Abstract
Statistical analysis was performed on physicochemical descriptors of ∼250 drugs known to interact with one or more SLC22 “drug” transporters (i.e., SLC22A6 or OAT1, SLC22A8 or OAT3, SLC22A1 or OCT1, and SLC22A2 or OCT2), followed by application of machine-learning methods and wet laboratory testing of novel predictions. In addition to molecular charge, organic anion transporters (OATs) were found to prefer interacting with planar structures, whereas organic cation transporters (OCTs) interact with more three-dimensional structures (i.e., greater SP3 character). Moreover, compared with OAT1 ligands, OAT3 ligands possess more acyclic tetravalent bonds and have a more zwitterionic/cationic character. In contrast, OCT1 and OCT2 ligands were not clearly distinquishable form one another by the methods employed. Multiple pharmacophore models were generated on the basis of the drugs and, consistent with the machine-learning analyses, one unique pharmacophore created from ligands of OAT3 possessed cationic properties similar to OCT ligands; this was confirmed by quantitative atomic property field analysis. Virtual screening with this pharmacophore, followed by transport assays, identified several cationic drugs that selectively interact with OAT3 but not OAT1. Although the present analysis may be somewhat limited by the need to rely largely on inhibition data for modeling, wet laboratory/in vitro transport studies, as well as analysis of drug/metabolite handling in Oat and Oct knockout animals, support the general validity of the approach—which can also be applied to other SLC and ATP binding cassette drug transporters. This may make it possible to predict the molecular properties of a drug or metabolite necessary for interaction with the transporter(s), thereby enabling better prediction of drug-drug interactions and drug-metabolite interactions. Furthermore, understanding the overlapping specificities of OATs and OCTs in the context of dynamic transporter tissue expression patterns should help predict net flux in a particular tissue of anionic, cationic, and zwitterionic molecules in normal and pathophysiological states.
Introduction
Organic anion transporter 1 (OAT1/SLC22A6), OAT3 (SLC22A8), organic cation transporter 1 (OCT1/SLC22A1), and OCT2 (SLC22A2), perhaps the best studied members of the SLC22 family of solute carriers, are responsible for the excretion of a wide variety of drugs, toxins, and metabolites in the kidney, liver, and other tissues (Nigam et al., 2007; Emami Riedmaier et al., 2012; Koepsell, 2013; Nigam, 2015; Nigam et al., 2015a,b). This family, originally proposed in 1997 on the basis of three family members (Lopez-Nieto et al., 1997), now consists of over 30 members in mammals (Lopez-Nieto et al., 1997; Eraly et al., 2004; Wu et al., 2009; Zhu et al., 2015). Although sharing overall sequence and predicted structural similarities, the four transporters have distinct preferences for interaction with ligands. As their names suggest, OATs, belonging to the “organic anion” transporter subfamily, mainly interact with anions, whereas OCTs, belonging to the “organic cation” transporter subfamily, mainly interact with cations (Popp et al., 2005). Nevertheless, the grouping of OATs and OCTs into two different transporter subfamilies, organic anions and organic cations, respectively, can be misleading when it comes to individual drugs, toxins, and metabolites. For example, OATs have the capacity to interact with cationic drugs (Ahn et al., 2009), and both OATs and OCTs appear to interact in vitro and in vivo with zwitterionic or mildly “cationic” metabolites such as creatinine and polyamines (Ahn et al., 2011; Imamura et al., 2011; Vallon et al., 2012). However, these studies were limited to a few interacting compounds. Moreover, evolutionary analysis also indicates that the SLC22 family is probably more complex than originally thought, as it appears to comprise at least six subgroups, including, apart from the “Oat” and “Oct” group, groups termed “Oat-like,” “Oat-related,” “Octn” (organic cation/carnitine transporter), and “Oct-related” (Zhu et al., 2015). Together these results raise certain questions about the simple conception of OATs as “organic anion” transporters and OCTs as “organic cation” transporters and demonstrate the need for deeper investigation of ligand interactions with the various SLC22 transporters.
Given that there are a large number of well established OAT1-, OAT3-, OCT1-, and OCT2-interacting drugs, we attempted to address this issue by performing a systematic computational and statistical analysis, as well as machine-learning analyses, on the basis of the physicochemical descriptors of drugs known to interact with one or more of these four transporters. Since the crystal structures of the four transporters are unknown at this time, ligand-based computational chemistry approaches were used here. Among these, one commonly used method is the development of quantitative structure-activity relationship (SAR and QSAR) models, which attempt to identify the correlation between the activity, or binding affinity of ligands and transporters, and the values of the physicochemical descriptors of the ligands. Previously developed QSAR models for OAT1, OAT3, and OAT6, which were built on the basis of inputs of approximately 10 descriptors, identified several physicochemical properties of ligands important for interaction with transporters (Kaler et al., 2007; Truong et al., 2008). In addition to QSAR models, another approach that has gained popularity is the application of machine-learning tools. Among these tools, the support vector machine method has been used to develop models for two ATP binding cassette (ABC) transporters, breast cancer resistance protein, and P-glycoprotein (Wang et al., 2011; Hazai et al., 2013); these models were mainly used for the in silico prediction of new substrates. Besides the support vector machine method, other powerful machine-learning tools, such as decision trees and random forests, have been used widely for different applications (Vaglio Laurin et al., 2014; Kim et al., 2015).
To investigate the functional differences between OAT1, OAT3, OCT1, and OCT2, a number of machine-learning methods were applied to understand which physicochemical properties within a set of ∼250 drugs affect the interactions between individual transporters and their ligands, as well as the relative importance of these properties in contributing to selectivity for interaction with a particular transporter. To obtain clear results, the analysis relied heavily on inhibition (Ki) data, since these data are available for almost all of the drugs studied. Actual transport (Km) data are much more limited; in a recent review on modeling of drug transporters, it was pointed out that the limited transport data (as opposed to inhibition data) is a general issue in the field of SLC and ABC drug transporters (Matsson and Bergstrom, 2015). Likewise, although it is generally assumed that inhibition data indicates competitive inhibition of transport of a characteristic substrate (e.g., para-aminohippurate; triethanolamine), competitive versus noncompetitive inhibition is rarely formally evaluated [in fact, this is part of a “wish-list of developments needed in the field” (Matsson and Bergstrom, 2015)]. Nevertheless, it is worth noting that, for those drugs for which transport data were available, it was generally consistent with inhibition data (Supplemental Table 1). Moreover, many of the general classes of compounds (e.g., antivirals, diuretics, antibiotics, metformin, zwitterions) have been studied in the SLC22 transporter knockout animals or tissues derived from them, and altered handling by the kidney and other tissues has been demonstrated in many of these cases (Eraly et al., 2006; Vanwert et al., 2007, 2008; Truong et al., 2008; Vallon et al., 2008b, 2012; Nagle et al., 2011, 2013).
The results of the machine-learning analyses were further supported by the generation of pharmacophore models of OAT and OCT ligands. Pharmacophore modeling studies aim to find the common features shared among the ligands in three-dimensional space, and previous studies using this method have built several pharmacophore models for relatively small subsets of various ligands of renal transporters, including drugs and metabolites (Ahn et al., 2009; Kouznetsova et al., 2011; Wikoff et al., 2011; Duan et al., 2012). To gain a much more comprehensive understanding of binding interactions and ligand selections, it was necessary to construct pharmacophore models on the basis of as many pharmaceutical drugs as possible (in our case, 253). This provided a more comprehensive chemical space to build complete pharmacophore models for the transporters, because each individual compound contributes some information to the broader representation of the whole chemical space of binding.
Our results indicate that, in addition to charge-related factors, OATs interact with planar structures, whereas OCTs interact with more three-dimensional structures, indicating that, in addition to charge, the topology of ligands is another important factor. In addition, subtle but important differences exist between OAT1 and OAT3; OAT3 has a propensity to bind some cations that structurally overlap with OCT ligands. This was experimentally confirmed by wet laboratory transport assays of ligands predicted by virtual screening.
Materials and Methods
The overall computational workflow is shown in Supplemental Fig. 1.
Water-soluble probenecid was purchased from Molecular Probes (Eugene, OR). The fluorescent tracers, 5-carboxyfluorescein and 6-carboxyfluorescein, and cationic drugs (loperamide hydrochloride, nebivolol hydrochloride, darifenacin hydrobromide, paliperidone, cisapride monohydrate, and halofantrine hydrochloride) were purchased from Sigma-Aldrich (St. Louis, MO).
Selection and Classification of Drugs Interacting with OAT1, OAT3, OCT1, and/or OCT2.
A comprehensive literature and internet search was performed to compile a list of pharmaceutical drugs and tracers that interact with any of the four SLC22 transporters investigated in this study (Supplemental Table 1). Approximately 250 drugs were analyzed and the inhibition affinity (Ki) and/or the substrate affinity (Km) were used as a measurement of a given drug’s interaction with a given transporter. This “interaction affinity” classified drugs as either high-affinity (i.e., Km or Ki ≤ 100 μM), mid-affinity (i.e., Km or Ki > 100 μM, but ≤ 1000 μM), low-affinity (i.e., Km or Ki > 1000 μM, but ≤ 2000 μM), or extremely low affinity (i.e., Km or Ki > 2000 μM, but ≤ 12000 μM). Since these transporters share a great deal of similarity and can interact with the same compounds, albeit usually with different affinities, drugs interacting with two or more transporters with similar interaction affinities (i.e., both interact with high affinity) were excluded from the subsequent data mining analysis aimed at defining the physicochemical descriptors that separate OAT1, OAT3, OCT1, and OCT2.
The net charge states of the drugs at physiologic pH (i.e., 7.4) were then determined in the computational environment of ICM software (a commercially available computational chemistry software; Molsoft LLC, San Diego, CA). Drugs were considered “cationic” if their net charge was greater than zero, “anionic” if their net charge was less than zero, “neutral” if their net charge was equal to zero and they contained no charged atoms, or “zwitterionic neutral” if their net charge was zero but they contained an equal number of positively charged and negatively charged atoms. However, since it is possible for a drug to have more than one charged species coexisting at a given pH, the percentage of each charge species was determined using pH/concentration curves created using the chemicalize software (www.chemicalize.org; ChemAxon, Cambridge, MA), and the species percentages were calculated at three different pH values, 7.2, 7.4, and 7.6. Finally, total positive species percentage, total negative species percentage, total neutral species percentage, and total zwitterionic neutral species percentage were calculated for individual charge–species bar diagrams. The results were then plotted.
