Abstract
Eomesodermin (Eomes) is a T-box transcription factor that has been implicated in the etiology of colorectal cancer and other human malignancies. We screened a panel of human primary colon cancers and patient-matched controls (n = 30) and detected Eomes overexpression at the mRNA and protein level. Similar results were obtained in a panel of rat colon tumors and adjacent normal-looking colonic mucosa (n = 24). In human colon cancer cells, forced overexpression of Eomes enhanced cell viability and protected against staurosporine-induced apoptosis. On the other hand, knocking down Eomes resulted in reduced cell viability, G2/M cell cycle arrest, and apoptosis induction. The apoptotic mechanism centered on the reciprocal downregulation of anti-apoptotic BIRC5 (Survivin) and upregulation of proapoptotic Bcl-2 modifying factor (BMF). In patients with colorectal cancer, high EOMES expression (n = 95) was associated with poor overall survival compared with individuals exhibiting low EOMES levels (n = 80). We conclude from the current investigation, and prior literature, that Eomes has a divergent role in cancer development, with evidence for tumor suppressor and oncogenic functions, depending on stage and tissue context. Further studies are warranted on the apoptotic mechanisms linked to the reciprocal regulation of BMF and BIRC5 in human colorectal cancers characterized by Eomes overexpression.
Keywords: Apoptosis, Bcl-2 modifying factor, Caspase, Heterocyclic amine, TCGA
Introduction
Eomes, also known as T-brain gene 2 (TBR2), was initially identified in Xenopus as a key early gene in mesoderm differentiation of gastrula stage embryos [1]. In mammals, Eomes regulates neurogenesis in the cortical subventricular zone [2], and is essential for hippocampal lineage progression from neural stem cells to intermediate progenitors and neurons [3]. Eomes also regulates CD8 T cell and natural killer (NK) cell differentiation and function, and is required for NK lineage cells to be maintained [4]. In CD8 T cells, Eomes is associated with the self-renewal of antigen-specific central memory cells. Eomes-deficient CD8 T cells undergo primary clonal expansion, but are defective in long-term survival, populating the bone marrow niche and re-expanding after re-challenge [5]. In human embryonic stem cells and mouse epiblast stem cells, EOMES is among the first genes induced upon endoderm differentiation and is regulated by pluripotency factors such as Nanog, Oct4 and Sox2 [6].
There is accumulating evidence for a role of Eomes in cancer etiology. In medulloblastoma, TBR1 (another T-box transcription factor) and Eomes/TBR2 were inversely associated and linked to the extent of DNA hypermethylation [7]. In bladder cancer, EOMES hypermethylation was one of several urinary biomarkers used for early detection of tumor recurrence [8]. Approximately 25% of patients with no EOMES methylation experienced a recurrence within 2 years, compared with recurrence in over 50% of subjects who were positive for EOMES methylation [8]. In human hepatocellular carcinoma, analyses of DNA methylation and hydroxymethylation identified EOMES as a candidate tumor suppressor [9]. In human colorectal cancer, EOMES expression was analyzed using in situ hybridization, identifying 46/88 (52%) and 42/88 (48%) of cases as being Eomes negative and Eomes positive, respectively [10]. A significant inverse correlation was noted for EOMES expression in colorectal cancers and the presence of lymph node metastases [10]. Another T-box transcription factor, T-bet, like Eomes, was implicated in the immunological control of tumor spread in colorectal cancer, via activated cytolytic CD8 T cells [11].
Given that both high and low Eomes expression has been observed in human colorectal cancer [10], we sought to clarify the role of Eomes using (i) human primary colon cancers and patient-matched controls, (ii) a preclinical model of colon tumor formation, and (iii) human colon cancer cells subjected to Eomes knockdown or overexpression. Mechanistic studies reported here implicated the reciprocal regulation of proapoptotic Bcl-2 modifying factor (Bmf) and antiapoptotic Survivin associated with Eomes overexpression in colorectal cancer.
Materials and methods
Source of tumors
Thirty pairs of frozen primary human colon adenocarcinoma specimens and patient-matched adjacent normal-looking tissues were kindly provided by Steven F. Moss and Lelia Simao (Rhode Island Hospital, Providence, RI). Human colon normal slides (ab4327) were obtained from Abcam (Cambridge, MA, USA), whereas colon cancer tissue microarrays (TMAs, BN05011) were purchased from US Biomax, Inc. (Rockville, MD, USA).
Colon tumors and matched adjacent normal-looking colonic mucosa samples were from one-year carcinogenicity bioassays in which male F344 rats were treated with a heterocyclic amine [12–16]. In some experiments, colonic mucosa from rats treated with no carcinogen was used as a negative control. The studies received prior approval from the Institutional Animal Care and Use Committee.
Quantitative real-time RT-PCR (qRT-PCR)
Frozen colon tumor samples and matched controls were thawed, and mRNA was extracted using the RNeasy kit (Qiagen, Valencia, CA, USA). RNA (2 μg) was reverse-transcribed using SuperScript III (Life Technologies, Grand Island, NY, USA). EOMES, BMF and BIRC5 mRNA levels were measured by qRT-PCR and normalized to ACTB. Forty cycles of PCR (95 °C/10 s, 58 °C/10 s, 72 °C/10 s) were run on a LightCycler 480 II system (Roche, Indianapolis, USA), in 20 μl total reaction volume containing cDNA, SYBR Green I dye (Roche), and target-specific primers. The amount of specific mRNA was quantified by determining the point at which the fluorescence accumulation entered the exponential phase (Ct), and the Ct ratio of the target gene to ACTB was calculated for each sample. At least three separate experiments were performed for each sample.
