Abstract
Our studies investigating the existence of tumor-initiating cell (TIC) populations in human esophageal squamous cell carcinoma (ESCC) had identified a subpopulation of cells isolated from ESCC patient-derived tumor specimens marked by an ALDHbri+ phenotype bear stem cell-like features. Importantly, KDM4C, a histone demethylase was enhanced in ALDHbri+ subpopulation, suggesting that strategies interfering with KDM4C may be able to target these putative TICs. In the present study, by genetic and chemical means, we demonstrated that, KDM4C blockade selectively decreased the ESCC ALDHbri+ TICs population in vitro and specifically targeted the TICs in ALDHbri+-derived xenograft, retarding engraftment. Subsequent studies of the KDM4C functional network identified a subset of pluripotency-associated genes (PAGs) and aldehyde dehydrogenase family members to be preferentially down-regulated in KDM4C inhibited ALDHbri+ TICs. We further supported a model in which KDM4C maintains permissive histone modifications with a low level of H3K9 methylation at the promoters of several PAGs. Moreover, ectopic expression of SOX2 restored KDM4C inhibition-dependent ALDHbri+ TIC properties. We further confirmed these findings by showing that the cytoplasmic and nuclear KDM4C staining increased with adverse pathologic phenotypes and poor patient survival. Such staining pattern of intracellular KDM4C appeared to overlap with the expression of SOX2 and ALDH1. Collectively, our findings provided the insights into the development of novel therapeutic strategies based on the inhibition of KDM4C pathway for the eliminating of ESCC TIC compartment.
Abbreviations: ESCC, Esophageal squamous cell carcinoma; TICs, Tumor-initiating cells; KDM4C, Lysine (K)-specific demethylase 4C
Introduction
TICs are thought to be resistant to current chemotherapeutic strategies due to their intrinsic stem-like properties and thus may provide the principal driving force behind recurrent tumor growth [1]. ESCC is one of the most aggressive types of human cancer, known for its high metastatic potential, enhanced heterogeneity and resistance to conventional therapy at advanced stages [2]. In this perspective, the aim of this study was to use human ESCC cell lines and patient-derived tumor specimens to identify and characterize putative ESCC TIC population. Further, to investigate the underlying molecular mechanism by which this population mediates ESCC maintenance.
Chromatin-associated proteins are essential for the specification and maintenance of cell identity. Histone demethylases constitute a large and diverse family of enzymes, which is regarded as an important type of histone modification during TICs maintenance [3]. The KDM4 subfamily (KDM4A-D) comprises the recently discovered Jumonji C (JmjC) domain containing (JMJD) histone demethylases, which demethylates mainly H3K9me2/3 or H3K36me3 [4]. A number of reports addressing the functions of the KDM4 demethylases showed that subunits are structurally similar and sequence homologous [4], [5] but may differ in their substrate specificity [6], [7], implying they may have widespread and redundant functions as well as unique targets and distinct biological roles. As there is increasing evidence that KDMs are associated with various disease states, they have emerged as attractive targets for the development of new therapeutic drugs [8], [9]. Efforts by many groups to find lysine demethylase inhibitors have led to the identification of several classes of inhibitors [10], [11], [12].
KDM4C, also known as JMJD2C/GASC1, is a member of the KDM4 subgroup of JmjC domain-containing proteins which catalyzes the demethylation of tri- and dimethylated lysine 9 and lysine 36 on histone H3. KDM4C was first identified as an oncogene that is amplified in esophageal cancer cell line, KYSE-150 [13]. Recent studies have implicated a role for KDM4C in self-renewal and pluripotency in preimplantation embryos and embryonic stem (ES) cells [14], [15], [16]. So far, the specific roles of KDM4C involved in maintenance of ESCC TICs or whether it can be used for TIC-directed therapy are largely unexplored.
Previous report identified ALDH1 as a marker of TICs in a broad spectrum of malignancies as assessed by the Aldefluor assay [17], [18], [19]. In this study, we used the similar techniques to identify cellular hierarchies in characterized ESCC cell line and patient-derived dissociated tumor specimens and demonstrated that a subset of cells contained ALDHbri+ components which endowed with stem cell properties as evidenced in clonogenicity self-renewal and are more tumorigenic in vivo. Further, we reported here that KDM4C blockade using either lentiviral-mediated gene silencing or its pharmacological inhibitor Caffeic acid (CaA) [20], preferentially reduced the proportion of ALDHbri+ cells, providing evidence for its roles in the maintenance of TICs in ESCC. Furthermore, we have identified several pluripotency-related genes and diverse members of aldehyde dehydrogenase family to be downstream targets of KDM4C.
Our study highlighted the role of the KDM4C in the maintenance of TICs in ESCC and probed the therapeutic potential of targeting KDM4C pathway activity in human ESCC.
Material and Methods
Fresh Clinical Tissue Specimen Collection and Processing
ESCC clinical samples were collected in the period between March 2008 and February 2010 from Anyang Cancer Hospital (Henan, China). All samples were approved by the committee for ethical review of research involving human subjects at The First Affiliated Hospital of Henan University of Science and Technology (Luoyang, China).
A portion of the samples (EC-1, 2, 3) were received in the laboratory immediately mechanically disaggregated and digested as described in Supplementary and subsequently incubated with Aldefluor® kit [17]. The resultant tumor cells were injected subcutaneously at different multiplicities, in the range 104–102(‘limiting dilution’), into NOD-SCID mice or seeded for the Esosphere formation assay (Supplemental Material and Methods).
Chromatin Immunoprecipitation Assay
ChIP assay was carried out as described previously [21]. Chromatin extracts were immunoprecipitated using 10 μg anti-KDM4C (A300-885 A; Bethyl Laboratories); 4 μg anti-trimethylated H3K9 (61,013; Active Motif), 4 μg anti-dimethylated H3K9 (39,683; Active Motif), and 2 μg anti-total H3 (39,763; Active Motif); Anti-GST (sc-459; Santa Cruz Biotechnology) antibodies were used as mock ChIP controls. Fold enrichments were calculated by determining the ratios of the amount of immunoprecipitated DNA to that of the input sample and were normalized to the level observed at a control region, which was defined as 1.0. The ChIP primers are available on request (Supplemental Table Primers).
Microarray Analysis and Gene Ontology Analysis
We performed transcriptome analysis by Agilent Human Whole Genome Microarrays in 48 hours KDM4C knockdown (or CaA-treated) and control ALDHbri+ ESCC TICs isolated from the ESCC clinical samples. The genes that decreased by ≥20% following KDM4C knockdown in primary ESCC TICs were analyzed by the DAVID Functional Annotation Tool, compared with a background of the total genes expressed in ALDHbri+ ESCC cells.
AlphaLISA Assay
The AlphaLISA Tri-Methyl-Histone H3 Lysine9 (H3K9me3) Cellular Detection Kit was obtained from PerkinElmer Life Sciences. Cells were cultured in 6-well tissue culture plates at 3 × 105/well in 3 ml of media for 12 h, then incubated with different concentrations of CaA (Sigma-Aldrich) for another 48 h. The same volume of DMSO was used as the vehicle control for CaA experiments at a final concentration of 0.1%. The luminescence signal was measured using the Envision (PerkinElmer Life Sciences) plate readers.
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism 5.0 software using t-test. Data are presented as the mean ± SEM. P ≤ .05 was regarded as being statistically significant.
Results
KDM4C Levels are Frequently Up-Regulated in a Subset of Patient-Derived Primary ESCC Cultures and Established Cell Lines
To investigate the role of KDM4C in the development of ESCC, we first examined KDM4C expression in well-characterized human ESCC cell lines, patient-derived ESCC tumors under conditions that permitted expansion in vitro and the normal human immortalized epithelial cells using Western blotting assay. Expression of KDM4C proteins were clearly detected in 3 of 5 established ESCC cell lines (EC9706, KYSE-150, KYSE-30, Figure S1A) and 3 of 5 primary ESCC cultures (Figure S1B, EC-1, 2, 3). However, it was documented at low level in one normal human Immortalized epithelial cells (SHEE, Figure S1A).
