Abstract
The genetic architecture of common traits, including the number, frequency, and effect sizes of inherited variants that contribute to individual risk, has been long debated. Genome-wide association studies have identified scores of common variants associated with type 2 diabetes, but in aggregate, these explain only a fraction of heritability. To test the hypothesis that lower-frequency variants explain much of the remainder, the GoT2D and T2D-GENES consortia performed whole genome sequencing in 2,657 Europeans with and without diabetes, and exome sequencing in a total of 12,940 subjects from five ancestral groups. To increase statistical power, we expanded sample size via genotyping and imputation in a further 111,548 subjects. Variants associated with type 2 diabetes after sequencing were overwhelmingly common and most fell within regions previously identified by genome-wide association studies. Comprehensive enumeration of sequence variation is necessary to identify functional alleles that provide important clues to disease pathophysiology, but large-scale sequencing does not support a major role for lower-frequency variants in predisposition to type 2 diabetes.
There is compelling evidence that individual risk of type 2 diabetes (T2D) is strongly influenced by genetic factors1. Progress in characterizing the specific T2D-risk alleles responsible has been catalyzed by the ability to perform genome-wide association studies (GWAS). Over the past decade, successive waves of T2D GWAS – featuring ever larger samples, progressively denser genotyping arrays supplemented by imputation against more complete reference panels, and richer ethnic diversity – have delivered >80 robust association signals2-8. However, in these studies, the alleles interrogated for association are predominantly common (minor allele frequency [MAF]>5%), and with limited exceptions7,9, the variants driving known association signals are also common, with individually-modest impacts on T2D risk 2-8,10. Variation at known loci explains only a minority of observed T2D heritability2,3,11.
Residual genetic variance is partly explained by a long tail of common variant signals of lesser effect2. However, the contribution to T2D risk attributable to lower-frequency variants remains a matter of considerable debate, not least because of the relevance of disease architecture to clinical application11. Next-generation sequencing enables direct evaluation of the role of lower-frequency variants to disease risk7,12,13. This paper describes the efforts of the coordinated, complementary strategies pursued by the Genetics of Type 2 Diabetes (GoT2D) and T2D-GENES (Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples) Consortia. GoT2D collected comprehensive genome-wide sequence data from 2,657 T2D cases and controls; T2D-GENES focused on exome sequence variation, assembling data (after inclusion of GoT2D exomes) from a multiethnic sample of 12,940 individuals. Both consortia used genotype data to expand the sample size available for association testing for a subset of the variants exposed by sequencing.
Analysis of genome-wide variation
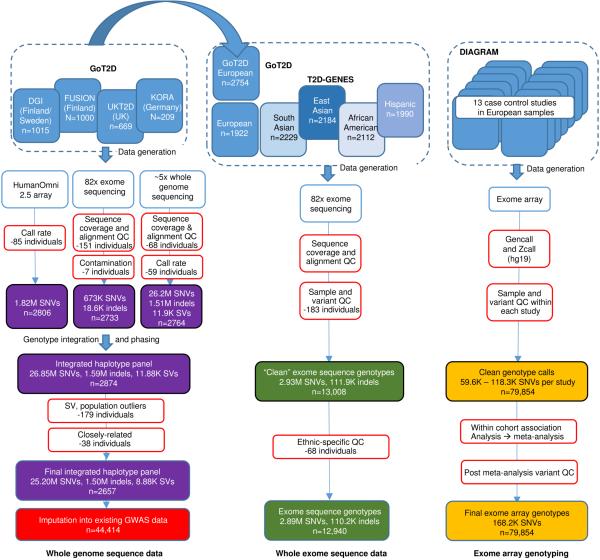
The GoT2D consortium selected for whole genome sequencing cases of type 2 diabetes (T2D) and ancestry-matched normoglycemic controls from northern and central Europe (Methods; Supplementary 1). To increase power to identify low-frequency (0.5%<MAF<5%) and rare (MAF<0.5%) T2D variants of large effect, we preferentially ascertained individuals from the extremes of genetic risk (Methods). The genome sequence of 1,326 cases and 1,331 control individuals was determined through joint statistical analysis of low-coverage whole-genome sequence (~5x), deep-coverage exome sequence (~82x), and array-based genotypes at 2.5M single nucleotide variants (SNVs) (Extended Data Fig. 1; Extended Data Table 2).
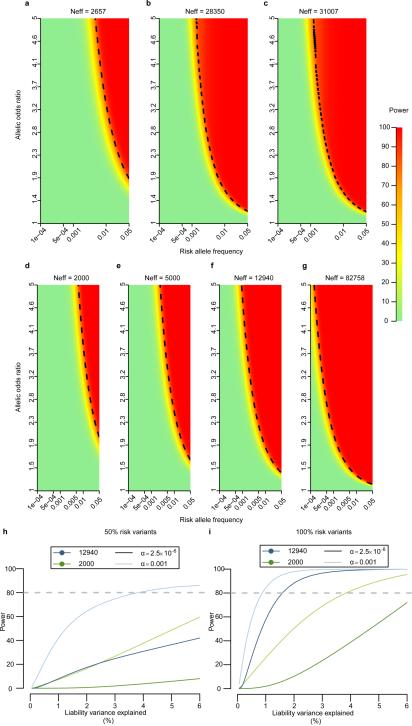
We detected, genotyped, and estimated haplotype phase for 26.7M genetic variants (Extended Data Fig. 1; Extended Data Table 3), including 1.5M short insertion-deletion variants (indels) and 8.9K large deletions. Individual diploid genomes carried a mean of 3.30M variants (range: 3.20M-3.35M), including 271K indels (262K-327K), and 669 (579-747) large deletions. These data include many variants not directly studied by previous GWAS, including all of the indels as well as 420K common and 2.4M low-frequency SNVs poorly tagged (r2≤0.30)3,4 by genotype arrays. We estimate near-complete ascertainment (98.2%) of SNVs with minor allele count >5 (MAF>0.1%), and high accuracy (>99.1%) at heterozygous genotypes (Methods; Fig. 1a). As half the sequenced individuals were T2D cases, ascertainment was enhanced for any rare or low-frequency variants that substantially increase T2D risk (Fig. 1a). Specifically, we estimate ≥80% power to detect (at genome-wide significance, α=5×10−8) T2D risk variants with MAF≥5% and OR≥1.87, or MAF≥0.5% and OR≥4.70 (Extended Data Fig. 4).
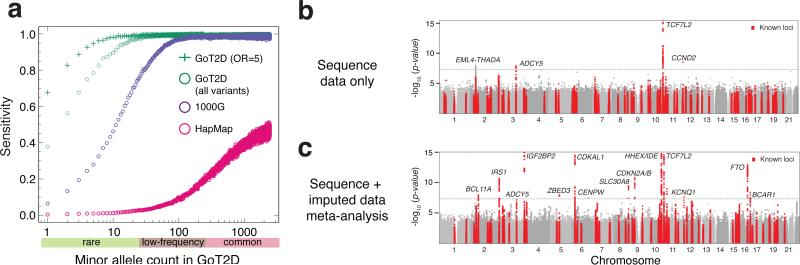
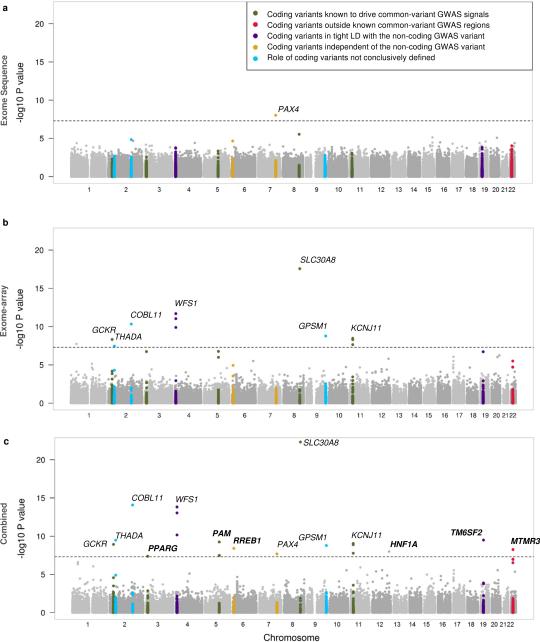
Figure 1. Ascertainment of variants and single-variant results.
a, Sensitivity of low-coverage genome sequence data to detect SNVs in the deep exome sequence data, relative to other variant catalogs. Points represent results for a specific minor allele count. All results assume OR=1 for all variants, unless stated otherwise. Manhattan plots of single-variant association analyses for: b, sequence data alone (1,326 cases and 1,331 controls) and c, meta-analysis of sequence and imputed data (total of 14,297 cases and 32,774 controls).
We tested all 26.7M variants for T2D association by logistic regression assuming an additive genetic model (Supplementary 2). Analyses using a mixed-model framework to account for population structure and relatedness generated almost identical results. At genome-wide significance, 126 variants at four loci were associated with T2D (Fig. 1b). This included two previously-reported common-variant loci (TCF7L2, ADCY5), a previously-reported low-frequency variant in CCND27 (rs76895963, MAF=2.6%, pseq=4.2×10−9), and a novel common-variant association near EML4 (MAF=34.8%, pseq=1.0×10−8). There was no significant evidence of T2D association for sets of low-frequency or rare variants within coding regions, nor within specified non-coding regulatory elements (Methods).
Power to detect association with low-frequency and rare variants of modest effect is limited in 2,657 individuals. To increase power for variants discovered via genome sequencing, we imputed sequence-based genotypes into 44,414 additional European-origin individuals (11,645 T2D cases, 32,769 controls; Methods) from 13 studies (Supplementary 3). We estimated power in the combined sequence plus imputed data, adjusting for imputation quality, to be ≥80% for variants with MAF≥5% and OR≥1.23, or MAF≥0.5% and OR≥1.92 (Extended Data Fig. 4). Meta-analysis combining results for the sequence and imputed data identified 674 variants across 14 loci associated with T2D at genome-wide significance (Fig. 1c). All were common except the CCND2 variant described above. We observed a novel association with a common variant near CENPW (rs11759026, MAF=23.2%, pmeta=3.5×10−8; Fig. 1c) and replicated this association in an additional 14,201 cases and 100,964 controls from the DIAGRAM consortium (p=2.5×10−4; pcombined=1.1×10−11; Methods). The EML4 signal detected in the sequence data did not replicate in the imputed data (p=0.59; pmeta=0.26; Fig. 1c).
To test for additional association signals we performed conditional analysis at loci previously associated with risk of T2D (Methods). We identified two novel association signals, both involving low-frequency variants, at a corrected significance threshold (α<1.8×10−6; Methods): one at the IRS1 locus (rs78124264, MAF=2.2%, pconditional=2.5×10−7) and one upstream of PPARG (rs79856023, MAF=2.2%, pconditional=9.2×10−7) (Extended Data Table 5). The PPARG signal overlaps regulatory elements in hASC pre-adipose and HepG2 cells, consistent with evidence that altered adipose regulation drives the primary PPARG signal14.
Analysis of coding variation
The T2D-GENES consortium adopted a complementary strategy, focused on variants in protein-coding sequence, and seeking to improve power to detect rare-variant association by exploiting the more robust functional annotation of coding variation and the potential to aggregate multiple alleles of presumed similar impact in the same gene12,15. We combined exome sequence data from 10,437 T2D cases and controls of diverse ancestry generated by T2D-GENES, with the equivalent data from GoT2D. This created a joint data set (after all QC) comprised of 12,940 individuals (6,504 cases; 6,436 controls) drawn from five ancestry groups: 4,541 of European origin, and ~2,000 [range: 1,943-2,217] each of South Asian, East Asian, Hispanic, and African American origin (Extended Data Fig. 1; Extended Data Table 2; Supplementary 4). Mean coverage was 82x across the coding sequence of 18,281 genes, identifying 3.04M variants (1.19M protein-altering) (Supplementary 5,6). Each diploid genome carried a mean of 9,243 (range: 8,423-11,487) synonymous, 7,636 (6,935-9,271) missense, and 250 (183-358) protein-truncating alleles (Supplementary 7).
We tested for T2D association within the five ancestral groups, assuming an additive genetic model, using mixed-model approaches that account for population structure and relatedness16, and combined ancestry-specific results via trans-ethnic meta-analysis (Methods). We estimate ≥80% power to detect (at genome-wide significance) T2D risk variants with MAF≥5% and OR≥1.36, or MAF≥0.5% and OR≥2.29 (Methods; Extended Data Fig. 4). Only one variant reached genome-wide significance (PAX4 Arg192His, rs2233580, p=9.3×10−9) (Table 1; Extended Data Figs. 6,7; Supplementary 8). This association was exclusive to East Asians, in whom the 192His allele is, in fact, common (MAF~10%) with a substantial effect size (allelic OR=1.79 [1.47-2.19]); 192His is virtually absent in other ancestries (MAF=0.014%). The rs2233580 association replicated in independent East Asian case-control data (n=3,301; p=5.9×10−7: Supplementary 9) and was distinct (r2<0.05) from previously-reported GWAS SNVs at the GCC1-PAX4 locus6,8. PAX4 encodes a transcription factor involved in islet differentiation and function17 (Supplementary 10), and PAX4 variants have been implicated in early-onset monogenic diabetes18. However, in East Asian cases, 192His was not associated with age of diabetes diagnosis (p=0.64), indicating this variant influences risk of type 2 rather than early-onset monogenic diabetes (Supplementary 9).
Table 1.
Nonsynonymous coding variants achieving genome-wide significance.
| Locus | Gene | Variant | RAF range | Eur MAF | Alleles | Exomes (N=12,940) | Exome-chip (N=79,854) | Combined (N=92,794) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| p-value | OR (95% CI) | p-value | OR (95% CI) | p-value | OR (95% CI) | ||||||
| Established common causal coding variant signals | |||||||||||
| GCKR | GCKR | rs1260326 Pro446Leu |
0.49-0.86 | 0.37 | C, T | 0.075 | 1.05 (0.99-1.11) | 4.8×10−9 | 1.07 (1.04-1.11) | 1.2×10−9 | 1.07 (1.04-1.10) |
| PPARG | PPARG | rs1801282 Pro12Ala |
0.86-0.99 | 0.14 | C, G | 0.0030 | 1.16 (1.06-1.27) | 1.8×10−7 | 1.10 (1.06-1.14) | 4.2×10−8 | 1.11 (1.07-1.15) |
| PAMI PPIP5K2 | PAM | rs35658696 Asp563Gly |
0.00-0.05 | 0.054 | G, A | 0.00045 | 1.36 (1.14-1.63) | 1.7×10−7 | 1.15 (1.08-1.23) | 5.7×10−10 | 1.17 (1.11-1.24) |
| PPIP5K2 | rs36046591 Ser1207Gly |
0.00-0.05 | 0.054 | G, A | 0.0099 | 1.34 (1.12-1.61) | 1.0×10−6 | 1.17 (1.10-1.25) | 3.3×10−8 | 1.19 (1.12-1.26) | |
| SLC30A8 | SLC30A8 | rs13266634 Asp325Trp |
0.58-0.91 | 0.33 | C, T | 2.9×10−6 | 1.15 (1.09-1.22) | 2.7×10−18 | 1.14 (1.11-1.17) | 4.8×10−23 | 1.14 (1.11-1.17) |
| KCNJ11/ABCC8 | KCNJ11 | rs5215 Val337Ile |
0.08-0.40 | 0.40 | C, T | 0.11 | 1.07 (1.01-1.13) | 3.4×10−9 | 1.07 (1.04-1.11) | 1.3×10−9 | 1.07 (1.05-1.10) |
| rs5219 Lys23Glu |
0.06-0.40 | 0.40 | T, C | 0.056 | 1.08 (1.02-1.14) | 5.1×10−9 | 1.07 (1.04-1.11) | 9.0×10−10 | 1.07 (1.05-1.10) | ||
| ABCC8 | rs757110 Ala1369Ser |
0.06-0.40 | 0.40 | C, A | 0.20 | 1.06 (1.00-1.12) | 2.3×10−8 | 1.07 (1.04-1.11) | 1.7×10−8 | 1.07 (1.04-1.10) | |
| Other coding variant associations within established common variant GWAS regions | |||||||||||
| THADA | THADA | rs35720761 Cys1605Tyr |
0.85-1.00 | 0.10 | C, T | 0.0021 | 1.12 (1.01-1.23) | 3.5×10−8 | 1.11 (1.07-1.16) | 3.3×10−10 | 1.12 (1.07-1.16) |
| COBLL1 | COBLL1 | rs7607980 Asn939Asp |
0.84-1.00 | 0.12 | T, C | 1.4×10−5 | 1.21 (1.11-1.33) | 4.7×10−11 | 1.14 (1.10-1.19) | 8.3×10−15 | 1.15 (1.11-1.19) |
| WFS1 | WFS1 | rs1801212 Val333Ile |
0.70-1.00 | 0.30 | A, G | 0.0026 | 1.14 (1.06-1.23) | 9.3×10−12 | 1.08 (1.04-1.12) | 9.0×10−14 | 1.09 (1.06-1.12) |
| rs1801214 Asn500Asn |
0.59-0.96 | 0.41 | T, C | 0.0019 | 1.08 (1.02-1.15) | 2.0×10−12 | 1.08 (1.05-1.11) | 1.5×10−14 | 1.08 (1.05-1.11) | ||
| rs734312 Arg611His |
0.11-0.85 | 0.47 | A, G | 0.12 | 1.05 (0.99-1.11) | 1.3×10−10 | 1.07 (1.03-1.10) | 6.9×10−11 | 1.06 (1.04-1.09) | ||
| RREB1 | RREB1 | rs9379084 Asp1171Asn |
0.87-0.98 | 0.11 | G, A | 2.2×10−5 | 1.19 (1.09-1.30) | 1.1×10−5 | 1.12 (1.06-1.17) | 4.0×10−9 | 1.13 (1.09-1.18) |
| PAX4 | PAX4 | rs2233580 Arg192His |
0.00-0.10 | 0.00 | T, C | 9.3×10−9 | 1.79 (1.47-2.19) | NA | NA | 9.3×10−9 | 1.79 (1.47-2.19) |
| GPSM1* | GPSM1* | rs60980157 Ser391Leu |
0.26 | 0.26 | C, T | NA | NA | 1.7×10−9 | 1.09 (1.06-1.12) | 1.7×10−9 | 1.09 (1.06-1.12) |
| CILP2 | TM6SF2 | rs58542926 Glu167Lys |
0.03-0.10 | 0.082 | T, C | 0.00015 | 1.22 (1.10-1.36) | 1.9×10−7 | 1.13 (1.08-1.18) | 3.2×10−10 | 1.14 (1.10-1.19) |
| Coding variant associations outside established common variant GWAS regions | |||||||||||
| MTMR3/ASCC2 | MTMR3 | rs41278853 Asn960Ser |
0.92-1.00 | 0.083 | A, G | 9.2×10−5 | 1.26 (1.12-1.42) | 3.2×10−6 | 1.12 (1.07-1.17) | 5.6×10−9 | 1.14 (1.09-1.19) |
| ASCC2 | rs11549795 Val123Ile |
0.92-1.00 | 0.083 | C, T | 0.00040 | 1.23 (1.10-1.38) | 2.0×10−5 | 1.11 (1.06-1.16) | 1.0×10−7 | 1.13 (1.08-1.18) | |
| rs28265 Asp407His |
0.92-1.00 | 0.083 | C, G | 0.00050 | 1.21 (1.08-1.36) | 1.9×10−5 | 1.11 (1.06-1.16) | 1.1× 10−7 | 1.12 (1.08-1.17) | ||
| rs36571 Pro423Ser |
0.92-1.00 | 0.083 | G, A | 0.0023 | 1.23 (1.08-1.40) | 2.0×10−5 | 1.11 (1.06-1.16) | 3.0×10−7 | 1.12 (1.08-1.17) | ||
These loci were identified through single-variant analyses of exome sequence data in 6,504 cases and 6,436 controls and exome-array in 28,305 cases and 51,549 controls. RAF: Risk allele frequency. Eur MAF: Minor allele frequency in Europeans. OR: odds-ratio. CI: confidence interval. N: Total number of individuals analysed. N: Total number of individuals analysed. Genome-wide significance defined as p < 5×10−8.
