Abstract
Pathogenic amino acid substitutions of the common E3 component (hE3) of the human alpha-ketoglutarate dehydrogenase and the pyruvate dehydrogenase complexes lead to severe metabolic diseases (E3 deficiency), which usually manifest themselves in cardiological and/or neurological symptoms and often cause premature death. To date, 14 disease-causing amino acid substitutions of the hE3 component have been reported in the clinical literature. None of the pathogenic protein variants has lent itself to high-resolution structure elucidation by X-ray or NMR. Hence, the structural alterations of the hE3 protein caused by the disease-causing mutations and leading to dysfunction, including the enhanced generation of reactive oxygen species by selected disease-causing variants, could only be speculated. Here we report results of an examination of the effects on the protein structure of ten pathogenic mutations of hE3 using hydrogen/deuterium-exchange mass spectrometry (HDX-MS), a new and state-of-the-art approach of solution structure elucidation. On the basis of the results, putative structural and mechanistic conclusions were drawn regarding the molecular pathogenesis of each disease-causing hE3 mutation addressed in this study.
Keywords: dihydrolipoamide dehydrogenase, pathogenic mutation, E3 deficiency, hydrogen/deuterium exchange, mass spectrometry
Graphical Abstract
1. Introduction
Human dihydrolipoamide dehydrogenase (hLADH, hE3) is the common third component enzyme in the mitochondrial 2-oxo acid dehydrogenase complex family comprising the pyruvate dehydrogenase complex (PDHc), the α-ketoglutarate (or 2-oxoglutarate) dehydrogenase complex (KGDHc or OGDHc), and the branched-chain α-keto acid dehydrogenase complex (BCKDHc); hE3 is also part of the glycine cleavage system [1–6]. The hE3 is an obligate homodimer of 474 amino acids in each monomer after cleavage of the mitochondrial signal peptide (35 amino acids); each monomer binds tightly, but non-covalently, a single FAD prosthetic group [7]. The overall stoichiometries (the numbers of E1, E2 and E3 subunits) of the above multienzyme complexes harboring hE3 are generally different and may also vary in different species owing to the specific reaction and the biochemical environment (e.g., substrate provision) [1, 3, 4, 8–12]; optimal stoichiometries of the KGDHc and the PDHc from human and E. coli origins were determined recently by us via reconstitution experiments using recombinant components [6]. The KGDHc catalyzes a rate-limiting step in the Krebs cycle facilitating the oxidative decarboxylation of α-ketoglutarate and generating succinyl-CoA and NADH, whereas PDHc, with a similar mechanism, converts pyruvate to acetyl-CoA and NADH [13]. The function of the common E3 component in the catalytic mechanism is to reoxidize the reduced dihydrolipoic acid moieties, covalently linked to the respective E2 components, while generating NADH. It has been demonstrated that in addition to their physiological catalytic activities, KGDHc and PDHc can generate significant amounts of the reactive oxygen species (ROS) superoxide and, its dismutation product, H2O2 under pathologically relevant conditions [6, 14–17]. ROS produced by the KGDHc was proved to be a major source of oxidative stress inside the mitochondrion [14, 15, 18, 19] which, together with the impaired activity of the KGDHc under certain pathological conditions, is heavily implicated in the progression of senescence/aging, neurodegenerative diseases (such as Alzheimer’s and Parkinson’s disease), ischemia-reperfusion, hypoxia- and glutamate-induced cerebral damage, infantile lactic acidosis, Friedreich’s ataxia, various atypical E3-deficiencies, among others [20–29]. The ROS-generating activity of the intact KGDHc (and the PDHc) had been attributed to the E3 component [6, 14–17]. However, both the isolated hE1 [6, 30] and the hE1-hE2 sub-complexes of hKGDHc [6] could also generate significant amounts of ROS; a feature which is unique to the human complex, and may become a predominant pathomechanism regarding ROS production in acidosis (where a partial detachment of E3 from KGDHc was proposed [16]), or in E2 deficiency [6, 30]. The hKGDHc, hPDHc, as well as the isolated hE3 component can consume NADH in the reverse E3 reaction while also generating superoxide and H2O2 from O2; this reaction dominates under elevated NADH/NAD+ ratios (also a pathologically relevant condition) [6, 15–17].
Pathogenic mutations of the dld gene (coding for hE3) lead to an inherited, often lethal disease known as E3-deficiency; the clinical course of E3-deficiency is greatly diversified and often involves cardiological and/or neurological symptoms [25, 31–42]. The following phenotypes, among many others, for hE3 pathogenic mutations commonly appear in clinical reports: failure to thrive, developmental delay, encephalopathy, hypotonia, seizure, hepatomegaly, liver dysfunction, lactic acidosis, hypoglycemia, microcephaly, and ataxia. The effects of several of the total of 14 clinically reported disease-causing amino acid substitutions of the hE3 component on various biochemical and biophysical parameters, including ROS-generating capacity, of the enzyme have previously been investigated by us and others. Among other findings, it was reported that four amino acid substitutions resulting from the respective pathogenic mutations, P453L, G194C, D444V, and E340K, significantly stimulated ROS generation by the isolated hE3 (and also by hKGDHc reconstituted with G194C-hE3 [6]) in the reverse catalytic direction; in all four cases, the sensitivity of ROS generation by hE3 to a more acidic pH was also augmented relative to the wild-type (wt) enzyme [17]. The E340K, D444V, and G194C substitutions of hE3 all triggered the oxidative deterioration of the lipoic acid (LA) cofactors of both PDHc-E2 and KGDHc-E2 in a yeast model and led to a great reduction in the respiratory function of the yeast cells [43]. Since the decreases in hE3 activity resulting from the pathogenic mutations do not, in general, correlate directly in magnitude to the severity of the respective pathogeneses [25, 35], the group at Semmelweis proposed that a missing link in the interpretation of selected pathogeneses is perhaps the enhanced ROS generation stimulated by the respective pathogenic mutations of hE3 [17]. To date, no report has been published regarding the high-resolution structure of any of the pathogenic mutant forms of hE3 determined by a direct structural method such as x-ray crystallography or NMR spectroscopy. Hence, the pathomechanisms of action, including that of the enhanced ROS generation by selected variants, could only be speculated based on the crystal structures of the hE3 [7, 44, 45], molecular modeling [46, 47] and biochemical/biophysical data (e.g. [17, 43, 48, 49]).
Hydrogen/deuterium-exchange mass spectrometry (HDX-MS) is a powerful methodology for structural investigation of biomacromolecules in solution, which nevertheless presents considerable methodological and technical challenges (primarily regarding the minimization of the “back exchange” of deuterium (2H) to protium (1H) on peptic peptides during analysis). The Rutgers laboratory is equipped with an efficiently performing HDX-MS instrument for this purpose, which has already been successfully used to investigate principally protein-protein interactions in related projects [50–53].
Here we report the results of an HDX-MS investigation performed on ten disease-causing hE3 variants to establish the structural alterations induced by the respective pathogenic substitutions relative to the wild-type protein structure. Based on our findings with the HDX-MS approach, we present plausible mechanisms for enzymatic dysfunction in case of the 10 disease-causing hE3 variants studied.
2. Results
In this study, deuterium incorporation of peptic peptides of hE3 was compared to those of the hE3 disease-causing variants at a peptide resolution to detect conformational perturbations induced by the implicated pathogenic amino acid substitutions; experiments for deuterium incorporation were carried out in solution under physiologically relevant buffer conditions at 25 °C.
Recombinant wild-type and variant proteins for this study were expressed in E. coli and purified to homogeneity; the purity of all proteins was greater than 95%, similarly to those reported from the Semmelweis group in previous studies on these hE3 variants [17, 54]. There are fourteen pathogenic mutations of hE3 documented in clinical reports to date which are associated with a single amino acid substitution in the mature hE3 monomer (and do not involve exon skipping or a frame shift); pathogenic mutations of the dld gene occur frequently in a compound heterozygous state [25, 41]. Our protein samples mimic the homozygous forms of the mutations owing to the characteristics of standard heterologous recombinant protein expression protocols. We were able to generate protein samples with adequate concentrations for the HDX-experiment for ten of the above mutations (see below); soluble expression of the remaining four variants (G101del, I12T, M326V, G426E) using all applicable codon synonyms, including the most favorable ones for E. coli expression, for each newly incorporated amino acid was also attempted, but proved to be quite inefficient, likely due to structural instability and/or insufficient folding.
It was demonstrated in [7] that each monomer of the catalytically active functional (obligate) hE3 homodimer comprises four domains: one for FAD-binding (residues 1–149), one for NAD+/NADH-binding (residues 150–282), one referred to as the central domain (residues 283–350), and one referred to as the interface domain (residues 351–474). A single FAD prosthetic group binds non-covalently to each monomer, and each monomer contributes amino acids to the active site of the partner monomer [7]. Of the ten successfully purified hE3 variants, the K37E substitution affects the FAD-binding domain, the G194(C) residue is located in the NAD+/NADH-binding domain, the E340(K) and the I318(T) residues reside in the central domain, and the remaining substitutions (I445M, R460G, R447G, P453L, D444V, I358T) are all associated with the interface domain.
