Abstract
Background
Dysfunction of long non-coding RNAs (lncRNAs) has been demonstrated to be involved in psychiatric diseases. However, the expression patterns and functions of the regulatory lncRNAs in schizophrenia (SZ) patients have rarely been systematically reported.
Material/Methods
The lncRNAs in peripheral blood mononuclear cells (PBMCs) were screened and compared between the SZ patients and demographically-matched healthy controls using microarray analysis, and then were validated by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) method. Three verified significantly dysregulated lncRNAs of PBMCs were selected and then measured in SZ patients before and after the antipsychotic treatment. SZ symptomatology improvement was measured by Positive And Negative Syndrome Scale (PANSS) scores.
Results
One hundred and twenty-five lncRNAs were significantly differentially expressed in SZ patients compared with healthy controls, of which 62 were up-regulated and 63 were down-regulated. Concurrent with the significant decrease of the PANSS scores of patients after the treatment, the PBMC levels of lncRNA NONHSAT089447 and NONHSAT041499 were strikingly decreased (P<0.05). Down-regulation of PBMC expression of NONHSAT041499 was significantly correlated to the improvement of positive and activity symptoms of patients (r=−0.444 and −0.423, respectively, P<0.05, accounting for 16.9% and 15.1%, respectively), and was also significantly associated with better outcomes (odds ratio 2.325 for positive symptom and 12.340 for activity symptom).
Conclusions
LncRNA NONHSAT089447 and NONHSAT041499 might be involved in the pathogenesis and development of SZ, and the PBMC level of NONHSAT041499 is significantly associated with the treatment outcomes of SZ.
MeSH Keywords: Drug Therapy; Microarray Analysis; RNA, Long Noncoding; Schizophrenia
Background
Schizophrenia (SZ) is one of the most severely disabling mental disorders, which usually begins in early adulthood and features disordered symptoms such as hallucinations, delusions, disturbed communication, reduced motivation, and blunted affect. SZ has been estimated to have a median lifetime prevalence of 4.0 per 1000 persons worldwide [1], and is a global devastating health and socioeconomic burden. Current evidence demonstrates that SZ is attributable to the interactions between environmental and genetic factors [2]; however, the mechanisms of the pathogenesis of SZ are still unclear. We currently lack reliable and simple biomarkers for the diagnosis of SZ and prognosis of antipsychotic treatment, which hampers the early diagnosis and effective treatment of SZ patients [3,4].
LncRNAs are non-coding transcripts of longer than 200 nucleotides, and were previously often considered to be transcriptional ‘noise’ [5]. Recently, accumulating evidence has revealed that a number of lncRNAs play critical roles in the regulation of gene expression, and cell proliferation and differentiation, and participate in the pathogenesis and development of various diseases [6–9], especially neuropsychiatric disorders and neurodegenerative diseases such as Alzheimer’s disease [10], major depressive disorder [11], Parkinson’s disease [12], and autism spectrum disorders [13]. Barry et al. showed that dysregulation of lncRNA Gomafu led to defective alternative splicing patterns which link to SZ, implying the role of Gomafu in SZ [8]. Rao et al. reported that lncRNA MIAT was significantly associated with paranoid SZ among the Chinese Han population [14]. Recently, Ren et al. demonstrated that 2 lncRNA modules were significantly associated with early-onset SZ [15]. However, few studies have investigated lncRNA expression profiles in the peripheral blood cells of SZ patients, and the response of lncRNAs to the antipsychotic medications is unclear.
The present study systematically screened the differentially expressed lncRNAs in the peripheral blood mononuclear cells (PBMCs) from SZ patients compared with healthy controls, using microarray method. We then observed the changes in the PBMC levels of 3 verified significantly dysregulated lncRNAs in response to antipsychotic treatment in SZ patients, and analyzed the association of the variation of the lncRNA expression with the improvement of symptoms. This study provides new insights into the mechanisms underlying the pathogenesis of SZ, and suggests that lncRNAs might be considered as novel biomarkers for the diagnosis of SZ and prognosis of the related treatment, and potentially as therapeutic targets.
Material and Methods
Patients
A total of 106 SZ patients aged 20–50 years who met the diagnostic criteria based on the Diagnostic and Statistical Manual of Mental Disorders by American Psychiatric Association [16] were prospectively enrolled between December 2013 and May 2015 at the No. 102 Hospital of the People’s Liberation Army (Changzhou, China). All patients were of Han ethnicity and did not take any antipsychotic medications for at least 3 months before the enrollment. Patients with history of severe medical diseases, structural brain disorders, cognitive disability, other psychiatric disorders, unstable psychiatric features, or movement disorders were excluded. In addition, patients who had post-traumatic amnesia for over 24 h or received blood transfusion therapy within 1 month or electroshock therapy within 6 months were also excluded from the study.
Forty-eight age-, gender-, and ethnicity-matched healthy control subjects without any family history of major psychiatric disorders (e.g., SZ, major depressive disorder and bipolar disorder) within the last 3 generations, and without any history of blood transfusion therapy or severe traumatic injury within 1 month, were recruited. All the control subjects were also of Han ethnicity.
The study was approved by the Institutional Review Board of No. 102 Hospital of the People’s Liberation Army. Written informed consent was obtained from each subject.
RNA extraction
Whole blood (5 ml) from each subject was collected in EDTA-containing anticoagulant tubes and processed within 1 h. PBMCs were isolated from the blood samples through density gradient centrifugation, collected and stored at −80°C until use. Total RNA was isolated from PBMCs using Trizol regent (Invitrogen, Carlsbad, CA, USA) and RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturers’ protocols, followed by Turbo DNase treatment (Life Technologies, Carlsbad, CA, USA), quantification by NanoDrop ND-2000 (Thermo Scientific, Delaware, ME, USA), RNA integrity detection by gel electrophoresis, and reverse transcription (Superscript III; Invitrogen).
LncRNA microarray
RNA samples from 3 SZ patients (patient #1: male, 23 years old; patient #2: male, 31 years old; patient #3: female, 28 years old) and 3 healthy controls (patient #1: male, 20 years old; patient #2: male, 33 years old; patient #3: female, 26 years old) were used for lncRNA microarray profiling. The RNA sample labeling, microarray hybridizing, and washing were conducted according to the manufacturer’s standard protocols. Briefly, total RNA was transcribed to double-stranded cDNA, synthesized into cRNA, labeled with Cyanine-3-CTP, and then hybridized onto the Agilent Human lncRNA array v4.0 (4X180 K, Design ID: 062918, Agilent Technologies, Santa Clara, CA, USA). After washing, the arrays were then scanned by the Agilent Scanner G2505C (Agilent Technologies). Array images were analyzed using Feature Extraction software 10.7.1.1 (Agilent Technologies) to extract the raw data, which were further normalized with the quantile algorithm using Genespring software (Version 12.5; Agilent Technologies). The probes that were all flagged as “P” in at least 1 out of the 2 groups were chosen for further data analysis. LncRNAs data were shown as fold-changes relative to the controls. Differentially expressed lncRNAs were identified by fold-changes and P values through t-test. The threshold for up- and down-regulated expression was a fold-change ≥2.0 and a P value ≤0.05. Finally, hierarchical clustering was conducted to display the distinguishable lncRNA expression patterns among samples.
