Abstract
Bacterial processes, such as stress responses and cell differentiation, are controlled at many different levels. While some factors, such as transcriptional regulation, are well appreciated, the importance of chromosomal gene location is often underestimated or even completely neglected. A combination of environmental parameters and the chromosomal location of a gene determine how many copies of its DNA are present at a given time during the cell cycle. Here, we review bacterial processes that rely, completely or partially, on the chromosomal location of involved genes and their fluctuating copy numbers. Special attention will be given to the several different ways in which these copy-number fluctuations can be used for bacterial cell fate determination or coordination of interdependent processes in a bacterial cell.
Keywords: DNA replication, chromosome organization, stress response, cell fate determination, gene dosage, Bacillus subtilis, Streptococcus pneumoniae, Vibrio cholerae
Trends
While well appreciated in eukaryotes, the relevance of gene copy numbers in bacteria is largely unexplored.
Copy number imbalance between two genes during DNA replication was found to be responsible for precisely timed activity pulses of the Bacillus subtilis sporulation initiator.
Besides the various ways in which chromosomal gene location has been shown to be involved in B. subtilis sporulation, more examples have recently been found in other bacteria as well.
Changing nutrient availability or malfunction of critical subcellular processes lead to distorted gene copy number distributions, with transcriptomic shifts as a result, for example accounting for the activation of pneumococcal competence.
Additionally, the discovery that mRNA diffusibility is unexpectedly low suggests that the chromosomal location of a gene may be in part determined by where in the cell its product is required.
How Genome Organization and Gene Function Are Connected
For decades, the importance of genome organization has been recognized. Virtually every process that interacts directly or indirectly with the chromosome has left its marks during the course of genome evolution. It has become clear that the order and orientation of features on a chromosome, as well as the three-dimensional structure of the chromosome, is of importance to a cell. Numerous examples of the interplay between genome organization and cellular processes are available. For example, essential genes tend to be located on the strand that is transcribed in the same direction as in which replication proceeds [1].
However, the importance of the genomic location of key elements is still often underestimated. In fact, very little attention is given to the many different ways in which genomic location can impact cell biology. We therefore review the various mechanisms by which the exact genomic location of a feature can play a role in the regulatory landscape and development of bacterial cells. More specifically, we focus on processes in which gene copy number or, more accurately, genome-wide copy number distributions play a role. It is a well established fact in eukaryotes that having an abnormal number of chromosomes (aneuploidy), leading to atypical gene copy numbers, can have detrimental effects, a well known example being Down syndrome (trisomy 21 in humans, [2]). Additionally, the need for female mammals to silence one of their two copies of the X-chromosome underlines the importance of DNA copy numbers 3, 4. Furthermore, amplification of specific nutrient transporter genes in Saccharomyces cerevisiae was observed to enhance fitness in nitrogen-limited conditions [5]. The correlation between copy number and gene expression implied by these examples was confirmed recently by Chen and Zhang, who showed that the timing of replication of a gene influences its final expression level in yeast [6]. Nevertheless, copy number effects are still only rarely considered in prokaryotes. During bacterial cell cycle progression, copy numbers around the chromosome fluctuate periodically. Both the periodicity [7] and the amplitude 8, 9 of this fluctuation can be employed to regulate certain processes in the cell. Furthermore, global or local (e.g., compartmentalization, see below) distortions of copy number fluctuations can be involved in bacterial ‘decision making’ and even play an important role during virulence [9].
Replication-Associated Copy Number Fluctuations
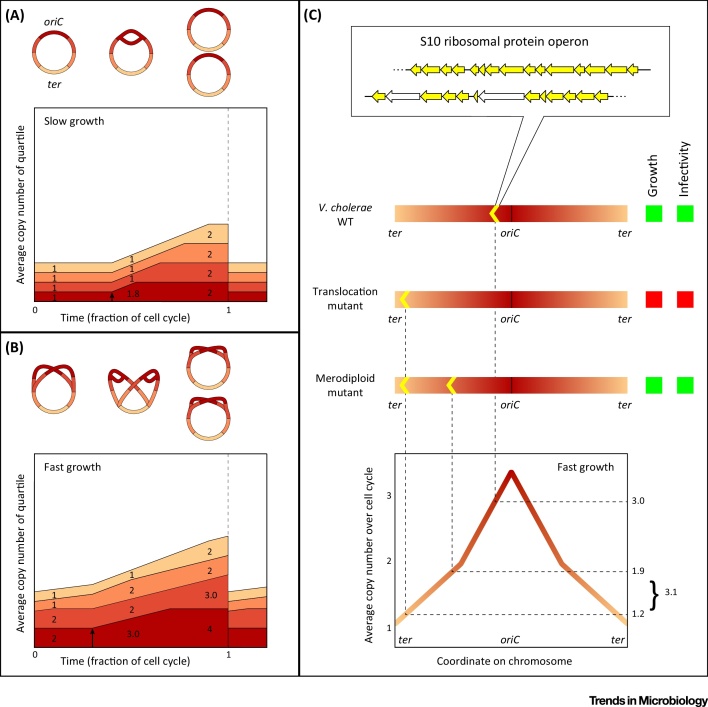
The majority of bacteria have their DNA organized on a single, circular chromosome, replication of which starts at a well defined origin of replication (oriC). From there, replication proceeds symmetrically in both directions around the chromosome and is terminated at the opposite end (the ter region) of the molecule, where both replication machineries (forks) meet. As a result, the various genes and other features on the chromosome are replicated in a fixed order, leading to periodic fluctuations of their copy numbers that are repeated every cell cycle. After termination of replication, cells still need a specific amount of time to finish cell division (the D-period [8]). The initiation of new rounds of replication is tightly regulated by a variety of factors 10, 11, 12, 13; this ensures that there is exactly one initiation event in each cell cycle, timed in such a way that replication and cell division are properly coordinated. When growth is sufficiently slow, cells have enough time to start and finish DNA replication within one cycle, and local copy numbers will generally only fluctuate between one and two copies of a certain region (Figure 1A). Some bacteria, however, have the capacity to grow so fast that replication of their entire chromosome cannot be completed within one cell cycle [14]. In this case, cells engage in multifork replication; before a replication fork has finished, a new replication initiation event takes place (still exactly once per cell cycle) at all (≥2) copies of oriC simultaneously, resulting in copy numbers of oriC-proximal regions of more than 2 (Figure 1B). For example, fast-growing Escherichia coli cells have been observed to contain up to 8 origins [15]. Since there is a clear correlation between gene copy number and gene expression 16, 17, 18, these fluctuations are relevant to a cell's transcriptome, as is exemplified by the various cases mentioned in this review.
