Abstract
Alzheimer's disease (AD) is a progressive neurodegenerative disorder that accounts for the most cases of dementia. AD affects more than 25 million people globally and is predicted to affect nearly one in 85 people worldwide by 2050. AD is characterized by the accumulation of dense plaques of β-amyloid peptide (Aβ) and neurofibrillary tangles of hyperphosphorylated tau that cause impairment in memory, cognition, and daily activities. Although early-onset AD has been linked to several mutations, reliable genetic markers for late-onset AD are lacking. Further, the diagnosis of AD biomarkers has its limitations and cannot detect early-stage AD. The identification of accurate, early, and non-invasive biomarkers for AD is, therefore, an unmet challenge. Recently, micro-RNAs (miRNAs) have emerged as a novel class of gene regulatory elements with conserved roles in development and disease. Recent discoveries have uncovered roles of miRNAs in several model organisms during aging and have identified potential miRNAs biomarkers of AD. Here we will discuss this emerging field of miRNAs associated with AD and prospects for the future.
Keywords: Alzheimer's disease, longevity, microRNAs, neurodegenerative diseases, stress
Introduction: Alzheimer's disease
Alzheimer's disease (AD) belongs in a class of incurable neurodegenerative diseases, such as Huntington's and Parkinson's, wherein proteotoxic aggregates accumulate with age, leading to progressive and widespread damage to the CNS and, ultimately, death (1). Although progress has been made in identifying genes associated with these devastating syndromes, the causes of these diseases are not well understood. AD is characterized by the formation of amyloid plaques, which are associated with the loss of neuronal function and neuronal cell death (2). The environmental or genetic factors that predis-pose towards the development of AD have been studied intensely for three decades and several genes have been linked with AD (3–6). Early-onset, familial AD is rare (estimated prevalence <1%) and has been associated with autosomal dominant mutations in the genes presenilin 1 (PSEN1), presenilin 2 (PSEN2), and the amyloid precursor protein gene (APP) (4). These mutations are believed to be responsible for the accumulation of the toxic Aβ amyloid peptide (Aβ1–42) which has a high propensity for aggregation and accumulates in the amyloid plaques that are a hallmark of AD. The Aβ1–42 peptide is generated by proteolytic cleavage of full-length APP by α, β, and γ-secretases (2, 4). The AD-associated mutations in PSEN1 and PSEN2 have been shown to impair the γ-secretase pathway and cause an increased production of Aβ1–42 (4). Similarly, at least 39 mutations in APP have been found which affect its processing and increase the production of Aβ1–42 (4). These findings support the “amyloid cascade hypothesis”, the idea that the incorrect processing of APP or clearance of Aβ1–42 is the key trigger of a cascade of events that lead to AD (4). While mutations in the APP, PSEN1, and PSEN2 genes are the most well-characterized examples of genes that are linked to familial early-onset AD, the major genetic determinant of late-onset AD has been identified as the ε4 allele of the apolipoprotein E (APOE) gene. The role of genetic vs. environmental risk factors in late-onset, sporadic AD are less well-understood but APOE ε4 is associated with a 5–20% overall increased risk in developing AD (5, –9). In support of the amyloid cascade hypothesis, APOE binds APP and affects the clearance of soluble Aβ1–42, but the pathogenic APOE ε4 allele appears to reduce the efficacy of this clearance mechanism (8–10). To highlight the complexity in the molecular pathology of AD, mutations of APP, PSEN1, and PSEN2 account for about 5–10% of early-onset AD while the APOE ε4 allele increases the risk of both early-onset as well as late-onset AD but is not sufficient to cause disease (6). Indeed, while 40–65% AD patients carry the APOE ε4 allele, 20–25% of the general population also carry one or both APOE ε4 alleles (6). Therefore, many studies have tried to identify novel genetic markers of AD and to date at least 21 additional genetic risk loci have been identified (6). In further support of the amyloid cascade hypothesis, Down Syndrome patients, which have trisomy of chromosome 21, where APP is located, develop Aβ plaques early in life and are at a higher risk of developing AD (4, 11).
Beyond genetics, research on AD has also focused on diagnostic approaches by neuropathology and neuro-imaging. Neuropathology aims at a definitive diagnosis of AD but is complicated by the fact that most patients with neuropathological disease show concomitant cerebrovascular pathology and significant overlap in AD and Lewy body dementia (LBD) pathology. Moreover, only few patients with definite AD show pathology exclusively associated with AD (4). Neuroimaging, computed tomography (CT), and magnetic resonance imaging (MRI) also help in the diagnosis of AD by excluding other causes of dementia, such as brain tumor and subdural hematoma (4). CT and MRI identify cerebral atrophy, which is visualized as enlarged ventricles and cortical sulci. However, the great overlap with normal aging and other dementias limit their diagnostic value. However, neuroimaging detects cerebral infarcts and white matter lesions, which are valuable to identify vascular dementia or mixed dementia (4). Neuroimaging in AD can be carried out by molecular imaging techniques. The main molecular imaging techniques used for imaging in dementia in humans are positron emission tomography (PET) and single photon emission computed tomography (SPECT) (12). The two main types of PET scanning have been used in AD diagnostics – FDG-PET, which uses the glucose analog 2-deoxy-2-[18F] fluoro-D-glucose (FDG) to measure brain glucose metabolism, and amyloid-PET, which measures Aβ accumulation in the brain (12). FDG-PET takes advantage of the observation that AD patients frequently present with abnormal glucose metabolism. As glucose is the main source of energy in the brain, reduced uptake of FDG (hypometabolism) is indicative of neuronal dysfunction and is associated with AD (12). A pattern of hypometabolism has characteristic spatial and temporal signatures with reduced FDG in the parieto-temporal, frontal, and posterior cingulate cortices in early AD patients. FDG-PET has high sensitivity (~90%) for the diagnosis of early AD but it has low specificity (71–73%) in differentiating AD from other dementia (12). For amyloid-PET, numerous 18F-labeled tracers have been tested to measure fibrillary Aβ present in neuritic amyloid plaques by PET. The 18F-labeled amyloid imaging agents that are approved by the Food and Drug Administration (FDA) to evaluate patients with cognitive decline are florbetapir, flutemetamol, and florbetaben (12, 13). Florbetaben PET has a high predictive value, sensitivity, and specificity that help in reliable detection of amyloid pathology (13). Florbetapir and flutemetamol also have high median sensitivities and specificities for the detection of Aβ plaques (12). Although amyloid-PET holds promise for differential diagnosis of dementia, technical considerations are crucial for proper analysis including proper consideration of brain atrophy, reader error, and consistent timing of sample imaging after tracer injection (12). In addition, both FDG-PET and amyloid-PET are expensive techniques. Although amyloid-PET gives spatial information about the localization of Aβ in the brain, the technique is partially redundant with measurements of Aβ1–42 from cerebrospinal fluid (CSF). Finally, Aβ1–42 expression increases with normal aging and overlaps with other dementias, such as Lewy body disease – therefore, a positive amyloid-PET result is not sufficient for AD diagnosis and other biomarkers and diagnostic tests are of critical importance. According to the National Institute of Neurological and Communicative Diseases and Stroke and the Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) criteria for the clinical diagnosis of AD mainly depends on the exclusion of other dementias which leads to low diagnostic accuracy especially in primary care settings and in patients with mild AD. These criteria also fail to address concomitant cerebrovascular disease – therefore, it is not always clear what extent of infarcts, lacunas, or white matter changes are considered normal before assessing it as a mixed dementia. Moreover, AD cannot be diagnosed until patients exhibit dementia (4). Based on the clinical suspicion or differential diagnosis from other neurological disorders, AD can be confirmed by analysis of total tau (t-tau), phosphorylated tau (p-tau), and Aβ42 in CSF. These difficulties in early diagnosis of AD emphasize the need for the identification of novel biomarkers for AD. The identification of blood-based biomarkers for AD is an emerging and promising new field. The discovery that miRNAs are expressed in the CSF and that miRNAs can affect longevity and stress resistance in several species suggests the possibility that miRNAs may also have important roles in diseases of aging such as AD. In this review, we will discuss the functions of miRNAs in aging and stress resistance, and consider their possible useful roles as biomarkers of AD.