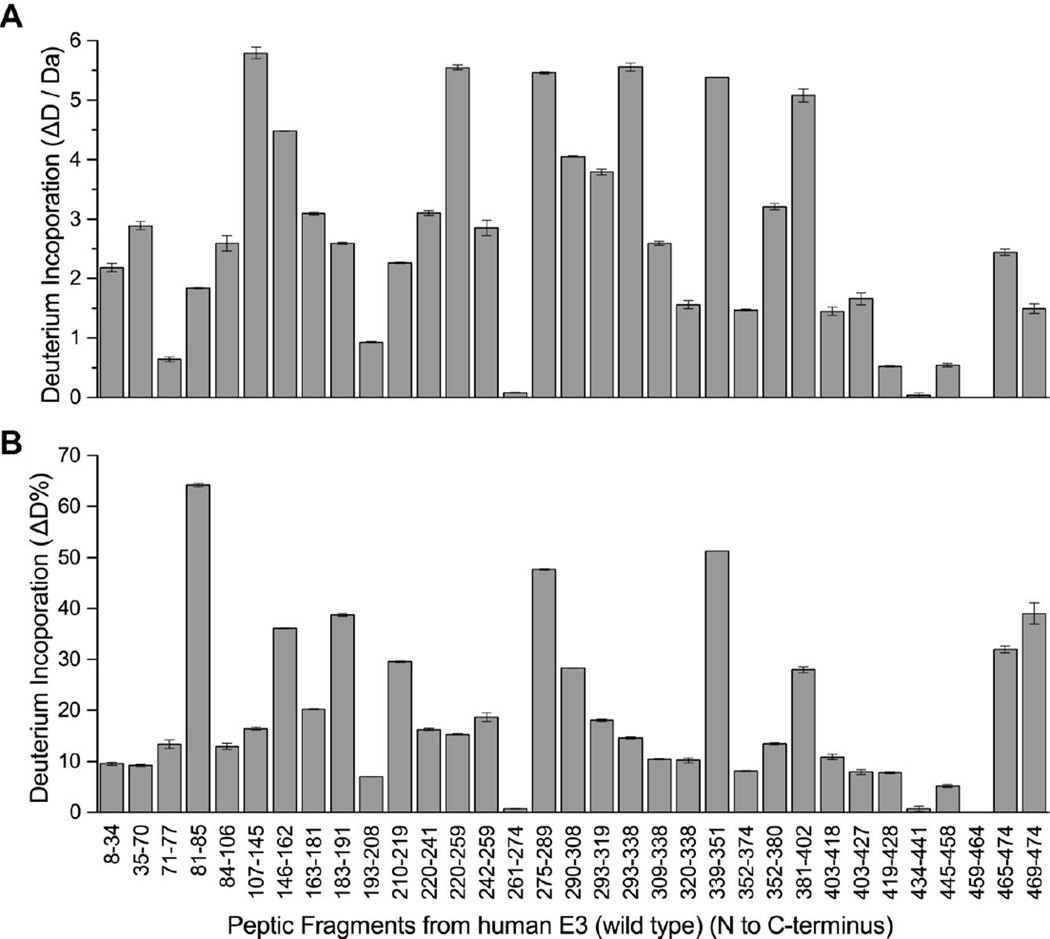
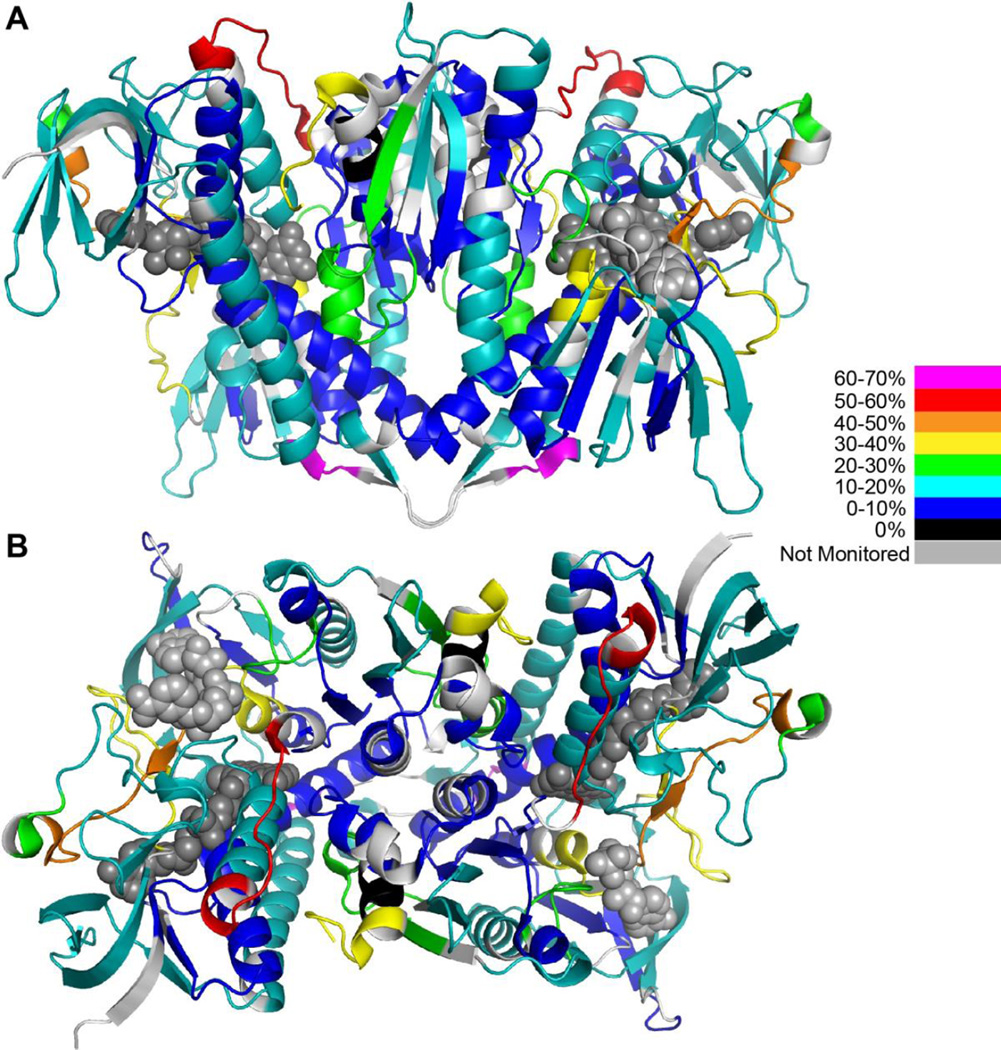
In this differential HDX-MS investigation, 33 peptic peptides from hE3 were selected for evaluation, constituting an overall sequence coverage of 95% (Figure S1). Additional peptides originating from the pathogenic hE3 variants were also used for evaluation, as listed in Table S1; these peptides usually contained single amino acid substitutions relative to the above mentioned 33 original peptides. The deuterium incorporation pattern of hE3 is shown in Figure 1. It is apparent that the regions 81–85 (purple in Figure 2), 275–289 (light brown in Figure 2), and 339–351 (red in Figure 2) display enhanced deuterium incorporation and hence can be stated to be flexible or exposed regions, while dimerization buries and stabilizes the interface domain, in particular fragments 434–441 (dark blue in Figure 2, with not exclusive coloring) and 459–464 (black in Figure 2). In fact, the entire interface domain, except the C-terminus, is rather unexposed and rigid in general, as expected. It is also noted that the interior of the central domain is also rather unexposed and rigid and possesses a conserved structure for hE3 and each of its variants studied here (see below and Figure 3).
Fig. 1.
Deuterium incorporation of peptic peptides of hE3. (A) Deuterium incorporation (ΔD, y axis) and (B) deuterium incorporation percentage (ΔD%, y axis, deuterons exchanged / maximum exchangeable sites × 100%) of peptic fragments from hE3 (x axis, listed from N to C terminus). Results represent mean ± S.D. from three independent measurements. Vertical error bars represent 1 SD in each direction.
Fig. 2.
Deuterium incorporation heatmap of hE3. Peptides with varying percentages of deuterium incorporation are highlighted with different colors. A and B represent two different orientations of the hE3 homodimer. FAD and NAD+ are depicted as dark and light grey space filling objects, respectively. For model building the hE3 structure with the PDB ID: 1ZMC and the Pymol program were used [7].
Fig. 3.
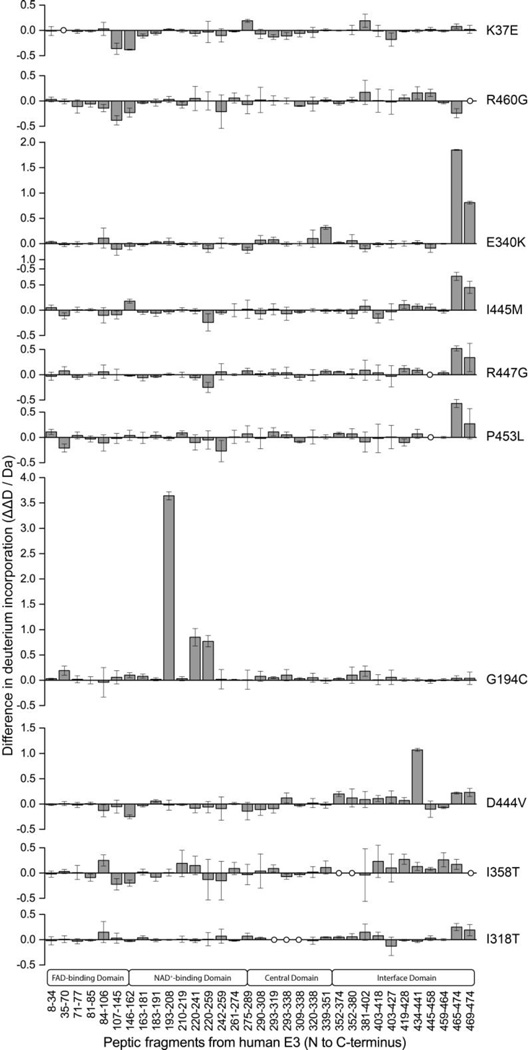
Modulation of deuterium incorporation by hE3 disease-causing variants. The difference plots display the deviations in deuterium incorporation by peptic fragments of hE3 variants (ΔΔD, y axis, deuterium incorporation by the hE3 variant minus deuterium incorporation by hE3). Results represent the mean ± S.D. from three independent experiments. Vertical error bars represent 1 SD in each direction.
The differential deuterium exchange profiles of the ten pathogenic hE3 variants relative to the deuterium exchange profile of the hE3 are shown in Figure 3; numerical data are displayed in Table S2. As seen, the disease-causing mutations triggered wide-ranging conformational alterations in hE3 in general, affecting not only amino acids in close proximity to the amino acid substituted, but also distant regions of hE3. This conclusion is still valid even though error ranges that signal statistically insignificant differences were also found. It is also evident from the results that there is no major overall alteration in the D-incorporation profile at the interface domain for any of the pathogenic variants, strengthening the notion that none of the pathogenic mutations examined here trigger monomerization of the functional hE3 homodimer. In four hE3 variants (E340K, I445M, R447G and P453L), a greatly enhanced exposure or dynamics of the C-terminus (fragments 465–474, 469–474) could be observed. In the symmetric hE3 homodimer both C-termini are partially embedded into the dimer interface and the exposure of this region to solvent as well as the intermonomeric dynamics of the dimer affect their mobility. The similar conformational effects resulting from the E340K or R447G substitutions could readily be rationalized as these two residues form an intermonomeric H-bond in hE3; in either scenario, the substitution results in considerable steric and charge modulations in close proximity to the C-terminus. The I445M and P453L substitutions are proximal in sequence to their most affected regions; these substitutions do not constitute major alterations in charge or size, but rather in overall shape. The linkage between H452, a catalytically important residue [55], and P453 is a cis peptide bond which most likely is converted to the trans orientation in P453L-hE3, perhaps inducing considerable distortions of the protein chain in this C-terminal region. The G194C substitution causes a major conformational anomaly in its surroundings in the NAD+/NADH-binding domain; G194 is part of an α-helix which might be considerably perturbed by the insertion of a large and polar Cys. The only other pathogenic variant that showed a major and rather local disturbance of the structure was the D444V-hE3; in the hE3 homodimer D444 and Y438’ form a hydrogen bridge the loss of which in D444V-hE3 may lead to enhanced flexibility at the C-terminus. In the case of the K37E, R460G, and I358T hE3 variants, structural alterations of a moderate magnitude are seen in various regions throughout the protein in each variant, the analysis of which requires more in-depth evaluations (see Discussion). The I318T substitution triggered major structural disturbance only at the C-termini; the explanation for this effect could be similar to the one suggested above for the E340K-hE3 variant.