Figure 1.
Replication-Associated Gene Copy Number Fluctuations. Simulated gene copy number distributions throughout the cell cycle (A and B). Each arm of the chromosome has been divided into four quartiles, which are color-coded based on their oriC-proximity. The height of each colored area in the graphs represents the average copy number within the corresponding quartile; as the replisome moves through a quartile, the corresponding graph area steadily increases in height until it is exactly doubled (i.e., the entire quartile is replicated), while the other areas maintain their height. The areas describing the copy number development of the four quartiles are stacked, so their combined height reflects the total DNA content of a cell. Average copy numbers of each quartile at 10%, 50%, and 90% of the cell cycle are shown in the plots. The script to run the simulations is available upon request. Replication initiation is indicated by black arrows. (A) During relatively slow growth (replication time/cell cycle = 0.5, D-period = 10% of cell cycle), only one replication fork is present at a time on each arm of the chromosome (top) and gene copy numbers will fluctuate between 1 and 2 (bottom). (B) During relatively fast growth (replication time/cell cycle = 1.6, D-period = 10% of cell cycle), multifork replication occurs (top) and gene copy numbers can exceed 2 (bottom). (C) The oriC-proximal location of the Vibrio cholerae S10 ribosomal protein operon is important for fitness [9]. Top: translocation of these genes to an oriC-distal site leads to lower gene copy numbers and therefore to a growth defect and attenuated infectivity. Merodiploid strains, with two copies of the S10 operon, show restored fitness and infectivity. Bottom: locus-dependent average copy number over the cell cycle for fast-growing cells (same parameters as in (B), closely matching the oriC-ter ratio observed by Soler-Bistué et al.[9]). Inspection of copy numbers at the varying loci of S10 operon placement shows that S10 gene dosage in the merodiploid strain is very similar to that in the wild-type strain.
Function-Associated Gene Order
The amplitude of a gene's copy number fluctuation will thus depend both on its genomic location, relative to oriC, and on growth rate. The impact of these dependencies is illustrated by the fact that translocations and chromosomal inversions preferentially occur in a copy-number-neutral fashion (i.e., symmetrical with respect to oriC) 19, 20, 21. Another example of the importance of gene order is the strong conservation of the oriC-proximal colocalization of important growth factors involved in replication, transcription and translation 14, 22, 23. The colocalization of these factors can be explained by a combination of the importance of their stoichiometry on the one hand and functional compartmentalization (see below) on the other. However, the fact that they are virtually always found close to the origin of replication rather reflects the cells’ need to correlate their expression with their requirement; when growth conditions improve, cells may switch to multifork replication, automatically boosting the expression of these essential growth factors due to the resulting dosage increase. Recent work by Soler-Bistué et al. demonstrates the relevance of the genomic position of ribosomal protein genes on the large chromosome of the human pathogen Vibrio cholerae, which harbors two circular chromosomes (Figure 1C) [9]. They showed that translocation of a locus bearing half of all ribosomal protein genes from oriC-proximal to various sites further away from the origin of replication results in significant defects in growth and host-invasion capacity. It is worth noting that these defects specifically occur during relatively fast growth, where the difference in copy number between oriC and ter, and therefore the relative effect of translocation of the ribosomal protein genes, is the largest. Both defects are relieved when, instead of one, two copies of the locus are present at an oriC-distal site, effectively restoring absolute ribosomal gene copy numbers and consequently ribosome production levels. The fact that these genes are then no longer colocalized with other important growth factors is, apparently, of lesser importance in this context.
Similarly, Sobetzko et al. demonstrated that nucleoid-associated proteins (NAPs) employed during exponential growth, together with their binding sites, show a tendency to be located closer to oriC than NAPs that act in (near–)stationary phase [23]. Simultaneously, they showed that genes with related functions have a propensity to be distributed at equal distances from oriC, without the necessity of being on the same arm of the chromosome [23]. Taken together, these observations underline that the variation in growth conditions encountered throughout evolution is directly reflected by the relative positioning on the chromosome of genes with related functions.
Chromosome Structure and Gene Expression
Of course, the correlation between gene copy numbers and final expression levels 16, 17, 18 represents only part of the puzzle. Bryant et al. showed that changing the chromosomal location of a reporter cassette in the Gram-negative model organism E. coli could lead to differences in expression level of several hundredfold [24]. Similar observations were made in the Gram-positive human pathogen Streptococcus pneumoniae (the pneumococcus) [25]. These variations cannot be explained by DNA copy number differences alone, especially since the experiments were executed in slow-growth conditions, with no multifork replication and hence with minimal copy number variation along the oriC-ter axis. Possible explanations for these large differences include gyrase activity, transcription-associated supercoiling and the presence of NAP binding sites 24, 26, 27, many of which play a role in determining chromosome structure 28, 29.