Collection and Preprocessing for Machine-Learning Analyses.
The pairwise comparison study employed in the machine-learning analyses was limited to nonoverlapping drugs (i.e., drugs which interacted with high affinity for only one transporter). The attributes to be compared in the machine-learning models were physicochemical properties of the drugs calculated using ICM (Molsoft), and tabulated in Konstanz information miner (KNIME), an open-source workflow platform for machine-learning (Beisken et al., 2013). Using ICM, about 50 physicochemical attributes of the drugs were calculated, including molecular quantum numbers, atom counts, bond counts, polarity counts, and topology counts. KNIME includes extensions capable of collecting data from three notable open source cheminformatics toolkits: RDKIT, Indigo, and CDK. Through the KNIME platform using RDKit, Indigo, and CDK, attributes were added to represent about 100 chemical features for each drug, such as molecular weight, molecular volume, Log P, Log S, polar surface area, etc. In addition to these physicochemical attributes, a class variable was also added to represent the transporter with which a given drug would interact.
After collecting the data, Weka, KNIME, and Excel were used to preprocess the data. Weka (cs.waikato.ac.nz/ml/weka) is an open source collection of machine-learning algorithms developed by the University of Waikato and is bundled together with tools for preprocessing data to make it more easily understood by the machine-learning algorithms. For example, the raw data extracted from KNIME and ICM contained some attributes that were overlapping, empty, or constant, and these were eliminated. The second step was to use Weka’s attribute-selection feature, Chi Square Evaluator, to rank the attributes according to their contribution to predicting the class variable. The Chi Square procedure is applied individually to each variable by first binarizing real-valued variables and then testing the expected-minus-observed counts with respect to the class, where the expected counts are assumed to be independent; larger counts result in a higher Chi Square statistic and suggest nonindependence (Agresti and Coull, 1996).
Machine-Learning Analyses.
After the data was compiled and preprocessed, machine-learning algorithms were employed to develop models. In this case, drugs that had a “high affinity” for the transporters OAT1, OAT3, OCT1, and OCT2 were treated as “instances,” and the physicochemical properties of the drugs were used as “attributes.” Six pairwise comparison studies were conducted: OAT1 versus OCT1, OAT1 versus OCT2, OAT3 versus OCT1, OAT3 versus OCT2, OAT1 versus OAT3, and OCT1 versus OCT2; as described above, in each comparison study, “overlapping” drugs, or ones that displayed high-affinity interaction with both of the transporters being compared, were eliminated from the analysis.
Several Weka machine-learning models were used: decision trees, decision rules, support vector machine, Bayesian models, and neural networks. Classification models that were well validated were obtained by several different techniques, but the preference was for those models that could help explain transporter binding/interaction data. For example, neural network models are a “black box” model, so, although they are not as useful as decision rule or decision tree models for defining distinguishing properties, they are still accurate classifiers. Comparable classification success rates (Supplemental Table 2) with several different algorithms demonstrate that there is a boundary related to transporter selectivity. Also of note, within a given model, depending on the features of the model selected, different decision trees were generated, probably owing in part to the overlap in molecular characteristics captured by various attributes. Multiple iterations of the algorithms and parameters were explored to arrive at models with the best validation scores.
In a decision tree, each node is a variable, and each branch represents a data split that depends on the value of the variable. An instance of the data determines a path down the tree, which ultimately leads to a leaf node that represents a class prediction. The decision tree is induced by ranking how well each variable can split the data at a decision node (starting with the root), splitting the data, and repeating the process for each branch. As the data gets split more and more, eventually each node mostly reflects one class or the other, and the branching stops. Typically, trees are induced and then lower levels are pruned back to improve performance in a cross-validation procedure. Compared with other techniques, a decision tree is more interpretable because the decisions are easily described.
A random forest is an ensemble of decision trees in which each tree is trained with different bootstrap samples (1000 in our case). The ensemble is averaged together to produce an aggregate classification. The trees are made slightly decorrelated by limiting the choice of variables during tree induction so that different combinations of variables can fill out the tree branches. An additional benefit of the bootstrap is that one can estimate the detrimental effect of variable permutations on predictions for each “left out of bag” sample. That effect is averaged and normalized over all trees, leading to a measure of variable importance. Because a decision tree is nonlinear in the way it partitions the input, the variable importance is potentially a measure of both interaction and main effects (Svetnik et al., 2003).
Statistical Analysis.
In addition to the machine-learning approach, statistical tests were used to study the significance of the calculated differences between ligand transporter interactions. In each of the pairwise comparison studies, t tests were performed on the physiologic properties to determine if the differences in the mean values for each were statistically significant between the two groups of drugs. Then, the physiologic properties were ranked according to their P values.
Creation of Pharmacophore Hypotheses.
Pharmacophore models were built in ICM, which performed clustering, alignment, and pharmacophore building on the basis of the atomic property field (APF) of the drug. APF considers the three-dimensional representation of atomic properties, such as hydrogen bond donors, hydrogen bond acceptors, SP2 hybridization, lipophilicity, size of large atoms, and positive and negative charges (Totrov, 2008). High affinity drugs were chosen as “actives.” Since the actives were diverse in their three-dimensional molecular structures, hierarchical clustering of actives on the basis of APF was first done to separate them into groups. Actives among the each group were then aligned, and a pharmacophore model was generated from the aligned drugs. To be included, each group needed to comprise a minimum of three drugs with dissimilarity score less than or equal to 0.25. The dissimilarity score is an indication of how similar two compounds are in APF and ranges from 0 to 1, where 0 = similarity and 1 = dissimilarity. Thus, clusters containing drugs that were too dissimilar would not be considered for pharmacophore model generation. APF properties were determined for each pharmacophore model using ICM, and the vectors of each APF property across all the models were added to calculate the total for that property. More extensive descriptions of this type of approach can be found elsewhere (Khan et al., 2012).
In Silico Screening and Uptake Assays.
Pharmacophore models were then used to virtually screen the DrugBank database with the ICM computational software. Some top hits were selected for further testing in an in vitro transport/uptake assay for interaction with selected transporters. Uptake assays, with probenecid serving as a negative control, were performed using Chinese hamster ovary cells constitutively expressing mouse Oat3 or Oat1, as previously described (Ahn et al., 2009; Wu et al., 2013, 2015; Zhu et al., 2015).
Results
The overall goal was to determine whether a formal systematic analysis of the physicochemical descriptors of drugs that interact with SLC22 transporters could: 1) identify properties, other than charge, that would help in predicting whether a ligand interacts with an OAT or OCT, and 2) uncover additional molecular properties of ligands predictive for interaction with prototypical members of these subfamilies (OAT1 versus OAT3 and OCT1 versus OCT2). A literature search identified a large number of pharmaceutical drugs and tracers with the ability to interact with OAT1, OAT3, OCT1, and OCT2 (i.e., 103, 105, 96, and 81, respectively) at all affinity levels (∼5 μM to 5 mM) (Supplemental Table 1); unless otherwise specified, machine-learning analysis, statistical analysis, and pharmacophore modeling were performed using drugs interacting with the transporters in the “high-affinity” range (i.e., ≤ 100 μM).
Because there is only limited direct transport data (Km) for these transporters compared with the amount of inhibition data (Ki), the analyses (Ki combined with Km) perforce is weighted toward inhibition data. The literature seems to assume competitive inhibition with a transported substrate (e.g., labeled para-aminohippurate or triethanolamine), but in nearly all cases the type of inhibition is not formally established by accepted biochemical criteria. This appears to be a general issue for most, if not all, solute carriers and ABC drug transporters (Matsson and Bergstrom, 2015). Nevertheless, we also carried out the decision tree analyses described below for those drugs with inhibition (Ki) data alone (excluding those drugs that had Km data), and generally similar results were obtained for these comparatively large datasets (Supplemental Figs. 2 and 3; Supplemental Table 3). We also tried to perform the analysis on the much smaller sets of drugs for which Km data were available; although a trend similar to the “Ki plus Km analysis” and the “Ki analysis” was often seen, there did not appear to be large enough samples to achieve clear results (Supplemental Fig. 4; Supplemental Table 3).
OAT3 Has Greater Capacity to Interact with Drugs of Positively-Charged Species and Zwitterionic-Neutral Species.
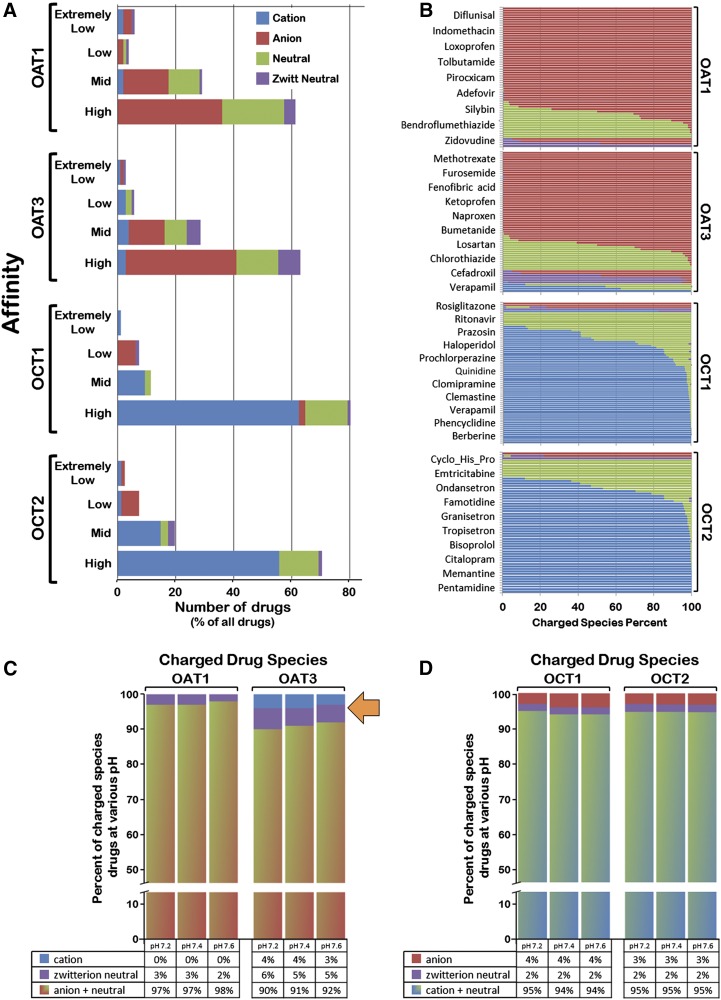
On the basis of the charge-species bar diagrams for individual transporters at pH 7.4 (Fig. 1, A and B), it was noted that the charged-species with which OAT1 and OAT3 mainly interacted were negatively charged (i.e., anionic), whereas OCT1 and OCT2 mainly interacted with positively charged species (i.e., cationic). Although the next most prevalent charged species with which both OATs and OCTs interacted were the neutral species, all four transporters interacted with zwitterionic-neutral species as well (Fig. 1). Notably, OAT3 (compared with OAT1) exhibited a greater ability to interact with zwitterionic-neutral species, as well as those with a charge opposite to that suggested by the name “organic anion transporter” (i.e., organic cations) (Fig. 1). At physiologic pH (i.e., pH 7.4), OAT1 does not interact with any positively charged species with high affinity; in contrast, OAT3 was able to interact with positively charged species at all affinities (which constituted 3.55% of species with which OAT3 interacts) (Fig. 1C). Both OCT1 and OCT2 interacted with negatively charged species, and the total negatively charged species percentages were 3.80% and 3.17%, respectively (Fig. 1D). Finally, the four transporters interacted with zwitterionic-neutral species to varying degrees; the total zwitterionic-neutral species percentages for OAT1, OAT3, OCT1, and OCT2 were the following: 2.75%, 5.44%, 1.78%, and 2.15%, respectively (Fig. 1). Thus, among these SLC22 transporters, OAT3 had the greatest ability to interact with zwitterionic-neutral species. To determine how well individual transporters interacted with “oppositely charged” and zwitterionic-neutral species together, we explored the total percentages of “oppositely charged” species percentage plus zwitterionic-neutral species percentage for each transporter (Fig. 1, C and D). Among the four transporters, OAT3 had a much higher total percentage than the rest of the transporters (the value for OAT3 was 8.98%, whereas the values for OAT1, OCT1, and OCT2 were 2.75%, 5.58%, and 5.33%, respectively) (Fig. 1). This began to suggest to us that, although OCT1 and OCT2 may be somewhat similar in their ligand specificities, OAT3 might be quite different than OAT1 especially with respect to the ability to interact with cations and zwitterions and may have more similarity (in terms of ligand preference) to OCTs than previously appreciated. This hypothesis was more formally explored in the studies below.