GPSM1 variant failed quality control in exome sequence: association p-values derive only from exome-array analysis. The synonymous variant Thr515Thr (rs55834942) in HNF1A also reached genome-wide significance (p=1.0×10−8) in the combined analysis. Alleles are aligned to the forward strand of NCBI Build 37 and represented as risk and other allele.
To increase power to detect association of rare variants that cluster in individual genes, we deployed gene-level variant aggregation tests15 across the exome sequence data (Methods; Supplementary 11). We observed no deviation from the null distribution of association statistics, and no single gene reached exome-wide significance (α=2.5×10−6) (Methods; Supplementary 12,13). When we focused on 634 genes mapping to known GWAS regions, only FES exceeded a reduced significance threshold of α=7.9×10−5 (psouthAsian=7.2×10−6, pmultiethnic=1.9×10−5) (Method; Supplementary 14). This aggregate signal was driven entirely by the South Asian-specific Pro536Ser variant (MAF=0.9%, OR=6.7 [2.6-17.3], p=7.5×10−6), indicating that FES is likely to be the effector gene at the PRC1 GWAS locus4.
To increase power to detect coding variant associations (Extended Data Fig. 4), we contributed early T2D-GENES exome data to the design of Illumina exome array9, and then collected genotypes from an additional 28,305 T2D cases and 51,549 controls of European-ancestry from 13 studies (Extended Data Fig. 1; Extended Data Table 2; Supplementary 15). Of 27,904 protein-altering variants with MAF>0.5% detected in exome sequence data from n=4,541 European individuals, variation at 81.6% was captured on the array (Supplementary 16).
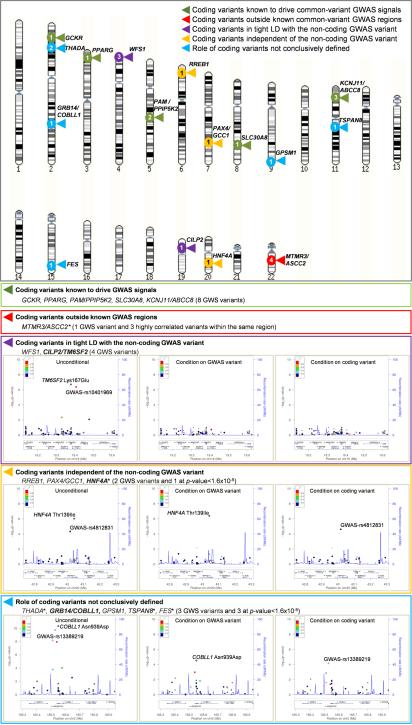
Association analysis in the combined sequence and array data from >90,000 individuals identified 18 coding variants (17 nonsynonymous), at 13 loci, which exceeded genome-wide significance (α=5×10−8) (Table 1; Extended Data Figs. 6,7). All of these were common (MAF>5%) and all but one mapped within established common-variant GWAS regions2,3. The exception, which we replicated in the INTERACT study19 (n=9,292; pINTERACT=2.4×10−4; pmeta=2.2×10−11), involved a common haplotype of four strongly-correlated coding variants in MTMR3 and ASCC2 (Table 1). Of these, MTMR3 Asn960Ser (MAF=8.3%) had the strongest residual association signal on conditional analysis, implicating MTMR3, encoding a phosphatidylinositol phosphatase20, as the probable effector transcript at this locus (Extended Data Table 5; Extended Data Figs. 6,7; Supplementary 10,17).
The remaining coding variant signals provided an opportunity to highlight causal alleles and effector transcripts for known GWAS signals. For five loci (SLC30A8, GCKR, PPARG, KCNJ11-ABCC8, PAM), the coding variants identified had previously been nominated as causal for their respective GWAS signals2,7,13. For the other seven loci, GWAS meta-analyses had previously highlighted a lead variant in non-coding sequence2,5,6. We (re)evaluated these relationships with conditional and credible set analyses, finding that, at most, the evidence supported a direct causal role for the coding variants concerned (Extended Data Table 5; Extended Data Figs. 6,7; Supplementary 10,17).
For example, at the CILP2 locus2, previous GWAS had identified the non-coding variant rs10401969 as the lead SNV. However, direct genotyping of TM6SF2 Lys167Glu on the exome array revealed complete linkage disequilibrium with rs10401969, and reciprocal signal extinction in conditional analyses (Extended Data Table 5; Extended Data Figs. 6,7). In previous GWAS, the association at Lys167Glu had been obscured by incomplete genotyping and poor imputation (Supplementary 18). The TM6SF2 Lys167 allele has been shown to underlie predisposition to hepatic steatosis21, and was associated with fasting hyperinsulinemia (p=1.0×10−4) in 30,824 non-diabetic controls from the present study. This combination of genetic and functional data, consistent with known mechanistic links between insulin resistance, T2D, and fatty liver disease22, implicates TM6SF2 Lys167Glu as the likely T2D-risk variant at this locus.
In contrast, the association at RREB1 Asp1171Asn represented a novel signal, conditionally independent of the adjacent common-variant GWAS signal. This association, together with that involving a second associated coding variant, Ser1554Tyr, which has a marked association with fasting glucose (p=2.7×10−9 in levels in 38,338 non-diabetic subjects from the present study) (Supplementary 19), establishes RREB123 as the probable effector gene at the SSR1 locus.
Given the concentration of coding-variant associations within established GWAS loci, we sought to nominate additional single-variant signals in 634 genes mapping to established T2D GWAS regions using a Bonferroni-corrected α=1.6×10−5 (Methods; Supplementary 14,20). At HNF4A, we confirmed a T2D association at Thr139Ile (European MAF range 0.7-3.8%, OR=1.15 [1.08-1.22], p=2.9×10−6)10 distinct both from the common non-coding lead GWAS SNV2,3,5, and multiple rare HNF4A variants implicated in monogenic diabetes24. Additional coding variant associations in TSPAN8 and THADA highlighted these two genes as probable effector transcripts in their respective GWAS regions (Supplementary 10,21).
Rare alleles in Mendelian genes
We extended gene-based tests for rare-variant associations to gene-sets implicated in monogenic or syndromic diabetes or in altered glucose metabolism24. Across 81 genes harboring rare alleles causal for monogenic or syndromic diabetes or related glycemic traits (‘Monogenic All’; Supplementary 22), the only variant or gene association genome-wide significance involved the previously-mentioned PAX4 Arg192His. However, across the entire gene-set, we observed a weak aggregate association with T2D-risk (p=0.023: Fig. 2a). The association was considerably stronger in two subsets of genes more directly implicated in monogenic and syndromic diabetes: a manually-curated set of 28 genes for which diabetes was the primary phenotype (‘Monogenic Primary’) and a partially-overlapping set of 13 genes reported in OMIM as causal for MODY or neonatal diabetes (‘Monogenic OMIM’) (Supplementary 22).
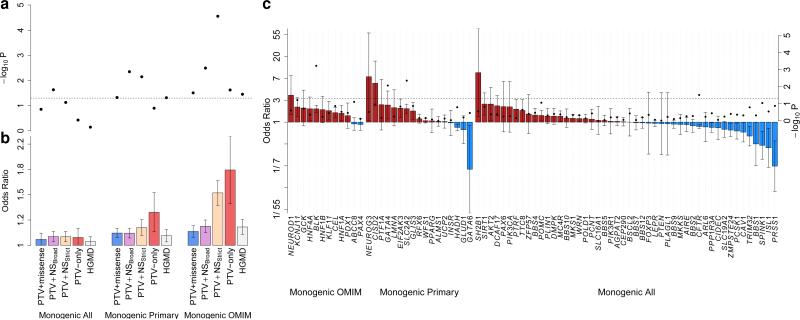
Figure 2. Association between T2D and variants in genes for Mendelian forms of diabetes.
a, p-values of aggregate association for variants from 6,504 T2D cases and 6,436 controls in three sets of Mendelian diabetes genes, for five variant “masks” (Methods). Dotted line: p=0.05. b, Estimated T2D odds ratio (OR) for carriers of variants in each gene-set and mask. Error bars: one standard error. c, Estimated ORs (bars, left axis) and p-values (dots, right axis) for carriers of variants in the PTV+NSstrict mask for each gene. Error bars: one standard error. Red: OR > 1; blue: OR < 1; dotted line: p=0.05.
The ‘Monogenic OMIM’ gene-set had a statistically robust signal of association (p=2.8×10−5, OR=1.51 [1.25-1.83]) driven by allelic burden of MAF<1% alleles. Effect size estimates tracked with increasing stringency of variant annotation and gene-set definition, consistent with progressive enrichment for functional over neutral alleles (Fig. 2b). This signal does not reflect inclusion among T2D cases of individuals who, in reality, had monogenic diabetes: the association was not concentrated among genes most frequently responsible for monogenic diabetes24 (Fig. 2c), and age of diabetes diagnosis was no younger in variant carriers than non-carriers (Supplementary 23). The association signal remained after all alleles listed as ’disease-causing’ within the Human Genetic Mutation Database were excluded (p=2.9×10−4, OR=1.50 [1.21-1.86]).
These analyses point to widespread enrichment for T2D association among rare coding alleles in genes causal for monogenic diabetes. In these genes, alleles of penetrance sufficient to drive familial segregation of early-onset diabetes coexist alongside those of more modest effect predisposing to later-onset T2D. No other compelling signals of rare-variant enrichment were detected using gene-set enrichment or protein-protein interaction analysis in other pre-defined gene-sets (Supplementary 24-26).
No evidence for synthetic association
In 2010, Goldstein and colleagues proposed that common-variant GWAS signals may be the consequence of low-frequency and rare variants that by chance cluster on common haplotypes25. While this hypothesis has been debated26,27 and assessed indirectly3,28, we used the near-complete ascertainment of genetic variation in 2,657 genome-sequenced individuals to directly test the importance of ‘synthetic’ associations29. We focused on the ten T2D GWAS loci at which our sample provided the strongest statistical evidence for association (p<0.001), implementing a conditional analysis procedure to assess whether combinations of SNVs within a 5Mb window could explain the common-variant signal (Extended Data Table 8; Methods).
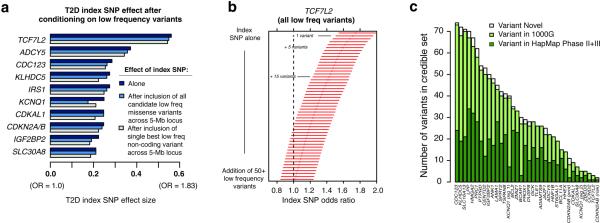
We first focused on missense variants, finding that none of the ten signals could be explained by low-frequency and rare variants within 2.5Mb of the common index SNV (Extended Data Fig. 9). For example, at the IRS1 locus, including the five observed missense IRS1 alleles in the model did not meaningfully diminish the index SNV association (punconditional=2.8×10−6, pconditional=4.3×10−6). With 99.7% ascertainment of low-frequency coding variants (Methods), these results rule out synthetic associations produced by missense variants at these ten loci.
We expanded the search to include all low-frequency and rare variants, non-coding and coding, within 2.5Mb of index SNVs. At no locus was a single low-frequency or rare variant sufficient to explain the GWAS signal (Extended Data Fig. 9). At 8 of the 10 loci, ≥10 low-frequency and rare variants were needed to reverse the direction of effect at the common index SNV; at TCF7L2, even 50 were insufficient (Extended Data Fig. 9). We note that the statistical procedure we developed and deployed is biased in favor of the synthetic association hypothesis, since it is highly prone to over-fitting. Nonetheless, at 8 of the 10 loci the data were indistinguishable from a null model of no synthetic association (Extended Data Table 8; Supplementary 27).
Nominating candidate functional alleles
Using the GoT2D whole genome sequence data, we constructed 99% ‘credible sets’ for each T2D GWAS locus on the assumption of one causal variant per locus (Methods)30. Across 78 published autosomal loci at which the reported index SNV had MAF>1%, 99% credible set sizes ranged from 2 (CDKN2AB) to ~1,000 (POU5F1) variants; at 71 loci, the credible set contained >10 variants (Extended Data Fig. 9; Supplementary 28). The GoT2D dataset provides near-complete ascertainment of common and low-frequency variants to support more comprehensive credible set analysis than studies based on genotyping or imputation alone3,31: of the credible set variants identified from whole genome sequence data, ~60% are absent from HapMap and ~5% from 1000G Phase 1 (Extended Data Fig. 9).
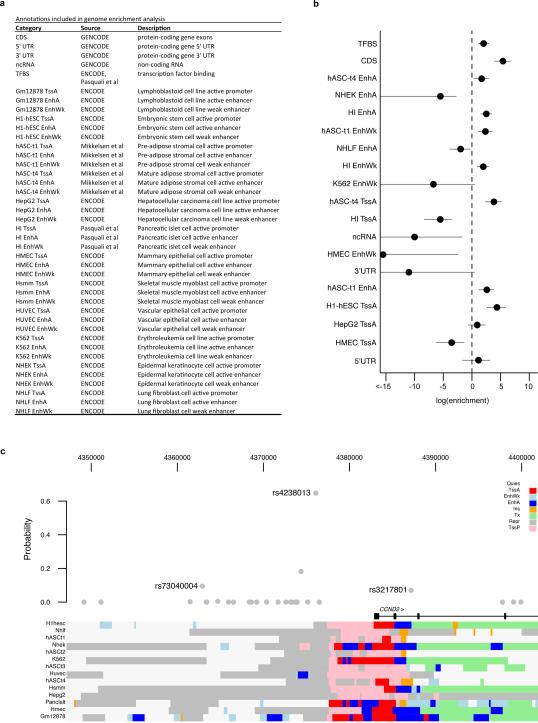
Genomic maps of chromatin state or transcription factor binding32-35 have been used to prioritize causal variants within credible sets36,37. We jointly modeled genetic association and genomic annotation data at T2D GWAS loci using fgwas38. Consistent with previous reports34,35, associated variants were enriched in coding exons, transcription factor binding sites, and enhancers active in pancreatic islets and adipose tissue (Extended Data Fig. 10). Overall, including the functional annotation data reduced credible set size by 35%. At several loci, access to complete sequence data prioritized variants that overlap relevant regulatory annotations and were previously overlooked. For example, at the CCND2 locus, three variants not present in HapMap Phase 2 have combined probability of 90.0% of explaining the common-variant signal2 (Extended Data Fig. 10); one of these (rs3217801) is a 2bp indel overlapping an islet enhancer element.
