Abstract
The interaction of Regulator of G protein Signaling 4 (RGS4) with the rat mu opioid receptor (MOR)/G protein complex was investigated. Solubilized MOR from rat brain membranes was immunoprecipitated in the presence of RGS4 with antibodies against the N‐terminus of MOR (anti‐MOR10–70). Activation of MOR with [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO) during immunoprecipitation caused a 150% increase in Goα and a 50% increase in RGS4 in the pellet. When 10 μM GTP was included with DAMGO, there was an additional 72% increase in RGS4 co‐immunoprecipitating with MOR (p = 0.003). Guanosine 5′‐O‐(3‐thiotriphosphate) (GTPγS) increased the amount of co‐precipitating RGS4 by 93% (compared to DAMGO alone, p = 0.008), and the inclusion of GTPγS caused the ratio of MOR to RGS4 to be 1 : 1 (31 fmoles : 28 fmoles, respectively). GTPγS also increased the association of endogenous RGS4 with MOR. In His6RGS4/Ni2+‐NTA agarose pull down experiments, 0.3 μM GTPγS tripled the binding of Goα to His6RGS4, whereas the addition of 100 μM GDP blocked this effect. Importantly, activation of solubilized MOR with DAMGO in the presence of 100 μM GDP and 0.3 μM GTPγS increased Goα binding to His6RGS4/Ni2+‐NTA agarose (p = 0.001).
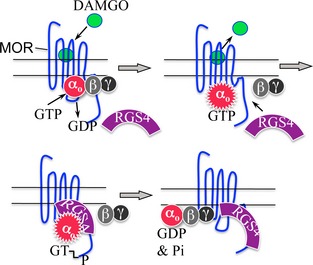
Regulators of G protein Signaling (RGS) shorten the time that G proteins are active. Activation of the mu opioid receptor (MOR) causes GTP to bind to and to activate Go (αoβγ). RGS4 then binds to the activated αo‐GTP/MOR complex and accelerates the intrinsic GTPase of αo. After αo dissociates from MOR, RGS4 remains bound to the C‐terminal region of MOR.
Keywords: adenylyl cyclase, cell signaling, G protein–coupled receptor, immunoprecipitation, opiate opioid, regulator of G protein signaling
Abbreviations used
- anti‐MOR10–70
antiserum against the N‐terminal region of MOR
- anti‐MOR349–398
antigen‐affinity purified antibody against the C terminal 50 amino acids of MOR
- anti‐RGS4170–205
antigen‐affinity purified antibody against the C terminal 36 amino acids of RGS4
- buffer A
50 mM Hepes (pH 7.4), 100 mM NaCl, 2 mM Tris.EDTA, 1 mM Tris.EGTA, 5 mM MgCl2, protease inhibitors, and phosphatase inhibitors (Sigma P0044)
- CHAPS
3‐[(3‐cholamidopropyl)‐dimethylammonio]‐1‐propanesulfonate
- DAMGO
[D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin
- GST‐RGS4170–205
glutathione‐S‐transferase fused to the C terminal 36 amino acids of RGS4
- MOR
mu opioid receptor
- RGS4
regulator of G protein signaling 4
- SHSY5Y cells
human neuroblastoma cell line expressing endogenous mu opioid receptors
- TEM buffer
50 mM Tris.HCl (pH 7.2), 2 mM EDTA, 1 mM EGTA, 5 mM MgCl2
Regulators of G protein signaling (RGS) accelerate the inactivation of G proteins that have been activated by their associated receptors by increasing the rate of GTP hydrolysis by the intrinsic GTPase of G proteins (Neubig and Siderovski 2002). Since the discovery of RGS proteins, attempts have been made to identify which RGS proteins modulate the signaling of known G protein–coupled receptors (GPCRs) and to determine the sequence of events that triggers the RGS protein's association with its GPCR and its cognate G proteins (Dohlman et al. 1995; Dohlman and Thorner 1997). This study investigates the interaction of RGS4 with the endogenous rat brain mu opioid receptor (MOR) and Go, one of its cognate G proteins.
The MOR is expressed in many regions of the brain and spinal cord and is essential for natural opiate reward and analgesia (Matthes et al. 1996; Cui et al. 2014). Activation of MOR by endogenous enkephalins or exogenous opiates leads to suppression of neuronal activities. At the molecular level, MOR is associated with Gi/o‐type G proteins that mediate inhibition of adenylyl cyclase activity, the opening of inwardly rectifying K+ channels and the closure of voltage‐gated Ca2+ channels (North 1986; Gosse et al. 1989; Chen and Yu 1994; Chalecka‐Franaszek et al. 2000). The activation of MOR results in the release of GDP from the α‐subunits of Gi and Go, and this allows GTP to bind to and activate these α subunits (Gilman 1987). The α‐GTP and the βγ subunits dissociate from the receptor and act on downstream effector systems. Signal termination is achieved by hydrolysis of GTP to GDP by an intrinsic GTPase of the α subunit of Gi and Go, and the rate of GTP hydrolysis is accelerated ≥ 40‐fold by the interaction of RGS proteins with α‐GTP (Berman et al. 1996a). Thus, the association of an RGS protein with a receptor shortens the time that the receptor's G proteins are active.
Multiple RGS proteins, including RGS2, RGS4, RGS6, RGS7, RGS8, RGS9‐2, RGS11, RGS19, and RGS20 have been proposed to regulate MOR signaling (Neubig and Siderovski 2002; Xie et al. 2007; Talbot et al. 2010; Psigfogeorgou et al. 2011; Traynor 2012; Wang and Traynor 2013). RGS9‐2, and to a lesser extent RGS4, appear to modulate the nociception pathway, whereas RGS4 modulates the reward pathway (Zachariou et al. 2003; Wang et al. 2009; Han et al. 2010). RGS4 also opposes morphine‐induced physical dependence in the locus coeruleus. In addition, RGS4 and RGS8 are enriched in the thalamus, a region of the brain where MOR modulates the relay of pain signals to the sensory cortex (Gold et al. 1997). A number of studies have demonstrated that exogenous RGS4 can attenuate MOR signaling (Xie et al. 2007; Talbot et al. 2010). Understanding the processes leading to the association of RGS proteins with MOR, and determining which RGS proteins modulate MOR functioning in the nociception pathway versus the reward pathway, may reveal targets for drugs that can reduce the influence of opiates on the reward pathway and enhance the influence of opiates as anti‐nociceptive agents.
It is not entirely clear if RGS4 can bind to endogenous MOR (and its cognate G proteins), if RGS4 is constitutively bound to MOR, or if activation of MOR can influence the binding of RGS4 to the MOR/G protein complex. Limited evidence suggests that GPCRs, like MOR, may play a role in attracting RGS proteins to their cognate G proteins (Bernstein et al. 2004; Hague et al. 2005; Georgoussi et al. 2006). In a study performed in a heterologous system, it was found that HA‐tagged RGS4 bound to myc‐tagged MOR in the presence of AMF (100 μM AlCl3, 2 mM MgCl2, 100 mM NaF) and that HA‐tagged RGS4 could bind to a peptide identical to the 70 amino acids of the C‐terminal region of MOR (MOR329–398). These authors also demonstrated membrane translocation of RGS4 following MOR activation in HEK293 cells (Leontiadis et al. 2009).
In this study, we investigated, in depth, the interactions among endogenous rat brain MOR, endogenous G proteins and recombinant and endogenous RGS4. Our ability to solubilize MOR in a functionally active state allowed us to investigate the roles of [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO), a MOR agonist, and guanine nucleotides in attracting RGS4 to the MOR/G protein complex. By immunoprecipitating MOR with an antibody directed against the N‐terminus of MOR, we were able to minimize any potential interference of the antibody with interactions among MOR, G proteins, and other proteins. This study demonstrates, for the first time, that GTP is required for maximal DAMGO‐stimulated RGS4 binding to the MOR/G protein complex. Furthermore, we were able to demonstrate that DAMGO activation of soluble MOR can stimulate the binding of Goα‐GTPγS to His6RGS4. In functional assays, RGS4 was found to cause a concentration‐dependent, non‐competitive attenuation of MOR‐mediated inhibition of adenylyl cyclase activity but did not affect MOR‐stimulated binding of GTPγS to G proteins.