Seeking the structural basis of pathogenicity for each disease-causing hE3 variant, we evaluated all of the statistically significant structural perturbations induced by the pathogenic hE3 substitutions as detected by HDX-MS, and assessed their plausible functional consequences in the Discussion.
3. Discussion
This appears to be the first study in the literature that experimentally addresses the structural alterations induced in the pathogenic hE3 variant enzymes. The effects of ten pathogenic amino acid substitutions on the overall protein structure of hE3 were investigated by HDX-MS. In the absence of high resolution structural data by X-ray crystallography or NMR spectroscopy on any of the pathogenic hE3 variants, the findings of this study provide the only structural information to date for a reliable evaluation of the molecular pathomechanisms of action in the related atypical forms of hE3 deficiency. Although HDX-MS does not provide residue-specific information and is restricted to a peptide-level resolution, it is able to analyze and map even the finest alterations in the structure that may lead to changes in protein-protein interactions or perhaps even to a catalytically dysfunctional protein conformation. The HDX-MS method is also capable of detecting local changes, due to various reasons (including a substitution), in the dynamics of the protein chain, which arguably also contributes to catalytic efficiency. The putative molecular pathogeneses of each disease-causing mutation of hE3 studied here are proposed below in light of the structural changes detected in the variants by HDX-MS. The contribution of the direct effects of the structural changes to the gross activity losses of hE3 or the harboring complexes in vivo upon the different mutations next to other potential auxiliary in vivo effects, such as suppression by DNA/RNA secondary structures, insufficient folding, structural instability, or accelerated degradation, cannot be judged by HDX-MS; most pathogenic hE3 variants, however, once successfully expressed to the soluble phase, are completely folded and stable under in vitro conditions [17].
D444V-hE3
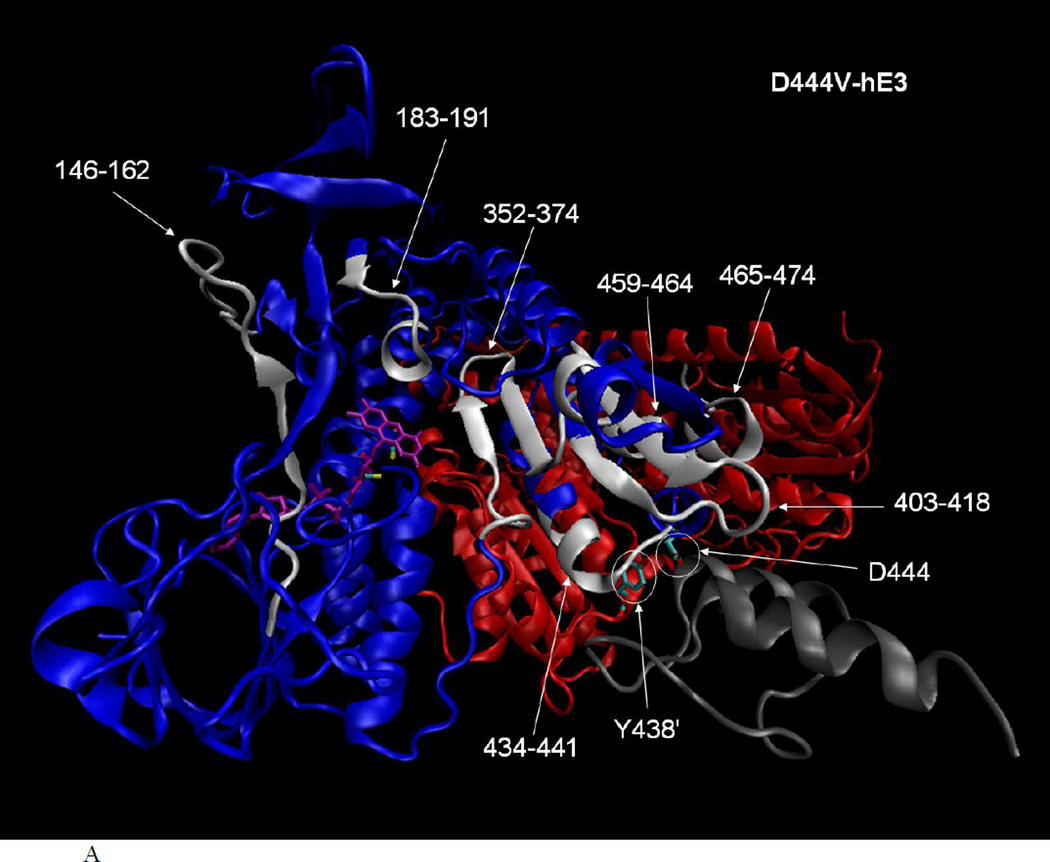
hLADH activity for this variant was measured (albeit under different assay conditions) to be 80% [43], 35% [17], 100% [44], 90% [49] of the control (hE3) using recombinant proteins and 15% of the control in the muscle homogenate of a patient homozygous for D444V (where immunoreactivity towards hE3 was reduced to 10%) [35]. In the latter study, the corresponding hKGDHc or hPDHc activities were 2 or 0% of the control, respectively, while the activity of hPDHc reconstituted from recombinant components using D444V-hE3 was 15% of the control [44]. According to previous studies, a H-bond between D444 and Y438’ (‘ for the other monomer), as well as the binding potential of hE3 towards the E3-binding domain of the E3-binding protein (E3BP) of hPDHc is lost [7, 44], but monomerization does not occur ([17, 44] and as also seen in Figure 3) upon this substitution. Regarding the loss in hLADH activity for isolated D444V-hE3, it can be seen in the crystal structure of hE3 (see Figure 4A) that three (146–162, 183–191, 352–374) of the seven (146–162, 183–191, 352–374, 403–418, 434–441, 459–464, 465–474) peptides with altered flexibility/exposure in D444V-hE3 according to HDX-MS are directly involved in FAD-binding. Yet, only ~5% FAD loss was measured for isolated D444V-hE3 [17], hence this effect cannot account for any greater activity loss and definitely not for the enhanced ROS-generating capacity of D444V-hE3 (30% increase compared to hE3) [17]. The same three peptides are in close proximity to the re face of the isoalloxazine ring of FAD where NAD+/NADH-binding occurs (see Figure 4A); there are no altered flexibility peptides in D444V-hE3 on the si face of the isoalloxazine ring, where the catalytic disulfide-exchange site and the LA-binding site are located. This implies disturbance in NAD+/NADH-binding in D444V-hE3, especially since the above three peptides altogether possess five binding residues for NADH (from the total of 13 binding residues for NADH in hE3, 38%) and two binding residues for NAD+ (from the total of 13 binding residues for NAD+ in hE3, 15%), according to structures with PDB IDs 1ZMD and 1ZMC, respectively. The other four affected peptides are farther from the active site and the LA-binding site and appear to be those regions more directly/locally affected by this substitution, and are instead likely to be implicated in E3BP-binding (see below). Thus, hLADH activity loss, as well as an increase in ROS-generating capacity of D444V-hE3 should be explained via the 146–162, the 183–191, and the 352–374 peptides. According to the HDX-MS results, it is plausible to hypothesize that FAD/NAD+/NADH-binding is in part compromised (via a propagating effect of the substitution), and this appears to be the primary cause for hLADH activity loss for D444V-hE3. The anomalies in the binding of the cofactors, however, do not seem to be of a sufficiently significant magnitude to negatively affect the relatively low-rate of ROS generation by this variant (0.24% of the catalytic rate in the reverse E3 reaction for isolated D444V-hE3 in the absence of NAD+ [17]). To the contrary, a 30% increase in ROS-generating capacity was detected, relative to hE3, for the isolated D444V-hE3 [17], a variant that does not induce a considerable conformational change in the LA-binding cavity, whence O2 most likely approaches FAD for the primary superoxide generation. Rather, ROS generation by this variant could be explained by a catalytic advantage for superoxide generation, likely involving the isoalloxazine ring of FAD, whose origins are not explained by HDX-MS. The conformation of FAD, and of the isoalloxazine ring, must be sensitive to changes of the protein locus where FAD is bound, and it may also manifest itself in alterations of the reactivity of the FAD moiety.
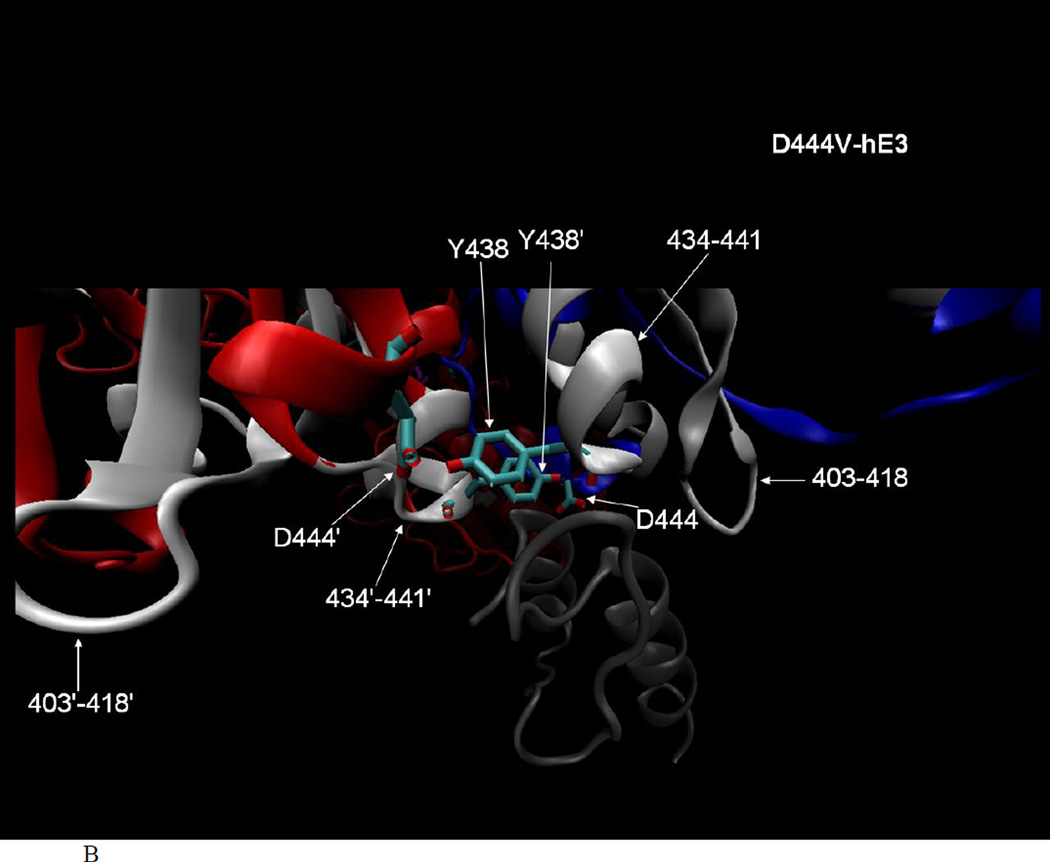
Fig. 4.
