subdatabase.
Discussion
An increased awareness of the role of transporters in the uptake and efflux of clinically relevant compounds has led to a growing interest in the development of PBPK models to investigate their influence on pharmacokinetics (Li et al., 2014; Rose et al., 2014; Posada et al., 2015; Snoeys et al., 2016). However, the lack of quantitative expression data for transporters in in vitro systems and human tissue has been a key limitation, meaning such PBPK models cannot be developed entirely from the “bottom up” and are typically built using both in vitro and clinical data in a “middle-out” approach. For example, if an approach such as that described by eq. 1 is used to scale in vitro CLint,j values to CLint,liver, the difficulty in obtaining a measured value of REF/RAF means that often this value can only be estimated by fitting clinical pharmacokinetic data. Recent proteomics techniques have provided a quantitative means of determining transporter expression in human samples (Ohtsuki et al., 2012; Prasad and Unadkat, 2014; Vildhede et al., 2015). These data facilitate the scaling of in vitro CLint,j values using an approach such as that described by eq. 2, with the advantage that this scaling approach may be more readily applied without the need for clinical data, assuming that the abundance of the transporter is correlated to its activity. Although a meta-analysis of the hepatic abundance of OATP transporters has previously been performed (Badée et al., 2015), our study aimed to expand the analysis to other hepatic transporters and to develop a database that was specific to a healthy adult Caucasian population. The information provided can be directly applied to the population-based scaling of rates of in vitro transport using the approach described by eq. 2, whereby virtual individuals can be created, each with a hepatic transporter abundance assigned on the basis of the mean and variability defined in our study.
A literature search was performed to gather quantitative abundance data for 24 hepatic transporter proteins to be used as a starting point for the incorporation of quantitative IVIVE scalars. A complete database of 1486 human liver measurements was established, from which a refined subdatabase of 431 measurements was obtained after exclusion of various data points on the basis of several criteria. Absolute abundance data were available from both crude and PM fractions. A purified PM fraction can be obtained from the CM by applying a gradient centrifugation step (Ohtsuki et al., 2012). However, data obtained from PMs were not included in the subdatabase, because corresponding protein yield values that enable the conversion of abundance values to picomoles of transporter per million hepatocytes are currently not reported in the literature (eq. 3) and there are concerns surrounding protein losses from this fraction (Harwood et al., 2014).
Although quantitative abundance data are essential for the bottom-up modeling of transporters, especially with respect to inhibition and induction studies, PBPK modeling should ultimately account for the relevant activity differences among transporters. In addition to protein that is active in the plasma membrane, measurements of transporter protein abundance in a CM will include protein that is in the plasma membrane but inactive and potentially protein residing in intracellular sites such as the endoplasmic reticulum, possibly acting as a reserve that can be trafficked to the plasma membrane on demand. A difference in the relative amounts of these forms of the same protein between an in vitro system and in vivo cells may lead to a difference in relative activity/abundance ratio. To account for this, the intersystem extrapolation factor for transporters (ISEF,T) is included in the scaling approach based upon transporter abundance (eq. 2) (Harwood et al., 2013). It is also imperative to account for activity differences where a clear link between activity and protein expression has been shown, such as OATP1B1 and OATP1B3, in which polymorphisms have been shown to result in different transporter activities in the Caucasian population (Seithel et al., 2008a; Niemi, 2010; Nies et al., 2013). Therefore, in this study, data were excluded for OATP1B1, MRP2, and BCRP transporters where a clear difference between abundance and activity has been reported (i.e., inclusion of abundance data from ET individuals only).
The observed heterogeneity of the OATP1B1 and OATP1B3 data that was present even after applying exclusion criteria was a key observation and suggests the involvement of additional factors, which may include tissue quality, the choice of peptide standards, or the sample preparation method. These observations emphasize the need to perform crosslaboratory comparisons and account for experimental variability in addition to population variability (Harwood et al., 2016).