Modelling disease architecture
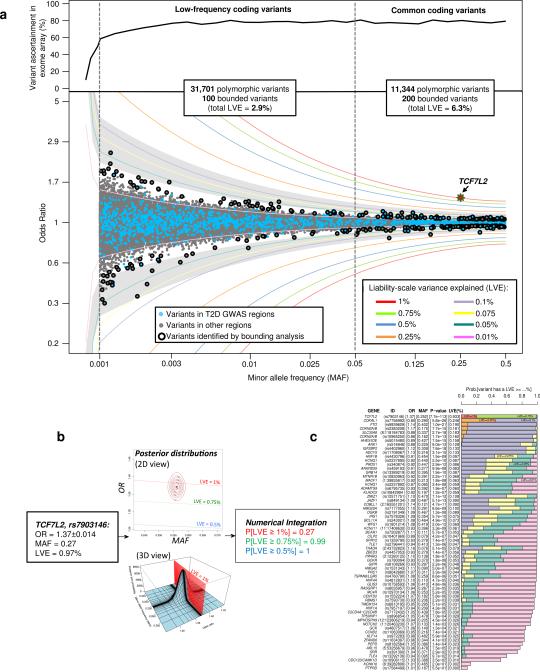
To evaluate the overall contribution of low-frequency coding variation to T2D risk, we estimated the proportion of variance in T2D-liability attributable to each such variant39 (Methods; Extended Data Fig. 11). We focused on exome array data to maximize sample size, and on variants with MAF>0.1%; sensitivity of variant ascertainment and accuracy of OR estimation decline below this threshold. Among the 31,701 variants on the exome array with 0.1%<MAF<5% there was a progressive increase in the maximum OR estimates with decreasing frequency. However, the liability variance explained for these variants rarely exceeded 0.05%, limiting power to detect association in the sample size available (Extended Data Fig. 11). We estimated (Methods) that the liability variance collectively attributable to coding variants in the 0.1%<MAF<5% range was 2.9%, compared to 6.3% for common variants.
Finally, we compared our whole genome T2D association results with predictions from population genetic simulations40 under twelve models that vary widely with respect to the proportion of heritability explained by common, low-frequency, and rare variants. We mirrored the GoT2D study design (with imputation) and performed in parallel the same association analysis on empirical and simulated data, focusing on variants with MAF>0.1% and allowing for power loss due to imperfect imputation (Methods).
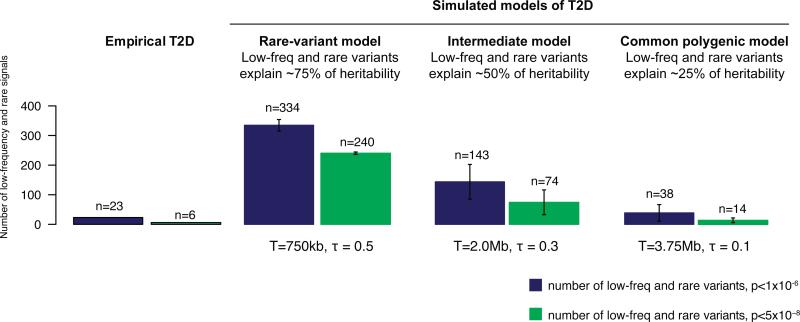
Figure 3 displays results for three representative models: a ‘purifying selection’ model in which low-frequency and rare variants explain ~75% of T2D heritability, an intermediate model in which low-frequency/rare and common variants both contribute substantially, and a ‘neutral’ model in which common variants explain ~75% of T2D heritability. Predictions of the first two models differ markedly in the numbers of low-frequency and rare risk variants that are associated with T2D. Specifically, these two models predict a larger number and greater effect size of low-frequency variants found in our whole genome sequencing study as compared to those observed in the empirical data. In contrast, empirical data are consistent with predictions under the ‘neutral’ common-variant model.
Figure 3. Empirical T2D association results compared to results under different simulated disease models.
Observed number of rare and low-frequency (MAF<5%) genetic association signals for T2D detected genome-wide after imputation compared to the numbers seen under three simulated disease models for T2D which were plausible given results (T2D recurrence risks, GWAS, linkage) prior to large-scale sequencing. Simulated models were defined by two parameters: disease target size T and degree of coupling τ between the causal effects of variants and the selective pressure against them40. Simulated data were generated to match GoT2D imputation quality as a function of MAF (Methods).
The century-old Mendelian-biometrician debate pitted those who attributed trait variation to rare variants of large effect against those who argued that trait variation is largely due to many common variants of small effect. The debate today is about whether the ‘missing heritability’ after GWAS is due largely to individually rare, highly-penetrant variants41 or to a large universe of common alleles of modest effect42. The results are of more than academic interest, since genetic architecture plays out powerfully in relation to the power of genetic diagnosis and the application of precision medicine.
Our data and analysis indicate that for T2D, nearly all common-variant associations detectable by whole genome sequencing were previously found by GWAS based on genotyping arrays and imputation: concerns about incomplete coverage due to ‘holes’ in HapMap11 coverage were, we show, unfounded. Of more lasting interest, the combination of genome and exome sequencing in large samples provides limited evidence of a role for lower-frequency variants — coding or genome wide — in T2D predisposition. Of course, rare risk alleles have long been known to contribute in families with early-onset forms of diabetes, and sequencing of Mendelian and GWAS genes has identified rare variants that influence disease risk43,44. Sequencing of T2D cases in much larger samples will undoubtedly uncover additional low-frequency and rare variants that provide biological and potentially clinical value. Nonetheless, our empirical and simulated data argue that these lower-frequency variants contribute much less to T2D heritability than do common variants. Moreover, the frequency spectrum of variant association signals is consistent with a model whereby limited selective pressure distributes most the genetic variance influencing T2D risk among common alleles40, consistent with the frequency distribution of inter-individual sequence variation. Similar large-scale sequencing-based exploration of other complex traits will be required to determine the extent to which the genetic architecture of T2D is representative of other late-onset diseases.
Our results further strengthen the case for sequencing of diverse samples: the population-enriched T2D risk variant in PAX4 dovetails with similar findings involving SLC16A1145 in East Asian and Native American populations and TBC1D446 in Greenland Inuits. Study of populations subject to bottlenecks and/or extreme selective pressures43,46,47 may be particularly fruitful.
Understanding the inherited basis of T2D will require much further progress in identifying the mechanisms whereby common, mostly non-coding, variants influence disease risk. The combination of global epigenetic measurements, genome editing48, and high-throughput functional assays49 make it increasingly practical to characterize large numbers of non-coding variants and the processes they impact. Genome sequencing in much larger numbers of individuals than included in the current study are needed and will no doubt provide foundational information to guide such experimentation and connect the results to human population variation, physiology, and disease. Integration of biological insights gleaned from common and rare variant associations to T2D into a unified picture of disease pathophysiology will be required to fully understand the basis of this common but challenging disease.
EXTENDED METHODS
Ethics statement
All human research was approved by the relevant institutional review boards and conducted according to the Declaration of Helsinki. All participants provided written informed consent.
1 Data generation
1.1 GoT2D integrated panel generation
1.1.1. GoT2D sequenced samples
Here we describe how we generated, processed, and carried out quality control (QC) on sequence and genotype data for the 2,891 individuals initially chosen for GoT2D from four studies, and how this resulted in 2,657 individuals (1,326 T2D cases and 1,331 non-diabetic controls) for analysis (Extended Data Figure 1). We preferentially sampled early-onset, lean, and/or familial T2D cases and overweight controls with low fasting glucose levels50. Specific details of selected samples are provided in Extended Data Table 2 and Supplementary 1.
1.1.2. DNA sample preparation
De-identified DNA samples were sent to the Broad Institute (DGI, FUSION), Wellcome Trust Centre for Human Genetics in Oxford (UKT2D), and Helmholtz Zentrum München (KORA) and prepared for genetic analysis. DNA quantity was measured by Picogreen (all), and samples with sufficient total DNA and minimum concentrations for downstream experiments were genotyped for a set of 24 SNVs using the Sequenom iPLEX assay (DGI, FUSION, UKT2D): one gender assay and 23 SNVs located across the autosomes. The genotypes for these SNVs were used as a quality filter to advance samples and a technical fingerprint for subsequent sequencing and genome-wide array genotypes.
1.1.3. Exome sequencing
Genomic DNA was sheared, end repaired, ligated with barcoded Illumina sequencing adapters, amplified, size selected, and subjected to in-solution hybrid capture using the Agilent SureSelect Human All Exon 44Mb v2.0 (DGI, FUSION, UK2T2D) and v3.0 (KORA) bait set (Agilent Technologies, USA). Resulting Illumina exome sequencing libraries were qPCR quantified, pooled, and sequenced with 76bp paired-end reads using Illumina GAII or HiSeq 2000 sequencers to ~82-fold mean coverage.
1.1.4. Genome sequencing
Whole-genome Illumina sequencing library construction was performed as described for exome capture above, except that genomic DNA was sheared to a larger target size and hybrid capture was not performed. Resulting libraries were size selected to contain fragment insert size of 380bp±20% (DGI, FUSION, KORA) and 420bp±25% (UKT2D) using gel electrophoresis or the SAGE Pippin Prep (Sage Science, USA). Libraries were qPCR quantified, pooled, and sequenced with 101bp paired-end reads using Illumina GAII or HiSeq 2000 sequencers to ~5-fold mean coverage.
1.1.5. HumanOmni2.5 array genotyping
Genotyping was performed by the Broad Genetic Analysis Platform. DNA samples were placed on 96-well plates and genotyped using the Illumina HumanOmni2.5-4v1_B SNV array.
1.1.6. Alignment and processing of exome and genome sequence data
1.1.6.1. Alignment of sequence reads to reference genome
Sequence data were processed and aligned to hg19 using the Picard (broadinstitute. github.io/picard/), BWA51, and GATK52,53 pipelines. Resulting BAM and VCF files were submitted to NCBI and are available in dbGaP (accession number phs000840.v1.p1, study name NIDDK_GoT2D).
1.1.6.2. Coverage and QC of aligned sequence reads
We excluded 151 exome samples with average coverage ≤20x in >20% of the target bases and 68 genome samples with average coverage ≤5x. After sequence alignment and post-processing, aligned sequence reads were screened based on multiple QC criteria, including number of mapped reads, number of mapped bases with <1% estimated base call error rate (>Q20), fraction of duplicate reads, fraction of properly paired reads, distribution of insert sizes, distribution of mean base quality with respect to sequencing cycles, and GC bias (Extended Data Figure 1).
1.1.6.3. Detecting and handling contamination of sequence reads
We assessed possible DNA contamination in the genome and exome sequence data using verifyBamID54 using two methods. First, we estimated the contamination level of sequenced samples using allele frequencies estimated from the HumanOmni2.5 array on a thinned set of 100,000 markers with minor allele frequency (MAF)>5%. Second, for samples with HumanOmni2.5 genotypes, we used these genotypes together with sequence data to estimate contamination and identify possible sample swaps. We excluded exome sequence data for 7 individuals and genome sequence data for 59 individuals with estimated contamination ≥2% using either method. Prior to variant calling, uncontaminated sample swaps were assigned to the correct sample label after searching for the matching pairs using the same method.
1.1.7. GoT2D integrated panel genotype calling
1.1.7.1. SNV identification
We processed whole-genome sequence reads across the remaining 2,764 QC-passed individuals by two SNV calling pipelines: GotCloud (www.gotcloud.org) and GATK UnifiedGenotyper55. We merged unfiltered SNV calls across the two call sets and then processed the merged site list through the SVM and VQSR filtering algorithms implemented by those pipelines. SNVs that failed both filtering algorithms were removed before genotyping and haplotype integration. For the 2,733 QC-passed exome sequenced individuals, we used GATK UnifiedGenotyper to call SNVs.
1.1.7.1.1. Illumina HumanOmni2.5 array genotyping
We used Illumina GenomeStudio v2010.3 with default clusters to call HumanOmni2.5 genotypes after comparing different clustering algorithms and observing that the default cluster resulted in highest concordance with sequence-based genotypes. Called genotypes were run through a standard QC pipeline; samples passing a call rate threshold of 95%, and genetic fingerprint (24 marker panel) and gender concordance were passed on to downstream GWAS QC. SNVs with GenTrain score<0.6, cluster separation score<0.4, or call rate<97% were considered technical failures at the genotyping laboratory and deleted before data release. We removed samples with call rate<98%, and SNVs monomorphic across all samples, failed by 1000G Omni 2.5 QC filter, or with Hardy-Weinberg equilibrium p<10−6 (Extended Data Figure 1). 85 samples were removed in this process.
1.1.7.2. Short insertion and deletion (indel) identification
For the whole-genome sequence data, we used the GATK UnifiedGenotyper to call short indels (<50bp). Because short indels are known to have high false positive rates due to systematic sequencing and alignment errors55, we used stringent filtering criteria in SVM and VQSR and excluded indels that failed either algorithm. For exome sequencing, we used GATK UnifiedGenotyper to call short indels, following best practices described elsewhere52.
1.1.7.3. Large deletion identification
We used GenomeSTRiP56 to call large (>100bp) deletions in the whole-genome sequence data. After initial discovery of large deletions in 2,764 QC-passed individuals, we merged the discovered sites with deletions identified in 1,092 sequenced individuals from the 1000G Project to increase sensitivity and then genotyped the merged site lists across the 2,764 individuals. After applying the default filtering implemented in GenomeSTRiP, pass-filtered sites variable in any of the samples were identified as candidate variant sites. Among these candidate sites, we excluded variants in known immunoglobin loci to reduce the impact of possible cell-line artifacts. We then excluded 136 more individuals owing to an unusually large number of variants per sample (>median+3×mean absolute deviation). Variants present only in these excluded individuals were removed from further analysis.
1.1.8. GoT2D integrated panel haplotype integration
1.1.8.1. Genotype likelihood calculation
We merged SNVs discovered from the three experimental platforms into one site list and calculated genotype likelihoods across all sites separately by platform. Because exome sequence data have substantial off-target coverage, we calculated likelihoods across the genome combining data from the genome and exome sequence experiments. For genome sequence, we calculated likelihoods using GotCloud; for exomes, we used GATK UnifiedGenotyper; for HumanOmni2.5 genotypes, we converted hard genotype calls into genotype likelihoods assuming a genotype error rate of 10−6. For indels, we calculated likelihoods in a similar way except the HumanOmni2.5 data could not be used. For structural variants (SVs), genotype likelihoods were calculated from GenomeSTRiP using the whole-genome sequence data.
1.1.8.2. Integration of genotype and sequence data
We calculated combined genotype likelihoods across each of the 2,874 individuals as the product of the corresponding genome, exome, and HumanOmni2.5 likelihoods assuming independent data across platforms (Extended Data Figure 1). We then phased the genotype data using the strategy developed for 1000G Phase 155. Specifically, we phased the integrated likelihoods using Beagle57 with 10,000 SNVs per chunk and 1,000 overlapping SNVs between consecutive chunks. We refined phased sequences using Thunder58 as implemented in GotCloud (genome.sph.umich.edu/wiki/GotCloud) with 400 states to improve genotype and haplotype quality.
1.1.9. GoT2D integrated panel QC
2,874 individuals were available in the integrated haplotype panel. To identify population outliers, we carried out principal components analysis (PCA). We computed PCs for each of the three variant types (SNVs, short indels, large deletions) using EPACTS on an LD-pruned (r2<0.20) set of autosomal variants obtained by removing large high-LD regions59,60, variants with MAF<0.01, and variants with Hardy-Weinberg equilibrium p<10−6. Inspecting the first ten PCs for each variant type, we identified 43 population outliers and 136 additional outliers for large deletions only; we excluded these 179 individuals. We excluded an additional 38 individuals based on close relationships (estimated genome-wide identity-by-descent proportion of alleles shared >0.20) with other study members. 2,657 individuals remained available for downstream analyses (Extended Data Figure 1).
1.1.10. GoT2D integrated panel evaluation of variant detection sensitivity
Since we had no external data to evaluate SNV and indel variant detection sensitivity and genotype accuracy for our integrated haplotype panel, we evaluated accuracy for the low-pass whole-genome sequence data using the exome sequence data as gold standard for variants at which exome sequence depth was ≥10. We consider the resulting sensitivity and accuracy estimates as lower bounds for the integrated panel, which combined information from the genome, exome, and HumanOmni2.5 data.
