Abstract
In regions of coendemicity for Plasmodium falciparum and Plasmodium vivax where mefloquine is used to treat P. falciparum infection, drug pressure mediated by increased copy numbers of the multidrug resistance 1 gene (pvmdr1) may select for mefloquine-resistant P. vivax. Surveillance is not undertaken routinely owing in part to methodological challenges in detection of gene amplification. Using genomic data on 88 P. vivax samples from western Thailand, we identified pvmdr1 amplification in 17 isolates, all exhibiting tandem copies of a 37.6–kilobase pair region with identical breakpoints. A novel breakpoint-specific polymerase chain reaction assay was designed to detect the amplification. The assay demonstrated high sensitivity, identifying amplifications in 13 additional, polyclonal infections. Application to 132 further samples identified the common breakpoint in all years tested (2003–2015), with a decline in prevalence after 2012 corresponding to local discontinuation of mefloquine regimens. Assessment of the structure of pvmdr1 amplification in other geographic regions will yield information about the population-specificity of the breakpoints and underlying amplification mechanisms.
Keywords: malaria, Plasmodium vivax, multidrug resistance, mdr1, copy number, mefloquine, Thailand
Outside of Africa, Plasmodium vivax has become the main cause of malaria-associated morbidity [1]. Previously considered benign, it is now widely acknowledged that this species is sometimes associated with severe and life-threatening disease, particularly in young children and pregnant women [2–8]. Concerted efforts are needed to contain the spread of emerging multidrug-resistant P. vivax parasites [9, 10].
Although mefloquine (MQ) is not generally used for the treatment of P. vivax infection, indirect drug pressure in regions of P. vivax and Plasmodium falciparum coendemicity may occur where it is used in artemisinin combination therapy (ACT) to treat P. falciparum malaria. The high frequency of mixed-species infections [11, 12] and the high risk of P. vivax infection recurrence following P. falciparum infection [13] likely impart significant selective pressure on P. vivax, particularly following administration of slowly eliminated drugs, such as MQ. The molecular basis of MQ resistance in P. falciparum has been evaluated comprehensively, with several clinical and in vitro studies implicating copy number (CN) amplification of the P. falciparum multidrug resistance 1 gene (pfmdr1) in resistant infection [14–18]. Investigation of the genetic architecture of pfmdr1 amplification in western Thailand highlighted the adaptive potential of P. falciparum parasites in response to MQ, demonstrating frequent amplification events; 5–15 independent amplification events were estimated among the isolates circulating in a single clinic [19]. Amplification of the orthologous pvmdr1 gene has been associated with a significant decrease in the ex vivo susceptibility of P. vivax to MQ [20]. However, in contrast to pfmdr1, the genetic structure of pvmdr1 amplification is poorly characterized. Our study sought to characterize the genetic architecture of pvmdr1 CN variants (CNVs) in P. vivax isolates from western Thailand.
MATERIALS AND METHODS
Ethics
Ethical approval for the study was provided by Mahidol University Faculty of Medical Technology Ethics Committee (MUTM 2011-043-03) and the Oxford Tropical Research Ethics Committee (OXTREC 45-10).
Sample Details
Samples were sourced from symptomatic patients with microscopy-confirmed monospecies P. vivax infection who were attending outpatient clinics in Mae Sod, western Thailand, between June 2003 and August 2015, and Timika, Papua Indonesia, between June 2004 and December 2006 (Supplementary Figure 1). MQ was not a recommended antimalarial or widely used in Papua Indonesia during the study period, and no pvmdr1 amplifications have been reported in the region previously [20, 21]; the Indonesian isolates therefore served as controls of MQ-unexposed P. vivax.
Whole-Genome Sequencing and Read Alignment
Leukocyte-depleted samples with ≥50 ng total DNA were subjected to whole-genome sequencing within the framework of a community study in the malaria Genomic Epidemiology Network [22]. Sequencing was undertaken on the Illumina GAII or Hi-Seq 2000 platform. Library preparation, cluster generation, and sequencing were undertaken as per the manufacturer's protocols for generating standard paired-end reads 75–150 base pairs long. Reads aligning to the human reference genome were removed before any analyses were undertaken. The remaining reads were aligned against the P. vivax SalI reference [23] (available at: http://plasmodb.org/common/downloads/release-10.0/PvivaxSal1/fasta/data/PlasmoDB-10.0_PvivaxSal1_Genome.fasta, Accessed 15 August 2016), using bwa [24], version 0.5.9-r16, with default parameters. Standard alignment metrics were generated for each sample using the bamcheck utility from samtools.