Another factor that must be taken into account, especially as more data become available in this field, is the possible linkage of hepatic transporters, metabolic enzymes, and/or cofactor proteins. To date, most of the available knowledge in this field is qualitative and requires further investigation before it can be incorporated into expression analyses. The correlation between OATP1B1 and OATP1B3 protein expression is the only example where colinearity has been quantitatively demonstrated (Nies et al., 2013). However, because the trends of the interstudy heterogeneity were maintained across several transporters in our study, a reliable assessment of correlation between transporters in matched livers was confounded (Fig. 6). The significant (P ≤ 0.05, rs > 0.5) correlations that were observed in the final subdatabase between all OATPs and between OATP1B1 and P-gp were not consistently observed in individual studies. An example of these correlations and their application to the power of studies is described in greater detail in another publication from our group (Emami Riedmaier et al., 2016).
It was ensured that only samples from healthy individuals were included in the final database, because disease has been associated with significant changes in the membrane expression of several transporters (Zollner et al., 2007, 2014; Ufer et al., 2009; Wojtal et al., 2009). In this study, we found that adult individuals with fatty liver disease had lower expression of OATP1B1 and OATP1B3 compared with individuals with normal liver appearance, which has not been previously identified. The lack of correlation between MRP2 and BCRP abundance in normal compared with fatty livers that was observed by Deo et al. (2012) and Prasad et al. (2013), respectively, was maintained when combined with the data from Li et al. (2009a,b), respectively, after application of exclusion criteria for adult, Caucasian, and ET samples.
In our subdatabase, a significant but weak correlation was observed between OATP2B1 abundance and age (rs = 0.268, P = 0.00418, n = 113) but not for any other transporter. This differs from the previous meta-analysis of OATP abundance, in which a significant correlation was found for OATP1B1 (rs = 0.33, P < 0.01, n = 80) (Badée et al., 2015); this is likely linked to the application of exclusion criteria, because a significant correlation for OATP1B1 was also identified in our complete database (rs = 0.177, P = 0.00769, n = 226). Peng et al. (2015) identified a significant (P = 0.028) correlation between OATP1B3 abundance and age, which was not observed in our study.
Several studies have observed differences in drug exposure in different ethnic populations, which could not be entirely explained by differences in physiologic parameters (i.e., liver size) or the effect of genetic variations (Tomita et al., 2013; Peng et al., 2015). Although the mechanisms underlying these ethnic differences are not yet fully understood, it is believed that they may at least partially be attributed to differences in the expression or intrinsic activity of transporters in different ethnic populations. As a result of these observations, 9% of the complete database was excluded from the subdatabase, of which most were Asian (72%). This analysis demonstrates the importance of characterizing tissue backgrounds in quantitative proteomic studies prior to study commencement or prospectively and clearly reporting these data in publications as performed by Prasad et al. (2014) and Peng et al. (2015).
By using the same exclusion criteria, a healthy adult Caucasian relationship between age and CM protein yield obtained from the extraction kit method (i.e., MPEK) was obtained. The mean relationship was comparable to the mean relationship between age and microsomal protein (MPPGL) (Barter et al., 2008), which can be reconciled by the fact that the resulting fraction is similar between methods. However, there was a considerably reduced variability in the yield from the MPEK method compared with the MPPGL data, which was also observed when the exclusion criteria were not applied to the MPEK yield data (data not shown). The MPPGL values determined by Barter et al. (2008) were corrected for losses in microsomal protein during the extraction procedure, which was not performed for the MPEK data. The lack of a correction for losses in the MPEK data would most likely result in increased variability rather than the decrease that was observed. The difference in variability between these extraction procedures could be linked to a significant reduction in experimental variability with the MPEK method, because the interindividual variability would be expected to be similar, given the similarity in the resulting fractions.
To our knowledge, this is the first in-depth analysis of current quantitative abundance data for a wide array of hepatic transporters with the aim of using these data for IVIVE. Similar studies will be warranted in other ethnic populations and for other organs.
Acknowledgments
The authors thank Dr. Bhagwat Prasad and Dr. Jashvant Unadkat (University of Washington), Dr. Michael Zhuo Wang (University of Kansas), and Dr. Yingying Guo-Avrutin (Eli Lilly) for providing the individual data for the CM yield and the associated demographics and disease background to liver samples (Deo et al., 2012; Prasad et al., 2013; Peng et al., 2015). The authors also thank Dr. Yurong Lai (Bristol-Myers Squibb, Princeton, NJ) for providing individual data for transporter abundances and associated demographics (Li et al., 2009a,b; Kimoto et al., 2012) and Eleanor Savill (Simcyp) and Jessica Waite (Simcyp) for assistance in formatting the manuscript for submission.