We estimated the sensitivity of low-pass genome sequence data to detect true SNVs by calculating the proportion of exome-sequencing-detected SNVs detected by low-pass genome sequencing in the 2,538 individuals with data for all three experimental platforms. For exome sequence allele counts <1,000, we merged adjacent allele count bins until the number of alleles was >1,000. We estimated the sensitivity of low-pass genome sequencing to detect common, low-frequency, and rare SNVs as 99.8%, 99.0%, and 48.2%, respectively. Similarly, we estimated the sensitivity of low-pass genome sequence to detect true short indels by calculating the proportion of exome sequencing-detected short indels detected by low-pass genome sequencing. Sensitivity estimates were >99.9%, 93.8%, and 17.9% for common, low-frequency, and rare short indels, respectively.
To estimate the sensitivity of the combined low-pass genome and exome sequence data, we focused on coding SNVs and calculated the proportion of HumanOmni2.5 SNVs detected by either sequencing platform. Because HumanOmni2.5 SNVs are enriched for common variants, we calculated a weighted averaged sensitivity at each allele count, weighted by the number of exome-detected variants given the allele count. Sensitivity estimates were 99.9%, 99.7%, and 83.9% for common, low-frequency, and rare variants.
1.1.11. GoT2D integrated panel evaluation of genotype accuracy
To evaluate genotype accuracy for SNVs, we focused on chromosome 20, and compared the concordance of low-pass whole-genome-sequence-based genotypes with those based on exome sequence. Overall genotype concordance was 99.86%. Homozygous reference, heterozygous, and homozygous non-reference concordances were 99.97%, 98.34%, and 99.72%. We also compared genotype concordance between exome sequence and HumanOmni2.5 genotypes. Overall concordance was 99.4%. When the HumanOmni2.5 genotypes were homozygous reference, heterozygous, and homozygous non-reference, concordances were 99.97%, 99.69%, and 99.88%. We evaluated genotype accuracy of indels for the 210 chromosome 20 indels that overlapped between those discovered by exome and genome sequencing. Overall genotype concordance was 99.4%. When the exome genotypes were homozygous reference, heterozygous, and homozygous non-reference, concordances were 99.8%, 95.8%, and 98.6%.
To evaluate the genotype accuracy of our low-pass genome sequence data to detect true structural variants, we took advantage of the 181 individuals in our study previously included in the WTCCC array-CGH based structural variant detection experiment61. Taking the WTCCC data as gold standard, we estimated genotype accuracy across 1,047 overlapping structural variants (with reciprocal overlap>0.8) genome-wide. The overall genotype concordance was 99.8%. When the WTCCC genotypes were homozygous reference, heterozygous, and homozygous non-reference, concordances were 99.9%, 99.6%, and 99.7%.
1.2. GoT2D+T2D-GENES multiethnic exome panel generation and QC
1.2.1. Samples
We considered 6,504 T2D cases and 6,436 controls from 14 studies of African American, East Asian, South Asian, Hispanic, and European ancestry. In contrast to the GoT2D whole-genome integrated panel, this data set also includes GoT2D individuals for whom whole genome data were not available. Sample characteristics are provided in Extended Data Table 2 and Supplementary 4. Sequence reads were processed and aligned to the reference genome (hg19) with Picard (http://picard.sourceforge.net). Polymorphic sites and genotypes were called with GATK, with filtering of sites performed using Variant Quality Score Recalibration (VSQR) for SNVs, and hard filters for indels. Genotype likelihoods were computed controlling for contamination.
Hard calls (the GATK-called genotypes but set as missing at a genotype quality (GQ)<20 threshold52) and dosages (the expected value of the genotype, defined as Pr(RX|data)+2Pr(XX|data), where X is the alternative allele) were computed for each sample at each variant site. Hard calls were used only for quality control, while dosages were used in all downstream association analyses. Multi-allelic SNVs and indels were dichotomized by collapsing alternate alleles into one category because downstream association analyses required bi-allelic variants.
Individuals were excluded from analysis if they were outliers on one of multiple metrics: poor array genotype concordance (where available), high number of variant alleles or singletons, high or low allele balance (average proportion of non-reference alleles at heterozygous sites), or excess mean heterozygosity or ratio of heterozygous to homozygous genotypes.
Within this reduced set of individuals, we then performed extended QC using ethnicity and T2D status to provide high-quality genotype data for downstream association analyses. Within each ethnicity, we excluded variants based on hard call rate (<90% in any cohort), deviation from Hardy-Weinberg equilibrium (p<10−6 in any ancestry group), or differential call rate between T2D cases and controls (p<10−4 in any ancestry group). We then considered autosomal variants that passed extended QC and with MAF>1% in all ancestry groups for trans-ethnic kinship analyses. We calculated identity-by-state (IBS) between each pair of samples based on independent variants (trans-ethnic r2<0.05) and constructed axes of genetic variation through PCA implemented in EIGENSTRAT62 to identify ethnic outliers (Supplementary 29). We also identified duplicates based on IBS, and excluded the sample from each pair with lowest call rate and/or mismatch with external information. The extended QC excluded 68 individuals, and 9.9% of SNVs and 90.8% of indels from the clean dataset.
2. Association analysis
2.1.1. Power calculation
We used the genetic power calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) to estimate power to detect T2D association assuming 8% prevalence. For the T2D-GENES+GoT2D exome sequence data set we assumed: (i) a fixed-effect across all five ancestry groups (12,940 individuals); and (ii) an effect specific to one group (2,000 individuals) (Extended Data Figure 4). We repeated our calculations for combined exome sequence and exome array data, assuming a fixed effect across all ethnicities, for an effective total sample size of 82,758 individuals (Extended Data Figure 4).
For the GoT2D integrated panel we allowed for incomplete variant detection by multiplying power by the estimated sensitivity to detect the variant as a function of MAF. For imputed variants, we first multiplied the sample size by the median imputation quality (rsq_hat) obtained from MaCH/Thunder or minimac63 for the corresponding MAF bin across the analyzed cohorts, and then multiplied the estimated power by the fraction of variants that passed the imputation quality cutoff for that MAF bin.
For gene-based tests in the T2D-GENES+GoT2D data, we made use of a Bonferroni correction for 20,000 genes, corresponding to p<2.5×10−6. We used a simulated haplotype dataset from the SKAT package (http://cran.r-project.org/web/packages/SKAT/vignettes/SKAT.pdf) and estimated the power of SKAT-O to detect association of variants within a gene at this threshold as a function of the phenotypic variance (1%) in a liability scale explained by additive genetic effects and the percentage of variants that were causal (50% and 100%). As for single-variant power calculations, we considered: (i) a fixed-effect across all ethnicities (12,940 individuals); and (ii) an effect specific to one ancestry group (2,000 individuals) (Extended Data Figure 4).
2.2. GoT2D integrated panel association analysis
2.2.1. Single-variant association analysis
We tested for T2D association in a logistic regression framework assuming an additive genetic model. We used the Firth bias-corrected likelihood ratio test64,65 as our primary analysis strategy; we repeated association analysis using the score test for inclusion in sample-size-weighted meta-analysis (Supplementary 2). Tests were adjusted for sex, the first two genotype-based PCs to account for population stratification, and an indicator function for observed temporal stratification based on sequencing date and center. PCs were calculated using linkage-disequilibrium (LD) pruned (r2<0.20) HumanOmni2.5M array variants with MAF>1% after removing large high-LD regions59,60.
2.2.2. Aggregate association analysis
To test for aggregate association within coding regions of the genome, we used the approach described in 2.3.6. For every gene and mask tested, p-values were greater than 2.5 × 10−4. We also tested for aggregate association among variants in non-coding regions of the genome. We aggregated variants in individual pancreatic islet enhancer elements (see 6.1), as these elements collectively demonstrated strongest genome-wide enrichment of T2D association. We performed both the burden and SKAT tests using genotypes from the integrated panel on variants with MAF<5% in each islet enhancer element. We used a Bonferroni threshold p<1.68×10−7 based on a nominal significance level of α=0.05 corrected for 298,240 elements with at least one variant. All elements tested in this manner had p-value greater than 2.5 × 10−6.
2.3. GoT2D+T2D-GENES multiethnic association analysis
2.3.1. Kinship analysis
Within each ancestry group, we considered autosomal variants that passed QC with MAF>1% for ethnic-specific kinship analyses. We calculated IBS between each pair of samples in the ancestry group based on independent variants (ethnic-specific r2<0.05) and constructed a kinship matrix to account for intra-ethnic population structure and relatedness in downstream mixed-model (EMMAX) based association analyses16. We also used IBS to identify pairs of related individuals within each ancestry group (defined by pi-hat>0.3). We then defined intra-ethnic related exclusion lists for downstream non-EMMAX association analyses using the following steps: (i) remove the control from each T2D-status discordant pair; and (ii) remove the sample with lowest call rate from each T2D-status concordant pair. We also constructed intra-ethnic axes of genetic variation through PCA implemented in EIGENSTRAT62. We identified axes of genetic variation in each ancestry group for inclusion as covariates in downstream non-EMMAX association analyses to account for intra-ethnic population structure that: (i) explain at least 0.5% genotypic variation; and/or (ii) demonstrate nominal association (p<0.05) with T2D in logistic regression analysis.
2.3.2. Single-variant association analysis
Within each ancestry group, we performed a score test of T2D association with each variant passing ethnic-specific QC in a linear regression framework under an additive model in EMMAX16. We also performed a Wald test of T2D association with each variant passing ethnic-specific QC in a logistic regression framework under an additive model with adjustment for ethnic-specific axes of genetic variation after exclusion of related samples (Supplementary 30). Within each ancestry group, we calculated genomic control inflation factors (score EMMAX and Wald) based on independent variants used for the ethnic-specific kinship analyses and corrected association summary statistics (p-value and SE) to account for residual population structure.
Subsequently, we performed trans-ethnic fixed-effects meta-analysis of ancestry-specific association summary statistics at each variant based on: (i) sample size weighting of score EMMAX directed p-values; and (ii) inverse-variance weighting of Wald beta/SE (to obtain unbiased estimates of allelic odds ratios and confidence intervals that cannot be constructed from EMMAX effect estimates). We also performed trans-ethnic meta-analysis of ancestry-specific association summary statistics (score EMMAX beta/SE) at each variant using MANTRA66, using pair-wise mean allele frequency differences at the subset of independent variants used for trans-ethnic kinship analyses as a prior for relatedness between ancestry groups.
2.3.3. Validation of PAX4 association signal in additional East Asian studies
We validated the PAX4 Arg192His (rs2233580) association signal in an additional 1,789 T2D cases and 1,509 controls of East Asian ancestry from Hong Kong, Korea, and Singapore (Supplementary 9). Within each study, we tested for association with T2D in a logistic regression model, and combined association summary statistics across studies through fixed-effects meta-analysis (Supplementary 9). Among T2D cases, we also tested for association with age of diagnosis in a linear regression model, and combined association summary statistics across studies through fixed-effects meta-analysis (Supplementary 9).
2.3.4. Admixture analysis
Admixed populations can offer greater statistical power to detect association because diverse ancestry increases genetic variation. However, admixture can also introduce false-positive signals due to population stratification and heterogeneity of effects because of differential LD67. To assess the contribution of ancestral background in the two admixed groups (African American and Hispanic), we inferred local ancestry based on SNVs in available GWAS data using two approaches. For African Americans, we ran HAPMIX68 using CEU and YRI haplotypes from HapMap as reference, and estimated the proportion of European ancestry at each genomic position. For Hispanics, we ran Multimix69 using European, West African, and Native American haplotypes from HapMap as reference, and estimated the proportion of European ancestry at each genomic position, since we observe only a very low West African contribution (1.1-3.2%, Supplementary 31). We then repeated our intra-ethnic EMMAX-based analyses within African American and Hispanic ancestry groups, this time adjusting for local ancestry by including the estimated proportion of European ancestry at each variant as a covariate. Adjustment for local ancestry resulted in numerically similar association statistics as those from unadjusted analyses in the African American and Hispanic samples.
2.3.5. Gene-based analysis
We generated four variant lists (‘masks’) based on MAF and functional annotation. We mapped variants to transcripts in Ensembl 66 (GRCh37.66). Using annotations from CHAoS v0.6.3, SnpEFF v3.1, and VEP v2.7, we identified variants predicted to be protein-truncating (e.g. nonsense, frameshift, essential splice site) denoted PTV-only or ‘Mask 1’; or protein-altering (e.g. missense, in-frame indel, non-essential splice site) in at least one mapped transcript (by at least one of the three algorithms) with MAF<1%, denoted PTV+missense or ‘Mask 2’. We additionally used the procedure described by Purcell et al.70 to identify subsets of missense variants with MAF<1% meeting ‘strict’ or ‘broad’ criteria for being deleterious, using annotation predictions from Polyphen2-HumDiv, PolyPhen2-HumVar, LRT, Mutation Taster, and SIFT; variants predicted deleterious by all five algorithms or by at least one algorithm were denoted PTV+NSstrict or ‘Mask 3’ and PTV+NSbroad or ‘Mask 4’, respectively. Indels predicted by CHAoS, SnpEFF, or VEP to introduce frameshifts were included in the ‘strict’ category. We calculated MAFs for each ancestry using high-quality genotype calls (GQ>20) for all samples passing extended QC. We considered a variant to have MAF<1% if MAF estimates for every ancestry group were <1%.
We used the MetaSKAT R package (v0.32)15 with the SKAT v0.93 library to perform SKAT-O71 analysis within each ancestry, and in meta-analysis. Within each ancestry group, we analyzed genotype dosages with adjustment for ethnic-specific axes of genetic variation after exclusion of 96 related individuals. We assumed homogenous allele frequencies and genetic affects for all studies within an ancestry group. We performed meta-analysis using genotype-level data, allowing for heterogeneity of allele frequencies and genetic effects between (but homogeneity within) ancestry groups. All analyses were completed using the recommended rho vector for SKAT-O: (0, 0.12, 0.22, 0.32, 0.52, 0.5, 1).
2.4. Imputed data
2.4.1. Samples
We carried out genotype imputation into 44,414 individuals (11,645 T2D cases and 32,769 controls) from 13 studies using the GoT2D integrated haplotypes as reference panel. Characteristics of the imputed studies are provided in Extended Data Table 2 and Supplementary 3.
2.4.2. Single-variant association meta-analysis
The one sequenced and thirteen imputed studies totaled 12,971 T2D cases and 34,100 controls. Each study performed its own sample- and variant-based QC. In each study, SNVs with minor allele count (MAC)≥1 passing QC were tested for T2D association assuming an additive genetic model adjusting for study-specific covariates. Association testing was performed using logistic regression Firth bias-corrected, likelihood ratio, or score tests as implemented in EPACTS (genome.sph.umich.edu/wiki/EPACTS) or SNPTEST72. To account for related samples in the Framingham Heart Study, generalized estimating equations (GEE) were used, as implemented in R. Residual population stratification for each study was accounted for using genomic control73. We then carried out fixed-effects sample-size weighted meta-analysis as implemented in METAL74.
2.4.3. Conditional analyses in established GWAS loci
We compiled a list of 143 previously-reported genome-wide significant SNVs in 81 T2D autosomal loci (a) from Morris et al.2 and Voight et al.4; (b) from papers they referenced; and (c) from references in the NHGRI GWAS catalog75. We LD pruned these SNVs (r2<0.95), yielding a list of 129 SNVs. We deleted the CILP2 locus (and two SNVs) from subsequent whole-genome analyses owing to large regions in which no variants passed QC, resulting in a list of 127 index SNVs at 80 autosomal loci. To identify additional T2D-associated variants within these 80 T2D autosomal loci in the genome-wide data, we repeated GWA analysis for 12 of the 13 studies (conditional analysis results for FHS were unavailable), conditioning on the 127 index SNVs. We performed fixed-effects inverse-variance meta-analysis to combine conditional analysis results from the studies totaling 12,298 cases and 26,440 controls. For each known locus, we analyzed all SNVs within 500kb of the known index SNVs; if there were multiple known index SNVs, we analyzed all SNVs within 500kb of the most proximal and distal index SNVs. We imposed a conditional-analysis significance threshold of α=1.8×10−6 based on a proportional number of multiple tests for ~83Mb of the ~3000Mb genome.
2.5. Exome array data
2.5.1. Samples
We considered 28,305 T2D cases and 51,549 controls from 13 studies of European ancestry, genotyped with the Illumina exome array. Characteristics of the studies are provided in Extended Data Table 2 and Supplementary 15.
2.5.2. Overlap of exome sequence variation with exome array
We assessed overlap of variants present on the exome array with those observed in our trans-ethnic exome-sequence data. Since exome array primarily contains SNVs that are predicted to be protein altering, we focused on nonsense, essential splice site, and missense variants. Only variants passing QC in both sequence and array data were included in our overlap assessment.
