Abstract
Background. Unlike tuberculosis, few studies have evaluated a host genetic basis for variability in susceptibility to latent Mycobacterium tuberculosis infection (LTBI). We performed a candidate gene association study of autophagy-related genes and LTBI.
Methods. We enrolled close contacts of individuals with pulmonary tuberculosis, assessed LTBI status, and determined clinical and sociodemographic risk factors for LTBI. In participants who self-identified as Asian or black, we compared haplotype-tagging single-nucleotide polymorphisms (SNPs) in ULK1 and GABARAP between cases (n = 143) and controls (n = 106). Using CRISPR/Cas9 in U937 monocytes, we investigated the effect of ULK1 deficiency on cytokine expression, autophagy, and M. tuberculosis replication.
Results. In Asian participants, we identified 2 ULK1 SNPs (rs12297124 and rs7300908) associated with LTBI. After adjustment for population admixture and clinical risk for LTBI, each rs12297124 minor allele conferred 80% reduction in LTBI risk (odds ratio, 0.18; 95% confidence interval, .07–.46). Compared with controls, ULK1-deficient cells exhibited decreased tumor necrosis factor secretion after stimulation with Toll-like receptor ligands and M. tuberculosis whole-cell lysate, increased M. tuberculosis replication, and decreased selective autophagy.
Conclusions. These results demonstrate a strong association of rs12297124, a noncoding ULK1 SNP, with LTBI and a role for ULK1 regulation of TNF secretion, nonspecific and M. tuberculosis–induced autophagy, and M. tuberculosis replication in monocytes.
Keywords: autophagy, candidate gene analysis, single nucleotide polymorphism, genetic susceptibility, tuberculosi
Mycobacterium tuberculosis infects one third of the world's population and is a leading cause of infectious disease–related death [1]. Infection with M. tuberculosis may result in several outcomes, including immediate bacillary multiplication resulting in primary tuberculosis, latent M. tuberculosis infection (LTBI), or reactivation tuberculosis. The mechanisms responsible for different tuberculosis outcomes are poorly understood. Several lines of evidence support the influence of host genetics on susceptibility to tuberculosis, including twin-based, candidate gene association, linkage, and genome-wide association studies [2–4]. The vast majority of studies that have evaluated a host genetic basis for variability in tuberculosis outcomes have assessed associations with active tuberculosis. However, the direct study of tuberculosis susceptibility in humans allows for unique insights not possible in animal models, including investigations into LTBI [5].
Despite a massive global burden, the host genetic basis for LTBI susceptibility is understudied [6]. The heritability of responses to LTBI tests—the tuberculin skin test (TST) and the interferon γ (IFN-γ)–release assay—is estimated to be high [7, 8]. Genome-wide linkage studies identified several loci associated with LTBI [9–11], and candidate gene association studies identified associations between polymorphisms in the genes encoding interleukin 10 and interleukin 4 and TST responses [12, 13]. Although these studies suggest genetic regulation of LTBI susceptibility, the mechanisms remain unknown. A better understanding of the impact of host genetics on LTBI is key to understanding tuberculosis pathogenesis.
The innate immune system, in particular alveolar macrophages, likely plays an important role in determining tuberculosis outcomes following inhalation of M. tuberculosis into the lungs and prior to T-cell priming. We and others have identified associations between common polymorphisms in innate immunity genes and susceptibility to tuberculosis [14–16]. Much of this work has focused on pattern-recognition receptors and their associated adaptor proteins [17–19]. In recent years there has been a growth in interest and understanding of autophagy, which protects the host by targeting invading pathogens for elimination (in addition to other functions) and has a major role in host tuberculosis defenses [20–23]. Autophagy is initiated by a ULK1-containing complex under regulation by mTOR and AMP kinase. GABARAP regulates later stages of autophagosome maturation. A number of candidate gene studies have demonstrated associations between polymorphisms in IRGM, an autophagy gene, and tuberculosis in various populations [24–26]. However, IRGM polymorphisms were not associated with the outcome of LTBI in one study [27]. To our knowledge, associations of genetic variants in autophagy genes with LTBI have not been identified.
We hypothesized that common variation in autophagy-related genes is associated with susceptibility to LTBI following tuberculosis exposure. With few exceptions [9], studies of LTBI susceptibility and genetic variation occurred in tuberculosis-endemic regions (geographic differences may affect responses to tuberculosis treatments) [28, 29] and did not adjust for epidemiologic risk factors, potentially leading to loss of study power. We enrolled close contacts of tuberculosis cases through a US tuberculosis program to examine whether variation in autophagy genes is associated with susceptibility to M. tuberculosis infection. To our knowledge, this is the first reported association of a ULK1 single-nucleotide polymorphism (SNP) with a tuberculosis-related outcome.
METHODS
Human Subjects
We recruited subjects through the Tuberculosis Control Program (TBCP), Public Health–Seattle and King County (Seattle, WA). Eligible study subjects were close contacts of patients with culture-confirmed pulmonary tuberculosis. Generally, close contact was defined as >8 hours of exposure to an index case in an enclosed space during the preceding 3 months. All participants were ≥18 years of age and without a prior history of tuberculosis. We excluded individuals who were immunocompromised, including known those with known human immunodeficiency virus (HIV) infection. HIV testing was not routinely offered to close contacts unless they received a diagnosis of active tuberculosis.