Abbreviations
- ABC
ATP binding cassette
- BCRP
breast cancer resistance protein
- CLint
intrinsic clearance
- CM
crude membrane fraction
- CV
coefficient of variation
- ET
extensive transporter
- HPGL
hepatocytes per gram of liver
- ISEF,T
intersystem extrapolation factor for transporters
- IT
intermediate transporter
- IVIVE
in vitro–in vivo extrapolation
- LC
liquid chromatography
- MATE
multidrug and toxin extrusion protein
- MPEK
membrane protein extraction kit
- MPPGL
microsomal protein per gram of liver
- MRP
multidrug resistance protein
- MS/MS
tandem mass spectrometry
- OATP
organic anion transporting polypeptide
- P450
cytochrome P450
- P-gp
P-glycoprotein
- PBPK
physiologically based pharmacokinetic
- PM
plasma membrane fraction
- PT
poor transporter
- RAF
relative activity factor
- REF
relative expression factor
- rs
rank Spearman correlation coefficient
- SLC
solute carrier family
- UT
ultrarapid transporter
Authorship Contributions
Participated in research design: Burt, Emami Riedmaier, Harwood, Gill, Neuhoff.
Performed data analysis: Burt, Emami Riedmaier, Harwood, Crewe, Gill, Neuhoff.
Wrote or contributed to the writing of the manuscript: Burt, Emami Riedmaier, Harwood, Crewe, Gill, Neuhoff.
Footnotes
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Achour B, Russell MR, Barber J, Rostami-Hodjegan A. (2014) Simultaneous quantification of the abundance of several cytochrome P450 and uridine 5′-diphospho-glucuronosyltransferase enzymes in human liver microsomes using multiplexed targeted proteomics. Drug Metab Dispos 42:500–510. [DOI] [PubMed] [Google Scholar]
- Badée J, Achour B, Rostami-Hodjegan A, Galetin A. (2015) Meta-analysis of expression of hepatic organic anion-transporting polypeptide (OATP) transporters in cellular systems relative to human liver tissue. Drug Metab Dispos 43:424–432. [DOI] [PubMed] [Google Scholar]
- Balogh LM, Kimoto E, Chupka J, Zhang H, Lai Y. (2012) Membrane protein quantification by peptide-based mass spectrometry approaches: studies on the organic anion-transporting polypeptide family. J Proteomics Bioinform S4:003. [Google Scholar]
- Barter ZE, Bayliss MK, Beaune PH, Boobis AR, Carlile DJ, Edwards RJ, Houston JB, Lake BG, Lipscomb JC, Pelkonen OR, et al. (2007) Scaling factors for the extrapolation of in vivo metabolic drug clearance from in vitro data: reaching a consensus on values of human microsomal protein and hepatocellularity per gram of liver. Curr Drug Metab 8:33–45. [DOI] [PubMed] [Google Scholar]
- Barter ZE, Chowdry JE, Harlow JR, Snawder JE, Lipscomb JC, Rostami-Hodjegan A. (2008) Covariation of human microsomal protein per gram of liver with age: absence of influence of operator and sample storage may justify interlaboratory data pooling. Drug Metab Dispos 36:2405–2409. [DOI] [PubMed] [Google Scholar]
- Bi YA, Kimoto E, Sevidal S, Jones HM, Barton HA, Kempshall S, Whalen KM, Zhang H, Ji C, Fenner KS, et al. (2012) In vitro evaluation of hepatic transporter-mediated clinical drug-drug interactions: hepatocyte model optimization and retrospective investigation. Drug Metab Dispos 40:1085–1092. [DOI] [PubMed] [Google Scholar]
- Bosgra S, van de Steeg E, Vlaming ML, Verhoeckx KC, Huisman MT, Verwei M, Wortelboer HM. (2014) Predicting carrier-mediated hepatic disposition of rosuvastatin in man by scaling from individual transfected cell-lines in vitro using absolute transporter protein quantification and PBPK modeling. Eur J Pharm Sci 65:156–166. [DOI] [PubMed] [Google Scholar]
- Choi MK, Shin HJ, Choi YL, Deng JW, Shin JG, Song IS. (2011) Differential effect of genetic variants of Na(+)-taurocholate co-transporting polypeptide (NTCP) and organic anion-transporting polypeptide 1B1 (OATP1B1) on the uptake of HMG-CoA reductase inhibitors. Xenobiotica 41:24–34. [DOI] [PubMed] [Google Scholar]