Real-time quantitative reverse-transcription PCR (qRT-PCR)
According to the microarray results, the 10 most dysregulated lncRNAs were chosen for further validation by qRT-PCR in 106 SZ patients versus 48 healthy controls. Blood samples were collected and PBMCs were isolated. Total RNAs were isolated from PBMCs using Trizol reagent (Invitrogen), and complementary DNA was synthesized using the Reverse Transcription TaqMan RNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s instructions. Real-time PCR was performed using a 7900HT Real-Time PCR System (Applied Biosystems). Data were analyzed using the SDS 2.3 software (Applied Biosystems) and DataAssist v3.0 software. The expression levels of lncRNAs were normalized to β-actin and were calculated using 2−ΔΔCt method.
Medical intervention and symptom assessment
According to the microarray and qRT-PCR results, 3 verified significantly dysregulated lncRNAs were further measured the expression variation in the PBMCs of 30 SZ patients before and after the antipsychotic treatment by qRT-PCR. Among these patients, 5 were treated with risperidone (starting dosage 2 mg, average dosage 3.9 mg, range 2–6 mg), 7 with ziprasidone (starting dosage 40 mg, average dosage 125 mg, range 40–140 mg), 8 with quetiapine (starting dosage 100 mg, average dosage 520 mg, range 100–800 mg), and 10 with olanzapine (starting dosage 5 mg, average dosage 12 mg, range 5–20 mg).
Positive and Negative Syndrome Scale (PANSS) is commonly used to evaluate the severity of symptoms of patients with SZ [17]. PANSS contains 33 items, including 3 for aggressiveness, 7 for positive symptoms, 7 for negative symptoms, and 16 for general psychopathological symptoms [17]. In this study, symptoms of patients were assessed at baseline and at 6 weeks after the antipsychotic treatment by experienced psychiatrists using the PANSS. The symptom improvement was reflected by the variation of the symptomatology scores and total score before and after the treatment. The reduction rate of symptomatology scores was calculated as the variation of symptomatology score before and after the medication treatment relative to the pre-medication symptomatology score.
Statistical analysis
Data are expressed as the mean ± standard deviation or percentages where appropriate, and were compared between SZ patient and healthy control groups using the Statistical Package for Social Sciences for Windows 22.0 (SPSS Inc. Chicago, IL, USA). The chi-square test was used to compare the categorical demographic variables, and the t test was used to compare quantitative demographic variables. The Mann-Whitney U test was used to compare the PBMC levels of the top 10 differentially expressed lncRNAs by microarray between SZ and healthy controls subjects. The paired-sample t test was for the comparison of the expression levels of lncRNAs in SZ patients between before and after the treatment. Pearson correlation analysis was performed to evaluate the correlation of change of the lncRNA expression level with the improvement of symptomatology scores. Regression analysis was then carried out using the variation of lncRNA NONHSAT041499 expression as independent variable and improvement of PANSS positive and activity symptoms as dependent variables. Stepwise regression analysis was to determine the lncRNA NHSAT041499 accountability of symptomatological improvement in SZ patients. ΔR2 was assessed to show the percentage of the variation of positive and activity subscales with the NHSAT041499 variation. Then, according to the reduction rate of symptomatology scores before and after the medication, SZ patients were divided into better (score reduction rate equal to or more than 50%) and worse (score reduction rate less than 50%) treatment outcome subgroups. Logistic regression analysis was then conducted to observe the association of NHSAT041499 with the treatment outcomes of patients, which was assessed by odds ratio (OR) and P values. P<0.05 (2-tailed) was considered statistically significant.
Results
Microarray analysis
Microarray analysis showed there were 125 lncRNAs significantly differentially expressed in SZ patients compared with healthy controls (fold change ≧2, P<0.05), among which 62 were up-regulated and 63 were down-regulated (Supplementary Table 1). The top 20 differentially expressed lncRNAs are shown in Table 1. In hierarchical clustering analysis, the normalized expression of the 125 significantly differentially expressed lncRNAs was recorded to generate a heat map, from which a general difference of the lncRNA expression in blood samples from SZ patients versus healthy control subjects were clearly displayed (Figure 1).
Table 1.
Top 20 aberrantly expressed lncRNAs in peripheral blood mononuclear cells from Schizophrenia patients versus healthy controls by microarray analysis.
| lncRNA | Fold-change | P-value | Style | Chromosome start end |
|---|---|---|---|---|
| ENST00000394742 | 5.6865215 | 0.001366189 | Down | Chr12 13100085 13106891 |
| TCONS_l2_00025502 | 5.130027 | 0.016324848 | Down | Chr6 143360562 143363461 |
| NONHSAT041499 | 5.1150675 | 0.004036811 | Up | Chr15 32806172 32819079 |
| NONHSAT098126 | 4.610542 | 0.047743667 | Up | Chr4 121606074 121631566 |
| NONHSAT003974 | 4.4428 | 0.006958588 | Up | Chr1 75428997 75430802 |
| ENST00000519337 | 4.2703557 | 0.020613242 | Up | Chr8 103866683 103868303 |
| ENST00000563823 | 3.775364 | 0.012858684 | Down | Chr1 672557231 72661585 |
| NONHSAT021545 | 3.6979194 | 0.023714427 | Up | Chr11 59570202 59573350 |
| ENST00000496491 | 3.6021621 | 0.026623152 | Up | Chr3 149095565 149104370 |
| ENST00000521622 | 3.5583007 | 0.00341083 | Down | Chr8 106810555 107072719 |
| NONHSAT 089447 | 3.514152 | 0.038919978 | Up | Chr3 46598887 46601178 |
| TCONS_l2_00021339 | 3.4050841 | 0.010671916 | Down | Chr4 147030377 147043080 |
| TCONS_l2_00024969 | 3.370955 | 0.017096879 | Down | Chr6 147182804 147240209 |
| NONHSAT104778 | 3.2426178 | 0.03928101 | Up | Chr5 157174512 157174715 |
| ENST00000581634 | 3.0800865 | 0.024413016 | Up | Chr18 76265165 76266410 |
| TCONS_l2_00000563 | 2.9737065 | 0.030129751 | Down | Chr1 143398221 143401768 |
| NONHSAT074892 | 2.9536893 | 0.03315027 | Down | Chr2 149666582 149685052 |
| NONHSAT005508 | 2.947432 | 0.030145906 | Up | Chr1 118628601 118629785 |
| TCONS_l2_00012438 | 2.896232 | 0.027099133 | Down | Chr19 28409545 28447647 |
| NONHSAT016970 | 2.8774061 | 0.04668065 | Up | Chr10 129849591 129850501 |
Figure 1.