We designed eight shRNAs directed against KDM4C and infected ESCC cells with lentiviral particles encoding shRNA sequences. KDM4C-shRNA-5 (shKDM4C) was employed for most studies, as it reliably yielded cells with >80% KDM4C knockdown at the protein levels. KDM4C-shRNA-7 was used as a second shRNA to confirm the KDM4C knockdown phenotype (Figure S1C).
KDM4C Expression is Enhanced in ALDHbri+ Subpopulation which is Critical for the Maintenance of ESCC TICs
To determine whether ESCC contains TIC population, cells from KYSE-150 cell line and patient-derived dissociated tumors of ESCC were isolated by virtue of ALDH activity. Representative cytograms are shown in Figure 1A, the ALDHbri+ populations, ranging from 3.92% to 15.6%, were detected in all cell preparations from freshly isolated ESCC specimens. The ALDHbri+ fraction of KYSE-150 cell line was approximately 0.97%. These experiments confirmed the presence of ALDHbri+ cells in human ESCC.
Figure 1.
A subset of ESCC cells contained ALDHbri+ components which endowed with stem cell properties as evidenced in clonogenicity and self-renewal in vitro. (A) Representative Aldefluor analysis in primary patient-derived ESCC cells and KYSE-150 cell line. Control samples incubated with the inhibitor, DEAB, were used to ensure identification of ALDHbri+ and ALDHlow- cells. (B) Sphere formation efficiency of fluorescence-activated cell sorting (FACS)-sorted ALDHbri+ and ALDHlow- cells isolated from fresh samples and cell lines. (C) Representative images of ALDHbri+ and ALDHlow- derived spheres of EC-2 (scale bar = 50 μm). (D) Representative Aldefluor analysis in dissociated esospheres of EC-2 in serial propagations. Control samples incubated with the inhibitor, DEAB, were used to ensure identification of ALDHbri+ and ALDHlow- cells. (E) Representative images of ALDHbri+ and ALDHlow- spheres of EC-2 in three passages (scale bar = 50 μm). (F). ALDHbri+ and unseparated subpopulations of EC-2 were capable of self-renewal in vitro, as shown by similar esosphere-initiating capacity in three passages.
To determine whether the subset defined by high ALDH activity (ALDHbri+) was enriched in TIC, we compared the ability of FACS-sorted ALDHbri+ ESCC cells and their ALDHlow- counterparts to generate esospheres in vitro and to initiate tumor formation in vivo.
Within 7 days of culture of ALDHbri+ and their ALDHlow- daughter cells in non-adherent, serum-free, growth factor-supplemented conditions. As previously reported, these culture conditions allowed the selection of undifferentiated cancer stem and progenitor cells, while serum-dependent differentiated tumor cells and non-transformed accessory cells were negatively selected [22]. The ALDHbri+ population was capable of generating more esospheres at a density of 5000 cells/ml than its ALDHlow- counterpart (Figure 1, B and C). Moreover, ALDHbri+ cells sorted from dissociated esospheres were capable of self-renewal in vitro as evidenced by similar ALDHbri+ population percentage (Figure 1D) and esosphere-initiating capacity in serial propagations (Figure 1, E and F), showing virtually unlimited growth potential.
Next, we investigated the ability of sorted cells to subcutaneously engraft and give rise to tumors by inoculation of limiting dilutions of cells (ranging from 102 to 104) into mammary fat pads of NOD/SCID mice.
As shown in Table I, using cells from patient EC-1, the sorted ALDHbri+ cells clearly showed increased tumor-forming capacity comparing with ALDHlow- and bulk counterparts when 104 or fewer cells were implanted, which was evident at extremely low multiplicities (i.e., 102 transplanted cells). Indeed, 104 ALDHbri+ cells formed tumors in 5/5 mice, compared to 2/5 and 4/5 mice with tumors from matched ALDHlow- and unsorted cells, respectively. 103 ALDHbri+ cells formed tumors in 5/5 mice, whereas 0/5 and 2/5 mice bear tumors when implanted with same matched ALDHlow- and unsorted cells, respectively. When mice were implanted with 102 cells, ALDHbri+ cells formed tumors in 2/5 mice, compared to no tumor formation from matched ALDHlow- and unsorted cells.
In addition to their increased tumor-forming capacity, the frequency of TICs was considerably higher in ALDHbri+ cells (1 in 32) than ALDHlow- cells (1 in 2704) or parental cells (1 in 979) (Table I). For comparison of tumor latency, the ALDHbri+ fraction was capable of generating tumors following injection of as few as 102 cells whereas about 104 ALDHlow- cells were needed for generating tumors in the same time period (Figure 2A). Tumor formation in the lower dilutions for ALDHlow- cells was either significantly delayed or undetected within the 2-month experimental time frame (Figure 2A). As indicated, tumor growth kinetics correlated with the latency and size of tumor formation correlated with the number of ALDHbri+ cells injected (Figure 2A).
Figure 2.
A subset of ESCC cells contained ALDHbri+ components which were tumorigenic in vivo. (A) Tumorigenesis of ALDHbri+ and ALDHlow- in NOD/SCID mice. Tumor growth curves were plotted for the numbers of engrafted cells (10,000, 1000, and 100 cells) and for each subpopulation (ALDHbri+ and ALDHlow-). Tumor growth kinetics correlated with the latency and size of tumor formation and the number of ALDHbri+ cells. No tumor was detected at the ALDHlow- cells' injection site (100 cells injected). Data represent mean ± SEM (n = 3). P < .05 compared with ALDHlow-. (B) Representative bioluminescent images of mice harboring tumors are shown. To evaluate the tumorigenesis of ALDHbri+ and ALDHlow-in vivo, we infected both cells with a lentivirus expressing luciferase, and inoculated 10,000, 1000 and 100 luciferase-tagged cells into NOD/SCID mice. Tumor formation was measured at weekly intervals following inoculation, revealed a statistically significant increase (P < .01) in tumoral formation in ALDHbri+ compared with ALDHlow- cells. (C) Differentiation of ALDHbri+ and ALDHlow- cells in vivo. Sorted ALDHbri+ and ALDHlow- cells from EC-2 and EC-3 were implanted into NOD/SCID mice and the xenografts derived from ALDHbri+ and those from ALDHlow- cells were analyzed by Aldefluor assay. (D) Histology and KDM4C IHC analysis of primary tumor and tumor xenograft generated from ALDHbri+ cells isolated from EC-2 and EC-3. The ALDHbri+ population was capable of regenerating the phenotypic heterogeneity of the initial tumor after a passage in NOD/SCID mice.
In subsequent experiments, we found that ALDHbri+ cells from samples EC-2 and EC-3 were also enriched for high tumorigenicity even at low cell input numbers. ALDHbri+ cells purified from EC-2 and EC-3 were tumorigenic at 100 cells, and resulted in tumor formation in 2/5 and 4/5 mice, respectively. In contrast, 1000 ALDHlow- cells from both EC-2 and EC-3 were essentially non-tumorigenic, with tumor formation in only 1/5 and 2/5 mice. We were unable to grow tumors with 100 or fewer ALDHlow- sorted cells from the primary EC-2 and EC-3 tumors (Table II and III). Using the IVIS imaging system, we monitored xenograft formation by measuring the change in bioluminescence signal and found that luciferase-tagged ALDHbri+ cells exhibited a greater frequency of tumor formation, particularly at low numbers of injected cells, compared to ALDHlow- counterparts (Figure 2B).
To investigate whether ALDHbri+ ESCC cells have long-term tumorigenic potential and self-renewing capacity, we evaluated their ability to generate tumors in serial transplantations. Tumors obtained by subcutaneous transplantation of both ALDHbri+ and ALDHlow- cells from primary xenografts were again reduced to single-cell suspension, sorted again by FACS into ALDHbri+ and ALDHlow- fractions, and again transplanted (104cells) into recipient mice. Table IV summarizes the results relative to a total of three rounds of serial transplantations. Clearly, whereas ALDHlow- rapidly lost their tumor-initiating capacity already after two rounds, ALDHbri+ cells demonstrate self-renewal by retaining their ability to engraft throughout the serial transplantation passages.