Immunoblotting
Proteins were immunoblotted using the methodologies reported [17–23]. Primary antibodies were from the following sources: Eomes/anti-TBR2 (Abcam); poly(ADP-ribose)polymerase (PARP), cleaved caspase 3, cleaved caspase 7, Survivin and Bmf (Cell Signaling, Beverly, MA, USA); β-actin (Sigma, St. Louis, MO, USA). Proteins were visualized and quantified as reported [18,19,23].
Immunohistochemistry
Tissues were sectioned at 4–5 μm, and Eomes antibody (Abcam, ab23345) was used at 1:25 dilution for human normal or colon cancer specimens, or 1:250 dilution for rat colon tumors and Swiss-rolled normal rat colonic tissue. Other details of the basic methodology were reported elsewhere [12,13].
Cell culture and transient transfection
Human HCT116, HT29, and other colon cancer cells (American Type Culture Collection, Manassas, VA, USA) were maintained in McCoy’s 5A medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone Laboratories, Logan, UT, USA), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. Cells (6 × 105) were seeded in 6-well plates overnight in antibiotic free media. For siRNA transfection, Eomes siRNA (SASI_Hs02_00339597, Sigma) or non-target siRNA control (siRNA universal negative control #1, SIC001, Sigma) at 100 nM final concentration was mixed with 12 μl Lipofectamine RNAiMax (Life Technologies) in serum reduced medium for 15 min at room temperature. For cDNA transfection, 1 μg Eomes cDNA construct (Origene Technologies, Rockville, MD, USA) or the corresponding pCMV6-AC-GFP empty vector was mixed with 2 μl X-tremeGENE HP transfection reagent (Roche) in serum reduced medium for 20 min, and then added to cell medium. Unless stated otherwise, cells were harvested 48 or 72 h after transfection.
In some experiments, 48 h after knockdown or overexpression of Eomes, staurosporine (STS, Abcam) was added to a final concentration of 0.5 μM in cell culture medium, with dimethylsulfoxide (DMSO) serving as vehicle control. Cells were examined 24 h later under the fluorescence microscope for signs of chromatin condensation, nuclear fragmentation, or membrane blebbing indicative of apoptosis. Cells also were harvested for immunoblotting, as described above.
Cell viability
Cells (3 × 103) in 100 μl media were seeded in 96-well plates overnight and transfected with siRNA or cDNA, as described above. At 24, 48, 72 and 96 h after transfection, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added and incubated for 3 h, followed by 100 μl of 10% SDS in 0.01 N HCl. Formation of formazan dye was assessed colorimetrically at 550 nm.
Cell cycle distribution
Cells treated with Eomes siRNA for 48 and 72 h, as described above, were harvested in cold PBS, fixed in 70% ethanol, and stored at 4 °C for 48 h. Fixed cells were washed with PBS and resuspended in propidium iodide/Triton X-100 staining solution containing RNase A. Samples were incubated in the dark for 30 min before determining the DNA content on an FC 500 Beckman Coulter flow cytometer. Cell cycle distribution was assessed using Multicycle Software (Phoenix Flow Systems, San Diego, CA).
Immunocytochemistry
Cells were grown on poly-L-lysine coated coverslips in 12-well plates. Forty-eight hours after transfection with Eomes expression construct or empty vector, and 24 h after STS or vehicle treatment (see above), cells were fixed with 5% buffered formalin phosphate, pH 7.2 for 10 min, washed with PBS, and incubated with methanol for 15 min. After treatment with 2.1% citric acid solution containing 0.5% Tween 20 for 10 min, cells were incubated with 1% BSA in PBS for 1 h. Primary antibody was incubated overnight at 4 °C, and after washing with PBS, Chromeo 642 goat anti-rabbit antibody (Active Motif, Carlsbad, CA, USA) was applied for 1 h at room temperature. ProLong® Gold Antifade with DAPI (Life Technologies) was applied and cells were examined by fluorescence microscope.