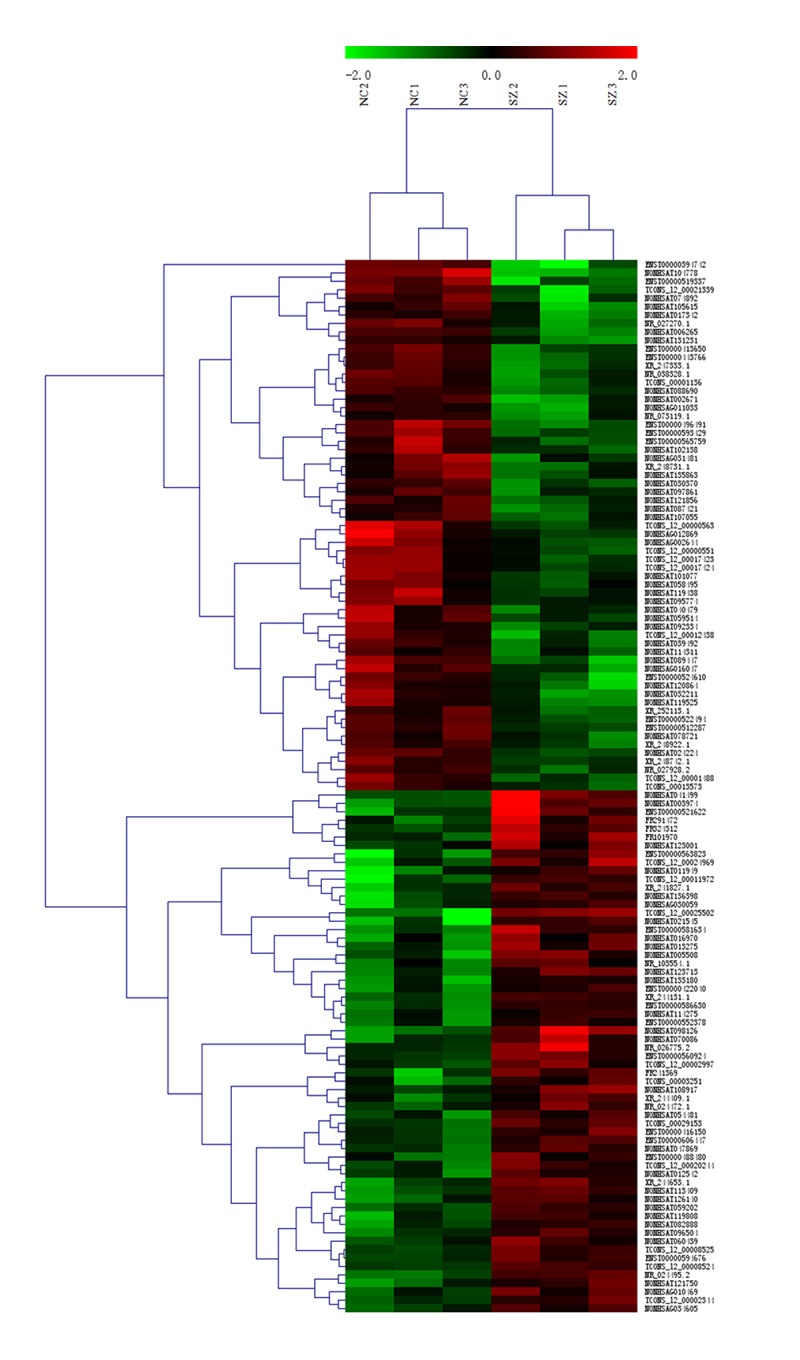
Hierarchical clustering analysis of differentially expressed lncRNAs in peripheral blood mononuclear cells from schizophrenia patients versus normal controls. Rows represent differentially expressed lncRNAs and columns represent blood samples. Color scale depicts the relative lncRNA expression; red shows up-regulation and green shows down-regulation. The 2.0, 0, and -2.0 indicate fold-changes in the corresponding spectrum. NC represents blood samples from normal controls and SZ represents samples from SZ patients. The differentially expressed lncRNAs were self-segregated into NC and SZ clusters.
Clinical characteristics of the patients
As shown in Table 2, the mean age of patients and healthy controls was 30.49±12.86 and 29.61±12.32 years, respectively. There was no significant difference in age, gender, residential location, sibling status, education, marital status, or family history of mental disorders between SZ patients and healthy controls (P>0.05, Table 2).
Table 2.
Demographic characteristics of patients with schizophrenia and healthy controls.
| Patients (n=106) | Controls (n=48) | t | P-value | |
|---|---|---|---|---|
| Age (years) | 30.49±12.86 | 29.61±12.32 | −0.379 | 0.705 |
| Gender (n/%) | 1.523 | 0.683 | ||
| Female | 52 (49.1%) | 25 (52.1%) | ||
| Male | 54 (50.9%) | 23 (47.9%) | ||
| Inhabitants (n/%) | 1.958 | 0.892 | ||
| Urban | 47 (44.3%) | 22 (45.8%) | ||
| Rural | 59 (55.7%) | 26 (54.2%) | ||
| Sibling status (n/%) | 2.123 | 0.096 | ||
| Only-child | 62 (58.5%) | 26 (54.2%) | ||
| Non-only child | 44 (41.5%) | 22 (45.8%) | ||
| Education (n/%) | 2.325 | 0.075 | ||
| Junior high school or below | 59 (55.7%) | 21 (43.8%) | ||
| Senior high school or above | 47 (44.3%) | 27 (56.2%) | ||
| Marital status (n/%) | 1.538 | 0.356 | ||
| Married | 50 (47.2%) | 28 (58.3%) | ||
| Unmarried | 56 (52.8%) | 20 (41.7%) | ||
| Family history of mental disorders (n/%) | 1.621 | 0.125 | ||
| With | 19 (17.9%) | 7 (14.6%) | ||
| Without | 87 (82.1%) | 41 (85.4%) |
qRT-PCR validation
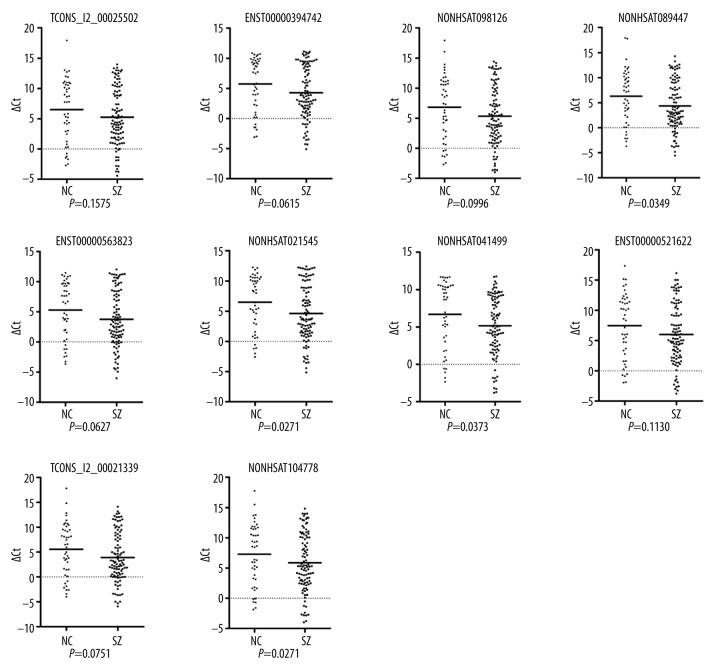
To validate the results of the microarray assay, 10 of the top 20 significantly differentially expressed lncRNAs (including 5 up-regulated lncRNAs: NONHSAT098126, NONHSAT089447, NONHSAT021545, NONHSAT041499, and NONHSAT104778, and 5 down-regulated lncRNAs: ENST00000394742, TCONS_l2_00025502, ENST00000563823, ENST00000521622, and TCONS_l2_00021339) were chosen for further validation in larger blood samples from 106 patients versus 48 healthy controls using qRT-PCR method. Results showed that the expression of lncRNAs NONHSAT089447, NONHSAT021545, NONHSAT041499, NONHSAT098126, and NONHSAT104778 was consistent with the microarray results, of which the first 3 lncRNAs exhibited significant difference of expression between patients and healthy controls (P<0.05) (Figure 2). These first 3 up-regulated lncRNAs were then chosen for further study.
