Abstract
Brd4 is an epigenetic reader protein and a member of the BET (bromodomain and extra terminal domain) family of proteins with two bromodomains that recognize acetylated lysine residues. Brd4 specifically binds to acetylated transcription factor NF-κB p65 and coactivates transcription. Polyomavirus JC (JCV) is regulated by a noncoding control region (NCCR) containing promoter/enhancer elements for viral gene expression including a binding site for NF-κB, which responds to proinflammatory cytokines such as TNF-α, the DNA damage response, calcium signaling and acetylation of the NF-κB p65 subunit on lysine residues K218 and K221. Earlier studies indicated that NF-κB is involved in the reactivation of persistent/latent JCV in glial cells to cause progressive multifocal leukoencephalopathy (PML), a severe demyelinating disease of the brain caused by replication of JCV in glial cells. To investigate the mechanism of action of NF-κB acetylation on JCV transcription, we examined Brd4 and found that JCV early transcription was stimulated by Brd4 via the JCV NF-κB site and that p65 K218 and K221 were involved. Treatment with the Brd4 inhibitor JQ1(+) or mutation of either K218 or K221 to glutamine (K218R or K221) inhibited this stimulation and decreased the proportion of p65 in the nucleus. We conclude that Brd4 is involved in the regulation of the activation status of JCV in glial cells.
Keywords: JC Virus, Progressive multifocal leukoencephalopathy, Viral persistence, Viral reactivation, Epigenetic regulation, protein acetylation
INTRODUCTION
Progressive multifocal leukoencephalopathy (PML) is a debilitating and often fatal demyelinating disease of the central nervous system (CNS) caused by the neurotropic polyomavirus JC (JCV), which replicates in glial cells causing cytolytic death of oligodendrocytes giving rise to expanding mutifocal lesions of myelin loss (Berger 2011). PML is a rare disease that occurs almost always in the context of immune system impairment, especially HIV-1/AIDS where it remains a complication despite the introduction of combination antiretroviral therapy (Tavazzi et al. 2012). No effective therapies for PML are available (Tavazzi et al. 2012; Clifford 2015). More recently, therapeutic immunomodulatory monoclonal antibodies used to treat autoimmune disorders such as natalizumab, rituximab and efalizumab have become recognized as another predisposing condition for PML (Berger 2010; Chahin and Berger 2015). Infection by JCV is very common throughout populations worldwide since most people acquire antibodies to the virus at an early age (White and Khalili 2011). However, the incidence of PML is very low suggesting that virus is restrained but then persists asymptomatically. Occasionally and in conditions of severe immune impairment, JCV undergoes reactivation in the glia of the brain to give PML (Jelcic et al. 2015; Wollebo et al 2015a). Defining the molecular mechanisms whereby this occurs is of paramount importance in understanding the JCV life cycle and the pathogenesis of PML.
Like other polyomaviruses, JCV is a small, non-enveloped, double-stranded DNA virus with a circular ~5 Kbp genome comprised of three regions, two protein-coding regions and a noncoding control region (NCCR) that lies between them (Padgett et al. 1971; DeCaprio et al. 2013). The early coding region contains large T-antigen (T-Ag) and small t-antigen (t-Ag), while the late coding region encodes the capsid proteins VP1, VP2 and VP3 and a regulatory protein know as agnoprotein (Padgett et al. 1971; DeCaprio et al. 2013). The NCCR contains the promoter/enhancer elements for expression of the early and late genes and the origin of viral DNA replication. Binding sites for a number of transcription factors are found within the NCCR and these regulate early and late transcription (White et al. 2009). In particular, we have reported an NF-κB site that activates JCV early and late transcription in response to NF-κB p65 expression (Romagnoli et al. 2009) or stimulation of the NF-κB pathway by proinflammatory cytokines such a TNF-α (Wollebo et al. 2011). We have proposed that proinflammatory cytokines such as those that occur in HIV-1/AIDS reactivate JCV leading to PML (White and Khalili 2011). In support of this, we found that TNF-α and its receptor TNFR1 are upregulated in clinical samples from HIV/PML and immunohistochemistry of PML brain tissue shows redistribution of NF-κB to the nucleus (Wollebo et al. 2016).
The JCV NF-κB site is also a nexus for the regulation of JCV by other pathways including DNA damage response signaling (White et al. 2014) and calcium signaling (Wollebo et al. 2012). Recently, we discovered that acetylation of NF-κB p65 regulates JCV. Thus, histone deacetylation inhibitors (HDACi) such as trichostatin A (TSA) strongly stimulated JCV transcription and mutations at the NF-κB site or siRNA to NF-κB p65 abrogated this effect (Wollebo et al. 2013). Furthermore, ectopic p65 expression, TNF-α stimulation or TSA treatment each elicited an identical gel shift with a probe containing the JCV NF-κB site sequence and this was supershifted by antibody to p65 in all cases (Wollebo et al. 2013). Site-directed mutagenesis analysis of the known lysine acetylation sites of NF-κB p65 (K218, K221 and K310) implicated K218 and K221 in transactivation of JCV transcription and showed that acetylation at these sites regulates NF-κB p65 activity toward JCV at the level of p65 binding to the JCV NCCR and transcriptional transactivation (Wollebo et al. 2015b).
To further understand the mechanism of NF-κB action on JCV transcription, we have examined the epigenetic reader protein Brd4, which is a member of the BET (bromodomain and extra terminal domain) family of proteins and contains two bromodomains that recognize acetylated lysine residues (Wu and Chiang 2007). Brd4 was first cloned from the mouse and designated MCAP or mitotic chromosome-associated protein (Dey et al. 2000). The human homolog has also been identified and the protein has been redesignated Brd4 (French et al. 2001). Brd4 specifically binds to acetylated NF-κB p65 and coactivates transcription in a number of ways (Huang et al. 2009). Brd4 serves as a scaffolding protein and is involved in physically linking chromatin remodeling proteins and transcriptional regulatory proteins (Devaiah and Singer 2013) including P-TEFb (Jang et al. 2005; Yang et al. 2005; Schröder et al. 2012; Diament and Dikstein 2013) and the SWI/SNF chromatin remodeling complex (Shi et al. 2013). Brd4 phosphorylates the carboxy-terminal domain of RNA polymerase II and this is abrogated by a small molecule inhibitor that prevents Brd4 binding to chromatin (Devaiah et al. 2012). Additionally, Brd4 may have upstream inhibitory effects on the NF-κB signaling pathway by interaction with IκB kinase (Ceribelli et al. 2014).