A. Altered flexibility peptides in the D444V-hE3 pathogenic variant studied by HDX-MS and represented in the hE3-E3BP structure (PDB ID: 1ZY8 [45]). The hE3 monomers are colored blue and red (chains G and H in 1ZY8, respectively), FAD is purple, the E3-binding domain of E3BP (44aa, residues 130–173, chain N in 1ZY8) is gray, important residues are circled, the sulfur atoms in the active center Cys residues are yellow, altered flexibility peptides are white and labeled. Structure features are shown in chain G only for clarity, but the analysis of the structure was performed using both monomers and one of the alternative locations of the E3-binding domain of E3BP (due to the symmetric alignment of the alternative coordinates of the E3-binding domain of E3BP relative to the hE3 dimer). B. Close-up representation to show the direct interactions between E3BP and the altered flexibility peptides of D444V-hE3 in hE3 (both monomers are labeled for altered flexibility peptides in B). The structural representations were created by VMD [72].
Regarding the loss of affinity of D444V-hE3 to E3BP and the resulting loss of the overall hPDHc (and hKGDHc) activity (see above), the crystal structures of the complex formed by hE3 and the E3-binding domain of E3BP [44, 45] revealed that D444, as well as its intermonomeric H-bridging partner Y438’ in hE3, make numerous direct contacts to the E3-binding domain of E3BP (D444 and Y438’ make five and seven interactions to E3BP, respectively). Substitution of Asp to Val in D444V-hE3 naturally abolishes the respective interactions towards E3BP. Moreover, this substitution triggers a significant displacement (or altered flexibility) of peptide 434–441 (see Figures 3 and 4B), which includes residue Y438, whose interactions with E3BP are likely to be also compromised. The two Y438 residues in the hE3 dimer form a crucial hydrophobic patch for E3BP binding [44, 45]. The peptide 434–441 contains additional already identified interaction partners towards E3BP, G439 and E437 [44, 45], the affinities of which to E3BP must also be altered upon the D444V substitution. In another altered flexibility peptide (403–418), two further interacting residues towards E3BP are found, T412 and D413, which are likely to be also affected by the D444V substitution. The D444V-hE3 is mostly dissociated from hPDHc (and likely hKGDHc), as dictated by the elevated Kd of the D444V-hE3–E3BP complex [44]. Hence, the LA-cofactors of these complexes are likely to be more prone to oxidative damage, in part by ROS generated via D444V-hE3 (see above), or the E1-E2 subcomplex itself for hKGDHc [6], which can further reduce the remaining overall activities of hPDHc and hKGDHc.
G194C-hE3
This is the most prevalent pathogenic hE3 variant (the carrier frequency for the mutation, e.g., in the Ashkenazi Jewish population is 1:94), which generally presents with an adult-onset; patients are usually homozygous for the respective mutation, however, compound heterozygosity also occurs. Biological samples of patients (originating from fibroblasts, lymphocytes, or muscle) showed E3-related activities in the following ranges: 6.5–30% (hLADH), 12.2–44% (hKGDHc), and 10.5–70% (hPDHc) of the respective controls [25, 34, 35, 38, 41, 56, 57]. The isolated G194C-hE3 exhibited hLADH activity of 100% [17], 75% [58] or 47% [43] of the control (hE3), while for enzyme complexes reconstituted with G194C-hE3 the following values were measured: 29% (hPDHc), and 61% (hKGDHc) [6]. The isolated G194C-hE3 displayed a 72% enhancement in ROS-generating capacity relative to hE3 in the reverse E3 reaction [17]. Reconstitution of hPDHc or hKGDHc with G194C-hE3 resulted in enhanced ROS generation (a 11% statistically significant increase relative to the control) only in case of the hKGDHc in its reverse E3 reaction [6]. The residue G194 is part of a helix whose selected residues bind to the cofactors of hE3 (FAD, NAD+/NADH) [7]. On substitution of Gly with Cys, a relatively large and polar side chain is introduced into this helix, which apparently perturbs cofactor-binding (28% FAD loss was measured for G194C-hE3 [17]). There are six peptides with modified flexibility in G194C-hE3 relative to hE3 according to HDX-MS (see Figures 3 and S2). The activity losses of the harboring complexes for this variant are not as detrimental as e.g., for the D444V substitution, suggesting a rather minor impact of this substitution on the binding of G194C-hE3 to E3BP. This conclusion is well justified by the distance of all affected peptides in G194C-hE3 from the E3BP-hE3 interface (see Figure S2). Although peptides 35–70, 146–162 and 163–181 present with flexibility modifications of moderate magnitudes in G194C-hE3, these three peptides together possess 17 of the 38 binding residues for FAD (45%) and one of the 13 for NADH (8%) in hE3 [7]. Therefore, any structural changes in these peptides would warrant their consideration as potential causes of the dysfunction (and FAD loss) of G194C-hE3. Contrary to this expectation, in our hands there was no functional loss, only FAD loss and elevated ROS-generating capacity, measured for isolated G194C-hE3 [17]). The other three peptides (193–208, 220–259, and 381–402), with considerable effects on flexibility for peptides 193–208 and 220–259, possess additional cofactor binding residues (altogether five for NAD+ (38%), two for NADH (15%), and none for FAD), whose perturbation must also contribute to the loss of FAD and potentially activity. There is flexibility alteration on the si face of the isoalloxazine ring of FAD and this is proposed to be the primary cause of the enhanced ROS-generating capacity of G194C-hE3, but it could possibly also contribute to an activity loss, especially flexibility alteration in peptide 35–70 that bears the disulfide-exchange site residues C45 and C50. There have been no experiments reported to measure the binding affinity of G194C-hE3 to E3BP, however, on the basis of HDX-MS results we propose that the activity loss of the hKGDHc or the hPDHc harboring G194C-hE3 is not due to an enhanced dissociation of G194C-hE3 from these complexes (see Figure S2). Rather, the underlying mechanism is suggested to be the prolonged oxidative deterioration of the LA cofactors (see above), which is potentially supplemented by an activity loss of the complexes due to the compromised activity of G194C-hE3 (based on hLADH activities measured for G194C-hE3 in [58] and [43]).
P453L-hE3
The peptide bond between P453 and H452, a catalytically important active site base, is in the cis configuration [7]; the O atom of this peptide bond is in a H-bond with N3-FAD. The P453L substitution was proposed to cause a considerable perturbation to the local structure near the disulfide-exchange site (of the partner monomer, see Figure S3), including the potential misplacement of the H452 side-chain and the elimination of the H-bond to N3-FAD, due to the preference for a trans peptide orientation by Leu and the greater size of the Leu side-chain [7]. No hLADH or overall PDHc activity was detectable upon this substitution in yeast [59], and the hLADH activity was only 6% of the control in cultured fibroblasts of a patient with compound heterozygosity (together with the K37E substitution) [31]. For an isolated P453L-hE3 component 4–9% hLADH activity and a 34% FAD loss were measured relative to hE3 [17]. The importance of P453 in the hE3 mechanism was confirmed also by a P453V variant that displayed a 0.06% hLADH activity [60]. The isolated P453L-hE3 component exhibited a 130% increase in ROS-generating capacity relative to hE3, which transformed this variant to be a very strong ROS-generating enzyme (catalytic activities for ROS-generation and the physiological forward reaction were approximately identical). The affinity loss of P453L-hE3 towards E3BP was measured to be 4-fold and believed insignificant [61]. There are numerous peptides in P453L-hE3, whose flexibility alterations relative to hE3 are considered significant (Figure S3). According to Figure S3, it is apparent that most of the conformational/flexibility changes in a hE3 monomer upon this substitution ought to be attributed instead to the distant effects of the P453L substitution in the other monomer. The HDX-MS data support the notion that the affinity of hE3 or P453L-hE3 towards E3BP should be similar (see above), hence dysfunctions of the complexes harboring P453L-hE3 are indeed unlikely to be the result of the dissociation of P453L-hE3 from the complexes, but rather the consequence of the greatly compromised hLADH activity and/or perhaps the greatly enhanced ROS generation by P453L-hE3. HDX-MS data justify the origin of the considerable FAD loss in P453L-hE3, as altered flexibility peptides in P453L-hE3 are much involved in the binding of FAD in hE3 (altogether with 26 of the 38 binding residues for FAD in hE3, 68%). NAD+/NADH-binding is also affected (four or two of the 13 binding residues for NAD+ (31%) or NADH (15%), respectively), but the rather ‘passive’ ~30% loss of cofactor-binding alone does not account for the observed deleterious activity loss. As seen in Figure S3, perturbation of the structure on both faces of the isoalloxazine ring is so extensive that it almost certainly distorts the structural integrity of the active site (residues C45, C50, H452’, FAD), which can account for the great activity loss as well as for the extreme elevation of the ROS-generating potential. Such a dramatic impact on the protein structure around the FAD prosthetic group must have effects on the FAD conformation itself, which may also contribute to the elevated ROS-generating capacity, as well as to the functional loss.
K37E-hE3
A 6% hLADH activity compared to control was found in cultured fibroblasts of a patient with compound heterozygosity for this substitution (together with the P453L substitution) [31]. In yeast, the K37E-hE3 exhibited 92% hLADH activity, 88% PDHc activity, and 68% KGDHc activity of the respective controls [59]. Isolated K37E-hE3 displayed 20% [61], 75% [62], or 70% [17] hLADH activity of the control. A negligible 1.7-fold increase in Kd was found for the K37E-hE3–E3BP complex relative to the hE3–E3BP complex [61]. A 24% [62] or 33% [17] FAD loss and an unaltered ROS-generating potential [17] were measured for K37E-hE3 relative to hE3. The methylene moieties of K37 stack on the adenine base of FAD, while the O2’ atom in the AMP moiety of FAD contacts the K37 side-chain via a H-bond in hE3 [7]. A Glu in place of a Lys would offer an opposite charge and a weaker stacking in an interaction with FAD. There are several altered flexibility peptides in K37E-hE3 according to HDX-MS (Figures 3 and S4A). As seen in Figure S4B there is an altered flexibility peptide (403–427) of K37E-hE3 in hE3 which is in close spatial proximity to E3BP in the hE3-E3BP complex. In fact, T412 and D413 of hE3, two residues of direct contact with E3BP [44, 45], are part of this peptide whose modulation could indeed account for the enhanced dissociation of the K37E-hE3– E3BP complex. There are 18 (50%), five (38%) and seven (54%) binding residues for FAD, NAD+ and NADH, respectively, in hE3 [7] affected by the K37E substitution according to HDX-MS (Figure S4A), which can account for the loss both in FAD and hLADH activity for K37E-hE3.