Figure 2.
Validation of the expression of lncRNAs by qRT-PCR analysis in the peripheral blood mononuclear cells from schizophrenia patients (n=106) and normal controls (n=48). The line represents the median value, and the plots were constructed by using GraphPad Prism 5 software. Statistical difference was analyzed using the Mann-Whitney U test.
LncRNA expression, and symptomatology scores and total score before and after the treatment in SZ patients
As shown in Table 3, ΔCT values of lncRNAs NONHSAT089447 and NONHSAT041499 were significantly increased in patients after the treatment (P<0.001), indicating the significant down-regulation of these lncRNA expression by the treatment. Consistently, the symptomatology scores and total score were significantly decreased after the treatment (P<0.001, Table 3).
Table 3.
LncRNA expression, symptomatology scores and total score before and after the antipsychotic treatment in schizophrenia patients.
| Item | Baseline (n=30) | After treatment (n=30) | t | P |
|---|---|---|---|---|
| NONHSAT089447 (ΔCT) | 3.23±4.26 | 5.13±3.51 | −4.577 | <0.001 |
| NONHSAT021545 (ΔCT) | 3.67±4.12 | 4.04±4.55 | −0.858 | 0.398 |
| NONHSAT041499 (ΔCT) | 4.56±3.92 | 6.77±3.13 | −5.056 | <0.001 |
| Positive subscale | 19.94±5.98 | 9.32±4.78 | 8.993 | <0.001 |
| Negative subscale | 21.29±8.24 | 10.58±4.56 | 8.778 | <0.001 |
| General psychopathology subscale | 40.90±10.85 | 19.71±5.05 | 11.29 | <0.001 |
| Total score | 81.84±18.13 | 39.61±7.64 | 13.80 | <0.001 |
| Lack of response | 10.10±4.57 | 6.00±2.42 | 5.507 | <0.001 |
| Disturbance of thought | 12.42±3.97 | 5.64±2.59 | 9.455 | <0.001 |
| Activity | 5.84±2.19 | 3.29±2.12 | 7.289 | <0.001 |
| Paranoid | 7.16±2.78 | 3.58±1.03 | 7.271 | <0.001 |
| Depression | 10.68±3.30 | 5.74±2.62 | 8.093 | <0.001 |
| Aggressiveness | 13.03±3.73 | 7.07±2.02 | 9.276 | <0.001 |
Down-regulation of lncRNA NONHSAT041499 expression was correlated with symptom improvement in SZ patients
Pearson correlation analysis revealed that the down-regulation of the lncRNA NONHSAT041499 expression was significantly correlated with the improvement of positive and activity symptoms after the treatment (r=−0.444 and −0.423, respectively, P<0.05, Table 4).
Table 4.
Correlation between the down-regulation of lncRNA expression and improvement of symptoms in schizophrenia patients.
| Item | NONHSAT089447 (r value) | NONHSAT041499 (r value) |
|---|---|---|
| Positive subscale | −0.122 | −0.444* |
| Negative subscale | −0.160 | −0.065 |
| General psychopathology subscale | −0.093 | −0.107 |
| Total score | −0.146 | −0.216 |
| Lack of response | −0.143 | −0.064 |
| Disturbance of thought | 0.139 | 0.116 |
| Activity | −0.193 | −0.423* |
| Paranoid | −0.172 | −0.016 |
| Depression | −0.035 | 0.149 |
| Aggressiveness | −0.323 | −0.110 |
P<0.05 represented significant difference for the correlation between the lncRNA with the symptoms.
LncRNA NONHSAT041499 down-regulation was significantly associated with improvement of treatment outcomes of SZ patients
Step-wise regression analysis revealed that the down-regulation of lncRNA NONHSAT041499 expression as independent variable accounted for 16.9% (ΔR2=0.169) and 15.1% (ΔR2=0.151) of the improvement of positive and activity symptoms, respectively (Table 5).
Table 5.
LncRNA NONHSAT041499 down-regulation accountability of symptomatology improvement of schizophrenia patients by step-wise regression analysis.
| Dependent variables | Regression model | Partial regression coefficient | Standard error | Standard coefficient | t | ΔR2 | P value |
|---|---|---|---|---|---|---|---|
| Δ Positive symptom | Constant | −7.965 | 1.464 | −5.440 | 0.000 | ||
| Δ NONHSAT041499 | −1.144 | 0.429 | −0.444 | −2.666 | 0.169 | 0.012 | |
| Δ Activity symptom | Constant | −1.800 | 0.438 | −4.106 | 0.000 | ||
| Δ NONHSAT041499 | −0.323 | 0.128 | −0.423 | −2.515 | 0.151 | 0.018 |
To further validate the association of NONHSAT041499 with the antipsychotic treatment outcomes, SZ patients were divided into better and worse treatment outcome subgroups according to the symptomatology score reduction rate. Taking the down-regulation of NONHSAT041499 expression as an independent variable, and the improvement of positive and activity symptoms as dependent variables, logistic regression analysis was carried out. The results showed that for positive symptom, the OR determined by NONHSAT041499 down-regulation (calculated as the increase of CT value, ΔCT) of better treatment outcome subgroup against worse treatment outcome subgroup was 2.325, and for activity symptom the OR was 12.340 (Table 6)
Table 6.
Association between lncRNA NONHSAT041499 down-regulation and treatment outcomes of schizophrenia patients by logistic regression analysis.
| Dependent variables | Regression model | B | Standard error | Wals | Odds ratio | Determination coefficient | P value |
|---|---|---|---|---|---|---|---|
| Δ Positive symptom | Constant | −1.912 | 0.800 | 5.717 | 0.148 | 0.017 | |
| Δ NONHSAT041499 | 0.844 | 0.319 | 7.018 | 2.325 | 0.366 | 0.008 | |
| Δ Activity symptom | Constant | −4.347 | 1.831 | 5.637 | 0.013 | 0.013 | |
| Δ NONHSAT041499 | 2.513 | 1.010 | 6.186 | 12.340 | 0.607 | 0.018 |
Discussion
Current treatment for SZ mainly comprises dopamine receptors system [18,19], 5-HT receptors [20], and GABA system [21] drugs, but the pharmacological mechanisms remain elusive. This makes it difficult for more effective treatment and prognosis of the outcomes. LncRNAs play important roles in various pathologic processes, including neuropsychiatric disorders and neurodegenerative diseases [11,13]. To date, only a few studies reported that lncRNAs were significantly associated with SZ [14,15]. There have been few reports on lncRNA expression profiling in SZ patients. Only a recent study demonstrated a microarray profiling of lncRNAs of SZ patients, with the focus on the analysis of co-expression network of lncRNAs and mRNAs and their correlation [15]. The association between these lncRNAs and the treatment outcomes of SZ patients is still unclear.