In this study, we examined the role of Brd4 in the regulation of JCV and found that Brd4 activates JCV early transcription and that this is blocked by mutations in the JCV NF-κB site or treatment with the small molecule Brd4 inhibitor JQ1(+). Furthermore wild-type NF-κB p65 was co-stimulatory with Brd4 but not when either p65 lysine 218 or 221 were mutated to arginine, which cannot be acetylated. JCV replication was increased by Brd4 expression and decreased by Brd4 inhibitor treatment. Thus Brd4 has a role in regulating JCV transcriptional status and replication via epigenetic changes in NF-κB.
METHODS
Cell culture and plasmids
Culture of the human TC620 oligodendroglioma cell line and SVG-A, a cell line originally derived from human glial cells transformed by origin-defective SV40 that expresses SV40 T-Ag was performed as we have previously described (Wollebo et al. 2011, 2015c). The reporter construct JCVE-LUC contains the JCV early promoter from the Mad-1 strain linked to the luciferase gene in the early orientation (Wollebo et al. 2011). Mutant versions of the JCV early promoter, m1 and m2, which contain mutations at two adjacent sites within the NF-κB binding site, have been described previously (Romagnoli et al. 2009). The expression plasmid pCMV-p65wt was previously described (Romagnoli et al. 2009) and expression plasmids for the p65 acetylation site Lys to Arg mutants (p65K218R, p65K221R and p65K310R) and Lys to Gln mutants (p65K218Q, p65K221Q and p65K310Q) have also been described (Wollebo et al. 2015b). Expression plasmid for Brd4 (pcDNA6B-Brd4) was made by cloning the coding region for human Brd4 (Addgene) into the BamHI/HindIII site of pcDNA6B.
Antibodies
The following antibodies were used for Western blot: Mouse monoclonal anti-p65 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-α-tubulin clone B512 (Sigma-Aldrich. St. Louis, MO) and anti-Lamin A/C (Cell Signaling, Danvers, MA). Purified mouse monoclonal anti-Grb2 (#610111) antibody was purchased from BD Biosciences, San Jose CA. Rabbit polyclonal antibody to Brd4 (sc-48772) was from Santa Cruz. Rabbit polyclonal antibodies against JCV agnoprotein and VP1 were previously described (Darbinyan et al. 2007).
Western blots
Western blot assays were performed as previously described (White et al. 2006). Briefly, 50 μg of protein was resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with primary antibody (1/1000 dilution) and secondary antibody (1/10000 dilution). Bound antibody was detected with the LI-COR system. Blots were incubated with IRDye® 680RD Goat Anti-Mouse Li-COR dyes and visualized with an Odyssey® CLx Imaging System (LI-COR, Inc., Lincoln, NE) using LI-COR Odyssey software. Band intensities were quantified using the Quantity One software (Bio-Rad, Hercules CA) and intensities normalized to loading control as previously described (Bellizzi et al. 2015).
Transient transfection and reporter assays
Co-transfection of reporter plasmids and expression plasmids were performed using Lipofectamine 2000 (Life Technologies Inc., Carlsbad CA) as we have previously described (Romagnoli et al. 2009; Wollebo et al. 2011). Briefly, TC620 cells were transfected with reporter constructs alone or in combination with expression plasmid(s) for 48 h prior to harvesting. The total amount of transfected DNA was normalized with empty vector DNA. For cytokine stimulation of transfected cells, TNF-α was used at a concentration of 10 ng/ml as described (Wollebo et al. 2011). Assay for luciferase was performed as previously described (Romagnoli et al. 2009; Wollebo et al. 2011).
Treatment with the Brd4 small molecule inhibitor JQ1(+)
JQ1(+) ((S)-tert-butyl 2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetate) is a potent inhibitor of the BET family of bromodomain proteins including BRD4 and is related to benzodiazepines (Filippakopoulos et al. 2010). It was used at a concentration of 2.5–10 μM and added to cells 24 hours prior to harvesting for luciferase or cell fractionation experiments. Parallel cultures treated with the same concentration of the inactive isomer JQ1(−) were used as negative controls.
MTT Assay
To assess cell viability after treatment, the MTT assay was performed with the Vybrant® MTT Cell Proliferation Assay Kit using a standard microplate absorbance reader according to the Manufacturer's protocol (Vybrant, Life Technologies, Inc).
Assay of JCV infection
Infection experiments were performed with SVG-A cells. Cells were transfected with Brd4 expression plasmid or treated with JQ1 and infected with Mad-1 JCV at an MOI of 1 as we have previously described (Radhakrishnan et al. 2003, 2004), harvested and analyzed after 7 days together with uninfected control cultures. Expression of the viral proteins VP1 and agnoprotein was measured in whole cell protein extracts by Western blot. In parallel, the growth medium of the cells was also collected to measure viral load by Q-PCR (Radhakrishnan et al. 2004).
Cell fractionation
TC620 cells were treated for 24 hours with 0, 2.5, 5 or 10 μg JQ1(+) or JQ1(−)(BD Biosciences) harvested for cell fractions using the NE-PER nuclear and cytoplasmic extraction kit according to the Manufacturer’s instructions (Thermo Scientific Pierce, Rockford IL; Cat. # PI-78835).