2.5.3. Data processing, QC, and kinship analysis
Within each study, exome array genotypes were initially called using GenCall (https://support.illumina.com/downloads/gencall_software.html) and Birdseed76. Sample and variant QC was then undertaken within each study based on several quality control filters. Criteria for sample exclusion included low call rate (<99%), mean heterozygosity, high singleton counts, non-European ancestry, sex discrepancy, GWAS discordance (where data were available), genotyping platform fingerprint discordance, and duplicate discordance. Variants were excluded based on call rate (<99%), deviation from Hardy-Weinberg equilibrium (p<10−6), duplicate, chromosome or allele mismatch, GenTrain score <0.6, Cluster separation score <0.4, and manual cluster checks. Missing genotypes were subsequently re-called using zCall, with a second round of QC to exclude poor quality samples (call rate <99% and mean heterozygosity) and variants (call rate <99%). Within each study, we considered independent autosomal variants that passed QC with MAF>1% for kinship analyses, and calculated IBS between each pair of samples. We used these statistics to: (i) identify non-European ancestry samples to be excluded from all downstream analyses; (ii) construct a kinship matrix to account for fine-scale population structure and relatedness in downstream EMMAX-based association analyses; (iii) identify related samples to be excluded from downstream non-EMMAX association analyses; and (iv) calculate axes of genetic variation for inclusion as covariates in downstream non-EMMAX association analyses to account for fine-scale population structure (if required).
2.5.4. Single-variant association analysis
Within each study, we performed a score test of T2D association with each variant passing QC in a mixed-model regression framework under an additive model in EMMAX16. We also performed a Wald test of T2D association with each variant in a logistic regression framework under an additive model with adjustment for axes of genetic variation after exclusion of related samples. For each test, we corrected SE and p-value for the genomic control inflation factor (if >1) calculated based on the independent autosomal variants used for kinship analysis.
Across studies, we performed fixed-effects meta-analysis of association summary statistics at each variant based on: (i) inverse-variance weighting of score EMMAX beta/SE; (ii) sample size weighting of score EMMAX directed p-values; and (iii) inverse-variance weighting of Wald beta/SE. For each of these meta-analyses, we applied a second round of correction of SE and p-value by genomic control, again calculated based on the independent autosomal SNVs used for kinship analyses.
2.5.5. Combined exome sequence and exome array single-variant analysis
We considered variants that were represented both in the exome sequence and on the exome chip. We began by performing fixed-effects meta-analysis of association summary statistics (after correction for genomic control, as described above) from the exome-chip meta-analysis and the European ancestry sequenced samples using: (i) inverse-variance weighting of score EMMAX beta/SE; (ii) sample size weighting of score EMMAX directed p-values; and (iii) inverse-variance weighting of Wald beta/SE. Subsequently, we performed trans-ethnic fixed-effects meta-analysis of ancestry-specific association summary statistics (after correction for genomic control, as described above) at each variant based on: (i) sample size weighting of score EMMAX directed p-values; and (ii) inverse-variance weighting of Wald beta/SE.
2.5.6. Gene-based analyses
We made use of the four variant masks defined for exome sequence gene-based analyses, but with MAF calculated across all exome array studies. Within each study, we performed SKAT-O analyses71, with adjustment for axes of genetic variation after exclusion of related samples. We combined p-values for association across studies via meta-analysis with Stouffer's method77.
2.5.7. Evaluating relationships between association signals for coding variants and previously reported lead SNVs at established GWAS loci
For coding variants mapping to established T2D susceptibility loci and achieving genome-wide significance in combined exome sequence and/or exome array analysis, we used complementary approaches with a range of available genetic data resources to evaluate their contribution to the association signals of previously reported lead SNVs. If the previously reported lead SNV (or a good proxy, r2≥0.8) was genotyped on the exome array, we performed reciprocal conditional analyses with the available exome array data. Within each study, we repeated EMMAX analyses in GWAS loci, including additively coded genotypes at the previously reported2 lead SNV or genome-wide significant coding variant as an additional covariate in the regression model. Across studies, we performed fixed-effects meta-analysis of association summary statistics at each variant based on: (i) inverse-variance weighting of score EMMAX beta/SE; (ii) sample size weighting of score EMMAX directed p-values. If the previously reported lead SNV (or a good proxy) was not genotyped on the exome array, we performed approximate reciprocal conditional analysis, implemented in GCTA78, using genome-wide meta-analysis association summary statistics from 12,971 T2D cases and 34,100 controls from the combined GoT2D integrated panel and imputed data. Patterns of LD between variants were estimated using a subset of the GoT2D integrated panel, restricted to 2,389 individuals with pairwise genetic relationship <0.025, as defined by the GCTA A statistic79. Finally, we interrogated 99% credible sets of variants at each GWAS locus, which together represent ≥99% of the probability of driving each association signal. We determined whether the coding variant at each locus was included in the credible set for the association signal for the previously reported lead SNV, and recorded its rank.
3. Enrichment of exome association signals in GWAS
To define T2D-associated intervals, we first identified all SNVs associated with T2D in published genome-wide association studies (GWAS) by searching literature and the NHGRI GWAS catalog (see also 2.4.3). We identified 143 autosomal SNVs, with some associated in more than one ancestry (167 SNV-ancestry pairs). For each SNV-ancestry pair, we identified the most distant pair of SNVs with r2>0.5 in 1000 Genomes Phase I data, using the appropriate continental subset of 1000 Genomes samples (EUR, AMR, or ASN). We used 1000 Genomes data, rather than our own exome sequence data, because most reported associations for T2D are with common, intergenic SNVs. We then extended each region of interest by moving out 0.02 cM from those two SNVs (to encompass nearby recombination hotspots), and added an additional 300kb upstream and downstream. We merged overlapping intervals, yielding 81 unique associated regions, and identified 634 genes completely or partially included within associated regions. In single-variant analyses, we analyzed 3,147 non-synonymous variants within these genes in the combined exome sequence and exome array datasets, using a Bonferroni corrected significance threshold of α=0.05/3,147=1.6×10−5. We considered gene-level association statistics from exome sequence for these 634 genes using a Bonferroni-corrected significance threshold of α=0.05/634=7.9×10−5.
We note that by reducing the stringency of the significance threshold for variants within GWAS loci, we increase the ‘experiment-wise’ type I error rate across the entire exome. Assuming that 3% of 100,000 coding variants interrogated in this study map to T2D GWAS loci, as defined above, we would need to change the threshold of significance outside of these regions to p<2.1×10−8 to maintain an ‘experiment-wise’ type I error rate of 5%.
4. Testing for ‘synthetic associations’ at T2D loci in GoT2D genome sequence data
To identify low-frequency or rare variants that could potentially define synthetic associations, we analyzed the ten T2D loci at which a previously-reported tag SNV achieved p<0.001 in our single-variant analysis of the genome sequence dataset. We defined as candidates at each locus all low-frequency or rare variants (excluding singletons) within a 5Mb window (centered on the prior GWAS signals) and tested for synthetic associations caused by either (1) a single low-frequency or rare variant or (2) multiple low-frequency or rare variants on a common haplotype.
To identify synthetic associations driven by a single low-frequency or rare variant at each of the ten loci, we performed a series of conditional analyses in which we tested for association between gene dosage at the previously reported GWAS index SNV and T2D risk via logistic regression, while including each candidate low-frequency or rare SNV (excluding singletons) as an additional covariate, one-by-one. If inclusion of the low-frequency or rare variant resulted in a conditional association p>0.05 for the tag SNV, we considered the common-variant association signal a potential synthetic association.
To identify synthetic associations based on sets of low-frequency or rare variants, we extended this approach. We (1) defined common haplotypes segregating at each T2D locus; (2) identified all low-frequency or rare (excluding singletons) variants occurring on T2D-associated haplotypes (haplotypes on which the T2D-associated GWAS index SNV minor allele is present); and (3) asked whether any combination of these low-frequency or rare variants could explain the effect observed at the T2D GWAS index SNV. We carried out these analyses restricting attention to protein-coding variants within the window and then again for all low-frequency and rare SNVs in the 5Mb window.
To define common haplotypes at each locus, we used the phased whole-genome sequence data. We first employed the phased genotypes for common (MAF>5%) variants segregating in the interval between recombination hotspots at the locus (to minimize the number of recombinant haplotypes identified). We next identified the haplotypes on which the T2D-associated (risk or protective) GWAS index SNV minor allele was present. We then assembled the set of low-frequency and rare variants from across the 5Mb interval which occurred on the background of these T2D-associated common-variant haplotypes. Due to recombination and imperfect phasing, low-frequency or rare (excluding singletons) variants are often observed on more than one haplotype background. We included all low-frequency or rare variants that occurred more frequently on a T2D-associated haplotype than on other haplotypes.
From this pool of low-frequency and rare variants, we considered only variants with the same direction of effect as the common GWAS index SNV minor allele, as required by the synthetic association hypothesis, which posits that low-frequency or rare variants of larger effect than the common SNV could induce a weaker association signal. We then used a greedy algorithm to select the low-frequency or rare variant which, when added to the index GWAS SNV's dosage in a logistic regression, most reduced the residual effect remaining at the index SNV, as measured by estimated conditional odds ratio. We repeated this process, adding variants to the model, until the estimated effect at the index SNV genotype or gene dosage changed sign, representing no residual effect of the index SNV. At each locus, we also counted the number of variants required to increase the association p-value at the GWAS index SNV beyond the nominal p=0.05 significance threshold (Extended Data Table 8).
5. Credible set analysis of GoT2D genome sequence data
At 78 of the 80 T2D GWAS loci (2.4.3), the previously reported index SNV had MAF>1% in our GoT2D genome-sequenced sample. At these 78 loci, we constructed credible sets of common variants that, with some minimum specified probability (e.g. ≥99%), contain the variant causal for the corresponding association signal. Our analysis assumes a single causal SNV per signal and that the SNV was genotyped30,31. We constructed credible sets for up to two independent association signals at each locus; at 5 loci with multiple independent (r2<0.10) GWAS index SNVs, we constructed two distinct credible sets.
For each GWAS index SNV, we identified the set of common variants with r2≥0.10 with the index SNV within a 5Mb window centered on the index SNV. For each variant in this set, we calculated the posterior probability of being causal31. We first calculated an approximate Bayes’ factor (ABF) for each variant as:
where r=0.04/[SE2+0.04], z=β/SE, and β and SE are the estimated effect size (log odds ratio) and its standard error from logistic regression. We then calculated the posterior probability for each variant as ABF/T, where T is the sum of the ABF values over all candidate variants across the interval. This calculation assumes a Gaussian prior with mean 0 and variance 0.04 for β, the same prior employed in the commonly used single-variant association program SNPTEST72.
We based the analysis on the genome-wide meta-analysis results, since most common variants were included in this analysis, and sample sizes were significantly larger than for the genome sequence data alone.
We calculated the effective imputed sample size for each variant in the meta-analysis data as , where is the imputation quality and is the effective sample size for imputation cohortj. To ensure approximately uniform sample size across variants, we considered to be well-imputed only those variants with effective imputed sample size (Neff)≥80% of the maximum observed across all variants in the window.
Indels were not imputed or meta-analyzed in this study, and <2% of common SNVs were not well-imputed by the above effective sample size criterion. To include these common variants while using the most precise estimates available, we calculated posterior probabilities separately from each genome-wide data source. Where an indel from the sequence dataset had a SNV proxy in high LD (r2≥0.80) in the meta-analysis dataset, we used the proxy's information instead. Where a common SNV that was poorly imputed had high-quality association data from the genome sequence data alone, the posterior probability from the genome sequence dataset was used instead. In each case, the final posterior probabilities for all SNVs were re-scaled such that their sum across a locus equaled one.
We used these final posterior probabilities to rank variants in decreasing order. To define credible sets of a specified level (e.g. 99%), we included variants with highest final posterior probabilities until their sum reached or exceeded that level (Supplementary 28).
6. Genome enrichment analyses of the GoT2D genome sequence data
6.1. Genomic annotation
We collected genome annotation data from several sources. First, we obtained gene transcript information from GENCODEv1480. For protein-coding genes, we included transcripts with a protein-coding tag that either were present in the conserved coding DNA sequence (CCDS) database or had experimentally confirmed mRNA start and end; we then included 5’ UTR, exon, and 3’ UTR regions from the resulting transcripts. For non-coding genes, we included transcripts with a lncRNA, miRNA, snoRNA, or snRNA tag.
Second, we defined regulatory chromatin states in 12 cell types. We collected sequence reads generated for the following assays: H3K4me1, H3K4me3, H3K27ac, H3K27me3, H3K36me3, and CTCF ChIP, in 9 ENCODE cell types (GM12878, K562, HepG2, Hsmm, HUVEC, NHEK, NHLF, hESC, HMEC)32, pancreatic islets35, and hASC (adipose stromal cell) pre- and mature adipocytes33. We mapped reads to hg19 using BWA51 and used the resulting mapped reads for all cell types to call regulatory states using ChromHMM81, assuming ten states. We then assigned names to the resulting state definitions: (1) H3K4me3, H3K27ac (active promoter); (2) H3K4me3, H3K27ac, H3K4me1 (active enhancer 1); (3) H3K27ac, H3K4me1 (active enhancer 2); (4) H3K4me1 (weak enhancer); (5) H3K27me3, H3K4me3, H3K4me1 (poised promoter); (6) H3K27me3 (repressed); (7) low/no signal 1; (8) CTCF (insulator); (9) low/no signal 2; and (10) H3K36me3 (transcription).
Third, we obtained transcription factor binding ChIP sites from three sources: 141 proteins from ENCODE32, 5 from Pasquali et al.35, and 1 from Mikkelsen et al.33.
From gene transcript data we defined CDS (protein coding transcript exons); ncRNA (non-coding RNA transcripts); and 3’ and 5’ UTR (UTR regions of coding transcripts). From chromatin state data for each of the 12 cell types we identified active enhancers (pooled active enhancer 1 and 2 elements); weak enhancers; and active promoters. From transcription factor binding sites we defined transcription factor binding sites (TFBS) (sites pooled across all factors). This resulted in a total of 41 annotation categories (Extended Data Figure 10).
6.2. Enrichment of genome annotation
We jointly modeled variants in credible sets using T2D association and the functional annotation classes using the method described by Pickrell38. First, we tested each annotation individually and identified the annotation that most improved the model likelihood. We then iteratively added annotations in this manner until the likelihood did not increase further. Using this set of annotations, we tested a range of penalized likelihoods (from 0-1 in .01 increments) using 10-fold cross-validation, and identified the penalty that gave the best cross-validation likelihood. Using this penalty, we then iteratively dropped annotations to identify the model with the maximal cross-validation likelihood. The resulting model included coding exons, TFBS, hASC mature adipose active enhancers and promoters, pancreatic islet active and weak enhancers and active promoters, hASC pre-adipose active and weak enhancers, NHEK active enhancers, NHLF active enhancers, K562 weak enhancers, HMEC weak enhancers and active promoters, H1-hESC active promoters, ncRNA, and 5’ and 3’ UTR (Extended Data Figure 10). Finally, we used this model to update posterior probabilities for each variant and re-calculate 99% credible sets.
7. Gene enrichment analyses in the GoT2D+T2D-GENES exome sequence data
We first used the SMP (statistics/matrix/permutation) gene-set enrichment procedure implemented in the PLINK/Seq package (http://atgu.mgh.harvard.edu/plinkseq/). This approach calculates enrichment statistics for large sets of genes to establish whether case-enrichment of rare variants is preferentially concentrated in a particular set of genes, controlling for any exome-wide/baseline difference in case and control rates. The procedure uses gene-based association statistics, and forms sums of these statistics over all genes in a set, the significance of which is evaluated by permutation. We considered the relative enrichment statistic, SSET/SEXOME, with significance evaluated empirically (10,000 replicates) based on the null distribution of this ratio. The reported effect sizes from the gene-set enrichment analysis are estimates of the unconditional odds ratio that do not take exome-wide differences in case/control rates into account70. We selected 18 ‘premium’ sets of genes (Supplementary 32) that reflect the current knowledge of pathways (N=15) involved in type 2 diabetes and the three sets of genes involved in monogenic form of diabetes defined above: ‘Monogenic All’ (N=81); ‘Monogenic Primary’ (N=28); and ‘Monogenic OMIM’ (N=13). We restricted these analyses to singleton and ultra-rare (MAF<0.1%) protein-truncating variants.
We then used biological knowledge to test for enrichment of association signal across established sets of genes from Gene Ontology, KEGG, Reactome, and Biocarta collections from MSigDB (version 4.0) as well as a number of hand-curated gene-sets (Supplementary 32) that had been generated for the SMP analyses. These analyses calculated measures of gene-set enrichment from gene-level association results (i.e. from SKAT-O) by means of a pre-ranked GSEA82 method (version 2.0.13), which consists of a weighted Kolmogorov-Smirnov (random bridge) statistic. In our analysis we performed 10,000 permutations on gene-set sizes from 5 to 5,000 genes.