This study systematically screened the differentially expressed lncRNAs in SZ patients in comparison to healthy controls and demonstrated that lncRNAs NONHSAT089447, NONHSAT021545, and NONHSAT041499 were significantly up-regulated in SZ patients. Down-regulation of NONHSAT089447 and NONHSAT041499 was concurrent with the improvement of symptoms of patients after the anti-psychotropic medication. These results suggest that these lncRNAs might be involved in the pathogenesis and development of SZ and could be considered as novel potential treatment targets. Reportedly, lncRNAs are transcribed in complex patterns (e.g., intergenic, overlapping, and antisense patterns) relative to the adjacent protein-coding genes [9], and participate in the regulation of the target gene expression by inducing chromatin remodeling and targeting transcription factors [7,22], suggesting the complexity of the regulatory pathways of lncRNAs. An integrated co-expression network analysis revealed significant correlation between lncRNAs and mRNAs, and that the lncRNAs, together with mRNAs, constructed co-expressed modules, some of which were associated with early-onset SZ [15]. Barry et al. showed that lncRNA Gomafu directly binds to the splicing factors, such as serine/arginine-rich splicing factor 1, to regulate the alternative splicing patterns whose defection is linked to SZ [8]. Ishizuka et al. reported that lncRNA Gomafu indirectly modulated RNA-binding protein Celf3 and other splicing factors to regulate the functions of the SZ-related genes, thus playing roles in SZ [23]. How these lncRNAs modulate these SZ-related genes to regulate SZ warrants further investigation.
Traditionally, diagnosis of SZ is based on the clinical symptoms [16,17]. Functional neuroimaging techniques have been developed to detect the neurotransmitters (e.g., dopamine and glutamate) implicated in SZ, and SZ-associated regional brain activity [24]. However, at present it is difficult to diagnose SZ because neither single clinical symptoms nor neurotransmitters are unique for SZ. Symptoms or SZ-related brain activity are usually manifested or detected when SZ is developed to a certain stage, which means such symptoms or brain activity-based diagnosis might lead to the delay of the SZ treatment. Accurate and early diagnosis of SZ is very important. This study demonstrates that lncRNAs NONHSAT089447, NONHSAT021545, and NONHSAT041499 were significantly up-regulated in SZ patients and that NONHSAT089447 and NONHSAT021545 down-regulation was significantly correlated with improvement of symptoms by the anti-psychotropic treatment, suggesting these lncRNAs could be used as new non-invasive biomarkers for the diagnosis of SZ. Particularly, based on the findings that lncRNAs play roles in SZ via upstream regulating the SZ-related genes [8,15,23], it is anticipated that lncRNAs could be even considered as early diagnostic biomarkers for SZ, which will be beneficial for the early treatment of SZ. Our next study, with larger patient numbers, will be carried out to validate the diagnostic value of NONHSAT041499 in SZ.
At present, although drugs (mainly dopamine receptor system) have been employed, SZ is difficult to treat. For more effective treatment, it is therefore necessary to predict the treatment outcomes. In this study, through Pearson correlation, and step-wise and logistic regression analysis, we revealed that NONHSAT041499 down-regulation was significantly correlated to the improvement of positive and activity symptoms of patients after the medication treatment, and was also significantly associated with better outcomes. This result implies NONHSAT041499 might be considered as a potent prognosis factor for the treatment outcome. To the best of our knowledge, similar studies have not been reported yet. Recently, we reported that down-regulation of plasma miRNA-181b predicted symptom improvement of SZ patients after antipsychotic treatment [25]. These results suggest potential non-invasive molecular markers for the prognosis of SZ patients. Further studies with larger sample sizes are needed to confirm the potential of NONHSAT041499 as a prognostic factor for the treatment of SZ.
LncRNAs closely interact with the regulated mRNAs. LncRNA NONHSAT089447, NONHSAT021545, and NONHSAT041499 were found to be co-expressed with many mRNAs that regulate various biological processes, including neuron apoptosis, learning, memory, behavior, sensory perception of sound, synapse organization and activity, layer formation in the cerebral cortex, stress-activated protein kinase signaling pathway, Ras protein signal transduction, and small GTPase-mediated signal transduction (unpublished data). The detailed bioinformatics study of the above-mentioned lncRNAs is under investigation, and molecular mechanisms whereby these lncRNAs participate in the pathogenesis and development of SZ need to be extensively explored.
A limitation of this study is that the sample size is relatively small for the regression analysis. Small patient numbers in the control group relative to the SZ group might decrease the statistical power for the comparison of the lncRNA expression levels between SZ and control groups. Further studies with more patients and more variables are needed to validate the present results.
Conclusions
We systematically screened the differentially expressed lncRNAs in the PBMCs of SZ patients, and demonstrated that down-regulation of NONHSAT041499 was significantly associated with the symptom improvement and better treatment outcomes of SZ patients. This study will be beneficial for the investigation of the mechanisms underlying the pathogenesis and development of SZ, and suggests the potential usefulness of lncRNA NONHSAT041499 as a novel biomarker for the diagnosis of SZ and prognosis of the treatment, as well as being a potential treatment target.
Supplementary materials
Supplementary Table 1.
Differentially expressed lncRNAs in peripheral blood mononuclear cells from Schizophrenia patients versus healthy controls by microarray.