MicroRNAs
MicroRNAs (miRNAs) are small, endogenous, non-protein coding RNAs, originally discovered in the nematode Caenorhabditis elegans (C. elegans) and found to be present in all plants and animals. miRNAs are synthesized either from introns or from the transcription of miRNA genes present at different loci in the genome. miRNA genes encode for long RNAs with hairpin structure, which are then processed by various RNase III enzymes (Drosha and Dicer) to form small miRNA duplexes of approximately 22 nucleotides in length. One of the strands of miRNA duplex binds to argonaute protein and forms the RNA-induced silencing complex (RISC). The RISC binds a target messenger RNA (mRNA) through miRNA:mRNA complementarity, whereby the miRNA pairs with the target mRNA either to cause mRNA cleavage and/or to induce translational repression. Full complementarity between a miRNA and the ORF of a target mRNA is more common in plants, whereas in animals the more typical mechanism involves translational repression by partial complementarity between a miRNA and the 3′-untranslated region (3′-UTR) of a target mRNA. The 5′ region of a miRNA, or the “seed region”, encompassing nucleotides 2–8, is essential for target specificity and is the most conserved region of a miRNA. Perfect or near perfect complementarity between a miRNA seed region and its target mRNA is both necessary and sufficient for target recognition. Mismatches in this core 5′ seed region of a miRNA lead to the loss of miRNA function due to compromised binding to its target mRNA. The loss of miRNA function due to mutations in a miRNA's seed sequence generally cause an upregulation of the target mRNA levels and/or target protein levels, reflecting the reduced levels of target mRNA cleavage and reduced translational repression. A seed region's relatively short length (6–8 nucleotides) is similar to the size of a typical transcription factor's binding site and suggests that miRNAs may have thousands of potential target mRNAs (14, 15).
miRNA functions in aging
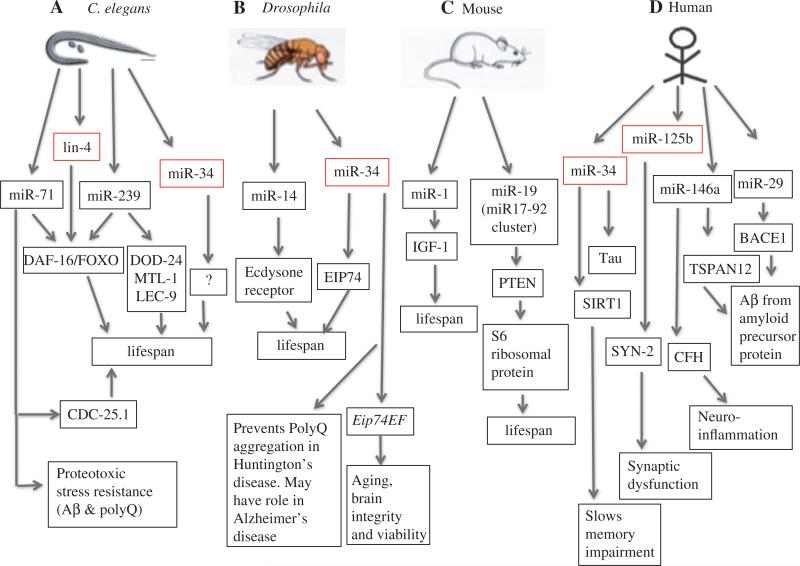
Just as miRNAs were first identified in C. elegans, the discovery of miRNAs that have adult-specific effects on longevity and stress resistance was also first made in nematodes. Early investigations into genetic pathways of aging lead to the discovery in the mid-1990s that genes such as age-1 and daf-2, conserved members of the IGF-1/insulin-like pathway (IIS), can dramatically affect C. elegans life span (16). In addition to the IIS pathway, many other pathways and environmental alterations have since been found to alter life span across phyla, such as genes in the TOR pathway, sirtuins, AMPK, cell respiratory genes, dietary/caloric restriction, and low-level stress (hormesis) (17). The genetic landscape of pathways that affect aging was further expanded by the discovery, in C. elegans, that mutations to lin-4, the founding member of the miRNA family, could have dramatic effects on C. elegans life span (18). Loss-of-function mutations of the lin-4 result in reduced life span while overexpression of lin-4 increase C. elegans life span and stress resistance (18). Today, several aging-associated miRNAs have been identified in C. elegans, Drosophila, and in vertebrates (Figure 1). A survey of miRNA genes with adult-specific functions identified four additional miRNAs – miR-71, miR-238, miR-246, and miR-239 – that function to alter C. elegans longevity as well as several novel miRNAs which appear to be expressed only during adulthood (30). Unlike lin-4, which is essential for proper development, most of these new aging-associated miRNAs had no previously identified functions (30). Both miR-71 and mR-239 mediate life span at least partially through the conserved IGF-1/Insulin pathway – miR-239 positively regulates expression of AGE-1/PDK-1 while miR-71 downregulates PDK-1 during C. elegans adulthood (30). Interestingly, miR-71 has also been shown to interact with the DNA damage response pathway by negatively regulating CDC-25.1 during adulthood (30). Importantly, these aging-associated miRNAs also have roles in the stress response of C. elegans. The short-lived miR-71, mir-238, miR-246 deletion mutants showed increased sensitivity to both heat and oxidative stress, while the long-lived miR-239 deletion mutants exhibited resistance to both heat and oxidative stress (30). Given that daf-2 in the IIS pathway has been shown to mediate the response to Aβ toxicity in C. elegans (26) and given the role of these aging-associated miRNAs in the stress response of C. elegans, it is possible that these miRNAs may also play vital roles in organismal response of aging-associated neurodegenerative diseases such as AD, which will be discussed further below.