Finally, to determine the abilities of the ALDHbri+ cells to re-establish tumor heterogeneity in vivo revealed ALDHbri+-derived xenografts contained a mixed population with 10.1% and 14.7% of ALDHbri+ cells similar to their original tumors of EC-2 and EC-3 (Figure 2C). Moreover, tumors formed by ALDHbri+ cells contained histological and antigenic (KDM4C) features closely resembling the original tumor, as shown by hematoxylin and eosin (H&E) staining and IHC analysis (Figure 2D). These suggested that a heterogeneous tumor phenotype was recapitulated following injection of a fraction of ALDHbri+ cells.
We subsequently tested the expression of KDM4C in sorted ALDHbri+ TICs and ALDHlow- tumor cells from three ESCC specimens by qPCR (Figure S2A) and Western blotting assay (Figure S2B). KDM4C expression was enriched in the ALDHbri+ subpopulation as compared with the ALDHlow- counterparts. In light of this commonality between sorted subpopulations, we hypothesized that KDM4C is involved in the maintenance of human ESCC ALDHbri+ cells.
KDM4C Inhibition Decreases the Clonogenicity and Self-Renewal of ALDHbri+ ESCC TICs
We next tested whether KDM4C function was involved in maintenance of the ALDHbri+ ESCC TICs. Silencing of KDM4C was achieved by lentiviral expression of shRNAs (Figure S2C). Aldefluor analysis revealed KDM4C knockdown led to a drastic decrease in the fraction of ALDHbri+ subpopulation from 9.8% to 2.05% in EC-2, from 15.6% to 3.93% in EC-3. The EC-1 had a small ALDHbri+ population that was reduced from 3.92% to 0.09% (Figure 3A). Moreover, repression of KDM4C expression in ALDHbri+ TICs significantly suppressed the ability of cells to initiate esospheres when grown in non-adherent serum-free conditions in vitro (Figure 3B) and expand in subsequent in vitro serial propagations (Figure 3C).
Figure 3.
KDM4C inhibition decreased the percentage, clonogenicity and self-renewal of ESCC ALDHbri+ TICs in vitro. (A) Quantification of the fraction of ALDHbri+ cells, determined by Aldefluor assay in patient-derived ESCC cells transduced with LV-c, LV-shKDM4C-5 or LV-shKDM4C-7. Significant reduction in the percentage of ALDHbri+ cells after silencing of KDM4C is shown. (B) Representative images of spheres formed by patient 2-derived ALDHbri+ TICs transduced with LV-c, LV-shKDM4C-5 or LV-shKDM4C-7. (Scale bar = 50 μm). Decreased Number and size of spheres are shown in ALDHbri+ cells after silencing of KDM4C. (C) ALDHbri+ cells were capable of self-renewal in vitro, as shown by similar esosphere-initiating capacity in three passages of EC-2. Significant reduction in the self-renewal of ALDHbri+ cells after silencing of KDM4C is shown. (D) Quantification of the fraction of ALDHbri+ cells, determined by Aldefluor assay in patient-derived ESCC cells treated with vehicle, 5 μM, 10 μM or 20 μM KDM4C inhibitor CaA. Significant reduction in the percentage of ALDHbri+ cells after CaA treatment is shown. (E) Representative images of spheres formed by patient 2-derived ALDHbri+ cells treated with vehicle, 5 μM, 10 μM or 20 μM CaA. (Scale bar = 50 μm). Decreased Number and size of spheres are shown in ALDHbri+ cells upon CaA treatment in a dose-dependent manner. (F) ALDHbri+ cells of EC-2 were capable of self-renewal in vitro, as shown by similar esosphere-initiating capacity in serial passages. Significant reduction in the self-renewal of ALDHbri+ cells after CaA treatment is shown. Data shown are the mean ± S.E.M. of at least three independent experiments. *P⩽0.05; **P⩽0.01.
As an additional test to investigate the requirement of KDM4C function, we inhibited it pharmacologically. The pharmacological inhibitor CaA treatment dramatically diminished the percentage of ALDHbri+ population, from 9.8% to 7.93% (5 μM) and 1.22% (20 μM) in EC-2 and from 15.6% to 9.3% (5 μM) and 0.92% (20 μM) in EC-3. The EC-1 had a small ALDHbri+ population that was reduced to 2.19% and 0.06% by 5 and 20 μM CaA, respectively (Figure 3D). We then examined the ability of CaA to block esospheres formation. ALDHbri+ TICs from primary cell cultures seeded and allowed to establish initial esospheres were then treated with vehicle, 5 μM, 10 μM, or 20 μM drug for an additional 7 days and the resulting esospheres were analyzed. Esospheres formation was inhibited in a dose-dependent manner, with a statistically significant 25% reduction in colonies grown in 5 μM CaA (P = .02) and an approximately 50% and 95% reduction in colonies grown in 10 μM and 20 μM CaA (P < .0001, respectively) (Figure 3E). After 7 days of CaA treatment, esospheres were dissociated and the ability of single cells to generate secondary spheres in absence of the drug was quantified. Consistently with shRNA approach, CaA treatment also led to a significant and dose-dependent reduction of the ability to form secondary spheres (Figure 3F). Taken together, our results indicate that KDM4C is required for the maintenance of the ALDHbri+ ESCC TICs in vitro.
Preferential Targeting of TICs by CaA in the ALDHbri+-Derived Xenograft Model In Vivo
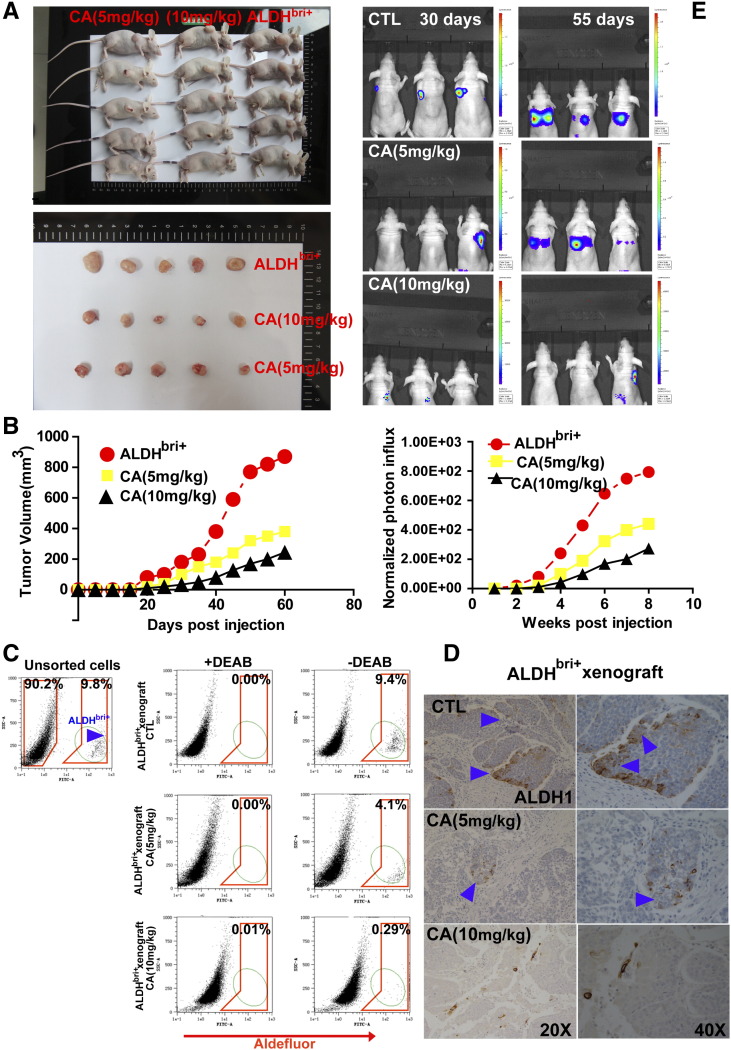
We subsequently determined whether KDM4C is required for TIC maintenance in vivo. ALDHbri+ TICs were injected subcutaneously in the mammary fat pads of NOD/SCID mice to initiate xenograft tumors. One week after the initial tumor inoculation, tumor-bearing animals were begun administered intraperitoneally with CaA (5 mg/kg or 10 mg/kg) or saline controls three times a week for a total of 8 weeks. For each xenotransplant, we observed significant inhibition of tumor initiation by the small-molecule inhibitor, as indicated by decreases in tumor size (Figure 4A) and increases in tumor latency (Figure 4B).