| lncRNA | Fold-change | P-value | Style |
|---|---|---|---|
| TCONS_l2_00008525 | 2.0184042 | 0.007859847 | Up |
| FR324312 | 2.5046852 | 0.019613983 | Up |
| FR241369 | 2.795938 | 0.026778212 | Up |
| NR_024495.2 | 2.6708856 | 0.001104883 | Up |
| NONHSAG012869 | 2.7815604 | 0.048556764 | Down |
| NONHSAG030059 | 2.4375992 | 0.04930478 | Up |
| NONHSAT119438 | 2.386957 | 0.042039905 | Down |
| XR_248742.1 | 2.0546277 | 0.013295304 | Down |
| TCONS_l2_00017424 | 2.2616987 | 0.039616782 | Down |
| NONHSAT123713 | 2.7456982 | 0.015772445 | Up |
| TCONS_l2_00017423 | 2.3137672 | 0.04239666 | Down |
| NONHSAT078721 | 2.090962 | 0.03983514 | Down |
| NONHSAT104778 | 3.2426178 | 0.03928101 | Up |
| NONHSAT016970 | 2.8774061 | 0.04668065 | Up |
| NONHSAT006265 | 2.74296 | 0.003577456 | Down |
| NONHSAT059514 | 2.416889 | 0.034944758 | Down |
| NONHSAT041499 | 5.1150675 | 0.004036811 | Up |
| NONHSAT092334 | 2.22519 | 0.045760788 | Down |
| TCONS_00029153 | 2.2907834 | 0.005757978 | Up |
| NONHSAT 089447 | 3.514152 | 0.038919978 | Up |
| ENST00000519337 | 4.2703557 | 0.020613242 | Up |
| NONHSAG016047 | 2.8309715 | 0.03872216 | Up |
| FR101970 | 2.8698637 | 0.029207746 | Up |
| ENST00000563823 | 3.775364 | 0.012858684 | Down |
| NONHSAT121750 | 2.3670413 | 0.028498909 | Up |
| NONHSAT135863 | 2.3467486 | 0.031746913 | Down |
| NONHSAT021545 | 3.6979194 | 0.023714427 | Up |
| NONHSAT011949 | 2.8012006 | 0.047748405 | Up |
| NR_038328.1 | 2.4504633 | 0.025065137 | Down |
| NONHSAT059202 | 2.1078913 | 0.003934796 | Up |
| TCONS_00013573 | 2.016616 | 0.021455377 | Down |
| XR_241827.1 | 2.519167 | 0.043524686 | Up |
| NONHSAT123001 | 2.2459533 | 0.04912253 | Up |
| NONHSAG034605 | 2.0021873 | 0.013193308 | Up |
| ENST00000581634 | 3.0800865 | 0.024413016 | Up |
| ENST00000522494 | 2.1406422 | 0.009605092 | Up |
| NONHSAT120864 | 2.6860943 | 0.047448784 | Down |
| NONHSAT108917 | 2.201703 | 0.025858361 | Up |
| NONHSAG010469 | 2.170598 | 0.02606947 | Up |
| NONHSAT040479 | 2.4519546 | 0.049309734 | Down |
| ENST00000488480 | 2.2488601 | 0.04426648 | Up |
| NONHSAT054481 | 2.1365309 | 0.033978045 | Up |
| NONHSAT131231 | 2.2493892 | 0.022191135 | Down |
| NONHSAT013275 | 2.7648053 | 0.026773857 | Up |
| ENST00000560924 | 2.2441285 | 0.008775448 | Up |
| TCONS_00003251 | 2.3368568 | 0.048376054 | Up |
| NONHSAT133180 | 2.255505 | 0.039863095 | Up |
| ENST00000422040 | 2.246702 | 0.034511745 | Up |
| NONHSAT113409 | 2.6231997 | 0.006450792 | Up |
| ENST00000552378 | 2.0455449 | 0.039099984 | Up |
| NONHSAT098126 | 4.610542 | 0.047743667 | Up |
| NONHSAT105615 | 2.6867616 | 0.037940864 | Down |
| NONHSAT119525 | 2.5023258 | 0.041274253 | Down |
| ENST00000565759 | 2.545508 | 0.038889498 | Down |
| TCONS_00001136 | 2.401271 | 0.017750448 | Down |
| NONHSAT003974 | 4.4428 | 0.006958588 | Up |
| NONHSAT101077 | 2.382952 | 0.025674935 | Down |
| TCONS_l2_00002344 | 2.021424 | 0.013821459 | Up |
| XR_244131.1 | 2.320134 | 0.005847468 | Up |
| NR_027928.2 | 2.035199 | 0.010459018 | Down |
| NR_026775.2 | 2.709865 | 0.037649963 | Up |
| TCONS_l2_00024969 | 3.370955 | 0.017096879 | Down |
| TCONS_l2_00021339 | 3.4050841 | 0.010671916 | Down |
| XR_252113.1 | 2.1277385 | 0.019082548 | Down |
| TCONS_l2_00001488 | 2.42787 | 0.016129335 | Down |
| ENST00000593429 | 2.664398 | 0.005580793 | Down |
| NONHSAT032211 | 2.8629696 | 0.03303966 | Down |
| NONHSAT039492 | 2.419247 | 0.012648112 | Down |
| NONHSAT121856 | 2.2077594 | 0.013875067 | Down |
| NONHSAT126140 | 2.4100482 | 0.026816849 | Up |
| ENST00000413650 | 2.50962 | 0.010103071 | Down |
| NONHSAG002644 | 2.5878246 | 0.046584073 | Down |
| ENST00000512287 | 2.095495 | 0.00460248 | Down |
| NONHSAT058495 | 2.0634727 | 0.04672545 | Down |
| ENST00000416150 | 2.0931425 | 0.029960617 | Up |
| XR_247333.1 | 2.482382 | 0.01222255 | Down |
| NR_027270.1 | 2.7393317 | 0.023086803 | Down |
| NONHSAT012542 | 2.0278077 | 0.03747045 | Up |
| ENST00000594676 | 2.0075424 | 0.007863165 | Up |
| FR291472 | 2.8738096 | 0.045134224 | Up |
| NR_103554.1 | 2.3072157 | 0.048855953 | Up |
| NONHSAT102138 | 2.3331935 | 0.035092298 | Down |
| NONHSAT095774 | 2.0473397 | 0.03872717 | Down |
| XR_244653.1 | 2.6912992 | 0.01806327 | Up |
| ENST00000521622 | 3.5583007 | 0.00341083 | Down |
| TCONS_l2_00002997 | 2.1203494 | 0.024146989 | Up |
| NONHSAT114311 | 2.045813 | 0.034777325 | Down |
| ENST00000586630 | 2.2811098 | 0.009588628 | Up |
| NONHSAT024224 | 2.280496 | 0.001247224 | Down |
| NONHSAT060439 | 2.0503778 | 0.03933156 | Up |
| ENST00000443766 | 2.5501273 | 0.006421687 | Down |
| NONHSAT017342 | 2.3088417 | 0.027970523 | Down |
| NONHSAG031481 | 2.6192472 | 0.04883049 | Down |
| NONHSAT097861 | 2.0948513 | 0.03657174 | Down |
| ENST00000524610 | 2.7294452 | 0.03419223 | down |
| NONHSAT047869 | 2.046003 | 0.014862789 | Up |
| ENST00000394742 | 5.6865215 | 0.001366189 | Down |
| TCONS_l2_00008524 | 2.0070338 | 2.13E-04 | Up |
| NONHSAT136398 | 2.4874713 | 0.046239804 | Up |
| NR_024472.1 | 2.0432649 | 0.027397072 | Up |
| NONHSAG011033 | 2.3757823 | 0.0297065 | Down |
| ENST00000496491 | 3.6021621 | 0.026623152 | Up |
| NONHSAT114275 | 2.109495 | 0.037710313 | Up |
| ENST00000606447 | 2.0563893 | 0.011615175 | Up |
| XR_244409.1 | 2.0452442 | 0.049372125 | Up |
| TCONS_l2_00000563 | 2.9737065 | 0.030129751 | Down |
| NONHSAT082888 | 2.0611625 | 0.02947335 | Up |
| NONHSAT005508 | 2.947432 | 0.030145906 | Up |
| TCONS_l2_00000551 | 2.3637156 | 0.031189756 | Down |
| NONHSAT070086 | 2.7337165 | 0.039751925 | Up |
| XR_248731.1 | 2.5972993 | 0.022589074 | Down |
| NONHSAT119808 | 2.2518048 | 0.03818275 | Up |
| TCONS_l2_00012438 | 2.896232 | 0.027099133 | Down |
| NONHSAT074892 | 2.9536893 | 0.03315027 | Down |
| TCONS_l2_00020244 | 2.2335203 | 0.027745022 | Up |
| NONHSAT107055 | 2.0738862 | 0.022926092 | Down |
| NONHSAT030370 | 2.3975239 | 0.006883643 | Down |
| NONHSAT087421 | 2.249703 | 0.021947894 | Down |
| NONHSAT096504 | 2.0448937 | 0.02514679 | Up |
| NONHSAT002671 | 2.6095362 | 0.02860338 | Down |
| TCONS_l2_00011972 | 2.8288274 | 0.034055557 | Up |
| NONHSAT088690 | 2.270206 | 0.006502805 | Down |
| TCONS_l2_00025502 | 5.130027 | 0.016324848 | Down |
| NR_073119.1 | 2.1553414 | 0.03583079 | Down |
| XR_248922.1 | 2.0368042 | 0.019383209 | Down |
Acknowledgement
We wish to thank all the individuals who participated in the study. We also acknowledge the great assistance we received from numerous mental health professionals in different clinical departments within and outside the No. 102 Hospital of PLA. In particular, we wish to convey sincere thanks to Gopath Global LLC., Chicago, USA for their professional laboratory services and No. 102 Hospital clinical laboratory of PLA for professional laboratory assistance.