Figure 1.
Association of miRNAs with aging and age-related diseases in different model organisms.
Model of the association between selected miRNAs, predicted targets, and phenotypes on life span or age-related diseases. The arrows denote functional or expression relationships that have identified between miRNAs and their targets or with specific phenotypes on life span or age-associated disease states. Note that the arrows only specify association but not the type of regulation (i.e. positive vs. negative regulation). This list is not exhaustive – see text and Table 1 for further details. Conserved miRNAs with already confirmed cross-species function in aging or age-related diseases are offset and highlighted in red.
The discovery that certain miRNAs affect life span in C. elegans and the abundance of miRNAs of unknown function across higher eukaryotes has led researchers to investigate the roles of miRNAs during adulthood in other species. Recent research in Drosophila has demonstrated functional roles for several miRNAs, including miR-8, miR-14, and miR-34 in adult life span. In Drosophila, the brain-expressed miRNAs, miR-14 and miR-34, are down-regulated and upregulated with age, respectively (31, 32). Interestingly, miR-34 was also identified as one of the five upregulated miRNAs during C. elegans aging where it appears to antagonize longevity (30, 33). In Drosophila, loss-of-function mutants of both miR-14 and miR-34 display shorter life span via ecdysone signaling. miR-14 affects life span through upregulation of the ecdysone receptor and miR-34 acts through EIP74, an ecdysone-induced transcription factor. The elevated ecdysone pathway in these mir-34 mutants causes neurodegeneration and early death (31, 32). miR-8 is also associated with modulating life span in flies through its target, the U-shaped gene, which inhibits PI3K in the IIS pathway (34). Loss of miR-8 also increases atrophin levels, leading to early-onset cell death in the brain and behavioral defects (32, 35). The fact that miR-8 absence, just like miR-34, leads to neurodegeneration and that it regulates IIS genes, implicates miR-8 as an important genetic modulator of neuronal homeostasis. Notably, miR-8, miR-14, and miR-34 are all highly conserved, suggesting important and possibly conserved roles, across evolution.
In mice, several miRNAs have also recently been associated with roles in regulating life span. It has been observed that several miRNAs such as miR-27a, miR-470, miR-669b, and miR-681, which function through insulin/IGF-signaling pathway, are upregulated in long-lived Ames dwarf mice (19). Studies of mouse brain have identified four upregulated miRNAs in aging mouse brain – miR-26b, let-7f, miR-17, and miR-10b (20). Interestingly, many more miRNAs (a total of 71 miRNAs) were downregulated with aging in mouse brain – a pattern that has also been noted during aging in other species (27, 30). Functionally, the increased expression of at least one miRNA – miR-1- in multiple tissues has been shown to cause premature aging in a mouse model of Hutchinson-Gilford progeria syndrome (32). miR-1 is known to be upregulated in both progeroid mice and in human Hutchinson-Gilford progeria cells (19). In progeroid mice, upregulated miR-1 suppresses liver IGF-1 synthesis even in the presence of increased levels of circulating growth hormone (19). The high levels of circulating growth hormone can contribute to premature aging of progeroid mice (19). The miR-17-92 cluster, represented by miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1, is conserved among vertebrates, and has been associated with roles in cell cycle, tumorigenesis, and aging. Humans contain two paralogues of the miR17-92 cluster – the miR-106a/363 and miR-106b/25 clusters (28). The miR17-92 cluster has several targets: miR-19a and miR19b-1 bind to the 3′UTR and regulate PTEN function which is involved in cell death. miR-20a targets the 3′UTR of both E2F2 and E2F3 while miR-17 and miR-20a regulate E2F1 expression. This suggests that miR-20a and miR-17 affect the cell cycle as all three E2Fs are involved in cell-cycle regulation (28). Members of the miR17-92 cluster also function in TGβ signaling. miR-17 and miR-20a directly target the TGβ receptor-II, while miR-18a regulates two members of TGβ signaling pathway – Smad2 and Smad4. As the TGβ signaling pathway mediates longevity in C. elegans and other organisms this offers another possible link between the miR17-92 cluster and aging-associated roles in the mouse. Finally, in several human replicative and organismal aging models, it has been shown that the expression of the members of the miR17-92 cluster is downregulated during aging and senescence, although the mechanism for the downregulation of this miRNA cluster is not yet known (36).
Interestingly, miR-19 also targets and represses PTEN, as observed in mouse primary B-cell culture (37). By downregulating PTEN, miR-19 activates PI3K pathway, thus promoting the phosphorylation of AKT and also the phosphorylation of the ribosomal S6 protein. This last effect is due to the activation of the mTOR (mammalian target of rapamycin) pathway, where activated AKT/mTOR pathway promotes cell survival (37). Notably, the AKT/mTOR pathway is also associated with regulating life span in various model organisms (17). The proper regulation of miR-19 expression might, therefore, be critical for maintenance of homeostasis in various organism or the cell response to stress (36).
Function of miRNAs in neurodegeneration
Previous studies on functions of the IIS pathway in a C. elegans model of Aβ1–42 toxicity have demonstrated that several genes of the IIS pathway, such as daf-2 and daf-16 (FOXO) have profound effects on the toxic effects of Aβ1–42 on C. elegans (26). Transgenic C. elegans animals that express the human form of Aβ1–42 show age-dependent paralysis due to the accumulation of proteotoxic Aβ1–42 aggregates (26). Knocking out daf-16 causes the rate of paralysis due to Aβ1–42 to accelerate, while knocking down daf-2 significantly retards paralysis (26). These results are consistent with previous observations that daf-2 and daf-16 antagonize and promote, stress resistance in C. elegans, respectively. Further, these results demonstrate that previously identified genetic pathways of stress resistance (such as IIS) are relevant to cellular responses to proteotoxicity such as those associated with various diseases of aging. As several miRNAs, such as lin-4, miR-71, and miR-239, have been shown to genetically interact with the IIS pathway and to function in the regulation of pathways of aging and stress resistance (18, 30), this suggests a possible link between miRNAs that function through IIS pathway and neurodegenerative diseases like AD (26).