Methods
Animals
See Supporting Information
Drugs and cell culture
[D‐Ala2, N‐Me‐Phe4, gly‐ol] enkephalin (DAMGO) was purchased from Sigma, St. Louis, MO, USA; [3H]cAMP from NEN Life Sciences, and [3H]DAMGO from Amersham Pharmacia (Piscataway, NJ, USA). [35S]GTPγS was obtained from Perkin Elmer Life Sciences (Boston, MA, USA).
Preparation of recombinant RGS4
His6RGS4 was expressed in JM109 E. coli, and His6RGS4 was extracted (in the presence of protease inhibitors, Sigma P 8849) on Ni2+‐NTA agarose resin. See Supporting Information Methods for details.
MOR and RGS4 Antibodies
See Supporting Information Methods for details.
Solubilization of rat brain membranes
Rat brain membranes were solubilized as previously described (Weems et al. 1996; Chalecka‐Franaszek et al. 2000). See Supporting Information for details.
MOR immunoprecipitation
Three polyclonal anti‐MOR antibodies were capable of immunoprecipitating MOR. Anti‐MOR340–398, anti‐MOR10–70, and anti‐MOR349–398 were bound to Protein A Sepharose resin and covalently cross‐linked to the resin with 20 mM dimethylpimelimidate (Chalecka‐Franaszek et al. 2000). A summary table of these antibodies and their potential uses is presented in Table S1.
Immunoblots
Routinely, immunoblots of material immunoprecipitated with anti‐MOR10–70 were cut into three portions: the portion from 90 to 52 kDa was used to detect MOR (a smear running between ~54 and 70 kDa); the middle portion of the blot between 52 and 31 kDa was used to screen for Goα (~40 kDa); and the bottom third of the blot between 31 and 12 kDa was used to screen for RGS4 (~26 kDa). See Supporting Information for details.
Detection of [3H]DAMGO binding activity in immunoprecipitated material
See Supporting Information.
His6RGS4/Ni2+‐NTA agarose pull‐down experiments
See Supporting Information for details. SHSY5Y cell cultures and adenylyl cyclase assays were performed as described in Supporting Information.
SHSY5Y cell cultures and adenylyl cyclase assays were performed
See Supporting Information for details.
Statistical analyses
The software ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/;, 1997–2014) was used to quantify the densities of protein bands on film that had been exposed to immunoblots. In experiments involving the IP of MOR with anti‐MOR10–70, the density of each band of co‐precipitated protein (Goα or RGS4) was divided by the density of the MOR band in the same lane (see Fig. 2). Also, the density of each Goα band was divided by the density of the RGS4 band in the same lane for comparisons in the Ni2+‐NTA/His6RGS4 pull down assay (see Fig. 4). Quantification of unknown amounts of RGS4 that co‐immunoprecipitated with MOR was accomplished by interpolation of nonlinear regression curves generated from known standard amounts of RGS4 that were processed with the unknown samples (see Fig. 1b). All statistical and curve‐fitting analyses were performed using PRISM v5.0 for Macintosh OS X (GraphPad Software, San Diego, CA, USA). Non‐linear regression analysis was used to determine the best fit of full concentration‐effect curves for adenylyl cyclase activity. The EC50 values and maximal effects were determined from best‐fit analyses. [3H]DAMGO receptor binding was analyzed using a one‐site binding hyperbola model. All data are expressed as mean ± standard error of the mean.
Figure 2.
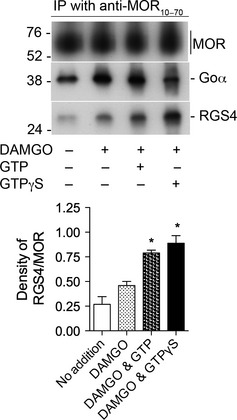
Activation of MOR with [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO) stimulates the association of RGS4 with MOR in a GTP‐dependent manner. Upper panel. Rat brain membranes were solubilized and processed as described in Methods (except in this experiment, intact membranes were not treated with DAMGO or guanine nucleotides prior to their solubilization). Soluble aliquots were incubated with 0.3 μM RGS4 alone, 0.3 μM RGS4, and 1 μM DAMGO, 0.3 μM RGS4, 1 μM DAMGO and 10 μM GTP or 0.3 μM RGS4, 1 μM DAMGO, and 1 μM GTPγS. Material immunoprecipitated by anti‐MOR10–70 was subjected to SDS–PAGE, and proteins were transferred to a nitrocellulose membrane. The top section of the blot was incubated with anti‐MOR349–398, the middle section with anti‐Goα, and the lower portion with anti‐RGS4. The densities of all bands were determined with ImageJ. Lower Panel. The experiment depicted in the upper panel was repeated two additional times. In each experiment, the density of each RGS4 band was divided by the density of the corresponding MOR band. The values of RGS4/MOR for each test group are displayed in the bar graph as mean ± SEM, n = 3. Of particular note, there was a 72% increase in the amount of RGS4 associated with MOR in the presence of DAMGO and GTP vs. DAMGO alone (unpaired, two‐tailed t‐test, * indicates p < 0.008). There was no difference in the densities of MOR among the four groups (two‐way anova, p = 0.60). There were differences in the levels of Goα among the groups: inclusion of DAMGO increased the amount of Goα (as was seen in Figure S2c) although the addition of GTP to DAMGO did not alter the amount of Goα associated with MOR (unpaired, two‐tailed t‐test, p = 0.79); and in six independent experiments, the addition of GTPγS decreased the association of Goα with MOR by 57 ± 7% (p = 0.02). These data are summarized in Table 1.
Figure 4.
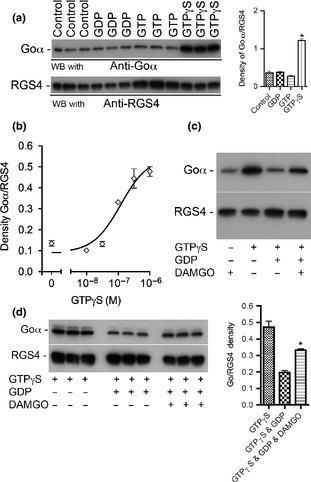
[D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO) stimulates GTPγS binding to Goα in solubilized rat brain membranes and causes GTPγS‐activated Goα to bind to RGS4. Solubilized rat brain membranes were used in these experiments (see Methods). (a) Guanine nucleotides were tested for their ability to activate Goα and to cause activated Goα to bind to 0.3 μM His6RGS4 pre‐bound to N2+‐NTA agarose. Neither 100 μM GDP nor 10 μM GTP affected Goα binding to RGS4. However, 1 μM GTPγS caused a 3‐fold increase in the density of the Goα band [right side of (a) unpaired, two‐tailed t‐test, p = 0.001 (the ratios of Goα/RGS4 in each lane were analyzed)]. In four independent experiments similar to that shown in (a), the addition of 1 μM GTPγS caused a 2.8‐fold increase in the amount of Goα bound to RGS4 (unpaired, two‐tailed t‐test, * indicates p = 0.0044). (b) GTPγS caused a concentration‐dependent increase in the binding of Goα to RGS4 (n = 3). The EC50 of GTPγS was 1.2 × 10−7 M. (c) The addition of 0.3 μM GTPγS caused an approximate 3‐fold increase in Goα binding to RGS4 whereas the combination of 0.3 μM GTPγS and 100 μM GDP caused Goα binding to RGS4 to return to the control level. However, when 1 μM DAMGO was added along with 0.3 μM GTPγS and 100 μM GDP, there was an increase in the binding of Goα to RGS4. (d) To confirm that activation of MOR with DAMGO could decrease the affinity of Goα for GDP and allow GTPγS to activate Goα, the experiment depicted in (c) was performed again in triplicate. Activation of MOR with DAMGO caused a 70% increase in the amount of Goα binding to RGS4 [(d) right side bar graph: GTPγS & GDP vs. GTPγS, GDP & DAMGO, unpaired, two‐tailed t‐test, * indicates p = 0.001].