Immunocytochemistry (ICC)
ICC was performed as we have recently described (White et al. 2014). TC620 cells were transfected with expression plasmids for p65 (wild-type or mutant) and Brd4, serum-starved overnight with 0.5% BSA and then either untreated or treated with 10 ng/ml TNF-α for 20 min. Cells were fixed in 4% paraformaldehyde in PBS for 10 min, washed, permeabilized for 5 min with 0.1% TritonX-100, blocked with 5% normal goat serum for 30 min and incubated for 3 h at 37°C with mouse anti-NF-κB p65 and rabbit anti-Brd4 at a 1:100 dilution in PBS. Cells were then washed, incubated for 2 h with secondary FITC-conjugated goat anti-rabbit and rhodamine-conjugated anti-mouse secondary antibodies at a 1:200 dilution, washed, mounted with DAPI-containing mounting medium (VECTASHIELD, Vector Laboratories Inc. Burlingame, CA) and viewed by fluorescence microscopy.
RESULTS
Expression of Brd4 stimulates the transcriptional activity of the JCV early promoter and this is mediated by the NF-κB binding site
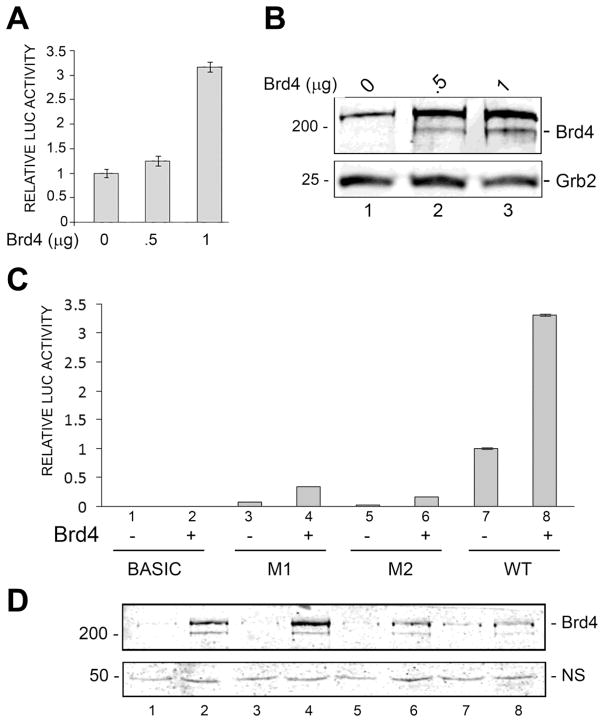
Our earlier studies have shown that in vivo acetylation of NF-κB p65 at lysines 218 and 221 is involved in the epigenetic regulation of JCV (Wollebo et al. 2015b). Brd4 is an epigenetic reader protein, which specifically binds to acetylated NF-κB p65 and coactivates transcription in a number of ways (Huang et al. 2009) and thus we investigated a role for Brd4 in the regulation of JCV by NF-κB p65. Co-transfection of Brd4 expression plasmid with luciferase reporter plasmid for the JCV early promoter into TC620 oligodendroglial cells showed a 3-fold stimulation of transcription (Fig. 1A). These data indicate a role for Brd4 in the regulation of JCV transcription. Expression of Brd4 was verified by Western blot (Fig. 1B).
Figure 1. Effect of Brd4 on JCV early transcription and effect of mutations in the NF-κB binding site.
A. TC620 cells were transfected with luciferase reporter plasmid for the JCV early promoter with or without expression plasmid for Brd4. After 48 h, cells were harvested and assayed for luciferase activity as described in Methods. Activities were normalized to control without expression plasmid. The bar represents one standard deviation. B. Western blot for Brd4 expression in the experiment in Panel A. Grb2 was the loading control. C. TC620 cells were transfected with luciferase reporter plasmid for the JCV early promoter (wild-type, m1 or m2) with or without expression plasmid for Brd4. After 48 h, cells were harvested and assayed for luciferase activity as described in Methods. Activities were normalized to control without expression plasmid. The bar represents one standard deviation. Where no error bar is seen, it is too small to be visible. D. Western blot for Brd4 expression in the experiment in Panel C. A nonspecific band (NS) on the same gel was the loading control.
To determine the site of action of Brd4 on the JCV early promoter, we employed mutants (m1 and m2) that contain base substitutions in the NF-κB binding site (Romagnoli et al. 2009). As shown in Fig. 1C, the stimulation of JCV early transcription is drastically reduced in the m1 and m2 mutants compared to wild-type in experiments where wild-type or mutant luciferase plasmids are transfected into TC620 cells with expression plasmid for Brd4. Expression of Brd4 was verified by Western blot (Fig. 1D). Thus, the stimulatory effect of Brd4 on JCV transcription is mediated through the NF-κB site and this is consistent with our hypothesis that Brd4 is exerting its effect through binding to NF-κB p65.