In Drosophila, the conserved miRNAs miR-34 has been shown to be essential for normal aging, as described above (38). Interestingly, miR-34 has essential roles in the maintenance of normal neuronal health in Drosophila brain during aging, indicating that miR-34 provides a link between aging and neurodegeneration in Drosophila (38). The neuronal brain expression of miR-34 increases during fly aging, in concordance with the upregulation of miR-34 in C. elegans during aging (30, 38). Studies have shown that miR-34 expression is low during development in flies, increases in young adult and becomes further upregulated with aging – similar to the temporal expression of miR-34 in C. elegans. Furthermore, loss of miR-34 causes no developmental abnormalities, but significantly shortens adult life span and causes aging-associated neurodegeneration (38). Null mir-34 flies (mir-34−/−) exhibit brain lesions, stress sensitivity, loss of brain integrity, accelerated brain aging, and various neurodegenerative phenotypes including locomotion defects. In an effort to identify the molecular mechanisms that mediate miR-34 function during adulthood in flies, researchers have identified Eip74EF as a candidate miR-34 target in Drosophila. The Eip74EF gene encodes two protein isoforms – E74A and E74B (referred as the E74A and E74B genes, respectively), both of which share the same 3′UTR and which are involved in the steroid hormone signaling pathways. Mutation of miR-34 predicted-binding sites in the 3′UTR of Eip74EF has confirmed that miR-34 negatively regulates Eip74EF by binding to its 3′UTR (38). In mir-34−/− flies E74A protein is upregulated leading to a negative impact on normal aging (38). Hence, adult-onset expression of miR-34 in adults may function at least in part to suppress E74A expression and thereby prevent the deleterious activity of E74A on brain integrity and viability in adults (38). Importantly, mir-34−/− mutant animals showed defects in protein misfolding as evidenced by an increase in the levels of inclusion bodies (38). As protein misfolding is a hallmark of many human neurodegenerative diseases, researchers tested whether miR-34 overexpression might reduce disease-associated protein misfolding by using animals that express PolyQ, a model for Huntington's disease in fly. Indeed, animals that overexpress miR-34 were shown to be protected from PolyQ aggregation, inclusion formation, and neuronal loss association with PolyQ expression (38). As AD is characterized by an accumulation of insoluble Aβ1–42 aggregates, it will be interesting to test if miR-34 overexpression will similarly protect from the toxic effects of Aβ1–42 in fly. Notably, miR-34 expression increases with age in both C. elegans and humans (30, 39) and it is misregulated in degenerative diseases in humans (40, 41).
Human disease: miRNAs in AD
Previous studies have demonstrated that specific miRNAs are expressed in the central nervous system (CNS), where they play vital roles in the molecular control of development and aging of the brain, neuronal development, and are associated with neurodegenerative diseases such as AD (22, 24, 25). miRNAs have been shown to regulate AD-related proteins in the brain, and miRNAs are stable in CSF and blood as they travel through biofluids within exosomes. Furthermore, miRNA levels in biofluids can be easily detected by standard molecular biology techniques such as quantitative PCR (qPCR). Hence, miRNAs are being considered as potential novel biomarkers in AD (24, 42, 43).
Schipper and colleagues first studied miRNAs as biomarkers in AD by microarray analysis and found that several miRNAs, such as miR-34a and miR-181b, were upregulated in peripheral blood mononuclear cells (PBMCs) in AD patients (44, 45). Numerous studies on the expression profiling of miRNAs in AD have been carried out in recent years from various biological sources such as peripheral blood, plasma, CSF, serum, brain tissue-derived extracellular fluid (ECF) using various techniques such as next generation sequencing, nanostring technology, miRNA PCR array, and qRT-PCR. These efforts, led by a variety of labs, have led to the identification of a number of dysregulated miRNAs in AD (25), which we discuss below.
miRNA profiling of AD
In a study of human brain tissue of AD patients, the expression of miR-9 and miR-128 was found to be elevated in the hippocampus of AD patients (46). Another study showed that miR-9, -26a, -132, and -146b expression is downregulated and miR-27, -29, -30, -34, and -125b were upregulated in the frontal gyrus of AD patients (47). An examination of the cortex of sporadic AD patients showed a downregulation of several miRNAs such as miR-181c, -15a, -9, -101, -29b, -19b, and -106b (48). Notably, miR-9 was shown as down-regulated here whereas it was shown to be upregulated in the hippocampus of AD patients (46–48). The relevance of this miRNA will be further explored below. Finally, measurements in human CSF have shown that miR-146b and miR-27a-3p are downregulated (47, 49) and miR-30 family members are upregulated in AD patients (25).
Below we discuss the relevance of miRNAs that were found to be dysregulated in the brain tissue of AD patients and discuss their possible involvement in the pathogenesis of AD.
miR-9
miR-9 is encoded by three different genes and expressed in the neocortex, retina, and fetal hippocampus. miR-9 is a retinoic acid-inducible miRNA that is regulated by NF-kB (25). It is associated with neurogenesis, morphogenesis, developmental patterning, brain cell proliferation, and glioblastoma (25). Other expression studies have demonstrated the altered expression of miR-9 in AD brains: Lukiw and colleagues showed an upregulation of miR-9 in the temporal cortex and hippocampus of AD patients (23, 46, 50). However, Hebert and coworkers reported a down-regulation of miR-9 in the cortex of sporadic AD patients (48). In a mouse model of AD (APPswe/PSΔE9), miR-9 expression was noted to be downregulated in the hippocampus of 6-month-old mice, but not 3-month-old mice (51). The predicted targets of miR-9 are fibroblast growth factor receptor 1 (FGFR1), CDK6, caudal-type home-obox 2 (CDX2), NFKB1, sirtuin 1 (SIRT1), and RE1 silence transcription factor (REST). miR-9 acts as a guardian of neuronal cells by preventing accumulation of lamin and toxic progerin in these cells, thereby offering Hutchinson-Gilford progeria syndrome patients some protection from neurodegeneration (21, 22). miR-9 also negatively regulates nuclear receptor TLX or nuclear receptor subfamily 2 group E member 1 (NR2E1), to promote neural differentiation and prevent stem cell proliferation (22, 24, 52).