Fig. 1.
(A) The distribution of charge states for pharmaceuticals that interacted with each of the transporters at various binding affinity ranges. The charge states of the pharmaceuticals were defined by considering the number of positive charges and negative charges calculated in ICM at the environment of pH = 7.4. The charge states are colored according to the legend in the upper right-hand corner of the graph. (B) The charge-species composition diagrams for the transporters. The charge-species composition for individual pharmaceuticals was measured on the basis of the pH/concentration curves found in chemicalize.org (an online compound database supported by ChemAxon), which were then grouped according to the transporters with which the pharmaceuticals interacted. The diagrams could indicate the capability of the transporters to interact with various charge species. The charge states are colored as defined in the legend in A. (C, D) Summary of the total percentage of various charge species for each transporter on the basis of the results of charge-species composition diagrams. The potential capability of OAT3 to interact with positively charged and zwitterionic species (arrow) was thereby clarified.
Effect of pH on the Ability of Transporters to Interact with Charged and Zwitterionic Neutral Drugs.
In addition to analyzing pH 7.4, we explored how varying the pH of the solution in silico might change the composition of charged species with which each of the transporters interacted. At different pH levels the percent composition of charged species for drugs considered to be anionic, cationic, or zwitterionic at pH 7.4 would be expected to vary. In a more acidic environment, drugs would be protonated and contain more positively charged species, whereas in a more basic environment, drugs would be deprotonated and contain more negatively charged species. This would affect the percent composition of the charged species for a particular drug (anion/cation/zwitterion) with which individual transporters potentially interacted at a particular pH (Fig. 1, C and D). The sum of the positively charged species percentage and zwitterionic-neutral species percentage for drugs that interact with the organic anion transporters OAT1 and OAT3 increased as pH decreased, and the sum of negatively charged and zwitterionic species of the organic cation transporters, OCT1 and OCT2, increased when pH shifted toward the basic direction. In addition, it was found that OAT3-interacting drugs (compared with drugs interacting with OAT1, OCT1, and OCT2) probably changed most dramatically throughout the pH range of 7.2–7.6; when pH was either lowered or increased, the sum of the total positively charged and zwitterionic-neutral species for OAT3-interacting drugs changed from 8.31% to 9.93% as the pH was lowered from 7.6 to 7.2. In contrast, the sum of those values for OAT1, OCT1, and OCT2 changed minimally (Fig. 1, C and D).
Ligand Overlap between OAT1 and OAT3 and between OCT1 and OCT2.
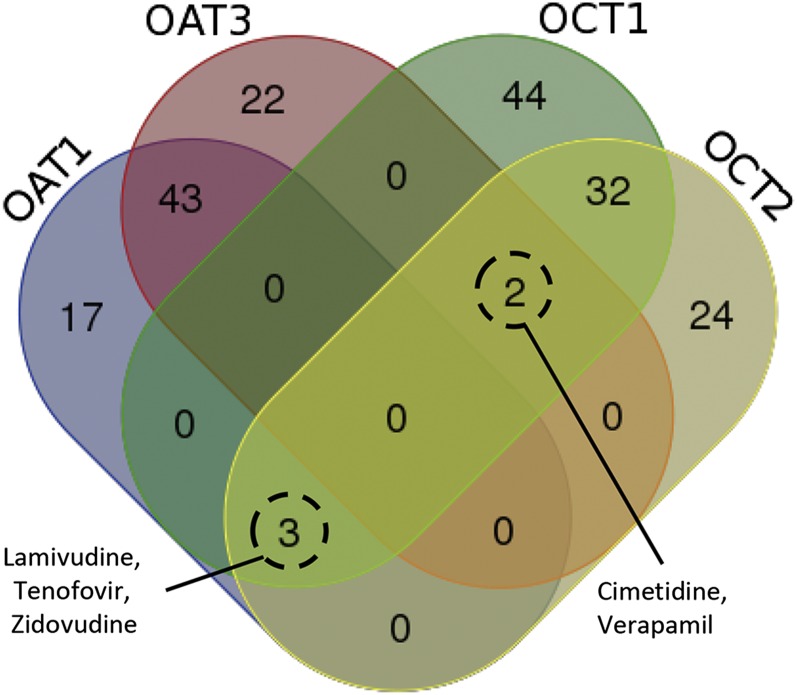
OAT1 and OAT3 were found to share a number of high-affinity ligands with ∼50% of the drugs showing affinities ≤100 μM for both organic anion transporters; likewise, OCT1 and OCT2 also shared many high-affinity ligands, with ∼35% of these drugs displaying high-affinity interactions for both organic cation transporters (Fig. 2). Comparisons of OAT high-affinity ligands with those of the OCTs revealed much less overlap, with only ∼1.8% of OAT1 and ∼1.2% of OAT3 high-affinity drugs also being able to interact with the organic cation transporters at affinities ≤100 μM (Fig. 2), which is consistent with known ligand differences between OATs and OCTs. To identify subtle differences in ligand specificity between transporters, the overlapping drugs (i.e., those interacting with two transporters with high affinity) were excluded from the subsequent data-mining analysis aimed at defining the physicochemical descriptors that separate OAT1, OAT3, OCT1, and OCT2.
Fig. 2.
Substrate overlap among transporters. The Venn diagram demonstrates the substrate specificity and substrate multispecificity between the transporters. Drugs found to be overlapping between the various transporters were excluded for the subsequent machine-learning analysis. Note: Although cimetidine and verapamil can bind OAT1 as well (Ahn et al., 2009), the affinity is roughly 10-fold less compared to OAT3.
Machine-Learning Analysis: Results of Exemplary Models for OAT1 versus OCT1.
We initially applied a number of machine-learning approaches. The overall results of applying classification algorithms using decision trees, neural networks, support vector machine, decision rules, and naïve Bayes for the comparison of OAT1 drugs and OCT1 drugs are presented in Supplemental Table 2. Although comparable results were generally obtained, decision trees developed using the J48 algorithm and random forest are discussed in detail for all the pairwise comparisons. Since in contrast to some of the other approaches, which are more like “black boxes” (e.g., neural networks), these models not only classified the data well, but provided a very logical way to demonstrate how physicochemical properties of the ligands affect the binding interaction between ligands and transporters. Of note, comparable classification success rates were obtained using different approaches (Supplemental Table 2), which suggests analyzable boundaries related to transporter selectivity.
Differences in Substrate Specificity Are More Probably between OAT1 and OAT3 Than between OCT1 and OCT2.
Table 1 shows the summary of weighted-average receiver operating characteristic (ROC) areas for the six decision tree models on the basis of high-affinity drugs when 10-fold cross-validation is performed. These were: OAT1/OCT1, OAT1/OCT2, OAT3/OCT1, OAT3/OCT2, OAT1/OAT3, and OCT1/OCT2. Most decision tree models were well validated, and only two trees had ROC areas less than 0.80, which were the trees for OAT1/OAT3 and OCT1/OCT2. This was probably owing to the fact that ligands for the two OATs and two OCTs were highly similar, and it is difficult to build a decision tree model to identify and predict differences. Nevertheless, the ROC areas for OAT1/OAT3 was 0.795, and that for OCT1/OCT2 was 0.639 (Table 1), indicating that the functional differences between OAT1 and OAT3 were more easily discriminated than those between OCT1 and OCT2. This is an important point for the analyses that follow.
TABLE 1 .
Weighted-average ROC areas: performance validation of various decision tree analyses
The table summarizes the results using 10-fold cross-validation of machine-learning decision tree models for: 1) high-affinity drugs (with affinity less than 100 μM), 2) high-affinity drugs without using charge as an attribute, and 3) mid-affinity drugs (with affinity between 100 and 1000 μM).
| Transporters Compared | High-Affinity Drugs |
Mid-Affinity Drugs |
||||
|---|---|---|---|---|---|---|
| Charge Included |
Charge Excluded |
Charge Included as an Attribute |
||||
| Correctly Classified | ROC Area | Correctly Classified | ROC Area | Correctly Classified | ROC Area | |
| OAT1/OCT1 | 86.57% | 0.905 | 80.60% | 0.823 | 82.50% | 0.874 |
| OAT1/OCT2 | 83.33% | 0.932 | 78.95% | 0.835 | 82.22% | 0.868 |
| OAT3/OCT1 | 86.33% | 0.880 | 77.70% | 0.764 | 80.00% | 0.880 |
| OAT3/OCT2 | 93.28% | 0.932 | 72.27% | 0.774 | 70.83% | 0.779 |
| OAT1/OAT3 | 69.77% | 0.795 | — | — | 86.37% | 0.722 |
| OCT1/OCT2a | 66.67% | 0.639 | — | — | 45.45% | 0.450 |
Note the poor results in the OCT1/OCT2 analysis are probably attributable to a small data set of six and five instances. —, results for OAT1/OAT3 and OCT1/OCT2 were inconclusive when charge was excluded. Please see text.