Clearly, a myriad of forces have been, and are, at play simultaneously in the evolution of chromosome organization. For that very reason, countless patterns have been, and doubtlessly will still be, discovered in both the order of chromosomal features and chromosome morphology. Although not the focus of this review, the impact of chromosome topology and overall structure on gene expression and chromosome organization cannot go unmentioned. It has been known for decades that a change in DNA supercoiling can affect gene expression [30], and it was recently shown by Sobetzko that preserving the gene-regulatory capacity of supercoiling has been a driving force in the evolution of chromosomal gene order [31]. Furthermore, the folding and compaction of the chromosome can bring genes that are located on very different parts of the DNA molecule into close proximity to each other 32, 33, 34. Interestingly, a periodic distribution of conserved gene pairs around the E. coli chromosome was observed with a period of 117 kb 35, 36. This periodicity possibly allows the chromosome to be folded such that these genes, and thereby their corresponding products, end up in close spatial proximity to each other. The surprisingly low diffusibility of some mRNA molecules from their production site suggests that the well defined organization of the chromosome within the cell might serve as a blueprint that determines where in the cell a certain protein is synthesized 37, 38, 39. However, recently, Moffitt et al. probed the localization of 75% of the E. coli transcriptome using a combination of fluorescence in situ hybridization and super-resolution microscopy, showing that the role of genome structure in mRNA localization is rather the exception than the rule [40].
As mentioned above, final production levels of a certain protein are determined by many more factors than gene dosage alone. For example, when Gerganova et al. moved the fis gene, encoding the E. coli global regulator FIS, from an oriC-proximal to a terminus-proximal location, cells were able to maintain the original protein level by upregulating transcription of fis [41]. Despite the nearly unchanged fis transcript and FIS protein levels, the mutant displayed a significant fitness loss. This was attributed to altered levels of some key NAPs that are directly regulated by FIS and that are, among other things, implicated in promoter binding and DNA supercoiling. While copy-number changes could not be blamed for this observation, the authors speculated that the limited diffusibility of both the fis transcript and FIS might be responsible, leading to different activation patterns of its regulon and thereby possibly affecting DNA topology and structure.
The Various Roles of Chromosome Organization in B. subtilis Sporulation
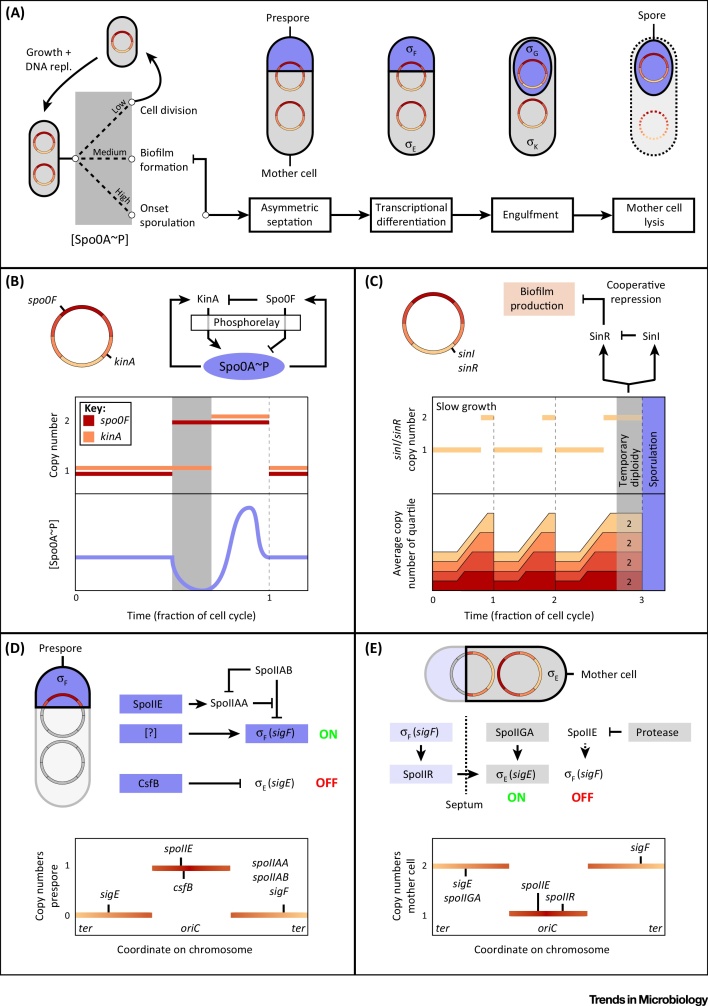
The Gram-positive model bacterium Bacillus subtilis is of great interest when studying culture heterogeneity and cell fate determination. When exposed to stress, it can behave in very different ways [42]; under mild nutritional stress B. subtilis can form biofilms, multicellular communities held together by an amyloid-like matrix, while more severe nutritional stress may lead to the formation of endospores, which can survive various extreme conditions. During sporulation a cell undergoes asymmetric division, which starts with asymmetric septum formation, dividing the cell into a prespore and a mother cell, followed by spore engulfment by the mother cell, spore maturation, and the subsequent lysis of the mother cell (Figure 2A). Here, we discuss the variety of ways in which chromosomal gene order is involved in the regulation of sporulation. For a more complete overview of this well studied phenomenon, several reviews are available (e.g., 43, 44, 45).
Figure 2.