Footnotes
Source of support: Departmental sources
References
- 1.Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2:e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–45. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz E, Bahn S. Biomarker discovery in psychiatric disorders. Electrophoresis. 2008;29:2884–90. doi: 10.1002/elps.200700710. [DOI] [PubMed] [Google Scholar]
- 4.Lakhan SE, Kramer A. Schizophrenia genomics and proteomics: Are we any closer to biomarker discovery. Behav Brain Funct. 2009;5:2. doi: 10.1186/1744-9081-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu P, Zuo X, Deng H, et al. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Barry G, Briggs JA, Vanichkina DP, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19(4):486–94. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 9.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arisi I, D’Onofrio M, Brandi R, et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J Alzheimers Dis. 2011;24:721–38. doi: 10.3233/JAD-2011-101881. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Li X, Sun N, et al. Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One. 2014;9:e93388. doi: 10.1371/journal.pone.0093388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sai Y, Zou Z, Peng K, Dong Z. The Parkinson’s disease-related genes act in mitochondrial homeostasis. Neurosci Biobehav Rev. 2012;36:2034–43. doi: 10.1016/j.neubiorev.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J Mol Neurosci. 2013;49(3):589–93. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SQ, Hu HL, Ye N, et al. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr Res. 2015;166(1–3):125–30. doi: 10.1016/j.schres.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 15.Ren Y, Cui Y, Li X, et al. A co-expression network analysis reveals lncRNA abnormalities in peripheral blood in early-onset schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2015;63:1–5. doi: 10.1016/j.pnpbp.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 16.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th ed. Text Revision American Psychiatric Association; Washington, DC: 2000. [Google Scholar]
- 17.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–76. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 18.Garzya V, Forbes IT, Gribble AD, et al. Studies towards the identification of a new generation of atypical antipsychotic agents. Bioorganic Med Chem Lett. 2007;17:400e5. doi: 10.1016/j.bmcl.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: Selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353e9. doi: 10.1038/nrd1346. [DOI] [PubMed] [Google Scholar]
- 20.Lewis DA, Gonzalez-Burgos G. Pathophysiologically based treatment interventions in schizophrenia. Nat Med. 2006;12:1016e22. doi: 10.1038/nm1478. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds GP, Beasley CL. GABAergic neuronal subtypes in the human frontal cortexddevelopment and deficits in schizophrenia. J Chem Neuroanatomy. 2001;22:95e100. doi: 10.1016/s0891-0618(01)00113-2. [DOI] [PubMed] [Google Scholar]
- 22.Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–68. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Ishizuka A, Hasegawa Y, Ishida K, et al. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cells. 2014;19(9):704–21. doi: 10.1111/gtc.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire P, Howes OD, Stone J, Fusar-Poli P. Functional neuroimaging in schizophrenia: diagnosis and drug discovery. Trends Pharmacol Sci. 2008;29(2):91–98. doi: 10.1016/j.tips.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Song HT, Sun XY, Zhang L, et al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J Psychiatr Res. 2014;54:134–40. doi: 10.1016/j.jpsychires.2014.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Differentially expressed lncRNAs in peripheral blood mononuclear cells from Schizophrenia patients versus healthy controls by microarray.
| lncRNA | Fold-change | P-value | Style |
|---|---|---|---|
| TCONS_l2_00008525 | 2.0184042 | 0.007859847 | Up |
| FR324312 | 2.5046852 | 0.019613983 | Up |
| FR241369 | 2.795938 | 0.026778212 | Up |
| NR_024495.2 | 2.6708856 | 0.001104883 | Up |
| NONHSAG012869 | 2.7815604 | 0.048556764 | Down |
| NONHSAG030059 | 2.4375992 | 0.04930478 | Up |
| NONHSAT119438 | 2.386957 | 0.042039905 | Down |
| XR_248742.1 | 2.0546277 | 0.013295304 | Down |
| TCONS_l2_00017424 | 2.2616987 | 0.039616782 | Down |
| NONHSAT123713 | 2.7456982 | 0.015772445 | Up |
| TCONS_l2_00017423 | 2.3137672 | 0.04239666 | Down |
| NONHSAT078721 | 2.090962 | 0.03983514 | Down |
| NONHSAT104778 | 3.2426178 | 0.03928101 | Up |
| NONHSAT016970 | 2.8774061 | 0.04668065 | Up |
| NONHSAT006265 | 2.74296 | 0.003577456 | Down |
| NONHSAT059514 | 2.416889 | 0.034944758 | Down |
| NONHSAT041499 | 5.1150675 | 0.004036811 | Up |
| NONHSAT092334 | 2.22519 | 0.045760788 | Down |
| TCONS_00029153 | 2.2907834 | 0.005757978 | Up |
| NONHSAT 089447 | 3.514152 | 0.038919978 | Up |
| ENST00000519337 | 4.2703557 | 0.020613242 | Up |
| NONHSAG016047 | 2.8309715 | 0.03872216 | Up |
| FR101970 | 2.8698637 | 0.029207746 | Up |