Statistics
Data were expressed as mean ± s.d. and comparisons were made between control and treatment groups (Student’s t-test). In the figures, significant results were indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Eomes is overexpressed in human and rat colon tumors
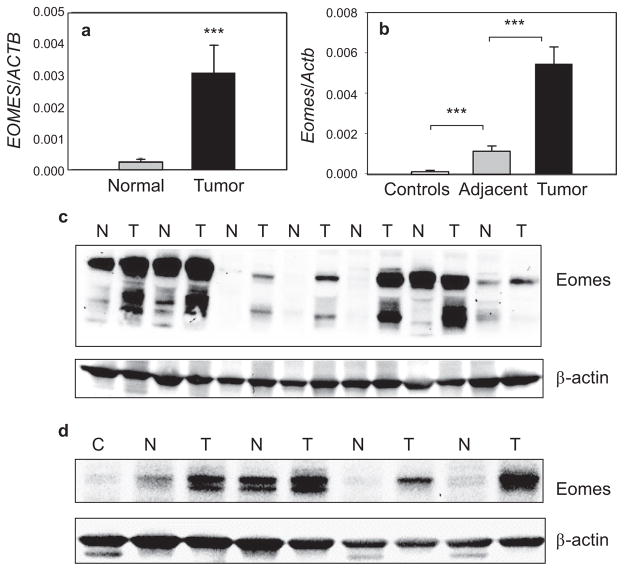
Human primary colon cancers had significantly elevated EOMES mRNA levels when compared with patient-matched controls (Fig. 1a). Specifically, EOMES had a relative expression level of 3.1 ± 1.2 × 10−3 in tumors compared with 0.26 ± 0.1 × 10−3 in normal-looking controls (***P < 0.001), representing a ~ 12-fold increase overall in human colon cancers expressing high EOMES mRNA levels. The thirty primary human colon cancers were characterized histopathologically as well-differentiated adenocarcinomas, and higher EOMES expression was detected in each case compared with the corresponding patient-matched control (30/30 = 100%).
Fig. 1.
Eomes overexpression in human primary colon cancers and rat colon tumors. (a) qRT-PCR data for EOMES normalized to ACTB in primary colon cancers and patient-matched controls, n = 30, mean ± s.d.; ***P < 0.001. (b) qRT-PCR data for Eomes normalized to Actb in carcinogen-induced rat colon tumors (n = 24), adjacent normal-looking colonic mucosa (n = 24), and colonic mucosa from untreated controls (n = 8), mean ± SD; ***P < 0.001. (c) Immunoblotting of primary human colon tumors (T) and patient-matched normal colon (N). (d) Rat colon tumors (T) and adjacent normal-looking colon (N). Immunoblots included normal colonic mucosa (C) from untreated rats as additional controls. Immunoblot data are from a single experiment and are representative of findings from two or more separate experiments. Note: EOMES = human, Eomes = rat gene designation.
In rat colon tumors archived from a previous study [12,13], Eomes mRNA expression was elevated 5-fold compared with adjacent normal-looking colon, or 78-fold compared with colonic mucosa from untreated controls (Fig. 1b). Normal-looking colonic mucosa adjacent to tumor also had 16-fold higher Eomes mRNA expression compared with colonic mucosa from untreated rats (Fig. 1b).
In human primary colon cancers, immunoblotting detected bands at ~75 kD and ~60 kD, corresponding to different Eomes isoforms (UniProtKB-O95936). In most cases, Eomes isoforms were elevated in tumors compared with patient-matched controls (Fig. 1c). Eomes protein expression also was consistently higher in rat colon tumors compared with adjacent normal-looking colon, whereas it was scarcely detectable in normal colonic mucosa from untreated controls (Fig. 1d).
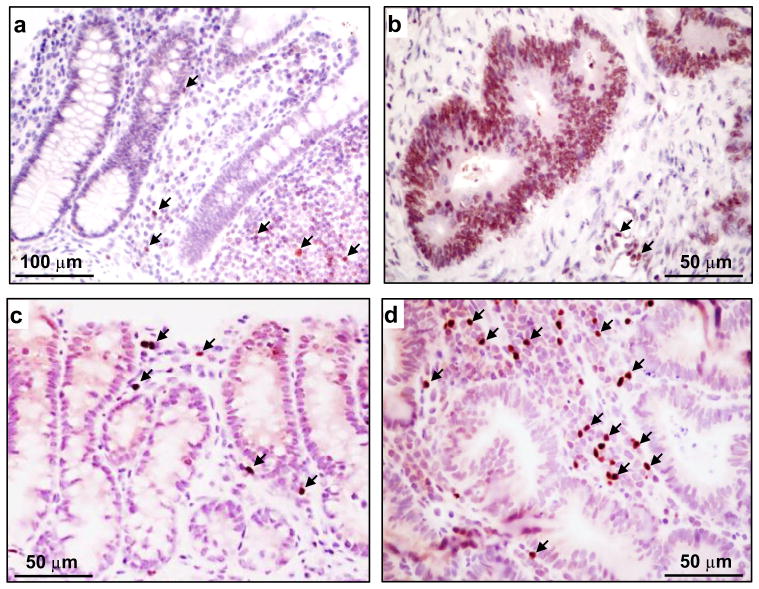
Human normal colon had Eomes mainly immunolocalized to lymphocytes (Fig. 2a), whereas human primary colon cancers had Eomes strongly expressed in epithelial cells, as well as in stroma lymphocytes (Fig. 2b). In rat colon tumors and adjacent normal-looking colonic mucosa, Eomes was immunolocalized mainly to lymphocytes (Figs 2c,d), with a staining pattern and lymphocytic infiltrate that tended to be more prominent within the tumors. The Human Protein Atlas also revealed that human colorectal cancers had Eomes localized to cytoplasmic and membrane compartments (HumanProteinAtlasEOMES: http://www.proteinatlas.org/ENSG00000163508-EOMES/cancer/tissue/colorectal+cancer).
Fig. 2.