Figure 1.
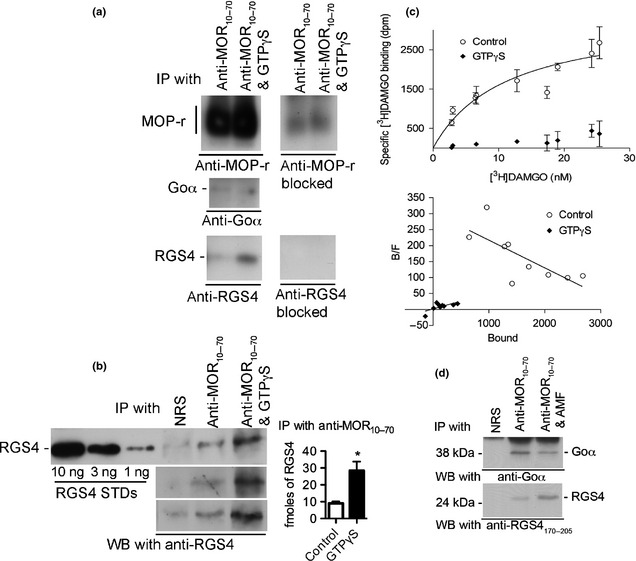
Activation of G proteins with GTPγS enhances the association of RGS4 with MOR. (a) MOR was immunoprecipitated with anti‐MOR10–70 in the presence of either 1 μM [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO) and 0.3 μM RGS4 or 1 μM DAMGO, 0.3 μM RGS4 and 1 μM GTPγS. The immunoprecipitate was subjected to SDS–PAGE (12% gel) and immunoblots were screened with the following antibodies: anti‐MOR349–398 (top panel), anti‐Goα (second panel from the top), and anti‐RGS4170–205 (third panel from the top). The inclusion of the appropriate blocking peptides selectively blocked recognition of MOR and RGS4 [right side of (a)]. All westerns in A were performed on a single nitrocellulose blot that was cut into sections. (b) The experiment shown in (a) was repeated three additional times. Anti‐RGS4170–205 detected RGS4 that was co‐immunoprecipated with MOR by anti‐MOR10–70 in the absence or presence of 1 μM GTPγS in three independent experiments. RGS4 standards (10, 3, and 1 ng) were included in each experiment. Values of co‐immunoprecipitated RGS4 were determined by extrapolation using nonlinear regression analysis of known RGS4 standards (Prism v5). In four independent experiments the amounts of RGS4 immunoprecipitated in the absence and in the presence of GTPγS were 9 ± 1.1 and 28.4 ± 5.4 fmoles, respectively [unpaired, two‐tailed t‐test, * indicates p = 0.013, see bar graph in right side of (b)]. (c) [3H]DAMGO saturation binding to MOR immunoprecipitated with anti‐MOR10–70 was used to quantify MOR. Aliquots of the immunoprecipitate identical to the aliquots that were loaded on to the gels depicted in (a) and (b), were used to measure [3H]DAMGO saturation binding (see Methods). Non‐specific binding was subtracted from total binding to calculate specific binding (open circles). Specific binding ranged from 80% of total binding at the lowest concentration of [3H]DAMGO to 55% of total binding at the highest concentration of [3H]DAMGO. Samples were incubated in triplicate. Data shown were combined from two independent experiments. Analysis of the data by non‐linear regression indicated that the Bmax was 3400 dpm of [3H]DAMGO (31.0 fmoles); the 95% confidence interval was 2240 dpm to 4559 dpm (20.4 fmoles to 41 fmoles of MO). The KD was determined to be 11 nM with a 95% confidence interval of 2.1–20 nM. The lower panel shows a Scatchard transformation of the data. (d) The inclusion of AMF (100 μM AlCl3, 2 mM MgCl2 and 100 mM NaF) during the immunoprecipitation with anti‐MOR10–70 also caused an increase in the co‐immunoprecipitated RGS4.
Results
Quantification of MOR and RGS4
Experiments were performed to determine if RGS4 associates with the MOR/G protein complex and, if it does, to determine the stoichiometry of MOR and RGS4 under condition where no guanine nucleotide was present or where 0.3 μM GTPγS was present. Rat brain membranes were incubated for 10 min at 30°C with 1 μM DAMGO (or 10 μM morphine instead of DAMGO when [3H]DAMGO binding was to be determined, Fig. 1c) and 0.3 μM His6RGS4, a concentration of RGS4 that had a near maximal effect in attenuating MOR signaling (see Figure S5). The rat brain membranes were solubilized, and solubilized MOR was immunoprecipitated with anti‐MOR10–70 as described in Methods. To determine the ratio of RGS4 to MOR, one portion of the immunoprecipitated material was used to quantify the amount of RGS4 by western blotting (Fig. 1b) and another portion was used to determine saturation [3H]DAMGO binding (Fig. 1c). GTPγS (1 μM) was included in some samples subjected to the immunoprecipitation procedure to determine if activation of the G proteins would affect His6RGS4 binding to the MOR/G‐protein complex (Fig. 1a and b). Interestingly, when 1 μM GTPγS was present, there was a 3‐fold increase in the amount of His6RGS4 co‐immunoprecipitating with MOR in spite of the fact that the amount of Goα was reduced by 57% (paired, two‐tailed t‐test, p = 0.02). In the absence of GTPγS, 9.0 ± 1.1 fmoles of RGS4 were co‐immunoprecipitated with MOR, whereas in the presence of 1 μM GTPγS, 28.4 ± 5.4 fmoles of RGS4 were co‐immunoprecipitated with MOR (Fig. 1b, bar graph). In four independent experiments, GTPγS caused a 3‐fold increase in the amount of RGS4 co‐immunoprecipitated with MOR (unpaired t‐test, p = 0.0128). The saturation [3H]DAMGO binding assay (Fig. 1c) indicated that the equivalent of 31 fmoles of MOR were present in the material that was loaded on each lane of the SDS gels shown in Fig. 1b. Thus, when G proteins were fully activated (with 1 μM GTPγS, the same concentration that maximally activates Go in inhibiting adenylyl cyclase activity, see Figure S6b), the ratio of MOR to RGS4 was approximately 1–1. The amounts of RGS4 co‐immunoprecipitated with MOR were determined by interpolation of the densities of the unknown bands from non‐linear regression curves generated from known His6RGS4 standards run in the same gel as the co‐immunoprecipitated His6RGS4 samples. The densities of bands were determined using the software ImageJ, and the curves were generated using the program Prism® v5.0. Also, the inclusion of AMF (100 μM AlCl3, 2 mM MgCl2 and 100 mM NaF) during the immunoprecipitation procedure resulted in an increase in His6RGS4 (Fig. 1d).
DAMGO‐stimulated binding of RGS4 to MOR
As activation of Goα with GTPγS increased the amount of RGS4 bound to the MOR/Goα complex, the influence of DAMGO and guanine nucleotides on the association of Goα and RGS4 with the MOR/G protein complex was further investigated. As is evident in Figure S2c, DAMGO alone caused an approximate 2‐fold increase in the association of Goα with MOR. In five independent experiments, the inclusion of DAMGO during the IP process increased the amount of Goα by 2.5‐fold (unpaired two‐tailed t‐test, p = 0.03). The inclusion of DAMGO also increased the amount of RGS4 co‐immunoprecipitating with MOR by 53% (Fig. 2). Interestingly, the combination of DAMGO and GTP resulted in an additional 71.6% increase in the amount of RGS4 that was co‐immunoprecipitated by anti‐MOR10–70 (unpaired two‐tailed t‐test, p = 0.003). As was shown in Fig. 1, the inclusion of GTPγS along with DAMGO during the IP process also significantly increased the amount of RGS4 co‐immunoprecipitated with the MOR/Go complex by 93% versus DAMGO alone (unpaired, two‐tailed t‐test, p = 0.008). The changes in Goα and RGS4 that resulted from the addition of DAMGO alone, or DAMGO in the presence of either GTP or GTPγS are shown in Table 1. In statistical analyses, the density of each RGS4 band was divided by the density of the corresponding MOR band (detected from approximately 54–70 kDa) to correct for any differences in sample loading. This experiment demonstrates that activation of MOR increases the association of RGS4 with the MOR/G protein complex in a GTP‐dependent manner.