Substrate Preferences between OATs and OCTs Appear to Be Mostly Attributable to Charge.
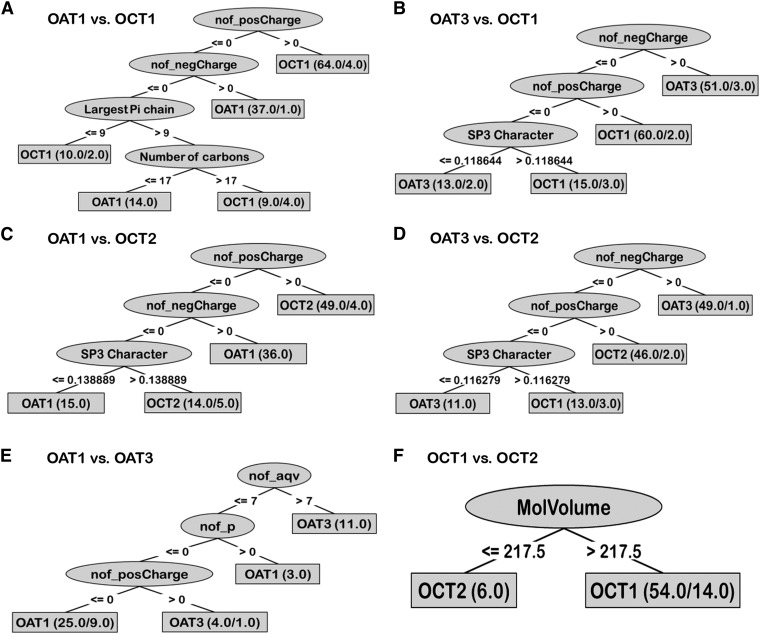
When an OAT was compared with an OCT in decision tree analysis, it was found that the first two physicochemical attributes that separated an OAT from an OCT were the number of negative (nof_negCharge) and positive charges (nof_posCharge) (Fig. 3). This is consistent with previous experimental data across mammalian species for many OAT1 and OCT1 ligands that include not only drugs, but also metabolites and toxins. Drugs that had the “number of negative charge greater than zero” were classified as OAT-interacting; in contrast, drugs that had the “number of positive charge greater than zero” interacted with OCTs (Fig. 3). Although this is compatible with the simple view that OATs transport anions and OCTs transport cations, as we describe elsewhere, a more complex picture emerged with further analysis. For example, even with pairwise comparisons, after charge, the next determinant attribute seen in most trees was SP3 character: Those drugs with greater SP3 values are classified as OCT drugs. This suggested that it was more probable that drugs with more three-dimensional and less planar character would turn out to be OCT ligands.
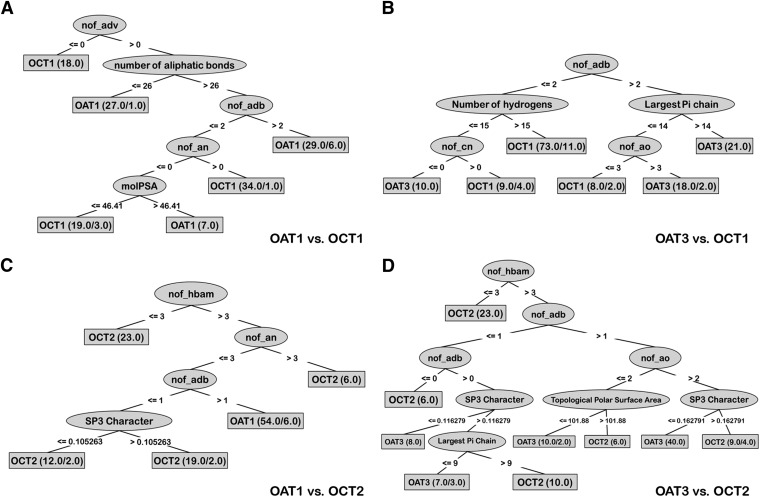
Fig. 3.
Decision trees generated on the basis of those drugs interacting with the transporters with high-affinity (i.e., ≤100 μM). The decision trees show that the main difference between OATs and OCTs are attributable to charge and charge-associated properties. Besides charge, the three-dimensionality versus planarity of the drug, indicated by SP3 character, was found to be another important factor in separating OAT and OCT drugs. In addition, some differences were found between two OATs, specifically in number of aqv, p, and posCharge (these attributes are further explained in the text).
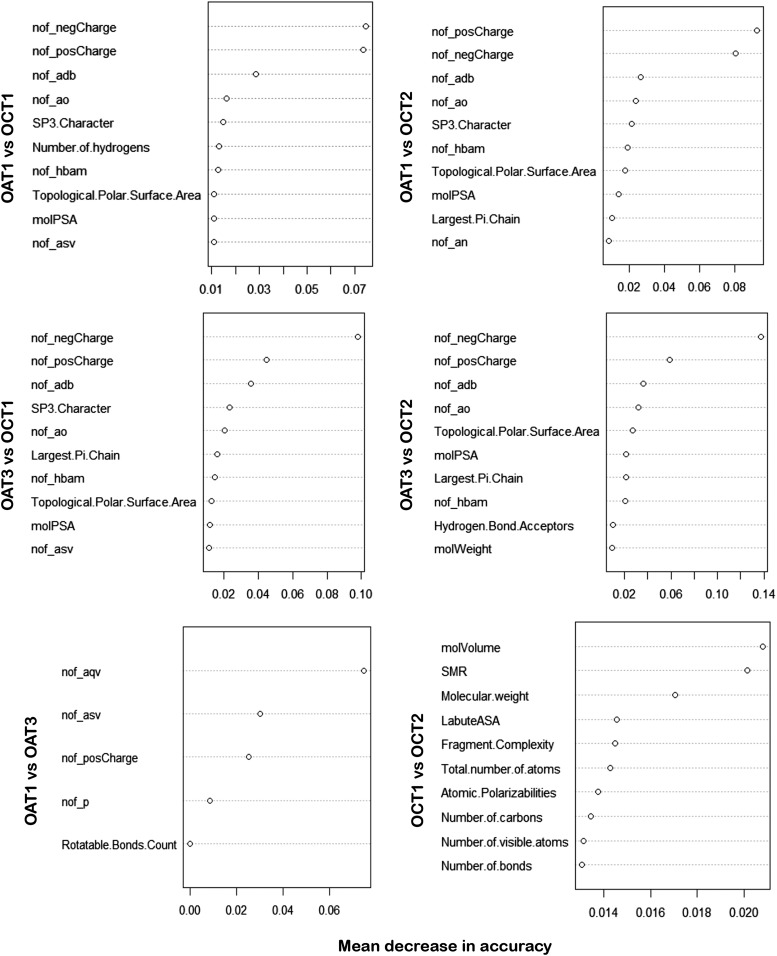
Random forest models were also used as an independent classification approach. In the variable importance plots derived from the random forest model for the pairwise OAT and OCT comparisons, the charge state information was also found to dominate in the ranking (Fig. 4). This supports the notion that the higher nodes in the decision tree are robustly important for classification across the bootstrap samples in the random forest. In addition, the variables found to be most important after the charge state were the number of acyclic double bonds (adb), acyclic oxygens (ao), followed by the “SP3 character.” After five or six variables, the importance levels drop off and little is gained by considering additional variables. For the pairwise comparison between OATs, the results also confirm and justify the decision tree interpretation. However, for OCT1 versus OCT 2, the results are not aligned, which is not surprising given that the classification performance is poor (Fig. 4).
Fig. 4.
Results on the basis of the random forest analyses. Variable importance plots showing comparisons for drug attributes predicting interaction with transporters. Importance of attributes are ranked from the upper right of the plot (most important) to the lower left (least important). As discussed in the text, these results are highly comparable to the results from decision trees.
Exclusion of Charge Reveals Potential Role of Physicochemical Properties Other Than Charge in Substrate Preference Differences.
The random forest models pointed to the potential role, in addition to charge, that other physicochemical features of the high-affinity drugs might play in separating OAT-interacting drugs from OCT-interacting drugs. Therefore, decision trees were constructed that excluded the properties of positive and negative charge (Fig. 5). The resulting trees split on a variety of other properties; the number of acyclic double bonds (“adb”), number of acyclic oxygens (“ao”), number of acyclic nitrogens (“an”), and the “SP3 character” were dominant.
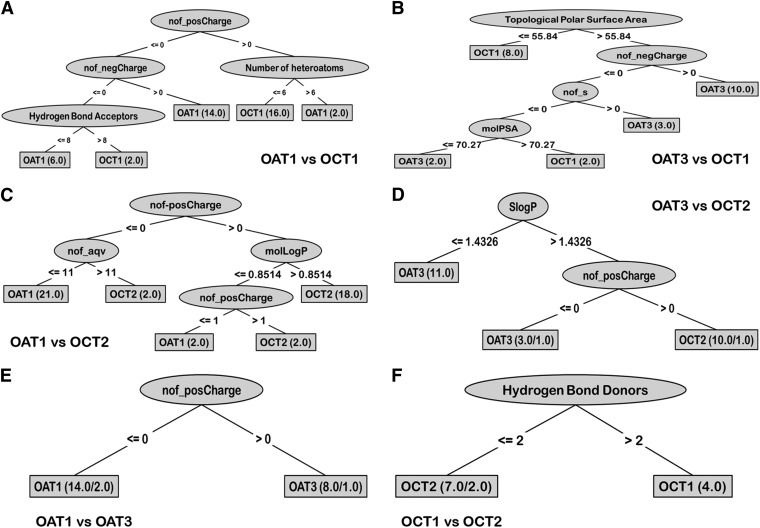
Fig. 5.
Decision trees excluding charge properties. The attributes of positive and negative charge were excluded in the building of the model so as to identify other important properties that potentially segregate OAT and OCT drugs. Again, it was found that some charge-associated attributes and the SP3 character were key determinants.