Eomes immunohistochemistry. (a) Human normal colon. (b) Human colon adenocarcinoma. (c) Rat normal-looking colonic mucosa adjacent to tumor. (d) Rat colon tumor. Arrows, Eomes-positive lymphocytes. Scale bar indicates 50 or 100 μm, as shown in the corresponding panel.
Eomes overexpression attenuates staurosporine-induced apoptosis
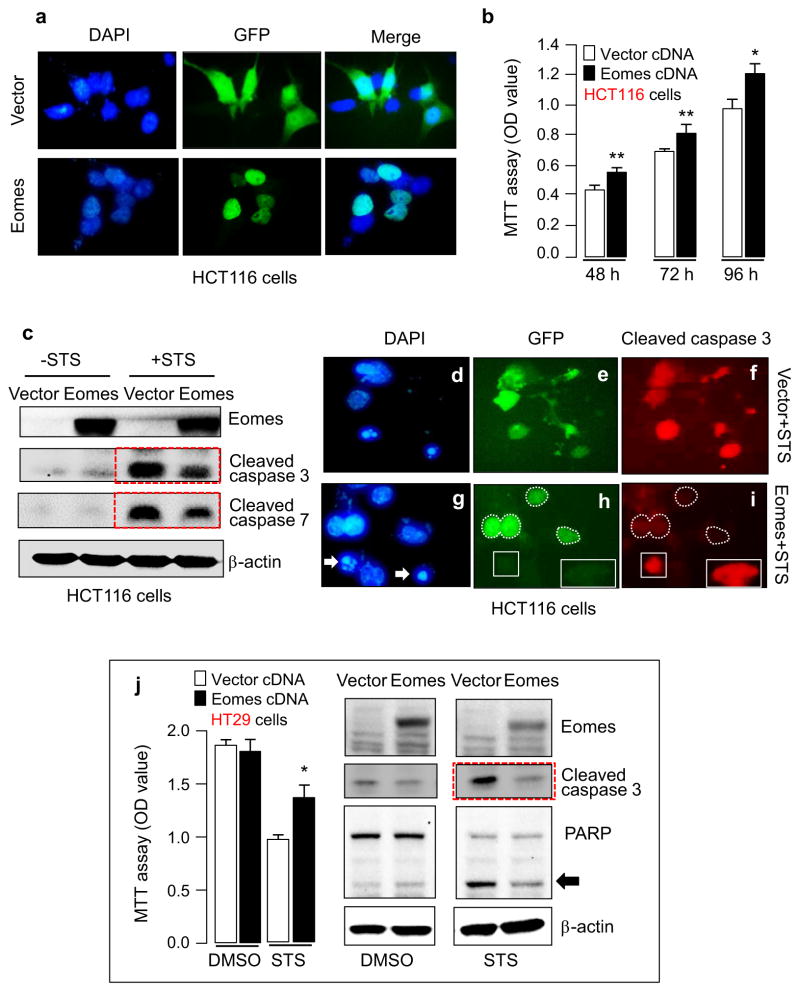
We screened a panel of human colon cancer and non-transformed colonic epithelial cell lines for EOMES mRNA expression; HCT116 cells had intermediate EOMES mRNA levels (data not shown) and were chosen for knockdown and overexpression experiments. Transient transfection of an Eomes expression construct revealed strong nuclear localization in cells that were GFP-positive, consistent with the transcription factor role of Eomes, whereas diffuse GFP expression was observed in cells transfected with the vector control (Fig. 3a). In cells transiently transfected with Eomes expression construct, cell viability was increased significantly between 48 and 96 h (Fig. 3b). Transient transfection of the Eomes construct for 48 h followed by 24 h incubation with staurosporine (STS) resulted in cleaved caspase levels that were noticeably reduced compared with the corresponding vector control treated with STS (Fig. 3c, red dashed boxes). Cells that overexpressed exogenously transfected Eomes (e.g., white dashed regions in Fig. 3h) were resistant to STS-induced apoptosis, as shown by the absence of cleaved caspase 3 in those cells (white dashed regions in Fig. 3i). On the other hand, cells that lacked exogenous Eomes (e.g., white boxed areas in Fig. 3h) had abundant cleaved caspase 3 (Fig. 3i) and nuclear condensation/ fragmentation indicative of apoptosis (e.g., Fig. 3g, arrows).
Fig. 3.
Forced expression of Eomes enhances cell viability and resists apoptosis induction. (a) HCT116 cells after transient transfection with green fluorescent protein (GFP)-vector control or GFP-Eomes expression construct, with 4′,6-diamidino-2-phenylindole (DAPI) as a nuclear counterstain. (b) MTT assays revealed increased cell viability after Eomes overexpression. Data = mean ± s.d., n = 3; *P < 0.05; **P < 0.01. (c) Immunoblotting revealed attenuated staurosporine (STS)-induced caspase activation after forced expression of Eomes. (d)–(i) STS-treated cells after transient transfection with vector or Eomes construct. Arrows, nuclear condensation/fragmentation in cells positive for cleaved caspase 3; white dashed lines, cells resistant to STS-induced apoptosis after forced Eomes expression; white boxes, cells lacking GFP (no exogenous Eomes) that underwent apoptosis, as evidenced by the presence of cleaved caspase 3. (j) Additional experiments with STS and forced expression of Eomes were conducted in HT29 cells. Data bars = mean ± s.d., n = 3; *P < 0.05. Arrow, cleaved poly(ADP-ribose)polymerase (PARP).