LTBI status was evaluated with tuberculin skin tests (TSTs) and/or QuantiFERON Gold-in-Tube (QFT) assays at the discretion of the TBCP. TSTs were performed using 5 tuberculin units (0.1 mL) of purified protein derivative (RT 23 solution, Sanofi-Pasteur) and read at 48–72 hours. A TST yielding an induration diameter of ≥5 mm was considered positive for LTBI [30]. The QFT assay was performed and interpreted according to manufacturer guidelines. Participants who were tested with both the TST and the QFT assay had blood collected before TST placement, and any positive result was considered evidence of LTBI. Per TBCP protocol, close contacts with a negative result of the initial test for LTBI were retested 8–10 weeks after the last tuberculosis exposure. We defined converters as participants who initially tested negative for LTBI but had a positive result on retesting.
SNP Selection
We selected 2 candidate autophagy genes, GABARAP and ULK1, and ancestry-informative markers [31]. We chose ULK1 on the basis of its central role in the initiation of autophagy and GABARAP because of its role in later stages of autophagosome maturation. Neither gene was examined in a previous study of tuberculosis genetic susceptibility [32]. Haplotype tagging SNPs were chosen using the 1000 Genomes Browser in Asian (CHB, CHS, and JPT) and African (ASW, LWK, and YRI) populations [33]. Selection criteria included a minor allele frequency of 2.5% (for GABARAP) or 5% (for ULK1), an R2 cutoff of 0.8 for linkage disequilibrium, and a region that included the gene ± 1 kb.
Genomic Techniques
We limited genotyping to participants who self-reported as Asian or black, to maximize study efficiency, as these ethnicities are the most frequent among individuals with tuberculosis in King County, Washington, and limit confounding due to population stratification. Included cases and controls were unrelated. Genomic DNA was prepared from saliva (Oragene Discover, DNA Genotek). By use of Illumina's GoldenGate Assay and a custom multiplex oligo pool, genomic DNA samples (250 ng) underwent allele-specific primer extension and amplification. Amplified products were then hybridized to Universal BeadChips and subsequently imaged on the Illumina iScan+. Cluster plots were visually inspected to ensure accurate genotyping calls. The call rate exceeded 95% for 92% of all genotyped SNPs. Genotypes were confirmed in a subset of individuals through genotyping with the Fluidigm BioMark microfluidic system. All GABARAP and ULK1 SNPs were in Hardy-Weinberg equilibrium (P > .001 by the exact test) based on the χ2 goodness-of-fit test [34].
CRISPR/Cas9 Gene Editing
Guide RNA targeting ULK1 (sequence GGACGCCTCCATGCTCAGCG[TGG]) was selected using ChopChop (available at: https://chopchop.rc.fas.harvard.edu/) [35]. The guide RNA and scaffold were cloned into a lentiviral plasmid, gRNA-Cas9-t2a-puromycin pRRL (a gift from Elizabeth Gray, University of Washington), using Clontech InFusion Cloning Kit (Takara Bio). Nontargeting control gRNA-CAs9-t2a-puromycin pRRL plasmids (a gift from Elizabeth Gray) were used as negative controls (sequences were obtained from the GeCKO v2 library, Addgene) [36]. Lentivirus was produced in HEK 293 T Lenti-X cells (Clontech) by transfecting ULK1-pRRL plasmid with packaging plasmids pRSV-Rev, pMD2.g, and pMDLg/pRRE (Addgene numbers 12253, 12259, and 12251, respectively), in Opti-MEM and TransiT-LT1 (Mirius Bio). Supernatants were filtered and incubated with U937 cells and 5 µg/mL polybrene (EMD Millipore). Transduced U937 cells were allowed to rest for 24 hours in fresh medium after incubation with lentiviral particles. After puromycin selection, Crispr-Cas9 targeting was evaluated by restriction fragment–length polymorphism (RFLP) screening, using restriction sites located within the Cas9 cut site. Genomic DNA was isolated and sequenced to confirm the presence of an insertion/deletion mutation (AGCCCCTCGCCCCCTGCCCACGCT[:/T]GAGCATGGAGGCGTCCT). To evaluate cell viability, cells were plated at 1 × 105 cells/well, removed daily, stained with trypan blue, and counted in a hemocytometer. Both viable and nonviable cells were recorded, and percentage viability calculated. Viability experiments were performed on 2 separate days, and data were pooled.
Cytokine Expression
U937 cells were plated at 5 × 104 cells/well in Roswell Park Memorial Institute (RPMI) medium with 10% fetal bovine serum, 2 mM L-glutamine, and HEPES, differentiated with 50 ng/mL phorbol myristate acetate (PMA; Invivogen, San Diego, CA) for 48 hours, washed with Hank's balanced salt solution (HBSS), and rested for 24 hours in complete medium. Cells were incubated with lipopolysaccharide (LPS; List Biological Labs, Campbell, CA), PAM2/PAM3 (Invivogen), or whole-cell lysate from M. tuberculosis strain H37Rv (Colorado State University, Fort Collins) for 24 hours. In addition, we used an NLRC4 ligand to activate the inflammasome with a chimeric protein of the type 3 secretion system needle protein of Burkholderia thailandensis fused to the binding domain of Bacillus anthracis lethal factor, which is used for cytoplasmic delivery when administered with B. anthracis protective antigen (a gift from Russell Vance, University of California, Berkeley). We evaluated supernatants for interleukin 1β (IL-1β) and tumor necrosis factor (TNF) levels, using the DuoSet enzyme-linked immunoassay (R&D Systems, Minneapolis), and for interferon β (IFN-β), using the TaqMan assay (Thermo Fisher Scientific, Waltham, MA) and primer Hs.PT.56a.39481063.g (IDT, Coralville, IA).