Figure 4.
The KDM4C inhibitor CaA preferentially abolished TIC in ALDHbri+-derived xenograft model in vivo. (A) Representative images of subcutaneous xenografts in nude mice taken at the same time for each group. For each xenograft, 10,000 cells were injected into the mammary fat pad of nude mice. There is a statistically significant size reduction of the tumors treated with 5 mg/kg or 10 mg/kg CaA compared with the control tumors (P < .01, respectively). (B) Tumor growth curves were plotted for the numbers of engrafted cells (10,000 FACS-sorted ALDHbri+ cells). There is a statistically significant size reduction of the tumor treated with 5 mg/kg or 10 mg/kg CaA compared with the controls during the course of each indicated time points (P < .01, respectively). (C) Mice bearing injection were treated with either vehicle or CaA (5 mg/kg or 10 mg/kg) three times a week for a total of 8 weeks. Cells were dissociated from tumors and subject to Aldefluor assay. Shown are cytograms of percent ALDHbri+ cells. (D) Xenotransplants from each group were collected, and ALDH1 IHC staining was done. Cytoplasmic ALDH1 expression was detected in the controls (arrowheads denote positive staining), whereas low expression was detected in the tumors treated with CaA. (E) Representative bioluminescent images of mice harboring tumors are shown (Upper panel). To evaluate the effect of CaA treatment on tumor initiation, we infected ALDHbri+ cells with a lentivirus expressing luciferase, and inoculated 10,000 luciferase-tagged cells into NOD/SCID mice. One week after the initial tumor inoculation, tumor-bearing animals were begun administered intraperitoneally with CaA (5 mg/kg or 10 mg/kg) or saline controls three times a week for a total of 8 weeks. Tumor formation was monitored using bioluminescence imaging. Quantification of the normalized photon flux (Lower panel), measured at weekly intervals following inoculation, revealed a statistically significant decrease (P < .01, respectively) in tumoral formation in CaA-treated compared with controls for mice inoculated with luciferase-tagged ALDHbri+ cells.
After 8 weeks of treatment, animals were sacrificed, residual xenograft tumors were harvested and dissociated into single cells and subjected to Aldefluor assay without further compound treatment. Results showed that residual controls contained an unchanged proportion of ALDHbri+ cells compared with parental tumor from which these ALDHbri+ cells have been derived. In contrast, CaA treatment caused significant reduction of the proportion of ALDHbri+ cells to 4.1% (5 mg/kg) and 0.29% (10 mg/kg), respectively (Figure 4C). These observations were confirmed by IHC of ALDH1 expression in the different xenotransplants, a decrease in ALDH1 staining was detected in CaA-treated tumors compared with controls (Figure 4D). We also monitored xenograft formation by in vivo imaging system (IVIS) and found that CaA treatment greatly hampered the tumor initiating capability of the luciferase-tagged ALDHbri+ cells (Figure 4E).
To further demonstrate that CaA treatment was able to reduce the ESCC TIC population, we used a functional in vivo assay consisting of re-implantation of equal cells from treated tumors into secondary recipients. Tumor cells derived from controls showed similar tumor re-growth at 104ALDHbri+ cells in secondary recipients. In contrast, when equal numbers of cells were injected, those from CaA-treated animals showed a 2- to 5-fold reduction in tumor incidence in secondary recipients (Table V).
Together, these studies demonstrate that CaA treatment specifically targets and reduces the ESCC ALDHbri+ TIC population.
KDM4C, Influences Unique Gene Signatures and Functional Networks in ALDHbri+ ESCC TICs
To understand the molecular basis of the KDM4C function in ALDHbri+ ESCC TICs maintenance, we performed transcriptomic analyses by Agilent Human Whole Genome Microarrays in 48 hours KDM4C knockdown and control ALDHbri+ ESCC TICs isolated from the ESCC clinical samples. By evaluating the overlap between down-regulated genes identified in CaA-treatment study and RNA interference screen, we identified 694 genes that were commonly down-regulated at least 2.0-fold between the screens (Figure 5A). Heatmap Figure 5B shows unsupervised clustering of transcripts obtained for the ALDHbri+ ESCC cells 48 h after KDM4C knockdown or mock transfection. GO analysis was used to functionally annotate differentially expressed genes and demonstrated that the overlapped down-regulated genes were generally enriched for functions in aldehyde dehydrogenase (NAD) activity, transcription factor binding/pluripotency maintenance, cell cycle regulation and differentiation. Selected genes that can be organized by function or family are illustrated in heatmap Figure 5C.
Figure 5.
Profiling of KDM4C knockdown and control ALDHbri+ ESCC TICs. (A) Venn diagrams for genes that are down-regulated by KDM4Csh knockdown or CaA treatment. ALDHbri+ cells were treated with shKDM4C or CaA. The mRNA levels in KDM4C-depleted cells were measured by Agilent Human Whole Genome Microarrays and compared with those in the controls. (B) Heatmap shows unsupervised clustering of down-expressed transcripts obtained for the ALDHbri+ ESCC cells after KDM4C knockdown (right) or mock transfection (left) using Agilent Human Whole Genome Microarrays. Colored spots indicate up-regulated (red) or down-regulated (blue) genes from microarray analysis. (C) Heatmap shows Gene ontology analysis of genes that are co-down-regulated by KDM4C knockdown and CaA treatment. Gene ontology analysis was performed using the program DAVID. (D) qPCR validation of differentially expressed genes. Expression levels were normalized to GAPDH. White and black and bars represent expression in controls and KDM4C knockdown ALDHbri+ cells, respectively (P < .01, respectively). (E) Immunostaining of control and KDM4C-shRNA ALDHbri+ cells. KDM4C-shRNA cells have decreased KDM4C, SOX2 and ALDH1 expression relative to control cells. Nuclei were stained with DAPI. (Scale bar = 50 μm).
Among these genes are a number of known pluripotency-associated genes, including Pou5f1, c-Myc, SOX2 and KLF4 which were chosen for further analysis because they had known functions for stemness maintenance, making them more desirable targets for later functional analysis. qPCR was carried out to validate this finding and results were concordant with the microarray results (Figure 5D1). Among these genes, SOX2, a key transcription factor important for the maintenance of pluripotency and self-renewal of ES cells was strikingly down-regulated and was chosen as a target for these studies. A representative example of TICs that were stained for both KDM4C and SOX2 is shown in Figure 5E1.
As depicted in the supervised hierarchical clustering (Figure 5B) and GO analysis heatmap (Figure 5C), we also confirmed a significant decrease in the expression of the diverse members of aldehyde dehydrogenase family in ALDHbri+ TICs, including ALDH1A1, ALDH1A3, ALDH16A1, ALDH6A1,ALDH7A1 and ALDH4A1 (Figure 5D2). A representative example of TICs that were stained for both KDM4C and ALDH1 is shown in Figure 5E2.