8. Investigation of genes implicated in Mendelian forms of diabetes in the exome data
We first curated a list of 81 genes termed the ‘Monogenic All’ gene-set (Supplementary 22), consisting of genes with pathogenic mutations reported to co-segregate with diabetes or a syndrome associated with an increased prevalence of diabetes. Two subsets of the ‘Monogenic All’ gene-set were then additionally defined: the ‘Monogenic Primary’ gene-set (N=28), consisting of genes with mutations leading to diabetes as a primary feature, and the ‘Monogenic OMIM’ gene-set (N=13), consisting of genes linked to Maturity Onset Diabetes of the Young (MODY) or Neonatal Diabetes in the OMIM catalog (entry #606391 and #606176). In addition to examining the significance of single-variant and gene-based tests within these gene-sets, we also performed an aggregate analysis of all variants in the gene-set. For each of the three gene-sets, we constructed five variant lists by applying the same four masks as in the exome-wide gene-level analysis (PTV-only, PTV+missense, PTV+NSbroad and PTV+NSstrict), as well as an additional mask containing all variants reported as ‘high confidence’ and ‘disease-causing’ in the Human Gene Mutation Database (HGMD), annotated using Biobase ‘GenomeTrax’ software (http://www.biobase-international.com/product/genome-trax). We then analyzed each of the fifteen variant lists with the SKAT-O test, using the same meta-analysis procedure and covariates as in the exome-wide gene-based analysis. To obtain effect-size estimates, for each variant list we applied a collapsing burden test, in which logistic regression of T2D status was performed on individual genotypes encoded as 0 (if they carried no variants in the list) or 1 (if they carried at least one variant in the list). Effect size estimates and standard errors were determined using the Firth penalized likelihood method. Analysis in the exome array dataset was performed by first generating fifteen variant lists based on the content of the exome array, computing the collapsing burden test for each cohort, and then combining associations and effect size estimates using an inverse variance weighted meta-analysis. To compare the age of diagnosis of variant carriers to those of non-carriers, we used a two-sided t-test.
9. Protein-protein interaction analyses in the exome data
We performed data-driven extraction of association signal enriched sub-networks (rather than relying on pre-defined gene-sets) from protein-protein interaction (PPI) data. We used two different approaches, both run using the curated PPI database InWeb383.
The first approach consists of two steps. First, the entire human PPI network was searched for protein complexes (clusters) using the algorithm implemented in clusterONE84, which identifies protein complexes with high cohesiveness. The method was run with default parameter settings (0.3 as density threshold, 0.8 as merging threshold, and 2 as the penalty-value node), and with the --fluff option activated, which allows the addition of highly connected boundary nodes to the cluster. Second, gene-based association p-values derived from SKAT-O analyses of the 12,940 multiethnic exome sequences were aggregated, using Fisher's method, for the genes encoding each of the proteins within a cluster to generate a ‘cluster association’ statistic.
An empirical p-value for the significance of these aggregated cluster association statistics was derived by comparing each cluster to a large number of complexes of the same topology, but composed of randomly sampled proteins. Specifically, a background distribution was obtained for each protein complex as follows: each protein in the cluster was randomly substituted by a different protein represented in the InWeb3 database, matched for number of minor allele carriers in the data set. SKAT-O p-values were assigned to each protein from the exome sequencing results, and an aggregated p-value was obtained for each pseudo-complex using Fisher's method, as above. This process was repeated 100,000 times, and the empirical p-value for each complex was calculated as the proportion of the iterations for which the Fisher's p-value of the observed complex was more significant than that of p-values for the pseudo-complexes. This procedure was repeated for all gene-level masks (PTV-only, PTV+missense, PTV+NSstrict and PTV+NSbroad).
To test the study-wide significance of apparently associated clusters, we used two permutation designs. In the first design, we generated 100,000 pseudo-complexes for each cluster, replacing each protein within each cluster with one protein from InWeb3, matched for the number of minor allele carriers in the data set. We calculated the number of permuted datasets which generated any ‘pseudocluster’ association p-value more significant than our most enriched cluster. In the second design, we used a Monte-Carlo algorithm to generate 10,000 random PPI networks, with the same degree as observed in the InWeb3 database, ran clusterONE on each, and once again compared the distribution of ‘best’ cluster association p-value with that observed in the real data.
The second approach uses the dense module searching algorithm (a heuristic ‘greedy’ method) described in dmGWAS85, where a module is defined as a sub-network within the whole network if it contains a locally increased proportion of low p-value genes. This method differs from the earlier method in using the association p-values, in combination with the PPI data, to construct the networks. The module is grown for each protein in the PPI by adding the neighboring nodes within a pre-defined distance (d=2) that can yield a maximum increment of the module score for module m, where k is the number of genes in the module and Zi is calculated from the p-value of exome gene-based tests using an inverse normal distribution function. The addition of neighborhood nodes is stopped when the increment is less than 10% of Z(k)m (that is, Z(k+1)m< +Z(k)m × 0.1). As with the clusterONE approach, this procedure was conducted for all four exome gene-based level masks.
To evaluate whether the top ranked-modules are significantly associated with T2D, we permuted case-control status across the 12,940 exomes (maintaining ethnic strata) 10,000 times and generated 10,000 SKAT-O gene-based association tests on all genes in the top 15 modules (once for each gene-based variant mask, 40,000 in total). During each permutation, Zm was re-calculated for each module, and a set of empirical p-values was obtained by comparing the p-value of the original module to these modules with the SKAT-O results from the swapped labels. Following the above procedure, all 15 top modules were found significantly enriched for the PTV+NSstrict and PTV+NSbroad gene-based variant masks (p<10−4, after the 10,000 case-control permutations).
10. Modelling disease architecture
10.1. T2D liability risk and architecture bounding in the exome array data
We used a Bayesian framework implemented in R to compute the probability that each variant explains more than a defined amount of the T2D-risk liability-scale variance (LVE). The joint distribution in the MAF-OR space is computed by assuming a T2D prevalence of 8% and beta and normal distributions for the MAF and the odds ratio (OR) respectively. The OR is calculated with reference to the minor allele. The MAF is adjusted to take account of apparent allele frequency heterogeneity between cohorts (subjects from missing cohorts are excluded from calculations). Analyses are restricted to variants with MAF>0.1% since the representation of variants with MAF below this threshold on the exome array is poor. The probability is obtained by numerically integrating over the joint distribution for MAF-OR combinations that explain more than the defined amount of liability-scale variance. For bounding the maximum number of variants that could contribute to T2D risk variance, we performed a sensitivity analysis on the 88 known T2D index SNVs present on the exome array to define the thresholded variance explained and the probability: this analysis shows that for a probability of >0.8 to explain 0.01% of the T2D risk variance, we were able to identify 91% of these known T2D SNVs. Ranges of OR and MAF consistent with 80% power to detect single-variant association in this dataset (for exome-wide significance, p<5×10−7) were calculated to reflect the fact that differences in sample size for individual variants (due to differences in allele frequency distribution and genotyping QC) also influence power. The relationship between power and LVE differs for risk and protective alleles because of unequal numbers of cases and controls.
10.2. Genetic architecture simulations based on GoT2D data and results
10.2.1. Range of simulated disease models
Following our previously published framework40, we conducted population genetic simulations of T2D architecture using the forward simulation program ForSim86. We assumed T2D prevalence 8% and heritability ~45%, and chose the mutation rate, recombination rate, a gamma distribution of selection coefficients, and other parameters of demographic history by fitting the simulated site frequency spectrum to empirical high coverage exome sequence data from GoT2D.
We then considered a wide range of disease models by varying two parameters: coupling parameter τ which regulates how strongly selection against a disease-causing allele depends on the per-allele disease risk87; and target size T, the summed lengths of the genomic regions within which mutations can influence T2D risk. Specifically, a variant's additive contribution to disease risk g is given by g=sτ(1+ε) where s is the selection coefficient under which the variant evolves and ε is drawn from a normal distribution40.
By varying τ and T, we generated a wide range of joint distributions for allele frequency and effect size. In total, we evaluated 12 models: τ=0, 0.1, 0.3, and 0.5 crossed with T=750kb, 2.0Mb, and 3.75Mb. Under models with higher selection against strongly deleterious alleles (larger τ), rare variants explain the bulk of heritability and can have large effects, while under models with weak dependence (smaller τ), common variants explain the bulk of heritability and rare variants collectively have weaker effects. Although we had previously excluded many models as producing predictions inconsistent with observed sibling relative risk, GWAS, and linkage results, prior work showed that models varying widely in the proportion of total heritability attributable to rare versus common variation were still plausible88. In this study, we explored whether the space of plausible disease models could be further constrained using whole genome sequence, imputation, and meta-analysis results.
10.2.2. Simulation procedure
ForSim enables simulation of variants across user-specified loci in large populations. Inputs include a demographic history (trained on European sequence data) and a gamma distribution of selection coefficients for a subset of variants under natural selection. We simulated genotypes for a current population of effective size 500,000 individuals40 and selected potential disease risk variants from those under selection appropriate to the intended target size. Each risk variant received a disease-specific effect size depending on the selection coefficient under which it evolved and the assumed degree of dependence between selection and effect size. Each individual was then designated as case or control depending on his/her cumulative genetic risk score plus a random environmental risk component chosen to achieve the estimated T2D heritability of ~45%. From this population simulated with both phenotypes and genotypes, we selected appropriate numbers of cases and controls and conducted single-variant association tests in order to compare the distribution of p-values from simulation to that observed in the current study. Results shown are the average of 25 independent simulation replicates for each disease model.
10.2.3. Comparison of simulated outcomes to empirical T2D results
We focused on comparing simulated outcomes under three disease models, each of which were previously found to be consistent with sibling relative risk, GWAS, and linkage results for T2D, but vary widely in causal variant properties (Fig. 3): a rare-variant model in which rare variants explain ~75% of T2D heritability (small target size T=750kb and moderate dependence between effect size and selection τ=0.5), an intermediate model in which rare, low-frequency, and common variants all contribute significantly to T2D heritability (T=2.0Mb and τ=0.3), and a common polygenic model in which common variants explain ~75% of T2D heritability (T=3.75Mb and weak dependence τ=0.1). We first compared the simulated outcomes of a whole-genome sequencing study in ~3K samples under each model. All three models predicted similar distributions of variant association test statistics using the sequenced individuals alone (data not shown).
However, the predictions began to diverge when we simulated imputation into GWAS samples and studied the distribution of test statistics after meta-analysis. For each simulated model, we sampled 14,175 cases and 14,175 controls (to match the effective sample size of the actual imputation cohorts used for meta-analysis). Because genotyping accuracy in simulated samples is perfect (unlike in imputation), we calculated average imputation quality as a function of MAC in the empirical data (using the r2 value reported by the imputation software that was used in each cohort). We then corrected, for each variant, the association test statistic in simulated data by multiplying the chi-squared value by the average imputation r2 for the variant MAC. We then re-computed association p-values from the corrected chi-squared statistics to compare p-value distributions in simulated versus empirical data. We plotted the distribution of association p-values for variants of different frequency classes in a quantile-quantile (QQ) plot, and compare these curves to the empirical T2D results (Fig. 3). Focusing on low-frequency variants, we also asked how many unique low-frequency signals achieved significant association to T2D risk under each simulated model, and compared these quantities to empirical observation (Fig. 3). These analyses demonstrate that the intermediate and rare-variant models produce an excess of association signal among low-frequency variants compared to observation, whereas the common polygenic model is consistent with the genome-wide distribution of association signals observed.
Extended Data
Extended Data Figure 1. Summary of samples and quality control procedures.
This figure summarises data generation for whole genome sequencing (GoT2D), exome sequencing (GoT2D and T2D-GENES) and exome array genotyping (DIAGRAM). In addition, GoT2D whole genome sequence data was imputed into GWAS data from 44,414 subjects of European descent.
Extended Data Figure 4. Power for single and aggregate variant association.
a-g. Power to detect single-variant association (α=5×10−8) at varying minor allele frequency (x-axis) and allelic odds-ratio (y-axis) for seven effective sample size (Neff) scenarios relevant to the genomes (a-c) and exomes (dg) component of this project. a. variant observed in 2,657 samples (the effective size of the GoT2D integrated panel); b. variant observed in 28,350 samples (the effective size of the imputed data set); c. variant observed in the GoT2D integrated panel and the imputed data set (effective sample size 31,007); d. ancestry-specific variant in 2,000 samples (the size of each of the non-European exome sequence data sets); e. European specific variant in 5,000 samples (the combined size of the European exome sequence data sets); f. variant observed with shared frequency across all ancestry groups in 12,940 samples (the size of the combined exome sequence data set); and g. variant observed in the combined exome array and sequencing data set (effective sample size 82,758). h-i. Power for gene based test of association (SKAT-O) according to liability variance explained. In h, 50% of the variants contribute to disease risk while the remaining 50% have no effect on disease risk; in i., 100% of the variants contribute to disease risk. For each, sample sizes considered are 2,000 (ancestry-specific effects; green) and 12,940 (ancestry-shared effects; blue). Power is shown for two levels of significance (α=2.5×10−6 and α=0.001). From these simulation studies, it is clear that under the optimistic model, where effects are shared across all ethnicities (blue line) and all variants contribute, power is >60% for 1% variance explained and α=2.5×10−6. However, power declines rapidly if either criterion is relaxed.
Extended Data Figure 6. Single variant analyses.
Manhattan plot of single-variant analyses generated from a. exome sequence data in 6,504 cases and 6,436 controls of African American, East Asian, European, Hispanic, and South Asian ancestry; b. exome array genotypes in 28,305 cases and 51,549 controls of European ancestry; and c. combined meta-analysis of exome array and exome sequence samples. Coding variants are categorized according to their relationships to the previously reported lead variant from GWAS region. Loci achieving genome-wide significance only in the combined analysis are highlighted in bold. The HNF1A variant reaching genome-wide significance in the combined analysis is a synonymous variant (Thr515Thr). The dashed horizontal line in each panel designates the threshold for genome-wide significance (p<5×10−8).
Extended Data Figure 7. Classification of coding variants according to their relationship to reported lead variants for each GWAS region.
The ideogram shows the location of 25 coding variant associations at 16 loci described in the text. The number in each circle corresponds to the number of associated variants at each locus. Variants are grouped into five categories based on inferred relationship with the GWAS lead variant. For some of these categories, the figure includes representative regional association plots based on exome array meta-analysis data from 28,305 cases and 51,549 controls. The locus displayed for each category is designated in bold. The first plot in each panel shows the unconditional association results; middle plot the association results after conditioning on the non-coding GWAS SNP; and the last plot the results after conditioning on the most significantly associated coding variant. Each point represents a SNP in the exome array meta-analysis, plotted with their p-value (on a –log10 scale) as a function of the genomic position (hg19). In each panel, the lead coding variant is represented by the purple symbol. The color-coding of all other SNPs indicates LD with the lead SNP (estimated by European r2 from 1000 Genomes March 2012 reference panel: red r2≥0.8; gold 0.6≤r2<0.8; green 0.4≤r2<0.6; cyan 0.2≤r2<0.4; blue r2<0.2; grey r2unknown). Gene annotations are taken from the University of California Santa Cruz genome browser. GWS: genome-wide significance. *Seven variants, three at ASCC2, and one each at THADA, TSPAN8, FES and HNF4A did not achieve genome-wide significance themselves, but are included because they fall into genes and/or regions with other significant association signals (see text).
Extended Data Figure 9. Exclusion of synthetic associations and construction of credible causal variant sets at T2D GWAS loci.
Ten T2D GWAS loci were selected for synthetic association testing (p<0.001; Methods). a, The effect size observed at the GWAS index SNV (sequence data) before (navy blue) and after (light blue, grey) conditioning on candidate rare and low-frequency (MAF<5%) variants which could produce synthetic association. b, Example of synthetic association exclusion at the TCF7L2 locus. c, Credible sets for T2D GWAS loci where credible set consisted of <80 variants displaying the proportion of credible set variants present in the HapMap and 1000G catalogs.
Extended Data Figure 10.
Genome enrichment analysis in GoT2D whole genome sequence data (n=2,657) a, Functional annotation categories were defined using transcription, chromatin state and transcription factor binding data from GENCODE, ENCODE and other studies. b, T2D association statistics for variants at each T2D locus were jointly modelled with functional annotation using fgwas. In the resulting model we identified enrichment of coding exons (CDS), transcription factor binding sites (TFBS), mature adipose active enhancers and promoters (hASC-t4 EnhA, TssA), pancreatic islet active and weak enhancers (HI EnhA, EnhWk), pre-adipose active and weak enhancers (hASC-t1 EnhA, EnhWk), embryonic stem cell active promoters (H1-hESC TssA) and 5’ UTR. Dots represent enrichment estimates and horizontal lines the 95% confidence intervals. c, At the CCND2 locus, three variants not present in HapMap2 have a combined 90% posterior probability of being causal (rs4238013, rs3217801, rs73040004). One of these variants, rs3217801, is a 2-bp indel that overlaps an islet enhancer element.