Transient transfection of the Eomes construct for 48 h followed by 24 h incubation with STS also was performed in HT29 cells (Fig. 3j). Forced expression of Eomes rescued HT29 cells from the reduced viability caused by STS treatment (*P < 0.05), and lowered STS-induced apoptotic markers, such as cleaved caspase 3 (Fig. 3j, red dashed box) and cleaved PARP (Fig. 3j, arrow). These results indicated that Eomes overexpression attenuated STS-induced apoptosis in HCT116 cells containing wild type p53, and in HT29 cells harboring mutant p53. Changes in cell density paralleled the findings from the MTT assays, although cell counts were not specifically recorded after Eomes overexpression or knockdown (see next).
Eomes knockdown causes reduced cell viability and activates apoptosis
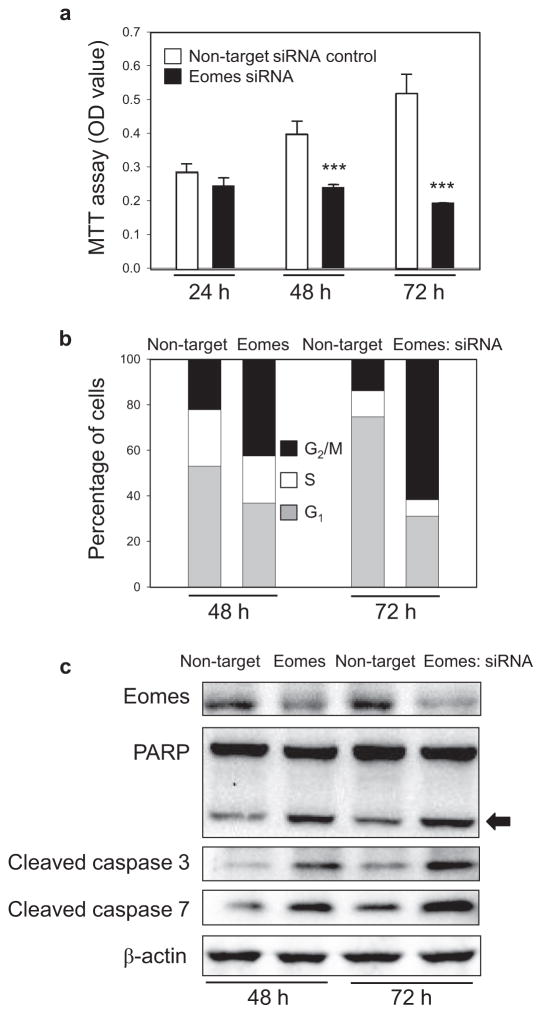
Subsequent experiments used Eomes-specific siRNA as a knockdown strategy (Fig. 4). In contrast to enhanced cell viability after Eomes overexpression (Fig. 3b), cell viability was inhibited significantly after 48 h in HCT116 cells transfected with Eomes-specific siRNA (Fig. 4a, solid bars). At 48 and 72 h post-transfection, 22% and 14% of the cells incubated with non-target siRNA occupied G2/M of the cell cycle, compared with 43% and 62% of the cells after Eomes knockdown (Fig. 4b, black bars), indicating G2/M cell cycle arrest. At the later time-point of 72 h, 15.90% of cells treated with Eomes-specific siRNA were present in the sub-G0/G1 compartment, compared with 6.63% of cells treated with non-target siRNA. Enhanced apoptosis due to Eomes knockdown was evident from morphological criteria, such as membrane blebbing and nuclear condensation (not shown), and from increased levels of cleaved caspases and PARP (Fig. 4c). Similar results were obtained in other colon cancer cell lines transfected with Eomes-specific siRNA (data not shown). We conclude that Eomes knockdown was synonymous with reduced cell viability and the activation of apoptosis.
Fig. 4.
Knockdown of Eomes reduces cell viability and triggers G2/M arrest and apoptosis. Human HCT116 cells treated with Eomes-specific siRNA exhibited (a) reduced viability, (b) G2/M arrest, and (c) apoptosis induction. Arrow, cleaved PARP. Results were from a single experiment, each repeated at least twice. Data in (a) indicate mean ± s.d., n = 3; ***P < 0.001.