KDM4C Inhibition is Involved in the Maintenance of H3K9me3 at the Pluripotency-Associated Genes Locus
To test the hypothesis that down-regulation of pluripotency-associated genes during KDM4C inhibition is linked to histone modifications, we investigated whether the demethylase inhibitor CaA affect selected global histone methylation states in ALDHbri+ cells, using AlphaLISA Assay. We observed that, treatment of ALDHbri+ cells with CaA for 48 h led to a dramatic increase of H3K9me3 levels dose-dependently, whereas H3K4me2 and H3K4me1 levels did not change significantly (Figure S3A). Subsequently, we used the technique of quantitative chromatin immunoprecipitation (qChIP) to probe histone changes at pluripotency-associated gene promoters during KDM4C inhibition. ChIP analysis was performed using antibodies that individually recognize either H3K9me2 or H3K9me3 and the primers amplifying the regions of SOX2 promoter as indicated in Figure 6A. KDM4C inhibition was found to cause substantial increases in the levels of H3K9me3 and H3K9me2 at the SOX2 promoter (Figure 6, A and B). To extend our study, we screened the promoter regions of other genes for KDM4C-dependent modulation of the H3K9 methylation levels and detected KDM4C bind the promoter of c-Myc and demethylated H3K9me3 and H3K9me2 at its promoter (Figure 6, C and D). KDM4C binds promoters of the core pluripotency regulators Pou5f1 providing additional evidence that KDM4C positively supports ALDHbri+ ESCC TICs pluripotency (Figure S3B) Representative agarose gel images showing PCR amplification products corresponding to the SOX2 (Figure S6A), c-Myc (Figure 6C) and Pou5f1 (Figure S3B) promoter regions are shown. The corresponding bands were detected in the samples from shKDM4C ALDHbri+ cells, but remained low in the control cells. Input DNA and anti-IgG antibody-precipitated DNA were used as positive and negative controls, respectively. Quantification of H3K9 methylation levels by qPCR at the promoters of SOX2 (Figure 6B), c-Myc (Figure 6D) and Pou5f1 (Figure S3B) were also shown. Subsequently, we performed a ChIP assay using two independently generated anti-KDM4C antibodies. The result showed that KDM4C was bound to the SOX2 promoter (Figure S3C). In contrast, Jmjd5 showed only a very low or near background level of binding (Figure S3D). Furthermore, the depletion of KDM4C using either shRNA construct abolished the KDM4C ChIP signal, indicating that the antibodies specifically recognized KDM4C (Figure S3E).
Figure 6.
Increased H3K9 methylation levels at the pluripotency-associated gene promoters in response to KDM4C inhibition in ALDHbri+ cells. ChIP assays were performed on ALDHbri+ cells following KDM4C knockdown (via transfection with control shRNA, KDM4CshRNA-5, KDM4CshRNA-7) or exposure or not exposure to CaA for various concentrations, as indicated. Cells were then collected for ChIP analyses using antibodies to the indicated H3K9 methylation forms to determine H3K9 methylation levels at the SOX2, c-Myc, and Pou5f1 promoters. (A) Representative agarose gels showing PCR amplification products corresponding to the SOX2 promoter region is shown. Specific antibodies that individually recognize either the di- (H3K9me2) or the trimethylated form of H3K9 (H3K9me3) were used. GST antibody was used as a control. (B) ChIP analysis of H3K9 methylation levels at the SOX2 promoter after KDM4C inhibition as quantified by real-time PCR. Relative promoter occupancies (% input) are shown with error bars based on standard errors (SEs) calculated from at least three replicates. The input signal is set as 100% (not depicted in graphs) for each set of assays. (C) Representative agarose gels showing PCR amplification products corresponding to the c-Myc promoter region is shown. Specific antibodies that individually recognize either the di- or the trimethylated form of H3K9 were used. (D) ChIP analysis of H3K9 methylation levels at the c-Myc promoter after KDM4C inhibition as quantified by real-time PCR. (E) ChIP analysis of H3K9 methylation levels at the SOX2 promoter after 48 h of exposure or not exposure to CaA for various concentrations, as indicated, as quantified by real-time PCR. Relative promoter occupancies (%input) by the indicated H3K9 methylated forms are shown with error bars based on SEs calculated from at least three replicates. (F) ChIP analysis of H3K9 methylation levels at the c-Myc promoter after 48 h of exposure or not exposure to CaA for various concentrations, as indicated, as quantified by real-time PCR. (G) Xenotransplants from each group were collected, Western blot analysis and IHC staining(H) were done. Representative immunoblots of H3K9me3 and H3K9me2 protein expression in whole tissue extracts from ALDHbri+-derived xenograft treated with or without CaA. Note the increase in H3K9me3 and H3K9me2 immunoreactivity in CaA-treated xenografts compared with controls. β-Actin was used as loading control. (H) Expression of SOX2 and H3K9me3 protein in paraffin sections of CaA-treated xenografts analyzed by IHC. Sections were counterstained with hematoxylin (blue nuclei). Note the increase in nuclear H3K9me3 methylation and decrease in SOX2 immunostaining in CaA-treated xenografts compared with controls (arrowheads denote positive staining).
To this end, we performed the ChIP assay using CaA. ALDHbri+ cells were exposed or not exposed to CaA at multiple concentrations of (5, 10, 20 μM) CaA for 48 h, and chromatin was prepared for analysis. Results revealed that treatment of ALDHbri+ cells with CaA led to a marked increase in levels of di-, and trimethylated H3K9 histones on both SOX2 (Figure 6E and S3F) and c-Myc promoters (Figure 6F and S3G), consistent with decreased SOX2 and c-Myc expression in these cells, whereas no effect was detected on a non-target, anti-GST. The data support a role of KDM4C in positively regulating a subset of pluripotency-associated genes by demethylation of H3K9me3 and H3K9me2 at their promoters.
To examine whether CaA blockage of the KDM4C demethylase function affects xenograft tumor growth via epigenetic modifications, we determine whether CaA affects H3K9 methylation globally in vivo. H3K9 methylation levels were analyzed in histone extracts prepared from ALDHbri+-derived xenograft tumors after exposure to CaA. Western blot analysis (Figure 6G) showed that global di- or the trimethylated forms of H3K9 were significantly increased in CaA-treated xenografts than in controls. To examine whether the effects reflect the expression of target genes involved in the repression of growth, we analyzed expression of SOX2 and H3K9me3 by using IHC analysis from xenograft tumors. The expression level of SOX2 was significantly decreased whereas the H3K9me3 was increased in CaA-treated xenografts than in controls (Figure 6H). To test whether CaA treatment promoted the alteration in epigenetic marks on SOX2 promoter, we examined the status of the H3K9me3 marks in xenografts. A substantial increase was found in the H3K9me3 at the SOX2 promoters (Figure S3H) These studies suggest that blockage of the KDM4C axis via CaA has potential to decrease ESCC TICs in vivo by epigenetic modification at promoters of target genes.
SOX2 is Involved in Mediating the Inhibitory Effects of KDM4C Inactivation on ESCC ALDHbri+ Cells
Studies employing in vitro culture of ALDHbri+ cells have shown that inhibition of SOX2 expression causes decreased clonogenicity and self-renewal (Figure 7, A and B), which is similar to the phenotype that was observed when KDM4C was depleted, suggesting that SOX2 serves as a primary and key direct target of KDM4C inactivation for growth inhibition in ESCC ALDHbri+ cells.
Figure 7.
SOX2 is the key downstream effector of KDM4C responsible for maintaining ALDHbri+ ESCC cells. (A) Representative images of spheres formed by ALDHbri+ cells transduced with LV-c or LV-shSOX2. (Scale bar = 50 μm, Upper panel). Esosphere formation analysis of ALDHbri+ cells reveal SOX2 knockdown lead to a reduction in clonogenic efficiency (Lower panel). (B) ALDHbri+ cells were capable of self-renewal in vitro, as shown by similar esosphere-initiating capacity in serial passages. Significant reduction in the self-renewal of ALDHbri+ cells upon SOX2 knockdown is shown. (C-E) Enforced SOX2 over-expression could rescue the differentiation phenotype induced by KDM4C depletion. ALDHbri+ cells were transfected with vehicle or a SOX2-overexpressing vector and challenged with shRNA directing against KDM4C. The cells were subjected to Aldefluor analysis, spheroid formation test and expand in subsequent serial propagations to examine self-renewal. Note the rescue and the maintenance of ALDHbri+ populations (C), spheroid formation (D) and self-renewal capability (E) in SOX2-overexpressing-KDM4C-shRNA treated cells. Little or no rescue was observed when the cells were challenged with vehicle. (F-G) Enforced SOX2 over-expression could rescue the differentiation phenotype induced by CaA treatment. ALDHbri+ cells were transfected with vehicle or a SOX2-overexpressing vector and treated with CaA. The cells were subjected to Aldefluor analysis and spheroid formation test. Note the rescue and the maintenance of ALDHbri+ populations (F) and spheroid formation capability (G) in SOX2-overexpressing-CaA treated cells. Little or no rescue was observed when the cells were challenged with vehicle. (H) Enforced SOX2 over-expression could rescue the engraftment capability upon CaA treatment. ALDHbri+ cells were transfected with vehicle or SOX2-overexpressing vector and subjected to engraftment capability analyses. Note the maintenance of engraftment capability in SOX2-overexpressing cells upon CaA treatment. Little or no rescue was observed when the cells were challenged with vehicle. Data shown are the mean ± S.E.M. of at least three independent experiments. *P⩽0.05; **P⩽0.01.