In the OAT1/OCT1 tree, the first attribute that split was “adv” (acyclic divalent nodes); drugs that had zero “adv” were classified as OCT1 drugs. The next attribute was “number of aliphatic bonds,” and drugs with the greater number of aliphatic bonds were classified as OCT1 drugs. When we examined the three other OAT versus OCT trees, they followed trends similar to the OAT1/OCT1 tree; OCT ligands generally had a higher number of “an” than OAT ligands, and OAT ligands had higher numbers of “adb” and “ao” than did OCT ligands. Interestingly, statistics from the accuracy of these decision tree models (which excluded charge) were not as strong as ones including charge but were still reasonable (Table 1). In addition, the attributes “ao,” “adb,” and “SP3 character” were confirmed as important attributes in the t test statistical analysis (below and Table 2).
TABLE 2 .
Pairwise comparisons of individual attributes for the four SLC22 transportersa
| Pairwise Comparison | Number of Attributes, by P Value |
Top Eight Attributes Ranked by P Valuea | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.005 | <0.01 | <0.05 | |||||||||
| OAT1 versus OCT1 | 27 | 29 | 37 | nof_posCharge (P = 5.95E-23) | nof_negCharge (P = 4.92E-13) | nof_adb (P = 2.4E-12) | nof_ao (P = 3.17E-09) | nof_hbam (P = 2.28E-08) | SP3 Character (P = 1.34E-07) | Number of hydrogens (P = 7.93E-07) | nof_asv (P = 1.11E-06) |
| OAT3 versus OCT1 | 17 | 18 | 23 | nof_negCharge (P = 1.10E-16) | nof_posCharge (P = 2.44E-15) | nof_adb (P = 4.98E-13) | nof_ao (P = 3.07E-10) | nof_hbam (P = 7.08E-09) | molPSA (P = 1.38E-06) | SP3 Character (P = 2.7E-06) | Topological polar surface area (P = 3.63E-06) |
| OAT1 versus OCT2 | 16 | 21 | 28 | nof_posCharge (P = 1.45E-19) | nof_negCharge (P = 1.31E-13) | nof_adb (P = 1.52E-11) | nof_ao (P = 1.15E-10) | nof_hbam (P = 1.08E-09) | SP3 Character (P = 4.59E-07) | molPSA (P = 8.03E-07) | nof_adv (P = 1.64E-06) |
| OAT3 versus OCT2 | 17 | 20 | 27 | nof_negCharge (P = 3.52E-17) | nof_posCharge (P = 8.12E-14) | nof_ao (P = 1.71E-11) | nof_adb (P = 3.24E-11) | nof_hbam (P = 1.27E-09) | Hydrogen Bond Acceptors (P = 1.03E-06) | molPSA (P = 1.32983E-06) | nof_adv (P = 1.66E-06) |
| OAT1 versus OAT3 | 1 | 2 | 18 | nof_aqv (P = 0.00257) | nof_posCharge (P = 0.00586) | nof_asv (P = 0.01339) | nof_asb (P = 0.01364) | nof_rbc (P = 0.01432) | molVolume (P = 0.01828) | Fragment Complexity (P = 0.01924) | Number of hydrogens (P = 0.01964) |
| OCT1 Versus OCT2 | 6 | 12 | 30 | nof_s (P = 0.00171) | molWeight (P = 0.00382) | LabuteASA (P = 0.00451) | Number of heavy atoms (P = 0.00495) | SMR (P = 0.00497) | nof_hac (P = 0.00514) | Vertex adjacency information magnitude (P = 0.00527) | nof_rings6 (P = 0.00545) |
The Student’s t tests calculated the P values for each attribute for each pairwise transporter comparison, and the level of significance is indicated. The results are found to be consistent with the results from the machine-learning analyses.
Pairwise Comparison between OAT1 and OAT3 Reveals Differences between the Two OATs.
When OAT1 and OAT3 were compared (Fig. 3E), the first attribute separating OAT1 and OAT3 ligands was the number of acyclic tetravalent nodes (“aqv”). Drugs that have the greater number of acyclic tetravalent nodes tended to be classified as interacting with OAT3. The next attribute separating the OAT ligands was the number of phosphorous atoms (“p”). Drugs that had at least one or more phosphorus atoms tended to be classified as OAT1-interacting. A third attribute that emerged from these comparisons of OAT1 and OAT3 ligands was the number of positive charges; drugs with a positive charge were associated with an OAT3 classification (Fig. 3E). (The aforementioned properties will be discussed in more detail below when we present wet laboratory support for the computational analysis.) In contrast to the comparison of the two OATs, the model generated for comparison of the two OCTs had poor validation performance; it appears that OCT ligands are too similar to be distinguished by the approaches we used; hence, the results for that decision tree model will not be discussed further.
Statistical Analysis Confirmed the Machine-Learning Analyses.
When performing t test analyses on individual attributes for each pairwise transporter comparison, we identified a number of attributes as statistically different between ligands interacting with each pair of transporters. The attributes that had the lowest P values for each comparison are summarized in Table 2 and are consistent with the machine-learning analyses. The two properties that had the lowest P values were the “number of positive charge” and the “number of negative charge,” corresponding to the results from the machine-learning analyses. After positive and negative charge, the next attributes that came out from the ranking were numbers of acyclic double bond (“adb”), acyclic oxygen (“ao”), hydrogen bond acceptor site (“hbam”), and SP3 character (Table 2). For the pairwise comparison of the two OATs, the two properties seen in the OAT1/OAT3 decision tree [i.e., the “number of acyclic tetravalent nodes” (“aqv”) and “number of positive charges”] were also found to have the lowest P values in the ranking. Again, the results from both decision trees and random forest are consistent with the statistical analysis.
Explanation of Properties Found to Be Relevant in Results.
As described above, on the basis of the results of machine-learning and statistical tests, we found that ligands of the OATs (either OAT1 or OAT3) generally had higher numbers of negative charge, acyclic double bonds, acyclic oxygen, and hydrogen bond acceptor sites than an OCT ligand (either OCT1 or OCT2). These properties tend to be associated with the anionic propensity. For example, most acyclic double bonds within the structures were in the forms of carbonyl (O=C), thial (S=C), sulfoxide (S=O), and the electronegative oxygen and sulfur within these double bonds are prominent hydrogen bond accepting sites. The “number of acyclic oxygen” is another property that expresses the anionic propensity, as the acyclic oxygen also serves as a potential hydrogen bond accepting site.
Importantly, in addition to having differences in properties associated with charges and ionization, ligands of OCTs and OATs are different in geometry-related properties, particularly with respect to the SP3 character value. SP3 character is defined as the number of SP3-hybridized carbons divided by the total number of atoms; it is one measure of the degree of three-dimensionality of a compound. If a drug has a higher SP3 character value, it is more three-dimensional; likewise, a lower SP3 character value is taken to imply that the drug is more planar (Lovering et al., 2009; Over et al., 2014). In machine-learning models and statistical analyses, drugs with a stronger affinity for the OCTs had a greater SP3 character value than those with a stronger affinity for the OATs, supporting the view that the “OCT-interacting drugs” are more three-dimensional than “OAT-interacting drugs.” As measured by SP3 character, compared with most other drugs in the data set, amantadine, nandrolone, and atropine are three OCT drugs that have highly three-dimensional structures, each with a SP3 character value of 0.357, 0.326, and 0.227, respectively. On the other hand, OAT drugs have much lower values of SP3 character, with none of the OAT drugs having SP3 character values greater than 0.300.
Some differences are also observed among the ligands of the two OATs; OAT3 tended to interact with drugs that have more acyclic tetravalent nodes and more positive charges, whereas OAT1 tended to interact with those that have more phosphorus atoms. An acyclic tetravalent node usually is composed of a carbon-forming tetravalent bond with four elements. In the decision tree model, 11 drugs were classified as OAT3 drugs from this node; among them were verapamil, pravastatin, enalapril, and methotrexate, and along with the higher number of acyclic tetravalent nodes, these drugs have longer and more hydrophobic chains. The next attribute separating OAT1 and OAT3 ligands was the number of phosphorous atoms (“p”). Drugs that had at least one or more phosphorus atoms were classified as interacting with OAT1; the three drugs in this category were cidofovir, tenofovir, and adefovir. The chemical structures of these drugs showed that the phosphorus atoms were in phosphate groups. Since the phosphate groups contain several oxygen atoms binding with phosphorus—some of which were deprotonated at the normal pH range—the phosphate group is highly anionic. Thus, the number of phosphorus atoms was directly correlated with the anionic propensity. In summary, even though both OAT1 and OAT3 were found to have functional overlap, there were some differences between their ligands identified in our analyses. OAT3 preferred to interact with drugs with more positive charge and long hydrophobic chains, and OAT1 ligands tended to be more anionic than OAT3.
Analysis of Mid-Affinity Drugs Supports the Results of High-Affinity Drugs.
Well described OAT ligands verified in vivo in knockouts include many compounds with an affinity greater than 100 μM (Eraly et al., 2006; Vallon et al., 2008a; Wikoff et al., 2011; Wu et al., 2013; Nigam, 2015; Nigam et al., 2015a,b). Thus, in addition to understanding the molecular interactions between transporters and drugs that bind with high affinity (≤100 μM), we also tried to study how OAT1, OAT3, OCT1, and OCT2 interact with drugs in the mid-affinity range (100–1000 μM). The decision trees constructed with mid-affinity drugs (Fig. 6) demonstrate that major factors involved in classifying a drug as an OAT or an OCT substrate were the result of charge, as in the high-affinity group. But the separation was less impressive than for the high-affinity (<100 μM) drugs. The decision tree comparing OAT1 and OAT3 in the mid-affinity range had only one node, which split on positive charge (Fig. 6). Drugs with a positive charge generally classified as OAT3-interacting.
Fig. 6.
The decision trees constructed with drugs that interact with the transporters at mid-affinity range (between 100 and 1000 μM). The trees show that, in the mid-affinity range, the main differences between OAT- and OCT-interacting drugs were still attributable to charges, but to a lesser degree than drugs in the high-affinity range.
Three-Dimensional Pharmacophore Models Showed Structural Similarities Corresponding to the Overlap in Functions for OATs and for OCTs.