Table 1.
Relative changes in the levels of Goα and RGS4 that co‐immunoprecipitated with MOR as a result of the presence of 1 μM DAMGO alone or 1 μM DAMGO in combination with either 10 μM GTP or 1 μM GTPγS
| Goα, % | RGS4, % | |
|---|---|---|
| No addition | 100 | 100 |
| DAMGO | 256 | 150 |
| DAMGO + GTP | 225 | 256 |
| DAMGO + GTPγS | 110 | 289 |
The relative amounts of Goα and RGS4 that were co‐immunoprecipitated with MOR in the absence of agonist, or in the presence of DAMGO alone, or DAMGO in combination with either GTP or GTPγS are shown. The values of Goα and RGS4 in the absence of either DAMGO or guanine nucleotides were set at 100%. Activation of MOR with DAMGO increases the association of Goα and RGS4 with MOR by 156% and 50%, respectively. Activation of MOR with DAMGO in the presence of 10 μM GTP did not significantly affect the amount of Goα associated with MOR but increased the amount of RGS4 associated with MOR by an additional 72% (vs. DAMGO alone). The combination of DAMGO & GTPγS caused a decrease in the level of Goα by 56% (vs. DAMGO alone) while causing an increase in the amount of RGS4 by 93% (vs. DAMGO alone). The data were compiled from 19 independent experiments where the densities of bands were quantified using ImageJ. Values of Goα and RGS4 were adjusted for the amount of MOR loaded in each lane of each gel.
As anti‐RGS4170–205 has not previously been characterized, we compared it to ABT17 (a.k.a. U1079, anti‐RGS41–205 from Millipore), an antibody known to be effective for detecting endogenous RGS4 from brain membranes (Krumins et al. 2004; Schwendt and McGinty 2007). In this experiment, the concentrations of protease and phosphatase inhibitors were increased (doubled) to improve the chances that ABT17 would detect endogenous RGS4. In this experiment, solubilized membranes were incubated with His6RGS4 and DAMGO and immunoprecipitated with anti‐MOR10–70 in the absence or presence of GTPγS. Both RGS4 antibodies not only detected an increase in His6RGS4 binding to MOR in the presence of GTPγS, but both antibodies detected an increase in the binding of endogenous RGS4 (lower band) in the presence of GTPγS (Fig. 3a). When the experiment was repeated with the addition of 10 μM MG132 but without any exogenous His6RGS4, only the lower molecular weight form of RGS4 (the endogenous form) was detected in the immunoprecipitate (Fig. 3b). An over‐exposure of the same blot reveals that both RGS4 antibodies recognized endogenous RGS4 in brain membranes (Figure S3b).
Figure 3.
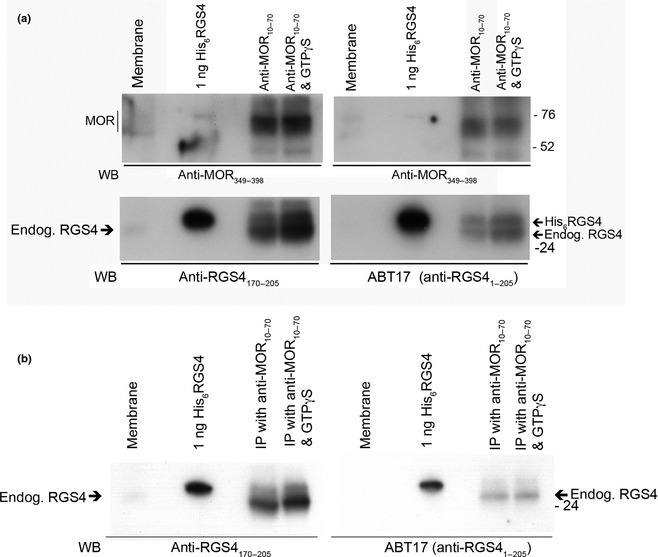
Comparison of anti‐RGS4170–205 to anti‐RGS41–205 A. In an experiment similar to the one depicted in Fig. 1a, solubilizes MOR was immunoprecipitated either in the presence of [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO) and 1 μM His6RGS4 or in the presence of DAMGO, 1 μM His6RGS4 plus 0.3 μM GTPγS. However, in an attempt to minimize proteolysis of endogenous RGS4, the routine concentrations of protease and phosphatase inhibitors were doubled, and the material immunoprecipitated by anti‐MOR10–70 was screened with anti‐RGS4170–205 (a.k.a. 1554) and anti‐RGS41–205 (a.k.a. ABT17 or U1079, an antibody capable of recognizing endogenous RGS4). The upper left and upper right panels were screened with 0.2 μg/mL of anti‐MOR349–398 to ensure the presence of equal amounts of MOR. The lower left and lower right panels were screened for the presence of RGS4 with 2 μg/mL of anti‐RGS4170–205 and a 1 : 2000 dilution of antiserum ABT17, respectively. Arrows to the right of lower panel mark the locations of His6RGS4 and endogenous RGS4. The inclusion of GTPγS appeared to increase the association of both His6RGS4 and endogenous RGS4 (lower band) with MOR. This experiment was repeated one time with nearly identical results. Endogenous RGS4 was also detected in brain membranes by both anti‐RGS4 antibodies. (b) When the same experiment depicted in A was repeated, but in the absence of exogenous RGS4, only the lower mol. weight band of RGS4 was detected in material co‐immunoprecipitated with MOR. The inclusion of GTPγS during the immunoprecipitation only modestly increased the association of RGS4 with MOR. When film was over‐exposed to this same blot, RGS4 was detected in brain membranes by both RGS4 antibodies (Figure S3b). Ten μM of the proteasome inhibitor, MG132, was included in this experiment.
Direct interaction of Goα with RGS4
The direct interaction of Goα with RGS4 was investigated using solubilized rat brain membranes (Weems et al. 1996) and His6RGS4 attached to Ni2+‐NTA agarose resin. In these experiments His6RGS4, pre‐bound to Ni2+‐NTA agarose resin, was incubated with solubilized rat brain membranes in the absence of guanine nucleotides or in the presence of 100 μM GDP, 10 μM GTP or 1 μM GTPγS for 1 h at 4°C in Buffer A with 4 mM Chaps (Fig. 4a). His6RGS4‐Ni2+‐NTA resin was separated from the soluble fraction by centrifugation, and the pellet was washed 4× with Buffer A containing 4 mM 3‐[(3‐cholamidopropyl)‐dimethylammonio]‐1‐propanesulfonate (CHAPS). The relative amounts of Goα and RGS4 in the pull down were estimated by immunoblotting followed by band density measurements with ImageJ. To correct for possible gel loading discrepancies, the density of Goα in each lane was divided by the density of RGS4 in the same lane, and the ratios of Goα/RGS4 were compared among treatment groups. The inclusion of GDP or GTP failed to affect the association of Goα with His6RGS4. However, the inclusion of GTPγS caused a significant increase (3‐fold) in the amount of Goα bound to His6RGS4/Ni2+‐NTA pull‐down (Fig. 4a, unpaired t‐test, p = 0.0001). In four independent experiments, the inclusion of GTPγS during the His6RGS4 pull‐down caused a 2.8‐fold increase in the amount of Goα in the pellet (p = 0.004). GTPγS caused a concentration‐dependent stimulation of Goα binding to RGS4 with an EC50 of 0.12 μM (Fig. 4b). The inclusion of 100 μM GDP prevented 0.3 μM GTPγS from activating Goα and thus prevented GTPγS from enhancing Goα binding to RGS4 (Fig. 4c). However, when 1 μM DAMGO was included during the incubation, there was a 70% increase in the binding of Goα to RGS4 (p = 0.003, Fig. 4d). The addition of 1 μM DAMGO alone (without guanine nucleotide) did not affect the binding of Goα to RGS4; nor did the addition of 1 μM DAMGO further increase the amount of Goα binding to RGS4 caused by 0.3 μM GTPγS alone (data not shown). GTPγS can also stimulate the binding of other Gα subunits to RGS4, and studies are underway to determine if MOR activation can stimulate the binding of GTPγS to these other Gα subunits and cause them to associate with RGS4 (Figure S4).