E340K-hE3
The missense mutation for this substitution was found together with a three nucleotide (single amino acid) deletion (G101del) on different alleles of a proband, and led to 3–11% hLADH, 12–14% hPDHc, and 1–6% hKGDHc activities of the controls in clinical samples [33]. In case of another carrier patient with compound heterozygosity (the other substitution was M326V) 3–14% hLADH and 3–44% hPDHc activities of the normal were measured [36]. A third patient (also a compound heterozygote resulting in the E340K and the I12T substitutions) displayed 9% hLADH, 59% hPDHc, 25% hKGDHc, and 62% hBCKDHc activities of the controls [25]. Isolated E340K-hE3 exhibited 64–71% [17], 0.2% [61], 100 % [44], 89% [43], or 71% [63] hLADH activities of the controls (hE3). The activity of hPDHc reconstituted with E340K-hE3 was 38% of the control [44]. The interaction of hE3 and E3BP was completely lost upon the E340K substitution as measured by either surface plasmon resonance (SPR) [61] or isothermal titration calorimetry (ITC) [44]. A 23% increase in ROS-generating capacity was detected for isolated E340K-hE3 [17], while the LA-cofactors of KGDHc and PDHc were oxidatively damaged to a great degree in yeast upon expression of E340K-hE3 [43]. The residue E340 forms a salt bridge to R447’ and the substitution of Lys for Glu results in an inversion of charges which obstructs the former interaction [7]; however, no monomerization results (see Figure 3 and [17, 44]). The majority of the studies on isolated recombinant protein showed 30% or less activity loss for E340K-hE3 (see above), which can potentially be ascribed to the six (NAD+, 46%) or three (NADH, 23%) cofactor binding residues (according to the PDB ID: 1ZMC/1ZMD structures [7]) which were affected upon the E340K substitution as measured by HDX-MS (see peptides 220–259 and 275–289 in Figure S5A). There were only three FAD binding residues (~8%) which were affected by this substitution (in the peptides 275–289 and 293–319) and this is what might be reflected in the modest 1% FAD loss measured elsewhere [17]. The elevated ROS-generating potential of E340K-hE3 cannot be explained via any conformational change on the si face of the isoalloxazine ring of FAD (see Figure S5A), but it could be accounted for via a potential effect of the altered flexibility peptide 275–289 in E340K-hE3 on the structure or reactivity of the isoalloxazine ring of FAD. The 62% loss in the overall hPDHc activity upon reconstitution with the E340K-hE3 variant, as well as the sometimes reported deleterious activity loss of the E340K-hE3-hPDHc in vivo (see above) could be explained by a perturbation of the direct interactions between V347 and G348 in hE3 and E3BP [44, 45] upon the E340K substitution via the altered flexibility peptide 339–351 (see Figure S5B). Interestingly, the flexibility of the R447 residue, another direct contact to E3BP, does not appear to be affected upon the loss of its salt bridge to E340 by this substitution.
R460G-hE3
In a compound heterozygote patient (the other mutation being a nonsense mutation on the other allele) 1.5–14% hLADH, 20% hKGDHc, and 11–26% hPDHc activities were measured [25, 32, 41]. For the isolated R460G-hE3 component the following hLADH activities were measured: 91% [61], 10% [44], 18–22% [17], 29% [64], and 40% [43] of the controls. The FAD content of R460G-hE3 is not reported in the literature whereas the ROS-producing potential was 74% of the control [17]. No overall hPDHc activity was detectable when the complex was reconstituted with the R460G-hE3 component [44]. The residue R460 forms a salt bridge with E333’ in hE3 [7]. The loss of this interaction upon the substitution of Gly for Arg can trigger proximal as well as distant conformational effects as reflected by the altered flexibility peptides in R460G-hE3 according to HDX-MS (see Figures 3 and S6A; in particular, peptide 309–338 affected in R460G-hE3 includes the E333 residue). No monomerization of hE3 due to this substitution was observed in previous studies [17, 44], nor was it detected by HDX-MS (Figure 3). Approximately 42% of the FAD binding residues in hE3 are perturbed in R460G-hE3 according to HDX-MS; NAD+ binding residues are not affected, while only 8% of the NADH binding residues are perturbed (see Figure S6A). As FAD binding residues appear to be greatly affected by this substitution, this is proposed to be the predominant cause of the impaired catalytic activity of isolated R460G-hE3, which is most likely accompanied by a FAD loss to an as yet unreported degree. As seen in Figure S6B, two altered flexibility peptides (434–441 and 445–458) are in close proximity to E3BP. Indeed, the residues E437, Y438, and G439 in the peptide 434–441, and R447 in the peptide 445–458 in hE3 were previously identified as direct contacts to E3BP [44, 45]. Perturbation of these interaction sites in R460G-hE3, as suggested by HDX-MS, is a plausible explanation for the orders of magnitude increase of the Kd of the R460G-hE3–E3BP complex [44, 61], as well as for the deleterious loss in hPDHc activity in vitro and in vivo (see above).
R447G-hE3
Patients homozygous for this substitution presented with the following E3-related activities in clinical samples: 20% hLADH (identical values were reported for two patients), 63% hPDHc (one patient), 0% hKGDHc (identical values were reported for two patients), and 56% hBCKDHc (one patient) activities of the normal. Substitution of R447 in a recombinant hE3 resulted in 92% (R447G) [44], 54% (R447G) [43], 85% (R447M) [61], and 110–122% (R447A) [65] hLADH activities of the controls. The hPDHc reconstituted with R447 variants displayed the following overall activities: 110% (R447A) [65] and 28% (R447G) [44] of the normal. A nearly 250-fold increase was observed for the Kd of the R447G-hE3–E3BP complex relative to the control by ITC [44]. For the R447M-hE3 variant the increase in Kd was nearly three-fold as measured by SPR [61], whereas the Kd was essentially unaltered for the R447A-hE3 variant by ITC [65]. Neither FAD loss nor ROS-generating potential has ever been evaluated directly for R447G-hE3, however, this variant also led to an oxidative deterioration of the LA-cofactors of both PDHc and KGDHc in yeast [43], which may imply an enhanced ROS-generating potential for the R447G-hE3 component. The residue R447 forms a salt bridge with E340’ in hE3 [7] and it is also a direct contact to E3BP [44, 45]. Upon the R447G substitution these direct interactions are lost, as clearly reflected by the flexibility enhancement of the E340 residue in R447G-hE3 according to HDX-MS (E340 being part of the altered flexibility peptide 339–351 in R447G-hE3, see Figures 3 and S7B), and also by the elevated Kd for the interaction of R447G-hE3 with E3BP (see above). As for the augmented Kd of the R447G-hE3–E3BP complex, several other direct contacts in hE3 towards E3BP are also affected upon the R447G substitution according to HDX-MS (V347, H348, E437, Y438, G439 [44, 45] in the altered flexibility peptides 339–351 and 434–441 in R447G-hE3, see Figure S7B), which can justify the results reported above in terms of affinity to E3BP and overall hPDHc activities (in vitro and in vivo). Cofactor binding of hE3 was also affected by this substitution according to HDX-MS: 11, 62 and 54% of the binding residues for FAD, NAD+ and NADH, respectively were affected in R447G-hE3 (in the peptides 183–191, 220–259, 275–289, 352–374, see Figure S7A). These results suggest a moderate FAD loss and a considerably altered NAD+/NADH-binding, which together can account for the loss in hLADH activity of the R447G-hE3 component (see above). Alterations of structural flexibility in the LA-binding pocket of R447G-hE3 were not observed, which would suggest a rather small if any effect of this substitution on the ROS-generating capacity of hE3. In contrast, however, the deleterious effects of this substitution on the oxidation status of the LA cofactors of KGDHc and PDHc [43] still suggest an elevated free radical production by R447G-hE3, which could perhaps be explained via the conformational effects of the altered flexibility peptides in the cofactor-binding domains of R447G-hE3 on the isoalloxazine ring of FAD and a resultant alteration in the reactivity of FAD towards superoxide production.
I318T-hE3
A compound heterozygote patient (the other mutation being a deletion, G101del) presented with 9–29% hLADH, 80% hPDHc, 24% hBCKDHc, and 107% hKGDHc activities of the controls [41]. There have been no reports on the FAD-content, the ROS-generating capacity, the affinity to E3BP, or the catalytic activity of the isolated variant. The replacement of Ile by Thr introduces a shorter and more polar side-chain into the local structure that must perturb the nearby structure, as well as the local charge distribution. It is important to note that there are two other pathogenic mutations of hE3 also leading to an Ile-to-Thr replacement (I12T, I358T) where the origin of the dysfunction should be based on similar local effects. HDX-MS provides four altered flexibility peptides in I318T–hE3 (see Figures 3 and S8A). The alteration in the flexibility of the peptides 146–162 and 352–374 in I318T-hE3 likely affects 14% of the FAD-binding residues and 15% of the NADH-binding residues (but none of the NAD+-binding residues) according to HDX-MS, probably accounting for the loss in hLADH activity, and likely reduction of the FAD content of I318T-hE3. Among the direct contacts of hE3 to E3BP, the residues V347 and H348 are located in the altered flexibility peptide 339–351 in I318T-hE3 (see Figure S8B). The perturbation of these interactions can account for the loss in the overall hPDHc activity (see above). No significant conformational changes could be observed on the si face of the FAD isoalloxazine ring (see Figure S8A), hence the ROS-generating capacity is unlikely to be affected by this substitution.
I445M-hE3
A patient homozygous for the relevant mutation presented with undetectable hLADH activity and 13–97% hPDHc activity of the (very variable) normal samples [40, 41]. There is no FAD-content, ROS-generating capacity, affinity to E3BP, or catalytic activity reported for the isolated variant. Replacement of Ile by Met introduces a side-chain with similar size, but different overall shape and somewhat higher polarity that likely triggers steric conflicts in the local structure and perhaps also long-range conformational changes. There are several altered flexibility peptides in I445M-hE3 according to HDX-MS (see Figures 3 and S9A) from which the following ones may affect the cofactor binding sites: 35–70, 146–162, 220–259, and 309–338. The implicated binding residues constitute 58, 23, and 8% of the total binding residues for FAD, NAD+, and NADH in hE3, respectively, which could by themselves account for the deleterious loss in hLADH activity, and likely also the FAD content of the I445M-hE3 component. Several direct contacts to E3BP are affected in I445M-hE3: T412 and D413 in the peptide 403–418 as well as E437, Y438, and G439 in the peptide 434–441 (see Figure S9B). Perturbation of these interacting sites in I445M-hE3 may justify the loss in overall hPDHc activity (see above) and suggests an increased Kd for the interaction of I445M-hE3 and E3BP. On the si face of the FAD isoalloxazine ring a considerable conformational disturbance can be seen (see Figure S9A) which might modulate the ROS-generating potential in I445M-hE3.