Extended Data Figure 11. Low frequency variants in exome array data.
Results from meta-analysis of 43,045 low-frequency and common coding variants on the exome array (assayed in 79,854 European subjects). a. Observed allelic ORs as a property of allele MAF. Variants missing in >8 cohorts or polymorphic in only one cohort were excluded. Colored lines represent contours for liability variance explained. Regions shaded grey denote ranges of OR and MAF consistent with 80% power (in this case, at α=5×10−7) to detect single-variant associations in this data set (given the observed range of missing data). Variants with a black collar are those highlighted by a bounding analysis as having a probability>0.8 of having LVE>0.1%; b. Distribution of each variant in the MAF/OR space was computed by assuming T2D prevalence of 8% and a beta and normal distribution for MAF and OR respectively. Probability is obtained by integrating the joint MAF-OR distributions over ranges of LVE; c. Single variant association, liability and bounding results for the known T2D GWAS variants on the exome array (Methods).
Extended Data Table 2.
Summary information for samples sets used in the association analyses.
| Ancestry | Study | Countries of Origin | Num. of Cases (% female) | Num. of Controls (% female) | Effective Sample Size |
|---|---|---|---|---|---|
|
Whole Genome Sequencing Studies
| |||||
| European | Finland-United States Investigation of NIDDM Genetics (FUSION) Study | Finland | 493 (41.5) | 486 (45.2) | 979 |
| European | Kooperative Gesundheitsforschung in der Region Augsburg (KORA) | Germany | 101 (44.5) | 104 (66.3) | 205 |
| European | Malmo-Botnia Study | Finland, Sweden | 410 (51.5) | 419 (44.1) | 829 |
| European | UK Type 2 Diabetes Genetics Consortium (UKT2D) | UK | 322 (46.2) | 322 (82.2) | 644 |
| Total Whole Genome Sequence | 1,326 | 1,331 | 2,657 | ||
|
Genome-Wide Array Studies | |||||
| European | INTERACT | France, Germany, Italy, Netherlands, Spain, Sweden, UK | 4624 (51.8) | 4668 (64.2) | 9292 |
| European | Wellcome Trust Case Control Consortium (WTCCC) | UK | 1586 (40.9) | 2938 (50.8) | 4120 |
| European | Kooperative Gesundheitsforschung in der Region Augsburg (KORA) | Germany | 993 (45.1) | 2985 (52.2) | 2980 |
| European | Framingham Heart Study (FHS) | US | 673 (42.6) | 7660 (55.1) | 2475 |
| European | Finland-United States Investigation of NIDDM Genetics (FUSION) Study | Finland | 1060 (43.1) | 1090 (51.3) | 2150 |
| European | Diabetes Genetics Initiative (DGI) | Finland, Sweden | 899 (46.6) | 1057 (49.6) | 1943 |
| European | Estonian Genome Center, University of Tartu (EGCUT-OMNI) | Estonia | 389 (58.6) | 6013 (54.2) | 1461 |
| European | Diabetes Gene Discovery Group (DGDG) | France, Canada | 677 (39.3) | 697 (59.7) | 1374 |
| European | Mt Sinai BioMe Biobank Platform (BioMe (Illumina)) | US | 255 (29.0) | 1647 (51.4) | 883 |
| European | Uppsala Longitudinal Study of Adult Men (ULSAM) | Sweden | 166 (0) | 953 (0) | 565 |
| European | Mt Sinai BioMe Biobank Platform (BioMe) | US | 132 (26.5) | 455 (34.7) | 409 |
| European | Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) | Sweden | 111 (41.4) | 838 (51.2) | 392 |
| European | Estonian Genome Center, University of Tartu (EGCUT-370) | Estonia | 80 (48.8) | 1768 (51) | 306 |
| Total Genome-Wide Array | 11,645 | 32,769 | 28,350 | ||
| Total Whole Genome Sequence + Genome-Wide Array | 12,971 | 34,100 | 31,007 | ||
|
Whole Exome Sequencing Studies | |||||
| African American | Jackson Heart Study | US | 500 (66.6) | 526 (63.3) | 1,026 |
| African American | Wake Forest School of Medicine Study | US | 518 (59.5) | 530 (56.0) | 1,048 |
| East Asian | Korea Association Research Project | Korea | 526 (45.6) | 561 (58.5) | 1,086 |
| East Asian | Singapore Diabetes Cohort Study; Singapore Prospective Study Program | Singapore (Chinese) | 486 (52.1) | 592 (61.3) | 1,068 |
| European | Ashkenazi | US, Israel | 506 (47.0) | 355 (56.9) | 834 |
| European | Metabolic Syndrome in Men Study (METSIM) | Finland | 484 (0) | 498 (0) | 982 |
| European | Finland-United States Investigation of NIDDM Genetics (FUSION) Study | Finland | 472 (42.6) | 476 (45.0) | 948 |
| European | Kooperative Gesundheitsforschung in der Region Augsburg (KORA) | Germany | 97 (44.3) | 90 (63.3) | 186 |
| European | UK Type 2 Diabetes Genetics Consortium (UKT2D) | UK | 322 (45.7) | 320 (82.8) | 642 |
| European | Malmo-Botnia Study | Finland, Sweden | 478 (54.8) | 443 (43.8) | 920 |
| Hispanic | San Antonio Family Heart Study, San Antonio Family Diabetes/Gallbladder Study, Veterans Administration Genetic Epidemiology Study, and the Investigation of Nephropathy and Diabetes Study Family Component | US | 272 (58.8) | 218 (58.7) | 484 |
| Hispanic | Starr County, Texas | US | 749 (59.7) | 704 (71.9) | 1,452 |
| South Asian | London Life Sciences Population Study (LOLIPOP) | UK (Indian Asian) | 531 (14.1) | 538 (15.8) | 1,068 |
| South Asian | Singapore Indian Eye Study | Singapore (Indian Asian) | 563 (44.4) | 585 (49.2) | 1,148 |
| Total Whole Exome Sequence | 6,504 | 6,436 | 12,892 | ||
|
Exome Array Studies | |||||
| European | ADDITION; Steno Diabetes Centre (SDC); Health06; Health08; Vejle Biobank; Inter99 | Denmark | 5813 (40.0) | 7987 (54.4) | 13,458 |
| European | Wellcome Trust Case Control Consortium (UK Type 2 Diabetes Consortium); Young Diabetics Study (YDX); Genetics of Diabetes and Audit Research Tayside Study (GoDARTS); Oxford Biobank; TwinsUK; 1958 Birth Cohort (BC58) | UK | 3576 (51.7) | 12675 (41.2) | 11,156 |
| European | Finland-United States Investigation of NIDDM Genetics (FUSION) Study; Finrisk2007; Metabolic Syndrome in Men Study (METSIM); Dose-Responses to Exercise Training (DR'sEXTRA); D2D2007 | Finland | 3593 (33.4) | 8222 (26.0) | 10,001 |
| European | Malmo Diabetes Cohort (MDC); All New Diabetics in Skane (ANDIS) | Sweden | 4633(41.0) | 5404 (59.5) | 9,978 |
| European | Prevalence, Prediction and Prevention of Diabetes (PPP); Diabetes Register in Vaasa (DIREVA) | Finland | 2910 (43.7) | 4596 (53.7) | 7,127 |
| European | Nurses’ Health Study (NHS) | US | 1413 (100.0) | 1695 (100.0) | 3,082 |
| European | Health Professionals Follow-up Study (HPFS) | US | 1184 (0.0) | 1287 (0.0) | 2,467 |
| European | The Exeter Family Study of Child Health (EFSOCH) | UK | 1446 (39.0) | 1567 (52.0) | 3,008 |
| European | Kooperative Gesundheitsforschung in der Region Augsburg (KORA) | Germany | 933 (45.3) | 2705 (51.7) | 2,775 |
| European | Estonian Genome Center at the University of Tartu (EGCUT) | Estonia | 882 (43.7) | 1506 (44.2) | 2,225 |
| European | Gene-Lifestyle Interactions and Complex Traits Involved in Elevated Disease Risk (GLACIER) | Sweden | 960 (47.6) | 957 (54.5) | 1,917 |
| European | Fenland cohort of the European Prospective Investigation of Cancer (Fen-EPIC) | UK | 691(47.0) | 1157 (54.5) | 1,730 |
| European | The Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS); Uppsala Longitudinal Study of Adult Men (ULSAM) | Sweden | 271(16.9) | 1791 (23.9) | 942 |
| Total Exome Array | 28,305 | 51,549 | 69,866 | ||
| Total Whole Exome Sequence + Exome Array | 34,809 | 57,985 | 82,758 | ||
Extended Data Table 3. Counts and properties of variants identified in sequenced subjects.
a. Variant numbers for the 2,657 individuals with whole genome sequence data passing QC and included in the association analysis data set; b. Variant numbers are provided for the 13,008 individuals passing initial rounds of QC from which further QC defined the 12,940 subjects included in the association analysis data set. Private refers to variants seen in only a single ancestral group; cosmopolitan to variants seen in all five major ancestral groups.
| a | |||
|---|---|---|---|
| Genomes integrated panel | |||
| SNV | Indel | SV | |
|
Variant
Type N (%total) |
25.2M (94%) | 1.50M (5.6%) | 8,876 (0.03%) |
| Coding | Non-coding | ||
|
Function N (%total) |
888K (3.3%) | 25.8M (97%) | |
| Rare (MAF<0.5%) | Low frequency (0.5<MAF<5%) | Common (MAF>5%) | |
|
Frequency
spectrum N (%total) |
6.26M (23%) | 4.16M (16%) | 16.3M (61%) |
| b137 | Novel | ||
|
dbSNV N (%total) |
14.6M (55%) | 12.1M (45%) | |
| b | ||||||
|---|---|---|---|---|---|---|
| Exome sequence data | ||||||
| All samples | African-American | East-Asian | European | Hispanic | South-Asian | |
| Samples: | 13,008 | 2,086 | 2,165 | 4,579 | 1,959 | 2,219 |
| T2D cases | 6,504 | 1,018 | 1,012 | 2,359 | 1,021 | 1,094 |
| T2D controls | 6,436 | 1,056 | 1,153 | 2,182 | 922 | 1,123 |
| Excluded from association analysis | 68 | 12 | 0 | 38 | 16 | 2 |
| Coverage: | ||||||
| Coding: | ||||||
| Mean (Mc) per gene | 81.7 ±23.7 | 83.2 ±24.0 | 84.6 ±23.8 | 78.6 ±23.3 | 83.8 ±24.1 | 78.2 ±23.2 |
| # of genes with Mc <20 | 368 | 302 | 302 | 351 | 269 | 325 |
| Non-coding: | ||||||
| Mean per gene | 59.0 ±21.0 | 60.9 ±21.5 | 62.2 ±21.6 | 57.5 ±20.6 | 59.2 ±21.2 | 55.4 ±20.3 |
| # of genes with Mc <20 | 1,150 | 738 | 731 | 1,102 | 804 | 945 |
| Variant annotations: | ||||||
| Synonymous SNV | 627,630 | 237,430 | 178,232 | 192,282 | 156,231 | 211,218 |
| Missense SNV | 1,110,897 | 354,797 | 296,707 | 327,049 | 231,351 | 344,191 |
| Start SNV | 2,055 | 593 | 523 | 639 | 384 | 583 |
| Nonsense SNV | 26,321 | 7,188 | 6,668 | 8,030 | 4,660 | 7,339 |
| Frameshift INDEL | 26,901 | 6,605 | 6,159 | 7,515 | 4,155 | 6,609 |
| Inframe INDEL | 11,090 | 3,471 | 2,963 | 3,145 | 2,068 | 3,165 |
| 3′UTR SNV, INDEL | 65,013 | 24,583 | 19,149 | 21,102 | 16,959 | 22,177 |
| 5′UTR SNV, INDEL | 43,965 | 16,920 | 13,520 | 15,562 | 11,634 | 15,595 |
| Intron SNV, INDEL | 931,449 | 352,398 | 270,564 | 296,970 | 243,139 | 314,810 |
| Essential splicing SNV, INDEL | 14,286 | 3,648 | 3,454 | 4,108 | 2,301 | 3,744 |
| Other splicing SNV, INDEL | 128,644 | 45,876 | 35,413 | 38,263 | 30,301 | 41,122 |
| Non-coding RNA SNV, INDEL | 18,113 | 7,247 | 5,996 | 6,715 | 5,084 | 6,706 |
| Intergenic SNV, INDEL | 37,345 | 14,335 | 11,498 | 13,614 | 10,700 | 12,937 |
| All | 3,043,709 | 1,075,091 | 850,846 | 934,994 | 718,967 | 990,196 |
| Coding frequency spectrum: | ||||||
| Rare (MAF<0.5%) | 95.79% | 83.30% | 90.06% | 89.19% | 84.56% | 89.89% |
| private | 77.93% | 53.79% | 65.47% | 51.80% | 37.26% | 61.55% |
| cosmopolitan | 0.35% | 1.80% | 3.02% | 1.88% | 2.24% | 1.73% |
| Low frequency (0.5<MAF<5%) | 2.57% | 10.36% | 4.61% | 5.52% | 8.21% | 5.10% |
| private | 0.17% | 1.43% | 1.10% | 0.26% | 0.52% | 1.02% |
| cosmopolitan | 0.60% | 1.50% | 1.54% | 1.94% | 2.74% | 1.62% |
| Common (MAF>5%) | 1.65% | 6.35% | 5.33% | 5.29% | 7.23% | 5.00% |
| private | 0.09% | 0.00% | 0.00% | 0.00% | 0.01% | 0.00% |
| cosmopolitan | 1.50% | 4.35% | 5.17% | 4.97% | 6.88% | 4.86% |
| Intron/UTR frequency spectrum: | ||||||
| Rare (MAF<0.5%) | 94.09% | 78.68% | 86.91% | 86.17% | 81.43% | 86.68% |
| private | 74.76% | 49.81% | 61.36% | 45.26% | 31.03% | 56.96% |
| cosmopolitan | 0.46% | 2.07% | 3.98% | 2.49% | 2.66% | 2.19% |
| Low frequency (0.5<MAF<5%) | 3.52% | 12.57% | 5.63% | 6.51% | 9.43% | 6.32% |
| private | 0.25% | 1.74% | 1.25% | 0.29% | 0.47% | 1.18% |
| cosmopolitan | 0.80% | 1.81% | 2.11% | 2.53% | 3.30% | 2.17% |
| Common (MAF>5%) | 2.39% | 8.76% | 7.46% | 7.32% | 9.14% | 7.00% |
| private | 0.15% | 0.00% | 0.00% | 0.01% | 0.00% | 0.00% |
| cosmopolitan | 2.17% | 5.94% | 7.26% | 6.93% | 8.77% | 6.81% |
Extended Data Table 5. Characterization of variant associations through conditional analysis.