CNV Detection and Characterization, Using Genomic Data
CNVs spanning the pvmdr1 region (PVX_080100; Pv_Sal1_chr10:361,701–366 095) were detected using pysamstats (available at: http://github.com/alimanfoo/pysamstats, Accessed 15 August 2016). For each sample, coverage in nonoverlapping 300–base pair bins was calculated and normalized by dividing by the median coverage across all bins with the same integer percentage GC content. CNVs were called using a hidden Markov model with the python package sklearn.hmm.GaussianHMM. All CNVs >3 kilobase pairs long encompassing pvmdr1 were recorded. Although the primary definition of genomic CN amplification was based on read depth at correctly mapping reads, assessment was also undertaken using face-away mapping reads [25].
Multilocus Genotype (MLG) Analysis
MLG analysis was conducted using Illumina genomic data on SNPs derived from a set of 291 clinical P. vivax isolates [21]. ENA accession codes for all samples are available at: https://www.malariagen.net/resource/17 (Accessed 15 August 2016). Prior to analysis, samples with a genome-wide within-isolate fixation index (FWS) score of <0.95, indicative of polyclonal infection, were excluded [26, 27]. MLG analysis in the regions flanking the pvmdr1 amplification was conducted using previously described genotype-calling definitions, whereby positions with <5 reads were assigned a missing genotype call, a minimum of 2 reference and 2 alternative alleles were required for heterozygote assignment, and all other calls were defined as homozygous for the major allele [27]. MLG analysis within the amplified region was conducted using allele frequencies derived from read counts. The relatedness between infections was illustrated for monoclonal infections with a single copy of pvmdr1 (CN1) or multiple copies of pvmdr1 (CN2+), using heat map plots constructed with the R heatmap.2 package.
Single-Nucleotide Polymorphism (SNP) Genotype Calling at pvmdr1 Y976F
The Y976F variant (Pv_Sal1_chr10:363169) did not pass the stringent criteria for inclusion in the 303K genome-wide SNP set but was included for analysis here. Genotype calls were defined as described above.
Pvmdr1 Amplification Breakpoint–Specific Polymerase Chain Reaction (PCR) Assay
Three PCR assays were designed to test for the presence or absence of pvmdr1 amplification with specific breakpoints. Primer pairs MDR1LF (5′-ACTGCGAAAGTCGCCTATTT-3′) and MDR1LR (5′-TCATCGTGTGGCACATTTTT-3′), and MDR1RF (5′- GGTGAAAAGGTCGAAGCAAA-3′) and MDR1RR (5′-GGGACACGTTCCTCAGAAGT-3′) were designed as positive controls, amplifying 408–base pair and 505–base pair products, respectively, in all isolates. The test assay comprised MDR1RF and MDR1LR, which amplify an approximately 600–base pair product spanning the junction between tandem copies in multicopy (CN2+) isolates with a specific amplification breakpoint. In isolates without the tandem amplification, MDR1RF and MDR1LR are positioned in opposite orientations and so will not yield a product (Supplementary Figure 2). Each assay was undertaken in a 20-µL volume comprising 0.3 units of Taq DNA polymerase (Qiagen), 0.25 µM each primer, 0.2 mM dNTPs (Qiagen), 2 mM MgCl2, and 2 µL of genomic DNA template. Thermocycling was undertaken as follows: 95°C for 3 minutes, followed by 35 cycles of 95°C for 30 seconds, 58°C for 40 seconds, and 72°C for 30 seconds, with a final step of 72°C for 5 minutes. The PCR products were assessed by standard agarose gel electrophoresis.
Quantitative PCR Data on pvmdr1
Details on pvmdr1 CN determined by SYBR Green–based quantitative PCR (qPCR) were sourced from a previous study, whereby CN amplification was defined as ≥1.5 ratio of pvmdr1 amplification against β-tubulin [20]. DNA was available for 48 Thai and 27 Indonesian isolates from the study.
STR Genotyping to Determine Multiplicity of Infection
Genotyping was undertaken at 3 short-tandem-repeat (STR) markers—Pv3.27, msp1F3, and MS16—that have demonstrated high genetic diversity in previous studies [28–33]. Amplification was undertaken using previously described methods [29]. The labeled PCR products were sized by denaturing capillary electrophoresis on an ABI 3100 Genetic Analyzer. Genotype calling was facilitated with GeneMapper, version 4.0. A minimum of 1 multiallelic locus was required to define an infection as polyclonal, and a minimum of 3 monoallelic loci were required to define an infection as monoclonal; all other infections were classified as genotype fails.