Brd4 inhibitor JQ1(+) causes a decline in the level of nuclear NF-κB p65
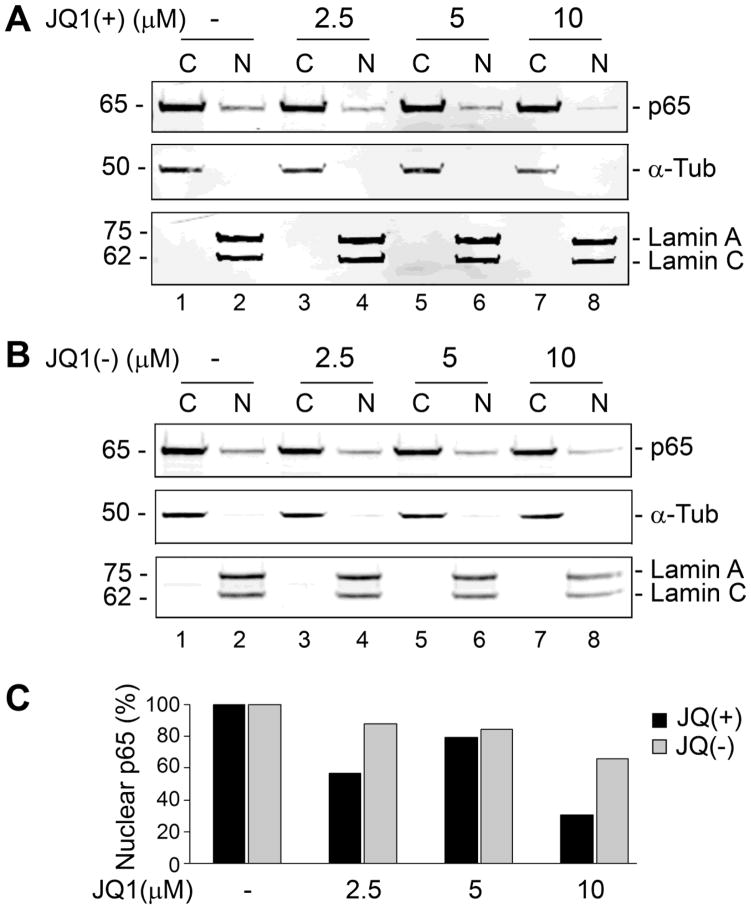
In the next series of experiments, we looked at the effect of JQ1(+), which is a thienotriazolodiazepine and a cell-permeable small molecule inhibitor of the BET family of bromodomain proteins including BRD4. JQ1(+) binds competitively to acetyl-lysine recognition motifs (bromodomains) and thereby provides highly potent and specific inhibition of Brd4 (Filippakopoulos et al. 2010). Since Brd4 specifically binds to acetylated NF-κB p65 in the nucleus (Huang et al. 2009), we investigated the effect of JQ1(+) on the level of nuclear NF-κB p65. TC620 cells were treated with various concentrations of JQ1(+) and harvested for isolation of nuclear and cytoplasmic fractions as described in Methods. As show in Figure 2, analysis of the fractions by Western blot revealed that there was a 70% decrease in nuclear NF-κB p65 after treatment with 10 μM JQ1(+)(A) compared to the biologically inactive isomer JQ1(−)(B). The nuclear and cytoplasmic p65 bands were quantified by densitometry and are shown a histogram of the percentage of p65 present in the nucleus, which decreases with JQ(+)(C). Thus Brd4 is involved in the nuclear localization of NF-κB p65. It is possible that interaction of Brd4 with p65 facilitates p65 binding to chromatin thus favoring p65 nuclear localization. Alternatively, it is known that IκB kinase activity is inhibited by small molecules targeting BET proteins and this might favor p65 retention in the cytoplasm bound to IκB (Ceribelli et al. 2014). Furthermore, depletion of Brd4 or treatment of cells with JQ1 has been shown to induce the ubiquitination and degradation of the active nuclear form of p65 (Zou et al. 2014).
Figure 2. Effect of Brd4 inhibitor JQ1 on NF-κB subcellular distribution.
TC620 cells were treated with or without (A) the active form of JQ1 (JQ1(+)) or (B) its inactive isomer (JQ1(−)) at concentrations of 2.5, 5 and 10 μM and cytoplasmic and nuclear fractions prepared as described in Methods. The distribution of NF-κB p65 in these fractions was measured by Western blot. Purity of fractions and equal loading was assessed by Western blot to the cytoplasmic protein α-Tubulin and the nuclear proteins Lamins A/C. The nuclear and cytoplasmic p65 bands were quantified by densitometry and are shown a histogram of the percentage of p65 present in the nucleus (C).
The Brd4 inhibitor JQ1(+) reduces JCV early promoter activity
TC620 cells were transfected with luciferase reporter plasmid for the JCV early promoter, treated with the Brd4 inhibitor JQ1(+) or its inactive isomer JQ(−), harvested and luciferase activity determined (Fig. 3A). Treatment with 10 μM JQ1(+) reduced JCV early promoter activity by about one third whereas JQ(−) was without significant effect. The expression of Brd4 was analyzed by Western blot (Fig. 3B) and viability by MTT assay (Fig. 3C). These data further implicate Brd4 in the regulation of JCV transcription.
Figure 3. Effect of Brd4 inhibitor JQ1 on JCV early transcription.
A. TC620 cells were transfected with luciferase reporter plasmid for the JCV early promoter together with vector plasmid or expression plasmid for Brd4 and treated with or without the active form of JQ1 (JQ1(+)) or its inactive isomer (JQ1(−)) at concentrations of 5 and 10μM. After 48 h, cells were harvested and assayed for luciferase activity as described in Methods. Activities were normalized to control without expression plasmid. The bar represents one standard deviation. B. Western blot for Brd4 expression in the experiment in Panel A. Grb2 was the loading control. C. To determine the effect of JQ1(+) and JQ1(−) on cell viability, cells were incubated with JQ1(+) at 5, 10 or 15μM for 24h (lanes 2, 4 and 6 respectively) or JQ1(−) at 5, 10 or 15μM for 24h (lanes 8, 10 and 12 respectively). An equal volume of DMSO was added to the odd numbered lanes corresponding to the amount for each following even numbered lane. Viabilty was measured by MTT assay.