To further determine whether reduced expression of SOX2 is responsible for the growth inhibition caused by KDM4C inactivation, we tried to co-transfected a SOX2 or vector control expression plasmid with KDM4Csh or shRNA constructs in ALDHbri+ cells. Ectopic expression of SOX2 led to a significant resistance of these cells to KDM4C inhibition as compared with controls as shown that ALDHbri+ cells co-transfected with a SOX2 vector plasmid and KDM4Csh constructs were found to retain TICs ALDH activity (Figure 7C), thus, cells could similarly initiate spheroid formation (Figure 7D) and expand in subsequent serial propagations (Figure 7E). The depletion of KDM4C was equally efficient in the control and SOX2-overexpressing ALDHbri+ cells (Data not shown). This excludes the possibility that the phenotypic differences were due to insufficient depletion of KDM4C.
Additionally, ectopic expression of SOX2 plasmid blocked the effects of CaA on the ALDHbri+ populations as shown by the maintenance of the ALDHbri+ populations (Figure 7F) and sphere-forming ability (Figure 7G) after treatment with the inhibitor. The rescued phenotype in SOX2-overexpressing ALDHbri+ cells was also supported by engraftment capability analyses. These cells maintained the ability to engraft when injected subcutaneously in immunodeficient animals as compared to control cells upon CaA treatment (Figure 7H). Taken together, the rescue result indicates that SOX2 can compensate for the loss of KDM4C.
KDM4C Pathway Correlates with Adverse Pathologic Phenotypes and Poor Overall Survival in ESCC Patients
We characterized the expression of KDM4C in patient ESCC by Quantitative real-time PCR (qPCR) analysis. As shown in Figure S4A, the overall expression level of KDM4C in ESCC was significantly higher than that in adjacent normal tissues (P = .001, Figure S4A). Afterward, Spearman rank correlation coefficient test and linear tendency test were conducted. It revealed that there is a statistically significant linear correlation between the expression of KDM4C and SOX2 (P < .001, r = 0.874) (Figure S4B and Table VI) as well as KDM4C and ALDH1 (P = .002, r = 0.644) (Figure S4C and Table VI) in ESCC.
To investigate the relationship between KDM4C pathway expression and clinicopathological characteristics, a series of archived ESCC specimens with multistage and differentiation were co-stained with specific antibodies for KDM4C, SOX2 and ALDH1 and the levels of expression were analyzed (Figure S4D). As shown in Figure S4D, an increase in KDM4C in the cytoplasmic and nuclear areas of ESCCs compared with normal mucosa was observed. Interestingly, the cancer cells at the tumor-host interface exhibited intense staining for the presence of KDM4C and ALDH1 in some cases (Figure S4E). These suggested that the KDM4C pathway-positive cells might be mainly localized at the invading tumor front thus endowed with adverse pathologic phenotypes.
As shown in Table VII, KDM4C protein was detectable with significantly higher frequency in advanced T-grade ESCCs than in tumors with early stage. Moreover, KDM4C increased concomitantly with tumor TNM stage. In addition, the numbers of sample positive for KDM4C were significantly greater in poorly-differentiated tumors compared with those well-differentiated ones (Table VII). There was no significant association between KDM4C expression and age, location, gross pathology and tumor size (Table VII). Kaplan–Meier analyses of overall survival showed that patients carrying tumors with increased expression of KDM4C had a significantly worse five-year overall survival than patients with tumors that had decreased expression (P = .0082, Figure S4F). Moreover, patients with co-expression of KDM4C and SOX2 had a poorer prognosis (P = .0006, Figure S4G).
Overall, our findings suggest that KDM4C signaling cascade contributes to adverse pathologic phenotypes and poor overall survival of ESCC.
Discussion
The KDM4/JMJD2 JmjC-containing histone lysine demethylases (KDM4A-KDM4D), which selectively remove the methyl group(s) from tri/dimethylated lysine 9/36 of H3 in a Fe2+ and α-KG-dependent manner and are involved in a plethora of cellular processes [3]. For example, several lines of evidence suggest that KDM4C, originally identified as a gene amplified in ESCC cell lines plays a role in malignant transformation [3]. Subsequent studies have shown amplifications of KDM4C in a metastatic lung sarcomatoid carcinoma, medulloblastomas, breast cancers, primary mediastinal B cell lymphomas, Hodgkin lymphomas, and acute myeloid leukemia [5], [9], [13], [23], [24], [25]. While KDM4C is considered an interesting drug target for cancer therapy, this demethylase has also been reported to fulfill vital functions during embryonic and postnatal development [14], [15], [16]. Our study described increased expression of KDM4C in the ESCC ALDHbri+ TICs and its involvement in TICs maintenance was rigorously demonstrated by a variety of standard assays including esospheres in vitro, tumorigenic potential in vivo and serial transplantations/passaging.
Based on sequence homologies and architectural similarities, these JmjC-domain containing demethylases shared similar enzymatic activities most likely exert overlapping functions roles at some developmental stages. Recently, it reported that KDM4A and KDM4C exert a strong overlap in the binding patterns in mESCs [26], neural stem cells [27] and transformed leukemic cells [5], where they exert redundant functions in preventing accumulation of H3K9me3 and H3K36me3. Likewise, our functional studies of KDM4C and the closely related KDM4A are complicated by the fact that the ALDHbri+ populations express both demethylases. Although, we failed to detect other JmjC domain-containing histone demethylases, including JMJD1A, KDM4B or KDM4D were up-regulated in this population. This raises the question as to whether functional redundancy exists within KDM4A and KDM4C in our cellular systems. Thus, a meaningful study of target gene specificity using shRNA technology was applied to investigate the relative contributions of the two demethylases to ALDHbri+ TICs maintenance. However, our results differ from the published observations in the following conceptually important ways. The first indication that KDM4C and KDM4A play non-redundant roles came from variations in expression levels of the subfamily members. Immunohistochemical studies of KDM4C and KDM4A in ESCC tissues have demonstrated tissue- and cell-specific expression, with very little overlap. This might reflect their distinct expression patterns and non-overlapping biological functions in different individuals. Second, KDM4A and KDM4C may not completely function redundantly but appear to regulate a distinct set of targets. We demonstrated that respective knockdown of the demethylases had distinct effects on gene expression, and depletion of both demethylases generally was not found to enhance the transcriptional effects. Nevertheless, Gene Ontology analyses showed that genes with deregulated expression upon loss of KDM4C were enriched for functions related to various cellular reprogramming. However, KDM4A does not appear to participate in maintaining the gene signature typical of pluripotent state. The fact [26] that this enzyme is dispensable for mESCs maintenance or mouse embryogenesis seems paradoxical and reveals cell context type-dependency of KDM4 enzymes effects. Third, several reports [3], [5], [26], [28] have described KDM4A-dependent modulation of H3K9me3 at selected tested promoter regions, and global increases in H3K9me3 have been reported for certain cell types. Surprisingly, KDM4A knockdown ALDHbri+ cells did not display comparable increases in H3K9me3 levels at regions analyzed by ChIP-qPCR. Thus, KDM4A appears to play a minor role in the regulation of H3K9 tri-methylation at KDM4C bound sites in ESCCs. Our data is consistent with a recently published report [29] describing binding specificity of KDM4C from that of the double Tudor domains of KDM4A and KDM4C in ESCC KYSE150 cell line and also consistent with the possibility that KDM4A has roles in transcriptional regulation that are uncoupled from H3K9me3 methylation [4]. Investigation of the relative roles of KDM4A and KDM4C in ESCC TICs maintenance provides additional evidence for distinct functions of the two enzymes. We indicate that KDM4C inhibition decreases the clonogenicity and self-renewal of ALDHbri+ ESCC TICs. However, we did not observe impaired esospheres-forming capacity or serial transplantations efficiency in vitro or substantial decrease in the population of ALDHbri+ cells upon genetic deletion of KDM4A in ESCC cells. Furthermore, a comparable phenotype was observed for ALDHbri+ cells in the absence of KDM4C and cells devoid of both KDM4A and KDM4C (our unpublished data). These data imply that KDM4C had vital functions in ESCC ALDHbri+ TICs maintenance that are not shared by KDM4A. Collectively, our results show that, there is less redundancy or interaction between KDM4C and KDM4A in our cellular context than previously anticipated. The discrepancy further highlights the importance of understanding in which cellular context and protein complexes these demethylases are acting and suggests that differences in genetic backgrounds, culture conditions and the use of knockdown constructs versus the study of knockout cells must be taken into account.