RGS4 attenuates DAMGO‐mediated inhibition of adenylyl cyclase activity RGS4 caused a concentration‐dependent attenuation of DAMGO‐mediated inhibition of adenylyl cyclase activity. Data from seven independent experiments (n = 28 for each data point) were combined to generate the curve shown in Figure S5. In the absence of RGS4, 1 μM DAMGO plus 10 μM GTP and 1 μM DAMGO plus 1 μM GTPγS caused a 60.4 ± 2.97 and 52.7% ± 2.57% inhibition of adenylyl cyclase activity, respectively. The addition of increasing concentrations of RGS4 caused a concentration‐dependent attenuation of DAMGO (plus GTP)‐mediated inhibition of adenylyl cyclase (EC50 of RGS4 = 83.6 ± 1.5 nM with a 95% confidence interval (CI): 43.2–162 nM). In the presence of 3 μM RGS4, DAMGO inhibited forskolin‐stimulated adenylyl cyclase activity by only 21.8% ± 2.27%. In contrast, RGS4 failed to attenuate GTPγS‐mediated inhibition of adenylyl cyclase activity.
Non‐competitive nature of RGS4 in attenuating MOR‐mediated inhibition of adenylyl cyclase activity DAMGO caused a concentration‐dependent inhibition of adenylyl cyclase activity in homogenates of SHSY5Y cells (Fig. 5, left side). The EC50s of DAMGO in the absence and in the presence of 1 μM RGS4 were 40 nM (95% CI from 26.9 to 59.5 nM) and 66 nM (95% CI from 31.8 to 137 nM), respectively. In the absence and presence of 1 μM RGS4, the maximal inhibitory effect of DAMGO was 53.7% ± 1.72% inhibition of adenylyl cyclase activity (95% CI from 50.3% to 57.2%) and 28.9 ± 1.83% inhibition of adenylyl cyclase activity (95% CI from 25.3% to 32.6% inhibition), respectively. Thus, RGS4 significantly diminished the maximal inhibitory effect (the efficacy) of DAMGO but did not significantly diminish the EC50 (the potency) of DAMGO. Figure 5, left side, was generated from four independent experiments, n = 16 for each data point. Both His6RGS4 and RGS4 (produced by cutting GST off GST‐RGS4 with thrombin, see Figure S6a) were equally effective in attenuating the efficacy of DAMGO in the adenylyl cyclase assay. Therefore, the presence of the His6 tag did not influence the effectiveness of RGS4. In contrast to the noncompetitive inhibition caused by RGS4, naltrexone (0.3 μM), an opioid receptor antagonist, caused a rightward shift in the concentration–response curve to DAMGO that is typical of competitive inhibition (Fig. 5, right side). The EC50s of DAMGO were 18.0 nM (95% CI from 10.5 to 30.8 nM) and 1.30 μM (95% CI from 0.351 to 4.74 μM) in the absence and presence of 0.3 μM naltrexone, respectively. The maximal inhibition of adenylyl cyclase activity was unaffected by naltrexone. All data in Fig. 5 were analyzed by non‐linear regression analysis using Prism® 5.
Figure 5.

RGS4 diminished the efficacy, but not the potency, of [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO) in inhibiting adenylyl cyclase activity (left panel); in contrast, naltrexone diminished the potency, but not the efficacy, of DAMGO (right panel). Left Panel. Adenylyl cyclase activity was measured in the presence of increasing concentrations of DAMGO in the absence (open circles) or the presence of 1 μM RGS4 (filled circles). The EC50s of DAMGO in the absence and presence of RGS4 were 40.0 nM (95% confidence interval 27–59 nM) and 66.0 nM (95% confidence interval 32–137 nM), respectively (EC50 values were not significantly different). In the absence and presence of 1 μM RGS4, DAMGO maximally inhibited adenylyl cyclase activity by 54% (95% confidence interval 50.3–57.2%) and 28.2% (95% confidence interval 25.3–32.6%), respectively (efficacies were significantly different p < 0.05). Data from four independent experiments were combined, n = 16 for each point. Right Panel. Adenylyl cyclase activity was measured in the presence of increasing concentrations of DAMGO in the absence (open squares) or presence of 0.3 μM naltrexone (filled squares). The EC50s of DAMGO in the absence and presence of naltrexone were 19 nM (95% confidence intervals 10.5–31 nM) and 1.30 μM (95% confidence intervals 0.351–4.74 μM), respectively (p < 0.05). The maximal inhibitions caused by DAMGO in the absence or presence of naltrexone were to 3.28 pmoles cAMP formed in 10 min (95% confidence intervals 2.44–4.01 pmoles) and 3.92 pmoles cAMP formed in 10 min (95% confidence intervals 1.28–6.56 pmoles), respectively (not significantly different). Data shown are from a single experiment (n = 4 for each point).
Aspects of the MOR/G protein complex functioning not affected by RGS4 RGS4 failed to affect GTPγS‐mediated inhibition of adenylyl cyclase activity (Figure 6b). In concentration–response curves, the EC50s of GTPγS were 31.2 nM (95% CI from 20.5 to 47.5 nM) and 33.2 nM (95% CI from 21.9 to 50.1 nM) in the absence and presence of 1 μM RGS4, respectively. GTPγS inhibited adenylyl cyclase activity to 43.7% of maximal activity (95% CI from 39.8 to 47.6% of maximal activity) and 39.4% of maximal activity (95% CI from 35.5 to 43.4% of maximal activity), in the absence and presence of 1 μM RGS4, respectively.
Figure 6.
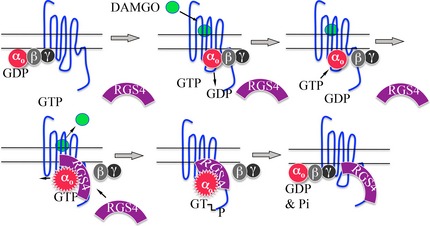
Proposed model of the interaction of RGS4 with MOR and Go It is proposed that the unoccupied MOR (blue serpentine line) is weakly associated with Gi/Go‐type G proteins (αo‐GDPβγ). Activation of MOR by [D‐Ala2, N‐Me‐Phe4, Gly5‐ol] enkephalin (DAMGO), a mu opioid agonist, causes αo‐GDPβγ to bind more tightly to MOR and causes GDP to dissociate from αo. GTP now binds to and activates αo (activated αo shown as serrated red circle bound with GTP). RGS4 is now attracted to ‘activated’ αo‐GTP. The interaction of RGS4 with αo‐GTP increases the intrinsic GTPase of αo resulting the hydrolysis of GTP to GDP and Pi. αo‐GDP re‐associates with βγ and, αo‐GDPβγ binds less tightly to MOR in the absence of agonist. We speculate the RGS4 may remain bound to the C terminal of MOR and remain situated in position to accelerate the inactivation of the next molecule of αo‐GTP.
Also, RGS4 did not significantly alter DAMGO‐stimulated binding of [35S]GTPγS to G proteins associated with SHSY5Y cell membranes (Figure S6c). This assay measures the ability of MOR activation by DAMGO to cause GDP to dissociate from cognate G proteins allowing [35S]GTPγS to bind to these G proteins. In this assay, the EC50s of DAMGO were 39.0 nM (95% CI from 15.7 to 96.5 nM) and 62.0 nM (95% CI from 22.6 to 170 nM) in the absence and presence of 1 μM RGS4, respectively. The maximal amounts of [35S]GTPγS bound in the absence and presence of 1 μM RGS4 were 1432 ± 58 dpm (95% CI from 1266 to 1598) and 1319 ± 82 dpm (95% CI from 1149 to 1490), respectively. Finally, RGS4 did not affect the binding of [3H]DAMGO to MOR in SHSY5Y cell membranes (Figure S6d).