The Various Roles of Gene Location in Bacillus subtilis Sporulation. (A) Overview of endospore development. Sporulation is initiated by high levels of Spo0A∼P, leading to a block of matrix formation. After asymmetric septation, transcriptional differentiation is directed by dedicated sigma-factors, eventually leading to spore maturation and mother cell lysis. (B) Cell-cycle-mediated pulsing controls onset of sporulation [7]. The pulsatile profile of the Spo0A∼P concentration during the cell cycle (bottom graph, bottom panel) is made possible by the asymmetrical placement (top left) and therefore copy number development of spo0F and kinA (bottom graph, top panel; red and orange lines, respectively), which are involved in the sporulation phosphorelay, leading to phosphorylation of Spo0A (top right). (C) Temporary diploidy preceding spore development leads to shut-down of matrix production [51]. Simulated copy number distributions throughout the cell cycle (same plotting and simulation parameters as in Figure 1A) demonstrate how gene dosage is affected prior to sporulation (bottom, orange-red areas). Due to the ter-proximal location of regulatory genes sinI and sinR (top left), unusual levels of their respective products are reached (bottom, light orange lines). Due to cooperativity in SinR-mediated repression of matrix production, biofilm formation is prevented (top right). (D) Transient overproduction of SpoIIE in the prespore leads to eventual release of active σF. Asymmetrical chromosome translocation (top left) leads to unusual copy number distributions (bottom). Initially, one-third of the chromosome is translocated into the prespore, leading to accumulation of oriC-proximally encoded proteins. These include SpoIIE, which is partially responsible for prespore-specific production of σF56, 57, and CsfB, which represses σE in the prespore [61]. (E) Prespore-localized expression of spoIIR and ter-proximal location of spoIIGA and sigE are required for activation of σE in the mother cell 57, 58. Asymmetric chromosome translocation (top) leads to increased levels of ter-proximally encoded proteins (bottom), including spoIIGA and sigE, resulting in σE activation in the mother cell. Most likely, σF is repressed because of mother-cell-specific degradation of SpoIIE.
Cell-Cycle-Mediated Pulsing Controls Onset of Sporulation
The decision to engage in sporulation is taken on a single-cell level and is controlled by a complex phosphorelay system, composed of several kinases and phosphotransferases [46], with master regulator Spo0A at the end of the line. When the amount of active, phosphorylated Spo0A (Spo0A∼P) reaches a threshold level, downstream genes are activated and the sporulation program begins [47]. It is, at that point, important for a cell to have completely copied its DNA, so that the spore can receive an intact chromosome and therefore proliferate 43, 44, 48. A number of studies showed that B. subtilis realizes the necessary coordination of sporulation activation with the cell cycle by producing a single sharp pulse of active Spo0A∼P every cycle, shortly after termination of DNA replication (Figure 2B) 7, 49, 50. Thereby, whenever the conditions are such that the threshold level of Spo0A∼P is reached, this will occur at a phase of the cell cycle in which two complete copies of the chromosome are present. Narula and coworkers recently showed that the pulse-like dynamics of Spo0A activation largely rely on two requirements, the first one being the presence of negative feedback between Spo0A∼P and one of the upstream phosphotransferases, Spo0F [7]. Crucially, this feedback is not instantaneous, but delayed since Spo0A∼P transcriptionally increases levels of Spo0F, which in turn is thought to interfere with substrate binding by KinA, the major sporulation kinase, lowering the phosphate flux towards Spo0A. The second requirement is that, during the replication cycle, there is a temporary imbalance between the copy numbers of spo0F and kinA. This imbalance is due to their different locations on the genome: since spo0F is located close to the origin, while kinA is located closer to ter, the kinA:spo0F ratio will temporarily drop after replication of spo0F, leading to a gradual decrease in Spo0A∼P. After kinA is also replicated, the Spo0A∼P level increases again, then overshoots (i.e., pulses), due to the aforementioned delayed negative feedback loop, and only then falls back to its equilibrium level. In accordance with this model, the researchers showed that translocating kinA and/or spo0F led to sporulation defects, stressing the importance of their chromosomal location for successful endospore formation.
Temporary Diploidy Ensures Mutual Exclusion of Sporulation and Biofilm Formation
Although biofilm formation and sporulation are activated by the same protein, phosphorylated Spo0A (Spo0A∼P), both states are mutually exclusive since matrix production is completely absent in cells that have started sporulating 51, 52. The regulation of matrix genes by Spo0A∼P is indirect, by activation of the gene encoding SinI, which counteracts SinR, a repressor of genes responsible for matrix production. Chai et al. showed that while the repression by SinR is cooperative, derepression by SinI takes place uncooperatively, that is, single SinI molecules bind SinR [51]. They argued that these binding characteristics provided an answer for the somewhat paradoxical mutual exclusion of sporulation and matrix formation. Part of the explanation lies in the difference in Spo0A∼P binding affinity between biofilm and sporulation gene promoters; while biofilm formation is mainly triggered by lower levels of Spo0A∼P, sporulation is activated only when levels are high [47]. More importantly though, the cells make use of the unique situation at the start of sporulation, when there are two complete copies of the chromosome present for a prolonged period of time (Figure 2C) [53]. Due to this temporary diploidy, copy numbers of terminus-proximal genes sinI and sinR are roughly doubled. Due to the cooperative nature of repression by SinR, this may definitively shift the balance towards complete absence of matrix gene expression.
Physical Compartmentalization Is Responsible for Transcriptional Differentiation between Mother Cell and Prespore
In the course of spore development the replicated chromosome of the predivisional cell is segregated such that both the mother cell and the spore receive one copy. Importantly, however, in the early stages of sporulation only the oriC-proximal one-third of the chromosome is translocated into the prespore (Figure 2D–E). The remainder of the molecule is gradually translocated over a time period of around 15 minutes 54, 55.