Eomes reciprocally regulates Bmf and Survivin in human colon cancer cells
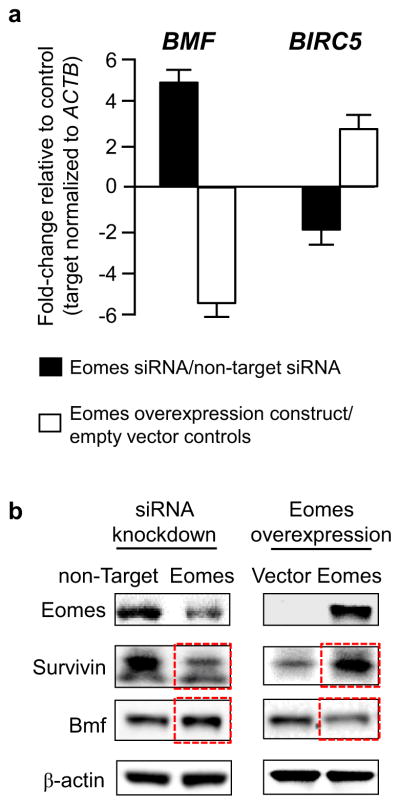
To investigate the apoptotic mechanism in more detail, multiple survival factors were profiled, as reported [17]. These experiments highlighted proapoptotic BMF and anti-apoptotic BIRC5 (the gene for Survivin) as key regulators of Eomes-mediated apoptosis in human colon cancer cells. Knockdown experiments using Eomes-specific siRNA induced BMF markedly while attenuating BIRC5 (Fig. 5a, solid bars), whereas transient overexpression of Eomes had the opposite effects, downregulating BMF and increasing BIRC5 (Fig. 5a, open bars). Immunoblotting confirmed the loss of Survivin with increased Bmf after Eomes knockdown, and increased Survivin with attenuated Bmf following Eomes overexpression (Fig. 5b).
Fig. 5.
BMF and BIRC5 (Survivin) are reciprocally regulated by Eomes. (a) Fold changes in BMF and BIRC5 mRNA levels after knockdown or overexpression of Eomes; qRT-PCR data (mean ± s.d., n = 3) were normalized to ACTB and then expressed relative to the corresponding non-target siRNA or vector control. (b) Immunoblots showing reciprocal changes in Survivin and Bmf protein expression (red boxes) after knockdown or overexpression of Eomes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Discussion
In this investigation, rat colon tumors and primary human colon cancers were found to overexpress Eomes at the mRNA and protein level. In human colon cancer cells, forced overexpression of Eomes increased cell viability and protected against STS-induced apoptosis, whereas Eomes knockdown resulted in reduced cell viability, G2/M arrest, and apoptosis induction. A key question for the future is whether the phenotypic changes observed in cell-based assays involving Eomes overexpression and knockdown might be recapitulated in vivo, for example, in mouse tumor xenograft experiments designed to track tumor growth rates and drug sensitivity. An array-based screen of multiple survival factors in human colon cancer cells implicated BIRC5 and BMF as critical players in the apoptotic mechanism of Eomes. In accordance with a possible direct transcriptional role, the BMF gene promoter has an Eomes binding site upstream of the transcription start site [6], in a region known to interact with transcription factors such as STAT3 and Sp3 [17]. Investigation of the corresponding region in BIRC5 identified no Eomes interactions (data not shown), consistent with prior work that detected Eomes on BIRC2, BIRC6, and BIRC7, but not on BIRC5 [6]. Post-transcriptional regulation of BMF and BIRC5 also might be an important mechanism, via the actions of one or more microRNAs [24–27], which also serve to regulate Eomes [28,29]. Further studies are warranted on the reciprocal regulation of BMF and BIRC5 in response to changes in Eomes expression.
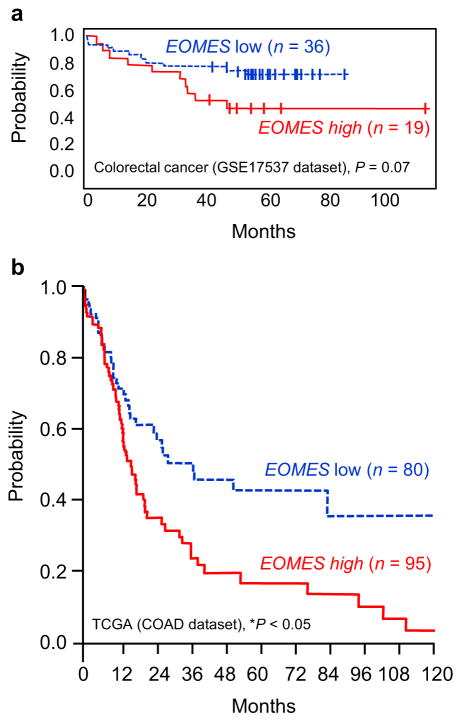
The results reported here imply an oncogenic role for Eomes in the large intestine. Interestingly, colorectal cancer dataset GSE17537 in PrognoScan [30] suggested worse overall survival in patients with high versus low EOMES expression (Fig. 6a). Mining of colorectal cancer data in the Cancer Browser indicated that patients in the upper quartile of EOMES expression had significantly worse overall survival than those in the lower quartile (Fig. 6b).
Fig. 6.
Survival curves for colorectal cancer patients in relation to EOMES expression. (a) Kaplan–Meier curves for colorectal cancer dataset GSE17537 in PrognoScan [30], stratified by high vs. low EOMES expression. (b) COADREAD results from The Cancer Genome Atlas (http://cancergenome.nih.gov/). Patients were stratified into upper and lower quartiles according to EOMES expression.