- Couvert P, Giral P, Dejager S, Gu J, Huby T, Chapman MJ, Bruckert E, Carrié A. (2008) Association between a frequent allele of the gene encoding OATP1B1 and enhanced LDL-lowering response to fluvastatin therapy. Pharmacogenomics 9:1217–1227. [DOI] [PubMed] [Google Scholar]
- de Graan AJM, Lancaster CS, Obaidat A, Hagenbuch B, Elens L, Friberg LE, de Bruijn P, Hu S, Gibson AA, Bruun GH, et al. (2012) Influence of polymorphic OATP1B-type carriers on the disposition of docetaxel. Clin Cancer Res 18:4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JW, Song IS, Shin HJ, Yeo CW, Cho DY, Shon JH, Shin JG. (2008) The effect of SLCO1B1*15 on the disposition of pravastatin and pitavastatin is substrate dependent: the contribution of transporting activity changes by SLCO1B1*15. Pharmacogenet Genomics 18:424–433. [DOI] [PubMed] [Google Scholar]
- Deo AK, Prasad B, Balogh L, Lai Y, Unadkat JD. (2012) Interindividual variability in hepatic expression of the multidrug resistance-associated protein 2 (MRP2/ABCC2): quantification by liquid chromatography/tandem mass spectrometry. Drug Metab Dispos 40:852–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly LA, Doney AS, Tavendale R, Lang CC, Pearson ER, Colhoun HM, McCarthy MI, Hattersley AT, Morris AD, Palmer CN. (2011) Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: a go-DARTS study. Clin Pharmacol Ther 89:210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami Riedmaier A, Burt H, Abduljalil K, Neuhoff S. (2016) More power to OATP1B1: an evaluation of sample size in pharmacogenetic studies using a rosuvastatin PBPK model for intestinal, hepatic and renal transporter-mediated clearances. J Clin Pharmacol 56 (Suppl 7):S132–S142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood MD, Achour B, Neuhoff S, Russell MR, Carlson G, Warhurst G, Rostami-Hodjegan A. (2016) In vitro-in vivo extrapolation scaling factors for intestinal P-glycoprotein and breast cancer resistance protein: part i: a cross-laboratory comparison of transporter-protein abundances and relative expression factors in human intestine and Caco-2 cells. Drug Metab Dispos 44:297–307. [DOI] [PubMed] [Google Scholar]
- Harwood MD, Neuhoff S, Carlson GL, Warhurst G, Rostami-Hodjegan A. (2013) Absolute abundance and function of intestinal drug transporters: a prerequisite for fully mechanistic in vitro-in vivo extrapolation of oral drug absorption. Biopharm Drug Dispos 34:2–28. [DOI] [PubMed] [Google Scholar]
- Harwood MD, Russell MR, Neuhoff S, Warhurst G, Rostami-Hodjegan A. (2014) Lost in centrifugation: accounting for transporter protein losses in quantitative targeted absolute proteomics. Drug Metab Dispos 42:1766–1772. [DOI] [PubMed] [Google Scholar]
- Ho RH, Choi L, Lee W, Mayo G, Schwarz UI, Tirona RG, Bailey DG, Stein CM, Kim RB. (2007) Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenet Genomics 17:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, Wang Y, Kim RB. (2006) Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology 130:1793–1806. [DOI] [PubMed] [Google Scholar]
- Howgate EM, Rowland Yeo K, Proctor NJ, Tucker GT, Rostami-Hodjegan A. (2006) Prediction of in vivo drug clearance from in vitro data. I: impact of inter-individual variability. Xenobiotica 36:473–497. [DOI] [PubMed] [Google Scholar]
- Iwai M, Suzuki H, Ieiri I, Otsubo K, Sugiyama Y. (2004) Functional analysis of single nucleotide polymorphisms of hepatic organic anion transporter OATP1B1 (OATP-C). Pharmacogenetics 14:749–757. [DOI] [PubMed] [Google Scholar]