Autophagy Studies
U937 cells were differentiated using PMA for 24 hours, then exposed to medium, starvation, or M. tuberculosis expressing mCherry at a multiplicity of infection (MOI) of 5 for 4 hours. Cells were washed (3 times) in phosphate-buffered saline and fixed in 100% methanol at −20°C for 5 minutes. Cover slips were incubated in Alexa488 anti-LC3 (clone D3U4C, Cell Signaling) at a dilution of 1:50 overnight at 4°C. After washing, the cover slips were mounted onto glass slides, using Prolong Gold anti-fade reagent (Molecular Probes). Images were acquired on a Nikon Eclipse Ti microscope (60× objective), corrected for background, and the number of LC3 puncta >1 µm in diameter were measured (Volocity 6.3, PerkinElmer, Waltham, MA) for at least 100 cells per condition. Each experiment was performed in triplicate. Overlaid fluorescent images were analyzed to determine the number of LC3-positive autophagosomes containing mycobacteria. A minimum of 100 phagosomes were analyzed for each cellular condition, with at least 3 independent experiments performed.
M. tuberculosis Infection
U937 cells with the ULK1 knockout, empty vector CRISPR control, or wild type were plated in sextuplet at a density of 75 000 cells/well and differentiated with PMA (50 ng/mL) overnight, washed twice with HBSS, and rested for 24 hours. A frozen aliquot of M. tuberculosis Erdman strain expressing lux (MTB-lux; a gift from Jeffrey Cox, University of California, San Francisco) was thawed, centrifuged, and resuspended in RPMI with 10% fetal calf serum. Luminescence from this organism strongly correlates with the number of M. tuberculosis colony-forming units on an agar plate (R2 = 0.99; data not shown). A separate set of wells was included with mLux in the absence of U937 cells as a control for background luciferase expression. After incubation for 4 hours (MOI, 10), wells were washed once with medium and then incubated with fresh medium for 7 days. Relative light units were measured at days 0, 1, and 4–7 (Synergy H4 reader, BioTek Instruments). The experiment was performed 3 separate times.
Statistical Methods
Differences were tested using the χ2 test, the Student t test, or the Wilcoxon rank sum test as appropriate and indicated in figure legends. We assessed associations between candidate SNPs and LTBI in genotypic exact models stratified by ethnicity, using Stata 11 (Stata, College Station, TX). SNPs with a significant association (P < .05) were evaluated using dominant, recessive, and additive models. To assess for population stratification, we performed principal components analysis using ancestry-informative markers and examined scree and score plots. Before candidate gene association analyses, we removed genetic outliers from each ethnicity on the basis of the first 2 principal components (n = 4). We evaluated significant SNP associations with LTBI, using multivariable logistic regression that adjusted for principal components and demographic and exposure-related variables that were associated with LTBI in the entire cohort. We chose demographic and exposure variables for evaluation, based on published and hypothesized associations with LTBI, including index case characteristics (acid-fast bacilli [AFB] smear burden, categorized as smear negative, low (1+/2+), or high (3+/4+); and radiographic evidence of bilateral disease or cavitations), exposure setting, and contact characteristics (sex, ethnicity, homelessness, education level, employment status, alcohol use, body mass index, smoking, foreign birth, BCG status, and tuberculosis history in first-degree relative). We included age in multivariable models regardless of bivariate significance. We compared logistic regression models, using partial likelihood ratio tests (nested models) or the Akaike information criterion (nonnested models). We report uncorrected P values throughout the article.
Expression Quantitative Trait Loci (eQTL) Analysis
We used publicly available data from the Genotype-Tissue Expression project (GTEx) to examine associations between SNPs of interest and messenger RNA levels in associated genes in a variety of cells and tissues [37]. GTEx generates eQTL P values based on linear regression analysis assuming an additive model, using a 2-tailed t test.
Ethics
This study was approved by the University of Washington Institutional Review Board, and informed consent was obtained from all participants.
RESULTS
Study Population
From 2010 to 2014, we enrolled 569 close contacts of tuberculosis index cases. Although we limited our candidate gene association study to individuals who self-identified as black or Asian, any close contact was eligible for enrollment to determine epidemiological and clinical characteristics associated with LTBI. In our cohort, 273 participants (48%) had LTBI (Table 1). Agreement between TST and QFT was moderate (κ = 0.54) [38]. LTBI test conversions were observed in 44 participants. In multivariable analysis of demographic, clinical, and exposure-related variables, radiographic evidence of bilateral disease (odds ratio [OR], 2.3; 95% confidence interval [CI], 1.5–3.5) and AFB smear grade (OR, 1.3; 95% CI, 1.1–1.5) in the index case and black ethnicity (compared with white; OR, 3.7; 95% CI, 1.7–8.0) and foreign birth (OR, 2.7; 95% CI, 1.5–5.0) in close contacts were independently associated with LTBI.
Table 1.