These findings are not in accordance with Eomes as a general protective factor in cancer etiology. In medulloblastoma, bladder cancer, and hepatocellular carcinoma, low Eomes expression and DNA hypermethylation coincided with a better prognosis [7–9]. However, in human colorectal cancer, in situ hybridization studies suggested a broad range of EOMES expression, with approximately half having low/no and half having high EOMES levels [10]. The current investigation of Eomes focused on colonic epithelial cells, rather than the tumor-infiltrating immune cells involved in metastasis [11]. Thus, for Eomes, stage of colorectal cancer may be an important consideration. The heterocyclic amine-induced rat colon carcinogenesis model used here is characterized by activation of β-catenin-dependent signaling [31–33], but does not typically involve invasion and metastasis [34–39]. Interestingly, in heterocyclic amine-treated mice, immunosuppressive effects were linked to the inhibition of T cell proliferation [40] and immunotoxicity in lymphoid tissues of the gut [41]. In these animal models, it might be instructive to assess immune cell infiltration and mechanisms governing the functional activity of T lymphocytes, via T-box transcription factors such as Eomes.
In summary, the current investigation and prior literature reports highlight a possible divergent role for Eomes in cancer etiology. Overexpression of EOMES in human and rat colon cancers prompted mechanistic work in human colon cancer cells that implicated a reciprocal regulation of BMF and BIRC5. Further studies are warranted on the timing of Eomes upregulation, from early stages through invasion and metastasis, and the precise apoptotic mechanisms involved.
Acknowledgments
Supported by NIH grants CA090890, CA122959, ES00210, and ES023512, the John S. Dunn Foundation, and a Chancellor’s Research Initiative from Texas A&M University.
Abbreviations
- ACTB
Actb (β-actin gene human, murine)
- Eomes
eomesodermin
- BIRC5
Survivin gene
- BMF
Bcl-2 modifying factor gene
- Bmf
Bmf protein
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethylsulfoxide
- GFP
green fluorescent protein
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NK
natural killer
- PARP
poly(ADP-ribose)polymerase
- qRT-PCR
quantitative real-time polymerase chain reaction
- STS
staurosporine
- TBR2
T-brain gene 2
- TMA
tissue microarray
Footnotes
Conflict of interest
The authors have nothing to disclose.
References
- 1.Ryan K, Garrett N, Mitchell A, Gurdon JB. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell. 1996;87:989–1000. doi: 10.1016/s0092-8674(00)81794-8. [DOI] [PubMed] [Google Scholar]
- 2.Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodge RD, Nelson BR, Kahoud RJ, Yang R, Mussar KE, Reiner SL, et al. Tbr2 is essential for hippocampal progression from neural stem cells to intermediate progenitors and neurons. J Neurosci. 2012;22:6275–6287. doi: 10.1523/JNEUROSCI.0532-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, et al. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, et al. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DT, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinert T, Borre M, Christiansen A, Hermann GG, Ørntoft TF, Dyrskøt L. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS ONE. 2012;7:e46297. doi: 10.1371/journal.pone.0046297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao F, Xia Y, Wang J, Lin Z, Ou Y, Liu X, et al. Integrated analyses of DNA methylation and hydroxymethylation reveal tumor suppressor roles of ECM1, ATF5, and EOMES in human hepatocellular carcinoma. Genome Biol. 2014;15:533. doi: 10.1186/s13059-014-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atreya I, Schimanski CC, Becker C, Wirtz S, Dornhoff H, Schnürer E, et al. The T-box transcription factor eomesodermin controls CD8 T cell activity and lymph node metastasis in human colorectal cancer. Gut. 2007;56:1572–1578. doi: 10.1136/gut.2006.117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Dashwood WM, Nian H, Löhr CV, Fischer KA, Tsuchiya N, et al. NADPH oxidase overexpression in human colon cancers and rat colon tumors induced by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Int J Cancer. 2011;128:2581–2590. doi: 10.1002/ijc.25610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang R, Löhr CV, Fischer K, Dashwood WM, Greenwood JA, Ho E, et al. Epigenetic inactivation of endothelin-2 and endothelin-3 in colon cancer. Int J Cancer. 2013;132:1004–1012. doi: 10.1002/ijc.27762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, Dashwood WM, Löhr CV, Fischer KA, Nakagama H, Williams DE, et al. Beta-Catenin is strongly elevated in rat colonic epithelium following short-term intermittent treatment with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and a high-fat diet. Cancer Sci. 2008;99:1754–1759. doi: 10.1111/j.1349-7006.2008.00887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Dashwood WM, Löhr CV, Fischer KA, Pereira CB, Louderback M, et al. Protective versus promotional effects of white tea and caffeine on PhIP-induced tumorigenesis and beta-catenin expression in the rat. Carcinogenesis. 2008;29:834–839. doi: 10.1093/carcin/bgn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parasramka M, Dashwood WM, Wang R, Abdelli A, Bailey GS, Williams DE, et al. MicroRNA profiling of carcinogen-induced rat colon tumors and the influence of dietary spinach. Mol Nutr Food Res. 2012;56:1259–1269. doi: 10.1002/mnfr.201200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang Y, Nian H, Rajendran P, Kim E, Dashwood WM, Pinto JT, et al. HDAC8 and STAT3 repress BMF gene activity in colon cancer cells. Cell Death Dis. 2014;5:e1476. doi: 10.1038/cddis.2014.