Statistical Tests
Proportions were examined using the Fisher exact test. Spearman rank correlation was used to test correlations in nonparametric data. All tests were performed using R software, version 2.12.1, and assuming a significance threshold of 0.05.
RESULTS
pvmdr1 CN Amplification Identified Using Genomic data
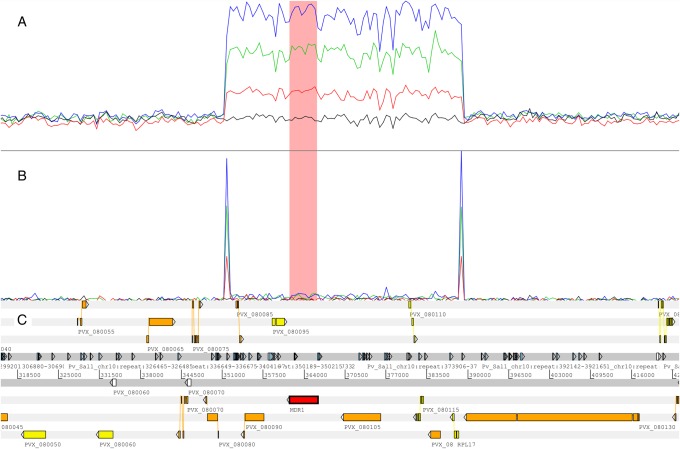
High-read-depth genomic data were available for 88 Thai P. vivax isolates. The median read depth across the 21.4–megabase pair core genome was 67.5 (range, 16–111) in these isolates. Seventeen isolates (19.3%) demonstrated increased read depth and face-away read profiles reflective of CN amplification in the region encompassing pvmdr1 (Supplementary Figure 3). Three additional infections (PD0173-C, PD0183-C, and PD0614-C) demonstrated evidence of amplification by face-away read mapping but not by read depth (Supplementary Figure 3). The majority of the amplified samples (14 [82%]) appeared to have 2 copies of the pvmdr1 region, whereas 2 (12%) had 3 copies, and 1 (6%) had 4 copies. Figure 1 illustrates the read coverage profiles of correctly mapping and face-away mapping reads in the region in each of a CN1, CN2, CN3, and CN4 isolate. Visual inspection of the alignments revealed a sequence of soft-clipped bases (ie, regions of the read that do not align in the same genomic region as the rest of the read) present on both the 5′ and 3′ ends of the amplified region, denoting the amplification breakpoints (Supplementary Figure 4). Common 5′ and 3′ breakpoint sequences consisting of 15–base pair and 18–base pair poly-A tracts, starting at Pv_Sal1_chr10:351600 and Pv_Sal1_chr10:389205, respectively, were observed in all CN2+ samples. In addition to pvmdr1, the 37.6–kilobase pair amplified region encompassed genes encoding 3 hypothetical proteins (PVX_080085, PVX_080105, and PVX_080120), a mitochondrial processing peptidase α protein (PVX_080095), a putative G10 protein (PVX_080110), a putative iron-sulfur cluster assembly accessory protein (PVX_080115), and a putative 50S ribosomal protein L17 (PVX_080125; Figure 1).
Figure 1.
Illustration of the breakpoint region and read depth variation in examples of isolates with no amplification and with 2, 3, and 4 predicted copies. A, Depth (height) of correctly mapping reads in examples of copy number (CN) 1 (black), CN2 (red), CN3 (green), and CN4 (blue) clonal isolates. B, Depth (height) of face-away mapping reads in the same samples, illustrating the breakpoint region. C, Genomic features in the region, including repeat regions. The pvmdr1 gene region is highlighted in pink in panels A and B.
MLG Patterns in the Regions Flanking the pvmdr1 CNV
Fifty-six of 88 isolates (63.6%), including 10 of 17 CN2+ isolates (58.8%), exhibited a genome-wide FWS of ≥ 0.95, consistent with monoclonal infections, and were included in MLG analysis. A total of 105 and 136 SNPs within 20 kilobase pairs of the 5′ and 3′ flanking regions, respectively, of the amplified region had at least 1 minor variant among the high-FWS samples. As illustrated in Supplementary Figure 5, a broad range of MLGs were observed, and there was no notable clustering of the CN2+ samples in either the 5′ or 3′ flanking regions.