Discussion
RGS proteins have been established as negative regulators of all G protein–coupled receptors (Neubig and Siderovski 2002) including MOR (Zachariou et al. 2003; Rodriguez‐Munoz et al. 2007; Leontiadis et al. 2009; Han et al. 2010; Talbot et al. 2010). This study focuses on the interaction of RGS4 with MOR and one of its cognate G proteins, Go, although Gi3, when activated by GTPγS, can also associate with RGS4 (Figure S4). We chose to study RGS4, because there is evidence that RGS4 acts as a GTPase activating protein for Gi/Go‐type G proteins, the type of G proteins that act as transducers for MOR and because RGS4 mRNA and protein are present in many brain regions that express MOR (Berman et al. 1996a,b; Gold et al. 1997; Hepler et al. 1997; Chalecka‐Franaszek et al. 2000; Krumins et al. 2004). Also, RGS4 has been implicated in the development of opiate physical dependence. RGS4 mRNA levels in the locus coeruleus doubled following 6 days of chronic morphine administration, and RGS4 knockout mice undergo a more severe withdrawal syndrome following acute withdrawal from morphine (Gold et al. 2003; Han et al. 2010). Surprisingly, RGS4−/− mice do not show any major phenotype related to the MOR signaling system (Grillet et al. 2005); the co‐expression of RGS8 and RGS4 in many regions of the brain suggests a functional redundancy between these two RGS proteins (Talbot et al. 2010). RGS4 is not the only RGS protein that modulates MOR activity, but RGS4 almost certainly modulates MOR activity in a number of brain regions. In this study, we show that endogenous RGS4 does indeed associate with MOR (Fig. 3). Future studies will focus on specific brain regions where RGS4 may regulate MOR signaling.
Association of RGS4 with the MOR/G protein complex
Earlier studies demonstrated that RGS4 binds directly to Gi/o‐type proteins, particularly in the presence of aluminum, magnesium, and fluoride (AMF) (Berman et al. 1996a; Watson et al. 1996). As MOR is associated with Gi and Go‐type G proteins, we investigated the interaction between RGS4 and the MOR/G protein complex from rat brain membranes. Previously, we developed a method to solubilize and immunoprecipitate active MOR in association with Gi1, Gi3, and Go; the ratio of MOR to G protein was approximately one to one when the immunoprecipitation was performed in the presence of morphine (Weems et al. 1996; Chalecka‐Franaszek et al. 2000). In this study, we followed the co‐immunoprecipitation of Goα as a representative of the G proteins that interact with MOR because Goα is the most abundant of G proteins in rat brain, and good Goα antibodies are available for immunoblotting. MORs were immunoprecipitated with three different anti‐MOR antibodies; each of these MOR antibodies caused the co‐immunoprecipitation of RGS4 (shown for anti‐MOR10–70 in Figs 1, 2, 3). The inclusion of the MOR agonist DAMGO during the immunoprecipitation caused a 150% increase in amount of Goα and a 50% increase in RGS4, whereas the addition of GTPγS (along with DAMGO) caused a three‐fold increase in RGS4 and a 50% decrease in the amount of Goα (Table 1). The concomitant increase in RGS4 and decrease in Goα caused by GTPγS suggests that activated G proteins attract RGS4 to the MOR/G protein complex and that a region of MOR may contribute to the continued binding of RGS4 to the complex following the dissociation of Goα. Leontiadis et al. (2009) have shown evidence that RGS4 binds directly to a portion of the C‐terminal region of MOR (MOR329–355), that is part of the 4th intracellular loop of MOR, without the assistance of G proteins. We speculate that G proteins that bind to the 3rd intracellular loop of MOR may occlude a portion of the 4th intracellular loop of MOR, the region of MOR that has been proposed to bind RGS4. When a GTP‐activated G protein dissociates from MOR, RGS4 is able to bind to the 4th intracellular loop of MOR positioning RGS4 to rapidly inactivate the next GTP‐activated G protein. Most importantly, in the current study, DAMGO stimulated the binding of RGS4 to the MOR/G protein complex in a GTP‐dependent manner. This finding clearly shows that activation of MOR causes GTP to bind to its cognate G proteins (including Go) and that the activated G proteins attract RGS4 to the MOR/G protein complex. Interestingly, using the procedures described in Methods, 28 fmoles of RGS4 (determined by western blotting) were co‐immunoprecipitated with 31 fmoles of MOR (determined by saturation [3H]DAMGO binding) in the presence of GTPγS. Thus, when G proteins (like Go) are fully activated, one molecule of RGS4 becomes associated with one molecule of MOR.
Association of endogenous RGS4 with the MOR/G proteins complex When the concentrations of protease inhibitors were double and when the proteasome inhibitor was included during the immunoprecipitation of MOR, endogenous RGS4 remained intact and associated with MOR (i.e., it was not degraded), and therefore GTPγS could only modestly increase the binding of endogenous RGS4 to MOR. We speculate that receptor activation (and subsequent G protein activation) causes RGS4 to bind to the MOR/G protein complex and that, after a period of time, RGS4 undergoes ubiquitination and proteolysis by the proteosome (Wang and Traynor 2011). The fact that His6RGS4 could be detected in material immunoprecipitated with MOR suggests that His6RGS4 is less susceptible to proteolysis than endogenous RGS4. In preliminary experiments performed with His6RGS4, we had observed a significant amount of ‘smearing’ in the location at and below ~26 kDa on western blots. We found that preparing our own cocktail of protease inhibitors that also included phosphatase inhibitors resulted in far less breakdown of His6RGS4 as is indicated by the presence of discrete bands of His6RGS4 around 26 kDa (as seen in Figs. 1 and 2). Clearly, even higher levels of inhibitors (2×) plus MG132 were needed to protect endogenous RGS4 from degradation (Fig. 3). Future studies will focus on the interactions of endogenous RGS proteins with MOR in distinct regions of the brain.
Association of Goα with RGS4 – influence of MOR
The interaction of RGS4 with Go was also studied in a preparation of solubilized rat brain membranes that had been passed through a SephadexG50 column pre‐equilibrated with 4 mM CHAPS in buffer A. Passage of the solubilized material through the SephadexG50 column removes all small molecules (including free peptides and nucleotides), reduces the CHAPS concentration to 4 mM, and maintains the MOR in a high affinity binding state that is sensitive to guanine nucleotides (Weems et al. 1996). In this soluble preparation, neither GTP nor GDP had an effect on the binding of Goα to His6RGS4‐Ni2+NTA agarose, but GTPγS (0.3 μM) caused a three‐fold increase in the binding of Goα to His6RGS4 (Fig. 4a). GTPγS caused a concentration‐dependent increase in the binding of Goα to RGS4 with an EC50 of 0.1 μM and a near maximal effect occurring at 0.3 μM (Fig. 4b). Interestingly, an excess of GDP (100 μM) blocked the activation of Goα GTPγS (0.3 μM) and diminished the subsequent binding of Goα to His6RGS4. However, activation of MOR with DAMGO during the incubation with GDP and GTPγS increased the binding of Goα to His6RGS4 (Fig. 4c and d). Presumably, 0.3 μM GTPγS activated the majority of solubilized Go‐type G proteins, just as 0.3 μM GTPγS caused a near maximal inhibition of adenylyl cyclase activity (Figure S6b). GDP (100 μM) blocks access of GTPγS to Goα and reduces the binding of Goα to RGS4, whereas activation of MOR by DAMGO decreases the affinity of 40–50% of the Goα molecules for GDP and resulted in the subsequent activation of these Goα molecules by GTPγS causing them (Goα‐GTPγS) to bind to His6RGS4‐Ni2+‐NTA agarose (Fig. 4d, right bar graph). Previously, we demonstrated that activation of MOR decreased the affinity of Gi/o‐type G proteins for GDP (and thus caused GDP to dissociate from G proteins) because GDP could prevent GTPγS from inhibiting adenylyl cyclase activity in the absence, but not in the presence of morphine (Gosse et al. 1989). Of course it has been accepted for decades that all GPCRs work by inducing the dissociation of GDP from their G proteins and allowing intracellular GTP to bind to, and to activate, their cognate G proteins (Gilman 1987). Indeed, in the experiment shown in Figure S6c, DAMGO stimulated the binding of [35S]GTPγS to SHSY5Y cell membranes in the presence of 10 μM GDP. Importantly, this experiment demonstrates that MOR activation of Goα caused GDP to dissociate from Goα, allowing GTPγS to bind to Goα and for Goα‐GTPγS to bind to RGS4. These findings clearly demonstrate that RGS4 is capable of binding to G proteins that have been activated by MOR.