For each locus, significantly associated SNVs are presented. Unconditional p-values are given in italics, and conditional p-values are shown for each pair of SNVs (p-values are for SNVs in the “Variant” column, with SNVs listed in header included as covariates in association analysis). The IRS1 and PPARG non-coding associations were characterized using exact conditional analysis in 38,738 samples from the GoT2D genome-wide imputed meta-analysis. Conditional analysis for coding variant associations was, for most loci, restricted to the exome array genotypes (28,305 cases, 51,549 controls). At THADA and RREB1, neither the non-coding lead GWAS SNVs nor close proxies were typed on the exome array, so approximate conditional analyses were undertaken using GCTA in 44,414 samples from the GoT2D genome-wide imputed meta-analysis (Methods). For several of these loci, unconditional association p-values for these loci do not reach genome-wide significance as sample sizes are smaller. At the GPSM1 locus, the previously reported GWAS SNV was not available on exome array and too poorly imputed in the GoT2D meta-analysis to allow meaningful inference
| Locus | Variant | MAF | Unconditional and conditional association p-values | Interpretation | |||
|---|---|---|---|---|---|---|---|
| Non-coding variant associations characterized in 38,738 samples in GoT2D genome wide imputed meta analysis | |||||||
| IRS1 | rs78124264 | rs7578326 | rs2943640 | rs2943641 | The association signal rs78124264 and the GWAS SNPs at this locus are distinct. Signals are not extinguished in reciprocal conditional analysis. Previous GWAS signals are not mediated through rs78124264, which represents a distinct association signal at this locus. | ||
| rs78124264 | 0.022 | 8.5×10−5 | 2.5×10−7* | 2.5×10−7* | 2.5×10−7* | ||
| rs7578326¶ | 0.35 | 1.2×10−7 | 1.1×10−5 | n.d. | n.d. | ||
| rs2943640¶ | 0.35 | 2.5×10−11 | n.d. | 4.5×10−10 | n.d. | ||
| rs2943641¶ | 0.36 | 9.0×10−12 | n.d. | n.d. | 1.5×10−10 | ||
| PPARG | rs79856023 | rs1801282 | The association signal rs79856023 and the GWAS SNP at this locus are distinct. Signals are not extinguished in reciprocal conditional analysis. Previous GWAS signal is not mediated through rs79856023, which represents a distinct association signal at this locus. | ||||
| rs79856023 | 0.022 | 1.2×10−4 | 9.2×10−7 | ||||
| rs1801282¶ | 0.13 | 1.6×10−6 | 1.2×10−5 | ||||
| Coding variant associations characterized in 28,305 cases and 51,549 controls typed on exome array | |||||||
| PAM | Asp563Gly (PAM) | Ser1207Gly (PPIP5K2) | Association signals for PAM Asp563Gly and PPIP5K2 Ser1207Gly are indistinguishable in reciprocal conditional analysis. Gene biology, as well as previous reports of additional PAM variants associated with T2D in Icelandic cohorts, highlights PAM as the probable transcript at this locus. | ||||
| Asp563Gly | 0.054 | 1.7×10−7 | 0.24 | ||||
| Ser1207Gly | 0.054 | 0.30 | 1.0×10−6 | ||||
| MTMR3-ASCC2 | Asn960Ser (MTMR3) | Val123Ile (ASCC2) | Asp407His (ASCC2) | Pro423Ser (ASCC2) | Association signals for the MTMR3 and ASCC2 coding variants are indistinguishable in reciprocal conditional analysis. The MTMR3 Asn960Ser variant has the strongest signal, and highlights MTMR3 as the most likely effector transcript at this locus. | ||
| Asn960Ser | 0.083 | 3.2×10−6 | 0.022 | 0.027 | 0.022 | ||
| Val123Ile | 0.083 | 0.15 | 2.0×10−5 | 0.066 | 0.76 | ||
| Asp407His | 0.083 | 0.18 | 0.99 | 1.9×10−5 | 0.88 | ||
| Pro423Ser | 0.083 | 0.18 | 0.67 | 0.98 | 2.0×10−5 | ||
| KCNJ11-ABCC8 | Val337Ile (KCNJ11) | Lys23Glu (KCNJ11) | Ala1369Ser (ABCC8) | Association signals for KCNJ11 Val337Ile and Lys23Glu and ABCC8 Ala1369Ser are indistinguishable in reciprocal conditional analysis. The relative causal contributions of the two genes, making up the two components of the sulfonlylurea-responsive potassium channel, are indistinguishable on statistical grounds. | |||
| Val337Ile | 0.40 | 3.4×10−9 | 0.17 | 0.049 | |||
| Lys23Glu | 0.40 | 0.48 | 5.1×10−9 | 0.082 | |||
| Ala1369Ser | 0.40 | 0.68 | 0.84 | 2.3×10−8 | |||
| WFS1 | Val333Ile | Asn500Asn | Arg611His | rs4689388 | Association signals for the WFS1 coding variants are indistinguishable from each other and the previously reported non-coding GWAS SNP at this locus in reciprocal conditional analysis. WFS1 is the likely effector transcript for the non-coding GWAS signal at this locus, although the causal variant in the gene is unclear. | ||
| Val333Ile | 0.30 | 9.3×10−12 | 0.024 | 0.00070 | 0.0030 | ||
| Asn500Asn | 0.41 | 0.0070 | 2.0×10−12 | 0.0049 | 0.027 | ||
| Arg611His | 0.47 | 0.020 | 0.62 | 1.3×10−10 | 0.19 | ||
| rs4689388¶ | 0.43 | 0.011 | 0.62 | 0.024 | 2.3×10−11 | ||
| CILP2-TM6SF2 | Glu167Lys (TM6SF2) | rs10401969 | Association signals for TM6SF2 Glu167Lys and the previously reported non-coding GWAS SNP at this locus are indistinguishable from each other in reciprocal conditional analysis. TM6SF2 is the probable effector transcript for the non-coding GWAS signal at this locus, with the effect mediated through Glu167Lys. | ||||
| Glu167Lys | 0.082 | 1.9×10−7 | 0.52 | ||||
| rs10401969¶ | 0.083 | 0.62 | 4.2×10−7 | ||||
| GRB14-COBLL1 | Asn939Asp (COBLL1) | rs13389219 | Association signals for COBLL1 Asn939Asp and the previously reported non-coding GWAS SNP at this locus are partially correlated. The association signal for the GWAS signal is not entirely extinguished in reciprocal conditional analysis. COBLL1 is a candidate effector transcript for the GWAS signal at this locus. | ||||
| Asn939Asp | 0.12 | 4.7×10−11 | 3.00×10−5 | ||||
| rs13389219¶ | 0.39 | 7.0×10−5 | 1.9×10−10 | ||||
| Coding variant associations characterized in 44,414 samples in GoT2D genome wide imputed meta analysis | |||||||
| THADA | Cys1605Tyr | rs10203174 | Association signals THADA Cys1605Tyr and the GWAS SNP are partially correlated. The association signal for the GWAS SNP is not entirely extinguished in reciprocal conditional analysis. THADA is a candidate effector transcript for the GWAS signal at this locus. | ||||
| Cys1605Tyr | 0.10 | 0.00035 | 0.92 | ||||
| rs10203174¶ | 0.10 | 0.0063 | 5.7×10−6 | ||||
| RREB1 | Asp1171Asn | rs9502570 | The association signals of RREB1 Asp1171Asn and the GWAS SNP at this locus are distinct. The association signal is not extinguished in reciprocal conditional analysis. Previous GWAS signal is not mediated through RREB1 Asp1171Asn. RREB1 Asp1171Asn represents a distinct association signal at this locus. | ||||
| Asp1171Asn | 0.11 | 0.0018 | 0.0017 | ||||
| rs9502570¶ | 0.28 | 0.0037 | 0.0042 | ||||
Conditional analysis was performed once for rs78124264 with all three previously known GWAS variants included as covariates.
Non-coding GWAS lead variant.
n.d. indicates “not determined.”
Extended Data Table 8. Testing for synthetic associations across GWAS-identified T2D loci.
Gene names refer to protein-coding transcript(s) closest to the index SNV. Reported index SNVs are the previously-reported GWAS variants (in European populations) with the strongest association signal in the GoT2D sequencing data (n=2,657). Relative likelihoods are based on causal models with only the chosen low-frequency and rare missense variants, relative to models with only the GWAS index SNV, assessed using the Akaike Information content (AIC) of each regression model, calculated as exp[(AICindex–AIClow-frequency or rare)/2]. n1 provides the number of low-frequency or rare variants required for the residual odds ratio at the GWAS index SNV, after joint conditioning on the low-frequency and rare variants, to switch direction of effect. n2 provides the number of low-frequency or rare variants required for the association p-value remaining at the GWAS index SNV, after joint conditioning on the low-frequency and rare variants, to exceed 0.05.
| Index SNV association signal |
Synthetic association by
missense variants |
Synthetic association by all
low- frequency and rare variants across 5Mb region |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index SNV association signal before inclusion of missense variants |
Index SNV association signal after inclusion of missense variants |
Index SNV association after inclusion of single best variant |
Testing groups of low- frequency and rare variants |
|||||||||||
| Gene | Index SNV | MAF | OR [95% interval] |
p-value | Number Missense Variants |
OR [95% interval] |
p-value | Relative likelihood of LF model |
Best LF Variant |
MAF | OR [95% interval] |
p-value | n1 | n2 |
| TCF7L2 | 10:114758349 | 0.27 | 1.75 [1.54-1.99] | 2.80×10−18 | 6 | 1.73 [1.52-1.97] | 2.33×10−17 | 1.8×10−17 | 10:114787948 | 1.6% | 1.72 [1.51-1.95] | 1.62×10−16 | >50 | 35 |
| ADCY5 | 3:123065778 | 0.19 | 0.69 [0.60-0.79] | 1.12×10−7 | 13 | 0.70 [0.61-0.81] | 9.00×10−7 | 9.7×10−8 | 3:123096056 | 2.5% | 0.71 [0.61-0.82] | 3.04×10−6 | 13 | 6 |
| IRS1 | 2:227093745 | 0.36 | 0.76 [0.68-0.86] | 2.80×10−6 | 5 | 0.77 [0.69-0.86] | 4.30×10−6 | 4.5×10−7 | 2:226993370 | 1.7% | 0.78 [0.70-0.88] | 2.19×10−5 | 12 | 6 |
| KCNQ1 | 11:2847069 | 0.45 | 0.78 [0.70-0.87] | 1.22×10−5 | >50 | 0.84 [0.75-0.94] | 2.07×10−3 | 1.0×10−7 | 11:2825279 | 4.7% | 0.81 [0.71-0.91] | 3.19×10−4 | 16 | 6 |
| CDC 123-CAMK1D | 10:12307894 | 0.25 | 1.33 [1.17-1.52] | 1.19×10−5 | 4 | 1.30 [1.13-1.50] | 2.06×10−4 | 7.1×10−5 | 10:12325477 | 3.8% | 1.29 [1.12-1.48] | 3.03×10−4 | 10 | 5 |
| CDKN2A-CDKN2B | 9:22137685 | 0.28 | 1.28 [1.14-1.45] | 4.52×10−5 | 4 | 1.27 [1.13-1.43] | 9.28×10−5 | 4.3×10−5 | 9:22133773 | 3.5% | 1.25 [1.10-1.41] | 5.98×10−4 | 22 | 7 |
| IGF2BP2 | 3:185511687 | 0.32 | 1.25 [1.11-1.41] | 1.65×10−4 | 14 | 1.21 [1.07-1.36] | 2.12×10−3 | 3.0×10−4 | 3:185550500 | 4.1% | 1.20 [1.07-1.36] | 2.91×10−3 | 8 | 3 |
| KLHDC5 | 12:27965150 | 0.17 | 0.76 [0.66-0.88] | 2.19×10−4 | 3 | 0.77 [0.66-0.89] | 4.45×10−4 | 1.2×10−3 | 12:27832062 | 2.0% | 0.80 [0.68-0.92] | 3.04×10−3 | 10 | 4 |
| SLC30A8 | 8:118184783 | 0.33 | 0.81 [0.72-0.91] | 2.95×10−4 | 2 | 0.81 [0.72-0.91] | 3.73×10−4 | 0.02 | 8:117964024 | 2.2% | 0.83 [0.73-0.93] | 1.23×10−3 | 17 | 6 |
| CDKAL1 | 6:20694884 | 0.18 | 1.28 [1.11-1.48] | 6.05×10−4 | 1 | 1.28 [1.11-1.48] | 7.57×10−4 | 0.007 | 6:20718780 | 2.8% | 1.23 [1.06-1.43] | 7.71×10−3 | 9 | 3 |
Supplementary Material
ACKNOWLEDGEMENTS
Grant support and acknowledgments are provided in supplementary information.
Footnotes
Current addresses (where relevant) are provided in supplementary information.
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
AUTHOR CONTRIBUTIONS
Author contributions are provided in supplementary information.
AUTHOR INFORMATION
Data deposition: Whole genome sequence data from the GoT2D project are available by application to the European Genotype Archive (EGAS00001001459) and from dbGAP (phs000840.v1.p1). Whole exome sequence data from the T2D-GENES project are available from the European Genotype Archive (EGAS00001001460) and from dbGAP (phs000847.v1.p1, phs001093.v1.p1, phs001095.v1.p1, phs001096.v1.p1, phs001097.v1.p1, phs001098.v1.p1, phs001099.v1.p1, phs001100.v1.p1, phs001102.v1.p1). Summary level data from the exome array component of this project (and from the exome and genome sequence) can be freely accessed at the Accelerating Medicines Partnership T2D portal (www.type2diabetesgenetics.org), and similar data from the GoT2D-imputed data at www.diagram-consortium.org.
COMPETING FINANCIAL INTERESTS
Ralph A DeFronzo has been a member of advisory boards for Astra Zeneca, Novo Nordisk, Janssen, Lexicon, Boehringer-Ingelheim, received research support from Bristol Myers Squibb, Boehringer- Ingelheim, Takeda and Astra Zeneca, and is a member of speaker's bureaus for Novo-Nordisk and Astra Zeneca.
Jose C Florez has received consulting honoraria from Pfizer and PanGenX.
Mark McCarthy has received consulting and advisory board honoraria from Pfizer, Lilly, and NovoNordisk.
Gilean McVean and Peter Donnelly are co-founders of Genomics PLC, which provides genome analytics.
REFERENCES
- 1.Willemsen G, et al. The concordance and heritability of type 2 diabetes in 34,166 Twin Pairs From International Twin Registers: The Discordant Twin (DISCOTWIN) Consortium. Twin Res Hum Genet. 2015;18:762–71. doi: 10.1017/thg.2015.83. [DOI] [PubMed] [Google Scholar]
- 2.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahajan A, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kooner JS, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–9. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YS, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinthorsdottir V, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–8. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 8.Ma RC, et al. Genome-wide association study in a Chinese population identifies a susceptibility locus for type 2 diabetes at 7q32 near PAX4. Diabetologia. 2013;56:1291–305. doi: 10.1007/s00125-013-2874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huyghe JR, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–25. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmueller KE, et al. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet. 2013;93:1072–86. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albrechtsen A, et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56:298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claussnitzer M, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–58. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, Teslovich TM, Boehnke M, Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet. 2013;93:42–53. doi: 10.1016/j.ajhg.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collombat P, et al. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooptiwut S, et al. Defective PAX4 R192H transcriptional repressor activities associated with maturity onset diabetes of the young and early onset-age of type 2 diabetes. J Diabetes Complications. 2012;26:343–7. doi: 10.1016/j.jdiacomp.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 19.InterAct Consortium et al. Design and cohort description of the InterAct Project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC Study. Diabetologia. 2011;54:2272–82. doi: 10.1007/s00125-011-2182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oppelt A, et al. Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 2013;14:57–64. doi: 10.1038/embor.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–6. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahdessian H, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913–8. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiagalingam A, Lengauer C, Baylin SB, Nelkin BD. RREB1, a ras responsive element binding protein, maps to human chromosome 6p25. Genomics. 1997;45:630–2. doi: 10.1006/geno.1997.5001. [DOI] [PubMed] [Google Scholar]
- 24.Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4:200–13. doi: 10.1038/ncpendmet0778. [DOI] [PubMed] [Google Scholar]
- 25.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson CA, Soranzo N, Zeggini E, Barrett JC. Synthetic associations are unlikely to account for many common disease genome-wide association signals. PLoS Biol. 2011;9:e1000580. doi: 10.1371/journal.pbio.1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wray NR, Purcell SM, Visscher PM. Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biol. 2011;9:e1000579. doi: 10.1371/journal.pbio.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sim X, et al. Transferability of type 2 diabetes implicated loci in multi-ethnic cohorts from Southeast Asia. PLoS Genet. 2011;7:e1001363. doi: 10.1371/journal.pgen.1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein DB. The importance of synthetic associations will only be resolved empirically. PLoS Biol. 2011;9:e1001008. doi: 10.1371/journal.pbio.1001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen TS, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–69. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46:136–43. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaulton KJ, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–9. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–73. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet. 1965;29:51–76. [Google Scholar]
- 40.Agarwala V, Flannick J, Sunyaev S. GoT2D Consortium & Altshuler, D. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet. 2013;45:1418–27. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–7. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flannick J, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–63. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnefond A, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44:297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigma Type 2 Diabetes Consortium et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moltke I, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–3. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 47.Sigma Type 2 Diabetes Consortium et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–14. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majithia AR, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A. 2014;111:13127–32. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
EXTENDED METHODS REFERENCES
- 50.Guey LT, et al. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet Epidemiol. 2011;35:236–46. doi: 10.1002/gepi.20572. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–48. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–76. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–51. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–5. doi: 10.1016/j.ajhg.2008.06.005. author reply 135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weale ME. Quality control for genome-wide association studies. Methods Mol Biol. 2010;628:341–72. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 61.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 63.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Firth D. Bias reduction of maximum-likelihood-estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 65.Ma C, Blackwell T, Boehnke M, Scott LJ, GoT2D investigators Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37:539–50. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–22. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12:523–8. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Churchhouse C, Marchini J. Multiway admixture deconvolution using phased or unphased ancestral panels. Genet Epidemiol. 2013;37:1–12. doi: 10.1002/gepi.21692. [DOI] [PubMed] [Google Scholar]
- 70.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–75. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 73.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 74.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–60. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rice WR. A Consensus Combined P-Value Test and the Family-Wide Significance of Component Tests. Biometrics. 1990;46:303–308. [Google Scholar]
- 78.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. S1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–16. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 84.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–2. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lambert BW, Terwilliger JD, Weiss KM. ForSim: a tool for exploring the genetic architecture of complex traits with controlled truth. Bioinformatics. 2008;24:1821–2. doi: 10.1093/bioinformatics/btn317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eyre-Walker A. Evolution in health and medicine Sackler colloquium: Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1752–6. doi: 10.1073/pnas.0906182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lyssenko V, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–32. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.