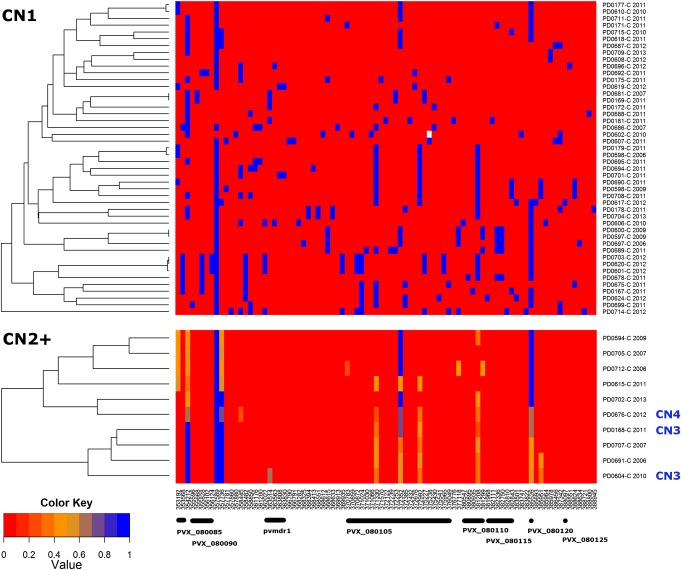
MLG Patterns Within the pvmdr1 CNV Region
Eighty-seven SNPs within the 37.6–kilobase pair amplified region had at least 1 minor variant among the high-FWS samples. With the exception of 1 genotype fail in 1 sample, all positions were successfully called. As illustrated in Figure 2, the large majority of positions in the CN1 isolates were homozygote for the reference or alternative allele, as expected in monoclonal samples. A broad range of MLGs were observed, with few isolates sharing identical or near-identical MLGs. The MLGs in the CN2+ isolates were more complex to interpret owing to multiple apparent heterozygote positions. Given the stringent FWS filtration to remove polyclonal infections, the heterozygote positions likely reflected allelic differences between copies. In accordance with this hypothesis, the median MAF of the heterozygote genotypes approximated the expected values of 50% (range, 42%–49%) in the 7 CN2 isolates, 33% in the 2 CN3 isolates (36% and 35%), and 25% in the CN4 isolate (29%; Supplementary Table 1). A total of 14 loci displayed at least 1 heterozygote genotype. The minor variants at these loci were not private to the CN2+ group; all 14 loci segregated among the CN1 samples. Furthermore, there was a significant correlation between the allele frequency at these loci in the CN1 vs CN2+ samples (ρ = 0.66, P = .011). These patterns suggest ongoing genetic exchange between CN1 and CN2+ isolates. The heterozygote positions clustered at the 5′ and 3′ ends of the amplified region, with pvmdr1 and the direct flanking sequence remaining largely invariable in the CN2+ samples, possibly reflecting recombination sites at the 5′ and 3′ ends of pvmdr1 (Figure 2). On visual inspection of the MLGs, we identified several combinations of CN1 MLGs that would produce heterozygote patterns observed at the 5′ or 3′ ends of pvmdr1 in CN2+ isolates; 2 examples are illustrated in Supplementary Figure 6.
Figure 2.
Heat map illustrating the relatedness of multilocus genotypes (MLGs) in the pvmdr1 region in CN1 (top) and CN2+ (bottom) isolates. MLGs were reconstructed using allele frequency data at 87 single-nucleotide polymorphisms in the amplified region. Color is used to represent the nonreference allele frequency (proportion of reads containing the nonreference allele within that sample). Homozygote reference (Sal1) alleles are presented in red (0.0) and alternative alleles in blue (1.0). Missing genotype calls are colored white. Copy number (CN) definitions are based on genomic read depth results. Samples with a FWS of < 0.95 have been excluded. MLGs are labeled with a sample identifier and year of collection.
Relationship Between pvmdr1 Duplication and Y976F Variation
Genotype calls were determined for the Y976F locus in the 88 isolates with genomic data. Among the 17 CN2+ isolates, the majority (94%) displayed the homozygote wild-type genotype (Supplementary Table 2). None (0%) of the CN2+ isolates exhibited a homozygous mutant genotype, compared with 11 of 71 CN1 isolates (15.5%), but the difference was not significant (P > .05).