In agreement with our findings, Leontiadis et al. (2009) demonstrated that when MOR and RGS4 were co‐transfected into HEK 293 cells, RGS4 could be co‐immunoprecipitated with MOR, and more interestingly, when aluminum, fluoride, and magnesium ions were included during the MOR immunoprecipitation, the amount of RGS4 in the immunoprecipitate tripled.
Influence of RGS4 on MOR Signaling
RGS4 caused a concentration‐dependent attenuation of MOR‐mediated inhibition of adenylyl cyclase (Figure S5). We assume that the ability of RGS4 to attenuate DAMGO‐mediated inhibition of adenylyl cyclase activity stems from its ability to accelerate the GTPase of Gi/o‐type G proteins since RGS4 has a similar EC50 in both of these systems (Berman et al. 1996b; Watson et al. 1996). The inhibitory effect of RGS4 was found to be primarily noncompetitive because RGS4 diminished the efficacy, but not the potency, of DAMGO in inhibiting adenylyl cyclase activity (Fig. 5, left panel). In contrast, naltrexone, an opioid receptor antagonist, diminished the potency, but not the efficacy of DAMGO (Fig. 5, right panel). The noncompetitive nature of the RGS4 inhibition is consistent with the hypothesis that RGS4 stabilizes the transition state of the intrinsic GTPase activity of Gi/o‐type G proteins as opposed to its having a direct effect on the ligand binding site on the receptor (Berman et al. 1996a,b; Watson et al. 1996; Tesmer et al. 1997). In contrast to its attenuating effect on MOR signaling, RGS4 had no effect on GTPγS‐induced inhibition adenylyl cyclase, a finding consistent with the resistance of GTPγS to hydrolysis by GTPase. Also, RGS4 did not affect DAMGO‐stimulated [35S]GTPγS binding to SHSY5Y membranes (Figure S6c). The rate‐limiting step in stimulating the binding of GTP to G proteins is the dissociation of GDP from the G protein (Gilman 1987). As DAMGO activation of MOR stimulates the release of GDP from Gi and Go in enhancing the binding of [35S]GTPγS to Gi and Go, we conclude that RGS4 has no major effect on the coupling of MOR to its G proteins or on the activation of these G proteins by the receptor and guanosine triphosphates. Also, RGS4 had no significant effect on either the affinity of MOR for [3H]DAMGO or on the maximal number of [3H]DAMGO binding sites (Figure 6d). Because high affinity agonist binding requires the association of the receptor with its G proteins, these results provide further evidence that RGS4 does not interfere with the coupling of MOR to its G proteins.
Proposed mechanism of RGS4 attenuation of MOR signaling
In the absence of an agonist, MOR (blue serpentine line) is associated weakly with its cognate G proteins, Gi1, Gi3, and Go (represented by αβγ) and perhaps is also temporarily associated with RGS4 (until RGS4 is proteolyzed) (Fig. 6). When DAMGO, a mu opioid agonist, binds to and activates MOR, Go (αβγ) binds more tightly to MOR, and MOR causes a decrease in the affinity of Goα for GDP. The dissociation of GDP from Goα allows GTP to bind to, and to activate, Goα. A Goα‐GTP r circle) dissociates slightly from Gβγ and slightly from MOR, and Goα‐GTP triggers downstream signaling (e.g., inhibition of adenylyl cyclase activity, opening of K+ channels, etc.). RGS4 binds to activated Goα‐GTP and perhaps simultaneously to the C‐terminal region of MOR (Leontiadis et al. 2009); the binding of RGS4 to Goα‐GTP increases the intrinsic GTPase of Goα [RGS4 stabilizes the transition state of Goα (Kimple et al. 2011)]. Once GTP is hydrolyzed to GDP, Goα‐GDP becomes weakly associated with MOR, and we speculate that RGS4 remains associated with the C‐terminal region of MOR where it is in position to rapidly inactivate the next round of Goα‐GTP produced by the activation of MOR. After a period of time, RGS4 is removed from the MOR/G protein complex by ubiquitination and proteolysis by the proteasome.
Supporting information
Figure S1. Anti‐MOR349–398 identifies MOR immunohistochemically in sections of rat brain.
Figure S2. Immunoprecipitation of the MOR/G protein complex Panel A.
Figure S3. Specificity of anti‐RGS4170–205 in western blots and ability of anti‐RGS4170–205 to recognize endogenous RGS4 A.
Figure 4. GTPγS stimulated the binding of Goα and Gi3α to RGS4.
Figure S5. RGS4 caused a concentration‐dependent attenuation of DAMGO‐mediated inhibition of adenylyl cyclase activity.
Figure S6. Influence of RGS4 on the signaling of MOR in SHSY5Y cell membranes.
Table S1. Antibodies used in the current study.
Table S2. Pre‐treatment of SHSY5Y cells with DAMGO enhanced the ability of RGS4 to attenuate DAMGO‐mediated inhibition of adenylyl cyclase activity in cell homogenates.
Acknowledgments and conflict of interest disclosure
We thank Susanne Mumby for the gift of RGS4 cDNA and Jeffrey Harmon and Fereshteh Nugent for helpful suggestions and for critically reviewing the manuscript. This work was supported by NRSA fellowship 102581; and USUHS grant RO75QU. The authors have no conflicts of interest to declare.
All experiments were conducted in compliance with the ARRIVE guidelines.