Characteristics of 273 Participants With and 296 Without Latent Mycobacterium tuberculosis Infection (LTBI)
| Characteristic | LTBI | No LTBI | P Value |
|---|---|---|---|
| Age, y, mean ± SD | 41.3 ± 15.9 | 41.0 ± 15.2 | .80 |
| Male sex | 153/273 | 153/296 | .30 |
| Ethnicity | <.001 | ||
| Asian | 113/273 | 82/296 | |
| Black | 58/273 | 45/296 | |
| Latino | 31/273 | 25/296 | |
| Pacific Islander | 35/273 | 21/296 | |
| White | 21/273 | 36/296 | |
| Other | 14/273 | 17/296 | |
| Foreign birth (n = 558) | 211/266 | 145/292 | <.001 |
| BCG vaccination (n = 563) | 165/270 | 118/293 | <.001 |
| BMI,a median | 25.0 | 25.8 | .17 |
| Homeless | 33/273 | 56/296 | .03 |
| Nocturnal exposure (n = 568) | 53/272 | 42/296 | .09 |
| Smoking (n = 562) | 63/272 | 70/290 | .79 |
| Alcohol use (n = 566) | 89/272 | 111/294 | .21 |
| Employed >20 h/wk | 121/273 | 137/296 | .61 |
| Highest education level (n = 559) | .04 | ||
| Less than grade school | 10/268 | 8/291 | |
| Grade school | 50/268 | 30/291 | |
| High school | 134/268 | 148/291 | |
| College | 55/268 | 77/291 | |
| >College | 19/268 | 28/291 | |
| Tuberculosis history in first-degree relative | 61/273 | 48/296 | .06 |
| Index characteristic | |||
| Bilateral disease detected by chest radiography (n = 461) | 120/237 | 78/224 | .001 |
| Cavitation(s) detected by chest radiography (n = 466) | 146/240 | 110/226 | .008 |
| AFB smear finding | <.001 | ||
| Negative | 11/236 | 36/232 | |
| 1+ or 2+ | 53/236 | 67/232 | |
| 3+ or 4+ | 172/236 | 129/232 | |
| LTBI diagnosis, participants, no. | |||
| TST (n = 256) | 105 | 151 | |
| IGRA (n = 358) | 171 | 187 | |
| Dual testing (n = 86) | 49 | 36 | |
Data are percentage of participants, unless otherwise indicated.
Abbreviations: AFB, acid-fast bacilli; IGRA, interferon γ release assay; TST, tuberculin skin test.
a Body mass index (BMI) is calculated as the weight in kilograms divided by the height in meters squared.
Genotyping Results by Ethnicity
To determine whether candidate polymorphisms in ULK1 and GABARAP were associated with susceptibility to M. tuberculosis infection, we performed genotyping in 143 close contacts with LTBI and 106 without LTBI. We evaluated 15 SNPs in ULK1 and 5 SNPs in GABARAP (Supplementary Table 1). Using genotypic models, we identified 2 SNPs in ULK1 in Asian participants that were significantly associated with LTBI (unadjusted P < .05): rs12297124 and rs7300908 (Table 2). No associations between GABRAP polymorphisms and LTBI were identified (Supplementary Table 1). We found that rs12297124 (OR, 0.36; 95% CI, .19–.72; P = .004) and rs7300908 (OR, 0.37; 95% CI, .18–.77; P = .007) had the strongest associations with LTBI in additive models (Table 3). As rs12297124 was in high linkage disequilibrium with rs7300908 (r2 = 0.88; Supplementary Figure 1), we focus the remainder of this article on understanding LTBI susceptibility with rs12297124, an intronic SNP that involves a G → T transversion.
Table 2.
2 ULK1 Single-Nucleotide Polymorphisms (SNPs) Are Associated With Latent Mycobacterium tuberculosis Infection (LTBI) in Asian Contacts
| SNP | Genotype Frequency in Controls |
Genotype Frequency in Cases With LTBI |
P Value |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 00 | 01 | 11 | Total | 00 | 01 | 11 | Total | Genotypic | Trend test | |
| ULK1 rs12297124 | 0.657 | 0.286 | 0.057 | 70 | 0.837 | 0.163 | 0.000 | 92 | .005 | .003 |
| ULK1 rs7300908 | 0.657 | 0.329 | 0.014 | 70 | 0.837 | 0.163 | 0.000 | 92 | .01 | .006 |
Abbreviations: 00, homozygous common allele; 01, heterozygous allele; 11, homozygous rare allele.
Table 3.
Multivariable Model of ULK1 rs12297124 Association With Latent Mycobacterium tuberculosis Infection in Asian Contacts
| Variable | Odds Ratio (95% CI) | P Value |
|---|---|---|
| Unadjusted additive model | ||
| ULK1 rs12297124 | 0.36 (.19–.72) | .004 |
| Additive model adjusted for first principal component | ||
| ULK1 rs12297124 | 0.30 (.15–.61) | .001 |
| Additive model adjusted for clinical variablesa | ||
| ULK1 rs12297124 | 0.18 (.07–.46) | .0004 |
| Bilateral radiographic disease | 1.89 (.78–4.54) | .16 |
| Smear burden (high, low, negative) | 2.09 (1.09–4.00) | .03 |
| Foreign birth | 0.82 (.16–4.14) | .81 |
| Age | 1.04 (1.01–1.06) | .01 |
Abbreviation: CI, confidence interval.
a Adjusted for PC1.