miR-29
The expression of both miR29a and miR29b has been reported as elevated in CSF but decreased in the cortex of AD patients (25). The altered expression of miR-29 in AD patients is particularly interesting as miR-29a targets beta-secretase 1 (BACE1), an enzyme which promotes the formation of Aβ from APP. Hence, the decrease in miR-29 expression in AD patients is associated with a concomitant increase of BACE1 in these patients, and therefore elevated levels of Aβ (25). The apparent discrepancy in measurements of miR-29 from different studies may stem from the fact that miR-29a levels in CSF have been shown to be strongly affected by blood contamination. This is a potential drawback to the use of this miRNA in that its levels are strongly correlated to the number of blood cells present in the CSF. Thus, its vulnerability to blood contamination warrants caution in its use as a biomarker in CSF especially for samples collected after a traumatic lumbar puncture. While consideration of these technical challenges will be essential in how miR-29a expression is interpreted, miR-29a remains a promising biomarker for AD (43).
miR-34
In humans, the miR-34 family consists of three members – miR-34a, miR-34b, and miR-34c. MiR-34a is transcribed from human chromosome 1p36, whereas miR-34b and miR-34c are co-transcribed from human chromosome 11q23. These miRNAs have different expression profiles, where miR-34a is mainly expressed in the brain and the highest expression of both miR-34b and miR-34c is observed in the lungs. As stated above, miR-34 is known to regulate aging in both C. elegans and Drosophila. Loss-of-function mutations of miR-34 increase life span in C. elegans by promoting autophagic flux (33). In Drosophila, the miR-34c is upregulated with age. Loss-of-function mutants of miR-34 in flies display shorter life span via ecdysone signaling and causes aging-associated neurode-generation (38). miR-34 is also a tumor suppressor miRNA that is associated with p53 expression in mouse and human (53). miR-34a and p53 activate each other, where miR-34a represses many p53 inhibitor genes and p53 induces expression of miR-34a (53). miR-34a is associated with p53-dependent processes, such as somatic cell reprogramming or inhibition of epithelial-mesenchymal transition (EMT) and metastasis, but not in p53-mediated stress response (53). Consistent with its role in aging-associated neurodegeneration in flies, miR-34 has also been associated with AD: (i) miR-34a is upregulated in brain tissue and blood mononuclear cells of AD patients, (ii) miR-34a regulates neuronal differentiation and neurite outgrowth, and (iii) miR-34a can inhibit the expression of human tau by binding to its long 3′UTR isoform (29). In addition, miR-34c also appears to be capable of binding tau 3′UTR and suppress its expression in gastric cancer cells (29). Further, and consistent with its upregulation during aging in flies (38), the miR-34c expression also increases in the aging mouse hippocampus, in a mouse model for AD and in the hippocampus of human AD patients (40). These results demonstrate a possible conserved functional role for miR-34c in neuronal health during aging. In mouse models of AD, miR-34c appears to have a negative effect on memory formation by suppressing hippocampal SIRT1 levels (40). Indeed, and as a preliminary exploration of possible miRNA therapeutic approaches for AD, injection of a miR-34 inhibitor in a mouse model of AD both restored SIRT1 levels as well as improved memory function in these mice (40). Together, these data suggest that miR-34 is a promising biomarker in AD and that miR-34c is a candidate target for AD therapeutics (40).
miR-106b
miR-106b belongs to the miR-20a microRNA family. miR-106b is highly expressed in human brain and its expression significantly decreases in AD patients (54). Studies have not determined whether decrease in miR-106b expression can be beneficial or detrimental to AD patients for two reasons: i) miR-106b decreases ATP-binding cassette transporter A1 (ABCA1) levels, which leads to impaired cholesterol efflux in neuronal cells and increase in Aβ levels (55) and ii) miR-106b suppresses the expression of APP, which may prevent Aβ accumulation (55). miR-106b is involved in neuronal differentiation and targets genes that control the cell cycle such as cyclin-dependent kinase inhibitor 1A (p21) and the E2F transcription factor (25, 55). Therefore, miR-106b might have an important role in the pathogenesis of AD.
miR-125b
miR-125b is an extensively studied miRNA and is found in human brain and retina. miR-125 is a particularly interesting candidate of AD-associated miRNAs as it is the human ortholog of lin-4 in C. elegans – the first miRNA shown to regulate organismal life span (18). In C. elegans, lin-4 promotes longevity via downregulation of its target, the transcription factor LIN-14, and concurrent derepression of the conserved pro-longevity and -stress resistance transcription factors DAF-16/FOXO and HSF-1 (18). Therefore, it is of particular interest that miR-125b expression is shown to be in upregulated in AD patients (50). In mammals, miR-125 has been shown to have an important role in the mammalian neuronal development and in neuronal differentiation during retinal development (52). Its regulatory role on the nervous system is partially due to the fact that miRNA-125b downregulates target genes that need to be turned off for proper neuronal development such as itchy E3 ubiquitin protein ligase, and diacylglycerol O-acyl-transferase 1 (22). It is known that elevated miR-125b downregulates synaptic protein synapsin-2 (SYN-2) and 15-lipoxygenase (15-LOX) thereby potentially causing pathogenic effects of AD such as synaptic and neurotrophic deficits and astrogliosis (50). As AD is associated with the innate immune system and the inflammatory response, it is also likely relevant that miR-125b appears to regulate factors involved in the innate immune system and in the pro-inflammatory response, such as complement factor-H protein (CFH) and inter-feron regulatory factor 4 (IRF4). miR-125b has predicted tandem binding sites on the 3′UTR of human CFH mRNA and it represses CFH expression in human primary astroglial cells and it also downregulates interferon regulatory factor 4 (IRF4) to activate immune response (52).
miR-146a
Another miRNA with possible links to AD via the inflammatory response is miR-146a which is a proinflammatory miRNA that is regulated by NF-kB (50). Previous studies have shown that miR-146a is highly upregulated in AD brain specifically in the temporal cortex and in the hippocampus (24, 50). The upregulated miR-146a in AD brain potentially targets inflammation-related and membrane-associated mRNAs, such as interleukin-1 associated kinase-1 (IRAK-1), a component of the innate immune response (50). Just like miR-125b, elevated miR-146a is known to have multiple binding sites in CFH mRNA 3′UTR and thus also potentially downregulates CFH (22, 50, 52). As a candidate AD biomarker, miR-146a is detectable in CSF but care must be taken in interpreting these values, because just like miR-29a, miR-146a levels are also influenced by blood contamination (43).
miR-155
A final miRNA that is associated with the inflammatory response is miR-155 which is a proinflammatory miRNA expressed in human neocortex and retina and is similarly regulated by NF-kB, as well as cytokines (52, 56). Studies have demonstrated that miR-155 is abundantly expressed in AD ECF and CSF (56). miR-155 is known to have binding sites in human CFH mRNA 3′UTR; miR-155 binding sites in CFH 3′UTR are, in fact, overlapping with miR-146a binding sites, which may be due to their related ribonucleotide sequence. miRNA-155 and miR146a are known to contribute to altered innate immune responses and inflammatory neuropathology (52, 56). Studies have shown that miR-155 regulates functions of T-cells during inflammation, where proper regulation of various T-cells can reduce AD-related pathologies. Therefore, not only is miR-155 a possibly valuable candidate AD biomarkers but a possible therapeutic target for AD (57).