Considering the significant implication of KDM4 demethylases in the development of various diseases, a thorough understanding of their molecular mechanism and effective therapeutic inhibition is of considerable interest. On the basis of the three-dimensional structures available and studies of their catalytic mechanisms, a number of KDM4 inhibitors have been identified and reported [10], [11], [12].
Caffeic acid (3,4-dihydroxycinnamic acid, CaA), a naturally occurring hydroxycinnamic acid derivatives, is an active component in the phenolic propolis extract and also in a wide variety of plants [30]. CaA was identified as several known JmjC histone demethylase inhibitors, which inhibits KDM4C with an IC50 value of about 10 μM [20]. Our studies reveal the identification of the novel inhibitors of KDM4C with therapeutic implications for ESCC, as demonstrated by dose-inhibition response of the proportion of ALDHbri+ cells in xenografts to this compound. Likewise, recent studies [30], [31], [32] published the use of CaA and its derivative, Caffeic acid phenethyl ester (CAPE) to inhibit CSCs-like properties in human breast cancer and malignant human keratinocyte. Although the molecular mechanisms underlying remain largely uninvestigated in this studies, the demonstration that CaA can inhibit the demethylation activity of KDM4C and KDM4C confers stem cell-like characteristics such as the ability to form mammospheres in breast CSCs [13] could be used as an argument to propose that this study provided some proof of concept for targeting the CSCs by KDM4C demethylase inhibitors.
One limitation of our study is that we cannot exclude additional targets of CaA based on its role as a pan-selective JmjC demethylases inhibitor [20]. For example, the fact that the microarray data are highly non-overlapping suggests that CaA hits other targets besides KDM4C. Indeed, CaA also exerts inhibitory effects on other pathways, such as DNA methylation, lipid synthesize and itself is an antioxidant [30]. Although it can be hard to reconcile, the discrepancy at least could be due to the fact that none of studies have been shown to be selective for the KDM4C demethylase thus far, although several mechanistic-based inhibitors toward the catalytic JmjC domain of the histone demethylases have been studied in recent years [11], [12], [33]. Based on the studies presented here, we hope that the available subfamily-specific chemical inhibitors will be refined potent and selective lysine demethylase inhibitors will be identified so that, in a few years, we can test the feasibility of using KDM4C-specific inhibitors for the treatment of ESCC.
Jin et al. [8] recently published the identification of a new chemical compound, SD70 by chemical library screening and obtained data that characterize it as a potential KDM4C inhibitor for cancer therapy. Likewise, Cheung et al. [9] provided the molecular and preclinical evidence for the potential clinical utility of SD70 in MLL leukemia. While our manuscript was being prepared for re-submission, we further demonstrated the therapeutic potentials of targeting KDM4C by SD70 in ESCC ALDHbri+ cells to rule out other inhibitory effects from CaA treatment. We provide evidence that SD70 causes pronounced changes in ESCC ALDHbri+ cell characteristics manifested by dose-dependent inhibition of progenitor formation and self-renewal in serum-free medium, reduction in clonal growth in soft agar and concurrent significant decrease in ALDHbri+ content, all signs of decreased malignancy potential. Furthermore, experiments showed that this small-molecule inhibitor could also remarkable lower tumor burdens and extend the disease latency, particularly in the ALDHbri+ population-derived xenograft in vivo model (our unpublished data), thus holding potential promise for ultimate use in ESCC therapy. Although we cannot exclude the possibility that CaA or SD70 also inhibits other JmjC demethylases, these results could also be faithfully reproduced using an independent KDM4C shRNA approach on the ESCC ALDHbri+ cells phenocopying effects of the two chemical inhibitors, and therefore suggesting that KDM4C might represent as at least one target mediating CaA and SD70 effects. Another important limitation is the focus on experiments presented mainly relied on the usage of shRNA, which could have the disadvantage of substantial off-target effects. Importantly, the conclusions presented in our study are based on the finding that KDM4C knockdown did not affect the expression of KDM4A.
A main finding of this study is that, we identified a number of KDM4C target genes in ESCC TICs that have important roles in the maintenance of pluripotency and are part of the core ES cell transcriptional circuitry (Figure S5), consistent with the previous study in early embryos and ES cells in which KDM4C is required for maintenance of pluripotency and occupies key pluripotency and epigenetic modifier genes [15]. Additionally, accumulating evidence has supported the existence of TICs share important characteristics with embryonic and normal tissue stem cells, including extensive self-renewal and stemness signatures [19], [34].
Interestingly, our result suggests that the specific demethylation of H3K9Me3 by KDM4C at the SOX2 promoter may be required in the ESCC TICs maintenance. However, we observed that the SOX2 rescue was only partial, as some differentiated phenotype were still observed. Thus it is possible that other SOX2-independent mechanisms exist for KDM4C in regulating the ESCC TICs. Thus, further investigation for other regulatory targets that also could be important in TIC physiology is warranted. Meanwhile, our finds demonstrated that, of a subset of isozymes in this family, a number of ALDH isozymes, including ALDH1A1, ALDH1A3 and ALDH3A1 were differentially expressed in ESCC ALDHbri+ cells with or without KDM4C inhibition. Different ALDH isoenzymes has been identified contribute to the elevated ALDH activity in TICs of varied origin thus has been employed as putative CSC markers in multiple tumor types, with focus on ALDH1 and ALDH3A1 [19]. In this regard, it is logical to hypothesize that KDM4C play roles in TIC physiology is proposed to, in part, through ALDH isozymes family. Thus, further investigations of the roles of ALDH1A isozymes in the KDM4C pathway are clearly warranted. These findings together provide insight into mechanisms by which KDM4C maintains the epigenetic state of specific target programs and highlight KDM4C-mediated epigenetic modifications as a means by which it is involved in orchestrating TIC physiology in ESCC.
To our knowledge, this is the first study to identify ALDH1 as a marker of ESCC TICs from freshly resected tumor specimens and, most importantly, is one of the few studies to associate KDM4C expression in ESCC with an adverse pathologic phenotypes and a poor prognosis. In addition, we first study to identify and characterize the molecular signatures and biological properties associated with KDM4C in ESCC ALDHbri+ TICs, which make KDM4C a novel and attractive therapeutic target for human ESCC. The characterization of molecular targets downstream of KDM4C in ESCC TICs in this study should provide not only a better understanding of the mechanisms regulating this specific subpopulation but also novel insights that could be used for the development of more effective cancer therapies for the treatment of this disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: Shegan Gao, Xiang Yuan, Gang Liu;
Development of methodology: Xiang Yuan, Jinyu Kong, Zhikun Ma, Na Li, Ruinuo Jia, Yiwen Liu;
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Fuyou Zhou, Xiang Yuan, Jinyu Kong, Zhikun Ma, Na Li, Ruinuo Jia, Yiwen Liu;
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Xiang Yuan, Jinyu Kong, Zhikun Ma, Na Li, Ruinuo Jia, Yiwen Liu;
Writing, review, and/or revision of the manuscript: Xiang Yuan, Gang Liu, Qimin Zhan;
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Qimin Zhan, Shegan Gao.