The transcriptional differentiation between the two compartments (prespore and mother cell) is largely initiated by two different sigma factors, σE and σF, that activate two separate gene expression programs in the mother cell and prespore, respectively [44].
As discussed by Hilbert and Piggot, σF activity is obstructed by anti-σ protein SpoIIAB (Figure 2D). The release of σF is promoted by the dephosphorylated form of SpoIIAA (an anti-anti-σ protein), which can, however, become phosphorylated through the kinase activity of SpoIIAB. Dephosphorylation of SpoIIAA is performed by protein phosphatase SpoIIE [44].
While σF, SpoIIAA, and SpoIIAB are encoded at a terminus-proximal site on a single transcriptional unit, spoIIE is located close to oriC. It was suggested that the transient overproduction of SpoIIE may be responsible for the eventual release of active σF in the prespore (Figure 2D)56, 57. Frandsen et al. showed that moving the coding sequence for σF to an origin-proximal site allowed spore production even in the absence of the normally required factors SpoIIAA and SpoIIE [56]. McBride et al. showed that displacement of spoIIE to origin-distal sites partially repressed prespore-specific genes, but did not affect overall sporulation efficiency [57]. These findings suggest that more factors play a role in initiating the prespore's transcriptional program.
In the mother cell, the precursor of σE (pro-σE), encoded by sigE, is inactive due to an N-terminal 27-amino-acid extension that can be cleaved off by SpoIIGA, resulting in active σE. Even though the N-terminal extension may be relevant for proper development of sporulation, it is the terminus-proximal location of the sigE-SpoIIGA operon that seems to be responsible for the initial accumulation in the mother cell [57]. To ensure that the mother cell only enters its transcriptional program when that of the prespore has fully initiated, σF-dependent, prespore-localized expression of another gene, spoIIR, is required for activation of σE in the mother cell (Figure 2E) 58, 59. SpoIIR is thought to function as a signal for the septum-localized SpoIIGA, which then processes pro-σE into σE [44]. The oriC-proximal location of spoIIR is required for sufficient expression levels in the prespore during asymmetrical segregation. This became clear when Khvorova et al. saw a drastic delay in timing and decrease in level of sporulation gene induction upon the translocation of spoIIR to a ter-proximal site [60].
In the mother cell, σF activity is prevented, perhaps in part due to lower copy number of SpoIIE, but probably mainly due to mother-cell-specific degradation of SpoIIE (Figure 2E) [57]. At the same time, σE activity in the prespore is repressed by binding to anti-σ protein CsfB, which is again encoded on the prespore-localized one-third of the chromosome (Figure 2D) [61].
B. subtilis sporulation thus provides us with many examples of the relevance of chromosome organization for key processes in a bacterial cell, through a variety of mechanisms.
Distortion of Natural Gene Dosage Fluctuation Induces Bacterial Competence
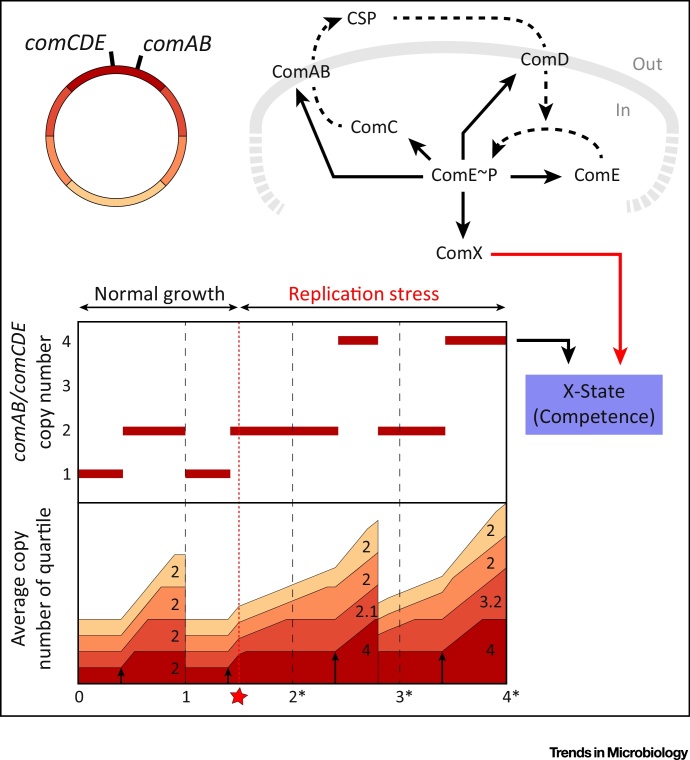
Whether or not a bacterium will perform multifork replication largely depends on the combination of its growth rate and its genome size. As discussed earlier, the oriC-proximal location of genes encoding important growth factors automatically correlates their production and requirement levels. A different way in which oriC-proximity is utilized is found in the pneumococcus (Figure 3). With its relatively small genome (∼2 Mb, [62]), multifork replication in rapidly dividing cells has not been observed [63]. This situation alters, however, when replication fork progression is directly or indirectly perturbed and slowed down. Since, as far as we know, there is no instantaneous feedback to the pneumococcal replication initiation system, new replication complexes may be loaded onto the genome before the stalled or slowed replication forks have finished, leading to increased dosage of oriC-proximal genes. Various factors can lead to this form of overinitiation: DNA damage (e.g., double-strand breaks induced by mitomycin C); insufficient functioning of type II topoisomerases, which are responsible for the relaxation of DNA required for replication forks to progress (e.g., induced by fluoroquinolone antibiotics); or limited nucleotide availability (e.g., induced by trimethoprim and hydroxyurea). S. pneumoniae makes use of this exceptional situation to activate competence or the so-called X-state, one of the most important stress response mechanisms it has at its disposal, allowing cells to take up exogenous DNA [64]. The activation of this system encompasses the expression of over a hundred genes 65, 66, 67, blocks cell division [68], and thus represents a significant burden for the cell. It is therefore important for the cell to somehow regulate the activation of this system. Despite the large number of genes eventually being activated, the on/off switch of the X-state is constituted by a positive feedback loop containing a set of only five genes organized into two operons [69], comAB and comCDE. Very-low-level basal expression occurs for both operons. ComC is a 41-residue peptide containing a double-glycine leader of 24 amino acids in length. Membrane-associated transporter complex ComAB exports ComC, cleaving off the leader peptide, and extracellularly releasing the 17-residue competence-stimulating peptide (CSP), which acts as a quorum-sensing autoinducer [70]. ComDE constitutes a typical two-component regulatory system; the membrane-bound histidine kinase ComD binds the extracellular CSP and consecutively transfers a phosphate group to the response regulator ComE, resulting in ComE∼P. ComE∼P then completes the positive feedback loop by enhancing expression of both comAB and comCDE [71]. Additionally, it induces the expression of comX, coding for the X-state-specific sigma factor σX, required for the activation of the entire