Since it was found that OAT3 ligands also possessed some cationic characteristics, on the basis of the machine-learning analyses, pharmacophore models for OAT3-, OCT1-, and OCT2-interacting drugs were built to compare the functional similarities/differences between the OAT and the OCTs in three-dimensional space (Fig. 7). The models showed that OAT3 and OCTs interacted with drugs that had hydrophobic and aromatic centers. However, a slight difference in compound backbone appeared, as the hydrophobic chains for OCT1 and OCT2 models would sometimes enclose cationic spheres (seen in OCT1 pharmacophore models 3, 4, 5, and 6), which is not observed in most OAT3 models. Overall, models of OAT3-interacting ligands were more anionic, and models of OCT-interacting ligands were more cationic. This can also be seen from Table 3, which shows the quantitative measurements of the seven properties for individual models; as measured by the mean, the table shows that ligands of the OATs had higher “hydrogen bond acceptors” and higher “negative charges”; in contrast, ligands of the OCTs had higher “hydrogen bond donors” and higher “positive charges.”
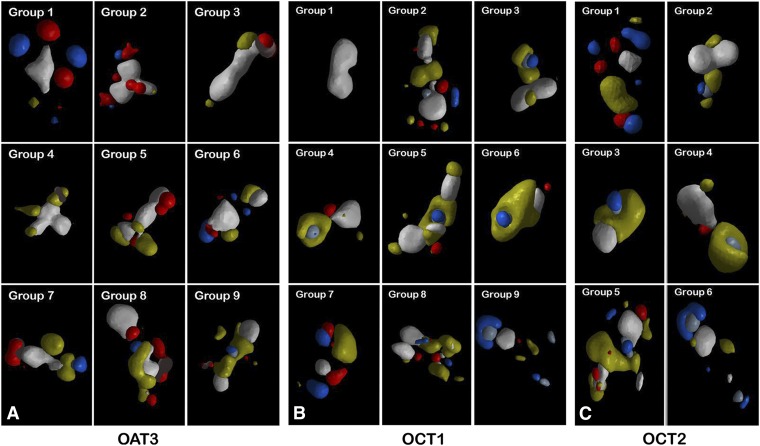
Fig. 7.
The pharmacophore models for OAT3, OCT1, and OCT2. Since the drugs interacting with each transporter were diverse in their three-dimensional structures, the drugs were clustered into groups on the basis of APF. The drugs within the same clustering groups were then aligned, and pharmacophore models for each group were created. In the pharmacophore models, different colors represent various APF properties: blue, hydrogen bond donor; red, hydrogen bond acceptor; white, aromaticity; yellow, hydrophobicity; light red, negative charges; light blue, positive charges. The OAT3 models and OCT1 models are found to be distinctive. With one notable exception, the OAT3 models for each group contained more characteristics of negative charges, electronegativity, and hydrogen bond acceptors, and vice versa for OCT1 models. However, the OAT3 model derived from group 9 was an exception as it contained several characteristics found largely in models from OCT groups.
TABLE 3 .
Quantitative APF property measurements
| Transporter | Pharmacophore Model | Hydrogen Bond |
Sp2 Hybridization | Lipophilic | Size (large) | Charge |
Electro |
Overall Space | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Donors | Acceptors | Positive | Negative | Positive | Negative | ||||||
| OAT3 | 1 | 143.204 | 154.117 | 839.695 | 101.509 | 213.651 | 0 | 0 | 219.724 | −117.089 | 47124 |
| 2 | 68.1087 | 484.381 | 1560.94 | 628.674 | 773.567 | 0 | −230.93 | 587.804 | −289.52 | 82173 | |
| 3 | 7.92777 | 175.642 | 1009.03 | 493.385 | 636.479 | 5.5641 | −193.954 | 529.816 | −102.394 | 80496 | |
| 4 | 88.2388 | 189.257 | 1441.67 | 925.382 | 1058.15 | 0 | −138.558 | 964.06 | −140.411 | 98000 | |
| 5 | 35.8698 | 387.069 | 1795.93 | 923.052 | 1148.9 | 0 | −197.94 | 987.326 | −151.581 | 120744 | |
| 6 | 220.122 | 169.155 | 894.646 | 580.978 | 656.921 | 0 | −38.4882 | 588.515 | −325.512 | 68894 | |
| 7 | 197.147 | 244.087 | 881.089 | 588.426 | 752.08 | 57.7324 | −173.197 | 773.25 | −176.682 | 66924 | |
| 8 | 123.888 | 427.116 | 1445.62 | 664.117 | 934.116 | 0 | −181.445 | 686.311 | −246.445 | 116550 | |
| >9a | 66.121 | 252.657 | 850.408 | 1164.04 | 1388.78 | 73.3208 | −175.956 | 1331.36 | −99.7383 | 149940 | |
| OCT1 | 1 | 48.1776 | 31.481 | 1056.59 | 570.64 | 704.827 | 102.635 | 0 | 823.36 | −23.6594 | 115248 |
| 2 | 90.1057 | 195.495 | 1020.17 | 716.752 | 937.245 | 46.1859 | 0 | 829.391 | −75.0204 | 92752 | |
| 3 | 89.3848 | 32.1592 | 845.243 | 850.424 | 951.403 | 110.846 | 0 | 997.719 | −20.1164 | 134160 | |
| 4 | 74.3945 | 95.9579 | 670.303 | 748.842 | 882.211 | 115.465 | 0 | 979.179 | −55.4599 | 93240 | |
| >5 | 80.1732 | 150.189 | 951.337 | 1241.55 | 1355.43 | 115.465 | 0 | 1323.47 | −66.1869 | 110124 | |
| >6 | 77.3553 | 68.1684 | 211.241 | 896.307 | 962.966 | 69.2789 | 0 | 1106.73 | −21.3618 | 80360 | |
| 7 | 216.391 | 229.452 | 556.192 | 339.69 | 495.407 | 17.6656 | −56.1539 | 503.882 | −61.267 | 62700 | |
| 8 | 193.59 | 217.192 | 1639.75 | 1511.09 | 1759.88 | 46.1859 | 0 | 1827.44 | −80.0704 | 164883 | |
| 9 | 554.411 | 58.0202 | 1109.79 | 377.711 | 472.479 | 346.394 | 0 | 887.07 | −48.0251 | 105408 | |
| OCT2 | 1 | 211.184 | 222.252 | 622.403 | 345.852 | 501.321 | 23.0927 | −69.2787 | 498.707 | −75.3364 | 115248 |
| 2 | 68.8115 | 29.6212 | 918.67 | 778.429 | 899.004 | 98.145 | 0 | 946.223 | −7.10932 | 92752 | |
| 3 | 101.393 | 47.913 | 287.96 | 825.327 | 881.816 | 79.382 | 0 | 992.614 | −23.1909 | 134160 | |
| 4 | 82.6908 | 86.1924 | 667.67 | 738.494 | 867.966 | 128.294 | 0 | 977.612 | −49.6171 | 93240 | |
| 5 | 133.952 | 214.602 | 1024.3 | 1186.25 | 1402.03 | 76.9765 | 0 | 1347.41 | −64.4698 | 110124 | |
| 6 | 502 | 39.793 | 1078.9 | 362.868 | 453.107 | 269.418 | 0 | 826.522 | −24.6827 | 80360 | |
OAT3 pharmacophore model 9 (arrowhead) was found to have higher value of positive charge and electropositive charge than the rest of OAT3 pharmacophore models. The APF property values of this model were also found to be comparable with several OCT1 models, such as OCT1 models 5 and 6 (arrowheads).
The Pharmacophore Models Revealed Structural Similarities between Ligands of OAT3 and OCT1.
Even though the majority of pharmacophore models for ligands of OAT3 had similar features, there was one clear exception, the pharmacophore model formed on the basis of group 9 for OAT3 (Fig. 7). Unlike other OAT ligand models, this model contained a hydrophobic chain that tended to enclose a sphere enriched with hydrogen bond donors and positive charges, which was a pattern shared among many OCT1 and OCT2 ligand models. Thus, this model (OAT3 pharmacophore model 9) was found to be very OCT-like, and the quantitative APF measurement of this model was found to have greater values of “positive charges” and “electropositive charges.”
Interestingly, the list of drugs used to construct this model from group 9 for OAT3 was found to be highly similar to the list of drugs that was independently separated on the basis of the first attribute or node in the OAT1/OAT3 decision tree (Fig. 3). Out of the nine drugs used to construct the pharmacophore model, six of them contained more than seven acyclic tetravalent nodes and were classified as “OAT3” drugs in the decision tree. This is important since it demonstrates that the results from the decision trees and the pharmacophore models identified the same differences found between ligands of OAT1 and OAT3, and the differences were attributable to the apparent capability of OAT3 to interact with OCT-like substrates.
Experimental Validation of In Silico Screening Results Identified New Cationic Drugs That Preferentially Interact with OAT3 But Not OAT1.