Combined effects of Brd4 and NF-κB p65 acetylation site mutants on JCV early promoter activity and TNF-α-mediated subcellular redistribution
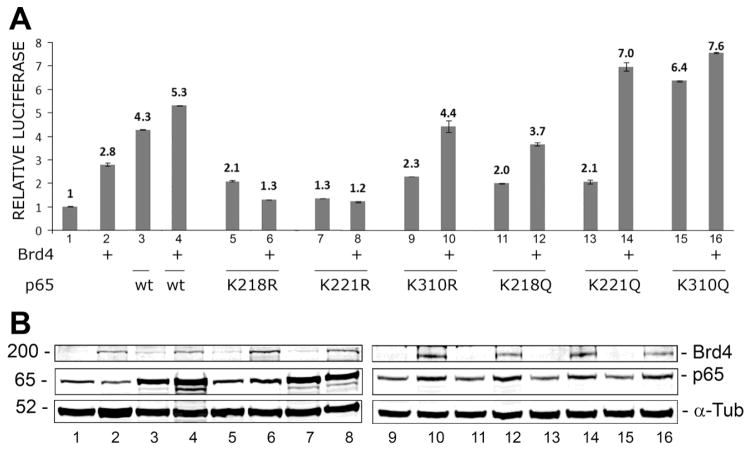
While expression of wild-type NF-κB p65 stimulated JCV early transcription 4-fold (Fig. 4A, lane 2) and Brd4 stimulated 3-fold (Fig. 4A, lane 3), together they stimulated the promoter 5.3-fold (Fig. 4A, lane 4). Previously, we had constructed a series of p65 acetylation site mutants where individual lysines (K) were mutated to arginine (R), which cannot be acetylated or glutamine (Q), which possesses a negative charge like acetyllysine (Wollebo et al. 2015b). Brd4 failed to costimulate with the K218R and K221R mutants (Fig. 4A, lanes 5–8) while K310R was only slightly reduced (Fig. 4A, lanes 9 and 10) indicating the importance of K218 and K221 in transcriptional co-activation and Brd4 binding. This was not the case for the K to Q mutants (Fig. 4A, lanes 11–16), which were still co-stimulated by Brd4 suggesting that Brd4 could bind to the negatively charged Q residues in K218Q and K221Q and co-activate JCV transcription. Expression of p65 and Brd4 in this experiment was confirmed by Western blot (Fig. 4B).
Figure 4. Effect of Brd4 and NF-κB p65 acetylation site mutants on JCV early transcription.
A. TC620 cells were transfected with vector or Brd4 expression plasmid with and without expression plasmid for NF-κB p65, wild-type or mutants K218R, K221R, K310R, K218Q, K221Q or K310Q. After 48 h, cells were harvested and assayed for luciferase activity as described in Methods. Activities were normalized to control without expression plasmid. The bar represents one standard deviation. B. The lysates from Panel A were analyzed by Western blot for expression of Brd4 and p65. The loading control was α-tubulin (α-Tub).
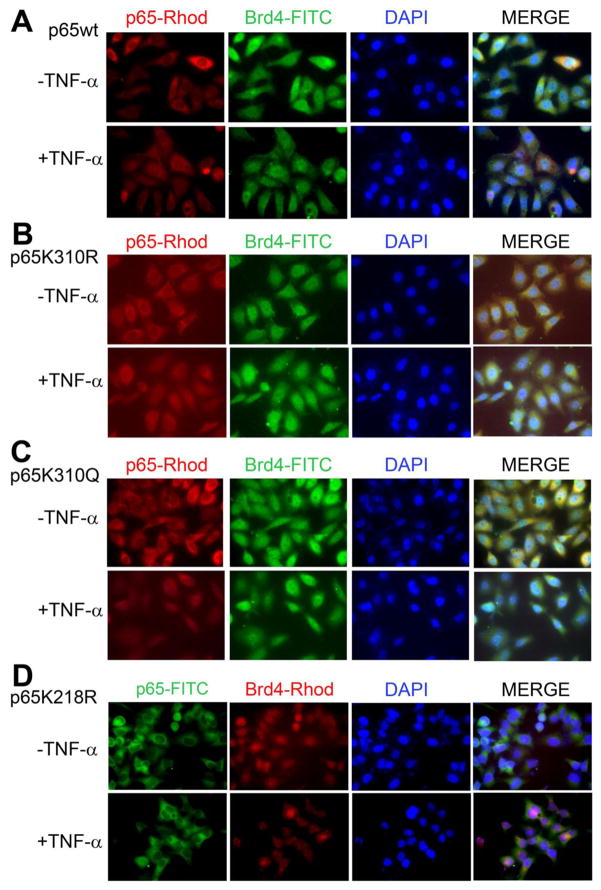
Immunocytochemistry showed that TNF-α treatment of TC620 cells transfected with wild-type p65 and Brd4 resulted in the expected cytoplasm to nucleus relocation of p65 (Fig. 5A). Similarly in the case of K310R (Fig. 5B) and K310Q (Fig. 5C), a comparable redistribution occurred but not for K218R (Fig. 5D) and K221R (data not shown), again pointing to the importance of K218 and K221 acetylation.
Figure 5. Effect of Brd4 and NF-κB p65 acetylation site mutants on redistribution of p65 in response to TNF-α stimulation.
TC620 cells were transfected with expression plasmid for Brd4 together with expression plasmid for NF-κB p65 either wild-type (A), K301R (B), K301Q (C) or K218R (D). Double-labeling immunofluorescence was performed for p65 (rhodamine), Brd4 (FITC) and nuclei (DAPI), except Panel D where FITC was used for p65 and rhodamine for Brd4 as described in Methods.
To assess the effect of Brd4 on JCV infection, we transfected SVGA cells with Brd4 expression plasmid or treated with JQ1 and infected with JCV. Viral infection was monitored by Western blot for viral proteins, VP1 and agnoprotein (Fig. 6A) and by Q-PCR for viral DNA in the culture supernatants (Fig. 6B). Expression of Brd4 enhanced viral infection while JQ1 was inhibitory. These data indicate that Brd4 has a crucial role in the JCV replicative life cycle.
Figure 6. Effect of Brd4 and Brd4 inhibitor JQ1 on JCV early replication.
A. SVG-A cells were transfected with Brd4 or treated with JQ1 and assayed for JCV infection (MOI = 1, 7 days post-infection). Viral infection was assessed by Western blot for VP1 and agnoprotein with α-tubulin as a loading control. Brd4 levels were also monitored by Western blot. B. Viral loads in the culture supernatants from the experiment in Panel A were quantified using Q-PCR and are shown as copy number/well.