competence regulon [72]. However, processes such as mRNA and protein degradation and dilution by growth will counteract this positive feedback loop and may prevent the X-state from switching on. Additionally, the autocatalytic efficiency of the system is dependent on medium parameters such as pH. Only when the local extracellular CSP concentration exceeds a certain threshold, the positive feedback may outcompete the counteracting forces and X-state gene expression may dramatically increase (possibly with several orders of magnitude). Hence, whether or not the X-state is activated depends on a complex set of parameters, including the copy numbers of comAB and comCDE; because of their oriC-proximal location on the chromosome (8˚ and –1˚, respectively), relative overinitiation (e.g., due to replication fork stalling) can push up the dosage of early X-state genes. It was shown that even a slight increase in dosage, of below twofold, can suffice to reach threshold CSP concentrations and lead to X-state activation [63]. Interestingly, it was recently shown that the production of pneumococcal bacteriocins (pneumocins) is also potentiated by X-state activation 73, 74. Since pneumocins play an important role in intra- and interspecies competition in their natural niche (the human nasopharynx), the gene-dosage-induced activation of the X-state may cause the composition of the nasopharyngeal flora to change, for better or for worse.
Figure 3.
Competence Activation in Streptococcus pneumoniae Due to Dosage Upshift of oriC-Proximal Regulator Genes. The oriC-proximal location of early competence genes allows the pneumococcus to activate this state in response to replication stress [63]. Simulated development of copy-number distribution during replication stress is shown in the bottom graph (bottom panel; same plotting parameters and (initially) same simulation parameters as in Figure 1A). Halfway in the second cell cycle, replication stress is applied (red star; new replication rate is one-third of original replication rate), while timing of replication initiation events is unaltered (black arrows). Note that time units indicated with an asterisk are multiples of the cell cycle time in the absence of replication stress. Due to the oriC-proximal location of comAB and comCDE, their expression levels increase (bottom graph, top panel) and once a certain threshold activity is reached, competence is activated via the positive feedback loop in its regulatory system (top right).
Implications for Future Research
mRNA Noise Prediction
Over the past 15 years it has become more and more clear that stochastic ‘noise’, which is intrinsically present in any process that takes place in a cell, can have tremendous effects on cell fate determination 75, 76. Phenotypic heterogeneity can arise when noisy expression of certain regulators only exceeds a threshold in a subpopulation of cells. To fully understand the impact of molecular noise on macroscopic properties, such as a bacterial phenotype, it is important to know all the relevant parameters that contribute to the absolute level of variation in a specific process. Recently, Peterson et al. showed that, for accurate modeling of mRNA distributions, it is imperative to account for DNA replication progressing and at some point doubling the copy number of the gene under study. Since replication is very tightly regulated, replication noise is negligible in this context, and incorporation into noise models should be relatively straightforward. Ignoring the effect of replication leads to an–in some cases very severe–overestimation of mRNA noise [77].
Normalization for Differential Gene Expression Analysis
With the upsurge of Next-Generation Sequencing (NGS) techniques, such as RNA-Seq, over the past decade, transcriptome studies have become much more attainable for many microbiologists around the globe. Analysis of the obtained datasets is often performed in automated pipelines that do not require much input from the user. The relative ease of use of these techniques has led to large amounts of highly valuable data. To our knowledge, however, none of the existing analysis packages take into account the possibility of a genome-wide copy number shift, which is expected in several conditions, including treatment with certain antibiotics [63] and changes in growth rate, temperature, or replication rate. As a result of such an altered gene-dosage pattern around the chromosome, it may become difficult to set a proper baseline of expression or differential expression; most normalization methods depend, at least in part, either on the assumption that most genes will have an unaltered expression level (e.g., upper-quartile or median normalization), or on the assumption that the total number of transcripts per cell remains roughly the same {e.g., transcripts per million (TPM, [78]) as a measure of expression}. In case of a global shift in copy-number distribution, neither of these assumptions is necessarily valid, and failing to acknowledge this may lead to overestimation of the number of truly differentially expressed genes, and simultaneously camouflage the changes of interest. The most accurate interpretation of data in these situations depends on the question one is trying to answer, but it is important to be aware of the role that copy-number changes have in these experiments.
Additionally, normalization methods for quantitative reverse-transcriptase-PCR (qRT-PCR) are usually based on the assumption that certain reference genes will keep a constant expression level in the various conditions that are being compared. This may be, even in the absence of copy-number effects, a very dangerous assumption, but it is especially so when gene-dosage distribution shifts are in play. It is therefore advisable to, in addition to using more than one reference gene, confirm that copy-number shifts are not responsible for the observed results. This can be accomplished by a qPCR experiment using chromosomal DNA as a template.