Validation of a pvmdr1 Amplification Breakpoint–Specific PCR Assay
The presence of a common breakpoint enabled the design of a targeted PCR assay to detect CN2+ isolates in samples of varying quality, including those not currently amenable to whole-genome sequencing. No amplification was observed in human and P. falciparum DNA. Among the 88 isolates with genomic data, 1 isolate (1%) with CN1 based on read depth was not defined as CN1 the PCR assay. All 17 isolates with CN2+ based on read depth were confirmed by the breakpoint-specific assay. Among the 71 infections with a CN1 read depth status, 13 (18.3%), including PD0173-C, PD0183-C, and PD0614-C, demonstrated evidence of a subpopulation of CN2+ clone(s), (30 CN2+ isolates [34.1%] in total; Table 1). STR-based analysis revealed that all 13 infections (100%) were polyclonal.
Table 1.
Concordance Between the Breakpoint-Specific PCR vs Read Depth and qPCR in Detecting pvmdr1 CN Variation
| Variable | Breakpoint-Specific PCR |
Concordance, % | |||
|---|---|---|---|---|---|
| CN1 | CN2+ | Fail | Total | ||
| Sequence read depth: Thailand | |||||
| CN1 | 57 (22a) | 13 (13) | 1 | 71 | … |
| CN2+ | 0 | 17 (7) | 0 | 17 | … |
| Total | 57 | 30 | 1 | 88 | 85 |
| qPCR: Thailand | |||||
| CN1 | 33 (11b) | 3 (2) | 1 | 37 | … |
| CN2+ | 1 (0) | 9 (4) | 1 | 11 | … |
| Total | 34 | 12 | 2 | 48 | 91 |
| qPCR: Indonesia | |||||
| CN1 | 25 | 0 | 2 | 27 | … |
| CN2+ | 0 | 0 | 0 | 0 | … |
| Total | 25 | 0 | 2 | 27 | 100 |
Number of polyclonal infections as defined by microsatellite genotyping are indicated in parentheses. Genotyping was not undertaken in the Indonesian isolates.
Abbreviation: CN, copy number.
a Two isolates with genotyping failure.
b One isolate with genotyping failure.
DNA was available for 48 Thai isolates with quantitative PCR (qPCR) data on pvmdr1 CN from a previous study [20]. Forty-six isolates (95.8%) yielded confident results with the breakpoint-specific assay. Ten of these isolates (21.7%) demonstrated evidence of increased pvmdr1 amplification, of which 9 (90%) were confirmed with the breakpoint-specific assay (Table 1). Among the 36 isolates defined as CN1 by qPCR, 3 (8%) demonstrated evidence of carrying at least 1 CN2+ clone (12 CN2+ isolates [26.1%] in total). Two of the 3 infections revealed evidence of polyclonality with STR analysis, versus 4 of 9 isolates with CN2+ status detected by both methods. In 27 Indonesian isolates with qPCR data and DNA available, 25 (93%) yielded confident results with the breakpoint-specific assay. No evidence (0%) of pvmdr1 amplification was observed by qPCR or with the breakpoint-specific assay (Table 1).
Temporal Trends in the Prevalence of pvmdr1 Duplication in Thailand Between 2003 and 2015
The breakpoint-specific assay was attempted on 226 Thai samples, including the genomic and qPCR samples. A total of 220 isolates (95.2%) yielded successful assays; 215 of these isolates had details on the date of blood sampling. Sampling dates ranged from 2003 to 2015, although sample size was low in 2008 (n = 1) and 2009 (n = 4) (Supplementary Table 3). As illustrated in Figure 3, with the exclusion of 2008–2009, a trend for declining prevalence of pvmdr1 CN2+ clones was observed over time. A significant difference was observed in CN2+ prevalence in the years before (2003–2011 [52%]) vs after (2012–2015 [17%]) MQ regimens were discontinued in the region (P = 1.635 × 10–5).
Figure 3.

Temporal trend in the prevalence of pvmdr1 amplification. Data from 2008 and 2009 were excluded owing to low sample size (1 and 4, respectively). Abbreviation: MQ, mefloquine.
DISCUSSION
This study provides the first in-depth description of the genetic architecture of pvmdr1 CN amplification. In contrast to the dynamics of pfmdr1 amplification in western Thailand, we observed the same pvmdr1 amplification breakpoints over a 12-year period. Capitalizing on the invariability of the breakpoint, we describe the application of a simple breakpoint-specific PCR assay to temporal surveillance of pvmdr1 amplification in western Thailand.