I358T-hE3
A compound heterozygote patient (the other mutation being at a consensus splice site of the dld gene, IVS9 +1G > A) presented with 29% and 14–100% of the normal hLADH and hPDHc activities, respectively [37]. The recombinant enzyme variant displayed 100% [66] or 30–41% [17] hLADH activity of the control. On the basis of the crystal structure of hE3 it was proposed that on substitution of Ile 358 by the polar Thr residue, primarily the NAD+/NADH-binding would be affected due to a disturbance of the residues near Ile 358 that bind to NAD+ or NADH [7]. However, HDX-MS results show only minor effects on the binding of these cosubstrates (0% and 8% of the NAD+ and NADH-binding residues were affected in I358T-hE3, respectively). FAD-loss is not reported in the literature for this variant. HDX-MS suggests a considerable perturbation of the FAD-binding site by this substitution (42% of all the FAD-binding residues were affected, see Figure S10A), which can account for the loss in hLADH activity and likely FAD-binding affinity of the I358T-hE3 component. The ROS-generating potential is not stimulated by the I358T substitution [17], which is underlined by the lack of alterations in flexibility at the si face of the FAD isoalloxazine ring according to HDX-MS (see Figure S10B). The altered flexibility peptides 434–441 and 445–458 in I358T-hE3 contain the E437, Y438, G439, and R447 residues which are direct contacts of hE3 towards E3BP and whose perturbation can also contribute to a loss in the overall hPDHc activity (see above and Figure S10B).
Our HDX-MS results provide a more detailed understanding of the effects of the disease-causing mutations of the human dld gene on the structure, dynamics and catalytic activities of the hE3 protein as an isolated enzyme. When compared to data published before on these hE3 variants (e.g. in [17]), the HDX-MS results presented here in most cases enabled identification of the peptides, or sometimes even the specific residues, in hE3 whose perturbation upon the respective mutations may indeed be responsible for specific changes in the biochemical functionality of hE3 or the multienzyme complexes harboring hE3. Based on the results, important insights could also be gained regarding the putative mechanistic explanations for the loss of functional integrity of the multienzyme complexes which harbor selected pathogenic hE3 variants. Concerning generalization with respect to disease phenotype and mechanistic conclusions, it appears according to the present and previous biochemical data, that each mutation leads to a specific mode or pathway for impairing hLADH activity and/or the activity of multienzyme complexes harboring hE3. In this respect, seeking generalized mechanisms (e.g., for certain groups of the 14 pathogenic mutations) is not likely to be beneficial or even feasible, rather the individual modes of action ought to be elucidated in future studies in even greater details. The structural information presented here, together with the suggested future mechanistic investigations, will help shed light also on the mechanism of enhanced ROS generation stimulated by selected pathogenic amino acid substitutions of hE3 and serve as a starting point for targeting this latter with potential future clinical benefits.
4. Materials and Methods
4.1. Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. Deuterium oxide (D2O, 99.9%) was obtained from Cambridge Isotope Laboratories (Tewksbury, MA, USA). Buffer components were obtained as molecular biology grade protease free chemicals from Sigma-Aldrich.
4.2. Cloning
The vector plasmid for the heterologous expression of the hE3 protein was generated by DNA2.0, Inc. (Menlo Park, CA, USA) using the pET-52b(+) plasmid from Novagen (Madison, WI, USA) and a DNA insert coding for the mature hE3 (474aa lacking the 35aa mitochondrial leader sequence) which was synthesized via codon-optimization for efficient E. coli expression. The gene of interest was preceded by a sequence coding for an N-terminal Strep-tag (for affinity purification) and a subsequent human rhinovirus (HRV) 3C protease cleavage site (between the Strep-tag and hE3); these sequences were also codon-optimized for E. coli expression. The expression host was BL21(DE3) (from Novagen) under resistance to ampicillin, which was conferred by the pET-52b(+) plasmid. The pathogenic mutations were introduced in-house by the QuikChange II site-directed mutagenesis kit (Stratagene; Cedar Creek, TX, USA) according to the manufacturer’s specifications. The primers for the mutations were designed by the QuikChange Primer Design program and were purchased from IBA (Gottingen, Germany) as HPLC purified oligonucleotides. Clones were confirmed by DNA sequencing (Agowa GmbH, Berlin, Germany). The forward and the reverse sequencing results always overlapped verifying both the sites of the mutations and the correctness of the entire gene constructs.
4.3. Protein expression and purification
Single colonies of BL21(DE3) plus hE3-pET-52b+ (always fresh transformants) from LB-agar plates supplemented with 0.1 mg/mL ampicillin (Amp) were used for overnight cultures in LB medium plus 0.1 mg/mL Amp (LB-Amp) at 37°C and 200 rpm. Overnight cultures were added to fresh 5 L LB-Amp in a 1:50 volumetric ratio and were left growing at 37 °C and 200 rpm. Protein expression was induced by 1 mM (final concentration) isopropyl β-D-1-thiogalactopyranoside (IPTG) at OD~0.5 and was allowed to proceed for 3 h at 25 °C. Cells were harvested at 6,500 g, 4 °C for 15 min, the pellet was frozen at −20 °C overnight, then the bacteria per 5 L of expression culture were lysed in 120 mL B-PER bacterial solubilizing solution (ThermoFisher Scientific, Waltham, MA, USA) in the presence of 24 mg lysozyme, 1.2 mL EDTA-free Halt protease inhibitor cocktail (ThermoFisher Scientific) and 12 µL Universal Nuclease (ThermoFisher Scientific) using a potter homogenizer and subsequently continuous gentle magnetic stirring at 4 °C for 30 min. Cell components were centrifuged at 15,000 g, 4 °C for 30 min. 70 µL 20 mM FAD and 33 µL 50 mg/mL avidin (IBA, Gottingen, Germany) were added to the cleared lysate. For affinity purification, a 5 mL Strep-Tactin Macroprep FPLC column (IBA, Gottingen, Germany) was applied on an AKTA Purifier 10 UPC FPLC system (GE Healthcare Biosciences AB, Uppsala, Sweden) according to our published protocol for hE3 purification [54, 67], which requires no further purification steps. Protein eluates were concentrated to 10–15 mg/mL using Amicon Ultracel centrifugation filtering tubes (MWCO=30 kDa; Millipore, Cork, Ireland) at 4 °C. Concentrated proteins were fast-frozen in liquid N2 and stored at −80 °C, until use.
4.4. Hydrogen/deuterium-exchange mass spectrometry (HDX-MS)
The HDX-MS analysis was carried out as described before [50, 52]. All 11 proteins (hE3 and the 10 variants, K37E, G194C, I318T, E340K, I358T, D444V, I445M, R447G, P453L and R460G hE3) were individually buffer-exchanged into 10 mM KH2PO4 (pH 7.0), 50 mM KCl, 2 µM FAD (Buffer A). Each protein was diluted to 40 µM concentration with Buffer A and was allowed to equilibrate for 1 h at 25 °C. The deuterium labeling reaction was initiated by diluting 2 µL of protein stock solution into 38 µL of the labeling buffer (10 mM KH2PO4, 50 mM KCl, 2 µM FAD, 99.9% D2O, pD 7.0; the pD value was obtained by adding 0.4 unit to the reading of a pH meter) at 25 °C, yielding a final concentration of 94.9% D2O. At 3 min of exchange, a 30 µL aliquot from the labeling reaction was rapidly quenched by an equal volume of ice-cold quench buffer (0.2 M KH2PO4, pH 2.6); a single exchange time point of 3 min was proved to be effective, as before [52, 53], to monitor all the significant structural alterations, relative to hE3, of the pathogenic hE3 variants studied here. The samples were immediately frozen in liquid nitrogen and stored at −80 °C until analysis, but for no more than two weeks. Non-deuterated samples were generated following the same procedure except that protein samples were diluted into Buffer A instead of the labeling buffer. The frozen deuterated sample was rapidly thawed and loaded into a 20 µL sample loop inside the refrigeration system using an ice-cold syringe. The protein sample (approximately 20 pmol in total) was carried by a 0.3 mL·min−1 digestion flow of 0.1% (v/v) formic acid into an immobilized pepsin column (Poroszyme Immobilized Pepsin Cartridge, 2.1 mm × 30 mm, ThermoFisher Scientific, placed in a customized column oven set to 15 °C) and digested for 20 s. The resultant peptides were immediately cooled to 0 °C through a heat exchanger and were concentrated and desalted on a peptide trap (Michrom peptide MacroTrap, 3 × 8 mm, Michrom Bioresources, Auburn, CA, USA). The peptides were eluted and separated in 15 min through a reversed-phase C18 HPLC column (Agilent Poroshell 300SB-C18, 2.1 mm × 75 mm, Agilent Technologies, Santa Clara, CA, USA) at a flow rate of 0.2 mL·min−1 applying a 2–40% acetonitrile gradient containing 0.1% (v/v) formic acid at 0 °C. ESI-FT-MS measurements were launched 5 min after the initiation of the elution process and lasted for 10 min. The time from initiation of digestion to elution of the last peptide was always less than 20 min. The mass spectrometer settings were as follows: ESI+ mode; capillary, 4,500 V; spray shield, 4,000 V; drying gas temperature, 190 °C; mass acquisition range, 400–2,000 m/z; scan rate, 0.5 scans per s (see Table S1 for the selected peptic peptides from each variant). All experiments were carried out in triplicate. Bruker Daltonics DataAnalysis 4.0 was used for spectrum analysis and data treatment. Peptides were identified against non-deuterated samples using the customized program DXgest, which matches experimental peptide mass with theoretically generated peptic peptide mass applying statistical considerations [68] for the pepsin cleavage pattern under H/D exchange conditions. Mass tolerance was set to <1.5 ppm for accurate identification. H/D exchange data for each individual peptide at various time points were processed using the program HX-Express [69]; back-exchange correction was not performed for the purposes of this comparative analysis. The number of exchangeable backbone amide protons (maxD) of a peptide was calculated to be the total number of amino acid residues in the peptide, excluding proline residues, minus the N-terminal residue that carries two fast-exchangeable amino protons [70, 71]. The percentage of deuterium incorporation (without the back-exchange correction) into each peptide was calculated using the following equation: ΔD%=ΔD/(maxD×1.0063×0.949) × 100%, where 1.0063 is the atomic mass difference between deuterium and hydrogen (protium), whereas 0.949 represents the D2O content in the labeling reaction. Plots were created by Origin (OriginLab, Northampton, MA, USA).
Supplementary Material
Highlights.
Study of common E3 of human α-ketoglutarate and pyruvate dehydrogenase complexes.
Effects of ten pathogenic mutations on human E3 structure examined by HDX-MS.
Structural changes identified account for loss in cofactor binding and activity.
Mutations lead to dysfunctional complexes and alter interactions in variants.
HDX-MS results suggest structural sources of elevated ROS-generating capacities.
Acknowledgments
We are grateful to Drs. Oliver Ozohanics, Karoly Vekey (both from the Research Centre for Natural Sciences, Hungarian Academy of Sciences, Budapest, Hungary) and Arpad Somogyi (Ohio State University, Columbus, OH, USA) for their contributions to the method development in mass spectrometry. Financial support is gratefully acknowledged from the Hungarian Academy of Sciences (MTA grant 02001 to A-V.V.), the Hungarian Scientific Research Fund (OTKA, grant 112230 to A-V.V.), the Hungarian Brain Research Program (grant KTIA_13_NAP-A-III/6. to A-V.V.), the Bolyai and the Fulbright Fellowships (to A.A.), NIH-R15GM116077 and NSF- CHE-1402675 (to F.J.).