ULK1 Genetic Associations
We genotyped ancestry-informative markers and performed a principal components analysis, which indicated that the cases and controls were genetically well matched and that clusters by principal components agreed with self-identified ethnicity (Supplementary Figure 2). After comparing models by using likelihood ratio tests, we included only the first principal component (PC1) in multivariate models. Among Asian participants, adjustment of rs12297124 for PC1 strengthened the association with LTBI (OR, 0.30; 95% CI, .15–.61; Table 3). In a multivariable model that included index case characteristics (bilateral disease on radiographs and smear grade), contact characteristics (foreign birth and age), and PC1, rs12297124 remained independently associated with LTBI (OR, 0.18; 95% CI, .07–.46; Table 3). Among black participants (50 cases and 36 controls), ULK1 rs12297124 was not significantly associated with LTBI in a model that included exposure-related variables and PC1 (OR, 0.67; 95% CI, .14–3.22). When we evaluated Asian and black participants together in a multivariable model, ULK1 rs12297124 remained independently associated with LTBI (OR, 0.32; 95% CI, .16–.66; P = .002). We also evaluated rs12297124 associations with LTBI as defined separately by QFT assay positivity and TST positivity in Asian participants: after adjustment for principal components, rs12297124 was associated with QFT assay–defined LTBI (OR, 0.31; 95% CI, .13–.73; P = .007) and TST-defined LTBI (OR, 0.08; 95% CI, .01–.64; P = .02).
eQTL Findings
Using the GTEx browser (available at: http://www.gtexportal.org/home/), we evaluated associations between ULK1 expression and haplotype-tagging SNPs. Of the 15 ULK1 SNPs that we genotyped, only rs12297124 was significantly associated with ULK1 expression in tissue from esophageal muscularis mucosae (P = .005; Supplementary Figure 3).
ULK1-Deficient Cells Are Associated With Increased M. tuberculosis Replication, Decreased TNF Secretion, and Decreased Autophagy
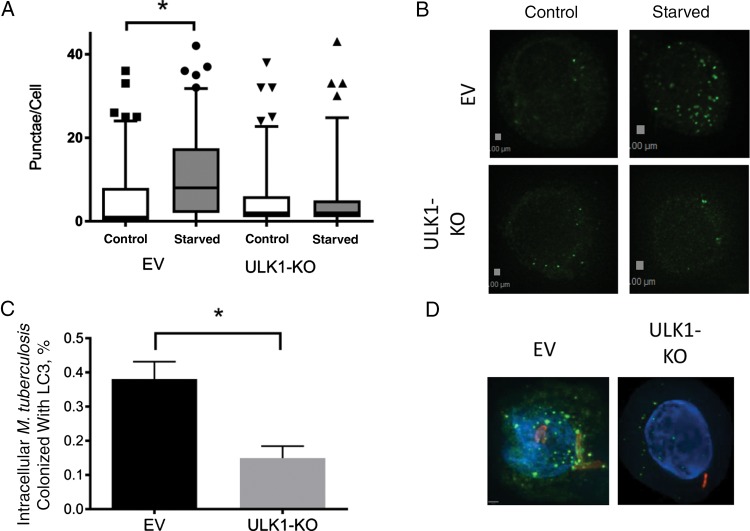
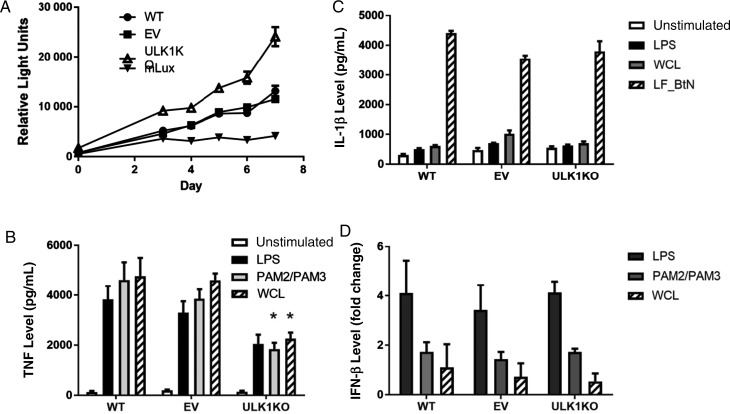
To evaluate potential mechanisms through which ULK1 affects LTBI susceptibility, we created an ULK1-deficient U937 cell line, using the CRISPR/Cas9 gene editing technique. We inserted a single base pair into ULK1 that generated a premature stop codon, which we verified by a RFLP assay (Supplementary Figure 4). We assessed nonselective autophagy to confirm that ULK1 function was impaired and found that starvation conditions did not induce autophagy in ULK1-deficient cells, as measured by the number of LC3-positive puncta per cell. This was in contrast to cells created with empty vector during CRISPR/Cas9 processing, in which starvation led to a significant increase in the number of LC3-positive puncta per cell (Figure 1A and 1B). Percentage cell viability did not differ between ULK1-deficient U937, empty vector, and wild-type cells. We infected ULK1-deficient, empty vector, and wild-type U937 cells with M. tuberculosis expressing the lux operon to assess differences in control of infection. Using a bioluminescent assay, we observed increased M. tuberculosis replication in ULK1-deficient cells compared to empty vector control and wild-type cells (P = .004 for days 3–7; Figure 2A).
Figure 1.