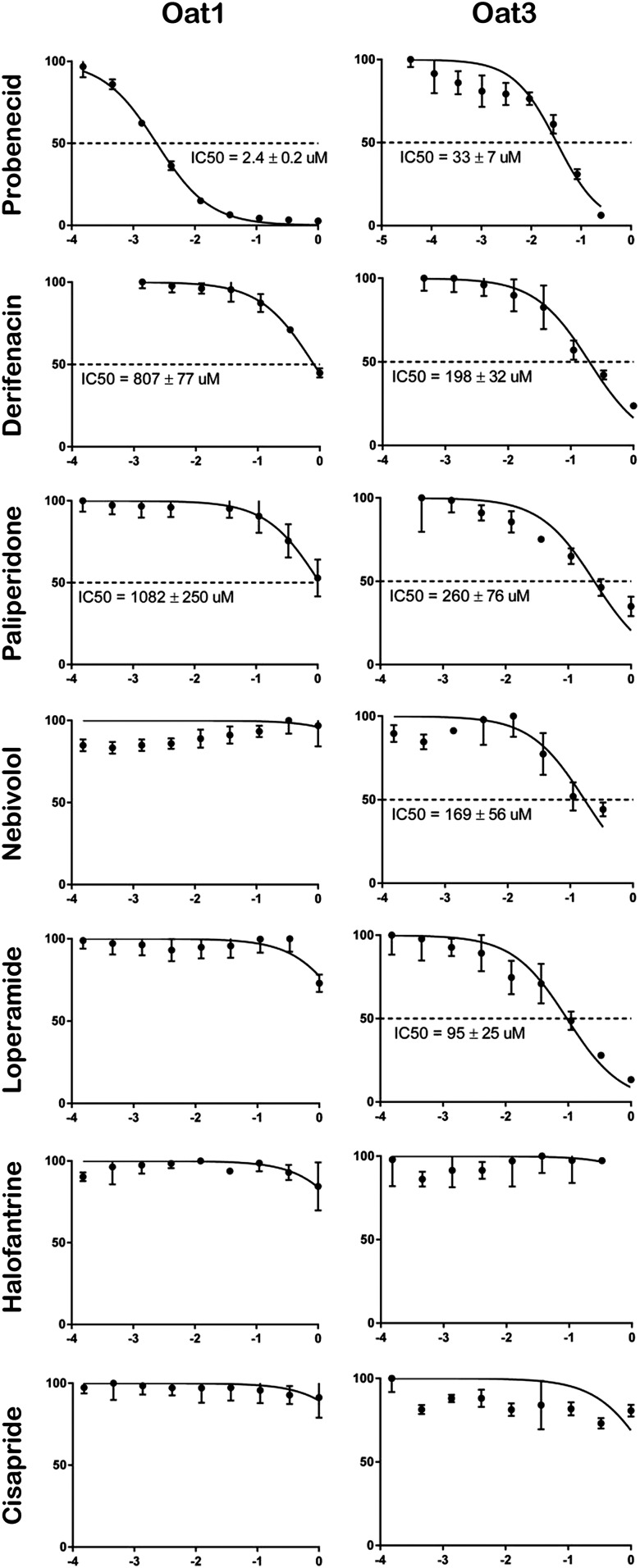
The finding that OAT3 prefers more cationic substrates than does OAT1 was thus consistent in decision tree and random forest analyses, and there was one (cationic) OAT3 pharmacophore model that was strikingly similar to OCT pharmacophore models. Thus, with the idea of trying to validate this experimentally, the OAT3 cationic model was used for virtual screening in silico. Using the pharmacophore model on the basis of group 9 of the OAT3 substrates, a virtual screen of the DrugBank database identified potential new OAT3 cationic ligands. Six top hits were selected for further wet laboratory validation. These hits were then tested for their ability to interact selectively with OAT3 using wet laboratory transport assays in OAT1-expressing or OAT3-expressing cells. Four of the ligands were found to interact with OAT3, with strong inhibition of tracer uptake. In marked contrast, when these six cationic drugs were tested in the OAT1 uptake assay, it was found that only two of them inhibited OAT1 function, and, importantly, with a much lower affinity (Fig. 8). The preference of these compounds for interaction with OAT3, but not OAT1, not only supports the validity of the pharmacophore model (model 9) but it is consistent with the machine-learning analysis indicating the capability of OAT3 to interact with cationic drugs. The measured IC50 values of tested compounds against OAT1 and OAT3 are summarized in Table 4.
Fig. 8.
Uptake inhibition assay on the basis of the virtual screening of the OAT3 pharmacophore model from group 9 (i.e., cationic pharmacophore) against the DrugBank database. For OAT1 inhibition assay, 10 μM 6-carboxyfluorescein was used as fluorescent tracer, and for OAT3 assay, 20 μM 5-carboxyfluorescein was used. Please see Materials and Methods and text for additional details.
TABLE 4 .
IC50 values of cationic drugs tested for interaction with OAT1 and OAT3
| Drug Name | IC50 (μM) |
|
|---|---|---|
| OAT1 | OAT3 | |
| Probenecida | 2.4 | 33 |
| Darifenacin | 807 | 198 |
| Paliperidone | 1082 | 260 |
| Loperamide | No significant inhibition | 95 |
| Nebivolol | No significant inhibition | 169 |
| Halofantrine | No inhibition | No inhibition |
| Cisapride | No inhibition | No inhibition |
The data for probenecid uptake inhibition is shown as a control.
Discussion
Recent knockout and in vitro data on a limited set of ligands suggest that the specificity of the OATs and OCTs of greatest clinical and pharmaceutical interest goes beyond whether the ligand is an anion or a cation (Ahn et al., 2009; Vallon, 2012; Nigam, 2015; Nigam et al., 2015a,b). Thus, molecular properties other than ligand charge need to be carefully addressed. To systemically examine this question, an extensive literature search was first done to build a complete-as-possible transporter-ligand database (nearly any compound found to interact with the transporters of interest was initially curated). Within this data (Supplemental Table 1), all drugs reliably known to interact with OAT1, OAT3, OCT1, and OCT2 were selected and used to study the functional differences and similarities between the transporters by applying machine-learning tools. Among the machine-learning tools (which included neural nets, support vector machines, and other methods as shown in Supplemental Table 2), decision trees and random forests were more helpful from the viewpoint of understanding this question of substrate specificity as opposed to simply fitting data (Figs. 3–6).
The results of the decision tree analyses were in agreement with the results of the random forest, and these results were further verified by conventional statistical tests (Table 2). The results indicated that, although the main difference between the ligand preferences of OATs and OCTs (with respect to physicochemical descriptors) was charge, the structure of ligands also affected the interaction with the transporters. Thus, in considering factors beyond charge, OCTs interacted with more three-dimensional structures (more SP3 character), whereas OATs interacted with planar compounds (Figs. 3–5). This may imply that the binding pockets of OCTs accommodate fewer planar compounds than those of OATs, which is worthy of further investigation once crystal structures of these transporters become available (Koepsell, 2013; Matsson and Bergstrom, 2015; Nigam et al., 2015a).
In addition to finding differences between OATs and OCTs, some differences among the submembers of these families were also identified. On the basis of machine-learning models and pharmacophore models, OAT1 and OAT3 were found to be different in that the latter possesses some ability to interact with cations, making it more functionally similar to OCT1 and OCT2 in this respect (Figs. 3 and 5). Among high-affinity drugs (<100 mM Km, Ki, or IC50), OAT3 could interact with ligands with more diverse structures (per machine-learning analysis of physicochemical descriptors and pharmacophore analysis) than did OAT1, again implying that OAT3 has different binding pockets than OAT1 and supporting the importance of obtaining structures for both transporters.
On the basis of the pharmacophore OAT3/OCT1 (Fig. 7) overlay, OAT3 binding pockets could have similarity to binding pockets of the OCTs, enabling OAT3 to bind some ligands with cationic characteristics. Our studies indicate that, although OAT1, OAT3, OCT1, and OCT2 are “multispecific” (or “polyspecific”), this multispecificity (polyspecificity) is restricted, and the actual interaction of each transporter with their ligands goes beyond conventional views about charge. This is our main finding, supported by machine-learning analysis, pharmacophore modeling, and wet laboratory transport assays. In particular, OAT3 stands out. OAT3 has overlapping ligands with OAT1, and like OAT1 it has a preference for planar anionic molecules, but OAT3 also accepts larger ligands and more cationic/zwitterionic ones—including those that might conventionally be viewed as OCT substrates. We support this conclusion with wet laboratory data using an OAT3 transport assay indicating that cationic drugs not previously reported (as far as we know) to be ligands indeed interact with OAT3. Together, the computational and wet laboratory analyses indicate that the boundary that separates OATs and OCTs is not as clear as the current literature suggests.
Thus, finding the differences and similarities between the transporters with respect to ligand preference can help to predict and identify new compounds that interact with the transporter (as we have done here), since the set of rules defined by decision trees can be further used for in-silico screening of new ligands/inhibitors (drugs, toxins, metabolites, signaling molecules). These rules can also be used to design new, potent, selective ligands that can target a particular transporter. These could be drugs that are aimed at targeting a particular tissue or body fluid, or alternatively, selective inhibitors of transport.
Expression of varying levels of OAT1, OAT3, OCT1, and OCT2 may thus help the cell alter the net ligand (drugs, toxins, metabolites, signaling molecules) taken up by kidney, liver, and other tissues in nonobvious ways. The potential relevance of this concept to normal physiology and pathophysiological states has been discussed in the Remote Sensing and Signaling Hypothesis (Kaler et al., 2006; Ahn and Nigam, 2009; Wu et al., 2011; Nigam, 2015; Nigam et al., 2015a). Our results should also be useful for predicting potential drug-drug interactions and drug-metabolite interactions.
As discussed throughout this article, the study may be somewhat limited owing to paucity of direct transport data and the reliance on inhibition data. As indicated in a recent review addressing ligand-based modeling of SLC and ABC drug transporters, the limited transport data available is an issue for the whole field (Matsson and Bergstrom, 2015); even with inhibition data, competitive versus noncompetitive inhibition is also generally not addressed, although the former is often assumed (Matsson and Bergstrom, 2015). However, at least for the drugs studied here the limited transport data were quite consistent with binding data. In support of this notion, one can also consider in vivo studies in the Oat and Oct knockout animals. A number of general classes of organic anion, organ cation, and organic zwitterion compounds analyzed here (e.g., antivirals, antibiotics, diuretics, metformin, zwitterions) have also been evaluated in the Oat1, Oat3, Oct1, and Oct2 knockout animals or in knockout tissues, and abnormalities in handling of these compounds consistent with inhibition affinities have been demonstrated (Eraly et al., 2006; Vanwert et al., 2007, 2008; Truong et al., 2008; Vallon et al., 2008b, 2012; Nagle et al., 2011, 2013). Indeed, the knockout data even seems to support the preference of Oat3 (compared with Oat1) for zwitterions such as creatinine (Vallon et al., 2012). Nevertheless, caution about relying entirely on inhibition data seems appropriate as there may be cases where high-affinity binding to transporters such as Oct1 may not necessarily correspond to physiologically relevant transport (He et al., 2016).
As discussed above, we also performed decision tree analyses on the set of drugs that had inhibition (Ki) data (not including drugs with transport data as indicated by Km values); in this analysis, results were obtained similar to those from the larger dataset consisting of both Ki and Km data (Supplemental Fig. 2). In addition, we attempted to obtain reliable decision trees for the considerably smaller set of compounds for which transport (Km) data had been found (Supplemental Fig. 3). Although similar trends (to the Ki plus Km decision trees) were found in some cases, clear, consistent, and significant results could not be generally obtained with this limited set of compounds with Km values. This again highlights the need for the field to obtain transport data for all the drugs and, with respect to inhibition data, the need to distinguish competitive from noncompetitive inhibition (Matsson and Bergstrom, 2015). In addition, it can be argued that ligand-based modeling for multispecific SLC drug transporters, which handle structurally diverse compounds, might be more difficult than for transporters that handle a single class of structurally similar compounds (Matsson and Bergstrom, 2015). This is one reason we believed it was reasonable to use as large a dataset as possible, despite the limitations described above—an approach that was partly experimentally validated. As more transport and other biochemical data becomes available, and as machine-learning and other data science approaches continue to improve, it may be possible to obtain an even clearer picture of the chemical features of drugs that enable transport by one or another SLC and/or ABC transporter.