The simplest explanation for the conservation of the pvmdr1 amplification breakpoint is that the amplifications have all descended from a single ancestral event. Given that isolates with 3 and 4 copies of the pvmdr1 region were observed, it seems there has been >1 amplification event, but the conservation of the breakpoint in these CN3+ isolates suggests that additional amplifications may have taken place on the background of a common ancestral duplication. One might expect to observe comparably high genetic relatedness in the region flanking the amplification in CN2+ versus CN1 isolates if the former have a recent common ancestor. MLG analysis in the regions flanking the amplification did not infer a single lineage of the CN2+ isolates. However, one cannot exclude the possibility that the genetic relatedness between the CN2+ isolates has been broken down by recombination with CN1 parasites over time. Indeed, de-amplification events might further obscure detection of a common lineage. Although the date at which a founding amplification event might have taken place remains unclear, we observed evidence of the same breakpoint in isolates from 2003—the earliest year tested.
The assumption of a single origin of the common breakpoint contrasts with the dynamics of pfmdr1 amplification in the same geographic region. A range of pfmdr1 breakpoints and amplicon sizes were observed in P. falciparum isolates collected from a single clinic between 2000 and 2003 [19]. Indeed, laboratory experiments infer high rates of pfmdr1 duplication (1 per 108 parasites) in drug selection experiments [34]. However, although P. vivax may be exposed to MQ in mixed-species infections, misdiagnoses, and recurrences following P. falciparum infection, the selective pressure of this concomitant drug exposure is not likely to be as strong as that for P. falciparum, possibly resulting in fewer founding amplification events. Differences in transmission dynamics between the 2 species are also likely to contribute to the contrasting pfmdr1 and pvmdr1 amplification patterns. The early development of the transmissible gametocyte stages in P. vivax may have further reduced the impact of drug selection [35]. In addition, the ability of P. vivax isolates to form dormant liver stages, which relapse weeks or months after inoculation, may facilitate longer persistence of certain lineages in P. vivax versus P. falciparum.
Although the degree of selective pressure on P. vivax parasites remains unclear, a recent study identified frequent duplications in the P. vivax Duffy binding protein 1 gene (pvdbp1) in Madagascar, which also had common breakpoints [25]. Sequence analysis inferred a single and recent ancestral origin of the pvdbp1 duplication in these isolates. The authors identified the same pvdbp1 breakpoints in non-Malagasy P. vivax infections but could not determine whether these isolates had a shared lineage with the Malagasy isolates. In combination with our results on pvmdr1, these findings suggest that common amplification breakpoints may not be unusual in P. vivax.
The possibility that multiple independent pvmdr1 amplification events have occurred using the same breakpoints cannot be excluded. It remains unclear, however, why the same breakpoints would be used, indeed in contrast to P. falciparum, where multiple breakpoints are evident [19]. The breakpoints of pfmdr1 and P. falciparum GTP-cyclohydrolase 1 amplifications contain low-complexity sequences such as monomeric A/T tracts [19, 36, 37]. Similarly, the breakpoints of the common pvdbp1 and pvmdr1 duplications contained long homopolymer tracts of T and A residues, respectively [25]. Although several repetitive sequences are observed in the regions flanking pvmdr1, it is possible that the DNA conformation or other mechanism(s) lead to preferential breakage at the poly-A tracts. Alternatively, amplifications may have occurred at other breakpoints but were associated with a fitness disadvantage, compared with those described in the current study, and thus have not reached high frequency.
We observed evidence of different allelic states between copies in isolates with pvmdr1 amplifications, as inferred by the presence of heterozygote calls in single-clone infections. Assuming that the mechanism of the original amplification event entailed duplication during mitosis, the ancestral duplication would have comprised identical copies and thus exhibited no heterozygote states. Over time, mutation and/or recombination events may have created different allelic states between copies in some lineages. Indeed, none of the polymorphisms among the CN2+ isolates were private to this group; rather, they were also variable in the CN1 group, possibly inferring occasional recombination between CN1 and CN2+ isolates. Alternatively, the original amplification(s) may have been produced during meiosis, with distinct maternal and paternal copies being retained. In this scenario, the ancestral amplification(s) could carry copies with different allelic states.