| ENST00000563823 | 3.775364 | 0.012858684 | Down |
| NONHSAT121750 | 2.3670413 | 0.028498909 | Up |
| NONHSAT135863 | 2.3467486 | 0.031746913 | Down |
| NONHSAT021545 | 3.6979194 | 0.023714427 | Up |
| NONHSAT011949 | 2.8012006 | 0.047748405 | Up |
| NR_038328.1 | 2.4504633 | 0.025065137 | Down |
| NONHSAT059202 | 2.1078913 | 0.003934796 | Up |
| TCONS_00013573 | 2.016616 | 0.021455377 | Down |
| XR_241827.1 | 2.519167 | 0.043524686 | Up |
| NONHSAT123001 | 2.2459533 | 0.04912253 | Up |
| NONHSAG034605 | 2.0021873 | 0.013193308 | Up |
| ENST00000581634 | 3.0800865 | 0.024413016 | Up |
| ENST00000522494 | 2.1406422 | 0.009605092 | Up |
| NONHSAT120864 | 2.6860943 | 0.047448784 | Down |
| NONHSAT108917 | 2.201703 | 0.025858361 | Up |
| NONHSAG010469 | 2.170598 | 0.02606947 | Up |
| NONHSAT040479 | 2.4519546 | 0.049309734 | Down |
| ENST00000488480 | 2.2488601 | 0.04426648 | Up |
| NONHSAT054481 | 2.1365309 | 0.033978045 | Up |
| NONHSAT131231 | 2.2493892 | 0.022191135 | Down |
| NONHSAT013275 | 2.7648053 | 0.026773857 | Up |
| ENST00000560924 | 2.2441285 | 0.008775448 | Up |
| TCONS_00003251 | 2.3368568 | 0.048376054 | Up |
| NONHSAT133180 | 2.255505 | 0.039863095 | Up |
| ENST00000422040 | 2.246702 | 0.034511745 | Up |
| NONHSAT113409 | 2.6231997 | 0.006450792 | Up |
| ENST00000552378 | 2.0455449 | 0.039099984 | Up |
| NONHSAT098126 | 4.610542 | 0.047743667 | Up |
| NONHSAT105615 | 2.6867616 | 0.037940864 | Down |
| NONHSAT119525 | 2.5023258 | 0.041274253 | Down |
| ENST00000565759 | 2.545508 | 0.038889498 | Down |
| TCONS_00001136 | 2.401271 | 0.017750448 | Down |
| NONHSAT003974 | 4.4428 | 0.006958588 | Up |
| NONHSAT101077 | 2.382952 | 0.025674935 | Down |
| TCONS_l2_00002344 | 2.021424 | 0.013821459 | Up |
| XR_244131.1 | 2.320134 | 0.005847468 | Up |
| NR_027928.2 | 2.035199 | 0.010459018 | Down |
| NR_026775.2 | 2.709865 | 0.037649963 | Up |
| TCONS_l2_00024969 | 3.370955 | 0.017096879 | Down |
| TCONS_l2_00021339 | 3.4050841 | 0.010671916 | Down |
| XR_252113.1 | 2.1277385 | 0.019082548 | Down |
| TCONS_l2_00001488 | 2.42787 | 0.016129335 | Down |
| ENST00000593429 | 2.664398 | 0.005580793 | Down |
| NONHSAT032211 | 2.8629696 | 0.03303966 | Down |
| NONHSAT039492 | 2.419247 | 0.012648112 | Down |
| NONHSAT121856 | 2.2077594 | 0.013875067 | Down |
| NONHSAT126140 | 2.4100482 | 0.026816849 | Up |
| ENST00000413650 | 2.50962 | 0.010103071 | Down |
| NONHSAG002644 | 2.5878246 | 0.046584073 | Down |
| ENST00000512287 | 2.095495 | 0.00460248 | Down |
| NONHSAT058495 | 2.0634727 | 0.04672545 | Down |
| ENST00000416150 | 2.0931425 | 0.029960617 | Up |
| XR_247333.1 | 2.482382 | 0.01222255 | Down |
| NR_027270.1 | 2.7393317 | 0.023086803 | Down |
| NONHSAT012542 | 2.0278077 | 0.03747045 | Up |
| ENST00000594676 | 2.0075424 | 0.007863165 | Up |
| FR291472 | 2.8738096 | 0.045134224 | Up |
| NR_103554.1 | 2.3072157 | 0.048855953 | Up |
| NONHSAT102138 | 2.3331935 | 0.035092298 | Down |
| NONHSAT095774 | 2.0473397 | 0.03872717 | Down |
| XR_244653.1 | 2.6912992 | 0.01806327 | Up |
| ENST00000521622 | 3.5583007 | 0.00341083 | Down |
| TCONS_l2_00002997 | 2.1203494 | 0.024146989 | Up |
| NONHSAT114311 | 2.045813 | 0.034777325 | Down |
| ENST00000586630 | 2.2811098 | 0.009588628 | Up |
| NONHSAT024224 | 2.280496 | 0.001247224 | Down |
| NONHSAT060439 | 2.0503778 | 0.03933156 | Up |
| ENST00000443766 | 2.5501273 | 0.006421687 | Down |
| NONHSAT017342 | 2.3088417 | 0.027970523 | Down |
| NONHSAG031481 | 2.6192472 | 0.04883049 | Down |
| NONHSAT097861 | 2.0948513 | 0.03657174 | Down |
| ENST00000524610 | 2.7294452 | 0.03419223 | down |
| NONHSAT047869 | 2.046003 | 0.014862789 | Up |
| ENST00000394742 | 5.6865215 | 0.001366189 | Down |
| TCONS_l2_00008524 | 2.0070338 | 2.13E-04 | Up |
| NONHSAT136398 | 2.4874713 | 0.046239804 | Up |
| NR_024472.1 | 2.0432649 | 0.027397072 | Up |
| NONHSAG011033 | 2.3757823 | 0.0297065 | Down |
| ENST00000496491 | 3.6021621 | 0.026623152 | Up |
| NONHSAT114275 | 2.109495 | 0.037710313 | Up |
| ENST00000606447 | 2.0563893 | 0.011615175 | Up |
| XR_244409.1 | 2.0452442 | 0.049372125 | Up |
| TCONS_l2_00000563 | 2.9737065 | 0.030129751 | Down |
| NONHSAT082888 | 2.0611625 | 0.02947335 | Up |
| NONHSAT005508 | 2.947432 | 0.030145906 | Up |
| TCONS_l2_00000551 | 2.3637156 | 0.031189756 | Down |
| NONHSAT070086 | 2.7337165 | 0.039751925 | Up |
| XR_248731.1 | 2.5972993 | 0.022589074 | Down |
| NONHSAT119808 | 2.2518048 | 0.03818275 | Up |
| TCONS_l2_00012438 | 2.896232 | 0.027099133 | Down |
| NONHSAT074892 | 2.9536893 | 0.03315027 | Down |
| TCONS_l2_00020244 | 2.2335203 | 0.027745022 | Up |
| NONHSAT107055 | 2.0738862 | 0.022926092 | Down |
| NONHSAT030370 | 2.3975239 | 0.006883643 | Down |
| NONHSAT087421 | 2.249703 | 0.021947894 | Down |
| NONHSAT096504 | 2.0448937 | 0.02514679 | Up |
| NONHSAT002671 | 2.6095362 | 0.02860338 | Down |
| TCONS_l2_00011972 | 2.8288274 | 0.034055557 | Up |
| NONHSAT088690 | 2.270206 | 0.006502805 | Down |
| TCONS_l2_00025502 | 5.130027 | 0.016324848 | Down |
| NR_073119.1 | 2.1553414 | 0.03583079 | Down |
| XR_248922.1 | 2.0368042 | 0.019383209 | Down |