Copy-Number Effects in Synthetic and Natural Systems
In this review we highlighted the role of copy-number fluctuations in bacterial processes. In synthetic constructs the copy number of an integrated DNA sequence can be critical for its proper functioning and in these cases the location of chromosomal integration should be carefully deliberated [18].
Surprisingly, beyond the well known replication-associated gene dosage, not too many examples of bacterial decision-making are available that have been ascribed to the genomic location of key factors involved. Over the past couple of years, however, several of these examples have emerged 7, 9, 51, 63. Combined with the fact that DNA replication is universally present in all living organisms, this is highly suggestive of the possibility that these effects are much more abundant in bacterial biology than currently acknowledged. Going further, the reviewed phenomena are not necessarily limited to bacteria; archaeal chromosomes typically contain no more than a few replication origins, and since bacterial and archaeal chromosomes share several organizational traits [79], some of the mechanisms discussed here may very well be active in archaea as well.
Concluding Remarks
Various aspects of chromosome organization, including chromosome structure and topology, have been described and are increasingly being studied. The growing pool of knowledge on properties related to the spatial organization of genes, including accessibility to transcription-related proteins, spatial colocalization of genes, and mRNA and protein diffusibility, will be very important for the understanding of bacterial gene regulation. However, while not unrelated, chromosomal gene order is yet another aspect that affects the regulatory landscape of a bacterial cell, and has not received as much attention as necessary. As described in this review, the exact position of a gene on the chromosome determines when, where, and how often its DNA is copied. Although gene order is not critical for cell survival in laboratory conditions [80], the significance of this facet of chromosome organization is emphasized by the several examples that have emerged of regulatory processes that depend on the dynamic copy number fluctuations during DNA replication 7, 9, 51, 63. Future research will have to determine how widespread these mechanisms are (see Outstanding Questions).
Additionally, regardless of the role of the chromosomal location of a gene under natural circumstances, it is important to keep in mind the potential impact certain experiments will have on copy number distributions in a cell; translocation of genes, antibiotic treatment, nutrient limitation, and other types of stress can each in their own way induce transcriptional changes by affecting gene dosage, either locally or genome-wide. A better understanding and increased awareness of the role of chromosomal gene order in the regulation of key processes is therefore paramount in understanding bacteria, and possibly also archaea, both in nature and in the laboratory.
Outstanding Questions.
Is Bacillus subtilis exceptional in its versatile use of gene location or does it actually represent the first step towards a more complete understanding of the bacterial regulatory landscape?
To what extent can gene order and chromosome structure function as a blueprint for the localization of various processes within a cell?
Can stress-induced distortions of gene copy number distributions more generally explain the activation of the accompanying bacterial stress response?
What is the effect of the variability of an organism's natural environment on its tendency to employ copy number changes for cell fate regulation?
Taking into account copy number effects during transcriptome analysis, can new information be obtained from already existing data sets?
Acknowledgments
We thank M. Kjos and L.E. Keller for comments on the manuscript. Work in the Veening laboratory is supported by the EMBO Young Investigator Program, a VIDI fellowship (864.12.001) from The Netherlands Organisation for Scientific Research, Earth and Life Sciences (NWO-ALW) and ERC starting grant 337399-PneumoCell.
References
- 1.Rocha E.P.C., Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 2.Kahlem P. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 2004;14:1258–1267. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno S. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp. Cell Res. 1959;18:415–418. doi: 10.1016/0014-4827(59)90031-x. [DOI] [PubMed] [Google Scholar]
- 4.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 5.Hong J., Gresham D. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet. 2014;10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X., Zhang J. The genomic landscape of position effects on protein expression level and noise in yeast. Cell Syst. 2016;2:347–354. doi: 10.1016/j.cels.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narula J. Chromosomal arrangement of phosphorelay genes couples sporulation and DNA replication. Cell. 2015;162:328–337. doi: 10.1016/j.cell.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper S., Helmstetter C.E. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 9.Soler-Bistué A. Genomic location of the major ribosomal protein gene locus determines Vibrio cholerae global growth and infectivity. PLoS Genet. 2015;11:e1005156. doi: 10.1371/journal.pgen.1005156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mott M.L., Berger J.M. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 11.Murray H., Koh A. Multiple regulatory systems coordinate DNA replication with cell growth in Bacillus subtilis. PLoS Genet. 2014;10:e1004731. doi: 10.1371/journal.pgen.1004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholefield G. DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 2011;21:188–194. doi: 10.1016/j.tcb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Wolański M. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2014;5:735. doi: 10.3389/fmicb.2014.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couturier E., Rocha E.P.C. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol. Microbiol. 2006;59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- 15.Fossum S. Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J. 2007;26:4514–4522. doi: 10.1038/sj.emboj.7601871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sousa C. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiol. Read. Engl. 1997;143:2071–2078. doi: 10.1099/00221287-143-6-2071. [DOI] [PubMed] [Google Scholar]
- 17.Block D.H.S. Regulatory consequences of gene translocation in bacteria. Nucleic Acids Res. 2012;40:8979–8992. doi: 10.1093/nar/gks694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer C. Effect of genome position on heterologous gene expression in Bacillus subtilis; an unbiased analysis. ACS Synth. Biol. 2016 doi: 10.1021/acssynbio.6b00065. Published online May 20, 2016. [DOI] [PubMed] [Google Scholar]
- 19.Campo N. Chromosomal constraints in Gram-positive bacteria revealed by artificial inversions. Mol. Microbiol. 2004;51:511–522. doi: 10.1046/j.1365-2958.2003.03847.x. [DOI] [PubMed] [Google Scholar]