Similar to P. falciparum, ex vivo investigation infers that, although pvmdr1 amplification reduces susceptibility to MQ, it increases susceptibility to chloroquine (CQ) [20]. Although requiring further confirmation, a point mutation conferring a Y976F change in pvmdr1 has been associated with low-level reduction in CQ susceptibility [38]. We were able to assess both pvmdr1 amplification and 976 variation in 88 isolates collected between 2006 and 2013. In accordance with a previous study undertaken in western Thailand [39], although 976-F variants were observed in our study, they were not observed in amplifications. This trend may reflect a significant fitness cost to the parasite of carrying both the mutation and amplification and lends suggestion that MQ pressure on the P. vivax population was possibly greater than CQ pressure during the sampling period.
Quantitative methods for detecting amplifications, such as read depth and quantitative PCR approaches, have the benefit that they are not constrained to specific amplicons. However, these approaches have limited sensitivity to detect low-frequency CN2+ clones in polyclonal infections—a caveat overcome by the breakpoint-specific PCR approach. In regions where polyclonal infections are common, quantitative approaches may miss a sizeable proportion of CN2+ clones. Indeed, relative to the breakpoint-specific PCR, 43% and 25% of infections with pvmdr1 amplification were missed by read depth (13 of 30) and qPCR approaches (3 of 12), respectively, in our study. The sensitivity, simplicity, and applicability to a range of sample qualities were advantageous features of the breakpoint-specific assay for surveillance of pvmdr1 amplification in western Thailand. Using this assay, we demonstrated significantly lower prevalence of CN2+ clones between 2012 and 2015 relative to earlier years tested. This trend is consistent with the local discontinuation of MQ plus artesunate in 2012 following documentation of its declining efficacy in the region [40]. However, the application of any assay using a specific breakpoint pattern is critically reliant on knowledge of the genetic background of the CN2+ clones circulating in the given population. Further investigation of the genetic background of pvmdr1 amplification is required in other geographic regions. These investigations will provide further insights into the evolutionary dynamics of pvmdr1 amplification and its adaptive potential.
In contrast to pfmdr1, the breakpoints of pvmdr1 CN amplification appear to be highly conserved in western Thailand. It remains unclear whether this dynamic reflects a single amplification origin and whether the same breakpoints are commonly observed in other geographic regions, permitting surveillance of reduced MQ susceptibility by using a rapid and cost-effective breakpoint-specific PCR approach.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the patients who contributed their samples to the study and the health workers and field teams who assisted with the sample collections.
Financial support. This work was supported by a Wellcome Trust Senior Research Fellowship in Clinical Science (grant 091625 to R. N. P.). Additional support for sequencing, data analysis, and project coordination was provided by the Wellcome Trust (098051, 090770/Z/09/Z), the Medical Research Council (G0600718), and the United Kingdom Department for International Development (M006212). D. S. was supported by the National Institutes of Health (grant R01 AI03228). SMRU is part of the Mahidol Oxford University Research Unit, supported by the Wellcome Trust of Great Britain.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. World malaria report 2014. Geneva: World Health Organization, 2014. [Google Scholar]
- 2.Barcus MJ, Basri H, Picarima H et al. . Demographic risk factors for severe and fatal vivax and falciparum malaria among hospital admissions in northeastern Indonesian Papua. Am J Trop Med Hyg 2007; 77:984–91. [PubMed] [Google Scholar]
- 3.Rodriguez-Morales AJ, Benitez JA, Arria M. Malaria mortality in Venezuela: focus on deaths due to Plasmodium vivax in children. J Trop Pediatr 2008; 54:94–101. [DOI] [PubMed] [Google Scholar]
- 4.Baird JK. Severe and fatal vivax malaria challenges ‘benign tertian malaria’ dogma. Ann Trop Paediatr 2009; 29:251–2. [DOI] [PubMed] [Google Scholar]
- 5.Nosten F, McGready R, Simpson JA et al. . Effects of Plasmodium vivax malaria in pregnancy. Lancet 1999; 354:546–9. [DOI] [PubMed] [Google Scholar]
- 6.Tjitra E, Anstey NM, Sugiarto P et al. . Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med 2008; 5:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poespoprodjo JR, Fobia W, Kenangalem E et al. . Vivax malaria: a major cause of morbidity in early infancy. Clin Infect Dis 2009; 48:1704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poespoprodjo JR, Fobia W, Kenangalem E et al. . Adverse pregnancy outcomes in an area where multidrug-resistant plasmodium vivax and Plasmodium falciparum infections are endemic. Clin Infect Dis 2008; 46:1374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price RN, Auburn S, Marfurt J, Cheng Q. Phenotypic and genotypic characterisation of drug-resistant Plasmodium vivax. Trends Parasitol 2012; 28:522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baird JK. Resistance to therapies for infection by Plasmodium vivax. Clin Microbiol Rev 2009; 22:508–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayxay M, Pukrittayakamee S, Newton PN, White NJ. Mixed-species malaria infections in humans. Trends Parasitol 2004; 20:233–40. [DOI] [PubMed] [Google Scholar]