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
References
- Berman D. M., Kozasa T. and Gilman A. G. (1996a) The GTPase‐activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J. Biol. Chem. 271, 27209–27212. [DOI] [PubMed] [Google Scholar]
- Berman D. M., Wilkie T. M. and Gilman A. G. (1996b) GAIP and RGS4 are GTPase‐activating proteins for the Gi subfamily of G protein a subunits. Cell 86, 445–452. [DOI] [PubMed] [Google Scholar]
- Bernstein L. S., Ramineni S., Hague C., Cladman W., Chidiac P., Levey A. I. and Hepler J. R. (2004) RGS2 binds directly and selectively to the M1 muscarinic acetylcholine receptor third intracellular loop to modulate Gq/11alpha signaling. J. Biol. Chem. 279, 21248–21256. [DOI] [PubMed] [Google Scholar]
- Chalecka‐Franaszek E., Weems H. B., Crowder A. T., Cox B. M. and Cote T. E. (2000) Immunoprecipitation of high‐affinity, guanine nucleotide‐sensitive, solubilized mu‐opioid receptors from rat brain: coimmunoprecipitation of the G proteins Go, Gi1, and Gi3. J. Neurochem. 74, 1068–1078. [DOI] [PubMed] [Google Scholar]
- Chen Y. and Yu L. (1994) Differential regulation by cAMP‐dependent protein kinase and protein kinase C of the mu opioid receptor coupling to a G protein‐activated K+ channel. J. Biol. Chem. 269, 7839–7842. [PubMed] [Google Scholar]
- Cui Y., Ostlund S. B., James A. S. et al (2014) Targeted expression of mu‐opioid receptors in a subset of striatal direct‐pathway neurons restores opiate reward. Nat. Neurosci. 17, 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G. and Thorner J. (1997) Minireview: RGS proteins and signaling by heterotrimeric G proteins. J. Biol. Chem. 272, 3871–3874. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Apaniesk D., Chen Y., Song J. and Nusskern D. (1995) Inhibition of G‐protein signalling by dominant gain‐of‐function mutants in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 1, 3635–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoussi Z., Leontiadis L., Mazarakou G., Merkouris M., Hyde K. and Hamm H. (2006) Selective interactions between G protein subunits and RGS4 with the C‐terminal domains of the mu‐ and delta‐opioid receptors regulate opioid receptor signaling. Cell. Signal. 18, 771–782. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. (1987) G proteins: transduction of receptor‐generated signals. Annu. Rev. Biochem. 56, 615–649. [DOI] [PubMed] [Google Scholar]
- Gold S. J., Ni Y. G., Dohlman H. G. and Nestler E. J. (1997) Regulators of G‐protein signalling (RGS) proteins: region specific expression of nine subtypes in rat brain. J. Neurosci. 17, 8024–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold S. J., Han M. H., Herman A. E. et al (2003) Regulation of RGS proteins by chronic morphine in rat locus coeruleus. Eur. J. Neurosci. 17, 971–980. [DOI] [PubMed] [Google Scholar]
- Gosse M. E., Frey E. A. and Cote T. E. (1989) Site of pertussis toxin‐induced ADP‐ribosylation of Gi is critical for receptor modulation of GDP interaction with Gi. Mol. Endocrinol. 3, 315–324. [DOI] [PubMed] [Google Scholar]
- Grillet N., Pattyn A., Contet C., Kieffer B. L., Goridis C. and Brunet J. F. (2005) Generation and characterization of Rgs4 mutant mice. Mol. Cell. Biol. 25, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hague C., Bernstein L. S., Ramineni S., Chen Z., Minneman K. P. and Hepler J. R. (2005) Selective inhibition of alpha1A‐adrenergic receptor signaling by RGS2 association with the receptor third intracellular loop. J. Biol. Chem. 280, 27289–27295. [DOI] [PubMed] [Google Scholar]
- Han M. H., Renthal W., Ring R. H. et al (2010) Brain region specific actions of regulator of G protein signaling 4 oppose morphine reward and dependence but promote analgesia. Biol. Psychiatry 67, 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler J. R., Berman D. M., Gilman A. G. and Kozasa T. (1997) RGS4 and GAIP are GTPase‐activating proteins for Gqa and block activation of phospholipase Cb by g‐thio‐GTP‐Gqa. Proc. Natl Acad. Sci. USA 94, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple A. J., Bosch D. E., Giguere P. M. and Siderovski D. P. (2011) Regulators of G‐protein signaling and their Galpha substrates: promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 63, 728–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumins A. M., Barker S. A., Huang C., Sunahara R. K., Yu K., Wilkie T. M., Gold S. J. and Mumby S. M. (2004) Differentially regulated expression of endogenous RGS4 and RGS7. J. Biol. Chem. 279, 2593–2599. [DOI] [PubMed] [Google Scholar]
- Leontiadis L. J., Papakonstantinou M. P. and Georgoussi Z. (2009) Regulator of G protein signaling 4 confers selectivity to specific G proteins to modulate mu‐ and delta‐opioid receptor signaling. Cell. Signal. 21, 1218–1228. [DOI] [PubMed] [Google Scholar]
- Matthes H., Maldondo R., Simonin F. et al (1996) Loss of morphine‐induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu‐opioid receptor gene. Nature 383, 819–823. [DOI] [PubMed] [Google Scholar]
- Neubig R. R. and Siderovski D. P. (2002) Regulators of G‐protein signalling as new central nervous system drug targets. Nat. Rev. Drug. Discov. 1, 187–197. [DOI] [PubMed] [Google Scholar]
- North R. (1986) Opioid receptor types and membrane ion channels. Trends Neurosci. 9, 114–117. [Google Scholar]
- Psigfogeorgou K., Terzi D., Papachatzaki M., Varidaki A., Ferguson D., Gold S. and Zachariou V. (2011) A unique role of RGS9‐2 in the striatum as a positive or negative regulator of opiate analgesia. J. Neurosci. 31, 5617–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Munoz M., de la Torre‐Madrid E., Sanchez‐Blazquez P. and Garzon J. (2007) Morphine induces endocytosis of neuronal mu‐opioid receptors through the sustained transfer of Galpha subunits to RGSZ2 proteins. Mol. Pain. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendt M. and McGinty J. F. (2007) Regulator of G‐protein signaling 4 interacts with metabotropic glutamate receptor subtype 5 in rat striatum: relevance to amphetamine behavioral sensitization. J. Pharmacol. Exp. Ther. 323, 650–657. [DOI] [PubMed] [Google Scholar]
- Talbot J. N., Roman D. L., Clark M. J., Roof R. A., Tesmer J. J., Neubig R. R. and Traynor J. R. (2010) Differential modulation of mu‐opioid receptor signaling to adenylyl cyclase by regulators of G protein signaling proteins 4 or 8 and 7 in permeabilised C6 cells is Galpha subtype dependent. J. Neurochem. 112, 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer J. J., Berman D. M., Gilman A. G. and Sprang S. R. (1997) Structure of RGS4 bound to AlF4‐activated G(i alpha1): stabilization of the transition state of GTP hydrolysis. Cell 89, 251–261. [DOI] [PubMed] [Google Scholar]
- Traynor J. (2012) mu‐Opioid receptors and regulators of G protein signaling (RGS) proteins: from a symposium on new concepts in mu‐opioid pharmacology. Drug Alcohol Depend. 121, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. and Traynor J. R. (2011) Opioid‐induced down‐regulation of RGS4: role of ubiquitination and implications for receptor cross‐talk. J. Biol. Chem. 286, 7854–7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. and Traynor J. (2013) Modulation of mu‐opioid receptor signaling by RGS19. Mol. Pharmacol. 83, 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu‐Chen L. Y. and Traynor J. R. (2009) Differential modulation of mu‐ and delta‐opioid receptor agonists by endogenous RGS4 protein in SH‐SY5Y cells. J. Biol. Chem. 284, 18357–18367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N., Druey K. M., Kehrl J. H. and Blumer K. J. (1996) RGS family members: GTPase‐activating proteins for heterotrimeric G‐protein alpha‐subunits. Nature 383, 172–175. [DOI] [PubMed] [Google Scholar]
- Weems H. B., Chalecka‐Franaszek E. and Cote T. E. (1996) Solubilization of high‐affinity, guanine nucleotide‐sensitive μ‐opioid receptors from rat brain membranes. J. Neurochem. 66, 1042–1050. [DOI] [PubMed] [Google Scholar]
- Xie Z., Li Z., Guo L. et al (2007) Regulator of G protein signaling proteins differentially modulate signaling of mu and delta opioid receptors. Eur. J. Pharmacol. 565, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou V., Georgescu D., Sanchez N. et al (2003) Essential role for RGS9 in opiate action. Proc. Natl Acad. Sci. USA 100, 13656–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Anti‐MOR349–398 identifies MOR immunohistochemically in sections of rat brain.
Figure S2. Immunoprecipitation of the MOR/G protein complex Panel A.
Figure S3. Specificity of anti‐RGS4170–205 in western blots and ability of anti‐RGS4170–205 to recognize endogenous RGS4 A.
Figure 4. GTPγS stimulated the binding of Goα and Gi3α to RGS4.
Figure S5. RGS4 caused a concentration‐dependent attenuation of DAMGO‐mediated inhibition of adenylyl cyclase activity.
Figure S6. Influence of RGS4 on the signaling of MOR in SHSY5Y cell membranes.
Table S1. Antibodies used in the current study.
Table S2. Pre‐treatment of SHSY5Y cells with DAMGO enhanced the ability of RGS4 to attenuate DAMGO‐mediated inhibition of adenylyl cyclase activity in cell homogenates.