- Jamei M, Bajot F, Neuhoff S, Barter Z, Yang J, Rostami-Hodjegan A, Rowland-Yeo K. (2014) A mechanistic framework for in vitro-in vivo extrapolation of liver membrane transporters: prediction of drug-drug interaction between rosuvastatin and cyclosporine. Clin Pharmacokinet 53:73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TN, Tucker GT, Tanner MS, Rostami-Hodjegan A. (2005) Changes in liver volume from birth to adulthood: a meta-analysis. Liver Transpl 11:1481–1493. [DOI] [PubMed] [Google Scholar]
- Jones HM, Barton HA, Lai Y, Bi YA, Kimoto E, Kempshall S, Tate SC, El-Kattan A, Houston JB, Galetin A, et al. (2012) Mechanistic pharmacokinetic modeling for the prediction of transporter-mediated disposition in humans from sandwich culture human hepatocyte data. Drug Metab Dispos 40:1007–1017. [DOI] [PubMed] [Google Scholar]
- Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. (2005) Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics 15:513–522. [DOI] [PubMed] [Google Scholar]
- Katz DA, Carr R, Grimm DR, Xiong H, Holley-Shanks R, Mueller T, Leake B, Wang Q, Han L, Wang PG, et al. (2006) Organic anion transporting polypeptide 1B1 activity classified by SLCO1B1 genotype influences atrasentan pharmacokinetics. Clin Pharmacol Ther 79:186–196. [DOI] [PubMed] [Google Scholar]
- Kimoto E, Yoshida K, Balogh LM, Bi YA, Maeda K, El-Kattan A, Sugiyama Y, Lai Y. (2012) Characterization of organic anion transporting polypeptide (OATP) expression and its functional contribution to the uptake of substrates in human hepatocytes. Mol Pharm 9:3535–3542. [DOI] [PubMed] [Google Scholar]
- Kunze A, Huwyler J, Camenisch G, Poller B. (2014) Prediction of organic anion-transporting polypeptide 1B1- and 1B3-mediated hepatic uptake of statins based on transporter protein expression and activity data. Drug Metab Dispos 42:1514–1521. [DOI] [PubMed] [Google Scholar]
- Lancaster CS, Bruun GH, Peer CJ, Mikkelsen TS, Corydon TJ, Gibson AA, Hu S, Orwick SJ, Mathijssen RHJ, Figg WD, et al. (2012) OATP1B1 polymorphism as a determinant of erythromycin disposition. Clin Pharmacol Ther 92:642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Palandra J, Nemirovskiy OV, Lai Y. (2009a) LC-MS/MS mediated absolute quantification and comparison of bile salt export pump and breast cancer resistance protein in livers and hepatocytes across species. Anal Chem 81:2251–2259. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang Y, Hua F, Lai Y. (2009b) Absolute difference of hepatobiliary transporter multidrug resistance-associated protein (MRP2/Mrp2) in liver tissues and isolated hepatocytes from rat, dog, monkey, and human. Drug Metab Dispos 37:66–73. [DOI] [PubMed] [Google Scholar]
- Li R, Barton HA, Yates PD, Ghosh A, Wolford AC, Riccardi KA, Maurer TS. (2014) A “middle-out” approach to human pharmacokinetic predictions for OATP substrates using physiologically-based pharmacokinetic modeling. J Pharmacokinet Pharmacodyn 41:197–209. [DOI] [PubMed] [Google Scholar]
- Maeda K, Sugiyama Y. (2013) Transporter biology in drug approval: regulatory aspects. Mol Aspects Med 34:711–718. [DOI] [PubMed] [Google Scholar]
- Neuhoff S, Yeo KR, Barter Z, Jamei M, Turner DB, Rostami-Hodjegan A. (2013) Application of permeability-limited physiologically-based pharmacokinetic models: part I-digoxin pharmacokinetics incorporating P-glycoprotein-mediated efflux. J Pharm Sci 102:3145–3160. [DOI] [PubMed] [Google Scholar]
- Niemi M. (2010) Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther 87:130–133. [DOI] [PubMed] [Google Scholar]
- Nies AT, Koepsell H, Winter S, Burk O, Klein K, Kerb R, Zanger UM, Keppler D, Schwab M, Schaeffeler E. (2009) Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology 50:1227–1240. [DOI] [PubMed] [Google Scholar]