ULK1 deficiency inhibits nonselective autophagy and Mycobacterium tuberculosis–selective autophagy in U937-derived monocytes and macrophages. A and B, Nonselective autophagy is inhibited without ULK1. ULK1-deficient (ULK1-knockout [KO]) or empty-vector (EV) U937 cells were incubated in nutrient-rich medium or starvation medium for 4 hours and then fixed, permeabilized, and stained for LC3. Images were obtained via fluorescent microscopy, and the number of LC3-positive puncta were measured per cell for each condition. A, Tukey plot of the number of puncta/cell in each condition. B, Deconvolved images of LC3 staining from individual cells in each condition, representative of each. Green images indicate LC3, and blue images indicate DAPI nuclear staining. Each experiment was performed with at least 100 individual observations and performed 3 times to ensure reproducibility. C and D, Selective autophagy of M. tuberculosis is inhibited in cells lacking ULK1. C, Bar graph of the proportion of intracellular bacteria colocalized with LC3-positive puncta. Bars indicate mean values, with error bars indicating standards error of the mean. D, Deconvolved images of intracellular M. tuberculosis colocalized with autophagosomes in an EV cell and intracellular M. tuberculosis not colocalized with LC3 in an ULK1-KO cell. Green images indicate LC3, red images indicate mycobacteria, and blue images indicate DAPI nuclear staining. To calculate statistics, at least 100 cells were measured for each condition, and each experiment was performed 3 times to ensure reproducibility. *P < .05 by the Wilcoxon rank sum test.
Figure 2.
Differential responses in ULK1-deficient monocyte derived macrophages. U937 cells with either the ULK1 knockout (KO), the empty-vector CRISPR control (EV), or wild type (WT). A, Mycobacterium tuberculosis replication in ULK1-deficient monocyte-derived macrophages. KO, EV, and WT cells infected with luciferase-expressing M. tuberculosis at a multiplicity of infection (MOI) of 10. A separate set of wells was included with luciferase-expressing M. tuberculosis (mLux) in the absence of U937 cells as a control for background luciferase expression. Relative light units were measured at the indicated time points. Each point represents the mean of 6 separate wells. P = .004 by the Wilcoxon rank-sum test on days 3–7 for ULK1-deficient cells as compared to cells with EV or WT. B, After stimulation with PAM2/PAM3 (250 ng/mL each), lipopolysaccharide (LPS; 100 ng/mL), and M. tuberculosis whole-cell lysate (WCL; 25 µg/mL) for 24 hours, tumor necrosis factor (TNF) was measured by enzyme-linked immunosorbent assay (ELISA). ULK1-deficient cells produced significantly less TNF in response to PAM2/PAM3 (*P = .004 by the Student t test) and WCL (P < .001) as compared to cells with WT or EV. C, After stimulating the inflammasome with Bacillus thailandensis needle protein fused to lethal factor for cytoplasmic delivery (LF_Btn), interleukin 1β (IL-1β) secretion was measured by ELISA . D, After stimulation with PAM2/PAM3 (250 ng/mL, each), LPS (100 ng/mL), and WCL (25 µg/mL) for 24 hours, interferon β (IFN-β) was measured by reverse transcription–polymerase chain reaction. Linearized transcription values are expressed as fold change as compared to unstimulated cells. Graph includes merged data from 2 independent experiments.
To determine a mechanism for ULK1-dependent replication, we assessed whether ULK1-deficient U937 cells displayed different cytokine responses to stimulation conditions as compared to empty vector and wild-type control cells. ULK1-deficient cells produced significantly less TNF in response to PAM2 and PAM3 (P = .004 and P = .001) and M. tuberculosis whole-cell lysate (P < .001 and P < .02), compared with wild-type and empty vector cells, respectively (Figure 2B). However, TNF responses to LPS stimulation were not significantly different between ULK1-deficient cells and control cells. When we stimulated the inflammasome with an NLRC4 ligand, IL-1β production was not different when comparing ULK1-deficient cells to empty vector or wild-type cells (Figure 2C). As ULK1 negatively regulates IFN-β [39], we assessed IFN-β responses in our cell lines to various stimulation conditions. Compared with empty vector and wild-type cells, ULK1-deficient cells had similar changes in IFN-β responses as compared to unstimulated states (Figure 2D).
We next investigated the impact of ULK1-deficiency on selective autophagy and infected cells with mCherry-expressing M. tuberculosis (Figure 1). In comparison to control cells, ULK1-deficient cells had significantly fewer intracellular bacteria colocalized with LC3-positive puncta (Figure 1C and 1D).
DISCUSSION
The primary finding of our study is that ULK1 polymorphisms were associated with LTBI in Asian individuals, with a reduction of >80% in the LTBI risk for each copy of the minor allele. ULK1-deficient monocytes had increased M. tuberculosis replication, decreased TNF response to stimulation with TLR ligands and M. tuberculosis whole-cell lysate, and impaired autophagy. Furthermore, variation in rs12297124 was associated with ULK1 expression in esophageal muscularis mucosa cells. Previous murine studies demonstrated that autophagy pathway genes regulate M. tuberculosis–specific pathways in monocytes and influence tuberculosis susceptibility during in vivo infection studies [21, 40–42]. Autophagy is initiated by an ULK1-containing complex under regulation by mTOR and AMP kinase. ULK1 activation leads to phosphorylation of beclin-1, which ultimately results in autophagosome formation [43]. Although polymorphisms in IRGM, an autophagy gene, have been associated with tuberculosis in diverse populations [24–26], a study of 22 autophagy genes in an Indonesian population did not identify any associations with tuberculosis [32]. To our knowledge, ULK1 is only the second autophagy gene associated with tuberculosis in genetic studies and the first to be associated with LTBI [24, 25, 27].