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajendran P, Delage B, Dashwood WM, Yu TW, Wuth B, Williams DE, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. doi: 10.1186/1476-4598-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajendran P, Kidane AI, Yu TW, Dashwood WM, Bisson WH, Löhr CV, et al. HDAC turnover, CtIP acetylation and dysregulated DNA damage signaling in colon cancer cells treated with sulforaphane and related dietary isothiocyanates. Epigenetics. 2013;8:612–623. doi: 10.4161/epi.24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parasramka M, Dashwood WM, Wang R, Saeed HH, Williams DE, Ho E, et al. A role for low-abundance miRNAs in colon cancer: the miR-206/Krüppel-like factor 4 (KLF4) axis. Clin Epigenetics. 2012;4:16. doi: 10.1186/1868-7083-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nian H, Bisson WH, Dashwood WM, Pinto JT, Dashwood RH. Alpha-keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis. 2009;30:1416–1423. doi: 10.1093/carcin/bgp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compounds, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008;29:1816–1824. doi: 10.1093/carcin/bgn165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajendran P, Dashwood WM, Li L, Kang Y, Kim E, Johnson G, et al. Nrf2 affects tumor growth, HDAC3 gene promoter interactions, and the response to sulforaphane in the colon. Clin Epigenetics. 2015;7:102. doi: 10.1186/s13148-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, et al. Mir-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem. 2009;23:347–358. doi: 10.1159/000218181. [DOI] [PubMed] [Google Scholar]
- 25.Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor mutlifocality. Clin Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, et al. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Wang S, Han F, Li J, Yu L, Zhou P, et al. MicroRNA-542-3p suppresses cellular proliferation of bladder cancer cells through post-transcriptionally regulating Survivin. Gene. 2016;579:146–152. doi: 10.1016/j.gene.2015.12.048. [DOI] [PubMed] [Google Scholar]
- 28.Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-29b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci USA. 2013;110:7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, et al. MicroRNA-29 regulates T-box transcription factors and interferon-g production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dashwood RH, Suzui M, Nakagama H, Sugimura T, Nagao M. High frequency of beta-catenin (Ctnnb1) mutation in the colon tumors induced by two heterocyclic amines in the F344 rat. Cancer Res. 1998;58:1127–1129. [PubMed] [Google Scholar]
- 32.Li Q, Dashwood WM, Zhong X, Nakagama H, Dashwood RH. Bcl-2 overexpression in PhIP-induced colon tumors: cloning of the Bcl-2 promoter and characterization of a pathway involving beta-catenin, c-Myc and E2F1. Oncogene. 2007;28:6194–6202. doi: 10.1038/sj.onc.1210438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Fageeh M, Li Q, Dashwood WD, Myzak MC, Dashwood RH. Phosphorylation and ubiquitination of oncogenic mutants of beta-catenin containing substitution at Asp32. Oncogene. 2004;23:4839–4846. doi: 10.1038/sj.onc.1207634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochiai M, Ogawa K, Wakabayashi K, Sugimura T, Nagase S, Esumi H, et al. Induction of intestinal adenocarcinomas by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in Nagase analbuminemic rats. Jpn J Cancer Res. 1991;82:363–366. doi: 10.1111/j.1349-7006.1991.tb01855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito N, Hasegawa R, Sano M, Tamano S, Esumi H, Takayama S, et al. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) Carcinogenesis. 1991;12:1503–1506. doi: 10.1093/carcin/12.8.1503. [DOI] [PubMed] [Google Scholar]
- 36.Guo D, Schut HA, Davis CD, Snyderwine EG, Bailey GS, Dashwood RH. Protection by chlorophyllin and indole-3-carbinol against 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-induced DNA adducts and colonic aberrant crypts in the F344 rat. Carcinogenesis. 1995;16:2931–2937. doi: 10.1093/carcin/16.12.2931. [DOI] [PubMed] [Google Scholar]
- 37.Guo D, Horio DT, Grove JS, Dashwood RH. Inhibition by chlorophyllin of 2-amino-3-methylimidazo[4,5-f]quinoline-induced tumorigenesis in the male F344 rat. Cancer Lett. 1995;95:161–165. doi: 10.1016/0304-3835(95)03882-w. [DOI] [PubMed] [Google Scholar]
- 38.Takeshita F, Ogawa K, Asamoto M, Shirai T. Mechanistic approach of contrasting modifying effects of caffeine on carcinogenesis in the rat colon and mammary gland induced with 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Lett. 2003;194:25–35. doi: 10.1016/s0304-3835(03)00051-x. [DOI] [PubMed] [Google Scholar]
- 39.Ochiai M, Watanabe M, Nakanishi M, Taguchi A, Sugimura T, Nakagama H. Differential staining of dysplastic aberrant crypt foci in the colon facilitates prediction of carcinogenic potentials of chemical in rats. Cancer Lett. 2005;220:67–74. doi: 10.1016/j.canlet.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Yun CH, Son CG, Jung U, Han SH. Immunosuppressive effect of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) through the inhibition of T-lymphocyte proliferation and IL-2 production. Toxicology. 2006;217:31–38. doi: 10.1016/j.tox.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Davis DA, Archuleta MM, Born JL, Knize MG, Felton JS, Burchiel SW. Inhibition of humoral immunity and mitogen responsiveness of lymphoid cells following oral administration of the heterocyclic food mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) to B6C3F1 mice. Fundam Appl Toxicol. 1994;23:81–86. doi: 10.1006/faat.1994.1082. [DOI] [PubMed] [Google Scholar]