In addition to the above-described miRNAs, several other miRNAs are associated with AD such as miR-107, miR-124, miR-132, miR-137, and miR-148a that are involved in tau phosphorylation, neuronal differentiation, APP processing, neuroinflammation, and cell cycle (22, 24).
RNA-based therapeutics in AD
As discussed here, miRNAs are an emerging class of bio-markers for AD due to their high abundance in the brain, CSF, and ECF. Additionally, the differential expression of several miRNAs in AD patients, and the identification of several miRNAs in model organisms with functions in stress resistance and neurodegeneration highlight likely critical roles for miRNAs in disease progression and possible new targets for therapeutics. Aberrant miRNA expression has been studied and analyzed using animal models of neurodegenerative disorders and, given the conservation of many miRNAs in pathological pathways, the results obtained from studies in model organisms are expected to inform on the pattern of expression and function of homologous miRNAs in human tissue (44, 58, 59). Further, the study of conserved genes and pathways that are regulated by AD-associated miRNAs will help in expanding the list of potential non-miRNA gene biomarkers. The study of dysregulated miRNAs in neuro-degenerative diseases and their associated targets may also aid in RNA-based therapeutics. As described above, one highly attractive therapeutic target is miR-34, which has been shown to be upregulated during aging in C. elegans, Drosophila, and in vertebrates (30, 38, 40). In addition, one specific miR-34 isoform, miR-34c, is associated with neurodegeneration in Drosophila and with memory deficits in a mouse model of AD (38, 40). In a proof-of-concept for a possible therapeutic approach for AD-associated miRNAs, researchers successfully reversed memory deficits in AD mice by injecting these animals with miR-34 seed inhibitors (40). While this experiment does not address challenges such as the specificity of miRNA silencing (for example, how to specifically target only one miR-34 isoform instead of all miR-34 family members) and delivery difficulties, it clearly establishes a proof-of-principle for approaches to possible therapeutics involving AD-associated miRNAs. Importantly, these approaches will benefit from similar approaches being tested in therapeutics of cancer-associated miRNAs (60). The progress of RNA-based therapies is more evident in hereditary neurodegenerative diseases compared to sporadic neurodegenerative disorders. In AD, RNA-based silencing of the amyloid and BACE1 pathways showed improvement in disease phenotypes (25). Miller et al. (61) designed small interfering RNAs (siRNAs) mainly against tau and APP because of their important role in the pathogenesis of inherited and sporadic AD. The tau protein is a key component of neurofibrillary tangles in the AD brain and cleavage of APP forms Aβ, the main constituent of senile plaques in AD brain. Both neurofibrillary tangles and senile plaques are hallmarks of AD. Researchers have reported that they have developed siRNAs that can be effective in silencing wild-type tau and APP and also mutant APP (Swedish double mutation APP/APPsw) and a tau mutant (V337M) associated with frontotemporal dementia with Parkinsonism linked to chromosome 17/FTDP-17. However, this RNAi therapy needs in vivo testing using animal models of AD. Also, the main challenge of siRNA therapy is the delivery of siRNA to the appropriate neurons. This can be overcome by the development of better expression plasmids, but the long-term safety of chronically co-opting the RNAi pathway to target genes remains uncertain. The in vivo RNAi studies using animal models of AD might provide answers to these questions (61). Another study has reported that RNAi silencing of APP adaptor proteins such as ShcA (SHC1), ShcC (SHC3), and Fe65 (APBB1) have effects on APP processing and Aβ production (62). These APP adaptor proteins bind through their phospho-tyrosine-binding domains with the conserved YENPTY motif in the APP-C terminus. APPC-terminal fragments – APP-C99 and APP-C83 are formed by cleavage of APP by BACE and α-secretase. These C-terminal fragments are cleaved further to form Aβ and p3 by γ-secretase. Aβ is an important component of senile plaques in AD brain and p3 promotes apoptosis. Studies have shown that the RNAi silencing of ShcC decreased APP C-terminal fragments and Aβ levels in H4 human neuroglioma cells overexpressing full-length APP (H4-FL-APP cells), but not in cells expressing APP-C99 (H4-APPC99 cells) (62). RNAi silencing of ShcC also reduced BACE levels in H4-FL-APP cells (62). While RNAi silencing of ShcA did not have an effect on Aβ levels in H4-FL-APP Cells, RNAi silencing of Fe65 did increase APP-CTF levels and decreased Aβ levels in H4-FL-APP cells. These results strongly suggest that pharmacologically blocking the interaction of APP with Fe65, ShcC, and other APP adaptor proteins might be a novel therapeutic approach for AD (62). Indeed, in a study of in vivo RNAi silencing, researchers used lentiviral vectors to deliver BACE1 siRNA in APP transgenic mice and observed a decrease in amyloid production and in neurodegeneration (63). This promising result, and others listed here, suggests that miRNAs, their targets, and RNAi-based technologies may hold promise as new avenues of treatment for AD.
Conclusion
miRNAs have significant roles in aging and stress resistance and are emerging as important in neurodegenerative diseases such as AD. miRNAs are abundant in brain and highly stable in biofluids and regulate several pathways that influence the onset and progression of neurodegenerative diseases. Hence, miRNAs serve as promising new biomarkers or as a base for future treatment of AD. Future studies will help to better understand molecular mechanisms behind the regulatory role of miRNAs in neurode-generative diseases, to validate their role as biomarkers and to develop novel pharmacological strategies for better clinical management of AD.
Table 1.