- Nies AT, Niemi M, Burk O, Winter S, Zanger UM, Stieger B, Schwab M, Schaeffeler E. (2013) Genetics is a major determinant of expression of the human hepatic uptake transporter OATP1B1, but not of OATP1B3 and OATP2B1. Genome Med 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa T, Minami H, Sugiura S, Tsuji A, Tamai I. (2005) Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: in vitro evidence and effect of single nucleotide polymorphisms. Drug Metab Dispos 33:434–439. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, et al. (2012) Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos 40:83–92. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Uchida Y, Kubo Y, Terasaki T. (2011) Quantitative targeted absolute proteomics-based ADME research as a new path to drug discovery and development: methodology, advantages, strategy, and prospects. J Pharm Sci 100:3547–3559. [DOI] [PubMed] [Google Scholar]
- Peng KW, Bacon J, Zheng M, Guo Y, Wang MZ. (2015) Ethnic variability in the expression of hepatic drug transporters: absolute quantification by an optimized targeted quantitative proteomic approach. Drug Metab Dispos 43:1045–1055. [DOI] [PubMed] [Google Scholar]
- Perrett HF, Barter ZE, Jones BC, Yamazaki H, Tucker GT, Rostami-Hodjegan A. (2007) Disparity in holoprotein/apoprotein ratios of different standards used for immunoquantification of hepatic cytochrome P450 enzymes. Drug Metab Dispos 35:1733–1736. [DOI] [PubMed] [Google Scholar]
- Posada MM, Bacon JA, Schneck KB, Tirona RG, Kim RB, Higgins JW, Pak YA, Hall SD, Hillgren KM. (2015) Prediction of renal transporter mediated drug-drug interactions for pemetrexed using physiologically based pharmacokinetic modeling. Drug Metab Dispos 43:325–334. [DOI] [PubMed] [Google Scholar]
- Prasad B, Evers R, Gupta A, Hop CECA, Salphati L, Shukla S, Ambudkar SV, Unadkat JD. (2014) Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos 42:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Lai Y, Lin Y, Unadkat JD. (2013) Interindividual variability in the hepatic expression of the human breast cancer resistance protein (BCRP/ABCG2): effect of age, sex, and genotype. J Pharm Sci 102:787–793. [DOI] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014) Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor NJ, Tucker GT, Rostami-Hodjegan A. (2004) Predicting drug clearance from recombinantly expressed CYPs: intersystem extrapolation factors. Xenobiotica 34:151–178. [DOI] [PubMed] [Google Scholar]
- Qiu X, Bi YA, Balogh LM, Lai Y. (2013) Absolute measurement of species differences in sodium taurocholate cotransporting polypeptide (NTCP/Ntcp) and its modulation in cultured hepatocytes. J Pharm Sci 102:3252–3263. [DOI] [PubMed] [Google Scholar]
- Ramsey LB, Bruun GH, Yang W, Treviño LR, Vattathil S, Scheet P, Cheng C, Rosner GL, Giacomini KM, Fan Y, et al. (2012) Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res 22:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RH, Neuhoff S, Abduljalil K, Chetty M, Rostami-Hodjegan A, Jamei M. (2014) Application of a physiologically based pharmacokinetic model to predict OATP1B1-related variability in pharmacodynamics of rosuvastatin. CPT Pharmacometrics Syst Pharmacol 3:e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland-Yeo K, Rostami-Hodjegan A, Tucker GT. (2004) Abundance of cytochromes P450 in human liver: a meta-analysis [Abstract]. Br J Clin Pharmacol 57:687–688. [Google Scholar]
- Seithel A, Glaeser H, Fromm MF, König J. (2008a) The functional consequences of genetic variations in transporter genes encoding human organic anion-transporting polypeptide family members. Expert Opin Drug Metab Toxicol 4:51–64. [DOI] [PubMed] [Google Scholar]
- Seithel A, Klein K, Zanger UM, Fromm MF, König J. (2008b) Non-synonymous polymorphisms in the human SLCO1B1 gene: an in vitro analysis of SNP c.1929A>C. Mol Genet Genomics 279:149–157. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270:414–423. [PubMed] [Google Scholar]