DISCUSSION
The transcription factor NF-κB is pivotal in the regulation of JCV activation and may mediate the reactivation of virus in response to extracellular proinflammatory cytokines, such as TNF-α, which turn on signaling pathways that lie upstream of NF-κB (Wollebo et al. 2011). Due to its many interactions with other proteins, this JCV NF-κB site also constitutes a nexus for JCV regulation by other signaling pathways including the DNA damage response (White et al. 2014) and calcium-activated signaling (Wollebo et al. 2012), which are mediated by direct interaction of NF-κB p65 with the proteins Rad51 and NFAT4 respectively. The pathological relevance of these proteins is indicated by the observations that Rad51 is barely detectable in normal brain but is robustly induced in HIV/PML brain sections (Darbinyan et al. 2007) and that NFAT4 preferentially localizes to the nuclei of oligodendrocytes and bizarre astrocytes in HIV/PML relative to what is observed in non-PML brain sections (Wollebo et al. 2016). A further level of complexity is added to this by the finding that NF-κB p65 is subject to epigenetic regulation of its stimulatory effect on JCV activity by acetylation. Thus, experiments demonstrated that histone deacetylation inhibitors (HDACi) such as trichostatin A (TSA) and sodium butyrate strongly activate JCV transcription via the NF-κB site in the JCV NCCR as revealed by analysis of JCV promoter mutants (Wollebo et al. 2013). We were further able to pinpoint this effect by site-directed mutagenesis analysis of lysine acetylation sites within NF-κB p65 and implicated K218R and K221R of p65 in transactivation of JCV transcription (Wollebo et al. 2015b). Gel shift studies revealed the importance of K218R and K221R in p65 binding to the JCV NF-κB site and activation of JCV transcription (Wollebo et al. 2015b). We have suggested that these multiple pathways of NF-κB regulation of JCV in glial cells may confer a switch-like response to the NF-κB signaling module whereby a threshold level of converging input signals triggers the switch between silent JCV and JCV reactivation (White et al. 2014) as suggested by the experimental evidence and mathematical modeling of NF-κB signaling by Shinohara et al. (2014).
The biological effects of acetylated lysine residues are mediated by epigenetic reader proteins, which recognize them through binding to their acetyl-lysine binding domains, known as bromodomains (Zeng and Zhou 2002). The ability of acetylated NF-κB to activate transcription is mediated by Brd4, a member of the BET family of proteins, which contains two bromodomains that recognize acetylated lysine residues (Dey et al. 2000). Brd4 specifically binds to acetylated NF-κB p65 and is involved in the co-activation of transcription through several mechanisms including forming a scaffold that physically links chromatin remodeling proteins, e.g., SWI/SNF, activation of transcription factors such as P-TEFb (Devaiah and Singer 2013; Diament and Dikstein 2013; Huang et al. 2009; Jang et al. 2005; Schröder et al. 2012; Shi et al. 2013; Yang et al. 2005) and phosphorylation of the carboxy-terminal domain of the RNA polymerase II through an atypical protein kinase possessed by Brd4, which is abrogated by a small molecule Brd4 inhibitor that prevents Brd4 binding to chromatin (Devaiah et al. 2012). Our data indicate that Brd4 is a transcriptional coactivator for NF-κB p65 at the JCV promoter since expression of Brd4 stimulated JCV transcription (Fig. 1A). The site of Brd4 stimulation was confirmed to be the JCV NF-κB site by mapping with JCV promoter mutants (Fig. 1C).
JQ1 is a potent and specific inhibitor of the BET family of proteins and a useful reagent to investigate the role of Brd4 (Filippakopoulos et al. 2010). In this study, we examined the effects of JQ1 on p65 levels in the nucleus and cytoplasm (Fig. 2), JCV transcription (Fig. 3) and JCV replication (Fig. 6). Brd4 specifically binds to acetylated NF-κB p65 in the nucleus (Huang et al. 2009) and the Brd4 inhibitor JQ1 (+) causes a substantial decrease in nuclear NF-κB p65 as shown in Figure 2. This may be because the interaction of Brd4 with p65 promotes p65 binding to chromatin and hence p65 nuclear localization. Alternatively, since JQ1(+) is known to inhibit upstream IκB kinase activity, this would favor the retention of p65 in the cytoplasm bound to unphosphorylated IκB (Ceribelli et al. 2014). It should also be noted that treatment of cells with JQ1 has been shown to induce the ubiquitination and degradation of nuclear p65 (Zou et al. 2014). The effects of JQ1(+) to inhibit transcription of a JCV early reporter construct (Fig. 3) and JCV replication in SVGA cells (Fig. 6) provide strong evidence for a role for Brd4 in JCV transcriptional activation and the advancement of the viral life cycle.
With regard to JCV DNA replication, it is clear that Brd4 has a positive effect since the inhibition of Brd4 by JQ1(+) results in decreased late protein expression and virion production during JCV infection of SVGA cells (Fig. 6). Interestingly, Maruyama et al reported that Brd4 interacts with replication factor C (RFC), inhibits RFC-dependent cellular DNA elongation reactions in vitro and retards cell cycle progression from G1 to S (Maruyama et al. 2002). In the case of JCV, replication of viral DNA is critically dependent on JCV T-Ag. In this case, Brd4 likely increases viral replication through its enhancing effects on NF-κB-stimulated transcription of the JCV early promoter and hence increased expression of T-Ag. Similarly in an earlier study, we found that NFAT4 inhibition decreases NF-κB-stimulated early transcription and also JCV replication (Wollebo et al. 2012). For other viruses, Wang et al. (2013) reported that Brd4 was required for human papillomavirus type 16 DNA replication and Brd4 is also involved in the replication of HIV-1 (Liu et al. 2014; Mbonye and Karn 2014) and Merkel cell polyomavirus (Wang et al. 2012) as described below.
Previously, our data with the panel of NF-κB p65 acetylation site lysine mutants had indicated that the lysine residues at positions 218 and 221 are involved in transactivation of JCV early promoter transcription and the effect of histone deacetylation inhibition (Wollebo et al. 2015b). The present data indicate that mutation of either of these two residues to arginine, which cannot be acetylated, abrogates the ability of p65 to mediate Brd4 stimulation of JCV early transcription (Fig. 4) and TNF-α stimulation of p65 translocation (Fig. 5). Thus lysines 218 and 221 are implicated in the mediation of acetylation of p65 in the context of JCV stimulation. This suggests that Brd4 is involved in the regulation of JCV latency/persistency and reactivation.