Abbreviations
- ABC
ATP binding cassette
- APF
atomic property field
- KNIME
Konstanz information miner
- OAT
organic anion transporter
- OCT
organic cation transporter
- QSAR
quantitative structure activity relationship
- ROC
receiver operating characteristic
- SLC
solute carrier
- Weka
Waikato environment for knowledge analysis
Authorship Contributions
Participated in research design: Nigam, Liu, Goldenberg.
Conducted experiments: Liu, Goldenberg, Wu, Chen, Lun.
Performed data analysis: Liu, Goldenberg, Rodriguez, Wu.
Contributed new reagents or analytic tools: Balac, Rodriguez, Abagyan.
Wrote or contributed to the writing of the manuscript: Liu, Goldenberg, Bush, Nigam.
Footnotes
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM098449, R01-GM104098], and by the National Institutes of Health Grants Eunice Kennedy Shriver National Institute of Child Health and Human Development [U54-HD07160]. The authors declare the following competing financial interests: Ruben Abagyan has significant financial interest in Molsoft LLC, San Diego, CA, or its products.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Agresti A, Coull BA. (1996) Order-restricted tests for stratified comparisons of binomial proportions. Biometrics 52:1103–1111. [PubMed] [Google Scholar]
- Ahn SY, Eraly SA, Tsigelny I, Nigam SK. (2009) Interaction of organic cations with organic anion transporters. J Biol Chem 284:31422–31430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SY, Jamshidi N, Mo ML, Wu W, Eraly SA, Dnyanmote A, Bush KT, Gallegos TF, Sweet DH, Palsson BO, et al. (2011) Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J Biol Chem 286:31522–31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SY, Nigam SK. (2009) Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol Pharmacol 76:481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisken S, Meinl T, Wiswedel B, de Figueiredo LF, Berthold M, Steinbeck C. (2013) KNIME-CDK: Workflow-driven cheminformatics. BMC Bioinformatics 14:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P, Li S, Ai N, Hu L, Welsh WJ, You G. (2012) Potent inhibitors of human organic anion transporters 1 and 3 from clinical drug libraries: discovery and molecular characterization. Mol Pharm 9:3340–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami Riedmaier A, Nies AT, Schaeffeler E, Schwab M. (2012) Organic anion transporters and their implications in pharmacotherapy. Pharmacol Rev 64:421–449. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Monte JC, Nigam SK. (2004) Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol Genomics 18:12–24. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, Nagle M, Monte JC, Rieg T, Truong DM, Long JM, et al. (2006) Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J Biol Chem 281:5072–5083. [DOI] [PubMed] [Google Scholar]
- Hazai E, Hazai I, Ragueneau-Majlessi I, Chung SP, Bikadi Z, Mao Q. (2013) Predicting substrates of the human breast cancer resistance protein using a support vector machine method. BMC Bioinformatics 14:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Garza D, Nigam SK, Chang G. (2016) Multispecific Organic Cation Transporter 1 (OCT1) from Bos taurus Has High Affinity and Slow Binding Kinetics towards Prostaglandin E2. PLoS One 11:e0152969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y, Murayama N, Okudaira N, Kurihara A, Okazaki O, Izumi T, Inoue K, Yuasa H, Kusuhara H, Sugiyama Y. (2011) Prediction of fluoroquinolone-induced elevation in serum creatinine levels: a case of drug-endogenous substance interaction involving the inhibition of renal secretion. Clin Pharmacol Ther 89:81–88. [DOI] [PubMed] [Google Scholar]
- Kaler G, Truong DM, Khandelwal A, Nagle M, Eraly SA, Swaan PW, Nigam SK. (2007) Structural variation governs substrate specificity for organic anion transporter (OAT) homologs. Potential remote sensing by OAT family members. J Biol Chem 282:23841–23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaler G, Truong DM, Sweeney DE, Logan DW, Nagle M, Wu W, Eraly SA, Nigam SK. (2006) Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem Biophys Res Commun 351:872–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, Wuxiuer Y, Sylte I. (2012) Binding modes and pharmacophore modelling of thermolysin inhibitors. Mini Rev Med Chem 12:515–533. [DOI] [PubMed] [Google Scholar]
- Kim SP, Gupta D, Israni AK, Kasiske BL. (2015) Accept/decline decision module for the liver simulated allocation model. Health Care Manage Sci 18:35–57. [DOI] [PubMed] [Google Scholar]
- Koepsell H. (2013) The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med 34:413–435. [DOI] [PubMed] [Google Scholar]
- Kouznetsova VL, Tsigelny IF, Nagle MA, Nigam SK. (2011) Elucidation of common pharmacophores from analysis of targeted metabolites transported by the multispecific drug transporter-Organic anion transporter1 (Oat1). Bioorg Med Chem 19:3320–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Nieto CE, You G, Bush KT, Barros EJ, Beier DR, Nigam SK. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J Biol Chem 272:6471–6478. [DOI] [PubMed] [Google Scholar]
- Lovering F, Kirincich S, Wang W, Combs K, Resnick L, Sabalski JE, Butera J, Liu J, Parris K, Telliez JB. (2009) Identification and SAR of squarate inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2). Bioorg Med Chem 17:3342–3351. [DOI] [PubMed] [Google Scholar]
- Matsson P, Bergström CA. (2015) Computational modeling to predict the functions and impact of drug transporters. In Silico Pharmacol 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle MA, Truong DM, Dnyanmote AV, Ahn SY, Eraly SA, Wu W, Nigam SK. (2011) Analysis of three-dimensional systems for developing and mature kidneys clarifies the role of OAT1 and OAT3 in antiviral handling. J Biol Chem 286:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle MA, Wu W, Eraly SA, Nigam SK. (2013) Organic anion transport pathways in antiviral handling in choroid plexus in Oat1 (Slc22a6) and Oat3 (Slc22a8) deficient tissue. Neurosci Lett 534:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK. (2015) What do drug transporters really do? Nat Rev Drug Discov 14:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK, Bush KT, Bhatnagar V. (2007) Drug and toxicant handling by the OAT organic anion transporters in the kidney and other tissues. Nat Clin Pract Nephrol 3:443–448. [DOI] [PubMed] [Google Scholar]
- Nigam SK, Bush KT, Martovetsky G, Ahn SY, Liu HC, Richard E, Bhatnagar V, Wu W. (2015a) The organic anion transporter (OAT) family: a systems biology perspective. Physiol Rev 95:83–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SK, Wu W, Bush KT, Hoenig MP, Blantz RC, Bhatnagar V. (2015b) Handling of Drugs, Metabolites, and Uremic Toxins by Kidney Proximal Tubule Drug Transporters. Clin J Am Soc Nephrol 10:2039–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Over B, McCarren P, Artursson P, Foley M, Giordanetto F, Grönberg G, Hilgendorf C, Lee MD, 4th, Matsson P, Muncipinto G, et al. (2014) Impact of stereospecific intramolecular hydrogen bonding on cell permeability and physicochemical properties. J Med Chem 57:2746–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Gorboulev V, Müller TD, Gorbunov D, Shatskaya N, Koepsell H. (2005) Amino acids critical for substrate affinity of rat organic cation transporter 1 line the substrate binding region in a model derived from the tertiary structure of lactose permease. Mol Pharmacol 67:1600–1611. [DOI] [PubMed] [Google Scholar]
- Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. (2003) Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci 43:1947–1958. [DOI] [PubMed] [Google Scholar]
- Totrov M. (2008) Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des 71:15–27. [DOI] [PubMed] [Google Scholar]
- Truong DM, Kaler G, Khandelwal A, Swaan PW, Nigam SK. (2008) Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J Biol Chem 283:8654–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaglio Laurin G, Cheung-Wai Chan J, Chen Q, Lindsell JA, Coomes DA, Guerriero L, Del Frate F, Miglietta F, Valentini R. (2014) Biodiversity mapping in a tropical West African forest with airborne hyperspectral data. PLoS One 9:e97910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon V, Eraly SA, Rao SR, Gerasimova M, Rose M, Nagle M, Anzai N, Smith T, Sharma K, Nigam SK, et al. (2012) A role for the organic anion transporter OAT3 in renal creatinine secretion in mice. Am J Physiol Renal Physiol 302:F1293–F1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon V, Eraly SA, Wikoff WR, Rieg T, Kaler G, Truong DM, Ahn SY, Mahapatra NR, Mahata SK, Gangoiti JA, et al. (2008a) Organic anion transporter 3 contributes to the regulation of blood pressure. J Am Soc Nephrol 19:1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon V, Rieg T, Ahn SY, Wu W, Eraly SA, Nigam SK. (2008b) Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am J Physiol Renal Physiol 294:F867–F873. [DOI] [PubMed] [Google Scholar]
- Vanwert AL, Bailey RM, Sweet DH. (2007) Organic anion transporter 3 (Oat3/Slc22a8) knockout mice exhibit altered clearance and distribution of penicillin G. Am J Physiol Renal Physiol 293:F1332–F1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwert AL, Srimaroeng C, Sweet DH. (2008) Organic anion transporter 3 (oat3/slc22a8) interacts with carboxyfluoroquinolones, and deletion increases systemic exposure to ciprofloxacin. Mol Pharmacol 74:122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chen Y, Liang H, Bender A, Glen RC, Yan A. (2011) P-glycoprotein substrate models using support vector machines based on a comprehensive data set. J Chem Inf Model 51:1447–1456. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. (2011) Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J Proteome Res 10:2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Baker ME, Eraly SA, Bush KT, Nigam SK. (2009) Analysis of a large cluster of SLC22 transporter genes, including novel USTs, reveals species-specific amplification of subsets of family members. Physiol Genomics 38:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Bush KT, Liu HC, Zhu C, Abagyan R, Nigam SK. (2015) Shared Ligands Between Organic Anion Transporters (OAT1 and OAT6) and Odorant Receptors. Drug Metab Dispos 43:1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Dnyanmote AV, Nigam SK. (2011) Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol Pharmacol 79:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Jamshidi N, Eraly SA, Liu HC, Bush KT, Palsson BO, Nigam SK. (2013) Multispecific drug transporter Slc22a8 (Oat3) regulates multiple metabolic and signaling pathways. Drug Metab Dispos 41:1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Nigam KB, Date RC, Bush KT, Springer SA, Saier MH, Jr, Wu W, Nigam SK. (2015) Evolutionary analysis and classification of OATs, OCTs, OCTNs, and other SLC22 transporters: structure-function implications and analysis of sequence motifs. PLoS One 10:e0140569. [DOI] [PMC free article] [PubMed] [Google Scholar]