- Snoeys J, Beumont M, Monshouwer M, Ouwerkerk-Mahadevan S. (2016) Mechanistic understanding of the nonlinear pharmacokinetics and intersubject variability of simeprevir: A PBPK-guided drug development approach. Clin Pharmacol Ther 99:224–234. [DOI] [PubMed] [Google Scholar]
- Tirona RG, Leake BF, Merino G, Kim RB. (2001) Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem 276:35669–35675. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Maeda K, Sugiyama Y. (2013) Ethnic variability in the plasma exposures of OATP1B1 substrates such as HMG-CoA reductase inhibitors: a kinetic consideration of its mechanism. Clin Pharmacol Ther 94:37–51. [DOI] [PubMed] [Google Scholar]
- Troutman MD, Thakker DR. (2003) Novel experimental parameters to quantify the modulation of absorptive and secretory transport of compounds by P-glycoprotein in cell culture models of intestinal epithelium. Pharm Res 20:1210–1224. [DOI] [PubMed] [Google Scholar]
- Tucker TGHA, Milne AM, Fournel-Gigleux S, Fenner KS, Coughtrie MWH. (2012) Absolute immunoquantification of the expression of ABC transporters P-glycoprotein, breast cancer resistance protein and multidrug resistance-associated protein 2 in human liver and duodenum. Biochem Pharmacol 83:279–285. [DOI] [PubMed] [Google Scholar]
- Ufer M, Häsler R, Jacobs G, Haenisch S, Lächelt S, Faltraco F, Sina C, Rosenstiel P, Nikolaus S, Schreiber S, et al. (2009) Decreased sigmoidal ABCB1 (P-glycoprotein) expression in ulcerative colitis is associated with disease activity. Pharmacogenomics 10:1941–1953. [DOI] [PubMed] [Google Scholar]
- Ulvestad M, Skottheim IB, Jakobsen GS, Bremer S, Molden E, Asberg A, Hjelmesæth J, Andersson TB, Sandbu R, Christensen H. (2013) Impact of OATP1B1, MDR1, and CYP3A4 expression in liver and intestine on interpatient pharmacokinetic variability of atorvastatin in obese subjects. Clin Pharmacol Ther 93:275–282. [DOI] [PubMed] [Google Scholar]
- Varma MV, Lai Y, Feng B, Litchfield J, Goosen TC, Bergman A. (2012) Physiologically based modeling of pravastatin transporter-mediated hepatobiliary disposition and drug-drug interactions. Pharm Res 29:2860–2873. [DOI] [PubMed] [Google Scholar]
- Vildhede A, Karlgren M, Svedberg EK, Wisniewski JR, Lai Y, Norén A, Artursson P. (2014) Hepatic uptake of atorvastatin: influence of variability in transporter expression on uptake clearance and drug-drug interactions. Drug Metab Dispos 42:1210–1218. [DOI] [PubMed] [Google Scholar]
- Vildhede A, Wiśniewski JR, Norén A, Karlgren M, Artursson P. (2015) Comparative proteomic analysis of human liver tissue and isolated hepatocytes with a focus on proteins determining drug exposure. J Proteome Res 14:3305–3314. [DOI] [PubMed] [Google Scholar]
- Wang L, Prasad B, Salphati L, Chu X, Gupta A, Hop CECA, Evers R, Unadkat JD. (2015) Interspecies variability in expression of hepatobiliary transporters across human, dog, monkey, and rat as determined by quantitative proteomics. Drug Metab Dispos 43:367–374. [DOI] [PubMed] [Google Scholar]
- Wojtal KA, Eloranta JJ, Hruz P, Gutmann H, Drewe J, Staumann A, Beglinger C, Fried M, Kullak-Ublick GA, Vavricka SR. (2009) Changes in mRNA expression levels of solute carrier transporters in inflammatory bowel disease patients. Drug Metab Dispos 37:1871–1877. [DOI] [PubMed] [Google Scholar]
- Zollner G, Thueringer A, Lackner C, Fickert P, Trauner M. (2014) Alterations of canalicular ATP-binding cassette transporter expression in drug-induced liver injury. Digestion 90:81–88. [DOI] [PubMed] [Google Scholar]
- Zollner G, Wagner M, Fickert P, Silbert D, Gumhold J, Zatloukal K, Denk H, Trauner M. (2007) Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver Int 27:920–929. [DOI] [PubMed] [Google Scholar]