- 12.Snounou G, White NJ. The co-existence of Plasmodium: sidelights from falciparum and vivax malaria in Thailand. Trends Parasitol 2004; 20:333–9. [DOI] [PubMed] [Google Scholar]
- 13.Douglas NM, Nosten F, Ashley EA et al. . Plasmodium vivax recurrence following falciparum and mixed species malaria: risk factors and effect of antimalarial kinetics. Clin Infect Dis 2011; 52:612–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson CM, Volkman SK, Thaithong S et al. . Amplification of pfmdr 1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol 1993; 57:151–60. [DOI] [PubMed] [Google Scholar]
- 15.Price RN, Uhlemann AC, Brockman A et al. . Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 2004; 364:438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price RN, Uhlemann AC, van Vugt M et al. . Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis 2006; 42:1570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A 1994; 91:1143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis 2006; 194:528–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair S, Nash D, Sudimack D et al. . Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol 2007; 24:562–73. [DOI] [PubMed] [Google Scholar]
- 20.Suwanarusk R, Chavchich M, Russell B et al. . Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis 2008; 198:1558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson RD, Amato R, Auburn S et al. . Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet 2016; 48:959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malaria Genomic Epidemiology Network. A global network for investigating the genomic epidemiology of malaria. Nature 2008; 456:732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlton JM, Adams JH, Silva JC et al. . Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 2008; 455:757–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menard D, Chan ER, Benedet C et al. . Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis 2013; 7:e2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auburn S, Campino S, Miotto O et al. . Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One 2012; 7:e32891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manske M, Miotto O, Campino S et al. . Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 2012; 487:375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koepfli C, Mueller I, Marfurt J et al. . Evaluation of Plasmodium vivax genotyping markers for molecular monitoring in clinical trials. J Infect Dis 2009; 199:1074–80. [DOI] [PubMed] [Google Scholar]
- 29.Abdullah NR, Barber BE, William T et al. . Plasmodium vivax population structure and transmission dynamics in Sabah Malaysia. PLoS One 2013; 8:e82553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Getachew S, To S, Trimarsanto H et al. . Variation in Complexity of Infection and Transmission Stability between Neighbouring Populations of Plasmodium vivax in Southern Ethiopia. PLoS One 2015; 10:e0140780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Auburn S, Cao J et al. . Genetic diversity and population structure of Plasmodium vivax in Central China. Malar J 2014; 13:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noviyanti R, Coutrier F, Utami RA et al. . Contrasting Transmission Dynamics of Co-endemic Plasmodium vivax and P. falciparum: Implications for Malaria Control and Elimination. PLoS Negl Trop Dis 2015; 9:e0003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray KA, Dowd S, Bain L et al. . Population genetics of Plasmodium falciparum and Plasmodium vivax and asymptomatic malaria in Temotu Province, Solomon Islands. Malar J 2013; 12:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preechapornkul P, Imwong M, Chotivanich K et al. . Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. Antimicrob Agents Chemother 2009; 53:1509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider KA, Escalante AA. Fitness components and natural selection: why are there different patterns on the emergence of drug resistance in Plasmodium falciparum and Plasmodium vivax? Malar J 2013; 12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triglia T, Foote SJ, Kemp DJ, Cowman AF. Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol Cell Biol 1991; 11:5244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair S, Miller B, Barends M et al. . Adaptive copy number evolution in malaria parasites. PLoS Genet 2008; 4:e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suwanarusk R, Russell B, Chavchich M et al. . Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One 2007; 2:e1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imwong M, Pukrittayakamee S, Pongtavornpinyo W et al. . Gene amplification of the multidrug resistance 1 gene of Plasmodium vivax isolates from Thailand, Laos, and Myanmar. Antimicrob Agents Chemother 2008; 52:2657–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrara VI, Lwin KM, Phyo AP et al. . Malaria burden and artemisinin resistance in the mobile and migrant population on the Thai-Myanmar border, 1999–2011: an observational study. PLoS Med 2013; 10:e1001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.