Summary on mRNA targets of AD-associated miRNAs.
| miRNA | Upregulated target(s) | Downregulated target(s) | Description | References |
|---|---|---|---|---|
| miR-34 | Tau, SIRT1, p53 inhibitor genes | Upregulated miR-34 family members in AD: – Enhanced miR-34a in AD inhibits the expression of tau – Increased expression of miR-34c suppresses SIRT1 and causes memory impairment |
(19–21) | |
| miR-125b | CDKN2A, SYN-2 & 15-LOX enzyme, IRF4, CFH, adapter-related protein complex 1, mu 1 subunit, itchy E3 ubiquitin protein ligase, and diacylglycerol O-acyl- transferase 1 | Upregulated miR-125b in AD: – Reduces cyclin-dependent kinase inhibitor 2A (CDKN2A), which is a negative regulator of astroglial cell growth, thus causing glial cell proliferation – Downregulates synaptic protein SYN-2 that is linked with the cytoplasmic surface of synaptic vesicles, which leads to synaptic failure – Suppresses 15-LOX, an enzyme required to convert omega-3 fatty acid DHA into the potent docosanoid neuroprotectin D1 (NPD1), resulting in lack of NPD1, and causing neurotrophic failure |
(22, 23) | |
| miR-9 | CFH, NR2E1 | Up-regulated miR-9 in AD: – Downregulates CFH, a key negative regulator of the innate immune and complement system, leading to neuroinflammation |
(22–24) | |
| miR-155 | CFH | Upregulated miR-155 in AD: – Downregulates CFH, a key negative regulator of the innate immune and complement system, leading to neuroinflammation |
(23) | |
| miR-146a | IRAK-2 | CFH, IRAK-1, TSPAN12 | Upregulated miR-146a in AD: – Represses CFH that leads to neuroinflammation – Downregulates IRAK-1 and increases IRAK-2, which supports proinflammatory signaling in brain cells – Decreases membrane spanning integral membrane protein tetraspanin-12 (TSPAN12) levels. This induces amyloidogenesis, vascular aberrations, and formation of Aβ42 peptide |
(22, 23) |
| miR-29a | BACE1 enzyme | Downregulated miR-29a in AD: – Increases BACE1, an enzyme that catalyzes the formation of Aβ protein from APP |
(6, 25) | |
| miR-106b | ABCA1, APP | Downregulated miR-106b in AD: – Represses ABCA1 leading to increase in Aβ levels – Suppresses the expression of APP that may prevent Aβ accumulation |
(25–29) |
See text for details.
Acknowledgements
The authors would like to thank Kaushik Muralidharan for help with the manuscript. This work was supported by an NIH grant to ADL (R15 AG051132-01).
List of abbreviations
- 15LOX
15-lipoxygenase
- 3′UTR
3′ Untranslated region
- ABCA1
ATP-binding cassette transporter A1
- AD
Alzheimer's disease
- Akt–mTOR
mammalian target of rapamycin
- APOE ε4
apolipoprotein E ε4 allele
- APP
amyloid precursor protein
- APPsw
Swedish double mutation APP
- Aβ42
amyloid-β42
- C. elegans
Caenorhabditis elegans
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- CDX2
caudal-type homeobox 2
- CFH
complement factor-H
- CNS
central nervous system
- CSF
cerebrospinal fluid
- ECF
extracellular fluid
- ELP4
elongator acetyltransferase complex subunit 4
- EMT
epithelial-mesenchymal transition
- FGFR1
fibroblast growth factor receptor 1
- FTDP-17
frontotemporal dementia with Parkinsonism linked to chromosome 17
- H4-FL-APP
cells H4 human neuroglioma cells overexpressing full-length APP cells
- HCF-1
host cell factor
- HSF1
Heat shock factor 1
- Insulin/IGF-1
insulin/insulin-like growth factor 1
- IRAK-1
interleukin-1 associated kinase-1
- IRF4
interferon regulatory factor 4
- LBD
Lewy-body dementia
- lgals3
galectin-3
- MIR17HG
miR-17/92 cluster host gene
- miRNAs/miR
microRNAs
- mRNA
messenger RNA
- mt-3
metallothionein-3
- NAD
nicotinamide adenine dinucleotide
- NDDs
neurodegenerative diseases
- NPD1
neuroprotectin D1
- NR2E1
nuclear receptor subfamily 2 group E member 1
- NRF-1
nuclear respiratory factor 1
- p-tau
phosphorylated tau
- PBMCs
peripheral blood mononuclear cells
- PDK-1
protein kinase-1
- PDK1-2
protein kinases B
- PSEN1
presenilin 1
- PSEN2
Presenilin 2
- qPCR
quantitative PCR
- REST
RE1 silence transcription factor
- RISC
RNA-induced silencing complex
- SIRT
sirtuin
- SYN-2
synapsin-2
- t-tau
total tau
- TSPAN12
tetraspanin-12
Footnotes
Conflict of interest statement: The authors declare no conflicts of interest.
Contributor Information
Manasa Basavaraju, Department of Biological Sciences, Quinnipiac University, Hamden, CT 06473, USA.
Alexandre de Lencastre, Department of Biological Sciences, Quinnipiac University, Hamden, CT 06473, USA.