Abbreviations
- KGDHc
alpha-ketoglutarate (also known as 2-oxoglutarate) dehydrogenase complex
- PDHc
pyruvate dehydrogenase complex
- BCKDHc
branched-chain α-keto acid dehydrogenase complex
- E3
dihydrolipoamide dehydrogenase, the E3 component of the 2-oxo acid dehydrogenase complexes shared by all complexes in a particular cell
- HDX-MS
hydrogen/deuterium-exchange mass spectrometry
- EDTA
ethylenediaminetetraacetic acid
- FAD
flavin adenine dinucleotide
- NAD+/NADH
nicotinamide adenine dinucleotide (oxidized/reduced forms)
- ROS
the reactive oxygen species superoxide anion and hydrogen peroxide
- Tris
2-amino-2-hydroxymethyl-propane-1,3-diol
- wt
wild-type
- h
human origin
- aa
amino acid
- Amp
ampicillin
- LB medium
Luria-Bertani medium
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- NMR
nuclear magnetic resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement. The authors declare no conflict of interest.
References
- 1.Massey V. The composition of the ketoglutarate dehydrogenase complex. Biochim. Biophys. Acta. 1960;38:447–460. doi: 10.1016/0006-3002(60)91280-4. [DOI] [PubMed] [Google Scholar]
- 2.Koike K, Hamada M, Tanaka N, Otsuka KI, Ogasahara K, Koike M. Properties and subunit composition of the pig heart 2-oxoglutarate dehydrogenase. J. Biol. Chem. 1974;249:3836–3842. [PubMed] [Google Scholar]
- 3.Reed LJ. Multienzyme complexes. Acc. Chem. Res. 1974;7:40–46. [Google Scholar]
- 4.Perham RN. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes - a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 5.Sheu KFR, Blass JP. Oxidative/Energy Metabolism in Neurodegenerative Disorders. Vol. 893. New York: New York Acad Sciences; 1999. The alpha-ketoglutarate dehydrogenase complex; pp. 61–78. [DOI] [PubMed] [Google Scholar]
- 6.Ambrus A, Nemeria NS, Torocsik B, Tretter L, Nilsson M, Jordan F, Adam-Vizi V. Formation of reactive oxygen species by human and bacterial pyruvate and 2-oxoglutarate dehydrogenase multienzyme complexes reconstituted from recombinant components. Free Radic. Biol. Med. 2015;89:642–650. doi: 10.1016/j.freeradbiomed.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. Crystal structure of human dihydrolipoamide dehydrogenase: NAD(+)/NADH binding and the structural basis of disease-causing mutations. J. Mol. Biol. 2005;350:543–552. doi: 10.1016/j.jmb.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Sanderson SJ, Khan SS, McCartney RG, Miller C, Lindsay JG. Reconstitution of mammalian pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase complexes: Analysis of protein X involvement and interaction of homologous and heterologous dihydrolipoamide dehydrogenases. Biochem. J. 1996;319:109–116. doi: 10.1042/bj3190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smolle M, Prior AE, Brown AE, Cooper A, Byron O, Lindsay JG. A new level of architectural complexity in the human pyruvate dehydrogenase complex. J. Biol. Chem. 2006;281:19772–19780. doi: 10.1074/jbc.M601140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brautigam CA, Wynn RM, Chuang JL, Chuang DT. Subunit and Catalytic Component Stoichiometries of an in Vitro Reconstituted Human Pyruvate Dehydrogenase Complex. J. Biol. Chem. 2009;284:13086–13098. doi: 10.1074/jbc.M806563200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel MS, Nemeria NS, Furey W, Jordan F. The Pyruvate Dehydrogenase Complexes: Structure-based Function and Regulation. J. Biol. Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa E, Oliver RM, Reed LJ. Alpha-Keto acid dehydrogenase complexes, V. Macromolecular organization of pyruvate and alpha-ketoglutarate dehydrogenase complexes isolated from beef kidney mitochondria. Proc. Natl. Acad. Sci. USA. 1966;56:534–541. doi: 10.1073/pnas.56.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed LJ, Oliver RM. The multienzyme alpha-keto acid dehydrogenase complexes. Brookhaven Symp. Biol. 1968;21:397–412. [PubMed] [Google Scholar]
- 14.Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J. Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tretter L, Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. J. Neurosci. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrus A, Tretter L, Adam-Vizi V. Inhibition of the alpha-ketoglutarate dehydrogenase-mediated reactive oxygen species generation by lipoic acid. J. Neurochem. 2009;109:222–229. doi: 10.1111/j.1471-4159.2009.05942.x. [DOI] [PubMed] [Google Scholar]
- 17.Ambrus A, Torocsik B, Tretter L, Ozohanics O, Adam-Vizi V. Stimulation of reactive oxygen species generation by disease-causing mutations of lipoamide dehydrogenase. Hum. Mol. Genet. 2011;20:2984–2995. doi: 10.1093/hmg/ddr202. [DOI] [PubMed] [Google Scholar]
- 18.Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J. Biol. Chem. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mailloux RJ, Gardiner D, O’Brien M. 2-Oxoglutarate dehydrogenase is a more significant source of O2.-/H2O2 than pyruvate dehydrogenase in cardiac and liver tissue. Free Radic. Biol. Med. 2016;97:501–512. doi: 10.1016/j.freeradbiomed.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Starkov AA, Adam-Vizi V. Calcium and mitochondrial reactive oxygen species generation: how to read the facts. J. Alzheim. Dis. 2010;20:S413–S426. doi: 10.3233/JAD-2010-100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson GE, Starkov A, Blass JP, Ratan RR, Beal MF. Cause and consequence: Mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases, Biochim. Biophys. Acta-Mol. Basis Dis. 2010;1802:122–134. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starkov AA. The Role of Mitochondria in Reactive Oxygen Species Metabolism and Signaling. In: Gibson GE, Ratan RR, Beal MF, editors. Mitochondria and Oxidative Stress in Neurodegenerative Disorders. Vol. 1147. 2008. pp. 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Droge W, Schipper HM. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell. 2007;6:361–370. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunik VI, Schloss JV, Pinto JT, Gibson GE, Cooper AJL. Enzyme-catalyzed side reactions with molecular oxygen may contribute to cell signaling and neurodegenerative diseases. Neurochem. Res. 2007;32:871–891. doi: 10.1007/s11064-006-9239-z. [DOI] [PubMed] [Google Scholar]
- 25.Cameron JM, Levandovskiy V, MacKay N, Raiman J, Renaud DL, Clarke JTR, Feigenbaum A, Elpeleg O, Robinson BH. Novel mutations in dihydrolipoamide dehydrogenase deficiency in two cousins with borderline-normal PDH complex activity. Am. J. Med. Genet. A. 2006;140A:1542–1552. doi: 10.1002/ajmg.a.31313. [DOI] [PubMed] [Google Scholar]
- 26.Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos. Trans. R. Soc. B-Biol. Sci. 2005;360:2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tretter L, Sipos I, Adam-Vizi V. Initiation of neuronal damage by complex I deficiency and oxidative stress in Parkinson’s disease. Neurochem. Res. 2004;29:569–577. doi: 10.1023/b:nere.0000014827.94562.4b. [DOI] [PubMed] [Google Scholar]
- 28.Gibson GE, Kingsbury AE, Xu H, Lindsay JG, Daniel S, Foster OJF, Lees AJ, Blass JP. Deficits in a tricarboxylic acid cycle enzyme in brains from patients with Parkinson’s disease. Neurochem. Int. 2003;43:129–135. doi: 10.1016/s0197-0186(02)00225-5. [DOI] [PubMed] [Google Scholar]
- 29.Gibson GE, Park LCH, Sheu K-FR, Blass JP, Calingasan NY. The [alpha]-ketoglutarate dehydrogenase complex in neurodegeneration. Neurochem. Int. 2000;36:97–112. doi: 10.1016/s0197-0186(99)00114-x. [DOI] [PubMed] [Google Scholar]
- 30.Nemeria NS, Ambrus A, Patel H, Gerfen G, Adam-Vizi V, Tretter L, Zhou J, Wang J, Jordan F. Human 2-Oxoglutarate Dehydrogenase Complex E1 Component Forms a Thiamin-derived Radical by Aerobic Oxidation of the Enamine Intermediate. J. Biol. Chem. 2014;289:29859–29873. doi: 10.1074/jbc.M114.591073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu TC, Kim H, Arizmendi C, Kitano A, Patel MS. Identification of two missense mutations in a dihydrolipoamide dehydrogenase-deficient patient. Proc. Natl. Acad. Sci. USA. 1993;90:5186–5190. doi: 10.1073/pnas.90.11.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong YS, Kerr DS, Craigen WJ, Tan J, Pan YZ, Lusk M, Patel MS. Identification of two mutations in a compound heterozygous child with dihydrolipoamide dehydrogenase deficiency. Hum. Mol. Genet. 1996;5:1925–1930. doi: 10.1093/hmg/5.12.1925. [DOI] [PubMed] [Google Scholar]
- 33.Hong YS, Kerr DS, Liu TC, Lusk M, Powell BR, Patel MS. Deficiency of dihydrolipoamide dehydrogenase due to two mutant alleles (E340K and G101del) - Analysis of a family and prenatal testing. Biochim. Biophys. Acta-Mol. Basis Dis. 1997;1362:160–168. doi: 10.1016/s0925-4439(97)00073-2. [DOI] [PubMed] [Google Scholar]
- 34.Shaag A, Saada A, Berger I, Mandel H, Joseph A, Feigenbaum A, Elpeleg ON. Molecular basis of lipoamide dehydrogenase deficiency in Ashkenazi Jews. Am. J. Med. Genet. 1999;82:177–182. [PubMed] [Google Scholar]
- 35.Shany E, Saada A, Landau D, Shaag A, Hershkovitz E, Elpeleg ON. Lipoamide dehydrogenase deficiency due to a novel mutation in the interface domain. Biochem. Biophys. Res. Commun. 1999;262:163–166. doi: 10.1006/bbrc.1999.1133. [DOI] [PubMed] [Google Scholar]
- 36.Cerna L, Wenchich L, Hansiková H, Kmoch S, Peskova K, Chrastina P, Brynda J, Zeman J. Novel mutations in a boy with dihydrolipoamide dehydrogenase deficiency. Med. Sci. Monitor. 2001;7:1319–1325. [PubMed] [Google Scholar]
- 37.Grafakou O, Oexle K, van den Heuvel L, Smeets R, Trijbels F, Goebel HH, Bosshard N, Superti-Furga A, Steinmann B, Smeitink J. Leigh syndrome due to compound heterozygosity of dihydrolipoamide dehydrogenase gene mutations. Description of the first E3 splice site mutation. Eur. J. Pediatr. 2003;162:714–718. doi: 10.1007/s00431-003-1282-z. [DOI] [PubMed] [Google Scholar]