The mechanism of how ULK1 variants regulate susceptibility to human M. tuberculosis infection is not known. Based on its well-described role in autophagy initiation, we speculated that human ULK1 deficiency leads to decreased autophagy and increased M. tuberculosis replication in monocytes. Our studies in ULK1-deficient monocytes support this model. However, additional mechanisms may be important, including ULK1 regulation of M. tuberculosis replication through effects on cell signaling. We showed decreased TNF responses, a key cytokine in control of M. tuberculosis infection, after stimulation of ULK1-deficient monocytes with M. tuberculosis whole-cell lysate, which is similar to previously reported TNF decreases in M. tuberculosis–exposed cells after 3MA-induced inhibition of autophagy [44, 45]. ULK1 phosphorylates STING [39] with negative regulation of IRF3-dependent IFN-β, which negatively regulates IL-1β, a cytokine that is protective in murine tuberculosis [46]. Autophagy has been shown to limit inflammasome activity, and inhibition of autophagy led to increased IL-1β secretion after infection with M. tuberculosis [44, 47]. In addition, ULK1 has been demonstrated to negatively regulate NLRP3 and caspase-1 activity with decreases in interleukin 18 expression [48]. Our data suggest that ULK1 mediates TLR and M. tuberculosis–induced TNF secretion, but not NLRC4-mediated IL-1β production. We currently do not know which ULK1 variants regulate gene function. Further insight into human ULK1 function will require fine-mapping and functional studies to discover which SNPs regulate ULK1 function and their effects on M. tuberculosis immunoregulation.
Our study has several limitations. Our small sample size for black participants may have limited our ability to detect small-to-moderate genetic relative risks in candidate genes. However, our use of controls who were exposed to infectious tuberculosis likely improved study power by reducing misclassification bias [49]. An additional limitation is that LTBI tests are imperfect and may lead to misclassification of study subjects. As with any candidate gene study, our findings should be replicated in a new population.
Infection with M. tuberculosis leads to a spectrum of outcomes that include sterilizing immunity, LTBI, subclinical disease, and symptomatic tuberculosis. Host genetic variation may impact tuberculosis pathogenesis at any stage in the spectrum. The majority of genetic studies of tuberculosis susceptibility has focused on the outcome of active tuberculosis and were unable to determine which steps in the tuberculosis spectrum were impacted. Given the orders of magnitude of difference in LTBI versus active tuberculosis prevalence, the specific genetic variations associated with susceptibility would be expected to differ between LTBI and active tuberculosis.
In summary, we found that a common ULK1 genetic variant was associated with protection against LTBI. To our knowledge, this is the first study to suggest a role for ULK1 in immunoregulation of M. tuberculosis. Our findings prompt further evaluation of this gene to validate our results and to further probe mechanistic explanations.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the staff from the TBCP, for their work; Rick Wells from the University of Washington, for his assistance; Dr Russell Vance, for providing the lethal factor and needle protein inflammasome stimulation system; and Dr Jeffrey Cox, for providing M. tuberculosis Erdman strain expressing lux.
D. J. H. and T. R. H. conceived and designed the study. D. J. H., A. D. G., J. A. S., M. N., and T. R. H. contributed to analyses and interpretation of analyses. All authors contributed to data acquisition and preparation and critical revision of the manuscript.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants 1K23AI85036 [to D. J. H.] and 1K24AI089794 [to T. R. H.]); the ATS Foundation and Merck, Inc. (to D. J. H.); the Firland Foundation (to D. J. H.); and the Burroughs Wellcome Foundation (to T. R. H.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Global tuberculosis report 2015. Geneva, Switzerland: WHO, 2015. http://www.who.int/tb/publications/global_report/en/. Accessed 2 February 2016. [Google Scholar]
- 2.Thye T, Vannberg FO, Wong SH et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 2010; 42:739–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisson-Dupuis S, Bustamante J, El-Baghdadi J et al. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol Rev 2015; 264:103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vannberg FO, Chapman SJ, Hill AV. Human genetic susceptibility to intracellular pathogens. Immunol Rev 2011; 240:105–16. [DOI] [PubMed] [Google Scholar]
- 5.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Ann Rev Immunol 2002; 20:581–620. [DOI] [PubMed] [Google Scholar]
- 6.Cobat A, Orlova M, Barrera LF, Schurr E. Host genomics and control of tuberculosis infection. Public Health Genomics 2013; 16:44–9. [DOI] [PubMed] [Google Scholar]
- 7.Cobat A, Gallant CJ, Simkin L et al. High heritability of antimycobacterial immunity in an area of hyperendemicity for tuberculosis disease. J Infect Dis 2010; 201:15–9. [DOI] [PubMed] [Google Scholar]
- 8.Jepson A, Fowler A, Banya W et al. Genetic regulation of acquired immune responses to antigens of Mycobacterium tuberculosis: a study of twins in West Africa. Infect Immun 2001; 69:3989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobat A, Poirier C, Hoal E et al. Tuberculin skin test negativity is under tight genetic control of chromosomal region 11p14–15 in settings with different tuberculosis endemicities. J Infect Dis 2015; 211:317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobat A, Gallant CJ, Simkin L et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med 2009; 206:2583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein CM, Zalwango S, Malone LL et al. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS One 2008; 3:e4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thye T, Browne EN, Chinbuah MA et al. IL10 haplotype associated with tuberculin skin test response but not with pulmonary TB. PLoS One 2009; 4:e5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zembrzuski VM, Basta PC, Callegari-Jacques SM et al. Cytokine genes are associated with tuberculin skin test response in a native Brazilian population. Tuberculosis (Edinb) 2010; 90:44–9. [DOI] [PubMed] [Google Scholar]