References
- 1.Förstl H, Kurz A. Clinical features of Alzheimer's disease. Eur Arch Psychiatry Clin Neurosci. 1999;249:288–90. doi: 10.1007/s004060050101. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 1991;12:383–8. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- 3.Waring SC, Rosenberg RN. Genome-wide association studies in Alzheimer disease. Arch Neurol. 2008;65:329–34. doi: 10.1001/archneur.65.3.329. [DOI] [PubMed] [Google Scholar]
- 4.Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet Lond Engl. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 5.Pericak-Vance MA, Bebout JL, Gaskell PC, Yamaoka LH, Hung W-Y, Alberts MJ, Walker AP, Bartlett RJ, Haynes CA, Welsh KA. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet. 1991;48:1034–50. [PMC free article] [PubMed] [Google Scholar]
- 6.Van Cauwenberghe C, Van Broeckhoven C, Sleegers K. The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med Off J Am Coll Med Genet. 2016;18:421–30. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–81. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser PS, Ryan RO. Impact of apolipoprotein E on Alzheimer's disease. Curr Alzheimer Res. 2013;10:809–17. doi: 10.2174/15672050113109990156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–13. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nistor M, Don M, Parekh M, Sarsoza F, Goodus M, Lopez GE, Kawas C, Leverenz J, Doran E, Lott IT, Hill M, Head E. Alpha- and beta-secretase activity as a function of age and beta-amyloid in Down syndrome and normal brain. Neurobiol Aging. 2007;28:1493–506. doi: 10.1016/j.neurobiolaging.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McConathy J, Sheline YI. Imaging biomarkers associated with cognitive decline: a review. Biol Psychiatry. 2015;77:685–92. doi: 10.1016/j.biopsych.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabri O, Sabbagh MN, Seibyl J, Barthel H, Akatsu H, Ouchi Y, Senda K, Murayama S, Ishii K, Takao M, Beach TG, Rowe CC, Leverenz JB, Ghetti B, Ironside JW, Catafau AM, Stephens AW, Mueller A, Koglin N, Hoffmann A, Roth K, Reininger C, Schulz-Schaeffer WJ. Florbetaben Phase 3 Study Group. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer's disease: Phase 3 study. Alzheimers Dement. 2015;11:964–74. doi: 10.1016/j.jalz.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Macfarlane L-A, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–61. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 18.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–7. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 19.Mariño G, Ugalde AP, Fernández AF, Osorio FG, Fueyo A, Freije JM, López-Otín C. Insulin-like growth factor 1 treatment extends longevity in a mouse model of human premature aging by restoring somatotroph axis function. Proc Natl Acad Sci USA. 2010;107:16268–73. doi: 10.1073/pnas.1002696107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inukai S, de Lencastre A, Turner M, Slack F. Watson M, editor. Novel m icroRNAs differentially expressed during aging in the mouse brain. PLoS One. 2012;7:e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nissan X, Blondel S, Navarro C, Maury Y, Denis C, Girard M, Martinat C, De Sandre-Giovannoli A, Levy N, Peschanski M. Unique preservation of neural cells in Hutchinson-Gilford progeria syndrome is due to the expression of the neural-specific miR-9 microRNA. Cell Rep. 2012;2:1–9. doi: 10.1016/j.celrep.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Van den Hove DL, Kompotis K, Lardenoije R, Kenis G, Mill J, Steinbusch HW, Lesch KP, Fitzsimons CP, De Strooper B, Rutten BP. Epigenetically regulated microRNAs in Alzheimer's disease. Neurobiol Aging. 2014;35:731–45. doi: 10.1016/j.neurobiolaging.2013.10.082. [DOI] [PubMed] [Google Scholar]
- 23.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–4. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 24.Femminella GD, Ferrara N, Rengo G. The emerging role of micro-RNAs in Alzheimer's disease. Front Physiol. 2015;6:40. doi: 10.3389/fphys.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 26.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–10. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 27.Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. Blagosklonny MV, editor. PLoS One. 2010;5:e10724. doi: 10.1371/journal.pone.0010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. J Neurochem. 2013;127:739–49. doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20:2159–68. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–82. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garg D, Cohen SM. miRNAs and aging: a genetic perspective. Ageing Res Rev. 2014;17:3–8. doi: 10.1016/j.arr.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Chen D, He Y, Meléndez A, Feng Z, Hong Q, Bai X, Li Q, Cai G, Wang J, Chen X. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age. 2013;35:11–22. doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved microRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The Conserved microRNA MiR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–45. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 36.Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501–6. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu N, Landreh M, Cao K, Abe M, Hendriks G-J, Kennerdell JR, Zhu Y, Wang LS, Bonini NM. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–23. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibáñez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:235–46. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 40.Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi F, Fischer A. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Björkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum Mol Genet. 2011;20:2225–37. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 42.Garza-Manero S, Arias C, Bermúdez-Rattoni F, Vaca L, Zepeda A. Identification of age- and disease-related alterations in circulating miRNAs in a mouse model of Alzheimer's disease. Front Cell Neurosci. 2015;9:53. doi: 10.3389/fncel.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller M, Jäkel L, Bruinsma IB, Claassen JA, Kuiperij HB, Verbeek MM. MicroRNA-29a is a candidate biomarker for Alzheimer's disease in cell-free cerebrospinal fluid. Mol Neurobiol. 2016;53:2894–9. doi: 10.1007/s12035-015-9156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grasso M, Piscopo P, Crestini A, Confaloni A, Denti MA. Circulating microRNAs in neurodegenerative diseases. EXS. 2015;106:151–69. doi: 10.1007/978-3-0348-0955-9_7. [DOI] [PubMed] [Google Scholar]
- 45.Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Biol. 2007;1:263–74. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 47.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 48.Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sala Frigerio C, Lau P, Salta E, Tournoy J, Bossers K, Vandenberghe R, Wallin A, Bjerke M, Zetterberg H, Blennow K, De Strooper B. Reduced expression of hsa-miR-27a-3p in CSF of patients with Alzheimer disease. Neurology. 2013;81:2103–6. doi: 10.1212/01.wnl.0000437306.37850.22. [DOI] [PubMed] [Google Scholar]
- 50.Lukiw WJ. NF-κB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol. 2012;235:484–90. doi: 10.1016/j.expneurol.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer's disease, inhibits bcl2 translation. Brain Res Bull. 2009;80:268–73. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Lukiw WJ, Andreeva TV, Grigorenko AP, Rogaev EI. Studying micro RNA function and dysfunction in Alzheimer's disease. Front Genet. 2012;3:327. doi: 10.3389/fgene.2012.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro F, Lieberman J. miR-34 and p53: new insights into a complex functional relationship. PLoS One. 2015;10:e0132767. doi: 10.1371/journal.pone.0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hébert SS, Horré K, Nicolaï L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–8. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Kim J, Yoon H, Ramírez CM, Lee S-M, Hoe H-S, Fernández-Hernando C, Kim J. miR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–83. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukiw WJ, Alexandrov PN, Zhao Y, Hill JM, Bhattacharjee S. Spreading of Alzheimer's disease inflammatory signaling through soluble micro-RNA. Neuroreport. 2012;23:621–6. doi: 10.1097/WNR.0b013e32835542b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song J, Lee JE. miR-155 is involved in Alzheimer's disease by regulating T lymphocyte function. Front Aging Neurosci. 2015;7:61. doi: 10.3389/fnagi.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. MicroRNA (miRNA) speciation in Alzheimer's disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int J Biochem Mol Biol. 2012;3:365–73. [PMC free article] [PubMed] [Google Scholar]
- 60.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller VM, Gouvion CM, Davidson BL, Paulson HL. Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32:661–8. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie Z, Dong Y, Maeda U, Xia W, Tanzi RE. RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Aβ generation. J Biol Chem. 2007;282:4318–25. doi: 10.1074/jbc.M609293200. [DOI] [PubMed] [Google Scholar]
- 63.Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8:1343–9. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]