- 38.Hong YS, Korman SH, Lee J, Ghoshal P, Qu Q, Barash V, Kang S, Oh S, Kwon M, Gutman A, Rachmel A, Patel MS. Identification of a common mutation (Gly194Cys) in both Arab Moslem and Ashkenazi Jewish patients with dihydrolipoamide dehydrogenase (E3) deficiency: Possible beneficial effect of vitamin therapy. J. Inherit. Metab. Dis. 2003;26:816–818. doi: 10.1023/b:boli.0000010004.12053.5b. [DOI] [PubMed] [Google Scholar]
- 39.Odievre MH, Chretien D, Munnich A, Robinson BH, Dumoulin R, Masmoudi S, Kadhom N, Rötig A, Rustin P, Bonnefont JP. A novel mutation in the dihydrolipoamide dehydrogenase E3 subunit gene (DLD) resulting in an atypical form of alpha-ketoglutarate dehydrogenase deficiency. Hum. Mutat. 2005;25:323–324. doi: 10.1002/humu.9319. [DOI] [PubMed] [Google Scholar]
- 40.Quintana E, Pineda M, Font A, Vilaseca MA, Tort F, Ribes A, Briones P. Dihydrolipoamide dehydrogenase (DLD) deficiency in a Spanish patient with myopathic presentation due to a new mutation in the interface domain. J. Inherit. Metab. Dis. 2010;33:S315–S319. doi: 10.1007/s10545-010-9169-4. [DOI] [PubMed] [Google Scholar]
- 41.Quinonez SC, Leber SM, Martin DM, Thoene JG, Bedoyan JK. Leigh Syndrome in a Girl With a Novel DLD Mutation Causing E3 Deficiency. Pediatr. Neurol. 2013;48:67–72. doi: 10.1016/j.pediatrneurol.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrozzo R, Torraco A, Fiermonte G, Martinelli D, Di Nottia M, Rizza T, Vozza A, Verrigni D, Diodato D, Parisi G, Maiorana A, Rizzo C, Pierri CL, Zucano S, Piemonte F, Bertini E, Dionisi-Vici C. Riboflavin responsive mitochondrial myopathy is a new phenotype of dihydrolipoamide dehydrogenase deficiency. The chaperon-like effect of vitamin B2. Mitochondrion. 2014;18:49–57. doi: 10.1016/j.mito.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Vaubel RA, Rustin P, Isaya G. Mutations in the Dimer Interface of Dihydrolipoamide Dehydrogenase Promote Site-specific Oxidative Damages in Yeast and Human Cells. J. Biol. Chem. 2011;286:40232–40245. doi: 10.1074/jbc.M111.274415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brautigam CA, Wynn RM, Chuang JL, Machius M, Tomchick DR, Chuang DT. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure. 2006;14:611–621. doi: 10.1016/j.str.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciszak EM, Makal A, Hong YS, Vettaikkorumakankauv AK, Korotchkina LG, Patel MS. How dihydrolipoamide dehydrogenase-binding protein binds dihydrolipoamide dehydrogenase in the human pyruvate dehydrogenase complex. J. Biol. Chem. 2006;281:648–655. doi: 10.1074/jbc.M507850200. [DOI] [PubMed] [Google Scholar]
- 46.Ambrus A, Adam-Vizi V. Molecular dynamics study of the structural basis of dysfunction and the modulation of reactive oxygen species generation by pathogenic mutants of human dihydrolipoamide dehydrogenase. Arch. Biochem. Biophys. 2013;538:145–155. doi: 10.1016/j.abb.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 47.Ambrus A, Mizsei R, Adam-Vizi V. Structural alterations by five disease-causing mutations in the low-pH conformation of human dihydrolipoamide dehydrogenase (hLADH) analyzed by molecular dynamics - Implications in functional loss and modulation of reactive oxygen species generation by pathogenic hLADH forms. Biochem. Biophys. Reports. 2015;2:50–56. doi: 10.1016/j.bbrep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klyachko NL, Shchedrina VA, Efimov AV, Kazakov SV, Gazaryan IG, Kristal BS, Brown AM. pH-dependent substrate preference of pig heart lipoamide dehydrogenase varies with oligomeric state - Response to mitochondrial matrix acidification. J. Biol. Chem. 2005;280:16106–16114. doi: 10.1074/jbc.M414285200. [DOI] [PubMed] [Google Scholar]
- 49.Babady NE, Pang YP, Elpeleg O, Isaya G. Cryptic proteolytic activity of dihydrolipoamide dehydrogenase. Proc. Natl. Acad. Sci. USA. 2007;104:6158–6163. doi: 10.1073/pnas.0610618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandrasekhar K, Wang JJ, Arjunan P, Sax M, Park YH, Nemeria NS, Kumaran S, Song JY, Jordan F, Furey W. Insight to the Interaction of the Dihydrolipoamide Acetyltransferase (E2) Core with the Peripheral Components in the Escherichia coli Pyruvate Dehydrogenase Complex via Multifaceted Structural Approaches. J. Biol. Chem. 2013;288:15402–15417. doi: 10.1074/jbc.M113.466789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arjunan P, Wang J, Nemeria NS, Reynolds S, Brown I, Chandrasekhar K, Calero G, Jordan F, Furey W. Novel Binding Motif and New Flexibility Revealed by Structural Analyses of a Pyruvate Dehydrogenase-Dihydrolipoyl Acetyltransferase Subcomplex from the Escherichia coli Pyruvate Dehydrogenase Multienzyme Complex. J. Biol. Chem. 2014;289:30161–30176. doi: 10.1074/jbc.M114.592915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Nemeria NS, Chandrasekhar K, Kumaran S, Arjunan P, Reynolds S, Calero G, Brukh R, Kakalis L, Furey W, Jordan F. Structure and Function of the Catalytic Domain of the Dihydrolipoyl Acetyltransferase Component in Escherichia coli Pyruvate Dehydrogenase Complex. J. Biol. Chem. 2014;289:15215–15230. doi: 10.1074/jbc.M113.544080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang JJ, Kumaran S, Zhou JY, Nerneria NS, Tao H, Kakalis L, Park YH, Birkaya B, Patel MS, Jordan F. Elucidation of the Interaction Loci of the Human Pyruvate Dehydrogenase Complex E2.E3BP Core with Pyruvate Dehydrogenase Kinase 1 and Kinase 2 by HID Exchange Mass Spectrometry and Nuclear Magnetic Resonance. Biochemistry. 2015;54:69–82. doi: 10.1021/bi5013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambrus A, Torocsik B, Adam-Vizi V. Periplasmic cold expression and one-step purification of human dihydrolipoamide dehydrogenase. Protein Expr. Purif. 2009;63:50–57. doi: 10.1016/j.pep.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Kim H, Patel MS. Characterization of 2 site specifically mutated human dihydrolipoamide dehydrogenases (His-452-]Gln and Glu-457-]Gln) J. Biol. Chem. 1992;267:5128–5132. [PubMed] [Google Scholar]
- 56.Sansaricq C, Pardo S, Balwani M, Grace M, Raymond K. Biochemical and molecular diagnosis of lipoamide dehydrogenase deficiency in a North American Ashkenazi Jewish family. J. Inherit. Metab. Dis. 2006;29:203–204. doi: 10.1007/s10545-006-0175-5. [DOI] [PubMed] [Google Scholar]
- 57.Brassier A, Ottolenghi C, Boutron A, Bertrand AM, Valmary-Degano S, Cervoni JP, Chretien D, Arnoux JB, Hubert L, Rabier D, Lacaille F, de Keyzer Y, Di Martino V, de Lonlay P. Dihydrolipoamide dehydrogenase deficiency: A still overlooked cause of recurrent acute liver failure and Reye-like syndrome. Mol. Genet. Metab. 2013;109:28–32. doi: 10.1016/j.ymgme.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 58.Yuan L, Cho YJ, Kim H. Characterization of Two Naturally Occurring Mutations Close to Cofactors in Human Dihydrolipoamide Dehydrogenase. Bull. Korean Chem. Soc. 2008;29:2327–2328. [Google Scholar]
- 59.Lanterman MM, Dickinson JR, Danner DJ. Functional analysis in Saccharomyces cerevisiae of naturally occurring amino acid substitutions in human dihydrolipoamide dehydrogenase. Hum. Mol. Genet. 1996;5:1643–1648. doi: 10.1093/hmg/5.10.1643. [DOI] [PubMed] [Google Scholar]
- 60.Kim H. Examination of the importance of Pro-453 in human dihydrolipoamide dehydrogenase predicted from the three-dimensional structure. Bull. Korean Chem. Soc. 2006;27:819–820. [Google Scholar]
- 61.Patel MS, Korotchkina LG, Sidhu S. Interaction of E1 and E3 components with the core proteins of the human pyruvate dehydrogenase complex. J. Mol. Catal. B-Enzym. 2009;61:2–6. doi: 10.1016/j.molcatb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu TC, Korotchkina LG, Hyatt SL, Vettakkorumakankav NN, Patel MS. Spectroscopic studies of the characterization of recombinant human dihydrolipoamide dehydrogenase and its side-directed mutants. J. Biol. Chem. 1995;270:15545–15550. doi: 10.1074/jbc.270.26.15545. [DOI] [PubMed] [Google Scholar]
- 63.Kim H. Characterization of Two Naturally Occurring Mutations (Gly-101 Deletion and Glu-340 to Lys Substitution) in Human Dihydrolipoamide Dehydrogenase of a Patient with Metabolic Acidosis. Bull. Korean Chem. Soc. 2012;33:2477–2478. [Google Scholar]
- 64.Yuan L, Kim H. Characterization of a Naturally Occurring Mutation (Arg-460 to Gly) Close to FAD in Human Dihydrolipoamide Dehydrogenase. Bull. Korean Chem. Soc. 2010;31:3511–3512. [Google Scholar]
- 65.Park Y-H, Patel MS. Characterization of interactions of dihydrolipoamide dehydrogenase with its binding protein in the human pyruvate dehydrogenase complex. Biochem. Biophys. Res. Commun. 2010;395:416–419. doi: 10.1016/j.bbrc.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan L, Kim H. Characterization of a Naturally Occurring Mutation (Ile-358 to Thr) in Human Dihydrolipoamide Dehydrogenase of a Patient with Leigh Syndrome. Bull. Korean Chem. Soc. 2012;33:1445–1446. [Google Scholar]
- 67.Ambrus A, Torocsik B, Adam-Vizi V. Refolding of the human dihydrolipoamide dehydrogenase. Biochem. Eng. J. 2009;45:120–125. doi: 10.1016/j.pep.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 68.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun. Mass Spectrom. 2008;22:1041–1046. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
- 69.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 70.Bai YW, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen-exchange. Proteins: Struct. Funct. Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kan Z-Y, Mayne L, Chetty PS, Englander SW. ExMS: Data Analysis for HX-MS Experiments. J. Am. Soc. Mass Spectrom. 2011;22:1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.