- 14.Randhawa AK, Shey MS, Keyser A et al. Association of human TLR1 and TLR6 deficiency with altered immune responses to BCG vaccination in South African infants. PLoS Pathog 2011; 7:e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berrington WR, Hawn TR. Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter? Immunol Rev 2007; 219:167–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abel L, El-Baghdadi J, Bousfiha AA, Casanova JL, Schurr E. Human genetics of tuberculosis: a long and winding road. Philos Trans R Soc Lond B Biol Sci 2014; 369:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davila S, Hibberd ML, Hari Dass R et al. Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis. PLoS Genet 2008; 4:e1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawn TR, Dunstan SJ, Thwaites GE et al. A polymorphism in Toll-interleukin 1 receptor domain containing adaptor protein is associated with susceptibility to meningeal tuberculosis. J Infect Dis 2006; 194:1127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khor CC, Chapman SJ, Vannberg FO et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet 2007; 39:523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manzanillo PS, Ayres JS, Watson RO et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013; 501:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar D, Nath L, Kamal MA et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 2010; 140:731–43. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753–66. [DOI] [PubMed] [Google Scholar]
- 23.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006; 313:1438–41. [DOI] [PubMed] [Google Scholar]
- 24.Intemann CD, Thye T, Niemann S et al. Autophagy gene variant IRGM -261 T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog 2009; 5:e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King KY, Lew JD, Ha NP et al. Polymorphic allele of human IRGM1 is associated with susceptibility to tuberculosis in African Americans. PLoS One 2011; 6:e16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Che N, Li S, Gao T et al. Identification of a novel IRGM promoter single nucleotide polymorphism associated with tuberculosis. Clin Chim Acta 2010; 411:1645–9. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, Li Q, Peng J et al. Association of autophagy-related IRGM polymorphisms with latent versus active tuberculosis infection in a Chinese population. Tuberculosis (Edinb) 2016; 97:47–51. [DOI] [PubMed] [Google Scholar]
- 28.Houben RM, Sumner T, Grant AD, White RG. Ability of preventive therapy to cure latent Mycobacterium tuberculosis infection in HIV-infected individuals in high-burden settings. Proc Natl Acad Sci USA 2014; 111:5325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mac Kenzie WR, Heilig CM, Bozeman L et al. Geographic differences in time to culture conversion in liquid media: Tuberculosis Trials Consortium study 28. Culture conversion is delayed in Africa. PLoS One 2011; 6:e18358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Targeted tuberculin testing and treatment of latent tuberculosis infection. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, July 1999. This is a Joint Statement of the American Thoracic Society (ATS) and the Centers for Disease Control and Prevention (CDC). This statement was endorsed by the Council of the Infectious Diseases Society of America. (IDSA), September 1999, and the sections of this statement. Am J Respir Crit Care Med 2000; 161:S221–47. [DOI] [PubMed] [Google Scholar]
- 31.Kosoy R, Nassir R, Tian C et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 2009; 30:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Songane M, Kleinnijenhuis J, Alisjahbana B et al. Polymorphisms in autophagy genes and susceptibility to tuberculosis. PLoS One 2012; 7:e41618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abecasis GR, Auton A, Brooks LD et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunetta KL. Genetic association studies. Circulation 2008; 118:96–101. [DOI] [PubMed] [Google Scholar]
- 35.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 2014; 42:W401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014; 11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med 2005; 37:360–3. [PubMed] [Google Scholar]
- 39.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013; 155:688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castillo EF, Dekonenko A, Arko-Mensah J et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A 2012; 109:E3168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakowski ET, Koster S, Portal Celhay C et al. Ubiquilin 1 Promotes IFN-gamma-Induced Xenophagy of Mycobacterium tuberculosis. PLoS Pathog 2015; 11:e1005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deretic V, Kimura T, Timmins G, Moseley P, Chauhan S, Mandell M. Immunologic manifestations of autophagy. J Clin Invest 2015; 125:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell RC, Tian Y, Yuan H et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinnijenhuis J, Oosting M, Plantinga TS et al. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response. Immunology 2011; 134:341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peral de Castro C, Jones SA, Ni Cheallaigh C et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol 2012; 189:4144–53. [DOI] [PubMed] [Google Scholar]
- 46.Novikov A, Cardone M, Thompson R et al. Mycobacterium tuberculosis triggers host type I IFN signaling to regulate IL-1beta production in human macrophages. J Immunol 2011; 187:2540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi CS, Shenderov K, Huang NN et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13:255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupfer C, Thomas PG, Anand PK et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol 2013; 14:480–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein CM. Genetic epidemiology of tuberculosis susceptibility: impact of study design. PLoS Pathog 2011; 7:e1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.