Brd4 has also been implicated in the reactivation of latent HIV-1 (Liu et al. 2014; Mbonye and Karn 2014). P-TEFb was first identified as a specific host cofactor required for HIV-1 transcription (Marshall and Price 1995). HIV-1 transcription utilizes a complex scheme to recruit P-TEFb and other transcriptional cofactors to the long terminal repeat (LTR) region. These include a P-TEFb/Brd4 complex where the interaction of Brd4 with acetylated histones recruits P-TEFb to the LTR chromatin (Wu and Chiang 2007). This interaction promotes basal HIV-1 transcription but inhibits Tat transactivation because Brd4 competes with Tat for binding to P-TEFb (Yang et al. 2005). Thus, in several systems, JQ1(+) can displace Brd4 from HIV-1 LTR chromatin and antagonize Brd4 inhibition of Tat transactivation resulting in HIV reactivation of latent HIV-1 (Li et al. 2013). Brd4 also has a role in the life cycle of Merkel cell polyomavirus (MCV), interacts with MCV large T-antigen (T-Ag) and plays a critical role in MCV DNA replication (Wang et al. 2012). Brd4 colocalizes with the MCV T-Ag in the viral replication origin complex in the nucleus and recruits replication factor C (RFC) to sites of viral DNA replication (Wang et al. 2012). For papillomavirus, Brd4 functions at several points in the viral life cycle through its ability to interact with the E2 protein, which is a multifunctional protein involved in papillomavirus transcription, maintenance and partitioning of extrachromosomal viral genomes and initiation of viral DNA replication (McBride and Jang 2013). Other viruses, e.g. herpesviruses, also exploit the cellular functions of Brd4 for their own purposes and these have been reviewed by Weidner-Glunde et al. (2010).
For cellular genes, Brd4 is involved in regulation of the promoter/enhancers of many genes, notably those that are involved in the inflammatory response and are inducible by NF-κB (Belkina et al. 2013; Brown et al. 2014; Xu and Vakoc 2014) and Brd4 binding is also a feature of so-called super-enhancers found at key cell identity genes (Amaral and Bannister 2014; Brown et al. 2014; Pott and Lieb 2015; Pelish et al. 2015). Thus Brd4 is a key regulatory protein responsible for the transcriptional response to the acetylation of transcription factors including NF-κB.
In conclusion, Brd4 is a cellular adaptor protein that functions to convey the effects of transcription factors such as NF-κB to downstream effects on transcription. Or data indicate that Brd4 is a key mediator of the effect of NF-κB acetylation on lysines 218 and 221 on JCV transcription. This may offer new insights on possible therapeutic interventions to counter the reactivation of JCV in the glial cells of the brain that leads to PML.
Acknowledgments
We thank past and present members of the Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. Anna Bellizzi was partially supported by postdoctoral fellowship for research Abroad 2014 from Institute Pasteur Cenci-Bolognetti Foundation. We thank Valeria Pietropaolo and Anna T. Palamara for their discussion and input into this study. This study utilized services offered by Temple University School of Medicine Comprehensive NeuroAIDS Center supported by NIH grant P30 MH092177. This work was supported by grant R01 AI077460 awarded by the NIH to MKW.
Footnotes
CONFLICT OF INTEREST
The authors Hassen S. Wollebo, Anna Bellizzi, Dominique H. Cossari, Mahmut Safak, Kamel Khalili and Martyn K. White declare that they have no conflicts of interest.
References
- Amaral PP, Bannister AJ. Re-place your BETs: the dynamics of super enhancers. Mol Cell. 2014;56:187–189. doi: 10.1016/j.molcel.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi A, White MK, Wollebo HS. Degradation of polyomavirus JC T-antigen by stress involves the LIP isoform of C/EBP. Cell Cycle. 2015;14:2075–2079. doi: 10.1080/15384101.2015.1042631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JR. Progressive multifocal leukoencephalopathy and newer biological agents. Drug Saf. 2010;33:969–983. doi: 10.2165/11537510-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Berger JR. The clinical features of PML. Cleve Clin J Med. 2011;78(Suppl 2):S8–12. doi: 10.3949/ccjm.78.s2.03. [DOI] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, Yang T, Wang H, Luscinskas FW, Croce KJ, Bradner JE, Plutzky J. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56(2):219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, Mathews Griner LA, Guha R, Shinn P, Keller JM, Liu D, Patel PR, Ferrer M, Joshi S, Nerle S, Sandy P, Normant E, Thomas CJ, Staudt LM. Blockade of oncogenic IκB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci USA. 2014;111:11365–11370. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahin S, Berger JR. A risk classification for immunosuppressive treatment-associated progressive multifocal leukoencephalopathy. J Neurovirol. 2015;21:623–631. doi: 10.1007/s13365-014-0303-1. [DOI] [PubMed] [Google Scholar]
- Clifford DB. Progressive multifocal leukoencephalopathy therapy. J Neurovirol. 2015;21:632–636. doi: 10.1007/s13365-014-0289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinyan A, White MK, Akan S, Radhakrishnan S, Del Valle L, Amini S, Khalili K. Alterations of DNA damage repair pathways resulting from JCV infection. Virology. 2007;364:73–86. doi: 10.1016/j.virol.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Imperiale MJ, Major EO. Polyomaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6. Lippincott, Williams & Wilkins; Philadelphia: 2013. pp. 6133–1661. [Google Scholar]
- Devaiah BN, Singer DS. Two faces of brd4: mitotic bookmark and transcriptional lynchpin. Transcription. 2013;4:13–17. doi: 10.4161/trns.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, 3rd, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci USA. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant G, Dikstein R. Transcriptional control by NF-κB: elongation in focus. Biochim Biophys Acta. 2013;1829:937–945. doi: 10.1016/j.bbagrm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Miyoshi I, Aster JC, Kubonishi I, Kroll TG, Dal Cin P, Vargas SO, Perez-Atayde AR, Fletcher JA. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19) Am J Pathol. 2001;159:1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jelcic I, Jelcic I, Faigle W, Sospedra M, Martin R. Immunology of progressive multifocal leukoencephalopathy. J Neurovirol. 2015;21:614–22. doi: 10.1007/s13365-014-0294-y. [DOI] [PubMed] [Google Scholar]
- Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41:277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RD, Wu J, Shao R, Xue YH. Mechanism and factors that control HIV-1 transcription and latency activation. J Zhejiang Univ Sci B. 2014;15:455–465. doi: 10.1631/jzus.B1400059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454–455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AA, Jang MK. Current understanding of the role of the Brd4 protein in the papillomavirus lifecycle. Viruses. 2013;5:1374–1394. doi: 10.3390/v5061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Zu Rhein GM, Walker DL, Echroade R, Dessel B. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet. 1971;1(7712):1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A, Fadeyi O, Christie AL, Du K, Banka D, Schneider EV, Jestel A, Zou G, Si C, Ebmeier CC, Bronson RT, Krivtsov AV, Myers AG, Kohl NE, Kung AL, Armstrong SA, Lemieux ME, Taatjes DJ, Shair MD. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol. 2003;77:10638–10644. doi: 10.1128/JVI.77.19.10638-10644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan S, Gordon J, Del Valle L, Cui J, Khalili K. Intracellular approach for blocking JC virus gene expression by using RNA interference during viral infection. J Virol. 2004;78:7264–7269. doi: 10.1128/JVI.78.13.7264-7269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli L, Wollebo HS, Deshmane SL, Mukerjee R, Del Valle L, Safak M, Khalili K, White MK. Modulation of JC virus transcription by C/EBPβ. Virus Res. 2009;146:97–106. doi: 10.1016/j.virusres.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnölzer M, Verdin E, Zhou MM, Ott M. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, Campbell AE, Kawaoka S, Shareef S, Zhu Z, Kendall J, Muhar M, Haslinger C, Yu M, Roeder RG, Wigler MH, Blobel GA, Zuber J, Spector DL, Young RA, Vakoc CR. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Behar M, Inoue K, Hiroshima M, Yasuda T, Nagashima T, Kimura S, Sanjo H, Maeda S, Yumoto N, Ki S, Akira S, Sako Y, Hoffmann A, Kurosaki T, Okada-Hatakeyama M. Positive feedback within a kinase signaling complex functions as a switch mechanism for NF-kB activation. Science. 2014;344:760–764. doi: 10.1126/science.1250020. [DOI] [PubMed] [Google Scholar]
- Tavazzi E, White MK, Khalili K. Molecular insights into polyomavirus JC and clinical updates on progressive multifocal leukoencephalopathy. Rev Med Virol. 2012;22:18–32. doi: 10.1002/rmv.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li J, Schowalter RM, Jiao J, Buck CB, You J. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog. 2012;8:e1003021. doi: 10.1371/journal.ppat.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Helfer CM, Pancholi N, Bradner JE, You J. Recruitment of Brd4 to the human papillomavirus type 16 DNA replication complex is essential for replication of viral DNA. J Virol. 2013;87:3871–3884. doi: 10.1128/JVI.03068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner-Glunde M, Ottinger M, Schulz TF. WHAT do viruses BET on? Front Biosci. 2010;15:537–549. doi: 10.2741/3632. [DOI] [PubMed] [Google Scholar]
- White MK, Skowronska A, Gordon J, Del Valle L, Deshmane SL, Giordano A, Khalili K. Analysis of a mutant p53 protein arising in a medulloblastoma from a mouse transgenic for the JC virus early region. Anticancer Res. 2006;26:4079–4092. [PubMed] [Google Scholar]
- White MK, Safak M, Khalili K. Regulation of gene expression in primate polyomaviruses. J Virol. 2009;83:10846–10856. doi: 10.1128/JVI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Khalili K. Pathogenesis of progressive multifocal leukoencephalopathy – revisited. J Infect Dis. 2011;203:578–586. doi: 10.1093/infdis/jiq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Kaminski R, Khalili K, Wollebo HS. Rad51 activates polyomavirus JC early transcription. PLoS One. 2014;9:e110122. doi: 10.1371/journal.pone.0110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Del Valle L, Safak M, Khalili K, White MK. Role for tumor necrosis factor-alpha in JC virus reactivation and progressive multifocal leukoencephalopathy. J Neuroimmunol. 2011;233:46–53. doi: 10.1016/j.jneuroim.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Melis S, Khalili K, Safak M, White MK. Cooperative Roles of NF-κB and NFAT4 in polyomavirus JC regulation at the KB control element. Virology. 2012;432:146–154. doi: 10.1016/j.virol.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Woldemichaele B, Khalili K, Safak M, White MK. Epigenetic regulation of polyomavirus JC. Virol J. 2013;10:264. doi: 10.1186/1743-422X-10-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, White MK, Gordon J, Berger JR, Khalili K. Persistence and pathogenesis of the neurotropic polyomavirus JC. Ann Neurol. 2015a;77:560–570. doi: 10.1002/ana.24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Bellizzi A, Cossari DH, Safak M, Khalili K, White MK. Epigenetic regulation of polyomavirus JC involves acetylation of specific lysine residues in NF-κB p65. J Neurovirol. 2015b;21:679–687. doi: 10.1007/s13365-015-0326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Bellizzi A, Kaminski R, Hu W, White MK, Khalili K. CRISPR/Cas9 system as an agent for eliminating Polyomavirus JC infection. PLoS One. 2015c;10:e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollebo HS, Cotto B, Adiga R, Langford D, White MK. Expression of Signaling Molecules in Progressive Multifocal Leukoencephalopathy. Curr HIV Res. 2016;14:47–53. doi: 10.2174/1570162x1401151102125319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Vakoc CR. Brd4 is on the move during inflammation. Trends Cell Biol. 2014;24:615–616. doi: 10.1016/j.tcb.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, Nair S, Chen LF. Brd4 maintains constitutively active NF-κB in cancer cells by binding to acetylated RelA. Oncogene. 2014;33:2395–2404. doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]