Study supervision: Gang Liu, Qimin Zhan, Shegan Gao.
Acknowledgments
We wish to thank Yinming Liang in Xinxiang Medical University for excellent technical help and for critical reading of an early version of the manuscript. We thank Dr. Shimada in Kyoto University for generously providing us with ESCC cell lines.
Footnotes
This work was supported in part by grants from the National Natural Science Foundation of China (81472234 for Shegan Gao), the National Natural Science Foundation of China (U1404817 for Xiang Yuan), and the Key Provincial Medical Sci-Tech Project of Henan (201402026 for Xiang Yuan), Henan Province Education Department Research item (152102310085 for Xiang Yuan).
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2016.08.005.
Contributor Information
Xiang Yuan, Email: yuanxiangdrive@163.com.
Jinyu Kong, Email: kongjinyu@126.com.
Zhikun Ma, Email: mazhikun1225@163.com.
Na Li, Email: linatec83@163.com.
Ruinuo Jia, Email: ruinuojia78@163.com.
Yiwen Liu, Email: liuyiwen615810@163.com.
Qimin Zhan, Email: qmzhan59@163.com.
Gang Liu, Email: liuga@karmanos.org.
Shegan Gao, Email: gsgvicepresident@163.com.
Appendix A. Supplementary Data
References
- 1.Rassouli FB, Matin MM, Saeinasab M. Cancer stem cells in human digestive tract malignancies. Tumour Biol. 2015 doi: 10.1007/s13277-015-4155-y. [DOI] [PubMed] [Google Scholar]
- 2.Yuan X, He J, Sun F, Gu J. Effects and interactions of MiR-577 and TSGA10 in regulating esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2013;6:2651–2667. [PMC free article] [PubMed] [Google Scholar]
- 3.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 4.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agger K, Miyagi S, Pedersen MT, Kooistra SM, Johansen JV, Helin K. Jmjd2/Kdm4 demethylases are required for expression of Il3ra and survival of acute myeloid leukemia cells. Genes Dev. 2016;30:1278–1288. doi: 10.1101/gad.280495.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ, Oppermann U. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J Biol Chem. 2011;286:41616–41625. doi: 10.1074/jbc.M111.283689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- 8.Jin C, Yang L, Xie M, Lin C, Merkurjev D, Yang JC, Tanasa B, Oh S, Zhang J, Ohgi KA. Chem-seq permits identification of genomic targets of drugs against androgen receptor regulation selected by functional phenotypic screens. Proc Natl Acad Sci U S A. 2014;111:9235–9240. doi: 10.1073/pnas.1404303111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung N, Fung TK, Zeisig BB, Holmes K, Rane JK, Mowen KA, Finn MG, Lenhard B, Chan LC, So CW. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell. 2016;29:32–48. doi: 10.1016/j.ccell.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotili D, Tomassi S, Conte M, Benedetti R, Tortorici M, Ciossani G, Valente S, Marrocco B, Labella D, Novellino E. Pan-histone demethylase inhibitors simultaneously targeting Jumonji C and lysine-specific demethylases display high anticancer activities. J Med Chem. 2014;57:42–55. doi: 10.1021/jm4012802. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T, Miyata N. Lysine demethylases inhibitors. J Med Chem. 2011;54:8236–8250. doi: 10.1021/jm201048w. [DOI] [PubMed] [Google Scholar]
- 12.Yu V, Fisch T, Long AM, Tang J, Lee JH, Hierl M, Chen H, Yakowec P, Schwandner R, Emkey R. High-throughput TR-FRET assays for identifying inhibitors of LSD1 and JMJD2C histone lysine demethylases. J Biomol Screen. 2012;17:27–38. doi: 10.1177/1087057111418228. [DOI] [PubMed] [Google Scholar]
- 13.Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene. 2009;28:4491–4500. doi: 10.1038/onc.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Zhang M, Zhang Y, Kou Z, Han Z, Chen DY, Sun QY, Gao S. The histone demethylase JMJD2C is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol Reprod. 2010;82:105–111. doi: 10.1095/biolreprod.109.078055. [DOI] [PubMed] [Google Scholar]
- 16.Katoh Y, Katoh M. Comparative integromics on JMJD2A, JMJD2B and JMJD2C: preferential expression of JMJD2C in undifferentiated ES cells. Int J Mol Med. 2007;20:269–273. [PubMed] [Google Scholar]
- 17.Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris DA. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30:2100–2113. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma I, AL A. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen AL, Kristensen LH, Stephansen KB, Kristensen JB, Helgstrand C, Lees M, Cloos P, Helin K, Gajhede M, Olsen L. Identification of catechols as histone-lysine demethylase inhibitors. FEBS Lett. 2012;586:1190–1194. doi: 10.1016/j.febslet.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol. 2011;31:3339–3350. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Bao Q, Schwarz B, Zhao L, Mysliwietz J, Ellwart J, Renner A, Hirner H, Niess H, Camaj P. Stem cell-like side populations in esophageal cancer: a source of chemotherapy resistance and metastases. Stem Cells Dev. 2014;23:180–192. doi: 10.1089/scd.2013.0103. [DOI] [PubMed] [Google Scholar]
- 23.Ehrbrecht A, Muller U, Wolter M, Hoischen A, Koch A, Radlwimmer B, Actor B, Mincheva A, Pietsch T, Lichter P. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol. 2006;208:554–563. doi: 10.1002/path.1925. [DOI] [PubMed] [Google Scholar]
- 24.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, Wright GW, Lenz G, Ngo VN, Shaffer AL. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18:590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Wary KK, Revskoy S, Gao X, Tsang K, Komarova YA, Rehman J, Malik AB. Histone Demethylases KDM4A and KDM4C Regulate Differentiation of Embryonic Stem Cells to Endothelial Cells. Stem Cell Rep. 2015;5:10–21. doi: 10.1016/j.stemcr.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cascante A, Klum S, Biswas M, Antolin-Fontes B, Barnabe-Heider F, Hermanson O. Gene-specific methylation control of H3K9 and H3K36 on neurotrophic BDNF versus astroglial GFAP genes by KDM4A/C regulates neural stem cell differentiation. J Mol Biol. 2014;426:3467–3477. doi: 10.1016/j.jmb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen MT, Kooistra SM, Radzisheuskaya A, Laugesen A, Johansen JV, Hayward DG, Nilsson J, Agger K, Helin K. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 2016;35:1550–1564. doi: 10.15252/embj.201593317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen MT, Agger K, Laugesen A, Johansen JV, Cloos PA, Christensen J, Helin K. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol Cell Biol. 2014;34:1031–1045. doi: 10.1128/MCB.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Jiang F, Chen L, Yang Y, Cao S, Ye Y, Wang X, Mu J, Li Z, Li L. Blockage of TGFbeta-SMAD2 by demethylation-activated miR-148a is involved in caffeic acid-induced inhibition of cancer stem cell-like properties in vitro and in vivo. FEBS Open Bio. 2015;5:466–475. doi: 10.1016/j.fob.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omene CO, Wu J, Frenkel K. Caffeic Acid Phenethyl Ester (CAPE) derived from propolis, a honeybee product, inhibits growth of breast cancer stem cells. Invest New Drugs. 2012;30:1279–1288. doi: 10.1007/s10637-011-9667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Li Y, Wang K, Wang Y, Yin W, Li L. P38/NF-kappaB/snail pathway is involved in caffeic acid-induced inhibition of cancer stem cells-like properties and migratory capacity in malignant human keratinocyte. PLoS One. 2013;8:e58915. doi: 10.1371/journal.pone.0058915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maes T, Carceller E, Salas J, Ortega A, Buesa C. Advances in the development of histone lysine demethylase inhibitors. Curr Opin Pharmacol. 2015;23:52–60. doi: 10.1016/j.coph.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 34.De Los AA, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.