- 20.Mackiewicz P. Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-interactions1004. interactions, 1004.1-1004.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darling A.E. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 2008;4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rocha E.P.C. The organization of the bacterial genome. Annu. Rev. Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 23.Sobetzko P. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E42–E50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant J.A. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014;42:11383–11392. doi: 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrándiz M-J. Role of global and local topology in the regulation of gene expression in Streptococcus pneumoniae. PloS ONE. 2014;9:e101574. doi: 10.1371/journal.pone.0101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vora T. Protein occupancy landscape of a bacterial genome. Mol. Cell. 2009;35:247–253. doi: 10.1016/j.molcel.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brambilla E., Sclavi B. Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli. G3 (Bethesda). 2015;5:605–614. doi: 10.1534/g3.114.016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dillon S.C., Dorman C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 29.Dorman C.J. Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation. J. Mol. Microbiol. Biotechnol. 2014;24:316–331. doi: 10.1159/000368850. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein E., Drlica K. Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc. Natl. Acad. Sci. U.S.A. 1984;81:4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobetzko P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res. 2016;44:1514–1524. doi: 10.1093/nar/gkw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le T.B.K. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marbouty M. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Wang X. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Képès F. Periodic transcriptional organization of the E. coli genome. J. Mol. Biol. 2004;340:957–964. doi: 10.1016/j.jmb.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 36.Wright M.A. Chromosomal periodicity of evolutionarily conserved gene pairs. Proc. Natl. Acad. Sci. U.S.A. 2007;104:10559–10564. doi: 10.1073/pnas.0610776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montero Llopis P. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roggiani M., Goulian M. Chromosome-membrane interactions in bacteria. Annu. Rev. Genet. 2015;49:115–129. doi: 10.1146/annurev-genet-112414-054958. [DOI] [PubMed] [Google Scholar]
- 39.Kuhlman T.E., Cox E.C. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol. Syst. Biol. 2012;8:610. doi: 10.1038/msb.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moffitt J.R. Spatial organization shapes the turnover of a bacterial transcriptome. Elife. 2016;5:e13065. doi: 10.7554/eLife.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerganova V. Chromosomal position shift of a regulatory gene alters the bacterial phenotype. Nucleic Acids Res. 2015;43:8215–8226. doi: 10.1093/nar/gkv709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López D. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 43.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 44.Hilbert D.W., Piggot P.J. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higgins D., Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burbulys D. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 47.Fujita M., Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hauser P.M., Errington J. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J. Bacteriol. 1995;177:3923–3931. doi: 10.1128/jb.177.14.3923-3931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine J.H. Pulsed feedback defers cellular differentiation. PLoS Biol. 2012;10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veening J-W. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chai Y. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López D. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Errington J. Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr. Opin. Microbiol. 2001;4:660–666. doi: 10.1016/s1369-5274(01)00266-1. [DOI] [PubMed] [Google Scholar]
- 54.Wu L.J., Errington J. Bacillus subtilis SpoIIIE Protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 55.Pogliano J. Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J. Bacteriol. 2002;184:1743–1749. doi: 10.1128/JB.184.4.1743-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frandsen N. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 1999;13:394–399. doi: 10.1101/gad.13.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBride S.M. Contributions of protein structure and gene position to the compartmentalization of the regulatory proteins σE and SpoIIE in sporulating Bacillus subtilis. Mol. Microbiol. 2005;57:434–451. doi: 10.1111/j.1365-2958.2005.04712.x. [DOI] [PubMed] [Google Scholar]
- 58.Eldar A. Partial penetrance facilitates developmental evolution in bacteria. Nature. 2009;460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karow M.L. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khvorova A. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J. Bacteriol. 2000;182:4425–4429. doi: 10.1128/jb.182.16.4425-4429.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serrano M. Dual-specificity anti-sigma factor reinforces control of cell-type specific gene expression in Bacillus subtilis. PLoS Genet. 2015;11:e1005104. doi: 10.1371/journal.pgen.1005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tettelin H. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 63.Slager J. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 64.Claverys J-P. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 65.Dagkessamanskaia A. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 2004;51:1071–1086. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 66.Peterson S. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 2000;182:6192–6202. doi: 10.1128/jb.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rimini R. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 2000;36:1279–1292. doi: 10.1046/j.1365-2958.2000.01931.x. [DOI] [PubMed] [Google Scholar]
- 68.Oggioni M.R. Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2004;48:4725–4732. doi: 10.1128/AAC.48.12.4725-4732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin B. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 2000;38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 70.Håvarstein L.S. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin B. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence) Mol. Microbiol. 2013;87:394–411. doi: 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 72.Lee M.S., Morrison D.A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kjos M. Expression of Streptococcus pneumoniae bacteriocins is induced by antibiotics via regulatory interplay with the competence system. PLoS Pathog. 2016;12:e1005422. doi: 10.1371/journal.ppat.1005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wholey W-Y. Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog. 2016;12:e1005413. doi: 10.1371/journal.ppat.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elowitz M.B. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 76.Jørgensen M.G. Noise and stochasticity in gene expression: a pathogenic fate determinant. In: Harwood C., Wipat A., editors. Microbial synthetic biology. 1st edn. Elsevier; 2013. pp. 157–176. [Google Scholar]
- 77.Peterson J.R. Effects of DNA replication on mRNA noise. Proc. Natl. Acad. Sci. U.S.A. 2015;112:15886–15891. doi: 10.1073/pnas.1516246112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wagner G.P. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 79.Vieira-Silva S., Rocha E.P.C. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 2010;6:e1000808. doi: 10.1371/journal.pgen.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hutchison C.A. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]